Abstract
The proportion of Staphylococcus aureus in the skin microbiome is associated with the severity of inflammation in the skin disease atopic dermatitis. Staphylococcus epidermidis, a commensal skin bacterium, inhibits the growth of S. aureus in the skin. Therefore, the balance between S. epidermidis and S. aureus in the skin microbiome is important for maintaining healthy skin. In the present study, we demonstrated that the heat-treated culture supernatant of Delftia acidovorans, a member of the skin microbiome, inhibits the growth of S. epidermidis, but not that of S. aureus. Comprehensive gene expression analysis by RNA sequencing revealed that culture supernatant of D. acidovorans increased the expression of genes related to glycolysis and the tricarboxylic acid cycle (TCA) cycle in S. epidermidis. Malonate, an inhibitor of succinate dehydrogenase in the TCA cycle, suppressed the inhibitory effect of the heat-treated culture supernatant of D. acidovorans on the growth of S. epidermidis. Reactive oxygen species production in S. epidermidis was induced by the heat-treated culture supernatant of D. acidovorans and suppressed by malonate. Further, the inhibitory effect of the heat-treated culture supernatant of D. acidovorans on the growth of S. epidermidis was suppressed by N-acetyl-L-cysteine, a free radical scavenger. These findings suggest that heat-resistant substances secreted by D. acidovorans inhibit the growth of S. epidermidis by inducing the production of reactive oxygen species via the TCA cycle.
Introduction
Atopic dermatitis (AD) is a chronic inflammatory skin disease characterized by variable clinical features, such as relapsing pruritus and eczema [1,2]. Genetic and environmental factors are associated with the onset and exacerbation of AD [3–5]. The skin of AD patients exhibits an elevated pH and barrier disruption. Moreover, the skin microbial composition is altered in AD patients compared with healthy humans. Microbial diversity on the human skin contributes to maintain healthy skin by modulating immune responses [6–8]. In the skin of AD patients, the microbial diversity is reduced due to an increased proportion of Staphylococcus aureus in the skin microbiome [4]. S. aureus exacerbates AD and induces Th2 cytokines and proteases in human skin [4,9,10]. On the other hand, Staphylococcus epidermidis, a coagulase-negative Staphylococcus, inhibits S. aureus growth by producing antimicrobial peptides and short-chain fatty acids [11,12]. The proportion of S. aureus in the skin of AD patients was decreased after transplantation of coagulase-negative Staphylococcus strains having antimicrobial activity [11]. Disrupting the balance between S. aureus and coagulase-negative Staphylococcus with antimicrobial activity against S. aureus in the skin microbiome is associated with exacerbation of AD [11,12]. The relative abundance of S. aureus is increased in the skin of patients with psoriasis (PS) as well as in those with AD [13,14]. S. epidermidis also inhibits the growth of Cutibacterium acnes, which is associated with acne vulgaris (AV) [15–17]. Therefore, S. epidermidis might play a key role in various skin disorders.
Delftia acidovorans, a gram-negative bacterium, is detected in the skin of AD patients by microbiome analysis [18–22]. D. acidovorans secretes a compound with antimicrobial activity against S. aureus, Enterococcus faecalis, Acinetobacter baumannii, Klebsiella pneumoniae, and Pseudomonas aeruginosa [23]. Thus, D. acidovorans may affect bacterial growth in the skin microbiome, but the effect of D. acidovorans on the growth of S. epidermidis remains unclear.
In the present study, we investigated the effect of D. acidovorans on the growth of S. epidermidis and elucidated its mechanisms. We found that the heat-treated culture supernatant (CS) of D. acidovorans inhibited S. epidermidis growth. Furthermore, we demonstrated that D. acidovorans heat-treated CS induced the production of reactive oxygen species (ROS) via the tricarboxylic acid cycle (TCA) cycle in S. epidermidis. Our findings provide important insight into how D. acidovorans affects the skin microbiome by inhibiting the growth of S. epidermidis, resulting in a skin microbiome imbalance related to various skin disorders.
Materials & methods
Reagents
Malonic acid, N-acetyl-L-cysteine (NAC), and gentamicin sulfate were purchased from Wako Pure Chemical Corporation (Osaka, Japan). Menadione was purchased from MilliporeSigma (St. Louis, MO, USA). 2,7-Dichlorodihydrofluorescein diacetate was purchased from Cayman Chemical (Ann Arbor, MI, USA).
Strain and growth conditions
The NBRC100911 strain of Staphylococcus epidermidis (S. epidermidis) and the NBRC100910 strain of Staphylococcus aureus (S. aureus) were obtained from the National Institute of Technology and Evaluation (Tokyo, Japan). S. epidermidis and S. aureus strains were spread on nutrient agar (5 g/L sodium chloride [Wako Pure Chemical Corporation], 5 g/L beef extract [Becton Dickinson, Franklin Lakes, NJ, USA], 10 g/L hipolypepton [Wako Pure Chemical Corporation], and 15 g/L agar powder [Wako Pure Chemical Corporation] before autoclaving) and grown overnight at 37°C. The JCM6218 strain of D. acidovorans was obtained from the Japan Collection of Microorganisms (Ibaraki, Japan). The D. acidovorans JCM6218 strain was spread on nutrient agar and grown overnight at 27°C.
