Abstract
Background
The replication-competent human immunodeficiency virus (HIV) reservoir is the major barrier to cure. The quantitative viral outgrowth assay (QVOA), the gold-standard method to quantify replication-competent HIV, is resource intensive, which limits its application in large clinical trials. The intact proviral DNA assay (IPDA) requires minimal cell input relative to QVOA and quantifies both defective and intact proviral HIV DNA, the latter potentially serving as a surrogate marker for replication-competent provirus. However, there are limited cross-sectional and longitudinal data on the relationship between IPDA and QVOA measurements.
Methods
QVOA and IPDA measurements were performed on 156 resting CD4 T-cell (rCD4) samples from 83 antiretroviral therapy-suppressed HIV-positive participants. Longitudinal QVOA and IPDA measurements were performed on rCD4 from 29 of these participants.
Results
Frequencies of intact, defective, and total proviruses were positively associated with frequencies of replication-competent HIV. Longitudinally, decreases in intact proviral frequencies were strikingly similar to that of replication-competent virus in most participants. In contrast, defective proviral DNA frequencies appeared relatively stable over time in most individuals.
Conclusions
Changes in frequencies of IPDA-derived intact proviral DNA and replication-competent HIV measured by QVOA are similar. IPDA is a promising high-throughput approach to estimate changes in the frequency of the replication-competent reservoir.
Keywords: HIV, reservoir, cure, IPDA, QVOA, longitudinal, decay
Decay of the HIV reservoir in resting CD4 T cells measured using the intact proviral DNA assay closely resembles that measured using the quantitative viral outgrowth assay.
Latent, replication-competent human immunodeficiency virus (HIV) proviruses in CD4 T cells are the major barrier to HIV cure; however, measuring these viruses is challenging due to their low frequency, quiescent state, and sequence variation [1–3].
The gold-standard for the determination of replication-competent virus frequency is the quantitative viral outgrowth assay (QVOA). Although the QVOA provides high certainty that the virus recovered can replicate, it requires large cell numbers and weeks of coculture, making its adaption to large clinical trials difficult [4–6]. Further, not all replication-competent HIV is induced from a single round of T-cell activation; therefore, while QVOA is a reliable measure of the true replication-competent reservoir that captures the natural decay of the HIV reservoir over time [4, 5], it is an underrepresentation of the absolute frequency of latent infection [7, 8].
In contrast to outgrowth assays, polymerase chain reaction (PCR) measurements of HIV DNA using single-probe assays greatly overestimate the frequency of the replication-competent HIV reservoir because in participants on long-term suppressive antiretroviral therapy (ART) the majority of HIV DNA is defective [7, 9–15]. The intact proviral DNA assay (IPDA) is a novel method of HIV DNA measurement designed to quantify provirus that is likely to be intact and replication competent, and requires minimal cell input relative to the QVOA [11]. The IPDA measures HIV DNA using digital droplet PCR (ddPCR) with 2 probes: one targeting the HIV packaging signal (PS), the other targeting the env region. When amplified together within the same provirus, this probe combination excludes over 95% of defective proviruses detectable by near full-length genome sequencing [11].
To inform the use of these assays in clinical studies designed to deplete the HIV reservoir, it is critical to evaluate the relationship between IPDA and QVOA measurements in diverse cohorts and longitudinally [6]. We performed the IPDA on stored resting CD4 T cells from a cohort of 83 ART-suppressed individuals in whom QVOA was also measured. In 29 of these individuals, longitudinal IPDA and QVOA measurements were also performed.
METHODS
Study Design and Human Subjects
This study was a retrospective analysis of samples from participants from a number of study protocols. Study participants were recruited through the University of North Carolina (UNC) Global HIV Clinical Trials Unit and the UNC Center For AIDS Research (CFAR) HIV Clinical Cohort. Resting CD4 T cells from leukapheresis samples were obtained via ongoing longitudinal collection protocols approved by the UNC Institutional Review Board, and all participants provided informed consent. All participants were stably suppressed on ART at the time of donation (HIV-1 RNA < 50 copies/mL). Some participants received single or multiple 400-mg doses of vorinostat, a dendritic cell HIV RNA vaccine, ex vivo expanded lymphocyte products, or a combination thereof during the longitudinal sampling period, as previously described [16–21]. Participants who received study interventions are designated in the figures. Individuals treated during acute or early HIV infection were assigned Fiebig staging, as described previously (Supplementary Table 2) [22].
Resting CD4 T-Cell Isolation
Lymphocytes were obtained by continuous-flow leukapheresis and isolation of resting CD4+ T cells (CD25−HLADR−CD69−) was performed as described elsewhere [23, 24].
Intact Proviral DNA Assay
The IPDA was performed as originally described by Bruner and colleagues [11], with a minor PCR annealing temperature modification (detailed in the Supplementary Methods) to improve droplet separation from the negative population in individuals with likely proviral polymorphisms in the primer/probe binding regions (Supplementary Figure 1). DNA shearing index values were similar to those reported previously (median, 0.34; interquartile range [IQR], 0.32–0.35) [10–12]. A median of 9.26 × 105 (IQR, 8.13 × 105–1.05 × 106) resting CD4 T-cell equivalents (measured assuming 2 RPP30 copies per genome) were assessed per IPDA across all samples tested, consistent with other reported IPDA measurements [12]. Gating for positive droplets was set using multiple DNA elution buffer, and HIV-seronegative CD4 T-cell DNA-negative control wells run on each digital PCR plate. IPDA proviral frequencies were left-censored at 5 copies/million cells based upon the occasional detection of low-amplitude false-positive droplets in negative control wells at a rate of 0–5 copies/million cells, depending on the run (Supplementary Figure 2). A detailed protocol, primer/probe sequence information, and example digital PCR plots are provided in the Supplementary Material.
