Abstract
Amyloid formation by amylin contributes to β-cell dysfunction in type-2 diabetes. The features which control the amyloidogenicity and toxicity of amylin are not understood. Not all species form islet amyloid and its presence or absence correlates with the in vitro behavior of the polypeptide. Rats do not develop type-2 diabetes or islet amyloid and rat amylin is non-amyloidogenic, except at very high concentrations. This has led to the notion that rodent amylins are non-amyloidogenic. Prairie vole amylin has an unusual sequence compared to human and rat amylin, including non-conservative Lys-1 to Glu and Asn-22 to Gly substitutions. The first reduces the net charge on the peptide, while the second disrupts a potential network of sidechain hydrogen bonds in the amyloid fiber, a so called Asn ladder. The prairie vole polypeptide forms amyloid more slowly than human amylin and is considerably less cytotoxic. An Asn-22 to Gly substitution in human amylin significantly reduces toxicity, increasing the effective concentration of amylin required to reach 50% toxicity by more than 7-fold, but has modest effects on the time to form amyloid. A Lys-1 to Glu replacement has less of an effect, but does reduce toxicity relative to human amylin, without having significant impact on the time to form amyloid. The effect of the Lys-1 Glu substitution on amyloid kinetics is more significant in Tris buffer than in phosphate buffered saline. The work demonstrates that the N-terminus of amylin plays a role in modulating toxicity and highlights the key role of position 22.
Keywords: Amylin, Amyloid, IAPP, Islet amyloid polypeptide, Type-2 Diabetes, Prairie Vole
Graphical Abstract
INTRODUCTION
Amylin, also known as islet amyloid polypeptide (IAPP), is a 37-residue pancreatic polypeptide hormone which is produced in the pancreatic β-cells and co-secreted with insulin (1–3). Human amylin (h-amylin) forms islet amyloid in type-2 diabetes and toxic species produced during amyloidogenesis lead to β-cell dysfunction and death (1, 2, 4–9). Not all species form islet amyloid and not all organisms develop type-2 diabetes (1, 10–13). To date there is a one-to-one correlation between the observation of type-2 diabetes and the presence of islet amyloid. For example, mice and rats do not form islet amyloid in vivo nor do they develop type-2 diabetes and mouse/rat amylin is non-amyloidogenic in vitro except at very high concentrations. The only known example of a rodent developing islet amyloid is the Degu, a South American rodent. However, Degu amyloid is composed of insulin rather than amylin (1). These observations have led to the view that rodent amylins are non-amyloidogenic and the reasons for the reduced amyloidogenicity of rat/mouse amylin have been investigated. The sequences of rat and mouse amylin are identical and differ from human amylin at only 6 positions (Figure 1). These include three prolines at residues 25, 28, and 29 and the replacement of His-18 in h-amylin with Arg in rat/mouse amylin. The three proline substitutions have been incorporated into h-amylin to produce a non-amyloidogenic form of the polypeptide, known as pramlintide, which is used clinically (14, 15).
Figure 1:
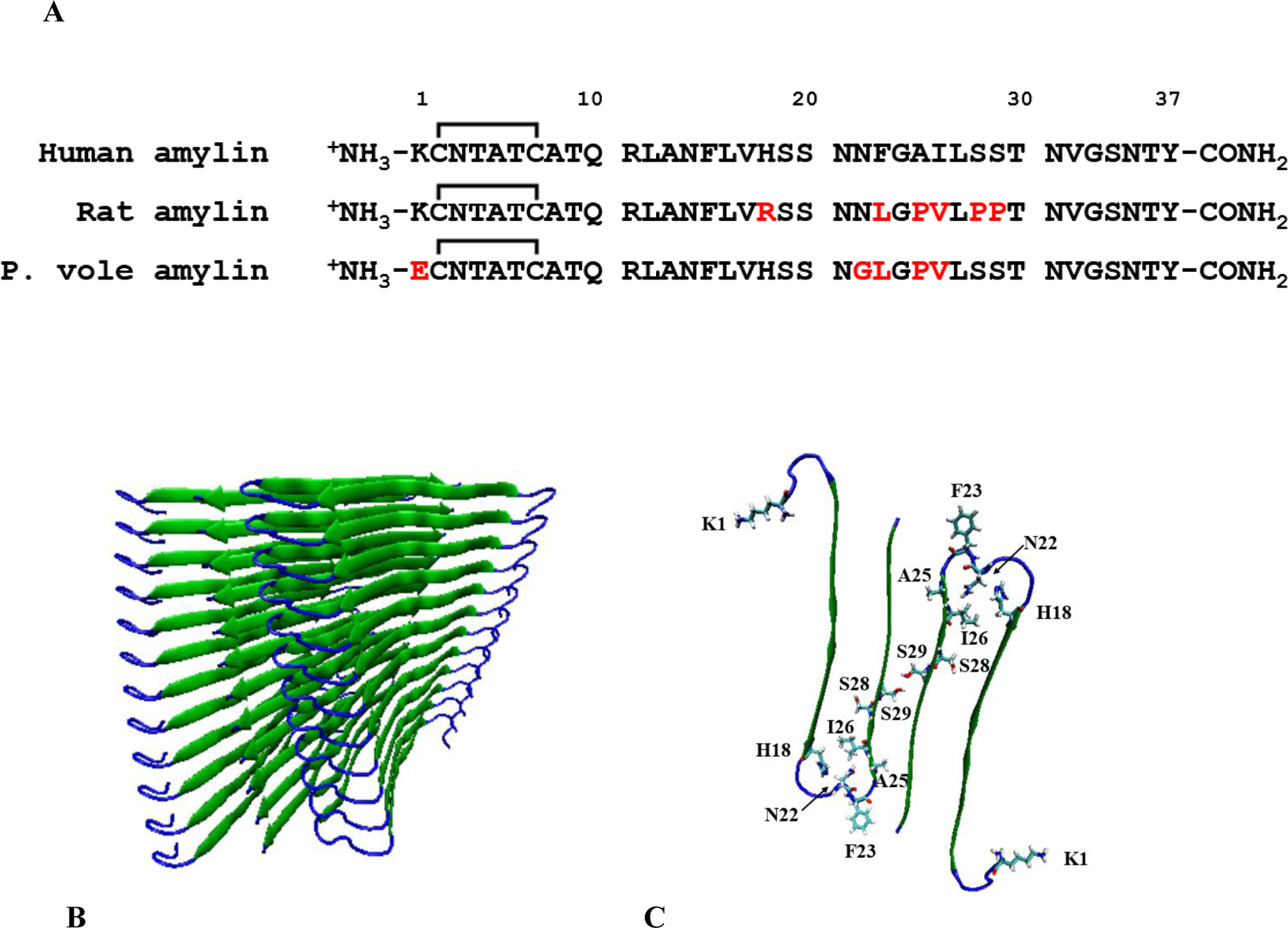
(A) The sequence of human, rat and prairie vole amylin. Residues which differ than human amylin are color-coded red. (B) A model of the human amylin amyloid fiber derived from x-ray structures of small peptides derived from human amylin (16). (C) One layer of the structure showing the residues in human amylin which differ in prairie vole amylin. The location of His-18, Ser-28 and Ser-29 are also shown.
