Abstract
In 2019, the SARS-CoV-2 beta-coronavirus, which caused a pandemic of severe acute respiratory viral infection COVID-19 (from COronaVIrus Disease 2019), was first detected. The susceptibility to SARS-CoV-2 and the nature of the course of the COVID-19 clinical picture are determined by many factors, including genetic characteristics of both the pathogen and the human. The SARS-CoV-2 genome has a similarity to the genomes of other coronaviruses, which are pathogenic for humans and cause a severe course of infection: 79% to the SARS-CoV genome and 50% to the MERS-CoV genome. The most significant differences between SARS-CoV-2 and other coronaviruses are recorded in the structure of the gene of the S protein, a key protein responsible for the virus binding to the receptor of the host organism cells. In particular, substitutions in the S protein of SARS-CoV-2, leading to the formation of the furin cleavage site that is absent in other SARS-like coronaviruses, were identified, which may explain the high pathogenicity of SARS-CoV-2. In humans, the genes that are significant for the initial stages of infection include ACE2, ANPEP, DPP4 (encode receptors for coronavirus binding); TMPRSS2, FURIN, TMPRSS11D, CTSL, CTSB (encode proteases involved in the entry of the coronavirus into the cell); DDX1 (the gene of ATP-dependent RNA helicase DDX1, which promotes replication of coronaviruses); and IFITM1, IFITM2, and IFITM3 (encode interferon-induced transmembrane proteins with an antiviral effect). These genes are expressed in many tissues (including those susceptible to the effects of SARS-CoV-2); rare and frequent variants that affect the structure of the encoded protein and its properties and expression level are described in them. A number of common genetic variants with proven functional significance are characterized by the variability in the allele frequency in the world’s populations, which can determine interpopulation differences in the prevalence of COVID-19 and in the clinical features of the course of this pathology. The expression level of genes that are important for the formation of the susceptibility to SARS-CoV-2 is affected by epigenetic modifications, comorbidities at the time of infection, taking medications, and bad habits.
Keywords: SARS-CoV-2 genome, COVID-19, candidate genes for susceptibility to SARS-CoV-2, SNV, eQTL, expression, DNA methylation
INTRODUCTION
The SARS-CoV-2 coronavirus, first identified in 2019, led to a pandemic of a severe acute respiratory viral infection known as COVID-19 (from COronaVIrus Disease 2019). According to official statistics, on June 21, 2020, the pandemic covered 215 countries (territories), more than 8.8 million people were infected with COVID-19, and more than 465 000 people died [1].
SARS-CoV-2 is an RNA beta-coronavirus (CoV). For humans, seven coronaviruses are known: alpha-coronaviruses HCoV-229E and HCoV-NL63 and beta-coronaviruses HCoV-OC43, HCoV-HKU1, SARS-CoV, MERS-CoV, and SARS-CoV-2 [2]. Coronaviruses differ in pathogenicity: 229E, OC43, NL63, and HKU1 cause mild forms of infectious disease, while SARS-CoV, MERS-CoV, and SARS-CoV-2 are highly pathogenic (SARS-CoV caused atypical pneumonia in 2002–2003; MERS-CoV caused Middle East respiratory syndrome in 2015) [2, 3]. The clinical picture of diseases caused by highly pathogenic coronaviruses has both general and specific features [4–6]. In particular, COVID-19 is characterized by greater contagiousness, but lower mortality, and significant heterogeneity of clinical symptoms, including in different age groups and in different countries of the world compared with the infectious disease caused by MERS-CoV [5, 7–9]. The most common symptoms of COVID-19 are cough, fever, and dyspnea; myalgia, diarrhea, nausea, and vomiting are also recorded [6, 8]. However, individual clinical symptoms vary widely: from an asymptomatic course and a mild cold to complications such as bronchitis, pneumonia, severe acute respiratory distress syndrome, and multiple organ failure [6].
The human susceptibility to coronavirus infection and the clinical picture of the disease depend on many causes, among which a certain role belongs to the genetic characteristics of both the viruses and the person [10–13]. The initial stage of infection of the body is the contact of the coronavirus with cells susceptible to it. In humans, about 100 genes whose products are involved in the process of virus entry into the cell are known; they are classified into five categories of Gene Ontology: viral entry into the host cell (GO: 0046718); virus receptor activity (GO: 0001618); fusion of the virus membrane with the host plasma membrane (GO: 0019064); clathrin-dependent endocytosis of the virus by the host cell (GO: 0075512); receptor-mediated endocytosis of the virus by the host cell (GO: 0019065) [14, 15]. As we study the interactions between viruses and host cells, the number of known genes that affect susceptibility to coronavirus infection will increase. At the same time, coronaviruses also differ in the structure of the genome and key molecules responsible for their entry into cells [11, 16, 17]. Therefore, not all human genes whose products are involved in the regulation of the entry of viruses into cells turn out to be significant in relation to the formation of the risk of developing the infection caused by various coronaviruses.
The present review analyzes the features of the SARS-CoV-2 genome and the structural and functional properties of human genes and proteins responsible for the susceptibility to this pathogen.
PATHOGENETICALLY SIGNIFICANT FEATURES OF THE SARS-COV-2 GENOME
The SARS-CoV-2 genome is structurally similar to the genomes of two bat coronaviruses bat-SL-CoVZC45 and bat-SL-CoVZXC21 (the identity is 88–96%) and has much less similarity to the genomes of SARS-CoV (79–80%) and MERS-CoV (about 50%) [17, 18]. The genomes of individual SARS-CoV-2 samples obtained from Chinese patients were characterized by the high similarity exceeding 99% [18], but estimates on genetic diversity are being refined as the number of studies on the structure of the SARS-CoV-2019 genomes from different regions of the world increases [19, 20].
The SARS-CoV-2 genome is characterized by lower values of the effective number of codons compared with SARS-CoV and MERS-CoV, which suggests a higher expression efficiency of genes, including genes encoding structural proteins: spike (S), envelope (E), membrane (M), nucleocapsid (N) [2, 21]. The S protein (spike protein), which is responsible for the attachment of SARS-CoV-2 to the membrane receptor, is considered critical for the entry into the host cell [22]. The SARS-CoV-2 coronavirus (like HCoV-NL63, SARS-CoV, MERS-CoV) uses membrane-bound angiotensin-converting enzyme 2 (ACE2) as a receptor [17, 23–25]. Accordingly, SARS-CoV-2 can penetrate into all cells expressing ACE2. However, coronaviruses can also use a number of other molecules as receptors to enter the cells of the host organism [25–27].
The structure of the SARS-CoV-2 S protein, as compared with other coronaviruses, has some features that are important for the entry into the host cell [16, 17, 28]. The identity between S proteins of SARS-CoV and SARS-CoV-2 is 72–75% [16, 17]. P. Zhou et al. [17] attributed three short inserts in the N-terminal domain and also changes in four of five key residues in the receptor-binding motif to the number of main differences in the structure of the gene sequence encoding the SARS-CoV-2 S protein. As compared with SARS-CoV, SARS-CoV-2 is characterized by a higher affinity for binding to the ACE2 receptor due to substitutions in the C-terminal domain of the S protein (there are more interactions with ACE2, H-bonds, etc.) [16, 28, 29], a better capability of fusion with the plasma membrane of the host cell due to amino acid substitutions in the HR1 domain of the S2 subunit of the S protein [30], and, in general, a higher activity [23].
