Abstract
Background:
Cardiovascular disease (CVD) is the first cause of world death, and myocardial infarction (MI) is one of the five primary disorders of CVDs which the patient electrocardiogram (ECG) analysis plays a dominant role in MI diagnosis. This research aims to evaluate some extracted features of ECG data to diagnose MI.
Methods:
In this paper, we used the Physikalisch-Technische Bundesanstalt database and extracted some morphological features, such as total integral of ECG, integral of the T-wave section, integral of the QRS complex, and J-point elevation from a cycle of normal and abnormal ECG waveforms. Since the morphology of healthy and abnormal ECG signals is different, we applied integral to different ECG cycles and intervals. We executed 100 of iterations on a 10-fold and 5-fold cross-validation method and calculated the average of statistical parameters to show the performance and stability of four classifiers, namely logistic regression (LR), simple decision tree, weighted K-nearest neighbor, and linear support vector machine. Furthermore, different combinations of proposed features were employed as a feature selection procedure based on classifier's performance using the aforementioned trained classifiers.
Results:
The results of our proposed method to diagnose MI utilizing all the proposed features with an LR classifier include 90.37%, 94.87%, and 86.44% for accuracy, sensitivity, specificity, respectively. Also, we calculated the standard deviation value for the accuracy of 0.006.
Conclusion:
Our proposed classification-based method successfully classified and diagnosed MI using different combinations of presented features. Consequently, all proposed features are valuable in MI diagnosis and are praiseworthy for future works.
Keywords: Biological signal processing, classification, cross-validation, electrocardiography, feature selection, linear support vector machine, myocardial infarction, simple tree, weighted K-nearest neighbor
Introduction
Myocardial infarction
The first cause of world death is cardiovascular disease (CVD), which causes more death than other diseases annually. In 2015, about 17.7 million people died from CVD, accounting for 31% of the world's total deaths, of which about 7.4 million people died from the stroke and 7.6 million deaths were because of coronary heart disease. Myocardial infarction (MI) is one of the five main complications of CVDs, which include persistent angina, unstable angina, heart failure, MI, and sudden death.[1,2] Consequently, early diagnosis of MI has critical importance in medical applications for the effective treatment of patients.
MI or “heart attack” occurs when one of the coronary arteries is completely blocked. A region of the myocardium fed by the coronary artery dies, and loses its blood and deprives of oxygen and other nutrients. The sudden and total occlusion of the artery, which precipitates the infarction, is usually due to coronary artery spasm or superimposed thrombosis.[3] As the ST-segment is the most widely used feature for MI detection, the onset of MI can be distinguished into two kinds of MI, namely an ST-segment elevation MI, which is caused by the blockage of an artery from the heart, and a non-ST-segment elevation MI, which depends on T-inversion or ST depression.[4,5,6,7]
Usually, in most cases of infarction, electrocardiogram (ECG) signal processing based on the investigation of the morphology of multi-lead ECG results in precise diagnosis.[8] Early ECG variations and specific electrocardiographic changes associated with MI occurred with the onset of myocardial dysfunctions.[3,7]
During acute MI, the ECG evolves through three stages:
T-wave peaking followed by T-wave inversion
ST-segment elevation
Emergence of a new Q-wave.
However, during an acute infarction, commonly, all three states can appear in the ECG, but it is possible to have any of these conditions without another one. For example, it is not unusual at all that the ST-segment elevation occurs without T-wave inversion. Considering all the three states, we can predict MI with the least error.[3,9] Some of these methods introduced are as follows:
If the T-wave peaks and then reverses, it represents myocardial ischemia. In addition, if an acute MI occurs, then T-wave can remain reverse about more than a few months to several years
If the ST-segment raises and joins with the T-wave, then it indicates a myocardial injury. Furthermore, the ST-segment usually recovers to baseline within a few hours when infarction happens
If a new Q-wave appears within hours to days, then it signifies MI. In most cases, the Q-wave persists for the rest of the patient life.[3]
The localization of infarction is essential since its prognostic and therapeutic effects are in the heart regions, which are already dead. Infarctions categorized into several general anatomic categories. These are inferior infarctions, lateral infarctions, anterior infarctions, and posterior infarctions. Combinations can also be realized, such as anterolateral infarctions, which are very common.[3]
Inferior infarction: The structural electrocardiographic changes of infarction reveal in the inferior leads such as II, III, and AVF
Lateral infarction: In this infarction, the occlusion of the left circumflex artery leading to changes in the left lateral leads AVL, I, V5, and V6
Anterior infarction: This infarction marked by specific changes in the precordial leads (V1–V6)
Posterior infarction: In this infarction, since there are no leads overlying the posterior wall of the heart, therefore, the anterior leads, especially V1, are needed to investigate for reciprocal changes, which are indeed essential to diagnose posterior MI.
Research background
According to all studies, MI diagnosing generally constitutes four major phases, including signal preprocessing step, ECG wave segmentation, feature extraction, and classification. In general, signal features extracted from the time-domain, the frequency-domain, and the transform-domain.[5,10] The importance of an efficient feature selection method is undeniable and affects the overall performance of classification methods.
There are several automated, semi-automated, or manual signal segmentation approaches for signal feature extraction. For example, Al Touma et al. proposed a system which utilized two algorithms, and the first algorithm could detect the critical points on the ECG waveforms and the second algorithm could detect possible MI based on the analysis of the aforementioned critical points.[4] Further, Gupta and Kundu applied a statistical index, namely dissimilarity factor “D” to classify normal and inferior MI data, without the need for any direct clinical feature extraction. Accordingly, the T-wave and the QRS sections of inferior MI datasets were automatically extracted from the leads II, III, and aVF and then compared with corresponding segments of healthy patients using the Physikalisch-Technische Bundesanstalt (PTB) database.[11]
Some research analyzes and investigates the effects and arrangements of each MI feature. Carley presented that the location of leads V1–V6 depends on historical convention, and there is clear evidence that ST-elevation, indicative of acute MI, exists outside of the standard 12-lead ECG.[12] Accordingly, we consider it as an essential factor to detect and localize MI in the current study. Besides, Muhammad Arif et al. employed time-domain features of each beat in the ECG signal, including T-wave amplitude, Q-wave, and ST-elevation using K-nearest neighbor (KNN) classifier in an automated scheme for diagnosis and localization of MI.[13] Similarly, Safdarian et al. applied two new time-domain features including total integral and T-wave integral as morphological signal features to detect and localize the MI in the left ventricle of the heart.[14]
On the other hand, some researches focused on the features in the frequency domain or the transform domain of the ECG signal. As an example, Nidhyananthan et al. proposed a wavelet-based method, which applied for detecting MI along with user-identity using a support vector machine (SVM) classifier to classify the normal and abnormal cases in these signals. In addition, the RR interval utilized to authenticate the ECG signal.[15] In addition, Sharma et al. represented a novel technique on multiscale energy and Eigen space approach to detect and localize MI from multi-lead ECG. Moreover, the nearest neighbor (KNN), and the SVM, along with both linear and radial basis function kernels applied as classifiers.[16]
Moreover, Noorian et al. proposed the radial basis function neural networks classifier with wavelet coefficient as features extracted from frank lead to diagnose and localize MI.[17] Moreover, Tripathy et al. proposed the multiscale convolutional neural network for automatic MI localization using the Fourier-Bessel (FB) series expansion-based empirical wavelet transform (EWT) for signal segmentation and feature extraction.[8] Concerning the use of neural networks, similarly, Zhang et al. proposed a multi-lead bidirectional-gated recurrent unit neural network, and their results show that the algorithm has higher sensitivity, positive predictivity, accuracy, and universality.[7]
In this study, we focused on the feature selection section as an essential part of a successful MI diagnosing and classification. We presented a classification-based feature selection method to analyze the worthiness of the extracted ECG morphological features and to tackle the problem of feature selection.