Here we examine amylin sequences from different rodents in order to search for factors that might modulate amyloidogenicity and toxicity. One sequence, that of prairie vole (p. vole, Microtus Ochrogaster), has unusual features including the substitution of Lys to Glu at the N-terminus. Residue one is a basic residue in h-amylin and in all of the known rodent sequences except for the p. vole sequence (Figure 2). The p. vole sequence also contains a very unusual Asn-22 to Gly replacement. Position 22 is Asn in h-amylin and in all but one other rodent sequence, where it is replaced by Ser (Figure 2). We demonstrate that the p. vole sequence forms amyloid and is cytotoxic under conditions where h-amylin is amyloidogenic and rat amylin is not, but also show that p. vole amylin is significantly less cytotoxic than h-amylin and is slower to form amyloid. Analysis of the consequences of substituting residues from p. vole amylin into h-amylin produces new insight into the role of the N-terminus of amylin in amyloid formation and toxicity, and highlights the importance of position 22 in modulating toxicity. The work also reveals that the sequence features which control amyloidogenicity can differ significantly from those that modulate toxicity.
Figure 2:

Sequence alignment of known rodent amylin sequences. Human amylin is included for comparison. Residues which differ from human amylin are colored red. Only a partial sequence has been reported for the western European hedgehog.
MATERIALS AND METHODS
Peptide Synthesis and Purification.
Human amylin, p. vole amylin, K1E h-amylin, and N22G h-amylin were synthesized using a CEM Liberty microwave peptide synthesizer on a 0.01 mmol scale with 9-fluorenylmethyloxycarbonyl (Fmoc) protected amino acids. Pseudoproline dipeptide (oxazolidine) derivatives of Ala-Thr and Leu-Ser were used as previously described (17, 18). 5-(4’-Fmoc-aminomethyl-3’,5’-dimethoxyphenol) valeric acid (PAL-PEG) resin was used to afford an amidated C-terminus. All β-branched residues, pseudoproline dipeptides, and the first amino acid (Tyr) attached to the resin were double coupled. Peptide coupling reactions were carried out for 2 minutes at 90 °C, except for Cys and His which were coupled at 55 °C. Peptides were cleaved using a cleavage solution of trifluoroacetic acid (TFA) (5.5 mL), triisopropylsilane (TIPS) (150 μL), 3,6-dioxa-1,8-octanedithiol (DODT) (150 μL), and H2O (150 μL). The peptides were stirred in the cleavage solution at room temperature for 3 hours, the volume reduced with N2 gas and crude peptide precipitated as a white solid using cold diethyl ether. Cleaved peptides were dissolved in 15% (v/v) acetic acid and lyophilized. The internal disulfide bond between residues 2 and 7 was formed by dissolving the dry peptide in 100% dimethyl sulfoxide (DMSO) at a concentration of 10 mg/mL and incubating at room temperature for 3 days. Peptides were purified using reverse-phase high performance liquid chromatography (HPLC) with a Higgins Analytical Proto 300 C18 preparatory column. Residual scavengers were removed using a 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) extraction procedure. Purified peptide was dissolved in neat HFIP to a concentration of 10 mg/mL, the solution was incubated at room temperature for 4 hours and then filtered using a 0.45 μM GHP membrane filter, and finally purified via HPLC. The mass of purified h-amylin, p. vole amylin, K1E h-amylin, and N22G h-amylin were measured using ESI MS; h-amylin expected: 3903.6, observed: 3902.9; p. vole amylin expected: 3825.2, observed 3825.2; K1E h-amylin expected: 3904.3, measured: 3903.8; N22G h-amylin expected: 3846.3, measured: 3845.8.