D. acidovorans culture supernatant preparation
A colony of the D. acidovorans JCM6218 strain was inoculated in 3 mL nutrient broth (NB) and incubated overnight at 32°C. The bacterial culture (3 mL) was inoculated into 300 mL NB and incubated overnight at 32°C with shaking (200 rpm, ZWY-240 Incubator Shaker, LABWIT Scientific, VIC, Australia). Bacterial cells were removed by centrifugation at 6300 rpm for 5 min (TOMY-MX305, TOMY Digital Biology Co. Ltd, Tokyo, Japan). The D. acidovorans CS was filtered using the Vacuum Filtration 500 rapid filter MAX (TPP, Schaffhausen, Switzerland). D. acidovorans CS incubated for 30 min at 100°C was used as D. acidovorans heat-treated CS in this study.
Bacterial growth inhibition assay
Bacterial growth inhibition assays were performed according to a previous report [24]. Samples including D. acidovorans CS and inhibitors were diluted with NB to the appropriate concentrations and dispensed in 50-μL aliquots into a 96-well plate (TPP). S. epidermidis and S. aureus suspensions (2 x 104 cells/ml) were prepared with NB and 50 μL was added to each well. After incubating at 37°C for 12 h or 32°C for 18 h, absorbance at 630 nm was measured using a microplate reader (iMark™ microplate reader; Bio-Rad Laboratories Inc., Hercules, CA, USA).
RNA sequencing analysis
RNA-sequencing (RNA-seq) analysis was performed according to a previous report [24]. An S. epidermidis suspension was prepared with NB to an absorbance of 1 at 630 nm. The suspension was diluted in the same volume of D. acidovorans CS and incubated at 37°C for 4 h. After incubation, RNA was extracted from the cells using EZ-Beads (AMR, Inc., Gifu, Japan) and a High Pure RNA Isolation kit (Roche, Basel, Switzerland) according to the manufacturer’s instructions. The RNA-seq library was prepared from the RNA using an NEBNext rRNA Depletion Kit and NEBNext Ultra II RNA Library Prep Kit for Illumina (New England Biolabs Japan Inc., Tokyo, Japan) and subjected to RNA-seq analysis using MiSeq (Illumina Inc., San Diego, CA, USA). Gene function information was examined using the GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and pathway information was analyzed using the Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.genome.jp/kegg/kegg_ja.html).
Time-kill assay
The S. epidermidis suspension (1×105 cells/ml) was prepared with phosphate-buffered saline (PBS) and treated with 20% heat-treated NB, 20% D. acidovorans heat-treated CS, or gentamicin (final concentration 10 μg/mL). Each sample was incubated at 37°C or 32°C. Aliquots were serially diluted in PBS and 100 μL of the diluted samples was spread on a nutrient agar plate at 0, 3, and 6 h. The viable cell number was determined by counting the colonies on the plate after incubation for 24 h at 37°C.
Measuring ROS production
Quantification of ROS production in S. epidermidis with 2’,7’-dichlorodihydrofluorescein diacetate) was performed according to previous reports [25,26]. S. epidermidis was suspended in 3 mL NB and incubated overnight at 37°C with shaking (150 rpm, ZWY-240 incubator shaker, LABWIT Scientific). The culture was suspended in NB to an absorbance of 0.001 at 630 nm and exposed to 100 μM 2’,7’-dichlorodihydrofluorescein diacetate (final concentration) for 1 h at 37°C. After incubation, the samples were centrifuged at 14,000 rpm for 5 min (TOMY-MX105) and the supernatants were removed. The bacterial pellets were suspended in fresh NB and treated with D. acidovorans heat-treated CS or menadione, an ROS inducer. Fluorescence (λ excitation = 485 nm, λ emission = 538 nm) was measured using the Fluoroskan Ascent™ (Thermo Fisher Scientific, Waltham, MA, USA). Fluorescence was determined by subtracting the background fluorescence.
Statistical analysis
Statistical differences between groups were analyzed by the Student t-test, Tukey-Kramer test, Dunnett test, and Williams test. All experiments were performed at least twice. Each experiment was performed in triplicate and error bars indicate the standard deviations of the means. A P value of less than 0.05 was considered statistically significant.