As a result of comparative analysis of the genomes of SARS-CoV-2 isolates from different regions of the world, multiple mutations, including in genes encoding structural proteins (including the S protein), were established [20]. For example, a mutation in the receptor-binding domain of the S protein at position 407, leading to a change in the secondary structure of the protein, since the positively charged amino acid arginine is replaced by the hydrophobic C-beta-branched amino acid isoleucine, was found in clinical isolates from India [19]. Amino acid substitutions in the S protein were found also in SARS-CoV-2 from other geographic regions [31]. Mutations that change the structure of key proteins responsible for the binding of the coronavirus to its receptor can affect the pathogenicity of the coronavirus.
After the S protein binding to the ACE2 receptor, the protein is cleaved into S1 and S2 subunits, which initiates fusion of the viral envelope with the cell membrane and the entry of HCoV into the cell [22]. For these purposes, coronaviruses (like other viruses) use various proteases of host cells, the choice of which is determined by the structural features and the availability of the site for proteolytic cleavage of the S protein [32]. For example, it was shown that, as compared with laboratory strains from 1966, modern clinical isolates of the HCoV-229E alpha-coronavirus use TMPRSS2, not cathepsin L for the entry, which is associated with two amino acid substitutions (R642M and N714K) in the S protein [33]. SARS-CoV-2 also uses TMPRSS2 for activation and cleavage of the S protein [34–36]. In addition, substitutions in the S protein gene leading to the formation of a furin-like cleavage site, which is absent in other SARS-like coronaviruses, were identified in SAR-CoV-2 [25, 37]. The presence of the furin cleavage site in SARS-CoV-2 determines the ability of virus–cell and cell–cell fusion, which ensures the high pathogenicity of SARS-CoV-2 compared with other beta-coronaviruses [38, 39].
Since the entry of coronaviruses into the cell occurs with the participation of protein products of the cells of the host organism, it can be expected that the features in the structure of genes encoding these proteins will determine individual differences in the susceptibility to coronaviruses, including SARS-CoV-2.
GENETIC FACTORS OF THE SUSCEPTIBILITY TO SARS-CoV-2 IN HUMANS
On the basis of a classical twin study, the heritability rates for some symptoms of COVID-19 were calculated: for fever, this parameter was 41 (95% confidence interval 12–70)%, for anosmia was 47 (27–67)%, and for delirium was 49 (24–75)%, which allowed us to estimate the predicted COVID-19 heritability rate at 50 (29–70)% [40]. These estimates indicate a significant genetic component in the formation of symptoms of this disease. The studies devoted to the identification of genetic markers of the susceptibility to SARS-CoV-2 are not numerous [12, 41, 42].
A number of genes whose products can participate in the process of human infection with coronaviruses, including SARS-CoV-2, are known (Table 1). These include the gene of ATP-dependent RNA helicase DDX1 (DDX1), which promotes replication of coronaviruses (as was shown for SARS-CoV) [56], as well as the genes IFITM1, IFITM2, and IFITM3 encoding interferon-induced transmembrane proteins [12, 57, 58] in addition to receptor genes (ACE2, ANPEP, DPP4) and protease genes (TMPRSS2, FURIN, TMPRSS11D, CTSL, CTSB), which contribute to the entry of the coronavirus into the cell.
Table 1.
Characteristics of candidate genes for the susceptibility to human SARS-CoV-2
| No. | Gene (alternative names#), chromosomal localization |
Protein | Number of LoF/missense variants with MAF ≤ 1% (according to [43]) |
Missense variants with MAF > 1%: rs (allele: min–max frequency) (according to [14, 43, 44]) |
Number of known eQTL variants (according to [45, 46]) |
Participation in processes of infection with coronavirus |
Coronaviruses that use products of given genes to enter host organism cells |
|---|---|---|---|---|---|---|---|
| 1 | ACE2 (ACEH), Xp22.2 | Angiotensin-converting enzyme 2 (ACE2) | 5/239 | None | 288 | Coronavirus receptor [25] | SARS-CoV-2; SARS-CoV; HCoV-NL63 [10, 16, 23, 24, 28, 35, 39] |
| 2 |
DPP4 (CD26), 2q24.2 |
Dipeptidyl peptidase 4 | 60/391 | rs1129599 (A: 0–4%) | 130 | Coronavirus receptor [47] |
SARS-CoV-2; hCoV-EMC (MERS-CoV) |
| 3 | ANPEP (APN, CD13), 15q26.1 | Aminopeptidase N | 47/547 |
rs8192297 (C: 16–37%) rs25651 (T: 23–51%) rs25653 (C: 10–61%) rs17240268 (A: 0–14%)* rs41276922 (A: 0–9%)* rs17240212 (A: 0–3%) |
594 | Coronavirus receptor [26] | HCV-229E [26, 50] |
| 4 | FURIN (FUR), 15q26.1 | Furin | 5/339 | rs16944971 (T: 0–9%) | 295 |
S protein cleavage; |
SARS-CoV-2 [29, 37, 51] |
| 5 |
TMPRSS11D (HAT), 4q13.2 |
Transmembrane protease serine 11D |
17/210 | 0 | 191 |
Cleavage of S protein and ACE2; promotes fusion of viral and cell membranes [35] |
SARS-CoV; HCoV-229E [35, 52] |
| 6 |
TMPRSS2, 21q22.3 |
Transmembrane protease serine 2 |
39/332 |
rs12329760 (T: 15–36%)* rs75603675 (A: 2–40%) |
203 |
S protein cleavage; |
SARS-CoV-2; SARS-CoV; MERS-CoV [34, 35, 39, 51, 53, 54] |
| 7 |
CTSL, 9q21.33 |
Cathepsin L | 14/192 | rs11541204 (A: 0–5%) | 258 | S protein activation; promotes entry of the virus into the cell [33, 52] |
SARS-CoV-2; |
| 8 |
CTSB, 8p23.1 |
Cathepsin B | 41/369 |
rs12338 (G: 49–67%)* rs1803250 (C: 0–15%)* rs17573 (T: 0–17%)* |
2553 |
Activates fusion |
SARS-CoV-2; SARS-CoV; MERS-CoV [14, 29, 55] |
| 9 |
DDX1, 2p24.3 |
ATP-dependent RNA helicase DDX1 | 14/305 | 0 | 463 |
Promotes coronavirus replication (in vitro) [56] |
SARS-CoV [56] |
| 10 |
IFITM1, 11p15.5 |
Interferon-induced transmembrane protein 1 |
4/72 | 0 | 201 | Restricting replication and entry of viruses into the host cell [14, 57] | SARS-CoV [14, 57] |
| 11 |
IFITM2, 11p15.5 |
Interferon-induced transmembrane protein 2 |
11/123 |
rs1059091 (A: 31–67%)* rs1058900 (T: 0–42%) rs14408 (T: 4–62%) |
220 | Restricting replication and entry of viruses into the host cell [14, 57] | SARS-CoV and other viruses [57, 58] |
| 12 |
IFITM3, 11p15.5 |
Interferon-induced transmembrane protein 3 |
6/92 | rs1136853 (T: 0–8%)* | 439 | Restricting replication and entry of viruses into the host cell; inactivation of new viruses releasing from the infected cell [59] | SARS-CoV-2; SARS-CoV and other viruses [12, 57, 58] |
LoF—loss of function variant; MAF—minor allele frequency. # Alternative names of genes and proteins found in the used literature sources are indicated. * Potentially pathogenic effect based on the results of using programs for predicting the functional effect of genetic variants built into the Ensemble browser.
