Abstract
Many enveloped viruses bud from cholesterol-rich lipid rafts on the cell membrane. Depleting cellular cholesterol impedes this process and results in viral particles with reduced viability. Viperin (Virus Inhibitory Protein, Endoplasmic Reticulum-associated, Interferon iNducible) is an endoplasmic reticulum membrane–associated enzyme that exerts broad-ranging antiviral effects, including inhibiting the budding of some enveloped viruses. However, the relationship between viperin expression and the retarded budding of virus particles from lipid rafts on the cell membrane is unclear. Here, we investigated the effect of viperin expression on cholesterol biosynthesis using transiently expressed genes in the human cell line human embryonic kidney 293T (HEK293T). We found that viperin expression reduces cholesterol levels by 20% to 30% in these cells. Following this observation, a proteomic screen of the viperin interactome identified several cholesterol biosynthetic enzymes among the top hits, including lanosterol synthase (LS) and squalene monooxygenase (SM), which are enzymes that catalyze key steps in establishing the sterol carbon skeleton. Coimmunoprecipitation experiments confirmed that viperin, LS, and SM form a complex at the endoplasmic reticulum membrane. While coexpression of viperin was found to significantly inhibit the specific activity of LS in HEK293T cell lysates, coexpression of viperin had no effect on the specific activity of SM, although did reduce SM protein levels by approximately 30%. Despite these inhibitory effects, the coexpression of neither LS nor SM was able to reverse the viperin-induced depletion of cellular cholesterol levels, possibly because viperin is highly expressed in transfected HEK293T cells. Our results establish a link between viperin expression and downregulation of cholesterol biosynthesis that helps explain viperin's antiviral effects against enveloped viruses.
Keywords: viperin, cholesterol regulation, interactome analysis, radical SAM enzyme, squalene monooxygenase, lanosterol synthase
Abbreviations: BHT, 2,6-di-tert-butyl-4-methylphenol; BIBB 515, (1-(4-chlorobenzoyl)-4-((4-(2-oxazolin-2-yl) benzylidene))piperidine); DAVID, Database for Annotation, Visualization and Integrated Discovery; ddhCTP, 3’-deoxy-3’,4’-didehydro-CTP; ER, endoplasmic reticulum; FPPS, farnesyl pyrophosphate synthase; HEK293T, human embryonic kidney 293T; HMGR, 3-hydroxy-3-methylglutaryl CoA reductase; IFN, interferon; LS, lanosterol synthase; PEI, polyethyleneimine; RSV, respiratory syncytial virus; SAINT, Significance Analysis of INTeractome; SM, squalene monooxygenase; TBS, Tris-buffered saline; TRAF6, TNF receptor–associated factor 6; UPLC, ultra performance LC; viperin, Virus Inhibitory Protein, Endoplasmic Reticulum-associated, Interferon iNducible
Cholesterol is a critical component of eukaryotic membranes and a precursor to many steroid hormones and bile acids (1). Cholesterol biosynthesis occurs on the cytosolic face of the endoplasmic reticulum (ER), primarily in the liver and intestines (2), and is one of the most extensively regulated biosynthetic pathways (3). The numerous regulation mechanisms include transcriptional upregulation through the sterol regulatory element-binding protein pathway (4); downregulation through the sterol- and isoprenoid-dependent degradation of 3-hydroxy-3-methylglutaryl CoA reductase (HMGR) (5, 6); the transcriptional regulation of HMGR (7, 8); the post-translational modification of HMGR (9); and, more recently, the direct sensing of cholesterol leading to proteasomal degradation of a key biosynthetic enzyme, squalene monooxygenase (SM), mediated by the E3 ubiquitin ligase, Membrane Associated Ring-CH-Type Finger 6 (10, 11, 12). Cholesterol is also an important component of lipid rafts in cell membranes (1, 13), which have been implicated in a variety of diseases, including Alzheimer's disease, prion diseases, and bacterial and viral infections (13). Notably, various enveloped viruses require cholesterol-rich lipid rafts to bud from the cell, thereby completing the viral replication cycle to produce infectious viruses (13, 14, 15, 16, 17, 18).
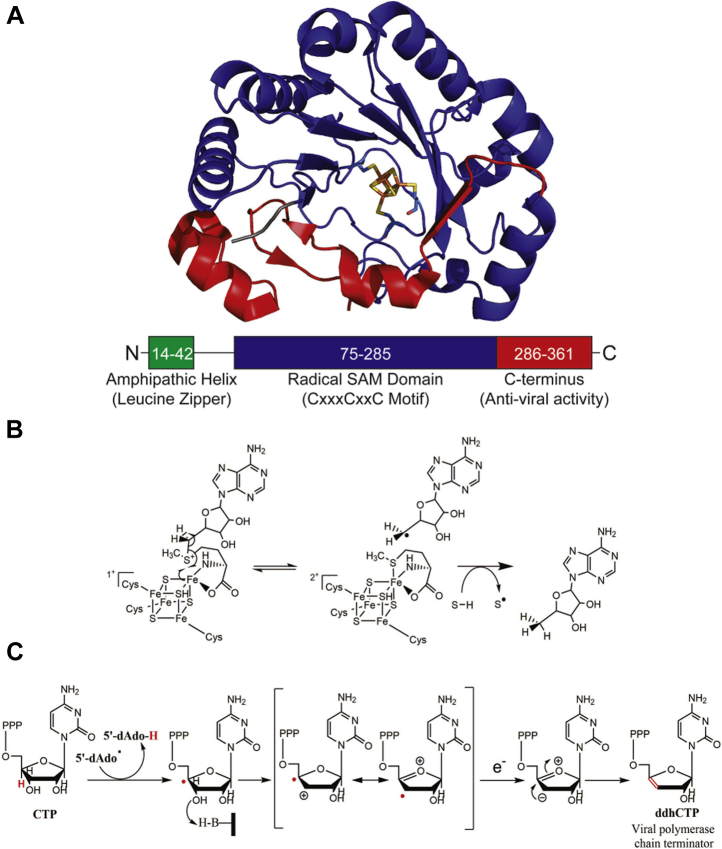
Eukaryotic cells exhibit a wide variety of defenses against viral infections (19). As part of the innate immune response, the first line of defense against infection, production of interferons (IFNs) promotes the upregulation of a wide range of genes to combat infection (20, 21). Viperin—Virus Inhibitory Protein, Endoplasmic Reticulum-associated, Interferon iNducible—(also known as cig5 and radical SAM domain–containing 2) is strongly induced by type I IFNs (16, 22, 23, 24) and is associated with a wide range of antiviral properties (22). In humans, viperin is a ∼42 kDa and 361-residue amino acid, ER-associated protein that, interestingly, is one of only eight radical-SAM enzymes identified in the human genome. The enzyme comprises three domains: an N-terminal amphipathic helix responsible for localizing the enzyme to the ER membrane, a conserved radical-SAM domain containing a canonical Fe4–S4 cluster-binding motif (CxxxCxxC), and a C-terminal domain, largely responsible for binding the substrate (Fig. 1) (23, 25, 26). Viperin catalyzes the dehydration of CTP to form 3’-deoxy-3’,4’-didehydro-CTP (ddhCTP) through a radical mechanism (27). This modified nucleotide has been shown to act as an effective chain-terminating inhibitor of some, but not all, viral RNA-dependent RNA polymerases (27) (Fig. 1).
Figure 1.
Structure and function of the viperin.A, crystal structure of mouse viperin (Protein Data Bank ID: 6Q2P) with the [Fe4–S4] cluster bound and its three domains highlighted. B, the general mechanistic scheme for the generation of 5’-deoxyadenosyl radical by radical SAM enzymes. C, proposed mechanism for the formation of ddhCTP catalyzed by viperin. ddhCTP, 3’-deoxy-3’,4’-didehydro-CTP; viperin, Virus Inhibitory Protein, Endoplasmic Reticulum-associated, Interferon iNducible.
Viperin is implicated in restricting a broad range of viruses including flaviviruses such as dengue virus (28, 29), hepatitis C virus (30, 31, 32), tick-borne encephalitis virus (33), West Nile virus (29), and Zika virus (33, 34, 35) and other types of viruses including human cytomegalovirus (22, 36, 37), HIV (38), and influenza A (18, 39). However, no common mechanism has emerged by which viperin inhibits viral replication. Depending on the virus, the C-terminal domain has been reported to primarily facilitate viperin's antiviral activity (21, 26, 28, 30, 40, 41, 42); in other cases, its antiviral effects have been attributed to the N-terminal helix or the radical-SAM domain (16, 18, 27, 30, 31, 41, 42, 43, 44, 45, 46, 47, 48, 49). The prevailing antiviral mechanisms proposed are localization to the ER and lipid droplets to inhibit viral budding (14, 16, 20, 31); inhibition of viral genome replication either by production of ddhCTP or interaction with viral replication complexes (27, 28, 30, 33, 40, 43, 44, 47, 50); stimulation of innate immune response pathways by interactions with intracellular signaling proteins (35, 49, 51, 52); and targeting of viral proteins for proteolytic degradation through the ubiquitination pathway (33, 43, 51, 53).
