Abstract
Polyploidy or whole-genome duplication (WGD) is widespread in plants and is a key driver of evolution and speciation, accompanied by rapid and dynamic changes in genomic structure and gene expression. The 3D structure of the genome is intricately linked to gene expression, but its role in transcription regulation following polyploidy and domestication remains unclear. Here, we generated high-resolution (∼2 kb) Hi-C maps for cultivated soybean (Glycine max), wild soybean (Glycine soja), and common bean (Phaseolus vulgaris). We found polyploidization in soybean may induce architecture changes of topologically associating domains and subsequent diploidization led to chromatin topology alteration around chromosome-rearrangement sites. Compared with single-copy and small-scale duplicated genes, WGD genes displayed more long-range chromosomal interactions and were coupled with higher levels of gene expression and chromatin accessibilities but void of DNA methylation. Interestingly, chromatin loop reorganization was involved in expression divergence of the genes during soybean domestication. Genes with chromatin loops were under stronger artificial selection than genes without loops. These findings provide insights into the roles of dynamic chromatin structures on gene expression during polyploidization, diploidization, and domestication of soybean.
Reorganization of chromatin structure was involved in expression divergence during polyploidization, diploidization, and domestication of soybean.
Introduction
Eukaryotic genomes are hierarchically organized at different levels in the nucleus (Sotelo-Silveira et al., 2018; Zheng and Xie, 2019). Chromatin folding brings long-range interactions between genomic elements that are tightly linked to regulation of gene expression (Lieberman-Aiden et al., 2009; Wang et al., 2015a). Recent advances in chromosome conformation capture (3C)-based methods, including Hi-C and Chromatin Interaction Analysis with Paired-End Tag, have provided a detailed genome-wide information on the 3D organization of genomes in animals (Fullwood et al., 2009; Lieberman-Aiden et al., 2009; Dixon et al., 2012; Zheng and Xie, 2019) and plants (Feng et al., 2014; Wang et al., 2015a; Dong et al., 2017; Liu et al., 2017; Sotelo-Silveira et al., 2018; Grob and Grossniklaus, 2019).
Mammalian genomes are partitioned into megabase-scale topologically associated domains (TADs), which are separated by regions enriched for CCCTC-binding factor (CTCF) protein and cohesion complex (Dixon et al., 2012; Phillips-Cremins et al., 2013; Sofueva et al., 2013). TADs are proposed to represent regulatory units that coordinate transcriptional regulation, chromatin states, and DNA replication (Beagan and Phillips-Cremins, 2020). Disruption of TAD boundaries is found to correlate with a wide range of diseases. For example, disrupting the TAD structure containing Epha4 enhancers leads to limb defects in human and mice (Lupianez et al., 2015). TAD structures are also characterized in maize (Zea mays), rice (Oryza sativa), and other plant species (Dong et al., 2017, 2018; Liu et al., 2017), although the canonical insulator protein CCCTC-binding factor (CTCF) has not been detected in plants. At a higher resolution, chromatin loops in plants are widely detected between genes and genes, or gene and distal regulatory elements, which are reported to be involved in stress response and organ development (Liu et al., 2016; Peng et al., 2019; Zhao et al., 2019; Sun et al., 2020a, 2020b). In maize, the KRN4 enhancer, located ∼33.1- to 49.9-kb downstream from the UB3 gene, could spatially interact with the UB3 promoter to affect UB3 expression and hence regulate inflorescence development (Du et al., 2020).
Soybean (Glycine max) and common bean (Phaseolus vulgaris) are two of the most important sources of vegetable protein for humans and livestock. These two legumes originated from a common ancestor, which underwent a whole-genome duplication (WGD) event at ∼56.5 million years ago (MYA), and diverged at ∼19.2 MYA (Lavin et al., 2005). After speciation, soybean experienced an independent WGD event at ∼10 MYA (Schmutz et al., 2010). Due to the relatively slow process of diploidization (Kim et al., 2009), most genes are present in multiple copies across the soybean genome (Schmutz et al., 2010; Kim et al., 2015b). Thus, soybean and common bean are commonly used to study genetic and epigenetic effects of polyploidization and diploidization on genome evolution.
