Summary
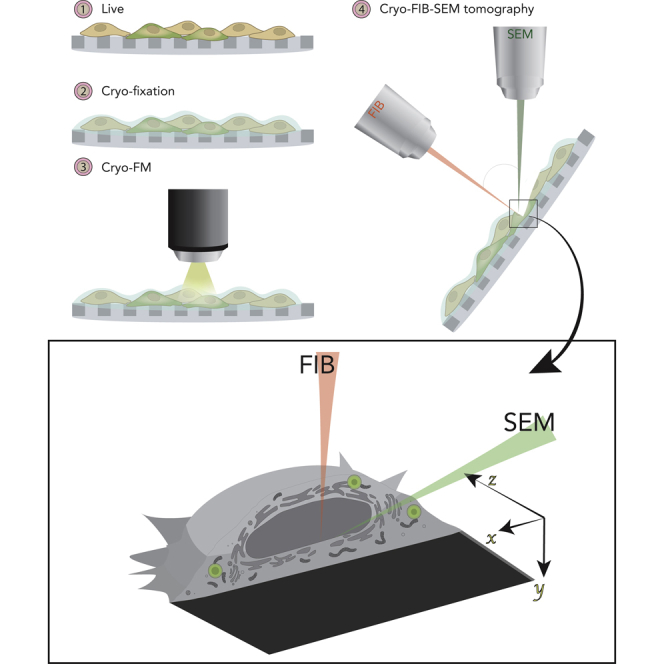
Imaging of cells and tissues has improved significantly over the last decade. Dual-beam instruments with a focused ion beam mounted on a scanning electron microscope (FIB-SEM), offering high-resolution 3D imaging of large volumes and fields-of-view are becoming widely used in the life sciences. FIB-SEM has most recently been implemented on fully hydrated, cryo-immobilized, biological samples. Correlative light and electron microscopy workflows combining fluorescence microscopy (FM) with FIB-SEM imaging exist, whereas workflows combining cryo-FM and cryo-FIB-SEM imaging are not yet commonly available. Here, we demonstrate that fluorescently labeled lipid droplets can serve as in situ fiducial markers for correlating cryo-FM and FIB-SEM datasets and that this approach can be used to target the acquisition of large FIB-SEM stacks spanning tens of microns under cryogenic conditions. We also show that cryo-FIB-SEM imaging is particularly informative for questions related to organelle structure and inter-organellar contacts, nuclear organization, and mineral deposits in cells.
Subject areas: Cell biology, Technical aspects of cell biology, Structural biology
Graphical abstract
Highlights
-
•
3D CLEM of cryo-FM and FIB-SEM datasets using fluorescently labeled lipid droplets
-
•
Cryo-FIB-SEM imaging of organelle-organelle interactions and nuclear organization
Cell biology; Technical aspects of cell biology; Structural biology
Introduction
Gaining a mechanistic understanding of biological processes often depends on developing the capability of visualizing cells and tissues in their native hydrated state at high resolution. Groundbreaking advances in volume electron microscopy and specimen preparation enable the 3D visualization of cells in unprecedented detail. These advances include adapting focused ion beam milling followed by scanning electron microscopy (FIB-SEM) to the life sciences (Heymann et al., 2006). FIB-SEM has since been used to gain previously inaccessible insights into both cells and tissues under physiological and pathological conditions (Heymann et al., 2006; Knott et al., 2008; Merchán-Pérez et al., 2009; Schneider et al., 2010, Schneider et al., 2011; Weiner et al., 2011, Weiner et al., 2016; Reznikov et al., 2013; Revach et al., 2015). The principle of FIB-SEM is that a biological sample is exposed to a focused ion beam (usually consisting of Ga ions) capable of removing thin layers of material by milling in a highly precise manner (5–10 nm). Between each sample milling, a scanning electron beam is used to image the newly exposed surface. By repeating this process hundreds or even thousands of times, a large sample volume can be acquired with an isotropic voxel reaching 3 nm (Wei et al., 2012; Xu et al., 2020), in resin and at room temperature. With the recent development of automated acquisition procedures, even relatively large volumes (>1,000 μm3) can be acquired within a few days, providing large ultrastructural datasets (Peddie and Collinson, 2014; Narayan and Subramaniam, 2015). The ability to acquire large volumes with high and isometric resolution holds the potential to look at the cellular environment in a more holistic manner.
One major drawback of conventional FIB-SEM imaging at room temperatures is that it requires dehydration and resin embedding, precluding the possibility to visualize cellular structures closer to their native hydrated state (Sviben et al., 2016; Vidavsky et al., 2016; Vidavsky et al., 2016; Kumar et al., 2020). Recent studies have shown that cellular membranes are particularly visible using in-column secondary electron detection (InLens SE) in scanning electron microscopy (SEM) (Schertel et al., 2013; Spehner et al., 2020). This finding was unexpected because without staining by heavy atoms to generate contrast based on back-scattered electrons from the flat surface, the sample should be electron transparent. It has since been suggested that contrast may be a product of low-energy type 1 secondary electrons, generated at the electron beam focal point at low voltage (<3 kV), which are sensitive to the local surface potential of different biological material (Schertel et al., 2013). Nevertheless, the extent to which three dimensional (3D) organellar structure and organization can be studied at high resolution using cryo-FIB-SEM needs to be explored further.
In parallel to the increasing popularity of FIB-SEM instruments, correlative light and electron microscopy (CLEM) workflows are becoming an important tool to study rare, dynamic, or undescribed cellular events by mapping information from fluorescence microscopy (FM) onto electron microscopy (EM) data of the exact same sample (Kukulski et al., 2011; Bykov et al., 2016; Karreman et al., 2016; Khalifa et al., 2016; Weiner et al., 2016; Weiner and Enninga, 2019; Scher and Avinoam, 2020). For correlative microscopy, visible features in both imaging modalities must be identified. This can be achieved either by using intrinsic features of the sample, such as the shape of cells, organelles, and other landmarks, or by adding fiducial markers that are both fluorescent and electron opaque. The latter is typically challenging under cryogenic conditions, even for cryosections (Masich et al., 2006), and even more so if a 3D correlation is needed, because the fiducials would have to be incorporated into the specimen volume. To overcome these challenges and develop a cryo-3D-CLEM approach, we examined whether organelles such as lipid droplets (LDs), which are relatively abundant in cells and well resolved by cryo-FIB-SEM imaging, can be used as internal fiducial markers to target the acquisition of large volumes in plunge-frozen cells grown on EM grids.
