The mixed-lineage leukemia (MLL) proto-oncogenic protein, as the founding member of human TrxG proteins, was originally identified through its association with both acute lymphoblastic leukemia and acute myeloid leukemia.1 MLL is a histone H3 lysine 4 (H3K4) methyltransferase that can execute methylation on a subset of target genes through its evolutionarily conserved SET domain, an activity that is essential for normal MLL function.2MLL is proteolytically cleaved into two distinct subunits: MLLC180 and MLLN320, which non-covalently interact to assemble an intramolecular complex involved in epigenetic transcriptional regulation.2
MLL is routinely regarded as a nuclear protein. Interestingly, however, our recent research revealed that the MLLC180 subunit alone can localize to cytoplasmic processing bodies (P-bodies),3,4 where microRNA (miRNA)-mediated gene silencing takes place,5 and affect the function of a subset of miRNA, as exemplified by the let-7a family.3,4 The dysregulated function of let-7a resulting from the reduced expression of MLLC180 was very important for maintaining a high level of MYC in MLL leukemia.4 Thus, our work uncovered an unexpected role for MLL in miRNA-mediated translational repression. However, how MLL participates in the regulation of miRNA function remains elusive. We therefore sought to uncover the underlying mechanisms of how MLL participates in miRNA-mediated translational repression. In this study, we demonstrated that MLL was required to recruit let-7a and miR-10a to the miRNA-induced silencing complex (miRISC), partly through its binding partner RAN. The methods and datasets are available as Online Supplementary Information files.
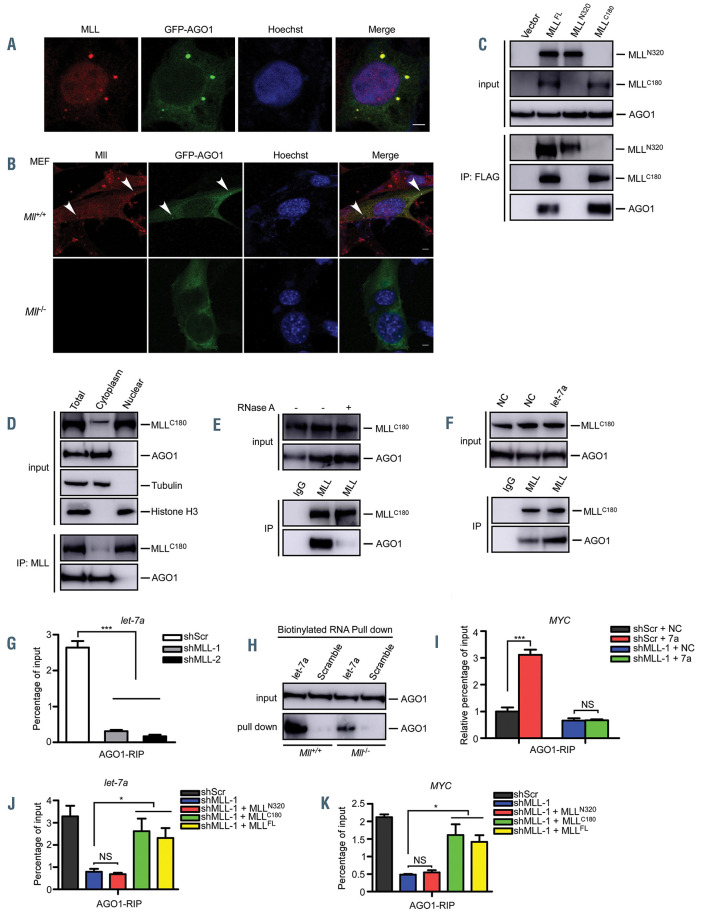
Most miRNA are loaded onto Argonaute (AGO) proteins in the miRISC and act as post-transcriptional regulators of their target mRNA.6 Unfortunately, how these miRNA are selectively loaded onto AGO proteins still remains poorly understood.6 Among miRISC-associated factors, AGO1 plays a predominant and specific role in miRNA-mediated translational repression.7 Our immunofluorescence results demonstrated that AGO1 and MLL were localized in the same cytoplasmic foci, which was disrupted upon MLL depletion (Figure 1A and B, Online Supplementary Figure S1A-C), suggesting an interaction between AGO1 and MLL. Using a specific P-body marker DCP1A, we further confirmed that MLL and AGO1 colocalized in the cytoplasmic P-bodies (Online Supplementary Figure S1D). Previous studies showed that Argonaute proteins could accumulate in stress granules in addition to P-bodies when cells were subjected to stress.8 We observed that upon arsenite treatment MLL, together with AGO1, could co-localize to stress granules, as indicated by the specific stress granule marker G3BP1 (Online Supplementary Figure S1E). These results are consistent with those of our previous study showing that MLL was present not only in P-bodies but also in stress granules.3 Co-immunoprecipitation experiments