Abstract
Previously, we isolated a novel strain of goose pegivirus (GPgV) that infects geese and shows high levels of lymphotropism. This novel pegivirus strain is phylogenetically distinct from previously known Pegivirus species, Pegivirus A-K, and qualifies as a candidate new Pegivirus species, GPgV. GPgV is tentatively named Pegivirus M. Here, to better understand the epidemic of GPgV infection and the coinfection of this virus with other viruses in Southwest China, 25 geese in poor health from Sichuan Province and 24 geese in poor health from the municipality of Chongqing were collected. The geese were tested for 9 types of goose viruses (goose hemorrhagic polyomavirus, GPgV, astrovirus, parvovirus, circovirus, reovirus, coronavirus, paramyxovirus, and avian influenza virus) by RT-PCR or nested RT-PCR. GPgV RNA was detected in 2 out of 25 monoinfections and 8 out of 25 coinfections with other viruses on Sichuan farms and 2 out of 24 monoinfections and 10 out of 24 coinfections on Chongqing farms. Overall, 22 of the 49 (44.9%) geese were positive for GPgV, which indicated a high infection rate. To the best of our knowledge, this is the first report of GPgV coinfection with other epidemic viruses. This study enhances our understanding of the emergence and epidemiology of Pegivirus.
Key words: pegivirus, goose, epidemiology, infection, coinfection
INTRODUCTION
All pegiviruses have positive-sense single-stranded RNA genomes. The pegivirus genome is translated into a single polyprotein, which subsequently undergoes co- and post-translational cleavage by cellular and viral proteases to form mature proteins.
The genus Pegivirus can be subdivided into phylogroup 1 (Pegivirus F, G, H, and J) and phylogroup 2 (Pegivirus A, B, C, D, E, I, and K) according to the phylogenetic relationships among its members. Pegivirus A includes GBV-A and other isolates from New World monkeys; Pegivirus B includes viruses derived from bats (GBV-D); Pegivirus C has been proposed as a new species, with the GBV-C/hepatitis V virus and related viruses isolated from Old World primates classified in this taxon; Pegivirus D and E are both derived from horses; Pegivirus F, G and I all include viruses derived from Old and New World bats; Pegivirus H includes viruses described as human pegivirus 2 and human hepegivirus; Pegivirus J includes viruses derived from rodents; and Pegivirus K comprises a virus isolated from pigs. Previously, we reported a new Pegivirus species, goose pegivirus (GPgV), which was isolated from dead geese in Southwest China. Experimental infection of GPgV in goslings via intravenous injection revealed robust replication and high lymphotropism (Wu et al., 2020).
Species of the Pegivirus genus (family Flaviviridae) are not commonly identified as disease-causing agents (Smith et al., 2016b; Stapleton et al., 2011); an exception is equine pegivirus, which causes persistent viremia (Chandriani et al., 2013). However, no clinical disease has been conclusively identified as associated with pegivirus infection in experimentally infected animals (Baechlein et al., 2016; Chandriani et al., 2013; Kapoor et al., 2013a; Kapoor et al., 2013b; Quan et al., 2013). Coinfection of pegivirus with other viruses affects the outcome of infection (Berg et al., 2015). This observation prompted us to speculate that GPgV might coinfect with other goose viruses. To date, no systematic study of GPgV infection and coinfection has been conducted. Thus, we detected GPgV infection and coinfection in goslings of poor health from local farms. Twenty-two of the 49 (44.9%) geese were positive for GPgV, and 18 of the 49 (36.7%) geese were coinfected with GPgV and another virus. Most GPgV infections occurred as coinfections with other viruses. To the best of our knowledge, this is the first report on the epidemic of GPgV infection and the first report of GPgV coinfection with a number of other goose viruses. Our study enhances our understanding of the emergence and epidemiology of Pegivirus-related diseases.
MATERIALS AND METHODS
Samples Collection
Some gosling-producing farms in Southwest China reported sporadic goose losses in past decades. Some dead adult geese showed typical symptoms of enteritis, including loss of appetite and weakness. Haemorrhages and hyperemia were observed in the small intestines of sick goslings, and enlarged spleens and thymuses were also obviously in detected 4 to 6 wk old goose. The dead goslings usually showed severe diarrhea and sudden death; in many cases, no obvious clinical symptoms were observed. In initial tests, the small and large intestines, livers, kidneys, spleens, and brains from the dead geese were sampled, and samples were negative for other viruses (e.g., goose parvovirus, goose circovirus, goose coronaviruses, goose paramyxovirus, and avian influenza virus) according to PCR or RT-PCR or Nest-PCR analysis (Table 1).
