Abstract
Formation of biomolecular condensates through liquid–liquid phase separation (LLPS) has been described for several pathogenic proteins linked to neurodegenerative diseases and is discussed as an early step in the formation of protein aggregates with neurotoxic properties. In prion diseases, neurodegeneration and formation of infectious prions is caused by aberrant folding of the cellular prion protein (PrPC). PrPC is characterized by a large intrinsically disordered N-terminal domain and a structured C-terminal globular domain. A significant fraction of mature PrPC is proteolytically processed in vivo into an entirely unstructured fragment, designated N1, and the corresponding C-terminal fragment C1 harboring the globular domain. Notably, N1 contains a polybasic motif that serves as a binding site for neurotoxic Aβ oligomers. PrP can undergo LLPS; however, nothing is known how phase separation of PrP is triggered on a molecular scale. Here, we show that the intrinsically disordered N1 domain is necessary and sufficient for LLPS of PrP. Similar to full-length PrP, the N1 fragment formed highly dynamic liquid-like droplets. Remarkably, a slightly shorter unstructured fragment, designated N2, which lacks the Aβ-binding domain and is generated under stress conditions, failed to form liquid-like droplets and instead formed amorphous assemblies of irregular structures. Through a mutational analysis, we identified three positively charged lysines in the postoctarepeat region as essential drivers of condensate formation, presumably largely via cation–π interactions. These findings provide insights into the molecular basis of LLPS of the mammalian prion protein and reveal a crucial role of the Aβ-binding domain in this process.
Keywords: liquid–liquid phase separation, neurodegenerative disease, prion protein, prion disease, protein self-assembly, aggregation, intrinsically disordered protein
Abbreviations: DLS, dynamic light scattering; FRAP, fluorescence recovery after photobleaching; LLPS, liquid–liquid phase separation; MBP, maltose-binding protein; PrP, prion protein; PrPC, cellular prion protein; TEV, tobacco etch virus
Various cellular processes occur in specialized compartments that can be membrane-surrounded organelles or biomolecular condensates formed by liquid–liquid phase separation (LLPS) (reviewed in (1, 2, 3)). On the molecular level, phase separation can be induced by intermolecular interactions between modular binding domains and cognate peptide motifs (4, 5). However, many phase-separating proteins contain large intrinsically disordered protein regions with low-complexity sequences, which do not form stable folded structures (6, 7). For those proteins, it has been described that multivalent (4, 8), that is, electrostatic (9, 10), π–π, cation–π (11, 12, 13, 14, 15), and hydrophobic (16) interactions can contribute to the formation of biomolecular condensates. Interestingly, many proteins implicated in neurodegenerative diseases have been shown to undergo LLPS, leading to the concept that altered LLPS can promote the formation of neurotoxic protein assemblies (reviewed in (17, 18, 19, 20)).
In mammals, a conformational transition of the cellular prion protein (PrPC) into the scrapie isoform leads to transmissible spongiform encephalopathies or prion diseases, such as Creutzfeldt–Jakob disease in humans, bovine spongiform encephalopathy in cattle, and scrapie in sheep and goat (21). PrPC shows an interesting modular structure: an extended intrinsically disordered N-terminal domain and a highly structured C-terminal domain of a similar length (Fig. 1A). Various approaches in transgenic mice and cultured cells, including primary neurons, revealed that the N-terminal domain modulates signaling activity of PrPC. Strikingly, this role is seemingly contradictory because a physiological function of PrPC to promote neuronal homeostasis under diverse stress conditions is linked to this domain as well as a pathophysiological activity of certain PrP mutants (reviewed in (22, 23, 24)). Moreover, neuronal PrPC mediates neurotoxic effects of scrapie prions (25, 26, 27, 28), Aβ (29, 30, 31, 32, 33), α-synuclein (34), and Tau (35) via an interaction of its N-terminal domain with beta-sheet–rich oligomeric conformers of the respective pathogenic proteins. A considerable fraction of mature PrPC is proteolytically processed (α-cleavage) under physiological conditions into a completely unstructured fragment, designated N1, and the corresponding C-terminal structured fragment C1. A second cleavage around amino acid position 90 (β-cleavage) is mainly observed under pathological conditions and generates the fragments N2 and C2 (36, 37, 38, 39) (Fig. 1A). Of note, N1 but not N2 contains the Aβ-binding domain (29, 40).
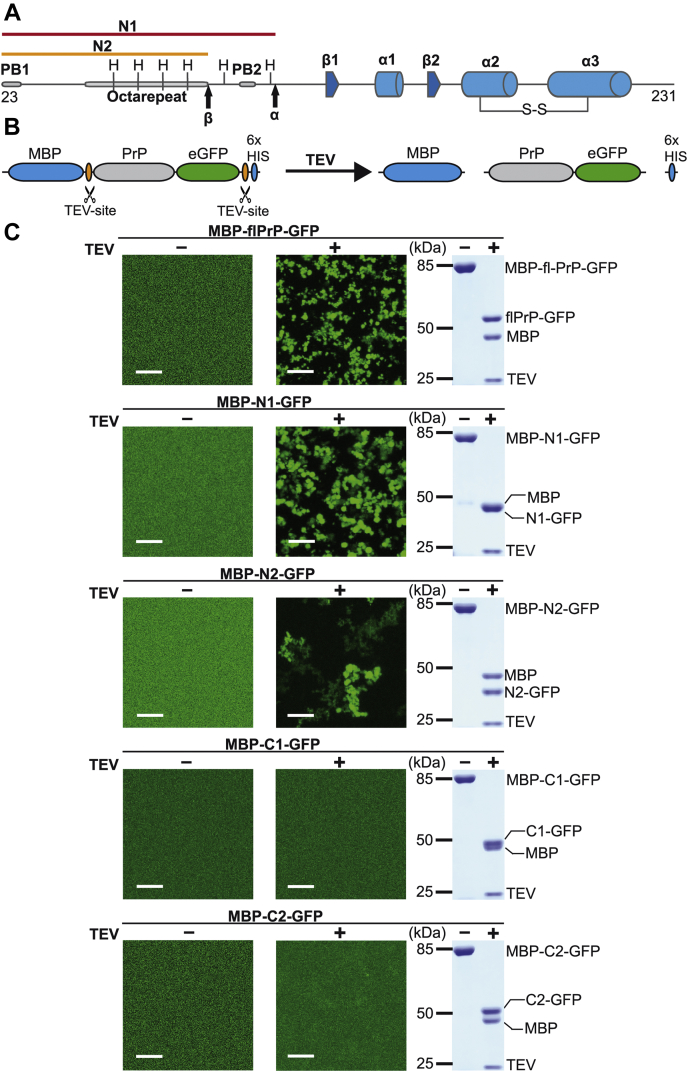
Figure 1.