Objectives
The main objective of our study is to evaluate the efficiency of the proposed extracted morphological features from ECG data in MI diagnosis and not necessarily diagnosing MI only. The higher the quality of ECG extracted features, the better the results in the early diagnosis of MI.
Materials and Methods
Signal acquisition
In this study, we proposed a classification-based feature selection procedure to evaluate the ECG morphological features for diagnosing MI. At this point, we collect our ECG data from PhysioNet. In the PTB database, the ECGs were derived from patients with various heart diseases and healthy volunteers.[18]
In our research, we have assigned two diagnostic classes, including 148 MI subjects and 52 standard control subjects, since we investigated MI types. Consequently, the sum of 59 healthy ECG records alongside 156 MI records forms the complete database of 215 signal records.
Signal preprocessing and feature extraction from electrocardiogram signals
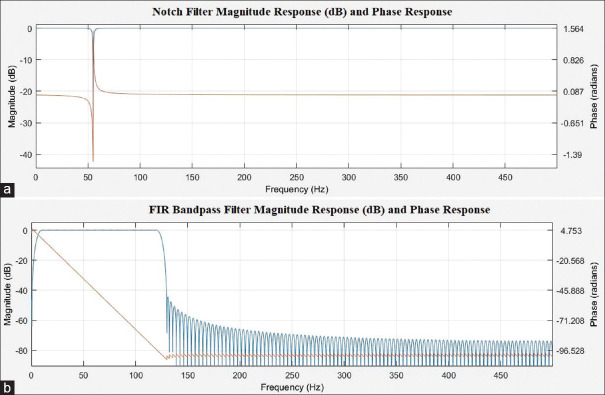
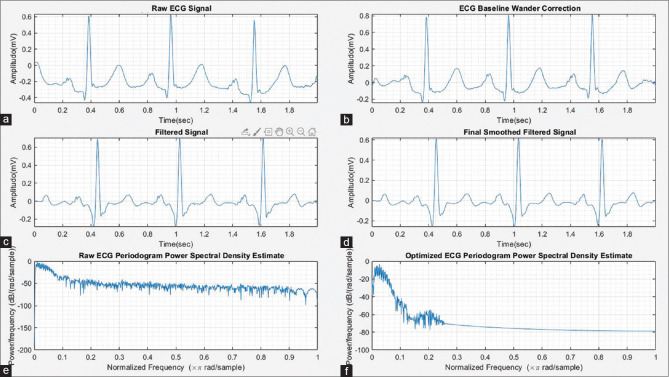
The raw ECG records contain various artifacts such as muscle noise, power line interference noise, and baseline wander.[4,10] In this article, we implemented two filters, namely the band-pass FIR filter (ECG band 0.5–150 Hz) and IIR notch filter (55 Hz) to remove noise from ECG signals. The notch filter removes the power line interference noise of 50–60 Hz. Moreover, the band-pass FIR filter is designed using a minimum order Kaiser window for the removal of ECG baseline wander. We utilized MATLAB filter design application and the design configuration summary of each filter presented and also, the Bode plot for the filters and also, signal optimization are shown in Figures 1 and 2.
Figure 1.
(a) Notch filter magnitude response (dB) and phase response; (b) FIR band-pass Filter magnitude response (dB) and phase response
Figure 2.
(a) Raw electrocardiogram signal, (b) electrocardiogram baseline wander correction, (c) filtered electrocardiogram signal, (d) final smoothed filtered electrocardiogram signal, (e) raw electrocardiogram periodogram power spectral density estimate, and (f) optimized electrocardiogram periodogram power spectral density estimate
IIR notch filter
Filter order: 2
Sampling frequency: 1000
Notch Bandwidth: 10 Hza
Passband ripple (Apass): 0.1 dB
FIR band-pass filter
Filter order: 124
Sampling frequency: 1000
Window: Kaiser
End of the first stopband (Fstop1): 0.5 Hz
Beginning of the passband (Fpass1): 10 Hz
End of the passband (Fpass2): 100 Hz
Beginning of the second stopband (Fstop2): 150 Hz
Passband ripple (Apass): 0.1 dB
Stopband attenuation for both stopbands (Astop1, Astop2): 40 dB.
In this review, we focused on ECG features and identified fundamental characteristics to extract, classify, and eventually determine exact MI types, which are presented below:
Peaking of the T-wave (hyperacute T-wave) followed by inversion of the T-wave
Elevation of the ST segment.
The idea is to integrate the ECG signal and extract the above features. Accordingly, we compute definite integrals of “ECG cycle,” “QRS complex,” and “T-wave” using “trapz” function in MATLAB (R2016b) software. As a result, we can extract morphology variations and hyperacute T-wave features of ECG signal in MI as well. Furthermore, we use ECG J-point elevation value to estimate ST-elevation and also extract it as a feature from three ECG leads, namely lead I, lead II, and lead V2.
According to the MATLAB software, the “trapz” operates numerical integration via the trapezoidal method, which can divide the area down into trapezoidal segments to approximate the integration over an interval. Therefore, the approximation is as follows:
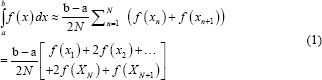
Where the spacing between each point is equal to the scalar value 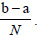 .
.
Thus, we obtain a general “features matrix” with 215 rows and 11 columns, which include features and labels. Consequently, the features extraction process of ECG signals accomplishes. The final features matrix arrays contain {F1, F2, F3, F4, F5, F6, F7}; features accompany with two class and four class labels.
F1: Integral of a complete ECG cycle
F2: Integral of ORS complex
F3: Integral of T-wave
F4: ST elevation in lead I
F5: ST elevation in lead II
F6: ST elevation in lead V2
F7: T-wave inversion (0 for no inversion and 1 for inversion).
