ABSTRACT
Ebola virus (EBOV) is a negative single-stranded RNA virus within the Filoviridae family and the causative agent of Ebola virus disease (EVD). Nonhuman primates (NHPs), including cynomolgus and rhesus macaques, are considered the gold standard animal model to interrogate mechanisms of EBOV pathogenesis. However, despite significant genetic similarity (>90%), NHP species display different clinical presentation following EBOV infection, notably a ∼1–2 days delay in disease progression. Consequently, evaluation of therapeutics is generally conducted in rhesus macaques, whereas cynomolgus macaques are utilized to determine efficacy of preventative treatments, notably vaccines. This observation is in line with reported differences in disease severity and host responses between these two NHP following infection with simian varicella virus, influenza A and SARS-CoV-2. However, the molecular underpinnings of these differential outcomes following viral infections remain poorly defined. In this study, we compared published transcriptional profiles obtained from cynomolgus and rhesus macaques infected with the EBOV-Makona Guinea C07 using bivariate and regression analyses to elucidate differences in host responses. We report the presence of a shared core of differentially expressed genes (DEGs) reflecting EVD pathology, including aberrant inflammation, lymphopenia, and coagulopathy. However, the magnitudes of change differed between the two macaque species. These findings suggest that the differential clinical presentation of EVD in these two species is mediated by altered transcriptional responses.
KEYWORDS: EBOV, transcriptomics, nonhuman primate, NHP, immunity, longitudinal
Introduction
Ebola virus (EBOV) is a single-stranded RNA virus and a member of the Filoviridae family. Infection with EBOV causes Ebola virus disease (EVD), which is characterized by excessive inflammation, coagulopathy, lymphopenia, and apoptosis that ultimately results in organ failure and death [1]. In the absence of antivirals and vaccines, case fatality rates range from 40% to 90% [1–3]. The recent 2013–2016 West Africa epidemic that incurred over 28,000 reported infections and the recent outbreaks of EBOV variants in the Democratic Republic of the Congo demonstrate a continued need for ongoing research into the mechanisms of EBOV pathogenesis [3–5].
EBOV preferentially infects antigen presenting cells notably dendritic cells (DCs) and monocytes/macrophages, which are believed to be major drivers of pathology [6,7]. Infection of monocytes/macrophages contributes to the massive induction of pro-inflammatory cytokines and chemokines that recruit additional myeloid cells to the site of infection, promote neutrophil-mediated immunity, and contribute to lymphocyte apoptosis [8–10]. In contrast, infection of DCs results in reduced expression of co-stimulatory molecules and impaired antigen presentation which in turn disrupts the mobilization of adaptive immunity [10–13]. This includes T cell-mediated and humoral immunity, both which are critical for controlling infection and conferring vaccine-mediated protection [2,14,15]. Fatal EVD cases are associated with a robust and sustained adaptive systemic cytokine storm and dramatic loss of lymphocytes, while survivors show early control of innate and immune responses [16–21].
Nonhuman primates (NHPs) are the gold standard animal model for EBOV pathogenesis studies as they accurately recapitulate pathobiology of EVD observed in humans [22–24]. Historically, cynomolgus macaques have been used to study response to EBOV infection and assess vaccine efficacy, while rhesus macaques are primarily utilized in therapeutic studies [25]. Both species are equally susceptible to EBOV infection; however, differences in the duration/severity of EVD and histopathological presentation exist. Importantly, time to euthanasia following infection with a lethal dose of EBOV-Mayinga or EBOV-Makona is shorter in cynomolgus macaques (5-6 days) compared to rhesus macaques (∼8 days) [26–29]. This is accompanied by earlier changes in inflammatory mediators, earlier onset of viremia and greater severity of cellular stress and coagulopathy (e.g. elevated BUN, CRE, and AST; reduced PLT) in rhesus macaques [26–29]. Interestingly, viral titres in lymphoid organs at the time of euthanasia are comparable between the two species despite the delayed viremia [26,27]. Other studies reported small variations in the kinetics of blood and serum markers of EVD between the two macaque species although extensive comparisons have not been conducted [27,28,30]. These differences in disease progression are not unexpected given that rhesus and cynomolgus macaques exhibit stark differences following infection with influenza A virus (IAV), severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), simian immunodeficiency virus (SIV), and simian varicella virus (SVV)[31–41]. However, no study to date has compared the host transcriptional responses to EBOV infection in these two species to elucidate molecular mechanisms explaining differences in disease pathogenesis.
To address this gap in knowledge, we compared transcriptional signatures in whole blood samples obtained from cynomolgus and rhesus macaques following infection with EBOV-Makona Guinea C07 infection. The EBOV-Makona C07 isolate was obtained early in the 2013–2016 West Africa EBOV epidemic [26,27]. Our analysis determined a shared core of antiviral, innate immune and adaptive immune genes induced in both species during infection that reflected canonical EVD pathology. In addition, we identified a more robust expression of genes that play roles in inflammation, apoptosis, coagulopathy and regulation of T lymphocytes in rhesus macaques. In contrast, expression of genes related to B cell-mediated immunity were more frequently dysregulated in cynomolgus macaques.
Materials and methods
Study cohorts
Historical whole blood RNA samples from rhesus macaques infected with Makona C07 (n = 3), as described in [26], were obtained from Dr. Marzi. Complete description of this study can be found in the original publication [26]. Briefly, whole blood samples were collected at days 0, 4 and 6 from rhesus macaques (n = 3) following intramuscular inoculation with 1,000 plaque-forming units (PFU) of EBOV-Makona C07 divided equally between both caudal thighs in the maximum containment laboratory at the Rocky Mountain Laboratories (RML), Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. RNA was extracted using the QIAmp viral Mini RNA kit (Qiagen) and shipped to the Messaoudi Laboratory [26]. Clinical signs of disease as well as blood chemistry values were reported in [26].
RNA sequencing data were obtained from our previous study ([29], PRJNA398558). Similar to the study with rhesus macaques, whole blood samples were collected at days 0, 4 and 6 from cynomolgus macaques (n = 5) following intramuscular inoculation with 1,000 PFU of EBOV-Makona Guinea C07 divided equally between both quadriceps. This study was conducted in the BSL4 at the University of Texas Medical Branch at Galveston (UTMB). Clinical signs of disease as well as blood chemistry values were reported in [29].