Results and discussion
Organelles are well resolved in cryo-FIB-SEM
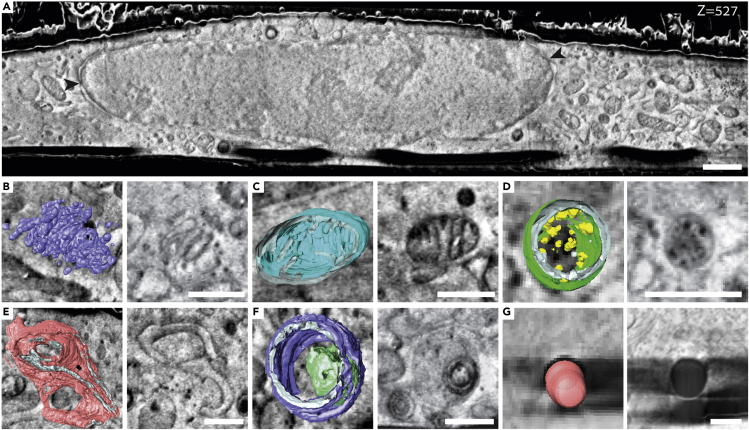
Plunge freezing of cells grown on EM grids is a well-established method to vitrify samples for several downstream cryo-EM techniques (Medalia et al., 2002, Medalia et al., 2007; Sartori et al., 2007; Mahamid et al., 2016; Spehner et al., 2020). In the present study, we grew mammalian cells on holey carbon-coated EM grids, cryo-immobilized them by plunge freezing, and imaged them using mixing of InLens SE and type 2 secondary electron (SE2) detection in cryo-FIB-SEM. Because the SE2 detector is less sensitive to charge accumulation on the cross section than the InLens SE detector, blending the two reduced the appearance of charging artifacts in the final image, resulting in a better signal to noise ratio. In all experiments (N = 5), cells appeared flat on the carbon film with the region close to the nucleus accounting for most of the volume (Figure 1A). The maximum volume acquired with an isometric voxel of 10 nm was ∼5,584μm3, which is approximately 45% of the volume of the cell. Much like in transmission electron microscopy (TEM) membranes and lipids generated a darker contrast compared with their surroundings, as previously shown (Schertel et al., 2013; Spehner et al., 2020). Hence, membranous organelles such as the endoplasmic reticulum, Golgi apparatus, multi-vesicular bodies, and mitochondria could be readily observed and segmented (Figures 1B–1G) (Schertel et al., 2013; Spehner et al., 2020).
Figure 1.
Cryo-FIB-SEM provides high-resolution information of sub-cellular compartments at their native state
(A) A representative slice from an FIB-SEM acquisition of a plunge frozen mammalian cell. Contrast is most likely generated from differences in surface potential of the different materials. Nuclear pores (black arrowheads) Voxel size: 10 × 10 × 10 nm. Scale bar: 1 μm.
(B–G) Overlays of representative micrographs and organelles segmentation (left) and a slice through the same organelle (right): Golgi apparatus (B), mitochondria (C), multi-vesicular body (D), endoplasmic reticulum (E), lysosome (F), and lipid droplet (G). Scale bar: 0.5 μm.
Lipid droplets can serve as fiducial markers for CLEM
Re-localizing cells of interest on EM grids is relatively straight forward (Figures 2A–2C). However, targeting specific regions of interest within the huge volume of the cell is still challenging. LDs have been recently used as fiducial markers post acquisition of 2D TEM data from cryo-lamella (Klein et al., 2021). Here, we tested whether fluorescently labeled LDs can be used for 3D correlative cryo-FM and FIB-SEM imaging.
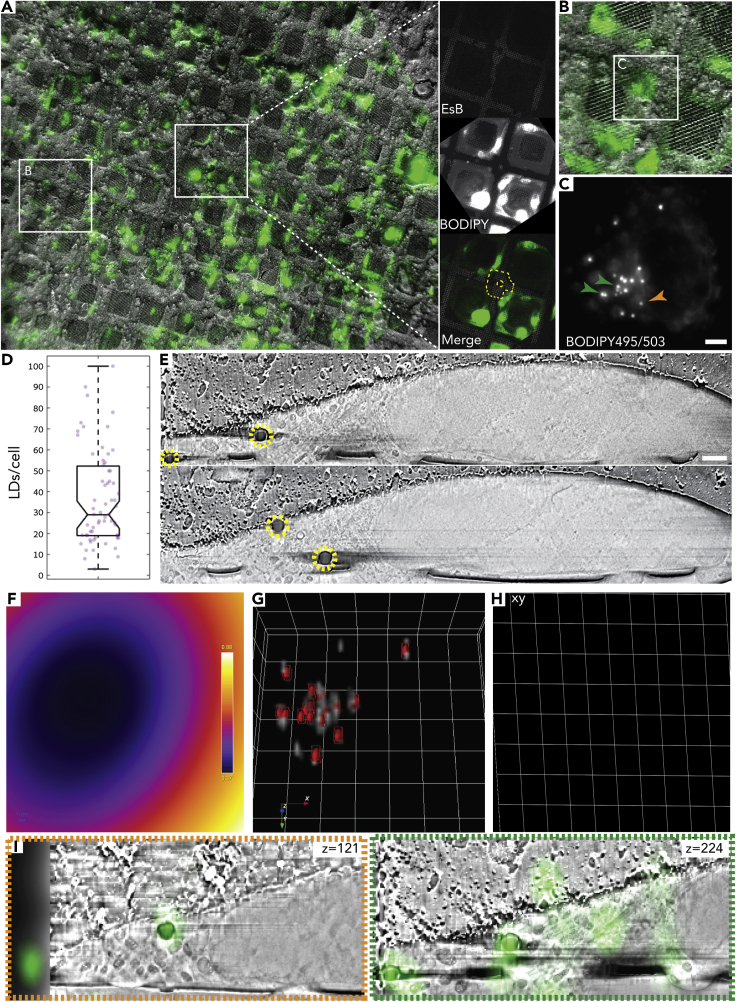
Figure 2.
3D correlation using lipid droplets as fiducial markers
(A) Overlay of a low-magnification cryo-FM and cryo-SEM (InLens SE/SE2 mixed detection) showing the same area on the EM grid. Cells of interest are identified in the cryo-SEM using an asymmetric mark at the center of the grid (white square and magnified on right) and other landmarks on the grid. Cells were labeled with BODIPY493/503 (green), which stains LDs. LDs are subsequently used as internal fiducial markers for CLEM.
