Abstract
A novel coronavirus, severe acute respiratory syndrome (SARS)–associated coronavirus (SARS-CoV), has been identified as the causal agent of SARS. Spike (S) protein is a major structural glycoprotein of the SARS virus and a potential target for SARS-specific cell-mediated immune responses. A panel of S protein–derived peptides was tested for their binding affinity to HLA-A*0201 molecules. Peptides with high affinity for HLA-A*0201 were then assessed for their capacity to elicit specific immune responses mediated by cytotoxic T lymphocytes (CTLs) both in vivo, in HLA-A2.1/Kb transgenic mice, and in vitro, from peripheral blood lymphocytes (PBLs) sourced from healthy HLA-A2.1+ donors. SARS-CoV protein-derived peptide-1 (SSp-1 RLNEVAKNL), induced peptide-specific CTLs both in vivo (transgenic mice) and in vitro (human PBLs), which specifically released interferon-γ (IFN-γ) upon stimulation with SSp-1–pulsed autologous dendritic cells (DCs) or T2 cells. SSp-1–specific CTLs also lysed major histocompatibility complex (MHC)–matched tumor cell lines engineered to express S proteins. HLA-A*0201–SSp-1 tetramer staining revealed the presence of significant populations of SSp-1–specific CTLs in SSp-1–induced CD8+ T cells. We propose that the newly identified epitope SSp-1 will help in the characterization of virus control mechanisms and immunopathology in SARS-CoV infection, and may be relevant to the development of immunotherapeutic approaches for SARS.
Introduction
The first recorded outbreak of severe acute respiratory syndrome (SARS) in late February 2003 has led to thousands of infected patients and hundreds of deaths. The etiologic agent of the syndrome, a novel coronavirus termed SARS-associated coronavirus (SARS-CoV), has since been identified and isolated,1, 2, 3, 4 and its genome sequenced.5,6 SARS is characterized by high fever, rigor, headache, nonproductive cough, or dyspnea and may progress to generalized, interstitial infiltrates in the lung, requiring intubation and mechanical ventilation.1,2,7, 8, 9 A common observation in patients with SARS is pronounced lymphopenia.1,10,11 A notable drop in CD4+ and CD8+ T lymphocyte counts occurs early in the course of the disease and is associated with adverse outcomes.12 To date, studies on SARS have generally been retrospective or limited to the description of initial clinical, hematologic, radiologic, and microbiologic findings. Further studies to evaluate the mechanisms of these manifestations may help us to better understand this disease.
Evidence suggests that CD8+ cytotoxic T lymphocytes (CTLs) play a pivotal role in both virus elimination and induction of immunopathology in respiratory syncytial virus (RSV) infection, following recognition of epitopes presented on target cells in the context of major histocompatibility complex (MHC) class I.13, 14, 15, 16, 17 Similarly, CTLs may participate in the clearance of virus in recovered SARS patients but also contribute to immunopathology in early stages of the disease; the molecular mechanisms underlying these CD8+ CTL-mediated effects remain poorly defined. HLA-A2 is the most common HLA-A allele in Asian populations, particularly in the Chinese, with an estimated frequency of more than 50%.18 As SARS affected many parts of Asia, and a reservoir of infection may persist in these regions, the identification of HLA-A*0201–restricted CTL epitopes of SARS-CoV is an important contribution to future studies concerning the role of CTLs in SARS-CoV pathogenesis and protection in at-risk populations.
Here we have studied a panel of SARS-CoV spike protein (SARS/S)–derived peptides to identify those with binding motifs for HLA-A*0201 molecules. We evaluated the ability of HLA-A*0201 binding peptides to provoke in vivo– and in vitro–specific CTL responses in HLA-A2.1/Kb transgenic (Tg) mice and peripheral blood lymphocyte (PBL) preparations from MHC-matched healthy donors, using dendritic cells (DCs) prepulsed with the peptides. Our findings show that anti–SARS-CoV CTLs generated from Tg mice and PBLs of healthy donors can elicit an antigen-specific, HLA-A2.1–restricted response, effectively killing peptide-pulsed T2 cells or HLA-A2.1+ tumor cell lines endogenously expressing S protein. To the best of our knowledge, our study is the first successful prospective identification of a novel HLA-A*0201–restricted CD8+ T-cell epitope from the SARS-CoV spike protein.
Materials and methods
Peptides
To identify potential HLA-A*0201 binding peptides within spike protein (SARS/S) of the SARS-CoV BJ01 strain19 (GenBank accession number AY278488), a computer-based program, accessed through the National Institutes of Health BioInformatics and Molecular Analysis Section (BIMAS) HLA Peptide Binding Predictions website (http://bimas.dcrt.nih.gov/molbio/hla_bind/index.html), was used.20,21 Peptides with high estimated half-time of dissociation (T1/2) were synthesized at GL Biochem (Shanghai, China) and purified to more than 95% by reverse phase high-performance liquid chromatography (HPLC), as confirmed by mass spectrometry. As a positive control for HLA-A*0201 binding ability, the HLA-A2.1–restricted carcinoembryonic antigen peptide-1 (CAP-1, YLSGANLNL) was used.22 The ovalbumin (OVA)–derived H-2b–restricted peptide OVA257-264 (SIINFKEL) was used as a negative control. Lyophilized peptides were dissolved in phosphate-buffered saline (PBS) at a concentration of 2 mg/mL and stored in aliquots at –80° C.
Animals and cell lines
HLA-A2.1/Kb transgenic (Tg) mice23 were purchased from The Jackson Laboratory (Bar Harbor, ME). Mice were bred and maintained in specific pathogen-free facilities. For experimental purposes, mice were used at 6 to 8 weeks of age. Cell surface HLA-A*0201 expression was assessed by flow cytometry using fluorescein isothiocyanate (FITC)–labeled HLA-A2–specific monoclonal antibody (mAb) BB7.2 (Serotec, Oxford, United Kingdom).
Human transporter associated with antigen processing (TAP)–deficient T2, embryonic kidney 293, colorectal carcinoma SW480 (HLA-A2.1+), and HT29 (HLA-A2.1–) cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and maintained in culture according to the supplier’s specifications.
Peptide binding assay
To determine whether synthetic peptides could bind to HLA-A*0201 molecules, peptide-induced HLA-A*0201 up-regulation on T2 cells was examined according to a protocol described previously.24,25 Briefly, T2 cells were incubated with 50 μM candidate peptides and 3 μg/mL human β2-microglobulin (Sigma, St Louis, MO) in serum-free Iscoves modified Dulbecco medium for 18 hours at 37° C in a 5% CO2 atmosphere. Expression of HLA-A*0201 on T2 cells was then determined by staining with FITC-conjugated anti–HLA-A2 mAb BB7.2 and data were analyzed using a FACSCalibur flow cytometer (Becton Dickinson, Mountain View, CA) and CellQuest software (Becton Dickinson). The fluorescence index (FI) was calculated as follows: FI = (mean FITC fluorescence with the given peptide – mean FITC fluorescence without peptide)/(mean FITC fluorescence without peptide). Peptides with FI more than 1 were regarded as high-affinity epitopes.