Peptide Stock and Sample Preparation.
Stock solutions were prepared by dissolving lyophilized peptide into 100% hexafluoroisopropanol (HFIP) to a target concentration of 1.6 mM. The concentration was determined by absorbance measurements at 280 nm, using an extinction coefficient of 1600 M−1cm−1 for all peptides as each contains a signal Tyr and no Trp. Aliquots for kinetic experiments were lyophilized for at least 12 hours prior to reconstitution.
Thioflavin-T Fluorescence Assays.
Aliquots of the HFIP stock solutions were lyophilized overnight and reconstituted in Tris buffer or PBS at pH 7.4 for kinetic assays. The final concentrations of the peptides and thioflavin-T were 16 μM and 32 μM, respectively. Black Corning 96-well nonbinding plates were used for thioflavin-T monitored kinetic assays. Plates were sealed with polyethylene sealing tape, and a clear lid was used. Wells at the edge of the plate were not used, but unused wells were filled with buffer. Fluorescence measurements were taken using a Beckmann Coulter DTX 880 plate reader and a Spectramax Gemini EM plate reader, with a multimode detector using an excitation wavelength of 450 nm and an emission of 485 nm. Readings were taken from the bottom of the wells at 10-minute intervals without agitation. Three to six parallel samples were tested for each experiment and average T50 values are reported. The uncertainties are the estimated standard deviations. All fluorescence assays were performed at 25 °C.
Sequence Alignment.
Rodent amylin sequences were retrieved from the NCBI data base. Many sequences are listed as the pro-form. The mature sequences were selected based upon the known prohormone processing sites, and the striking conserved disulfide bond and C-terminal aromatic residue.
Transmission Electron Microscopy.
Transmission electron microscopy (TEM) images were recorded at the Central Microscopy Imaging Center at Stony Brook University. 15 μL of peptide solutions were collected from sample wells at the end of the thioflavin-T binding kinetic assays and blotted on a carbon-coated Formvar 300-mesh copper grid for 1 minute and then negatively stained with 2% uranyl acetate for 1 minute.
Cytotoxicity Assays and Measurement of EC50 Values.
INS-1 cells were purchased from AddexBio and cultured with optimized RPMI-1640 (AddexBio, #C0004–02) medium supplemented with 10% ultra-low IgG FBS (Gibco, #16250078). CellTox Green (Promega, # G8741) and CellTiter-Glo 2.0 (Promega, #G9242) assays were used to evaluate the cytotoxicity of h-amylin, h-amylin mutants, and p. vole amylin towards INS-1 cells. Cells were seeded at ~50 % confluence in 96-well half-area clear bottom white plates (Greiner, #675083) and incubated for 36 hrs in a 5 % CO2 humidified incubator at 37 °C. Serial dilutions of the peptides were freshly prepared before use from lyophilized aliquots. Cells were then exposed to varying concentrations of peptide diluted in fresh complete medium for 24 hours. For the CellTox Green assay the cells were exposed to the peptide in presence of the assay dye (1:5000 dilution) for 24 hours before fluorescence intensity was read at 480 nm excitation with 525 nm emission using a Clariostar plate reader. For the CellTiter-Glo 2.0 assays, culture plates were cooled to room temperature and then an equal volume of the assay reagent was added. The plates were shaken for 1 min at 700 rpm and the luminescence intensity was read. Statistical analysis and calculation of EC50 values were conducted with Graph Pad Prism 5.
In Situ Photochemical Cross-Linking Experiments.
Peptides were cross-linked using tris (bipyridyl) Ru(II) chloride (Ru(bpy)3), in the presence of ammonium persulfate (APS). Samples were dissolved in PBS buffer at pH 7.4, and were centrifuged for 10 minutes at 17,800 g before the reaction. In a typical reaction, 2.5 µL of Ru(bpy)3 and APS were added to a 15 µL aliquot of 40 µM peptide, and then irradiated for 10 s with a 140W incandescent bulb. The ratios of Ru(bpy)3, APS, and peptide were 3.5 : 70 : 1, respectively. Samples were quenched using 10 µl of Tricine sample buffer containing 5 % β-mercaptoethanol. The samples were then heated for five minutes at 85 °C before loading on the gel. Silver staining was used to visualize the gels (19).
RESULTS
Figure 2 displays a sequence alignment of known rodent amylin sequences and includes h-amylin for comparison. Rodent amylins, like all other reported amylin sequences, contain several conserved features. All known amylin sequences have a disulfide bridge between residues two and seven and an amidated C-terminal tyrosine. P. vole amylin is unique in that it contains a negatively charged Glu at the first position, whereas all other rodent sequences and h-amylin contain a Lys or an Arg. The Lys to Glu substitution will reduce the net charge on the polypeptide from between 2 to 4 to between 0 to 2 at physiological pH depending on the exact pKa of His-18 and the N-terminus (20). Most rodent sequences contain an Arg at position 18, however, p. vole amylin, like h-amylin, contains a His. Replacement of His-18 in h-amylin with Arg has been shown to lead to a reduction in the rate of amyloid formation and toxicity (20, 21). The p. vole polypeptide contains a single proline at position 25 while rat/mouse amylin contains three at positions 25, 28 and 29 (Figure 1). The p. vole sequence also includes an unusual Asn to Gly replacement at residue 22. Asn residues have been proposed to play a role in stabilizing amyloid fibers by forming so called “Asn ladders” that lead to a network of sidechain-sidechain hydrogen bonds along the long axis of the fiber. Asn-22 is part of the partially ordered loop linking the two β-strands, but is not part of the core β-sheet structure in either of the high resolution models of the h-amylin amyloid fiber (16, 22). The effect of the Lys-1 to Glu and Asn-22 to Gly substitutions have not been reported, indeed there is very little work on the potential role of the N-terminal region of amylin. Consideration of the differences between rat/mouse and p. vole amylin led us to examine the ability of p. vole amylin to form amyloid in vitro and its effect on cultured β-cells, and to test the consequences of the Lys-1 to Glu and Asn-22 to Gly substitutions in the context of h-amylin.
