Abstract
Androgen receptor (AR) signaling continues to drive castration-resistant prostate cancer (CRPC) in spite of androgen deprivation therapy (ADT). Constitutively active shorter variants of AR, lacking the ligand binding domain, are frequently expressed in CRPC and have emerged as a potential mechanism for prostate cancer to escape ADT. ARv7 and ARv567es are 2 of the most commonly detected variants of AR in clinical samples of advanced, metastatic prostate cancer. It is not clear if variants of AR merely act as weaker substitutes for AR or can mediate unique isoform-specific activities different from AR. In this study, we employed LNCaP prostate cancer cell lines with inducible expression of ARv7 or ARv567es to delineate similarities and differences in transcriptomics, metabolomics, and lipidomics resulting from the activation of AR, ARv7, or ARv567es. While the majority of target genes were similarly regulated by the action of all 3 isoforms, we found a clear difference in transcriptomic activities of AR versus the variants, and a few differences between ARv7 and ARv567es. Some of the target gene regulation by AR isoforms was similar in the VCaP background as well. Differences in downstream activities of AR isoforms were also evident from comparison of the metabolome and lipidome in an LNCaP model. Overall our study implies that shorter variants of AR are capable of mediating unique downstream activities different from AR and some of these are isoform specific.
Keywords: prostate cancer, androgen receptor, splice variants, ARv7, ARv567es, omics data
The androgen receptor (AR) is a ligand activated transcription factor that plays a major role in the growth and development of normal prostate gland as well as prostate cancer (1). Treatment for metastatic prostate cancer consists of reducing the levels of circulating androgens chemically or surgically to inhibit AR signaling. This strategy known as androgen deprivation therapy (ADT) works initially to curb the growth of prostate cancer. However, prostate cancer eventually develops resistance to ADT and progresses into the lethal castration-resistant prostate cancer (CRPC) (2); this is the second leading cause of cancer-related mortality among men in the United States. CRPC has been shown to depend on AR signaling in spite of ADT, and several means for reactivation of AR have been identified (3-6).
Androgen receptor splice variants have emerged as an important possible mechanism for metastatic prostate cancer to escape ADT (7-10). AR splice variants lack the carboxy terminal ligand binding domain of AR and hence their transcriptional activity is not regulated by androgens. Expression of AR variants is enriched in CRPC (11, 12). More than 20 different splice variants have been identified (13). ARv7 is the best characterized and most widely expressed AR variant in prostate cancer cell lines and metastatic patient samples (10). It consists of exons 1 to 3 and an additional 16 amino acids from a cryptic exon at the C terminal end (Fig. 1A). The presence of ARv7 is positively correlated with resistance to ADT and reduced overall patient survival (14).
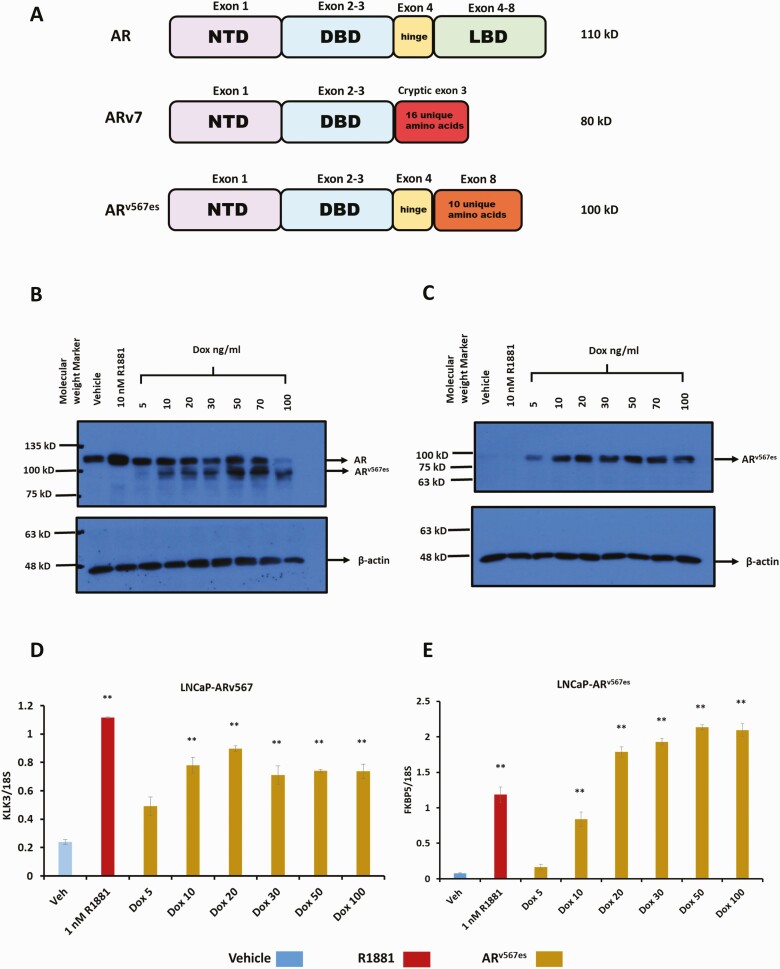
Figure 1.
Characterization of LNCaP-ARv567es cell line. (A) Structure of AR isoforms. Schematic showing the domain architecture of AR, ARv7 and ARv567es as well as the exons contributing to each of those domains and their molecular weight as detected on a western blot. NTD, amino terminal domain; DBD, DNA binding domain; LBD, ligand binding domain. The domains depicted in the schematic are not proportional to actual size. LNCaP-ARv567es cells were treated with vehicle (0.1% ethanol), 10 nM R1881 or 5, 10, 20, 30, 50 or 100 ng/mL doxycycline for 24 hours in medium containing 10% steroid depleted, charcoal stripped serum (CSS). Following treatment, protein and RNA were isolated for Western blots and RT-qPCR, respectively. (B) Western blot probed with AR441 (AR) antibody (upper), followed by antibody for β-actin (lower) using 10 µg protein extract per lane, (C) Western blot first probed with Abcam’s rabbit antibody specific for ARv567es (upper), followed by antibody for β-actin (lower). (D) RT-qPCR for canonical AR target gene, KLK3. (E) RT-qPCR for another AR target gene, FKBP5 (**P < .01).
Most of the splice variants contain exons 1 to 3 and some amount of unique sequence. In contrast, ARv567es, another AR splice variant frequently detected in human prostate cancer metastases (8) consists of exons 1 to 4 and 10 unique amino acids derived from out-of-frame translation of exon 8 (Fig. 1A). Exons 5, 6, and 7 of AR are missing in ARv567es. Thus, it contains the hinge region, which plays a role in nuclear localization among other activities and contains multiple post-translational modifications (15-17). Unlike other AR splice variants that were discovered in prostate cancer cell lines and result from alternative splicing of mRNA, ARv567es was discovered in LuCaP xenografts (8) as a result of an AR genomic rearrangement (18). AR genomic amplifications and rearrangements are very common in prostate cancer and contribute to its elevated expression as well as expression of variants (18, 19). ARv567es has been shown to lead to de novo tumorigenesis in a mouse model (20). Since AR splice variants coexist with AR in prostate cancer cell lines such as 22Rv1 and LN95 (9, 10, 21), it is challenging to delineate the differences in the transcriptional program and downstream activities mediated by AR variants versus AR. AR variants have the capacity to interact with each other and with AR (22), but the contribution of these interactions to variant function is unresolved, with some investigators failing to detect interactions in their studies (23) or requirements for AR in variant-mediated gene expression. Multiple studies primarily in the 22Rv1 cell line have shown that depleting ARv7 or a combination of variants expressed in the line alters gene expression, that some of these genes differ from AR-regulated genes (23-26), and unique ARv7 chromatin binding sites can be detected. Tindall’s group (26) compared gene expression in cells depleted of AR with those depleted of all AR isoforms and found substantial variant-dependent gene expression independent of AR. Thus, although the 2 AR isoforms can dimerize, AR is not required for variant activity. The differences in activity have not been consistent across cell lines or studies and seem to be cell context specific (13). ARv567es is not endogenously expressed at significant levels in any of the commonly studied prostate cancer cell lines. To address the activity of ARv567es, Nyquist et al. (18) generated a TALEN engineered R1-AD1 cell line (a monoclonal subline isolated from CWR-R1 prostate cancer cells) expressing ARv567es (18) without AR. This line did not show convincing unique activities of ARv567es relative to AR in the parental line, but rather a subset of AR activities. However, the protein level of ARv567es was significantly lower than AR in the parental line (18). Previous work done in our laboratory had shown that inducible expression of ARv7 in the LNCaP cell model led to isoform-specific differences in target gene regulation and downstream metabolic activities (27, 28). In this study, our goal was to compare the activities of ARv567es with those of AR and ARv7 in the same cellular background and to extend the analysis of downstream activities for all 3 isoforms to changes in lipids. We found that while many actions were common to all 3 AR isoforms some of the activities mediated by ARv7 and ARv567es are not only different from AR but also different from each other.
Materials and Methods
Cell culture
Parental LNCaP and VCaP cells as well as HEK 293T/17 cells were purchased from the American Type Culture Collection (Manassas, VA). LNCaPAR-V7/pHage (LNCaP-ARv7) (28) and VCaPAR-V7/pHage (VCaP-ARv7) (28) cell lines were derived from LNCaP and VCaP cell lines by infection by a lentivirus encoding ARv7 followed by antibiotic selection of a pool of resistant virally infected cells. They express androgen receptor splice variant ARv7 in response to doxycycline (Dox). The VCaPAR-V7/pHage cells were re-derived for this study using the same approach and their characterization is shown in Fig. 1 (for supplementary figures and tables see (29)) pCMV-3xFlag ARv567es vector, a kind gift from Dr. Stephen Plymate (University of Washington) was used to generate LNCaP-ARv567es and VCaP-ARv567es cell lines that express the variant ARv567es upon addition of dox as previously described for the ARv7 lines in 28. The LNCaP empty vector cell line was derived using the vector backbone. LNCaP cell lines were maintained in RPMI-1640 medium with 10% fetal bovine serum (FBS). VCaP cell lines were maintained in DMEM/F-12 (1:1) medium with 10% FBS. G418 (600 µg/mL and 1400 µg/mL) (Thermo Fisher Scientific Waltham MA, #10131-035) was used to select and maintain LNCaP and VCaP cell lines, respectively. All experiments were carried out in medium supplemented with 10% charcoal stripped serum (CSS) depleted of endogenous androgens. Cells were subjected to short tandem repeat analysis and stocks were frozen. Fresh stocks of cells were brought up every 3 months and routinely tested for mycoplasma.