(B) Overlay of a higher magnification cryo-FM and cryo-SEM showing a cell of interest.
(C) Maximum intensity projection of a focus series showing BODIPY distribution in a cell of interest. Arrows (green and yellow) correspond to LDs shown in the micrographs in (I). Scale bar: 5 μm.
(D) Quantification of the number of LDs per cell among potential cells for CLEM. The average number is 37 ± 22 LDs/cell (N = 63, median = 29 LDs/cell).
(E) Two representative cryo-FIB-SEM micrographs of the correlated cell; LDs (yellow circles) are clearly visible. Voxel size: 10 × 10 × 10 nm. Scale bar: 1 μm.
(F) Average z-projection of the error prediction map per image after affine image transformation using 17 LDs. The error prediction ranges from 370 (purple) to 880 nm (white).
(G) The transformed FM micrograph (gray) rendered as a volume including spheres (red) indicating the space in which the actual fiducial is with 95% confidence.
(H) Transformed grid slice of xy-plane based on the affine transformation showing the slight stretch of the image in the y and z directions (scaling of 27% and 39%, respectively, z deformation is not shown). This is probably caused by artifact induced from tilt correction and line averaging (on the y axis) and minute differences in the slice thickness and stack alignment (on the z axis).
(I) Two representative micrographs showing the overlay of the transformed FM on top of the FIB-SEM dataset (magnified ×1.5 from E). The affine transformation allowed for correlation of both LDs in the high-LDs-density area (green) as well as the low-LDs areas (orange).
LDs, also called lipid bodies, are organelles that store neutral lipids including triglycerides and cholesterol esters (Tauchi-Sato et al., 2002; Olzmann and Carvalho, 2019). To use LDs as internal fiducials for correlation, we stained mammalian cells grown on grids with the fluorescent neutral lipid dye 4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene (BODIPY 493/503) before plunge freezing and visualized the cells by cryo-FM (Schorb et al., 2017). We observed that 100% of the cells displayed some BODIPY staining (Figure 2) and selected cells for acquisition based on their LDs distribution. Cells suitable for acquisition showed an average of 37 ± 22 LDs/cell (N = 63, median = 29 LDs/cell) (Figure 2D), which assured having at least 15 LDs for correlation within a sub-volume of the cell. Whole grid landmarks such as symbols or missing grid squares were used to relocate cells in the FIB-SEM (Figure 2A). One or two cells were acquired in one acquisition session, which lasted 36–60 h (Figure 2E). Correlation of both datasets was performed under the ICY software (De Chaumont et al., 2012). We initialized the correlation by manually marking the centers of each LD in the FIB-SEM stack. In parallel, the centroids of the LDs were automatically defined by wavelet spot detection in the FM stack. We subsequently used AutoFinder in the eC-CLEM image registration plugin (Paul-Gilloteaux et al., 2017) to automatically match the centers of LDs in FIB-SEM and the centroids of LDs in FM, which were independently identified. AutoFinder then computed and applied the rigid transformation to the image. Correlation precision was estimated using a leave-one-out approach (Kukulski et al., 2011) with 17 of 18 LDs, yielding an error estimation on the omitted LD of 3.5 μm. Use of a validated statistical approach (Paul-Gilloteaux et al., 2017; Potier et al., 2021) yielded an average expected error for every point in the volume of 2–4.7 μm (Figure S1A), for the rigid registration. Since the rigid registration did not give an accurate enough correlation (Figures S1 and S2A), we used an affine transformation on the rough registration from the AutoFinder using the same 17 LDs, yielding an error estimation on the omitted LD of 730 nm and an average expected error estimation of 370–880 nm (Figures 2F–2I and S2C). On analysis of the resulting image transformation (Figures 2F–2I), we observed image stretching in the yz direction of the FIB-SEM coordinate system (Figure 2H). This distortion can be explained by uncompensated drift during stack acquisition. We used fast scanning with line averaging to provide an acceptable compromise between the signal to noise ratio and charging artifacts. However, some charging of the surface occurs, which likely causes the slight image distortion in the xy plane. FIB-SEM data are also collected at a 90° with respect to the FM data, which means that the axis with the lowest resolution (z) is different in FM and FIB-SEM. However, the contribution of this factor to the overall correlation precision is accounted for in the error estimation calculation. Although in theory more fiducials should increase the correlation precision, this is not always the case as it also depends on their localization accuracy and on how they are distributed in the volume (Paul-Gilloteaux et al., 2017; Potier et al., 2021). To test the robustness of the correlation with less fiducials, we repeated the refinement process using only nine well-distributed LDs, some of which were from a cluster of LDs in the center of the cell. This approach reduced the average expected error to 270–600 nm (Figures S2C and S2D). Measuring the discrepancy between the assigned and transformed positions for all 18 or only the 9 left-out LDs yielded an error estimation of 220 ± 164 and 247 ± 203 nm, respectively. This improvement in accuracy is likely because local inaccuracies in picking LD centers within a tight cluster disproportionately increases the error (Figures S2C and S2D). These experiments show that LDs are suitable and attractive as internal fiducial markers for 3D correlation.