Recombinant adenovirus preparation and infection
The open reading frame (ORF) of SARS/S cDNA was amplified by polymerase chain reaction (PCR) using forward (5′-GCCTCGAGACCATGTTTATTTTCTTATTATTTCTTACTCTCACTAGTGGTAGTGACCT-3′) and reverse (5′-TTTATGTGTAATGTAATTTGACACCCTT-3′) primers from a plasmid containing the SARS/S cDNA sequence obtained from SARS-CoV BJ01 strain.19 The PCR product was digested by XhoI and inserted into pShuttle-CMV vector (Stratagene, La Jolla, CA). After confirmation by DNA sequencing, the linearized shuttle vector was cotransformed into Escherichia coli strain BJ5183 together with pAdEasy-1, the supercoiled viral DNA plasmid (Stratagene). The recombinant adenovirus plasmid DNA containing SARS/S or β-galactosidase (β-gal) ORF was designated as pAd-SARS/S or pAd-LacZ, respectively. Recombinant adenoviruses were propagated in 293 cells, the titers of which were determined by plaque assay on 293 cells as previously described by us.26 For infection, SW480 (HLA-A2.1+) and HT29 (HLA-A2.1–) cells were incubated with pAd-LacZ or pAd-SARS/S at a multiplicity of infection (MOI) of 50:1 for 24 hours at 37° C in a 5% CO2 atmosphere and then washed twice with complete RPMI 1640 medium. Under these conditions, more than 90% of incubated target cells were infected with the recombinant adenoviruses as detected by β-gal staining, and SARS/S expression was confirmed by reverse transcriptase (RT)–PCR and Western blot using antibody to SARS/S (IMGENEX, San Diego, CA).
Generation of CTLs in HLA-A2.1/Kb transgenic mice
HLA-A2.1 transgenic mice are a well-established model for the study of HLA-A2.1–restricted CTL epitopes and vaccine development.25,27, 28, 29, 30, 31, 32, 33, 34 In this study, bone marrow–derived DCs were generated from transgenic mice as previously described by us.35 On day 7, DCs were harvested and washed. For induction of peptide-specific CTLs, DCs were pulsed with 20 μg/mL peptides in the presence of 3 μg/mL β2-microglobulin. After incubation at 37° C for 3 hours, cells were washed 3 times in PBS and resuspended in PBS for injections. HLA-A2.1/Kb transgenic mice were immunized intraperitoneally with 1 × 106 DCs/mouse weekly, for 3 weeks. At 7 days after the last immunization, splenic single-cell suspensions from primed mouse spleens were cultured in 6-well plates at a density of 1 × 107 cells/well, in the presence of 1 × 106 10 μg/mL peptide-loaded irradiated syngeneic DCs. On day 6 of culture, induced cells were harvested and tested by enzyme-linked immunospot (ELISPOT), cytotoxicity, and tetramer staining assays.
Generation of CTLs in healthy donors
Dendritic cells are characterized by the unique capacity to activate naive T cells and initiate primary T-cell response.36, 37, 38 Antigen-specific T-cell responses from peripheral blood mononuclear cells (PBMCs) can be elicited with antigenic peptide- or protein-pulsed autologous DCs.25,39, 40, 41, 42, 43 Here, PBMCs were isolated from whole blood of 11 healthy HLA-A2.1+ volunteer donors by Ficoll/Hypaque (Sigma) density gradient centrifugation. Human peripheral blood monocyte-derived DCs were generated as previously described by us.44 On day 5 of culture, 10 ng/mL recombinant human tumor necrosis factor α (TNF-α; Peprotech, Rocky Hill, NJ) was added to the medium to induce phenotypic and functional maturation. Then, 48 hours later, DCs were pulsed with 20 μg/mL peptide in the presence of 3 μg/mL β2-microglobulin at 37° C for 3 hours and irradiated at 30 Gy before use. Peripheral blood lymphocytes (PBLs, 2 × 106) were cocultured with 2 × 105 peptide-pulsed irradiated autologous DCs in a 24-well plate in the presence of 10 ng/mL recombinant human interleukin-7 (IL-7; Peprotech). The next day, recombinant human IL-10 (Peprotech) was added to the culture medium, to give a final concentration of 10 ng/mL. After 7 days, lymphocytes were restimulated with peptide-pulsed irradiated autologous DCs in medium containing 10 ng/mL IL-7 and 10 ng/mL IL-10, and then supplemented with 20 IU/mL IL-2 (Sigma) 24 hours later. Lymphocytes were restimulated each week in the same manner. At 7 days after the fourth round of restimulation, cells were harvested and tested by ELISPOT assay, cytotoxicity assay, and tetramer staining. For ELISPOT assay and cytotoxicity assays, CD8+ T lymphocytes were purified by CD4+ cell–negative depletion using human CD4 microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany).
Cytotoxicity assay
Cultured CTLs were tested for cytotoxicity in a standard 4-hour 51Cr release assay.45,46 T2 cells were loaded with 20 μg/mL peptide at 37° C for 1 hour. Peptide-pulsed T2 cells and SARS/S-expressing tumor cell lines were labeled with 51Cr sodium chromate (100 μCi/106 [3.7 MBq/106] cells) for 90 minutes at 37° C. 51Cr-labeled target cells were washed 3 times and mixed with graded doses of effectors in 96-well round-bottom tissue culture plates. After incubation at 37° C for 4 hours, a total of 100 μL supernatant was collected from each well and radioactivity was counted with a gamma counter. Each assay was performed in triplicate. Percent specific lysis was determined according to the following formula: percent specific lysis = [(mean experimental cpm – mean spontaneous cpm)/(mean maximum cpm – mean spontaneous cpm)] × 100%. Spontaneous and maximum release were determined by incubating the labeled targets with medium alone and 2% Triton X-100, respectively. Spontaneous release was always less than 15% of maximum release. The standard deviation (SD) of triplicate wells was less than 15%.
