Abstract
This study aims to build a radiological model based on standard MR sequences for detecting methylguanine methyltransferase (MGMT) methylation in gliomas using texture analysis. A retrospective cross‐sectional study was undertaken in a cohort of 53 glioma patients who underwent standard preoperative magnetic resonance (MR) imaging. Conventional visual radiographic features and clinical factors were compared between MGMT promoter methylated and unmethylated groups. Texture analysis extracted the top five most powerful texture features of MR images in each sequence quantitatively for detecting the MGMT promoter methylation status. The radiomic signature (Radscore) was generated by a linear combination of the five features and estimates in each sequence. The combined model based on each Radscore was established using multivariate logistic regression analysis. A receiver operating characteristic (ROC) curve, nomogram, calibration, and decision curve analysis (DCA) were used to evaluate the performance of the model. No significant differences were observed in any of the visual radiographic features or clinical factors between different MGMT methylated statuses. The top five most powerful features were selected from a total of 396 texture features of T1, contrast‐enhanced T1, T2, and T2 FLAIR. Each sequence’s Radscore can distinguish MGMT methylated status. A combined model based on Radscores showed differentiation between methylated MGMT and unmethylated MGMT both in the glioblastoma (GBM) dataset as well as the dataset for all other gliomas. The area under the ROC curve values for the combined model was 0.818, with 90.5% sensitivity and 72.7% specificity, in the GBM dataset, and 0.833, with 70.2% sensitivity and 90.6% specificity, in the overall gliomas dataset. Nomogram, calibration, and DCA also validated the performance of the combined model. The combined model based on texture features could be considered as a noninvasive imaging marker for detecting MGMT methylation status in glioma.
Keywords: glioma, magnetic resonance imaging, methylguanine methyltransferase, texture analysis
A retrospective cross‐sectional study was undertaken in a cohort of 53 glioma patients who underwent standard preoperative magnetic resonance (MR) imaging. Conventional visual radiographic features and clinical factors were compared between methylguanine methyltransferase (MGMT) promoter methylated and unmethylated groups. Texture analysis extracted the top five most powerful texture features of MR images in each sequence quantitatively for detecting the MGMT promoter methylation status.
Abbreviations
- CE
contrast‐enhanced
- CNS
central nervous system
- DCA
decision curve analysis
- GLCM
gray level co‐occurrence matrix
- MGMT
Methylguanine methyltransferase
- mRMR
Minimum‐redundancy maximum‐relevance
- PCR
polymerase chain reaction
- qMSP
quantitative methylation‐specific PCR
- ROC
receiver operating characteristic
- ROI
region of interest
1. INTRODUCTION
Methylguanine methyltransferase is an enzyme that repairs DNA. When the MGMT promoter is methylated, the MGMT gene is silenced/inactivated epigenetically, which suppresses DNA repair activity. 1 The methylation of the MGMT promoter is associated with sensitivity to TMZ, as tumor cells lacking MGMT activity are significantly more sensitive to the cytotoxic effects of TMZ. 2 Determining the MGMT promoter status before treatment is very important for high‐grade gliomas, especially for GBM, for which TMZ is considered part of the standard treatment protocol. Furthermore, it has also been suggested that the survival benefit of MGMT promoter methylation is not exclusive to patients treated with TMZ and radiation therapy 3 ; prolonged survival has also been reported in patients with methylated MGMT promoters irrespective of treatment method. Consequently, the methylation of the MGMT promoter has been widely accepted as a strong prognostic and predictive molecular marker of gliomas, whether high‐grade or low‐grade. 4 , 5 The MGMT promoter status is typically determined by methylation‐specific PCR, pyrosequencing, or a methylation chip based on a specimen, which could be hindered by the relatively long detection period, tumor heterogeneity, or the unavailability of surgery or biopsy.
A preoperative noninvasive method of evaluating the MGMT promoter methylation status would be of benefit and could help guide more efficacious decisions to be made on treatment. Although promising results have been reported with the use of conventional and functional MR to predict MGMT promoter methylation status, 6 , 7 limited studies have focused on the use of texture analysis for this prediction. 8 Given that MGMT promoter methylation status is an independent prognostic biomarker both for high‐ and low‐grade gliomas, and is also a predictive biomarker for treatment response to chemotherapy with an alkylating agent regardless of histological classification, 5 , 9 the ability to predict MGMT promotor methylation status using texture analysis could have great utility in clinical practice.
This study retrospectively investigated the imaging characteristics from preoperative conventional MR images of gliomas using a texture analysis approach, aiming to build a reliable radiological model to noninvasively predict the MGMT promoter methylation status in glioma patients, especially in GBM patients.