The role of ACE2 as a SARS-CoV-2 receptor is considered proven [16, 23, 25, 28, 39, 60]. The data on the possibility of using SARS-CoV-2 other receptors known for coronaviruses are ambiguous. A number of researchers [27, 48] attributed dipeptidyl peptidase 4 (DPP4, also known as CD26) to potential SARS-CoV-2 receptors, acting as a receptor for MERS-CoV, on the basis of modeling the structural interactions between the S protein and receptors of other coronaviruses [49]. Moreover, SARS-CoV-2 and MERS-CoV are characterized by the presence of identical DPP4 regions that are critical for the binding to the S protein [47]. At the same time, P. Zhou et al. [17] in an in vitro study found that SARS-CoV-2 does not use DPP4 as a receptor. The same authors did not confirm the significance of aminopeptidase N (ANPEP, also known as APN and CD13) as a SARS-CoV-2 receptor. Interestingly, ANPEP is a receptor for HCoV-229E, but does not have such properties for HCoV-OC43 [26]. However, ANPEP and DPP4 are characterized by expression patterns similar to those of ACE2 in 13 tissues, which allowed F. Qi et al. [61] to consider them as potential receptors for SARS-CoV-2. Further studies are needed to resolve the controversy regarding the receptor properties of DPP4 and clarify the possible receptor role of ANPEP for SARS-CoV-2.
After SARS-CoV-2 attaches to the ACE2 receptor, proteases, including TMPRSS2 and FURIN, are required to ensure the entry of the coronavirus into the cell [29, 37, 39]. D. Bestle et al. [51] found that, for SARS-CoV-2 to enter the cells, its S protein must be cleaved at two different sites by host cell proteases: by TMPRSS2 at the S2 site and by FURIN at the S1/S2 site.
The TMPRSS2 transmembrane protease and related proteases (such as TMPRSS11D) promote the entry of the coronavirus into the cell by two mechanisms: by cleaving the ACE2 receptor (which can stimulate the coronavirus uptake) and by cleaving the S protein of the coronavirus (which leads to the protein activation and fusion of the viral envelope with the membrane of the host organism cell) [35, 39]. The study by S. Bertram et al. [52] showed that TMPRSS2 activates cathepsin-independent entry of HCoV-229E into the host cell, and TMPRSS2 activation protects 229E-S-dependent entry into the cell from inhibition by interferon-induced transmembrane proteins. In model animals, it was found that the deficiency of this protease in the cells of the respiratory tract reduces the severity of SARS-CoV and MERS-CoV, which indicates the importance of TMPRSS2 in the infection process [62].
The entry of SARS-CoV-2 into the cell is also activated by lysosomal cathepsins [29]. Unlike TMPRSS2, cathepsins promote the entry of coronaviruses into the cell through endocytosis (lysosomal pathway); the choice of a protease for the entry is determined by the structural features of the S protein of the viruses [33]. The TMPRSS2 protease and lysosomal cathepsins have a cumulative effect with furin on the activation of the SARS-CoV-2 entry into the cells of the host organism [29].
Interferon-induced transmembrane proteins (IFITM1, IFITM2, IFITM3) are able to restrict replication of coronaviruses and their entry into cells and inactivate new coronaviruses when they are released from infected cells (shown for SARS-CoV and other viruses such as influenza A virus, Dengue virus, and West Nile virus) [57–59, 63]. Since IFITM proteins are induced by type I and II interferons, they are believed to be crucial for the antiviral action of interferon [63]. In the study by Y. Zhang et al. [12], it was found that the structural features of the IFITM3 gene are associated with the severity of the course of COVID-19.
Thus, most of the genes included in Table 1 are categorized as those having proven or potential significance in terms of the involvement of their protein products in infection of the cells with SARS-CoV-2. Five genes (TMPRSS11D, ANPEP, DDX1, IFITM1, IFITM2) can be considered as likely significant, since the products encoded by them are involved in the entry processes of other coronaviruses (SARS-CoV, HCoV-229E) and a number of other viruses into the cells of the host organism [35, 52, 56–58] (see Table 1). Further in the paper, all these genes will be designated as candidate genes for the susceptibility to SARS-CoV-2.
Among the genes involved in the consideration, only ACE2 is localized on the X chromosome, which may act as a possible cause of gender differences in the susceptibility to SARS-CoV-2 [64–66]; other genes are localized on eight different chromosomes (Table 1).
The candidate genes for the susceptibility to SARS-CoV-2 are expressed in many organs/tissues, including those sensitive to this coronavirus (according to clinical observations [7–9]). However, there are differences in the level of their expression in different organs (Fig. 1). On the basis of the expression level of the candidate genes for the susceptibility to SARS-CoV-2, tissues are grouped into several clusters (Fig. 1). One cluster combines tissues of the lungs, aorta, coronary artery, thyroid gland, and visceral and subcutaneous adipose tissues; the second cluster unites tissues of other organs involved in the comparison; the greatest differences in the expression level were observed in blood cells.
Fig. 1.
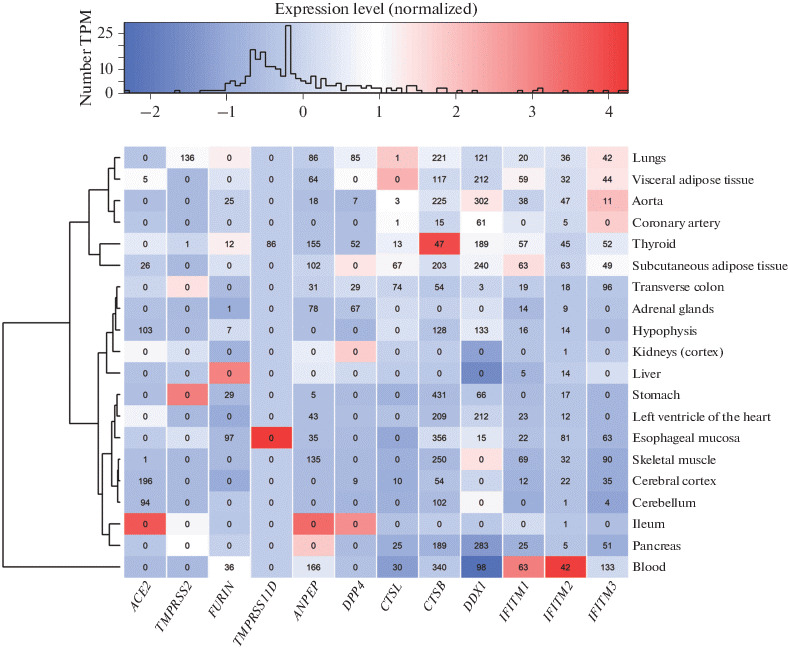
The heat map reflecting the expression level of the candidate genes for the susceptibility to SARS-CoV-2 in various human organs/tissues (constructed from median values of the number of transcripts per million (TPM) according to [45]). The expression level is normalized according to the genes for which zero corresponds to the average level of gene expression. The values in the wells reflect the number of statistically significant eQTLs affecting the level of gene expression in the respective tissues.
No differences in the expression level of the genes ACE2, TMPRSS2, CTSB, and CTSL between men and women were found [67]. Protein coexpression is an important condition for the entry into the cells and the spread of viruses in the human body [68], while organs and tissues where these genes are expressed can potentially be susceptible to coronavirus infection [69].
Individual differences in the susceptibility to SARS-CoV-2 may be affected by variants in the candidate genes that alter protein structure or expression. In addition, gene expression may depend on the methylation level of their promoter regions.
