Abstract
Background and purpose
Computed tomography (CT) is one of the most common medical imaging modalities in radiation oncology and radiomics research, the computational voxel-level analysis of medical images. Radiomics is vulnerable to the effects of dental artifacts (DA) caused by metal implants or fillings and can hamper future reproducibility on new datasets. In this study we seek to better understand the robustness of quantitative radiomic features to DAs. Furthermore, we propose a novel method of detecting DAs in order to safeguard radiomic studies and improve reproducibility.
Materials and methods
We analyzed the correlations between radiomic features and the location of dental artifacts in a new dataset containing 3D CT scans from 3211 patients. We then combined conventional image processing techniques with a pre-trained convolutional neural network to create a three-class patient-level DA classifier and slice-level DA locator. Finally, we demonstrated its utility in reducing the correlations between the location of DAs and certain radiomic features.
Results
We found that when strong DAs were present, the proximity of the tumour to the mouth was highly correlated with 36 radiomic features. We predicted the correct DA magnitude yielding a Matthews correlation coefficient of 0.73 and location of DAs achieving the same level of agreement as human labellers.
Conclusions
Removing radiomic features or CT slices containing DAs could reduce the unwanted correlations between the location of DAs and radiomic features. Automated DA detection can be used to improve the reproducibility of radiomic studies; an important step towards creating effective radiomic models for use in clinical radiation oncology.
Keywords: Radiomics, Computed tomography, Metal artifact detection, Deep learning
1. Introduction
Computed tomography (CT) images are a commonly-used medical imaging modality in radiation oncology. Recent advances in machine learning and deep learning have led to the development of advanced image processing techniques for medical imaging applications, including CT scans [1]. CT-derived quantitative features (also referred to as radiomic features) have shown promising results in personalized medicine [2], and when combined with machine learning, have potential utility in diagnostic and prognostic applications. For these features to be predictive of radiotherapy response, they must be highly reproducible and safeguards for data corruption must be put in place [3]. Unfortunately, radiomic features may be highly sensitive to high-density materials such as metal prosthesis or dental fillings [4]; the latter commonly causes dental artifacts, which pose a problem for imaging of head and neck patients. The metal in dental fillings has a much larger atomic number than soft tissues, resulting in a significantly higher attenuation for x-ray beams passing through the metal. As a result, these dental artifacts (DA) present as bright and dark streaks on the reconstruction images. These artifacts not only obscure large portions of the image’s reconstructed pixels, but studies have also shown that dental artifacts alter features computed by radiomics computational platforms in CT images [4], [5]. They also affect target volume delineation [6], and radiation therapy dose calculation accuracy [7]. There is a need to account for artifacts during image data processing.
Several studies have tried to address this data processing challenge by removing slices affected by DAs [4] or by using metal artifact reduction (MAR) algorithms [8]. Recently, a convolutional neural network (CNN) [9] and hand-crafted radiomic feature-based model [10] have been developed to detect the presence of DAs in CT volumes. However, to the best of our knowledge, no studies differentiated between DAs of different magnitudes or quantified how the location of these artifacts could affect quantitative imaging features used to train radiomic models. Furthermore, previous DA detection studies have classified hand-drawn regions of interest (ROI) as DA positive or DA negative [10] but have not examined the correlation between radiomic features in a given ROI and its distance from the DA source. These methods, even if effective at screening datasets for artifacts, could cause vast amounts of data to be unnecessarily marked as unclean, even if the artifacts do not homogeneously affect radiomic features in the patient’s image volume.
Robustness and reproducibility of radiomic studies requires an understanding of imaging artifacts, and their influence on extracted features. This study explores the relationships between a tumour's radiomic features and its proximity to dental artifacts present in the images. Furthermore, we introduce Artifact Labelling Tool for Artifact Reduction (ALTAR), a novel methodology to identify patient images that are at risk of impact from dental artifacts. This methodology will assist in safeguarding radiomic studies and has been made openly available for usage by the radiomic community.