In soybean, WGD genes display higher expression and higher CG gene-body methylation level than single-copy genes (Kim et al., 2015b). Cultivated soybean (G. max) was domesticated from wild species (Glycine soja) in east Asia 6,000–9,000 years ago for higher seed yield and wider geographical distribution; these characteristics were accompanied by extraordinary morphological changes including loss of pod shattering, erect and compact stem architecture, and reduction in photoperiod sensitivity (Kim et al., 2012; Sedivy et al., 2017). Quantitative trait locus analysis and genome-wide association study have identified a handful of genes that play important roles in soybean domestication, including SHAT1-5 and GmTfl1 (Liu et al., 2010; Tian et al., 2010; Dong et al., 2014; Zhou et al., 2015). Furthermore, epigenetic regulation is proposed to play an important role in soybean domestication (Shen et al., 2018). Compartments and TAD-like domain structures are reorganized after interspecific hybridization and polyploidization in Arabidopsis thaliana and cotton (Gossypium hirsutum) (Wang et al., 2017; Zhu et al., 2017; Zhang et al., 2019). However, how chromatin architecture reorganization shapes transcriptome patterns during post-polyploid diploidization (G. max versus P. vulgaris) and soybean domestication (G. max versus G. soja) remains largely unknown.
Here we generated high-resolution (∼2 kb) in situ Hi-C maps for G. max, G. soja, and P. vulgaris to uncover the 3D chromatin structure of these legume species. By integrating Hi-C, ChIP-seq, MethylC-seq, RNA-seq, and ATAC-seq data, we revealed relationships between chromatin interaction, epigenetic marks, chromatin accessibilities, and transcriptional abundance during processes of polyploidization and diploidization. We demonstrated that divergence of chromatin interactions may contribute to expression bias of WGD genes in soybean. In addition, chromatin loop reorganization was associated with expression variation of the genes during domestication. The results shed light on unique roles of dynamic chromatin structures in gene expression during evolution and domestication of paleopolyploid soybean.
Results
Conserved Rabl-like organization of chromosomes in legume species
To uncover the roles of 3D chromatin structure in transcription regulation during polyploidization, diploidization, and domestication of legume species, we performed in situ Hi-C experiments with two biological replicates using young leaves of G. max, G. soja, and P. vulgaris. We obtained a total of 857–1,717 million sequencing reads and identified 293–741 million valid interaction pairs for each species, generating 1.5- to 2.2-kb high-resolution (defined as the resolution at which 80% of loci have 1,000 or more contacts with any other locus) in situ Hi-C maps (Supplemental Data Set S1). The high ratio of intra to interchromosomal interactions (>2) indicated high quality of Hi-C data in all species (Supplemental Data Set S1). We also performed ATAC-seq for chromatin accessibilities, ChIP-seq for histone modifications, MethylC-seq for DNA methylation, and RNA-seq for transcriptomes to further dissect relationships between chromatin interaction, chromatin states and gene expression during soybean evolution and domestication (Supplemental Data Set S1).
Consistent with other plant species (Dong et al., 2017), the Hi-C map at the chromosomal level showed intense interactions along the main diagonal and much weaker signals for interchromosomal interactions in all legume species (Figure 1A;Supplemental Figure S1). Strong interaction signals were also detected on the anti-diagonal lines for intrachromosomal interactions, which may reflect the Rabl-like chromosome configuration in which telomeric and subtelomeric regions cluster in the interphase nuclei (Figure 1A;Supplemental Figure S1) (Mascher et al., 2017).
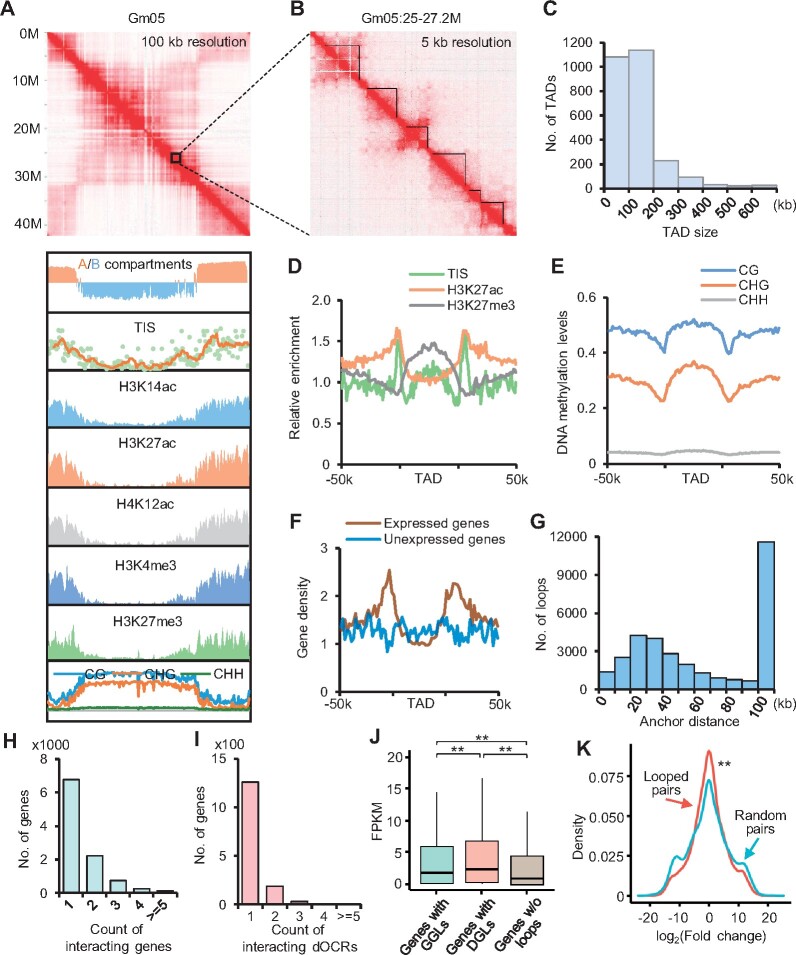
Figure 1.