Viperin interacts with a diverse array of cellular and viral proteins. For example, viperin binds to the nonstructural proteins of various flaviviruses, leading to inhibition of genome replication and/or viral assembly (28, 30, 33, 40, 50). In addition, viperin facilitates innate immune signaling in the Toll-like receptor-7/9 pathways through interactions with interleukin receptor–associated kinase 1 and E3 ubiquitin ligase, TNF receptor–associated factor 6 (TRAF6), to promote K63-linked polyubiquitination of interleukin receptor–associated kinase 1 by TRAF6, eventually leading to upregulation of IFN expression (49, 51, 52). Although primarily localized to the ER and lipid droplets, viperin can be translocated to the mitochondria where it binds the mitochondrial trifunctional protein β-subunit (Hydroxyacyl-CoA Dehydrogenase Trifunctional Multienzyme Complex Subunit Beta) and inhibits the thiolysis of β-ketoacyl-CoA esters, thereby inhibiting fatty acid catabolism and decreasing cellular ATP levels (36, 37, 54).
Here, we report studies that identify enzymes in the cholesterol biosynthetic pathway as novel targets for viperin. Our studies point to a new role for viperin in regulating cholesterol biosynthesis, which may explain the enzyme's previously observed effect of inhibiting the budding of influenza A and other enveloped viruses from cells.
Results
Viperin expression reduces cholesterol biosynthesis
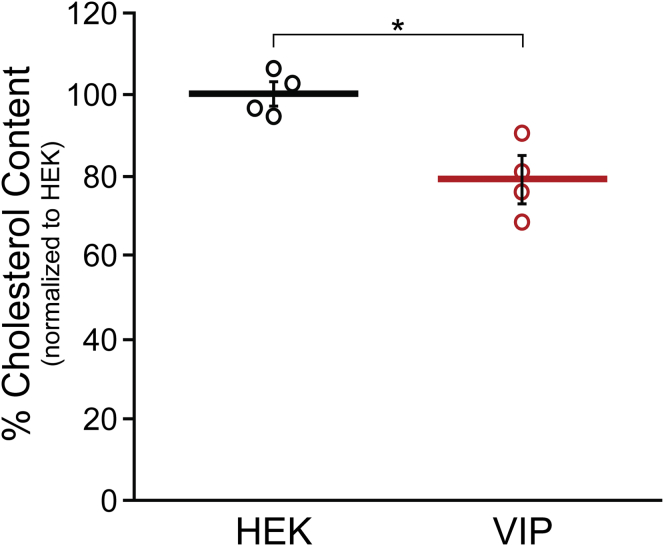
Various studies had previously linked viperin to the downregulation of cholesterol biosynthesis (39, 46, 55), but the effect of viperin expression on cellular cholesterol biosynthesis had not been investigated. To examine this question, human embryonic kidney 293T (HEK293T) cells were grown in reduced serum media containing lipoprotein-depleted serum to minimize uptake of exogenous cholesterol (56). Cells were transfected with either viperin or an empty vector control and grown for a further 36 h before harvesting. Lipids were extracted from the cell pellets and total cellular cholesterol levels quantified by LC–MS, with the results normalized to total cellular protein. The cholesterol content of the control cells was 79.6 ± 2.1 nmol/mg of cellular protein, whereas for cells overexpressing viperin, the cholesterol content was 63.1 ± 3.6 nmol/mg (Fig. 2). These values represent the average of four biological replicates with three technical replicates of each measurement. This represents a 21% decrease in cholesterol, which is both statistically significant, p = 0.008, and potentially biologically significant given that the statin-induced reduction of cholesterol biosynthesis is reported to substantially impair the replication of enveloped viruses such as influenza A and respiratory syncytial virus (RSV) (57, 58). Cells cultured in media containing serum that was not lipoprotein depleted exhibited no significant change in cellular cholesterol levels in response to viperin expression, implying that viperin alters cholesterol biosynthesis rather than cholesterol uptake.
Figure 2.
HEK293T cells expressing viperin exhibit reduced cholesterol levels. HEK293T cells were grown in reduced serum media containing lipoprotein-depleted serum and transfected with empty vector (HEK) or viperin (VIP) as indicated. Cholesterol was extracted from cell pellets, and total cholesterol levels were quantitated via LC–MS using a standard curve generated using pure cholesterol. Results were normalized against total cellular protein content. Values presented are the average of four biological replicates ± SEM. ∗Denotes p < 0.01. HEK293T, human embryonic kidney 293T cells; viperin, Virus Inhibitory Protein, Endoplasmic Reticulum-associated, Interferon iNducible.
The interactome of viperin includes several cholesterol biosynthetic enzymes
Viperin and many of the enzymes involved in cholesterol biosynthesis are associated with the ER membrane (16, 59), leading us to consider whether viperin may downregulate cholesterol levels by inhibiting one or more of these ER-bound enzymes. To examine which, if any, of the cholesterol biosynthetic enzymes may interact with viperin, we undertook a proteomic analysis to map the interactome of viperin and in the process obtain a more comprehensive inventory of the cellular proteins that interact with viperin. To accomplish this, a viperin construct bearing an N-terminal 3x-FLAG tag was transiently expressed in HEK293T cells, as described in the Experimental procedures section. About 42 h post-transfection, the expressed viperin was immunoprecipitated using anti-FLAG antibody-conjugated magnetic beads. The immunoprecipitated proteins were subjected to on-bead reduction, alkylation, and tryptic digestion followed by MS of the alkylated tryptic peptides. An empty 3x-FLAG-tagged pcDNA3.1(+) construct was transfected in HEK293T cells to serve as a control. The MS analyses were performed on three biological replicates for both the viperin and empty vector control samples.
For the initial list of identified proteins, containing 3670 hits, the fold change in protein abundance in the viperin samples over the empty vector control was calculated, with protein abundance estimated using spectral counts (i.e., the total number of peptide-to-spectrum matches, per protein). The shortened list was then analyzed using the Significance Analysis of INTeractome (SAINT) software (60) via the Contaminant Repository for Affinity Purification (CRAPome; www.crapome.org) web resource (61) to exclude common and nonspecifically bound proteins. For each protein identified in the dataset SAINT computes, using statistical modeling of spectral counts across bait purifications and the controls, a confidence score (probability) that the particular protein interacts with the bait protein. By setting the SAINT probability cutoff to be ≥0.9, we obtained a list of 100 possible hits (Table S1). It was apparent from this initial list that proteins involved in steroid biosynthesis and lipid metabolism were heavily represented.
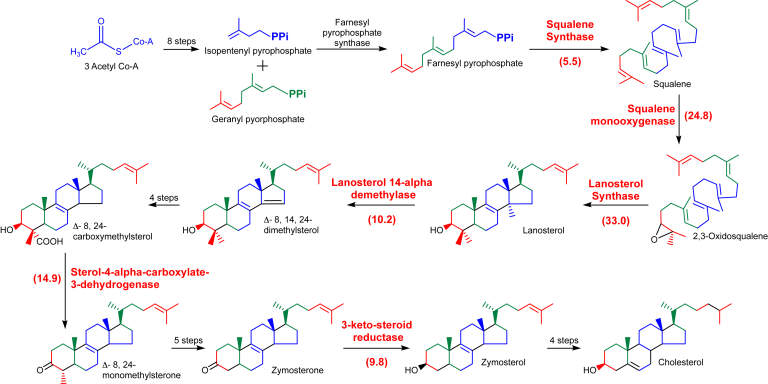
Therefore, we next performed a pathway enrichment analysis on these hits using the Database for Annotation, Visualization and Integrated Discovery (DAVID), version 6.8, software. Functional annotation of these proteins through Kyoto Encyclopedia of Genes and Genomes pathway analysis showed that the highest enrichment scores, 3.59, in DAVID at high stringency were for enzymes in sterol biosynthesis pathways (Table 1 and Fig. 3). This observation provided initial support for the hypothesis that viperin inhibits one or more of the enzymes in cholesterol biosynthesis.
Table 1.
Cholesterol biosynthetic enzymes identified by the proteomic analysis of proteins that were coimmunoprecipitated by viperin
| UniProtKB | Gene name | Enzyme | Enrichment (fold) | SAINT score | Sequence coverage (%) | Unique peptides |
|---|---|---|---|---|---|---|
| P48449 | LSS | LS | 33.0 | 1 | 59.01 | 35 |
| Q14534 | SQLE | SM | 24.8 | 1 | 53.5 | 23 |
| Q15738 | NSDHL | sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | 14.9 | 1 | 67.3 | 19 |
| Q16850 | CYP51A1 | lanosterol 14-alpha demethylase | 10.2 | 1 | 27.2 | 11 |
| P56937 | HSD17B7 | 3-ketosteroid reductase | 9.8 | 1 | 32.6 | 10 |
| P37268 | FDFT1 | squalene synthase | 5.5 | 0.97 | 40.0 | 14 |
The fold enrichment represents the factor by which each protein was enriched in the sample with respect to the empty vector control.
Figure 3.
Enzymes from the cholesterol biosynthetic pathway identified in the interactome of viperin. An overview of the pathway is presented with enzymes identified as interacting with viperin highlighted in red, with enrichment factors given in parentheses. viperin, Virus Inhibitory Protein, Endoplasmic Reticulum-associated, Interferon iNducible.
Viperin interacts with SM and LS to form a ternary complex
Many of the cholesterol biosynthetic enzymes have been shown to interact with each other in a functional complex (59), so it is likely that some of the enzymes identified by the screen are enriched through indirect interactions, rather than binding directly to viperin. Therefore, we focused on the two most highly enriched enzymes: SM and lanosterol synthase (LS). Notably, SM and LS were both substantially enriched over other cholesterol biosynthetic enzymes and were the two most highly enriched proteins identified in the proteomic screen (Table S1). SM catalyzes the conversion of squalene to 2,3-oxidosqualene, and LS catalyzes the subsequent cyclization of 2,3-oxidosqualene to lanosterol (Fig. 2), which are key steps in sterol biosynthesis (2).