showed that MLLC180 but not MLLN320 interacts with AGO1 (Figure 1C). Additionally, we demonstrated that the interaction between MLLC180 and AGO1 preferentially occurs in the cytoplasm, and not in the nucleus (Figure 1D). Interestingly, the interaction between MLL and AGO1 decreased dramatically after RNase A treatment, as revealed by co-immunoprecipitation assays, indicating that this interaction was an RNA-dependent indirect interaction, rather than a direct protein-protein interaction (Figure 1E, Online Supplementary Figure S1F). Indeed, the interaction between MLL and AGO1 was enhanced by co-transfected let-7a (Figure 1F, Online Supplementary Figure S1G), indicating that miRNA might play a critical role in the MLL and AGO1 axis.
miRNA-mediated gene silencing requires miRNA to associate with AGO proteins and other silencing factors to form a functional miRISC to repress target mRNA.6 Given that miRNA may fully function in mediating gene silencing even without the existence of microscopically visible P-bodies,9 functional miRISC may still be formed upon MLL depletion. We thus further examined whether the depletion of MLL would impair the recruitment of miRNA to form the functional miRISC. We focused on let-7a and miR-10a, which were two MLL-binding miRNA reported in our previous studies.3 We performed anti-AGO1 RNA immunoprecipitation (RIP) experiments and the results showed that MLL depletion resulted in the loss of binding of let-7a and miR-10a to AGO1 (Figure 1G, Online Supplementary Figure S1H-K). A pull-down assay using biotinylated let-7a further validated that the binding of AGO1 to let-7a was reduced in Mll knockout (Mll-/-) murine embryo fibroblasts (MEF) (Figure 1H). In addition, the recruitment of let-7a and miR-10a target mRNA, MYC, HRAS and HOXA1, to AGO1 was largely impaired in the MLL-depleted cells (Figure 1I and Online Supplementary Figure S1L and S1M). Notably, AGO1 expression was not affected by the knockdown of MLL (Online Supplementary Figure S1H), suggesting that this impaired recruitment of miRNA and its target mRNA to AGO1 was likely not caused by the reduced AGO1 protein levels. Together with the above-mentioned data showing that the interaction between MLL and AGO1 was RNA-dependent, these results indicated that MLL and miRNA may require each other in order to be efficiently recruited by AGO1 and form a functional miRISC.
To further investigate the role of MLL in the recruitment of miRNA to miRISC, we reintroduced shRNAresistant MLLN320, MLLC180 or full-length MLL (MLLFL) into MLL knockdown 293T cells or Mll knockout (Mll-/-) MEF cells and found that the recruitment of let-7a and miR-10a to miRISC was rescued by exogenous MLLC180 (Figure 1J and K, Online Supplementary Figure S1P-S). Collectively, these results indicated that MLL plays a causal role in targeting miRNA and their target mRNA to AGO1 to form a translationally repressed miRISC complex, highlighting the importance of MLL in the control of miRNA-mediated expression.