Protein extraction and Western blotting
The cells were washed with 1× ice cold phosphate-buffered saline (PBS) and harvested using a manual scraper. Cells in ice cold PBS were first spun at top speed in a microcentrifuge for 2 minutes to collect the cell pellet and cold PBS supernatant was discarded. TESH buffer (1×; 10 mM Tris, 1 mM EDTA, 12 mM monothioglycerol, pH adjusted to 7.7 with HCl) with 0.4 M NaCl and protease inhibitors (Sigma-Aldrich St. Louis MO, #S8820) was added to the cell pellets and they were subjected to 3 cycles of incubation in a dry ice ethanol bath, followed by thawing in a 37°C water bath and vortexing. After 3 cycles, extracts were spun at top speed for 2 minutes in a microcentrifuge, and protein in the resultant supernatant was collected for further analysis through Western blotting. Protein extracts were quantified using Bio-Rad protein assay dye (Bio-Rad Hercules CA, #5000006), resolved on a 7.5% gel using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane. The membrane was probed with anti-AR antibody, AR441 (30) at 1:1000 concentration, or antibody specific for ARv567es (31) at 1:1000 concentration in 2.5% milk-1× PBS after blocking at room temperature for 1 hour in 5% milk-1× PBS. Equal protein loading across lanes was demonstrated using an antibody for β actin (32).
RNA extraction and RT-qPCR
Total RNA was extracted from the cells using RNAsol reagent (GenDepot, Barker TX #R6101-020) according to the manufacturer’s instructions. A 500-ng bolus of RNA was used to synthesize cDNA using amfiRivert cDNA Synthesis Platinum Master Mix (GenDepot, #R5600-100). Target gene expression was measured using SYBR green polymerase chain reaction (PCR) master mix (Applied Biosystems, Waltham MA #4334973) on Fast Real-Time PCR System (ABI 7500). RT-qPCR data were analyzed using the relative standard curve method. 18S qPCR was used to normalize quantity of cDNA used as input. The primer sequences used to quantify RNA by RT-qPCR are listed in Table 1.
Table 1.
Primer sequences used to amplify genes through RT-qPCR
| Gene name | Forward primer | Reverse primer |
|---|---|---|
| 18S | ACCGCAGCTAGGAATAATGGA | GCCTCAGTTCCGAAAACCA |
| PSA (KLK3) | ACCTGCACCCGGAGAGCT | TCACGGACAGGGTGAGGAAG |
| FKBP5 | GGATATACGCCAACATGTTCAA | CCATTGCTTTATTGGCCTCT |
| SGK1 | CCTTGACGCTGGCTGTGA | TGACCCCGAGTTTACCGA |
| EDN2 | AGGGACATTTCCACAGTCAAG | TCCAGCTCACGACACTATCT |
| CTGF | CATTCTCCAGCCATCAAGAGAC | CCACAAGCTGTCCAGTCTAATC |
| P3H2 | AGAAGGTGGTCCTCTACTCTATG | GTTCTTCCGACAGGACGTTATC |
| TMEM100 | GTTAGAGGCTCTTTCCCAGAAG | GAGAGGCAATCCATTACCAGAG |
| OR5B2 | CTAACCAGTGTCCCAGAACTAC | GACAAGAGTCCATCAGGATCAG |
| NPR3 | CTATGGAGATGGCTCATGGAAG | CAGGTTTCACTGTCCTCAGTAG |
RNA sequencing
LNCaP-ARv567es cells were plated at 250 000 cells per well in a 6-well plate in RPMI-1640 medium with 10% FBS. Cells were allowed to attach overnight and the next day the medium was switched to RPMI 1640 with 10% CSS followed by treatment with vehicle (0.1% ethanol), 10 nM R1881 or 30 ng/mL of Dox. After 24 hours, mRNA was extracted using Qiagen’s RNeasy mini kit (Germantown MD, #74104) and Qiashredder spin columns (#79654) were used for sample homogenization. Two vehicle and 2 Dox-treated samples from 2 independent experiments performed in different weeks were submitted for RNA sequencing. TruSeq Stranded mRNA from Illumina (San Diego, CA) was used for library preparation and samples were sequenced on NextSeq 500 machine, also by Illumina.
RNA-sequencing data analysis
Reads were quality trimmed using TrimGalore!, a tool that employs Cutadapt and FastQC to apply trimming and quality controls to fastq files. After post-trimming quality assessment using FastQC, they were aligned to the UCSC hg19 genome build using HISAT2 (33-35). Expression of protein coding genes was quantified using featureCounts (36) and the GENCODE gene model. Low expression genes were removed, and gene expression was normalized using the trimmed mean of M-values algorithm. The R-package edgeR was used to determine the differential gene expression (37). Principle component analysis and heatmap visualization of differential gene expression were performed as quality control measures for the biological replicates (data not shown). Genes that were included in a gene signature for a given comparison were considered statistically significant for a fold change of at least 1.5 and a false discovery rate (FDR) of no more than 0.05. Venn diagrams were generated using the VennDiagram R-package using gene signatures from the differential gene expression analyses. Gene set enrichment analysis was performed using GSEA v3.0 against the HALLMARK, REACTOME, and KEGG pathway collections. Pathways were considered significant for an FDR less than 0.05 (38).
Seahorse Glycolytic rate assay
Prior to starting the assay, a 96 well Seahorse plate (Agilent, Santa Clara, CA) was coated with 0.01% poly-D-lysine (Sigma, St. Louis MO, #P7280), for 1 hour to promote better attachment of cells. Wells were washed twice with 1× PBS before plating 7500 LNCaP-ARv7 or LNCaP-ARv567es cells per well in RPMI medium with 10% CSS to a total volume of 80 µL per well. A manual multichannel pipette was used to gently pipette cells up and down to ensure uniform spread of cells in all wells. The background wells only received medium without cells. The cells were allowed to attach overnight in an incubator at 37°C and 5% CO2. The next day either vehicle (0.1% ethanol), 10 nM R1881, 20 ng/mL Dox (for LNCaP-ARv7), or 30 ng/mL Dox (LNCaP-ARv567es) were added to the wells. The vehicle, hormone or Dox treatments were first mixed at 5× the concentration in RPMI media with 10% CSS and 20 µL of 5× treatments was added to wells to bring up the final volume in each well to 100 µL as well as to dilute the final treatments to 1×. Cells were again placed in an incubator for 24 hours. The next day, cells were gently washed 3 times with a manual multichannel pipette using Seahorse XF RPMI (pH adjusted) medium, supplemented with XF glucose, XF pyruvate and XF L-glutamine to a final concentration of 10 mM, 1 mM, and 2 mM respectively according to the manufacturer’s instructions. The components in the Seahorse XF Glycolytic Rate Assay Kit (Agilent, #103344-100) such as Rotenone and Antimycin A as well as 2-deoxy-D-glucose were also reconstituted in XF RPMI media and the 96-well plate was further processed and run on Agilent’s XFe96 machine according to the manufacturer’s instructions. Results were analyzed using the accompanying Wave software. The experiment was performed 3 independent times and a representative experiment is shown. Each experiment consisted of 6 to 8 wells/technical replicates for each cell line and treatment.
Sample preparation for metabolomics
Two T75 flasks each containing 6 million LNCaP empty vector cells along with 1 flask each with the same number of LNCaP-ARv7, and LNCaP-ARv567es cells were plated in RPMI-1640 medium with 10% FBS. The next day, the medium was switched to 10% CSS and 1 flask of LNCaP empty vector cells was treated with vehicle (0.1% ethanol) and the other 1 with 10 nM R1881. LNCaP-ARv7 cells were treated with 20 ng/mL Dox and LNCaP-ARv567es cells were treated with 30 ng/mL Dox. All treatments were for 48 hours, after which the medium was aspirated and cells were trypsinized, quantified using a coulter counter and separated into aliquots of 5 million cells, which were washed twice in cold PBS before being pelleted and stored at –80°C. Three independent biological replicates were collected from independent experiments for each of the 4 treatment groups. Samples were extracted and analyzed using liquid chromatography/mass spectrometry (LC/MS) in the CPRIT Cancer Proteomics and Metabolomics core as described previously in 39-43.
Citrate assay
Three T75 flasks of LNCaP-ARv7 and 3 of LNCaP-ARv567es cells containing 6 million cells/flask were plated in RPMI-1640 medium with 10% FBS. After 24 hours, the medium was switched to RPMI-1640 with 10% CSS and the 3 flasks from each cell line were treated with Vehicle (0.1% ethanol), 10 nM R1881, 20 ng/mL Dox (LNCaP-ARv7), or 30 ng/mL Dox (LNCaP-ARv567es). After 48 hours, medium was collected from all 6 plates. An empty T75 flask with no cells and only medium was also incubated for 48 hours to be used as a negative control for the experiment. Abcam’s Citrate assay kit (Cambridge MA, #ab83396) was used to determine the amount of citrate released by the cells into 10 µL of medium. For each sample, absorbance was measured with and without enzyme for background correction. The average of 3 independent experiments is shown.
Oil red O staining
LNCaP-ARv567es cells were plated at 100 000 cells per well in a 6-well plate in RPMI-1640 medium with 10% FBS. The next day medium was changed to RPMI-1640 with 10% CSS and cells were treated with either vehicle (0.1% ethanol), 10 nM R1881, or 30 ng/mL Dox (LNCaP-ARv567es). Treatments continued for a total of 6 days. Media with 10% CSS and treatments were replenished after 3 days. On Day 6, cells were washed with cold PBS and fixed with 4% paraformaldehyde for 20 minutes followed by staining with 0.3% Oil red O solution in 60% isopropanol (Sigma, #O1391) for 10 minutes. Cells were further washed 3 more times with PBS and imaged with a Nikon Eclipse TE300 microscope.