ELISPOT assay
ELISPOT assays were performed using a commercially available kit (R&D Systems, Minneapolis, MN). pAd-SARS/S–transfected tumor cell lines, or peptide-pulsed DCs and T2 cells were used as stimulator cells. Effector cells (1 × 105) and stimulator cells (1 × 105) were seeded into 96-well polyvinylidene fluoride (PVDF)–backed microplates coated with monoclonal antibody specific for mouse or human interferon γ (IFN-γ). After incubation at 37° C for 16 hours, cells were removed and plates processed according to the manufacturer’s instructions. Resulting spots were counted with a stereomicroscope (Carl Zeiss, Thornwood, NY) under magnifications of × 20 to × 40. Only brown colored spots with fuzzy borders were scored as spot-forming cells (SFCs). The data in the figures refer to the mean of triplicate assays. SDs were generally within 10% of the mean.
Tetramer production and staining
Tetrameric HLA-A*0201 peptide complexes (tetramers) were constructed as previously described by us.26 Briefly, DNA encoding the 14–amino acid residues recognized by the biotinylation enzyme Bir A was inserted to the COOH terminus of the extracellular region of HLA-A*0201 heavy chain and the entire construct cloned into pET 21d plasmid (Novagen, Madison, WI) as an NheI/BamHI fragment. Recombinant HLA-A*0201 heavy chain containing the Bir A site and human β2-microglobulin was refolded in vitro together with the peptides corresponding to SARS-CoV protein-derived peptide-1 (SSp-1, RLNEVAKNL) or CAP-1 (YLSGANLNL). The HLA-A*0201 peptide–β2-microglobulin complexes were then purified through a Sephacryl S-200 column (Amersham Pharmacia Biotech, Piscataway, NJ) and biotinylated using Bir A enzyme (Avidity, Boulder, CO). The biotinylated complexes were further purified by gel filtration, and tetramerization was accomplished by mixing biotinylated HLA-A*0201 peptide–β2-microglobulin and phycoerythrin (PE)–conjugated streptavidin (Biosource, Camarillo, CA) at a molar ratio of 4:1. Peptide-induced lymphocyte cultures were first incubated with PE-conjugated tetramers for one hour at 37° C, and then FITC-labeled antihuman/mouse CD8 mAb (PharMingen, San Diego, CA) was added and cells were incubated for an additional 30 minutes at 4° C. After washing with PBS, stained cells were fixed with 0.5% paraformaldehyde and analyzed by flow cytometry.24,27,47, 48, 49
Results
Selection of potential HLA-A2.1 binding peptides from SARS-CoV spike protein
Based on computer software predictions, the 20 candidate nonameric peptides with the highest estimated half-time of dissociation from HLA-A*0201 were selected and synthesized (Table 1 ). To evaluate the binding affinity of these peptides to HLA-A*0201 molecules, we used a T2 cell–peptide binding test. This assay measures the increase of HLA-A*0201 molecules induced on T2 cells following exposure to exogenous HLA-A*0201 binding peptides, with high-affinity peptides inducing HLA-A*0201 up-regulation more strongly than low-affinity peptides.
Table 1.
T2-HLA-A*0201 binding affinity of SARS-Spike protein-derived peptides
| Rank | Start position | Sequence | Score* | FI† |
|---|---|---|---|---|
| 1 | 1042 | VVFLHVTYV | 484.238 | 0.5 |
| 2 | 982 | RLQSLQTYV | 382.536 | 0.3 |
| 3 | 958 | VLNDILSRL | 342.461 | 0.6 |
| 4 | 897 | VLYENQKQI | 232.693 | 0.4 |
| 5 | 1174 | NLNESLIDL | 201.447 | 1.1 |
| 6 | 734 | LLLQYGSFC | 171.868 | 0.4 |
| 7 | 411 | KLPDDFMGC | 135.453 | 0.4 |
| 8 | 787 | ILPDPLKPT | 119.463 | 0.3 |
| 9 | 151 | MIFDNAFNC | 102.176 | 0.4 |
| 10 | 2 | FIFLLFLTL | 94.987 | -0.1 |
| 11 | 596 | VLYQDVNCT | 93.239 | 0.4 |
| 12 | 1096 | IITTDNTFV | 89.418 | 0.3 |
| 13 | 1167 | RLNEVAKNL | 87.586 | 1.7 |
| 14 | 846 | LLTDDMIAA | 79.642 | 0.4 |
| 15 | 735 | LLQYGSFCT | 73.477 | 0.2 |
| 16 | 1214 | ILLCCMTSC | 71.872 | 0.3 |
| 17 | 803 | LLFNKVTLA | 71.872 | 0.3 |
| 18 | 106 | TMNNKSQSV | 50.232 | 0.2 |
| 19 | 940 | ALNTLVKQL | 49.134 | 0.3 |
| 20 | 131 | ELCDNPFFA | 48.732 | 0.4 |
Estimated half-time of dissociation (T1/2) of HLA-A*0201 peptide complexes calculated using the website http://bimas.dcrt.nih.gov/molbio/hla_bind/index.html.
Increase of HLA-A*0201 molecules on T2 cells. FI = (mean FITC fluorescence with the given peptide-mean FITC fluorescence without peptide)/(mean FITC fluorescence without peptide). FI more than 1 indicates high-affinity peptides; FI of 1 or less, low-affinity peptides. Positive control peptide CAP-1 (YLSGANLNL) and negative control peptide OVA257-264 (SIINFKEL) had FI 1.9 and 0.1, respectively.
Of the 20 candidate peptides, only no. 5 and no. 13 were high-affinity epitopes (FI = 1.1 and 1.7, respectively); all other peptides were of low affinity (FI ≤ 1; Table 1). The positive control CAP-1 bound HLA-A*0201 strongly (FI = 1.9), whereas no binding was associated with the negative control (FI = 0.1).
In vivo induction of peptide-specific CD8+ CTLs in HLA-A2.1/Kb transgenic mice
To determine the in vivo immunogenic potential of peptides, we immunized HLA-A2.1/Kb transgenic mice with syngeneic DCs loaded with these high-affinity peptides. After 3 rounds of weekly in vivo stimulation, splenocytes from primed mice were restimulated in vitro for an additional 6 days and tested for IFN-γ production and CTL reactivity. T2 cells were pulsed with various concentrations (1-100 μg/mL) of either peptide no. 5 or peptide no. 13 and used as stimulators in ELISPOT and targets in cytotoxicity assays. No specific reactivity could be detected in cell lines generated from Tg mice immunized with peptide no. 5 upon incubation with T2 cells loaded with any concentration of peptide no. 5 in the ELISPOT assay or at any effector-target ratio in the CTL assay (data not shown). Peptide no. 5 was thus excluded from further study. Bulk CTLs from Tg mice responding to peptide no. 13 (referred to as SSp-1) showed strong IFN-γ production, indicative of CTL reactivity, to T2 cells loaded with SSp-1 in a dose-dependent fashion but not to T2 cells alone or those loaded with the irrelevant peptide CAP-1 (Figure 1A ). CTLs from SSp-1–immunized mice efficiently lysed SSp-1–pulsed T2 target cells but did not lyse T2 cells alone or CAP-1–pulsed T2 cells at any effector-target ratio (Figure 1B). These results indicated that IFN-γ production and cytotoxicity of induced CTLs was SSp-1 peptide specific.