Genetic Variants Affecting Protein Structure
Nonsynonymous substitutions are recorded in all candidate genes for the susceptibility to SARS-CoV-2, but in most cases they are extremely rare, with low polymorphism, or are not recorded in all ethnoterritorial groups (Table 1; [44]). This may be associated with the functional significance of proteins and enzymes encoded by these genes, as well as their localization in the cell: all the considered proteins, except DDX1, are localized in the cell membrane [14]. At the same time, a large number of rare variants which are important for the entry of SARS-CoV-2 into the cells of the host organism and the development of infection were registered in the ACE2 gene, including S19P, I21T/V, E23K, A25T, K26R, T27A, E35D/K, E37K, Y50F, N51D/S, M62V, N64K, K68E, F72V, E75G, M82I, T92I, Q102P, G220S H239Q, G326E, E329G, G352V, D355N, H378R, Q388L, P389H, E467K, H505R, R514G/*, and Y515C; interracial differences in the frequency of variants were recorded for some of them (in bold) [67]. Since the ACE2 gene is localized on the X chromosome, the presence of such variants in men in the hemizygous state may act as an unfavorable factor for SARS-CoV-2 infection. In the candidate genes for the susceptibility to SARS-CoV-2, a large number of variants leading to amino acid substitutions or loss of protein function were identified with the detection rate of less than 1% (see Table 1). It is possible that just such rare variants both in the ACE2 gene [67] and in other considered genes may act as genetic factors that determine individual susceptibility to SARS-CoV-2.
The highly polymorphic missense variants in the candidate genes for the susceptibility to SARS-CoV-2 in all territorial groups characterized within the 1000 Genomes Project (Phase 3) include rs12329760 (p.Val160Met) of the TMPRSS2 gene; rs25653 (p.Arg86Gln), rs8192297 (p.Ile603Met), and rs25651 (p.Ser752Asn) of the ANPEP gene; rs12338 (p.Leu26Met, p.Leu26Val) of the CTSB gene; and rs1059091 (p.Ile121Leu) of the IFITM2 gene (Table 1). Moreover, rs12329760 of the TMPRSS2 gene, rs12338 of the CTSB gene, and rs1059091 of the IFITM2 gene are classified as potentially pathogenic according to the programs SIFT, PolyPhen, Mutation Assessor, and MetaLR, built into Ensemble [44].
The frequency of allele registration for most missense variants differs by 2 or more times between the studied ethnoterritorial groups (Table 1). SNPs, which significantly differ in the level of polymorphism between regions of the world (in some cases, the frequency of rare alleles varies from 0 to 42%), are also of interest for consideration. Thus, according to the 1000 Genomes Project [44], the A allele for rs75603675 (p.Gly8Asp) of the TMPRSS2 gene is recorded with the frequency of 2% in populations of East Asia, 22% in populations of South Asia, 0% in Africa, 27% in America, and 40% in European populations; the frequency of the T allele for rs1058900 (p.Val33Ala) of the IFITM2 gene is 0, 7, 4, 20, and 42%, respectively; the frequency of the T allele for rs14408 (p.Met41Arg) of the IFITM2 gene is 4, 27, 24, 33 and 60%, respectively.
Missense substitutions in the functionally significant region of the protein molecule (as, for example, in the case of rs12329760 of the TMPRSS2 gene, the p.Val160Met substitution is in the functional SRCR domain of the protein (mediates protein-protein interactions and ligand binding) [14]) can played an important role from the point of view of the formation of the susceptibility to coronaviruses. In the ANPEP gene, variants including those leading to nonsynonymous substitutions were identified in the region critical for binding to HCoV-229E (amino acids 260–353) [14, 50]. Such substitutions can affect the properties of proteins and, consequently, the differences in the degree of the susceptibility to coronavirus infection of the owners of different genotypes for these variants.
Among the polymorphic variants localized in exons of the candidate genes for the susceptibility to SARS-CoV-2 involved in the consideration, pathogenetic significance in relation to the risk of developing infectious diseases was previously established only for one SNP, rs12252 of the IFITM3 gene (OMIM: 605579, predisposition to severe course of influenza) [70, 71]. An association with a higher risk of severe H1N1/09 influenza was recorded for individuals with the C allele for rs12252 [71]. The authors of the cited study showed in vitro that cell lines with the CC genotype were characterized by a lower level of IFITM3 expression and a higher susceptibility to infection than lines with the TT genotype for this SNP. Despite the fact that the rs12252 polymorphic variant leads to the synonymous substitution (p.Ser14Ser), for some transcripts, this substitution is located in the region of splicing or 5'UTR [44]. Cells expressing the protein shortened by 22 amino acids are unable to restrict viral replication as compared with the wild-type IFITM3 protein [71]. The frequency of the unfavorable C allele varies from 4% in European populations to 53% in South Asian populations [44].
It was shown that carriers of the CC rs12252 genotype of the IFITM3 gene also have a risk of a more severe course of COVID-19 (the proportion of carriers of this genotype among patients with the average severity of the disease is 28.57%, with the severe course is 50%, and among the dead is 66.7%) [12]. Despite the fact that this study was performed on a small sample, taking into account the previously obtained data on the association of rs12252 with the severe course of H1N1/09 influenza [70, 71], on the lightning-fast development of viral pneumonia with the severe clinical picture in mice with knockout of the Ifitm3 gene under infection with low pathogenicity influenza virus [71], on the inability of CC homozygotes to limit replication of influenza H1N1/09 virus in the case of synthesis of truncated IFITM3 protein [71], and on the critical role of the 21 N-terminal amino acids of IFITM3 (as well as the C-terminal transmembrane region of the protein) for antiviral activity against vesicular stomatitis virus in vitro [72], it can be assumed that this variant has a high predictive value in assessing the risk of developing COVID-19 and for determining the nature of the course of infection caused by SARS-CoV-19.
Genetic Variants with Regulatory Potential
Numerous SNPs, including variants that can affect the level of gene expression, are recorded in the candidate genes for the susceptibility to SARS-CoV-2. eQTLs, the number and spectrum of which are different for different organs and tissues, are known for the candidate genes for the susceptibility to coronavirus infection (see Table 1, Fig. 1; see also [45]). For example, a higher expression level of the TMPRSS2 gene in the lungs is typical of the owners of the AA genotype for rs35074065, the TT genotype for rs34783969, the AA genotype for rs463727, etc., compared with other genotypes. In individuals with the AA genotype for rs1622599, a higher expression level of the CTSB gene was observed in lung tissues, blood cells, pancreas, and tibial artery, but a lower level was in the mucous membrane of the esophagus and thyroid gland [45].
The eQTL category for the TMPRSS2 gene includes rs2070788 and rs383510, whose GG and TT genotypes, respectively, are characterized by a higher level of this gene expression (including in the lung tissue) [45]. The same genotypes are associated with the susceptibility to influenza A (H7N9) virus and with the risk of severe influenza A (H1N1) [73]. Differences in the frequency of unfavorable alleles and, consequently, genotypes of the eQTL series are recorded between territorial groups of the population [44]. For example, the unfavorable G allele for rs2070788 of the TMPRSS2 gene occurred with the frequency of 27% in African populations, 36% in East Asian populations, and more than 46% in European, South Asian, and American populations.
Interestingly, the TMPRSS2 gene is located in close proximity to the genes MX1 and MX2, which encode two interferon-induced dynamin-like GTPases. MX1 has antiviral activity against a wide range of RNA viruses and some DNA viruses; MX2 is characterized by a strong antiviral effect against human immunodeficiency virus type 1. Markers rs2070788 and rs383510 are also eQTLs for the MX1 gene [14, 45, 74].
The A allele for rs17514846 located in the intron of the FURIN gene [75–77], associated with diseases of the cardiovascular system (hypertension, ischemic heart disease, atherosclerosis of the coronary arteries), leads to a higher transcriptional activity of this gene in the endothelial cells of the vessels than the alternative allele [45, 78]. These data are in good agreement with clinical observations according to which a higher susceptibility to SARS-CoV-2 infection and a more severe course of COVID-19 are observed in people with diseases of the cardiovascular system [79]. The frequency of registration of the unfavorable A allele for rs17514846 varies from 16% in populations of East Asia to 88% in African populations; in European populations, the frequency of this allele is 46% [44].
