Abstract
Objectives
Exosomes are small lipid bilayer vesicles that are defined by their endocytic origin and size range of 30–140 nm. They are constantly produced by different cell types, by both healthy and abnormal cells, and can be isolated from almost all body fluids.
Little information exists in isolating exosomes from plasma due to the complexity of its content and the presence of contaminating plasma proteins.
Design and methods
We carried-out liquid chromatography-mass spectrometry (LC-MS/MS) analyses of plasma-derived vesicles from 4 healthy donors obtained by 2 coupled methodologies: Ultracentrifugation (UC) coupled with size-exclusion chromatography (SEC) to isolate and subsequently enrich exosomes.
We compared the proteins detected by UC alone and UC coupled with SEC.
Results
In the coupled UC + SEC methodology we found 52.25% more proteins enriched in exosomes as CD9, Annexins, YWHAZ (14-3-3 family) and others, than by using UC alone. There is also a reduction of 98.8% of contaminating plasma proteins by coupling UC and SEC in comparison to using UC alone.
Conclusions
We conclude that exosomes can be successfully isolated from plasma using a very simple combination of standard methods, which could largely improve the proteomics profiling of plasma exosomes.
Keywords: Exosomes, Size-exclusion chromatography, Ultracentrifugation, Mass spectrometry, Human plasma
Abbreviations: AGO2, Argonaute protein; cfNAs, Cell-free nucleic acids; CSF, cerebrospinal fluid; ctDNA, circulating tumor DNA; miRNA, microRNA; EVs, extracellular vesicles; FDR, false discovery rate; ILVs, intraluminal vesicles; MVBs, multivesicular bodies; SEC, size exclusion chromatography; UC, ultracentrifugation
Highlights
-
•
Isolating exosomes with the least possible contaminants is very important for their clinical application.
-
•
Isolating exosomes from human plasma faces considerable challenges.
-
•
There is no ideal single isolation technique until this date.
-
•
Exosomes can be successfully isolated from plasma using a combination of Ultracentrifugation and SEC.
1. Introduction
Exosomes, the smallest category of extracellular vesicles (EVs), are small, lipid bilayer membrane vesicles (30–100 nm) that are released from all cell types into the extracellular space and are present in almost all biological fluids [1,2]. Blood, urine, cerebrospinal fluid (CSF), breast milk, ascites fluid, amniotic fluid, bile, semen, saliva and sputum all contain thousands to billions of exosomes per microliter of sample [3]. Exosomes are highly heterogeneous and likely reflect the phenotypic state of the cell that generates them [4,5]. Depending on the cell or tissue of origin, many different roles and functions have been attributed to exosomes [3]. EVs represent an important mode of intercellular communication and play key roles in many physiological and pathological processes [[6], [7], [8]]. They deliver macromolecular messages that enable cell-to-cell communication and signaling [3,9,10]. These tiny membrane vesicles transmit EV-mediated signals by proteins, lipids, nucleic acids and sugars, and the unique molecular pattern of this package direct the type of extracellular signal to be transmitted to target cells [11]. Cell-free nucleic acids (cfNAs) have been used as a minimally invasive detection method for molecular biomarkers in body fluids. Particularly, cell-free circulating tumor DNA (ctDNA) and microRNA (miRNA) are targets of interest for various diagnostic applications [12,13].
In the circulating blood, miRNAs are either bound by protective proteins, such as.
Argonaute protein AGO2 and Nucleoplasmin, loaded into high-density lipoprotein (HDL), or encapsulated within exosomes. Given the transportability of vesicles, the role of miRNAs in exosomes is gaining increasing attention. MiRNAs are involved in many biological activities as cell proliferation, differentiation and migration and in disease initiation and progression [2,14] conveying information via circulating vesicles as a way of intercellular communication [2,15,16].
Interest in exosomes range from their mode of action and various functions in the body to more practical applications such as development of biomarkers based on analysis of their RNA and protein content and their use in clinical diagnostics [3].
In general, exosomes are derived from the luminal membrane of multivesicular bodies (MVBs), which are constitutively released by fusion with the cell membrane [1,17]. During the biogenesis of exosomes and prior to their secretion, various molecules are uploaded into their lumen suggesting that the composition of exosomes is not random and is not a mere reflection of the cell. The selection of exosomal cargo encapsulated into intraluminal vesicles (ILVs) is a selectively regulated process [1,11,18]. This process distinguishes exosomes from microvesicles (MV, diameter 100–1000 nm), which are formed directly via outward budding of the plasma membrane [4,19].
Exosomes, therefore, are selectively enriched in tetraspanins (e.g. CD9 and CD81), Annexins, heat-shock proteins, 14-3-3 proteins, integrins, ALIX, TSG101 and others [1,4,7,11,17,18,[20], [21], [22], [23]].
Databases, including ExoCarta (www.exocarta.org), EVpedia (www.evpedia.info), Vesiclepedia (www.microvesicle.org) and Plasma Proteome Database (www.plasmaproteomedatabase.org), have cataloged the protein, lipid, and RNA content of exosomes, which have been identified in several EVs preparations. In this study we concentrated on the 100 most identified proteins in the ExoCarta database [1,11].