Figure 1.
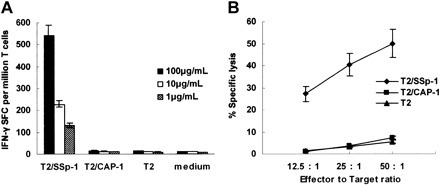
Immunogenicity of SARS-S–derived peptide SSp-1 in HLA-A2.1/Kb transgenic mice. IFN-γ release by bulk CTLs from immunized mice was detected by ELISPOT assay (A) and lytic activity was tested using a standard 4-hour chromium release assay (B). (A) Bulk CTLs from SSp-1–immunized mice released IFN-γ in response to T2 cells pulsed with SSp-1 (T2/SSp-1) in a dose-dependent manner, but not those with irrelevant peptide CAP-1 (T2/CAP-1) or T2 cells alone. (B) Bulk CTLs from SSp-1–immunized mice lysed T2 cells loaded with SSp-1, but not T2 cells pulsed with the irrelevant peptide CAP-1 or T2 cells alone. Data represent means ± SD.
In vitro generation of peptide-specific CD8+ CTLs from healthy human donor PBLs
To investigate the capacity of SSp-1 to mobilize a human CTL repertoire, SSp-1–specific CTLs were generated by in vitro sensitization of healthy HLA-A2.1+ donor PBLs with autologous DCs prepulsed with SSp-1. In more than 60% (7/11) of healthy HLA-A2.1+ individuals tested, SSp-1–specific CD8+ T cells were generated. To determine the antigen specificity of in vitro–induced CTLs, T2 cells and autologous DCs were pulsed with SSp-1 and used as stimulators and/or targets in ELISPOT and cytotoxicity assays. Human CTLs produced IFN-γ in response to SSp-1 in a concentration-dependent manner, but not to the irrelevant peptide CAP-1 (Figure 2A-B ). In vitro–generated effectors lysed T2 cells pulsed with SSp-1 but did not lyse target cells alone or CAP-1–pulsed T2 cells (Figure 2C).
Figure 2.
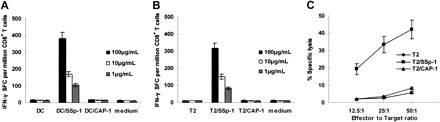
Specific reactivity of human CTLs induced by SSP-1–loaded DCs in vitro. SSp-1–specific CTLs were generated from the PBMCs of 7 of 11 HLA-A2.1+ healthy donors through 4 sequential rounds of stimulation with SSp-1–pulsed DCs. Resulting CTLs were tested for IFN-γ release (A-B) and SSp-1–specific lysis (C) using an ELISPOT assay and a standard 4-hour chromium release assay. Autologous DCs (A) and T2 cells (B) were pulsed with indicated concentrations of SSp-1 (DC/SSp-1, T2/SSp-1) or irrelevant peptide CAP-1 (DC/CAP-1, T2/CAP-1), and then used as stimulators in an IFN-γ release assay. (C) CTLs lysed T2 cells loaded with SSp-1, but not T2 cells loaded with the irrelevant peptide CAP-1 or T2 cells alone. Results are representative of 3 independent experiments. Data represent means ± SD.
Recognition of SSp-1 as a naturally processed peptide
To further address whether the immunogenic epitope SSp-1 is naturally processed and presented, CTLs generated by in vitro sensitization of healthy donor PBLs were tested for the capacity to recognize HLA-A2.1+ tumor cell line SW480 cells genetically modified to endogenously express SARS/S. CTLs were capable of killing pAd-SARS/S–transfected HLA-A2.1+ SW480 cells that expressed both HLA-A*0201 molecules and SARS-S. However, no lysis of pAd-LacZ–transduced SW480 cells (HLA-A2.1+, SARS-S–), pAd-SARS/S–transduced HT29 cells (HLA-A2.1–, SARS-S+), or pAd-LacZ–transduced HT29 cells (HLA-A2.1–, SARS-S–) was observed (Figure 3A ). These results indicate that SSp-1 is not only naturally processed but is also an HLA-A2.1–restricted CTL epitope. This was further confirmed by measuring IFN-γ secretion of T cells following contact with SARS/S-expressing SW480 cells. CTLs produced IFN-γ in response to pAd-SARS/S–transduced SW480 cells, but pAd-SARS/S–transduced HT29 cells and pAd-LacZ–transduced SW480 and HT29 cells did not induce IFN-γ secretion (Figure 3B).
Figure 3.
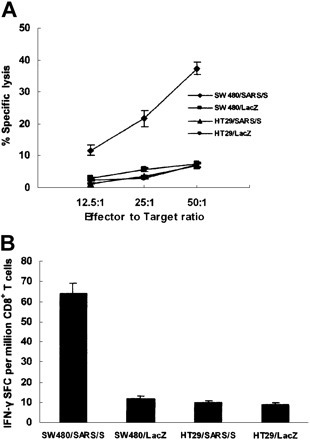
Identification of SSp-1 as a naturally processed and presented HLA-A2.1–restricted epitope. DC-induced human CTLs against SSp-1 were tested for specific lytic activity (A) and IFN-γ release (B) in response to genetically modified cell lines. (A) CTLs lysed pAd-SARS/S–transduced HLA-A2.1+ SW480 cells (SW480/SARS/S) but did not lyse pAd-SARS/S–transduced HLA-A2.1– HT29 cells (HT29/SARS/S) or pAd-LacZ–transduced SW480 (SW480/LacZ) and HT29 cells (HT29/LacZ). (B) CTLs elicited strong IFN-γ production in response to pAd-SARS/S–transduced SW480 cells, but only very low levels were elicited by pAd-SARS/S–transduced HT29 cells or pAd-LacZ–transduced cell lines. Results are representative of 3 independent experiments. Data represent means ± SD.
Taken together, these results indicate that the SARS/S-derived peptide, SSp-1 (RLNEVAKNL), is not only able to induce specific CTL responses in HLA-A2.1/Kb transgenic mice but can also mobilize a specific CTL repertoire in PBLs from healthy HLA-A2.1+ human donors. Moreover, SSp-1 is a naturally processed immunogenic peptide and is recognized by CTLs in an antigen-specific and HLA-A*0201–restricted manner.