The genotypes rs3118869 and rs2378757 affect the expression level of the CTSL gene [45, 80]. For the first specified eQTL, the dependence of the expression level on genotypes differed in organs/tissues (a higher level of CTSL expression was recorded in carriers of the C allele—in the skin, tibial nerve, and colon; in carriers of homozygous AA and CC genotypes—in subcutaneous adipose tissue and tibial artery; in carriers of the heterozygous genotype—in the blood). N. Mbewe-Campbell et al. [80] showed that the CC genotype of rs3118869, which is located in the CTSL gene promoter, is associated with arterial hypertension. It is possible that patients with arterial hypertension and the CC rs3118869 genotype of the CTSL gene are more susceptible to SARS-CoV-2 and damage to those organs, where the carriage of the CC genotype leads to a higher level of expression of this gene. For rs2378757, in 11 organs/tissues (including the lungs, aorta, subcutaneous adipose tissue), the level of CTSL gene expression decreased depending on the number of the A alleles: AA > AC > CC.
The specificity of expression profiles of the candidate genes for the susceptibility to SARS-CoV-2 (including because of the genetic characteristics of individuals) can determine interpopulation differences in the spread of COVID-19. In particular, some researchers associate different clinical scenarios of the course of COVID-19 in the regions of the world (in particular, in China and Italy) with the peculiarities of the expression level of the FURIN gene (one of the key genes; its product is involved in the process of the coronavirus entry into the host cell) [81]. For the FURIN gene, about 300 eQTLs are known (Table 1), but their number (Fig. 1) and spectrum differ in different organs and tissues [45].
The given examples of functionally significant polymorphic variants do not reflect their entire diversity. A. Paniri et al. [82], using in silico analysis of predicting the functional significance of the TMPRSS2 gene variability, identified 21 SNPs that affect the function and structure of the protein encoded by it: folding, posttranslational modification, splicing, and the regulatory potential of microRNA. In particular, rs12329760 changes the shape of the protein in such a way that a pocket of the protein is formed de novo; rs875393 creates a new donor site and sites in silencers and destroys three sites in the enhancer motif. A number of SNPs (rs12627374, rs456142, rs462574, rs456298) lead to the destruction/creation of binding sites and weakening or enhancement of the effects of various microRNAs (hsa-miR-548c-3p, hsa-miR-127-3p, hsa-miR1324, hsa-miR-5089, hsa-miR-204-5p, hsa-miR-211-5p, hsa-miR-4685-3p, hsa-miR-4716-5p). All these structural and functional features of the TMPRSS2 gene, like other candidate genes for the susceptibility to SARS-CoV-2, may underlie individual differences in the susceptibility to this infectious disease and determine the nature of its course.
Factors Affecting Expression of Candidate Genes for the Susceptibility to SARS-CoV-2
Clinical observations show that the presence of pathological conditions (diseases of the cardiovascular system and kidneys, endocrine disorders) and old age act as risk factors for the development, as well as the severe course of COVID-19 [8, 64, 83, 84]. In some diseases and with increasing age, changes in the expression level of the candidate genes for the susceptibility to SARS-CoV-2, including because of epigenetic modifications, may be recorded.
Expression of the ACE2 and TMPRSS2 genes in the sustentacular (accessory) cells of the olfactory epithelium increases with age (shown in the model objects) [85]. Older people have higher DPP4 activity [86] and CTSB level (CTSL level did not change) in various tissues and organs [87]. These observations may explain a higher susceptibility to COVID-19 in elderly people [88].
In many diseases, a change in the expression level of the candidate genes for the susceptibility to SARS-CoV-2 is recorded. In patients with idiopathic dilated cardiomyopathy and ischemic cardiomyopathy, expression of the ACE2 gene in the ventricular myocardium is increased [66]. At the same time, a higher risk of developing COVID-19 and complications from the cardiovascular system is shown in persons suffering from diseases of this system [83, 84, 88].
It should be noted that the level of ACE2 expression in scrapings of epithelial cells of the nose and bronchi in patients with bronchial asthma and respiratory allergies was reduced and showed an inverse relationship to the severity of the clinical picture (ACE2 expression was the lowest in patients with a high level of allergic sensitization and with allergic asthma), but nonatopic asthma was not associated with decreased expression of this gene [89]. The authors note that these data may explain such an unexpected clinical observation as a milder course of COVID-19 in patients with respiratory allergies and at the same time emphasize that ACE2 expression is one of the factors that may influence the response of these individuals to SARS-CoV-2 infection. Indeed, an increase in expression of the DPP4 and ANPEP genes was observed in the bronchi in allergic asthma [90], which may also affect the susceptibility to SARS-CoV-2. In addition, expression of all the candidate genes for the susceptibility to SARS-CoV-2 involved in the consideration is recorded in the lungs, and eQTLs affecting the expression level in the lungs were established for many genes (Fig. 1). Perhaps, the inconsistency of the results in relation to the effect of asthma on the risk of development and the nature of the course of COVID-19 is associated with the genetic characteristics of territorial population groups. Thus, Z. Zhu et al. [91] note that adults with asthma have a higher risk of severe COVID-19, and an increased risk was observed precisely among patients with nonallergic asthma.
An increase in the expression level of the DPP4 gene was registered in the alveolar epithelium and alveolar macrophages in patients with chronic obstructive pulmonary disease (COPD), with cystic fibrosis [92], with insulin resistance (shown in model animals and in cell culture) [93], and under hypoxic conditions [47]. An increased level of the DPP4 protein is considered as a predictor for developing metabolic syndrome [86]. In persons with type 2 diabetes mellitus (T2DM), a higher level and activity of DPP4 was observed in the plasma compared with healthy individuals; an increased level of this protein (but not activity) was also recorded in patients with T2DM and obesity compared with T2DM without obesity [94].
Drugs can have modifying effects on the regulation of the work of the candidate genes for the susceptibility to SARS-CoV-2 and the synthesis of proteins encoded by them. Thus, the expression level of the TMPRSS11D gene in lung biopsies of patients with COPD depended on the drug administration (fluticasone and salmeterol, a combination of inhaled steroids and long-acting beta-agonists) [95]. There were no differences in the expression level of the ACE2 and TMPRSS2 genes in the ileum and colon between patients with inflammatory bowel diseases and healthy individuals, but drug therapy in patients (in particular, administration of tumor necrosis factor blockers) led to a decrease in the expression level of ACE2 [96].
The level of expression of the candidate genes for the susceptibility to SARS-CoV-2 can be modified by the presence of other viruses in the body, and the direction of the change in expression depends on the infectious agent. Somewhat unexpectedly, some viruses (in particular, H5N1 avian influenza virus) increase expression of the IFITM1, IFITM2, and IFITM3 genes [97], which have antiviral effect [57–59, 63]. At the same time, human papillomaviruses of high oncogenic risk (hrHPV) suppress expression of the IFITM1 gene in the keratinocytes as early as 48 h after infection [98]. It is possible that a change in the expression level under the influence of viruses depends not only on the infectious agent but also on the duration of the infectious process, as well as on the genetic characteristics of the organism and its functional state at the time of infection.
Other exogenous factors can also act as modifiers of the functioning of the candidate genes for the susceptibility to SARS-CoV-2. In the lungs of smokers, an increase in expression of the ACE2 and FURIN genes was found, but smoking had no effect on the expression level of the TMRPSS2 gene [99]. Smoking is associated with increased expression of the ACE2 gene in adipose tissue [100]. Some researchers suggest that adipose tissue in obese individuals may act as a source for a wider spread of viruses, their increased release, immune activation, and amplification of cytokines [64].