2. MATERIALS AND METHODS
2.1. Dataset
Fifty‐three consecutive patients with histopathologically validated gliomas (44 high‐grade gliomas and nine low‐grade gliomas) (33 men and 20 women, aged 46.547 ± 13.580, [range 18‐72] years) who underwent standard MRI for presurgical planning at our hospital between September 2013 and September 2018 were included in this retrospective cross‐sectional study. Our study was approved by our institutional review board, and the requirement of written informed consent was waived. The inclusion criteria were as follows: (a) adults with histopathology verified primary gliomas who had no history of central nervous system malignancy; (b) underwent MGMT promoter methylation status testing; (c) underwent preoperative MRI including routine diagnostic protocol; (d) MR images were obtained without artifacts that affect image observation and post‐processing; and (e) had not undergone radiotherapy, chemotherapy, or other treatment prior to MRI acquisition and surgery. The gender, age, histopathologic types (GBM, diffusely infiltrating astrocytomas [including NOS], oligodendroglioma [including NOS], diffuse midline glioma, and other astrocytomas), and tumor grade (high and low) were considered as clinical factors associated with MGMT status. According to the WHO guidelines, gliomas are graded I‐IV using the following histological criteria: cytological atypia, mitotic activity, cellularity, microvascular proliferation, and/or necrosis. Gliomas graded I and II were classified as low‐grade gliomas, whereas gliomas graded III and IV were classified as high‐grade gliomas. 10
2.2. Histopathological protocols
Tumor tissue specimens were fixed in 10% formalin and embedded in paraffin wax using routine processing. For each subject, H&E‐stained sections were examined to classify the tumors according to the WHO International Histological Classification of Tumours. The MGMT promoter methylation status was determined by qMSP. The qMSP process involves the treatment of genomic DNA, denatured by sodium hydroxide with hydroquinone and sodium bisulfite to convert unmethylated C into T. The target gene was amplified by primers designed according to the gene sequence after C‐to‐T conversion. Because the C in the methylated CPG island cannot be converted into T, the target gene will not be amplified with the above primers. Therefore, the purpose of identifying genomic DNA methylation can be achieved by the above methods. To detect the MSP amplification products, if the fragments can be amplified with the primers for the methylated DNA chain, it means that there is methylation at the detected site. If the primers for the unmethylated DNA chain after treatment are used to amplify the fragments, it means that there is no methylation at the detected sites. To reduce the influence of tumor heterogeneity, we selected the solid components of the tumor for amplification, avoiding necrotic or hemorrhagic areas. DNA extracted from unstained tissue slides made from paraffin‐embedded blocks of tumor tissues was subjected to bisulfite modification by a Simlex OUP FFPE DNA extraction kit (TIB), according to the manufacturer’s instructions. Polymerase chain reaction was carried out with a DRR007 kit (Takara) using the Verity 96‐Well Thermal Cycler (Thermo Fisher Scientific), and pyrosequencing was done using the PyroMark Q96 system (Qiagen) in the 5 CpG island region within the MGMT promoter. A glioma was defined as “methylated” if the average methylation rate of the CpG regions was 8% or higher, otherwise, the tumor was classified as “unmethylated” (Figure 1). 11
FIGURE 1.
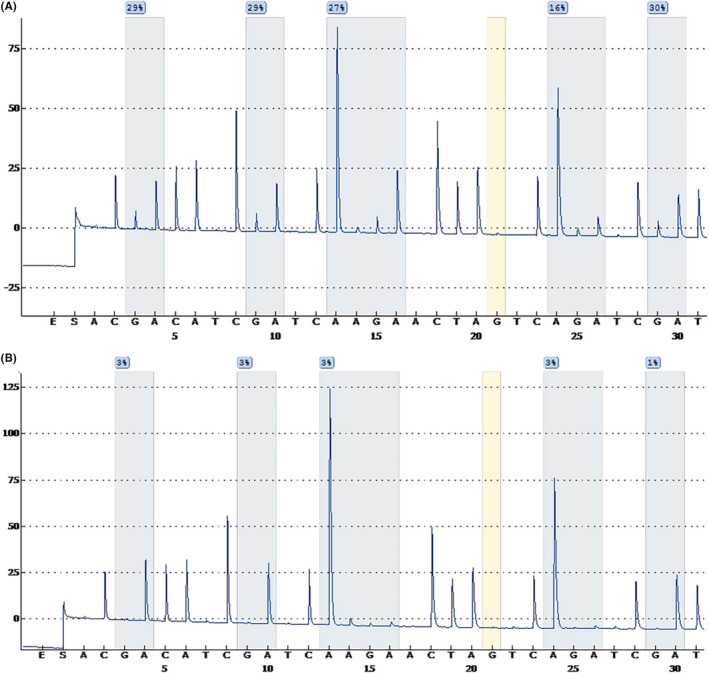
Sequencing peak map of methylguanine methyltransferase (MGMT) promoter methylation in glioma tumor tissue specimens. A, MGMT promoter methylated. B, MGMT promoter unmethylated
2.3. Magnetic resonance imaging protocols
For all patients, MR imaging was acquired within 72 hours before tumor resection. The MR scans were acquired on a 3T MR scanner (Verio; Siemens) using a 16‐channel head coil. The routine diagnostic protocol included T1w imaging, CE T1w imaging, T2w imaging, and T2‐FLAIR imaging. Axial T1w scanning was acquired at spin‐echo sequence with the following parameters: TR/TE of 400 ms/15 ms, matrix of 320 × 192, FOV of 24 cm, and a voxel size of 0.4 × 0.4 × 6 mm3. T2 weighted scanning was acquired at a fast spin‐echo sequence with a TR/TE of 6000 ms/99 ms, matrix of 320 × 192, FOV of 24 cm, and a voxel size of 0.4 × 0.4 × 6 mm3. T2‐FLAIR was acquired at a TR/TE of 9000 ms/94 ms, matrix of 320 × 192, FOV of 24 cm, and a voxel size of 0.5 × 0.5 × 6 mm3. Contrast‐enhanced T1w scanning was acquired at a 3‐D MP RAGE sequence with a TR/TE of 2100 ms/2.299 ms, matrix of 512 × 512, FOV of 24 cm, and a voxel size of 0.9 × 0.9 × 0.9 mm3, after administration of the contrast agent gadodiamide (0.1 mmol/kg; Omniscan, GE Medical Systems) at a rate of 2.5‐3 mL/s, followed by a 20 cm3 saline chaser at the same flow rate.