Frequencies of SSp-1–specific CD8+ T cells determined by tetramer staining
To visualize and enumerate SSp-1–specific CD8+ T cells from among lymphocytes induced either in vitro from healthy donor PBLs or in vivo in Tg mice, we constructed phycoerythrin (PE)–labeled HLA-A*0201 peptide tetramers containing the nonameric peptide SSp-1. These tetramers were designed to specifically bind to SSp-1–reactive T-cell clones but not unreactive clones. Virtually no specific tetramer staining could be detected in splenocyte preparations from unimmunized Tg mice or among unstimulated frozen PBLs from healthy HLA-A2.1+ donors (data not shown). As a negative control, HLA-A*0201 tetramers containing the peptide CAP-1 were prepared. Cells were simultaneously labeled with FITC-conjugated anti-CD8 mAb to distinguish CD8+ T cells. The frequency of SSp-1–specific CD8+ T cells present following in vitro stimulation of healthy HLA-A2.1+ donor PBLs with SSp-1–loaded DCs was 0.50% by HLA-A*0201–SSp-1 tetramer staining, with only 0.01% labeled by HLA-A*0201–CAP-1 tetramers (Figure 4A-B ). SSp-1–specific CD8+ T cells arising in Tg mice immunized with SSp-1–pulsed DCs made up 8.50% of the total number of CD8+ T cells by HLA-A*0201–SSp-1 tetramer staining, with only 0.01% labeled by HLA-A*0201–CAP-1 tetramers (Figure 4C-D).
Figure 4.
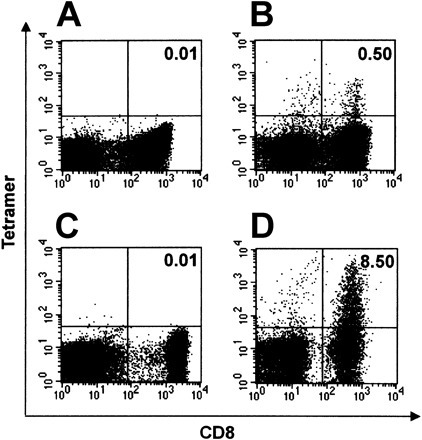
Frequency of SSp-1–specific CD8+ T cells. SSp-1–specific CD8+ T-cell frequency among DC-induced human CTLs against SSp-1 (A-B) and splenocytes derived from immunized mice and restimulated for 6 days in vitro (C-D). Tetramer-binding CD8+ T cells are shown in the upper-right quadrant and are labeled with the percentage of total CD8+ T cells. SSp-1–specific CTLs generated from human PBLs were stained with control HLA-A*0201/CAP-1 tetramers (A) and HLA-A*0201/SSp-1 tetramers (B). Results are representative of 3 independent experiments. Splenocytes from Tg mice immunized with SSp-1–pulsed DCs were stained with control HLA-A*0201/CAP-1 tetramers (C) and HLA-A*0201/SSp-1 tetramers (D).
Discussion
In this study, we identify a SARS-CoV spike protein–derived epitope, SSp-1 (RLNEVAKNL), with high affinity for HLA-A*0201 molecules, designed using computer algorithms and verified via MHC peptide binding assay. The peptide is further characterized by being immunogenic in vivo in HLA-A2.1/Kb transgenic mice, inducing in vitro–specific CTL responses from HLA-A*0201–matched healthy donor PBLs and being endogenously processed and presented in the context of HLA-A*0201 molecules on the surface of cell lines. CTLs generated in response to SSp-1 immunization and/or stimulation secrete IFN-γ and kill target cells in a peptide-specific and HLA-A*0201–restricted manner. MHC-peptide tetramer staining not only permitted the visualization of SSp-1–specific CD8+ T cell frequency in induced T-cell lines but also further confirmed strong binding of HLA-A*0201/SSp-1 tetramers with T-cell receptors (TCRs). Another candidate peptide, no. 5, failed to induce significant peptide-specific CTLs in Tg mice, despite its high binding affinity for HLA-A*0201, indicating the need for peptides predicted by computer algorithms to be biofunctionally validated. Peptide-MHC tetramers should be able to bind more than one TCR on a specificT cell, and thus have correspondingly slower dissociation rates. CD8 binding significantly enhances the avidity as well as the stability of interactions between TCR and cognate tetramers.50,51 It is notable that human HLA-A2/peptide tetramers can detect high-affinity TCR only on murine CTLs27,52,53 since murine CD8 does not bind human HLA-A2 molecules and therefore cytotoxicity assays would be more sensitive than HLA-A2–restricted tetramer binding for murine CTLs in the HLA-A2 transgenic mice.
Spike protein is a major structural glycoprotein of coronaviruses. The mature spike protein is inserted into the viral envelope, with most of the protein exposed on the surface of viral particles. Spike protein molecules associate in groups of 3 to form the characteristic corona-like structures of this virus family.6 In some coronaviruses, the spike protein has been identified as the protein responsible for both entry of the virus into susceptible cells and also for eliciting neutralizing antibody from the host.54, 55, 56, 57 In this study, an endogenously processed and presented peptide derived from the SARS-CoV spike protein could induce significant CTL responses in PBLs from healthy human donors, suggesting that CTL precursor populations against SARS/S exist within the T-cell repertoire of healthy individuals. CTL reactivity is postulated to be important in control of virus replication and in pathogenesis of SARS, but the exact mechanisms remain unclear.
In addition to CTL-mediated immunopathology, host genes that affect the viral receptor, virus production, and immune responses to infection can determine the outcome of coronavirus infections.58 The death of some SARS patients may have resulted from host factors that exacerbated the disease and enhanced antibody production.1,2,11,13 Pronounced macrophage infiltration was observed in the alveoli and the interstitium of the lung tissues obtained from the autopsy of SARS patients.2,9 Proinflammatory cytokines released by sensitized macrophages in the alveoli may play a prominent role in the pathogenesis of SARS, resulting in cytokine dysregulation.