Both RML and UTMB studies obtained EBOV-Makona Guinea C07 from the same source (PMID24738640) and passaged only 1–2 times on Vero cells. Both labs confirmed the sequence of the virus before use and determined identicality with the published sequence (KJ660347).
Both RML (OLAW# A4149-01) and UTMB (IBC# 201-1581, OLAW# A3314-01) are AAALAC accredited institutions. All procedures followed standard operating procedures (SOPs) approved by the respective Institutional Biosafety Committee (IBC). Animal work was performed in strict accordance with the recommendations described in the Guide for the Care and Use of Laboratory Animals of the National Institute of Health, the Office of Animal Welfare and the Animal Welfare Act, United States Department of Agriculture. The studies were approved by the Institutional Animal Care and Use Committees (IACUC). Procedures were conducted in animals anesthetized by trained personnel under the supervision of veterinary staff. The humane endpoint criteria for euthanasia were specified and approved by the IACUC. All efforts were made to ameliorate animal welfare and minimize animal suffering in accordance with the Weatherall report on the use of nonhuman primates in research (https://royalsociety.org/policy/publications/2006/weatherall-report/).
Library generation, sequencing, and Bioinformatic analysis
Quality and quantity of whole blood RNA obtained from rhesus macaques were determined using an Agilent 2100 Bioanalyzer. cDNA libraries were constructed using the TruSeq stranded total RNA LT-LS kit. RNA was treated with RNase H and DNase I following depletion of ribosomal RNA (rRNA). Adapters were ligated to cDNA products and the subsequent ∼300 base pair (bp) amplicons were PCR-amplified and selected by size exclusion. Quality and concentration of the cDNA libraries were assessed using Agilent 2100 Bioanalyzer and subjected to 150 bp single-end sequencing using the Illumina NovaSeq platform.
Preliminary data analysis was performed with RNA-Seq workflow module of systemPipeR, developed by Backman et. al. RNA-Seq reads were demultiplexed, quality-filtered and trimmed using Trim Galore (average Phred score cut-off of 30, minimum length of 50 bp). FastQC was used to generate quality reports. Reads were aligned using hisat2 to the reference genome Macaca mulatta (Macaca_mulatta.Mmul_8.0.1.dna.toplevel.fa) and the Macaca_mulatta.Mmul_8.0.1.97.gtf was used for annotation. Raw expression values (gene-level read counts) were generated using the summarizeOverlaps function and normalized (read per kilobase of transcript per million mapped reads, rpkm) using the edgeR package. Statistical analysis with edgeR was used to determine DEGs meeting the following criteria: genes with median rpkm of ≥5, a FDR corrected p-value ≤ 0.05 and a log2fold change ≥ 1 compared to day 0.
Construction, sequencing and bioinformatics analysis of cDNA libraires from blood samples obtained from cynomolgus macaques were previously described in detail in [29]. Briefly, cDNA libraries were constructed using the TruSeq stranded total RNA LT-LS kit and subjected to 100 bp (rhesus) single-end sequencing using the Illumina NextSeq platform. This data set is available through NCBI SRA under project PRJNA398558. Analysis was carried out exactly as described above with the exception of using reference genome Macaca fascicularis (Macaca_fascicularis.Macaca_fascicularis_5.0.dna.-toplevel.fa) and annotation file Macaca_fascicularis.Macaca_fascicularis_5.0.94.gtf.
To identify commonly regulated genes during infection with adjustments for species-specific differences (Figure 3), edgeR was performed with GLM capabilities. To identify DEGs between infected rhesus macaques at d6 and infected cynomolgus macaques at d4, the GLM approach was used with makeContrasts function to account for average species-related effects. Sparse partial least squares discrimination analysis (SPLS-DA) was performed using the mixOmics R package for classification and validation. Models were initially built with 10 components and validated with three-fold cross-validation and 10 repetitions.
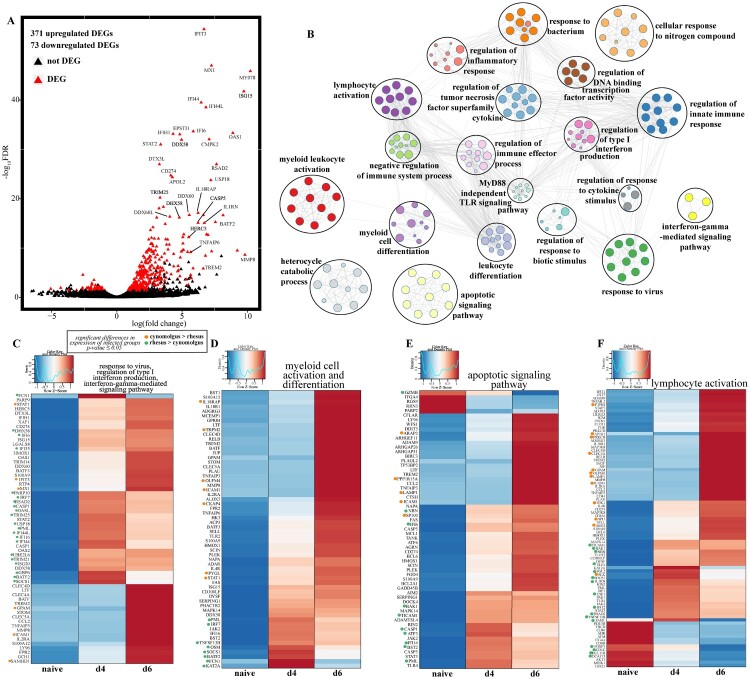
Figure 3.