The laptop we used had Windows 8.1 as the operating system, a Core i7 CPU, and 12 GB of RAM.
In this study, initially, we create a database from ECG-extracted features. Subsequently, we can classify MI and obtain the results using a 5-fold cross-validation technique to detect MI from ECG data. The 5-fold cross-validation divides the ECG data into four training datasets and a validation dataset as the inputs of four classifiers, namely the fine decision tree, the linear SVM (L-SVM), the weighted KNN, and the logistic regression (LR). We suggest the diagnosis of MI by evaluating ECG data using two procedures, including the two-class and the four-class classification. The dataset in the two-class classification method contains healthful records and MI records, however; in the four-class classification procedure, we have healthful records, inferior, anterior, and posterior MI records.
Statistical analysis
Statistical analyses to investigate the relationship between the features
To test the normality of the data (features extracted from the ECG signals), the test of the normality of the following features was performed in SPSS software (Version 20, IBM Corp. Released 2011. IBM SPSS Statistics for Windows, Version 20.0. Armonk, NY: IBM Corp.).
Features include F1, F2, F3, F4, F5, F6, and F7; Table 1 shows the test results for the above features.
Table 1.
Tests of normality of the features
| Features | Kolmogorov-Smirnova | Shapiro-Wilk | ||||
|---|---|---|---|---|---|---|
| Statistic | df | P | Statistic | df | P | |
| F1 | 0.094 | 215 | 0.000 | 0.879 | 215 | 0.000 |
| F2 | 0.056 | 215 | 0.093 | 0.981 | 215 | 0.006 |
| F3 | 0.051 | 215 | 0.200* | 0.975 | 215 | 0.001 |
| F4 | 0.513 | 215 | 0.000 | 0.424 | 215 | 0.000 |
| F5 | 0.435 | 215 | 0.000 | 0.585 | 215 | 0.000 |
| F6 | 0.402 | 215 | 0.000 | 0.616 | 215 | 0.000 |
| F7 | 0.442 | 215 | 0.000 | 0.577 | 215 | 0.000 |
*This is a lower bound of the true significance, aLilliefors significance correction
As shown in Table 1, and according to P value and the skewness results for all seven sets of extracted features, it indicates that the features are abnormal. Furthermore, Table 1 represents the comparison of variance status and skewness values of all three groups of the above characteristics.
Since the data of all seven groups of features do not have a normal distribution, parametric tests (such as t-test or ANOVA test) cannot be used to analyze these features. Therefore, we must use nonparametric analyses to investigate the independence of features. We then examine whether there is a significant difference between the seven sets of extracted features using the Friedman nonparametric test.
The results of Friedman nonparametric test show that there is a significant difference between the seven groups of different features due to the P < 0.01. In other words, the Chi-square value of Friedman's test is confirmed with an error of <1% and a confidence level of 99%. According to Tables 2 and 3, because of the significant difference between the different groups of extracted features, we can use them to enter classification algorithms. Finally, let us look at the accuracy, sensitivity, and specificity values used by classifying algorithms on these data.
Table 2.
Statistical analysis for the features
| Statistics | F1 | F2 | F3 | F4 | F5 | F6 | F7 |
|---|---|---|---|---|---|---|---|
| Valid (n) | 215 | 215 | 215 | 215 | 215 | 215 | 215 |
| Missing (n) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mean | 0.0001 | 0.0056 | 0.0063 | 0.1488 | 0.3163 | 0.3814 | 0.3023 |
| SD | 0.00050 | 0.01447 | 0.01238 | 0.35676 | 0.46611 | 0.48686 | 0.46034 |
| Variance | 0.000 | 0.000 | 0.000 | 0.127 | 0.217 | 0.237 | 0.212 |
| Skewness | 0.816 | 0.186 | −.428 | 1.987 | 0.796 | 0.492 | 0.867 |
| SE of skewness | 0.166 | 0.166 | 0.166 | 0.166 | 0.166 | 0.166 | 0.166 |
| Kurtosis | 10.122 | 1.281 | 2.195 | 1.967 | −1.380 | −1.775 | −1.260 |
| SE of kurtosis | 0.330 | 0.330 | 0.330 | 0.330 | 0.330 | 0.330 | 0.330 |
SE – Standard error; SD – Standard deviation
Table 3.
Friedman test
| Ranks | Mean rank |
|---|---|
| F1 | 3.47 |
| F2 | 4.09 |
| F3 | 4.42 |
| F4 | 3.52 |
| F5 | 4.17 |
| F6 | 4.47 |
| F7 | 3.87 |
| Test statisticsa | |
| n | 215 |
| χ2 | 49.306 |
| df | 6 |
| Asymptotic P | 0.000 |
aFriedman test
Statistical analysis to examine the relationship between data sets of extracted features from healthy and myocardial infarction classes
To test the normality of the data (extracted features from ECG signals of healthy and patient data), the following test of the normality of the following features was performed in SPSS software (Version 20). Review classes are given below.
Healthy
Patients with MI.
Moreover, the features examined in the above have seven independent classes including F1, F2, F3, F4, F5, F6, and F7.
In the previous section, the normality test was examined. At this stage, since the two healthy and MI classes represent two completely independent nonparametric groups, we use Mann–Whitney and Kolmogorov–Smirnov tests.
First, the tests above are performed on all seven features for two class labels (only healthy and only MI). Table 4 shows the results of the above two tests and reveals that all the features extracted from healthy ECG and MI samples (except for feature 4) in both groups (with label 1 for healthy and label 2 for MI, respectively) have a P < 0.05 value for both tests, which means that there is a significant difference between the data of healthy individuals and MI for these extracted features (except for the feature number 4, of course).
Table 4.
Mann-Whitney and Kolmogorov-Smirnov test
| Mann-Whitney test | F1 | F2 | F3 | F4 | F5 | F6 | F7 |
|---|---|---|---|---|---|---|---|
| Mann-Whitney test statisticsa | |||||||
| Mann-Whitney U | 3066.000 | 3102.000 | 1058.000 | 4303.000 | 3563.500 | 3365.500 | 2792.000 |
| Wilcoxon W | 15312.000 | 15348.000 | 13304.000 | 6073.000 | 5333.500 | 5135.500 | 4562.000 |
| Z | −3.774 | −3.685 | −8.707 | −1.192 | −3.168 | −3.611 | −5.590 |
| Asymptotic P (two-tailed) | 0.000 | 0.000 | 0.000 | 0.233 | 0.002 | 0.000 | 0.000 |
| Two-sample Kolmogorov-Smirnov test statisticsa | |||||||
| Most extreme differences, absolute | 0.330 | 0.294 | 0.655 | 0.065 | 0.226 | 0.269 | 0.393 |
| Positive | 0.330 | 0.294 | 0.655 | 0.000 | 0.000 | 0.000 | 0.000 |
| Negative | −0.024 | −0.013 | 0.000 | −0.065 | −0.226 | −0.269 | −0.393 |
| Kolmogorov-Smirnov Z | 2.156 | 1.923 | 4.284 | 0.425 | 1.476 | 1.758 | 2.573 |
| Asymptotic P (two-tailed) | 0.000 | 0.001 | 0.000 | 0.994 | 0.026 | 0.004 | 0.000 |
aGrouping variable: Label_2_class
Then, analyses are performed on all seven features for four class labels (healthy samples and three types of MI patient samples). At this stage, since one healthy class and three patient classes represent four completely independent groups, we use Kruskal–Wallis and median tests because these tests can be performed on more than two independent nonparametric groups.