Results
Growth inhibitory effect of substances secreted by D. acidovorans against S. epidermidis
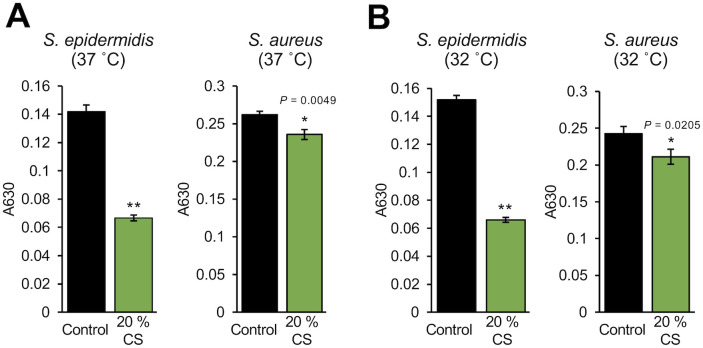
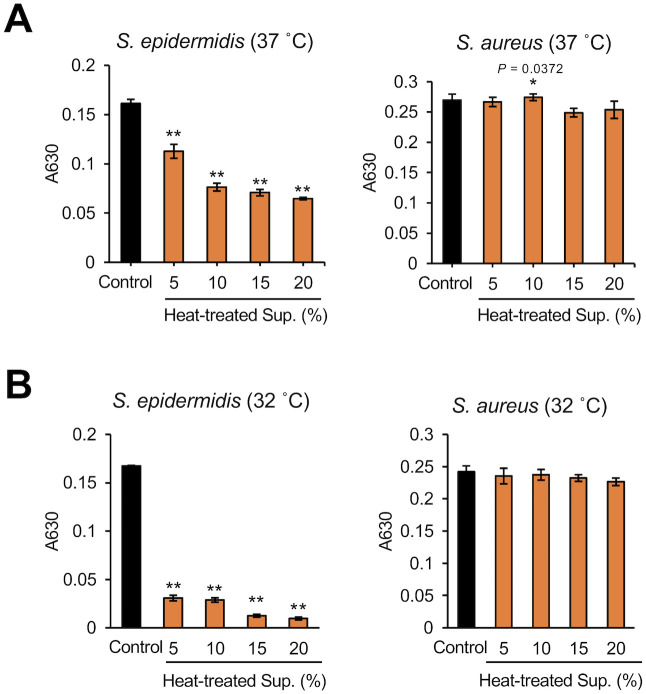
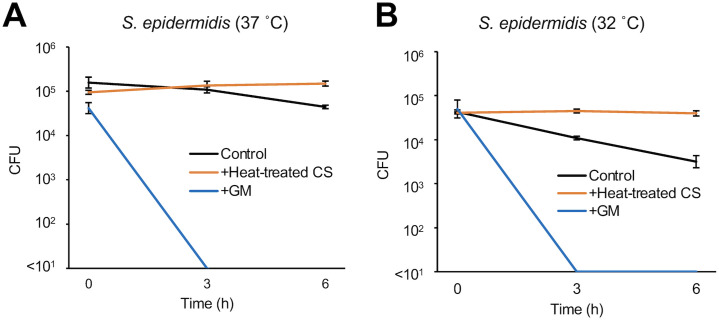
We investigated whether D. acidovorans produces substances that affect the growth of S. epidermidis. Compared with S. aureus, the growth of S. epidermidis at 37°C or 32°C was significantly inhibited by D. acidovorans CS (Fig 1). D. acidovorans heat-treated CS also inhibited S. epidermidis growth in a dose-dependent manner (Fig 2). We next examined whether the inhibitory activity of D. acidovorans heat-treated CS against S. epidermidis is bactericidal or bacteriostatic. D. acidovorans heat-treated CS did not decrease the viable number of S. epidermis in PBS (Fig 3). On the other hand, gentamicin, which has bactericidal activity, decreased the number of viable bacteria within 3 h (Fig 3). These results suggest that heat-stable substances secreted by D. acidovorans exhibit bacteriostatic activity, but not bactericidal activity, against S. epidermidis.
Fig 1. Inhibitory activity of the culture supernatant of D. acidovorans against S. epidermidis growth.
S. epidermidis (1×104 cells/mL) and S. aureus (1×104 cells/mL) in NB were mixed with 20% D. acidovorans CS. After incubating at 37°C for 12 h (A) or 32°C for 18 h (B), absorbance at 630 nm was measured using a microplate reader. Error bars indicate the standard deviations (SD) of the means (n = 3). Statistical differences between groups were analyzed by the Student t-test. (*: P < 0.05, **: P < 0.001).
Fig 2. Dose dependency of the inhibitory effect of heat-treated culture supernatant of D. acidovorans against S. epidermidis growth.
S. epidermidis (1×104 cells/mL) and S. aureus (1×104 cells/mL) in NB were mixed with 5% to 20% D. acidovorans heat-treated CS (Heat-treated CS). After incubating at 37°C for 12 h (A) or 32°C for 18 h (B), absorbance at 630 nm was measured using a microplate reader. Error bars indicate the SD of the means (n = 3). Statistical differences between groups were analyzed by the Williams test. (*: P < 0.05, **: P < 0.001).
Fig 3. Time-kill assay of heat-treated culture supernatant of D. acidovorans against S. epidermidis.
S. epidermidis (1×105 cells/mL) in PBS was mixed with 20% NB (Control), 20% D. acidovorans heat-treated CS (Heat-treated CS), and 10 μg/mL gentamicin (GM) at 37°C (A) or 32°C (B). Each line shows the number of viable cells of S. epidermidis at 0, 3, and 6 h. Error bars indicate the SD of the means (n = 3).