Table 1.
Detection of goose pegivirus in the samples from the farm located in the southwest of China.
| Coinfection with |
||||||||
|---|---|---|---|---|---|---|---|---|
| Region | Sample | Monoinfection | GPV | GoCV | GAV | GPV and GoCV | GoCV and GAV | GPV and GAV |
| Chongqing farms | 24 | 2 | 8 | 3 | 4 | 2 | 1 | 2 |
| Guanghan farms | 21 | 2 | 1 | 4 | 1 | 0 | 1 | 0 |
| Leshan farms | 4 | 0 | 3 | 3 | 0 | 3 | 0 | 0 |
| Sum | 49 | 4 | 12 | 10 | 5 | 5 | 2 | 2 |
| Infection rate | 8.20% | 24.50% | 20.40% | 10.20% | 10.20% | 4.10% | 4.10% | |
Abbreviations: GAV, goose astrovirus; GPV, goose parvovirus; GoCV, goose circovirus.
Virus Culture In Vitro
Virus culture in vitro had been described before (Wu et al., 2020). Breifly, 5 eggs were used for each passage and only the allantoic fluid from the egg with the highest virus load, determined using qRT-PCR, was used for the next inoculation step. Virus loads were determined using qRT-PCR.
Metagenomic Sequencing and Bioinformatics
To investigate the possible causative pathogen or pathogens causing this disease, viruses isolated from an intestine and liver homogenate were blindly passed into 9-day-old embryonated goose eggs through allantoic cavity injection. After 10 passages, the allantoic fluid of infected eggs was collected and centrifuged at 8000 g/min for 20 min to remove cell debris. The supernatant was collected and further ultracentrifuged at 45,000 g/min for 2 h, and the pellet was resuspended with PBS and used for viral DNA/RNA extraction. Because the initial identification attempts yielded negative results, we performed a pathogen discovery protocol by using a next-generation sequencing instrument. The viral DNA and RNA were extracted separately using a QIAamp DNA extraction kit and QIAN Viral RNA Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. Concentrations of the extracted RNA were quantified by using a Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA). The extracted samples were used to prepare the libraries by using NEB Next Ultra II RNA Library Prep Kit (NEB, Beijing, China). Similarly, we prepared paired-end DNA sequencing libraries from the DNA samples using a Nextera XT DNA Library Prep Kit (Illumina).
Phylogenetic Analysis
Phylogenetic analysis was conducted based on amino acids sequences of the pegivirus. Multiple alignments were constructed using the ClustalW method of the software MEGA (Version 7). The Genbank accession numbers of Goose pegivirus used in this study were Goose pegivirus1 (MW091548) and Goose pegivirus2 (MW365447). The phylogenetic tree was drawn using the neighbor-joining method of the MEGA program (Version 7.0) with absolute distances following 200 bootstrap replicates (Kumar et al., 2016).
Nested RT-PCR
Primers derived from the E1-E2 region of the GPgV genome (Table 1) were designed. Two sets of nested PCR primers were used, including the outer primer pair with a 1554-base span and the inner primer pair with a 194-base span. We tested 49 geese in poor health from 16 farms randomly in Sichuan Province and the municipality of Chongqing (Table 2). Various tissues of geese and goslings, including the large intestine, small intestine, thymus, spleen and bursa of Fabricius, were combined into pooled samples. The PCR mix containing 0.4 μL of 10 μM mix PCR primer, 5 μL 2 × Taq Master Mix (Vazyme, Nanjing, China) and 0.5 ul template cDNA was brought to 10 ul with Rnase-free water. The thermal cycling program involved an initial denaturation at 94°C for 5 min, followed by 30 cycles, each consisting of denaturation at 94°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 20 s, with a final extension for 10 min at 72°C. The second reaction contained 0.1 μL of the first PCR product as template and in the same condition as first round.
Table 2.