Sample preparation for untargeted lipidomics
LNCaP-ARv7 cells were treated with vehicle, 10 nM R1881, or 20 ng/mL Dox along with LNCaP-ARv567es cells treated with 30 ng/mL Dox and samples were prepared exactly as described for metabolomics except that 4 biological replicates were prepared from 4 independent experiments. Lipids were extracted using a modified Bligh–Dyer method (44). The extraction was carried out using 2:2:2 ratio of water:methanol:dichloromethane at room temperature after isotopic labeled standards internal standards (ISTDs) were added to the cell line samples and the quality control pool as described earlier (45). After homogenization of the samples, the organic layer was collected and completely dried under vacuum. Before MS analysis, the dried extract was resuspended in 100 μL of buffer containing 10 mmol/L NH4Ac and subjected to LC/MS. The lipidome was separated using reverse-phase chromatography. To monitor the lipid extraction process, we used an aliquot from a standard pool of liver tissue. The data acquisition of each sample was performed in both positive and negative ionization modes using a TripleTOF 5600 equipped with a Turbo VTM ion source. The instrument performed 1 TOF MS survey scan (150 ms) and 15 MS/MS scans with a total duty cycle time of 2.4 ms. The mass range in both modes was 50 to 1200 m/z. We controlled the acquisition in both MS and MS/MS spectra by data-dependent acquisition function of the Analyst TF software (AB Sciex) described previously (45-48).
Replicates and statistical analysis
Unless otherwise noted, experimental values are presented as an average of 3 separate wells/plates from the same experiment and standard error of mean. Experiments with LNCaP cell lines were repeated a minimum of 3 times independently and a representative experiment shown. Experiments with VCaP cell lines were repeated a minimum of 2 times independently and a representative experiment shown. A 2-tailed t-test was used to calculate P value vehicle-treated cells in the same cell line.
Statistical analysis for targeted metabolomics and untargeted lipidomics
MassHunter (Agilent Technologies) was used to find the peak area for each metabolite. Targeted metabolite analysis was performed in 3 separate runs. In the first run, carnitines, nucleotides, amino acids, amino sugars and tricarboxylic acid cycle (TCA) metabolites were analyzed. Coenzyme A metabolites and fatty acids were analyzed in the second and third runs respectively. For each experiment the metabolite peak area was normalized by the peak area of an internal standard. Carnitines, nucleotides, and amino acids were normalized using thymine-d4. Fatty acids, coenzyme A metabolites, and TCA metabolites were normalized using L-zeatine. Amino sugars were normalized using 15 N glutamic acid.
Multi quant (SCIEX) was used to find the peak area for each lipid. Lipids identified using untargeted lipidomics were also normalized using an internal standard, PG 34:0 or lysoPC 17:0, depending on the detection method. The normalized relative peak values were then log2-transformed. A compound-by-compound 1-way analysis of variance was performed on each study (the 3 separate targeted metabolite runs or the untargeted lipidomic experiment) to determine the metabolites that were differentially expressed under the conditions tested (ARv7 or ARv567es expression, R1881 treatment and vehicle controls). Differential metabolites were identified for each experiment by adjusting the p-values for multiple hypothesis testing using the Benjamini–Hochberg procedure. An FDR threshold of less than 0.25 was used to determine significance. To find which lipid classes were differentially expressed due to AR and AR variant expression the relative peak values for each lipid in a class were summed before log-2 transformation. A pairwise t-test was then performed for each lipid class between the test conditions (ARv567es or ARv7 expression, AR (R1881) treatment and vehicle) to determine if the test condition altered the relative abundance of that lipid class between the different test conditions. The P values for each pairwise combination of test conditions for a lipid class was corrected for multiple hypothesis testing using the Benjamini–Hochberg procedure. *denotes P value ≤ .05 and ** denotes P value of ≤ .01. Heatmaps were generated by mapping the z-score for each metabolite or lipid to a color scale for each sample. z-Scores were determined by subtracting the mean of the log2 transformed normalized expression of a metabolite or lipid from the log2 transformed normalized expression value of that metabolite or lipid for a given sample and then dividing by the standard deviation of the log2 transformed normalized expression value for that metabolite or lipid. Means and standard deviation were determined across all samples, such that the z-scores represents the number of standard deviations away from the mean for a given metabolite or lipid, thereby placing all metabolite or lipids on a similar scale.
Results
Generation of an LNCaP cell line with inducible expression of ARv567es
In order to study the capabilities and downstream activities of ARv567es and compare it with AR and ARv7, we generated an LNCaP inducible cell line in which ARv567es protein is expressed upon addition of Dox. This cell model allows for the activation of AR (using the synthetic androgen, R1881) or ARv567es (using Dox) in the same system with comparable protein expression for both AR and ARv567es. Figure 1B shows a Western blot of the protein levels of both AR and ARv567es in LNCaP-ARv567es cells treated with R1881 or with increasing concentrations of Dox. The same protein extracts were also used for a western to probe with an antibody specific for the ARv567es variant (Fig. 1C). At lower levels of Dox, we noticed a dose dependent increase in protein levels of ARv567es. Doses of Dox higher than 20 to 30 ng/mL, seemed to only modestly increase protein levels of ARv567es (Fig. 1C). Next using RT-qPCR, we determined that in our model, ARv567es induces canonical AR target genes of such as KLK3 (Fig. 1D) and FKBP5 (Fig. 1E) in agreement with previous studies (8, 18). Increasing doses of Dox led to a corresponding increase in induction of both KLK3 and FKBP5, although relative PSA (KLK3) levels stabilized around 20 to 30 ng/mL Dox and the further increase in KLK3 levels was very modest with higher treatments of Dox (Fig. 1D). Based on the preliminary results in Fig. 1, we selected the dose of 30 ng/mL Dox treatment for induction of ARv567es protein in LNCaP-ARv567es cells for all further experiments.
Unique gene targets of ARv567es and ARv7
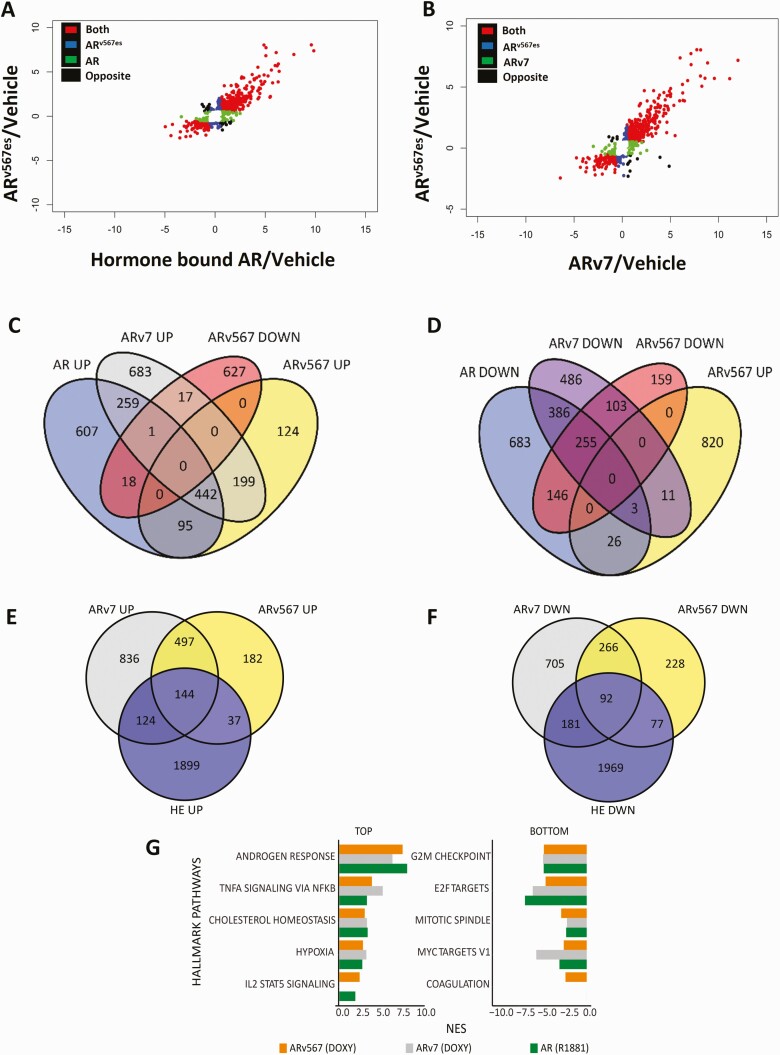
We employed RNA-sequencing to identify genes up- or downregulated by ARv567es and compared the resulting data with similar data sets for AR and ARv7. Although the majority of ARv567es regulated target genes (up- or downregulated) were also regulated and in the same direction by AR, there were also genes uniquely regulated by AR or ARv567es and even some that were regulated in the opposite direction (Fig. 2A). A similar comparison of RNA-sequencing data for ARv567es and ARv7 showed that the majority of the genes are regulated by both variants in the same direction with a smaller proportion of genes uniquely regulated by each variant (Fig. 2B). Further bioinformatics analysis of the combined RNA-sequencing data showed that 442 genes were upregulated by AR and both variants (Fig. 2C) whereas 255 were downregulated by all 3 isoforms (Fig. 2D). Total ARv7 and ARv567es regulated genes were also compared with variant-dependent genes in 22Rv1 cells (Fig 2E and 2F). The 22Rv1 genes were previously identified by simultaneously depleting multiple variants (24). There was minimal overlap between the 2 models. Of the upregulated genes, 17% of ARv7 genes (268) and 21% of ARv567es genes (181) overlapped with 22Rv1 genes (Fig. 2E), but there was a much higher overlap between the variants in the LNCaP background; 75% of ARv567es genes (663) overlapped with ARv7 genes. The extent of overlap was similar for downregulated genes with 22% of ARv7 (273) and 25% of ARv567es (169) overlapping with 22RV1 genes (Fig. 2F). Again, the overlap within the LNCaP line was higher; 54% of ARv567es (663) genes overlapped with ARv7 genes.
Figure 2.
Common and unique gene targets of AR and its splice variants identified by RNA-sequencing. (A) Scatter plot of the log2 fold change showing genes regulated by ARv567es and AR in the same direction, in the opposite direction, and genes unique for each isoform compared to vehicle controls. (B) Scatter plot of the log2 fold change showing genes regulated by ARv567es and ARv7 in the same direction, the opposite direction and unique target genes for each isoform compared to vehicle controls. (C) Venn diagram depicting numbers of genes commonly or uniquely upregulated by the 3 isoforms: AR, ARv7, and ARv567es. (D) Venn diagram depicting numbers of genes commonly or uniquely downregulated by all 3 isoforms: AR, ARv7 and ARv567es. (E) Venn diagram depicting the overlap of genes upregulated by ARv567es or ARv7 with the 22Rv1 data set from He et al. (24). (F) Venn diagram depicting numbers of genes downregulated by ARv567es or ARv7 with 22Rv1 data set from He et al. For these analyses, only genes with fold change exceeding 1.5× and adjusted P ≤ .05, compared with vehicle treatment were included. (G) The 5 Hallmark pathways most upregulated or downregulated by ARv567es (based on the normalized enrichment score/NES; FDR <0.05). For comparison, we provide the NES for the same pathways (when significantly enriched) regulated by AR and ARv7.