For all subjects, the entire tumor 3‐D ROI was segmented manually using ITK‐SNAP software (version 3.4.0; http://www.itksnap.org) 12 in each sequence. The ROIs include the entire tumor avoiding blood vessels and discernible peritumoral edema. This was carried out by a clinical radiologist (LW with 4 years of experience) and verified by a senior radiologist (WH with 11 years of experience), who were blinded to molecular information.
2.4. Imaging analysis: Visual radiographic features
Two radiologists blinded to patients’ molecular data (LW And WH) reviewed the MR images and assessed the following tumor visual radiographic features by consensus: tumor location, crossing the midline (+/−), enhancement degree (none, mild, marked), necrosis/cyst change (none, mild, marked), and presence of peritumoral edema (none, mild, marked). Necrosis was defined as an area located within the CE region of the tumor that had no or little discernable contrast enhancement. Cystic regions were defined as homogeneous areas isointense to cerebrospinal fluid on T1w and T2w images with a thin enhancing rim on T1 postcontrast images. Classification criteria were evaluated by the VASARI scoring system. 13
2.5. Imaging analysis: Texture analysis
Image processing was applied before feature extraction, including image resampling to 1 × 1 × 1 mm3 voxel size using linear interpolation and image gray normalization to uniform grayscale of 0‐255. A total of 396 texture features were extracted using AK software (Analysis Kit; GE Healthcare). The feature set included histogram features, GLCM features, gray level run‐length matrix features, form factor features, and gray level size‐zone matrix features. These features could characterize intratumor heterogeneity.
To eliminate the differences in the value scales of extraction features, feature normalization was carried out before feature selection, and each feature for all patients was normalized with Z‐scores, subtracting the mean value and dividing by SD. Minimum redundancy maximum relevance was used to eliminate the redundant and irrelevant features. The top five features 14 among 396 texture features with the greatest correlation with the MGMT methylation status were screened in. The Radscore of each sequence was then calculated for each patient using a linear combination of the five selected features weighted by their respective coefficients. The data texture analysis flow is shown in Figure 2.
FIGURE 2.
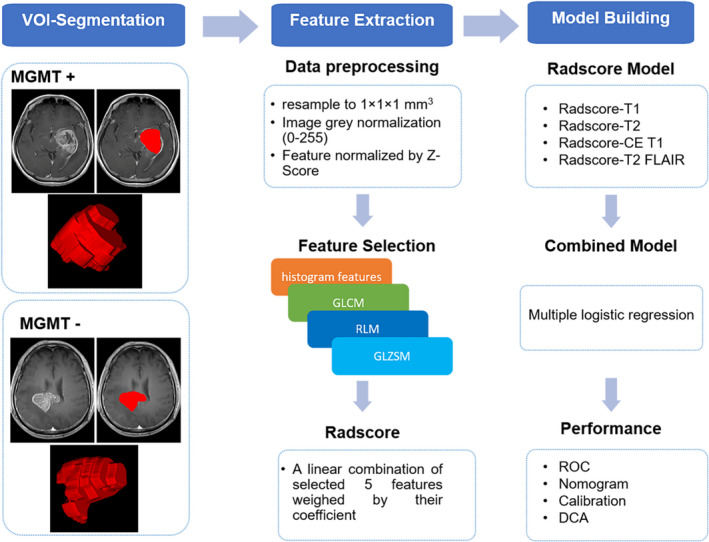
Workflow of texture analysis to detect methylguanine methyltransferase (MGMT) methylation in glioma. CE, contrast‐enhanced; DCA, decision curve analysis; FLAIR, fluid attenuated inversion recovery; GLCM, gray level co‐occurrence matrix; GLZSM, gray level size‐zone matrix; RLM, gray level run‐length matrix; Radscore, radiomic signature; ROC, receiver operating characteristic; VOI, volume of interest
2.6. Radiological model building and validation
The Radscore of each sequence was considered a variable. A radiological combined model was built based on Radscores of four sequences using multiple logistic regression. To estimate the prediction error and confidence interval, we further tested the proposed model using a 1000‐iteration bootstrap analysis on the overall dataset. The performance of the radiological model of four sequences was validated by a ROC curve. The radiological combined model was validated by a ROC curve, nomogram, calibration, and DCA.