Cryo-FIB-SEM as a tool to investigate cellular ultrastructure in 3D
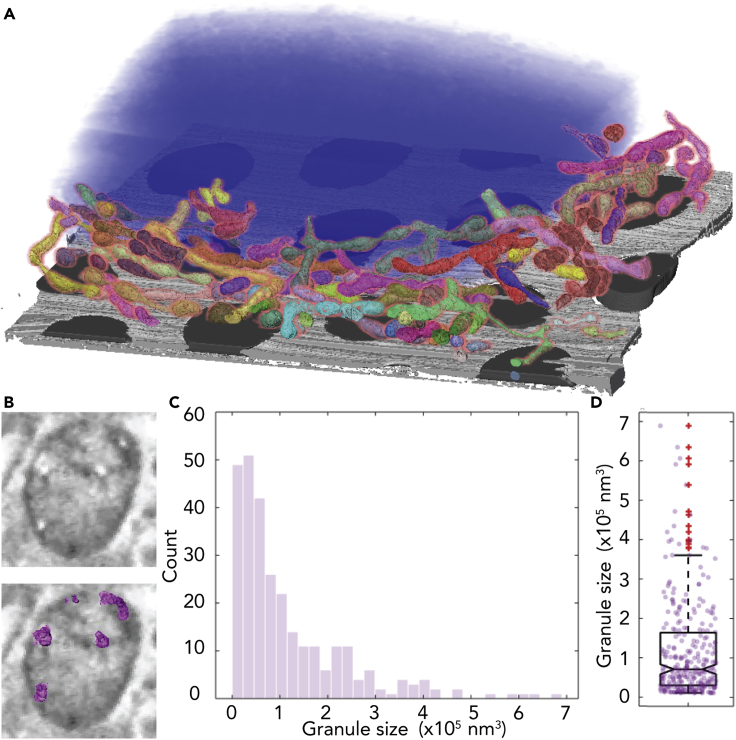
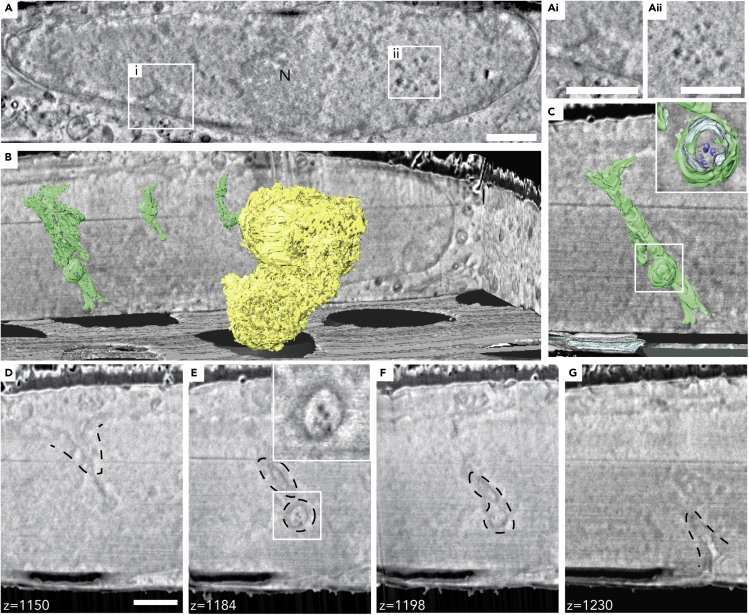
From the volumes that we obtained in cryo-FIB-SEM, we segmented a large fraction of the mitochondrial network (Figure 3A, Video S1). We observed that interactions between adjacent mitochondria and the mitochondrial cristae could be readily resolved as well as small granules inside the mitochondria matrix (Figures 1C and 3B, Video S1). Since it is not possible to determine the elemental composition of such small objects in the cryo-FIB-SEM, we could only speculate based on their size distribution (Figures 3C and 3D) that they were calcium phosphate granules as observed by cryo-scanning transmission electron microscopy (Wolf et al., 2017). The nuclear envelope (NE) and its nuclear pores were also clearly visible. The nucleus contrast was not uniform, and structures such as the heterochromatin in the periphery of the nucleus, the nucleoli, and nuclear speckles were also clearly visible (Figure 4A). Occasionally, we observed NE invaginations into the nucleus (Figure 4B). We segmented the NE invaginations and observed that they often followed a sinuous path (Figure 4B, Video S1). On one occasion the NE invagination formed a tube approximately 3.4 μm in length and ∼200 nm in diameter that crosses the nucleus (Figures 4C–4G, Video S1). We also observed small particles 42 ± 7 nm in diameter (N = 5) within the NE invagination (Figures 4B, 4E, and 4F). Together, these observations demonstrate that cryo-FIB-SEM is a particularly informative approach to study the 3D organization of membrane and membrane-less organelles in intact hydrated cells and highlights the need to combine cryo-FIB-SEM with fluorescence information.
Figure 3.
Inter- and intra-mitochondrial organization as visualized by cryo-FIB-SEM
(A) Segmentation of part of the mitochondrial network (assorted colors). Nucleus (blue), Holey carbon support film (gray).
(B) A cross section of one mitochondrion showing the white particles (upper image) and the same cross section with the segmented particles (magenta; bottom image).
(C) Histogram of the size distribution of the particles inside the mitochondrial matrix shows similarity to size distribution of amorphous calcium phosphate granules (N = 287).
(D) Box plot of the same dataset; the average volume of the granules is 1.16 x 105 ± 1.22 × 105 nm3.
Figure 4.
Nuclear organization as visualized by cryo-FIB-SEM
(A) A representative slice from a cryo-FIB-SEM acquisition highlighting the nucleus. Differences in nuclear content is visible based on contrast change. N, nucleolus; Ai, heterochromatin; Aii, nuclear speckles. Scale bar: 1 μm.
(B) Segmentation of nucleoli (yellow) and nuclear envelope invaginations (green). One of the observed invaginations passed through the whole nucleus and was less sinuous than others. Holey carbon support film (gray).
(C) The nuclear invagination crossing the entire nucleus spanning approximately 3.4 μm in length and approximately 200 nm in diameter. Inset shows the particles inside the invagination (blue).
(D–G) Representative slices through the nuclear envelope invagination showing the particles inside the spherical appendage (E and F). Inset shows a 1.5× magnification of the boxed area. The small particles measured are 42 ± 7 nm in diameter (N = 5). Scale bar: 1 μm.
Conclusions
The cryo-FM and FIB-SEM correlative imaging workflow presented here can be used to study any cell that has LDs dispersed around the region of interest. It is particularly useful for studying cellular structures that are not well preserved by conventional FIB-SEM. The resolution and correlation precision can be further improved by using cryo-confocal or super-resolution FM (Arnold et al., 2016; Wolff et al., 2016; Hoffman et al., 2020). Further improving the correlation precision will be important if specific regions need to be targeted for milling of thin lamella suitable for TEM. Much can also be done to enhance the success rate of data acquisition, alignment, image processing, and analysis.
The workflow will be particularly useful for pinpointing specific regions of the mitochondrial network, unambiguously identifying organelles or sub-structures within the nucleolus, to resolve their underlying ultrastructure in 3D. It would also be beneficial for identifying specific cells or sub-cellular regions in more complex samples such as tissues. Cryo-FIB-SEM imaging appears highly suited for questions related to organelle remodeling and nuclear organization because it provides sufficient resolution and a more complete view of larger objects than what can often be gauged from thin sections.
