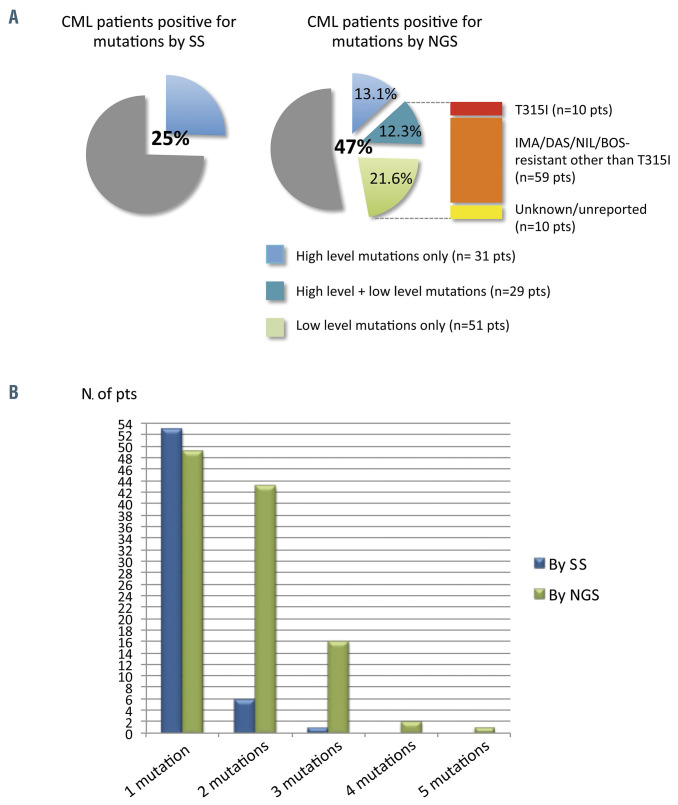
Figure 3.
Comparison between Sanger sequencing and next-generation sequencing – the NEXT-in-CML study. (A) Percentage of patients positive for mutations, as determined by Sanger sequencing (SS) and by next-generation sequencing (NGS). Among patients positive for mutations by NGS, 31 (13.1%) had high-level mutations only (≥20%; detectable by SS too); 29 (12.3%) had both ≥1 high-level mutations and ≥1 low-level mutations (≤20%; detectable by NGS only); 51 (21.6%) had only lowlevel mutations. A low-level T315I mutation was detected in ten patients; 59 additional patients had ≥1 low-level mutations known to be associated with resistance to imatinib or second-generation tyrosine kinase inhibitors other than the T315I mutation (Y253H; E255K/V; V299L; F317L/V/I/C; F359V/I/C). The remaining ten patients had only low-level mutations with an unknown resistance profile and/or not listed in the COSMIC database. (B) Patients positive for one or multiple mutations as assessed by SS versus NGS. CML: chronic myeloid leukemia; pts: patients; IMA: imatinib; DAS: dasatinib; NIL: nilotinib; BOS: bosutinib. (Adapted, with permission, from Soverini S et al.30).