Role of the S. epidermidis TCA cycle in the inhibitory effect of heat-stable substances secreted by D. acidovorans
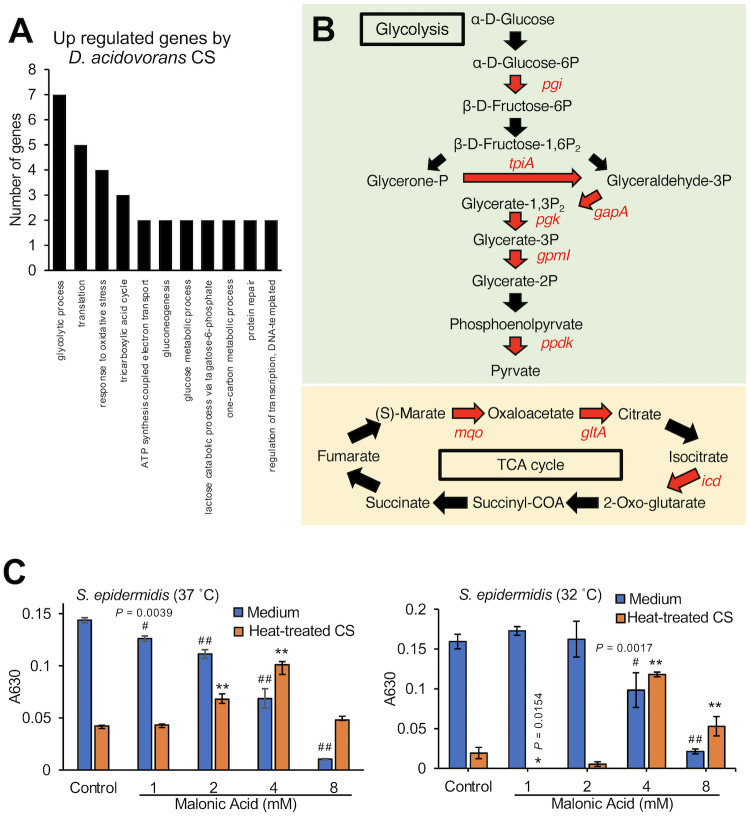
To understand the mechanism of action of the substances secreted by D. acidovorans, we analyzed gene expression in S. epidermidis in response to the D. acidovorans culture supernatant (CS). Expression of 100 genes in S. epidermidis was increased more than 2-fold by the D. acidovorans CS and expression of 7 genes was decreased to less than half (S1 and S2 Tables in S1 File). Gene ontology term analysis revealed that S. epidermidis genes related to glycolysis, the TCA cycle, and the response to oxidative stress were upregulated by the D. acidovorans CS (Fig 4A). Pathway analysis showed that expression of the 6 genes related to glycolysis and 3 genes related to the TCA cycle in S. epidermidis was increased more than 2-fold by the D. acidovorans CS (Fig 4B). Because pyruvate produced by glycolysis triggers TCA cycle activation, we focused on the relationship between activation of the TCA cycle, which is downstream of glycolysis, and the inhibitory activity of D. acidovorans heat-treated CS against S. epidermidis. Malonic acid (4 mM), a TCA cycle inhibitor, suppressed the inhibitory effect of the D. acidovorans heat-treated CS against S. epidermidis growth (Fig 4C). These results suggest that substances secreted by D. acidovorans inhibit the growth of S. epidermidis via TCA cycle regulation.
Fig 4. Suppressive effect of a TCA cycle inhibitor on the inhibitory effect of heat-treated culture supernatant of D. acidovorans against S. epidermidis growth.
(A) Gene expression analysis of S. epidermidis (1×109 cells/mL) mixed with 50% D. acidovorans CS at 37°C for 4 h. The graphs show the number of upregulated genes in S. epidermidis after D. acidovorans CS treatment. (B) Upregulated genes related to glycolysis and the TCA cycle. Red letters represent genes that increased more than 2-fold after treatment with D. acidovorans CS in S. epidermidis. (C) S. epidermidis (1×104 cells/mL) in NB was mixed with 1–4 mM malonic acid and 15% D. acidovorans heat-treated CS (Heat-treated CS). After incubating at 37°C for 12 h or 32°C for 18 h, absorbance at 630 nm was measured using a microplate reader. Error bars indicate the SD of the means (n = 3). Statistical differences of each group were analyzed by the Dunnett test. (#: P < 0.05 (Medium), # #: P < 0.001 (Medium), *: P < 0.05 (Heat-treated CS), **: P < 0.001 (Heat-treated CS)).
Heat-stable substances secreted by D. acidovorans induce ROS production in S. epidermidis
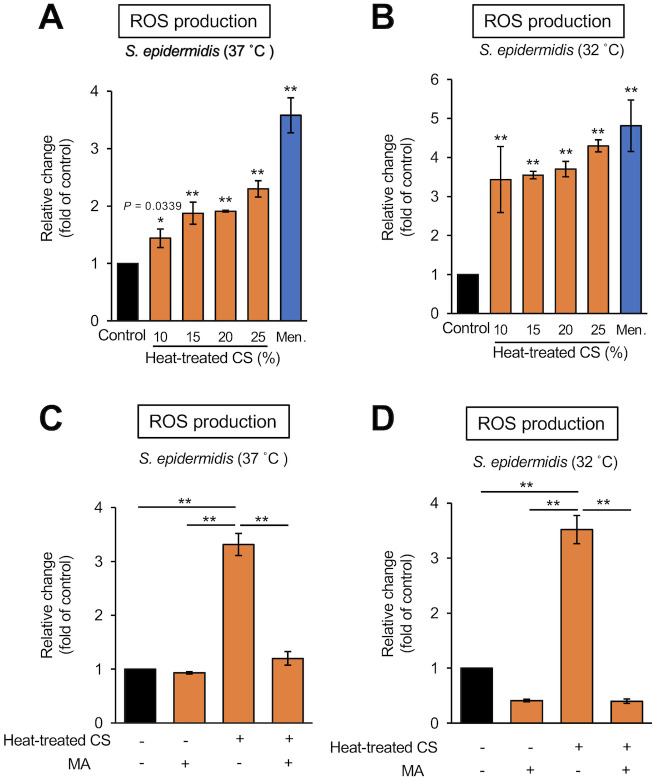
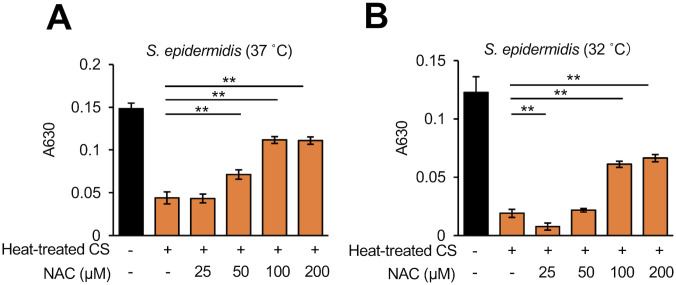
The TCA cycle is linked to ROS production in S. epidermidis [27]. ROS production in S. epidermidis was induced by exposure to the D. acidovorans heat-treated CS (Fig 5A and 5B). The addition of malonic acid inhibited the ROS production in S. epidermidis induced by the D. acidovorans heat-treated CS (Fig 5C and 5D). Addition of a free radical scavenger, NAC, suppressed the S. epidermidis growth inhibition in a dose-dependent manner (Fig 6). These results suggest that substances secreted by D. acidovorans inhibit the growth of S. epidermidis via TCA cycle-triggered ROS production (Fig 7).