The process of isolating exosomes is one of the most challenging approaches, especially from human plasma [24]. In studies documented in the ExoCarta database, exosomes have been mostly isolated from the supernatants of cultured (cancer) cells. This is because of the relatively simple chemical composition of most culture media which enables isolation of exosomes almost lacking contaminating plasma proteins. Many other studies were performed on body fluids with also relatively low protein concentrations as urine, CSF and ascites fluid [3].
However, isolating exosomes from human plasma faces considerable challenges mainly due to the complex composition of plasma and the unavoidable contamination of exosome preparations with plasma proteins, protein aggregates and other extracellular vesicles, such as apoptotic bodies [25]. In addition, plasma exosomes are ‘coated’ with proteins, glycoproteins or glycolipids which are likely to cause their aggregation and a potential loss upon subsequent isolation steps [26].
Considerable attention should be dedicated to isolation of exosomes from plasma, with minimal invasive medical procedures, to serve as biomarkers for cancer and other diseases [20].
Differential centrifugation, density gradient isolation, ultrafiltration, size exclusion chromatography (SEC) and immuno-affinity are frequent methods used for EVs isolation [11,[27], [28], [29]]. However, each of these methods has its own limitations ranging from co-isolating contaminants, including non-vesicular proteins and lipids, to low EV recovery [30].
To date, there is no ideal single isolation technique and the development of novel methodologies to increase EV recovery and purity will highly benefit the clinical application of EVs as disease biomarkers [30]. In an attempt to overcome some of the limitations, coupling two or more methods has been proposed [[31], [32], [33], [34]]. Koh et al. studied the outcomes after comparing different methods of isolation of exosomes from bovine plasma. They pointed to the conclusion that isolating exosomes by ultracentrifugation (UC) with their subsequent enrichment by SEC provided the most consistent yield of plasma exosomes based upon the particle yield, exosome morphology and presence of exosome markers which they identified by immunoblotting [32]. An et al. combined one cycle of UC with SEC and found that this method provided improved results relative to the SEC method, although the blood protein contamination was slightly higher than that of their optimized UC method [35].
In this study, we compared the direct use of UC, for the isolation of exosomes from human plasma, with using UC followed by SEC for the enrichment of exosomes. In order to get a good view on the content of exosomes isolated, we analysed our preparations by LC-MS serving as an objective method for characterization of proteins rich in exosomes.
2. Materials and methods
2.1. Blood collection and plasma processing
Peripheral blood of 4 healthy volunteers was collected following standard procedures that minimize contamination by platelet and platelet-derived vesicles [36]. All work was done in accordance with the Mannheim University Hospital Ethics Committee guidelines based on German law and the Declaration of Helsinki. Written informed consents were obtained by all participants. Briefly, after venous puncture, blood was collected with EDTA pretreated tubes, samples were gently inverted 8–10 times and processed within 30 min of collection by 2 consecutive centrifugation steps; 1600 g for 10 min and at 3000 g for 10 min at room temperature. Centrifugation is done without brakes. Whenever possible, plasma samples were processed for exosome isolation before being frozen at −20 °C.
2.2. Ultracentrifugation
Ultracentrifugation was performed according to Raposo et al. [37] with some modifications.
In brief, frozen plasma samples were thawed for the first time after freezing, centrifuged at 2000 g for 10 min at room temperature and then at 10,000 g for 20 min to pellet cellular debris. After each centrifugation step, the supernatant is transferred into a new test tube while the generated pellets are being discarded. The plasma was then diluted 1:1 with PBS (Dulbecco's Phosphate Buffered Saline (Sigma) to decrease their viscosity. This increases the purity of EVs by decreasing the co-isolated contaminants, such as protein aggregates. Moreover it can improve the efficiency of EV isolation since higher viscosity resulted in lower sedimentation efficiency [38]. The sample is then inserted in Ultra-Clear centrifuge tubes 16 × 102 mm (Beckman Coulter) and centrifuged at 14,000 g for 35 min at 4 °C. to precipitate microvesicles. Supernatant was inserted in clean tubes and centrifuged at 100,000 g, 4 °C for 1.5 h to precipitate exosomes (Surespin 630). Supernatant was discarded and the subsequent pellet was re-suspended with PBS for washing. A second UC run was performed at 100,000 g, 4 °C for 1.5 h. The washed pellet was resuspended with PBS to a final volume of 200 μl, which was split into equal halves to proceed in 2 directions (with/without SEC).
2.3. Exosome isolation by SEC
For identification of exosome containing fractions of SEC, 0.5 ml of human plasma were loaded onto the mini-PURE EVs Size Exclusion Chromatography mini Columns for Exosome and microvesicle isolation (Hansa BioMed), and 12 fractions (160 μl each) were collected. Elution volumes were reduced by the speed vacuum Concentrator plus (Eppendorf) at 60 °C and RIPA buffer was added to be then examined by WB.
For obtaining a single exosome containing fraction, 0.5 ml of human plasma were loaded onto a SEC mini column and 3 fractions (~700 μl each) were collected. Elution volumes were reduced by speed vacuum at 60 °C and RIPA buffer was added.
Exosomes isolated by Ultracentrifugation were completed with PBS to 0.5 ml and were applied to a SEC mini column and collection of the exosomal fraction (700 μl) using PBS as the elution buffer was done. The collected fraction was reduced by speed vacuum at 60 °C. RIPA buffer was then added and samples were either used directly or frozen at −20 °C until analysis by LC-MS for protein profiling. For confirmation of the presence of exosomes in these preparations, 3–5 μL of the sample were examined by WB.