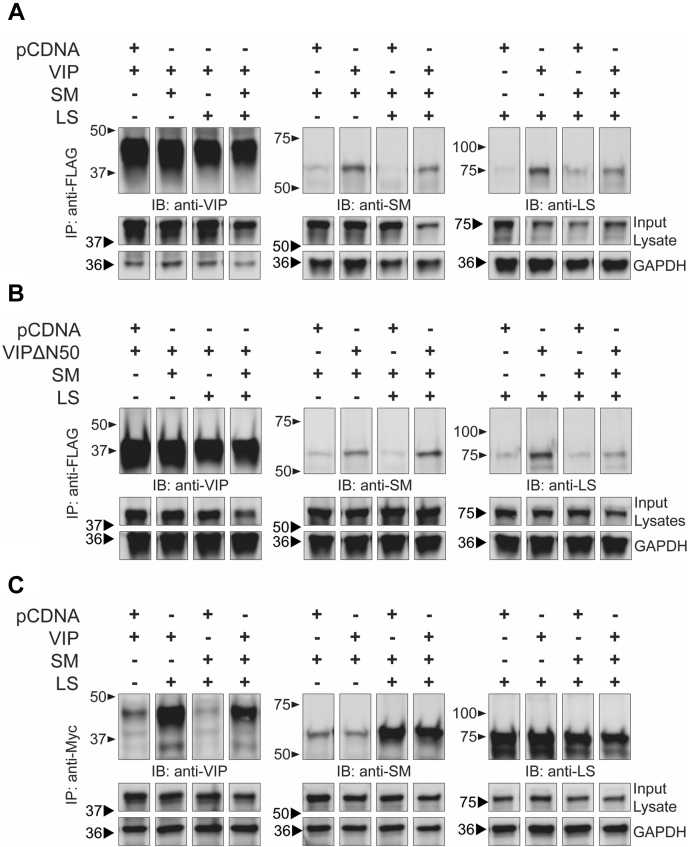
To validate the interactions between LS and SM with viperin, we transiently expressed these proteins in HEK293T cells and subjected them to coimmunoprecipitation analysis. Viperin, SM, and LS were transiently expressed with N-terminal 3x-FLAG, C-terminal V5, and C-terminal Myc epitope tags, respectively. Furthermore, because the N terminus of viperin has proven important for its interactions with various other proteins (16, 30, 43, 47), we also coexpressed a truncated viperin construct lacking the N-terminal 50 residues (viperin-ΔN50) that localize viperin to the ER membrane to examine whether ER localization was important in this instance. Coimmunoprecipitation experiments were performed using either viperin or viperin-ΔN50 as bait proteins with SM and/or LS serving as the prey proteins. Both viperin and viperin-ΔN50 were found to coprecipitate SM and LS when coexpressed individually. Consistent with this observation, viperin and viperin-ΔN50 also coprecipitated both SM and LS when all three enzymes were coexpressed (Fig. 4, A and B).
Figure 4.
Viperin forms a ternary complex with SM and LS.A, HEK293T cells were transfected with empty vector (pCDNA), viperin (VIP), SM, and LS as indicated. Viperin was immunoprecipitated using anti-FLAG magnetic beads and blots probed with antiviperin, anti-SM, or anti-LS polyclonal antibodies. Viperin is shown to pull down both SM and LS. B, the experiment described in A was repeated using a viperin construct lacking the ER membrane–localizing N-terminal domain (VIPΔ50). Similar results were obtained, demonstrating that the interaction of viperin with SM and LS does not depend on ER localization. C, a complementary experiment was performed in which LS was immunoprecipitated using anti-Myc magnetic beads. Consistent with the results in A, LS pulls down both SM and viperin. ER, endoplasmic reticulum; HEK293T, human embryonic kidney 293T cells; LS, lanosterol synthase; SM, squalene monooxygenase; Viperin, Virus Inhibitory Protein, Endoplasmic Reticulum-associated, Interferon iNducible.
To further validate these interactions, a second set of coimmunoprecipitation experiments was performed using LS as the bait protein and viperin and/or SM as the prey proteins. LS coprecipitated both SM and viperin when each coexpressed independently with LS. LS also coprecipitated both viperin and SM when all three enzymes were coexpressed (Fig. 4C). Taken together, these experiments indicate that viperin binds both SM and LS, and also that SM and LS interact with each other, suggesting that the three enzymes form a ternary complex in vivo. These results also indicate that the N-terminal ER-localizing domain of viperin is not required for it to bind either SM or LS.
We note that in these experiments there was some nonspecific binding of each enzyme to the anti-FLAG or anti-Myc beads used to precipitate complexes, as is evident from Figure 4. This indicates that there may be some intrinsic stickiness to these ER-associated proteins. Nevertheless, the significant differences in band intensity between the control and experimental lanes, combined with the large enrichment factors for SM and LS observed in the interactome screening described previously, provide confidence that the interactions are genuine.
Viperin expression reduces LS activity but not SM activity
Previous studies have shown that viperin exerts a range of effects on the proteins it interacts with. Depending on the enzyme, viperin may inhibit or enhance catalytic activity; in other cases, it may target the protein for proteolytic degradation (33, 51, 54, 55). Given the novel interaction between viperin, SM, and LS, and the important role SM and LS play in catalyzing the initial committed steps in sterol biosynthesis, we next examined whether viperin altered the enzymatic activity of either SM or LS.
SM activity assays were performed in triplicate using lysates prepared from HEK293T cells transfected with SM and the results compared with lysates prepared from cells cotransfected with viperin and/or LS. The growth medium was supplemented after transfection with BIBB 515 (1-(4-chlorobenzoyl)-4-((4-(2-oxazolin-2-yl) benzylidene))piperidine), a potent LS inhibitor to prevent conversion of 2,3-oxidosqualene to lanosterol during the assay (62). Assays containing 0.1 mM flavin adenine dinucleotide, 1.0 mM NADPH, and 20 μM squalene were incubated for 2 h at 37 °C, after which time 2,3-oxidosqualene was extracted with ethyl acetate and quantified using LC–MS. LS assays were conducted similarly to the SM assays except that cells were not treated with BIBB 515. In this case, assays contained 50 μM 2,3-oxidosqualene as substrate and were incubated for 2 h at 37 °C prior to extraction and LC–MS analysis. The relative amounts of the various enzymes in the assays were quantified by Western blotting, as described in the Experimental procedures section.
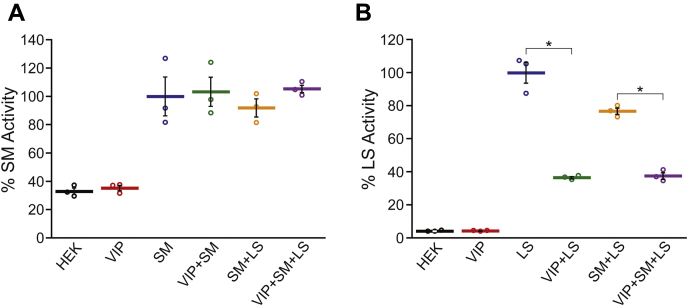
The control SM assays exhibited an appreciable level of background activity because of endogenous SM. However, cells transfected SM exhibited an approximately threefold increase in SM activity to 7.4 ± 0.7 nmol h−1 mg−1 of total protein, consistent with the expression of active enzyme (Fig. 5A). Coexpression of viperin with SM resulted in a modest but statistically significant decrease in the level of SM expression by 29 ± 3% (n = 3, p = 0.002; Fig. S1) compared with expression of SM alone. This resulted in a corresponding decrease in the SM activity measured. But after normalizing for the changes in SM expression levels, no significant difference in the specific activity of SM was observed when it was coexpressed with either viperin or LS or viperin and LS together (Fig. 5A).
Figure 5.
The effect of viperin expression on the enzymatic activity of SM and LS. Lysates were prepared from HEK293T cells transfected with either empty vector (HEK), viperin (VIP), SM, and LS as indicated. A, activity of SM. The amount of 2,3-oxidosqualene produced after 2 h was determined and normalized to the amount of SM expressed in the lysate. The SM activity measured in the HEK and VIP samples arises from endogenously expressed SM. The SM-only sample is arbitrarily set as 100%. B, activity of LS. The amount of lanosterol produced after 2 h was determined and normalized to the amount of LS expressed in the lysate. The LS-only sample is arbitrarily set as 100%. Values presented are the average of three biological replicates ± SEM. ∗Indicates p < 0.001. HEK293T, human embryonic kidney 293T cells; LS, lanosterol synthase; SM, squalene monooxygenase; viperin, Virus Inhibitory Protein, Endoplasmic Reticulum-associated, Interferon iNducible.
In contrast to SM, the endogenous levels of LS activity were much lower, and consequently, transfection with LS resulted in a large, ∼20-fold, increase in the LS activity of the cell lysates, with the LS activity = 6.7 ± 0.7 nmol h−1 mg−1 of total protein (Fig. 5B). In this case, coexpression of viperin modestly increased the expression levels of LS by 29 ± 3% (n = 3, p = 0.007; Fig. S2), but despite this, the LS activity was substantially reduced. After normalizing for expression levels, LS activity was reduced by 60%. The reduction in LS activity because of viperin coexpression was independent of whether SM was coexpressed. Coexpression of SM with LS in the absence of viperin resulted in a slight decrease of ∼20% in LS activity that was not statistically significant (p > 0.05).
