Abstract
Plants are subjected to fluctuations in light intensity, and this might cause unbalanced photosynthetic electron fluxes and overproduction of reactive oxygen species (ROS). Electrons needed for ROS detoxification are drawn, at least partially, from the cellular glutathione (GSH) pool via the ascorbate–glutathione cycle. Here, we explore the dynamics of the chloroplastic glutathione redox potential (chl-EGSH) using high-temporal-resolution monitoring of Arabidopsis (Arabidopsis thaliana) lines expressing the reduction–oxidation sensitive green fluorescent protein 2 (roGFP2) in chloroplasts. This was carried out over several days under dynamic environmental conditions and in correlation with PSII operating efficiency. Peaks in chl-EGSH oxidation during dark-to-light and light-to-dark transitions were observed. Increasing light intensities triggered a binary oxidation response, with a threshold around the light saturating point, suggesting two regulated oxidative states of the chl-EGSH. These patterns were not affected in npq1 plants, which are impaired in non-photochemical quenching. Oscillations between the two oxidation states were observed under fluctuating light in WT and npq1 plants, but not in pgr5 plants, suggesting a role for PSI photoinhibition in regulating the chl-EGSH dynamics. Remarkably, pgr5 plants showed an increase in chl-EGSH oxidation during the nights following light stresses, linking daytime photoinhibition and nighttime GSH metabolism. This work provides a systematic view of the dynamics of the in vivo chloroplastic glutathione redox state during varying light conditions.
Monitoring the daily in vivo dynamics of the chloroplastic GSH redox state in light-stressed wild-type plants versus photoprotective mutants provides insight into the photosynthesis-dependent production of oxidants.
Introduction
As sessile organisms that grow under highly variable light intensities, plants must constantly sense, respond, and adapt to the instantaneous modulation of daytime photon fluxes. This regulation is critical when the absorption of light energy, and the resulting production of NADPH and ATP, exceed the capacity of downstream reactions. This may occur either during the dark-to-light transition, under high light (HL), or under fluctuating light (FL) intensities (Kono and Terashima, 2014; Tikhonov, 2015). Under these circumstances, overreduction of the photosynthetic electron transport chain can produce deleterious reactive oxygen species (ROS) in chloroplasts.
Several photoprotective mechanisms have evolved in plants to achieve homeostasis and optimal photosynthetic performance under HL and fluctuating environmental conditions, thereby protecting the photosynthetic machinery from light stresses. Among them are energy dissipation via heat, known as non-photochemical quenching (NPQ; Demmig-Adams, 1990; Horton and Ruban, 1992), cyclic electron flow (CEF; Arnon et al., 1954; Tagawa et al., 1963), and water–water cycle (WWC; Mehler, 1951). Plants also cope with ROS overproduction by generating low molecular weight antioxidants such as glutathione (GSH) and ascorbate as well as antioxidative enzymes such as ascorbate peroxidase (APX) and peroxiredoxins (PRX; Mittler et al., 2004; Takahashi and Badger, 2011; Kono and Terashima, 2014).
The rapid NPQ component, known as qE, is induced by increasing acidification of the thylakoid lumen and involves the activity of violaxanthin de-epoxidase (VDE), which catalyzes the conversion of violaxanthin to zeaxanthin (Niyogi et al., 1998). Analysis of the VDE null mutant (npq1) demonstrated that this mechanism increases plant tolerance to variation in light intensity by protecting photosystem II (PSII) under FL conditions in the field (Külheim et al., 2002).
CEF and WWC play a role in balancing the stromal ATP/NADPH ratio according to the requirements of primary metabolism and the need for protecting photosystem I (PSI) from photoinhibition (Avenson et al., 2005; Shikanai, 2007; Takahashi et al., 2009; Miyake, 2010). CEF and WWC thereby serve as electron sinks under excessive light conditions (Asada, 1999; Ort and Baker, 2002). In higher plants, two CEF pathways are known to exist: the PROTON GRADIENT REGULATION 5 (PGR5)-dependent pathway, which is believed to be the main pathway in C3 plants and the NADH dehydrogenase-like (NDH)-complex-dependent pathway (Shikanai, 2016). Although PGR5 is a key player in protecting PSI functionality by regulating CEF (Munekage et al., 2002; Suorsa et al., 2012; 2013), the mechanism for its action is not yet clear. When grown under HL or FL, the Arabidopsis thaliana (Arabidopsis) PGR5 null mutant (pgr5) shows stunted growth, impaired NPQ, a lower electron transport rate (ETR) relative to the wild type (WT) (Munekage et al., 2002) and also shows severe damage to PSI (Munekage et al., 2004; Suorsa et al., 2012). Therefore, exposure of pgr5 to increased light intensities has served as an experimental model for assessing the physiological effects of PSI photoinhibition (Tiwari et al., 2016).
During the WWC, electrons are donated from PSI to molecular oxygen in the Mehler reaction, yielding superoxide radicals (), which are then dismutated to molecular oxygen (O2) and H2O2 in a reaction catalyzed by superoxide dismutase (SOD; Mehler, 1951; Rizhsky et al., 2003). Subsequent reduction of H2O2 to water is mediated mainly by APXs and PRXs (Awad et al., 2015). APX relies on electrons from reduced glutathione (GSH), channeled to APX through the ascorbate–glutathione cycle (Foyer and Noctor, 2011). In addition, ascorbate can also be directly regenerated by ferredoxin or by NAD(P)H via MDA reductase activity (Asada, 1999). Thus, electrons for the detoxification of WWC-derived H2O2 are drawn, at least partially, from the cellular glutathione pool. Notably, the production of H2O2 through the WWC can play a regulatory role, allowing communication between the photosynthetic light-harvesting reactions and downstream metabolic processes by transmitting oxidative signals to redox-regulated proteins (Dangoor et al., 2012; Eliyahu et al., 2015).
In the past 15 years, redox-sensitive green fluorescent proteins (roGFPs) have allowed for in vivo monitoring of the glutathione redox potential (EGSH) at high spatiotemporal resolution (Meyer et al., 2007; Rosenwasser et al., 2010; Nietzel et al., 2019). In vitro characterization of roGFP showed that roGFP reduction and oxidation are mediated by glutaredoxins (GRXs), which catalyze the reversible electron flow between glutathione and target proteins (Meyer et al. 2007; Gutscher et al., 2008; Trnka et al., 2020). In addition, the redox status of roGFP was not significantly affected by thioredoxin (Trx) or other redox-active compounds such as NADPH and ascorbate (Meyer et al. 2007; Gutscher et al. 2008). The pronounced preference of roGFP for EGSH was further corroborated by using GSH biosynthesis inhibitors and Arabidopsis mutants compromised in GSH biosynthesis and reduction of glutathione disulfide (GSSG) (Meyer et al. 2007; Meyer and Dick, 2010; Marty et al., 2019). Importantly, recording of the roGFP fluorescence and its dynamics appears to reflect the transmission of redox signals, ultimately leading to modulation of the redox state of native target proteins of GRXs (Meyer, 2008; Rosenwasser, 2014b).
Due to their high sensitivity, reversibility, and insensitivity to pH alterations in the physiological range (Schwarzländer et al., 2008), roGFP-based redox sensors are powerful tools for investigating redox dynamics in subcellular compartments. Recently, roGFP-expressing plants and algae have been tested under various biotic and abiotic stresses and were shown to develop organelle-specific roGFP oxidation patterns (Rosenwasser et al., 2010; van Creveld et al., 2015; Bratt et al., 2016; Volpert et al., 2018; Mizrachi et al., 2019; Nietzel et al., 2019).
The balance between ROS production and their scavenging by the chloroplastic GSH pool was suggested to play a significant role in sensing and responding to light stress because a disruption of this balance by elevating either GSH or ROS levels results in oxidative stress (Foyer et al., 1997; Karpinski et al., 1997; Creissen et al., 1999; Mullineaux et al., 2018). However, the diurnal dynamics of the chloroplastic glutathione redox state in response to changing light conditions have not been systematically explored. Here, we examined diurnal fluctuations in chloroplastic EGSH under different light conditions by developing an automated system for monitoring the roGFP2 oxidation state throughout the day and night. We discovered unique patterns of chloroplast-specific EGSH under normal, HL, and FL conditions. Moreover, by examining these patterns in Arabidopsis lines that are mutated in key proteins involved in photoprotective mechanisms, we revealed their interaction with the chloroplast GSH redox state.
Results
The chloroplastic EGSH redox state is directly affected by photosynthetic electron transport
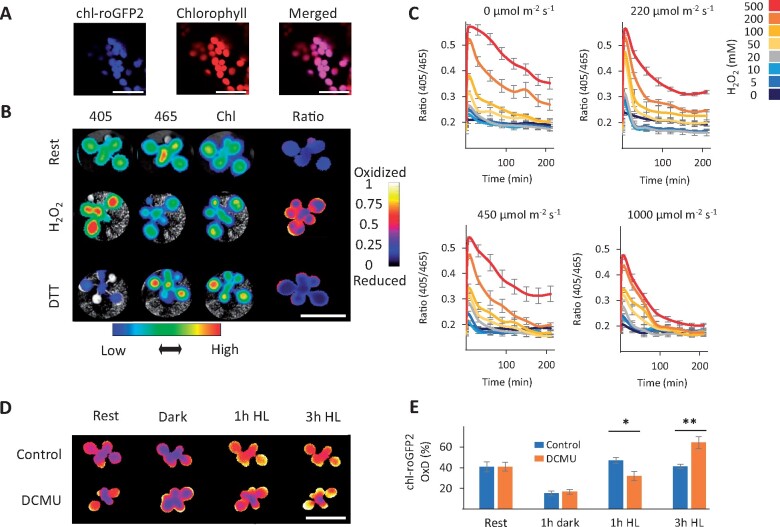
In order to map the in vivo temporal alterations in the chloroplastic EGSH (chl-EGSH) under “steady state” and light treatment conditions, we used a chloroplast-targeted redox-sensitive GFP 2 sensor (chl-roGFP2; Figure 1). Chloroplast targeting was achieved by using either the transketolase target peptide (chl-TKTP-roGFP2, Schwarzländer et al., 2008) or a 2-Cys PRX A target peptide (chl-PRXaTP-roGFP2, this study), as both target proteins to the chloroplast stroma (König et al., 2002; Schwarzländer et al., 2008). Chloroplast targeting was verified by the overlap of the chl-roGFP2 fluorescence signal with the chlorophyll fluorescence signal (Figure 1, A and Supplemental Figure S1). To monitor the response of the entire Arabidopsis shoot rosette to imposed redox changes, 3-week-old plantlets with ubiquitous expression of chl-roGFP2 were examined. Whole-plant in vivo fluorescence images showed that external application of H2O2 caused an increased emission after excitation at 405 nm fluorescence intensity and decreased when excited at 465 nm (Figure 1, B). Conversely, treatment of plants with DTT caused a less intense signal with 405 nm excitation and increased brightness with 465 nm excitation (Figure 1, B). Ratiometric analysis of the raw images (405/465 nm) resulted in informative false-color images showing the oxidation and reduction of the probe after application of H2O2 and DTT, respectively (Figure 1, B), as well as a positive correlation between the concentration of H2O2 applied and chl-roGFP2 oxidation (Supplemental Figure S2). These results validated the sensitivity of chl-roGFP2 to redox alterations and are consistent with probe performance, previously observed in plant and mammalian cells (Hanson et al., 2004; Dooley et al., 2004; Jiang et al., 2006; Meyer et al., 2007; Schwarzländer et al., 2008).
