Introduction
Seminal papers over the past four decades have described phenotypic driver mutations, such as BCR-ABL1, JAK2, MPL, CALR, KIT and CSF3R, in subsets of myeloproliferative neoplasms (MPN). These mutations affect cytokine signaling or regulation, and result in malignant hematopoiesis.1-3 Such discoveries and the accompanying biological insights have resulted in successful therapeutic approaches for many people diagnosed with MPN, in particular chronic myeloid leukemia (CML), myelofibrosis (MF) and systemic mastocytosis (SM).4-6 The greatest advance has been in CML, with a significant proportion of patients being able to achieve a major molecular response (MMR or MR3; BCR-ABL1 ≤0.1% on the International Scale) following treatment with an ABL1-tyrosine kinase inhibitor (TKI), leading to these patients having lifespans indistinguishable from those of the general population, although the time by which this response milestone should be reached remains controversial.7 Qualified success with significant symptomatic benefit and modest gains in survival have also been achieved in people with MF and SM, following treatment with JAK inhibitors and KIT inhibitors, respectively.6 Some of these achievements have been facilitated by the rational integration of next-generation sequencing (NGS) assays, high-sensitive polymerase chain reaction (PCR) assays on DNA or RNA (sensitivity 0.01%- 0.1%), and single-cell analyses, in efforts to improve personalized treatment approaches.8 Such efforts have opened a new era of precision medicine for diverse malignancies in which relatively non-specific and often toxic drugs are being replaced by safer and better tolerated agents whose mechanism of action is precisely defined, and for which the treatment algorithm is guided by individual genetic information. Here we examine how molecular testing in MPN can shape diagnosis, monitoring, and treatment algorithms and enable more precise early identification of targeted therapy resistance, particularly in CML (Figure 1). We also discuss the potential impact of persistent or new clonal hematopoiesis on the molecular testing of individuals with MPN and the potential impact on measurable residual disease. This manuscript describes some of the current highlights and challenges related to genetic testing in MPN in 2021.
Clinically validated tests for detecting and monitoring BCR-ABL1 and KIT mutations
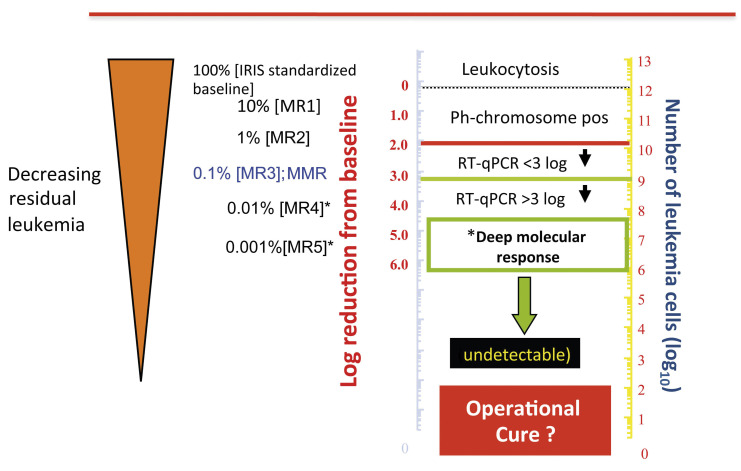
For patients with CML, a qualitative reverse transcriptase PCR, conducted at diagnosis on peripheral blood cells, enables precise identification of BCR-ABL1 transcripts. Once TKI therapy has commenced, quantitative reverse transcriptase PCR, which adheres to the well-established International Scale, is used for sensitive and accurate monitoring of the levels of BCR-ABL1. However, this has inherent limitations with regard to its lower limit of detection and quantification of BCR-ABL1 transcripts, problems which became important as TKI that were more powerful, compared to imatinib, were developed.9 This, in turn, affected the definition and identification of deep molecular response (DMR), defined as a greater than 4 (MR4), 4.5 (MR4.5) or 5 (MR5) log reduction in BCR-ABL1 transcripts on the International Scale, below the standardized baseline (Figure 2). DMR is now recognized to be of considerable clinical importance to prospectively identify patients likely to remain in remission after discontinuing TKI therapy. Indeed, several current CML guidelines advocate the use of a sustained DMR, for at least 2 years, to select patients for consideration of TKI cessation.10,11
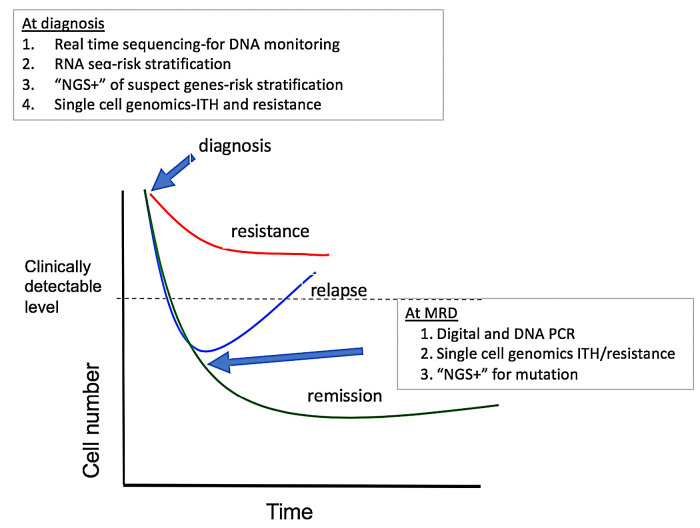
Figure 1.