LS coexpression inhibits ddhCTP synthesis by viperin
Our previous studies have shown that viperin's enzymatic activity can be significantly altered by its interactions with other enzymes; in some cases, viperin is activated, whereas in others, it is inhibited (43, 51). To examine whether either SM or LS potentially modulated viperin's ddhCTP-forming activity, cell lysates were prepared under anaerobic conditions from HEK293T cells expressing either viperin alone or coexpressing SM and/or LS. Viperin activity was assayed anaerobically using 300 μM CTP and 300 μM SAM as substrates. The reaction was analyzed by quantifying the formation of 5’-deoxyadenosine, and the concentration of viperin in the cell extracts was determined by immunoblotting (Fig. S3) as previously described (43).
In cell lysates expressing only viperin, the observed turnover number, kobs was 4.3 ± 0.5 h−1, which agreed well with values we previously reported (43). In lysates coexpressing SM, no significant change in viperin activity was observed, with kobs = 3.7 ± 0.3 h−1. However, coexpression of viperin with LS resulted in a significant reduction in the specific activity of viperin, with kobs = 2.2 ± 0.2 h−1. When all three proteins were coexpressed, viperin's specific activity was reduced further, kobs = 1.0 ± 0.2 h−1. These data suggest that the decrease in viperin's enzymatic activity arises from inhibition by LS that is potentiated by the interaction of viperin and LS with SM. Although these represent statistically significant decreases in viperin activity, it is unclear if the inhibitory effect of LS has any biological significance.
Effect of LS and SM coexpression on the viperin-induced reduction in cholesterol biosynthesis
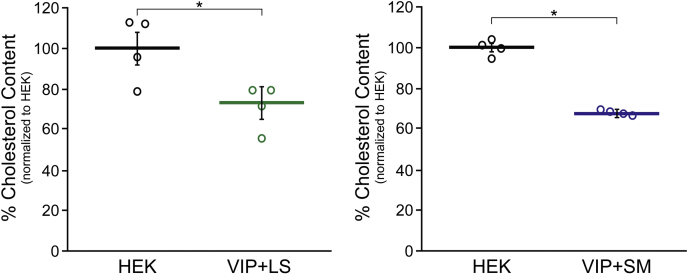
Finally, we examined whether coexpression of SM or LS with viperin would reverse the decrease in cholesterol levels observed in cells transfected with viperin alone. HEK293T cells were cultured in lipoprotein-depleted media as described previously and transfected with viperin, or viperin and LS, or viperin and SM, or empty vector as a control. The cells were harvested 36 h post-transfection, as before, and total cellular cholesterol levels quantified by LC–MS. As shown in Figure 6, coexpression of either LS or SM with viperin failed to reverse the viperin-induced decrease in cholesterol levels. In the viperin + LS cells, cholesterol levels were 72 ± 10% that of the empty vector control. In the viperin + SM cells, cholesterol levels were 68 ± 2% that of the empty vector control. Control experiments established that transfection of HEK293T cells with either SM or LS did not change cellular cholesterol levels. A further control experiment in which the viral protein, NS5A, which similarly localizes to the ER membrane, was transfected into HEK293T cells also had no effect on cellular cholesterol levels. These experiments confirm that the observed reduction in cholesterol biosynthesis is specific to the expression of viperin.
Figure 6.
Viperin-induced reduction in cholesterol levels in HEK293T cells is not reversed by coexpression of SM or LS. HEK293T cells were grown in reduced serum media containing lipoprotein-depleted serum and transfected with empty vector (HEK) or cotransfected with viperin and SM (VIP + SM) or viperin and LS (VIP + LS) as indicated. Cholesterol was extracted from cell pellets, and total cholesterol levels were quantitated via LC–MS using a standard curve generated using pure cholesterol. Results were normalized against total cellular protein content. In both cases, statistically significant reduction in cholesterol levels was observed (cf., Fig. 3): VIP + LS p = 0.05; VIP + SM p = 0.001. Values presented are the average of four biological replicates ± SEM. HEK293T, human embryonic kidney 293T cells; LS, lanosterol synthase; SM, squalene monooxygenase; viperin, Virus Inhibitory Protein, Endoplasmic Reticulum-associated, Interferon iNducible.
Discussion
Cholesterol has been shown to be essential for the viability of a number of enveloped viruses that bud from cholesterol-rich lipid rafts, including influenza A, RSV, and parainfluenza viruses (18, 39, 63, 64, 65, 66). For example, lowering cellular cholesterol levels by 20 to 40% in human alveolar epithelial (A549) cells through treatment with either gemfibrozil and/or lovastatin significantly impaired the budding of human parainfluenza virus. In this case, the titer of infective virions released was reduced by 88 to 98% (67). For influenza A and RSV, a similar statin-induced reduction in cholesterol levels resulted in viral titers that were reduced less dramatically, by ∼10-fold. But in this case, the resulting viral particles, which contained reduced amounts of cholesterol, were both less stable and less infectious (57). These observations demonstrate the potential importance of downregulating cholesterol biosynthesis as an antiviral response to enveloped viruses. Our results show that viperin expression lowers cellular cholesterol levels by between 200 and 30%, depending upon the experimental conditions, which, based on these prior studies, would substantially impair viral budding and viability.
The initial indication that viperin may downregulate cholesterol biosynthesis came from studies in which viperin was found to retard influenza A virus particles from budding from infected cells, although changes in cholesterol levels were not reported (18, 39). Using a yeast two-hybrid screen, viperin was identified as interacting with farnesyl pyrophosphate synthase (FPPS), which catalyzes an early step in the cholesterol biosynthetic pathway (18). However, more recent studies from our laboratory failed to identify a physical interaction between FPPS and viperin. Rather, it appears that viperin may indirectly downregulate FPPS expression, likely by increasing its rate of proteolytic degradation (55). Consistent with these earlier results, FPPS was not identified in the proteomic screen described here. Nevertheless, viperin-induced downregulation of FPPS may contribute to overall decrease in cholesterol biosynthesis we observed.
Multiple studies have provided evidence for viperin's interaction with a remarkably wide range of proteins (19, 21, 68). Many of the protein partners for viperin have been inferred by following changes in cellular physiology upon infection with various viruses. Here, we sought to achieve a broader picture of the proteins that may interact with viperin using a proteomics approach. The identification of several proteins that have previously been documented to interact with viperin, such as Hydroxyacyl-CoA Dehydrogenase Trifunctional Multienzyme Complex Subunit Alpha and Hydroxyacyl-CoA Dehydrogenase Trifunctional Multienzyme Complex Subunit Beta (fatty acid β-oxidation) (37), Cytosolic Iron-Sulfur Assembly Component 1 and MMS19 Homolog, Cytosolic Iron-Sulfur Assembly Component (iron-sulfur cluster installation) (69) serves to validate the screening approach.
However, we note that protein partners that are in low cellular abundance may have been missed by the screen, as would proteins that are only expressed upon viral infection. Also, the location of viperin at the ER membrane may bias the analysis toward enrichment of other ER-resident proteins. Arguing against this latter source of bias is the fact that, with the exception of one protein (ribophorin 1: dolichyldiphospho-oligosaccharide protein glycosyltransferase subunit 1), none of the 20 most abundant ER proteins (70) were identified in our interactome analysis. Therefore, we feel confident that our interactome analysis has identified a genuine interaction of viperin with SM and LS, as opposed to their association simply arising from their colocation at the ER membrane.
In some respects, our results reflect the consensus in the literature that viperin appears to be fairly promiscuous in its interactions with other proteins (36, 40, 49), as evidenced by the wide variety of proteins represented in the list of potential interaction partners. However, in addition to cholesterol biosynthetic enzymes, our analyses using DAVID also identified that a significant number of proteins involved in membrane lipid metabolism interact with viperin (Table S1). This observation suggests that, beyond lowering cholesterol levels, viperin may alter lipid metabolism, and hence, the lipid composition of cell membranes more extensively in response to viral infections.
The two enzymes that viperin appears to interact with most strongly, based on their enrichment factors, are LS and SM. SM catalyzes the committed step in sterol biosynthesis and is highly regulated by both transcriptional and post-translational mechanisms (10, 11, 12). Indeed most, if not all, cholesterol biosynthetic enzymes appear to be regulated at the transcriptional level in response to sterol levels through the action of sterol regulatory element-binding proteins (3, 4, 9, 71). Although the regulation of LS is less well studied, the fact that SM and LS form a complex with other cholesterol biosynthetic enzymes (59) suggests that the effects of regulating SM may be propagated to other enzymes in the complex. The cholesterol biosynthetic enzymes reside in the ER membrane and are therefore likely to be in close proximity to viperin, which is also localized to the ER membrane. It therefore seems reasonable that these enzymes could be subject to regulation by viperin.
We consistently observed that coexpression of viperin reduced the levels of SM accumulating in cells by ∼30%. Although this is not a large change in protein levels, it does suggest that viperin may reduce SM levels by increasing its rate of proteasomal degradation. In support of this idea, the ubiquitin-dependent degradation of SM is known to be regulated in response to cellular cholesterol levels through the action of the E3 ubiquitin ligase, Membrane Associated Ring-CH-Type Finger 6 (11), and it is generally considered that viperin exerts some of its antiviral effects by increasing the rate of proteasomal degradation of its target proteins (15, 68). Although the evidence for viperin's interaction with proteasomal degradation machinery is indirect, viperin has recently been shown to activate TRAF6 (72), which is an E3 ligase involved in the K63-linked polyubiquitination of proteins involved in immune signaling (49, 51).