In Table 5, the results of the above two tests indicate that all the extracted features from the ECG signals of healthy and patient samples (except for F4 in the Kruskal–Wallis test, and F2 and F4 in the median test for all four healthy and patient classes, label 1, label 2, label 3, and label 4, respectively) have valued P < 0.05 for both tests. It means that there is a significant difference between the data of healthy people and MI patients for all the extracted features, except for the two features mentioned above. Therefore, choosing these features to distinguish between healthy data and three MI patient groups is rational and performs acceptable discrimination between the classes as well.
Table 5.
Kruskal-Wallis and median tests
| F1 | F2 | F3 | F4 | F5 | F6 | F7 | |
|---|---|---|---|---|---|---|---|
| Kruskal-Wallis test statisticsa | |||||||
| χ2 | 45.620 | 13.594 | 82.702 | 2.374 | 11.126 | 59.767 | 36.221 |
| df | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| Asymptotic P | 0.000 | 0.004 | 0.000 | 0.499 | 0.011 | 0.000 | 0.000 |
| Median test statisticsa | |||||||
| n | 215 | 215 | 215 | 215 | 215 | 215 | 215 |
| Median | 0.0001 | 0.0049 | 0.0057 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| χ2 | 35.824b | 4.324b | 70.553b | 2.385c | 11.178d | 60.047e | 36.390f |
| df | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| Asymptotic P | 0.000 | 0.229 | 0.000 | 0.496 | 0.011 | 0.000 | 0.000 |
aGrouping variable: Label_4_Class, b2 cells (25.0%) have expected frequencies <5. The minimum expected cell frequency is 2.5, c2 cells (25.0%) have expected frequencies <5. The minimum expected cell frequency is 0.7, d2 cells (25.0%) have expected frequencies <5. The minimum expected cell frequency is 1.6, e2 cells (25.0%) have expected frequencies <5. The minimum expected cell frequency is 1.9., f2 cells (25.0%) have expected frequencies <5. The minimum expected cell frequency is 1.5
According to Table 5, the significant difference between the different classes of extracted features is noticeable, and we can use them to enter classification algorithms.
Classification
In this section, we compute integral of filtered ECG signal to extract aforementioned features, and next, we choose J-point elevation value in mv unit manually for 215 data samples and use these characteristics to create feature matrix. Furthermore, we apply 5-fold and 10-fold cross-validation for each of the classifiers and calculate the sensitivity and specificity using these two formulas below.[7,10,19,20]


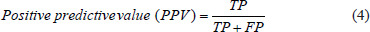
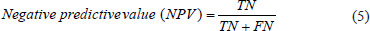
Moreover, we executed 100 iterations for 5-fold and 10-fold cross-validation on the 2-class and 4-class classification to show the stability of the proposed models.
K-fold cross-validation
The cross-validation estimate of accuracy equals the overall number of correct classifications divided by the number of samples in the dataset. Accordingly, assume that the inducer J maps a given dataset into a classifier and D(i) is the test set that includes sample xi = (vi, yi). Note that δ is the mean square error function, then the cross-validation estimate of accuracy is:[21]
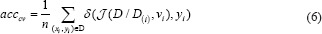
Logistic regression
LR is used to describe the data and explain the relationship between a dependent binary variable and one or more independent variables.[22,23] The regression analysis uses the log odds to compute output Y. Further, it tackles the problem of probabilities limitation and uses a cost function to compute the individual cost of each observation Yp. The LR equation is:
β = A0+A1(x) (7)
Further,
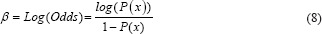
We define a cost function threshold of 0.5 for decision-making, so we have:
if Y ≥ 0.5: cost = -Log(Yp) (9)
if Y ≤ 0.5: cost = -Log(1-Yp) (10)
Where β is the log odds and A0 and A1 are logistic coefficients with initial values determined by optimization, which must minimize the cost function. Hence, the probability Y using a sigmoid function is:

Linear support vector machine
In machine learning, the SVMs include supervised learning models, which are a set of points in the n-dimensional data that specify the boundaries of the classes to classify the data. Consider a collection of training samples that each sample belongs to one or the other of two classes, the SVM finds the optimal hyperplane, which splits all data points of one class from those of the other class.[10,24]
This discussion follows Hastie et al.[24] and Cristianini and Shawe-Taylor.[25] The training data define as a set of points (vectors) xi along with their groups yi. For some dimension d, the xi ∊ Rd, and the yi = ±1. The equation of a hyperplane is:
f(x) = βx' + b(12)
Where β ∊ Rd and b is a real number.
The following problem defines the optimal separating hyperplane (i.e., the decision boundary). Find β and b, which minimize ||β|| such that for all data points (xi,yi), yjf (xj) ≥ 1–ξi. Hence, for i ∊ {1,…, n}, we define ξi = max (0, 1–yjf (xj)). The support vectors are the xi on the boundary for those which yjf (xj) = 1. For mathematical ease, the problem is usually assumed as the equivalent problem of minimizing ||β||.[24,26] In our work and according to MATLAB, the length of beta is equal to the number of predictors used to train the model so for more information check MATLAB help. Finally, we have:
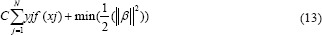
Where if C grows too large, the algorithm will reduce ||β||, leading to a hyperplane that classifies each training sample correctly. Conversely, if C becomes too small, then the algorithm increases ||β|| causing large training error.[26]
Decision tree
The decision tree learning is one of the predictive modeling methods applied in statistics, which maps observations about an item (presented in the branches) to conclude on the target value of the item (shown in the leaf). In tree classification, the target variable can accept a limited set of values; in these tree structures, leaves and branches, respectively, represent class labels and a combination of features, which leads to those class labels.[10,27]
K-nearest neighbors' algorithm
One of the nonparametric methods in pattern recognition is the KNN algorithm utilized for classification, which the term nearest is the foundation of the KNN so that each new instance compares to all prior cases and then assigns to the group with closer samples to each other. Since the KNN output is a class membership, therefore, the KNN assigns the sample to the class most common among its k nearest neighbors (k is a positive integer, typically small). The sample assigns to a class of that single nearest neighbor if k = 1.[10,22]
In the weighted KNN classifier, we assign a weight 1/k to the k nearest neighbors and all others a weight zero. That is, where the ith nearest neighbor assigns a weight ωni with 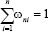 .[23]
.[23]
Let 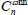 denotes the weighted nearest classifier with weights
denotes the weighted nearest classifier with weights 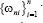 . With regard to regularity conditions on class distributions, the excess risk has the following asymptotic expansions
. With regard to regularity conditions on class distributions, the excess risk has the following asymptotic expansions 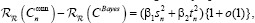
for constants β1 and β2, where 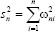 and
and 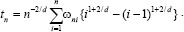 .