Primers used for the detection of goose virus in this study.
| Primers | Primer sequence (5’-3’) | Size (bp) |
|---|---|---|
| Goose pegivirus F1 | GGTCTACGCCTACAACCAC TTTCTCCTCAATGGCTGTGC |
1554 |
| Goose pegivirus R1 | ||
| Goose pegivirus F2 | CGACAAGGGTGCCTGCTGAG CCGTAGGTCGCATAGGT |
194 |
| Goose pegivirus R2 | ||
| Goose parvovirus F | TATAGATAGCCTCCAACGGG CATATACATCCGACGG |
778 |
| Goose parvovirus R | ||
| Goose circovirus F | CBATTAATAACCCTACCTTTGA GACCAATCAGAACGATGACC |
462 |
| Goose circovirus R | ||
| Goose hemorrhagic polyomavirus F | TGTTGCTGGATGAGAATGGG AACCCGTACTTCCTCAACCT |
275 |
| Goose hemorrhagic polyomavirus R | ||
| Goose astrovirus F1 | ATGAATTRKATTBKYACAGCAG KTCCATCHAYARTACACCA |
277 |
| Goose astrovirus R1 | ||
| Goose astrovirus F2 | GTGAAAACATCTGTGTTCGC GTTTTGGAAGTTGGCATCCC |
179 |
| Goose astrovirus R2 | ||
| Goose reovirus F | GTTCCATTCTGCTCCCCGG CGTCGAACACCATGTCAACC |
634 |
| Goose reovirus R | ||
| Goose coronavirus F | ACTCARWTGAATTTGAAATAYGC TCACAYTTWGGATARTCCCA |
251 |
| Goose coronavirus R | ||
| Goose paramyxovirus F | TCACTCCTCTTGGCGACTC CAAACTGCTGCATCTTCC |
282 |
| Goose paramyxovirus R | ||
| Avian influenza virus F | TTCTAACCGAGGTCGAAAC AAGCGTCTACGCTGCAGTCC |
229 |
| Avian influenza virus R |
RESULTS
Phylogenetic Analysis
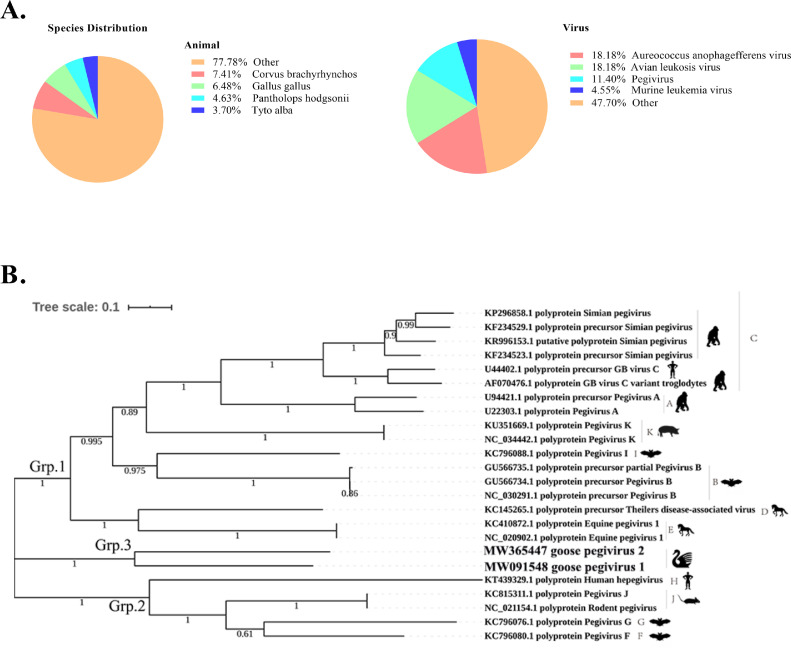
By performing high-throughput sequencing, we obtained a total of 6.79 Mb of raw reads, which were assembled using Trinity software after removing the host sequences (Haas et al., 2013). Contiguous sequences (contigs) were assembled by de novo assembly. Finally, we obtained 202 copies of unigenes. We compared the 202 assembled unigenes with the nr database by using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Considering that an assembled sequence may contain the RNA sequence of the host, the assembled unigenes were also compared with the sequences of animals. The sequencing data showed that the samples contained a great number of avian RNA fragments (Figure 1A). In the sequencing results, many avian and algal viruses were found. We excluded all algal viruses from further analysis. Among the avian viruses, avian leukemia virus may be integrated into the genome as an endogenous retrovirus. Among the viral sequences, 11.7% were from pegiviruses. Statistics are provided in supplement Table 1. The DNA contigs failed to identify any known virus. According to the DNA-based data assembly result annotations, no viral sequences were found.