We first analyzed the sequence of human, prairie vole, and rat amylin using several popular amyloid prediction programs. These include TANGO, PASTA, ZipperDB, Aggrescan, and Waltz (23–27). All predict that human amylin is the most amyloidogenic polypeptide. Aggrescan, TANGO, and ZipperDB predict that p. vole is more amyloidogenic than rat amylin while WALTZ and PASTA predict that rat and p. vole amylin have similar amyloidogenicity (Supporting Tables S1–S4, Supporting Figure S1).
Prairie vole amylin forms amyloid in vitro.
We monitored amyloid formation using fluorescence detected thioflavin-T binding assays. Thioflavin-T is a small molecule whose quantum yield increases upon binding to amyloid fibers. The dye offers a simple probe of amyloid formation and has been widely used in studies of amyloid formation by amylin (28, 29). Thioflavin-T is an extrinsic probe and there are examples of thioflavin-T assays leading to false negative results and there are examples of thioflavin-T binding to non-amyloid structures, thus we also use transmission electron microscopy (TEM) to directly confirm the presence or absence of amyloid (30).
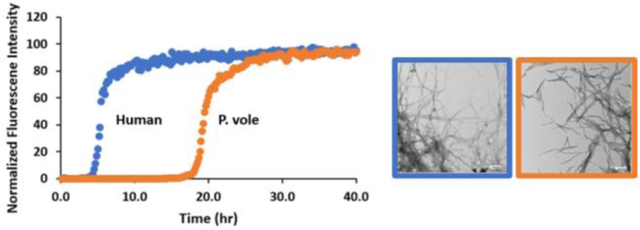
We first examined the two polypeptides in phosphate buffered saline (PBS, 10mM phosphate 140mM KCl). PBS has been widely employed in biophysical studies of amylin amyloid formation. T50, the value required to reach 50% of the final intensity in a thioflavin experiment is commonly used to compare relative times of amyloid formation. P. vole amylin forms amyloid in PBS (pH 7.4, 25 °C), but does so approximently four fold more slowly than h-amylin with a T50 of 19.2 ± 0.4 (hours) vs 5.1 ± 0.2 (hours) for h-amylin (Figure 3, Table 1, supporting Figure S2). TEM images confirm the presence of amyloid fibers for both polypeptides. The data in figure-3 is plotted as normalized fluorescence intensity for ease of comparison. Un-normalized data from multiple measurements are shown in the supporting information (Supporting Figure S2). The final thioflavin-T intensities are all similar to each other for a given peptide, and between peptides, although the p vole peptide shows higher final intensity (supporting information). The final thioflavin-T intensity cannot be directly interpreted in terms of the amount of amyloid formed since it depends on the affinity of the dye for different fibers and the quantum yield of the bound dye (which can vary between fibers). In addition, the lateral association of fibers, which can differ between fibers formed by different peptides, can occlude thioflavin-T binding sites.
Figure 3:

h-Amylin and p vole-amylin p.vole amylin form amyloid. (A) Thioflavin-T assays of amyloid formation conducted for wild type h-amylin (Blue) and p. vole amylin (Orange) in PBS buffer (pH 7.4, 10 mM with 140 mM KCl). The vertical axis was normalized to 100 for both peptides. Representative curves are shown and non-normalized data is shown in the supporting information. (B) TEM images recorded for samples collected at the end of the kinetic experiments: wild type h-amylin (Blue) and p. vole amylin (Orange). Scale bars represent 100 nm. (C) Thioflavin-T assays of amyloid formation conducted with wild type h-amylin (Blue) and p. vole amylin (Orange) in Tris buffer (pH 7.4, 20 mM) representative curves are shown. (D) TEM images recorded for samples collected at the end of the kinetic experiments: wild type h-amylin (Blue) and p. vole amylin (Orange). Scale bars represent 100 nm.
Table 1:
Summary of T50 values measured in PBS and Tris buffer at pH 7.4, 25°C, 16µM peptide. Quoted uncertainties are the apparent standard deviations.
| Peptide | T50 (hrs) |
|---|---|
| h-amylin in PBS | 5.1 ± 0.2 |
| p. vole amylin in PBS | 19.2 ± 0.4 |
| K1E h-amylin in PBS | 4.4 ± 0.2 |
| N22G h-amylin in PBS | 8.0 ± 2.0 |
| h-amylin in Tris | 19.3 ± 0.8 |
| p. vole in Tris | 178.8 ± 15.5 |
| K1E h-amylin in Tris | 7.8 ± 0.3 |
We next conducted experiments in Tris buffer as it has also been widely used in biophysical studies of amylin and because the rate of amyoid formation by h-amylin is known to be slower in Tris relative to PBS (20). h-Amylin and p. vole amylin both form amyloid in Tris as judged by thioflavin-T assays and TEM (Figure 3, Table 1). The difference in the T50 values of p. vole amylin and h-amylin are larger in Tris than in PBS. The T50 of p. vole amylin in Tris is 178.8 ± 15.5 (hours) which is 9 fold larger than the value for h-amylin in Tris, 19.3 ± 0.8 hours (Table 1). TEM images of both peptides reveal dense matts of amyloid fibers. Rat/mouse amylin does not form amyloid under these conditions for at least 10 days. The slower rate of amyloid formation in Tris vs PBS is consistent with prior reports and likely arises because phosphate binds to fibers and/or pre-fiber intermediates and promote amyloid formation (20, 30, 31).
