Abstract
The standard rapid approach for the diagnosis of coronavirus disease 2019 (COVID-19) is the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA. The detection of specific anti-SARS-CoV-2 immunoglobulins is crucial for screening people who have been exposed to the virus, whether or not they presented symptoms. Recent publications report different methods for the detection of specific IgGs, IgMs, and IgAs against SARS-CoV-2; these methods mainly detect immunoglobulins in the serum using conventional techniques such as rapid lateral flow tests or enzyme-linked immunosorbent assay (ELISA). In this article, we report the production of recombinant SARS-CoV-2 spike protein and the development of a rapid, reliable, cost-effective test, capable of detecting immunoglobulins in serum and saliva samples. This method is based on interferometric optical detection. The results obtained using this method and those obtained using ELISA were compared. Owing to its low cost and simplicity, this test can be used periodically for the early detection, surveillance, detection of immunity, and control of the spread of COVID-19.
Keywords: SARS-CoV-2, Immunoglobulins, Serum, Saliva, Interferometric optical detection method, Biosensing
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an RNA virus responsible for the pandemic that has infected over 120 million people and caused over 2.1 million deaths worldwide [1]. The infectivity of the virus, coupled with the extent of globalization has been responsible for its rapid spread, reaching most countries within a couple of months [2]. The infectivity and virulence also posed a problem for many countries where a large percentage of sanitary workers were infected, presenting an obstacle to the functioning of healthcare systems [3]. Therefore, countries need to be able to test the population in a fast, cheap, convenient, and reliable manner. In fact, since January 2020, the World Health Organization has encouraged each country with three words: “test, test, test” [4]. Frequent testing is also important to gain an understanding of the transmission, infectivity, and morbidity of the virus, and the herd immunity in the population.
In Spain and other European countries, most of the tests conducted are serum-based. These are generally of two types: the first test, which is more commonly used, analyzes total immunoglobulin (Ig) levels; the second test analyzes the presence of anti-SARS-CoV-2 IgM and IgG antibodies. The third diagnostic test is the polymerase chain reaction (PCR)-based method used for the detection of viral RNA in a nasopharyngeal sample. Usually, the first type of test is used in patients and the second and/or third tests are conducted only if the first yields positive results [1]. Although this strategy has several advantages, it has been found to produce a large number of false positives, require repetitions (which increases the possibility of cross-contamination), and is more invasive and uncomfortable for the patient [2].
By September 2020, over 150 tests have been carried out per 1000 people in Spain [1]; these showed a prevalence of SARS-CoV-2 of 8.9‰. However, many of these tests were repetitions or were conducted in conjunction with other tests [3].
Data from epidemiological studies conducted around the world have shown the necessity to develop new types of tests (as mentioned above), which use samples that are easier to obtain, such as saliva. Saliva has previously been used for the detection of infections [4]. IgA levels in saliva samples have previously been reported for patients testing positive for SARS-CoV-2 [5]; testing these levels would also facilitate the population-based mass screening for COVID-19 [6].
The spike protein is a glycosylated protein present on the outer surface of SARS-CoV-2; it plays a key role in viral entry into host cells [7,8]. Cryogenic electron microscopy studies have shown that trimeric arrangement and structural changes are necessary for the fusion of host and viral membranes [9]. The S1 and S2 subunits of the spike protein are affected to different extents by these changes, which ultimately allow the receptor-binding domain to access the target. The nature of the function of the spike protein is the basis of many studies, which target the protein with the goal of neutralizing the virus. Antibodies detected in patients infected with SARS-CoV-2 have also been reported to target the spike protein [[10], [11], [12]].
While these characteristics of the spike protein drive its use in test systems, different complications arise. Each monomer of the spike protein is 180 kDa and is composed of two subdomains, which are folded in a complicated manner. The protein is heavily glycosylated and can undergo other post-translational modifications, including acylation and phosphorylation [13]. The safety, yield, and reproducibility, among other factors, dictate the use of the recombinant protein, as opposed to the spike protein isolated from the natural source. However, not all systems are adequate for the production of proteins with these characteristics. Pichia pastoris has previously been used for the expression of difficult proteins with high yields, at cheap prices, using scalable protocols (including industries) [14]. In fact, a domain of the SARS-CoV spike protein was successfully expressed in P. pastoris in 2009 [15].
Immunoassay technology for in vitro diagnostics is of relevance in healthcare, clinical, agro-food, environmental, and pharmaceutical research, as well as for drug control and other fields. The field of optical techniques has experienced significant developments in biochemical sensing, which can be divided into approaches based on interferometry and resonance, absorption, and fluorescence. The latter two are based on labeled technologies such as enzyme-linked immunosorbent assay (ELISA) or lateral flow-based systems [16]. However, the significant advantages of optical biosensing systems based on combined interferometry and resonance, used with or without optical waveguides, are worth mentioning [17,18]. The applications of surface plasmon resonance-based biosensors [[19], [20], [21]], diffraction grating coupled interferometry [22], photonic crystals [23,24], ring resonators [[25], [26], [27]], Mach–Zehnder [28,29], guided-mode resonance biosensors [30,31], Young interferometers [32], and resonant nanopillars [33], among others, have been widely reported.
In order to detect specific anti-SARS-CoV-2 immunoglobulins in the serum or saliva samples of patients, immune responses must be emulated in vitro in a biosensor. To achieve this, we immobilized the recombinant SARS-CoV-2 spike protein in the sensing area of photonic biosensors. These photonic biosensors are based on biophotonic sensing cells (BICELLs), which have recently been reported [[34], [35], [36]]. Not only are the biosensors important, but also is the manner in which the transducing signals are readout in them. The interferometric optical detection method has been reported [37,38] to be a reliable system to archive the demanding limit of detection required for most in vitro diagnostic systems. It also has capacity for high-throughput screening. Both of these features are essential for the detection of antibodies specific to SARS-CoV-2.
Therefore, the work described here intends to present an in vitro diagnostic system consisting of a truncated and recombinant SARS-CoV-2 spike protein immobilized in BICELL photonic transducers. This system aims to detect anti-SARS-CoV-2 IgG, IgM, and IgA antibodies in serum and saliva samples of patients testing PCR-positive, using the interferometric optical detection method (IODM); the results obtained were compared and correlated with those obtained by ELISA.
ELISA is a gold standard laboratory technique that allows for the specific detection of a large variety of specific analytes in various types of samples. Therefore, it has diverse applications in the fields of clinical diagnostics, food quality, and biotechnology, among others. However, it has several disadvantages: it is a laborious procedure, requires a large sample volume, has limited options for multiplexing, and has a detection limit just lower than the nanomolar range [39,40]. The success of the IODM has already been proven. Its ability to conduct high throughput screening and its cost-effectiveness could also be beneficial in health systems.
2. Materials and methods
2.1. Production of recombinant truncated spike protein using Pichia pastoris
SARS-CoV-2 complementary DNA was kindly donated by Isabel Solá (Consejo Superior de Investigaciones Científicas – Centro Nacional de Biotecnología, CSIC-CNB, Spain). The truncated region of the spike gene was first amplified by PCR using specific primers. After cloning into the pPICZalpha plasmid, which served the vector, P. pastoris Bg11 cells were electro-transformed, and the protein was produced as per the manufacturer’s instructions. A fragment corresponding to the largest epitope was produced, as previously described [41].