Cynomolgus and rhesus macaques share a core of EVD-related genes. (A) Volcano plot of global gene expression changes shared between infected cynomolgus and rhesus macaques. DEGs (average rpkm ≥5) are denoted in red. Exemplar DEGs are labelled. (B) GO network depicting functional enrichment of DEGs expressed by both infected rhesus and cynomolgus using Metascape. Clustered nodes of identical colour correspond to one GO term. Node size represents the number of DEGs associated with the GO term. Grey lines represent shared interactions between GO terms, with density and number indicating the strengths of connections between closely related GO terms. Heatmaps representing DEGs enriching to GO terms (C) “response to virus,” “regulation of production of type I interferon,” “interferon-gamma-mediated signaling pathway,” (D) “lymphocyte activation,” (E) “myeloid cell activation,” “myeloid cell differentiation,” (F) “apoptotic signaling pathway” depicted in part B. Where GO terms consisted of more than 60 DEGs, only 60 are represented. Each column represents the median of the normalized rpkm of samples. Range of colours is based on scaled and centred rpkm values of the represented DEGs. Red represents upregulated; blue represents downregulated. DEGs expressed to a greater extent in cynomolgus (orange) or rhesus (green) macaques are coloured (two-tailed T test).
Functional enrichment of DEGs was performed using Metascape to identify relevant GO biological process terms. Heatmaps, bubbleplots, Venn diagrams and violin plots were generated using R packages ggplot2 and VennDiagrams. GO network plots were generated in Cytoscape (Version 3.5.1). Graphs were generated using GraphPad Prism software (version 8).
Statistical analysis
Differences in transcript abundance between the two macaque species was determined with a two-tail independent T test using the genefilter R package. PERMANOVA was performed using the vegan R package to assess contributions of variance from each principal component in Figure S1. *p-value≤0.05, **p-value≤0.01, ***p-value≤0.001.
Supplementary Material
Acknowledgements
AP, KM, IM and AM designed the experiment, conducted the experiments, analyzed the data, and wrote the manuscript. All authors approved the manuscript.
Correction Statement
This article was originally published with errors, which have now been corrected in the online version. Please see Correction (http://dx.doi.org/10.1080/22221751.2022.2036036)
Funding Statement
This work was supported by the National Institutes of Health (NIH) [grant numbers R21AI143301 and U19A109945], and in part by the Intramural Research Program National Institute of Allergy and Infectious Diseases, NIH.
Data availability
Sequencing data for rhesus macaques is available at BioProject PRJNA718880. Sequencing data for cynomolgus macaques is available at BioProject PRJNA398558.
Results
EBOV infection of cynomolgus and rhesus macaques results in distinct transcriptional profiles that have comparable biological function
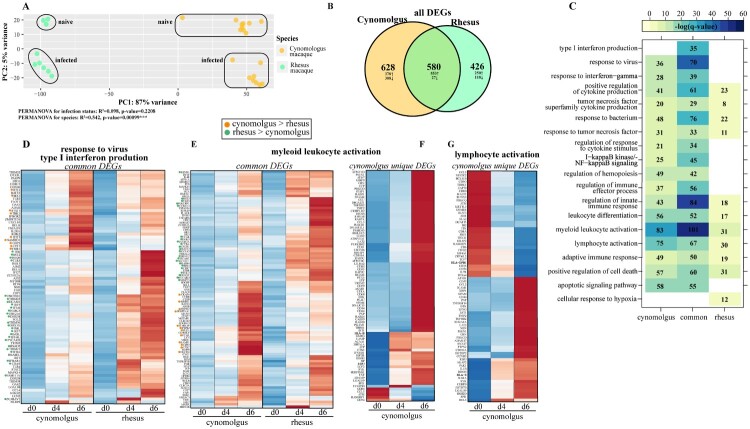
We leveraged access to an existing RNA sequencing data set from cynomolgus macaques (PRJNA398558) and historical whole blood RNA samples from rhesus macaques infected with 1,000 PFU of EBOV-Makona C07 to determine the impact of the species on the host transcriptional response to EBOV infection [26,29]. Data from these two studies showed that cynomolgus macaques exhibited an earlier onset of viremia (3 days post infection [DPI]) than rhesus macaques, who were viremic at 5–6 DPI ([26,29], Figure S1A). We performed a comparative bivariate analysis of transcriptional profiles obtained at 4 and 6 DPI to capture peak changes in disease prior to euthanasia. Principal component analysis (PCA) revealed substantial transcriptional differences between cynomolgus and rhesus macaques regardless of infection, with species (PC1) accounting for 87% of the variance while infection (PC2) accounted for 5% of the variance (Figure 1A).
Figure 1.
Bivariate analysis identifies species-specific transcriptomes prior to and following infection. (A) Principal component analysis of cynomolgus or rhesus macaques uninfected or infected with EBOV-Makona Guinea C07. (B) Venn diagram of DEGs detected in infected macaque species (4 and 6 days post infection) relative to uninfected counterparts. (C) GO enrichment depicting functional enrichment of DEGs found in Figure S1E using Metascape. The colour intensity represents the statistical significance, as shown by the negative log of the FDR-adjusted p-value (-log(q-value)), with the range of colours based on the GO terms with lowest and highest -log(q-value) values for the entire set of GO terms. Numbers of DEGs enriching to each GO term per column are represented in each box; blank boxes represent no statistical significance. Gene expression heatmaps representing DEGs from GO terms found in part C: (D) “response to virus” and “type I interferon production,” (E,F) “myeloid leukocyte activation” and (G) “lymphocyte activation.” Heatmaps represent DEGs either common to both or unique to one species. Each column represents the median of the normalized rpkm of samples. Range of colours is based on scaled and centred rpkm values of the represented DEGs. Red represents upregulated; blue represents downregulated. DEGs expressed to a greater extent in cynomolgus (orange) or rhesus (green) macaques are coloured (two-tailed T test).