Treatment response and potential uses of emerging technologies for diagnostics, monitoring and mutation testing in chronic myeloproliferative malignancies. At diagnosis, methods can be used to quickly identify breakpoints useful for designing monitoring assays, as well as other mutations that might influence the initial response to treatment with a tyrosine kinase inhibitor. RNAsequencing can be used to assay specific genes and pathways associated with early response. Single-cell genetics can be used to identify potentially troublesome complex heterogeneity and populations with a resistance signature. During therapy, digital and DNA polymerase chain reaction analyses can be used as more sensitive determinants of deep molecular response, and next-generation sequencing can be used to identify mutations in patients without a deep response who appear to be relapsing. RNA-seq: RNA-sequencing; NGS: nextgeneration sequencing; MRD: measurable residual disease; ITH: intratumoral heterogeneity; PCR: polymerase chain reaction. (Adapted with permission from Radich JP et al.7)
At present, there are several tests in the clinic for the detection and monitoring of BCR-ABL1 transcripts, some of which have not been validated robustly; furthermore, they show considerable interlaboratory variations and variable levels of sensitivity. With the current focus on treatmentfree remission (TFR), the importance of using analytically and clinically well-validated tests, preferably approved by regulatory bodies, is being recognized. As an illustration, three recently US Food and Drug Administration (FDA)- approved tests appear to perform better than the ‘standard’ quantitative reverse transcriptase PCR tests and may have greater appeal for monitoring very low levels of BCR-ABL1 transcripts. Indeed, it is of interest that over a decade ago, learning from the earlier lessons following the introduction of DNA PCR for BCR-ABL1 and the harmonizing the International Scale BCR-ABL1 transcript measurements, Goldman and colleagues in London, instigated a patientspecific DNA-based method of detection and quantification of an individual patient’s CML clone.12,13 This method involved the rapid identification of BCR-ABL1 fusion junctions by targeted NGS, coupled with the use of a dPCR platform, in patients with very low-level molecular residual disease.. The first FDA-approved test (Asuragen Inc., Austin, TX, USA) is performed with a manufactured kit that can be used on several thermal cyclers, although the FDA approval specifies a specific machine.14 The second approved test is the Cepheid cartridge technique (Cepheid, Sunnyvale, CA, USA), which is attractive given its simplicity and short turnaround time.15 More recently, a water-oil emulsion droplet technology, developed by Bio-Rad (Bio-Rad Laboratories Inc., Hercules, CA, USA), known as digital droplet PCR, was approved by the FDA.16 The digital droplet PCR assay has recently been tested in studies evaluating the feasibility of discontinuing TKI therapy safely in patients with CML who had been in DMR for >2 years.17 The studies documented that, compared with quantitative reverse transcriptase PCR, digital droplet PCR was better at forecasting the success of TKI discontinuation. Digital droplet PCR technology has several advantages over quantitative reverse transcriptase PCR tests that rely on exponential amplification to estimate the target amount. The digital droplet PCR assay relies on a binary endpoint (yes or no), which is much more lenient to poor RNA quality and inhibitors. The BCRABL1 transcript level is estimated using Poisson distribution based on the number of positive droplets. The Cepheid test is particularly suitable for analysis of smaller sample batches analyzed frequently; for larger batches of samples tested infrequently, the Asuragen or Bio-Rad platforms might be more cost-effective. The use of DNA for CML monitoring is difficult, specifically because the breakpoint on chromosome 9 is vast compared with the ‘small’ breakpoint cluster region on chromosome 22. There are techniques to find the breakpoint using a series of BCR and ABL primers. Once an amplification product has been generated, it can be sequenced, with patient-specific primers generated for subsequent PCR amplification of the BCR-ABL1 breakpoint. However, there may be new technologies that will make breakpoint detection much faster. Breakthroughs in ‘realtime’ sequencing, such as Pacific Bioscience and Nanopore technology, which can read exceeding long sequences at breathtaking speeds, potentially allow BCR-ABL1 breakpoint detection to be performed with a single sequencing run.8