Recombinant SARS-CoV-2 spike protein (rS1) was purified from the supernatant of the yeast culture and isolated by chromatography. Briefly, supernatants were dialyzed against 0.1 M ammonium acetate (pH 6.8) for 8 h at 4 °C (Spectrum Labs Spectra/6–8 kD MWCO RC Dry dialysis membrane, Fisher Scientific, Waltham, MA, USA). After freeze-drying, the dialyzed supernatant was fractionated by size exclusion chromatography on a Sephacryl S-200 High Resolution system (GE Healthcare, Chicago, IL, USA) in 0.1 M ammonium acetate (pH 6.8) (1 mL/min, 5 mL fractions). Fractions were quantified by bicinchoninic acid test (Thermo Scientific, Waltham, MA, USA) and analyzed by Coomassie staining and immunoassays using specific antibodies against the SARS-CoV-2 spike protein (Invitrogen, Thermo Fisher, Carlsbad, CA, USA).
The protein quality was assessed by matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry, and its identity was verified by peptide-mass fingerprint analysis, using an Ultraflex workstation (Bruker Daltonics, Bremen, Germany) equipped with a 337 nm nitrogen laser by standard methods. FlexControl Software version 2.4. (Bruker Daltonics) was used for sample analysis and for control of the analytical method parameters.
2.2. Human samples
To evaluate the immune activity of rS1, ELISA was performed using saliva samples collected during the voluntary and experimental pilot trial for the early detection, surveillance, and control of COVID-19 of the Universidad Politécnica de Madrid carried out at the Center of Biomedical Technology (http://www.ctb.upm.es/blog/).
The presence of SARS-CoV-2 antigens in the serum samples collected from patients with and without SARS-CoV-2 infection was confirmed by PCR. These samples were supplied by the Health Research Institute biobank, belonging to the Hospital Clínico San Carlos of Madrid (B.0000725; PT17/0015/0040; ISCIII-FEDER). Clinical studies were approved by the local ethics committee of the Hospital Clínico San Carlos (20/404-E_COVID). The samples were classified as per the clinical diagnostic criteria, as moderate, severe, or mild. Once the serum samples were received, they were thawed and treated at 56 °C to achieve complement deactivation.
2.3. Activity assays
Briefly, polystyrene 96-well microtiter plates (Costar 3590, Corning) were coated with 50 μL purified rS1 (5 μg/mL) and incubated for 2 h at 37 °C. After blocking, the plates were washed and incubated with saliva samples at 1:10 dilution (ON, 4 °C). The presence of specific IgA antibodies was detected by incubation with polyclonal horseradish peroxidase (HRP)-labelled anti-IgA antibodies (Thermo Fisher) for 1 h at 25 °C. The plates were washed again and then developed by treatment with 50 μL peroxidase substrate buffer (Ultra-TMB, Thermo Scientific). After 30 min, the reaction was stopped by treatment with 50 μL 2 N HCl, and the optical density was measured at 450 nm. Phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) was used as a negative control. Assays were performed in triplicate.
When the recombinant protein activity was studied, 98 % of patients who tested PCR-positive for SARS-CoV-2 (n = 54; March–June 2020) showed positive titers for IgA, IgG, and IgM antibodies. The test results of patients who had not been in contact with the virus (n = 14) were negative. Therefore, even though the percentage similarity between the activity of the natural protein and the recombinant fragment is not known, we can confirm that it is a good bioreceptor for use in the detection of anti-SARS-CoV-2 antibodies
In ELISA, the negative control was coated with casein blocking buffer; signals greater than 0.1 absorbance units were not obtained in any of the cases. The final data shown were obtained by subtracting the value of the negative control from the values obtained for the recombinant protein.
2.4. Use of multiplexed biosensors on a kit
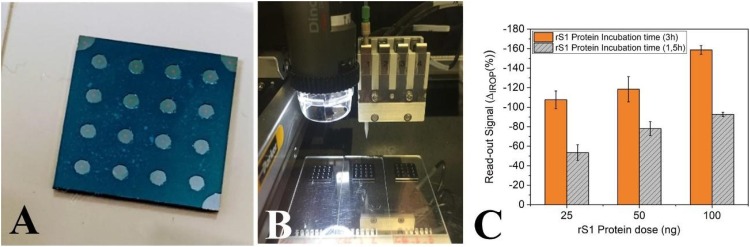
In this study, 16 independent BICELLs, transduced on chips, packaged on a kit, were used, since they were easy to handle. Each BICELL is based on two Fabry-Perot interferometers: one layer of SiO2 and a thin SU-8 polymeric film which exhibits reliable optical biosensing [42,43].
For the fabrication of the BICELLs, SU-8 2000.5 (MicroChem Corp., Newton, MA, USA) was spin-coated on a silicon substrate with a thin layer of SiO2. It was then soft-baked at 115 °C for 30 s. An ultraviolet light-exposure process was then conducted, followed by a post-baking step at 115 °C for 4 min and a developing step for 2 min (Laguna et al., 2015). Finally, the BICELL surface was activated by means of an O2 plasma process [44], to immobilize the recombinant SARS-CoV-2 spike protein onto the sensing surface. We tested and verified that the biofunctionalized biosensors can maintain their stability and activity for at least 3 months.
2.5. Biosensor readout
All immunoassay measurements were carried out using the IODM to read the 16 multiplexed BICELLs on a single kit (Fig. 1 A). The read-out used to evaluate biosensing was based on the increased relative optical power (IROP) signal; this has been reported previously [38]. In brief, IROP is the unit of measurement used for the quantification of changes in the optical readout for the two interferometric signals. When one of the interferometers changes its response due to biological recognition, the IROP signal changes. We define ΔIROP (%) as the difference between the reference IROP and the IROP after recognition of the biological event. Therefore, this methodology allows us to detect the concentration of the target biomolecules in an analyzed sample after specific binding to the biosensing response, as previously reported [37,43].
Fig. 1.
Biofunctionalization of biophotonic sensing cells (BICELLs). (A) Sixteen 200 μm BICELLs integrated into one biokit. (B) BioDot dispensing 1 μL of reagent per BICELL in three biokits. (C) Sensing recognition curve for 200 μm BICELLs. A calibration curve of ΔIROP (%) vs concentration of rS1 protein (25–100 ng/cell) after incubation for 1.5 and 3 h. n = 4 BICELLs for each concentration. Abbreviations: ΔIROP, difference in the increased relative optical power signal between the reference and after recognition of the biological event; rS1, recombinant SARS-CoV-2 spike protein.
2.6. Biofunctionalization
The whole batch of immunoassays was performed using rS1, produced in P. pastoris and purified as described in Section 2.1. The rS1 protein was automatically dropped over each BICELL (1 μL, as a 300 μg/mL solution in PBS [pH 7.5]) (Sigma-Aldrich, St. Louis, MO, USA) using an automated liquid dispensing platform, the BioDot AD1520TM (Fig. 1B). The BICELLs were then incubated for 3 h in a humid environment at 37 °C. After incubation, the biokits were washed by directing a stream of ultrapure water onto the surface of each BICELL and dried using filtered air. The ΔIROP (%) signal of the rS1 biofilm was then measured and the values were confirmed to be within our admissible limits of tolerance. A dose-response curve (concentration-ΔIROP [%]) was used to determine the concentration of protein to be biofunctionalized and to establish the tolerance limits (Fig. 1C). All kits were blocked with casein hydrolysate 1x (Sigma-Aldrich) for 1 h with agitation. Finally, the BICELLs were washed with 20 mL ultrapure water, shaken with ultrapure water for 45 s, and dried using clean air. The BioDot dispensing system can accurately dispense small volumes of reagent (in the order of 100 nL), allowing excellent reproducibility in the biofunctionalization stage.