Compared with other cryo-CLEM workflows that integrate volume EM approaches such as cryo-soft X-ray tomography, cryo-electron tomography, and cryo-serial sections TEM (Schorb et al., 2017; Bharat et al., 2018; Kounatidis et al., 2020), cryo-FIB-SEM provides a good compromise between spatial resolution and throughput (Scher and Avinoam, 2020). Although TEM provides higher spatial resolution in 2D, the throughput and resolution in the axial direction (z) is significantly lower than that of cryo-FIB-SEM. Cryo-soft X-ray tomography provides comparable spatial resolution in 2D and higher throughput owing to the faster acquisition time, albeit at the expense of total imaging volume (Carzaniga et al., 2014). The disadvantage of soft X-ray tomography might be the lower availability of relevant instrumentation.
In conclusion, we demonstrated the use of organelles as fiducial markers for cryo-CLEM as an alternative for external fiducial markers to the specimen. Specifically, we show the usefulness of LDs as fiducials for correlation. Although we used BODIPY staining for this study, other neutral lipid dyes in the red and far-red wavelengths can be used. In combination with fluorescent reporters highlighting specific transient processes or states, this workflow can be used to address different questions in cell biology, where resolving the ultrastructural organization of the cell at its native state is important. Since cryo-confocal and super-resolution FM have already been realized and developed, they can be applied when higher resolution or thicker high-pressure-frozen samples are needed. Cryo-FIB-SEM tomography is clearly a promising imaging approach that is not commonly used.
Limitations of this study
We identify three main avenues by which cryo-fluorescence and FIB-SEM imaging can be further improved. First, the use of BODIPY staining to generate in situ fiducial markers for correlation will inevitably limit the number of available wavelengths that can be used for labeling other cellular features. Although other neutral lipid dyes that emit in red and far red can be used, it would also be beneficial to explore the use of other dyes for different organelles. Second, future developments in hardware and software will undoubtfully increase the success rate of data acquisition, FIB-SEM stack alignment, image analysis, segmentation, and correlation. Third, an integrated cryo-confocal or a cryo-super resolution fluorescence microscope in the cryo-FIB-SEM would greatly facilitate the acquisition and improve the correlation precision that can be obtained.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| BODIPY™ 493/503 (4,4-Difluoro-1,3,5,7,8-Pentamethyl-4-Bora-3a,4a-Diaza-s-Indacene) | Thermo Fisher Scientific | Cat#D3922 |
| Experimental models: cell lines | ||
| Mouse: C2C12 cell line | ATCC | Cat# CRL-1772 |
| Software and algorithms | ||
| Amira (v2019.3) | Thermo Fisher Scientific |
http://www.fei.com/software/amira-3d-for-life-sciences/ RRID:SCR_007353 |
| eC-CLEM (v.2.1.0) | Paul-Gilloteaux et al., 2017 | http://icy.bioimageanalysis.org/plugin/ec-clem/- Used release: https://github.com/anrcrocoval/ec-clem/raw/master/binary/ec_clem-2.1.0-SNAPSHOT.jar |
| ICY | De Chaumont et al., 2012 |
http://icy.bioimageanalysis.org/ RRID:SCR_010587 |
| Matlab 2019b | Mathworks |
http://www.mathworks.com/products/matlab/ RRID:SCR_001622 |
| Fiji | Schindelin et al., 2012 |
https://fiji.sc/ RRID:SCR_002285 |
| Python 3.7 | Python software foundation |
http://www.python.org/ RRID:SCR_008394 |
| Pycharm – community version | JetBrains |
https://www.jetbrains.com/pycharm/ RRID:SCR_018221 |
| Image processing python script | Spehner et al., 2020 | |
| Adobe Illustrator | Adobe |
http://www.adobe.com/products/illustrator.html RRID:SCR_010279 |
| Other | ||
| Quantifoil R 3.5/1 200 Au mesh | Quantifoil Microtools GmbH | IMS Cat#Q250AR-35 |
| C-Clip Ring | Thermo Fisher Scientific | Cat#1036173 |
| C-Clips | Thermo Fisher Scientific | Cat#1036171 |
| DMEM, high glucose | Thermo Fisher Scientific | Cat#41965039 |
| HEPES 1M | Biological Industries | Cat#03-025-1B |
| Sodium Pyruvate 100mM | Biological Industries | Cat#03-042-1B |
| Penicillin-streptomycin solution | Biological Industries | Cat#03-031-1B |
| FBS | Thermo Fisher Scientific | Cat#12657-029 |
| DPBS | Biological Industries | Cat#02-020-1A |
| DPBS without calcium and magnesium | Biological Industries | Cat#02-023-1A |
| Trypsin EDTA solution B | Biological Industries | Cat#02-052-1B |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ori Avinoam (ori.avinoam@weizmann.ac.il).
Materials availability
This study did not generate new unique reagents.
Data and code availability statement
Data and code would be made available upon request.
Experimental model and subject details
Murine C2C12 were cultured in a growth medium composed of Dulbecco’s Modified Eagle Medium (DMEM; Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific), 1% sodium pyruvate (Biological industries, Israel), 1% penicillin-streptomycin (Biological industries) and 20mM HEPES (Biological industries), and maintained at 37°C, 5% CO2. Cell splitting has been done through 2 washes of Dulbecco’s Phosphate Buffer Saline (DPBS; Biological industries) without calcium and magnesium followed by trypsin (Biological industries) incubation for 5 minutes at 37°C, 5% CO2.
Method details
Cryo-immobilization and cryo-FM
For Cryo-FM, Cells were seeded on Quantifoil R3.5/1 Au 200 mesh EM grids (Quantifoil Micro Tools, Germany) at a cell density of 340 cells/mm2. After 10-20 hours cells were stained with BODIPY 493/503 (1:1000 in DPBS; Thermo Fisher Scientific) at 37°C for 20 min followed by three washes in growth medium.
Grids were plunge-frozen between 45-90 min after staining using a Leica EM GP (Leica Microsystems, Vienna, Austria) set to 37°C, 90% humidity and following 2.5s back-blotting. To facilitate their handling, grids were loaded into c-clip rings (Thermo Fisher Scientific) under liquid nitrogen prior to imaging. Imaging was performed using a cryo-CLEM microscope (Leica Microsystems) as previously described (Schorb et al., 2017). Samples were mounted onto a costume-made cartridge (EMBL, Heidelberg, Germany) in a liquid N2-cooled cryo-CLEM shuttle and transferred to the microscope. Samples were imaged under HCX PL APO 50× CLEM cryo-objective and images were acquired using an ORCA-flash4.0 (Hamamatsu Photonics, Hamamatsu city, Japan) with GFP (ex450-490/em500-550) filter. Initially, a spiral scan was acquired in brightfield (50ms exposure, 100% lamp intensity). To screen the grid and identify cells of interest, a grid map was by acquiring (250ms exposure, 100% lamp intensity) and stitching images with step size of 1-2μm over 50-100μm. We chose between seven and fifteen cells per grid based on their LD signal. For cells of interest, focus series were acquired using 25ms exposure (brightfield) and 200ms exposure (GFP), and a step size between 150-250nm over a 10μm range.