Figure 1.
Light-dependent redox modification of chloroplast-targeted roGFP2. A, Confocal images of chl-roGFP2 Arabidopsis plants. Bars = 20 µm. B, Ratiometric analysis of chl-roGFP2 during rest, under oxidized (500 mM H2O2) and reduced (100 mM DTT) conditions using whole-plant imaging. The plants were excited with 405 nm and 465 nm LEDs light sources and fluorescence emission was collected using a 510-nm emission filter. For chlorophyll detection, a 405-nm LED light source and a 670-nm emission filter were used. Color-coded keys represent fluorescence intensities (below image) or 405/465 ratio values (to right of image). Bars = 2 cm. C, The effect of light intensities on chl-roGFP2 reduction after application of H2O2 as examined by whole-plant imaging. Values represent means of six plants ± SE. D and E, The effect of DCMU on chl-roGFP2 oxidation using whole-plant imaging and plate-reader approaches. Oxidation of chl-roGFP2 was detected using whole-plant imaging. Bar = 2 cm. (D) or a plate reader (E). Values for the degree of chl-roGFP2 oxidation (OxD) are presented as means of seven to eight plants ± SE. Asterisks (*) mark significant differences between the ratio of the control/DCMU treatment (t test, P < 0.05, Supplemental File S1). The chl-roGFP2 data were acquired using the chl-TKTP-roGFP2 line.
To examine the dependence of the redox state of chl-roGFP2 on photosynthetic activity, we treated plants with various concentrations of H2O2 to induce oxidation, and we subsequently measured the recovery of chl-roGFP2 from oxidation under different light intensities. Regardless of the applied H2O2 concentration, the chl-roGFP2 fluorescence ratio immediately declined, resulting in distinct concentration-dependent peaks (Figure 1, C). Surprisingly, the decline was light-dependent and became faster and more efficient with increasing light intensities (Figure 1, C). Almost full reduction of chl-roGFP2 was only achieved when plants were illuminated with HL (1000 µE m−2 s−1). In this case, ratios for plants treated with 500 mM H2O2 approached resting levels with low fluorescence ratios after about 200 min (Figure 1, C). Although light-dependent chl-roGFP2 reduction was observed following H2O2 treatment, probe oxidation was observed in plants exposed to 1 h of HL conditions (700 µmol m−2 s−1), but not in plants pretreated with 150 µM 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), as determined by whole-plant imaging (Figure 1, D) and quantitative fluorometry (Figure 1, E). Compared with control plants, a higher level of oxidation was observed in DCMU-treated plants after 3 h under HL. Taken together, these results suggest that the chl-roGFP2 redox state is directly affected by photosynthetic activity.
Distinct patterns of chloroplastic EGSH during a diurnal cycle
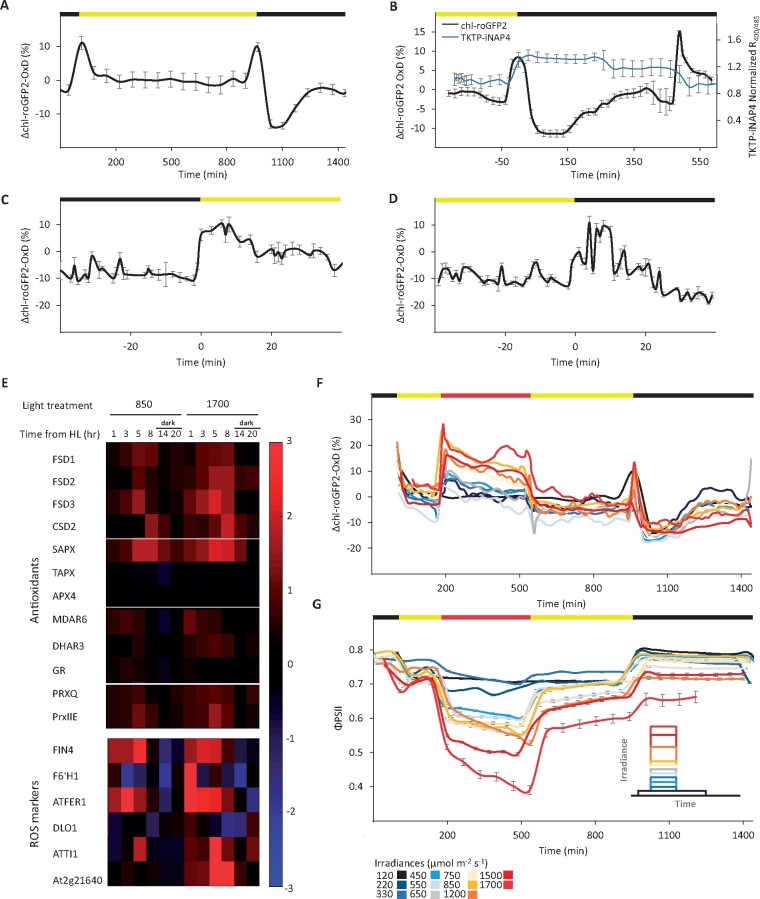
The observed relationship between photosynthesis activity and chl-roGFP2 redox state motivated us to systematically monitor its oxidation patterns under normal growth light and under changing light conditions. To this end, we developed an automated system that allows for continuous measurement of the degree of oxidation (OxD) of chl-roGFP2 and chlorophyll-fluorescence-derived photosynthetic parameters from many plants under dynamic environmental conditions (see the “Materials and methods” section). Examination of chl-roGFP2 oxidation under typical laboratory growth conditions (16-h light at 120 µmol m−2 s−1/8-h dark) showed a stable chl-roGFP2 state during the day of approximately OxD = 25%. Taking into account a stromal pH of 8, this OxD would correspond to an EGSH = −347 mV. These results are in agreement with previous calculations of chloroplastic EGSH under steady-state conditions (Schwarzländer et al., 2008; Rosenwasser et al., 2010). The average daytime OxD was then taken as a “steady-state” reference point and pronounced deviations in OxD that occurred during the transition from dark to light and vice versa were expressed as ΔOxD. A peak of oxidation with an increase in OxD of about 10% (ΔOxD = ∼10%) occurred during the dark–light transition (Figure 2, A).
Figure 2.
Diurnal changes in the degree of chl-roGFP2 oxidation under normal and HL growth conditions. chl-roGFP2 fluorescence was monitored over a 24-h period and chl-roGFP2 OxD values were calculated. Oxidation values were normalized to steady-state values (as observed during the day at a constant light intensity of 120 µmol m−2 s−1). Horizontal bars above graphs show light (yellow) and dark (black) periods. A, Changes in chl-roGFP2 OxD over time from the light onset under normal growth light conditions. Values are presented as means of eight plants ± SE. B, Changes in chl-roGFP2 OxD and normalized R400/485 of TKTP-iNAP4 over time during the nighttime. Results of TKTP-iNAP4 were normalized using the pH control sensor (TKTP-iNAPc) according to Lim et al. (2020) and values are presented as means of 12 plants ± SE. C, Oxidation and rereduction of chl-roGFP2 during dark-to-light transitions. Values are shown as means of 16–24 plants ± SE. D, Oxidation and rereduction of chl-roGFP2 during light-to-dark transitions, shown as means of 16–45 plants ± SE. E, Temporal changes in expression of chloroplast-targeted genes encoding antioxidant proteins and established ROS markers. Gene expression levels in plants exposed to HL (850 or 1700 µmol m−2 s−1) were normalized to the gene expression in plants grown under normal light conditions (120 µmol m−2 s−1) at each indicated time point after start of the HL illumination. All data are displayed as the log2 ratio. The red and blue colors represent high or low expression levels, respectively. F, Diurnal changes in chl-roGFP2 OxD in WT plants exposed to HL. For each treatment, between 21 and 24 plants, divided into three independent plates, were tested. For clear presentation of the redox changes of roGFP2 over time, a “sliding window” approach for the data along the x-axis was taken, in which each data point represents the average of three plates (n = 3). roGFP2 data were acquired using the chl-TKTP-roGFP2 line. For a depiction of absolute roGFP2 OxD values and illustration of SE values see Supplemental Figures S15, S16. G, Diurnal changes in chl-roGFP2 ΦPSII values in plants exposed to the same conditions as in (F). ΦPSII values were derived from chlorophyll fluorescence analysis and represent means of 12 plants ± SE. Inset: Experimental design for the HL experiments.
Surprisingly, a very similar peak of transient oxidation with the same amplitude occurred during the light–dark transition. In this case, however, OxD did not return to the steady-state level but rather became even more reduced as ΔOxD = −13%. This reduction was then followed by gradual oxidation, which after about 6 h stabilized near the steady-state daytime reference value (Figure 2, A). To better interpret chloroplast redox-metabolism-alterations during the nighttime, we estimated stromal NADPH availability using Arabidopsis plants expressing the recently developed NADPH sensor (TKTP-iNAP4; Tao et al., 2017; Lim et al., 2020). The observed increase in the TKTP-iNAP4 ratio (Figure 2, B) obtained by dividing the emissions at 510 nm after sequential excitation at 400 and 485 nm suggests an increase in NADPH levels at the beginning of the night, implying that the corresponding reduction of EGSH is governed by the higher reduction rate of the GSH pool by glutathione reductase (GR).
To further characterize the oxidation occurring during dark-to-light transitions, we collected data from 20 experiments, cumulatively involving approximately 2600 plants. This large data set allowed for a high-temporal resolution analysis of the chl-roGFP2 OxD values that surround the transitions points. Within the first minute of the light phase, we observed a rapid burst in chl-roGFP2 OxD levels that remained stable for 10–15 min, and this was followed by chl-roGFP2 reduction toward steady-state daytime values determined after about 20 min (Figure 2, C). Contrastingly, chl-roGFP2 oxidation in the light–dark transition occurred approximately 5 min after lights were switched off. This oxidation of ΔOxD = ∼10% was followed by a gradual reduction, reaching values that were detected during the day after 20 min (Figure 2, D). These results demonstrate significant oxidation of stromal EGSH during darkness–light transitions and imply the transmission of oxidative signals during the induction and termination phases of photosynthesis.