2.7. Statistical analysis
All statistical analyses were undertaken with R software (version 3.5.2, https://www.rproject.org), SPSS (version Windows 25, https://www.spss.com), and Medcalc (version 19.1.6, https://www.medcalc.org). For discrimination between MGMT methylated or MGMT unmethylated using visual radiographic features and clinical factors, the χ2 test was used to compare categorical variables, and an independent‐sample t test (conforming to a normal distribution)/Mann‐Whitney U test (not conforming to the normal distribution) was used to compare continuous variables (SPSS). The ROC analysis was carried out with Medcalc. Logistic regression, nomogram, calibration, and DCA were carried out with the glm, PredictABEL, and DecisionCurve packages of R software.
3. RESULTS
3.1. Basic information
Methylguanine methyltransferase methylation status was defined based on qMSP results (Figure 1). Of the 53 patients, 21 (39.62%) belonged to the MGMT methylated group and 32 patients (60.38%) belonged to the MGMT unmethylated group. There were 32 GBM patients in our overall dataset; 11 (34.38%) belonged to the MGMT methylated group and 21 patients (65.62%) belonged to the MGMT unmethylated group. None of the five visual radiographic features and four clinical factors showed a significant difference between the MGMT methylated and unmethylated groups. The clinical information of the patients is shown in Table 1. Representative cases of MR images of MGMT methylated and unmethylated groups are shown in Figure 3.
TABLE 1.
Clinical factors of patients with glioma (n = 53)
| MGMT methylated (n = 21) | MGMT unmethylated (n = 32) | P value | |
|---|---|---|---|
| Age, years; mean ± SD | 47.43 ± 12.55 | 45.66 ± 15.10 | .658 |
| Gender | |||
| Male | 11 (52.38) | 22 (68.75) | .229 |
| Female | 10 (47.62) | 10 (31.25) | |
| Grade | |||
| High (WHO III‐IV) | 17 (80.95) | 25 (78.12) | .806 |
| Low (WHO I‐II) | 4 (19.05) | 7 (21.88) | |
| Histopathological type | |||
| Glioblastoma | 8 (38.10) | 15 (46.88) | .827 |
| Astrocytoma/oligoastrocytoma | 11 (52.38) | 12 (37.50) | |
| Oligodendroglioma | 2 (9.52) | 2 (6.25) | |
| Others | 0 (0.00) | 3 (9.38) | |
Data are shown as n (%) unless otherwise indicated.
MGMT, methylguanine methyltransferase.
FIGURE 3.
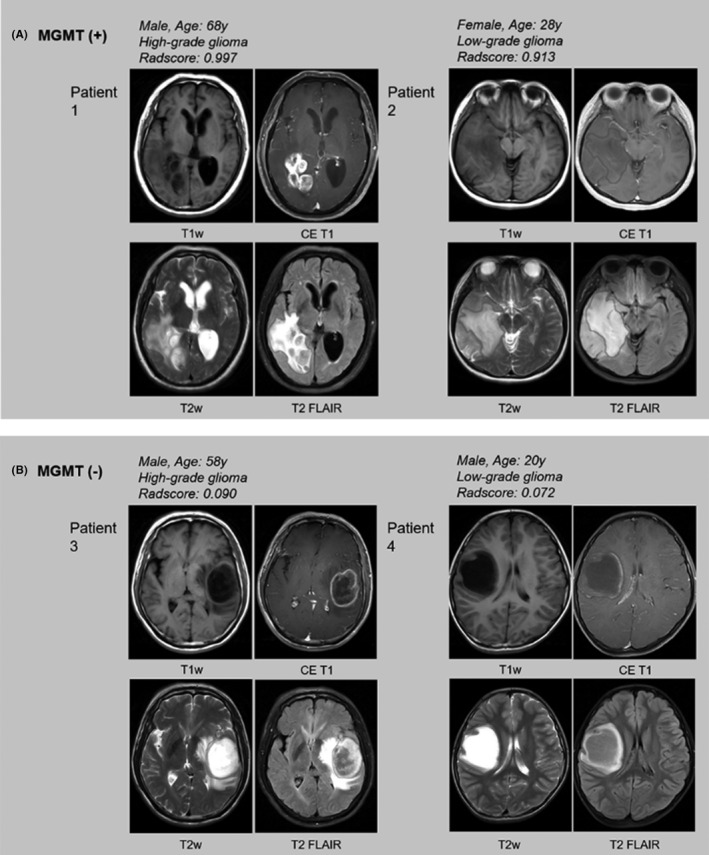
Representative images of glioma patients with or without methylguanine methyltransferase (MGMT) methylation. A, Patient 1, a 68‐y‐old man with MGMT promoter methylated high‐grade glioma. The combined model radiomic signature (Radscore) is 0.997. Patient 2, a 28‐y‐old woman with MGMT promoter methylated low‐grade glioma. The combined model Radscore is 0.913. B, Patient 3, a 58‐y‐old man with MGMT promoter unmethylated high‐grade glioma. The combined model Radscore is 0.090. Patient 4, a 20‐y‐old man with MGMT promoter unmethylated low‐grade glioma. The combined model Radscore is 0.072. CE, contrast‐enhanced; FLAIR, fluid attenuated inversion recovery; T1w, T1 weighted; T2w, T2 weighted
3.2. Texture analysis
After the selection of the minimum redundancy maximum relevance algorithm and elimination of redundancies, the five texture features with the greatest correlation with MGMT methylation and the least redundancy between features were selected in each group. All of these texture features are list in Table 2. For T1w images, only Radscore (P = .011) was significantly different between the MGMT methylated group and unmethylated groups. For CE T1w images, Radscore (P < .001), Compactness2 (P = .011), SphericalDisproprotion (P = .01), and Sphericity (P = .007) had a significant difference between the MGMT methylated and unmethylated groups. For T2w images, Radscore (P = .009), GLCMEntropy_angle45_offset7 (P = .023), GLCMEntropy_angle90_offset4 (P = .027), GLCMEntropy_angle90_offset7 (P = .019), and GLCMEntropy_angle135_offset7 (P = .033) had a significant difference between the MGMT methylated group and unmethylated group. Only Radscore (P = .011) can discriminate against the MGMT methylated status in FLAIR images.