Although we observed that viperin coexpression reduced the enzymatic activity of LS in cell lysates and the expression levels of SM, coexpression of either LS or SM with viperin did not reverse the viperin-induced decrease in cellular cholesterol levels. This observation was somewhat surprising, given the strong interactions between these enzymes and viperin and their important roles in cholesterol biosynthesis. However, it is possible that under the conditions of these experiments, in which LS, SM, and viperin were expressed at artificially high levels, that there was still sufficient viperin present to titrate out the additional LS and SM. It is also possible that viperin downregulates cholesterol biosynthesis by restricting the flux through other points in the pathway, for example, by reducing FPPS levels, or through a more circuitous route not captured in these studies. Further experiments will be needed to better establish how viperin downregulates this important biosynthetic pathway.
Conclusions
We observed that transient expression of viperin results in a 20 to 30% decrease in cellular cholesterol biosynthesis, which is sufficient to explain previous observations that viperin retards virus budding from the cell membrane. Consistent with this observation, the interactome of viperin includes a number of enzymes involved in the later stages of cholesterol biosynthesis. Of these, SM and LS were the most highly enriched proteins identified, and their interactions with viperin were validated by coimmunoprecipitation. Coexpression of viperin reduced the cellular levels of SM by ∼30% and inhibited the enzymatic activity of LS by over 60%. However, overexpression of either SM or LS in HEK293T cells failed to reverse the effects of viperin expression on cellular cholesterol levels. This observation suggests that viperin's downregulation of cholesterol biosynthesis is a complex phenomenon and not simply an effect of its interactions with these two biosynthetic enzymes.
Experimental procedures
Cell lines
The HEK293T cell line was obtained from American Type Culture Collection.
Plasmids
The expression constructs for viperin and viperin-ΔN50, lacking the N-terminal 50 residues, in pcDNA3.1(+) were as described previously (43). The genes encoding SM and LS were synthesized and cloned into the pcDNA3.1(+) vector commercially (GenScript). SM was inserted between the BamHI and XbaI restriction sites and included a C-terminal V5 tag. LS was inserted between the HindIII and BamHI restriction sites and included a C-terminal Myc tag. A Kozak consensus sequence (5’-GCCACC-3’) was included upstream of each protein to allow for protein expression in mammalian (HEK293T) cells.
Antibodies
Rabbit polyclonal viperin (11833-1-AP), mouse monoclonal viperin (MABF106), goat anti-rabbit Ig secondary (170-6515), and goat antimouse Ig secondary (626520) antibodies were used as described previously (43). Rabbit polyclonal SM (12544-1-AP), rabbit polyclonal LS (13715-1-AP), and rabbit polyclonal GAPDH (10494-1-AP) antibodies were purchased from ProteinTech. Mouse monoclonal GAPDH (6C5) antibody (CB1001) was purchased from EMD Millipore.
Reagents
For cell culture, Dulbecco's modified Eagle's medium, Opti-MEM reduced serum medium, 0.05% trypsin–EDTA, and PBS were obtained from Gibco (Thermo Fisher Scientific); lipoprotein-deficient serum from fetal calf was purchased from Millipore Sigma. For transfection of DNA into HEK293T cells, transfection grade linear polyethyleneimine (PEI) HCl was purchased from Polysciences, Inc, and Fugene transfection agent was purchased from Promega. Phenylmethylsulfonyl fluoride and Roche complete EDTA-free protease inhibitor cocktail tablets were purchased from Thermo Fisher Scientific and Millipore Sigma, respectively. For coimmunoprecipitation, anti-FLAG M2 magnetic beads and Pierce anti-c-Myc magnetic beads were purchased from Millipore Sigma and Thermo Fisher Scientific, respectively. S-(5′-adenosyl)-l-methionine p-toluenesulfonate salt was purchased from Millipore Sigma, and CTP disodium salt hydrate was purchased from Acros Organics. For the SM and LS assays, NADPH tetrasodium salt, flavin adenosine dinucleotide disodium salt hydrate, 2,3-oxidosqualene, lanosterol, and 2,6-di-tert-butyl-4-methylphenol (BHT) were purchased from Millipore sigma. Squalene was purchased from Thermo Fisher Scientific, and BIBB 515 was purchased from Cayman Chemical.
Cell culture, transfection, and harvesting
HEK293T cells were cultured, transfected, and harvested as described previously (43). Briefly, HEK293T cells were grown to 50 to 60% confluency after which transfection with the desired construct(s) (in pcDNA3.1(+)) was carried out using a PEI transfection agent. Transfections (and cotransfections) were performed with each 20 μg plasmid, using a 1:2 ratio of plasmid to PEI. DNA and PEI were mixed and incubated for 10 min at room temperature and then added to HEK293T cells (at the desired confluency) in a 10-cm culture plate. Typically, cells were allowed to grow for a further 24 to 48 h before harvesting by gentle centrifugation and stored at −80 °C. For cells used in SM assays, the LS-specific inhibitor, BIBB 515, 1 μM was added 24 h after transfection to prevent further conversion of 2,3-oxidosqualene to lanosterol by endogenous LS (62).
The cells used for cholesterol analyses were first gradually acclimated to increasing Opti-MEM serum-free media in four passages at 10, 25, 50, and 75% Opti-MEM media, to allow for normal growth with minimal serum. Then cells were passed into 90% Opti-MEM medium and 10% medium comprising Dulbecco's modified Eagle's medium supplemented with lipoprotein-depleted fetal calf serum; grown to 50 to 60% confluency and transfected with either an empty pCDNA3.1 (+) vector as a control or viperin in pCDNA3.1 (+) as described earlier. Cells were grown for 36 h post-transfection, harvested, and stored at −80 °C.
Proteomic screening
HEK293T cells overexpressing either 3x-FLAG-viperin or 3x-FLAG-tagged empty vectors (3x-FLAG-pcDNA3.1) as a control were harvested in PBS from 10-cm plates. Cells were incubated on ice for 20 min with 0.5 ml lysis buffer (20 mM Tris [pH 7.5], 500 mM NaCl, 0.1% Tween-20, 1 mM phenylmethylsulfonyl fluoride, and Complete Protease Inhibitor Cocktail) and lysed using a Fisher Scientific handheld sonicator at 10% amplitude (25 pulses; 1 s on, 5 s off). Cell lysates were cleared by centrifugation at 14,000 rpm for 30 min at 4 °C, and the pellets were discarded. Anti-FLAG M2 Magnetic Beads (M8823; Sigma–Aldrich) were washed and pre-equilibrated with ice-cold 20 mM Tris (pH 7.5), 500 mM NaCl, and 0.1% Tween-20. Total protein concentration of the supernatant was measured by Protein Detergent-Compatible Assay (Bio-Rad) and subjected to immunoprecipitation using anti-Flag M2 magnetic beads. Supernatant (total protein concentration ∼7 μg/μl) was incubated with the pre-equilibrated anti-FLAG magnetic beads at 50:1 (w/v) ratio for 2 h at 4 °C. Beads were washed three times with 20× bead volume of Tris-buffered saline (TBS) using a magnetic tube rack (MagRack 6; GE Life Sciences).
The beads were resuspended in 50 μl of 0.1 M ammonium bicarbonate buffer (pH ∼8); cysteine residues were reduced by adding 50 μl of 10 mM DTT and incubating at 45 °C for 30 min. Samples were cooled to room temperature and cysteine alkylated by incubation with 65 mM 2-chloroacetamide for 30 min at room temperature in the dark. Overnight digestion with 1 μg sequencing grade, modified trypsin was carried out at 37 °C with constant shaking. Digestion was stopped by acidification, and peptides were desalted using SepPak C18 cartridges (Waters). Samples were dried, and the resulting peptides were redissolved in 8 μl of 0.1% formic acid/2% acetonitrile solution. About 2 μl of the peptide solution were chromatographed on a nanocapillary reverse phase column (Acclaim PepMap C18; 2 micron, 50 cm, Thermo Scientific) using a 0.1% formic acid/2% acetonitrile (buffer A) and 0.1% formic acid/95% acetonitrile (buffer B) gradient at 300 nl/min over a period of 180 min (2–22% buffer B in 110 min, 22–40% in 25 min, 40–90% in 5 min followed by holding at 90% buffer B for 5 min and re-equilibration with buffer A for 25 min). Eluent was directly introduced into Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific) using an Easy-Spray source. MS1 scans were acquired at a resolution of 120 K (automatic gain control target = 1 × 106; maximum injection time = 50 ms). Data-dependent collision–induced dissociation MS–MS spectra were acquired using top speed method (3 s) following each MS1 scan (normalized collision energy = ∼32%; automatic gain control target = 1 × 105; and maximum injection time = 45 ms).
Proteins were identified by comparing the MS–MS data against Homo sapiens protein database (UniProt; 42,054 entries, November 30, 2016) using Proteome Discoverer (version 2.1; Thermo Scientific). Search parameters included MS1 mass tolerance of 10 ppm and fragment tolerance of 0.2 Da; two missed cleavages were allowed; carbamidimethylation of cysteine was considered a fixed modification and oxidation of methionine, deamidation of asparagine and glutamine, phosphorylation of serine, threonine, and tyrosine, ubiquitination of lysine (diglycine signature) were considered as potential modifications. False discovery rate was determined using percolator, and proteins/peptides with a false discovery rate of ≤1% were retained for further analysis.