Figure 1.
(A) Next-generation sequencing was used to identify the taxa represented in the samples. The proportional distributions of animal and viral species are shown. The unigenes were compared with the nr database through BLAST. (B) Phylogenetic analysis of pegiviruses. Maximum-likelihood evolutionary analyses were conducted in MEGA7 (Kumar, Stecher and Tamura, 2016). The analysis was conducted with 24 amino acid sequences of pegivirus polyprotein. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (200 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. GenBank accession numbers are given for reference virus sequences.
The genomic organization of goose pegivirus was similar to those of other pegiviruses: NH2-S-E1-E2-X-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH. The GPgV 5’UTR is longer than the 5′UTRs of other pegiviruses. The conserved GNRZ and PPT motifs were observed in the GPgV 5’UTR. GPgV lacks the microRNA (miR)-122-binding site in the 5′UTR that has been detected in the hepaciviruses. No polyU/UC was identified in the 3′UTR. The GPgV amino acid sequence was submitted to http://www.cbs.dtu.dk/services/, and the signal peptidase sites were found at nt 19, 211, 578 and 650. Of these signalase sites, the 578 site along with the downstream 650 site creates an S protein corresponding to that observed in Theiler's disease-associated virus (TDAV) *, HCV p7, and bat pegvirus (BPgV) X. Many motifs are extremely conserved among members of the Hepacivirus and Pegivirus genera and were also found in GPgV. The NS2 protein of GPgV contains a motif for NS2 cysteine proteases (histidine-1067, glutamic acid-1095 and cysteine-1108), responsible for cleavage at the NS2/NS3 site. The catalytic triad (histidine-1199, aspartic acid-1223, and serine-1281) and RNA helicase that contains the zinc-binding residues (cysteine-1239, cysteine-1286 and histidine-1290) of the N-terminal portion of GPgV NS3, walker A (GSGK at positions 1347-1350) and walker B (DEGH at positions 1434-1438) in its C-terminal are conserved among pegiviruses. GPgV NS5B is 574 aa long and contains functional RNA-dependent RNA polymerase motifs as palm, fingers, and thumb subdomains. There are also 5 conserved motifs in the palm subdomain (A to E) in GPgV compared to those in HCV. GPgV motif A contains the D-X4-D region (DATCFD), motif B contains GX2TTX3N (GVLTTSSSN), and motif C contains the highly conserved GDD active site (GDD). Phylogenetic analysis of the complete coding regions of 2 identified GPgV strains revealed that the Pegivirus genus was divided into 2 groups (group 1 and group 2), whereas GPgV formed group 3 and did not cluster with any known pegiviruses (Figure 1B).
Virus Pathogenicity