Cryo-FIB-SEM
Grid in c-clip rings were mounted onto a custom-made cryo-sample holder inside a modified Leica EM-VCM500 (Leica Microsystems) and transferred to a pre-cooled Zeiss Crossbeam 550 FIB-SEM (-150°C, Carl Zeiss Microscopy, Oberkochen, Germany) using Leica EM-VCT100 shuttle (Leica Microsystems). The Leica EM-VCM500 is closed to form a glove box and dried using constant flow of clean N2 gas (output pressure ∼1.5psi) in order to reduce exposure to humidity. The cryo-sample holder has a pre-tilt of -25° and the cryo-stage was tilted to 35°-40° during acquisition. Pt precursor deposition was performed by opening and closing the nuzzle of the multi-port gas-injection system in 3 cycles of 30s, without heating the source, while working distance was 7-8mm. FIB milling was executed by SmartFIB software (Carl Zeiss) using 30kV and probe current of 100pA to cut 10nm-thick sections, ion beam dose factor was 8, and dwell time 160μs. SEM micrographs were acquired at 1.6kV acceleration voltage with a probe current of 40pA and InLens SE/SE2 mixed detection with the smartSEM (Carl Zeiss; mixing ratio between 0.67-0.72). InLens SE detectors are efficient in detecting SE of type 1, that are generated close to or at the impact point of the primary beam. These electrons are derived from the upper part of the interaction volume and therefore supply direct information from the surface. Hence, InLens SE detectors are more sensitive than in-chamber SE2 detectors to differences in surface potential and charge accumulation over the cross section. Therefore, combining the signal of both increases the signal to noise ratio. Scanning speed of SEM was 1 and noise reduction was performed by line averaging (Figures 1, 3, and 4: N=104, Figure 2: N=140). The voxels sizes in data showed are 10x10x10 nm. During the first 10 hours of the session, temperature was maintained by manually refilling the dewar attached to the microscope every two hours. For nighttime acquisition, liquid N2 was replenished using an automatic 50 liters liquid N2 handling system (Norhof LN2 microdosing systems, Ede, The Netherlands), the microdosing system was filled prior to its connection. The level of liquid N2 in the microdoser was checked and refilled as needed.
Image processing and segmentation
Images were filtered using Fourier filtering approach with either Fiji (Schindelin et al., 2012) or a Fourier filtering Python 3.7 script kindly provided by Dr. Luca Bertinetti in order to reduce curtaining artifacts (Spehner et al., 2020). Stack alignment was performed either by cross-correlation alignment using Align Slices module in Amira 2019.3 (Thermo Fisher Scientific) or based on Fourier shift theorem as described in Spehner et al. (Spehner et al., 2020), Python 3.7 script was kindly provided by Dr. Luca Bertinetti. Both alignment methods allowed only for translation of the slices and not rotation or scaling. To enhance contrast, unsharp mask filter was applied to the stacks in Fiji software (Schindelin et al., 2012). In order to reduce charging artefact, charge correction was performed as described in Spehner et al. with a Python 3.7 script kindly provided by by Dr. Luca Bertinetti (Spehner et al., 2020).
Data segmentation was performed manually using Amira 2019.3 with the threshold tool on every 3rd or 4th slice. The rest of the slices were interpolated. Because intra mitochondrial granules span over ∼3 slices, owing of their small size, they were segmented without interpolation. All the segmented labels were converted into surfaces using Generate Surface module and presented using a Surface View object. To represent the volume of the nucleus in Figure 3, volume rendering was applied to the whole stack and the excess volume was cropped out using Volume Edit module.
Quantification and statistical analysis
Quantifications
Intra-mitochondrial granules quantification was performed based on the segmented particles. Volume of each particle was calculated by Amira using Material Statistics feature per particle. To avoid manual errors in the segmentation and exclude objects at the edge of the resolution, objects smaller than 10 voxels were left out of the quantification. The histogram and box plot chart were plotted using MATLAB 2019b (MathWorks).
LDs count per cell was made using the following processing steps in Fiji (Schindelin et al., 2012): Cells of interest were cropped and an intensity z-projection was calculated based on standard deviation. The image was then threshold to form a binary image and using the watershed tool. The LDs were counted using the analyze particles tool and corrected manually for false positive and false negatives. Box plot was plotted using MATLAB 2019b.
Correlation
For 3D correlation, the processed cryo-FIB-SEM was binned in Fiji (Schindelin et al., 2012). The center off mass was manually picked in the FIB-SEM dataset and the BODIPY signal was segmented using spot detector (parameters: detect bright spots, scale 2, sensitivity 10, export to ROI) in Icy (De Chaumont et al., 2012). Rough, rigid transformation was applied using Auto Finder feature in eC-CLEM (Paul-Gilloteaux et al., 2017) (parameters: FM with detected ROI spots as source image, FIB-SEM with manually picked ROI as target image, pre-aligned data, max error 10 microns, percentage of point to kept 70%, “already in the same orientation” option) following by either rigid or affine transformation with either 9 or 17 LDs identified manually under ec-CLEM v2.1.0. Error estimation heatmaps and 95% interval confidence spheres were calculated by eC-CLEM v2.1.0.
Acknowledgments
We thank Dr. Luca Bertinetti for providing scripts for image processing, Dr. Martin Schorb from the EMBL electron microcopy core facility for assistance in optimizing the cryo-FM image acquisition, the EM unit of the Weizmann Institute of Science, Dr. Neta Varsano and Dr. Mattia Morandi for critical reading of the manuscript, and members of the Avinoam team for fruitful discussions. P.P.-G. is part of MicroPICell facility (BioGenouest), member of the national infrastructure France-BioImaging (ANR-10-INBS-04), and acknowledges funding from ANR-18-CE45-0015. This research was supported by the Minerva Foundation with funding from the Federal German Ministry for Education and Research, the David Barton Center for Research on the Chemistry of Life, and the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement No 851080) to O.A. O.A is the Miriam Berman Presidential Development Chair.