TABLE 2.
Features selected based on texture analysis
| Sequences | Texture features |
MGMT methylated n = 21 |
MGMT unmethylated n = 32 |
P value |
|---|---|---|---|---|
| T1w | ClusterShade_angle135_offset7 | −0.05 ± 1.38 | 0.03 ± 0.66 | .791 |
| Compactness1 | −0.27 ± 0.98 | 0.18 ± 0.98 | .106 | |
| GLCMEntropy_AllDirection_offset1_SD | 0.25 ± 1.46 | −0.16 ± 0.48 | .140 | |
| LowGreyLevelRunEmphasis_AllDirection_offset1_SD | 0.22 ± 1.58 | −0.14 ± 0.10 | .202 | |
| LongRunHighGreyLevelEmphasis_AllDirection_offset7_SD | −0.10 ± 0.23 | 0.07 ± 1.28 | .542 | |
| Radscore‐T1 | −0.03 ± 1.14 | −0.69 ± 0.67 | .011 | |
| CE T1w | GLCMEntropy_AllDirection_offset4_SD | −0.27 ± 0.50 | 0.24 ± 1.25 | .086 |
| GLCMEntropy_angle135_offset7 | 0.28 ± 1.23 | −0.23 ± 0.86 | .083 | |
| Compactness2 | 0.49 ± 1.09 | −0.28 ± 0.88 | .010 | |
| SphericalDisproportion | 0.49 ± 1.09 | −0.28 ± 0.88 | .010 | |
| Sphericity | −0.48 ± 1.05 | 0.28 ± 0.90 | .007 | |
| Radscore‐CET1 | 0.68 ± 2.07 | −1.30 ± 1.16 | <.001 | |
| T2w | ClusterProminence_angle0_offset1 | −0.21 ± 0.90 | 0.28 ± 1.07 | .089 |
| GLCMEntropy_angle45_offset7 | 0.37 ± 1.16 | −0.29 ± 0.89 | .023 | |
| GLCMEntropy_angle90_offset4 | 0.35 ± 1.18 | −0.28 ± 0.85 | .027 | |
| GLCMEntropy_angle90_offset7 | 0.38 ± 1.17 | −0.30 ± 0.86 | .019 | |
| GLCMEntropy_angle135_offset7 | 0.33 ± 1.15 | −0.29 ± 0.90 | .033 | |
| Radscore‐T2 | −0.12 ± 0.97 | −0.74 ± 0.69 | .009 | |
| T2‐FLAIR | ClusterShade_AllDirection_offset4 | −0.10 ± 0.93 | 0.07 ± 1.07 | .548 |
| ClusterShade_angle0_offset7 | −0.09 ± 0.94 | 0.07 ± 1.06 | .586 | |
| ClusterShade_angle45_offset4 | −0.10 ± 0.93 | 0.08 ± 1.07 | .53 | |
| ClusterShade_angle90_offset4 | −0.10 ± 0.92 | 0.08 ± 1.07 | .531 | |
| ClusterShade_angle90_offset7 | −0.09 ± 0.91 | 0.07 ± 1.08 | .573 | |
| Radscore‐FLAIR | −0.20 ± 0.93 | −0.68 ± 0.66 | .034 |
CE, contrast‐enhanced; FLAIR, fluid attenuated inversion recovery; GLCM, gray level co‐occurrence matrix; MGMT, methylguanine methyltransferase; Radscore, radiomic signature; T1w, T1 weighted; T2w, T2 weighted.
3.3. Model development and analysis
Among the four sequences’ Radscores, the Radscore of CE T1 accounted for the highest weight in the radiological combined model built by multivariate logistic regression with an odds ratio value of 2.712 (Figure 4A). The ROC results of the four sequences of radiological models based on Radscore and the radiological combined model are shown in Table 3 and Figure 4B. The radiological combined model (proposed model) showed the best performance for discrimination of MGMT methylation status, with sensitivity, specificity, and AUC of 72.7%, 90.6%, and 0.833 (95% confidence interval, 0.706‐0.922), building by multivariate logistic regression (Enter) greater than the forward model. The density distribution map from the bootstrap analysis regarding the distributions of AUCs (0.85 ± 0.04) (Figure 4C) showed that the combined model lacked overfitting. We also verified the model performance in the GBM dataset using the ROC curve for consideration of GBM’s clinical and imaging characteristics. The combined model has an AUC of 0.818, with 90.5% sensitivity and 72.7% specificity (95% confidence interval, 0.672‐0.947), for discrimination of MGMT methylation in GBMs (Figure 4D). No significant difference was observed in the ROC performance of the combined model compared to the overall glioma dataset over GBM datasets (P = .89).