Given the significant contribution of species differences to transcriptional responses (PERMANOVA, p-value=0.00099), we first investigated species-specific differences prior to infection (Fig. S1B-F). Differentially expressed genes (DEG) were identified as protein-coding genes reaching statistical significance (|log2(fold-change)| ≥ 1, FDR-adjusted p-value ≤ 0.05, average rpkm ≥ 5). Over 2,000 genes DEGs were detected between the two species, with most (1,872) being upregulated in cynomolgus macaques compared to rhesus macaques (Fig. S1B). We performed functional enrichment of these DEGs to understand their biological relevance. DEGs enriched to immunological signatures related to both innate and adaptive immunity, including “LPS- vs ctrl-treated monocyte DN [differentiation],” “Naïve vs memory T cell DN” and “Plasma cell vs memory B cell DN” (Fig. S1C). DEGs belonging to innate immunity signatures exhibited a mixture of up- and down-regulation in cynomolgus macaques relative to rhesus macaques, including those involved in chemotaxis (e.g. CCR2, CD37, CD44, CD47, CSF1R, CXCR4), complement regulation (e.g. CD55) and neutrophil-mediated immunity (e.g. S100A6) (Fig. S1D). On the other hand, DEGs integral for cellular (e.g. CD246, CD28, CD40LG, CD86) and humoral (e.g. BCL6, LYN) immunity were primarily upregulated in cynomolgus (Fig. S1E). Additionally, we detected a notable number of DEGs related to antiviral defense and inflammation, such as DDX60L, JAK2 and IKBKB, that were expressed to a greater extent in cynomolgus macaques (Fig. S1F).
We next investigated the transcriptional response to EBOV-Makona C07 infection. Approximately 1,000 DEGs were identified in both species, with 580 DEGs shared between the two (Figure 1B). Most DEGs were upregulated in both cynomolgus (931 upregulated, 277 downregulated) and rhesus (861 upregulated, 145 downregulated) macaques. DEGs in both rhesus and cynomolgus macaques enriched to GO terms related to antiviral immunity (e.g. “response to virus,” “type I interferon production”), immune activation (e.g. “myeloid leukocyte activation” and “lymphocyte activation”), and cytokine signalling/inflammation (e.g. “regulation of cytokine production” and “cytokine mediated signaling pathways”) (Figure 1C). Only DEGs detected in rhesus macaques belonged to “cellular response to hypoxia” (e.g. ADAM8, VHL) (Figure 1C). Notable common DEGs enriching to GO terms related to antiviral immunity included key transcription factors (e.g. STAT1, STAT2, IRF7), interferon-stimulated genes (ISGs) (e.g. HERC5, IFIT3, OAS1, TRIM56, BST2), immune mediators/receptors (e.g. CCL4, IL2RA,), and pattern recognition receptors (TLR2-4, NLRP3). In both macaque species, the expression of these genes progressively increased, with highest level of expression observed 6 DPI (Figure 1D). Some ISGs were highly expressed only in rhesus (e.g. ISG20, TLR3, TRIM25) or cynomolgus macaques (e.g. MX1, STAT1, TBK1) (Figure 1D). Additionally, a large number of common DEGs enriched to GO terms indicative of myeloid cell activation and inflammation including genes encoding key transcription factors (e.g. RELB, NFKB), inflammatory factors (e.g. S100A12, S100A9, MMP25, IL18RAP, IL4R), and chemokine receptor (e.g. CXC1 CD47) (Figure 1C, E). Some of these genes were more highly expressed to a greater extent in rhesus (e.g. CD47, CXCR1, PYCARD, TICAM1) and others in cynomolgus macaques (e.g. CD93, IL18RAP) (Figure 1E).
Some DEGs that were only detected in cynomolgus macaques were important for monocyte activation and inflammation (e.g. CD14, ITGAM, MMP9, TGFBR2); while some of the genes that were unique to rhesus macaques played a role in NFKB-mediated inflammation and complement activation (e.g. CCR2, IL6, NFKB1, C3AR1, C5AR1) (Figure 1F). Finally, DEGs related to adaptive immunity were shared between both species (e.g. CD274, PDCD1LG2, RIPOR2). Genes that play a role in T cell-mediated immunity (e.g. MALT1, TCF7, ZBTB7B, TRBC1, ZAP70) were only downregulated in cynomolgus macaques (Figure 1G).
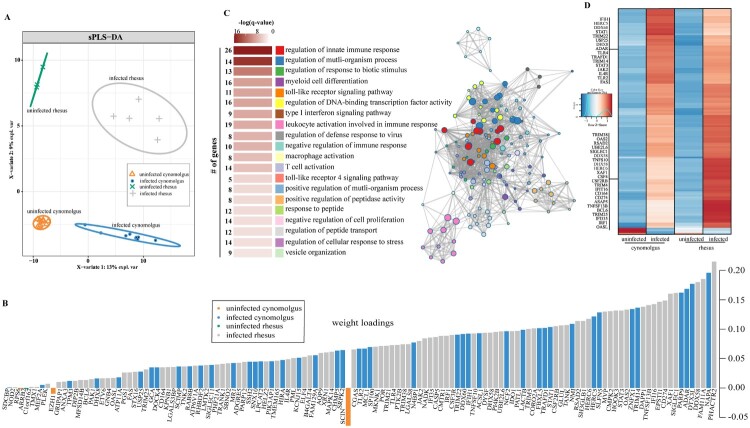
To better define the genes that explain the separation between infected and uninfected animals, we used a sparse partial least-squares discrimination analysis (sPLS-DA) (Figure 2A). Unlike principal component analysis, sPLS-DA preserves maximum covariance between defined groups and assumes sparsity, permitting identification of the minimum number of features responsible for driving a biological event. This analysis identified 130 genes that enriched to GO terms related to antiviral defense (e.g. “type I interferon signaling pathway”), mobilization of myeloid cell-mediated immunity (e.g. “myeloid cell differentiation”), inflammation (e.g. “TLR4-signaling pathway”), and T cell activation (e.g. “T cell activation”) (Figure 2B, C). The overwhelming majority of these 130 genes were upregulated (Figure 2D). Expression of genes related to antiviral defense were elevated in either rhesus (e.g. CGAS, IFI16, IRF1, JAK2, OAS2) or cynomolgus macaque (e.g. HERC5, STAT1, TRIM22) infection. Additionally, genes with roles in inflammation/apoptosis (e.g. CASP5, TNFSF13B, TLR4, XAF1) or intracellular trafficking (e.g. CHMP5, NAPA, SNX10) were more highly induced in infected rhesus or cynomolgus macaques, respectively. The three genes associated with uninfected cynomolgus are involved in epigenetic regulation (EZH1), translation (RPS6) and exocytosis (SCIN) (Figure 2B).