Author contributions
N.S. and O.A. conceived and designed the experiments and analyzed the data. N.S. with help from K.R. carried out the experiments. N.S. with help of P.P.-G. correlated the data. N.S. and O.A. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community.
Published: July 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102714.
Supplemental information
References
- Arnold J., Mahamid J., Lucic V., De Marco A., Fernandez J.J., Laugks T., Mayer T., Hyman A.A., Baumeister W., Plitzko J.M. Site-pecific cryo-focused ion beam sample preparation guided by 3D correlative microscopy. Biophys. J. 2016;110:860–869. doi: 10.1016/j.bpj.2015.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat T.A.M., Hoffmann P.C., Kukulski W. Correlative microscopy of vitreous sections provides insights into BAR-domain organization in situ. Structure. 2018;26:879–886.e3. doi: 10.1016/j.str.2018.03.015. Elsevier Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykov Y.S., Cortese M., Briggs J.A., Bartenschlager R. ‘Correlative light and electron microscopy methods for the study of virus–cell interactions’. FEBS Lett. 2016;590:1877–1895. doi: 10.1002/1873-3468.12153. [DOI] [PubMed] [Google Scholar]
- Carzaniga R., Domart M.C., Collinson L.M., Duke E. Cryo-soft X-ray tomography: a journey into the world of the native-state cell. Protoplasma. 2014;251:449–458. doi: 10.1007/s00709-013-0583-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Chaumont F., Dallongeville S., Chenouard N., Hervé N., Pop S., Provoost T., Meas-Yedid V., Pankajakshan P., Lecomte T., Le Montagner Y., Lagache T. Icy: an open bioimage informatics platform for extended reproducible research. Nat. Methods. 2012;9:690–696. doi: 10.1038/nmeth.2075. [DOI] [PubMed] [Google Scholar]
- Heymann J.A., Hayles M., Gestmann I., Giannuzzi L.A., Lich B., Subramaniam S. Site-specific 3D imaging of cells and tissues with a dual beam microscope. J. Struct. Biol. 2006 doi: 10.1016/j.jsb.2006.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman D.P., Shtengel G., Xu C.S., Campbell K.R., Freeman M., Wang L., Milkie D.E., Pasolli H.A., Iyer N., Bogovic J.A., Stabley D.R. Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science. 2020;367 doi: 10.1126/science.aaz5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreman M.A., Mercier L., Schieber N.L., Solecki G., Allio G., Winkler F., Ruthensteiner B., Goetz J.G., Schwab Y. Fast and precise targeting of single tumor cells in vivo by multimodal correlative microscopy. J. Cell Sci. 2016;129:444–456. doi: 10.1242/jcs.181842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalifa G.M., Kirchenbuechler D., Koifman N., Kleinerman O., Talmon Y., Elbaum M., Addadi L., Weiner S., Erez J. ‘Biomineralization pathways in a foraminifer revealed using a novel correlative cryo-fluorescence–SEM–EDS technique’. J. Struct. Biol. 2016;196:155–163. doi: 10.1016/j.jsb.2016.01.015. Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Klein S., Wimmer B.H., Winter S.L., Kolovou A., Laketa V., Chlanda P. Post-correlation on-lamella cryo-CLEM reveals the membrane architecture of lamellar bodies. Commun. Biol. 2021;4:1–12. doi: 10.1038/s42003-020-01567-z. Springer US. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott G., Marchman H., Wall D., Lich B. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J. Neurosci. 2008 doi: 10.1523/JNEUROSCI.3189-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kounatidis I., Stanifer M.L., Phillips M.A., Paul-Gilloteaux P., Heiligenstein X., Wang H., Okolo C.A., Fish T.M., Spink M.C., Stuart D.I., Davis I. 3D correlative cryo-structured illumination fluorescence and soft X-ray microscopy elucidates reovirus intracellular release pathway. Cell. 2020;182:515–530.e17. doi: 10.1016/j.cell.2020.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukulski W., Schorb M., Welsch S., Picco A., Kaksonen M., Briggs J.A. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J. Cell Biol. 2011;192:111–119. doi: 10.1083/jcb.201009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Rechav K., Kaplan-Ashiri I., Gal A. Imaging and quantifying homeostatic levels of intracellular silicon in diatoms. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aaz7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahamid J., Pfeffer S., Schaffer M., Villa E., Danev R., Cuellar L.K., Förster F., Hyman A.A., Plitzko J.M., Baumeister W. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science. 2016;351:969–972. doi: 10.1126/science.aad8857. [DOI] [PubMed] [Google Scholar]
- Masich S., Östberg T., Norlén L., Shupliakov O., Daneholt B. A procedure to deposit fiducial markers on vitreous cryo-sections for cellular tomography. J. Struct. Biol. 2006;156:461–468. doi: 10.1016/j.jsb.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Medalia O., Beck M., Ecke M., Weber I., Neujahr R., Baumeister W., Gerisch G. Organization of actin networks in intact Filopodia. Curr. Biol. 2007;17:79–84. doi: 10.1016/j.cub.2006.11.022. [DOI] [PubMed] [Google Scholar]
- Medalia O., Weber I., Frangakis A.S., Nicastro D., Gerisch G., Baumeister W. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 2002;298:1209–1213. doi: 10.1126/science.1076184. [DOI] [PubMed] [Google Scholar]
- Merchán-Pérez A., Rodriguez J.R., AlonsoNanclares L., Schertel A., DeFelipe J. Counting synapses using FIB/SEM microscopy: a true revolution for ultrastructural volume reconstruction. Front. Neuroanat. 2009;3:1–14. doi: 10.3389/neuro.05.018.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan K., Subramaniam S. Focused ion beams in biology. Nat. Methods. 2015;12:1021–1031. doi: 10.1038/nmeth.3623. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann J.A., Carvalho P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019;20:137–155. doi: 10.1038/s41580-018-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul-Gilloteaux P., Heiligenstein X., Bell M., Domart M.C., Larijani B., Collinson L., Raposo G., Salamero J. EC-CLEM: Flexible multidimensional registration software for correlative microscopies. Nat. Methods. 2017;14:102–103. doi: 10.1038/nmeth.4170. [DOI] [PubMed] [Google Scholar]