2.4. Protein quantification
We used Qubit protein assay (Invitrogen) according to the manufacturer. Samples of isolated material before and after SEC were adjusted to an equal starting volume to be directly comparable. 1 μl (before SEC) or 10 μl (after SEC) of samples were added to 199 or 190 μl working solution respectively (end volume 200 μl). Samples were then incubated at room temperature in the dark for 15 min and then measured with respect to a standard curve.
2.5. Western blot analysis
Eluates obtained from SEC were mixed with 4 × non-reducing SDS sample buffer, then heated at 95 °C for 5 min and loaded onto a 1.0 mm × 12 well 4%–12% Bis -Tris gel (Novex). Benchmark prestained protein ladder (Thermo Scientific) was added to one well as a control to monitor the molecular weight of the protein samples. The gel was run under denaturing conditions at 120 V for 1.5 h then transferred to a Nitrocellulose blotting membrane (GE Healthcare Life Sciences) using a semi-dry blotting instrument (Biometra). After transfer, the membranes were blocked for 1 h using Block buffer (5% Skimmed milk in TBS 0.1% Tween 20), then incubated overnight with CD9 antibody (Clone/PAD: TS9, Invitrogen) in a dilution of 1:500 with the Block buffer, at 4 °C. The membrane was then incubated with the secondary antibody (Polyclonal goat anti-mouse immunoglobulins/HRP, Dako). The ECL Prime Western Blotting Detection Reagent (GE Health care) was utilized to label the membrane. Membranes were then exposed to the imager ChemiDoc XRS+ with image lab software (Bio Rad) for 1–3 min and the image was analysed.
For the step of identification of exosome containing fractions of SEC, we stripped the Nitrocellulose membrane with mild stripping buffer. After stripping the membrane was blocked with Block buffer for 1 h, then incubated overnight with CD63 antibody (Clone/PAD: TS63, Invitrogen) in a dilution of 1:500 with the Block buffer, at 4 °C and proceeded as with CD9 antibody.
In parallel, another gel was prepared by the same way and stained with Coomassie blue stain.
2.6. Nanoparticle tracking analysis
The Nanoparticle tracking analysis (NTA) was performed using the ZetaView device (Particle Metrix, Meerbusch; Germany). 1 μl of isolated extracellular vesicles (from fraction 2 after SEC) were 1:2000–1:5000 diluted in PBS 1:1000 and measured. The ZetaView settings were adjusted to sensitivity 80%, shutter 100, 11 positions, and 2 cycles.
2.7. Electrophoresis, in-gel digestion
To the RIPA exosomes lysates an appropriate amount of 4x loading buffer was added, the samples were heated to 95 °C for 5 min and cooled on ice prior loading onto NuPAGE 10% Bis-Tris Gels (life technologies). The whole exosome lysate was loaded onto the gel, this way the four samples were directly comparable since the starting volume was the same. SDS polyacrylamide gelelectrophoresis (SDS-PAGE) was performed according to the manufacturer's specification. Proteins were fixed within the polyacrylamide matrix by incubating the entire gel in 5% acetic acid in 1:1 (vol/vol) water:methanol for 30 min. After Coomassie staining (60 min) the gel slab was rinsed with water (60 min) and each lane was excised and cut into small pieces. Subsequently the proteins were in-gel destained (100 mM ammonium bicarbonate/acetonitrile 1:1 (vol/vol)), reduced (10 mM DTT), alkylated (50 mm Iodoacetamide) and finally Trypsin digested by overnight incubation at 37 °C. The generated peptides were collected from the gel pieces, which were further subjected to a peptide extraction step with an acidic (1.5% formic acid) acetonitrile (66%) solution. Both peptides containing samples are combined and dried down in a vacuum centrifuge.
2.8. Mass spectrometry (LC-MS/MS)
Dried peptides were re-dissolved in 0.1% trifluoroacetic acid and loaded on a C18 precolumn (Acclaim; Dionex) using an RSLCnano HPLC system (Dionex). Peptides were then eluted with an aqueous-organic gradient, resolved on a C18 column (Acclaim; Dionex) with a flow rate of 300 nl/min and electrosprayed into a LTQ Orbitrap XL mass spectrometer (Thermo Scientific). A Triversa Automate (Advion biosciences) was used as ion source. Each scan cycle consisted of one FTMS full scan and up to seven ITMS dependent MS/MS scans of the seven most intense ions. Dynamic exclusion (30 s), mass width (10 ppm) and monoisotopic precursor selection were enabled. All analyses were performed in positive ion mode. Extracted MS/MS spectra were searched against the Uniprot/Swissprot database using the PEAKS search engine (Bioinformatics Solutions Inc.) accepting common variable modifications and one missed tryptic cleavage. Peptide tolerance was ±10 ppm and MS/MS tolerance was ±0.5 Da. All protein identification experiments were carried out using the corresponding decoy database and a false discovery rate (FDR) of 1%.