2. Materials and methods
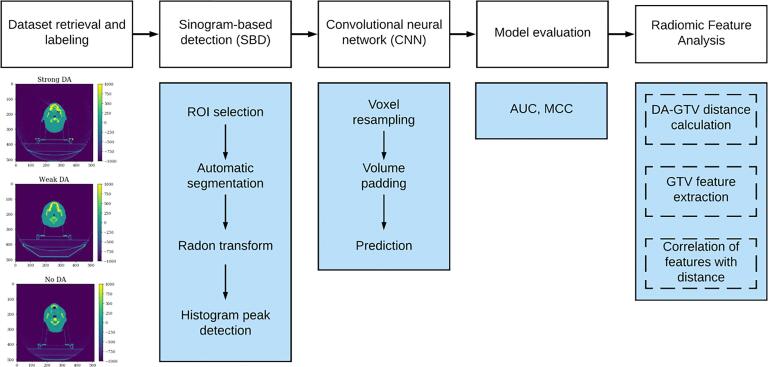
The design of our study is represented in Fig. 1. To train our new DA detection model, we manually labelled a new CT dataset containing 3D axial scans of 3211 cancer patients for the presence of metal dental artifacts. We then developed a novel sinogram-based detection algorithm to classify images with the strongest artifacts present, and we combined this with a pretrained binary DA detection CNN. We then evaluated the models and compared their classification performance to human annotators. Finally, we performed a statistical analysis of radiomic features in order to determine the impact of DAs on features in the gross tumour volume (GTV) (Fig. 2).
Fig. 1.
The study design includes five main steps: (1) retrieval of head and neck CT imaging volume dataset and labelling of DA; (2) initial classification of DA using a sinogram-based detection (SBD) method; (3) secondary classification of SBD-classified dental artifacts using a previously trained CNN; (4) model evaluation; and (5) exploration of the effect of DA magnitude and its distance from the GTV on radiomic features.
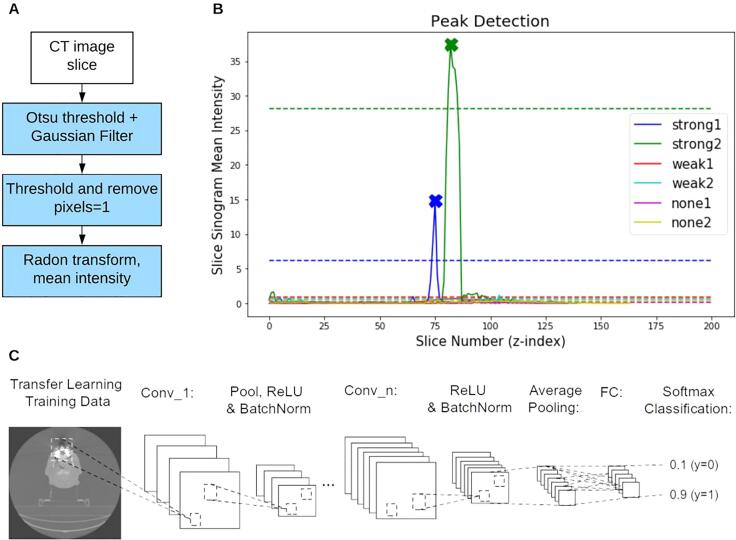
Fig. 2.