Quantitative Viral Outgrowth Assay
Recovery and quantification of replication-competent virus was performed as described elsewhere [23–25]. The number of resting CD4+ T cells in infectious units per million (IUPM) was estimated using a bias-corrected maximum likelihood method [26]. In 3 individuals, no p24-positive wells were recovered. IUPM for these individuals was calculated using an imputed 1 positive well at the top of the limiting dilution, and are designated as censored measurements in the figures.
Statistical Analysis
Association of IPDA parameters with QVOA parameters was assessed with several different approaches. First, Spearman correlations were used to assess the cross-sectional association of each IPDA parameter (intact, 5′-deleted, 3′-deleted/hypermutated, and total DNA) with the frequency of replication-competent virus (IUPM) measured by QVOA. For individuals with longitudinal sampling, the earliest time point was used for this analysis. Second, the association between QVOA and IPDA for all 156 resting CD4 T-cell (rCD4) measurements (cross-sectional and longitudinal) were assessed with linear mixed effects models. Models were fit using log10-transformed values with QVOA (IUPM rCD4) as the outcome and total, 5′-deleted, 3′-deleted/hypermutated, and intact DNA (copies per million rCD4) as the covariates of interest in separate models, with a random effect for individuals.
Longitudinal changes in QVOA and IPDA measurements were also compared. First, percent change per year in each of 5 frequency measurements (IUPM, intact, 5′-deleted, 3′-deleted/hypermutated, and total DNA) were calculated using the first and last time point available for each participant. Differences in percent change per year between each IPDA parameter and QVOA were evaluated using linear mixed effects models with percent change per year as the outcome, assay type (QVOA = 1, IPDA parameter = 0) as the covariate of interest, and a random effect for individuals. The estimated slope for the assay type in the model represents the mean difference in percent change per year values between the 2 assays.
Second, for each participant decay slopes (log10-transformed QVOA or IPDA parameter as the outcome, years of ART-suppression as the covariate of interest) for each assay were calculated using linear regression models. Decay slopes from these models for QVOA and IPDA parameters were compared using Spearman correlation. All reported P values are unadjusted for multiple testing.
RESULTS
Cohort Characteristics and Assay Performance
The IPDA and QVOA were conducted using resting CD4 T cells from 83 participants on suppressive ART. IUPM rCD4 T-cell frequencies were derived from QVOA, and the frequency of intact, 5′-deleted, and 3′-deleted/hypermutated proviruses per million rCD4 T cells were derived from IPDA. Total HIV DNA frequency per million rCD4 T cells was also estimated using the sum of intact, 5′-deleted, and 3′-deleted/hypermutated frequency measurements, with the caveat that proviruses with both 5′- and 3′-defects will be missed (estimated 4% of proviruses) [12].
For 7 of 83 individuals in the cohort, there was an IPDA PCR amplification failure for either the PS (3 individuals) or env probe (4 individuals), likely due to proviral polymorphisms in the primer/probe binding regions (Supplementary Figure 2). This represents an overall amplicon failure rate of 8.4%, consistent with recent reports of amplicon failure rates in large cohorts of individuals with subtype B infection [12]. Individuals with amplicon failures were excluded from the analyses.
Of the remaining 76 individuals, 16 participants were treated during acute HIV infection (AHI) or early HIV infection (Fiebig stages III–VI; Supplementary Table 2) with a median ART-suppression time of 0.81 years (IQR, 0.77–0.87) at the first time point of sampling. Sixty individuals were treated during chronic HIV infection (CHI), with a median ART-suppression time of 4.4 years (IQR, 2.7–7.0) at the first time point of sampling (Table 1). Some participants received study interventions designed to modulate the reservoir (vorinostat, HIV-expanded T cells, and/or an autologous dendritic cell HIV RNA vaccine) [16–21]. No study intervention had a significant impact on viral outgrowth frequencies, as previously reported [16–21].
Table 1.
Participant Demographic and Clinical Characteristics
| Characteristic | ART Initiation During Acute or Early HIV Infection | ART Initiation During Chronic HIV Infection |
|---|---|---|
| Sample size | 16 | 60 |
| Race/ethnicity, No. (%) | ||
| Black | 7 (44) | 29 (48) |
| Non-Hispanic white | 8 (50) | 27 (45) |
| Hispanic white | 1 (6) | 2 (3) |
| Other | 0 (0) | 3 (5) |
| Biological sex, No. (%) | ||
| Male | 16 (100) | 39 (65) |
| Female | 0 (0) | 21 (35) |
| Age, y, median (IQR) | 32 (27–46) | 49 (37–54) |
| Years on therapy, median (IQR) | 0.95 (0.89–0.99) | 7.06 (3.97–15.60) |
| Years suppressed, median (IQR) | 0.81 (0.77–0.87) | 4.41 (2.69–6.96) |
| Current CD4, cells/μL, median (IQR) | 867 (586–1068) | 742 (635–942) |
| Estimated Pre-ART nadir CD4a (IQR) | 515 (314–667) | 257 (25–464) |
| No. missing data points | 1 | 11 |
Abbreviations: ART, antiretroviral therapy; HIV, human immunodeficiency virus; IQR, interquartile range.
aFor some participants, insufficient CD4 count information was available to estimate pre-ART nadir CD4; the number of missing CD4 nadir estimates is indicated.