Clinical tests show that a notable drop in CD4+ and CD8+ T-lymphocyte counts occurs early in the course of the syndrome.1,10,11 Probable explanations for this rapid decline in peripheral blood T-cell numbers in SARS patients include: (1) The SARS virus infects T cells and damages them directly. (2) SARS-CoV infection of pulmonary tissues causes cells present in these tissues, such as epithelia cells, pneumocytes, matrix cells, and infiltrating antigen-presenting cells (APCs), to release certain cytokines or chemokines that attract T cells into the lungs, leading to an observed decrease in T-cell frequency in the peripheral blood. (3) As with any critical illness, SARS is accompanied by the activation of the hypothalamic-pituitary-adrenal axis, resulting in increased levels of adrenocorticotropic hormone (ACTH) and cortisol, aimed at maintaining the integrity of the vasculature and modulating the actions of proinflammatory and anti-inflammatory cytokines.59 Both glucocorticoids and stimulation of the hypothalamic-pituitary-adrenal axis lead to lymphocyte margination,10 the initial step involved in lymphocyte migration out of circulating blood; SARS patients not exhibiting lymphopenia may have adrenal insufficiency. Glucocorticoids administered to treat SARS may thus induce, or exacerbate, T-lymphocyte movement out of the intravascular compartment. However, treatment with glucocorticoid drugs does not account for lymphopenia in all patients, and certainly not for lymphopenia observed at the initial presentation. (4) Our results show the existence of functional anti–S protein CTL precursors within the peripheral T-cell repertoire of healthy human donors. The SARS virus or its components may thus act as an antigen or superantigen and polyclonally activate T cells, leading to activation-induced cell death (AICD). Peptide- or antigen-loaded DC numbers decline as functional effector CTLs appear,60, 61, 62 which may in turn have a negative effect on the persistence of functional effector CTLs. (5) Apoptosis may also contribute to SARS-associated lymphopenia.63 Respiratory syncytial virus (RSV) disease, measles, and sepsis show parallels to SARS, in that they also cause lymphopenia. In SARS, apoptotic cells, including pneumocytes, lymphocytes, and monocytes, and CD68+ monocytes are observed in abundance in the lung, spleen, and lymph nodes at postmortem examination.64 In models of sepsis, inhibitors of apoptosis ameliorate illness and prevent death,65 pointing to important therapeutic implications for patients with SARS. Interestingly, the mean B-lymphocyte count remains stable in the peripheral blood of SARS patients,8,12 with 100% of patients exhibiting antibody responses to SARS-CoV, with typical IgM and IgG profiles, during the convalescent phase.58,66
In pulmonary reovirus infection in athymic mice, a lower plaque-forming value of 106 is associated with pathologic changes consistent with bronchiolitis obliterans with organizing pneumonia, whereas a higher inoculum of 107 is associated with development of the more serious acute respiratory distress syndrome (ARDS).67 To lessen the risk of progression to ARDS, an effective antiviral strategy to reduce the viral load may be important; decreasing the initial cytolytic damage and viral load at the onset may in turn result in decreased immunopathologic damage.8 Thus, we must seek to maintain a fine balance between CTL-induced immune protection and pathology in the treatment of SARS disease. The HLA-A*0201–SSp-1 tetramers we constructed may be useful for monitoring the fluctuation of SSp-1–specific CD8+ T-cell numbers, and by extrapolation, SARS-specific CD8+ T-cell numbers as a whole, in the peripheral blood of SARS patients during clinical or therapeutic treatments, providing an additional indicator for treatment efficacy and short/long-term prognosis.
In conclusion, our study has identified a novel HLA-A*0201–restricted, immunogenic CD8+ T-cell epitope of the SARS-CoV spike protein, SSp-1. We propose that, as a representative CTL antigen, SSp-1 could be used for evaluation of SARS-CoV–specific CD8+ T-cell responses during the course of SARS infection and treatment. Furthermore, this newly identified epitope should help in the characterization of mechanisms of virus control and immunopathology in SARS-CoV infection, and may be relevant to the development of immunotherapeutic approaches for this sometimes lethal respiratory disease.
Acknowledgments
The contribution of Dr Jane Rayner in critically reading the manuscript is sincerely acknowledged. Dr Shuxun Liu is acknowledged for her guidance with generation of DCs and CTLs. We are grateful to Dr Zhenhong Guo, Ms Rui Zhang, and Ms Chunfang Luo for their excellent technical assistance with flow cytometry analysis.
Footnotes
Prepublished online as Blood First Edition Paper, March 11, 2004; DOI 10.1182/blood-2003-11-4072.
Supported by grants from the National Key Basic Research Program of China (2003CB514108, 2001CB510002, and 2001CB510009), the Natural Science Foundation of Shanghai (03DZ14104), and the National Natural Science Foundation of China (30121002).
B.W. and H.C. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
- 1.Lee N, Hui D, Wu A. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 2.Ksiazek TG, Erdman D, Goldsmith CS. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 3.Peiris J, Lai S, Poon L, Guan Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drosten C, Gunther S, Preiser W. Indentification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 5.Rota PA, Oberste MS, Monroe SS. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 6.Marra MA, Jones SJ, Astell CR. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 7.Kuiken T, Fouchier R, Schutten M. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peiris JS, Chu CM, Cheng VC. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholls JM, Poon LL, Lee KC. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panesar NS. Lymphopenia in SARS. Lancet. 2003. 1985;361:361. doi: 10.1016/S0140-6736(03)13557-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Booth CM, Matukas LM, Tomlinson GA. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA. 2003;289:2801–2809. doi: 10.1001/jama.289.21.JOC30885. [DOI] [PubMed] [Google Scholar]
- 12.Wong R, Wu A, To KF. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. Brit Med J. 2003;326:1358–1362. doi: 10.1136/bmj.326.7403.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Openshaw PJ. Immunopathological mechanisms in respiratory syncytial virus disease. Springer Semin Immunopathol. 1995;17:187–201. doi: 10.1007/BF00196165. [DOI] [PubMed] [Google Scholar]