.
The optimal weighting scheme 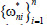 , which moderates the two terms in the aforementioned above, is given as follows:
, which moderates the two terms in the aforementioned above, is given as follows:
set 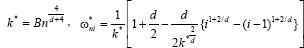 for i = 1,2 …,k* and
for i = 1,2 …,k* and 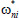 for i = k* + 1,…,n.[19]
for i = k* + 1,…,n.[19]
Results
According to Table 6, in two-class classification, the LR represents optimal accuracy, sensitivity, and specificity considering the standard deviation (SD) value and is more reliable caparisoning the other classifiers to diagnose MI. The comprehensive analysis of two-class classification reveals that the LR with 90.37% of precision has a better performance in comparison with the others. In the four-class classification, we should estimate sensitivity and specificity for each class individually. Therefore, in Table 7, we only report accuracy and related SD values for the models. The true-positive rate (TPR) (sensitivity), the false-negative rate (FNR), the positive predictive value (PPV), and the false predictive rate (FPR) for each classifier are determined in Figures 3-6.
Table 6.
5-fold and 10-fold cross-validation for two-class classification using different classifiers
| Method | Classifier | Mean accuracy | SD | Sensitivity | Specificity | FPR | NPV | PPV |
|---|---|---|---|---|---|---|---|---|
| 5-fold cross validation for two-class classification | Logistic regression | 90.344 | 0.00793 | 94.872 | 86.441 | 13.55 | 86.44 | 94.87 |
| Linear SVM | 88.809 | 0.00980 | 95.59 | 78.407 | 21.59 | 87.05 | 92.13 | |
| Weighted KNN | 85.772 | 0.01041 | 94.231 | 69.492 | 30.50 | 82 | 89.09 | |
| Simple tree | 82.772 | 0.02041 | 98.718 | 83.051 | 16.94 | 96.07 | 93.90 | |
| 10-fold cross validation for two-class classification | Logistic regression | 90.372 | 0.00618 | 94.872 | 86.441 | 13.559 | 86.441 | 94.872 |
| Linear SVM | 89.06 | 0.00769 | 95.622 | 78.407 | 21.593 | 87.142 | 92.133 | |
| Weighted KNN | 86.181 | 0.00823 | 94.231 | 69.492 | 30.508 | 82 | 89.091 | |
| Simple tree | 83.823 | 0.01455 | 98.718 | 83.051 | 16.949 | 96.078 | 93.902 |
SD – Standard deviation; PPV – Positive predictive value; NPV – Negative predictive value; FPR – False predictive rate; SVM – Support vector machine; KNN – K-nearest neighbor
Table 7.
5-fold and 10-fold cross-validation for four-class classification using different classifiers
| Method | Classifier | Mean accuracy | SD |
|---|---|---|---|
| 5-fold cross validation for four-class classification | Simple tree | 63.96 | 0.0243 |
| Linear SVM | 73.09 | 0.0115 | |
| Weighted KNN | 68.07 | 0.0172 | |
| 10-fold cross validation for four-class classification | Simple tree | 63.93 | 0.0210 |
| Linear SVM | 73.44 | 0.0072 | |
| Weighted KNN | 68.32 | 0.0116 |
SD – Standard deviation; SVM – Support vector machine; KNN – K-nearest neighbor
Figure 3.
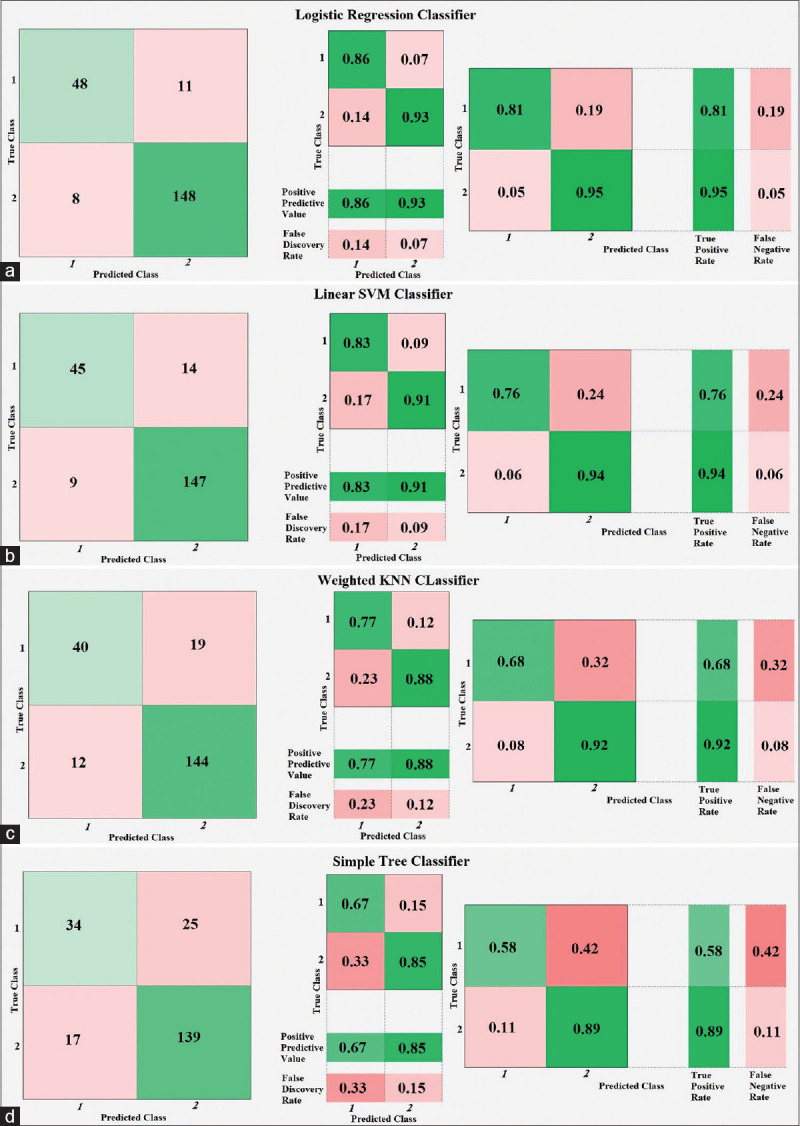
Two-class classification confusion matrices using 10-fold cross-validation for (a) logistic regression, (b) linear support vector machine, (c) weighted K-nearest neighbor, and (d) simple tree classifiers
Figure 6.