For 29 participants (9 treated during AHI/early HIV infection, 20 treated during CHI), longitudinal QVOA and IPDA measurements were conducted. For individuals with longitudinal sampling, the earliest time point available was used for the cross-sectional analysis and for the assessment of demographic and clinical characteristics (Table 1).
Cross-sectional Comparison of QVOA and IPDA Measurements
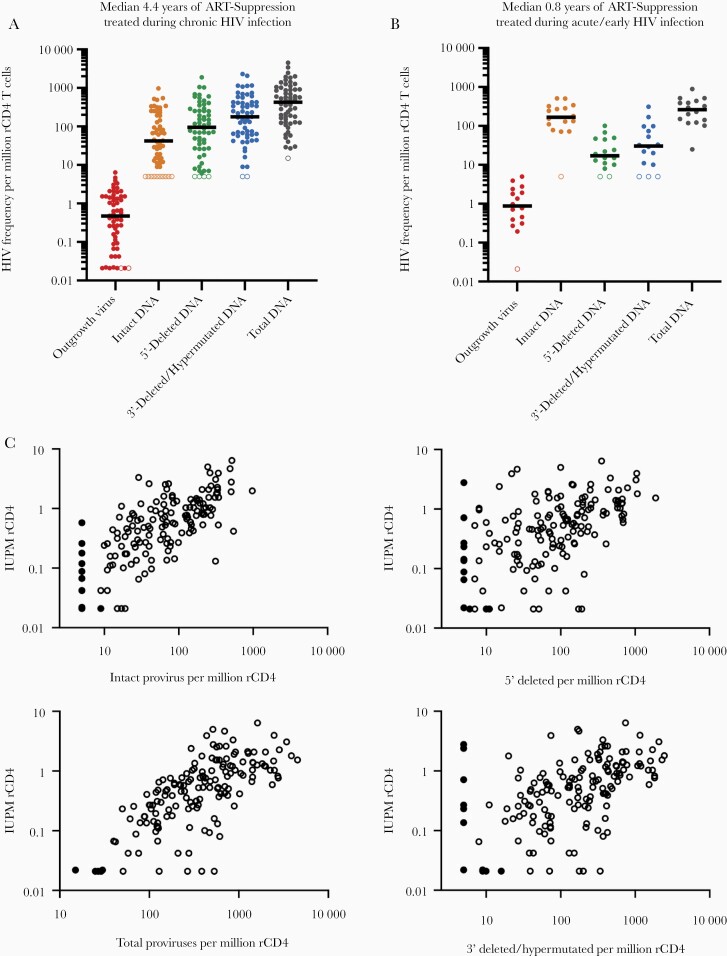
We first compared proviral DNA and outgrowth virus frequencies within individuals treated in either CHI (Figure 1A) or in AHI/early HIV infection (Figure 1B). For individuals treated in CHI, outgrowth virus was recovered at the lowest frequencies (median, 0.471 IUPM rCD4 cells), followed by intact HIV DNA (median, 42 copies/million rCD4 cells), 5′-deleted HIV DNA (median, 95 copies/million), 3′-deleted/hypermutated HIV DNA (median, 177 copies/million rCD4 cells), and total HIV DNA (median, 428 copies/million rCD4 cells) (Figure 1A). Intact DNA made up a median of 11.6% (IQR, 6.2%–27.8%) of total HIV proviruses in this cohort of CHI-treated individuals.
Figure 1.
Comparison of IPDA and QVOA HIV reservoir measurements. A, Frequency per million rCD4 T cells for each assay for individuals treated (A) during CHI on long-term (median 4.4 years) suppressive ART; and (B) during acute or early HIV infection on short-term (median 0.8 years) suppressive ART. Left-censored values are indicated with open symbols for each assay parameter. A,B, Black bars represent median values. C, Relationship between IPDA parameters and QVOA-derived IUPM for 156 resting CD4 T-cell samples from 76 individuals. Black, closed circles represent left-censored values for an IPDA and/or QVOA parameter. Mixed effects models of log10(IUPM) indicated associations between QVOA and log10(intact DNA) (estimated slope, 0.74; 95% CI, .64–.84), log10(total DNA) (estimated slope, 0.97; 95% CI, .80–1.14), log10(3′ deleted/hypermutated DNA) (estimated slope, 0.42; 95% CI, .24–.60), and log10(5′-deleted DNA) (estimated slope, 0.34; 95% CI, .17–.52). Abbreviations: ART, antiretroviral therapy; CHI, chronic HIV infection; CI, confidence interval; HIV, human immunodeficiency virus; IPDA, intact proviral DNA assay; IUPM, infectious units per million; QVOA, quantitative viral outgrowth assay; rCD4, resting CD4.