3. Results
3.1. Identification of exosomes containing fractions of SEC
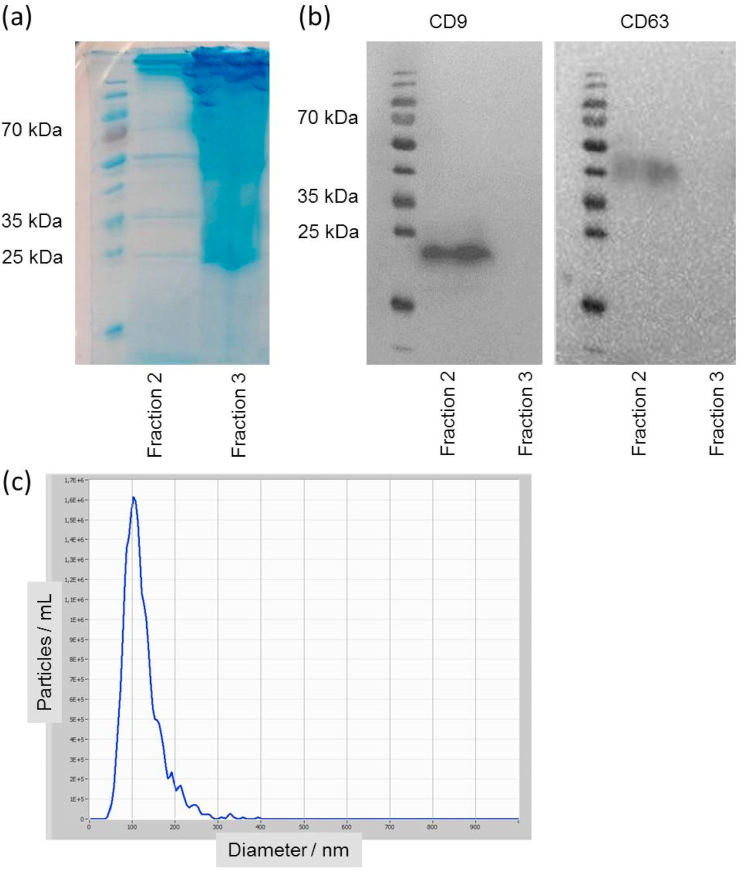
As a preliminary test, we simply tested the size exclusion columns using 0.5 ml of plasma in order to identify exosomes containing fractions. We then used anti-CD9 and anti- CD63 antibodies to identify the presence or absence of exosomal markers in each fraction by Western blot. Specifically, using 0.5 ml of plasma, we found that out of 12 fractions gained after SEC and each having a volume of around 160 μl, fractions 5–9 contained the highest concentrations of exosomal marker proteins, peaking in fraction 7, while in the later fractions (10–12) markers were absent. We limited the subsequent analysis to fractions 5–8, the bulk of the proteins were eluted in later fractions (data not shown). We then modified this experiment by collecting only three (instead of twelve) fractions but with larger volumes (700 μl). They were tested for protein by Coomassie blue stain (Fig. 1a) and for anti-CD9 and anti-CD63 antibodies by Western blot (Fig. 1b). The first fraction is devoid of any exosomes. The second fraction is the part collected for investigation of exosomes. The third fraction is also devoid of any markers. In contrast, the main bulk of the contaminating plasma proteins were eluted starting with this fraction. This preliminary work was done since correct identification of the right fraction after SEC is essential to obtain robust and reproducible results.
Fig. 1.
(a) Coomassie blue staining of the second and third eluted fractions from SEC of 0.5 ml human plasma after SDS-PAGE. (b) Western blot for the exosomal marker CD9 for the same SEC fractions as in (a). We also used the exosomal marker CD63 after stripping the Nitrocellulose membrane with mild stripping buffer. (c) Exemplary Nanoparticle Tracking Analysis of fraction 2 of one sample after SEC.
To further prove the presence of exosomes in fraction 2 nanoparticle tracking analysis was performed to visualize the size distribution of the vesicles and the particle count. Here we detected mean size values between 94 and 113 nm, an exemplary graph is shown in Fig. 1c.
In a standardized way, we only used fraction 2 (corresponding to elution volume from 700 μl to 1400 μl) when using SEC columns with a starting material volume of 500 μl plasma, in isolation and enrichment of exosomes for further preparation steps.