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (73) partner repository with the dataset identifier PXD023999.
Immunoblotting
Immunoblotting of HEK293T cell lysates was performed as previously described (51, 55). Primary antibodies were used at the following dilutions: rabbit polyclonal and mouse monoclonal antiviperin diluted 1:3000, rabbit polyclonal anti-SM diluted 1:2000, rabbit polyclonal anti-LS diluted 1:1000, and rabbit polyclonal and mouse monoclonal anti-GAPDH diluted 1:5000. Both goat anti-rabbit and goat antimouse Ig secondary antibodies were diluted at 1:5000. All dilutions were done using 5% w/v skim milk dissolved in 20 mM Tris (pH 7.5), 500 mM NaCl, and 0.1% Tween-20. All quantitative protein measurements presented represent the average of at least three independent biological replicates in coordination with GAPDH controls.
Coimmunoprecipitation assays
Immunoprecipitation experiments were performed essentially as described previously (43, 51). When using viperin as the bait protein, Anti-FLAG M2 Magnetic Beads (M8823; Sigma–Aldrich) were pre-equilibrated with 20 mM Tris/Cl (pH 7.5), 500 mM NaCl, and 0.1% Tween-20 (wash buffer). About 50 μl of 50% slurry (per culture) was added to cleared cell lysate in a 1.5 ml Eppendorf tube and incubated by end-to-end mixing for 2 h at 4 °C. The flow through was removed by placing the tube in a magnetic tube rack for 1 min, waiting for all beads to migrate to the side of the tube, and removing the residual solution. The beads were washed with wash buffer three times for 5 min each by end-to-end mixing at 4 °C. Protein complexes were eluted by adding 40 μl of 4× sample buffer supplemented with 2 mM 2-mercaptoethanol to each sample and subsequent incubation at 95 °C for 5 min. The samples were centrifuged to remove the magnetic beads and analyzed by immunoblotting. When using LS as the bait protein, the same protocol was used except that 40 μl of 50% slurry (per culture) of pre-equilibrated and chilled Pierce Anti-c-Myc Magnetic Beads (Thermo Fisher Scientific) were substituted for anti-FLAG beads.
Viperin activity assays
Viperin activity assays were performed as previously described (43). All assays were carried out using at least three biological replicates in an anaerobic chamber (COY) with O2 content <50 ppm. Briefly, HEK293T cells containing viperin and/or SM and/or LS were harvested from one 10-cm diameter tissue culture plate for each assay and resuspended in 0.5 ml anoxic lysis buffer (aforementioned). The cell suspension was lysed by sonication and centrifuged for 15 min at 14,000 rpm in an Eppendorf centrifuge at 4 °C. The supernatant was used for the activity assay. 5 mM DTT, 5 mM sodium dithionite, and 300 μM CTP were added to the cleared lysate and incubated at room temperature for 30 min. The reaction was initiated by the addition of 200 μM (final concentration) SAM and allowed to proceed for 1 h at room temperature. The reaction was quenched by incubation at 95 °C for 5 min. After chilling the solution to 4 °C, it was centrifuged for 30 min at 14,000 rpm, and the supernatant was removed. The organic component of the solution was extracted with acetonitrile and analyzed in triplicate by ultra performance LC (UPLC)–MS–MS as previously described (74).
SM activity assays in HEK293T cell lysates
HEK293T cells overexpressing different combinations of viperin, SM, and LS (which had been incubated post-transfection with 1 μM BIBB 515) were harvested from a 10-cm diameter tissue culture plate; the cells from one plate were sufficient for one assay. HEK293T cells transfected with an empty pcDNA3.1(+) vector were used as a negative control. SM activity was assayed by measuring the conversion of squalene to 2,3-oxidosqualene. Cells were resuspended in lysis buffer (TBS, 0.1% Tween-20, and Complete Protease Inhibitor Cocktail) and lysed as described previously. All assays were performed in lightproof tubes and performed as previously described (72), but with minor modifications. Briefly, assays contained 200 μl cleared HEK cell lysate prepared from cells expressing the protein(s) of interest, 0.1 mM flavin adenine dinucleotide, and 1 mM NADPH. Assays were initiated by the addition of squalene (from a 500 μM stock solution prepared in ethanol) to a final concentration of 20 μM in a final reaction volume of 250 μl. Assays were incubated at 37 °C for 2 h and quenched with 1 ml ethyl acetate supplemented with 0.2 mg/ml BHT purged with nitrogen followed by vortexing at room temperature for 30 min. The organic layer was isolated by centrifugation for 10 min at 14,000 rpm. About 300 μl of the organic layer was removed and evaporated to complete dryness under a nitrogen atmosphere. The resulting film was reconstituted with 50 μl acetonitrile supplemented with 0.2 mg/ml BHT.
LC–MS analysis was performed as previously described (75) with minor differences. Briefly, 2 μl of sample was injected into an Agilent 1290 Infinity Series HPLC equipped with a Waters Acquity UPLC BEH C18 column (1.7 μm, 2.1 × 50 mm) in line with an Agilent Q-TOF Mass Spectrometer (model G6520B). The HPLC separation was done on a Waters Acquity UPLC BEH C18 column (1.7 μm, 2.1 × 50 mm). Mobile phase A comprised water with 0.1% formic acid, whereas mobile phase B comprised 20% isopropanol, 80% acetonitrile, and 0.1% formic acid. Analytes were eluted isocratically at 0.6 ml/min using 2% mobile phase A and 98% mobile phase B over 3.75 min. Q-TOF spectra were acquired using an atmospheric pressure chemical ionization ion source in positive ion mode. The mass spectrometer was scanned from m/z 50 to m/z 1000 with one spectrum per second, and 2,3-oxidosqualene was detected by monitoring a mass transition pair of m/z 427.38 to 409.38. The LC–MS chromatograms were integrated, and resulting peaks were analyzed using Mass Hunter software (Agilent).
LS activity assays from HEK293T cell lysate
LS activity assays were carried out in the same manner as the SM assays except that cells were not treated with BIBB 515. HEK293T cells transfected with an empty pcDNA3.1(+) vector were used as a negative control. For each assay, cells were harvested from a single 10-cm diameter tissue culture plate, and cleared lysates were prepared. LS activity was measured by formation of lanosterol from 2,3-oxidosqualene. All reactions were carried out in lightproof tubes and contained 200 μl cleared HEK cell lysate and were initiated by the addition of 50 μM 2,3-oxidosqualene, final concentration (500 μM stock, all solutions prepared in ethanol), in a final reaction volume of 250 μl. Assays were incubated at 37 °C for 2 h and then worked up as described previously except that the reactions were quenched with chloroform. LC–MS analysis was performed as described previously, but the ion analyzed was that of lanosterol minus the OH group having an m/z of 409.38.
Cholesterol analysis of HEK293T cells
HEK293T cell pellets were resuspended in 500 μl TBS supplemented with 0.1% Tween-20 and lysed by sonication. Cholesterol was extracted from 50 μl of the resulting lysates with 1.2 ml ethyl acetate supplemented with 0.2 mg/ml BHT purged with nitrogen and vortexing at room temperature for 30 min. The organic component was isolated by centrifugation for 10 min at 14,000 rpm. About 500 μl of the organic layer was removed and evaporated to dryness under a nitrogen atmosphere. The resulting film was redissolved in 500 μl acetonitrile containing 0.2 mg/ml BHT. LC–MS analysis was performed as described previously, except the ion analyzed was that of the cholesterol radical without hydroxide at an m/z of 369.25. Cholesterol content was determined with respect to a standard curve constructed from pure cholesterol samples. Cholesterol concentrations were normalized with respect to the total protein concentration of the cell determined by bicinchoninic acid assay.
Statistical analyses
All measurements of enzyme activities and cholesterol levels made in HEK298T cells or cell lysates represent the average of at least three biological replicates, with three technical replicates of each. Results from all studies were compared with unpaired two-tailed Student's t test using GraphPad Prism v5.0 software, (GraphPad Software, Inc.). p Values less than 0.05 were considered significant.
Data availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD023999. All other data are contained within the article or supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
T. J. G., S. G., A. M. P., K. S., V. B., and Y. K. investigation; T. J. G., S. G., and E. N. G. M. writing–original draft; T. J. G., A. I. N., R. T. K., and E. N. G. M. writing–review and editing; T. J. G., S. G., J. W., V. B., Y. K., R. T. K., and A. I. N. methodology; V. B. and A. I. N. formal analysis; A. I. N. resources; R. T. K. and E. N. G. M. conceptualization; R. T. K. and E. N. G. M. funding acquisition; E. N. G. M. supervision; E. N. G. M. project administration.