FIGURE 4.
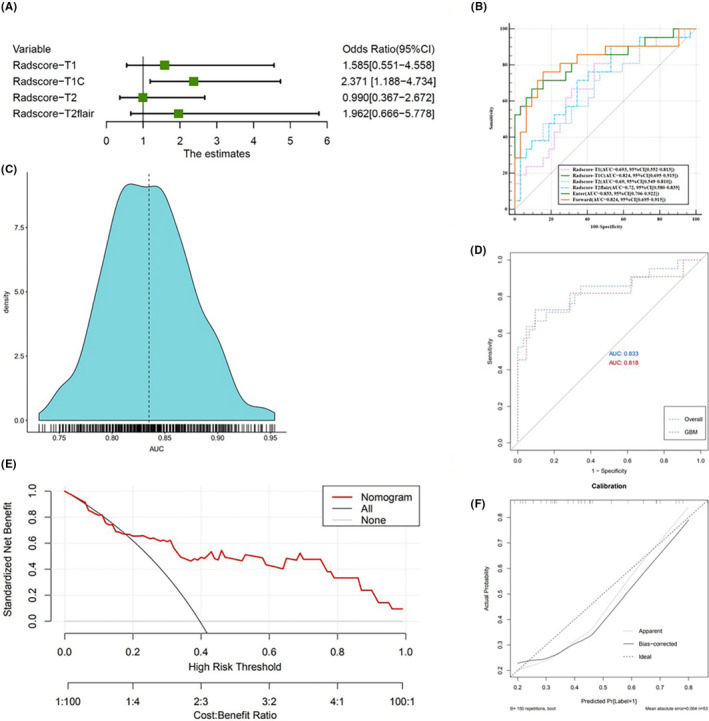
A, Forest plot of independent predictors of methylguanine methyltransferase (MGMT) in glioma, with odds ratios. B, Receiver operating characteristic (ROC) analysis for different sequence models and the combined model. C, Density distribution map regarding the distribution of area under the ROC curves (AUCs) from the bootstrap analysis for the combined model. D, Efficacy of the combined model in recognizing MGMT promoter methylation in gliomas was similar to that in glioblastomas (GBM), showing no significant difference. E, Decision curve for the nomogram. F, Calibration plot of the nomogram. CI, confidence interval; FLAIR, fluid attenuated inversion recovery
TABLE 3.
Receiver operating characteristic (ROC) results of models to detect methylguanine methyltransferase methylation in glioma using texture analysis
| Variable | AUC | 95% CI | Sensitivity | Specificity | Cut‐off | P value |
|---|---|---|---|---|---|---|
| Radscore‐T1 | 0.693 | 0.552‐0.813 | 81.0 | 56.2 | >−0.662 | .009 |
| Radscore‐CE T1 | 0.824 | 0.695‐0.915 | 76.2 | 84.5 | >−0.495 | <.001 |
| Radscore‐T2 | 0.690 | 0.549‐0.810 | 47.6 | 84.5 | >−0.064 | .012 |
| Radscore‐T2 FLAIR | 0.720 | 0.580‐0.835 | 90.5 | 46.9 | >−0.697 | .002 |
| Combined Radscore model (Enter) | 0.833 | 0.706‐0.922 | 72.7 | 90.6 | >0.454 | <.001 |
| Combined Radscore model (Forward) | 0.824 | 0.695‐0.915 | 76.2 | 84.5 | >0.389 | <.001 |
AUC, area under the ROC curve; CE, contrast‐enhanced; CI, confidence interval; FLAIR, fluid attenuated inversion recovery; Radscore, radiomic signature.
The performance of the combined model were as follows: calibration curve and decision curve. Calibration evaluates whether the nomogram (combined model) is over the fitting quantitative index Hosmer–Lemeshow test; the closer the P value is to 1, the better the effect of the model. The difference between the probabilities discriminated by the combined model and the observed probabilities was insignificant (P = .7099). Decision curve analysis was used to evaluate whether this nomogram would assist in differentiating MGMT methylation status, when the threshold probability ranged from 0 to 1 according to the decision curve analysis, the nomogram obtained the greatest benefit compared with a “treat all” strategy, a “treat none” strategy. The combined model is shown as a nomogram in Figure 3C and its discrimination and calibration plots are presented in Figure 3D. On the decision curve (Figure 3C), the black line represents the assumption that all glioma patients have MGMT methylation. The thick gray line represents the assumption that no glioma patients have MGMT methylation. The red line is the net benefit of the prediction of MGMT methylation on the basis of the combined model. The decision curve showed that if the threshold probability of a patient or doctor is higher than 20%, using the combined model nomogram in the current study to predict MGMT promoter status adds more benefit than the treat‐all or treat‐none strategy. The combined model is shown as a nomogram in Figure 4E and its discrimination and calibration plots are presented in Figure 4F.