Global 3D genome organization in soybean. A, Hi-C interaction matrix (100-kb resolution), A/B compartments (100-kb bins), chromatin accessibility and epigenetic marks (200-kb bins) throughout chromosome Gm05. TIS indicates frequency of Tn5 insertion sites. B, Representative TAD structures in chromosome Gm05. Black contours indicate the TADs and boundaries. C, Size distributions of TADs in whole genome. D and E, Chromatin accessibility, histone modifications (D), and DNA methylation (E) around TADs. F, Distributions of expressed (FPKM ≥ 1) and unexpressed (FPKM = 0) genes around TADs. G, Size distributions of intrachromosomal loops. H and I, Count of genes anchored with different numbers of genes (H) and dOCRs (I). J, Expression levels of genes with GGLs, DGLs, or without loops. **Indicates P < 1.2e−5 (Wilcoxon rank-sum test). K, Kernel density of log2(Fold change) for gene pairs linked by a loop or randomly selected. **Indicates P < 3.5e−16 (Kolmogorov–Smirnov test).
Previous studies reported that the genome could be partitioned into A/B compartments in animals and plants (Lieberman-Aiden et al., 2009; Dong et al., 2017; Liu et al., 2017). In legume species, chromatin regions were also segregated into A/B compartments wherein the A compartments were mainly located in chromosome arms with higher chromatin accessibility and histone modifications involved in transcriptional regulation, but the B compartments were located in centromeric and pericentromeric regions showing higher DNA methylation levels (Figure 1A;Supplemental Figure S2; Supplemental Data Set S2). These data indicate conserved 3D-spatial chromatin structure at the chromosomal-level among these legume species.
Long-range chromatin loops contribute to gene co-expression
We next determined higher-order chromatin organization and the effect of chromatin interaction on gene expression in cultivated soybean. We identified 2,621 TADs with median length of 105 kb at 5-kb resolution, representing 35% of the soybean genome (Figure 1, B and C; Supplemental Data Set S3). Boundaries of TADs were marked by higher levels of chromatin accessibility and activated histone modifications including H3K27ac (Figure 1D;Supplemental Figure S3A), but void of repressed epigenetic marks such as H3K27me3 (Figure 1D) and DNA methylation (Figure 1E). Expressed genes were enriched in TAD boundaries (Figure 1F), suggesting open chromatin and active gene expression may be involved in TAD establishment. Moreover, Teosinte branched1/Cycloidea/Proliferating (TCP)-binding motifs were enriched at TAD boundaries, which was also found in other species (Supplemental Figure S3B; E-value < 3.7e-5) (Liu et al., 2017; Karaaslan et al., 2020), implying the roles of TCP proteins in TAD establishment.
At 2-kb resolution, we identified 32,181 intrachromosomal loops with median size of 54 kb (Figure 1G;Supplemental Data Set S4). Approximately 34.7% (11,155/32,181) of intrachromosomal loops had genes at both anchors, which were termed as gene–gene loops (GGLs). A total of 10,133 genes were connected by GGLs, most of which mainly interacted with one locus or two other loci (Figure 1H). To gain insights into effects of distal enhancer-like elements on gene expression, we identified 11,974 distal open chromatin regions (dOCRs) that were >2-kb upstream of transcriptional start site (TSS) and 500-bp downstream of transcriptional termination site (TTS; Supplemental Figure S3C; Supplemental Data Set S5). Compared with adjacent sequences, dOCRs were DNA hypomethylated and did not show enrichment of histone modifications (Supplemental Figure S3, D and E).