- 14.Ostler T, Davidson W, Ehl S. Virus clearance and immunopathology by CD8+ T cells during infection with respiratory syncytial virus are mediated by IFN-gamma. Eur J Immunol. 2002;32:2117–2123. doi: 10.1002/1521-4141(200208)32:8<2117::AID-IMMU2117>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 15.Brandenburg AH, de Waal L, Timmerman HH, Hoogerhout P, de Swart RL, Osterhaus AD. HLA class I-restricted cytotoxic T-cell epitopes of the respiratory syncytial virus fusion protein. J Virol. 2000;74:10240–10244. doi: 10.1128/jvi.74.21.10240-10244.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostler T, Ehl S. Pulmonary T cells induced by respiratory syncytial virus are functional and can make an important contribution to long-lived protective immunity. Eur J Immunol. 2002;32:2562–2569. doi: 10.1002/1521-4141(200209)32:9<2562::AID-IMMU2562>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 17.Culley FJ, Pollott J, Openshaw PJ. Age at first viral infection determines the pattern of T cell-mediated disease during reinfection in adulthood. J Exp Med. 2002;196:1381–1386. doi: 10.1084/jem.20020943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shieh DC, Lin DT, Yang BS, Kuan HL, Kao KJ. High frequency of HLA-A*0207 subtype in Chinese population. Transfusion. 1996;36:818–821. doi: 10.1046/j.1537-2995.1996.36996420761.x. [DOI] [PubMed] [Google Scholar]
- 19.Qin E, Zhu Q, Yu M. A complete sequence and comparative analysis of a SARS-associated virus (Isolate BJ01). Chinese Science Bulletin. 2003;48:941–948. doi: 10.1007/BF03184203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor R. BioInformatics and Molecular Analysis Section (BIMAS) HLA peptide binding predictions. Available at: http://bimas.dcrt.nih.gov/molbio/hla_bind/index.html. 2000 [Google Scholar]
- 21.Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994;152:163–175. [PubMed] [Google Scholar]
- 22.Morse MA, Deng Y, Coleman D. A phase I study of active immunotherapy with carcinoembryonic antigen peptide (CAP-1)-pulsed, autologous human cultured dendritic cells in patients with metastatic malignancies expressing carcinoembryonic antigen. Clin Cancer Res. 1999;5:1331–1338. [PubMed] [Google Scholar]
- 23.Vitiello A, Marchesini D, Furze J, Sherman LA, Chesnut RW. Analysis of the HLA-restricted influenza-specific cytotoxic T lymphocyte response in transgenic mice carrying a chimeric humanmouse class I major histocompatibility complex. J Exp Med. 1991;173:1007–1015. doi: 10.1084/jem.173.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuzushima K, Hayashi N, Kimura H, Tsurumi T. Efficient identification of HLA-A*2402-restricted cytomegalovirus-specific CD8+ T-cell epitopes by a computer algorithm and an enzyme-linked immunospot assay. Blood. 2001;98:1872–1881. doi: 10.1182/blood.v98.6.1872. [DOI] [PubMed] [Google Scholar]
- 25.Passoni L, Scardino A, Bertazzoli C. ALK as a novel lymphoma-associated tumor antigen: identification of 2 HLA-A2.1-restricted CD8+ T-cell epitopes. Blood. 2002;99:2100–2106. doi: 10.1182/blood.v99.6.2100. [DOI] [PubMed] [Google Scholar]
- 26.Ju D, Tao Q, Lou G. Interleukin 18 transfection enhances antitumor immunity induced by dendritic cell-tumor cell conjugates. Cancer Res. 2001;61:3735–3740. [PubMed] [Google Scholar]
- 27.La Rosa C, Wang Z, Brewer JC. Preclinical development of an adjuvant-free peptide vaccine with activity against CMV pp65 in HLA transgenic mice. Blood. 2002;100:3681–3689. doi: 10.1182/blood-2002-03-0926. [DOI] [PubMed] [Google Scholar]
- 28.Ishioka GY, Fikes J, Hermanson G. Utilization of MHC class I transgenic mice for development of minigene DNA vaccines encoding multiple HLA-restricted CTL epitopes. J Immunol. 1999;162:3915–3925. [PubMed] [Google Scholar]
- 29.Mateo L, Gardner J, Chen Q. An HLA-A2 polyepitope vaccine for melanoma immunotherapy. J Immunol. 1999;163:4058–4063. [PubMed] [Google Scholar]
- 30.Firat H, Garcia-Pons F, Tourdot S. H-2 class I knockout, HLA-A2.1-transgenic mice: a versatile animal model for preclinical evaluation of antitumor immunotherapeutic strategies. Eur J Immunol. 1999;29:3112–3121. doi: 10.1002/(SICI)1521-4141(199910)29:10<3112::AID-IMMU3112>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 31.Vissers JL, De Vries IJ, Schreurs MW. The renal cell carcinoma-associated antigen G250 encodes a human leukocyte antigen (HLA)-A2.1-restricted epitope recognized by cytotoxic T lymphocytes. Cancer Res. 1999;59:5554–5559. [PubMed] [Google Scholar]
- 32.Arichi T, Saito T, Major ME. Prophylactic DNA vaccine for hepatitis C virus (HCV) infection: HCV-specific cytotoxic T lymphocyte induction and protection from HCV-recombinant vaccinia infection in an HLA-A2.1 transgenic mouse model. Proc Natl Acad Sci U S A. 2000;97:297–302. doi: 10.1073/pnas.97.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Cheuk E, D’Souza C, Hu N, Liu Y, Lang H, Chamberlain JW. Human MHC class I transgenic mice deficient for H2 class I expression facilitate identification and characterization of new HLA class I-restricted viral T cell epitopes. J Immunol. 2002;169:5571–5580. doi: 10.4049/jimmunol.169.10.5571. [DOI] [PubMed] [Google Scholar]
- 34.Bullock TN, Mullins DW, Engelhard VH. Antigen density presented by dendritic cells in vivo differentially affects the number and avidity of primary, memory, and recall CD8+ T cells. J Immunol. 2003;170:1822–1829. doi: 10.4049/jimmunol.170.4.1822. [DOI] [PubMed] [Google Scholar]
- 35.Cao X, Zhang W, He L. Lymphotactin genemodified bone marrow dendritic cells act as more potent adjuvants for peptide delivery to induce specific antitumor immunity. J Immunol. 1998;161:6238–6244. [PubMed] [Google Scholar]
- 36.Janeway CA, Bottomly K. Signals and signs for lymphocytes responses. Cell. 1994;76:275–285. doi: 10.1016/0092-8674(94)90335-2. [DOI] [PubMed] [Google Scholar]
- 37.Mellman I, Steinman RM. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106:255–258. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 38.Celluzzi CM, Mayordomo JI, Storkus WJ, Lotze MT, Falo LD. Peptide-pulsed dendritic cells induce antigen-specific, CTL-mediated protective tumor immunity. J Exp Med. 1996;183:283–287. doi: 10.1084/jem.183.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang S, Linette GP, Longerich S, Haluska FG. Antimelanoma activity of CTL generated from peripheral blood mononuclear cells after stimulation with autologous dendritic cells pulsed with melanoma gp100 peptide G209-2M is correlated to TCR avidity. J Immunol. 2002;169:531–539. doi: 10.4049/jimmunol.169.1.531. [DOI] [PubMed] [Google Scholar]