The chloroplastic EGSH dynamically responds to HL conditions
Next, we focused on investigating the dynamics of chl-EGSH under HL conditions. First, we assessed the induction of the WWC and the ascorbate–glutathione cycle under HL conditions by quantifying the level of expression of chloroplast-targeted genes involved in these pathways and several established marker genes for oxidative stress (Figure 2, E; the complete list of genes is described in Supplemental Data Set S1). Plants were exposed to 3 h of normal growth conditions (120 µmol m−2 s−1) at the beginning of the day, followed by a phase of different HL conditions at 850 or 1700 µmol m−2 s−1 for a 6-h period. After the HL phase, the light was dimmed again to the normal intensity for an additional 7-h period, followed by 8 h of darkness. Samples for RNA extraction were collected at different time points during the HL phase, after returning to normal growth conditions and during the subsequent dark phase. Control plants were exposed to normal growth conditions of 120 µmol m−2 s−1 for the entire 16-h light period.
We observed induction in four genes encoding for chloroplastic superoxide dismutase (FSD1, FSD2, FSD3, and CSD2) and the gene for stromal ascorbate peroxidase (sAPX) during the HL phase. Moderate induction was also observed for monodehydroascorbate reductase (MDAR6), dehydroascorbate reductase (DHAR3) and two PRXs (PRXQ and PrxIIE). No changes in the expression level of GR2 were observed. We also observed the induction of two H2O2-specific markers FER1 and FIN4 (op den Camp et al., 2003; Gadjev et al., 2006) as early as 1 h into the HL phase in both examined HL irradiances and the induction of two general oxidative stress markers, TI1 and At2g21640, mainly in response to 1700 µmol m−2 s−1. These results point to the induction of the WWC and ascorbate–glutathione cycle under the examined HL conditions.
We thus investigated chl-roGFP2 OxD patterns under HL conditions by exposing plants to various HL intensities during a 24-h cycle. Plants were exposed to 3 h of normal growth light conditions (120 µmol m−2 s−1) at the beginning of the day, after which the light irradiance was raised to various intensities (220, 330, 450, 550, 650, 750, 850, 1200, 1500, or 1700 µmol m−2 s−1) for a 6-h period. After the HL phase, the light was dimmed to the normal growth light conditions for an additional 7-h period, followed by 8 h of darkness (Figure 2, G inset). In addition to chl-roGFP2 oxidation (Figure 2, F), the PSII operating efficiency (ΦPSII) was continuously monitored based on chlorophyll fluorescence imaging (Figure 2, G).
We observed immediate chl-roGFP2 oxidation, followed by a gradual reduction, when plants were shifted from 120 µmol m−2 s−1 to any higher light intensity, indicating a rapid chl-EGSH response to unpredicted increases in the light input (Figure 2, F). No significant changes in the TKTP-iNAP4 ratio were detected upon shifting of plants from normal growth light conditions to HL of 850 µmol m−2 s−1, implying that higher ROS production rates dominated the increase in chl-roGFP2 oxidation (Supplemental Figure S3). Further reduction and stabilization of the chl-roGFP2 redox state were observed when plants were returned to normal growth light conditions after the HL period (Figure 2, F). Reduction at the onset of darkness, followed by gradual oxidation, as observed during normal growth conditions, was observed for all light treatments (Figure 2, F). Similar chl-roGFP2 diurnal patterns were observed when plants were exposed to similar light treatments under elevated CO2 levels (850 ppm), despite slightly higher reduction rates during the HL phase (Supplemental Figure S4). These light-induced chl-roGFP2 oxidation patterns were not observed in DCMU-treated plants, which exhibited gradual oxidation of the roGFP2 probe, further pointing to the direct regulation of roGFP2 dynamics by photosynthesis (Supplemental Figure S5).
To ensure that the observed HL responses were not affected by the availability of endogenous GRXs, which catalyze the reversible electron flow between GSH and roGFP2, we examined oxidation patterns in plants expressing chloroplast-targeted Grx1-roGFP2 (TKTP‐Grx1‐roGFP2). In this probe, the human glutaredoxin-1 (Grx1) is fused to roGFP2; hence, its equilibrium with the glutathione redox couple is not dependent on endogenous GRXs (Marty et al., 2009; Park et al., 2013). Similar diurnal oxidation patterns to those observed with chl-roGFP2 under HL were detected using TKTP Grx1-roGFP (Supplemental Figure S6). Furthermore, the examination of the expression level of seven genes coding for chloroplast-targeted GRXs under HL conditions showed that their expression levels were either unchanged or slightly upregulated (Supplemental Figure S7). These results suggest that both probes are suitable for monitoring the chloroplastic EGSH under dynamic light conditions.
Examination of the correlation between chl-roGFP2 oxidation and light irradiance revealed a binary response rather than a quantitative correlation (Figure 2, F). While irradiance values of 220–650 µmol m−2 s−1 triggered a similar initial chl-roGFP2 ΔOxD of approximately 10%, equivalent to an increase in EGSH of approximately 6 mV, irradiances of 750–1700 µmol m−2 s−1 triggered an initial oxidation of approximately 20%, equivalent to an increase in EGSH of approximately 13 mV. In contrast, the decrease in ΦPSII correlated with the light intensity (Figure 2, G). This binary response suggests a light-dependent threshold of chl-roGFP2 oxidation near 750 μE m−2 s−1, which matched the light saturation area, as indicated by light-response curves generated based on chlorophyll fluorescence measurements (Supplemental Figure S8). Overall, the data show the differential response of photosynthesis efficiency and redox metabolism to HL and suggest a regulatory role of chl-EGSH during the shift from light-unsaturated to light-saturated photosynthesis.
FL triggers oscillations in stromal EGSH
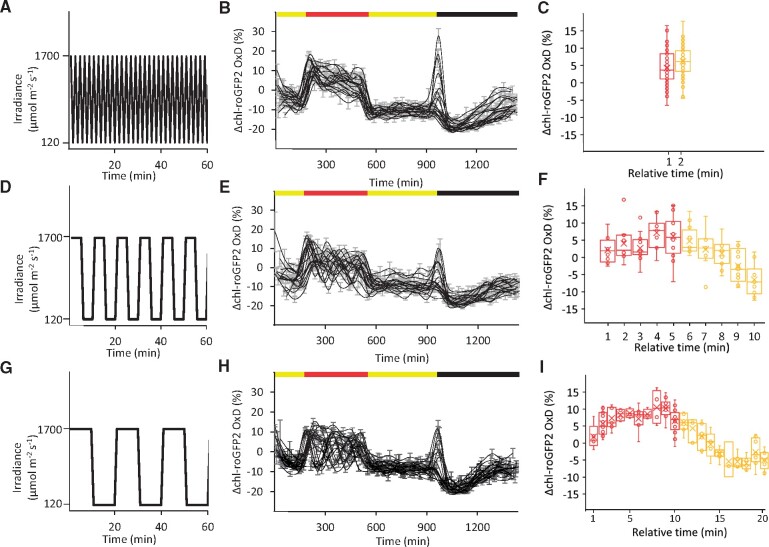
As plants in the field are inevitably confronted with varying light intensities, we further examined chl-roGFP2 redox dynamics under conditions of FL (Figure 3). To this end, after an initial illumination phase at 120 µmol m−2 s−1, plants were exposed, in the middle of the day, for 6 h of FL alternating between 1700 and 120 µmol m−2 s−1, in intervals of 1, 5, or 10 min (Figure 3, A, D, and G). Under all tested frequencies, a gradual and partial loss of photosynthetic activity was observed, as indicated by decreasing ΦPSII values, reaching a minimum at the end of the FL period (Supplemental Figure S9). Shifting plants back to normal light conditions resulted in partial ΦPSII recovery, with values still lower than those measured at the beginning of the day (Supplemental Figure S9). Plants exposed to the 1-min intervals showed a pattern that resembled HL treatments, including initial chl-roGFP2 oxidation (ΔOxD = ∼20%, Figure 3, B), followed by a gradual reduction when plants were moved back to normal growth light conditions. In contrast, oscillations in the chl-roGFP2 oxidation were observed in plants exposed to intervals of 5 and 10 min (Figure 3, E and H).
Figure 3.
Diurnal changes in chl-roGFP2 OxD under FL conditions. A, D, and G, Light irradiance during time over a 60-min period, extracted from the FL phase, is presented for the applied intervals of 1, 5, and 10 min. B, E, and H, Changes in chl-roGFP2 OxD values (ΔOxD) over time in Arabidopsis plants expressing chl-TKTP-roGFP2. After the start of the illumination period (yellow bar, 120 µmol m−2 s−1 for 3 h), the illumination regime was switched to FL alternating between HL (1700 µmol m−2 s−1) and normal light (120 µmol m−2 s−1) with different intervals. After 6 h FL (red bar), illumination continued continuously at 120 µmol m−2 s−1 for 7 h before the diurnal cycle ended with 8 h darkness (black bar). Oxidation values were normalized to steady-state values (as observed during the day at constant light conditions of 120 µmol m−2 s−1). Each black line represents the oxidation dynamics recorded for an independent well plate which consists of eight plants. Values represent means of these eight plants ± SE. C, F, and I, ΔOxD as a function of relative time (min) within each FL cycle. Values measured during the HL (1700 µmol m−2 s−1) phases within each light cycle are presented in red. Values measured during the phases with normal light intensities (120 µmol m−2 s−1) during each light cycle are presented in yellow. The presented box spans the interquartile range, and whiskers represent the minimum and maximum values (not including outliers). The median and mean of at least six plants are marked by a vertical line and x sign inside the box, respectively.
To make sense of the original data, we simplified the visual analysis and extracted all values recorded during the 6-h FL treatment and combined these data into a single apparent FL cycle, i.e. all ΔOxD values were sorted according to their relative time in the FL cycle (e.g. from the 10-min FL cycle experiment depicted in Figure 3, H, values for both 20 and 40 min were considered the 20th min each for their respective cycle and thus pooled together). This analysis resulted in a surprisingly clear picture showing an oscillation between higher and lower chl-roGFP2 oxidation states during the 5- and 10-min FL cycles, whereas no oscillations could be observed for the 1-min FL cycle (Figure 3, C, F, and I). For the 5- and 10-min FL cycles, we observed a gradual increase in oxidation during the HL phase and a decrease during the subsequent normal light phase. Importantly, these patterns were conserved in all FL cycles and were not altered despite the gradual decrease in ΦPSII during the FL phase (Supplemental Figure S9). It should be noted that the fact that no oscillations were observed in plants exposed to 1-min intervals does not necessarily mean that that chl-EGSH is constant under these conditions because equilibration of redox potentials between roGFP2 and the stromal glutathione pools involves a GRX-catalyzed biochemical reaction and is thus not instantaneous. Taken together, these results point to frequency-dependent oscillations of the chl-EGSH under FL.