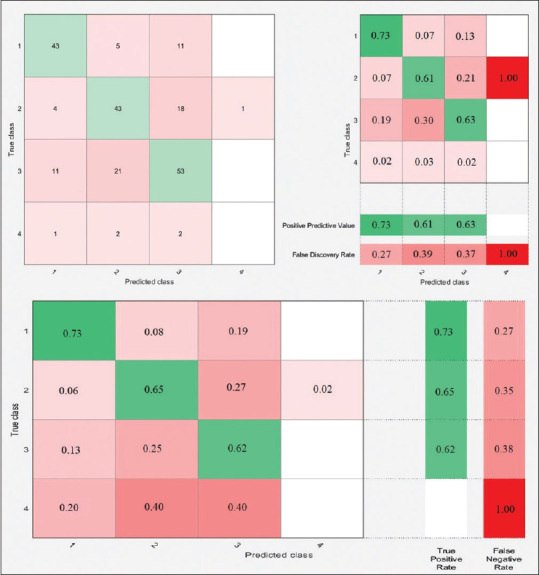
Four-class classification confusion matrices using 10-fold cross-validation for simple tree classifier
Figure 5.
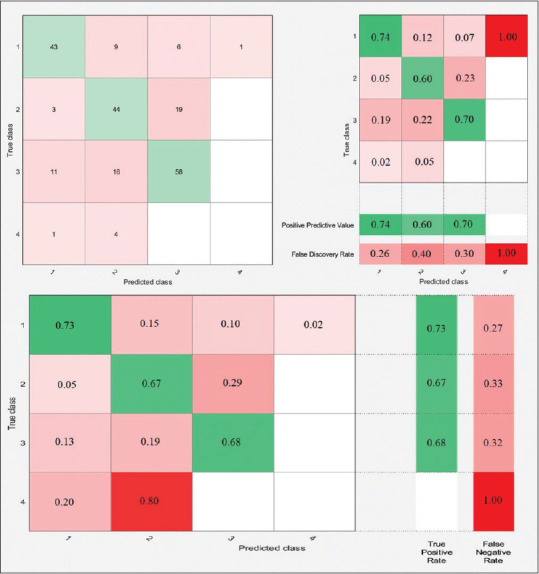
Four-class classification confusion matrices using 10-fold cross-validation for weighted K-nearest neighbor classifier
Discussion
There are representative performance schematics of all classifiers and their confusion matrices, as shown in Figures 3-6. Let us define each class to analyze the two-class and four-class classifications.
-
Two-class classification
Class 1: healthy records, Class 2: MI records.
-
Four-class classification
Class 1: healthy records, Class 2: anterior MI records, Class 3: inferior MI records, and Class 4: posterior MI records.
For example, in Figure 3a, the left-sided image represents the number of observations for the LR classifier. Moreover, the middle schematic indicates PPV and FPR. Furthermore, the right-sided schematic shows the TPR and FNR for each class in the two-class classification using 10-fold cross-validation. According to the aforementioned right-sided image, TPR for the first class is 81%, declaring that 81% of normal records correctly classified as healthy ECG, and FNR for the first class is 19%, demonstrating that 19% of healthy ECG records misclassified and predicted as MI records. Accordingly, the same comprehension analytic procedure can be applied for the four-class classification in, Figures 4-6 for different classifiers.
Figure 4.
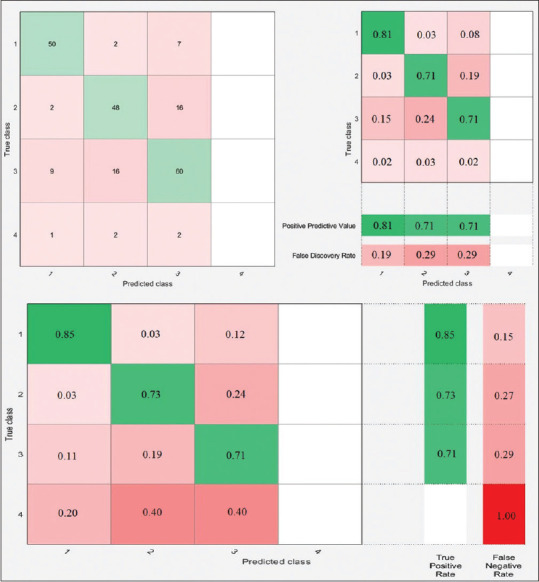
Four-class classification confusion matrices using 10-fold cross-validation for linear support vector machine classifier
According to Tables and Figures, the performance of L-SVM with 73.44% of accuracy is considerable in four-class classification. Note that all classifiers failed to predict the fourth class successfully just because of minimal sample volume.
As our main objective is to show how valuable is the strength of extracted features in the successful diagnosis of the MI, it is more satisfying to choose a different combination of signal features as an efficient method to observe how the accuracy of each classifier changes. According to the feature matrix, it consists of seven key features, namely F1, F2, F3, F4, F5, F6, and F7. In the proposed method, three signal features remain as default. Since F1, F2, and F3 extracted from lead II, we consider them as homogenous features and should always persist.
Based on Table 8, the precision of all classifiers with the set of {F1, F2, F3, F5, F6, F7} is more reliable in comparison with other sets of selected features in two-class classification. Furthermore, the F4, which is the set of J-point elevation values of ECG signals of lead I in mv, decreases the accuracy of classifiers. On the other hand, it seems that ignoring F4 from all features leads to a significant increase in the precision of classifiers.
Table 8.