3.2. Analysis of plasma exosomes by LC-MS
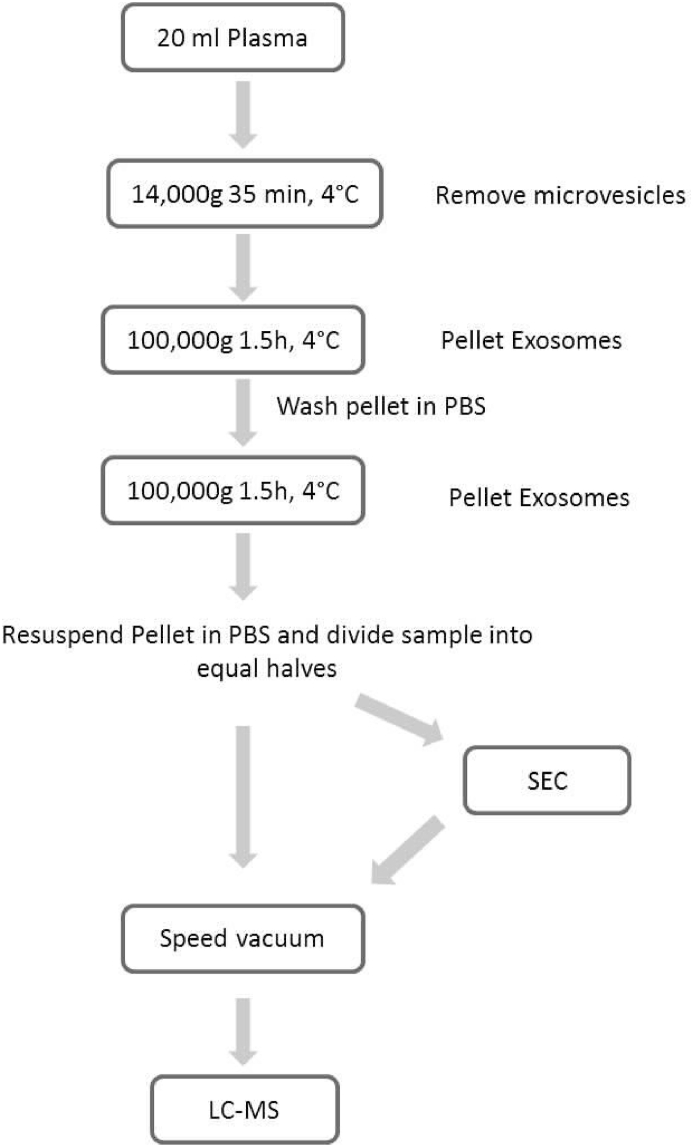
Our approach was to isolate exosomes by UC, resuspend the pellet in PBS and divide the sample into equal halves. One half was subjected to subsequent enrichment by SEC, while the other half was not, for comparison. The workflow for isolation of exosomes is presented in Fig. 2.
Fig. 2.
Workflow demonstrating the methodologies used in exosomes isolation and detection.
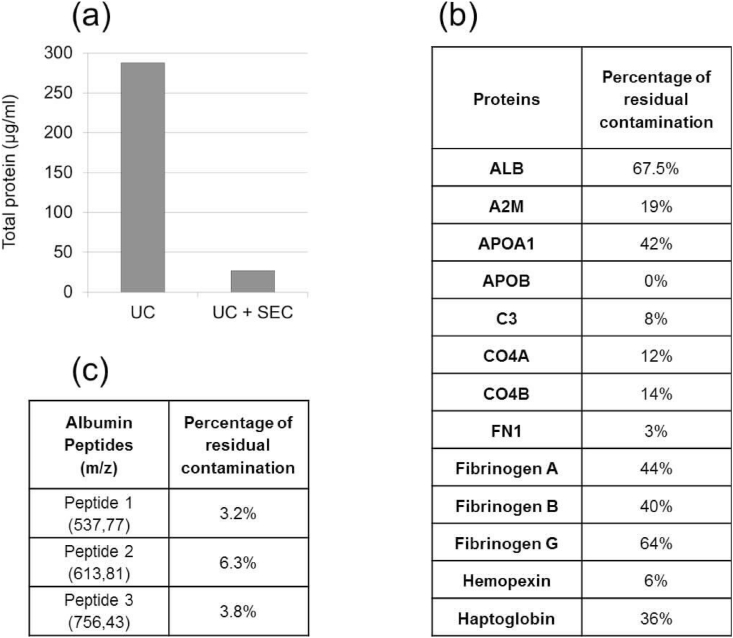
We quantified the protein concentration after UC both before and after SEC by Qubit protein assay. Mean protein concentration after UC and before SEC was 288 μg/ml (s = 31), and mean protein concentration after UC and after SEC (fraction 2) was 26.4 μg/ml (s = 13.4). Protein concentration was 11 times (90.8%) less by using SEC (Fig. 4a).
Fig. 4.
Protein quantification and analysis of contaminating plasma proteins (a) Diagram showing the difference in concentration of total protein after UC and SEC. (b). Table showing the percentage of residual peptides in some of the major contaminating plasma proteins when comparing isolating exosomes using ultracentrifugation (UC) alone or using UC coupled with size exclusion chromatography (SEC), using the mean from four different samples.(c)Table showing the percentage of residual contamination of 3 Albumin peptides, regarding signal intensity, when comparing isolating exosomes using ultracentrifugation coupled with size exclusion chromatography (SEC) compared to UC alone, using the mean from four different samples.