Funding and additional information
This work was supported in part by the National Institutes of Health grants GM 093088 to E. N. G. M., DK 046960 to R. T. K., and GM 094231 to A. I. N. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Peter Cresswell
Supporting information
References
- 1.Incardona J.P., Eaton S. Cholesterol in signal transduction. Curr. Opin. Cell Biol. 2000;12:193–203. doi: 10.1016/s0955-0674(99)00076-9. [DOI] [PubMed] [Google Scholar]
- 2.Cerqueira N.M.F.S.A., Oliveira E.F., Gesto D.S., Santos-Martins D., Moreira C., Moorthy H.N., Ramos M.J., Fernandes P.A. Cholesterol biosynthesis: A mechanistic overview. Biochemistry. 2016;55:5483–5506. doi: 10.1021/acs.biochem.6b00342. [DOI] [PubMed] [Google Scholar]
- 3.Espenshade P.J., Hughes A.L. Regulation of sterol synthesis in eukaryotes. Annu. Rev. Genet. 2007;41:401–427. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- 4.Brown M.S., Goldstein J.L. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 5.Faust J.R., Luskey K.L., Chin D.J., Goldstein J.L., Brown M.S. Regulation of synthesis and degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase by low density lipoprotein and 25-hydroxycholesterol in UT-1 cells. Proc. Natl. Acad. Sci. U. S. A. 1982;79:5205–5209. doi: 10.1073/pnas.79.17.5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein J.L., DeBose-Boyd R.A., Brown M.S. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 7.Osborne T.F., Gil G., Goldstein J.L., Brown M.S. Operator constitutive mutation of 3-hydroxy-3-methylglutaryl coenzyme A reductase promoter abolishes protein binding to sterol regulatory element. J. Biol. Chem. 1988;263:3380–3387. [PubMed] [Google Scholar]
- 8.Rajavashisth T.B., Taylor A.K., Andalibi A., Svenson K.L., Lusis A.J. Identification of a zinc finger protein that binds to the sterol regulatory element. Science. 1989;245:640–643. doi: 10.1126/science.2562787. [DOI] [PubMed] [Google Scholar]
- 9.Goldstein J.L., Brown M.S. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 10.Gill S., Stevenson J., Kristiana I., Brown A.J. Cholesterol-dependent degradation of squalene monooxygenase, a control point in cholesterol synthesis beyond HMG-CoA reductase. Cell Metab. 2011;13:260–273. doi: 10.1016/j.cmet.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 11.Zelcer N., Sharpe L.J., Loregger A., Kristiana I., Cook E.C.L., Phan L., Stevenson J., Brown A.J. The E3 ubiquitin ligase MARCH6 degrades squalene monooxygenase and affects 3-hydroxy-3-methyl-glutaryl coenzyme A reductase and the cholesterol synthesis pathway. Mol. Cell. Biol. 2014;34:1262–1270. doi: 10.1128/MCB.01140-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howe V., Chua N.K., Stevenson J., Brown A.J. The regulatory domain of squalene monooxygenase contains a re-entrant loop and senses cholesterol via a conformational change. J. Biol. Chem. 2015;290:27533–27544. doi: 10.1074/jbc.M115.675181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simons K., Ehehalt R. Cholesterol, lipid rafts, and disease. J. Clin. Invest. 2002;110:597–603. doi: 10.1172/JCI16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzgerald K.A. The interferon inducible gene: Viperin. J. Interf. Cytokine Res. 2011;31:131–135. doi: 10.1089/jir.2010.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helbig K.J., Beard M.R. The role of viperin in the innate antiviral response. J. Mol. Biol. 2014;426:1210–1219. doi: 10.1016/j.jmb.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 16.Hinson E.R., Cresswell P. The N-terminal amphipathic α-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J. Biol. Chem. 2009;284:4705–4712. doi: 10.1074/jbc.M807261200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 18.Wang X., Hinson E.R., Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe. 2007;2:96–105. doi: 10.1016/j.chom.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 19.Mattijssen S., Pruijn G.J.M. Viperin, a key player in the antiviral response. Microbes Infect. 2012;14:419–426. doi: 10.1016/j.micinf.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 20.Duschene K.S., Broderick J.B. Viperin: A radical response to viral infection. Biomol. Concepts. 2012;3:255–266. doi: 10.1515/bmc-2011-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo J.Y., Yaneva R., Cresswell P. Viperin: A multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe. 2011;10:534–539. doi: 10.1016/j.chom.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chin K.C., Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 2001;98:15125–15130. doi: 10.1073/pnas.011593298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaveta G., Shi J., Chow V.T.K., Song J. Structural characterization reveals that viperin is a radical S-adenosyl-l-methionine (SAM) enzyme. Biochem. Biophys. Res. Commun. 2010;391:1390–1395. doi: 10.1016/j.bbrc.2009.12.070. [DOI] [PubMed] [Google Scholar]
- 24.Duschene K.S., Broderick J.B. The antiviral protein viperin is a radical SAM enzyme. FEBS Lett. 2010;584:1263–1267. doi: 10.1016/j.febslet.2010.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haldar S., Paul S., Joshi N., Dasgupta A., Chattopadhyay K. The presence of the iron-sulfur motif is important for the conformational stability of the antiviral protein, viperin. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fenwick M.K., Li Y., Cresswell P., Modis Y., Ealick S.E. Structural studies of viperin, an antiviral radical SAM enzyme. Proc. Natl. Acad. Sci. U. S. A. 2017;114:6806–6811. doi: 10.1073/pnas.1705402114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gizzi A.S., Grove T.L., Arnold J.J., Jose J., Jangra R.K., Garforth S.J., Du Q., Cahill S.M., Dulyaninova N.G., Love J.D., Chandran K., Bresnick A.R., Cameron C.E., Almo S.C. A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature. 2018;558:610–614. doi: 10.1038/s41586-018-0238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helbig K.J., Carr J.M., Calvert J.K., Wati S., Clarke J.N., Eyre N.S., Narayana S.K., Fiches G.N., McCartney E.M., Beard M.R. Viperin is induced following Dengue virus type-2 (DENV-2) infection and has anti-viral actions requiring the C-terminal end of viperin. PLoS Negl. Trop. Dis. 2013;7 doi: 10.1371/journal.pntd.0002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang D., Weidner J.M., Qing M., Pan X.-B., Guo H., Xu C., Zhang X., Birk A., Chang J., Shi P.-Y., Block T.M., Guo J.-T. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and Dengue virus infections. J. Virol. 2010;84:8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helbig K.J., Eyre N.S., Yip E., Narayana S., Li K., Fiches G., Mccartney E.M., Jangra R.K., Lemon S.M., Beard M.R. The antiviral protein viperin inhibits hepatitis C virus replication via interaction with nonstructural protein 5A. Hepatology. 2011;54:1506–1517. doi: 10.1002/hep.24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hinson E.R., Cresswell P. The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic α-helix. Proc. Natl. Acad. Sci. U. S. A. 2009;106:20452–20457. doi: 10.1073/pnas.0911679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helbig K.J., Lau D.T.Y., Semendric L., Harley H.A.J., Beard M.R. Analysis of ISG expression in chronic hepatitis C identifies viperin as a potential antiviral effector. Hepatology. 2005;42:702–710. doi: 10.1002/hep.20844. [DOI] [PubMed] [Google Scholar]
- 33.Panayiotou C., Lindqvist R., Kurhade C., Vonderstein K., Pasto J., Edlund K., Upadhyay A.S., Överby A.K. Viperin restricts Zika virus and tick-borne encephalitis virus replication by targeting NS3 for proteasomal degradation. J. Virol. 2018;92 doi: 10.1128/JVI.02054-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Der Hoek K.H., Eyre N.S., Shue B., Khantisitthiporn O., Glab-Ampi K., Carr J.M., Gartner M.J., Jolly L.A., Thomas P.Q., Adikusuma F., Jankovic-Karasoulos T., Roberts C.T., Helbig K.J., Beard M.R. Viperin is an important host restriction factor in control of Zika virus infection. Sci. Rep. 2017;7:4475–4481. doi: 10.1038/s41598-017-04138-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanwalscappel B., Tada T., Landau N.R. Toll-like receptor agonist R848 blocks Zika virus replication by inducing the antiviral protein viperin. Virology. 2018;522:199–208. doi: 10.1016/j.virol.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seo J.Y., Cresswell P. Viperin regulates cellular lipid metabolism during human cytomegalovirus infection. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seo J.Y., Yaneva R., Hinson E.R., Cresswell P. Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science. 2011;332:1093–1097. doi: 10.1126/science.1202007. [DOI] [PubMed] [Google Scholar]
- 38.Nasr N., Maddocks S., Turville S.G., Harman A.N., Woolger N., Helbig K.J., Wilkinson J., Bye C.R., Wright T.K., Rambukwelle D., Donaghy H., Beard M.R., Cunningham A.L. HIV-1 infection of human macrophages directly induces viperin which inhibits viral production. Blood. 2012;120:778–788. doi: 10.1182/blood-2012-01-407395. [DOI] [PubMed] [Google Scholar]
- 39.Waheed A.A., Freed E.O. Influenza virus not cRAFTy enough to dodge viperin. Cell Host Microbe. 2007;2:71–72. doi: 10.1016/j.chom.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 40.Wang S., Wu X., Pan T., Song W., Wang Y., Zhang F., Yuan Z. Viperin inhibits hepatitis C virus replication by interfering with binding of NS5A to host protein hVAP-33. J. Gen. Virol. 2012;93:83–92. doi: 10.1099/vir.0.033860-0. [DOI] [PubMed] [Google Scholar]