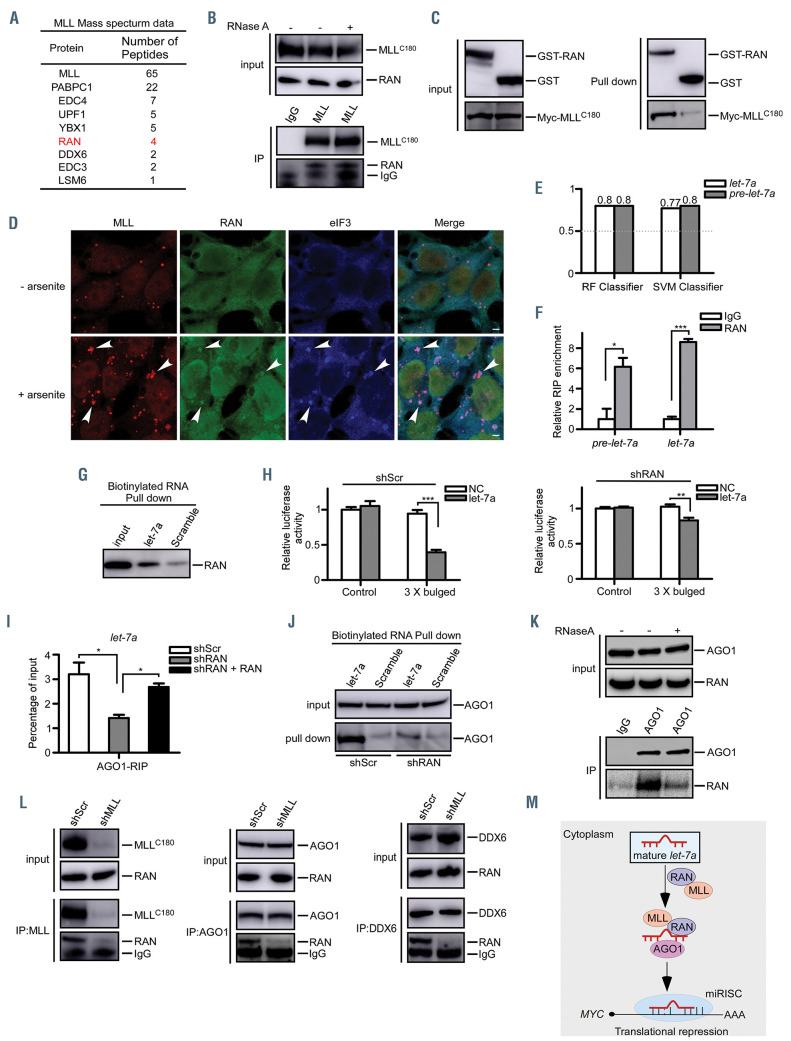
MLLC180 itself does not possess any predictable RNA recognition motif, so we reasoned that MLL might recruit RNA components indirectly through its binding partners. Our proteomics data showed that RAN, a small GTPase involved in the import of cargo through nuclear pore complexes,10 was one of the proteins displaying strong interactions with MLL in the cytoplasm (Figure 2A). In line with a previous study,11 we found that RAN interacted with MLL in an RNA-independent manner (Figure 2B). We further confirmed that RAN could pull down MLLC180, indicating a direct interaction between MLLC180 and RAN (Figure 2C). Moreover, immunofluorescence data showed that upon arsenite treatment, MLL together with RAN co-localized to stress granules, as revealed by a stress granule marker eIF3 (Figure 2D), suggesting a potential role of RAN in regulating mRNA translation besides involving the import of cargo. RAN and XPO5 can form a complex which plays a critical role in nucleocytoplasmic transport of pre-miRNA molecules.10 Unlike XPO5, which dissociates from pre-miRNA in the cytoplasm, RAN could still associate with pre-miRNA in the cytoplasm.12 According to the RPISeq online tool,13 RAN could bind the pre-miRNA and the mature miRNA (Figure 2E, Online Supplementary Figure S2A). Next, our RIP assay confirmed that RAN could bind not only the pre-miRNA but also the mature miRNA (Figure 2F, Online Supplementary Figure S2B). Moreover, RNA pull-down results showed a higher RAN expression in the let-7a or miR-10a-biotinylated group compared to that in the control group (Figure 2G, Online Supplementary Figure S2C). These results were consistent with a previous finding that RAN was an RNA-binding protein,14 suggesting that RAN may be involved in the later steps of miRNA processing and function.
Figure 1.