The effect of smoking on the methylation level of the cg23161492 site located in intron 1 of the ANPEP gene (smoking reduces the level of DNA methylation) in blood leukocytes was shown in a number of studies [101–104]. A change in the methylation level of this CpG site affects expression of the ANPEP gene in lung tissue [105]. Interestingly, six genetic variants (rs28565347, rs12442778, rs12440213, rs8030857, rs8031576, and rs748508), which can affect the methylation pattern of CpG sites (mQTLs) in blood leukocytes, were identified for cg23161492 [106]. The methylation level of another site cg05312779, localized in 3'UTR of the ANPEP gene, is statistically significantly associated with the lung function; in particular, an increase in DNA methylation was associated with a decrease in the Tiffeneau index in adolescent girls [107]. These findings are consistent with clinical observations that smokers have a higher risk of developing COVID-19 symptoms when infected with SARS-CoV-2 [100]. However, assessments of the impact of smoking on the risk of developing COVID-19 are ambiguous (see, for example, [65]), which once again emphasizes the complexity of regulating the response of the host organism to infection with SARS-CoV-2.
Disturbance of the level of DNA methylation in other genes is also considered as a factor modulating the response to SARS-CoV-2 infection [108]. It is assumed that hypomethylation of the regulatory regions of the ACE2 gene in CD4+ T cells in patients with systemic lupus erythematosus, owing to oxidative stress caused by viral infection, can lead to an increase in expression of the ACE2 gene [108]. Hypomethylation of CpG sites and a high level of expression of the ACE2 gene are typical of many cancers [109], and this may explain the high risk of complications in patients with COVID-19 and such comorbidities [83].
Thus, despite the short period of research, to date, data indicating that the susceptibility and nature of the course of infection caused by SARS-CoV-2 may be determined by the genetic characteristics of the coronavirus and the human body have been accumulated. The structural and functional features of the SARS-CoV-2 S protein and human genes encoding receptors, proteases, and other proteins and enzymes that are important for the entry and replication of the coronavirus, as well as factors modifying the functioning of these genes (including the presence of concomitant diseases, harmful habits, infection with other viruses, drug administration, etc.), can determine the susceptibility and clinical picture of COVID-19 at the individual level and the features of the epidemiological situation at the population level.
COMPLIANCE WITH ETHICAL STANDARDS
Conflict of interest. The authors declare that they have no conflict of interest.
Statement on the welfare of animals. This article does not contain any studies involving animals performed by any of the authors.
Statement of compliance with standards of research involving humans as subjects. This article does not contain any studies involving humans as subjects of research.
Footnotes
Translated by D. Novikova
REFERENCES
- 1.WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/. Accessed June 23, 2020.
- 2.Malik Y.A. Properties of coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020;42:3–11. [PubMed] [Google Scholar]
- 3.de Wit E., van Doremalen N., Falzarano D. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J., Zheng X., Tong Q. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J. Med. Virol. 2020;92:491–494. doi: 10.1002/jmv.25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tu Y.F., Chien C.S., Yarmishyn A.A. Review of SARS-CoV-2 and the ongoing clinical trials. Int. J. Mol. Sci. 2020;21:E2657. doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmermann P., Curtis N. Coronavirus infections in children including COVID-19: an overview of the epidemiology, clinical features, diagnosis, treatment and prevention options in children. Pediatr. Infect. Dis. J. 2020;39:355–368. doi: 10.1097/INF.0000000000002660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park S.E. Epidemiology, virology, and clinical features of severe acute respiratory syndrome—coronavirus-2 (SARS-CoV-2; Coronavirus Disease-19) Clin. Exp. Pediatr. 2020;63:119–124. doi: 10.3345/cep.2020.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goyal P., Choi J.J., Pinheiro L.C. Clinical characteristics of Covid-19 in New York City. N. Engl. J. Med. 2020;382:2372–2374. doi: 10.1056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu K., Chen Y., Lin R., Han K. Clinical features of COVID-19 in elderly patients: a comparison with young and middle-aged patients. J. Infect. 2020;80:e14–e18. doi: 10.1016/j.jinf.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W., Zhang C., Sui J. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X., Geng M., Peng Y. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020;10:102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, Y., Qin, L., Zhao, Y., et al., Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with disease severity in COVID-19, J. Infect. Dis., 2020, pii: jiaa224. 10.1093/infdis/jiaa224 [DOI] [PMC free article] [PubMed]
- 13.Clohisey S., Baillie J.K. Host susceptibility to severe influenza A virus infection. Crit. Care. 2019;23:303. doi: 10.1186/s13054-019-2566-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.QuickGO. https://www.ebi.ac.uk/QuickGO/. Accessed June, 2020.
- 16.Chen Y., Guo Y., Pan Y., Zhao Z.J. Structure analysis of the receptor binding of 2019-nCoV. Biochem. Biophys. Res. Commun. 2020;525:135–140. doi: 10.1016/j.bbrc.2020.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou P., Yang X.-L., Wang X.-G. Pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu R., Zhao X., Li J. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins Wenjie and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saha P., Banerjee A.K., Tripathi P.P. A virus that has gone viral: amino-acid mutation in S protein of Indian-isolate of Coronavirus COVID-19 might impact receptor-binding, thus infectivity. Biosci. Rep. 2020;40:BSR20201312. doi: 10.1042/BSR20201312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin, C., Genotyping coronavirus SARS-CoV-2: methods and implications, Genomics, 2020. pii: S0888-7543(20)30318-9. 10.1016/j.ygeno.2020.04.016 [DOI] [PMC free article] [PubMed]
- 21.Kandeel M., Ibrahim A., Fayez M., Al-Nazawi M. From SARS and MERS CoVs to SARS-CoV-2: moving toward more biased codon usage in viral structural and nonstructural genes. J. Med. Virol. 2020;92:660–666. doi: 10.1002/jmv.25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pillay T.S. Gene of the month: the 2019-nCoV/SARS-CoV-2 novel coronavirus spike protein. J. Clin. Pathol. 2020;73:366–369. doi: 10.1136/jclinpath-2020-206658. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Y., Yu D., Yan H. Design of potent membrane fusion inhibitors against SARS-CoV-2, an emerging coronavirus with high fusogenic activity. J. Virol. 2020;94:e00635–20. doi: 10.1128/JVI.00635-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofmann H., Pyrc K., van der Hoek L. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walls, A.C., Park, Y.J., Tortorici, M.A., et al., Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein, Cell, 2020, vol. 181, no. 2, pp. 281—292. e6. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed]
- 26.Yeager C.L., Ashmun R.A., Williams R.K. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Y., Zhang, Z., Yang, L., et al., The MERS-CoV receptor DPP4 as a candidate binding target of the SARS-CoV-2 spike, iScience, 2020, vol. 23, no. 6, p. 101160. 10.1016/j.isci.2020.101160 [DOI] [PMC free article] [PubMed]
- 28.Wang, Q., Zhang, Y., Wu, L., et al., Structural and functional basis of SARS-CoV-2 entry by using human ACE2, Cell, 2020, vol. 181, no. 4, pp. 894—904. e9. 10.1016/j.cell.2020.03.045 [DOI] [PMC free article] [PubMed]
- 29.Shang J., Wan Y., Luo C. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia S., Liu M., Wang C. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang T.J., Yang D.M., Wang M.L. Genomic analysis and comparative multiple sequence of SARS-CoV2. J. Chin. Med. Assoc. 2020;83:537–543. doi: 10.1097/JCMA.0000000000000335. [DOI] [PubMed] [Google Scholar]
- 32.Garten W., Braden C., Arendt A. Influenza virus activating host proteases: identification, localization and inhibitors as potential therapeutics. Eur. J. Cell. Biol. 2015;94:375–383. doi: 10.1016/j.ejcb.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 33.Shirato K., Kanou K., Kawase M., Matsuyama S. Clinical isolates of human coronavirus 229E bypass the endosome for cell entry. J. Virol. 2016;91:e01387–16. doi: 10.1128/JVI.01387-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shulla A., Heald-Sargent T., Subramanya G. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heurich A., Hofmann-Winkler H., Gierer S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014;88:1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinke L.M., Spiegel M., Plegge T. Different residues in the SARS-CoV spike protein determine cleavage and activation by the host cell protease TMPRSS2. PLoS One. 2017;12:e0179177. doi: 10.1371/journal.pone.0179177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coutard B., Valle C., de Lamballerie X. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabaan A.A., Al-Ahmed S.H., Haque S. SARS-CoV-2, SARS-CoV, and MERS-CoV: a comparative overview. Infez. Med. 2020;28:174–184. [PubMed] [Google Scholar]
- 39.Hoffmann M., Kleine-Weber H., Schroeder S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams, F.M.K., Freydin, M., Mangino, M., et al., Self-reported symptoms of covid-19 including symptoms most predictive of SARS-CoV-2 infection, are heritable, medRxiv, 2020. https://www.medrxiv.org/. 10.1101/2020.04.22.20072124 [DOI] [PubMed]
- 41.Zietz, M. and Tatonetti, N.P., Testing the association between blood type and COVID-19 infection, intubation, and death, medRxiv, 2020. https://www.medrxiv.org/. 10.1101/2020.04.08.20058073 [DOI] [PMC free article] [PubMed]
- 42.Zaidi F.Z., Zaidi A.R.Z., Abdullah S.M., Zaidi S.Z.A. COVID-19 and the ABO blood group connection. Transfus. Apheresis Sci. 2020;59:102838. doi: 10.1016/j.transci.2020.102838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Genome Aggregation Database (gnomAD). https://gnomad.broadinstitute.org/. Accessed June, 2020.