In CML, there are two variants of the BCR-ABL1 transcript, depending on whether the break in BCR occurs in the intron between exons e13 and e14, or in the intron between exons e14 and e15.6 A break in the former intron yields an e13a2 mRNA junction and a break in the latter intron yields an e14a2 junction. Most patients have transcripts with features of either e13a2 or e14a2, but occasional patients have both transcripts present in their leukemia cells. The prognostic significance of the precise type of BCR-ABL1 transcript is now being increasingly recognized in efforts to achieve DMR and potential TFR following successful TKI treatment. Earlier studies in patients treated with imatinib had suggested that patients with e14a2 transcripts had deeper and faster responses compared to those with e13a2 transcripts.18 Subsequently, Baccarani and colleagues observed lower response rates and inferior outcomes following nilotinib treatment in patients with e13a2 transcripts.19 These investigators also noted a relationship of transcript type with age and gender. More recently, a French study, led by Genthon and colleagues, also documented deeper and faster responses, in terms of achieving MR3 and MR4, in patients expressing e14a2 compared with those expressing e13a2 transcripts.20 The differences in clinical outcomes based on transcript subtype has so far only been investigated in small studies and this issue needs to be assessed prospectively in larger cohorts receiving any of the five currently licensed TKI treatments for CML. A study of 20 patients who were in TFR for longer than 1 year, having achieved sustained MR4.5 following treatment with TKI, suggested the potential importance of the lineage of measurable residual disease as a potential predictive biomarker of TFR outcome.21 The investigators, Pagani and colleagues, used fluorescence-activated cell sorting of CML cells known to express BCR-ABL1 mRNA, granulocytes, monocytes, B cells T cells and natural killer cells; BCR-ABL1 DNA PCR was used to investigate the lineage of these residual CML cells. The observation of BCR-ABL1 DNA being present only in B- and T-lymphocytes begs the question of the CML cell lineage contributing to the success of TFR. The study could not, however, establish the absence of CML stem cells (CD34+38–26+) in a cohort of patients in TFR with undetectable BCR-ABL1 mRNA transcripts, which are considered to account for the loss of TFR in such patients.22 Regardless, larger studies assessing the lineage of measurable residual disease in TFR patients are warranted.
Figure 2.
The International Scale for quantitative reverse transcriptase polymerase chain reaction analysis of BCR-ABL1 transcripts. IRIS: International Randomized Study of Interferon and STI571; MMR: major molecular response; MR4, MR4.5 and MR5: 4-, 4.5- and 5-log reductions, respectively, from the IRIS standardized baseline; Ph: Philadelphia chromosome; RT-qPCR: quantitative reverse transcriptase polymerase chain reaction.
In patients with SM, the identification and quantification of the KIT D816V mutation in hematopoietic stem and progenitor cells, as well as mature mast cells, by highly sensitive (sensitivity <0.01%) and specific assays, such as allelespecific oligonucleotide quantitative PCR and digital droplet PCR, allows an accurate diagnosis.23-26 This information is also useful for risk stratification, complementing conventional biomarkers such as serum tryptase level and percentage of bone marrow infiltration by mast cells, and may be used for monitoring patients receiving treatment. More recently, the use of targeted NGS panels for the characterization of associated gene mutations, such as SRSF2, ASXL1 and RUNX1, has improved prognostication and has led to the development of novel prognostic scoring systems to optimize clinical management.27