The unstructured N1 fragment of PrP is necessary and sufficient for liquid–liquid phase separation.A, schematic representation of the mammalian prion protein. B, scheme of the experimental approach. The N-terminal maltose-binding protein (MBP) and the C-terminal His tag (6xHis) can be cut off from the PrP–GFP fusion protein by incubation with TEV protease. C, fusion proteins (30 μM in 10 mM Tris, pH 7.4) composed of full-length (fl) PrP or the indicated PrP fragments were incubated in the absence (TEV−) or presence (TEV+) of TEV protease for 1 h, and then, fluorescent image data were recorded by using a microscope. The scale bar represents 10 μm. An aliquot of each sample (4.5 μg) was analyzed in parallel by SDS-PAGE and Coomassie brilliant blue staining (right panels). The positions of the respective proteins are indicated. β, cleavage site that generates N2 and C2; α, cleavage site that generates N1 and C1; β1,2, beta strand; α1-3, alpha helices; H, histidine; PB, polybasic cluster; PrP, prion protein; S–S, disulfide bridge; TEV, tobacco etch virus.
The ability of full-length PrP to form biomolecular condensates has been described recently (41, 42, 43, 44, 45); however, the underlying molecular mechanisms that drive the formation of liquid-like droplets remain unknown. In the present study, we analyzed phase separation of full-length PrP and its major proteolytic fragments N1, N2, C1, and C2. Our experiments revealed that N1 is necessary and sufficient to drive phase separation of full-length PrP. Within N1, positively charged residues located in the Aβ-binding domain are essential for the formation of biomolecular condensates, most likely via cation–π interactions.
Results and discussion
N1–PrP but not N2–PrP undergoes LLPS
To study LLPS of PrP in vitro, we used maltose-binding protein (MBP)–PrP–eGFP fusion proteins (Fig. 1B). Phase separation was induced by tobacco etch virus (TEV) protease-mediated liberation of PrP–eGFP from MBP and monitored by laser scanning microscopy. Full-length PrP rapidly formed droplets upon TEV protease cleavage, indicative of LLPS (Fig. 1C, TEV+). A key feature of proteins undergoing condensate formation is the presence of intrinsically disordered and low-complexity regions (6), which are important drivers of phase separation. To study the role of the unstructured domain of PrP in LLPS, we cloned N1 and N2, the two disordered N-terminal fragments of PrP that are generated in vivo through proteolytic processing by an α- and β-cleavage, respectively (Fig. 1A). Indeed, N1 underwent phase separation similarly to full-length PrP, indicating that the structural elements in the C-terminal domain are not required for the formation of biomolecular condensates. In contrast, N2 formed irregular assemblies instead of liquid-like droplets (Fig. 1C, TEV+), which were not present before TEV cleavage (Fig. 1C, TEV−). These findings raised the question of whether the globular domain can undergo LLPS in the absence of the unstructured domains. Consequently, we analyzed C1 and C2, the C-terminal fragments generated after α- or β-cleavage, in parallel to N1 or N2 (Fig. 1A). Both before and after TEV cleavage, GFP fluorescence was evenly distributed, indicating that neither C1 nor C2 formed biomolecular condensates under these conditions, corroborating recent results (41). Of note, the lack of LLPS was not due to inefficient TEV cleavage (Fig. 1C, right panels).
To study the droplets/assemblies formed by N1 and N2 in more detail, we performed confocal laser scanning microscopy and recorded Z-stacks with a depth of 10 μm. Representative volumetric three-dimensional reconstructions supported the notion that N1 formed droplets, similarly to full-length PrPs, whereas the N2 assemblies were of irregular structures (Fig. 2A, upper panels). A signature of biomolecular condensates are highly dynamic molecules that rapidly exchange both within the condensate and with the surrounding environment. We recorded fluorescence recovery after photobleaching (FRAP) to test for a dynamic behavior of the PrP droplets. Full-length PrPs as well as N1 molecules in the droplets were highly dynamic, characteristic of a liquid-like state. In contrast, fluorescence recovery of the irregular N2 structures was greatly delayed, indicating formation of a gel-like or aggregated state (Fig. 2A, lower panels). Because we recorded FRAP 1 h after TEV protease-mediated cleavage, we considered the possibility that the N2 assemblies initially had liquid-like properties but then rapidly converted into a gel-like or aggregated state. Consequently, we did the FRAP analysis immediately after addition of TEV protease. Already under this condition, the N2 assemblies turned out to be nondynamic (Fig. S1A).