Figure 2.
EBOV infection results in species-specific transcriptomes. (A) Sparse partial least squares discrimination analysis (SPLS-DA) of EBOV-infected and uninfected cynomolgus or rhesus macaques. (B) 130 gene sufficient to resolve transcriptional differences in part A. Weight loadings are coloured by group, where greater weight represents greater correlation with the infection state. (C) GO network depicting functional enrichment of 130 genes identified in part B. Clustered nodes of identical colour correspond to one GO term. Node size represents the number of genes associated with the GO term. Grey lines represent shared interactions between GO terms, with density and number indicating the strengths of connections between closely related GO terms. Accompanying graph illustrates the significance (negative log of the FDR-adjusted p-value (-log(q-value))) and number of genes associated with each GO term. (D) Heatmap representing 130 genes identified in part B. Each column represents the median of the normalized rpkm of samples. Range of colours is based on scaled and centred rpkm values of the represented DEGs. Red represents upregulated; blue represents downregulated.
A conserved core of genes reflecting EVD pathology is induced in cynomolgus and rhesus macaques
Given the significant difference between the transcriptomes of uninfected rhesus and cynomolgus macaques (PERMANOVA, p-value=0.00099), we employed generalized linear model (GLM) to identify core genes that are differentially expressed with infection while correcting for species differences (Figure 3A). The majority of the DEGs were upregulated (n = 371), while 73 genes were downregulated (Figure 3A). In line with previous analyses and studies, these DEGs enriched to GO terms reflecting EVD pathology, including antiviral defense (e.g. “regulation of type I interferon production”), immune activation (e.g. “regulation of immune effector process”), inflammation (e.g. “regulation of inflammatory response”), and apoptosis (e.g. “apoptotic signaling pathway”) (Figure 3B).
DEGs that enriched to GO terms associate with antiviral responses and IFN signalling were significantly upregulated 4 DPI and played a role in viral nucleic acid detection (e.g. DDX58, DDX60, OAS1) and antagonism of viral processes (e.g. ISG20, HERC5, MX1) (Figure 3C). On the other hand, DEGs that enriched to GO terms “myeloid cell activation and differentiation” were predominantly upregulated at 6 DPI and included genes involved in NFKB signalling (e.g. RELB, TLR2), inflammation (e.g. IL-18R1, S100A12, TNFAIP3), and cell migration (e.g. CCL2, CLEC5A, MMP8) (Figure 3D). DEGs that enriched to apoptosis included FAS and OLFM4, BIRC3, and CASP1 and were highly upregulated DPI6 (Figure 3E). Additionally, upregulated DEGs that enriched to “lymphocyte activation” encoded proteins important for inflammatory and antiviral processes (e.g. IL-1RN, JAK2, STAT3, CD44, and BST-2) while down-regulated genes played a role in T cell signalling (e.g. CD8, FOXP1, CD84) (Figure 3F). CD274 which encodes PDL1, a marker of T cell exhaustion, was upregulated at 6 DPI (Figure 3F).
We next investigated potential differences in the induction of these genes between the two species. Several antiviral genes were expressed to a greater extent in rhesus macaques (e.g. IRF7, GBP6, IFI35, ISG20, BST2) (Figure 3C, D). Additionally, genes related to pro-apoptotic processes (e.g. BAK1, CASP1), and regulation of leukocyte-mediated immunity (e.g. FCN, KAT2A, TNFSF13B) were expressed to significantly greater levels in rhesus macaques while those promoting T cell activation (e.g. ARG2, CELC4A, PTPRJ) and adhesion (e.g. ICAM1, OLFM4) were more elevated in cynomolgus macaques (Figure 3D-F).
Since rhesus macaques exhibit a delayed disease progression compared to cynomolgus macaques infected with EBOV-Makona C07, we compared whole blood transcriptional profile obtained from rhesus macaques at 6 DPI with those obtained from cynomolgus macaque 4 DPI using GLM to account for species effect (Fig. S2). We identified ∼400 DEGs, with most (336) being upregulated in rhesus macaques at 6 DPI compared to cynomolgus macaques 4 DPI (Fig.S2A). A large number of DEGs enriched to GO terms relating to innate processes such as myeloid activity (e.g. CD63, CD74, CSTA, CST7), stress response (e.g. FRX1, HSF1, TGFBR3, UPS1), and inflammation (e.g. IFNAR1, C1QB, IKBKE) (Fig. S2A, B). While the majority of DEGs in these GO terms were upregulated in rhesus macaques at 6 DPI, DEGs involved in translation (e.g. EIF3A, FMR1) and organelle mobilization (e.g. KIF5A, SEPTIN9, VPS39) were downregulated compared to cynomolgus macaques at 4 DPI (Fig. S2A).
Regression analysis identifies differences in the regulation of innate and adaptive immune genes
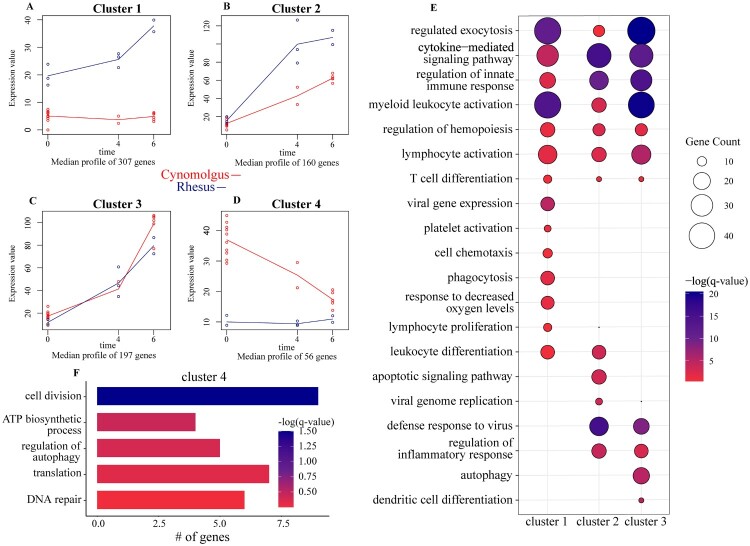
Our previous analyses used a bivariate strategy, focusing on genes differentially expressed with infection relative to uninfected counterparts regardless of days post challenge. To identify clusters of genes with similar and different kinetics of gene expression between the two species following EBOV-Makona C07 infection, we applied a two-ways forward regression model using maSigPro (Figure 4) [42]. Genes that were considered significant in at least 16 comparisons were retained and clustered by temporal expression patterns, resulting in four clusters (Figure 4A-D). Genes in cluster 1 (n = 307) were progressively upregulated in rhesus macaques throughout infection. These genes enriched primarily to “myeloid leukocyte activation” (e.g. CD47, JAK2, S100A9), “cytokine-mediated signaling pathway” (e.g. IKBKE, IL1RN, IL18BP, IL22RA), “cell chemotaxis” (e.g. CCL20, CCL23, CCL25, CXCR4), and “platelet activation” (e.g. TREML1, FCGER1, ADM) (Figure 4E, S3A).