An illustration of the two binary DA classifiers used in this study. (A) Two steps in the sinogram-based detection (SBD). First, one slice from a CT volume is thresholded and blurred, before being thresholded again to remove pixels in the body of the patient. The remaining pixels are thresholded again, revealing the streaks outside the patient’s body. The image is then transformed to the sinogram domain and the mean sinogram pixel intensity is computed. (B) An example of the ‘mean sinogram intensity’ for each slice in six CT volumes (each image represented with a different colour). A peak detection algorithm is applied to this plot for a given patient to detect slices likely to contain DAs. We annotate the detected slices with Xs to show that the algorithm detected one peak from each of the green and blue curves (both images labelled as ‘strong DA’). The dashed lines represent the peak detection threshold for each patient. (C) The CNN architecture used in the study. The network consisted of 5 convolutional layers (conv_1 to conv_5) creating a total of 64 filters. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.1. Dataset retrieval and labelling
The dataset used for this study consists of 3211 head and neck cancer axial CT image volumes collected from 2005-07-26 to 2017-08-17 at the University Health Network (UHN) in Toronto, Canada (REB approval #17-5871). This dataset is referred to as RADCURE. We developed Artifact Labelling Tool for Artifact Reduction (ALTAR), an open-source algorithm and web-application enabling the review of large sets of images and the annotation of the magnitude and location of the dental artifacts. Using this tool, each 3D image in RADCURE was labelled by human annotators as either containing a “strong,” “weak,” or no dental artifact. The location of the slice with the strongest artifact was also labelled, or for images with no DA present, the location of the most central axial slice in the mouth was labelled. Full details of this labelling process are described in the Supplementary methods.
2.2. Radiomic feature analysis
The relationship between quantitative imaging features and the existence and location of dental artifacts was studied. 1547 radiomic features were extracted from each 3D image in RADCURE which contained a gross tumour volume (2490 images) using the default settings of the open-source Python package, Pyradiomics (version 2.1.2) [11], [12]. A Wilcoxon rank-sum test between image features from volumes with strong DAs and image volumes with no DAs was performed, applying the Bonferroni correction to the p-values to adjust for multiple tests, and considering p < 0.05 significant. Next, the partial Spearman correlation (adjusted for tumour volume) between the feature values and DA-GTV distances was computed independently for each DA magnitude (1039 strong, 751 weak, 877 none). Finally, we removed 3D images where the GTV overlapped with the strongest DA slice and we performed the same Wilcoxon rank sum test between radiomic features from strong-DA and no-DA images from this smaller group of 1006 patients (529 strong, 477 no DA).
2.3. Automated classification
We created an automated three-class DA classification and location pipeline using a sequential combination of three algorithms. We developed a thresholding and sinogram-based detection (SBD) algorithm to create the first classifier in the pipeline which predicts a binary DA class for each patient’s 3D CT image volume. This was done by applying the Otsu threshold [13] to each axial slice and in order to segment the patient’s head. The head was then removed from the image and the remaining background pixels are transformed to the sinogram domain. We then apply a peak detection (Python Scipy version 1.4.1) algorithm on the mean sinogram intensities in the vertical stack of 2D images for a patient. If a peak was detected, the entire 3D volume was classified “DA positive”.
The original images were then passed to a pre-trained convolutional neural network (CNN) for binary classification. This network was developed and trained by Welch et al. [9] and a detailed description of how it was implemented in our study is provided in the Supplementary methods. We combined the binary predictions from the SBD algorithm and the CNN for each 3D patient scan to create a 3-class classifier using the decision tree architecture shown in Fig. 3.
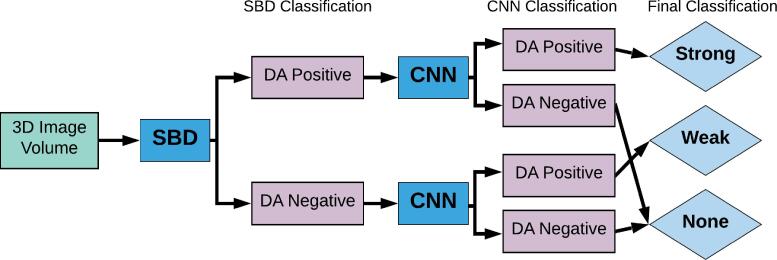
Fig. 3.