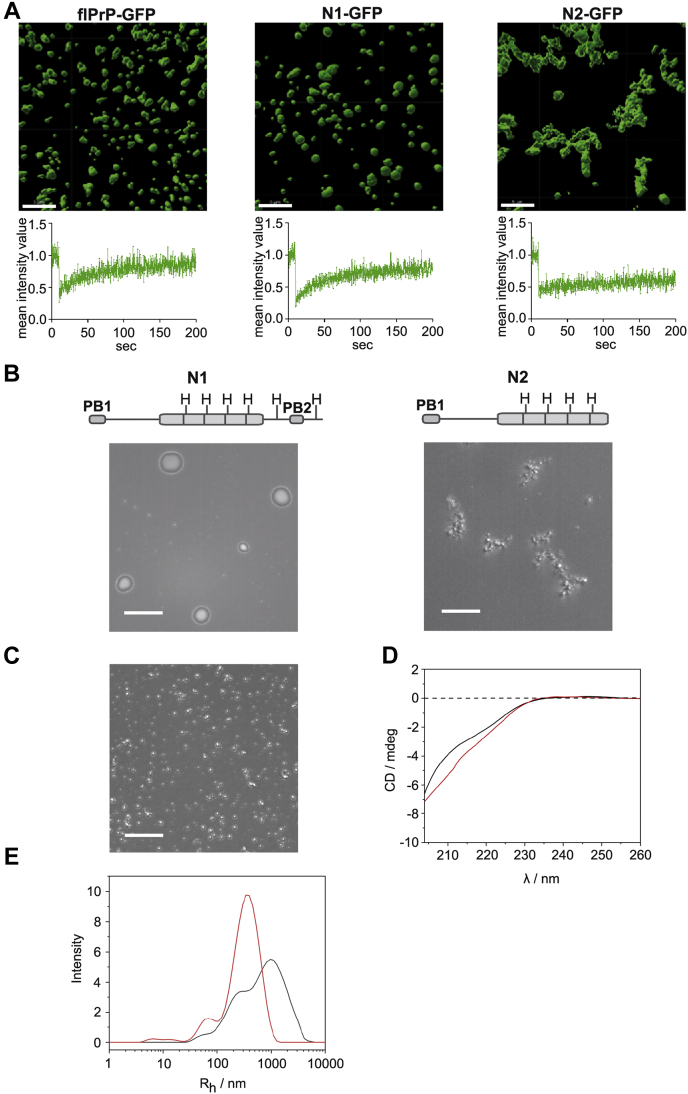
Figure 2.
Loss of the postoctarepeat domain interferes with the formation of condensates with liquid-like properties.A, volumetric three-dimensional reconstitution of the indicated proteins (10 μM in 10 mM Tris, pH 7.4) after TEV-mediated cleavage (1 h). Reconstitution was performed using Z-stack images (volume of 67.5 × 67.5 × 10 μm) produced by confocal laser scanning microscopy. The scale bar represents 5 μm (upper panels). Protein mobility within the droplets was measured by fluorescence recovery after photobleaching (FRAP). After 10 s of baseline recording (prebleach), a small area of interest (AOI) was photobleached. The average normalized fluorescence intensity of three AOIs was plotted over time (lower panels). B, MBP-N1 and MBP-N2 (10 μM in 10 mM Tris, pH 7.4) were analyzed by bright-field microscopy 1 h after TEV cleavage. A schematic representation of the fragments is shown above the images. C and D, recombinant N1 (64 μM in 20 mM Tris HCl, pH 7.4) was analyzed by bright-field microscopy (the scale bar represents 10 μm) (C) and CD spectroscopy (D). Shown are CD spectra in the absence (black line) and the presence of 4 M urea (red line). E, hydrodynamic radius (Rh) distribution functions for a 50 μM solution of N1 in 20 mM Tris HCl, pH 7.4 (black line) and in a 4 M urea-containing buffer (red line). MBP, maltose-binding protein; TEV, tobacco etch virus.
To address the possibility that the GFP tag influences LLPS, we purified MBP-N1 and MBP-N2 fusion proteins without GFP. Images taken by using bright-field microscopy confirmed that after TEV cleavage, MBP-N1 rapidly formed droplets, whereas MBP-N2 formed assemblies of irregular structures (Fig. 2B).
Next, we analyzed the conformation of phase-separated N1 after purifying recombinant N1 lacking both the MBP and GFP tag. N1(64 μM in 20 mM Tris HCl, pH 7.4) spontaneously formed liquid-like droplets, corroborating our findings with the TEV-induced cleavage of MBP-fusion proteins (Fig. 2C, Fig. S1B). At the same conditions, the CD spectrum of N1 is characterized by a negative CD band below 210 nm, indicating that the protein displays a fully disordered secondary structure at this condition (46). In the presence of 4 M urea, which prevents phase separation, only minor changes in the CD spectrum of N1 were observed. These findings show that the formation of liquid-like droplets by N1 is not accompanied by a significant conformational change (Fig. 2D). Finally, dynamic light scattering (DLS) experiments were carried out to determine the size of the N1 droplets (Fig. 2E). Based on the intensity distribution functions of the hydrodynamic radius, the N1 samples exhibited a largely polydisperse size distribution pattern, indicative of droplet formation of sizes ranging from 40 to several 1000 nm. Large hydrodynamic radius values beyond 1000 nm, that is, sizes in the μm range, indicate droplet formation as detectable by light microscopy. Interestingly, even in the urea-containing buffer, where macroscopic phase separation does not occur, N1 is not only present in its monomeric state but also forms larger oligomeric assemblies.
In sum, these experiments revealed that the unstructured domain of PrP is necessary and sufficient for LLPS. Furthermore, the comparative analysis of N1 and N2 has shown that the postoctarepeat region is crucial in driving LLPS of PrPs.