Interestingly, when IPDA and QVOA measurements were performed on samples from AHI/early treated participants after a median of 0.8 years on suppressive ART, a different pattern emerged: intact DNA comprised a much larger portion of the reservoir (median, 177 copies/million rCD4 cells), while 5′-deleted (median, 17 copies/million rCD4 cells) and 3′-deleted/hypermutated (median, 31 copies/million rCD4 cells) DNA species were markedly less frequent (Figure 1B).
Intact, Defective, and Total HIV DNA Frequency Measurements from the IPDA are Positively Associated with QVOA Measurements
Next, the correlation between IPDA parameters (intact, 5′-deleted, 3′-deleted/hypermutated, and total HIV DNA per million rCD4 T cells) and QVOA (outgrowth virus, IUPM rCD4 T cells) was assessed using Spearman correlations for all independent measurements (n = 76 individuals at the earliest time point of sampling). As previously reported in an overlapping cohort, all IPDA parameters positively associated with QVOA measurements (Supplementary Figure 3) [25].
In addition, in a linear mixed model analyses of 156 observations (see “Methods”) all IPDA parameter frequencies were positively associated with IUPM measured using QVOA (Figure 1C). Intact and total HIV DNA appeared to be better predictors of viral outgrowth frequencies than measures of defective HIV DNA frequency, as expected given the replication deficiency of defective DNA (Figure 1C).
Changes in Intact HIV DNA Frequency Closely Resemble Changes in Viral Outgrowth Frequency for AHI/Early Treated Participants
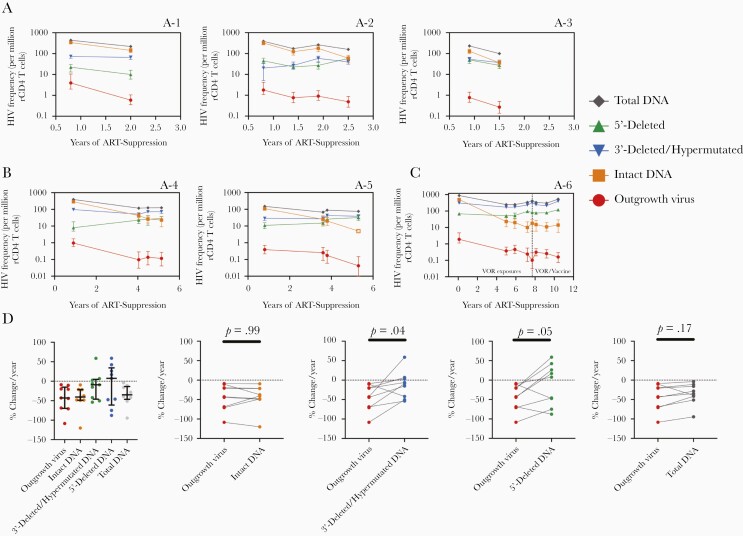
Longitudinal QVOA and IPDA measurements were performed in 9 AHI/early treated participants. For these participants, longitudinal sampling spanned a median of 1.3 (IQR, 1.0–4.2) years with a median of 2 (IQR, 2–4) time points per participant.
In 6 participants (A-1 to A-3 Figure 2A and A-7 to A-9, Supplementary Figure 4) with sampling from 0.5 to 3 years post ART suppression, decreases in intact proviral DNA frequency paralleled decreases in outgrowth virus frequency over time. In contrast, changes in defective or total HIV DNA frequencies were more variable, with apparent increases and decreases over time depending on the individual and defective proviral species (Figure 2A and Supplementary Figure 4).
Figure 2.
Longitudinal comparison of IPDA and QVOA measurements in participants treated during acute/early HIV infection. A-1 to A-6 indicates individual participants that were treated during acute/early HIV infection. Changes in HIV reservoir frequency over time for IPDA and QVOA measurements in (A) 3 participants with sampling 0.5–2.5 years post-ART suppression; (B) 2 participants with sampling 0.5–6 years post-ART suppression; and (C) 1 participant with sampling over 11 years of ART suppression. Open symbols indicate left-censored values. Time periods when participant received a study intervention (VOR and/or an autologous dendritic cell HIV vaccine as specified in the graph) are indicated by dotted line. Errors bars for QVOA represent 95% CIs from the maximum likelihood method [26]. Errors bars for IPDA measurements represent 95% CIs for the proviral frequency estimates using the total error from the QuantaSoft analysis software, which takes into account technical and subsampling error associated with ddPCR [27]. D, Percent change per year for each HIV reservoir frequency for all acute/early treated individuals (n = 9). Black lines represent the median for each parameter with the IQR. Mean difference between each IPDA measure and QVOA was assessed using linear mixed effect models (see “Methods”). Estimate mean differences versus QVOA were −0.1 (95% CI, −11.3 to 11.2) for intact DNA, −31.3 (95% CI, −56.1 to −6.6) for 3′-deleted/hypermutated DNA, −32.1 (95% CI, −59.4 to −4.9) for 5′-deleted DNA, and −8.1 (95% CI, −18.7 to 2.4) for total DNA. Abbreviations: ART, antiretroviral therapy; CI, confidence interval; ddPCR, digital droplet polymerase chain reaction; HIV, human immunodeficiency virus; IPDA, intact proviral DNA assay; IQR, interquartile range; QVOA, quantitative viral outgrowth assay; rCD4, resting CD4; VOR, vorinostat.