2.7. In vitro detection of total Ig, IgG, IgM, and IgA in serum
Once the kits with the rS1 virus protein were biofunctionalized, serum samples were diluted 1:10 and 1.5 μL of the sample was added per cell. This was then incubated for 3 h at 37 °C in a humid chamber to measure the total Ig levels. Washing was performed using 20 mL ultrapure water, followed by shaking for 10 min with PBS (Sigma-Aldrich), and two syringes of 20 mL ultrapure water, before drying with filtered air. The IgG, IgM, and IgA titers were measured simultaneously, allowing the total Ig to be determined directly. To separate the titers from the antibodies, the kits were incubated with secondary antibodies (αIgG, αIgM, αIgA). The kits from the washing step were incubated with 1.5 μL of αIgG (1:250, Sigma-Aldrich), αIgM (1:20, Sigma-Aldrich), and αIgA (1:10, Sigma-Aldrich) for 3 h at 37 °C in a humid environment. Then, the washing step was performed using 20 mL ultrapure water and shaking for 2 min in Milli-Q water. The kit was then dried using clean and dry air. IROP (%) were measured after each stage of incubation.
2.8. In vitro detection of IgA in saliva
First, saliva samples were centrifuged for 10 min at 15,000 rpm and the supernatant was collected. The sensing surface was biofunctionalized with the rS1 viral protein, as described in Section 2.6. To measure the total Ig levels directly, 2 μL of the saliva samples were incubated in each well overnight at 4 °C in a humid chamber. The washing step was performed with 20 mL ultrapure water and shaking for 45 s in Milli-Q water, before drying with clean air. To quantify the concentration of IgA in saliva and corroborate the results previously obtained, the kits were incubated with specific antibodies (αIgA). They were treated with 2 μL αIgA (1:10, Sigma-Aldrich) and incubated for 3 h at 37 °C in a humid environment. Next, the washing step was performed using 20 mL ultrapure water and shaking for 2 min in Milli-Q water. The chip was dried with clean and dry air. IROP (%) values were measured after each stage of incubation.
To increase the speed of the measurements, the assay was conducted using the same protocol, changing only the saliva incubation time. In this manner, the samples were incubated for 1, 3, and 6 h, and overnight as a control; the duration of incubation with αIgA (1:10, Sigma-Aldrich) was not changed.
2.9. Correlation assays
To determine the correlation between the two techniques (BICELL-based ELISA and IODM), ELISA plates were coated with 50 μL of purified rS1 (5 μg/mL) and incubated for 2 h at 37 °C. After blocking, the plates were washed and incubated with serum samples (1:40 dilution) and saliva samples (1:10 dilution) in blocking buffer:PBS (1:4 dilution) overnight at 4 °C. The presence of specific IgG, IgM, and IgA antibodies was detected by incubation with HRP-labelled polyclonal antibodies against IgG (1:25000), IgM (1:20000), and IgA (1:10000) (Thermo Fisher) for 1 h at 25 °C. Plates were washed again with 0.05 % PBS-Tween and then developed with 50 μL peroxidase substrate buffer (Ultra-TMB, Thermo Scientific). After 30 min, the reaction was stopped with 50 μL of 2 N HCl, and the optical density was measured at 450 nm. PBS containing 1% BSA was used as a negative control. Assays were performed in triplicate.
3. Results and discussion
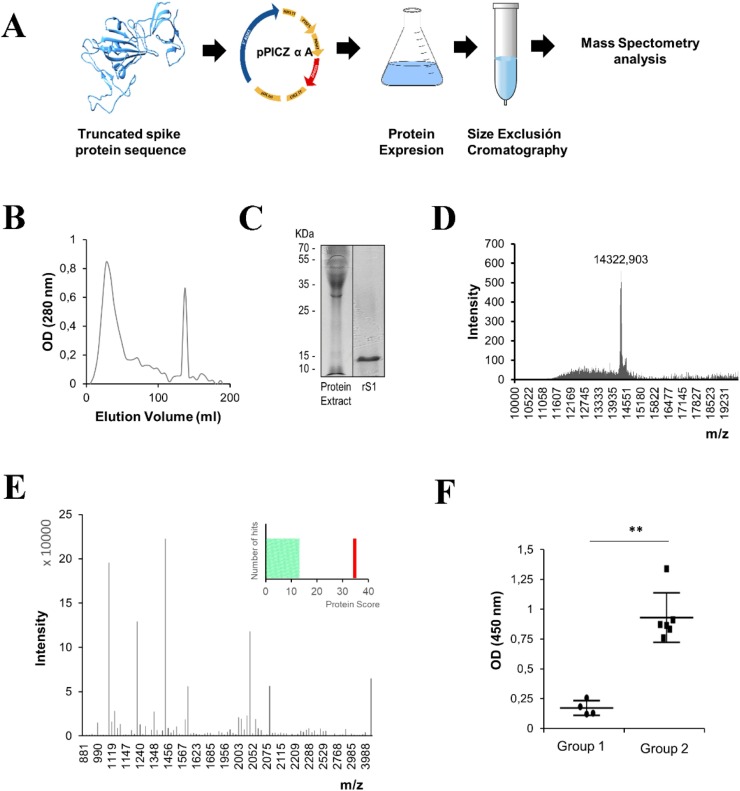
The domain of the SARS-CoV-2 spike protein (rS1) was expressed as a recombinant protein and purified from the media culture by size exclusion chromatography. It was then analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (Fig. 2 A–C). To assess the purity and identity of the protein, two different mass spectrometry analyses were carried out. Molecular weight analysis of the protein by MALDI-TOF revealed the presence of a unique peak representing a mass of 14322 Da (Fig. 2D), while peptide-mass fingerprinting verified the identity of the protein (Fig. 2E). After confirming the purity and nature of the protein, the activity was evaluated using saliva samples obtained from volunteers who had tested PCR-positive (Fig. 2F). The specificity of recognition was evaluated by ELISA. The results showed that the IgA antibodies present only in the patients who tested PCR-positive for SARS-CoV-2 (Group 2) were able to recognize rS1.
Fig. 2.
(A) Scheme of the production of truncated recombinant SARS-CoV-2 spike protein (rS1). (B) The protein extract was fractionated by size exclusion chromatography on a Sephacryl S-200 High Resolution system. The elution profile of the extract was detected at 280 nm (shown above). (C) The protein extract and purified rS1 (5 μg) were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis and Coomassie staining. (D) Molecular weight determination by matrix-assisted laser desorption/ionization-time of flight mass spectrometry of purified rS1. Measurements were performed in the linear positive mode, operating in the range of m/z = 4000–60000. (E) Peptide mass fingerprinting spectrum and Mascot scores (p < 0.05) of the purified rS1. (F) Activity assay to analyze the binding of rS1 with specific IgA in the saliva samples obtained from the volunteers. Group 1: saliva from volunteers who tested negative for SARS-CoV-2 by PCR and serological tests. Group 2: saliva samples from volunteers who tested positive for SARS-CoV-2 by PCR and serological tests. Statistical significance (**p < 0.01) determined by Mann-Whitney test for unpaired samples (n = 6). Abbreviations: OD, optical density.
In this work, we report the detection of specific anti-SARS-CoV-2 immunoglobulins in the serum and saliva of volunteers, using 16 BICELL-multiplexed kits. We demonstrated the in vitro detection of specific total Ig in patient samples, while avoiding the nonspecific adsorption of the serum and saliva complex matrix.