Testing for BCR ABL kinase domain mutations
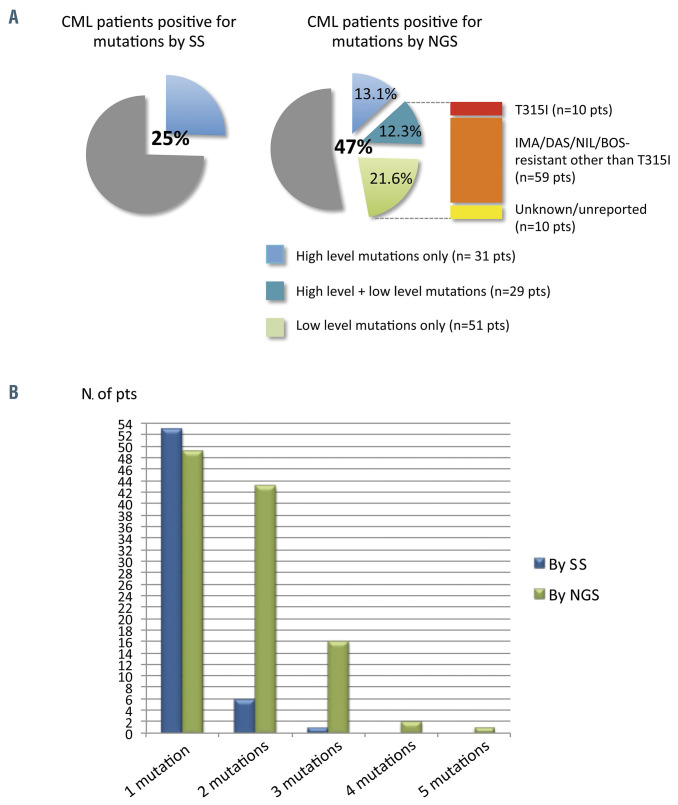
Mutation testing for BCR-ABL1 kinase domain mutations and the co-existence of subclones in general can be assessed by a variety of technologies. In general, the more sensitive the test, the more complex and expensive it is. Sanger sequencing has a low error rate but a poor sensitivity of only 10-20%. In contrast, NGS has a better sensitivity of roughly 1% and is useful for the identification of compound mutations.28 However, the error rate associated with the library amplification and preparation can be up to one in a 1,000 base pairs, particularly when sequencing from mRNA, which relies on the error-prone reverse transcriptase. Barcode correction techniques may improve this, but the best technique may be so-called duplex sequencing, which is novel in that it sequences both DNA strands, dramatically reducing the error rate since mutations are only called if the complementary base change is seen on the other strand. Indeed, recent reports suggest that low frequency mutations that are detected by NGS but have unpredictable clinical courses (e.g., disappear spontaneously in some patients) may represent artifacts from the error rate inherent in NGS.29 Soverini and colleagues recently conducted the first prospective, ‘real-world’ assessment of NGS-based BCR-ABL1 knockdown mutation testing compared with Sanger sequencing in a large cohort of consecutively studied CML patients in whom TKI had failed or who were in the ‘warning’ category described by the European Leukemia Net (ELN) guidelines.30 The researchers demonstrated the importance of low-level mutations, defined as mutations with a variant allele frequency of 3-20%, and their clinical relevance (Figure 3). These observations are the basis for guiding genomic-based personalized therapies for CML further, in particular through the identification of high-risk individuals prior to the ELN ‘warning’ stage. These tests should also improve the efficiency and safety of clinical trials designed to reduce the risk of blast transformation in patients who respond suboptimally to TKI. Important challenges now are to improve sensitivity further, reduce the turnaround time and lower the costs, which remain high.
Intratumoral heterogeneity
Studies of hematologic malignancies as well as diverse solid tumors have revealed a surprising amount of intratumor heterogeneity, i.e., the finding of multiple, related clones rather than one uni-clonal monolith. Thus, the kinetics of disease progression, response and relapse follow the rules of Darwinian selection. Many neoplasms, including CML, MF and advanced SM, have been found to exhibit greater clonal complexity than previously thought, as new myeloid mutations have been found in these diseases.31 For example, Jawaher and colleagues identified the emergence of the KIT D816V mutation as a distinct and late event in patients with multi-mutated advanced SM.32
Trying to map intratumor heterogeneity by the sequencing of bulk populations is limited by the simple fact that one is sequencing the average mutation frequency of all the cells from various clones. New technologies allow for the sequencing of RNA (e.g., 10x Genomics) or DNA (e.g., Mission Bio) from single cells. Major advantages of singlecell technologies include the higher resolution offered to understand the types of cells present, to detect rarer cell populations and to study their function (inferred by RNA expression) or clonal structure (inferred by mutation pattern). Disadvantages, other than the financial costs, are that each cell is a ‘one and done’ experiment. Furthermore, it can be difficult to determine real signal versus experimental noise, which is especially problematic with RNA, for which simple factors such as time from sample acquisition to experiment can influence gene expression.