4. DISCUSSION
In this study, we investigated the relationship between MGMT promoter methylation and radiological characteristics of gliomas based on the texture analysis of conventional MR images. Each sequence model and the combined model based on the Radscore of texture features can be used to differentiate the MGMT methylated and MGMT unmethylated tumor groups. Between the sequence models, the combined model could be the most promising. No significant correlation was found between visual radiographical features and clinical factors, and MGMT methylated status.
Most previous investigations have used CE T1w images to target volume definition of glioma tumors. 8 We used each sequence to outline the ROI of tumors for texture feature extraction of each sequence, trying to enable a lower bias for the extraction of features among the different sequences.
None of the top five texture features extracted from T1w and T2‐FLAIR showed the ability to differentiate MGMT methylated from MGMT unmethylated tumors, which is not surprising for T1w images; except in instances of hemorrhage within a tumor, the tumor tissue showed a similar signal intensity with the normal brain tissue on T1w images. The texture extracted from T1w images has limited ability to distinguish the MGMT methylated status. For T2‐FLAIR, it could be difficult to distinguish the area of the tumor and peritumoral edema. Kanazawa et al’s 15 results also indicated that none of the texture features extracted from T2‐FLAIR were associated with MGMT promoter methylation. We use Radscore, which integrated the information of the top five texture features for further analysis. The Radscore of both T1w and T2‐FLAIR can distinguish MGMT promoter methylation in glioma; this suggests that Radscore is more powerful than conventional texture features in the determination of the MGMT methylated status. For T2w and CE T1, some of the top five texture features showed a significant difference between MGMT methylated and MGMT unmethylated groups. However, Radscore integrated information from the top five texture features showed the best performance in differentiating methylated MGMT from unmethylated MGMT.
The combined model demonstrated better performance in differentiating MGMT methylated from MGMT unmethylated glioma than any other single sequence model with a 70.2% sensitivity and 90.6% specificity. Regarding GBM clinical and imaging characteristics, we also verified the efficacy of the combined model in the only GBM dataset. Our result showed that the combined model has a 90.4% sensitivity and 72.7% specificity in the GBM dataset. Furthermore, the bootstrap analysis confirmed that our combined model lacked overfitting. Jiang et al 16 compared the performance between the 3D‐CE‐T1w single radiomics model, T2w single radiomics model, and fusion radiomics model (based on the selected features from both the 3D‐CE‐T1w and T2w modalities) in predicting MGMT promoter methylation of low‐grade glioma. They also demonstrated that the fusion radiomics model was more powerful than the 3D‐CE‐T1w single or T2w single radiomics model. In our combined model, CE T1 was weighted and contributes the most.
We also studied several visual radiographic features (tumor location, crossing the midline, enhancement degree, necrosis/cyst degree, and presence of peritumoral edema) and clinical factors (age, gender, grade, and histological type), but none were correlated with MGMT methylation status, which is in agreement with previous results. 8 , 17 The enhancement mode judged by the radiologist with the naked eye cannot distinguish the status of MGMT methylation. However, texture features extracted from contrast T1 images can predict MGMT methylation preoperatively. With the application of a filtration histogram‐based approach, texture analysis can evaluate the distribution of gray levels in an image; the filtration step extracts and enhances features of different sizes corresponding to fine, medium, and coarse texture scales. This is then followed by quantification with histogram‐based statistical metrics. 18 Thus, this yields a potentially promising imaging biomarker to assess the microstructure and heterogeneity of tumors. The application of texture analysis to determine the mutation of IDH1 has also been confirmed. 19 This study demonstrates the potential for texture analysis to distinguish between MGMT methylated and MGMT unmethylated tumors by quantifying heterogeneity, without additional nonroutine imaging.
This study has several limitations. First, the study design was retrospective, and the patients with WHO grade I‐IV tumors were all included in the study, which caused relatively complicated tumor biological characteristics with notable heterogeneity. However, we verified the effectiveness of the combined model in the GBM dataset to overcome this disadvantage. We therefore show that the combined model, based on texture analysis, performed well to discriminate MGMT methylation status both in the GBM and the overall dataset. Second, our sample size was not large enough to undertake radiomics analyses to validate the performance of our model in this study. Therefore, further larger cohort studies are needed to validate the effectiveness of the Radscore model of texture analysis in the prediction of MGMT methylation.
Our results add new information to the imaging features of gliomas based on the MGMT methylation status using conventional MR. We have shown that the combined model based on texture features could be considered as a noninvasive imaging marker for detecting MGMT promoter methylation in glioma patients.
DISCLOSURE STATEMENT
The authors have no conflicts of interest to disclose.
ACKNOWLEDGMENTS
This study was funded by the Natural Science Foundation of Hainan Province (819QN351) and the Key Research and Development Program of Hainan Province (ZDYF2019173).