Fig 5. ROS production induced in S. epidermidis by heat-treated culture supernatant of D. acidovorans.
(A, B) ROS production in S. epidermidis (1×106 cells/mL) mixed with NB (Control), 10–25% D. acidovorans heat-treated CS (Heat-treated CS), or 70 μg/mL menadione (Men.) at 37°C (A) or 32°C (B) for 24 h. Menadione was used as a positive control. Fluorescence (λ excitation = 485 nm, λ emission = 538 nm) was measured using the Fluoroskan Ascent™. Error bars indicate the SD of the means (n = 3). Statistical differences between groups were analyzed by the Dunnett test. (*: P < 0.05, **: P < 0.001). (C, D) ROS production in S. epidermidis (1×106 cells/mL) mixed with NB (Control), 4 mM malonic acid (MA), 25% D. acidovorans heat-treated CS (Heat-treated CS), or 4 mM MA + 25% heat-treated CS at 37°C (C) or 32°C (D) for 24 h. Fluorescence (λ excitation = 485 nm, λ emission = 538 nm) was measured using the Fluoroskan Ascent™. Error bars indicate the SD of the means (n = 3). Statistical differences between groups were analyzed by the Tukey-Kramer test. (*: P < 0.05, **: P < 0.001).
Fig 6. Suppressive effect of N-acetyl-L-cysteine on the inhibitory activity of heat-treated culture supernatant of D. acidovorans against S. epidermidis growth.
S. epidermidis (1×104 cells/mL) in NB was mixed simultaneously with 15% D. acidovorans heat-treated CS (Heat-treated CS) and 25–200 μM N-acetyl-cysteine (NAC). After incubating at 37°C for 12 h (A) or 32°C for 18 h (B), absorbance at 630 nm was measured using a microplate reader. Error bars indicate the SD of the means (n = 3). Statistical differences between groups were analyzed by the Tukey-Kramer test. (*: P < 0.05, **: P < 0.001).
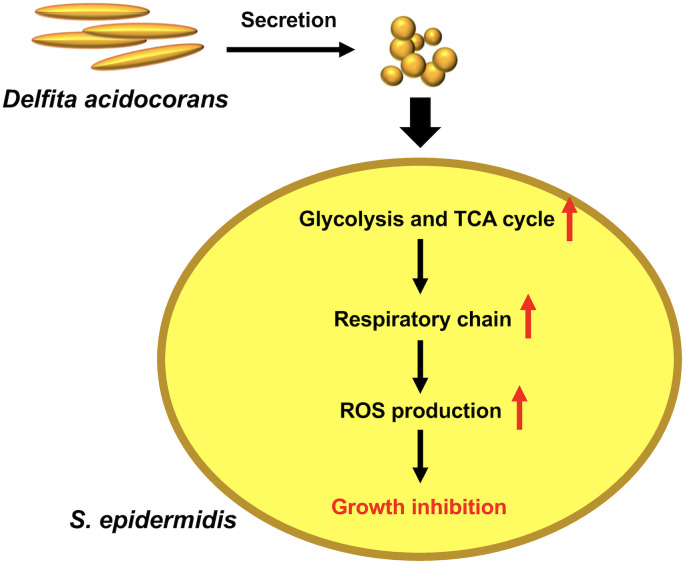
Fig 7. Model of the action of heat-stable substances secreted by D. acidovorans against S. epidermidis.
Heat-stable substances secreted by D. acidovorans induce the expression of the genes related to glycolysis and the TCA cycle and subsequently increase ROS production in S. epidermidis. The induced ROS production leads to growth inhibition in S. epidermidis.
Discussion
The findings of the present study demonstrated that heat-stable substances secreted by D. acidovorans inhibited the growth of S. epidermidis, but not that of S. aureus. The heat-stable substances induced ROS production through the TCA cycle in S. epidermidis. Moreover, the inhibitory effect against S. epidermidis growth by the heat-stable substances was suppressed by NAC, a radical scavenger. Our findings suggest that D. acidovorans secretes heat-stable substances that inhibit S. epidermidis growth by TCA cycle-triggered ROS production.