We then identified 2,132 intrachromosomal loops with one anchor in dOCRs and another anchor in genes, which were named dOCR-gene (DG) loops (DGLs). Similar to GGLs, most genes mainly interacted with no more than two dOCRs (Figure 1I). The expression levels of the genes with loops, especially those interacted with dOCRs, were significantly higher than the genes without loops (Figure 1J). Accordingly, the genes with loops showed higher levels of chromatin accessibility and active histone modifications but lower DNA methylation levels than the genes without loops (Supplemental Figure S3, F–N).
These chromatin loops may contribute to co-expression of anchored genes. To test this, we assayed expression variation of the genes paired with chromatin loops. Strikingly, gene pairs linked by chromatin loops showed significantly lower expression variation than randomly selected gene pairs (P < 3.5e−16, Kolmogorov–Smirnov test) (Figure 1K), suggesting a role of chromatin loops in gene co-expression.
Interchromosomal interactions enriched in B compartments
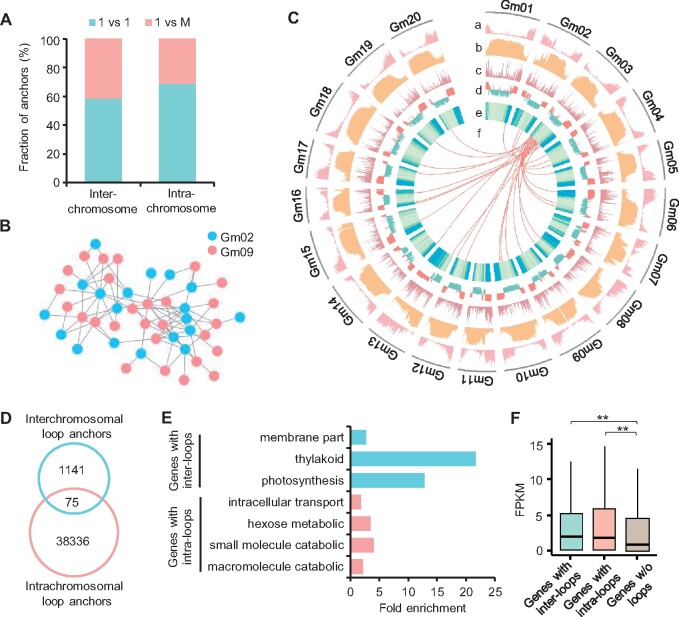
Interchromosomal interactions occurred at much lower frequency than intrachromosomal interactions (Supplemental Figure S1). To explore potential roles of interchromosomal interactions, a total of 1,589 interchromosomal loops were identified using strict criteria (see “Methods” section) (Supplemental Data Set S6), of which 26 (1.6%) interchromosomal loops linked whole-genome duplicated block pairs. Furthermore, only two WGD gene pairs were linked by interchromosomal loops, suggesting interchromosomal interactions mainly occurred in noncollinear regions/genes.
Compared to intrachromosomal loop anchors (∼1.7 loops per anchor), interchromosomal anchors tended to interact with more loci (∼2.6 loops per anchor) (Figure 2A) (P < 2e−65, Fisher’s exact test). Interestingly, we identified 96 interchromosomal interaction networks consisting of at least 3 loop anchors (Supplemental Figure S4A). For example, anchors in interchromosomal loops between Gm02 and Gm09 could be linked with each other to establish an interaction network (Figure 2B). In contrast to the enrichment of intrachromosomal loops in the A compartments, interchromosomal loops were frequently located in the B compartments (Figure 2C;Supplemental Figure S4B). Accordingly, 69% of intrachromosomal loops overlapped with genes, but only 40% of interchromosomal loops linked genes. Consistent with the above notion, few loci were shared between inter and intrachromosomal loop anchors (Figure 2D).
Figure 2.