Huang W‐Y, Wen L‐H, Wu G, et al. Radiological model based on the standard magnetic resonance sequences for detecting methylguanine methyltransferase methylation in glioma using texture analysis. Cancer Sci. 2021;112:2835–2844. 10.1111/cas.14918
Clinical Trial Registration information: This retrospective cross‐sectional study was approved by our institutional review board, and the requirement of written informed consent was waived.
Funding information
This study was funded by the Natural Science Foundation of Hainan Province (819QN351) and the Key Research and Development Program of Hainan Province (ZDYF2019173).
Contributor Information
Feng Chen, Email: fenger0802@163.com.
Jian‐nong Zhao, Email: zhaojnhngh@gmail.com, Email: zjn@vip.163.com.
REFERENCES
- 1. Chen X, Zhang M, Gan H, et al. A novel enhancer regulates MGMT expression and promotes temozolomide resistance in glioblastoma. Nat Commun. 2018;9(1):2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Villalva C, Cortes U, Wager M, et al. O6‐Methylguanine‐methyltransferase (MGMT) promoter methylation status in glioma stem‐like cells is correlated to temozolomide sensitivity under differentiation‐promoting conditions. Int J Mol Sci. 2012;13(6):6983‐6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Melguizo C, Prados J, González B, et al. MGMT promoter methylation status and MGMT and CD133 immunohistochemical expression as prognostic markers in glioblastoma patients treated with temozolomide plus radiotherapy. J Transl Med. 2012;10(1):250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen X, Yan Y, Zhou J, et al. Clinical prognostic value of isocitrate dehydrogenase mutation, O‐6‐methylguanine‐DNA methyltransferase promoter methylation, and 1p19q co‐deletion in glioma patients. Ann Transl Med. 2019;7(20):541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hegi ME, Liu L, Herman JG, et al. Correlation of O6‐methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26(25):4189. [DOI] [PubMed] [Google Scholar]
- 6. Rundle‐Thiele D, Day B, Stringer B, et al. Using the apparent diffusion coefficient to identifying MGMT promoter methylation status early in glioblastoma: importance of analytical method. J Med Radiat Sci. 2015;62(2):92‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bahrami N, Hartman S, Chang Y‐H, et al. Molecular classification of patients with grade II/III glioma using quantitative MRI characteristics. J Neurooncol. 2018;139(3):633‐642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Korfiatis P, Kline TL, Coufalova L, et al. MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med Phys. 2016;43(6):2835‐2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bell EH, Zhang P, Fisher BJ, et al. Association of MGMT promoter methylation status with survival outcomes in patients with high‐risk glioma treated with radiotherapy and temozolomide: an analysis from the NRG Oncology/RTOG 0424 trial. JAMA Oncol. 2018;4(10):1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Louis DN, Perry A, Reifenberger G. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 11. Yoon R, Kim H, Paik W, Shim W, Kim S, Kim J. Different diagnostic values of imaging parameters to predict pseudoprogression in glioblastoma subgroups stratified by MGMT promoter methylation. Eur Radiol. 2017;27(1):255‐266. [DOI] [PubMed] [Google Scholar]
- 12. Yushkevich PA, Piven J, Hazlett HC, et al. User‐guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage. 2006;31(3):1116‐1128. [DOI] [PubMed] [Google Scholar]
- 13. Su CQ, Lu SS, Han QY, Zhou MD, Hong XN. Intergrating conventional MRI, texture analysis of dynamic contrast‐enhanced MRI, and susceptibility weighted imaging for glioma grading. Acta Radiol. 2019;60(6):777‐787. [DOI] [PubMed] [Google Scholar]
- 14. Gao C, Xiang P, Ye J, Pang P, Wang S, Xu M. Can texture features improve the differentiation of infiltrative lung adenocarcinoma appearing as ground glass nodules in contrast‐enhanced CT? Eur J Radiol. 2019;117:126‐131. [DOI] [PubMed] [Google Scholar]
- 15. Kanazawa T, Minami Y, Takahashi H, et al. Magnetic resonance imaging texture analyses in lower‐grade gliomas with a commercially available software: correlation of apparent diffusion coefficient and T2 skewness with 1p/19q codeletion. Neurosurg Rev. 2019;43:1211‐1219. [DOI] [PubMed] [Google Scholar]
- 16. Jiang C, Kong Z, Liu S, et al. Fusion radiomics features from conventional MRI predict MGMT promoter methylation status in lower grade gliomas. Eur J Radiol. 2019;121:108714. [DOI] [PubMed] [Google Scholar]
- 17. Gupta A, Omuro A, Shah A, et al. Continuing the search for MR imaging biomarkers for MGMT promoter methylation status: conventional and perfusion MRI revisited. Neuroradiology. 2012;54(6):641‐643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Skogen K, Schulz A, Dormagen JB, Ganeshan B, Helseth E, Server A. Diagnostic performance of texture analysis on MRI in grading cerebral gliomas. Eur J Radiol. 2016;85(4):824‐829. [DOI] [PubMed] [Google Scholar]
- 19. Jakola AS, Zhang Y‐H, Skjulsvik AJ, et al. Quantitative texture analysis in the prediction of IDH status in low‐grade gliomas. Clin Neurol Neurosurg. 2018;164:114‐120. [DOI] [PubMed] [Google Scholar]