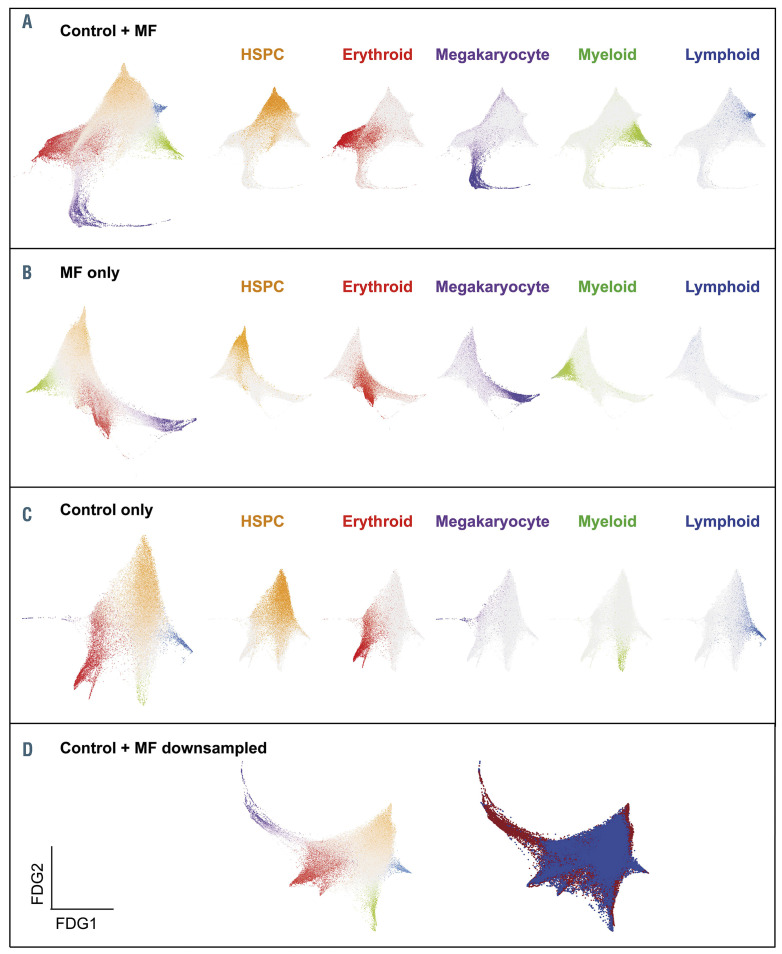
Recent work using single-cell RNA-sequencing has garnered considerable novel insight into normal and aberrant hematopoiesis, cell-cell interactions, characterization of bone marrow and immune (non-clonal) cells as well as tissue stroma and leukemia-initiating cells.33-36 This technology enables detection and characterization of intratumor heterogeneity and provides much needed granularity to key issues, including acquisition of individual or specific combinations of somatic mutations or proteins expressed by cellular subtypes and mechanisms of relapse or treatment resistance. As an illustration, the use of single-cell-level sequencing in a study of MF, conducted by Psaila and colleagues, revealed megakaryocyte-biased hematopoiesis with megakaryocyte progenitors demonstrating distinct inflammatory and metabolic signatures, and increased expression of the surface antigen G6B (MPIG6B) on MF stem cells and progenitors (Figure 4).36 These findings raise the possibility of new therapies for MF, which could target the MF clone and MF-associated fibrosis. In CML, insights into the intratumoral heterogeneity of CML stem cells has revealed subgroups with distinct molecular signatures that are resistant to TKI.34 Furthermore novel molecular pathways related metabolism and the bone marrow microenvironment are being deciphered. Patients with advanced SM, in particular those with an associated hematologic malignancy, often have multilineage involvement by KIT and multimutated clones, which are associated with a poorer prognosis.27 Additionally, through single-cell analysis, Grootens and colleagues identified KIT 816V in early SM stem cells and progenitors, suggesting that this biomarker may not be restricted to the mast cell lineage.37 In the same study it was observed that the mutation frequency was 100% in mature CD45RA+ mast cells.
Figure 3.
Comparison between Sanger sequencing and next-generation sequencing – the NEXT-in-CML study. (A) Percentage of patients positive for mutations, as determined by Sanger sequencing (SS) and by next-generation sequencing (NGS). Among patients positive for mutations by NGS, 31 (13.1%) had high-level mutations only (≥20%; detectable by SS too); 29 (12.3%) had both ≥1 high-level mutations and ≥1 low-level mutations (≤20%; detectable by NGS only); 51 (21.6%) had only lowlevel mutations. A low-level T315I mutation was detected in ten patients; 59 additional patients had ≥1 low-level mutations known to be associated with resistance to imatinib or second-generation tyrosine kinase inhibitors other than the T315I mutation (Y253H; E255K/V; V299L; F317L/V/I/C; F359V/I/C). The remaining ten patients had only low-level mutations with an unknown resistance profile and/or not listed in the COSMIC database. (B) Patients positive for one or multiple mutations as assessed by SS versus NGS. CML: chronic myeloid leukemia; pts: patients; IMA: imatinib; DAS: dasatinib; NIL: nilotinib; BOS: bosutinib. (Adapted, with permission, from Soverini S et al.30).
Figure 4.