10-fold cross-validation for two-class and four-class classification with different classifiers and also various combinations of selected features
| Method | 10-fold cross-validation | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Four-class classification | Feature selection | F1 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| F2 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| F3 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| F4 | ✓ | ✓ | ✓ | ✓ | - | - | ✓ | ✓ | - | ✓ | - | - | ✓ | ||
| F5 | ✓ | ✓ | ✓ | - | ✓ | - | - | ✓ | - | - | ✓ | ✓ | - | ||
| F6 | ✓ | ✓ | - | ✓ | ✓ | ✓ | - | - | - | - | ✓ | - | ✓ | ||
| F7 | ✓ | - | ✓ | ✓ | ✓ | ✓ | ✓ | - | ✓ | - | - | ✓ | - | ||
| Classifier accuracy | Simple tree (%) | 64.7 | 64.7 | 63.7 | 64.7 | 64.7 | 64.7 | 63.7 | 63.7 | 63.7 | 63.7 | 64.7 | 63.7 | 64.7 | |
| Linear SVM (%) | 73.0 | 72.6 | 59.1 | 70.2 | 74.0 | 70.7 | 64.2 | 60.5 | 61.4 | 63.7 | 73.5 | 61.9 | 70.7 | ||
| Weighted KNN (%) | 67.9 | 66.0 | 54.9 | 69.8 | 69.8 | 70.7 | 58.1 | 53.0 | 57.2 | 58.1 | 69.8 | 59.5 | 68.8 | ||
| Two-class classification | Feature selection | F1 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| F2 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| F3 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| F4 | ✓ | ✓ | ✓ | ✓ | - | - | ✓ | ✓ | - | ✓ | - | - | ✓ | ||
| F5 | ✓ | ✓ | ✓ | - | ✓ | - | - | ✓ | - | - | ✓ | ✓ | - | ||
| F6 | ✓ | ✓ | - | ✓ | ✓ | ✓ | - | - | - | - | ✓ | - | ✓ | ||
| F7 | ✓ | - | ✓ | ✓ | ✓ | ✓ | ✓ | - | ✓ | - | - | ✓ | - | ||
| Classifier accuracy | Simple tree (%) | 83.7 | 83.7 | 82.8 | 83.7 | 83.7 | 83.7 | 82.8 | 82.8 | 82.8 | 82.8 | 83.7 | 84.7 | 83.7 | |
| Linear SVM (%) | 89.8 | 89.8 | 84.7 | 89.3 | 89.8 | 89.3 | 81.9 | 84.2 | 82.3 | 81.9 | 89.3 | 80.9 | 88.8 | ||
| Weighted KNN (%) | 87.9 | 87.0 | 86.0 | 87.0 | 88.8 | 88.8 | 84.2 | 87.0 | 83.7 | 84.7 | 86.5 | 85.1 | 87.4 | ||
| Logistic regression (%) | 90.7 | 90.2 | 84.2 | 89.3 | 91.2 | 89.3 | 83.3 | 85.1 | 81.9 | 82.3 | 91.2 | 82.8 | 90.2 | ||
SD – Standard deviation; SVM – Support vector machine; KNN – K-nearest neighbor
As noted in the Statistical Analysis section, in the two-class analysis, the F4 feature did not differ significantly between healthy and MI samples. According to Tables 4 and 5, in four-class analysis, the same F4 feature did not make a significant difference between healthy and three MI samples, which confirms the best results of all the classifiers in distinguishing between extracted features. Note that other extracted features ({F1, F2, F3, F5, F6, F7}), following statistical analysis, and the results of the accuracy of all the classifiers presented in this article, have a significant difference, both in two-class labeling and in four-class labeling.
In MI diagnosis studies, various approaches applied including using wavelet, applying a similarity factor between normal, MI record-keeping, employing eigenvalue, and the energy of ECG signal as extracted features. Since our method applied mentioned classifiers, a similar approach should be compared to our work to achieve a reasonable assessment. Accordingly, Table 9 presents a comparison of our results to other research works. In the proposed method, it is clear that in 10-fold cross-validation with the LR classifier, the accuracy, sensitivity, and specificity achieved an acceptable result in comparison with the others. According to Tables 6-9, we executed 100 iterations for 5-fold and 10-fold cross-validation and calculating the statistical average and SD on the two-class and four-class classification to show that the proposed method along with the “Statistical Analysis” section has acceptable and reliable results and the compatibility of both results is not accidental.
Table 9.
The comparative analysis of the proposed method with other methods for two-class classification
| Two-class classification of MI | |||||
|---|---|---|---|---|---|
| Methods | Extracted features | Classifier | Accuracy (%) | Sensitivity (%) | Specificity (%) |
| Proposed method | Integral of ECG and J-point value | Weighted KNN | 87.9 | 94 | 73 |
| Linear SVM | 89.8 | 95 | 76 | ||
| Logistic regression | 90.37 | 94 | 83 | ||
| L. N. Sharma et al.[16] | Multiscale wavelet energies and eigenvalues of multiscale covariance matrices | KNN | 82.28 | 81.98 | 82.32 |
| Linear SVM | 88.02 | 87.23 | 88.53 | ||
| S. Selva Nidhyananthan et al.[15] | Multiscale wavelet energies and eigenvalues of multiscale covariance matrices | Linear SVM | 90.42 | 89 | 87 |
| Naser Safdarian et al.[14] | Integral of ECG and T-wave | KNN | 89.47 | NA | NA |
SVM – Support vector machine; KNN – K-nearest neighbor; MI – Myocardial infarction
Conclusion
In this study, we examined different extracted features from ECG data in a classification-based feature selection method, and the results show that they are more efficient in the two-class classification in comparison with the four-class. The ECG J-point belonging to all leads is a valuable feature, but in case of better accuracy, it is recommended to extract it by precise automatic algorithms. In addition, the results show that eliminating the F4 feature, which consists of the values of J-point in lead I, approximately increases the precision of all classifiers in the proposed combination of features space. It is essential to point out the more accurate extraction algorithms, the better classification, and results. Finally, the proposed features are valuable for the diagnosis and localization of MI and are recommended for future works.
Suggestions and future studies
There are several suggestions for future studies. To improve the performance of classifiers, the accuracy of the feature extraction technique should increase. As a whole, these are our suggestions for future research:
A robust and precise algorithm to detect all parts of ECG correctly
Detect pathologic Q wave and extract it as a feature
Using other classifiers and especially the nonlinear classifiers.
Compliance with ethical standards
Disclosure of potential conflicts of interest
Since data have obtained from PhysioNet Database free of charge, this study has not funded by any real person or legal institution.
Informed consent and research involving human participants and/or animals
This article does not contain any studies with human participants and/or animals performed by any of the authors but as we have obtained human participant's data from PhysioNet Database so, there have been no practical experiments on human by any of the present authors.
Financial support and sponsorship
None.
Conflicts of interest
There are no conflicts of interest.
BIOGRAPHIES

Seyed Ataddin Mahmoudinejad received the B.Eng. Degree in biomedical engineering from the Islamic Azad University of Dezful, Dezful, Iran, in 2013. In 2015, he graduated from the M.Sc. Eng. in biomedical engineering from Islamic Azad University of Kazeroon, Kazeroon, Iran. Since then, he has been with the Technology Development Centre, Dezful University of Medical Sciences, Dezful, Iran. Also, he is currently a member of the Intellectual Property Committee of the Medical University of Dezful. In 2019, he became a PhD student at the medical university of Tehran, Tehran, Iran. His researches interests are biomedical signal processing artificial intelligence and medical robotic. He studies biomedical engineering in the field of medical robotic until now.