Damage of PSI suppresses oscillations of EGSH in FL
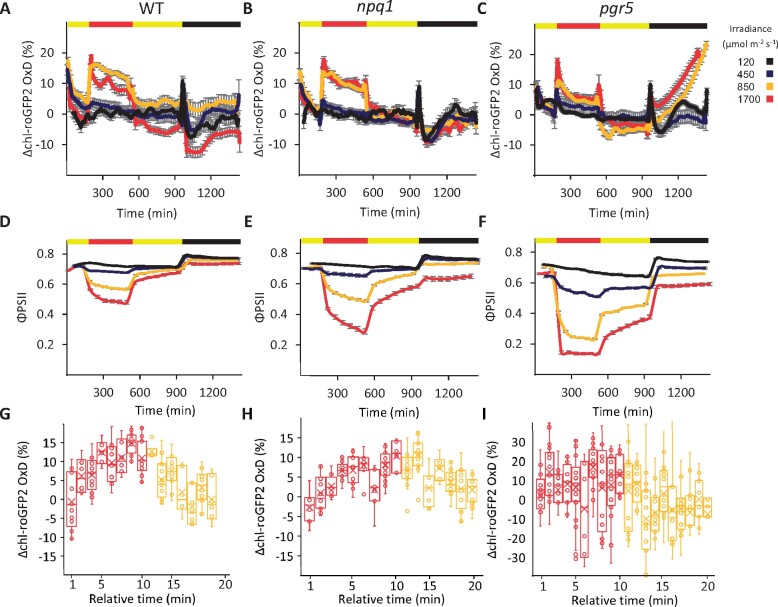
To examine the cross-talk between chl-EGSH patterns and light photoprotection mechanisms, the chl-roGFP2 probe was expressed in npq1 and pgr5 plants, and chl-roGFP2 OxD was measured in parallel with ΦPSII under the same HL regimens (Figure 4). Despite the clear decrease in ΦPSII in npq1, when compared with WT plants under identical HL conditions (e.g. reaching approximately 0.3 and 0.5 under 1700 µmol m−2 s−1, for npq1 and WT plants, respectively; Figure 4, D and E), nearly identical chl-roGFP2 oxidation trends were observed in both lines (Figure 4, A and B). Severely impaired photosynthetic efficiency was observed in pgr5 plants exposed to HL, reaching ΦPSII values of approximately 0.15 in plants illuminated at 1700 µmol m−2 s−1 (Figure 4, F). As in WT, shifting of pgr5 to HL resulted in fast oxidation, but chl-roGFP2 OxD values recovered much quicker than in WT (Figure 4, A and C, respectively). These results suggest that photoinhibition of PSII does not necessarily result in stronger chl-EGSH oxidation and that PSI photoinhibition results in faster reduction rates of chl-EGSH.
Figure 4.
Diurnal changes in chl-roGFP2 OxD and ΦPSII under HL and FL conditions in mutants with defects in photoprotective mechanisms. A–C, Daily changes in chl-roGFP2 OxD under different light regimes in WT (A), npq1 (B), and pgr5 (C) plants expressing chl-PRXaTP-roGFP2. The respective absolute ratio (400/485) data are provided in Supplemental Figure S17. Oxidation values were normalized to steady-state oxidation levels under normal growth light. The illumination regime was: 3 h normal light (yellow bar, 120 µmol m−2 s−1), followed by 6 h HL (red bar, intensities as indicated by line color), 7 h normal light (120 µmol m−2 s−1), and 8 h darkness. Each treatment involved between 24 and 32 plants divided into four independent plates that were consolidated in a “sliding window” (n = 4) display. Values represent means ± SE. D, E, and F, ΦPSII for WT (D), npq1 (E), and pgr5 (F) plants exposed to the same light regimes as in A–C. ΦPSII values were derived from chlorophyll fluorescence measurements and represent means of 12 plants ± SE. G, H, and I, Changes in chl-roGFP2 OxD during FL cycles with 10-min HL (1700 µmol m−2 s−1) and 10-min normal light (120 µmol m−2 s−1) for WT (G), npq1 (H), and pgr5 (I) plants. Values measured during the HL (1700 µmol m−2 s−1) phases within each light cycle are presented in red. Values measured during the phases with normal light intensities (120 µmol m−2 s−1) during each light cycle are presented in yellow. The presented box spans the interquartile range, and whiskers represent the minimum and maximum values (not including outliers). The median and mean of at least six plants are marked by a vertical line and x sign inside the box, respectively.
As PGR5 was found to be essential for photoprotection, specifically under FL conditions (Munekage et al., 2002; Suorsa et al., 2013; Yamori et al., 2016; Yamamoto and Shikanai, 2019), we examined chl-roGFP2 oxidation in WT, npq1, and pgr5 plants under FL conditions with 1- and 10-min intervals, as explained earlier. As shown for HL, FL also resulted in an apparent decrease in ΦPSII in npq1 and pgr5 compared with WT (Supplemental Figure S10). Recording of ΔOxD over an entire diurnal cycle with an FL phase of 1-min intervals did not significantly impact the overall oxidation pattern compared with plants exposed to continuous HL (Supplemental Figure S11, A). Accordingly, no significant difference in oxidation patterns was observed between the HL phase and the normal-light phases of each cycle in all three lines (Supplemental Figure S12). Under FL condition of 10-min intervals, the chl-roGFP2 redox state oscillated between relatively reduced and relatively oxidized values according to the relative timing within the FL cycle in WT and npq1 plants (Figure 4, G and H). Intriguingly, the fluctuation in ΔOxD during the FL cycle could not be observed in pgr5 plants (Figure 4, I). Taken together, these results demonstrate that the chl-EGSH response to FL conditions is dependent on PGR5 activity and is impaired under PSI photodamage.
Gradual relaxation of chloroplastic EGSH oxidation and ΦPSII impairment in pgr5 plants
Interestingly, a significant increase in chl-roGFP2 OxD was observed in pgr5 plants during the night following high and FL conditions, denoting an increase in chl-EGSH (Figure 4, C and Supplemental Figure S11, A and B). As shown, oxidation levels gradually increased during the night, reaching their maximum level at the end of the night period. The observed shift in OxD of approximately 25% (compared with the end-of-day values) in the night following HL phase of 1700 µmol m−2 s−1 denote an increase in chl-EGSH of approximately 16 mV. To ensure that the observed light response in pgr5 is not affected by the availability of endogenous GRXs, we examined the expression level of chloroplast targeted GRXs at the end of the night. No significant changes in the expression level of most chloroplast targeted GRXs were observed in pgr5 plants exposed to HL compared with those exposed to normal light or WT (Supplemental Figure S13), suggesting no alterations in the availability of endogenous GRXs at this time point. At the end of the night, chl-roGFP2 OxD values returned to steady-state OxD levels immediately when the light was turned on. In agreement with the chl-EGSH oxidation, we detected an induction of several oxidative stress marker genes (F6′H1, DLO1, and TI1) at the end of the night exclusively in pgr5 plants expose to HL (Supplemental Figure S13).
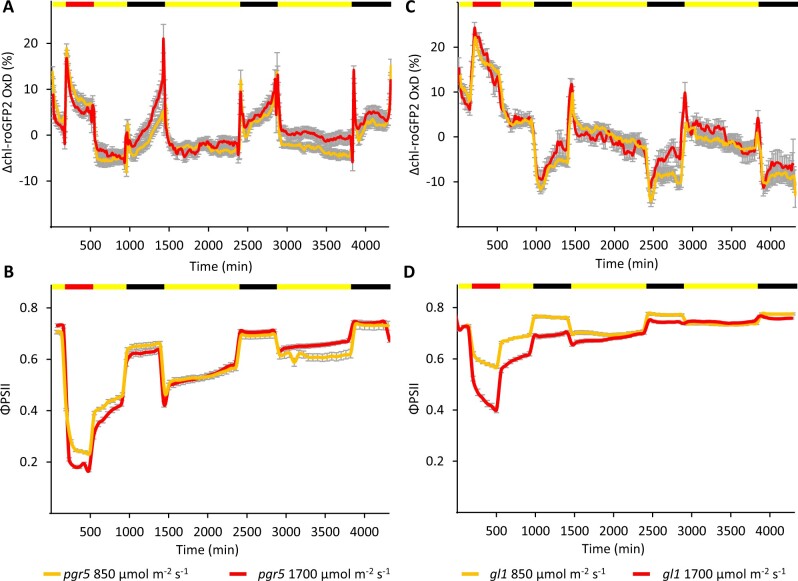
To further elucidate this phenomenon, we subjected pgr5 plants to 72-h experiments consisting of 24-h HL experiments (containing a 6-h HL period of 850 or 1700 µmol m−2 s−1), followed by 2 days with normal light (Figure 5, A and B; data of gl1, which served as the background line for the pgr5 mutation, are presented in Figure 5, C and D). During the first day of recovery under normal-light conditions, ΦPSII values were still lower than in gl1 plants (0.5–0.56 in pgr5 versus approximately 0.7 in gl1), and night chl-roGFP2 OxD oxidation clearly occurred again, albeit to a lower degree. Complete relaxation of the night chl-roGFP2 OxD oxidation was observed during the third night after HL treatment, reaching chl-roGFP2 OxD and ΦPSII values comparable to those observed under normal light conditions (Supplemental Figure S14). During the days, despite night oxidation, the chl-roGFP2 values were comparable on all the examined days. This night oxidation effect, which appeared hours after the HL stress, suggests a link between photoinhibition during the day and alterations in redox metabolism during the nights following the stress conditions.
Figure 5.
Diurnal changes in chl-roGFP2 OxD and ΦPSII during 1 day of HL regime followed by 2 days of normal light conditions in pgr5 and gl1 plants. A and C, Changes in chl-roGFP2 OxD over time from the experiment onset. chl-roGFP2 fluorescence was monitored throughout the experiment and chl-roGFP2 OxD values were calculated. Oxidation values were normalized to steady-state oxidation levels. Each treatment involved 32 plants divided into 4 independent plates that were consolidated in a “sliding window” (n = 4) display. Values represent means ± SE for pgr5 (A) and gl1 (C). roGFP2 data were acquired using the chl-PRXaTP-roGFP2. B, ΦPSII over time from the experiment onset. ΦPSII values were derived from chlorophyll fluorescence measurements and represent means of 12 plants ± SE for pgr5 (B) and gl1 (D). The color bar denotes the light conditions: black, night; yellow, normal light; and red, HL.
Discussion
The light-dependent flow of electrons from water to ferredoxin, the photosynthetic linear electron flow, produces the reducing power required for diverse downstream metabolic events, including carbon, nitrogen, and sulfur assimilation as well as for antioxidant activity. On the other hand, reduction of molecular oxygen at the acceptor site of PSI is the primary source of superoxide anion radical () and H2O2 in photosynthesizing chloroplasts (Asada, 1999; Badger et al., 2000). Thus, solar energy is the source for reducing power and oxidizing agents, subsequently requiring tight regulation to avoid imbalances between them.