- 44.Ensemble Genome Browser 88. https://www.ensembl.org/. Accessed May, 2020.
- 45.Genotype—Tissue Expression (GTEx) project. https://www.gtexportal.org/home/. Accessed May, 2020.
- 46.GTEx Consortium, The Genotype—Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y., Yang L., Dong L. Crosstalk between the Akt/mTORC1 and NF-κB signaling pathways promotes hypoxia-induced pulmonary hypertension by increasing DPP4 expression in PASMCs. Acta Pharmacol. Sin. 2019;40:1322–1333. doi: 10.1038/s41401-019-0272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vankadari N., Wilce J.A. Emerging COVID-19 coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg. Microbes Infect. 2020;9:601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raj V.S., Mou H., Smits S.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kolb A.F., Maile J., Heister A., Siddell S.G. Characterization of functional domains in the human coronavirus HCV 229E receptor. J. Gen. Virol. 1996;77:2515–2521. doi: 10.1099/0022-1317-77-10-2515. [DOI] [PubMed] [Google Scholar]
- 51.Bestle, D., Heindl, M.R., Limburg, H., et al., TMPRSS2 and furin are both essential for proteolytic activation and spread of SARS-CoV-2 in human airway epithelial cells and provide promising drug targets (preliminary report), bioRxiv, 2020. https://www.biorxiv.org/. 10.1101/2020.04.15.042085
- 52.Bertram S., Dijkman R., Habjan M. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J. Virol. 2013;87:6150–6160. doi: 10.1128/JVI.03372-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsuyama S., Nao N., Shirato K. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. U.S.A. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shirato K., Kawase M., Matsuyama S. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 2013;87:12552–12561. doi: 10.1128/JVI.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simmons G., Gosalia D.N., Rennekamp A.J. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu L., Khadijah S., Fang S. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J. Virol. 2010;84:8571–8583. doi: 10.1128/JVI.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang I.C., Bailey C.C., Weyer J.L. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7:e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang D., Weidner J.M., Qing M. Identification of five interferon-induced cellular proteins that inhibit west Nile virus and dengue virus infections. J. Virol. 2010;84:8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amini-Bavil-Olyaee S., Choi Y.J., Lee J.H. The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe. 2013;13:452–464. doi: 10.1016/j.chom.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wrapp D., Wang N., Corbett K.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020;526:135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iwata-Yoshikawa N., Okamura T., Shimizu Y. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J. Virol. 2019;93:e01815–18. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brass A.L., Huang I.C., Benita Y. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ryan P.M., Caplice N.M. Is adipose tissue a reservoir for viral spread, immune activation and cytokine amplification in COVID-19. Obesity. 2020;28:1191–1194. doi: 10.1002/oby.22843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gaibazzi, N., Tuttolomondo, D., Guidorossi, A., et al., Smoking prevalence is low in symptomatic patients admitted for COVID-19, medRxiv, 2020. https://www.medrxiv.org/. 10.1101/2020.05.05.20092015
- 66.Goulter A.B., Goddard M.J., Allen J.C., Clark K.L. ACE2 gene expression is up-regulated in the human failing heart. BMC Med. 2004;2:19. doi: 10.1186/1741-7015-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Darbani B. The expression and polymorphism of entry machinery for COVID-19 in human: juxtaposing population groups, gender, and different tissues. Int. J. Environ. Res. Public Health. 2020;17:3433. doi: 10.3390/ijerph17103433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bertram S., Heurich A., Lavender H. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS One. 2012;7:e35876. doi: 10.1371/journal.pone.0035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fu J., Zhou B., Zhang L. Expressions and significances of the angiotensin-converting enzyme 2 gene, the receptor of SARS-CoV-2 for COVID-19. Mol. Biol. Rep. 2020;47:4383–4392. doi: 10.1007/s11033-020-05478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Online Mendelian Inheritance in Man. http:www.omim.org/. Accessed June, 2020.
- 71.Everitt A.R., Clare S., Pertel T. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weidner J.M., Jiang D., Pan X.-B. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J. Virol. 2010;84:12646–12657. doi: 10.1128/JVI.01328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheng Z., Zhou J., To K.K. Identification of TMPRSS2 as a susceptibility gene for severe 2009 pandemic A (H1N1) influenza and A (H7N9) influenza. J. Infect. Dis. 2015;212:1214–1221. doi: 10.1093/infdis/jiv246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.EBI search. https://www.ebi.ac.uk/ebisearch/overview.ebi/about. Accessed 06.2020.