In addition, this is a method for direct detection and does not require any chemical developers or signal amplification (Fig. 3, Fig. 5AA). The use of O2 plasma etching of SU8 epoxy resist to immobilize rS1 improved the specific detection of immunoglobulins, since the activation of the open epoxy rings is sustained for the first hour, during which the immobilization of rS1 takes place. We observed that the dose of the protein required to properly cover the sensing area of BICELLs was 100 ng, and the required incubation time was 3 h (Fig. 1C). This result was obtained by dropping 1 L of 100 g mL−1 protein and comparing two incubation times (1.5 h and 3 h); saturation of the surface was observed at 3 h. Moreover, to prevent the non-specific adsorption of serum and saliva, we blocked the surface with casein hydrolysate.
Fig. 3.
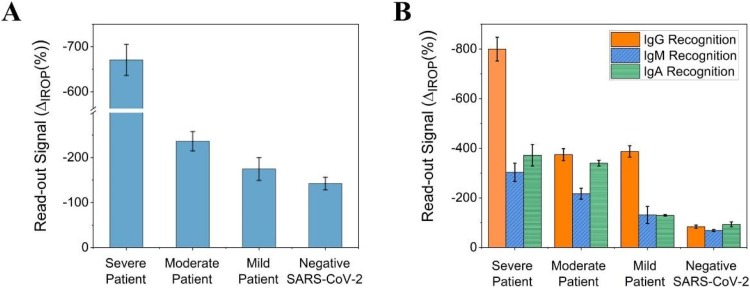
In vitro detection of Ig total, IgG, IgM, and IgA in serum. (A) The vertical axis represents the ΔIROP (%) corresponding to the total Ig levels detected in the sera of patients with severe, moderate, mild, and no SARS-CoV-2 infection. (B) The vertical axis represents the ΔIROP (%) corresponding to the levels of specific IgG, IgM, and IgA antibodies in the same group of patients. Abbreviations: ΔIROP, difference in the increased relative optical power signal between the reference and after recognition of the biological event; Ig, immunoglobulin.
Fig. 5.
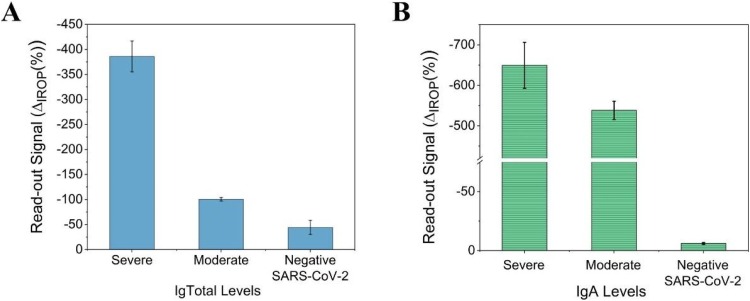
In vitro detection of IgA in saliva. (A) Ig total levels in saliva determined by a direct detection method. The antibody titers in patients with severe, moderate, and no disease are represented. (B) The ΔIROP (%) corresponding to the specific anti-rS1 IgA antibody levels. Abbreviations: ΔIROP, difference in the increased relative optical power signal between the reference and after recognition of the biological event; rS1, recombinant SARS-CoV-2 spike protein; Ig, immunoglobulin.
To determine the type of immunoglobulins — IgG, IgM, and IgA in the serum and IgA in saliva — the corresponding secondary antibodies were used in the second step of recognition.
First, total specific immunoglobulins to SARS-CoV-2and IgG, IgM, and IgA titers in serum samples were measured. Stronger read-out signals of total Ig levels were observed in patients with severe disease; the signals decreased in patients with moderate or mild disease (Fig. 3A). The titers differed based on the clinical classification (severe, moderate, and mild). Upon comparison of the individual immunoglobulin titers, we found that the level of IgGs was significantly elevated in patients with severe disease, compared to those with moderate disease (Fig. 3B). The levels of IgMs and IgAs were also found to be higher in patients who were classified as severe and moderate, in comparison with those who had mild symptoms (Fig. 3B). Finally, patients who tested negative for SARS-CoV-2 infection showed a low signal, representative of the background signal of the in vitro detection system.
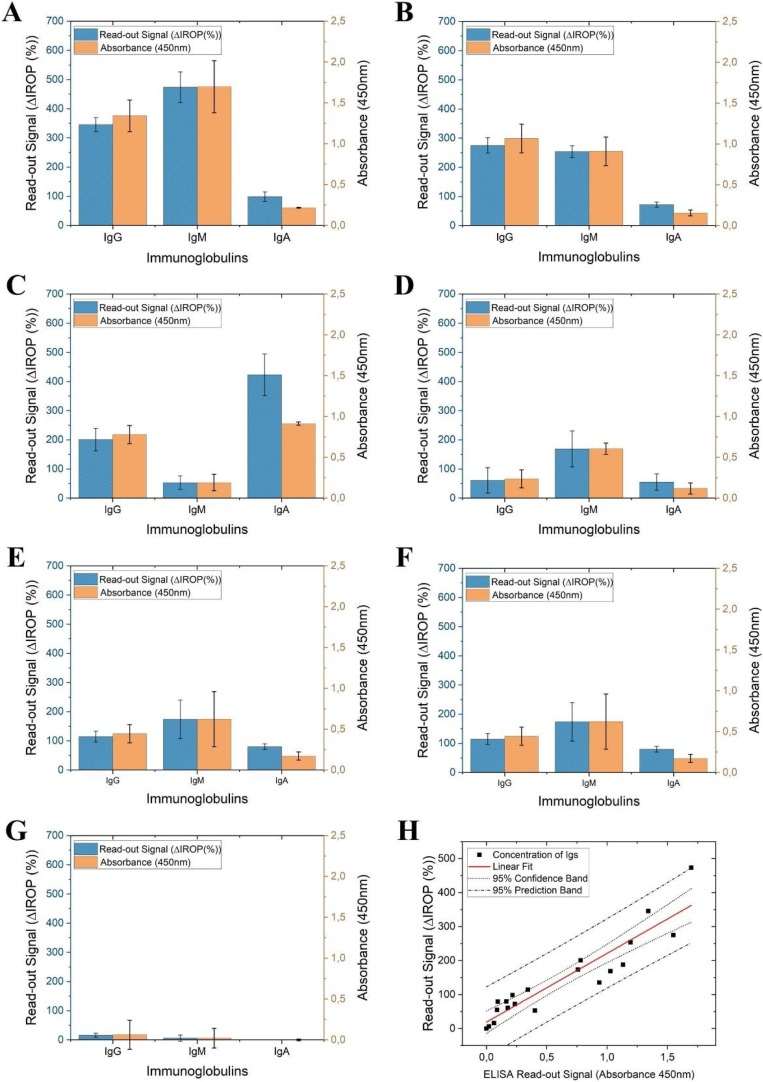
To validate this method, these results obtained using the BICELL-based IODM were correlated with those obtained by ELISA, for serum samples of patients with severe, moderate, mild, and no disease. The levels of individual specific antibodies (IgG, IgM, and IgA) against SARS-CoV-2 were measured. The antibody titers of three patients with severe disease (Fig. 4 A–C) and three with moderate disease (Fig. 4D–F) were measured, and similar results were observed using both techniques for all the patients. For both protocols, samples from a volunteer who tested negative for SARS-CoV-2 by PCR and tested by ELISA was used as a negative control (Fig. 4G).
Fig. 4.
Detection of IgG, IgM, and IgA in serum samples by measurement of ΔIROP and correlation of the results with those determined by ELISA. (A–C) Immunoglobulins levels in the sera of three patients with severe disease, measured by IODM and ELISA. (D–F) IgG, IgM, and IgA levels in the sera of three patients with moderate disease. (G) Samples of sera from healthy volunteers, who tested negative for SARS-CoV-2 infection, were used as a negative control. (H) Linear fitting between the results obtained by ELISA and the IODM, with a 95 % confidence interval and a 95 % confidence band. Abbreviations: ΔIROP, difference in the increased relative optical power signal between the reference and after recognition of the biological event; ELISA, enzyme-linked immunosorbent assay; IODM, interferometric optical detection method; Ig, immunoglobulin.