Single-cell -omics demonstrate a trajectory for megakaryocyte-biased hematopoiesis in myelofibrosis. (A–D) Force-directed graphs (FDG) for aggregates of all control + myelofibrosis (MF) cells (A), MF only (B), control only (C), and control + down-sampled MF dataset (D). In (D), the left graph shows the lineage signature gene score and in the right graph cells are colored according to the donor type (healthy donors, blue; MF patient, red). Gene expression trajectories are visualized by superimposing the expression scores of lineage signature gene sets on the FDG. Gray cells represent uncommitted hematopoietic stem and progenitor cells or cells with expression of more than one lineage signature. (Published, with permission, from Psalia B, et al.36)
Screening for clonal hematopoiesis of indeterminate potential
NGS-based genetic analysis of large unselected populations assessing acquisition of somatic mutations has provided evidence of age-related clonal expansion of recurrent mutations in known oncogenes in hematopoietic stem and progenitor cells in the absence of overt hematologic malignancies, a condition termed clonal hematopoiesis.38 The somatic mutations that define clonal hematopoiesis tend to be similar to those found in hematologic malignancies, such as ASXL1, DNMT3A, RUNX1, JAK2, TET2, SF3B1 and others, with a variant allele frequency >2% in subjects without cytopenias or a history of a myeloid neoplasm. The mutations are typically present in circulating granulocytes, monocytes, and natural killer cells; they can also affect B cells and, rarely, T cells. Clonal hematopoiesis is considered to represent the early steps of leukemogenesis and is associated with an increase in the risk of myeloid and lymphoid malignancies of 0.5% to 1.0% per year.39 Evidence from retrospective case-control studies supports the role of clonal hematopoiesis in therapy-related myeloid neoplasms, too.40 The mutations can also be associated with acquired drug resistance. Somewhat surprisingly, clonal hematopoiesis is also associated with a pro-inflammatory state and an increased risk of various non-hematologic diseases, in particular cardiovascular disease, attributed to genes that are involved in regulating inflammation and accelerating atherosclerosis. 41,42 Interestingly, Hameisterr and colleagues recently investigated whether clonal hematopoiesis might affect the course of COVID-19 in hospitalized older patients who had tested positive for SARS-CoV-2 infection and found no significant association.43
Although it is recognized that people with clonal hematopoiesis develop MPN at a higher rate than those without mutations, the precise impact of clonal hematopoiesis on the prognosis and treatment of MPN is an enigma.44 An important challenge is, therefore, to assess the clinical relevance of clonal hematopoiesis at the time of diagnosis of MPN and assess its potential prognostic value, in particular in the transformation to acute leukemia and in treatment resistance. The presence of clonal hematopoiesis may also affect the ‘real-world’ situation in people with CML, MF and SM who have been treated ‘successfully’, including those who have undergone allogeneic stem cell transplantation, when such mutations could be donorderived and influence the assessment of measurable residual disease.45 Genomic studies, in particular those involving single-cell sequencing, are being increasingly integrated into the investigation of MPN at diagnosis and transformation, which also raises the question of which genes should ideally be included in the NGS panel. Furthermore, we are beginning to fathom the complexity of the cancer tissue ecosystem and how this is affected by different features, such as the cells’ metabolism and how it co-opts normal genes, stromal and immune cells within the microenvironment, among other variables. In this regards the recent work of Van Etten, Krause and others on the specific, targetable interactions with the microenvironment in people with imatinib-resistant CML is important.46 Despite the therapeutic advances in CML, MF and SM, the outlook for people whose disease transforms remains bleak and highlights the need for suitable prognostic scores to identify those at high risk of progression who may benefit from more intensive initial treatments, including allogeneic stem cell transplantation.
Future prospects
We have clearly made significant progress by examining MPN through genetics and physiology, by the unprecedented application of ‘-omics’ technology, ultrasensitive sequencing technology and single-cell genomics. Such approaches have arguably enhanced our understanding of chronic myeloproliferative malignancies, and the characterization of the underlying intratumor heterogeneity and the ability of the neoplastic clone to evolve and adapt has been recognized as a principal challenge for targeted treatments and immunotherapies.47 The different genetic tests in MPN clinics have undoubtedly improved our ability to monitor patients effectively and have refined diagnostic risk stratification. 48 For CML patients, they also enhance the probability of TKI cessation and achieving TFR; for patients with MF and SM, they are complementary to the World Health Organization 2016 diagnostic criteria and help in navigating treatment decisions. These tests also allow for a better selection of targeted agents to be tested in subgroups with variant somatic mutations. Nevertheless, much work remains. For example, we have little understanding of the cell-intrinsic and -extrinsic mechanisms underpinning the transformation of MPN into acute leukemia, or the mechanisms of resistance to the newer inhibitors, to mention a few.49 The emerging picture is complex but has created a platform upon which to build novel therapeutic approaches.
Acknowledgments
The authors would like to thank Dr Alpa Parmar and participants of the XIV Post ASH MPN workshop for their assistance.
Funding Statement
Funding: The authors would like to thank Alpine Oncology Foundation for funding.