Two AHI/early treated participants (A-4 and A-5) had sampling from 0.5 to 6 years after ART suppression (Figure 2B). Intact DNA and viral outgrowth frequencies decreased markedly, while defective DNA species remained relatively stable over time. In an AHI/early treated individual (A-6) with sampling from over 11 years of ART suppression, intact and viral outgrowth frequencies decreased markedly, while defective and total DNA species appeared to expand (Figure 2C). Taken together, these data suggest that intact proviral DNA selectively decays over years of ART suppression in AHI/early treated individuals.
To test the longitudinal association between QVOA and IPDA parameters in AHI/early treated individuals, percent change per year for each IPDA parameter was compared to QVOA. Percent change for intact proviral DNA frequencies most closely resembled the percentage change observed with QVOA. In contrast, defective and total DNA frequencies appeared to decrease at a slower rate relative to viral outgrowth frequencies in the majority of individuals (Figure 2D).
Changes in Intact HIV DNA Frequency Closely Resemble Changes in Viral Outgrowth Frequency for Participants Treated During Chronic HIV Infection
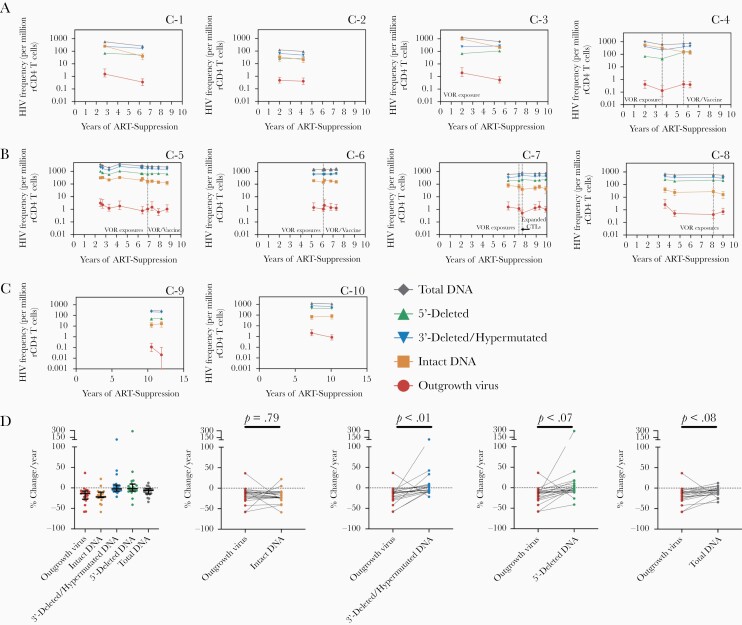
QVOA and IPDA measurements were also performed longitudinally in 20 CHI-treated participants. Longitudinal sampling spanned a median of 2.9 (IQR, 2.1–3.6) years with a median of 4 (IQR, 2–6) time points per participant.
The direction of change from the first to last time point of sampling for viral outgrowth and intact proviral DNA frequencies was the same for 17/20 participants (Figure 3 and Supplementary Figure 5). In addition, changes in intact proviral frequencies were generally concordant with changes in IUPM on a time point to time point basis. Of all the IPDA parameters (intact, 5′-deleted, 3′-deleted/hypermutated, and total DNA), intact DNA frequencies most closely resembled changes in outgrowth frequencies in individuals suppressed both during the first 5 years of ART suppression (Figure 3A and Supplementary Figure 5) and during very long-term (5–10 years) suppression (Figure 3B and Supplementary Figure 5). In 3/20 participants, the direction of intact proviral DNA frequency changes were opposite that of changes in viral outgrowth frequencies (Figure 3C and Supplementary Figure 5), which may be explained by assay variance. For instance, in 2 participants on very long-term therapy (C-9 and C-10; Figure 3C), intact DNA frequency was stable whereas QVOA declined; however, the decline in QVOA was within the previously reported occasional 6-fold longitudinal variance in the assay [4].
Figure 3.
Longitudinal comparison of IPDA and QVOA measurements in participants treated during chronic HIV Infection. C-1 to C-10 indicates individual participants that were treated during chronic HIV infection. Changes in HIV reservoir frequency over time for IPDA and QVOA measurements in (A) 4 participants with sampling during the first 5 years post-ART suppression; (B) 4 participants with sampling following 5–10 years post-ART suppression; and (C) participants where the direction of change for intact DNA and QVOA were not concordant. Time periods when participants received a study intervention (VOR, CTLs, and/or an autologous dendritic cell HIV vaccine as specified in the graphs) are indicated by dotted lines. Errors bars for QVOA represent the 95% CIs from the maximum likelihood method [26]. Errors bars for IPDA measurements represent 95% CIs for the proviral frequency estimates using the total error from the QuantaSoft analysis software, which takes into account technical and subsampling error associated with ddPCR [27]. D, Percent change per year for each HIV reservoir frequency for all individuals treated during CHI (n = 20). Black lines represent the median for each parameter with the IQR. Mean difference between each IPDA measure and QVOA was assessed using a linear mixed effects model (see “Methods”). Estimate mean differences versus QVOA were 1.6 (95% CI, −10.2 to 13.4) for intact DNA, −23.4 (95% CI, −38.2 to −8.5) for 3′-deleted/hypermutated DNA, −31.0 (95% CI, −62.1 to 0.2) for 5′-deleted DNA, and −9.3 (95% CI, −19.1 to 0.4) for total DNA. Abbreviations: CTLs, expanded cytotoxic lymphocytes; ART, antiretroviral therapy; CI, confidence interval; ddPCR, digital droplet polymerase chain reaction; HIV, human immunodeficiency virus; IPDA, intact proviral DNA assay; IQR, interquartile range; QVOA, quantitative viral outgrowth assay; rCD4, resting CD4; VOR, vorinostat.