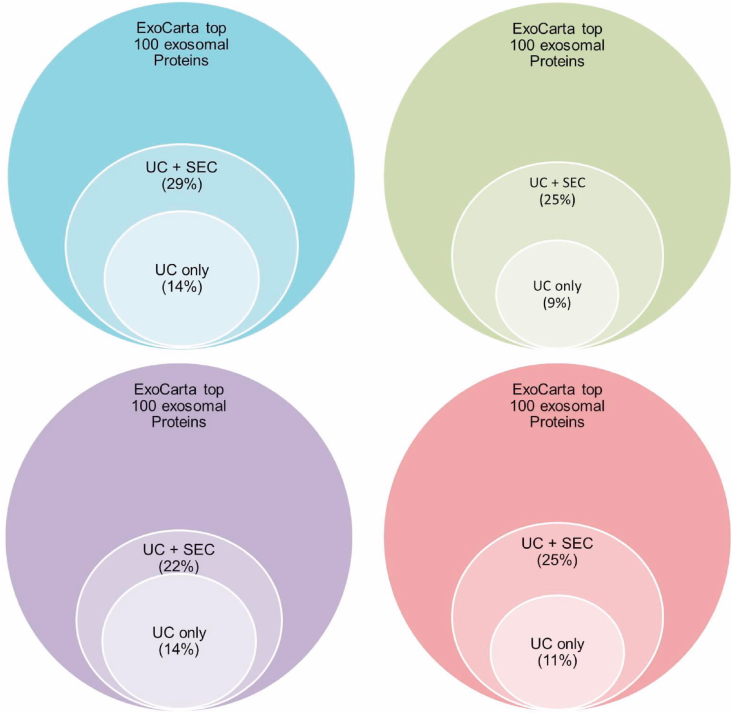
We continued by analyzing with LC-MS. We used the ExoCarta database of the top 100 human proteins mostly found in exosomes as a reference. UC + SEC was capable of enriching exosomal proteins, as by analyzing these preparations, we could detect approx. 30% of the proteins enriched in exosomes referring to the top 100 protein list in ExoCarta.
By proteomic analysis of exosomal preparations by this coupled methodology we identified one of the tetraspanins; CD9, Heat shock proteins as HSPA8 and HSPA1A, integrins as ITGA6, Annexins as ANXA2 and ANXA1, 14-3-3 proteins e.g. YWHAZ, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), tubulins e.g. TBA1A, TBA1B and TBA1C, Ras-related protein Rap-1b (RAP1B), Alpha Enolase (ENO1), Moesin (MSN), Ezrin (EZR), Stomatin (STOM), Thrombospondin 1 (THSB1), Gelsolin (GSN), Peptidyl-prolyl cis-trans isomerase A (PPIA), Filamin A (FLNA), Peroxiredoxin-2 (PRDX2) and cell division cycle 42 protein (CDC42) (Suppl. Table S1). The difference in percentage of exosomal proteins between the 2 methods used, using the ExoCarta top 100 proteins enriched in exosomes as a reference, is presented in Fig. 3.
Fig. 3.
Proteins enriched in exosomes were isolated by 2 methods; UC alone and UC coupled with SEC, and analysed by LC-MS. Proteins detected were compared using the ExoCarta database as a reference. Four different samples are shown.
Our results were detection of twice the amount of EV-specific proteins (average 25) by this coupled mechanism when compared to using UC alone (average 12). UC alone lead to identification of only 47.75% (mean of all four samples) of the exosomal proteins found when coupling UC + SEC (see Fig. 3).
We used LC-MS to compare marker peptides of 13 of the major contaminating plasma proteins. Based on the number of their peptides and by comparing them before and after SEC, we found that SEC greatly adds to the reduction of contamination as seen in Fig. 4b.
Albumin however is very resistant and the residual contamination is still significant after SEC. This led us to further analysis of albumin peptides. We selected 3 albumin peptides that were present in all 4 samples, had no posttranslational modifications and had no miscleavage. We analysed the signal intensity of each, and there was a reduction of ~95% after using UC + SEC when compared to using UC alone (Fig. 4c).
4. Discussion
Isolation of EVs from human plasma for downstream analysis is still challenging, due to the difficulty of obtaining pure EV isolates. Processing of plasma-derived EVs is commonly associated with co-isolation of contaminants. Supernatants of cultured cells have been the most common material used in isolation of exosomes in many studies. They have a relatively simple chemical composition when compared to plasma which facilitates isolation of exosomes almost lacking contaminating proteins. Moreover, the origin of exosomes is determined and easily controlled [26].
The need for a time efficient, highly reproducible and robust method of isolating exosomes from plasma, for their application in different varieties of clinical diagnostics, is highly essential. This method should provide an appropriate yield with the lowest possible degree of contamination.
Ultracentrifugation alone cannot achieve absolute separation of exosomes due to co-sedimentation of other macromolecules. The pellet from a high-speed spin will contain extravesicular proteins, protein aggregates, lipoprotein particles, and other contaminants. Resuspending and recentrifuging each pellet in PBS may aid in removing some of these impurities, but absolute separation is impossible by this method alone [36].
Correspondingly, using SEC as a stand-alone methodology for isolating exosomes, de Menezes-Neto, A., et al. could not detect tetraspanins by MS, even though they confirmed their presence by a bead-exosome FACS assay. Very few exosome enriched proteins were detectable by MS using this method alone [39].
By coupling UC with SEC, we gained the advantage of UC in the ability to process large volumes of plasma with better yields of exosomes due to the capability of SEC to get rid of a great fraction of contaminating plasma proteins. The protein profile of the exosome preparations (isolated by UC or UC + SEC) was determined by LC-MS with the analysis of both the membrane bound and internalized proteins. The overall number of identified proteins by either method was lower than that compared to other studies, being only around 150. This is highly likely due to the fact that the mass spectrometry device used for this work is rather dated.
In principle, detection of proteins enriched in exosomes, such as CD9 and ANXA proteins, and the absence of proteins such as the endoplasmic reticulum protein calnexin, is an indication that the exosome enriched pellet is indeed exosomes and not contaminating vesicles from other compartments of the cell [40].