A previous study demonstrated that D. acidovorans secreted heat-stable antimicrobial substances that inhibit the growth of S. aureus [23]. Therefore, we assessed the inhibitory activity of D. acidovorans heat-treated CS against S. epidermidis. We demonstrated that heat-stable substances secreted by D. acidovorans induced the expression of genes related to glycolysis and the TCA cycle, and the inhibitory effect against growth of S. epidermidis was suppressed by malonic acid, an inhibitor of succinate dehydrogenase in the TCA cycle. On the other hand, 4 mM malonic acid alone inhibited S. epidermidis growth, but this inhibitory activity was suppressed by the heat-stable substances of D. acidovorans. Therefore, we assumed that the inhibitory effect of malonic acid on the TCA cycle competed with the increased gene expression in the TCA cycle induced by the heat-stable substances secreted by D. acidovorans. The finding also suggests that the heat-stable substances of D. acidovorans affect the TCA cycle in S. epidermidis. We also assumed that the effect heat-stable substances of D. acidovorans on the TCA cycle of S. epidermidis led to an increase in the ROS production because the ROS production induced by the heat-stable substances of D. acidovorans was inhibited by malonic acid. Furthermore, the suppressive effect of NAC suggested that the induced ROS production in S. epidermidis caused the inhibitory effect of the heat-stable substances of D. acidovorans against the growth of S. epidermidis. S. aureus, on the other hand, produces staphyloxanthin, an antioxidant that confers resistance to ROS [28,29]. We speculated that the heat-stable substances of D. acidovorans exhibit no inhibitory effects against S. aureus growth because of differences in the resistance to ROS.
D. acidovorans is often isolated and detected by microbiome analysis from clinical samples, including from the skin of patients with AD [18,19,30–34]. We found that heat-stable substances secreted by D. acidovorans inhibited the growth of S. epidermidis, but not the growth of S. aureus. In the skin of patients with AD, the relative abundance of S. aureus is significantly increased, and enterotoxin or protease derived from S. aureus exacerbates inflammation of AD [4,9,10]. The relative abundance of S. aureus is also increased in the skin of patients with PS and Th17 cytokines are induced by S. aureus [13,14]. S. epidermidis inhibits the growth of S. aureus, which might be associated with AD and PS [11,12]. Moreover, S. epidermidis inhibits the growth of C. acnes, which is associated with AV [15–17]. We therefore hypothesized that the inhibitory activity of substances secreted by D. acidovorans against S. epidermidis is indirectly associated with exacerbation of AD, PS, and AV. D. acidovorans might be related to various skin disorders. Additional studies are needed to elucidate the relationship between D. acidovorans and AD, PS, and AV.
Previous studies reported that D. acidovorans secretes delftibactin, which has antimicrobial activity against S. aureus [23,35]. Delftibactin reacts in a solution containing AuCl3 and forms a gold precipitation, resulting in detoxification by chelating Au3+ [35]. We demonstrated that the D. acidovorans heat-treated CS used in this study did not form a gold precipitation by mixing it with a solution containing AuCl3 (S1 Fig in S1 File). Furthermore, gold toxicity against S. epidermidis was not inhibited by D. acidovorans heat-treated CS (S2 Fig in S1 File). Therefore, we assumed that the D. acidovorans heat-treated CS used in this study may not contain a sufficient amount of delftibactin to inhibit the growth of S. aureus. We considered that differences in both the strain and culture conditions between previous studies and the present study could affect the composition of delftibactin. Tejman-Yarden et al. indicated the existence of a fraction other than the delftibactin fraction that exerts antimicrobial activity in the culture supernatant of D. acidovorans. In addition, a substance characterized as C39H68N14O17, which is similar to the composition of delftibactin is present in the delftibactin fraction [23]. It is possible that the active substances focused on in this study are in these fractions. Identifying the active substances in the D. acidovorans heat-treated CS is an important topic for future studies.
Conclusion
D. acidovorans secretes heat-stable substances that have inhibitory activity against S. epidermidis growth through TCA cycle-triggered-ROS production. How D. acidovorans affects the proportion of S. aureus and S. epidermidis in the human skin microbiome is an important topic for future studies.
Supporting information
(DOCX)
(XLSX)
Data Availability
All data identified in the present study are provided in the paper and its Supplementary information (S1 Dataset). The RNA seq data have been deposited at DDBJ/ENA/GenBank: Submission ID = mpu_microbiology-0001, BioProject = PRJDB11464, BioSample = SAMD00294166, SAMD00294165, Accession number = DRA011812.