Assessment of percent change per year of ART-suppression for each assay demonstrated that percent change for intact proviral DNA frequencies closely matched the percent change observed with QVOA for CHI participants (Figure 3D). Percent change values for intact proviral DNA frequencies did not appear to be different from viral outgrowth frequencies. In contrast, the rate of change was lower for 3′-deleted/hypermutated and 5′-deleted proviral frequencies relative to viral outgrowth frequencies. Total DNA appeared to decrease more quickly than defective species but still decreased more slowly than viral outgrowth (Figure 3D).
Finally, as an additional metric to assess the relationship between longitudinal changes in QVOA and changes in IPDA parameters, QVOA- and IPDA-decay slopes were calculated using linear regression models for all participants, and then compared using Spearman correlation. Slopes for intact DNA and QVOA correlated (Spearman r = 0.47, P = .01) whereas slopes for QVOA and defective DNA did not (Spearman r = 0.21, P = .28 for 5′-deleted and Spearman r = 0.20, P = .31 for 3′-deleted/hypermutated DNA; Supplementary Figure 6). Consistent with previous reports, intact DNA rate of change did not correlate with rate of change for 3′- or 5′-defective DNA [10].
DISCUSSION
To inform on the use of IPDA and QVOA in clinical studies designed to characterize or deplete the HIV reservoir, this study assessed the relationship between IPDA and QVOA measurements in ART-suppressed individuals in cross-sectional and longitudinal analyses.
In the cross-sectional analysis, intact proviral DNA frequency positively associated with the frequency of replication-competent HIV measured by QVOA in resting CD4 T cells from a cohort of 76 ART-suppressed individuals. This is consistent with data from a separate cohort in which Bruner and colleagues reported a positive association of IPDA with QVOA measurements in CD4 T cells [11]. In CHI-treated participants on a median of 4.4 years of suppressive ART, frequencies and patterns of intact, 5′-deleted, 3′-deleted/hypermutated, and total proviruses for CHI-treated participants were similar to those observed in other cohorts, both in absolute number and in relationship to one another (QVOA < intact DNA < 5′-deleted DNA ≈ 3′-deleted/hypermutated DNA < total DNA) [10–12]. In addition, for CHI-treated participants the median percentage of total proviruses that were intact in this study (11.6%) was similar to other reports of ART-suppressed CHI-treated individuals (mean 8% in [12], median 9.8% in [28]).
To date, no study has assessed the longitudinal relationship between IPDA and QVOA measurements in CHI-treated participants. This is a critical evaluation if the IPDA is to be used to evaluate interventions seeking to reduce the frequency of persistent infection. We observed that percent changes in QVOA frequencies were similar to percent changes in intact HIV DNA, in contrast to defective DNA species which changed at slower rate or in some cases appeared to expand. This is consistent with recent studies reporting that intact proviral DNA decays more rapidly than defective or total proviral DNA [10, 28–31]. This study confirms and extends those findings to demonstrate that outgrowth virus decreases at a similar proportional rate to intact DNA measured in the same samples, and at a slower proportional rate to defective and total HIV DNA. Defining the mechanisms underlying the selective decay of intact proviruses is a critical endeavor to inform cure strategies [28].
This study also incorporated a subset of 16 individuals treated during AHI/early HIV infection with sampling at a median of 0.81 (IQR, 0.77–0.87) years of ART suppression. Interestingly, for AHI/early treated individuals, we observed that intact proviruses made up a much larger fraction of the total proviruses at these early time points compared to CHI-treated individuals on several years of suppressive ART. There are several potential explanations for this phenomenon, which are not mutually exclusive. First, this may reflect higher starting levels of intact DNA in AHI/early treated versus CHI-treated individuals that selectively decay with longer times of ART suppression [10, 28, 32]. Supporting this model, analysis of longitudinal IPDA data from AHI/early treated individuals in this study revealed that defective proviruses became more predominant with longer times of viral suppression. This is consistent with reports of high proportions of intact DNA in AHI-treated participants sampled immediately following ART suppression [32], but low proportions of intact DNA in AHI-treated participants sampled predominantly years after ART suppression [9].