- Peddie C.J., Collinson L.M. Exploring the third dimension: volume electron microscopy comes of age. Micron. 2014 doi: 10.1016/j.micron.2014.01.009. [DOI] [PubMed] [Google Scholar]
- Potier G., Lavancier F., Kunne S., Paul-Gilloteaux P. arXiv preprint; 2021. A Registration Error Estimation Framework for Correlative Imaging; pp. 1–11. [Google Scholar]
- Revach O.Y., Weiner A., Rechave K., Sabanay I., Livne A., Geiger B. Mechanical interplay between invadopodia and the nucleus in cultured cancer cells. Sci. Rep. 2015;5:1–13. doi: 10.1038/srep09466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznikov N., Almany-Magal R., Shahar R., Weiner S. Three-dimensional imaging of collagen fibril organization in rat circumferential lamellar bone using a dual beam electron microscope reveals ordered and disordered sub-lamellar structures. Bone. 2013;52:676–683. doi: 10.1016/j.bone.2012.10.034. Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Sartori A., Gatz R., Beck F., Rigort A., Baumeister W., Plitzko J.M. Correlative microscopy: Bridging the gap between fluorescence light microscopy and cryo-electron tomography. J. Struct. Biol. 2007;160:135–145. doi: 10.1016/j.jsb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Scher N., Avinoam O. 50 Shades of CLEM: how to choose the right approach for you. Methods Cell Biol. 2020 doi: 10.1016/bs.mcb.2020.08.001. Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Schertel A., Snaidero N., Han H.M., Ruhwedel T., Laue M., Grabenbauer M., Möbius W. Cryo FIB-SEM: volume imaging of cellular ultrastructure in native frozen specimens. J. Struct. Biol. 2013;184:355–360. doi: 10.1016/j.jsb.2013.09.024. Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider P., Meier M., Wepf R., Müller R. Towards quantitative 3D imaging of the osteocyte lacuno-canalicular network. Bone. 2010;47:848–858. doi: 10.1016/j.bone.2010.07.026. Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Schneider P., Meier M., Wepf R., Müller R. Serial FIB/SEM imaging for quantitative 3D assessment of the osteocyte lacuno-canalicular network. Bone. 2011;49:304–311. doi: 10.1016/j.bone.2011.04.005. Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Schorb M., Gaechter L., Avinoam O., Sieckmann F., Clarke M., Bebeacua C., Bykov Y.S., Sonnen A.F.P., Lihl R., Briggs J.A. New hardware and workflows for semi-automated correlative cryo-fluorescence and cryo-electron microscopy/tomography. J. Struct. Biol. 2017;197:83–93. doi: 10.1016/j.jsb.2016.06.020. The Authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spehner D., Steyer A.M., Bertinetti L., Orlov I., Benoit L., Pernet-Gallay K., Schertel A., Schultz P. Cryo-FIB-SEM as a promising tool for localizing proteins in 3D. J. Struct. Biol. 2020;211:107528. doi: 10.1016/j.jsb.2020.107528. Elsevier. [DOI] [PubMed] [Google Scholar]
- Sviben S., Gal A., Hood M.A., Bertinetti L., Politi Y., Bennet M., Krishnamoorthy P., Schertel A., Wirth R., Sorrentino A., Pereiro E. A vacuole-like compartment concentrates a disordered calcium phase in a key coccolithophorid alga. Nat. Commun. 2016;7:1–9. doi: 10.1038/ncomms11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauchi-Sato K., Ozeki S., Houjou T., Taguchi R., Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J. Biol. Chem. 2002;277:44507–44512. doi: 10.1074/jbc.M207712200. © 2002 ASBMB. Currently published by Elsevier Inc; originally published by American Society for Biochemistry and Molecular Biology. [DOI] [PubMed] [Google Scholar]
- Vidavsky N., Addadi S., Schertel A., Ben-Ezra D., Shpigel M., Addadi L., Weiner S. Calcium transport into the cells of the sea urchin larva in relation to spicule formation. Proc. Natl. Acad. Sci. U S A. 2016;113:12637–12642. doi: 10.1073/pnas.1612017113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidavsky N., Akiva A., Kaplan-Ashiri I., Rechav K., Addadi L., Weiner S., Schertel A. Cryo-FIB-SEM serial milling and block face imaging: large volume structural analysis of biological tissues preserved close to their native state. J. Struct. Biol. 2016;196:487–495. doi: 10.1016/j.jsb.2016.09.016. Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Wei D., Jacobs S., Modla S., Zhang S., Young C.L., Cirino R., Caplan J., Czymmek K. High-resolution three-dimensional reconstruction of a whole yeast cell using focused-ion beam scanning electron microscopy. BioTechniques. 2012;53:41–48. doi: 10.2144/000113850. [DOI] [PubMed] [Google Scholar]
- Weiner A., Dahan-Pasternak N., Shimoni E., Shinder V., von Huth P., Elbaum M., Dzikowski R. 3D nuclear architecture reveals coupled cell cycle dynamics of chromatin and nuclear pores in the malaria parasite Plasmodium falciparum. Cell Microbiol. 2011;13:967–977. doi: 10.1111/j.1462-5822.2011.01592.x. [DOI] [PubMed] [Google Scholar]
- Weiner A., Enninga J. The pathogen–host interface in three dimensions: correlative FIB/SEM applications. Trends Microbiol. 2019;27:426–439. doi: 10.1016/j.tim.2018.11.011. Elsevier Ltd. [DOI] [PubMed] [Google Scholar]
- Weiner A., Mellouk N., Lopez-Montero N., Chang Y.Y., Souque C., Schmitt C., Enninga J. Macropinosomes are key players in early shigella invasion and vacuolar escape in epithelial cells. PLoS Pathog. 2016;12:1–24. doi: 10.1371/journal.ppat.1005602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S.G., Mutsafi Y., Dadosh T., Ilani T., Lansky Z., Horowitz B., Rubin S., Elbaum M., Fass D. 3D visualization of mitochondrial solid-phase calcium stores in whole cells. eLife. 2017;6:1–18. doi: 10.7554/eLife.29929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G., Hagen C., Grünewald K., Kaufmann R. Towards correlative super-resolution fluorescence and electron cryo-microscopy. Biol. Cell. 2016;108:245–258. doi: 10.1111/boc.201600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C.S., Pang S., Shtengel G., Müller A., Ritter A.T., Hoffman H.K., Takemura S.Y., Lu Z., Pasolli H.A., Iyer N., Chung J. Isotropic 3D electron microscopy reference library of whole cells and tissues. bioRxiv. 2020 doi: 10.1101/2020.11.13.382457. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and code would be made available upon request.