Prairie vole amylin is considerably less cytotoxic than human amylin.
We next compared the cytotoxicity of p. vole and h-amylin using INS-1 β-cells, a standard pancreatic cell line. Two independent assays were used, one monitors cellular ATP levels (the CellTiter-Glo assay) and the second is an assay which monitors permeabilization of the cell membrane to a small dye (CellTox Green). The p. vole polypeptide is considerably less cytotoxic than h-amylin. The EC50 values for h-amylin, the peptide concentration required to achieve 50% effect, were found to be 40.0 ± 1.9 μM using the assay which monitors ATP production and 32.6 ± 2.8 μM via the cell permeabilization assay. The EC50 values for p. vole amylin were 7.0 to 8.2-fold larger: 341± 8.9 μM as judged by the ATP assay and 235.8 ± 19 μΜ as measured using the permeability assay (Figure 4, Table 2). The different EC50 values obtained for the two assays applied to the same amylin peptide demonstrate that detectable membrane permeabilization occurs at peptide concentrations where no significant change in ATP level is observed. This is consistent with prior studies of h-amylin (31).
Figure 4:

Cell viability assays for wild type h-amylin (Blue), wild type p. vole amylin (Orange), K1E h-amylin (Red) N22G h-amylin (Green). (A) The effect of the different amylin variants on cellular ATP levels as measured using the CellTiter-Glo assay. (B) The effect of the different amylin variants on membrane permeability as measured using the CellTox Green assay. The X-axis in both plots is plotted on a logarithmic scale.
Table 2:
EC50 values determined for human and p. vole amylin and human amylin variants.
| CellTiter-Glo | CellTox Green | |||
|---|---|---|---|---|
| EC50 (μM) | ±SEM | EC50 (μM) | ±SEM | |
| h-amylin | 40.0 | 1.9 | 32.6 | 2.8 |
| p. vole amylin | 341.3 | 8.9 | 235.8 | 19.0 |
| K1E h-amylin | 100.1 | 2.4 | 79.0 | 9.2 |
| N22G h-amylin | 302.5 | 14.0 | 190.9 | 20.3 |
Analysis of the consequences of the Lys-1 to Glu substitution and the Asn-22 to Gly substitution in h-amylin.
The Glu-1 and Gly-22 substitutions found in p. vole amylin are unusual compared to other rodent sequences and to h-amylin, and both represent non-conservative changes. Thus, we examined the consequences of the Glu-1 and Asn-22 replacements in the context of wild type human amylin. The N-terminal 7 residues of h-amylin are thought to be outside of the ordered cross β-sheet core of the amylin amyloid fiber as the disulfide between residue 2 and 7 constrains this portion of the chain into a conformation which is not compatible with β-sheet structure. Asn-22 is in the loop region which links the two β-strands in the high-resolution models of the h-amylin amyloid fiber. There are limited studies of the consequences of substitutions in the N-terminal region of amylin, although a 2–37 variant which lacks Lys-1 has been studied (31). Thioflavin-T assays and TEM studies show that K1E h-amylin forms amyloid on a similar time scale to wild type h-amylin in PBS, but does so more rapidly in Tris (Figure 5, Table 1, Supporting Figure S3). In PBS the T50 value of the K1E variant is 4.4 ± 0.2 hours and is 5.1 ± 0.2 hours for wild type (Figure 5). The T50 for the K1E h-amylin in Tris buffer is 7.8 ± 0.3 hours compared to the value of 19.3 ± 0.8 hours for wild type h-amylin (Supporting Figure S3). The fact that the relative rate of amyloid formation by wild type h-amylin and K1E h-amylin are similar in PBS, but differ in Tris, highlights the difficulties in objectively defining the amyloidogenicity of a polypeptide since the concept is dependent upon solution conditions.
Figure 5:

(A) Thioflavin-T assays of amyloid formation conducted for wild type h-amylin (Blue), wild type p. vole amylin (Orange), K1E h-amylin (Red), and N22G h-amylin (Green) in PBS buffer (pH 7.4, 10 mM with 140 mM KCl). The vertical axis is normalized to 100. Representative curves are shown. (B) TEM images recorded for samples collected at the end of the kinetic experiments: wild type h-amylin (Blue), wild type p. vole amylin (Orange), K1E h-amylin (Red), and N22G h-amylin (Green). Scale bars represent 100 nm.
Cell viability assays indicate that the K1E variant is less cytotoxic than wild type h-amylin with an EC50 of 100.1 ± 2.4 μM, determined via monitoring ATP production and 79.0 ± 9.2 μM from the permeabilization assay. In both cases, the EC50 values are approximately 2.5 fold larger than measured for wild type h-amylin. These results indicate that the Glu replacement contributes to the reduced toxicity of p. vole amylin, but has complicated effects on the time scale of amyloid formation.