Figure 4.
Regression analysis determines longitudinally regulated genes distinctly regulated by each species. (A-D) Gene expression of the four uniquely-regulated gene clusters identified by MaSigPro using a two-ways forward regression strategy. (E) Bubbleplot representing functional enrichment of genes belonging to clusters 1–3 during infection (4 and 6 DPI). Colour intensity of each bubble represents the negative log of the FDR-adjusted p-value (-log(q-value)) and the relative size of each bubble represents the number of DEGs belonging to the specified Gene Ontology (GO) term. (F) GO term bar graph representing functional enrichment of genes belong to cluster 4 during infection. Colour intensity of each bar represents the -log(q-value) for the corresponding GO term.
Genes in cluster 2 (n = 160) were upregulated by both species, albeit to greater levels in rhesus compared to cynomolgus macaques (Figure 4E). These genes enriched to “defense response to virus” (e.g. IFI16, IRF7, ISG15, OAS2, RSAD2), and “apoptotic signaling pathway” (e.g. BAK1, CASP1, TNFRSF15A) (Figure 4E, S3B). In contrast, cluster 3 (197) genes were upregulated to comparable levels throughout infection by both species. These genes enriched to GO terms “defense response to virus” (e.g. HERC5, IFIT3, MX1, MYD88, STAT1, TBK1), “lymphocyte activation” (e.g. B2M, PRKCB, LAMP1, TREML2) in addition to “autophagy” (e.g. RAB8A, TECPR2) and “dendritic cell differentiation” (e.g. AZI2, TRPM2) (Figure 4E, S3C). Genes in cluster 4 (n=56) were downregulated with infection in cynomolgus macaques only (Figure 4E, S3D). These genes are mostly involved in nucleic acid metabolism, such as “DNA repair” and “translation” (e.g. UWAF1, RFC1, SHPRH), as well as “cell division” (e.g. KIZ, KIF2A, PDS5B) (Figure 4E, S3D).
Discussion
NHPs, primarily cynomolgus and rhesus macaques, are considered the gold standard animal models for EBOV infection given their accurate recapitulation of human pathobiology [22–24]. Here we compared transcriptional responses to infection with a lethal dose of EBOV-Makona Guinea C07, an early isolate from the 2013–2016 West African EBOV epidemic, in both cynomolgus and rhesus macaques. Although both rhesus and cynomolgus macaques succumb to lethal intramuscular infection with EBOV, cynomolgus macaques succumb to disease ∼ 2 days earlier than rhesus macaques and display smaller changes in clinical indicators (e.g. AST, BUN, CRE) and earlier onset of viremia [26,29]. Differences in disease severity have also been noted between these two species for influenza A virus infection, and, more recently, COVID-19 [32,36]. Rhesus macaques are the model of choice for SIV and SHIV infections due to higher levels of viremia [33]. In SVV infection, rhesus macaques best capitulate establishment of viral latency normally seen with VZV infection in humans [39,40]. However, the molecular underpinnings of these differences in viral pathogenesis between the two species have not been thoroughly unexplored.
Our analysis identified a shared core of DEGs similar to those reported in following EBOV-Kikwit challenge in cynomolgus macaques as well as human infections [12,29,43–45]. In both macaque species, genes encoding ISGs, inflammatory cytokines, and apoptotic molecules, as well as genes associated with myeloid cell activation, were significantly upregulated. This transcriptional response is a hallmark of EVD and believed to be a key driver of disease progression. However, a subset of these genes was upregulated to greater extent in rhesus macaques, such as IRF1, BST-1/2, TLR4 and BCL6. Considering the important role these genes play in restricting viral spread, this distinct innate immune transcriptome could potentially explain the delayed kinetics of disease manifestation and viral replication seen in rhesus macaques. Additionally, genes relating to coagulation and hypoxia were upregulated to a greater extent in this species (e.g. ADAM8, TREML1, FCGER1). These transcriptional findings are consistent with exaggerated coagulopathy and hepatocellular damage (e.g. more severe loss of platelets and higher levels of AST) in rhesus compared to cynomolgus macaques reported in previous studies [28,29].
Both B cell- and T cell-mediated responses are believed to be correlates of protection against EBOV infection, as indicated by high levels of neutralizing antibodies and antiviral T cells in human EVD survivors [2,14,15,46,47]. On the other hand, lymphopenia is a characteristic of EVD [16,19–21]. Molecular indicators of T cell loss were more evident in cynomolgus macaques as indicated by the significant downregulation of key genes (CD3G, CD3E, ZAP70, CD8B, IL7R), providing a potential explanation for the shorter disease course observed in cynomolgus macaques. T cell exhaustion resulting from over-activation is associated with poor prognosis and fatal EVD in humans [47–50]. Although expression of PDL1, whose upregulation is also associated with fatal human infection [48] was comparable in both species, expression of FOXP1 which promotes B cell expansion was downregulated to a greater extent while expression VSIR, an inhibitor of T cell activation, was only upregulated in infected rhesus macaques. These observations suggest that dysregulation of the adaptive branch maybe less pronounced in rhesus macaques. Interestingly, some genes that played a role in cell division and nucleic acid metabolism (e.g. CCNY, CHD9, SHPRH, TPI1) were more significantly downregulated in cynomolgus macaques throughout infection indicative of greater cellular damage and stress and in line with more rapid disease progression.