Funding Statement
This study was supported by the Japan Society for the Promotion of Science, JSPS, https://www.jsps.go.jp/english/index.html, (grant number JP20K07208 to O.C., JP16K08384, and JP19K07176 to T.S.). The funders did not play any role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Katayama I, Aihara M, Ohya Y, Saeki H, Shimojo N, Shoji S, et al. Japanese guidelines for atopic dermatitis 2017. Allergol Int. 2017;66: 230–247. doi: 10.1016/j.alit.2016.12.003 [DOI] [PubMed] [Google Scholar]
- 2.Eckert L, Gupta S, Amand C, Gadkari A, Mahajan P, Gelfand JM. The burden of atopic dermatitis in US adults: Health care resource utilization data from the 2013 National Health and Wellness Survey. J Am Acad Dermatol. 2018;78: 54–61.e1. doi: 10.1016/j.jaad.2017.08.002 [DOI] [PubMed] [Google Scholar]
- 3.Meylan P, Lang C, Mermoud S, Johannsen A, Norrenberg S, Hohl D, et al. Skin Colonization by Staphylococcus aureus Precedes the Clinical Diagnosis of Atopic Dermatitis in Infancy. J Invest Dermatol. 2017;137: 2497–2504. doi: 10.1016/j.jid.2017.07.834 [DOI] [PubMed] [Google Scholar]
- 4.Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. Cold Spring Harbor Lab; 2012;22: 850–859. doi: 10.1101/gr.131029.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kennedy EA, Connolly J, Hourihane JO, Fallon PG, McLean WHI, Murray D, et al. Skin microbiome before development of atopic dermatitis: Early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J Allergy Clin Immunol. 2017;139: 166–172. doi: 10.1016/j.jaci.2016.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naik S, Bouladoux N, Linehan JL, Han S-J, Harrison OJ, Wilhelm C, et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. Nature Publishing Group; 2015;520: 104–108. doi: 10.1038/nature14052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linehan JL, Harrison OJ, Han S-J, Byrd AL, Vujkovic-Cvijin I, Villarino AV, et al. Non-classical Immunity Controls Microbiota Impact on Skin Immunity and Tissue Repair. Cell. 2018;172: 784–796.e18. doi: 10.1016/j.cell.2017.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai Y, Cogen AL, Radek KA, Park HJ, Macleod DT, Leichtle A, et al. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J Invest Dermatol. 2010;130: 2211–2221. doi: 10.1038/jid.2010.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams MR, Nakatsuji T, Sanford JA, Vrbanac AF, Gallo RL. Staphylococcus aureus Induces Increased Serine Protease Activity in Keratinocytes. J Invest Dermatol. 2017;137: 377–384. doi: 10.1016/j.jid.2016.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Syed AK, Reed TJ, Clark KL, Boles BR, Kahlenberg JM. Staphlyococcus aureus phenol-soluble modulins stimulate the release of proinflammatory cytokines from keratinocytes and are required for induction of skin inflammation. Camilli A, editor. Infect Immun. 2015;83: 3428–3437. doi: 10.1128/IAI.00401-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med. 2017;9: eaah4680. doi: 10.1126/scitranslmed.aah4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Traisaeng S, Herr DR, Kao H-J, Chuang T-H, Huang C-M. A Derivative of Butyric Acid, the Fermentation Metabolite of Staphylococcus epidermidis, Inhibits the Growth of a Staphylococcus aureus Strain Isolated from Atopic Dermatitis Patients. Toxins (Basel). Multidisciplinary Digital Publishing Institute; 2019;11: 311. doi: 10.3390/toxins11060311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Totté JEE, van der Feltz WT, Bode LGM, van Belkum A, van Zuuren EJ, Pasmans SGMA. A systematic review and meta-analysis on Staphylococcus aureus carriage in psoriasis, acne and rosacea. Eur J Clin Microbiol Infect Dis. 2016;35: 1069–1077. doi: 10.1007/s10096-016-2647-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang H-W, Yan D, Singh R, Liu J, Lu X, Ucmak D, et al. Alteration of the cutaneous microbiome in psoriasis and potential role in Th17 polarization. Microbiome. BioMed Central; 2018;6: 154–27. doi: 10.1186/s40168-018-0533-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Kuo S, Shu M, Yu J, Huang S, Dai A, et al. Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: implications of probiotics in acne vulgaris. Appl Microbiol Biotechnol. Springer Berlin Heidelberg; 2014;98: 411–424. doi: 10.1007/s00253-013-5394-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christensen GJM, Scholz CFP, Enghild J, Rohde H, Kilian M, Thürmer A, et al. Antagonism between Staphylococcus epidermidis and Propionibacterium acnes and its genomic basis. BMC Genomics. BioMed Central; 2016;17: 152–14. doi: 10.1186/s12864-016-2489-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura K, O’Neill AM, Williams MR, Cau L, Nakatsuji T, Horswill AR, et al. Short chain fatty acids produced by Cutibacterium acnes inhibit biofilm formation by Staphylococcus epidermidis. Sci Rep. Nature Publishing Group; 2020;10: 21237–12. doi: 10.1038/s41598-020-77790-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandwein M, Fuks G, Israel A, Sabbah F, Hodak E, Szitenberg A, et al. Skin Microbiome Compositional Changes in Atopic Dermatitis Accompany Dead Sea Climatotherapy. Photochem Photobiol. 2019;95: 1446–1453. doi: 10.1111/php.13119 [DOI] [PubMed] [Google Scholar]