GSEA analysis of Hallmark pathways for the LNCaP models showed that androgen response along with TNFα signaling and cholesterol homeostasis pathways were the most upregulated pathways by expression of ARv567es (Fig. 2G). The top 4 pathways up- or downregulated by ARv567es were also similarly regulated by the other 2 isoforms. The fifth highest upregulated pathway (IL2 STAT signaling) was not regulated by ARv7 and the fifth most downregulated pathway (coagulation) was regulated only by ARv567es (Fig. 2G).
Validation of target gene regulation by AR and its variants
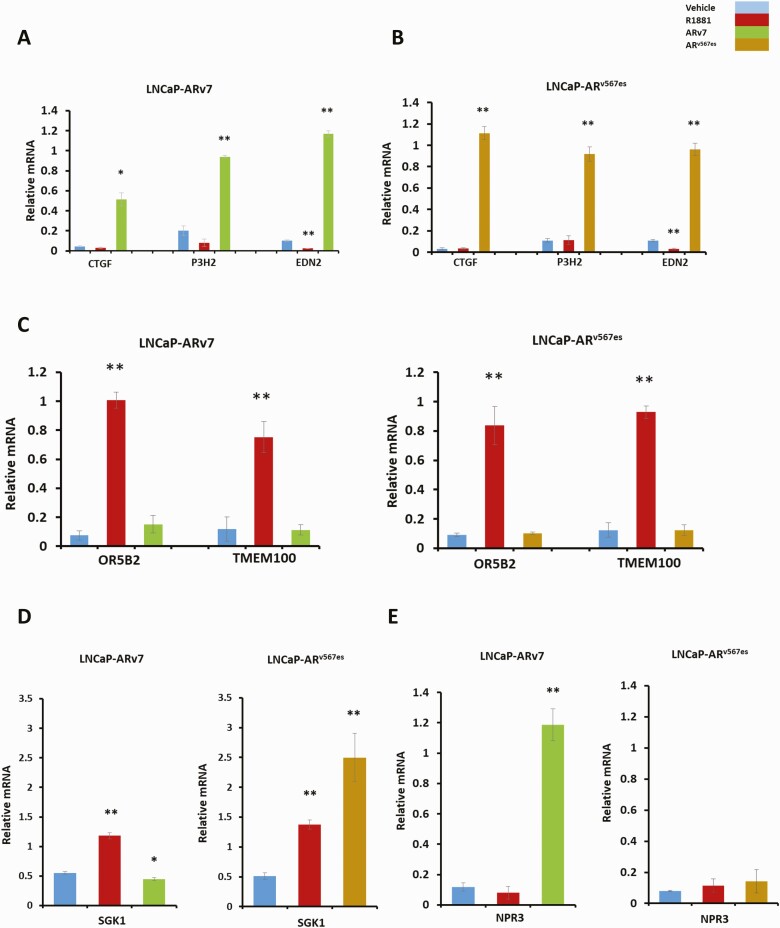
The RNA-sequencing data suggested that many of the genes upregulated by ARv567es are also upregulated by ARv7 (Fig. 2E). This was confirmed by RT-qPCR analysis of selected targets, which showed that the variant ARv567es induced EDN2 (Fig. 3B), a gene we had previously found to be highly induced by ARv7, but not by AR (27, 28) (Fig. 3A). We also confirmed 2 additional candidate genes CTGF and P3H2 that were also only highly induced by AR variants (Fig. 3A and 3B). There are also genes such as TMEM100 and OR5B2, induced by AR but not by its variants (Fig. 3C). We found examples of genes such as SGK1 which show induction by both AR and ARv567es, but not by ARv7 (Fig. 3D). We also noted some others that were very weakly induced by ARv567es relative to AR, but not induced by ARv7 (data not shown). Finally, at least 1 gene, NPR3, was only induced by ARv7 (Fig. 3E). Although our testing was not exhaustive, we did not confirm induction of any gene exclusively by ARv567es. Most genes induced by ARv567es were also induced by AR or ARv7 to some extent.
Figure 3.
Validation of isoform-specific target genes using RT-qPCR. LNCaP-ARv7 and LNCaP-ARv567es cells were treated with vehicle (0.1% ethanol), 1 nM R1881, 20 ng/mL doxycycline (LNCaP-ARv7), or 30 ng doxycycline (LNCaP-ARv567es) in triplicates in medium containing 10% CSS for 24 hours, after which RNA was extracted and converted into cDNA for qPCR analysis. qPCR results relative to 18S for CTGF (connective tissue growth factor), P3H2 (prolyl 3-hydroxylase 2), and EDN2 (endothelin 2) in LNCaP-ARv7 A) and (B) LNCaP-ARv567es cell lines. (C) qPCR results relative to 18S for genes specifically induced only by hormone bound AR but not the variants, OR5B2 (Olfactory Receptor Family 5 Subfamily B Member 2) and TMEM100 (Transmembrane Protein 100). (D) qPCR for SGK1 gene induced by AR and ARv567es but not ARv7 E) qPCR for NPR3 (Natriuretic Peptide Receptor 3), a unique gene target of ARv7. (*P < .05, **P < .01)
Generation of a VCaP cell line with inducible expression of ARv567es
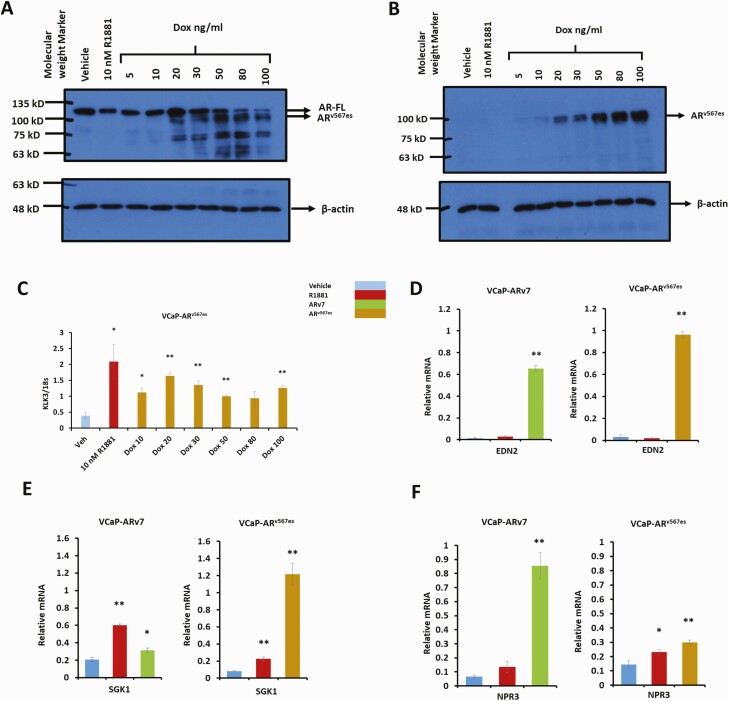
In order to study the variant ARv567es in a different cell model and to test similarities in regulation of target genes, we generated a VCaP cell line with Dox inducible expression of ARv567es. We had previously described a VCaP cell line that expressed ARv7 in response to Dox (28). That line required relatively high levels of Dox to induce ARv7. Thus, we again infected VCaP cells with the ARv7 lentivirus. The dose response of ARv7 in this new line is shown in Fig. S1A (29). Note that VCaP cells endogenously express a small amount ARv7 as reported previously (10). Figure 1B and 1C (29) show the dose dependent induction of KLK3 and EDN2, a variant-specific gene. Although the endogenous ARv7 presumably contributes to basal expression, the difference in gene induction for these genes is at least 10-fold suggesting that endogenous ARv7 is a minor contributor to the gene expression and also would provide minimal background relative to ARv567es activity. Figure 4A shows protein levels of both AR and ARv567es in VCaP-ARv567es cells treated with R1881 or with increasing concentrations of Dox. The same protein extracts were also used for Western blot analysis using an antibody specific for the ARv567es variant (Fig. 4B). ARv567es induced KLK3 (PSA) in the VCaP background (Fig. 4C), similar to its behavior in LNCaP cells (Fig. 1C).
Figure 4.
Characterization of the VCaP-ARv567es cell line and common gene targets of the variants in LNCaP and VCaP models. VCaP-ARv567es cells were treated with vehicle (0.1% ethanol), 10 nM R1881, or the indicated doses of doxycycline for 24 hours in CSS medium. Protein and RNA were extracted for Western blot and qPCR. (A) Western blot probed with AR441 antibody (upper), followed by antibody for β-actin (lower) using 5 µg protein extract per lane. (B) Western blot first probed with Abcam’s rabbit antibody specific for ARv567es (upper), followed by antibody for β-actin (Lower). (C) qPCR for canonical AR target gene, KLK3 VCaP-ARv7 and VCaP-ARv567es cells were treated with vehicle (0.1% ethanol), 10 nM R1881, 30 ng/mL Dox (VCaP-ARv567es), or 100 ng/mL Dox (VCaP-ARv7) in CSS medium for 24 hours in triplicates. After 24 hours of treatment, mRNA was extracted and converted to cDNA for qPCR analysis shown in Fig. 4D-G. qPCR data for (D) EDN2 (E) SGK1 (F) NPR3 relative to 18S (*P < .05, **P < .01)
Similar to its behavior in the LNCaP background, ARv567es induced genes such as EDN2 and SGK1 in the VCaP background (Fig. 4D and 4E). EDN2 is only induced by the variants and not by hormone bound AR in the LNCaP and VCaP backgrounds (Figs. 3A, 3B, and 4D). Similarly, in both cell types SGK1 is induced by AR and ARv567es (Fig. 3D and 4E). However, there is a very weak induction by ARv7 in VCaP cells whereas ARv567es induced SGK1 10-fold and was much more effective than R1881 in inducing SGK1 in the VCaP background (Fig. 4E). Finally, NPR3 was highly induced (about 10 fold) by ARv7 in VCaP cells with no more than a 2-fold increase by ARv567es (Fig. 4F).