Finally, as previously reported for the detection of interferon gamma [45], linear fitting, correlating the results from the BICELL-based IODM and ELISA was prepared; good correlation between both techniques was observed (see Fig. 4H).
After analyzing the serum samples, we tested whether this system could produce accurate results for other types of samples, such as saliva, since it is less invasive and simpler to obtain. Therefore, we measured the levels of anti-rS1 IgA antibodies (an early biomarker of COVID-19) in the saliva of patients who tested positive for SARS-CoV-2 infections and had different antibody titers (severe and moderate) (see Fig. 5 ).
A correlation was observed between the total Ig levels in serum and saliva samples (Figs. 3A and 5 A). A higher concentration of specific antibodies was observed in patients with a higher severity of disease, while those with moderate disease showed lower levels of these antibodies. It was observed that the specific IgA titer of both samples increased as the severity of the disease increased. The IgA titers in saliva were found to be higher than those in the serum samples, since it is a characteristic antibody of the mucosa (Figs. 3B and 5 B). The background signals for both total Ig and IgA titers (antibody titers for volunteers who tested negative for SARS-CoV-2 infection) were very low Fig. 5.
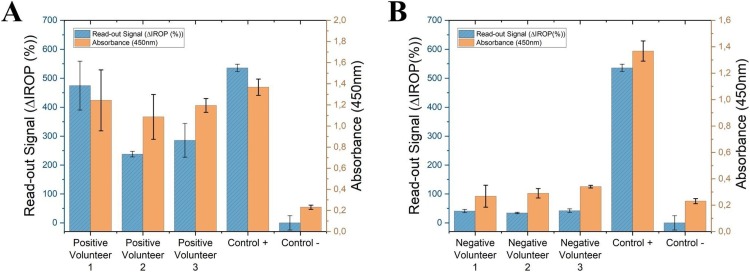
Finally, to verify these results, saliva samples from three volunteers who tested positive and three who tested negative for SARS-CoV-2 infection were measured by ELISA and IODM. IgA recognition values measured by the two techniques showed good correlation. The same positive and negative controls (confirmed by PCR) were used for both techniques (see Fig. 6 ).
Fig. 6.
Detection IgA in saliva by IODM and ELISA. (A) ΔIROP (%) and absorbance values for IgA in the saliva samples of volunteers who tested positive for SARS-CoV-2 infection. (B) IgA values measured by IODM and ELISA of volunteers who tested negative for SARS-CoV-2 infection. In all cases, the same PCR-confirmed positive and negative controls were used. Abbreviations: ΔIROP, difference in the increased relative optical power signal between the reference and after recognition of the biological event; ELISA, enzyme-linked immunosorbent assay; IODM, interferometric optical detection method; Ig, immunoglobulin.
To shorten the waiting time to obtain the results, the saliva of a patient with a high antibody titer was incubated for 1, 3, 6, and 24 h. After 3 h of incubation, the sensing response was not significantly improved. It was also observed that the response at this incubation time was similar to that for the direct detection of Ig total and the specific recognition of IgA. Therefore, for further experiments, saliva samples were incubated for only 3 h.
4. Conclusions
We report, for the first time, the specific detection of immunoglobulins in serum and saliva samples, using BICELLs biofunctionalized with a truncated SARS-CoV-2 spike protein with an IODM readout, without the need for any chemical developer. Moreover, we measured and compared the titers for different types of anti-rS1 immunoglobulins (IgGs, IgMs, and IgAs) to evaluate the in vitro detection system for the diagnosis of SARS-CoV-2 infection using serum and saliva samples. This technique was found to be suitable for use with both types of samples and the presence of specific antibodies against SARS-CoV-2 could be detected. Furthermore, the results showed a good correlation with those obtained by ELISA; the method produced results in the linear range of the correlation curve. It is worth mentioning that, here, we report only the features of this in vitro detection system for detecting specific immunoglobulins for SARS-CoV-2; the clinical implications of these results are beyond the scope of this article. Based on our findings, we can conclude that the in vitro detection system and the assays described in this paper present promising alternatives for the detection of SARS-CoV-2 infection, and open up the possibility of a new, cost-effectiveness, high throughput technique for patient screening. Additionally, this method allows the use of saliva samples, the collection of which is less invasive and traumatic than for other types of samples. This could facilitate the periodical use of this technique for early detection, surveillance, and control of the spread of SARS-CoV-2 infections.
Author contributions
A.M.M. Murillo: writing-original draft, investigation, methodology of the measurement system and immunoassay procedure, formal analysis and data curation, visualization; J. Tomé-Amat: production of the SARS-CoV-2 rS1 protein; Y. Ramírez: investigation, methodology of the measurement system, immunoassay procedure, formal analysis; Maria Garrido-Arandia: investigation and methodology of production of the SARS-CoV-2 protein; L.G. Valle: design of the dropping procedure for the biofunctionalization, biofunctionalization of the diagnostic kits; G. Hernández-Ramírez: production of the SARS-CoV-2 rS1 protein; L. Tramarin and B. Santamaría: design and micron-nano fabrication of the bio-detection kits, including protein immobilization; P. Herreros: diagnostic kit packaging, biofunctionalization of diagnostic kits, design of the protocol for the micro-dropping system; Araceli Diaz-Perales: conceptualization and supervision, methodology, writing-review and editing Miguel Holgado: conceptualization and supervision, methodology, writing- review and editing, project administration, funding acquisition.
Declaration of Competing Interest
None.
Acknowledgments
Authors acknowledge the support of Universidad Politécnica de Madrid under the project reference: VIMPACTO20MHB. This work was also supported in part by the Spanish Ministry (Ministerio de Economía y Competitividad) under project HERON under Grant TEC2017-84846-R, and grants IND2018/BIO9650, IND2019/IND-17207 PEJD-2019-PRE/BIO-15988 from Comunidad de Madrid.
Authors also acknowledge Drs. Luis Enjuanes and Isabel Sola from the Coronavirus laboratory, Centro Nacional de Biotecnología belonging to the Spanish Research Council (CNB-CSIC), who provided the DNA of the SARS-CoV-2 S gene used to express the recombinant protein included in the detection system; the Biobank of the Hospital Clínico San Carlos-IdISSC for the patient’s samples; and Bio Optical Detection for the support on the readout platform. Mass spectrometry analysis were performed on the Centro de Apoyo a la Investigación (CAI) de Espectrometría de Masas of Universidad Complutense de Madrid.
We wish also to thank the Unidad de Innovación (UI); Unidad de Investigación Clínica y Ensayos Clínicos (UICEC); Unidad de Apoyo Metodológico a la Investigación (UAMI); Biobanco; Unidad de Apoyo a los Comités de Etica (UACE; Unidad de Apoyo a Farmacia: gestión de medicación de investigación (UFAR); and Laboratorios Gabinetes y Servicios Clínicos de Apoyo a la Investigación (LABSERCLI) for their most valuable efort.