We also determined significant transcriptional differences between uninfected species. Interestingly, we identified processes related to innate, adaptive and antiviral immunity. The greater expression of genes characteristic of B and T cells in uninfected cynomolgus macaques relative to rhesus macaques suggests a possible mechanism for more pronounced adaptive immune dysregulation during infection, which may permit aberrant viral replication and early onset of viremia. This may occur in conjunction with baseline levels of transcriptional activity reflecting inflammation and hyperactivation of innate immune cells. Future studies will seek to further understand these differences at the cellular level to determine predisposition for different immune responses.
There are several caveats to this study. First, while our transcriptional findings provide insights into molecular differences in host response to EBOV infection, these findings should be followed up by functional studies that explore differences at the protein-level. Second, studies in cynomolgus and rhesus macaques were performed at two different BSL-4 facilities. Although inoculation titre and route, as well as immune monitoring procedures were identical, potential variations in experimental procedures could have influenced our results, including variation in precise virus stock composition. Nevertheless, data reported in this manuscript show distinct but overlapping transcriptional responses to EBOV-Makona infection in cynomolgus and rhesus macaques. Differences in the regulation of EVD-associated genes, notably those important for type I IFN responses, apoptosis, and T cell adaptive immunity may explain differences in disease progression. These findings provide critical insights for viral pathogenesis studies by facilitating the choice of the most appropriate NHP species for experiments.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Rivera A, Messaoudi I.. Molecular mechanisms of Ebola pathogenesis. J. Leukoc. Biol. 2016;100:889–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinski AN, Messaoudi I.. To B or Not to B: mechanisms of protection conferred by rVSV-EBOV-GP and the roles of innate and adaptive immunity. Microorganisms. 2020;8(10):1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coltart CEM, Lindsey B, Ghinai I, et al. . The Ebola outbreak, 2013–2016: old lessons for new epidemics. Philos. Trans. R. Soc. B Biol. Sci. 2017;372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.2014-2016 Ebola Outbreak in West Africa | History | Ebola (Ebola Virus Disease) | CDC. Available from: https://www.cdc.gov/vhf/ebola/history/2014-2016-outbreak/index.html.
- 5.2020 Democratic Republic of the Congo, Equateur Province | Democratic Republic of Congo | Outbreaks | Ebola (Ebola Virus Disease) | CDC. Available from https://www.cdc.gov/vhf/ebola/outbreaks/drc/2020-june.html.
- 6.Geisbert TW, Young HA, Jahrling PB, et al. . Pathogenesis of Ebola hemorrhagic fever in primate models: evidence that hemorrhage is not a direct effect of virus-induced cytolysis of endothelial cells. Am. J. Pathol. 2003;163:2371–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geisbert TW, Hensley LE, Larsen T, et al. . Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am. J. Pathol. 2003;163:2347–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wahl-Jensen V, Kurz S, Feldmann F, et al. . Ebola virion attachment and entry into human macrophages profoundly effects early cellular gene expression. PLoS Negl. Trop. Dis. 2011;5:e1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falasca L, Agrati C, Petrosillo N, et al. . Molecular mechanisms of Ebola virus pathogenesis: focus on cell death. Cell Death Differ. 2015;22:1250–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geisbert TW, Hensley LE, Gibb TR, et al. . Apoptosis induced in vitro and in vivo during infection by Ebola and Marburg viruses. Lab. Investig. J. Tech. Methods Pathol. 2000;80:171–186. [DOI] [PubMed] [Google Scholar]
- 11.Iampietro M, Younan P, Nishida A, et al. . Ebola virus glycoprotein directly triggers T lymphocyte death despite of the lack of infection. PLOS Pathog. 2017;13:e1006397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menicucci AR, Versteeg K, Woolsey C, et al. . Transcriptome analysis of Circulating immune cell subsets highlight the role of monocytes in Zaire Ebola Virus Makona pathogenesis. Front. Immunol. 2017;8:1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Younan P, Santos RI, Ramanathan P, et al. . Ebola virus-mediated T-lymphocyte depletion is the result of an abortive infection. PLOS Pathog. 2019;15:e1008068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thom R, Tipton T, Strecker T, et al. . Longitudinal antibody and T cell responses in Ebola virus disease survivors and contacts: an observational cohort study. Lancet Infect. Dis. 2020;21(4):507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakabe S, Sullivan BM, Hartnett JN, et al. . Analysis of CD8+ T cell response during the 2013–2016 Ebola epidemic in West Africa. Proc. Natl. Acad. Sci. 2018;115:E7578–E7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baize S, Leroy EM, Georges AJ, et al. . Inflammatory responses in Ebola virus-infected patients. Clin. Exp. Immunol. 2002;128:163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McElroy AK, Erickson BR, Flietstra TD, et al. . Ebola hemorrhagic fever: novel biomarker correlates of clinical outcome. J. Infect. Dis. 2014;210:558–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villinger F, Rollin PE, Brar SS, et al. . Markedly elevated levels of interferon (IFN)-gamma, IFN-alpha, interleukin (IL)-2, IL-10, and tumor necrosis factor-alpha associated with fatal Ebola virus infection. J Infect Dis 1999;179(Suppl 1):S188–S191. [DOI] [PubMed] [Google Scholar]
- 19.Wauquier N, Becquart P, Padilla C, et al. . Human fatal Zaire ebola virus infection is associated with an aberrant innate immunity and with massive lymphocyte apoptosis. PLoS Negl. Trop. Dis. 2010;4(10):e837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez A, Lukwiya M, Bausch D, et al. . Analysis of human peripheral blood samples from fatal and nonfatal cases of Ebola (Sudan) hemorrhagic fever: cellular responses, virus load, and nitric oxide levels. J. Virol. 2004;78:10370–10377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta M, Spiropoulou C, Rollin PE.. Ebola virus infection of human PBMCs causes massive death of macrophages, CD4 and CD8 T cell sub-populations in vitro. Virology. 2007;364:45–54. [DOI] [PubMed] [Google Scholar]