Flowchart of the SBD-CNN hybrid algorithm for dental artifact detection. Images were annotated manually and then first binned using SBD (Sinogram based detection) based on the average intensity of the corresponding sinogram. Subsequently, the original images were classified using the CNN model. Images that were labelled as artifact positive by both the SBD and CNN were categorized as having strong dental artifacts. Images labelled as artifact negative by both methods were labelled as having no artifacts. This way our hybrid model is capable of labelling images based on the strength of artifact presence.
Finally, we passed any images that were classified as “strong” or “weak” to the DA location detection algorithm. For this, we developed a thresholding-based algorithm to detect the axial slice containing a DA in a given patient CT image volume. The thresholding-based algorithm works by first clipping the HU values between the maximum intensity in one patient’s CT image volume and 200 HU above that maximum. The standard deviation of each axial slice is then computed and peak detection is performed on the standard deviations of each axial slice using the scipy find_peaks function in a similar manner to the SBD peak detection step. The specific parameters chosen for the peak detection function are described in detail in the Supplementary. If any peaks are detected, the algorithm simply returns the indices of those peaks for that patient. Otherwise, the lower bound of the clipping range is decreased by 50 HU and the process is repeated for the patient until at least one peak is found.
2.4. Performance assessment
A subset containing 2319 patient image volumes was set aside for model evaluation. The remaining 892 images were used to develop the three-class hybrid algorithm and the DA location detection algorithm. The test set was chosen by removing any images that were used by Welch et al. in the training and evaluation of the CNN. Since this study used a cohort of patients from the same institution as RADCURE, we ensured that no images used for model evaluation in our study were used by Welch et al. Furthermore, since the thresholding location detection algorithm is intended to be used on DA-positive images, we tested this method on the 1551 patients who had a DA from the 2319 patient test set.
We primarily used the Matthews correlation coefficient (MCC) to assess the accuracy of our prediction models. The MCC is equivalent to the Chi-square coefficient for binary labels and accounts for the potential class imbalance. We found the MCC to be a useful metric for DA detection, as an effective dataset cleaning tool should have a high accuracy and a low false-negative rate (the rate at which a classifier fails to detect DAs when they are present). The MCC can also be generalized to multiclass cases, allowing us to compare the performance of our binary and three-class classifiers. In the Supplementary we also provide the performance of the CNN on its own as a binary classifier using the Area Under the Receiver Operating Characteristic Curve (AUC). For all AUC and MCC values we also estimated a p-value from 5000 iterations of a randomized permutation test.
2.5. Research reproducibility
The application we created to manually annotate images, Artifact Labelling Tool for Artifact Reduction (ALTAR) can be downloaded from our GitHub repository (https://github.com/bhklab/ALTAR). The code for the SBD, CNN, and thresholding location detection is open source (Creative Commons Non-Commercial) and freely available from on our GitHub repository (https://github.com/bhklab/DA-Detection). To ensure full reproducibility of our study we created a Code Ocean capsule to allow users to easily run and reuse our analysis pipeline (https://codeocean.com/capsule/2097894/tree).
3. Results
3.1. Dataset labelling
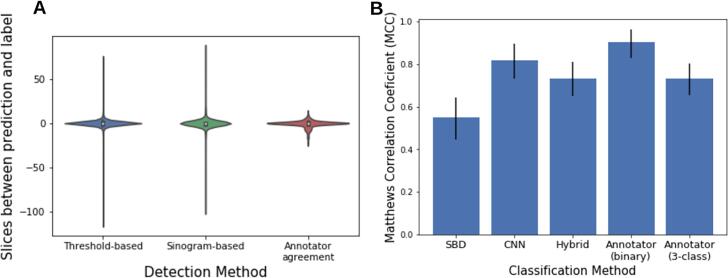
After reviewing 3211 image sets using our ALTAR web-application, we identified 2180 volumes containing artifacts (1289 strong and 891 weak) and 1031 volumes with no dental artifacts. The manual labelling was consistent in the set of 482 images that were labelled by two researchers, yielding three-class Matthews correlation coefficient (MCC) of 0.73 (p << 0.01, 95% CI [0.66, 0.81]), and 0.91 (p << 0.01, 95% CI [0.83, 0.96]) for binary classes (strong/weak vs none; Supplementary Table 2). The annotators labelled the same slice as containing the “strongest DA,” or the patient’s mouth in DA negative cases, in 46% of patients (Fig. 5A). The two annotators labelled the strongest DA slice to within 5 slices of each other 82% of the time.