Investigators of IdISSC-COVID-TASKFORCE and COVID-19_ URG-HCSC Register: Carlos Javier Llamas, Laura Matilla, Mª Luisa Bretón, Beatriz Rojano, José Antonio Bustamante, Eric Jorge García Lamberechts, Manuel Maroto, Álvaro Martin Ruiz, María Martínez Agüero, Arturo Corbatón, Cesario Fernández Alonso, Jaime Abelaira, Pablo Matías, Raúl Perales, María Rosario Blázquez, M. Carmen Muñoz, Alejandra Ortega, Carlota Clemente, Sara Laínez, Antonio Trino Salto Ariza, Pedro Villarroel González Elipe, Juan Manuel Algarra, Javier Candel, Encarnación Fernández del Palacio, Rosa Moreno Rodríguez, Daniel Muñoz Jiménez, Vicente Estrada, María Jesús Téllez, Ángel Molino, María José Núñez, Noemí Cabello, Nieves Martel, Myriam Calle, Miguel Holgado, María Luaces, Germán Seara, Joan Cháfer, Ángel Luis del Rey, Mercedes Martínez-Novillo, Alberto Delgado-Iribarren, Mª Ángeles Cuadrado Cenzual, María José Torrejón Martínez, Fernando Ataúlfo González Fernández, Silvia Sánchez Ramón, Asunción Mora, Ana Arribi Vilella, Miguel Fernández Armero, José Manuel Martínez Sesmero, Rocío Manzano, Virginia Puebla, Juan Arrazola, Ana Bustos, Laura Galván, Ana Mañas, Ángel Nava Muñoz, Sara Gómez Peña, Beatriz Cabeza Martínez, Irene Martín López, Joaquín López Herraiz, Alfonso Calle, Miguel Ángel Rubio Herrera, Isabel Runckle, Eduardo Anguita Mandley, Blanca García Reneses, David Fraguas, Marina Díaz Marsa, José Luis Carrasco, Julia Sevilla, Iabel Ramos, Marta Navas, Aránzazu Álvarez de Arcaya, José Ramón Núñez, Mar García Arenillas, Lourdes Cabrera, Carlos Macaya, Fernando Macaya, Iván Nuñez-Gil, Jorge Matías-Guiu, Jesús Porta, Miguel Sánchez, Fernando Martínez Sagasti, Leonor Laredo, Emilio Vargas, Alberto Mariano Lázaro, Carmen de Burgos, Manuel Fuentes Ferrer, Gloria Mato Chian, Beatriz Peláez Ros, María Ángeles Valcárcel de la Iglesia, Náyade del Prado, José Mª Leal Pozuelo, José Luis Fernández Rueda, Víctor Hernández Martín-Romo, Elena Urcelay, Antonio Portolés, and Julio Mayol.
Acknowledgments
Funding
This work was supported by the Research and Innovation Program of the Universidad Politécnica de Madrid (https://www.upm.es/Investigacion/Programa_Propio_UPM) VIMPACTO20MHB. Projects IND2018/BIO-9650, IND2019/IND-17207, and BIOPIELTEC-CM (Ref S2018/BAA-4480) were of the Madrid Regional Research Program, and project TEC2017-84846-R was supported by the Spanish Ministry of Economy and Competitiveness. AllerScreening (H2020-NMBP-X-KET-2017 – 768641) was supported by the European Commission. Instituto de Salud Carlos III (ISCIII) was co-founded by FEDER Thematic Networks and Cooperative Research Centers: ARADYAL (RD16/0006/0003).
Biographies
Ana María Martín Murillo. I received my bachelor’s degree in Biology from Universidad Autónoma de Madrid (UAM) (2015), Master’s degree in Immunology research from Universidad Complutense de Madrid (UCM) and Superior’s degree of Technician in laboratory of clinical and biomedical analysis from I. E. S Las Musas (2018). I am currently studying a doctorate in Mechanical Engineering in the Industrial Engineering School (ETSII-UPM), inside the group of the Optics, Photonics and Biophotonics at the Center for Biomedical Technology CTB-UPM. I’m basing mi PhD in surface silanization. I`ve been granted with a “Doctorado Industrial” (Industrial PhD) by CAM. Taking part into some of the projects in which the group is involved including regional and national projects.
Jaime Tome-Amat. I received my Bachelor’s degree in Biochemistry in 2007 and my Ph.D. in Biochemistry and Molecular Biology in 2012. After I worked as a Post-Doctoral researcher at Cornell University and in the Mount Sinai Hospital (2013–2017). Currently, I work as Post-Doc at Araceli Diaz-Perales’s Lab in the Centre for Biotechnology and Plant Genomics in the area of food allergy, developing in vitro and in vivo models to study mucosal inflammation, with a special interest in sensitization process.
Yolanda Ramírez. I received my Bachelor’s degree in Chemical engineering from Industrial Engineering School (ETSII-UPM). I am a doctoral student in the Optics, Photonics and Biophotonics group of the Polytechnic University of Madrid at the Biomedical Technology Centre. I am basing my PhD in the screening of drugs for autoimmune diseases using technology developed by the group. In addition, I am focusing my research on the validation of this technology with standard techniques. I`ve been granted with a “Doctorado Industrial” (Industrial PhD) by CAM. I am coauthor of scientific publications and I’ve attended several congresses.
María Garrido-Arandia. Graduated in Biochemistry and assistant professor in Universidad Politécnica de Madrid (UPM). I have devoted my research career to the computational study of allergens, trying to unveil molecular patterns that make these proteins become allergenic. Furthermore, I have explored allergen interactions with their ligands and other molecules, such as antibodies or cell receptors, involved within the immune response triggered when they are put into contact with allergic patients.
Luis González Valle. Predoctoral student hired at the Universidad Politécnica de Madrid, in Optics, Photonics and Biophotonics Group, where I am participating in projects for SARS-COV-2 detection in real sample using Optical Biosensors, and in projects related to High Throughput Screening in Optical Biosensors of molecules related to Alzheimer's disease using Nanoparticles. I studied the Degree in Biochemistry at the Complutense University of Madrid, where I presented my End of Degree work at the National Center of Biotechnology, based on cell culture techniques, cytometry and confocal microscopy. In addition, I studied a Master in Neuroscience at the Complutense University of Madrid where I presented my master’s thesis, in which I used biochemical techniques and RT-Q-PCR.
Guadalupe Hernandez-Ramirez. Degree in Biotechnology at Universidad Politécnica de Madrid (2017) and MSc in Agroforestry Biotechnology at UPM (2018). Currently, I am developing my doctoral thesis in the Prof. Araceli Díaz-Perales’s research line, “Plant Allergens”, at Centre for Plant Biotechnology and Genomics (INIA-UPM). The Plant Allergens group is focused on the study of molecular basis of plant allergens, characterizing allergens and improving their diagnosis and treatment. In this context, my PhD research aims the description of the immunological role of the Alt a 1, the major allergen of the fungus Alternaria alternata.
Luca Tramarin. Bachelor’s degree (2016) at the University of Pavia (UNIPV) and master’s degree (2018) at the Technical University of Madrid (UPM) in Biomedical Engineering. PhD candidate at the Industrial Engineering School (ETSII) of the UPM, part of the Group of Optics, Photonics and Biophotonics at the Center of Biomedical Technology (CTB-UPM). Postgraduate researcher for micro- and nano- fabrication tasks by Nanoimprint Lithography for biosensing application at the CTB-UPM during 8 months. Master’s work with title “Design of a nanophotonic structure for label free biosensing” which led to a scientific publication as first author, currently participating in 3 research projects (European, National and Regional).
Pedro Herreros. In 2017 I received my degree in Biotechnology at Technical University of Madrid and in 2018 I completed a M.Sc in Regenerative Medicine and Cell Therapy at CEU San Pablo University. I am a PhD student in the group of Optics, Photonics and Biophotonics located in the Center of Biomedical Technology of the Technical University of Madrid. My PhD work is based on the design and development of organ-on-a-chip devices as a new cell culture method. Taking part into some of the projects in which the group is involved including regional, national, and European projects such as BIOPIELTEC-CM, HERON, and Aller-Screening.