The polybasic motif in the postoctarepeat region drives phase separation of PrPs
To gain insight into mechanisms underlying phase separation of N1, we had a closer look at amino acids that can drive phase separation via intermolecular interactions. The scheme in Figure 3A depicts the amino acid composition of the N-terminal domain of PrPs. The unstructured domain is devoid of any negatively charged amino acids and contains only nine amino acids with positively charged side chains, which are in either an N- or C-terminal cluster (PB1 and PB2; see also Fig. 1A). In the “sticker and spacer” model that is frequently used to describe LLPS by intrinsically disordered domains, glycine is generally considered a spacer (47). Thus, it is interesting that N1 contains numerous glycines. Hydrophobic residues are evenly distributed over the entire domain with all but two of them being prolines. This is worth mentioning based on a possible specific role they are playing in phase separation. The partially positively charged faces of the pyrrolidine ring has been described to mediate intermolecular interactions with the negatively charged π faces of aromatic residues (48). The aromatic amino acids are also rather evenly distributed within the N1 domain without obvious cluster formation. The location of the six histidine residues is indicated because copper binding to these histidines seems to influence the structure and physiological function of PrPC as well as the formation of the scrapie isoform of prion protein (49, 50, 51, 52).
Figure 3.
The polybasic cluster of the Aβ-binding domain drives phase separation.A, schematic drawing of selected amino acids in the intrinsically disordered N-terminal domain. The domain that is missing from N2 (aa 90–111) is indicated in gray. B, schematic representation of the constructs. The positively charged residues (+) and the histidines (H) in the octarepeat domain are illustrated. C, the indicated proteins (10 μM in 10 mM Tris, pH 7.4) were analyzed 1 h after TEV cleavage. Fluorescent image data (upper panels) and FRAP (lower panels) were recorded as described in Figures 1 and 2. FRAP, fluorescence recovery after photobleaching; TEV, tobacco etch virus.
Comparing N2 with N1, the most striking difference is the lack of the polybasic cluster in the postoctarepeat domain (PB2), which serves as a binding domain for neurotoxic Aβ oligomers (29, 40). Conceptually, the positively charged lysines may contribute to phase separation via intermolecular cation–π interactions (11, 12, 13, 14). To test this hypothesis experimentally, we mutated the three lysines in PB2 to alanines (Fig. 3B) and analyzed phase separation. Indeed, the loss of the three lysine residues in the postoctarepeat region significantly impaired condensate formation of N1•PB2>A. The microscopic analysis revealed the formation of irregular structures, similarly to N2 (Fig. 3C, Figs. S2 and S3). Importantly, the FRAP recording supported the notion that N1•PB2>A lost its liquid-like properties. To support our concept that lysines are effective through their positive charge, we generated N1•PB2>R in which the three lysine residues in PB2 were mutated to positively charged arginines. Indeed, N1•PB2>R underwent LLPS similarly to N1 (Fig. S4A). In contrast, mutation of the aromatic residues (N1•W|Y > G) impaired LLPS, emphasizing the important role of cation–π interactions between positively charged residues and aromatic residues in driving LLPS of N1 (Fig. S4B). Based on these findings, we were wondering whether the other polybasic motif, located at the very N terminus, has a similar function in LLPS. N1•PB1>A did also not undergo LLPS and formed assemblies with irregular structures, indicating a gel-like or aggregated state, as judged by FRAP recordings (Fig. 3C, Figs. S2 and S3). As expected, deleting both the N- and C-terminal polybasic motifs (N1•PB1+2 > A) aggravated this phenotype. Finally, we mutated the four histidines within the octarepeat region. However, phase separation of N1•OR-H>G was not impaired (Fig. 3C, Figs. S2 and S3).
In conclusion, our study provided new mechanistic insights into the propensity of the mammalian prion protein to undergo LLPS. Our experiments revealed that the intrinsically disordered N1 fragment of PrP is necessary and sufficient for the formation of biomolecular condensates. On the molecular level, phase separation is governed primarily by basic residues within the Aβ-binding domain, most likely through intermolecular cation–π interactions of the lysines with neighboring aromatic side chains (Fig. 4). These findings are relevant for at least two major aspects in prion protein biology. First, binding of Aβ oligomers or other neurotoxic β-sheet–rich conformers to the polybasic motif is expected to interfere with LLPS of full-length PrPs and the N-terminal fragments, similarly to the deletion of the lysines. In support of this notion, it has been reported that binding of Aβ oligomers modulates phase separation and alters the conformation of full-length PrPs (44, 53). Second, it is tempting to speculate that LLPS of PrPs is linked to both its conversion into prions or neurotoxic conformers and its physiological function. Notably, this applies not only for the signaling competence of the full-length PrPC but also for the biological activities of N1 after its liberation from the PrPC by α-cleavage.
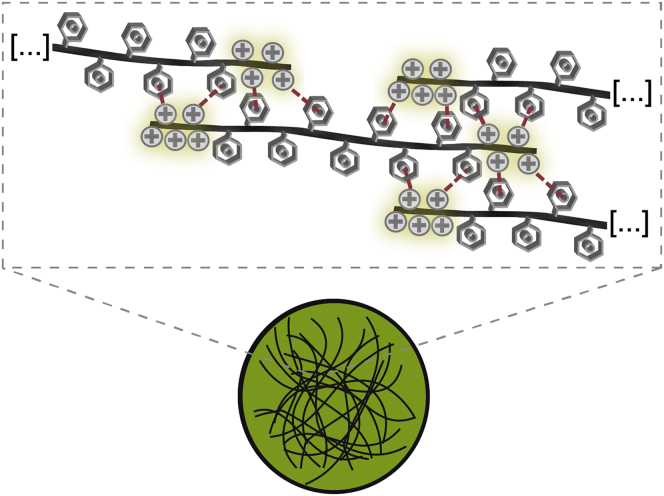
Figure 4.