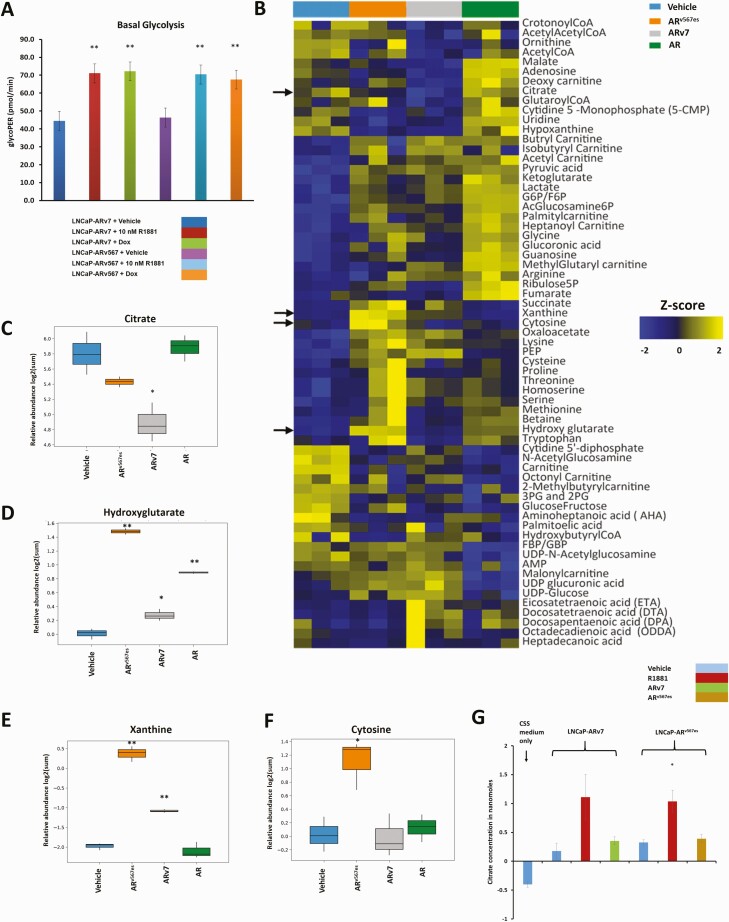
Metabolic changes mediated by androgen receptor isoforms
We have shown, previously, that AR and ARv7 not only alter gene expression, but induce both common and unique changes in metabolism (27). Previous studies (27) had shown that both AR and ARv7 increased glycolysis in the LNCaP model. Using the Seahorse platform, we found that expression of ARv567es led to an increase in basal rate of glycolysis in LNCaP cells, similar to the action of hormone bound AR and ARv7 (Fig. 5A). Targeted metabolomics analyses (liquid chromatography and mass spectrometry analyses) to evaluate steady state levels of metabolites including amino acids, nucleotides, CoA carnitines, fatty acids, amino sugars, and TCA cycle metabolites in control LNCaP cells or cells with activated AR, ARv7, or ARv567es were performed. A heat map showing the z-score of the log2 transformed normalized expression value for metabolites whose levels changed significantly upon treatment (Fig. 5B) reveals that many of the metabolites are regulated in the same direction by all isoforms although the extent of regulation differs. In some cases, AR has a greater effect on metabolites whereas in other cases 1 or both of the variants show more extensive changes. There are also a group of compounds whose levels increase when cells are treated with R1881, but that decrease when variants are expressed. Lastly, there were differences between the 2 variants supporting the concept that the AR isoforms have unique as well as overlapping actions. We confirmed a previous finding that ARv7 reduced citrate levels compared to hormone bound AR or vehicle-treated cells (27) (Fig. 5C). ARv567es was the most effective isoform in increasing relative levels of the onco-metabolite hydroxyglutarate with AR also showing a good increase and ARv7 a much more modest increase (Fig. 5D). ARv567es increased xanthine levels to substantially higher levels than did ARv7 whereas AR had no effect on levels (Fig. 5E). ARv567es was unique in that it led to a large increase in the steady state levels of cytosine, the base required for cytidine synthesis, compared to all of the other treatment groups (Fig. 5F).
Figure 5.
Metabolic differences between the actions of AR and the variants. (A) The basal rate of glycolysis was determined using LNCaP-ARv7 and LNCaP-ARv567es cells treated with vehicle, 10 nM R1881, 20 ng/mL Dox (LNCaP-ARv7), or 30 ng/mL Dox (LNCaP-ARv567es) for 24 hours in 10% CSS medium measured using the Seahorse platform. (B) Heat map showing Z scores for metabolites whose levels were changed as a result of the action of AR and its variants. LNCaP empty vector cells treated with vehicle (0.1% ethanol) or 10 nM R1881 were compared with an equal number of LNCaP-ARv7 cells treated with 20 ng/mL Dox or LNCaP-ARv567es cells treated with 30 ng/mL Dox for 48 hours in 10% CSS media. Three biological replicates from 3 independent experiments were used. Arrows indicate metabolites shown in subsequent box plots. Relative abundance (log2 scale of the peak area of the metabolite normalized to the internal standard) of (C) citrate, (D) hydroxyglutarate, (E) xanthine, and (F) cytosine from the experiment in Fig. 5B. Comparisons were performed between each of the 3 isoforms and vehicle, respectively. (G) Measurement of citrate secreted into the medium by LNCaP-ARv7 and LNCaP-ARv567es cells in CSS medium, treated with vehicle, 10 nM R1881, 20 ng/mL Dox (LNCaP-ARv7), or 30 ng/mL Dox (LNCaP-ARv567es) for 48 hours. Average of 3 independent experiments is shown. (t-test comparison with respect to vehicle-treated cells, *P < .05, **P < .01)
In our previous studies, we found that ARv7 reduced steady state citrate levels similarly to CRPCs and that this drop was not due to a decrease in synthesis (27). Either the citrate was secreted, or it was more rapidly metabolized to products such as lipids. To distinguish between these 2 alternatives, we measured the amount of citrate secreted into the medium by control cells or cells with 1 of the 3 AR isoforms activated. Hormone bound AR led to a higher amount of citrate being secreted into the surrounding cell culture medium, whereas neither ARv7 nor ARv567es increased the levels of secreted citrate beyond that of vehicle-treated cells (Fig. 5G). Thus, the reduced level of citrate in variant expressing cells is not due to enhanced secretion.
Androgen receptor isoforms differentially regulate lipid composition and metabolism
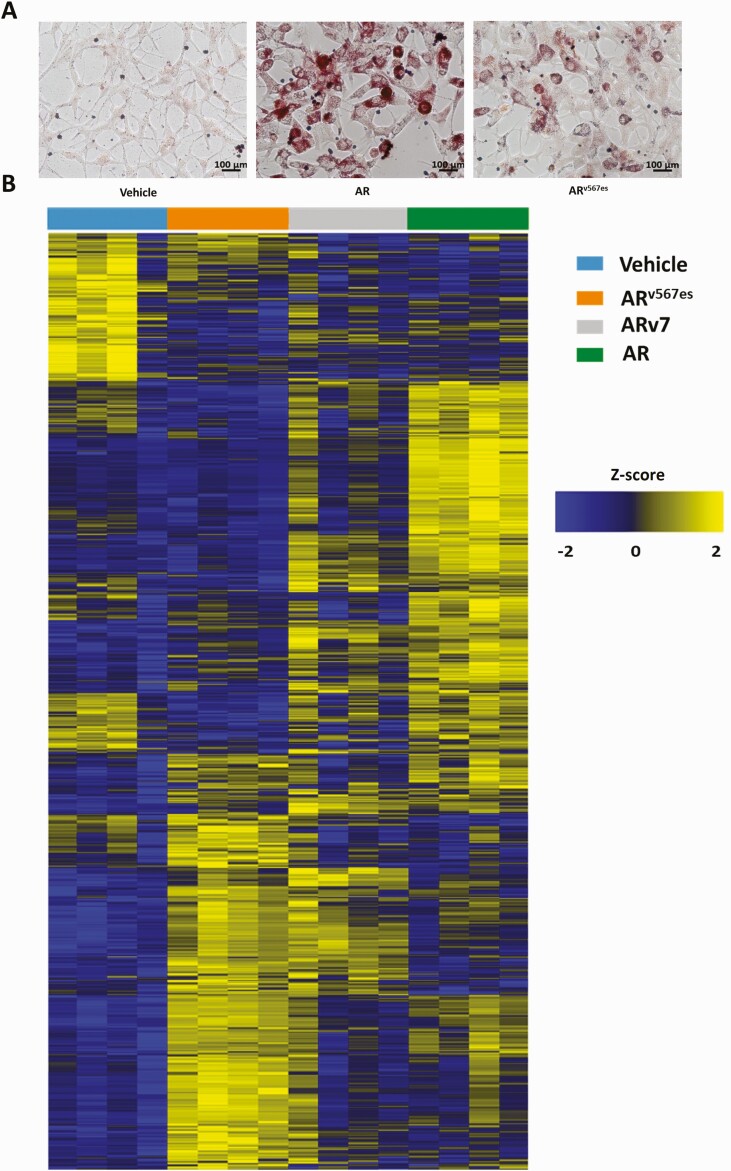
Hormone bound AR has been shown to stimulate accumulation of neutral lipids (49, 50) and ARv7 was less effective in inducing lipid droplet formation than AR (27). In order to compare the ability of ARv567es to induce lipid droplet formation with AR, we performed Oil red O staining in LNCaP cells expressing ARv567es and found that that ARv567es induces some accumulation of lipid droplets but seems to be less efficient than AR (Fig. 6A).
Figure 6.
Differences in lipid content resulting from the action of AR and the variants. (A) Phase contrast images of Oil red O staining for lipid accumulation in LNCaP-ARv567es cells treated with vehicle, 10 nM R1881, or 30 ng/mL Dox for 6 days in medium containing CSS. (B) Heat map representing Z scores of individual lipids whose level changes as a result of the action of AR, ARv7, and ARv567es. LNCaP-ARv7 cells were treated with vehicle (0.1% ethanol), 10 nM R1881 or 20 ng/mL Dox along with LNCaP-ARv567es cells that were treated with 30 ng/mL Dox for 48 hours in CSS medium. Four biological replicates collected from 4 independent experiments were used. Five million cells from each treatment were subjected to liquid chromatography and mass spectrometry to identify the lipids.