MLL is required for the loading of let-7a onto AGO1. (A) 293T cells were transfected with GFP-AGO1. Immunofluorescence experiments were performed to visualize the localization of GFP-AGO1 and MLL. MLL-CT antibody, which recognizes MLLC180 (aa2829-2883), was used to detect MLL. Scale bar, 5 mm. (B) Mll wild-type (Mll+/+) and Mll knockout (Mll-/-) MEF cells were transfected with GFP-AGO1. Immunofluorescence experiments were performed to visualize the localization of GFP-AGO1 and MLL. Arrowheads show the localization of MLL with the GFP-AGO1. Scale bar, 5 mm. (C) 293T cells were transfected with FLAGtagged full-length MLL (MLLFL), MLLC320, MLLC180 or empty vector. Cell lysates were prepared and subjected to anti-FLAG immunoprecipitation assays. The interaction between MLL and AGO1 was analyzed by western blot assays using indicated antibodies. (D) The cytosolic and nuclear fractions of 293T cells were separated and subjected to immunoprecipitation using anti-MLL antibodies. Co-purified proteins were examined by immunoblots using the indicated antibodies. (E) 293T cell lysates were treated with RNase A followed by anti-MLL immunoprecipitation. Western blots were performed using the indicated antibodies. (F) The interaction between MLL and AGO1 was assessed after let-7a transfection. Anti-MLL immunoprecipitation assays were performed, results were analyzed by immunoblots with indicated antibodies. (G) Extracts of 293T-shScr and 293T-shMLL cells were subjected to RNA immunoprecipitation (RIP) analysis using anti- AGO1 antibody, and pulled down RNA were analyzed by quantitative reverse transcription polymerase chain reaction (qRT-PCR) using specific primers for let-7a. (H) Mll+/+ and Mll-/- MEF cellular lysates were subjected to a biotinylated- let-7a RNA pull-down assay. Then let-7a-immunoprecipitated AGO1 proteins were subjected to western blot analysis. Scrambled miRNA were used as a negative control. (I) 293T-shScr and 293T-shMLL cells were transfected with Agomir-negative control (NC) and Agomir- let-7a mimic (let-7a) followed by anti-AGO1 RIP experiments at 24 h post-transfection. Total RNAs were isolated to analyze the MYC mRNA level by qRT-PCR. (J, K) 293T-shScr and 293T-shMLL cells with the latter being rescued by exogenous shRNA-resistant MLLN320, MLLC180 or MLLFL were performed with anti-AGO1 RIP experiments at 24 h after transfection. Total RNA were isolated to analyze the let-7a (J) and MYC (K) levels by qRT-PCR using specific primers. NS, no significant difference. *P<0.05, **P<0.01, ***P<0.001. Data represent mean and standard error fo mean of three independent experiments.
Figure 2.
MLL contributes to the loading of let-7a onto AGO1 through interacting with RAN. (A) List of MLL-associated proteins identified by mass spectrometric analysis. 293T cells transfected with MLL were harvested and subjected to the nuclear-cytoplasmic fractionation. The cytoplasmic fractions were prepared for the immunoprecipitation assays followed by mass spectrometric analysis. (B) 293T cell lysates were treated with RNase A followed by anti-MLL immunoprecipitation. Western blots were performed using the indicated antibodies. (C) Direct interaction between MLLC180 and GST-RAN was examined. Left panels: western blots showing the inputs of purified GST-RAN and Myc-MLLC180. Right panels: the pull-down immunoblots were shown with GST-RAN as the bait and the pulled MLLC180 detected by an anti-Myc antibody. (D) 293T cells were untreated (upper panels) or treated with arsenite (0.5 mM, 45 min) (lower panels), then fixed and stained with the indicated antibodies. Note that eIF3 is specific for stress granules. Arrowheads show the localization of MLL with RAN and eIF3. Scale bar, 5 mm. (E) The RPISeq tool was used to predict the interactions between RAN and let-7a or pre-let-7a. The random forest (RF) classifier and support vector machine (SVM) classifier represent the confidence of the prediction. In performance evaluation experiments, predictions with probabilities >0.5 were considered “positive”. (F) 293T cellular lysates were prepared and anti-RAN RIP experiments were performed. Pulled down RNA were isolated, pre-let-7a and mature let-7a were analyzed by qRT-PCR using specific primers. (G) 293T