Fig. 5.
Performance of DA classification. (A) Distributions of how close the predicted slice index is to the labelled index for the threshold-based and sinogram based-detection methods (e.g. ). The difference in slice label between two human annotators for a set of 482 CT volumes is also shown. (B) Performance (MCC) of the DA magnitude classification techniques used in this study. The p-value of the MCC for all classifiers was <0.001). The sinogram-based detection (SBD) and convolutional neural network (CNN) are both binary classifiers. The SBD was tested on 3211 CT image volumes and the CNN binary classifier was tested on a subset of 2319 image volumes. The SBD-CNN hybrid algorithm is a three-class classifier and the three-class MCC is therefore displayed here.
3.2. Analysis of radiomic features and dental artifacts
To test for differences between features from DA positive and DA negative image volumes, we performed a Wilcoxon rank sum test between features from each group. We found that 442 features were significantly different between the DA and no DA groups (Wilcoxon rank sum test corrected p-value <0.05), while 55 features varied significantly between strong DA and no DA patients when the artifact was 40 mm–80 mm from the GTV. No features were significantly different between strong DA and no DA for patients with a DA more than 80 mm from the GTV.
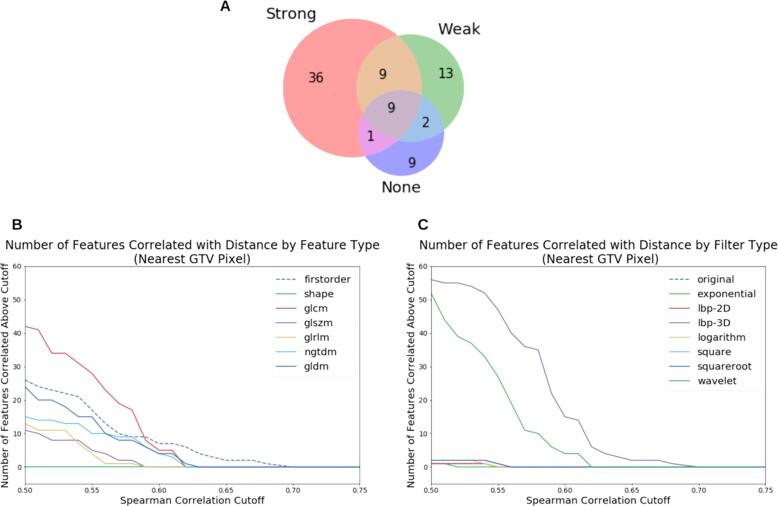
To assess the correlation between the radiomic features and distance of the GTV from the DA slice, we computed the partial Spearman correlation between DA-GTV distance and radiomic feature value, controlling for tumour volume (Fig. 4). 36 features were correlated with distance only in images with strong DAs (e.g. those same features were not correlated with distance when computed from weak and no-DA images). All but two of these 36 features were found to use the “lbp-3D-k” filter. Nine of these 36 features were also found to be significantly different between strong-DA and no-DA images in the Wilcoxon rank sum test.
Fig. 4.