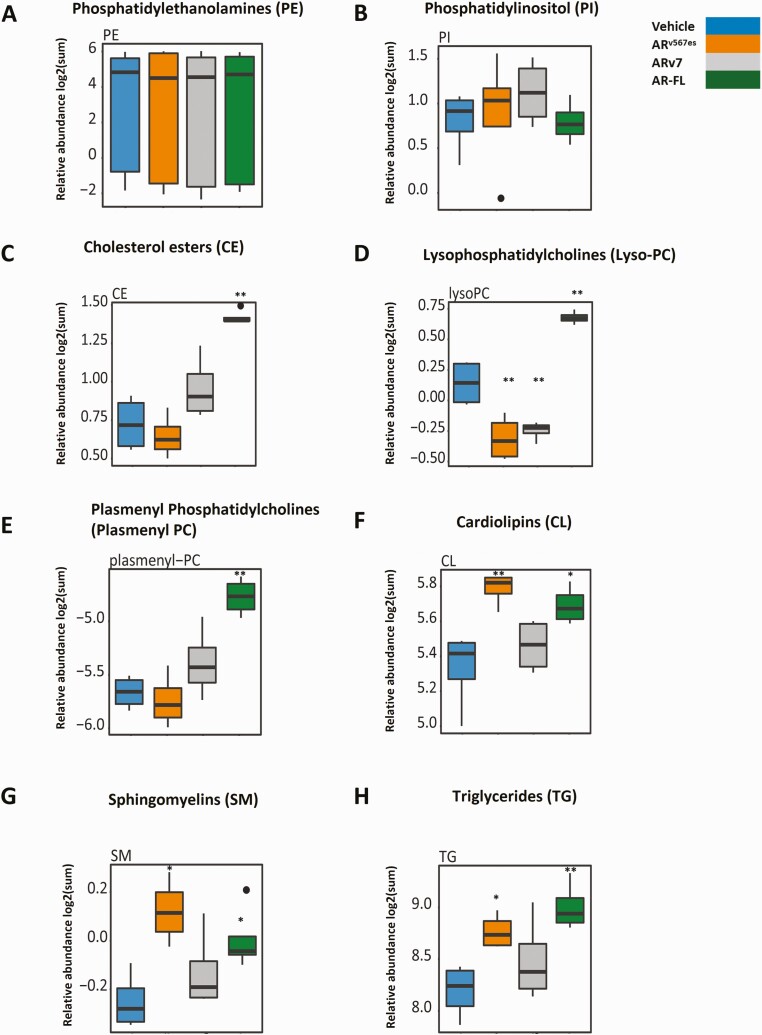
In order to further investigate differences in lipid profiles resulting from the action of AR, ARv7, or ARv567es in LNCaP cells, we employed untargeted shotgun lipidomics to examine the relative abundance of about 1170 lipids spanning 18 different lipid classes including phosphatidylserines, phosphocholines (PCs), ceramides, cardiolipins (CLs), triglycerides (TGs), etc. A heat map representation of our untargeted lipidomics analysis is shown in Fig. 6B. The z scores for log2 transformed normalized data are depicted. There are some lipids whose levels are changed by all 3 isoforms. Both ARv567es and AR strongly induce isoform-specific changes. ARv7 is more similar to AR in some aspects and more similar to ARv567es in others, but in general has a more modest effect on the levels of individual lipids. To examine classes of lipids, the abundance of all lipids within each class of lipids was determined by summing the individual lipids abundances and the log2 transformed normalized levels as a result of various treatments were compared. We did not observe any difference in the action of the 3 isoforms in altering the overall levels of some of the lipid classes such as phosphatidylethanolamines and phosphatidylinositol (Fig. 7A and 7B). However, we observed clear differences between vehicle and hormone treated samples in relative abundance of lipids from several other classes such as cholesterol esters (CEs), lysosomal PCs (lyso-PCs), plasmenyl phosphatidylcholines (plasmenyl PCs), CLs, sphingomyelins (SM), and triglycerides (TGs) (Fig. 7C-H). For lyso-phosphatidylcholines (lyso-PC), the variants reduced levels whereas hormone-bound AR increased levels (Fig. 7D). ARv567es expression led to an increase in the relative abundance of lipids in classes such as CLs, SMs, and TGs, similar to the action of hormone bound AR (Fig. 7F-H). Metabolism is very complex with multiple alternative pathways leading to the same products. Regulation of the enzymes is not only at the transcriptional level, but at several other levels including post-translational modifications, and through regulation by small molecule binding. We sought to determine whether any of these changes in cholesterol esters and in lyso-PC could be explained by changes in gene expression. Cholesterol homeostasis was 1 of the major Hallmark pathways upregulated by all 3 isoforms (Fig. 2G). Of the 74 genes attributed to this Hallmark pathway, 12 are regulated by the AR isoforms in our RNA-seq analyses (Table 1 (29) and GSE143907 and GSE151429). Some of these are directly involved in cholesterol synthesis, and others have fewer direct roles. Of the 12 regulated genes, 2 (HMGCS1 and LSS) are enzymes involved in cholesterol synthesis with HMGCS1 performing the first step in the mevalonic acid pathway. Interestingly, they are both induced by all 3 isoforms, but to a substantially higher extent by ARv7 and R1881. The metabolite pattern shows that R1881 induces much higher levels of cholesterol esters relative to control with a suggestion that ARv7 increases them as well (Fig. 7C). Cholesterol esters are produced when cholesterol levels are increased and these can be sequestered in lipid droplets. As shown in Fig. 6A, lipid droplet formation is highest in R1881 treated cells consistent with higher levels of cholesterol esters. Cholesterol is also the precursor of steroids and CYP11A1, the enzyme that catalyzes the first step in steroid biosynthesis is somewhat higher in ARv7 expressing cells (Table 2 (29)). Multiple enzymes involved in steroid synthesis and metabolism are found in our data (Table 2 (29)). One intriguing possibility for the difference in cholesterol ester levels between AR and the variants is the induction of CYP3A5 by variants. Knockout of CYP3A results in increased cholesterol and androgens in mouse prostate (51)
Figure 7.
Alterations in lipid classes induced by AR, ARv7, and ARv567es. Box plots of the relative abundance (log2 summed normalized peak area) evaluating the effect of AR and the variants on various lipid classes compared to the vehicle treatment group. (A) Phosphatidylethanolamines. (B) Phosphatidylinositol. (C) Cholesterol esters. (D) Lysophosphatidylcholines. (E) Plasmenyl phosphatidylcholines. (F) Cardiolipins. (G) Sphingomyelins. (H) Triglycerides (t-test comparison with respect to vehicle-treated cells, *P < .05 and **P < .01).
LysoPC levels were enhanced in R1881 treated cells, but reduced in cells with activated variants (Fig. 7D). LysoPC is produced by hydrolysis of PC by PLA2 (phospholipase A2) enzymes and the reaction is reversed by LPCAT enzymes. According to GeneCards, the various PLA2 enzymes show varied preferences for different phospholipids and in some cases for the specific fatty acid side chain. Although some of the enzymes in both classes are regulated by AR isoforms (Table 3 (29)), there is no simple pattern that might explain the altered overall abundance.
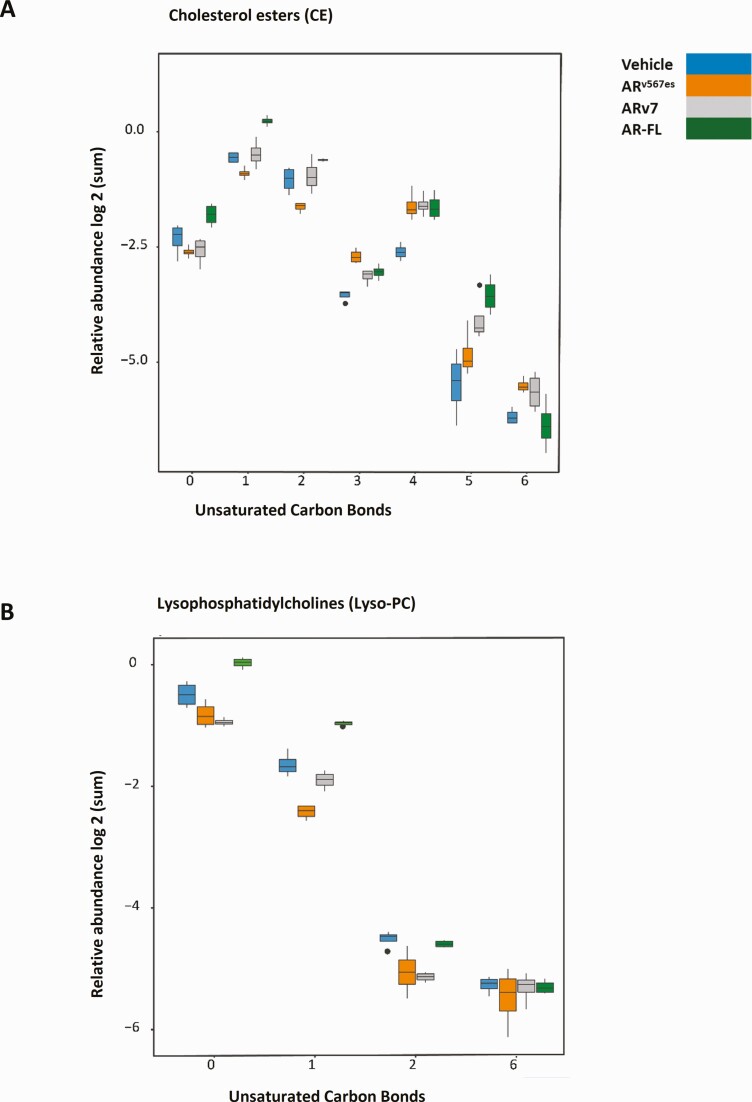
One added complication is the heterogeneity within classes of lipids. Figure 7 shows variation by classes, but within the class there are multiple lipid side chains and degrees of saturation of the individual lipids adding further to the diversity. Figure 8A shows the relative abundance of cholesterol esters with various numbers of unsaturated bonds. The groups represent the number of unsaturated bonds in the fatty acid. Note that R1881, which is most abundant overall, is most abundant for the saturated (0) and minimally unsaturated classes, whereas the expression is more equivalent for the minor highly unsaturated classes (statistical comparisons within the groups can be found in Table 4 (29)). In addition, chain length of the lipids can vary. Figure 2 (29) shows examples of the relative abundance of individual CE with different length side chains and different degrees of unsaturation. We also examined the abundance of Lyso PC with respect to saturation (Fig. 8B) (statistical comparisons are in Table 5 (29)) and levels of individual lipids (Fig. 3) (29). Again, the R1881 treated cells contained the highest level of the relatively saturated lipids, but levels were more similar for the minor more unsaturated forms.
Figure 8.