Model for phase separation of N1. In addition to favorable hydrophobic interactions, N1 mainly uses intermolecular cation–π interactions to form liquid droplets. Depicted are the aromatic rings of aromatic amino acid residues and the positive charge of lysine and arginine (+). Red dashed lines indicate the formation of cation–π interactions.
Experimental procedures
Constructs/plasmids
Plasmid maintenance and amplification was carried out using Escherichia coli TOP10 (Thermo Fisher Scientific). All PrP constructs were generated by standard PCR cloning techniques and are based on the coding region of mouse PrP gene (Prnp; GenBank accession number M18070) modified to express PrP-L108M/V111M (54), allowing detection by the mAb 3F4 (55). Full-length PrP: aa 23 to 230; N1: aa 23 to 114; N2: aa 23 to 89; C1: aa 111 to 230; C2: aa 90 to 230; N1•PB1>A: the polybasic motif K-K-R-P-K (aa 23–27) was changed to A-A-A-P-A; N1•PB2>A: the polybasic motif K-P-S-K-P-K (aa 100–105) was changed to A-P-S-A-P-A; N1•PB2>R: the polybasic motif K-P-S-K-P-K (aa 100–105) was changed to R-P-S-R-P-R; N1•PB1+2>A: both polybasic motifs were changed; N1•OR H>G: the four H in the octarepeat region (aa 49–89) were changed to G; N1•W|Y>G: all aromatic amino acid residues were changed to G. To generate the MBP-TEV-PrP-eGFP-TEV-His6 plasmids (Fig. 1B), we exchanged the FUS coding region from the pMal-TEV-FUS-eGFP-TEV-His6 plasmid, kindly provided by Dorothee Dormann (56), by those of the respective PrP variants.
Protein expression and purification
MBP-PrP-eGFP constructs were transformed into either BL21-DE3 strains for N-terminal constructs or Origami B (DE3) competent cells (Novagen) for full-length PrP and C1 and C2 constructs. For protein expression, 1-l bacterial culture of the lysogeny broth medium was inoculated and grown to an absorbance (600 nm) of 0.9 to 1.0. For BL21-DE3 cultures, 30-min incubation on ice was performed before IPTG induction. Then, the expression was induced with 100 μM IPTG and the culture was incubated over night at 12 °C, 120 rpm. For Origami B (DE3) cultures, the expression was induced with 0.5 mM IPTG and incubated over night at 25 °C, 120 rpm. Bacteria were harvested by centrifugation (5,000g, 4 °C, 20 min), and the pellet was washed with 20 ml Millipore water and centrifuged again (2,000g, 4 °C, 20 min). Pellets were stored at −20 °C until further use. For purification, the bacterial pellet was resuspended in the lysis buffer (50 mM Na2HPO4/NaH2PO4 (pH 8.0), 500 mM NaCl, 0.01 mM ZnCl2, 10% glycerol). Protein lysis was performed via SLM AMINCO French Press (Thermo Fisher Scientific), and the protein solution was centrifuged (40,000g, 45 min, 4 °C). The supernatant was loaded on a His-Trap FF column (GE Healthcare) equilibrated with the lysis buffer and washed with three CV lysis buffer containing 20 mM imidazole and in case of the N-terminal constructs washed again with three CV lysis buffer containing 50 mM imidazole. Proteins were eluted with the lysis buffer containing 200 mM imidazole and dialyzed over night at 4 °C in the dialysis buffer (50 mM Na2HPO4/NaH2PO4 (pH 8.0), 500 mM NaCl, 0.01 mM ZnCl2, 5% glycerol). The protein concentration was determined by NanoDrop 2000 (Thermo Scientific) and aliquoted and stored at −80 °C until further use.
To study N1-PrP without an MBP tag, N1 was cloned into a pGATEV vector and expressed in and purified from E. coli BL21-DE3.
Sample preparation
Protein aliquots were thawed on ice and centrifuged at 20,000g for 10 min at 4 °C to remove aggregates. Using Vivaspin 500 columns with 30-kDa molecular weight cut off (Sartorius Stedim biotech), the buffer was exchanged to 10 mM Tris, pH 7.4, by centrifuging five times for 7 min at 12,000g at 4 °C. After buffer exchange, the final protein concentration was again determined by NanoDrop 2000. For LLPS induction, TEV protease was added to the sample and incubated 1 h for complete cleavage before microscopy.
Laser scanning microscopy
As described previously (57), fluorescent imaging laser scanning microscopy on a microscope (ELYRA PS.1; Carl Zeiss) with an imaging detector (LSM 880; Carl Zeiss) was performed. For z-stack scanning, a 63× numerical aperture 1.4 oil-immersion objective was used to record a stack of 67.5 × 67.5 × 10 μm and 0.330 μm for each optical section. The argon laser power was set to 0.006% at 488 nm with pixel dwell time of 5.71 μs. During all measurements, laser power, gain, and field of view were kept constant. Data were imported into Imaris 9.3.1 for three-dimensional analysis of the z-stack images, and the surface module was used for reconstruction of the surfaces. For FRAP experiments, ZEN2.1 bleaching and region software module and Plan-Apochromat 100× numerical aperture 1.46 oil differential interference contrast M27 objective was used. For regions of interest, three circular areas with a 12-pixel diameter were chosen. One region was bleached with 100% laser power and a pixel dwell time of 8.71 ms, with a scan time of 111.29 ms and a pixel dwell time of 1.61 ms, and the other two regions were used as the reference signal and background signal. Data calculation was performed in Excel 2016, and diagrams were made with GraphPad Prism.