cellular lysates were subjected to biotinylated- let-7a RNA pull-down assays. Then let-7a-immunoprecipitated RAN proteins were subjected to western blot analysis. Scrambled miRNA were used as negative controls. (H) 293T-shScr and shRAN cells transfected with Agomir-negative control (NC) or Agomir- let- 7amimic (let-7a) were subjected to dual luciferase reporter assays. The ratio of luciferase activity was measured and normalized to the value of the cells transfected with the control reporter and NC. (I) Extracts of 293T-shScr and shRAN cells, with the latter being rescued by shRNA-resistant RAN, were subjected to anti-AGO1 RIP assays. Pulled-down RNA were analyzed by qRT-PCR using specific primers for let-7a. (J) 293T-shScr and 293T-shRAN cellular lysates were subjected to biotinylated- let-7a RNA pull-down assays. Then let-7a-immunoprecipitated AGO1 proteins were subjected to western blot analysis. Scrambled miRNA were used as negative controls. (K) 293T cell lysates were treated with RNase A followed by anti-AGO1 immunoprecipitation. Western blots were performed using the indicated antibodies. (L) Extracts of 293T-shScr and 293T-shMLL cells were collected and co-immunoprecipitation assays were performed and analyzed using the indicated antibodies. (M) The proposed mechanism through which MLL and RAN are involved in the loading of let-7a onto AGO1. MLL is required for the loading of let-7a onto AGO1 via a direct interaction with RAN. Thus, RAN serves as a molecular adaptor for the assembly of MLL-associated miRISC. NS, no significant difference. *P<0.05, **P<0.01, ***P<0.001. Data represent the mean and standared error of mean of three independent experiments.
We next probed whether RAN is required to mediate gene silencing of miRNA targets. As shown in Figure 2H and Online Supplementary Figure S2D,E, luciferase activity in RAN-depleted cells was increased compared with that in control cells, indicating that the loss of RAN impaired the let-7a and miR-10a silencing functions. RIP experiments showed that the binding of both let-7a, miR-10a and MYC, HOXA1 to AGO1 was decreased in RANdepleted cells, an effect that could be recovered by the reintroduction of RAN (Figure 2I, Online Supplementary Figure S2F-I). Our previous studies demonstrated that MLLC180 plays a causal role in the miRNA functional deficiency, 3,4 so we then investigated the role of RAN-binding in MLLC180-regulated miRNA function. We found that MLLC180 failed to rescue the miRNA activity when Ran was depleted, an effect that could be recovered by MLLC180 together with reintroduction of RAN, suggesting that RAN was required for the MLLC180-mediated miRNA regulation (Online Supplementary Figure S2J-K). Given the fact that RAN is a small GTPase involved in nucleocytoplasmic transport,10 we determined whether the GTPase activity of RAN is required for the functional interaction of the MLL-miRISC complex. As revealed in Online Supplementary Figure S2L, both wild-type RAN (RANWT) and GTPase-deficient mutant (RANQ69L) could partially reverse the deficits in the binding of let-7a to AGO1 caused by loss of endogenous RAN. We also observed that depletion of RAN significantly impaired the interaction between MLL and AGO1, which could be recovered by RANWT or RANQ69L re-expression (Online Supplementary Figure S2M), suggesting that the GTPase activity of RAN was not required for the function of the MLL-miRISC complex. Additionally, the binding of AGO1 to let-7a or miR-10a was decreased in RAN-depleted cells as revealed by a pull-down assay using biotinylated let-7a or miR-10a (Figure 2J, Online Supplementary Figure S2N). These results indicated that RAN, beyond pre-miRNA export, was required for miRNA-mediated gene silencing.
To decipher the role of RAN in the function of miRISC, we tested the interaction between RAN and AGO1. We observed that AGO1 had an RNA-dependent indirect interaction with RAN (Figure 2K). Importantly, coimmunoprecipitation experiments revealed that besides AGO1, DDX6 a key P-body protein specifically involved in miRNA-mediated translational repression,15 interacts with RAN, but these interactions decreased significantly upon MLL depletion (Figure 2L), indicating that MLL is accountable for these interactions.