Correlation between GTV-DA distance and feature values, based on the partial correlation using Spearman correlation. (A) Venn diagram showing the number of features with |r| > 0.55 calculated from patients from each DA class. This diagram only includes significant correlations (p < 0.05). For instance, 36 features had |r| > 0.55 and were found in patients with strong DAs (pink region), but those features had |r| < 0.55 when calculated from weak or no-DA images). Nine features had |r| > 0.55 when calculated for all three DA groups (grey region). (B) The number of features with DA-GTV distance correlation above a given cutoff, grouped by feature type. (C) These correlations grouped by filter type. (B) and (C) only include significant features (p < 0.05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In order to validate the effect of removing “bad” images on radiomic features, we removed all images where the centre slice of the DA overlapped with any pixel in the GTV. We found that only 123 features were significantly different between the groups (p < 0.05). Repeating the test with randomly selected samples of the same size, this number of 123 features was in the bottom 4.1% of repeated test results.
3.3. Classifier performance
The combined SBD and CNN DA three-class classifier yielded an MCC of 0.73 (p-value = 0.0002, 95% CI [0.65, 0.81]) on 2319 images (945 strong, 606 weak, and 768 without artifacts). This was identical to the three-class agreement between human annotators (MCC = 0.73, p << 0.01, 95% CI [0.65, 0.81] Fig. 5B). This hybrid algorithm was able to make use of two different binary DA detection algorithms which independently performed worse than human labelling. Together, the two methods complement each other and are able to stratify images into three distinct DA magnitude classes.
The thresholding-based DA location detection algorithm was tested on 1551 images with artifacts (1231 strong, 856 weak). The algorithm identified the exact slice which was labelled in 36% of cases. In 92% of cases, the algorithm identified a slice within 5 slices above or below the label (Fig. 5A).
4. Discussion
The main goal of this study consists of investigating potential spurious correlations in radiomic features due to metal artifacts and creating an automated pipeline for DA classification in large imaging datasets to safeguard against these risks and improve reproducibility. Using a subset of 2490 patients, we extracted 1547 radiomic features and investigated statistical differences in these features based on their DA locations and magnitude. Then we used a large dataset of 3211 head and neck cancer CT image volumes to build a DA location and magnitude classifier.
Analysis of radiomic features between strong DA and no DA patients revealed that approximately a third of features varied significantly with dental artifact status when the DA was less than 40 mm from the GTV. We found that the number of features associated with dental artifact status decreased significantly as the distance between the GTV and DA increased. This suggests that the location plays an important role in the effect of DAs on radiomic features. To further investigate this distance dependence, we examined the correlation between DA-GTV distance and the radiomic feature values. We found that, among the correlated features (Spearman correlation >0.55), only 0.5% of features (9 out of 1547 radiomic features) were found in DA negative volumes only. However, a larger set of 36 correlated features were specifically found in the strong DA volumes. Interestingly, 34 features in this set are exclusively composed of radiomic features which used the “LocalBinaryPattern3D-kurtosis” (‘lbp-3D-k’) filter, suggesting that their lack of robustness in the presence of DA makes them unsuitable for radiomics modeling. lbp-3D-k filter computes the kurtosis (a measure of the tailedness of a distribution) [14] from the local binary pattern, a rotationally invariant measure of texture in three dimensions [15]. We hypothesize that DAs are altering the width of the distribution of specific texture metrics in three dimensions. These strong correlations between DA-GTV distance and specific radiomic features highlights the need for robust data curation pipelines for DAs in radiomic studies.
We were then able to further motivate the use of DA location detection in dataset cleaning. By removing images with their GTV in the same slice as the DA, the number of features significantly different between strong-DA and no-DA images was significantly reduced (123 DA-affected feature vs an average of 188 features by randomly selecting 1006 patients; p-value = 0.041). This highlights the need for a DA location detector in dataset cleaning. By removing only the images with a GTV overlapping with a DA, we were able to significantly improve the robustness of the features extracted from the dataset.
Interestingly, the fact that only nine of those 36 distance-correlated features were significantly different between strong-DA and no-DA images (based on the Wilcoxon rank sum test) suggests that these two analyses are detecting different types of dependency. In particular, using the Spearman correlation with GTV-DA distance may be a more strict criterion by which to select features to exclude from a radiomics study. We suggest using both analyses in order to select features robust to DAs in future radiomics studies.