Second, the IPDA probes were designed using sequences from predominantly CHI-treated individuals, and their ability to identify full-length intact proviruses may differ in AHI/early treated participants [11]. There is some evidence of differential rates of proviral hypermutation and deletion in AHI/early treated individuals that could potentially affect the performance of the IPDA [9]. However, despite probe design based on sequences mostly from CHI-treated individuals, the IPDA still appears to measure a selective decay of intact proviruses that tracks with QVOA over time in AHI/early treated individuals. Additional work is clearly needed to elucidate the dynamics of the intact versus defective HIV reservoir immediately following ART suppression and further explore the utility of the IPDA in AHI/early treated participants.
This study has several limitations. First, longitudinal sampling was limited in terms of the time range of sampling and in number of participants sampled (n = 20 CHI-treated and n = 9 AHI/early treated). Second, many individuals received interventions designed to perturb or deplete the HIV reservoir. Receipt of study interventions in some participants precluded a formal assessment of natural reservoir decay. Third, neither the IPDA nor QVOA are able to capture clonal proliferation of infected cells, which heavily influences reservoir dynamics during suppressive ART [33]. Longitudinal evaluations of HIV reservoir clonality will depend on near full-length genome sequencing approaches with integration site analysis. Fourth, unintegrated PS+env+ 2-long terminal repeat (2-LTR) circles may have contributed to IPDA measurements at some of the earlier time points examined for AHI/early treated individuals in this study [34]. However, the contribution of 2-LTR circles to IPDA measurements for individuals suppressed for greater than 1 year is likely minimal, as most studies indicate 2-LTR circles rapidly decay upon ART initiation and are either undetectable or represent a minority of total HIV DNA following 1 year of ART [9, 35, 36].
Further, the IPDA is a quantitative, high-throughput assay that requires minimal cell input to estimate the frequency of HIV DNA that is likely to be intact; however, it does not provide sequence confirmation of intactness for proviruses positive for the PS and env probes. Therefore, another limitation of the current study is that the proportion of proviruses called intact by the IPDA probes may have defects elsewhere in the genome that render them replication defective. This is estimated to occur for approximately 30% of PS+env+ proviruses [11]. It is conceivable that this percentage may vary across individuals, though sequencing of PS+env+ proviruses in droplets is necessary to confirm this limitation [13]. Near full-length genome sequencing approaches for reservoir measurement, compared to IPDA, have a lower misidentification rate of defective proviruses as intact because they provide near–full-length sequence information. However, they are currently lower-throughput and may be limited in quantitative accuracy by long-range PCR efficiency [6, 10, 12]. Misidentification of defective proviruses as intact may partially explain why IPDA-intact proviral frequencies are approximately 50- to 100-fold higher than QVOA measurements in this study and others [11, 12]. Proviruses that are difficult to induce ex vivo also contribute significantly to this difference [7, 8]. Nonetheless, the intact proviral frequency estimate from the IPDA provides a much more accurate upper limit on the frequency of replication-competent virus than single-probe assays, and appears to track well with changes in QVOA over time.
Overall, this study adds to the growing body of evidence that intact provirus selectively decays [10, 28–31]. Importantly, percent decreases in intact, but not defective, proviral frequencies were similar to percent decreases in QVOA measurements tracked over time in the same samples. These findings lend strong support for the use of the IPDA in the arsenal of assays to assess potential replication-competent reservoir depletions in clinical studies designed to deplete the HIV reservoir.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the participants who made this study possible. We thank Y. Park, M. Cohen, M. Napier, S. Peterson, and the staff of the University of North Carolina (UNC) HIV Cure Center, UNC Blood Bank, and UNC Clinical and Translational Research Center (CTRC) for clinical support.
Disclaimer . The contents of this publication are solely the responsibility of the authors and do not represent the official views of the National Institutes of Health.
Financial support. This work was supported by the National Institutes of Health (grant numbers UM1AI126619 to D. M. M., R01AI134363 to N. M. A., P30-AI050410 to J. J. E. and M. G. H., and F30AI145588 to S. D. F.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: Conference on Retroviruses and Opportunistic Infections, 8–11 March 2020, Boston, MA.