CD spectroscopy
CD spectroscopy experiments were carried out at the temperature of 25 °C by means of a Jasco J-715 spectropolarimeter from JASCO Corporation. Far-UV spectra of N1 were recorded at the concentration of 64 μM by using a quartz cuvette with a path length of 0.01 cm. The experiments were performed in 20 mM Tris HCl buffer, pH 7.4, in the absence and in the presence of 4 M urea. From each sample, an appropriate background spectrum was subtracted. All the experiments were performed with the following instrumental parameters: scan rate of 50 nm min−1, response time of 2 s, and bandwidth of 5 nm. It is important to note that owing to the strong absorbance of urea, it is not possible to record the CD spectra down to 190 nm.
DLS
DLS measurements were performed by means of a Zetasizer Nano S from Malvern Instruments Limited. The instrument is equipped with a He–Ne laser operating at 633 nm. Each size distribution function is the result of 15 accumulations. The concentration of the N1 sample was 50 μM. The experiments were performed in 20 mM Tris HCl buffer, pH 7.4, in the absence and in the presence of 4 M urea at the temperature of 25 °C.
Data availability
All data are contained within the article and the supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Prof. Dorothee Dormann for providing the pMal-FUS plasmids. We are grateful to Petra Goldmann, Barbara Kachholz, and Andrea Roth-Sturm for technical support.
Confocal laser scanning and SR-SIM microscopy was funded by the German Research Foundation and the State Government of North Rhine-Westphalia (INST 213/840-1 FUGG).
Author contributions
J. K., Y.-H. L., R. O., V. B., R. W., K. F. W., and J. T. formal analysis; J. K., Y.-H. L., R. O., and V. B. investigation; J. K., Y.-H. L., R. O., and V. B. visualization; J. K., Y.-H. L., R. O., and V. B. writing–review and editing; R. W., K. F. W., and J. T. conceptualization; R. W., K. F. W., and J. T. supervision; R. W., K. F. W., and J. T. funding acquisition; R. W., K. F. W., and J. T. writing–original draft; R. W., K. F. W., and J. T. project administration.
Funding and additional information
This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy–EXC 2033–390677874–RESOLV; TA 167/6-3 and TA 167/11-1 (to J. T.); and WI/2111-6 (to K. F. W.).
Edited by Paul Fraser
Supporting information
References
- 1.Shin Y., Brangwynne C.P. Liquid phase condensation in cell physiology and disease. Science. 2017;357 doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 2.Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Flynn B.G., Mittag T. The role of liquid-liquid phase separation in regulating enzyme activity. Curr. Opin. Cell Biol. 2021;69:70–79. doi: 10.1016/j.ceb.2020.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li P., Banjade S., Cheng H.C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J.V., King D.S., Banani S.F., Russo P.S., Jiang Q.X., Nixon B.T., Rosen M.K. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banjade S., Rosen M.K. Phase transitions of multivalent proteins can promote clustering of membrane receptors. Elife. 2014;3 doi: 10.7554/eLife.04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darling A.L., Liu Y., Oldfield C.J., Uversky V.N. Intrinsically disordered proteome of human membrane-less organelles. Proteomics. 2018;18 doi: 10.1002/pmic.201700193. [DOI] [PubMed] [Google Scholar]
- 7.Brangwynne C.P., Tompa P., Pappu R.V. Polymer physics of intracellular phase transitions. Nat. Phys. 2015;11:899–904. [Google Scholar]
- 8.Pierce W.K., Grace C.R., Lee J., Nourse A., Marzahn M.R., Watson E.R., High A.A., Peng J., Schulman B.A., Mittag T. Multiple weak linear motifs enhance recruitment and processivity in SPOP-mediated substrate ubiquitination. J. Mol. Biol. 2016;428:1256–1271. doi: 10.1016/j.jmb.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pak C.W., Kosno M., Holehouse A.S., Padrick S.B., Mittal A., Ali R., Yunus A.A., Liu D.R., Pappu R.V., Rosen M.K. Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell. 2016;63:72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin Y.H., Forman-Kay J.D., Chan H.S. Sequence-specific polyampholyte phase separation in membraneless organelles. Phys. Rev. Lett. 2016;117:178101. doi: 10.1103/PhysRevLett.117.178101. [DOI] [PubMed] [Google Scholar]
- 11.Kang J., Lim L., Lu Y., Song J. A unified mechanism for LLPS of ALS/FTLD-causing FUS as well as its modulation by ATP and oligonucleic acids. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.3000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das S., Lin Y.H., Vernon R.M., Forman-Kay J.D., Chan H.S. Comparative roles of charge, pi, and hydrophobic interactions in sequence-dependent phase separation of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U. S. A. 2020;117:28795–28805. doi: 10.1073/pnas.2008122117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nott T.J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T.D., Bazett-Jones D.P., Pawson T., Forman-Kay J.D., Baldwin A.J. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S., Huang J., Lee Y., Dutta S., Yoo H.Y., Jung Y.M., Jho Y., Zeng H., Hwang D.S. Complexation and coacervation of like-charged polyelectrolytes inspired by mussels. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E847–E853. doi: 10.1073/pnas.1521521113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherrill C.D. Energy component analysis of pi interactions. Acc. Chem. Res. 2013;46:1020–1028. doi: 10.1021/ar3001124. [DOI] [PubMed] [Google Scholar]