Email: a-mahmoudinejad@razi.tums.ac.ir

Naser Safdarian received M.Sc in Biomedical Engineering from Science and Research Branch, Islamic Azad University, Tehran, Iran, in 2011. His researches interests are Biomedical Signal Processing, especially ECG and EMG Signal Processing, Intelligence Algorithms, Optimization, and Medical Image Processing. He is currently a PhD student in Biomedical Engineering.
Email: naser.safdarian@yahoo.com
References
- 1.Cardiovascular Diseases (CVDs), Fact Sheet. World Health Organization, Media Centre. [Updated May, 2017].
- 2.Mendis S, Thygesen K, Kuulasmaa K, Giampaoli S, Mähönen M, Ngu Blackett K, et al. World Health Organization definition of myocardial infarction: 2008-09 revision. Int J Epidemiol. 2011;40:139–46. doi: 10.1093/ije/dyq165. [DOI] [PubMed] [Google Scholar]
- 3.Thaler M. The only EKG Book you'll Ever Need. Lippincott Williams & Wilkins (LWW) 2017 [Google Scholar]
- 4.Al Touma A, Tafreshi R, Khan M. Detection of Cardiovascular Abnormalities Through 5-Lead System Algorithm. IEEE: 2016 IEEE-EMBS International Conference on Biomedical and Health Informatics (BHI) 2016 [Google Scholar]
- 5.Lu H, Ong K, Chia P. An automated ECG classification system based on a neuro-fuzzy system. Comput Cardiol. 2000;27:387–90. [Google Scholar]
- 6.de Bliek EC. ST elevation: Differential diagnosis and caveats. A comprehensive review to help distinguish ST elevation myocardial infarction from nonischemic etiologies of ST elevation. Turk J Emerg Med. 2018;18:1–0. doi: 10.1016/j.tjem.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Li R, Dai H, Liu Y, Zhou B, Wang Z. Localization of Myocardial Infarction with Multi-lead Bidirectional Gated Recurrent Unit Neural Network. IEEE Access. 2019 [Google Scholar]
- 8.Tripathy RK, Bhattacharyya A, Pachori RB. Localization of Myocardial Infarction from Multi-lead ECG signals using multiscale analysis and convolutional neural network. IEEE Sensors J. 2019;19:11437–48. [Google Scholar]
- 9.Lines GT, de Oliveira BL, Skavhaug O, Maleckar MM. Simple t-wave metrics may better predict early ischemia as compared to st segment. IEEE Trans Biomed Eng. 2016;64:1305–9. doi: 10.1109/TBME.2016.2600198. [DOI] [PubMed] [Google Scholar]
- 10.Ansari S, Farzaneh N, Duda M, Horan K, Andersson HB, Goldberger ZD, et al. A review of automated methods for detection of myocardial ischemia and infarction using electrocardiogram and electronic health records. IEEE Rev Biomed Eng. 2017;10:264–98. doi: 10.1109/RBME.2017.2757953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta R, Kundu P. Dissimilarity Factor Based Classification of Inferior Myocardial Infarction ECG. IEEE:IEEE First International Conference on Control, Measurement and Instrumentation (CMI) 2016 [Google Scholar]
- 12.Carley SD. Beyond the 12 lead: Review of the use of additional leads for the early electrocardiographic diagnosis of acute myocardial infarction. Emerg Med (Fremantle) 2003;15:143–54. doi: 10.1046/j.1442-2026.2003.00431.x. [DOI] [PubMed] [Google Scholar]
- 13.Arif M, Malagore IA, Afsar FA. Detection and localization of myocardial infarction using K-nearest neighbor classifier. J Med Syst. 2012;36:279–89. doi: 10.1007/s10916-010-9474-3. [DOI] [PubMed] [Google Scholar]
- 14.Safdarian N, Dabanloo NJ, Attarodi G. A new pattern recognition method for detection and localization of myocardial infarction using T-wave integral and total integral as extracted features from one cycle of ECG signal. J Biomed Sci Eng. 2014;7:818. [Google Scholar]
- 15.Nidhyananthan SS, Saranya S, Kumari RS. Myocardial Infarction Detection and Heart Patient Identity Verification. IEEE:International Conference on Wireless Communications, Signal Processing and Networking (WiSPNET) 2016 [Google Scholar]
- 16.Sharma LN, Tripathy RK, Dandapat S. Multiscale energy and eigenspace approach to detection and localization of myocardial infarction. IEEE Trans Biomed Eng. 2015;62:1827–37. doi: 10.1109/TBME.2015.2405134. [DOI] [PubMed] [Google Scholar]
- 17.Noorian A, Dabanloo NJ, Parvaneh S, editors. Wavelet based method for localization of myocardial infarction using the electrocardiogram. Comput Cardiol. 2014;2014:645–8. [Google Scholar]
- 18.Goldberger AL, Amaral LA, Glass L, Hausdorff JM, Ivanov PC, Mark RG, et al. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation. 2000;101:E215–20. doi: 10.1161/01.cir.101.23.e215. [DOI] [PubMed] [Google Scholar]
- 19.Samworth RJ. Optimal weighted nearest neighbour classifiers. Ann Statist. 2012;40:2733–63. [Google Scholar]
- 20.Wu J, Bao Y, Chan SC, Wu H, Zhang L, Wei XG, editors. Myocardial Infarction Detection and Classification-A New Multi-Scale Deep Feature Learning Approach. IEEE:IEEE International Conference on Digital Signal Processing (DSP) 2016 [Google Scholar]
- 21.Kohavi R. A Study of Cross-Validation and Bootstrap for Accuracy Estimation and Model Selection. Montreal, Canada: Ijcai; 1995. [Google Scholar]
- 22.Altman NS. An introduction to kernel and nearest-neighbor nonparametric regression. Am Statist. 1992;46:175–85. [Google Scholar]
- 23.Stone CJ. Consistent nonparametric regression. Ann Statist. 1977;5:595–620. [Google Scholar]
- 24.Hastie T, Tibshirani R, Friedman J, Franklin J. The elements of statistical learning: Data mining, inference and prediction. Mathem Intelligencer. 2005;27:83–5. [Google Scholar]
- 25.Cristianini N, Shawe-Taylor J. An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods. London: Cambridge University Press; 2000. [Google Scholar]
- 26.Cortes C, Vapnik VN. Support vector networks. Machine Learning. 1995;20:273–95. [Google Scholar]
- 27.Quinlan JR. Induction of decision trees. Machine Learning. 1986;1:81–106. [Google Scholar]