- 19.Li W, Xu X, Wen H, Wang Z, Ding C, Liu X, et al. Inverse Association Between the Skin and Oral Microbiota in Atopic Dermatitis. J Invest Dermatol. 2019;139: 1779–1787.e12. doi: 10.1016/j.jid.2019.02.009 [DOI] [PubMed] [Google Scholar]
- 20.Leung DYM, Calatroni A, Zaramela LS, LeBeau PK, Dyjack N, Brar K, et al. The nonlesional skin surface distinguishes atopic dermatitis with food allergy as a unique endotype. Sci Transl Med. 2019;11: eaav2685. doi: 10.1126/scitranslmed.aav2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tay ASL, Li C, Nandi T, Chng KR, Andiappan AK, Mettu VS, et al. Atopic dermatitis microbiomes stratify into ecological dermotypes enabling microbial virulence and disease severity. J Allergy Clin Immunol. 2020. doi: 10.1016/j.jaci.2020.09.031 [DOI] [PubMed] [Google Scholar]
- 22.Byrd AL, Deming C, Cassidy SKB, Harrison OJ, Ng W-I, Conlan S, et al. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci Transl Med. 2017;9: eaal4651. doi: 10.1126/scitranslmed.aal4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tejman-Yarden N, Robinson A, Davidov Y, Shulman A, Varvak A, Reyes F, et al. Delftibactin-A, a Non-ribosomal Peptide With Broad Antimicrobial Activity. Front Microbiol. Frontiers; 2019;10: 2377. doi: 10.3389/fmicb.2019.02377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto Y, Nakashima T, Cho O, Ohkubo T, Kato J, Sugita T. Pyruvate-triggered TCA cycle regulation in Staphylococcus aureus promotes tolerance to betamethasone valerate. Biochem Biophys Res Commun. 2020;528: 318–321. doi: 10.1016/j.bbrc.2020.05.035 [DOI] [PubMed] [Google Scholar]
- 25.Van Acker H, Gielis J, Acke M, Cools F, Cos P, Coenye T. The Role of Reactive Oxygen Species in Antibiotic-Induced Cell Death in Burkholderia cepacia Complex Bacteria. Kaufmann GF, editor. PLoS ONE. Public Library of Science; 2016;11: e0159837. doi: 10.1371/journal.pone.0159837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langlois J-P, Millette G, Guay I, Dubé-Duquette A, Chamberland S, Jacques P-É, et al. Bactericidal Activity of the Bacterial ATP Synthase Inhibitor Tomatidine and the Combination of Tomatidine and Aminoglycoside Against Persistent and Virulent Forms of Staphylococcus aureus. Front Microbiol. Frontiers; 2020;11: 805. doi: 10.3389/fmicb.2020.00805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas VC, Chittezham Thomas V, Kinkead LC, Janssen A, Schaeffer CR, Woods KM, et al. A dysfunctional tricarboxylic acid cycle enhances fitness of Staphylococcus epidermidis during β-lactam stress. Gilmore MS, editor. mBio. American Society for Microbiology; 2013;4: 233. doi: 10.1128/mBio.00437-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nasser A, Moradi M, Jazireian P, Safari H, Alizadeh-Sani M, Pourmand MR, et al. Staphylococcus aureus versus neutrophil: Scrutiny of ancient combat. Microb Pathog. 2019;131: 259–269. doi: 10.1016/j.micpath.2019.04.026 [DOI] [PubMed] [Google Scholar]
- 29.Clauditz A, Resch A, Wieland K-P, Peschel A, Götz F. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect Immun. American Society for Microbiology Journals; 2006;74: 4950–4953. doi: 10.1128/IAI.00204-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yildiz H, Sünnetçioğlu A, Ekin S, Baran Aİ, Özgökçe M, Aşker S, et al. Delftia acidovorans pneumonia with lung cavities formation. Turgut E, editor. Colomb Med. 2019;50: 215–221. doi: 10.25100/cm.v50i3.4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawamura I, Yagi T, Hatakeyama K, Ohkura T, Ohkusu K, Takahashi Y, et al. Recurrent vascular catheter-related bacteremia caused by Delftia acidovorans with different antimicrobial susceptibility profiles. J Infect Chemother. 2011;17: 111–113. doi: 10.1007/s10156-010-0089-x [DOI] [PubMed] [Google Scholar]
- 32.Mahmood S, Taylor KE, Overman TL, McCormick MI. Acute infective endocarditis caused by Delftia acidovorans, a rare pathogen complicating intravenous drug use. J Clin Microbiol. 2012;50: 3799–3800. doi: 10.1128/JCM.00553-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bilgin H, Sarmis A, Tigen E, Soyletir G, Mulazimoglu L. Delftia acidovorans: A rare pathogen in immunocompetent and immunocompromised patients. Can J Infect Dis Med Microbiol. Hindawi; 2015;26: 277–279. doi: 10.1155/2015/973284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen A, Fegan M, Hayward C, Chakraborty S, Sly LI. Phylogenetic relationships among members of the Comamonadaceae, and description of Delftia acidovorans (den Dooren de Jong 1926 and Tamaoka et al. 1987) gen. nov., comb. nov. Int J Syst Bacteriol. Microbiology Society; 1999;49 Pt 2: 567–576. doi: 10.1099/00207713-49-2-567 [DOI] [PubMed] [Google Scholar]
- 35.Johnston CW, Wyatt MA, Li X, Ibrahim A, Shuster J, Southam G, et al. Gold biomineralization by a metallophore from a gold-associated microbe. Nat Chem Biol. Nature Publishing Group; 2013;9: 241–243. doi: 10.1038/nchembio.1179 [DOI] [PubMed] [Google Scholar]