Beatriz Santamaría. I received my Bachelor's from Salamanca University (2012), Master's degree in Laser Technology from Technical University of Madrid (UPM) (2014) and Doctoral degree (Ph.D.) in 2020. I am currently Assistant professor in Chemical, Mechanical and Industrial Design Engineering Department at UPM and post-doctoral researcher member of Group of Optics, Photonics and Biophotonics at the Center for Biomedical Technology. I have participated in 2 European and in 7 National research projects. I am author and co-author of 11 JCR scientific publications published in several areas such metrology, instrumentation, nanotechnology, optics, and biotechnology.
Araceli Díaz-Perales. Graduated in Biological Sciences in Universidad Complutense de Madrid and PhD in Universidad Politécnica de Madrid. Professor of Immunology and Molecular Pathology at Universidad Politécnica de Madrid and Principal Investigator of Plant Allergens groups at Centre for Plant Biotechnology and Genomics (UPM-INIA). Her scientific career has been developed within the fields of food safety, health and plant biotechnology. She has contributed to the development of different technologies towards the improvement of clinical handling of allergic diseases and she has authored numerous original scientific papers, patents and scientific communications.
Miguel Holgado. I received my bachelor’s and Master’s degree in Electrical Engineering from Technical University of Madrid (UPM) (1996), and Doctoral degree (Ph.D.) at the Institute of Material Science (ICMM) belonging to the Spanish National Research Council CSIC (2000). I am group leader of the Optics, Photonics and Biophotonics at the Center for Biomedical Technology CTB-UPM, and associate professor at the Applied Physics and Material Engineering Department of Industrial Engineering School (ETSII-UPM). I am author/co- author of more than 150 scientific contributions. I am also the inventor of 7 patents applications. In addition, I am also founder of Bio Optical Detection; a spin-off company (BIOD S.L.) from the UPM, which develops optical Point-of Care devices.
References
- 1.Ritchie H., Ortiz-Ospina E., Beltekian D., Mathieu E., Hasell J., Macdonald B., Giattino C., Roser M. Glob. Chang. Data Lab. (2020) 2020. Total COVID-19 tests per 1,000 people, Sep 28.https://ourworldindata.org/grapher/full-list-cumulative-total-tests-per-thousand-map?time=2020-02-20..atest®ion=Europe (Accessed September 28, 2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.E.C. for D.P, C. ECDC . 2020. Population-wide Testing of SARS-CoV-2: Country Experiences and Potential Approaches in the EU/EEA and the United Kingdom European Commission Request Definition.https://www.ecdc.europa.eu/sites/default/files/documents/covid-19-population-wide-testing-country-experiences.pdf (Accessed September 28, 2020) [Google Scholar]
- 3.E.C. for D.P, C. ECDC . 2020. Epidemiology of COVID-19, Eur. Cent. Dis. Prev. Control.https://www.ecdc.europa.eu/en/covid-19/latest-evidence/epidemiology (Accessed September 28, 2020) [Google Scholar]
- 4.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., Geng B., Muenker M.C., Moore A., Vogels C., Petrone M., Ott I., Lu P., Lu-Culligan A., Klein J., Venkataraman A., Earnest R., Simonov M., Datta R., Handoko R., Naushad N., Sewanan L., Valdez J., White E., Lapidus S., Kalinich C., Jiang X., Kim D., Kudo E., Linehan M., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Weizman O.-E., Wong P., Yang Y., Bermejo S., Odio C., Omer S., Dela Cruz C., Farhadian S., Martinello R., Iwasaki A., Grubaugh N., Ko A. 2020. Saliva Is More Sensitive for SARS-CoV-2 Detection in COVID-19 Patients Than Nasopharyngeal Swabs. [DOI] [Google Scholar]
- 5.Azkur A.K., Akdis M., Azkur D., Sokolowska M., van de Veen W., Brüggen M.C., O’Mahony L., Gao Y., Nadeau K., Akdis C.A. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy Eur. J. Allergy Clin. Immunol. 2020;75:1564–1581. doi: 10.1111/all.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu T., Hsiung J., Zhao S., Kost J., Sreedhar D., Hanson C.V., Olson K., Keare D., Chang S.T., Bliden K.P., Gurbel P.A., Tantry U.S., Roche J., Press C., Boggs J., Rodriguez-Soto J.P., Montoya J.G., Tang M., Dai H. Quantification of antibody avidities and accurate detection of SARS-CoV-2 antibodies in serum and saliva on plasmonic substrates. Nat. Biomed. Eng. 2020 doi: 10.1038/s41551-020-00642-4. [DOI] [PubMed] [Google Scholar]
- 7.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sternberg A., Naujokat C. Structural features of coronavirus SARS-CoV-2 spike protein: targets for vaccination. Life Sci. 2020;257:118056. doi: 10.1016/j.lfs.2020.118056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. 2020. Structural Basis for the Recognition of SARS-CoV-2 by Full-length Human ACE2 Downloaded From.http://science.sciencemag.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petherick A. Developing antibody tests for SARS-CoV-2. Lancet (London, England) 2020;395:1101–1102. doi: 10.1016/S0140-6736(20)30788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y., Yang C., feng Xu X., Xu W., wen Liu S. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fung T.S., Liu D.X. Post-translational modifications of coronavirus proteins: roles and function. Future Virol. 2018;13:405–430. doi: 10.2217/fvl-2018-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Looser V., Bruhlmann B., Bumbak F., Stenger C., Costa M., Camattari A., Fotiadis D., Kovar K. Cultivation strategies to enhance productivity of Pichia pastoris: a review. Biotechnol. Adv. 2014;33:1177–1193. doi: 10.1016/j.biotechadv.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Chuck C.P., Wong C.H., Chow L.M.C., Fung K.P., Waye M.M.Y., Tsui S.K.W. Expression of SARS-coronavirus spike glycoprotein in Pichia pastoris. Virus Genes. 2009;38:1–9. doi: 10.1007/s11262-008-0292-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chin C.D., Linder V., Sia S.K. Commercialization of microfluidic point-of-care diagnostic devices. Lab Chip. 2012;12:2118–2134. doi: 10.1039/c2lc21204h. [DOI] [PubMed] [Google Scholar]
- 17.Zanchetta G., Lanfranco R., Giavazzi F., Bellini T., Buscaglia M. Emerging applications of label-free optical biosensors. Nanophotonics. 2017;6:627–645. doi: 10.1515/nanoph-2016-0158. [DOI] [Google Scholar]
- 18.Casquel R., Holgado M., Laguna M.F., Hernández A.L., Santamaría B., Lavín Á., Tramarin Luca, Herreros P. Engineering vertically interrogated interferometric sensors for optical label-free biosensing. Anal. Bioanal. Chem. 2020;412:3285–3297. doi: 10.1007/s00216-020-02411-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Homola J. Surface plasmon resonance sensors for detection of chemical and biological species. Chem. Rev. 2008;108:462–493. doi: 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- 20.Nair S., Escobedo C., Sabat R.G. Crossed surface relief gratings as nanoplasmonic biosensors. ACS Sens. 2017;2:379–385. doi: 10.1021/acssensors.6b00696. [DOI] [PubMed] [Google Scholar]