To automate DA classification, we propose a DA magnitude and location classifier to help reduce confounding correlations relating to DAs. Rather than using whole-CT volume DA classifiers to remove samples or patients from training sets, we suggest using a DA location detection algorithm to only remove images where the DA is close to the CT region of interest. The binary DA CNN classifier from Welch et al. was found to perform just as well on this larger dataset (AUC = 0.97, p << 0.01, 95% CI [0.94, 0.99]) as it had in the original study (AUC = 0.91 ± STD 0.01) and outperformed our sinogram-based detection method. However, we were able to leverage the SBD’s ability to discriminate between strong and weak artifacts (90% true positive rate (95% CI [for strong, 25% for weak DAs) to create a three-class DA classifier. The three-class classifier also performed as well as two human annotators, with both the three-class MCC of the annotators and the algorithm being 0.73 (p << 0.01, 95% CI [0.65, 0.80]).
In addition to developing a novel multi-class dental artifact detection method, we developed an algorithm to detect the slice containing the artifact. We found that both the sinogram-based and the thresholding-based location detection methods agreed with a human-annotator as well as two humans would agree with one another. This was determined by comparing 482 images in our dataset that were annotated twice by different human observers (see Supplementary). The algorithms predicted the DA to be within 5 slices of the human label in 80–90% of cases, while two humans agreed on the label to within 5 slices in 82% of cases. Due to the simplicity of the thresholding-based algorithm, it could be used as an efficient add-on to any DA detection algorithm, or used to annotate the locations of DAs in datasets where the DA status of images is known, but not their location. This could be useful for removing data corrupted by DAs from a dataset without having to exclude the entire CT volume of DA positive patients.
In general, our results show that it is crucial to quantify the image quality of datasets used in radiomic studies. We have shown that this can either be done using this DA detection tool to find images containing strong DAs, or manually labelling the data in order to understand the severity of DAs in the dataset.
Although there are currently no widely-accepted tools for reducing the effects of DAs in radiomic studies, this work paves the way for future investigation of metal artifact reduction models, specifically targeted at improving reproducibility in radiomics. While multiple DA reduction approaches have been explored in clinical settings [16], [17], these methods have not been investigated for their applications to reducing the impact of DAs on radiomic features (hand-engineered as with PyRadiomics or otherwise). This is an important area of research as our work highlights the strong links between DAs and radiomic features.
Our study has several potential limitations. The analysis in this study has largely focused on the vertical location of DAs and their vertical distance from the GTV. This ignores any potential relationship between DA distance and radiomic features in the x, y plane (within a slice). We also acknowledge the inherent subjectivity of our manual labelling process, as individual researchers and clinicians may have widely varying definitions of strong, weak and no artifacts. Although our analysis of annotator agreement shows that this was not a major problem in our study, it does mean that our work could be difficult to reproduce with different data and researchers.
In conclusion, we have developed a novel dental artifact detection algorithm which when combined with a convolutional neural network, created a three-class classifier for CT images with strong, weak, and no DAs. We then created a simple thresholding-based algorithm to detect the location of DAs in DA positive CT volumes. These new tools have been made open-source to be used in future studies to assess and account for the effects of DAs on radiomic models. We stress that our findings suggest that radiomic features are affected not only by the presence of DAs, but also by their location in the images.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by the Canadian Institutes for Health Research (CIHR) radiomics grant (# 426366). We would like to thank the head and neck group at the Princess Margaret Cancer Centre for their contributions to this study and their invaluable work to create the dataset used in this study. We would also like to acknowledge the contributions of the Radiomics Medicine Program at the University of Toronto and the University Health Network. Finally, we would like to thank Dean Zhu and Aleesha Masud for their help annotating the data and helping develop ALTAR.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2021.04.001.
Contributor Information
Scott Bratman, Email: scott.bratman@uhnresearch.ca.