- 16.Yeo G.C., Keeley F.W., Weiss A.S. Coacervation of tropoelastin. Adv. Colloid Interf. Sci. 2011;167:94–103. doi: 10.1016/j.cis.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Alberti S., Dormann D. Liquid-liquid phase separation in disease. Annu. Rev. Genet. 2019;53:171–194. doi: 10.1146/annurev-genet-112618-043527. [DOI] [PubMed] [Google Scholar]
- 18.Babinchak W.M., Surewicz W.K. Liquid-liquid phase separation and its mechanistic role in pathological protein aggregation. J. Mol. Biol. 2020;432:1910–1925. doi: 10.1016/j.jmb.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darling A.L., Shorter J. Combating deleterious phase transitions in neurodegenerative disease. Biochim. Biophys. Acta Mol. Cell Res. 2021;1868:118984. doi: 10.1016/j.bbamcr.2021.118984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zbinden A., Perez-Berlanga M., De Rossi P., Polymenidou M. Phase separation and neurodegenerative diseases: A disturbance in the force. Dev. Cell. 2020;55:45–68. doi: 10.1016/j.devcel.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 21.Prusiner S.B. Prions. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beland M., Roucou X. The prion protein unstructured N-terminal region is a broad-spectrum molecular sensor with diverse and contrasting potential functions. J. Neurochem. 2012;120:853–868. doi: 10.1111/j.1471-4159.2011.07613.x. [DOI] [PubMed] [Google Scholar]
- 23.Iraci N., Stincardini C., Barreca M.L., Biasini E. Decoding the function of the N-terminal tail of the cellular prion protein to inspire novel therapeutic avenues for neurodegenerative diseases. Virus Res. 2014;207:62–68. doi: 10.1016/j.virusres.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 24.Resenberger U.K., Winklhofer K.F., Tatzelt J. Neuroprotective and neurotoxic signaling by the prion protein. Top. Curr. Chem. 2011;305:101–119. doi: 10.1007/128_2011_160. [DOI] [PubMed] [Google Scholar]
- 25.Brandner S., Isenmann S., Raeber A., Fischer M., Sailer A., Kobayashi Y., Marino S., Weissmann C., Aguzzi A. Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature. 1996;379:339–343. doi: 10.1038/379339a0. [DOI] [PubMed] [Google Scholar]
- 26.Mallucci G., Dickinson A., Linehan J., Klohn P.C., Brandner S., Collinge J. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 2003;302:871–874. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- 27.Chesebro B., Trifilo M., Race R., Meade-White K., Teng C., LaCasse R., Raymond L., Favara C., Baron G., Priola S., Caughey B., Masliah E., Oldstone M. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science. 2005;308:1435–1439. doi: 10.1126/science.1110837. [DOI] [PubMed] [Google Scholar]
- 28.Rambold A.S., Müller V., Ron U., Ben-Tal N., Winklhofer K.F., Tatzelt J. Stress-protective activity of prion protein is corrupted by scrapie-prions. EMBO J. 2008;27:1974–1984. doi: 10.1038/emboj.2008.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lauren J., Gimbel D.A., Nygaard H.B., Gilbert J.W., Strittmatter S.M. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-beta oligomers. Nature. 2009;457:1128–1132. doi: 10.1038/nature07761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nygaard H.B., Strittmatter S.M. Cellular prion protein mediates the toxicity of beta-amyloid oligomers: Implications for Alzheimer disease. Arch. Neurol. 2009;66:1325–1328. doi: 10.1001/archneurol.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith L.M., Strittmatter S.M. Binding sites for amyloid-beta oligomers and synaptic toxicity. Cold Spring Harb. Perspect. Med. 2017;7 doi: 10.1101/cshperspect.a024075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Resenberger U.K., Harmeier A., Woerner A.C., Goodman J.L., Muller V., Krishnan R., Vabulas R.M., Kretzschmar H.A., Lindquist S., Hartl F.U., Multhaup G., Winklhofer K.F., Tatzelt J. The cellular prion protein mediates neurotoxic signalling of beta-sheet-rich conformers independent of prion replication. EMBO J. 2011;30:2057–2070. doi: 10.1038/emboj.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purro S.A., Nicoll A.J., Collinge J. Prion protein as a toxic acceptor of amyloid-beta oligomers. Biol. Psychiatry. 2018;83:358–368. doi: 10.1016/j.biopsych.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 34.Ferreira D.G., Temido-Ferreira M., Miranda H.V., Batalha V.L., Coelho J.E., Szego E.M., Marques-Morgado I., Vaz S.H., Rhee J.S., Schmitz M., Zerr I., Lopes L.V., Outeiro T.F. Alpha-synuclein interacts with PrPC to induce cognitive impairment through mGluR5 and NMDAR2B. Nature Neurosci. 2017;20:1569–1579. doi: 10.1038/nn.4648. [DOI] [PubMed] [Google Scholar]
- 35.Ondrejcak T., Klyubin I., Corbett G.T., Fraser G., Hong W., Mably A.J., Gardener M., Hammersley J., Perkinton M.S., Billinton A., Walsh D.M., Rowan M.J. Cellular prion protein mediates the disruption of hippocampal synaptic plasticity by soluble tau in vivo. J. Neurosci. 2018;38:10595–10606. doi: 10.1523/JNEUROSCI.1700-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen S.G., Teplow D.B., Parchi P., Teller J.K., Gambetti P., Autilio-Gambetti L. Truncated forms of the human prion protein in normal brain and in prion diseases. J. Biol. Chem. 1995;270:19173–19180. doi: 10.1074/jbc.270.32.19173. [DOI] [PubMed] [Google Scholar]
- 37.Mange A., Beranger F., Peoc'h K., Onodera T., Frobert Y., Lehmann S. Alpha- and beta- cleavages of the amino-terminus of the cellular prion protein. Biol. Cell. 2004;96:125–132. doi: 10.1016/j.biolcel.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 38.Harris D.A., Huber M.T., van Dijken P., Shyng S.L., Chait B.T., Wang R. Processing of a cellular prion protein: Identification of N- and C-terminal cleavage sites. Biochemistry. 1993;32:1009–1016. doi: 10.1021/bi00055a003. [DOI] [PubMed] [Google Scholar]