- 21.Gomez-Cruz J., Nair S., Manjarrez-Hernandez A., Gavilanes-Parra S., Ascanio G., Escobedo C. Cost-effective flow-through nanohole array-based biosensing platform for the label-free detection of uropathogenic E. coli in real time. Biosens. Bioelectron. 2018;106:105–110. doi: 10.1016/j.bios.2018.01.055. [DOI] [PubMed] [Google Scholar]
- 22.Jankovics H., Kovacs B., Saftics A., Gerecsei T., Tóth É., Szekacs I., Vonderviszt F., Horvath R. Grating-coupled interferometry reveals binding kinetics and affinities of Ni ions to genetically engineered protein layers. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-79226-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Algorri J.F., Zografopoulos D.C., Tapetado A., Poudereux D., Sánchez-Pena J.M. Infiltrated photonic crystal fibers for sensing applications. Sensors (Switzerland) 2018;18:1–32. doi: 10.3390/s18124263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee M.R., Fauchet P.M. Two-dimensional silicon photonic crystal based biosensing platform for protein detection. Opt. Express. 2007;15:4530. doi: 10.1364/oe.15.004530. [DOI] [PubMed] [Google Scholar]
- 25.Taniguchi T., Hirowatari A., Ikeda T., Fukuyama M., Amemiya Y., Kuroda A., Yokoyama S. Detection of antibody-antigen reaction by silicon nitride slot-ring biosensors using protein G. Opt. Commun. 2016;365:16–23. doi: 10.1016/j.optcom.2015.11.068. [DOI] [Google Scholar]
- 26.Ksendzov A., Lin Y. Integrated optics ring-resonator sensors for protein detection. Opt. Lett. 2005;30:3344. doi: 10.1364/ol.30.003344. [DOI] [PubMed] [Google Scholar]
- 27.Flueckiger J., Schmidt S., Donzella V., Sherwali A., Ratner D.M., Chrostowski L., Cheung K.C. Sub-wavelength grating for enhanced ring resonator biosensor. Opt. Express. 2016;24:15672. doi: 10.1364/oe.24.015672. [DOI] [PubMed] [Google Scholar]
- 28.Dong X., Du H., Sun X., Luo Z., Duan J. A novel strain sensor with large measurement range based on all fiber mach-zehnder interferometer. Sensors (Switzerland) 2018;18 doi: 10.3390/s18051549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bastos A.R., Vicente C.M.S., Oliveira-Silva R., Silva N.J.O., Tacão M., da Costa J.P., Lima M., André P.S., Ferreira R.A.S. Integrated optical Mach-Zehnder interferometer based on organic-inorganic hybrids for photonics-on-a-chip biosensing applications. Sensors (Switzerland) 2018;18:1–11. doi: 10.3390/s18030840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim W.J., Kim B.K., Kim A., Huh C., Ah C.S., Kim K.H., Hong J., Park S.H., Song S., Song J., Sung G.Y. Response to cardiac markers in human serum analyzed by guided-mode resonance biosensor. Anal. Chem. 2010;82:9686–9693. doi: 10.1021/ac101716p. [DOI] [PubMed] [Google Scholar]
- 31.Wei X., Weiss S.M. Grating coupled waveguide biosensor based on porous silicon. Mater. Res. Soc. Symp. Proc. 2011;1301:219–224. doi: 10.1557/opl.2011.553. [DOI] [Google Scholar]
- 32.Cross G.H., Reeves A.A., Brand S., Popplewell J.F., Peel L.L., Swann M.J., Freeman N.J. A new quantitative optical biosensor for protein characterisation. Biosens. Bioelectron. 2003;19:383–390. doi: 10.1016/S0956-5663(03)00203-3. [DOI] [PubMed] [Google Scholar]
- 33.Hernández A.L., Casquel R., Holgado M., Cornago I., Fernández F., Ciaurriz P., Sanza F.J., Santamaría B., Maigler M.V., Laguna M.F. Resonant nanopillars arrays for label-free biosensing. Opt. Lett. 2016;41:5430. doi: 10.1364/ol.41.005430. [DOI] [PubMed] [Google Scholar]
- 34.Holgado M., Sanza F.J., López A., Lavín Á., Casquel R., Laguna M.F. Description of an advantageous optical label-free biosensing interferometric read-out method to measure biological species. Sensors (Switzerland) 2014;14:3675–3689. doi: 10.3390/s140203675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavín Á., Casquel R., Sanza F.J., Laguna M.F., Holgado M. Efficient design and optimization of bio-photonic sensing cells (BICELLs) for label free biosensing. Sens. Actuators B Chem. 2013;176:753–760. doi: 10.1016/j.snb.2012.09.058. [DOI] [Google Scholar]
- 36.Sanza F.J., Holgado M., Ortega F.J., Casquel R., López-Romero D., Bañuls M.J., Laguna M.F., Barrios C.A., Puchades R., Maquieira A. Bio-photonic sensing cells over transparent substrates for anti-gestrinone antibodies biosensing. Biosens. Bioelectron. 2011;26:4842–4847. doi: 10.1016/j.bios.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 37.Maigler M.V., Holgado M., Laguna M.F., Sanza F.J., Santamaria B., Lavin A., Espinosa R.L. A new device based on interferometric optical detection method for label-free screening of C-Reactive protein, IEEE trans. Instrum. Meas. 2018:1–7. doi: 10.1109/TIM.2018.2876073. [DOI] [Google Scholar]
- 38.Holgado M., Maigler M.V., Santamaría B., Hernandez A.L., Lavín A., Laguna M.F., Sanza F.J., Granados D., Casquel R., Portilla J., Riesgo T. Towards reliable optical label-free point-of-care (PoC) biosensing devices. Sens. Actuators B Chem. 2016;236:765–772. doi: 10.1016/J.SNB.2016.06.047. [DOI] [Google Scholar]
- 39.Hosseini S., Vázquez-Villegas P., Rito-Palomares M., Martinez-Chapa S.O. Enzym. Immunosorbent Assay From A to Z. Springer; Singapore, Singapore: 2018. Advantages, disadvantages and modifications of conventional ELISA; pp. 67–115. [DOI] [Google Scholar]
- 40.Satija J., Punjabi N., Mishra D., Mukherji S. Plasmonic-ELISA: expanding horizons. RSC Adv. 2016;6:85440–85456. doi: 10.1039/c6ra16750k. [DOI] [Google Scholar]
- 41.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;80(367):1260–1263. doi: 10.1126/science.aax0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laguna M., Holgado M., Hernandez A.L., Santamaría B., Lavín A., Soria J., Suarez T., Bardina C., Jara M., Sanza F.J., Casquel R. Antigen-antibody affinity for dry eye biomarkers by label free biosensing. Comparison with the ELISA technique. Sensors (Switzerland) 2015;15:19819–19829. doi: 10.3390/s150819819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Espinosa R.L., Laguna M.F., Fernández F., Santamaria B., Sanza F.J., Maigler M.V., Álvarez-Millán J.J., Canalejas-Tejero V., Holgado M. A proof-of-concept of label-free biosensing system for food allergy diagnostics in biophotonic sensing cells: performance comparison with immunoCAP. Sensors (Switzerland) 2018;18 doi: 10.3390/s18082686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grimaldi I.A., Testa G., Persichetti G., Loffredo F., Villani F., Bernini R. Plasma functionalization procedure for antibody immobilization for SU-8 based sensor. Biosens. Bioelectron. 2016;86:827–833. doi: 10.1016/j.bios.2016.07.090. [DOI] [PubMed] [Google Scholar]
- 45.Laguna M., Ramirez Y., Fernandez Martinez C., Espinosa R.L., Lavín A., Holgado M. A point-of-care based on label-free interferometric optical detection method to evaluate interferon gamma (IFN- γ): a correlation with the ELISA technique. Sensors. 2020;20 doi: 10.3390/s20174776. [DOI] [PMC free article] [PubMed] [Google Scholar]