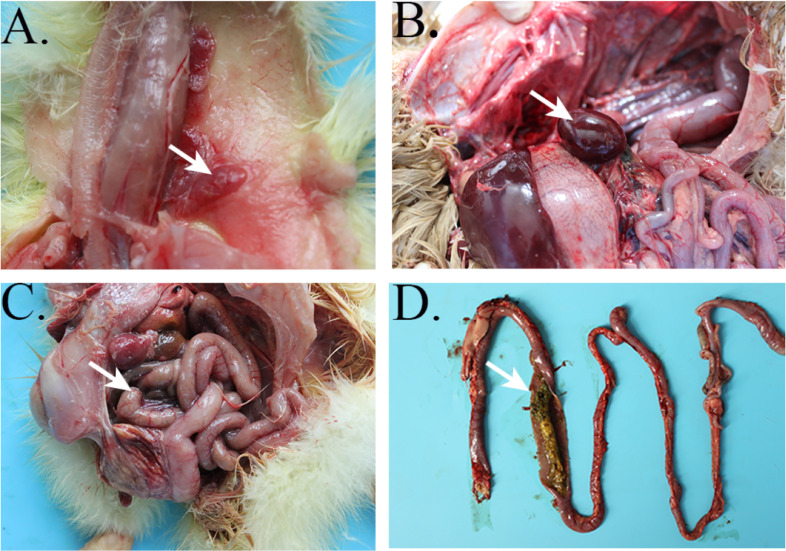
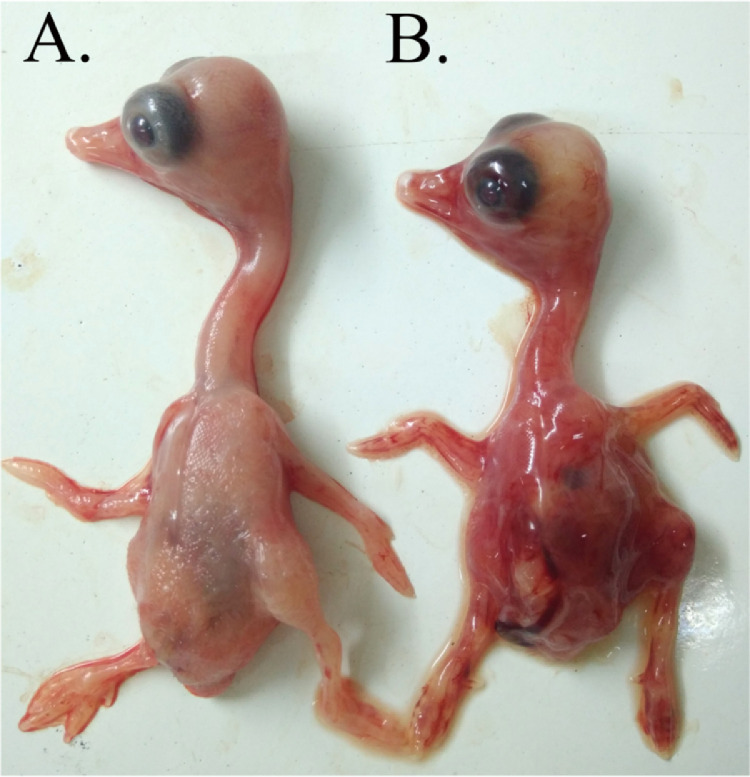
Anatomical analysis revealed that the infected geese had similar and typical visible lesions in the thymus, spleen, and intestine. In the dead goslings, the thymus was enlarged and red with congestion (Figure 2A). The spleen showed serious splenectasis and was reddish brown with congestion (Figure 2B). Most of the intestinal tract exhibited embolism, and ecchymosis hemorrhages were visible (Figure 2C and 2D). The isolated GPgVs were purified and passaged for further study. Viruses from the 9th passage were used to infect goose embryos, which showed severe retardation and cutaneous hemorrhages, in contrast to embryos in the PBS group (Figure 3A and 3B).
Figure 2.
Gross findings of tissues of diseased geese. (A) Thymus with hyperemia and hemorrhage. (B) A dead goose with swollen spleen. (C, D) Intestine showing swelling and embolism.
Figure 3.
Goose embryos infected with GPgV. (A) Mock-infected embryos and (B) GPgV-infected embryos. Goose embryos were inoculated with 0.2 mL of allantoic fluid from the fifth passage of the virus. Severe retardation and cutaneous hemorrhages were observed in the embryos at 5 d postinoculation.
Epidemiological Survey
Coinfection of people with human immunodeficiency virus (HIV) and human pegivirus can reduce HIV disease (Lefrere et al., 1999; Nunnari et al., 2003; Tillmann et al., 2001; Williams et al., 2004). But, simian pegivirus preinfection does not reduce pathological immune activation during acute simian immunodeficiency virus infection (Bailey et al., 2017). We want to know which goose virus is coinfected with GPgV in the case of natural infection, so as to provide ideas for subsequent coinfection research. We investigated the GPgV epidemic in southwest China, nested RT-PCR for GPgV detection was conducted. GPgV RNA was detected in 12 of 24 goslings (50%) from Chongqing farms and in 10 of 25 goslings (40%) from Sichuan farms; these results were confirmed by sequencing. To better understand the coinfection of this pegivirus with other viruses, we tested for most types of viruses that can infect geese, including goose parvovirus, circovirus, reovirus, hemorrhagic polyomavirus, astrovirus, coronavirus, paramyxovirus and avian influenza virus (the primers used are listed in Table 2). All the geese had been injected with goose parvovirus egg yolk antibody before. The infection rates of goose parvovirus, circovirus and astrovirus were 36.7%, 38.8% and 26.5%, respectively. The tests for other viruses were negative. The infection rate of GPgV was 44.9%, with 36.7% coinfections and 8.2% mono-infections. The highest rate of coinfection of GPgV was with goose parvovirus, at 24.5% (Table 1).