To further strengthen our findings, we explored how RAN behaves in a leukemic context. Co-immunoprecipitation assays performed in three leukemia cell lines, JM1, REH and U937, showed that MLL interacts with RAN (Online Supplementary Figure S2O). In REH and U937 cells, MLL together with RAN co-localized to stress granules following arsenite treatment, as illustrated by immunofluorescence assay (Online Supplementary Figure S2P). Additionally, we found that the binding of let-7a to AGO1 was decreased in RAN-depleted REH and U937 cells, an effect that could be restored by the reintroduction of RAN (Online Supplementary Figure S2Q-R). Consistent with the results obtained from 293T cells, we observed that AGO1 had an RNA-dependent indirect interaction with RAN in REH cells (Online Supplementary Figure S2S). Moreover, the interaction between AGO1 and RAN was impaired in MLL-depleted REH cells (Online Supplementary Figure S2T-V). As expected, the binding of RAN to AGO1 was reduced in MLL leukemic cells due to the downregulation of MLLC180 (Online Supplementary Figure S2W).
Collectively, in the present study, we demonstrated that MLL was required for recruiting let-7a and its target mRNA to the miRISC, partly through its direct binding partner RAN (Figure 2M), unraveling an unexpected role for RAN in the loading of miRNA onto AGO1. Our findings provide an alternate mechanism and expanded the functional scope of RAN in the miRNA processing pathway. Thus, the discovery of interplay between MLL and miRNA represents a new regulatory layer, and an additional level of complexity, in the control of gene expression.
Supplementary Material
Acknowledgments
We would like to thank all the members of Liu’s laboratory for their technical assistance. We also thank the Core Facility and Technical Service Center (Shanghai Institute of Hematology) for generous support with cell imaging. We apologize for not citing all the relevant references due to space limitations.
Funding Statement
Funding: this work was supported by the National Key Research and Development Program of China (2018YFA0107802), the National Natural Science Foundation of China (81973996, 81900107 and 81570119), the Program of Shanghai Academic/Technology Research Leader (19XD1402500), the Shanghai Municipal Education Commission Gaofeng Clinical Medicine grant (20161304), the Shanghai Municipal Health Commission (2019CXJQ01), the Shu Guang project (14SG15), the Collaborative Innovation Center of Hematology, and the Samuel Waxman Cancer Research Foundation.
References
- 1.Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71(4):691-700. [DOI] [PubMed] [Google Scholar]
- 2.Dou Y, Milne TA, Tackett AJ, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121(6):873-885. [DOI] [PubMed] [Google Scholar]
- 3.Zhu S, Chen Z, Wang R, et al. MLL is required for miRNA-mediated translational repression. Cell Discov. 2019;3(5):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu S, Cheng X, Wang R, et al. Restoration of microRNA function impairs MYC-dependent maintenance of MLL leukemia. Leukemia. 2020;34(9):2484-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005; 7(6):633-636. [DOI] [PubMed] [Google Scholar]
- 6.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509-524. [DOI] [PubMed] [Google Scholar]
- 7.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell. 2007;26(5):611-623. [DOI] [PubMed] [Google Scholar]
- 8.Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci U S A. 2006;103(48):18125-18130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leung AKL. The whereabouts of microRNA actions: cytoplasm and beyond. Trends Cell Biol. 2015;25(10):601-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003; 17(24):3011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura T, Mori T, Tada S, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10(5):1119-1128. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Xu X, Ma Z, et al. Dynamic mechanisms for pre-miRNA binding and export by Exportin-5. RNA. 2011;17(8):1511-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muppirala UK, Honavar VG, Dobbs D. Predicting RNA-protein interactions using only sequence information. BMC Bioinformatics. 2011;12:489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brannan KW, Jin W, Huelga SC, et al. SONAR discovers RNA-binding proteins from analysis of large-scale protein-protein interactomes. Mol Cell. 2016;64(2):282-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16(7):421-433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.