The reduction of H2O2 via the ascorbate–glutathione pathway is thought to be the major mechanism leading to GSSG accumulation in plants (Rahantaniaina et al., 2013). Although the direct role of the GSH pool in detoxifying photosynthetically produced ROS has not been directly proven and the involvement of DHARs has been debated (Polle, 2001; Rahantaniaina et al., 2017), a recent study demonstrated the role of GSH in recycling ascorbate under HL conditions either nonenzymatically or via DHAR activity (Terai et al., 2020). Accordingly, light-dependent alterations in chl-EGSH likely reflect the balance between photosynthesis-dependent H2O2 production and NADPH availability for GR activity.
By applying whole-plant fluorescence imaging, we resolved the probe’s spatial fluorescence patterns and validated probe expression in the entire tissue (Figure 1, B). This is of interest as partial lack of the fluorescence signals resulting from gene silencing is frequently reported for plant protein-based biosensors (Jones et al., 2014; Exposito-Rodriguez et al., 2017). As silencing of the genetically encoded probe increases over generations, working with earlier generations of the desired line is typically recommended (Schwarzländer et al., 2015). Specifically, we have found chl-roGFP2 expression to be stable across generations in WT and the mutant lines used in this study. Moreover, our measurements with chl-roGFP2 provided the same readout regardless of whether or not a catalyzing GRX was fused to the probe or not. This suggests that the amounts of GRXs capable of catalyzing the redox equilibration of roGFP2 with EGSH are sufficient in chloroplasts for reliable measurements of chl-EGSH even with free roGFP2. It is generally assumed that endogenous GRX-target proteins with redox-sensitive thiols will respond to changes in chl-EGSH in a similar form as roGFP2 (Meyer, 2008). Thus, changes in chl-EGSH monitored with roGFP2 might indicate a light-induced modulation in the activity of such stromal proteins.
By measuring chl-roGFP2 oxidation at high temporal resolution over several consecutive days and in response to diverse light conditions, we revealed the dynamic response of chl-EGSH to changing light conditions. The presented results showed oxidation peaks during darkness–light and light–darkness transitions (Figure 2, C and D). During the former, the discrepancy between the light-dependent reactions, which are activated immediately upon light exposure, and the Calvin–Benson–Bassham cycle reactions, which involve a 15-min activation phase, resulted in a limited overflow of electrons from PSI to NADP+ (Wirtz et al., 1982; Tikhonov, 2015) and transient diversion of electrons to O2 reduction. The subsequent reduction of the chl-roGFP2 oxidation state may result from the full induction of CO2 assimilation activity and a decrease in O2 reduction in parallel with H2O2 detoxification. These results align with the suggested induction of the WWC during the induction phase of photosynthesis (Radmer and Kok, 1976; Miyake, 2010; Volpert et al., 2018).
The light-dependent increase in oxidants, which triggers changes in chl-EGSH, may transmit oxidative signals to redox-sensitive proteins during the photosynthesis induction phase (Buchanan and Balmer, 2005; Rahantaniaina et al., 2013). Indeed, the reception of oxidative signals by chloroplastic atypical TRXs (which regulate NPQ induction and starch biosynthesis) was demonstrated shortly after illumination (Dangoor et al., 2012; Eliyahu et al., 2015). Similarly, peaks in oxidant production during light–dark transitions can play a regulatory role in oxidizing redox-sensitive proteins at night, allowing transmission of reducing signals when the light is turned on again. Oxidative signals transmitted from H2O2 to target proteins, mediated by 2-Cys PRX at the onset of the dark period, have been demonstrated (Eliyahu et al., 2015; Ojeda et al., 2018; Vaseghi et al., 2018). Similarly, oxidation of key proteins in photosynthesis, including the ATP synthase CF1-γ subunit, fructose 1,6-bisphosphatase (FBPase) and sedoheptulose 1,7-bisphosphatase (SBPase), has been observed after the light was switched off (Yoshida et al., 2018). Dynamic response of the EGSH upon light-to-dark transition was also recently reported in the model moss Physcomitrella patens (Müller-Schüssele et al., 2020). However, the molecular features and biophysical mechanism underlying the induction of these oxidation peaks during light–dark transitions, as well as the possible involvement of the GSH/GRXs pathway in oxidizing target proteins during the photosynthesis induction and termination phases, are still to be resolved.
As opposed to a relatively reduced “steady-state” under normal growth conditions, chl-roGFP2 underwent considerable oxidation under HL conditions followed by gradual reduction (Figure 2, F). Interestingly, light irradiances of 220–650 µmol m−2 s−1 induced a lower chl-roGFP2 oxidation than irradiances of 750–1700 µmol m−2 s−1, whereas within the two ranges, there did not seem to be a significant difference. This binary effect, together with the contrasting nonbinary behavior of the drop in ΦPSII during the HL period, suggests a regulatory role for chl-EGSH, rather than a safety valve for energy dissipation through the WWC. Furthermore, it also implies the existence of a threshold near the light saturation point, which separates between two distinct states of oxidant production and, consequently, the chl-EGSH. We hypothesize that these two redox states signal distinct metabolic responses, in accordance with the severity of the HL conditions. The similarity in chl-EGSH OxD patterns between WT and npq1 plants, despite the significantly lower PSII efficiency in the mutant line (Figure 4), further supports this signaling role of chl-EGSH dynamics, as NPQ is a major mechanism for dissipating excess photons in higher plants. The combination of increased antioxidant activity, as demonstrated by the induction of antioxidant genes (Figure 2, E) as well as enhanced consumption of reducing equivalents, may explain the gradual reduction observed during the HL phase (Figure 2, F).
A frequency-dependent shift between two oxidation states was observed under FL conditions (Figure 3), suggesting its role in photosynthesis regulation under natural conditions, in which the solar flux varies due to the diurnal cycle, canopy structure, and varying cloud coverage. Interestingly, although a decrease in photosynthesis efficiency was observed during FL (Supplemental Figure S9), the ability to shift between the two chl-roGFP2 oxidation states was not impaired throughout the FL period (Figure 3). The fact that the observed fluctuating oxidation patterns were not induced in pgr5 plants may point toward a connection between PSI photoinhibition and rates of ROS production under these conditions. This link is further displayed by the faster reduction rates observed in pgr5 plants (Figure 4, C). It has recently been demonstrated that pgr5 plants are highly sensitive to HL-induced damage of iron–sulfur (Fe–S) clusters of PSI. This damage provides an additional photoprotective mechanism by inducing a nonphotochemical photoprotective energy quenching state (Tiwari et al., 2016). Based on the data observed here, we suggest that this PSI quenching state results in impaired regulation of the chloroplast EGSH. This results are in line with the proposed role of PSI photoinhibition in preventing excessive ROS production (Lima-Melo et al., 2019a) and the increased antioxidant capacity and decreased production of superoxide and H2O2 in pgr5 plants (Suorsa et al., 2012). Accordingly, alterations in ROS production and downstream redox signaling may affect the sensitivity of pgr5 to FL conditions.
Although focusing on light-dependent oxidant production, continuous diurnal chl-roGFP2 OxD measurement uncovered a unique phenotype in pgr5 plants during the nights following the HL and FL conditions. Pronounced gradual oxidation was observed on the first night following the HL stress, reaching the highest observed OxD from all experiments by the end of the night (Figure 4, C). Furthermore, nighttime oxidation was observed again the next night and typical night redox patterns seen in control plants were only observed in pgr5 plants on the third night after treatment (Figure 5, A). This effect, which only surfaced hours after exposure to light stress, was not directly caused by photoinhibition and may be related to the slow recovery time of PSI (Sonoike, 2011; Zhang et al., 2011). Accordingly, restoration of CO2 assimilation in HL-treated pgr5 plants was achieved during the third day in normal light conditions and was attributed to the recovery of PSI parameters, such as maximum oxidizable P700 and maximal reduction state of ferredoxin (Lima-Melo et al., 2019b).
It is possible that chl-EGSH oxidation, observed during the two nights after HL stress, results from starch starvation caused by PSI photoinhibition in pgr5 plants (Lima-Melo et al., 2019b). It has been reported that starch accumulates in chloroplasts during the day and is metabolized during the night as an NADPH source via the pentose phosphate pathway (Weise et al., 2004; Kirchsteiger et al., 2009). Therefore, the observed nighttime oxidation in pgr5 plants following HL stress may be due to a deficiency in NADPH resulting from starch deficiency. Accordingly, the contrasting effect observed in WT, in which reduction of chl-EGSH was observed at the beginning of the night, likely results from higher NADPH availability during the night, as observed using the TKTP-iNAP4 sensor (Figure 2, B). However, the demand for NADPH to maintain the highly reduced state of chl-EGSH during the night assumes GSH oxidation at night.
Interestingly, earlier work demonstrated that in addition to their role in thiol-redox maintenance, GSH and GRXs participate in assembling Fe–S clusters and transferring them to acceptor proteins, linking GSH and iron metabolism (Rouhier et al., 2008; Mühlenhoff et al., 2010; Rouhier et al., 2010; Kumar et al., 2011). Accordingly, repair and biogenesis of PSI after HL-induced damage of Fe–S clusters may require the function of GSH and GRXs. It is possible that the observed oxidation of chl-EGSH several nights after HL or FL stress in pgr5 plants (Figures 4, C, 5, A and Supplemental Figure S11), in which PSI centers are susceptible to photoinhibition, mirrors the diversion of reduced GSH for the slow process of biogenesis and repair of PSI Fe–S clusters. In view of that, the gradual reduction and oxidation of chl-EGSH at night, as observed during normal growth conditions as well as during HL and FL conditions in WT plants (Figures 2, A, F, 3, B, E, and H), may reflect the natural repair daily cycle of PSI. Whether this repair machinery operates exclusively at nights, or that a shortage of photosynthetically produced reducing power in the dark allowed to uncover this GSH requirement is not yet clear. An increasing demand for GSH may occur under chilling stress conditions, in which PSI is the major site of photoinhibition (Terashima et al., 1994), resulting in slow repair of PSI Fe–S clusters that will be further aggravated when GR activity is suppressed (Shu et al., 2011). Further work will be required to characterize the molecular mechanisms regulating nighttime chl-EGSH oxidation and the possible connection to PSI recovery.
In conclusion, by continuously monitoring roGFP2 oxidation patterns over several days, we provide a comprehensive view of photosynthetically related alteration in the chloroplastic EGSH under various light conditions and uncover an uncharacterized link between PSI photoinhibition and nighttime GSH metabolism. Monitoring of compartment-specific redox metabolism under varying environmental conditions will further expand the fundamental understanding of the role of redox signaling in light acclimation of higher plants.