- 75.van der Harst P., Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of Coronary Artery Disease. Circ. Res. 2017;122:433–443. doi: 10.1161/circresaha.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nikpay M., Goel A., Won H.H. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takeuchi F., Akiyama M., Matoba N. Interethnic analyses of blood pressure loci in populations of East Asian and European descent. Nat. Commun. 2018;9:5052. doi: 10.1038/s41467-018-07345-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang X., Yang W., McVey D.G. FURIN expression in vascular endothelial cells is modulated by a coronary artery disease–associated genetic variant and influences monocyte transendothelial migration. J. Am. Heart Assoc. 2020;9:e014333. doi: 10.1161/JAHA.119.014333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li B., Yang J., Zhao F. Prevalence and impact of cardiovascular metabolic diseases on COVID-19 in China. Clin. Res. Cardiol. 2020;109:531–538. doi: 10.1007/s00392-020-01626-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mbewe-Campbell N., Wei Z., Zhang K. Genes and environment: novel, functional polymorphism in the human cathepsin L (CTSL1) promoter disrupts a xenobiotic response element (XRE) to alter transcription and blood pressure. J. Hypertens. 2012;30:1961–1969. doi: 10.1097/HJH.0b013e328356b86a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Afsar C.U. 2019-nCoV-SARS-CoV-2 (COVID-19) infection: cruciality of Furin and relevance with cancer. Med. Hypotheses. 2020;140:109770. doi: 10.1016/j.mehy.2020.109770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Paniri, A., Hosseini, M.M., and Akhavan-Niaki, H., First comprehensive computational analysis of functional consequences of TMPRSS2 SNPs in susceptibility to SARS-CoV-2 among different populations, J. Biomol. Struct. Dyn., 2020, pp. 1—18. 10.1080/07391102.2020.1767690 [DOI] [PMC free article] [PubMed]
- 83.Mason, K.E., McHale, P., and Pennington, A., Age-adjusted associations between comorbidity and outcomes of COVID-19: a review of the evidence, medRxiv, 2020. https://www.medrxiv.org/. 10.1101/2020.05.06.20093351 [DOI] [PMC free article] [PubMed]
- 84.Lippi G., Wong J., Henry B.M. Hypertension in patients with coronavirus disease 2019 (COVID-19): a pooled analysis. Pol. Arch. Int. Med. 2020;130:304–309. doi: 10.20452/pamw.15272. [DOI] [PubMed] [Google Scholar]
- 85.Bilinska K., Jakubowska P., von Bartheld C.S., Butowt R. Expression of the SARS-CoV-2 entry proteins, ACE2 and TMPRSS2, in cells of the olfactory epithelium: identification of cell types and trends with age. ACS Chem. Neurosci. 2020;11:1555–1562. doi: 10.1021/acschemneuro.0c00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bassendine M.F., Bridge S.H., McCaughan G.W., Gorrell M.D. Covid-19 and co-morbidities: a role for dipeptidyl peptidase 4 (DPP4) in disease severity? J. Diabetes. 2020;12:649–658. doi: 10.1111/1753-0407.13052. [DOI] [PubMed] [Google Scholar]
- 87.Kakimoto Y., Sasaki A., Niioka M. Myocardial cathepsin D is downregulated in sudden cardiac death. PLoS One. 2020;15:e0230375. doi: 10.1371/journal.pone.0230375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Siordia J.A. Epidemiology and clinical features of COVID-19: a review of current literature. J. Clin. Virol. 2020;127:104357. doi: 10.1016/j.jcv.2020.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jackson D.J., Busse W.W., Bacharier L.B. Association of respiratory allergy, asthma, and expression of the SARS-CoV-2 receptor ACE2. J. Allergy Clin. Immunol. 2020;146:203–206. doi: 10.1016/j.jaci.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van der Velden V.H., Wierenga-Wolf A.F., Adriaansen-Soeting P.W. Expression of aminopeptidase N and dipeptidyl peptidase IV in the healthy and asthmatic bronchus. Clin. Exp. Allergy. 1998;28:110–120. doi: 10.1046/j.1365-2222.1998.00198.x. [DOI] [PubMed] [Google Scholar]
- 91.Zhu, Z., Hasegawa, K., Ma, B., et al., Association of asthma and its genetic predisposition with the risk of severe COVID-19, J. Allergy Clin. Immunol., 2020, vol. 146, no. 2, pp. 327—329. e4. 10.1016/j.jaci.2020.06.001 [DOI] [PMC free article] [PubMed]
- 92.Meyerholz D.K., Lambertz A.M., McCray P.B. Dipeptidyl peptidase 4 distribution in the human respiratory tract: implications for the Middle East Respiratory Syndrome. Am. J. Pathol. 2016;186:78–86. doi: 10.1016/j.ajpath.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheng F., Yuan G., He J. Dysregulation of DPP4 is associated with the AMPK/JAK2/STAT3 pathway in adipocytes under insulin resistance status and liraglutide intervention. Diabetes Metab. Syndr. Obes. 2019;12:2635–2644. doi: 10.2147/DMSO.S229838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sarkar J., Nargis T., Tantia O. Increased plasma dipeptidyl peptidase-4 (DPP4) activity is an obesity-independent parameter for glycemic deregulation in type 2 diabetes patients. Front. Endocrinol. 2019;10:505. doi: 10.3389/fendo.2019.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van den Berge M., Steiling K., Timens W. Airway gene expression in COPD is dynamic with inhaled corticosteroid treatment and reflects biological pathways associated with disease activity. Thorax. 2014;69:14–23. doi: 10.1136/thoraxjnl-2012-202878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Burgueno J.F., Reich A., Hazime H. Expression of SARS-CoV-2 entry molecules ACE2 and TMPRSS2 in the gut of patients with IBD. Inflamm. Bowel. Dis. 2020;26:797–808. doi: 10.1093/ibd/izaa085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang H., Chen L., Luo J., He H. NP and NS1 proteins of H5N1 virus significantly upregulated IFITM1, IFITM2, and IFITM3 in A549 cells. Afr. Health Sci. 2019;19:1402–1410. doi: 10.4314/ahs.v19i1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ma W., Tummers B., van Esch E.M. Human papillomavirus downregulates the expression of IFITM1 and RIPK3 to escape from IFNγ- and TNFα-mediated antiproliferative effects and necroptosis. Front. Immunol. 2016;7:496. doi: 10.3389/fimmu.2016.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cai G., Bossé Y., Xiao F. Tobacco smoking increases the lung gene expression of ACE2, the receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020;201:1557–1559. doi: 10.1164/rccm.202003-0693LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hopkinson, N.S., Rossi, N., El-Sayed Moustafa, J., et al., Current tobacco smoking and risk from COVID-19: results from a population symptom app in over 2.4 million people, medRxiv. 2020. https://www.medrxiv.org/. 10.1101/2020.05.18.20105288
- 101.Zhang X., Hu Y., Aouizerat B.E. Machine learning selected smoking-associated DNA methylation signatures that predict HIV prognosis and mortality. Clin. Epigenet. 2018;10:155. doi: 10.1186/s13148-018-0591-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li S., Wong E.M., Bui M. Causal effect of smoking on DNA methylation in peripheral blood: a twin and family study. Clin. Epigenet. 2018;10:18. doi: 10.1186/s13148-018-0452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zeilinger S., Kühnel B., Klopp N. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One. 2013;8:e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sayols-Baixeras S., Lluís-Ganella C., Subirana I. Identification of a new locus and validation of previously reported loci showing differential methylation associated with smoking: the REGICOR study. Epigenetics. 2015;10:1156–1165. doi: 10.1080/15592294.2015.1115175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee M.K., Hong Y., Kim S.-Y. DNA methylation and smoking in Korean adults: epigenome-wide association study. Clin. Epigenet. 2016;8:103. doi: 10.1186/s13148-016-0266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gao X., Thomsen H., Zhang Y. The impact of methylation quantitative trait loci (mQTLs) on active smoking-related DNA methylation changes. Clin. Epigenet. 2017;9:87. doi: 10.1186/s13148-017-0387-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sunny S.K., Zhang H., Rezwan F.I. Changes of DNA methylation are associated with changes in lung function during adolescence. Respir. Res. 2020;21:80. doi: 10.1186/s12931-020-01342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sawalha A.H., Zhao M., Coit P., Lu Q. Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. Clin. Immunol. 2020;215:108410. doi: 10.1016/j.clim.2020.108410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chai P., Yu J., Ge S. Genetic alteration, RNA expression, and DNA methylation profiling of coronavirus disease 2019 (COVID-19) receptor ACE2 in malignancies: a pan-cancer analysis. J. Hematol. Oncol. 2020;13:43. doi: 10.1186/s13045-020-00883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