DISCUSSION
The International Committee on the Taxonomy of Viruses indicate that Pegivirus species are phylogenetically distinct from the assigned genera within the Flaviviridae (Hepacivirus, Flavivirus and Pestivirus), forming a separate monophyletic cluster based on analysis of homologous genes (Smith et al., 2016a; Stapleton et al., 2011). Here, we report a goose pegivirus that is phylogenetically distinct from all present pegiviruses, thus forming the third clade/phylogroup. The genetic distance and nucleotide identities of the concatenated coding genes of goose pegivirus suggested that it is a new species, Pegivirus M.
GB-agent was originally identified as a coinfection pathogenesis (Williams et al., 2004). According to our epidemiological investigation data, a high ratio of coinfection in pegivirus-positive birds was shown. This finding indicated that pegivirus may play a role in the progression of infectious disease, secondary infection or inflammatory mechanisms. The etiology, pathobiology, and epidemiology of pegivirus were largely unknown because of a lack of experimental animal models and in vitro culture systems. For the first time, a pegivirus was identified in a nonmammal, a bird, which is important for understanding the evolution and epidemic of Pegivirus. On the other side, GPgV may cause damage to immune organs. Our epidemiological data revealed a very high rate of coinfection in pegivirus-positive birds. Additional attention should be given to this emerging pathogen.
Acknowledgments
ACKNOWLEDGMENTS
This work was funded by grants from, the National Key Research and Development Program of China (2017YFD0500800), the Sichuan-International Joint Research for Science and Technology (2018HH0098), the China Agricultural Research System (CARS-42-17), and the Program Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (SCCXTD-2020-18).
DISCLOSURES
This manuscript has not been simultaneously submitted for publication in another journal and been approved by all co-authors. The authors declare that they do not have any conflict of interest.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2021.101251.
Appendix. Supplementary materials
REFERENCES
- Baechlein C., Grundhoff A., Fischer N., Alawi M., Hoeltig D., Waldmann K.H., Becher P. Pegivirus infection in domestic pigs, Germany. Emerg. Infect Dis. 2016;22:1312–1314. doi: 10.3201/eid2207.160024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A.L., Buechler C.R., Matson D.R., Peterson E.J., Brunner K.G., Mohns M.S., Breitbach M., Stewart L.M., Ericsen A.J., Newman C.M., Koenig M.R., Mohr E., Tan J., Capuano S., 3rd, Simmons H.A., Yang D.T., O'Connor D.H. Pegivirus avoids immune recognition but does not attenuate acute-phase disease in a macaque model of HIV infection. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg M.G., Lee D., Coller K., Frankel M., Aronsohn A., Cheng K., Forberg K., Marcinkus M., Naccache S.N., Dawson G., Brennan C., Jensen D.M., Hackett J., Jr., Chiu C.Y. Discovery of a novel human pegivirus in blood associated with hepatitis C virus co-infection. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandriani S., Skewes-Cox P., Zhong W., Ganem D.E., Divers T.J., Van Blaricum A.J., Tennant B.C., Kistler A.L. Identification of a previously undescribed divergent virus from the Flaviviridae family in an outbreak of equine serum hepatitis. PNAS. 2013;110:E1407–E1415. doi: 10.1073/pnas.1219217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M., MacManes M.D., Ott M., Orvis J., Pochet N., Strozzi F., Weeks N., Westerman R., William T., Dewey C.N., Henschel R., LeDuc R.D., Friedman N., Regev A. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Simmonds P., Cullen J.M., Scheel T.K., Medina J.L., Giannitti F., Nishiuchi E., Brock K.V., Burbelo P.D., Rice C.M., Lipkin W.I. Identification of a pegivirus (GB virus-like virus) that infects horses. J. Virol. 