References
- 1.Vainchenker W, Kralovics R. Genetic basic and molecular pathophysiology of classical myeloproliferative neoplasms. Blood. 2017; 129(6):667-679. [DOI] [PubMed] [Google Scholar]
- 2.Grinfeld J, Nangalia J, Baxtyer EJ, et al. Classification and personalized prognosis in myeloproliferative neoplasms. N Engl J Med. 2018;379(15):1416-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mughal TI, Gotlib J, Mesa R, et al. Recent advances in the genomics and therapy of BCR/ABL1-positive and -negative chronic myeloproliferative neoplasms. Leuk Res. 2018;67:67-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verstovsek S, Mesa RA, Gotlib J, et al. A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N Engl J Med. 2012;366(9):799-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arock M, Sotlar K, Gotlib J, et al. New developments in the field of mastocytosis and mast cell activation syndromes: a summary of the Annual Meeting of the European Competence Network on Mastocytosis (ECNM) 2019. Leuk Lymphoma. 2020;61(5): 1075-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mughal TI, Radich JP, Deininger MW, et al. Chronic myeloid leukemia: reminiscences and dreams. Haematologica. 2016;101(5): 541-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith G, Apperley J, Milojkovic D, et al. A British Society for Haematology Guideline on the diagnosis and management of chronic myeloid leukaemia. Br J Haematol. 2020;191(2):171-193. [DOI] [PubMed] [Google Scholar]
- 8.Radich J, Yeung C, Wu D. New approaches to molecular monitoring in CML (and other diseases). Blood. 2019;134(19):1578-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mughal TI, Pemmaraju N, Radich JP, et al. Emerging translational science discoveries, clonal approaches, and treatment trends in chronic myeloproliferative neoplasms. Hematol Oncol. 2019;37(3):240-252. [DOI] [PubMed] [Google Scholar]
- 10.Hochhaus A, Baccarani M, Silver RT, et al. European Leukemia Net 2020 recommendations for treating chronic myeloid leukemia. Leukemia. 2020;34(4):1495-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deininger MW, Shah NP, Altman JK, et al. Chronic myeloid leukemia - NCCN guidelines -version 2.2021. J Natl Compr Canc Netw. 2020;18(10):1385-1415. [DOI] [PubMed] [Google Scholar]
- 12.Alikian M, Ellery P, Forbes M, et al. Next generation sequencing-assisted DNA-based digital PCR for a personalized approach to the detection and quantification of residual disease in chronic myeloid patients. J Mol Diagn. 2016;18(2):176-189. [DOI] [PubMed] [Google Scholar]
- 13.Hughes T, Deininger M, Hochhaus A, et al. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: review and recommendations for harmonizing current methodology for detecting BCRABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;108(1):28-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown JT, Laosinchai-Wolf W, Hedges JB, et al. Establishment of a standardized multiplex assay with the analytical performance required for qualitative measurement of BCR-ABL1 on the international reporting scale. Blood Cancer J. 2011;1(3):e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radich JR. Chronic myeloid leukemia: Global impact from a local laboratory. Cancer. 2017;123(14):2594-2596. [DOI] [PubMed] [Google Scholar]
- 16.Yan D, Pomicter AD, O’Hare T, Deininger MW. ddeeper than deep: can ddPCR predict successful imatinib cessation? Clin Cancer Res. 2019;25(22):6561-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hochhaus A, Breccia M, Saglio G, et al. Expert opinion- management of chronic myeloid after resistance to second-generation tyrosine kinase inhibitors. Leukemia. 2020;34(6):1495-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ercaliskan A, Eskazan AE. The impact of BCR-ABL1 transcript type on tyrosine kinase inhibitor responses and outcomes in patients with chronic myeloid leukemia. Cancer. 2018;124(19):3806-3818. [DOI] [PubMed] [Google Scholar]
- 19.Baccarani M, Castagnetti F, Gugliotta G, et al. The proportion of different BCR-ABL1 transcript types in chronic myeloid leukemia. An international overview. Leukemia. 2019;33(5):1173-1183. [DOI] [PubMed] [Google Scholar]
- 20.Genthon A, Nicolini FE, Huguet F, et al. Infleunce of major BCR-ABL1 transcript subtype on outcome in patients with chronic myeloid leukemia in chronic phase treated frontline with nilotinib. Oncotarget. 2020;11(26):2560-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pagani IS, Dang P, Saunders V, et al. Lineage of measureable residual disease in patients with chronic myeloid leukemia in treatment- free remission. Leukemia. 2020;34(4): 1052-1061. [DOI] [PubMed] [Google Scholar]