- 39.Altmeppen H.C., Puig B., Dohler F., Thurm D.K., Falker C., Krasemann S., Glatzel M. Proteolytic processing of the prion protein in health and disease. Am. J. Neurodegener. Dis. 2012;1:15–31. [PMC free article] [PubMed] [Google Scholar]
- 40.Chen S., Yadav S.P., Surewicz W.K. Interaction between human prion protein and amyloid-beta (abeta) oligomers: Role OF N-terminal residues. J. Biol. Chem. 2010;285:26377–26383. doi: 10.1074/jbc.M110.145516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tange H., Ishibashi D., Nakagaki T., Taguchi Y., Kamatari Y.O., Ozawa H., Nishida N. Liquid-liquid phase separation of full-length prion protein initiates conformational conversion in vitro. J. Biol. Chem. 2021;296:100367. doi: 10.1016/j.jbc.2021.100367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang J.J., Li X.N., Liu W.L., Yuan H.Y., Gao Y., Wang K., Tang B., Pang D.W., Chen J., Liang Y. Neutralizing mutations significantly inhibit amyloid formation by human prion protein and decrease its cytotoxicity. J. Mol. Biol. 2020;432:828–844. doi: 10.1016/j.jmb.2019.11.020. [DOI] [PubMed] [Google Scholar]
- 43.Matos C.O., Passos Y.M., do Amaral M.J., Macedo B., Tempone M.H., Bezerra O.C.L., Moraes M.O., Almeida M.S., Weber G., Missailidis S., Silva J.L., Uversky V.N., Pinheiro A.S., Cordeiro Y. Liquid-liquid phase separation and fibrillation of the prion protein modulated by a high-affinity DNA aptamer. FASEB J. 2020;34:365–385. doi: 10.1096/fj.201901897R. [DOI] [PubMed] [Google Scholar]
- 44.Kostylev M.A., Tuttle M.D., Lee S., Klein L.E., Takahashi H., Cox T.O., Gunther E.C., Zilm K.W., Strittmatter S.M. Liquid and hydrogel phases of PrP(C) linked to conformation shifts and triggered by Alzheimer's amyloid-beta oligomers. Mol. Cell. 2018;72:426–443.e412. doi: 10.1016/j.molcel.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Passos Y.M., do Amaral M.J., Ferreira N.C., Macedo B., Chaves J.A.P., de Oliveira V.E., Gomes M.P.B., Silva J.L., Cordeiro Y. The interplay between a GC-rich oligonucleotide and copper ions on prion protein conformational and phase transitions. Int. J. Biol. Macromol. 2021;173:34–43. doi: 10.1016/j.ijbiomac.2021.01.097. [DOI] [PubMed] [Google Scholar]
- 46.Kelly S.M., Jess T.J., Price N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 47.Wang J., Choi J.M., Holehouse A.S., Lee H.O., Zhang X., Jahnel M., Maharana S., Lemaitre R., Pozniakovsky A., Drechsel D., Poser I., Pappu R.V., Alberti S., Hyman A.A. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018;174:688–699.e616. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zondlo N.J. Aromatic-proline interactions: Electronically tunable CH/pi interactions. Acc. Chem. Res. 2013;46:1039–1049. doi: 10.1021/ar300087y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown D.R., Qin K., Herms J.W., Madlung A., Manson J., Strome R., Fraser P.E., Kruck T., von Bohlen A., Schulz-Schaeffer W., Giese A., Westaway D., Kretzschmar H. The cellular prion protein binds copper in vivo. Nature. 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 50.Millhauser G.L. Copper and the prion protein: Methods, structures, function, and disease. Annu. Rev. Phys. Chem. 2007;58:299–320. doi: 10.1146/annurev.physchem.58.032806.104657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eigenbrod S., Frick P., Bertsch U., Mitteregger-Kretzschmar G., Mielke J., Maringer M., Piening N., Hepp A., Daude N., Windl O., Levin J., Giese A., Sakthivelu V., Tatzelt J., Kretzschmar H. Substitutions of PrP N-terminal histidine residues modulate scrapie disease pathogenesis and incubation time in transgenic mice. PLoS One. 2017;12 doi: 10.1371/journal.pone.0188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen X.T.A., Tran T.H., Cojoc D., Legname G. Copper binding regulates cellular prion protein function. Mol. Neurobiol. 2019;56:6121–6133. doi: 10.1007/s12035-019-1510-9. [DOI] [PubMed] [Google Scholar]
- 53.Konig A.S., Rosener N.S., Gremer L., Tusche M., Flender D., Reinartz E., Hoyer W., Neudecker P., Willbold D., Heise H. Structural details of amyloid beta oligomers in complex with human prion protein as revealed by solid-state MAS NMR spectroscopy. J. Biol. Chem. 2021;296:100499. doi: 10.1016/j.jbc.2021.100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winklhofer K.F., Heller U., Reintjes A., Tatzelt J. Inhibition of complex glycosylation increases formation of PrPSc. Traffic. 2003;4:313–322. doi: 10.1034/j.1600-0854.2003.00088.x. [DOI] [PubMed] [Google Scholar]
- 55.Kascsak R.J., Rubenstein R., Merz P.A., Tonna-DeMasi M., Fersko R., Carp R.I., Wisniewski H.M., Diringer H. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J. Virol. 1987;61:3688–3693. doi: 10.1128/jvi.61.12.3688-3693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hofweber M., Hutten S., Bourgeois B., Spreitzer E., Niedner-Boblenz A., Schifferer M., Ruepp M.D., Simons M., Niessing D., Madl T., Dormann D. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell. 2018;173:706–719.e713. doi: 10.1016/j.cell.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 57.Ahlers J., Adams E.M., Bader V., Pezzotti S., Winklhofer K.F., Tatzelt J., Havenith M. The key role of solvent in condensation: Mapping water in liquid-liquid phase-separated FUS. Biophys. J. 2021;120:1266–1275. doi: 10.1016/j.bpj.2021.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and the supporting information.