Characters of interchromosomal contacts in soybean. A, Fraction of anchors showing “1 versus 1” or “1 versus M” in intra and interchromosomal loops. “1 versus 1” indicates a locus only interacted with another locus, whereas “1 versus M” indicates a locus interacted with multiple loci (≥2). B, A representative interchromosomal contact network between Gm02 and Gm09. C, Circle plot of H3K27ac (a), CG methylation (b), density of intrachromosomal loop anchors (c), A/B compartments (d), gene density (e), and interchromosomal loops between Gm03 and other chromosomes (f). D, Venn diagram showing the overlap between inter and intrachromosomal loop anchors. E, Significantly enriched GO terms of genes with interchromosomal loops (inter-loops) or intrachromosomal loops (intra-loops). F, Expression levels of genes with inter-loops, intra-loops, and without (w/o) loops. **Indicates P < 0.01 (Wilcoxon rank-sum test).
Gene ontology (GO) analysis showed that the genes linked by interchromosomal loops were enriched in photosynthesis-related terms, whereas the genes with intrachromosomal loops were overrepresented in macromolecule catabolic, small molecular catabolic, and hexose metabolic processes (Figure 2E). Similar with the genes linked by intrachromosomal loops, the genes with interchromosomal loops displayed significantly higher expression levels than genes without loops (P < 0.01, Wilcoxon rank-sum test) (Figure 2F), indicating that both inter and intrachromosomal loops were positively associated with gene expression.
Chromatin interactions involved in expression bias of WGD genes
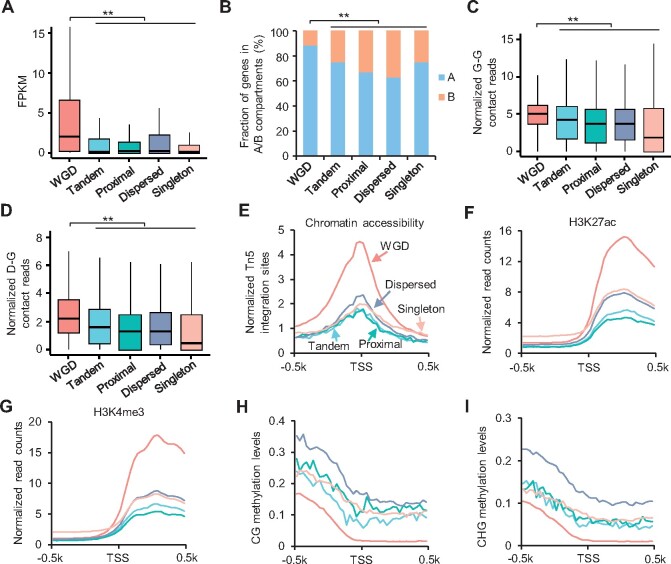
Due to the relatively slow process of diploidization after the WGD ∼10 MYA, nearly 75% of genes are present in multiple copies in the soybean genome (Schmutz et al., 2010). To uncover the role of chromatin interactions in expression divergence of genes during diploidization, we split genes into WGD genes, small-scale duplicated genes (tandem, proximal, and dispersed duplicated genes), and single-copy genes (Kim et al., 2015b). We found that WGD genes showed higher expression levels than small-scale duplicated genes and single-copy genes (Figure 3A), as observed previously (Kim et al., 2015b). Consistent with previous studies that higher frequency of retention of WGD genes occurs in chromosomal arms than pericentromeric regions (Du et al., 2012; Zhao et al., 2017), WGD genes were highly enriched in the A compartments (Figure 3B). Interestingly, WGD genes contained more gene–gene (G–G) and D–G contacts than other types of genes (Figure 3, C and D; Supplemental Figure S5, A and B), showing higher levels of chromatin accessibilities and activated histone modifications but depleted of DNA methylation around TSSs (Figure 3, E–I; Supplemental Figure S5, C–F). By comparison, no obvious epigenetic changes except DNA methylation around TTSs were observed in WGD genes compared with small-scale duplicated and single-copy genes (Supplemental Figure S5, G–O).
Figure 3.
More chromatin interactions in WGD genes than small-scale duplicate and single-copy genes. A, Expression levels of WGD, tandem, proximal, dispersed, and singleton genes. **Indicates P < 8e−100 (Wilcoxon rank-sum test). B, Percentage of WGD, small-scale duplicate, and single-copy genes in A/B compartments. **Indicates P < 2e−45 (Fisher’s exact test). C and D, Normalized G–G (C) and D-G (D) contacts of WGD, small-scale duplicate and single-copy genes. Y-axis indicates normalized contacts normalized by ICE method and gene length. **Indicates P < 2.5e−27 (Wilcoxon rank-sum test). E–I, Distributions of chromatin accessibility (E), H3K27ac (F), H3K4me3 (G), and DNA methylation (H, I) around TSSs of WGD, small-scale duplicate and single-copy genes. Different types of genes were indicated by different colors of lines.