2013;87:7185–7190. doi: 10.1128/JVI.00324-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Simmonds P., Scheel T.K., Hjelle B., Cullen J.M., Burbelo P.D., Chauhan L.V., Duraisamy R., Sanchez Leon M., Jain K., Vandegrift K.J., Calisher C.H., Rice C.M., Lipkin W.I. Identification of rodent homologs of hepatitis C virus and pegiviruses. mBio. 2013;4 doi: 10.1128/mBio.00216-13. e00216-00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrere J.J., Roudot-Thoraval F., Morand-Joubert L., Petit J.C., Lerable J., Thauvin M., Mariotti M. Carriage of GB virus C/hepatitis G virus RNA is associated with a slower immunologic, virologic, and clinical progression of human immunodeficiency virus disease in coinfected persons. J. Infect. Dis. 1999;179:783–789. doi: 10.1086/314671. [DOI] [PubMed] [Google Scholar]
- Nunnari G., Nigro L., Palermo F., Attanasio M., Berger A., Doerr H.W., Pomerantz R.J., Cacopardo B. Slower progression of HIV-1 infection in persons with GB virus C co-infection correlates with an intact T-helper 1 cytokine profile. Ann Intern Med. 2003;139:26–30. doi: 10.7326/0003-4819-139-1-200307010-00009. [DOI] [PubMed] [Google Scholar]
- Quan P.L., Firth C., Conte J.M., Williams S.H., Zambrana-Torrelio C.M., Anthony S.J., Ellison J.A., Gilbert A.T., Kuzmin I.V., Niezgoda M., Osinubi M.O., Recuenco S., Markotter W., Breiman R.F., Kalemba L., Malekani J., Lindblade K.A., Rostal M.K., Ojeda-Flores R., Suzan G., Davis L.B., Blau D.M., Ogunkoya A.B., Alvarez Castillo D.A., Moran D., Ngam S., Akaibe D., Agwanda B., Briese T., Epstein J.H., Daszak P., Rupprecht C.E., Holmes E.C., Lipkin W.I. Bats are a major natural reservoir for hepaciviruses and pegiviruses. PNAS. 2013;110:8194–8199. doi: 10.1073/pnas.1303037110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.B., Becher P., Bukh J., Gould E.A., Meyers G., Monath T., Muerhoff A.S., Pletnev A., Rico-Hesse R., Stapleton J.T., Simmonds P. Proposed update to the taxonomy of the genera Hepacivirus and Pegivirus within the Flaviviridae family. J. Gen. Virol. 2016;97:2894–2907. doi: 10.1099/jgv.0.000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.B., Simmonds P., Izopet J., Oliveira-Filho E.F., Ulrich R.G., Johne R., Koenig M., Jameel S., Harrison T.J., Meng X.J., Okamoto H., Van der Poel W.H., Purdy M.A. Proposed reference sequences for hepatitis E virus subtypes. J. Gen. Virol. 2016;97:537–542. doi: 10.1099/jgv.0.000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton J.T., Foung S., Muerhoff A.S., Bukh J., Simmonds P. The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J. Gen. Virol. 2011;92:233–246. doi: 10.1099/vir.0.027490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillmann H.L., Heiken H., Knapik-Botor A., Heringlake S., Ockenga J., Wilber J.C., Goergen B., Detmer J., McMorrow M., Stoll M., Schmidt R.E., Manns M.P. Infection with GB virus C and reduced mortality among HIV-infected patients. N. Engl. J. Med. 2001;345:715–724. doi: 10.1056/NEJMoa010398. [DOI] [PubMed] [Google Scholar]
- Williams C.F., Klinzman D., Yamashita T.E., Xiang J., Polgreen P.M., Rinaldo C., Liu C., Phair J., Margolick J.B., Zdunek D. Persistent GB virus C infection and survival in HIV-infected men. N. Engl. J. Med. 2004;350:981. doi: 10.1056/NEJMoa030107. [DOI] [PubMed] [Google Scholar]
- Wu Z., Wu Y., Zhang W., Merits A., Simmonds P., Wang M., Jia R., Zhu D., Liu M., Zhao X., Yang Q., Wu Y., Zhang S., Huang J., Ou X., Mao S., Liu Y., Zhang L., Yu Y., Tian B., Pan L., Rehman M.U., Chen S., Cheng A. The first nonmammalian pegivirus demonstrates efficient in vitro replication and high lymphotropism. J. Virol. 2020;94 doi: 10.1128/JVI.01150-20. e01150-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.