Alterations in individual lipids induced by AR, ARv7, and ARv567es. Box plots show relative abundance (log2 summed normalized peak area) of lipids in (A) cholesterol esters (CE) group and (B) lysophosphatidyl (LysoPC) groups differing in degree of unsaturation of carbon bonds due to action of AR and the variants.
Discussion
One of the major challenges in comparing isoform activities is the variations in gene expression even for AR and variants in different cell lines (23). Some of the variation certainly is due to secondary actions of long term treatment of cells with hormone or siRNA or, in our case Dox. Whereas hormone immediately activates AR, induction of the variant requires many hours and thus we are limited to longer term actions. However, these activities presumably are what would be relevant in tumors. In order to eliminate variations due to cell line background, we used LNCaP cell lines with inducible expression of the variants ARv7 and ARv567es. We found that ARv7 and ARv567es both regulate many genes in common with AR, but they also regulate unique genes compared to AR. Both variants induced EDN2 (Fig. 3A and 3B), a previously identified ARv7 preferential target gene (27, 28). Only the variants led to induction of connective tissue growth factor, implicated in angiogenesis and prostate cancer tumorigenesis (52) in the LNCaP model (Fig. 3A and 3B). While it is formally possible that increasing expression of an isoform might enable it to induce genes that are not induced at lower levels of the isoform, we sought to compare similar amounts of the isoforms. In the case of AR and ARv7 in the LNCaP background, we had shown previously that 0.1 to 0.3 nM R1881 was sufficient to maximally activate KLK3 and TMPRSS2 (28) (see Figs. 2C and 3E). Even 10 nM R1881 was insufficient to activate EDN2 (Fig. 3E). Moreover, increasing ARv7 expression increased EDN2, but did not cause induction of RASSF3 or EXTL2, 2 AR specific genes (Fig. 3A,B in (28) ). For this study, we chose genes that are highly regulated. Thus, it is unlikely that the observed differences are a reflection of conditions used.
Although ARv7 has been studied by numerous labs, much less has been done to compare the activities of the different variants. Previous work done in our laboratory comparing ARv7 with an AR construct terminating at amino acid 660 (AR N-terminal domain), which lacks the hormone binding domain and much of the hinge (28) showed no difference in regulation of the select target genes examined, hence indicating that short unique variant-specific sequences at the carboxy terminal may not contribute towards transcriptional specificity of AR variants. Transient transfection of ARv7 or ARv567es in LNCaP cells led to a distinctly different transcriptional program compared to AR (53), but no discernable differences were observed between transcriptional changes mediated by ARv7 and ARv567es. However, employing our LNCaP and VCaP models with Dox inducible expression of ARv7 and ARv567es, we have observed a few differences between the 2 variants as well. ARv567es seems to have a phenotype intermediate between AR and ARv7. For example, SGK1, a known AR responsive gene, was robustly induced by ARv567es, but not by ARv7. On the other hand, ARv567es induces the ARv7 target EDN2. However, ARv567es does not mimic ARv7 in all respects. NPR3 was strongly induced by ARv7 compared to AR and ARv567es. Although the comparison of gene expression showed that about 14% of genes upregulated by ARv567es were specific (Fig. 2C), when expression was tested by qPCR, genes were not highly induced or the differences between ARv567es and ARv7 were either not detected or were found to be minimal suggesting that the “unique” ARv567es genes were in many cases a result of the choice of fold cut-off for the comparison. Our studies, however, provided a strong confirmation for genes regulated only by AR or the variants.
Although others have sought to determine whether AR isoforms differ in their transcriptional programs, little has been done to examine downstream effects. We previously reported (27) that activation of AR and ARv7 differentially influenced steady state levels of metabolites. Here, we sought to determine whether there are differences among the 3 AR isoforms (Fig. 5B) and extended our studies to include untargeted analysis of lipids (Fig. 6B). Among the changes observed was an inverse correlation between levels of xanthine and hypoxanthine in our metabolomics data (Fig. 5B). ARv7 and ARv567es action led to lower levels of hypoxanthine and higher levels of xanthine compared to vehicle-treated cells, whereas AR action led to no significant change in relative abundance of hypoxanthine or xanthine. Interestingly, higher levels of xanthine appear to be present in metastatic prostate cancer compared with primary tumors (54). Hence it is possible that ARv7 and ARv567es action leads to higher levels or activity of xanthine dehydrogenase/oxidase (XDH), the enzyme that catalyzes the conversion of hypoxanthine to xanthine. Our RNA Sequencing data shows that induction of ARv7 led to a 4-fold increase in XDH mRNA levels, compared to vehicle-treated cells, although we have not verified this using qPCR.
Citrate is usually either oxidized in the mitochondria as part of the TCA cycle for energy production or transported to the cytosol to serve as a precursor for fatty acid synthesis (55). Prostate epithelial cells have a unique metabolic feature in that rather than oxidizing citrate for energy production, it is secreted as a component of prostatic fluid (56). Prostate cancer is characterized by reduced citrate levels, marking a shift from citrate secretion to citrate oxidation (56). Our previous studies showed that the rate of synthesis of citrate was actually enhanced by ARv7 induction (27), suggesting increased utilization or secretion of citrate. However, neither variant increased levels of citrate in the medium (Fig. 5G) indicating that there is increased metabolism of citrate. ARv567es induced the accumulation of lipid droplets (Fig. 6A) although not to the same extent as AR and our previous study also found that ARv7 was less effective in inducing lipid droplet formation than AR (27). Prostate cancer progression is marked by increased de novo lipogenesis (57-59). Increased concentrations of phospholipids such as PC and glycerophosphocholines have also been reported in high grade prostate cancer (60). The combination of our lipidomics data and transcriptomics shows that there is increased cholesterol metabolism leading in the case of AR to increased levels of cholesterol esters and lipid droplets. The gene expression profiles suggest that variants are also likely to increase synthesis of cholesterol, but the cholesterol not only is used to synthesize steroids, but also may be further metabolized by enzymes such as CYP3A5 leading to a reduced accumulation of cholesterol esters and lipid droplets. As a limitation, we must note that these hypotheses are drawn from RNA-Seq data. Although it is formally possible that some of the metabolite differences detected in cells expressing ARv567es are due to it being a separate LNCaP cell line that was compared to vehicle, hormone, and Dox treated ARv7 cells, we think that is for the most part unlikely since all cells were propagated in the absence of variant, so that there was no selective pressure for change. Because of the complex regulation of metabolic enzymes, further studies of rates of synthesis using flux analyses or enzyme assays are needed. Depletion of the candidate enzymes would be insufficient to support these concepts.
In conclusion, we find strong evidence that although the variants mimic many of the actions of AR, there are clear examples of isoform-specific regulation. While this would be expected for AR, which has the Ligand Binding Domain (LBD) and thus has additional interaction surfaces, it was not a foregone conclusion that the variants would have unique activities. The recent cryogenic electron microscopy (cryo-EM) structural study of AR bound to DNA and interacting with 2 coactivators (61) shows that the amino-terminal domains wrap around the LBD interacting in multiple sites within AR and that only 1 molecule of SRC-3 is bound to the dimer. Although structures of the variants are not available yet, the loss of the LBD interactions will cause the N terminal domain of the variants to be in different conformations that could facilitate unique interactions with other proteins bound to DNA as well as interactions with novel coregulators. Our studies show that while ARv567es mimics many transcriptional activities of ARv7, there are at least some instances where it is unable to induce an ARv7 target gene (NPR3) and others where it can mimic AR (SGK1). Unlike ARv7, ARv567es contains exon 4, which encodes the hinge region and a portion of the LBD. Thus, there may be unique interaction sites in this region including some of the post-translational modifications that contribute to the unique activities. We also detected a variety of downstream differences in steady state levels of metabolites and lipids supporting our belief that the differences in gene expression change downstream actions. However, additional studies will be required to confirm these changes, the functional significance, and the molecular basis for these differences.
Acknowledgments
We thank W.E. Bingman III for his technical assistance and Dr. David Rowley for the use of his microscope. We thank the Molecular and Cellular Biology tissue culture core for supplying media for maintenance of cell culture and also for providing HEK293T/17 cells for lentivirus production as well as the Baylor college of Medicine (BCM) protein and monoclonal antibody production core directed by Dean Edwards and supported by the P30 Cancer Center support grant (NCI-CA125123) for AR441 antibody. This project was also supported by the CPRIT Proteomics and Metabolomics Core Facility (RP170005), and Dan L. Duncan Cancer Centerthrough the P30 Cancer Center support grant (NCI-CA125123).
We thank Prof. Preethi Gunaratne and Dr. Sujash Chatterjee for RNA sequencing at the University of Houston sequencing core.
Financial Support: This project was supported by Cancer Prevention and Research Institute of Texas (CPRIT) RP150648 and the CPRIT Proteomics and Metabolomics Core Facility RP170005, National Institutes of Health (NIH)/National Cancer Institute (NCI) P30 CA125123, National Institutes of Health (NIH)/National Institute of Environmental Health Sciences (NIEHS) P30 ES030285, and the Dan L. Duncan Cancer Center.
Glossary
Abbreviations
- ADT
androgen deprivation therapy
- AR
androgen receptor
- CE
cholesterol ester
- CL
cardiolipin
- CRPC
castration-resistant prostate cancer
- CSS
charcoal stripped serum
- FBS
fetal bovine serum
- LC/MS
liquid chromatography/mass spectrometry
- PBS
phosphate-buffered saline
- PC
phosphocholine
- PCR
polymerase chain reaction
- TG
triglyceride
Additional Information
Disclosures: The authors have nothing to disclose.