References
- 1. Wang Z, Simonetti FR, Siliciano RF, Laird GM. Measuring replication competent HIV-1: advances and challenges in defining the latent reservoir. Retrovirology 2018; 15:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Falcinelli SD, Ceriani C, Margolis DM, Archin NM. New frontiers in measuring and characterizing the HIV reservoir. Front Microbiol 2019; 10:2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Margolis DM, Archin NM, Cohen MS, et al. Curing HIV: seeking to target and clear persistent infection. Cell 2020; 181:189–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Crooks AM, Bateson R, Cope AB, et al. Precise quantitation of the latent HIV-1 reservoir: implications for eradication strategies. J Infect Dis 2015; 212:1361–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Siliciano JD, Kajdas J, Finzi D, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 2003; 9:727–8. [DOI] [PubMed] [Google Scholar]
- 6. Abdel-Mohsen M, Richman D, Siliciano RF, et al. ; BEAT-HIV Delaney Collaboratory to Cure HIV-1 Infection . Recommendations for measuring HIV reservoir size in cure-directed clinical trials. Nat Med 2020; 26:1339–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ho YC, Shan L, Hosmane NN, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013; 155:540–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hosmane NN, Kwon KJ, Bruner KM, et al. Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: potential role in latent reservoir dynamics. J Exp Med 2017; 214:959–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bruner KM, Murray AJ, Pollack RA, et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 2016; 22:1043–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peluso MJ, Bacchetti P, Ritter KD, et al. Differential decay of intact and defective proviral DNA in HIV-1-infected individuals on suppressive antiretroviral therapy. JCI Insight 2020; 5:e132997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bruner KM, Wang Z, Simonetti FR, et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 2019; 566:120–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Simonetti FR, White JA, Tumiotto C, et al. Intact proviral DNA assay analysis of large cohorts of people with HIV provides a benchmark for the frequency and composition of persistent proviral DNA. Proc Natl Acad Sci U S A 2020; 117:18692–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gaebler C, Lorenzi JCC, Oliveira TY, et al. Combination of quadruplex qPCR and next-generation sequencing for qualitative and quantitative analysis of the HIV-1 latent reservoir. J Exp Med 2019; 216:2253–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Imamichi H, Dewar RL, Adelsberger JW, et al. Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc Natl Acad Sci U S A 2016; 113:8783–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hiener B, Horsburgh BA, Eden JS, et al. Identification of genetically intact HIV-1 proviruses in specific CD4+ T cells from effectively treated participants. Cell Rep 2017; 21:813–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gay CL, Kuruc JD, Falcinelli SD, et al. Assessing the impact of AGS-004, a dendritic cell-based immunotherapy, and vorinostat on persistent HIV-1 infection. Sci Rep 2020; 10:5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Archin NM, Liberty AL, Kashuba AD, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 2012; 487:482–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Archin NM, Kirchherr JL, Sung JA, et al. Interval dosing with the HDAC inhibitor vorinostat effectively reverses HIV latency. J Clin Invest 2017; 127:3126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Archin NM, Bateson R, Tripathy MK, et al. HIV-1 expression within resting CD4+ T cells after multiple doses of vorinostat. J Infect Dis 2014; 210:728–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sung JA, Lam S, Garrido C, et al. Expanded cytotoxic T-cell lymphocytes target the latent HIV reservoir. J Infect Dis 2015; 212:258–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sung JA, Patel S, Clohosey ML, et al. HIV-specific, ex vivo expanded T cell therapy: feasibility, safety, and efficacy in ART-suppressed HIV-infected individuals. Mol Ther 2018; 26:2496–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stekler JD, Tapia K, Maenza J, et al. No time to delay! Fiebig stages and referral in acute HIV infection: Seattle primary infection program experience. AIDS Res Hum Retroviruses 2018; 34:657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Archin NM, Eron JJ, Palmer S, et al. Valproic acid without intensified antiviral therapy has limited impact on persistent HIV infection of resting CD4+ T cells. AIDS 2008; 22:1131–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lehrman G, Hogue IB, Palmer S, et al. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet 2005; 366:549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Falcinelli SD, Shook-Sa BE, Dewey MG, et al. Impact of biological sex on immune activation and frequency of the latent HIV reservoir during suppressive antiretroviral therapy. J Infect Dis 2020; 222:1843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trumble IM, Allmon AG, Archin NM, et al. SLDAssay: a software package and web tool for analyzing limiting dilution assays. J Immunol Methods 2017; 450:10–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tzonev S. fundamentals of counting statistics in digital PCR: I just measured two target copies—what does it mean? Methods Mol Biol 2018; 1768:25–43. [DOI] [PubMed] [Google Scholar]
- 28. Gandhi RT, Cyktor JC, Bosch RJ, et al. Selective decay of intact HIV-1 proviral DNA on antiretroviral therapy [published online ahead of print 21 August 2020]. J Infect Dis 2021;223(2):225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Antar AA, Jenike KM, Jang S, et al. Longitudinal study reveals HIV-1-infected CD4+ T cell dynamics during long-term antiretroviral therapy. J Clin Invest 2020; 130:3543–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garcia-Broncano P, Maddali S, Einkauf KB, et al. Early antiretroviral therapy in neonates with HIV-1 infection restricts viral reservoir size and induces a distinct innate immune profile. Sci Transl Med 2019; 11:eaax7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pinzone MR, VanBelzen DJ, Weissman S, et al. Longitudinal HIV sequencing reveals reservoir expression leading to decay which is obscured by clonal expansion. Nat Commun 2019; 10:728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee GQ, Reddy K, Einkauf KB, et al. HIV-1 DNA sequence diversity and evolution during acute subtype C infection. Nat Commun 2019; 10:2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cohn LB, Chomont N, Deeks SG. The biology of the HIV-1 latent reservoir and implications for cure strategies. Cell Host Microbe 2020; 27:519–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pinzone MR, Bertuccio MP, VanBelzen DJ, Zurakowski R, O’Doherty U. Next-generation sequencing in a direct model of HIV infection reveals important parallels to and differences from in vivo reservoir dynamics. J Virol 2020; 94:e01900-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Besson GJ, Lalama CM, Bosch RJ, et al. HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin Infect Dis 2014; 59:1312–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ananworanich J, Chomont N, Eller LA, et al. ; RV217 and RV254/SEARCH010 Study Groups . HIV DNA set point is rapidly established in acute HIV infection and dramatically reduced by early ART. EBioMedicine 2016; 11:68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.