Materials and methods
Plant material, growth conditions, and experimental setup
Arabidopsis thaliana WT (ecotype Columbia-0), npq1 (CS3771, At1g08550, obtained from ABRC), and pgr5 (EMS mutant line, At2g05620, obtained from Prof. T. Shikanai) lines were used throughout this research. Columbia ecotype glabrous 1 (gl-1, At3g27920, obtained from Prof. T. Shikanai [Kyoto University]), which served as the reference ecotype for the pgr5 line, was also used. Plants were sown on soil, placed at 4°C for 2 days to ensure uniform germination, and then grown under 16-h/8-h light/dark cycles with a photosynthetic photon flux density of 120 µmol m−2 s−1 (21°C, 60%–70% RH, ambient CO2) for 2–3 weeks. For roGFP2 and PAM analysis, plants were transferred to 12-well cell-culture multiwell plates in solid peat plugs, and the plugs were covered (with holes for plants) with black plastic to prevent autofluorescence. For DCMU experiments, 2–3-week-old plants were sprayed with 150 µM DCMU (D2425-100G, Sigma), kept for 1 h in the dark, and then exposed to HL conditions of 700 µmol m−2 s−1.
In all experiments, 2–3-week-old plants were incubated in 21°C, 60%–70% RH, and ambient CO2, unless stated otherwise. The following light conditions were applied: Normal growth experiments were carried out under 16-h light (120 µmol m−2 s−1)/8-h dark per 24-h cycle. For HL experiments, plants were exposed to 3 h of normal light conditions (120 µmol m−2 s−1), and then light irradiance was elevated during 6 h to the following HL intensities: 220, 330, 450, 550, 650, 750, 850, 1200, 1500, or 1700 µmol m−2 s−1. After the HL phase, plants were grown again under normal light conditions for an additional 7 h and finally were under an 8-h darkness period. For FL experiments, plants were exposed to 3 h of 120 µmol m−2 s−1, followed by 6 h of FL between 1700 (HL) and 120 (normal light) µmol m−2 s−1 in frequencies of 1, 5, or 10 min (i.e. 1, 5, or 10 min of 1700 µmol m−2 s−1 followed by 1, 5, or 10 min 120 µmol m−2 s−1 and vice versa). Afterward, plants were grown again for 7 h under 120 µmol m−2 s−1 and finally under an 8-h darkness period.
Generation of chl-roGFP2 expressing lines
Chloroplast targeting was achieved by using either the transketolase target peptide (Schwarzländer et al., 2008) or a 2-Cys PRX A (PRXa; Q96291) target peptide (this study), both of which will target proteins to the chloroplast stroma (König et al., 2002; Schwarzländer et al., 2008). For the generation of the chl-roGFP2 line, the full gene sequence was chemically synthesized. The first 74 amino acids from PRXa were used as a signal peptide. The chl-roGFP2 gene was cloned into the plant cloning vector pART7 using XhoI and HindIII restriction enzymes. The whole construct including the CaMV 35S promoter and ocs terminator was then cloned into the binary vector pART27 using the restriction enzyme NotI. The pART27 plasmid, which contains the chl-roGFP2 construct, was transformed into GV3101 Agrobacterium tumefaciens. Transformation of A. thaliana was performed by floral dip (Clough and Bent, 1998). Transformed lines were selected based on kanamycin resistance and the chl-roGFP2 fluorescence signal.
Confocal microscopy
Images were acquired with a Leica TCS SP8 confocal system (Leica Microsystems) and the LAS X Life Science Software, while using a HC PL APO ×40/1.10 objective. All images were acquired at a resolution of 4096 × 4096 pixels. Images were acquired with emission at 500–520 nm following excitation at 488 nm for chl-roGFP2 fluorescence and emission at 670 nm following excitation at 488 nm for chlorophyll fluorescence. Merged images were generated using Fiji (Image J) software (Schneider et al., 2012).
chl-roGFP2 fluorescence measurements and analysis
Whole-plant chl-roGFP2 fluorescence imaging was detected using an Advanced Molecular Imager HT (Spectral Ami-HT, Spectral Instruments Imaging, LLC., USA) and AMIview software for image acquisition. This analysis was used to screen for fluorescent plants and for qualitative analysis. For chl-roGFP2 fluorescence detection, excitation by 405 ± 10 nm or 465 ± 10 nm LED light sources and 510 ± 20 nm emission filter were used. Oxidized chl-roGFP2 produces greater fluorescence at 405 nm compared to the reduced state, whereas highly reduced chl-roGFP2 shows a stronger signal at 465 nm compared to the oxidized state. Therefore, the 405/465 ratio is a reliable indicator of the degree of oxidation. For chlorophyll detection, a 405 nm ± 10 LED light source and a 670 ± 20 nm emission filter were used.
Quantitative chl-roGFP2 analysis was mainly carried out by the fluorometer method (Rosenwasser et al., 2010) using a multimode microplate reader (Tecan Spark, Tecan, Switzerland). For all plate reader experiments, roGFP2 fluorescence was excited at 400 ± 10 nm and 485 ± 10 nm, respectively, and emission recorded at 520 ± 5 nm. Chlorophyll autofluorescence was excited at 400 ± 10 nm and recorded in the 670 ± 20 nm band. For extended times, plants were grown in 12-well plates and placed in a growth chamber (Fytoscope FS-SI-4600, Photon Systems Instruments, Czech Republic) for illumination at defined light intensities. For recording of chl-roGFP2 fluorescence, the well plates were automatically inserted into the plate reader using a robot (KiNEDx KX.01467, Peak Analysis and Automation [PAA], Hampshire, UK) controlled by a self-compiled program (Overlord, PAA).
For the plate-reader analysis, a 9 × 9-pixel matrix was defined for each well and chlorophyll autofluorescence was detected, to generate a chlorophyll mask. This mask was then used to select pixels, which returned a positive chlorophyll fluorescence signal. Only those pixels were subsequently considered for the roGFP2 analysis. For background correction, the average signal of WT plants without chl-roGFP2 was determined and the values for this background autofluorescence subtracted from the values detected in the chl-roGFP2 fluorescence analysis. A similar experimental setup was used to measure the fluorescence emitted from the TKTP-iNAP4 and TKTP-iNAPc expressing lines. The pH-corrected ratio for TKTP-iNAP4 was calculated by dividing the fluorescence ratio values obtained from the TKTP-iNAP4 line by those obtained from the TKTP-iNAPc line, according to Lim et al. (2020). The degree of oxidation (the relative quantity of oxidized roGFP proteins, OxD) of roGFP2 was calculated based on the fluorescence signal according to Equation (1) (Meyer et al., 2007):
where R represents the ratio (400/485) at each point in the experiment, Rred represents the ratio under fully reduced conditions, Rox represents the ratio under fully oxidized conditions, I485min represents the fluorescence emitted at 520 nm when excited at 485 nm under fully oxidized conditions, and I485max represents the fluorescence emitted at 520 nm when excited at 485 nm under fully reduced conditions. In order to be able to calculate chl-roGFP2 OxD, we treated chl-roGFP2 Arabidopsis plants and measured the 400/485 nm fluorescence ratio under fully oxidized and fully reduced conditions by using 1–4 M H2O2 and 100 mM DTT, respectively, by immersing plants in the respective solutions. A custom-written Matlab script (available on request) was used to allow a robust analysis of multiple files, which contained the measurements from each experiment. The scripts collect the roGFP2 values from each plant into one matrix. In addition, experimental metadata such as the start and endpoint, plant lines, light irradiance, temperature, relative humidity (RH, %), and CO2 concentration were also collected and added to the output file. EGSH values were calculated according to Schwarzländer et al. (2008).
Chlorophyll fluorescence measurements
The ΦPSII of each plant was detected in 60-min intervals using a Walz PAM IMAGING PAM M-series IMAG-K7 (MAXI) fluorometer. The ETR was calculated as the multiplication of ΦPSII by the quantity of photons emitted by the light source (PAR), by the average percentage of the photons absorbed by the photosynthetic machinery (0.84), and by the amount of photons allocated to PSII (0.5; assuming that PSI and PSII contain the same amount of reaction centers, therefore each photosystem receives 50% of the photon influx). Accordingly, ETR = Y(II) × PAR × 0.84 × 0.5. Full ETR light induction curves’ data are in Supplemental Figure S8. A custom-written Matlab script was used to allow a robust analysis of multiple files (available upon request).
Gene expression analysis
RNA was extracted from 3-week-old rosettes using the Spectrum Plant RNA Kit (STRN50; Sigma). For each time point, three (for the 1 and 3 h in HL) or two (for later time points) independent biological replicates were analyzed. The extracted RNA was digested and cleaned with the TURBO DNase kit (Ambion) according to the manufacturer’s instructions and was reverse-transcribed to cDNA with the ThermoScript RT-PCR system (Invitrogen). The qPrimerDB (Lu et al., 2018) was used for selecting gene-specific primers for reverse-transcription quantitative-PCR (RT-qPCR). The BioMark HD system was used for high-throughput qPCR, using standard fast-cycling conditions and melt-curve analysis, following the manufacturer’s instructions (Fluidigm). Cycle threshold (Ct) values for each reaction (which correspond to the logarithm of the fold change in gene expression) were used for analysis and expression data were calculated using the ddCT approach (Livak and Schmittgen, 2001). The ACT2 gene (At3g18780) was used as a reference for equalizing the levels of RNA.
Statistics
Values in graphs mainly represent means of 8 plants per plate for chl-roGFP2 OxD and 12 plants per plate for ΦPSII, and error bars represent the respective standard errors. For many experiments, three to four plates were analyzed, and for a clear presentation, a “sliding window” approach was taken, in which each data point represents the average of three to four plates (24–32 plants). Statistical significance was tested using a two-tail Student’s t test and analysis of variance (ANOVA) at 95% confidence level and was indicated by asterisks when shown (Supplemental File S1).
Accession numbers
Germplasm used included npq1 (CS3771, At1g08550, obtained from ABRC), pgr5 (EMS mutant line, At2g05620, obtained from Prof. T. Shikanai), and gl1 (At3g27920, obtained from Prof. T. Shikanai), which served as the reference ecotype for the pgr5 line. Accession numbers for genes used in gene expression analyses are available in Supplemental Data Set S1.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Confocal images of the chl-PRXaTP-roGFP2 line.
Supplemental Figure S2. Dose-dependent chl-roGFP2 oxidation in response to H2O2.
Supplemental Figure S3. Changes in chl-roGFP2 oxidation and NADPH levels under high-light conditions.
Supplemental Figure S4. Diurnal changes in chl-roGFP2 OxD and ΦPSII under high light and elevated CO2.