- 40.Subklewe M, Chahroudi A, Schmaljohn A, Kurilla MG, Bhardwaj N, Steinman RM. Induction of Epstein-Barr virus-specific cytotoxic T-lymphocyte responses using dendritic cells pulsed with EBNA-3A peptides or UV-inactivated, recombinant EBNA-3A vaccinia virus. Blood. 1999;94:1372–1381. [PubMed] [Google Scholar]
- 41.Wang QJ, Huang XL, Rappocciolo G. Identification of an HLA A*0201-restricted CD8+ T-cell epitope for the glycoprotein B homolog of human herpesvirus 8. Blood. 2002;99:3360–3366. doi: 10.1182/blood.v99.9.3360. [DOI] [PubMed] [Google Scholar]
- 42.Stott LM, Barker RN, Urbaniak SJ. Identification of alloreactive T-cell epitopes on the Rhesus D protein. Blood. 2000;96:4011–4019. [PubMed] [Google Scholar]
- 43.Brossart P, Heinrich KS, Stuhler G. Identification of HLA-A2-restricted T-cell epitopes derived from the MUC1 tumor antigen for broadly applicable vaccine therapies. Blood. 1999;93:4309–4317. [PubMed] [Google Scholar]
- 44.Liu S, Yu Y, Zhang M, Wang W, Cao X. The involvement of TNF-alpha-related apoptosis-inducing ligand in the enhanced cytotoxicity of IFN-beta-stimulated human dendritic cells to tumor cells. J Immunol. 2001;166:5407–5415. doi: 10.4049/jimmunol.166.9.5407. [DOI] [PubMed] [Google Scholar]
- 45.Brossart P, Bevan MJ. Selective activation of Fas/FasL mediated cytotoxicity by a self-peptide. J Exp Med. 1996;183:2449–2458. doi: 10.1084/jem.183.6.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keogh E, Fikes J, Southwood S, Celis E, Chesnut R, Sette A. Identification of new epitopes from four different tumor-associated antigens: recognition of naturally processed epitopes correlates with HLA-A*0201-binding affinity. J Immunol. 2001;167:787–796. doi: 10.4049/jimmunol.167.2.787. [DOI] [PubMed] [Google Scholar]
- 47.Kuzushima K, Hayashi N, Kudoh A. Tetramer-assisted identification and characterization of epitopes recognized by HLA A*2402-restricted Epstein-Barr virus-specific CD8+ T cells. Blood. 2003;101:1460–1468. doi: 10.1182/blood-2002-04-1240. [DOI] [PubMed] [Google Scholar]
- 48.Altman JD, Moss PA, Goulder PJ. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 49.Lechner F, Wong DK, Dunbar PR. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med. 2000;191:1499–1512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi EM, Chen, Wooldridge L. High avidity antigen-specific CTL identified by CD8-independent tetramer staining. J Immunol. 2003;171:5116–5123. doi: 10.4049/jimmunol.171.10.5116. [DOI] [PubMed] [Google Scholar]
- 51.Kerry SE, Buslepp J, Cramer LA. Interplay between TCR affinity and necessity of coreceptor ligation: high-affinity peptide-MHC/TCR interaction overcomes lack of CD8 engagement. J Immunol. 2003;171:4493–4503. doi: 10.4049/jimmunol.171.9.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hernandez J, Lee PP, Davis MM, Sherman LA. The use of HLA A2.1/p53 peptide tetramers to visualize the impact of self tolerance on the TCR repertoire. J Immunol. 2000;164:596–602. doi: 10.4049/jimmunol.164.2.596. [DOI] [PubMed] [Google Scholar]
- 53.Schell TD, Lippolis JD, Tevethia SS. Cytotoxic T lymphocytes from HLA-A2.1 transgenic mice define a potential human epitope from simian virus 40 large T antigen. Cancer Res. 2001;61:873–879. [PubMed] [Google Scholar]
- 54.Bonavia A, Zelus BD, Wentworth DE, Talbot PJ, Holmes KV. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HcoVE-229E. J Virol. 2003;77:2530–2538. doi: 10.1128/JVI.77.4.2530-2538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arbour N, Ekande S, Cote G. Persistent infection of human oligodendrocytic and neuroglial cell lines by human coronavirus 229E. J Virol. 1999;73:3326–3337. doi: 10.1128/jvi.73.4.3326-3337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wessner DR, Shick PC, Lu Mutational analysis of the virus and monoclonal antibody binding sites in MHVR, the cellular receptor of the murine coronavirus mouse hepatitis virus strain A59. J Virol. 1998;72:1941–1948. doi: 10.1128/jvi.72.3.1941-1948.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saeki K, Ohtsuka N, Taguchi F. Identification of spike protein residues of murine coronavirus responsible for receptor-binding activity by use of soluble receptor-resistant mutants. J Virol. 1997;71:9024–9031. doi: 10.1128/jvi.71.12.9024-9031.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holmes KV. SARS-associated coronavirus. N Engl J Med. 2003;348:1948–1951. doi: 10.1056/NEJMp030078. [DOI] [PubMed] [Google Scholar]
- 59.Thompson BT. Glucocorticoids and acute lung injury. Crit Care Med. 2003;31:S253–S257. doi: 10.1097/01.CCM.0000057900.19201.55. [DOI] [PubMed] [Google Scholar]
- 60.Lu W, Wu X, Lu Y, Guo W, Andrieu JM. Therapeutic dendritic-cell vaccine for simian AIDS. Nat Med. 2003;9:27–32. doi: 10.1038/nm806. [DOI] [PubMed] [Google Scholar]
- 61.Hermans IF, Ritchie DS, Yang J, Roberts JM, Ronchese F. CD8+ T cell-dependent elimination of dendritic cells in vivo limits the induction of antitumor immunity. J Immunol. 2000;164:3095–3101. doi: 10.4049/jimmunol.164.6.3095. [DOI] [PubMed] [Google Scholar]
- 62.Norbury CC, Malide D, Gibbs JS, Bennink JR, Yewdell JW. Visualizing priming of virus-specific CD8+ T cells by infected dendritic cells in vivo. Nat Immunol. 2002;3:265–271. doi: 10.1038/ni762. [DOI] [PubMed] [Google Scholar]
- 63.O’Donnell R, Tasker RC, Roe MF. SARS: understanding the coronavirus: apoptosis may explain lymphopenia of SARS. Br Med J. 2003;327:620. doi: 10.1136/bmj.327.7415.620-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang QL, Ding YQ, He L. Detection of cell apoptosis in the pathological tissues of patients with SARS and its significance. Di Yi Jun Yi Da Xue Xue Bao. 2003;23:770–773. [PubMed] [Google Scholar]
- 65.Hotchkiss RS, Chang KC, Swanson PE. Caspase inhibitors improve survival in sepsis: a critical role of the lymphocyte. Nat Immunol. 2000;1:496–501. doi: 10.1038/82741. [DOI] [PubMed] [Google Scholar]
- 66.Li G, Chen X, Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N Engl J Med. 2003;349:508–509. doi: 10.1056/NEJM200307313490520. [DOI] [PubMed] [Google Scholar]
- 67.Majeski EI, Harley RA, Bellum SC, London SD, London L. Differential role for T cells in the development of fibrotic lesions associated with reovirus 1/L-induced bronchiolitis obliterans organizing pneumonia versus acute respiratory distress syndrome. Am J Respir Cell Mol Biol. 2003;28:208–217. doi: 10.1165/rcmb.4891. [DOI] [PubMed] [Google Scholar]