Benjamin Haibe-Kains, Email: bhaibeka@uhnresearch.ca.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Ha S. Perspectives in radiomics for personalized medicine and theranostics. Nucl Med Mol Imaging. 2019;53:164–166. doi: 10.1007/s13139-019-00578-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welch M.L., McIntosh C., Haibe-Kains B., Milosevic M.F., Wee L., Dekker A. Vulnerabilities of radiomic signature development: the need for safeguards. Radiother Oncol. 2018;130:2–9. doi: 10.1016/j.radonc.2018.10.027. [DOI] [PubMed] [Google Scholar]
- 4.Ger R.B., Craft D.F., Mackin D.S., Zhou S., Layman R.R., Jones A.K. Practical guidelines for handling head and neck computed tomography artifacts for quantitative image analysis. Comput Med Imaging Graph. 2018;69:134–139. doi: 10.1016/j.compmedimag.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leijenaar R.T.H., Carvalho S., Hoebers F.J.P., Aerts H.J.W.L., van Elmpt W.J.C., Huang S.H. External validation of a prognostic CT-based radiomic signature in oropharyngeal squamous cell carcinoma. Acta Oncol. 2015;54:1423–1429. doi: 10.3109/0284186X.2015.1061214. [DOI] [PubMed] [Google Scholar]
- 6.Hansen C.R., Christiansen R.L., Lorenzen E.L., Bertelsen A.S., Asmussen J.T., Gyldenkerne N. Contouring and dose calculation in head and neck cancer radiotherapy after reduction of metal artifacts in CT images. Acta Oncol. 2017;56:874–878. doi: 10.1080/0284186X.2017.1287427. [DOI] [PubMed] [Google Scholar]
- 7.Kim Y., Tomé W.A., Bal M., McNutt T.R., Spies L. The impact of dental metal artifacts on head and neck IMRT dose distributions. Radiother Oncol. 2006;79:198–202. doi: 10.1016/j.radonc.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 8.Gjesteby L., De Man B., Jin Y., Paganetti H., Verburg J., Giantsoudi D. Metal artifact reduction in CT: where are we after four decades? IEEE Access. 2016;4:5826–5849. [Google Scholar]
- 9.Welch M.L., McIntosh C., Purdie T.G., Wee L., Traverso A., Dekker A. Automatic classification of dental artifact status for efficient image veracity checks: effects of image resolution and convolutional neural network depth. Phys Med Biol. 2020;65:015005. doi: 10.1088/1361-6560/ab5427. [DOI] [PubMed] [Google Scholar]
- 10.Wei L., Rosen B., Vallières M., Chotchutipan T., Mierzwa M., Eisbruch A. Automatic recognition and analysis of metal streak artifacts in head and neck computed tomography for radiomics modeling. Phys Imaging Radiat Oncol. 2019;10:49–54. doi: 10.1016/j.phro.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aerts H.J.W.L., Velazquez E.R., Leijenaar R.T.H., Parmar C., Grossmann P., Carvalho S. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Griethuysen J.J.M., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern. 1979;9:62–66. [Google Scholar]
- 14.Kokoska S., Zwillinger D. 0 ed. CRC Press; 2000. CRC standard probability and statistics tables and formulae, student edition. [Google Scholar]
- 15.Banerjee J., Moelker A., Niessen W.J., van Walsum T. Computer vision – ACCV 2012 workshops. Springer; Berlin Heidelberg: 2013. 3D LBP-based rotationally invariant region description; pp. 26–37. [Google Scholar]
- 16.Zhang Y., Yu H. Convolutional neural network based metal artifact reduction in X-Ray computed tomography. IEEE Trans Med Imaging. 2018;37:1370–1381. doi: 10.1109/TMI.2018.2823083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakao M., Imanishi K., Ueda N., Imai Y., Kirita T., Matsuda T. Regularized three-dimensional generative adversarial nets for unsupervised metal artifact reduction in head and neck CT images. IEEE Access. 2020;8:109453–109465. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.