Supplemental Figure S5. Changes in chl-roGFP2 oxidation in DCMU-treated plants under high-light conditions.
Supplemental Figure S6. Diurnal changes in chloroplast-targeted Grx1-roGFP2 oxidation under high-light conditions.
Supplemental Figure S7. Temporal changes in expression levels of chloroplast-targeted GRXs under high-light conditions.
Supplemental Figure S8. ETR light-response curves in WT, npq1, and pgr5 plants.
Supplemental Figure S9. Diurnal changes in ΦPSII during illumination with fluctuating light.
Supplemental Figure S10. Diurnal changes in ΦPSII during fluctuating light experiments in WT, npq1, and pgr5 plants.
Supplemental Figure S11. Diurnal changes in chl-roGFP2 OxD during fluctuating light experiments in WT, npq1, and pgr5 plants.
Supplemental Figure S12. Changes in chl-roGFP2 OxD during fluctuating light cycles in WT, npq1, and pgr5 plants.
Supplemental Figure S13. Temporal changes in expression levels of chloroplast-targeted GRXs, antioxidant genes, and ROS gene markers in WT and pgr5 plants at the end of night.
Supplemental Figure S14. Diurnal changes in chl-roGFP2 OxD and ΦPSII during the third day of recovery-from-high-light experiments in pgr5 plants.
Supplemental Figure S15. Diurnal changes in chl-roGFP2 OxD during high-light experiments in wild type (WT) plants.
Supplemental Figure S16. Diurnal changes in chl-roGFP2 OxD during high-light experiments in wild type (WT) plants.
Supplemental Figure S17. Diurnal changes in chl-roGFP2 fluorescence ratios under high-light conditions in wild type WT and mutants with defects in photoprotective mechanisms.
Supplemental Data Set S1. Temporal changes in the expression levels of chloroplast-targeted genes encoding antioxidant proteins, GRXs, and established ROS markers in response to HL conditions.
Supplemental File S1. Statistical analysis tables.
Supplementary Material
Acknowledgments
The authors thank Avihai Danon, Zach Adam, and Assaf Vardi for critical comments on the manuscript. They also thank Prof. Toshiharu Shikanai, who kindly provided the pgr5 and gl1 seeds for this research, Prof. Wallace Boon Leong Lim, who kindly provided the TKTP-iNAP4 and TKTP-iNAPc Arabidopsis lines and Ms. Charis Chan for her help in shipping the seeds, and Daniel Waiger for his help in acquiring the confocal images. Finally, they thank David Pilzer for his support in performing the gene expression analysis.
Z.H. and S.R. designed the research and analyzed the data. Z.H., N.L., E.Z., M.H., and S.R. performed the research. A.J.M. contributed new plant lines. Z.H. and S.R. wrote the manuscript with inputs from N.L. and A.J.M.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Shilo Rosenwasser (shilo.rosenwaser@mail.huji.ac.il).
Funding
This research was supported by the Israel Science Foundation (Grant Nos. 826/17 and 827/17) to S.R.
Conflict of interest statement. The authors declare that there is no conflict of interest.
References
- Arnon DI, Allen MB, Whatley FR (1954) Photosynthesis by isolated chloroplasts. Nature 174: 394–396 [DOI] [PubMed] [Google Scholar]
- Asada K (1999) The water–water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol 50: 601–639 [DOI] [PubMed] [Google Scholar]
- Avenson TJ, Cruz JA, Kanazawa A, Kramer DM (2005) Regulating the proton budget of higher plant photosynthesis. Proc Natl Acad Sci USA 102: 9709–9713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad J, Stotz HU, Fekete A, Krischke M, Engert C, Havaux M, Berger S, Mueller MJ (2015) 2-Cysteine peroxiredoxins and thylakoid ascorbate peroxidase create a water–water cycle that is essential to protect the photosynthetic apparatus under high light stress conditions. Plant Physiol 167: 1592–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR, von Caemmerer S, Ruuska S, Nakano H (2000) Electron flow to oxygen in higher plants and algae: rates and control of direct photoreduction (Mehler reaction) and rubisco oxygenase. Phil Trans R Soc Lond B Biol Sci 355: 1433–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratt A, Rosenwasser S, Meyer A, Fluhr R (2016) Organelle redox autonomy during environmental stress. Plant Cell Environ 39: 1909–1919 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56: 187–220 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Creissen G, Firmin J, Fryer M, Kular B, Leyland N, Reynolds H, Pastori G, Wellburn F, Baker N, Welburn A, et al. (1999) Elevated glutathione biosynthetic capacity in the chloroplasts of transgenic tobacco plants paradoxically causes increased oxidative stress. Plant Cell 11: 1277–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangoor I, Peled-Zehavi H, Wittenberg G, Danon A (2012) A chloroplast light-regulated oxidative sensor for moderate light intensity in Arabidopsis. Plant Cell 24: 1894–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams B (1990) Carotenoids and photoprotection in plants a role for the xanthophyll zeaxanthin. Biochim Biophys Acta 1020: 1–24 [Google Scholar]
- Dooley CT, Dore TM, Hanson GT, Jackson WC, Remington SJ, Tsien RY (2004) Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J Biol Chem 279: 22284–22293 [DOI] [PubMed] [Google Scholar]
- Eliyahu E, Rog I, Inbal D, Danon A (2015) ACHT4-driven oxidation of APS1 attenuates starch synthesis under low light intensity in Arabidopsis plants. Proc Natl Acad Sci U S A 112: 12876–12881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito-Rodriguez M, Laissue PP, Yvon-Durocher G, Smirnoff N, Mullineaux PM (2017) Photosynthesis-dependent H2O2 transfer from chloroplasts to nuclei provides a high-light signalling mechanism. Nat Commun 8: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Lopez-Delgado H, Dat JF, Scott IM (1997) Hydrogen peroxide- and glutathione-associated mechanisms of acclimatory stress tolerance and signalling. Physiol Plant 100: 241–257 [Google Scholar]
- Foyer CH, Noctor G (2011) Ascorbate and glutathione: the heart of the redox hub. Plant Physiol 155: 2–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Breusegem FV (2006) Transcriptomic footprints disclose specificity of reactivated oxygen species signaling in Arabidopsis. Plant Physiol 141: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutscher M, Pauleau A-L, Marty L, Brach T, Wabnitz GH, Samstag Y, Meyer AJ, Dick TP (2008) Real-time imaging of the intracellular glutathione redox potential. Nat Methods 5: 553–559 [DOI] [PubMed] [Google Scholar]
- Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ (2004) Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem 279: 13044–13053 [DOI] [PubMed] [Google Scholar]
- Horton P, Ruban AV (1992) Regulation of photosystem II. Photosynth Res 34: 375–385 [DOI] [PubMed] [Google Scholar]
- Jiang K, Schwarzer C, Lally E, Zhang S, Ruzin S, Machen T, Remington SJ, Feldman L (2006) Expression and characterization of a redox-sensing green fluorescent protein (reduction–oxidation-sensitive green fluorescent protein) in Arabidopsis. Plant Physiol 141: 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Danielson JÅH, ManojKumar SN, Lanquar V, Grossmann G, Frommer WB (2014) Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. eLife 3: e01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux PM (1997) Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9: 627–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchsteiger K, Pulido P, González M, Cejudo FJ (2009) NADPH thioredoxin reductase c controls the redox status of chloroplast 2-cys peroxiredoxins in Arabidopsis thaliana. Mol Plant 2: 298–307 [DOI] [PubMed] [Google Scholar]
- König J, Baier M, Horling F, Kahmann U, Harris G, Schürmann P, Dietz K-J (2002) The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of photosynthetic electron flux. Proc Natl Acad Sci U S A 99: 5738–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono M, Terashima I (2014) Long-term and short-term responses of the photosynthetic electron transport to fluctuating light. J Photochem Photobiol B Biol 137: 89–99 [DOI] [PubMed] [Google Scholar]
- Külheim C, Ågren J, Jansson S (2002) Rapid regulation of light harvesting and plant fitness in the field. Science 297: 91–93 [DOI] [PubMed] [Google Scholar]
- Kumar C, Igbaria A, D’Autreaux B, Planson A-G, Junot C, Godat E, Bachhawat AK, Delaunay-Moisan A, Toledano MB (2011) Glutathione revisited: a vital function in iron metabolism and ancillary role in thiol-redox control. EMBO J 30: 2044–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S-L, Voon CP, Guan X, Yang Y, Gardeström P, Lim BL (2020) In planta study of photosynthesis and photorespiration using NADPH and NADH/NAD+ fluorescent protein sensors. Nat Commun 11: 3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-Melo Y, Alencar VTCB, Lobo AKM, Sousa RH V, Tikkanen M, Aro E-M, Silveira JAG, Gollan PJ (2019a) Photoinhibition of photosystem I provides oxidative protection during imbalanced photosynthetic electron transport in Arabidopsis thaliana. Front Plant Sci 10: 916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-Melo Y, Gollan PJ, Tikkanen M, Silveira JAG, Aro E-M (2019b) Consequences of photosystem-I damage and repair on photosynthesis and carbon use in Arabidopsis thaliana. Plant J 97: 1061–1072 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lu K, Li T, He J, Chang W, Zhang R, Liu M, Mengna Y, Fan Y, Ma J, Sun W,. et al. (2018) qPrimerDB: a thermodynamics-based gene-specific qPCR primer database for 147 organisms. Nucleic Acids Res 46: D1229–D1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty L, Bausewein D, Müller C, Bangash SAK, Moseler A, Schwarzländer M, Müller-Schüssele SJ, Zechmann B, Riondet C, Balk J (2019) Arabidopsis glutathione reductase 2 is indispensable in plastids, while mitochondrial glutathione is safeguarded by additional reduction and transport systems. New Phytol 224: 1569–1584. [DOI] [PubMed] [Google Scholar]
- Marty L, Siala W, Schwarzländer M, Fricker MD, Wirtz M, Sweetlove LJ, Meyer Y, Meyer AJ, Reichheld J-P, Hell R (2009) The NADPH-dependent thioredoxin system constitues a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc Natl Acad Sci U S A 106: 9109–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler AH (1951) Studies on reactions of illuminated chloroplasts: I. Mechanism of the reduction of oxygen and other hill reagents. Arch Biochem Biophys 33: 65–77 [DOI] [PubMed] [Google Scholar]
- Meyer AJ (2008) The integration of glutathione homeostasis and redox signaling. J Plant Physiol 165: 1390–1403 [DOI] [PubMed] [Google Scholar]
- Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot J-P, Hell R (2007) Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J 52: 973–986 [DOI] [PubMed] [Google Scholar]