Using UC coupled with SEC aided in enriching exosomal proteins and reducing protein contamination from highly abundant proteins like albumin or immunoglobulins. The percentages of residual contamination are shown in Fig. 4b. A significant amount of contaminating plasma proteins and lipoproteins was eliminated by this second step of SEC, as evidence by the significant decrease in the number of peptides of most of the popular contaminating plasma proteins; e.g. Albumin, Alpha 2 macroglobulin, apolipoprotein A1 and B, complement components (C3, CO4A and CO4B), Fibrinogen, Haptoglobin and Fibronectin as demonstrated in Fig. 4b. It has to be mentioned that SEC does not completely eliminate contaminating proteins like albumin or immunoglobulins but it reduces their amount to such a degree that low abundance exosomal proteins can then be detected.
The overall number of residual albumin peptides was reduced but not as efficient as other proteins. However, when we analysed 3 different albumin peptides to compare signal intensity (million counts) between the 2 methodologies used, the signal intensities for the 3 peptides were greatly reduced (~95%) by UC + SEC pointing to the great effect SEC has on reducing even the most resistant plasma proteins.
The comparison shows that adding the SEC step after UC enhances the purity of exosomes and therefore enables a higher identification rate than using UC alone.
Other proteins enriched in exosomes, that were not a part of the top 100 proteins in the ExoCarta database e.g. integrins as ITGA2B, ITGB3 and ITGA6, talin 1 (TLN1), SLC4A1 (Band 3 anion transport protein), Keratins (e.g. KRT1, KRT2, KRT9 and KRT10), ras-related C3 botulinum toxin substrate 2 (RAC2), Myosins (e.g. MYOVA and MYO9, Clusterin (CLU) and Radixin (RDX) were significantly detectable using the coupled methodology when compared to UC alone (see supplementary file).
By doubling the amount of starting material (plasma) we were able to detect even more proteins enriched in exosomes (in the ExoCarta top 100) e.g. PDCD6IP, PGK1, other 14-3-3 proteins (YWHAE, YWHAG..) and Valosin-containing protein (VCP) (data not shown). However, this large amount of plasma used as the starting material is not easily applicable.
5. Conclusions
This study demonstrates that a two-step isolation methodology, combining UC followed by SEC, isolates EVs from human plasma, and efficiently separates EVs from the main contaminating plasma proteins and lipoproteins, as evidenced by characterization of protein content by proteomic characterization by LC-MS/MS. Twice the number of exosomal proteins can be identified by using UC + SEC when compared to UC alone.
Funding
Sara Alameldin has received a governmental scholarship from the Cultural Affairs and Missions Sector, Ministry of Higher education, Arab Republic of Egypt.
Declaration of interest
The authors declare no competing interests.
Author contributions
Sara Alameldin: performed the experiments. participated in the analyses of data and wrote the manuscript. designed the experiments. All authors discussed the results and context of the manuscript; Victor Costina: was responsible for LC-MS analyses. All authors discussed the results and context of the manuscript; Hesham A. Abdel-Baset: All authors discussed the results and context of the manuscript; Katja Nitschke: performed NTA. All authors discussed the results and context of the manuscript; Phillip Nuhn: performed NTA. All authors discussed the results and context of the manuscript; Michael Neumaier: designed the experiments. All authors discussed the results and context of the manuscript; Maren Hedtke: designed the experiments. participated in the analyses of data and wrote the manuscript. All authors discussed the results and context of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.plabm.2021.e00241.
Contributor Information
Sara Alameldin, Email: dr.saraalameldin@aun.edu.eg.
Victor Costina, Email: victor.costina@umm.de.
Hesham A. Abdel-Baset, Email: heshama1964@yahoo.com.
Katja Nitschke, Email: Katja.Nitschke@medma.uni-heidelberg.de.
Phillip Nuhn, Email: Phillip.Nuhn@umm.de.
Michael Neumaier, Email: michael.neumaier@umm.de.