- 22.Bennett RS, Huzella LM, Jahrling PB, et al. . Nonhuman Primate Models of Ebola Virus disease. Curr. Top. Microbiol. Immunol. 2017;411:171–193. [DOI] [PubMed] [Google Scholar]
- 23.Geisbert TW, Pushko P, Anderson K, et al. . Evaluation in Nonhuman primates of vaccines against Ebola virus. Emerg Infect Dis 2002;8:503–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bente D, Gren J, Strong JE, et al. . Disease modeling for Ebola and Marburg viruses. Dis. Model. Mech. 2009;2:12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geisbert TW, Strong JE, Feldmann H.. Considerations in the Use of Nonhuman Primate Models of Ebola Virus and Marburg virus infection. J. Infect. Dis. 2015;212(Suppl 2):S91–S97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marzi A, Chadinah S, Haddock E, et al. . Recently identified mutations in the Ebola virus-Makona genome Do Not alter Pathogenicity in animal models. Cell Rep. 2018;23:1806–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marzi A, Feldmann F, Hanley PW, et al. . Delayed disease progression in cynomolgus macaques infected with Ebola Virus Makona strain. Emerg. Infect. Dis. 2015;21:1777–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warren T, Zumbrun E, Weidner JM, et al. . Characterization of Ebola Virus Disease (EVD) in Rhesus Monkeys for development of EVD therapeutics. Viruses. 2020;12(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Versteeg K, Menicucci AR, Woolsey C, et al. . Infection with the Makona variant results in a delayed and distinct host immune response compared to previous Ebola virus variants. Sci. Rep. 2017;7:9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marzi A, Robertson SJ, Haddock E, et al. . VSV-EBOV rapidly protects macaques against infection with the 2014/15 Ebola virus outbreak strain. Science. 2015;349:739–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munster VJ, Feldmann F, Williamson BN, et al. . Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rockx B, Kuiken T, Herfst S, et al. . Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haaft PT, Almond N, Biberfeld G, et al. . Comparison of early plasma RNA loads in different macaque species and the impact of different routes of exposure on SIV/SHIV infection. J. Med. Primatol. 2001;30:207–214. [DOI] [PubMed] [Google Scholar]
- 34.Skinner JA, Zurawski SM, Sugimoto C, et al. . Immunologic characterization of a Rhesus Macaque H1N1 challenge model for candidate influenza virus vaccine assessment. Clin. Vaccine Immunol. CVI. 2014;21:1668–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marriott AC, Dennis M, Kane JA, et al. . Influenza a virus challenge models in Cynomolgus Macaques using the authentic inhaled aerosol and intra-nasal routes of infection. PLoS ONE. 2016;11(6):e0157887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mooij P, Koopman G, Mortier D, et al. . Pandemic swine-origin H1N1 influenza virus replicates to higher levels and induces more fever and acute inflammatory cytokines in Cynomolgus versus Rhesus monkeys and can replicate in common marmosets. PloS One. 2015;10:e0126132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White TM, Mahalingam R, Traina-Dorge V, et al. . Simian varicella virus DNA is present and transcribed months after experimental infection of adult African Green monkeys. J. Neurovirol. 2002;8:191–203. [DOI] [PubMed] [Google Scholar]
- 38.White TM, Mahalingam R, Traina-Dorge V, et al. . Persistence of simian varicella virus DNA in CD4(+) and CD8(+) blood mononuclear cells for years after intratracheal inoculation of African Green monkeys. Virology. 2002;303:192–198. [DOI] [PubMed] [Google Scholar]
- 39.Messaoudi I, Barron A, Wellish M, et al. . Simian varicella virus infection of rhesus macaques recapitulates essential features of varicella zoster virus infection in humans. PLoS Pathog. 2009;5(11):e1000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahalingam R, Messaoudi I, Gilden D.. Simian varicella virus pathogenesis. Curr. Top. Microbiol. Immunol. 2010;342:309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reimann KA, Parker RA, Seaman MS, et al. . Pathogenicity of simian-human immunodeficiency virus SHIV-89.6P and SIVmac is attenuated in cynomolgus macaques and associated with early T-lymphocyte responses. J. Virol. 2005;79:8878–8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nueda MJ, Tarazona S, Conesa A.. Next maSigPro: updating maSigPro bioconductor package for RNA-seq time series. Bioinformatics. 2014;30:2598–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eisfeld AJ, Halfmann PJ, Wendler JP, et al. . Multi-platform ‘omics analysis of human Ebola virus disease pathogenesis. Cell Host Microbe. 2017;22:817–829. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, Speranza E, Muñoz-Fontela C, et al. . Transcriptomic signatures differentiate survival from fatal outcomes in humans infected with Ebola virus. Genome Biol. 2017;18:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kash JC, Walters K-A, Kindrachuk J, et al. . Longitudinal peripheral blood transcriptional analysis of a patient with severe Ebola virus disease. Sci. Transl. Med. 2017;9(385):eaai9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koch T, Rottstegge M, Ruibal P, et al. . Ebola virus disease survivors show more efficient antibody immunity than vaccinees despite similar levels of circulating immunoglobulins. Viruses. 2020;12(9):915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Speranza E, Ruibal P, Port JR, et al. . T-Cell receptor diversity and the control of T-cell homeostasis mark Ebola virus disease survival in humans. J. Infect. Dis. 2018;218:S508–S518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruibal P, Oestereich L, Lüdtke A, et al. . Unique human immune signature of Ebola virus disease in Guinea. Nature. 2016;533:100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agrati C, Castilletti C, Casetti R, et al. . Longitudinal characterization of dysfunctional T cell-activation during human acute Ebola infection. Cell Death Dis. 2016;7:e2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiedemann A, Foucat E, Hocini H, et al. . Long-lasting severe immune dysfunction in Ebola virus disease survivors. Nat. Commun. 2020;11:3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data for rhesus macaques is available at BioProject PRJNA718880. Sequencing data for cynomolgus macaques is available at BioProject PRJNA398558.