- 41.Dukhovny A., Shlomai A., Sklan E.H. The antiviral protein viperin suppresses T7 promoter dependent RNA synthesis-possible implications for its antiviral activity. Sci. Rep. 2018;8:8100. doi: 10.1038/s41598-018-26516-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fenwick M.K., Su D., Dong M., Lin H., Ealick S.E. Structural basis of the substrate selectivity of viperin. Biochemistry. 2020;59:652–662. doi: 10.1021/acs.biochem.9b00741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh S., Patel A.M., Grunkemeyer T.J., Dumbrepatil A.B., Zegalia K., Kennedy R.T., Marsh E.N.G. Interactions between viperin, vesicle-associated membrane protein A, and hepatitis C virus protein NS5A modulate viperin activity and NS5A degradation. Biochemistry. 2020;59:780–789. doi: 10.1021/acs.biochem.9b01090. [DOI] [PubMed] [Google Scholar]
- 44.Ebrahimi K.H., Howie D., Rowbotham J.S., McCullagh J., Armstrong F.A., James W.S. Viperin, through its radical-SAM activity, depletes cellular nucleotide pools and interferes with mitochondrial metabolism to inhibit viral replication. FEBS Lett. 2020;594:1624–1630. doi: 10.1002/1873-3468.13761. [DOI] [PubMed] [Google Scholar]
- 45.Honarmand Ebrahimi K., Vowles J., Browne C., McCullagh J., James W.S. ddhCTP produced by the radical-SAM activity of RSAD2 (viperin) inhibits the NAD + -dependent activity of enzymes to modulate metabolism. FEBS Lett. 2020;594:1631–1644. doi: 10.1002/1873-3468.13778. [DOI] [PubMed] [Google Scholar]
- 46.Mikulecky P., Andreeva E., Amara P., Weissenhorn W., Nicolet Y., Macheboeuf P. Human viperin catalyzes the modification of GPP and FPP potentially affecting cholesterol synthesis. FEBS Lett. 2018;592:199–208. doi: 10.1002/1873-3468.12941. [DOI] [PubMed] [Google Scholar]
- 47.Xu C., Feng L., Chen P., Li A., Guo S., Jiao X., Zhang C., Zhao Y., Jin X., Zhong K., Guo Y., Zhu H., Han L., Yang G., Li H. Viperin inhibits classical swine fever virus replication by interacting with viral nonstructural 5A protein. J. Med. Virol. 2020;92:149–160. doi: 10.1002/jmv.25595. [DOI] [PubMed] [Google Scholar]
- 48.Jiang X., Chen Z.J. Viperin links lipid bodies to immune defense. Immunity. 2011;34:285–287. doi: 10.1016/j.immuni.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saitoh T., Satoh T., Yamamoto N., Uematsu S., Takeuchi O., Kawai T., Akira S. Antiviral protein viperin promotes toll-like receptor 7- and toll-like receptor 9-mediated type i interferon production in plasmacytoid dendritic cells. Immunity. 2011;34:352–363. doi: 10.1016/j.immuni.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 50.Foster T.L., Belyaeva T., Stonehouse N.J., Pearson A.R., Harris M. All three domains of the hepatitis C virus nonstructural NS5A protein contribute to RNA binding. J. Virol. 2010;84:9267–9277. doi: 10.1128/JVI.00616-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dumbrepatil A.B., Ghosh S., Zegalia K.A., Malec P.A., Hoff J.D., Kennedy R.T., Marsh E.N.G. Viperin interacts with the kinase IRAK1 and the E3 ubiquitin ligase TRAF6, coupling innate immune signaling to antiviral ribonucleotide synthesis. J. Biol. Chem. 2019;294:6888–6898. doi: 10.1074/jbc.RA119.007719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J.J., Kim K.S., Eom J., Lee J.B., Seo J.Y. Viperin differentially induces interferon-stimulated genes in distinct cell types. Immune Netw. 2019;19 doi: 10.4110/in.2019.19.e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan Y., Miao Y., Qian L., Zhang Y., Liu C., Liu J., Zuo Y., Feng Q., Guo T., Zhang L., Chen X., Jin L., Huang F., Zhang H., Zhang W. Targeting UBE4A revives viperin protein in epithelium to enhance host antiviral defense. Mol. Cell. 2020;77:734–747. doi: 10.1016/j.molcel.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 54.Dumbrepatil A.B., Zegalia K.A., Sajja K., Kennedy R.T., Marsh E.N.G. Targeting viperin to the mitochondrion inhibits the thiolase activity of the trifunctional enzyme complex. J. Biol. Chem. 2020;295:2839–2849. doi: 10.1074/jbc.RA119.011526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Makins C., Ghosh S., Román-Meléndez G.D., Malec P.A., Kennedy R.T., Marsh E.N.G. Does viperin function as a radical S-adenosyl-L-methioninedependent enzyme in regulating farnesylpyrophosphate synthase expression and activity? J. Biol. Chem. 2016;291:26806–26815. doi: 10.1074/jbc.M116.751040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blattmann P., Henriques D., Zimmermann M., Frommelt F., Sauer U., Saez-Rodriguez J., Aebersold R. Systems pharmacology dissection of cholesterol regulation reveals determinants of large pharmacodynamic variability between cell lines. Cell Syst. 2017;5:604–619. doi: 10.1016/j.cels.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bajimaya S., Frankl T., Hayashi T., Takimoto T. Cholesterol is required for stability and infectivity of influenza A and respiratory syncytial viruses. Virology. 2017;510:234–241. doi: 10.1016/j.virol.2017.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gower T.L., Graham B.S. Antiviral activity of lovastatin against respiratory syncytial virus in vivo and in vitro. Antimicrob. Agents Chemother. 2001;45:1231–1237. doi: 10.1128/AAC.45.4.1231-1237.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mo C., Bard M. A systematic study of yeast sterol biosynthetic protein-protein interactions using the split-ubiquitin system. Biochim. Biophys. Acta. 2005;1737:152–160. doi: 10.1016/j.bbalip.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 60.Choi H., Larsen B., Lin Z.Y., Breitkreutz A., Mellacheruvu D., Fermin D., Qin Z.S., Tyers M., Gingras A.C., Nesvizhskii A.I. SAINT: Probabilistic scoring of affinity purificationg-mass spectrometry data. Nat. Methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mellacheruvu D., Wright Z., Couzens A.L., Lambert J.P., St-Denis N.A., Li T., Miteva Y.V., Hauri S., Sardiu M.E., Low T.Y., Halim V.A., Bagshaw R.D., Hubner N.C., Al-Hakim A., Bouchard A. The CRAPome: A contaminant repository for affinity purification-mass spectrometry data. Nat. Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eisele B., Budzinski R., Müller P., Maier R., Mark M. Effects of a novel 2,3-oxidosqualene cyclase inhibitor on cholesterol biosynthesis and lipid metabolism in vivo. J. Lipid Res. 1997;38:564–575. [PubMed] [Google Scholar]
- 63.Rabbani M.A.G., Ribaudo M., Guo J.-T., Barik S. Identification of interferon-stimulated gene proteins that inhibit human parainfluenza virus type 3. J. Virol. 2016;90:11145–11156. doi: 10.1128/JVI.01551-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li W., Li J., Sun M., Yang L., Mao L., Hao F., Liu M., Zhang W. Viperin protein inhibits the replication of caprine parainfluenza virus 3 (CPIV 3) by interaction with viral N protein. Antiviral Res. 2020;184:104903. doi: 10.1016/j.antiviral.2020.104903. [DOI] [PubMed] [Google Scholar]
- 65.McGillivary G., Jordan Z.B., Peeples M.E., Bakaletz L.O. Replication of respiratory syncytial virus is inhibited by the host defense molecule viperin. J. Innate Immun. 2013;5:60–71. doi: 10.1159/000342473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jumat M.R., Huong T.N., Ravi L.I., Stanford R., Tan B.H., Sugrue R.J. Viperin protein expression inhibits the late stage of respiratory syncytial virus morphogenesis. Antiviral Res. 2015;114:11–20. doi: 10.1016/j.antiviral.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 67.Bajimaya S., Hayashi T., Frankl T., Bryk P., Ward B., Takimoto T. Cholesterol reducing agents inhibit assembly of type I parainfluenza viruses. Virology. 2017;501:127–135. doi: 10.1016/j.virol.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ghosh S., Marsh E.N.G. Viperin: An ancient radical SAM enzyme finds its place in modern cellular metabolism and innate immunity. J. Biol. Chem. 2020;295:11513–11528. doi: 10.1074/jbc.REV120.012784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Upadhyay A.S., Stehling O., Panayiotou C., Rösser R., Lill R., Överby A.K. Cellular requirements for iron-sulfur cluster insertion into the antiviral radical SAM protein viperin. J. Biol. Chem. 2017;292:13879–13889. doi: 10.1074/jbc.M117.780122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Itzhak D.N., Tyanova S., Cox J., Borner G.H.H. Global, quantitative and dynamic mapping of protein subcellular localization. Elife. 2016;5 doi: 10.7554/eLife.16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brown M.S., Goldstein J.L. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patel A.M., Marsh E.N.G. The antiviral enzyme, viperin, activates protein ubiquitination by the E3 ubiquitin ligase, TRAF6. J. Am. Chem Soc. 2021;143:4910–4914. doi: 10.1021/jacs.1c01045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wong J.M.T., Malec P.A., Mabrouk O.S., Ro J., Dus M., Kennedy R.T. Benzoyl chloride derivatization with liquid chromatography-mass spectrometry for targeted metabolomics of neurochemicals in biological samples. J. Chromatogr. A. 2016;1446:78–90. doi: 10.1016/j.chroma.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Padyana A.K., Gross S., Jin L., Cianchetta G., Narayanaswamy R., Wang F., Wang R., Fang C., Lv X., Biller S.A., Dang L., Mahoney C.E., Nagaraja N., Pirman D., Sui Z. Structure and inhibition mechanism of the catalytic domain of human squalene epoxidase. Nat. Commun. 2019;10:97. doi: 10.1038/s41467-018-07928-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD023999. All other data are contained within the article or supporting information.