Data Availability
The metabolomics and lipidomics data is available at https://www.metabolomicsworkbench.org/. AR and ARv7 RNA-sequencing data have been deposited to Gene Expression Omnibus (GEO) with the accession number GSE143907. ARv567 RNA Sequencing data has also been deposited to the GEO database with the accession number GSE151429. Supplementary material can be accessed at https://datadryad.org/stash/share/4Wn7ppkOqY-oMXqzNR_N4KY7HsUsARuJmjYCPtTUFTk
References
- 1.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25(2):276-308. [DOI] [PubMed] [Google Scholar]
- 2.Wadosky KM, Koochekpour S. Molecular mechanisms underlying resistance to androgen deprivation therapy in prostate cancer. Oncotarget. 2016;7(39):64447-64470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montgomery RB, Mostaghel EA, Vessella R, et al. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res. 2008;68(11):4447-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Locke JA, Guns ES, Lubik AA, et al. Androgen levels increase by intratumoral de novo steroidogenesis during progression of castration-resistant prostate cancer. Cancer Res. 2008;68(15):6407-6415. [DOI] [PubMed] [Google Scholar]
- 5.Heemers HV, Tindall DJ. Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr Rev. 2007;28(7): 778-808. [DOI] [PubMed] [Google Scholar]
- 6.Shafi AA, Yen AE, Weigel NL. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol Ther. 2013;140(3):223-238. [DOI] [PubMed] [Google Scholar]
- 7.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68(13):5469-5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun S, Sprenger CC, Vessella RL, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120(8):2715-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Z, Yang X, Sun F, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69(6):2305-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu R, Dunn TA, Wei S, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69(1):16-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hörnberg E, Ylitalo EB, Crnalic S, et al. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS One. 2011;6(4):e19059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Morrissey C, Sun S, et al. Androgen receptor variants occur frequently in castration resistant prostate cancer metastases. PLoS One. 2011;6(11). Published online November 17, 2011. doi: 10.1371/journal.pone.0027970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu J, Van der Steen T, Tindall DJ. Are androgen receptor variants a substitute for the full-length receptor? Nat Rev Urol. 2015;12(3):137-144. [DOI] [PubMed] [Google Scholar]
- 14.Antonarakis ES, Lu C, Wang H, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371(11):1028-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou ZX, Kemppainen JA, Wilson EM. Identification of three proline-directed phosphorylation sites in the human androgen receptor. Mol Endocrinol. 1995;9(5):605-615. [DOI] [PubMed] [Google Scholar]
- 16.Zhou ZX, Sar M, Simental JA, Lane MV, Wilson EM. A ligand-dependent bipartite nuclear targeting signal in the human androgen receptor. Requirement for the DNA-binding domain and modulation by NH2-terminal and carboxyl-terminal sequences. J Biol Chem. 1994;269(18):13115-13123. [PubMed] [Google Scholar]
- 17.Fu M, Rao M, Wang C, et al. Acetylation of androgen receptor enhances coactivator binding and promotes prostate cancer cell growth. Mol Cell Biol. 2003;23(23):8563-8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nyquist MD, Li Y, Hwang TH, et al. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proc Natl Acad Sci U S A. 2013;110(43):17492-17497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Yang R, Henzler CM, et al. Diverse AR gene rearrangements mediate resistance to androgen receptor inhibitors in metastatic prostate cancer. Clin Cancer Res. 2020;26(8):1965-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu G, Sprenger C, Sun S, et al. AR variant ARv567es induces carcinogenesis in a novel transgenic mouse model of prostate cancer. Neoplasia. 2013;15(9):1009-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcias G, Erdmann E, Lapouge G, et al. Identification of novel truncated androgen receptor (AR) mutants including unreported pre-mRNA splicing variants in the 22Rv1 hormone-refractory prostate cancer (PCa) cell line. Hum Mutat. 2010;31(1):74-80. [DOI] [PubMed] [Google Scholar]
- 22.Zhan Y, Zhang G, Wang X, et al. Interplay between cytoplasmic and nuclear androgen receptor splice variants mediates castration resistance. Mol Cancer Res. 2017;15(1):59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Z, Wu D, Thomas-Ahner JM, et al. Diverse AR-V7 cistromes in castration-resistant prostate cancer are governed by HoxB13. Proc Natl Acad Sci U S A. 2018;115(26):6810-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He Y, Lu J, Ye Z, et al. Androgen receptor splice variants bind to constitutively open chromatin and promote abiraterone-resistant growth of prostate cancer. Nucleic Acids Res. 2018;46(4):1895-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai L, Tsai YH, Wang P, et al. ZFX mediates non-canonical oncogenic functions of the androgen receptor splice variant 7 in castrate-resistant prostate cancer. Mol Cell. 2018;72(2):341-354.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu J, Lonergan PE, Nacusi LP, et al. The cistrome and gene signature of androgen receptor splice variants in castration resistant prostate cancer cells. J Urol. 2015;193(2): 690-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shafi AA, Putluri V, Arnold JM, et al. Differential regulation of metabolic pathways by androgen receptor (AR) and its constitutively active splice variant, AR-V7, in prostate cancer cells. Oncotarget. 2015;6(31):31997-32012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krause WC, Shafi AA, Nakka M, Weigel NL. Androgen receptor and its splice variant, AR-V7, differentially regulate FOXA1 sensitive genes in LNCaP prostate cancer cells. Int J Biochem Cell Biol. 2014;54:49-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagandla H, , Robertson MJ, , Putluri V, et al. Supplementary data for Isoform-specific Activities of androgen receptor and its splice variants in prostate cancer cells. ProMED-mail website. https://datadryad.org/stash/share/4Wn7ppkOqY-oMXqzNR_N4KY7HsUsARuJmjYCPtTUFTk. Accessed November 24, 2020. [DOI] [PMC free article] [PubMed]
- 30.RRID:AB_11000751. Accessed November 24, 2020.
- 31.RRID:AB_2861276. Accessed November 24, 2020.
- 32.RRID:AB_11004139. Accessed November 24, 2020.
- 33.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9): 1650-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. Published online May 2, 2011. 2011. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 35.Andrews S. FastQC – a quality control tool for high throughput sequence data. Babraham Bioinforma. 2010. doi: citeulike-article-id:11583827. ProMED-mail website. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed August 15, 2020.
- 36.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923-930. [DOI] [PubMed] [Google Scholar]
- 37.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terunuma A, Putluri N, Mishra P, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest. 2014;124(1):398-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Putluri N, Maity S, Kommagani R, et al. Pathway-centric integrative analysis identifies RRM2 as a prognostic marker in breast cancer associated with poor survival and tamoxifen resistance. Neoplasia. 2014;16(5):390-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Putluri N, Shojaie A, Vasu VT, et al. Metabolomic profiling reveals a role for androgen in activating amino acid metabolism and methylation in prostate cancer cells. PLoS One. 2011;6(7):e21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Putluri N, Shojaie A, Vasu VT, et al. Metabolomic profiling reveals potential markers and bioprocesses altered in bladder cancer progression. Cancer Res. 2011;71(24):7376-7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhowmik SK, Ramirez-Peña E, Arnold JM, et al. EMT-induced metabolite signature identifies poor clinical outcome. Oncotarget. 2015;6(40):42651-42660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911-917. [DOI] [PubMed] [Google Scholar]
- 45.Vantaku V, Dong J, Ambati CR, et al. Multi-omics integration analysis robustly predicts high-grade patient survival and identifies CPT1B Effect on fatty acid metabolism in bladder cancer. Clin Cancer Res. 2019;25(12):3689-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Breil C, Abert Vian M, Zemb T, Kunz W, Chemat F. “Bligh and Dyer” and Folch methods for solid–liquid–liquid extraction of lipids from microorganisms. Comprehension of solvatation mechanisms and towards substitution with alternative solvents. Int J Mol Sci. 2017;18(4):708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piyarathna DWB, Rajendiran TM, Putluri V, et al. Distinct lipidomic landscapes associated with clinical stages of urothelial cancer of the bladder. Eur Urol Focus. 2018;4(6):907-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Purwaha P, Gu F, Piyarathna DWB, et al. Unbiased lipidomic profiling of triple-negative breast cancer tissues reveals the association of sphingomyelin levels with patient disease-free survival. Metabolites. 2018;8(3):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swinnen JV, Van Veldhoven PP, Esquenet M, Heyns W, Verhoeven G. Androgens markedly stimulate the accumulation of neutral lipids in the human prostatic adenocarcinoma cell line LNCaP. Endocrinology. 1996;137(10): 4468-4474. [DOI] [PubMed] [Google Scholar]
- 50.Swinnen JV, Esquenet M, Goossens K, Heyns W, Verhoeven G. Androgens stimulate fatty acid synthase in the human prostate cancer cell line LNCaP. Cancer Res. 1997;57(6): 1086-1090. [PubMed] [Google Scholar]
- 51.Hashimoto M, Kobayashi K, Yamazaki M, et al. Cyp3a deficiency enhances androgen receptor activity and cholesterol synthesis in the mouse prostate. J Steroid Biochem Mol Biol. 2016;163:121-128. [DOI] [PubMed] [Google Scholar]
- 52.Yang F, Tuxhorn JA, Ressler SJ, McAlhany SJ, Dang TD, Rowley DR. Stromal expression of connective tissue growth factor promotes angiogenesis and prostate cancer tumorigenesis. Cancer Res. 2005;65(19):8887-8895. [DOI] [PubMed] [Google Scholar]
- 53.Hu R, Lu C, Mostaghel EA, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72(14):3457-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sreekumar A, Poisson LM, Rajendiran TM, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457(7231):910-914. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 55.Liu Y. Fatty acid oxidation is a dominant bioenergetic pathway in prostate cancer. Prostate Cancer Prostatic Dis. 2006;9(3):230-234. [DOI] [PubMed] [Google Scholar]
- 56.Costello LC, Franklin RB. The intermediary metabolism of the prostate: a key to understanding the pathogenesis and progression of prostate malignancy. Oncology. 2000;59(4):269-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rossi S, Graner E, Febbo P, et al. Fatty acid synthase expression defines distinct molecular signatures in prostate cancer. Mol Cancer Res. 2003;1(10):707-715. [PubMed] [Google Scholar]
- 58.Swinnen JV, Roskams T, Joniau S, et al. Overexpression of fatty acid synthase is an early and common event in the development of prostate cancer. Int J Cancer. 2002;98(1):19-22. [DOI] [PubMed] [Google Scholar]
- 59.Swinnen JV, Vanderhoydonc F, Elgamal AA, et al. Selective activation of the fatty acid synthesis pathway in human prostate cancer. Int J Cancer. 2000;88(2):176-179. [DOI] [PubMed] [Google Scholar]
- 60.Zhou X, Mao J, Ai J, et al. Identification of plasma lipid biomarkers for prostate cancer by lipidomics and bioinformatics. PLoS One. 2012;7(11):e48889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu X, Yi P, Hamilton RA, et al. Structural insights of transcriptionally active, full-length androgen receptor coactivator complexes. Mol Cell. 2020;79(5):812-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The metabolomics and lipidomics data is available at https://www.metabolomicsworkbench.org/. AR and ARv7 RNA-sequencing data have been deposited to Gene Expression Omnibus (GEO) with the accession number GSE143907. ARv567 RNA Sequencing data has also been deposited to the GEO database with the accession number GSE151429. Supplementary material can be accessed at https://datadryad.org/stash/share/4Wn7ppkOqY-oMXqzNR_N4KY7HsUsARuJmjYCPtTUFTk