- Meyer AJ, Dick TP (2010) Fluorescent protein-based redox probes. Antioxid Redox Signal 13: 621–650 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Miyake C (2010) Alternative electron flows (water–water cycle and cyclic electron flow around PSI) in photosynthesis: molecular mechanisms and physiological functions. Plant Cell Physiol 51: 1951–1963 [DOI] [PubMed] [Google Scholar]
- Mizrachi A, Graff van Creveld S, Shapiro OH, Rosenwasser S, Vardi A (2019) Light-dependent single-cell heterogeneity in the chloroplast redox state regulates cell fate in a marine diatom. eLife 8: e47732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenhoff U, Molik S, Godoy JR, Uzarska MA, Richter N, Seubert A, Zhang Y, Stubbe J, Pierrel F, Herrero E, et al. (2010) Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab 12: 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Schüssele SJ, Wang R, Gütle DD, Romer J, Rodriguez-Franco M, Scholz M, Buchert F, Lüth VM, Kopriva S, Dörmann P (2020) Chloroplasts require glutathione reductase to balance reactive oxygen species and maintain efficient photosynthesis. Plant J 103: 1140–1154 [DOI] [PubMed] [Google Scholar]
- Mullineaux PM, Exposito-Rodriguez M, Laissue PP, Smirnoff N (2018) ROS-dependent signalling pathways in plants and algae exposed to high light: comparisons with other eukaryotes. Free Radic Biol Med 122: 52–64 [DOI] [PubMed] [Google Scholar]
- Munekage Y, Hashimoto M, Miyake C, Tomizawa K-I, Endo T, Tasaka M, Shikanai T (2004) Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429: 579–582. [DOI] [PubMed] [Google Scholar]
- Munekage Y, Hojo M, Meurer J, Endo T, Tasaka M, Shikanai T (2002) PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110: 361–371 [DOI] [PubMed] [Google Scholar]
- Nietzel T, Elsässer M, Ruberti C, Steinbeck J, Ugalde JM, Fuchs P, Wagner S, Ostermann L, Moseler A, Lemke P, et al. (2019) The fluorescent protein sensor roGFP2-Orp1 monitors in vivo H2O2 and thiol redox integration and elucidates intracellular H2O2 dynamics during elicitor-induced oxidative burst in Arabidopsis. New Phytol 221: 1649–1664 [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Grossman AR, Björkman O (1998) Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10: 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojeda V, Pérez-Ruiz JM, Cejudo FJ (2018) 2-Cys peroxiredoxins participate in the oxidation of chloroplast enzymes in the dark. Mol Plant 11: 1377–1388 [DOI] [PubMed] [Google Scholar]
- op den Camp RGL, Przbyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg É, Göbel C, Feussner I,. et al. (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ort DR, Baker NR (2002) A photoprotective role for O2 as an alternative electron sink in photosynthesis. Curr Opin Plant Biol 5: 193–198 [DOI] [PubMed] [Google Scholar]
- Park S-W, Li W, Viehhauser A, He B, Kim S, Nilsson AK, Andersson MX, Kittle JD, Ambavaram MMR, Luan S, et al. (2013) Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc Natl Acad Sci U S A 110: 9559–9564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polle A (2001) Dissecting the superoxide dismutase–ascorbate–glutathione-pathway in chloroplasts by metabolic modeling. Computer simulations as a step towards flux analysis. Plant Physiol 126: 445–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radmer RJ, Kok B (1976) Photoreduction of O2 primes and replaces CO2 assimilation. Plant Physiol 58: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahantaniaina M-S, Li S, Chatel-Innocenti G, Tuzet A, Mhamdi A, Vanacher H, Noctor G (2017) Glutathione oxidation in response to intracellular H2O2: key but overlapping roles for dehydroascorbate reductases. Plant Signal Behav 12: e1356531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahantaniaina M-S, Tuzet A, Mhamdi A, Noctor G (2013) Missing links in understanding redox signaling via thiol/disulfide modulation : how is glutathione oxidized in plants. Front Plant Sci 4: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Mittler R (2003) The water–water cycle is essential for chloroplast protection in the absence of stress. J Biol Chem 278: 38921–38925 [DOI] [PubMed] [Google Scholar]
- Rosenwasser S, Graff van Creveld S, Schatz D, Malitsky S, Tzfadia O, Aharoni A, Levin Y, Gabashvili A, Feldmesser E, Vardi A (2014b) Mapping the diatom redox-sensitive proteome provides insight into response to nitrogen stress in the marine environment. Proc Natl Acad Sci 111: 2740 LP–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwasser S, Rot I, Meyer AJ, Feldman L, Jiang K, Friedman H (2010) A fluorometer-based method for monitoring oxidation of redox-sensitive GFP (roGFP) during development and extended dark stress. Physiol Plant 138: 493–502 [DOI] [PubMed] [Google Scholar]
- Rouhier N, Couturier J, Johnson MK, Jacquot J-P (2010) Glutaredoxins: roles in iron homeostasis. Trends Biochem Sci 35: 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Lemaire SD, Jacquot J-P (2008) The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation. Annu Rev Plant Biol 59: 143–166 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzländer M, , Dick TP, , Meyer AJ, , Morgan B (2015) Dissecting Redox Biology Using Fluorescent Protein Sensors. Antioxid Redox Signal 24: 680–712 [DOI] [PubMed] [Google Scholar]
- Schwarzländer M, Fricker MD, Müller C, Marty L, Brach T, Novak J, Sweetlove LJ, Hell R, Meyer AJ (2008) Confocal imaging of glutathione redox potential in living plant cells. J Microsc 231: 299–316 [DOI] [PubMed] [Google Scholar]
- Shikanai T (2007) Cyclic electron transport around photosystem I: genetic approaches. Annu Rev Plant Biol 58: 199–217 [DOI] [PubMed] [Google Scholar]
- Shikanai T (2016) Regulatory network of proton motive force: contribution of cyclic electron transport around photosystem I. Photosynth Res 129: 253–260 [DOI] [PubMed] [Google Scholar]
- Shu D-F, Wang L-Y, Duan M, Deng Y-S, Meng Q-W (2011) Antisense-mediated depletion of tomato chloroplast glutathione reductase enhances susceptibility to chilling stress. Plant Physiol Biochem 49: 1228–1237 [DOI] [PubMed] [Google Scholar]
- Sonoike K (2011) Photoinhibition of photosystem I. Physiol Plant 142: 56–64 [DOI] [PubMed] [Google Scholar]
- Suorsa M, Grieco M, Järvi S, Gollan PJ, Kangasjärvi S, Tikkanen M, Aro E-M (2013) PGR5 ensures photosynthetic control to safeguard photosystem I under fluctuating light conditions. Plant Signal Behav 8: e22741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suorsa M, Järvi S, Grieco M, Nurmi M, Pietrzykowska M, Rantala M, Kangasjärvi S, Paakkarinen V, Tikkanen M, Jansson S, et al. (2012) PROTON GRADIENT REGULATION5 is essential for proper acclimation of Arabidopsis photosystem I to naturally and artificially fluctuating light conditions. Plant Cell 24: 2934–2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa K, Tsujimoto HY, Arnon DI (1963) Role of chloroplast ferredoxin in the energy conversion process of photosynthesis. Proc Natl Acad Sci U S A 49: 567–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Badger MR (2011) Photoprotection in plants: a new light on photosystem II damage. Trends Plant Sci 16: 53–60 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Milward SE, Fan D-Y, Chow WS, Badger MR (2009) How does cyclic electron flow alleviate photoinhibition in Arabidopsis. Plant Physiol 149: 1560–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Zhao Y, Chu H, Wang A, Zhu J, Chen X, Zou Y, Shi M, Liu R, Su N, et al. (2017) Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism. Nat Methods 14: 720–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai Y, Ueno H, Ogawa T, Sawa Y, Miyagi A, Kawai-Yamada M, Ishikawa T, Martua T (2020) Dehydroascorbate reductases and glutathione set a threshold for high-light-induced ascorbate accumulation. Plant Physiol 183: 112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima I, Funayama S, Sonoike K (1994) The site of photoinhibition in leaves of Cucumis sativus L. at low temperatures is photosystem I, not photosystem II. Planta 193: 300–306 [Google Scholar]
- Tikhonov AN (2015) Induction events and short-term regulation of electron transport in chloroplasts : an overview. Photosynth Res 125: 65–94 [DOI] [PubMed] [Google Scholar]
- Tiwari A, Mamedov F, Grieco M, Suorsa M, Jajoo A, Styring S, Tikkanen M, Aro E-M (2016) Photodamage of iron–sulphur clusters in photosystem I induces non-photochemical energy dissipation. Nat Plants 2: 16035. [DOI] [PubMed] [Google Scholar]
- Trnka D, Engelke AD, Gellert M, Moseler A, Hossain MF, Lindenberg TT, Pedroletti L, Odermatt B, de Souza JV, Bronowska AK,. et al. (2020) Molecular basis for the distinct functions of redox-active and FeS-transfering glutaredoxins. Nat Commun 11: 3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Creveld SG, Rosenwasser S, Schatz D, Koren I, Vardi A (2015) Early perturbation in mitochondria redox homeostasis in response to environmental stress predicts cell fate in diatoms. ISME J 9: 385–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseghi M-J, Chibani K, Telman W, Liebthal MF, Gerken M, Schnitzer H, Mueller SM, Dietz K-J (2018) The chloroplast 2-cysteine peroxiredoxin functions as thioredoxin oxidase in redox regulation of chloroplast metabolism. eLife 7: e38194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpert A, Graff van Creveld S, Rosenwasser S, Vardi A (2018) Diurnal fluctuations in chloroplast GSH redox state regulate susceptibility to oxidative stress and cell fate in a bloom-forming diatom. J Phycol 54: 329–341 [DOI] [PubMed] [Google Scholar]
- Weise SE, Weber APM, Sharkey TD (2004) Maltose is the major form of carbon exported from the chloroplast at night. Planta 218: 474–482 [DOI] [PubMed] [Google Scholar]
- Wirtz W, Stitt M, Heldt HW (1982) Light activation of calvin cycle enzymes as measured in pea leaves. FEBS Lett 142: 223–226 [Google Scholar]
- Yamamoto H, Shikanai T (2019) PGR5-dependent cyclic electron flow protects photosystem I under fluctuating light at donor and acceptor sides. Plant Physiol 179: 588–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamori W, Makino A, Shikanai T (2016) A physiological role of cyclic electron transport around photosystem I in sustaining photosynthesis under fluctuating light in rice. Sci Rep 6: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Hara A, Sugiura K, Fukaya Y, Hisabori T (2018) Thioredoxin-like2/2-Cys peroxidoxin redox cascade supports oxidative thiol modulation in chloroplasts. Proc Natl Acad Sci US A 115: E8296–E8304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Jia Y, Gao H, Zhang L, Li H, Meng Q (2011) Characterization of PSI recovery after chilling-induced photoinhibition in cucumber (Cucumis sativus L.) leaves. Planta 234: 883–889 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.