We next examined the effect of the Asn-22 to Gly substitution in the context of h-amylin. This residue takes part in an Asn ladder in the model of the h-amylin amyloid fiber based on peptide crystal structures, but is not part of either of the β-strands. The mutant peptide forms amyloid as judged by thioflavin-t assays and TEM measurements, with a T50 (8.0 ± 2.0 hours) that is only slightly larger than the T50 of h-amylin in PBS (Figure 5). In striking contrast, the substitution dramatically reduces the cytotoxicity of amylin towards cultured β-cells. The EC50 value of the N22G variant measured using the ATP levels is 302.5 ± 14.0 μM and is 190.9 ± 20.3 μM based on the cell membrane permeabilization assay (Table 2). These values of EC50 are 6 to 7.5 larger than those measured for h-amylin.
Finally, we tested if p. vole amylin and the human mutants form oligomers, using the in situ photochemical cross-linking method introduced by Teplow et.al (19). Amyloid formation by wild type h-amylin is well established to proceed with the formation of low order oligomers. Analysis of the results of the in situ cross-linking studies show that h-amylin, p. vole amylin, and the h-amylin mutants all form oligomers, and up to hexamers can be detected for each peptide (Supporting Figure S4).
CONCLUSIONS
The data presented here provides an example of an amyloidogenic rodent amylin and thus shows that not all rodent amylins are non-amyloidogenic. To the best of our knowledge, there are no reported studies designed to detect islet amyloid in the p. vole, nor is it known if the p. vole develops type-2 diabetes, hence it is not possible to deduce if the p. vole peptide represents a breakdown in the correlation between the in vitro amyloidogenicity of amylin and the propensity to develop diabetes. This work highlights the unanticipated role of the extreme N-terminus in amylin cytotoxicity and the unexpected role of residue 22.
The analysis of the Glu-1 variant of human amylin provides further evidence that there is not a direct one-to-one relationship between the net charge of the polypeptide and the rate of amyloid formation, since the K1E replacement reduces the net charge by 2 units, but amyloid formation in PBS occurs on the same time scale as wild type h-amylin.
The dependence of the time scale of amyloid formation by h-amylin, p. vole amylin, and K1E h-amylin on the choice of buffer highlights the difficulties in objectively defining the amyloidogenicity of a polypeptide, since the concept is dependent upon solution conditions. For example, amyloid formation by the K1E variant occurs on the same time scale as h-amylin in PBS, but is faster than h-amylin in Tris (Table 1). These effects likely reflect competing interactions; the K1E mutation reduces the net charge on the peptide in the region of highest charge density and this could lead to faster aggregation. However, the rate of amylin amyloid formation is known to depend on the type of the anion in solution and is known to be faster in phosphate (20). In Tris the dominate effect is likely the reduction of electrostatic repulsion, but the substitution likely causes two effects in PBS. First reduction of charge should lead to faster aggregation, but secondly the replacement is expected to reduce phosphate binding which would, by itself, slow amyloid formation. The net result is that the substitution causes only small effects in PBS. The observed buffer dependent effects reveal the importance of studying amyloid formation under different conditions.
The modest effect of the Asn-22 to Gly substitution on the time scale of amyloid formation shows that the network of sidechain hydrogen bonds at residue 22, the “Asn ladder”, is not required for rapid amyloid formation by h-amylin. This is consistent with early studies that demonstrated that an Asn-22 to Leu replacement did not alter the T50 of amyloid formation by a truncated construct of amylin composed of residues 8–37 (32). More recent work has also shown that an amide sidechain is also not required at position 21 as an Asn-21 to Ala substitution still forms amyloid (33).
Both the Lys-1 to Glu and the Asn-22 to Gly substitutions have measurable effects on the toxicity of amylin towards cultured cells with the Asn-22 replacement having a particularly striking effect. The data indicates that residues outside of the cross β-sheet core impact cytotoxicity. Prior work has reported that replacement of Asn-21 with Gly reduced toxicity; although an EC50 value was not reported, no cytotoxicity was observed at the highest concentration studied (50 μM). In addition, the Asn-21 to Gly substitution, like the Asn-22 to Gly variant studied here, had modest effects on the time course of amyloid formation (33). In contrast, a Ser-20 to Gly mutation is known to enhance the rate of amyloid formation in vitro and to lead to enhanced toxicity in a cultured islet model (34–37). Collectively these independent studies highlight the importance of the Ser-20 to Asn-22 region in modulating toxicity.
The most striking observation with the substitutions studied here is the significant decrease in toxicity observed for Asn-22 Gly h-amylin in the absence of significant effects on the time scale of amyloid formation. Toxicity is reduced by a factor of 6 to 7.5-fold, while the values of T50 differ by only a factor of 1.2 to 2-fold. This result shows that the features which control the rate of amyloid formation and cytotoxicity are not always linked, in the sense that some replacements can have dramatic effects on cytotoxicity, but small effects on the time scale of in vitro amyloid formation. Prior studies have shown that pre amyloid h-amylin oligomers are the toxic species produced during amyloidogenesis while amyloid fibers are non-toxic (38). Thus, the effect of the N22G substitution on toxicity is likely exerted at the level of the oligomers. This makes the N22G mutant an interesting candidate for in depth studies of the properties of oligomers. The observation of the effects of the N-terminal substitution on toxicity also reveals an unexpected role for this region of amylin in modulating cytotoxicity.
Supplementary Material
ACKNOWLEDGEMENTS.