- 22.Bocchia M, Sicuranza A, Abruzzese E, et al. Residual peripheral blood CD26+ leukemic stem cells in chronic myeloid leukemia patients during TKI therapy and during treatment-free remission. Front Oncol. 2018;8:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arock M, Sotlar M, Akin C, et al. KIT mutation analysis in mast cell neoplasms: recommendations of the European Competence Network on Mastocytosis. Leukemia. 2015;29(6):1223-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greiner G, Gurbisz M, Ratzinger F, et al. Digital PCR : a sensitive and precise method for KIT D816V quantification in mastocytosis. Clin Chem. 2018;64(3):547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reiter A, George TI, Gotlib J. New developments in diagnosis, prognostication, and treatment of advanced systemic mastocytosis. Blood. 2020;135(16):1365-1376. [DOI] [PubMed] [Google Scholar]
- 26.Martelli M, Monaldi C, De Santis S, et al. Recent advances in the molecular biology of systtemic mastocytosis: implications for diagnosis, prognosis, and therapy. Int J Mol Sci. 2020;21(11):3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jawhar M, Schwaab J, Hausmann D, et al. Splenomegaly, elevated alkaline phosphatase and mutations in SRSF2/ASXL1/ RUNXL1 gene panel are strong adverse prognostic markers in patients with systemic mastocytosis. Leukemia. 2016;30(12): 2342-2350. [DOI] [PubMed] [Google Scholar]
- 28.Wu D, Sherwood A, Fromm JR, et al. Highthroughput sequencing detects minimal residual disease in acute T lymphoblastic leukemia. Sci Transl Med. 2012;4(134): 134ra63. [DOI] [PubMed] [Google Scholar]
- 29.Sasine JP, Schiller GJ. Acute myeloid leukemia: how do we measure success? Curr Hematol Malig Rep. 2016;11(6):528-536. [DOI] [PubMed] [Google Scholar]
- 30.Soverini S, Bavaro L, De Benedittis C, et al. Prospective assessment of NGS-detectable mutations in CML patients with nonoptimal response: the NEXT-in-CML study. Blood. 2020;135(8):534-541. [DOI] [PubMed] [Google Scholar]
- 31.Lee J, Godfrey AL, Nangalia J. Genomic heterogeneity in myeloproliferative neoplasms and applications to clinical practice. Blood Rev. 2020;42:100708. [DOI] [PubMed] [Google Scholar]
- 32.Jawhar M, Schwaab J, Schnittger S, et al. Molecular profiling of myeloid progenitor cells in multi-mutated advanced systemic mastocytosis identifies KIT D816V as a distinct and late event. Leukemia. 2015;29(5):1115-1122. [DOI] [PubMed] [Google Scholar]
- 33.Tang F, Barbacioru C, Wang Y, et al. mRNASeq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377-382. [DOI] [PubMed] [Google Scholar]
- 34.Psaila B, Mead AJ. Single-cell approaches reveal novel cellular pathways for megakaryocyte and erythroid differentiation. Blood. 2019;133(13):1427-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris V, Marion W, Hughes T, et al. Singlecell analysis reveals mechanisms of plasticity of leukemia initiating cells. bioRxiv. 2020; April 30. [Epub ahead of print] [Google Scholar]
- 36.Psaila B, Wang G, Rodriguez-Meira A, et al. Single-cell analyses reveal megakaryocytebiased hematopoiesis in myelofibrosis and identify mutant clone-specific targets. Mol Cell. 2020;78(3):477-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grootens J, Ungerstedt JS, Ekoff M, et al. Single-cell analysis reveals the KIT D816V mutation in haematopoietic stem and progenitor cells in systemic mastocytosis. EBioMedicine. 2019;43:150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mead A. Single cell genomics in chronic myeloid leukemia. Hemasphere. 2018;2(S2): 54-55. [Google Scholar]
- 39.Steensma DP, Bejar R, Jaiswal S, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hasserjian RH, Steensma DP, Graubert TA, Ebert BL. Clonal hematopoiesis and measureable disease assessment in acute myeloid leukemia. Blood. 2020;135(20): 1729-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warren JT, Link DC. Clonal hematopoiesis and risk for hematologic malignancy. Blood. 2020;136(14):1599-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaiswal S, Natarajan P, Ebert BL. Clonal hematopoiesis and atherosclerosis. N Engl J Med. 2017;377(14):1401-1402. [DOI] [PubMed] [Google Scholar]
- 43.Hameister E, Stolz SM, Fuhrer Y, et al. Clonal hematopoiesis in hospitalized elderly patients with COVID-19. Hemasphere. 2020;4(4):e453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jaiswal S. Clonal hematopoiesis and nonhematologic disorders. Blood. 2020;136(14): 1606-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boettcher S, Wilk CM, Singer J, et al. Clonal hematopoiesis in donors and long-term survivors of related allogeneic hematopoietic stem cell transplantation. Blood. 2020;135(18):1548-1559. [DOI] [PubMed] [Google Scholar]
- 46.Kumar R, Pereira RS, Zanetti C, et al. Specific, targetable interactions with the microenvironment influence imatinib-resistant chronic myeloid leukemia. Leukemia. 2020;34(8):2087-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nangalia J, Mitchell E, Green AR. Clonal approaches to understand the impact of mutations on hematologic disease development. Blood. 2019;133(13):1436-1455. [DOI] [PubMed] [Google Scholar]
- 48.Branford S, Kim DDH, Apperley JA, et al. Laying the foundation for genomicallybased risk assessment in chronic myeloid leukemia. Leukemia. 2019;33(8):1835-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunbar A, Rampal R, Levine R. Leukemia secondary to myeloproliferative neoplasms. Blood. 2020;136(1):61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]