Maren Hedtke, Email: maren.hedtke@umm.de.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Guo W. Exosomes: new players in cancer (review) Oncol. Rep. 2017;38:665–675. doi: 10.3892/or.2017.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J. Exosome and exosomal microRNA: trafficking, sorting, and function. Dev. Reprod. Biol. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li M. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Phil. Trans. Roy. Soc. Lond. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalluri R. The biology and function of exosomes in cancer. J. Clin. Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kowal J. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. U.S.A. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardiner C. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J. Extracell. Vesicles. 2016;5:32945. doi: 10.3402/jev.v5.32945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raposo G., Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yanez-Mo M. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valadi H. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 10.Taylor D.D., Gercel-Taylor C. The origin, function, and diagnostic potential of RNA within extracellular vesicles present in human biological fluids. Front. Genet. 2013;4:142. doi: 10.3389/fgene.2013.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosa-Fernandes L., Rocha V.B., Carregari V.C., Urbani A., Palmisano G. A perspective on extracellular vesicles proteomics. Frontiers in chemistry. 2017;5:102. doi: 10.3389/fchem.2017.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarzenbach H., Hoon D.S., Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Canc. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 13.van Ginkel J.H. Cell-free nucleic acids in body fluids as biomarkers for the prediction and early detection of recurrent head and neck cancer: a systematic review of the literature. Oral Oncol. 2017;75:8–15. doi: 10.1016/j.oraloncology.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Png K.J., Halberg N., Yoshida M., Tavazoie S.F. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2011;481:190–194. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 15.Kuroiwa T. CD40 ligand-activated human monocytes amplify glomerular inflammatory responses through soluble and cell-to-cell contact-dependent mechanisms. J. Immunol. 1999;163:2168–2175. [PubMed] [Google Scholar]
- 16.Harvey S., Martinez-Moreno C.G., Luna M., Aramburo C. Autocrine/paracrine roles of extrapituitary growth hormone and prolactin in health and disease: an overview. Gen. Comp. Endocrinol. 2015;220:103–111. doi: 10.1016/j.ygcen.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Colombo M., Raposo G., Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 18.Mentkowski K.I., Snitzer J.D., Rusnak S., Lang J.K. Therapeutic potential of engineered extracellular vesicles. AAPS J. 2018;20:50. doi: 10.1208/s12248-018-0211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bortoluzzi S., Lovisa F., Gaffo E., Mussolin L. Small RNAs in circulating exosomes of cancer patients: a minireview. High-throughput. 2017;6 doi: 10.3390/ht6040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munson P., Shukla A. Exosomes: potential in cancer diagnosis and therapy. Medicines (Basel) 2015;2:310–327. doi: 10.3390/medicines2040310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Toro J., Herschlik L., Waldner C., Mongini C. Emerging roles of exosomes in normal and pathological conditions: new insights for diagnosis and therapeutic applications. Front. Immunol. 2015;6:203. doi: 10.3389/fimmu.2015.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Runz S. Malignant ascites-derived exosomes of ovarian carcinoma patients contain CD24 and EpCAM. Gynecol. Oncol. 2007;107:563–571. doi: 10.1016/j.ygyno.2007.08.064. [DOI] [PubMed] [Google Scholar]
- 23.Ciravolo V. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J. Cell. Physiol. 2012;227:658–667. doi: 10.1002/jcp.22773. [DOI] [PubMed] [Google Scholar]
- 24.Mateescu B. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J. Extracell. Vesicles. 2017;6:1286095. doi: 10.1080/20013078.2017.1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jablonska J., Pietrowska M., Ludwig S., Lang S., Thakur B.K. Challenges in the isolation and proteomic analysis of cancer exosomes-implications for translational research. Proteomes. 2019;7 doi: 10.3390/proteomes7020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller L., Hong C.S., Stolz D.B., Watkins S.C., Whiteside T.L. Isolation of biologically-active exosomes from human plasma. J. Immunol. Methods. 2014;411:55–65. doi: 10.1016/j.jim.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li P., Kaslan M., Lee S.H., Yao J., Gao Z. Progress in exosome isolation techniques. Theranostics. 2017;7:789–804. doi: 10.7150/thno.18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeringer E., Barta T., Li M., Vlassov A.V. Strategies for isolation of exosomes. Cold Spring Harb. Protoc. 2015:319–323. doi: 10.1101/pdb.top074476. 2015. [DOI] [PubMed] [Google Scholar]
- 29.Lobb R.J. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles. 2015;4:27031. doi: 10.3402/jev.v4.27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carpintero-Fernandez P., Fafian-Labora J., O'Loghlen A. Technical advances to study extracellular vesicles. Frontiers in molecular biosciences. 2017;4:79. doi: 10.3389/fmolb.2017.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onodi Z. Isolation of high-purity extracellular vesicles by the combination of iodixanol density gradient ultracentrifugation and bind-elute chromatography from blood plasma. Front. Physiol. 2018;9:1479. doi: 10.3389/fphys.2018.01479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koh Y.Q., Almughlliq F.B., Vaswani K., Peiris H.N., Mitchell M.D. Exosome enrichment by ultracentrifugation and size exclusion chromatography. Front Biosci (Landmark Ed) 2018;23:865–874. doi: 10.2741/4621. [DOI] [PubMed] [Google Scholar]
- 33.Mathias R.A., Lim J.W., Ji H., Simpson R.J. Isolation of extracellular membranous vesicles for proteomic analysis. Methods Mol. Biol. 2009;528:227–242. doi: 10.1007/978-1-60327-310-7_16. [DOI] [PubMed] [Google Scholar]
- 34.Wei R. Combination of size-exclusion chromatography and ultracentrifugation improves the proteomic profiling of plasma-derived small extracellular vesicles. Biol. Proced. Online. 2020;22:12. doi: 10.1186/s12575-020-00125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.An M., Wu J., Zhu J., Lubman D.M. Comparison of an optimized ultracentrifugation method versus size-exclusion chromatography for isolation of exosomes from human serum. J. Proteome Res. 2018;17:3599–3605. doi: 10.1021/acs.jproteome.8b00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Witwer K.W. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raposo G. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J., He X., Deng Y., Yang C. An update on isolation methods for proteomic studies of extracellular vesicles in biofluids. Molecules. 2019;24 doi: 10.3390/molecules24193516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Menezes-Neto A. Size-exclusion chromatography as a stand-alone methodology identifies novel markers in mass spectrometry analyses of plasma-derived vesicles from healthy individuals. J. Extracell. Vesicles. 2015;4:27378. doi: 10.3402/jev.v4.27378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lotvall J. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.