We thank the members of the Raleigh lab for helpful discussions. We thank Mr. Zachary Ridgway for assistance with the cross-linking studies. This work was supported by US NIH grant GM078114 and by Wellcome Trust Award 107927/Z/15/Z.
Abbreviations:
- EC50
the concentration of peptide to reach 50% of the effective response in an amylin cytotoxicity assay
- Fmoc
Fluorenylmethyloxycarbonyl
- h-amylin
human amylin
- HFIP
hexafluro isopropanol
- HPLC
high performance liquid chromatography
- p. vole amylin
prairie vole amylin
- PBS
phosphate buffered saline
- T50
the time required to achieve 50% of the maximum Thioflavin- T intensity in an amyloid assay
- TEM
transmission electron microscopy
Footnotes
ASSOCIATED CONTENT
Supporting Information
Supporting Information is available free of charge on the ACS publications website at DOI:
Amyloidogenicity prediction data for h-amylin, p. vole amylin, and rat amylin. Non-normalized thioflavin-T assays showing multiple curves for h-amylin, K1E h-amylin, N22G h-amylin and p. vole amylin. Thioflavin-T assays and TEM images comparing amyloid formation by wild type h-amylin and K1E amylin in Tris buffer. The results of in situ cross linking experiments conducted with all four polypeptides.
REFERENCES
- 1.Westermark P, Andersson A, and Westermark GT (2011) Islet amyloid polypeptide, islet amyloid, and diabetes mellitus. Physiol Rev 91, 795–826. [DOI] [PubMed] [Google Scholar]
- 2.Akter R, Abedini A, Ridgway Z, Zhang XX, Kleinberg J, Schmidt AM, and Raleigh DP (2017) Evolutionary adaptation and amyloid formation: does the reduced amyloidogenicity and cytotoxicity of ursine amylin contribute to the metabolic adaption of bears and polar bears? Isr J Chem 57, 750–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hay DL, Chen S, Lutz TA, Parkes DG, and Roth JD (2015) Amylin: pharmacology, physiology, and clinical potential. Pharmacol Rev 67, 564–600. [DOI] [PubMed] [Google Scholar]
- 4.Cooper GJS, Willis AC, Clark A, Turner RC, Sim RB, and Reid KBM (1987) Purification and characterization of a peptide from amyloid-rich pancreases of type-2 diabetic-patients. P Natl Acad Sci USA 84, 8628–8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westermark P, Wernstedt C, Wilander E, Hayden DW, Obrien TD, and Johnson KH (1987) Amyloid fibrils in human insulinoma and islets of langerhans of the diabetic cat Are derived from a neuropeptide-like protein also present in normal islet cells. P Natl Acad Sci USA 84, 3881–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper GJS, Leighton B, Dimitriadis GD, Parrybillings M, Kowalchuk JM, Howland K, Rothbard JB, Willis AC, and Reid KBM (1988) Amylin found in amyloid deposits in human type-2 diabetes-mellitus may be a hormone that regulates glycogen-metabolism in skeletal-muscle. P Natl Acad Sci USA 85, 7763–7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Opie EL (1901) On the relation of chronic interstitial pancreatitis to the islands of langerhans and to diabetes mellitus. J Exp Med 5, 397–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao P, Abedini A, and Raleigh DP (2013) Aggregation of islet amyloid polypeptide: from physical chemistry to cell biology. Curr Opin Struc Biol 23, 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abedini A, and Schmidt AM (2013) Mechanisms of islet amyloidosis toxicity in type 2 diabetes, FEBS Lett 587, 1119–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betsholtz C, Christmansson L, Engstrom U, Rorsman F, Svensson V, Johnson KH, and Westermark P (1989) Sequence divergence in a specific region of islet amyloid polypeptide (IAPP) explains differences in islet amyloid formation between species. Febs Letters 251, 261–264. [DOI] [PubMed] [Google Scholar]
- 11.Westermark P, Engstrom U, Johnson KH, Westermark GT, and Betsholtz C (1990) Islet amyloid polypeptide - pinpointing amino-acid-residues linked to amyloid fibril formation. P Natl Acad Sci USA 87, 5036–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburn TT, and Lansbury PT (1993) Interspecies sequence variations affect the kinetics and thermodynamics of amyloid formation - peptide models of pancreatic amyloid. Journal of the American Chemical Society 115, 11012–11013. [Google Scholar]
- 13.Akter R, Cao P, Noor H, Ridgway Z, Tu LH, Wang H, Wong AG, Zhang X, Abedini A, Schmidt AM, and Raleigh DP (2016) Islet amyloid polypeptide: structure, function, and pathophysiology. J Diabetes Res 2016, 2798269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryan G, Briscoe TA, and Jobe L (2009) Review of pramlintide as adjunctive therapy in treatment of type 1 and type 2 diabetes. Drug Des Devel Ther 2, 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isaacs D, Yager S, Parker M, Wolfe L, Luxenburg J, and Lekic S (2019) Adjunct antihyperglycemic agents in overweight and obese adults with type 1 diabetes. The Annals of Pharmacotherapy 53, 371–384. [DOI] [PubMed] [Google Scholar]
- 16.Wiltzius JJ, Sievers SA, Sawaya MR, Cascio D, Popov D, Riekel C, and Eisenberg D (2008) Atomic structure of the cross-beta spine of islet amyloid polypeptide (amylin). Protein Sci 17, 1467–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marek P, Woys AM, Sutton K, Zanni MT, and Raleigh DP (2010) Efficient microwave-assisted synthesis of human islet amyloid polypeptide designed to facilitate the specific incorporation of labeled amino acids. Organic Letters 12, 4848–4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abedini A, and Raleigh DP (2005) Incorporation of pseudoproline derivatives allows the facile synthesis of human IAPP, a highly amyloidogenic and aggregation-prone polypeptide, Org Lett 7, 693–696. [DOI] [PubMed] [Google Scholar]
- 19.Bitan G, and Teplow DB (2004) Rapid photochemical cross-linking - A new tool for studies of metastable, amyloidogenic protein assemblies, Accounts Chem Res 37, 357–364. [DOI] [PubMed] [Google Scholar]
- 20.Marek PJ, Patsalo V, Green DF, and Raleigh DP (2012) Ionic strength effects on amyloid formation by amylin are a complicated interplay among Debye screening, ion selectivity, and Hofmeister effects. Biochemistry 51, 8478–8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abedini A, and Raleigh DP (2005) The role of His-18 in amyloid formation by human islet amyloid polypeptide. Biochemistry 44, 16284–16291. [DOI] [PubMed] [Google Scholar]
- 22.Luca S, Yau WM, Leapman R, and Tycko R (2007) Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid-state NMR. Biochemistry 46, 13505–13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldschmidt L, Teng PK, Riek R, and Eisenberg D (2010) Identifying the amylome, proteins capable of forming amyloid-like fibrils. P Natl Acad Sci USA 107, 3487–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maurer-Stroh S, Debulpaep M, Kuemmerer N, de la Paz ML, Martins IC, Reumers J, Morris KL, Copland A, Serpell L, Serrano L, Schymkowitz JWH, and Rousseau F (2010) Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nature Methods 7, 237–42. [DOI] [PubMed] [Google Scholar]
- 25.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, and Serrano L (2004) Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nature Biotechnology 22, 1302–1306. [DOI] [PubMed] [Google Scholar]
- 26.Trovato A, Seno F, and Tosatto SCE (2007) The PASTA server for protein aggregation prediction. Protein Engineering Design & Selection 20, 521–523. [DOI] [PubMed] [Google Scholar]
- 27.Conchillo-Solé O, de Groot NS, Avilés FX, Vendrell J, Daura X, and Ventura S (2007) AGGRESCAN: a server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinformatics 8, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LeVine H (1999) Quantification of beta-sheet amyloid fibril structures with thioflavin T. Method Enzymol 309, 274–284. [DOI] [PubMed] [Google Scholar]
- 29.Tu L-H, and Raleigh DP (2013) Role of aromatic interactions in amyloid formation by islet amyloid polypeptide. Biochemistry 52, 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong AG, Wu C, Hannaberry E, Watson MD, Shea JE, and Raleigh DP (2016) Analysis of the amyloidogenic potential of pufferfish (Takifugu rubripes) islet amyloid polypeptide highlights the limitations of thioflavin-T assays and the difficulties in defining amyloidogenicity. Biochemistry 55, 510–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee K, Zhyvoloup A, Raleigh DP (2019) Amyloidogenicity and cytotoxicity of des-Lys-1 human amylin provides insight into amylin self-assebly and highlights the difficulties of defining amyloidogenicity. Protein Engineering, Design, and Selection, 1–7 doi: 10.1093/protein/gzz036. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koo BW, Hebda JA, and Miranker AD (2008) Amide inequivalence in the fibrillar assembly of islet amyloid polypeptide. Protein Eng Des Sel 21, 147–154. [DOI] [PubMed] [Google Scholar]
- 33.Godin E, Nguyen PT, Zottig X, and Bourgault S (2019) Identification of a hinge residue controlling islet amyloid polypeptide self-assembly and cytotoxicity. J Biol Chem 294, 8452–8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakagashira S, Sanke T, Hanabusa T, Shimomura H, Ohagi S, Kumagaye KY, Nakajima K, and Nanjo K (1996) Missense mutation of amylin gene (S20G) in Japanese NIDDM patients. Diabetes 45, 1279–1281. [DOI] [PubMed] [Google Scholar]
- 35.Sakagashira S, Hiddinga HJ, Tateishi K, Sanke T, Hanabusa T, Nanjo K, and Eberhardt NL (2000) S20G mutant amylin exhibits increased in vitro amyloidogenicity and increased intracellular cytotoxicity compared to wild-type amylin. American Journal of Pathology 157, 2101–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao P, Tu LH, Abedini A, Levsh O, Akter R, Patsalo V, Schmidt AM, and Raleigh DP (2012) Sensitivity of amyloid formation by human islet amyloid polypeptide to mutations at residue 20. J Mol Biol 421, 282–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meier DT, Entrup L, Templin AT, Hogan MF, Mellati M, Zraika S, Hull RL, and Kahn SE (2016) The S20G substitution in hIAPP is more amyloidogenic and cytotoxic than wild-type hIAPP in mouse islets. Diabetologia 59, 2166–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abedini A, Plesner A, Cao P, Ridgway Z, Zhang J, Tu LH, Middleton CT, Chao B, Sartori DJ, Meng F, Wang H, Wong AG, Zanni MT, Verchere CB, Raleigh DP, and Schmidt AM (2016) Time-resolved studies define the nature of toxic IAPP intermediates, providing insight for anti-amyloidosis therapeutics. Elife 5, e12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