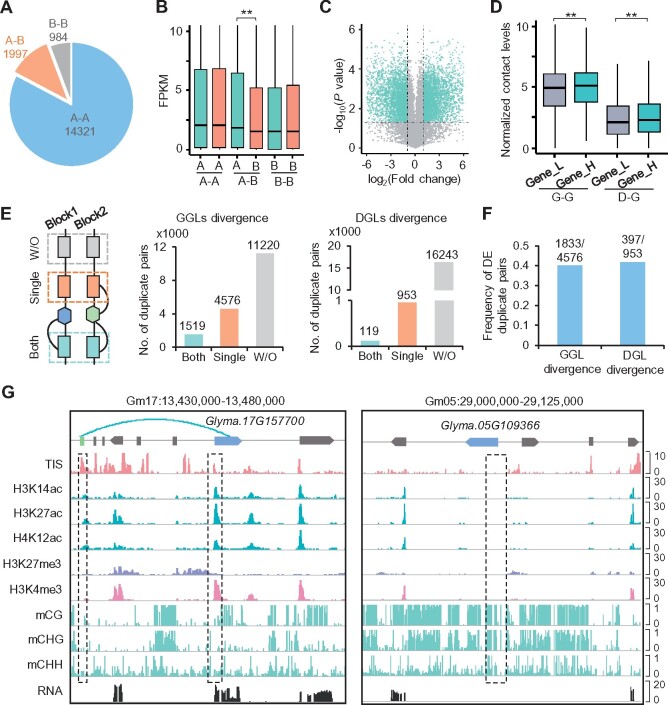
In total, 11.5% (1,997/17,302) of WGD gene pairs have two homoeologous genes located in different types of A/B compartments (Figure 4A). Across these WGD gene pairs, genes in compartment A showed significantly higher expression levels than genes in compartment B (Figure 4B). Whereas, no significant expression bias was observed in WGD genes located in the same type of A/B compartments (Figure 4B). We further identified 7,031 WGD gene pairs showing significantly differential expression levels between two homoeologs (Figure 4C). Compared with genes with lower expression levels in WGD gene pairs, higher-expressed genes were coupled with more G–G and D–G contacts as well as higher levels of chromatin accessibilities and activated histone modifications, but lower DNA methylation levels (Figure 4D;Supplemental Figure S6). In contrast, the WGD gene pairs that were not differentially expressed showed similar levels of chromatin contacts, chromatin accessibilities, and epigenetic modifications between two homoeologs (Supplemental Figure S7).
Figure 4.
Chromatin interactions contribute to expression bias of WGD genes. A, Number of WGD genes with or without A/B compartment divergence. A–A or B–B, respectively, indicates both WGD genes were located in A or B compartments. A–B indicates WGD genes were located in different types of compartments. B, Expression levels of WGD genes with or without A/B compartment divergence. **Indicates P < 0.01 (Wilcoxon rank-sum test). C, Volcano plot showing differentially expressed WGD genes. Dots in baby blue indicate differentially expressed WGD genes with P < 0.05 and abs (log2(Fold change)) ≥1. D, Normalized G–G or D–G contact levels in differentially expressed WGD genes. Gene_H and gene_L indicate genes with higher expression levels or lower expression levels in each pair, respectively. Y-axis indicates contacts normalized by ICE method and gene length. **Indicates P < 9e−06 (Wilcoxon rank-sum test). E, Divergence of intrachromosomal GGLs and DGLs between WGD genes. Both, both WGD genes with loops; Single, only one of WGD genes with loops; W/O, both WGD genes without loops. F, Frequency of differentially expressed (DE) duplicate pairs in WGD genes with GGL or DGL divergence. G, Representative IGV snapshots showing biased expression of WGD genes with divergence of D–G interaction, chromatin accessibility, histone modifications, and DNA methylation. Rectangle blocks indicate dOCRs and arrowed blocks indicate genes.
In addition, we identified 4,576 and 953 WGD gene pairs wherein only one homoeolog overlapped with GGLs or DGLs, respectively (Figure 4E). Notably, ∼40% of them showed expression bias between the two homoeologs (Figure 4F). For example, Glyma.17G157700 with DGLs showed higher levels of chromatin accessibilities, active histone marks, and expression, but lower DNA methylation levels than the homoeologous gene Glyma.05G109366 without loops (Figure 4G). These results suggest that chromatin interactions may integrate histone modifications and DNA methylation to maintain expression divergence between retained WGD genes.
Chromatin architecture alteration during polyploidization and diploidization
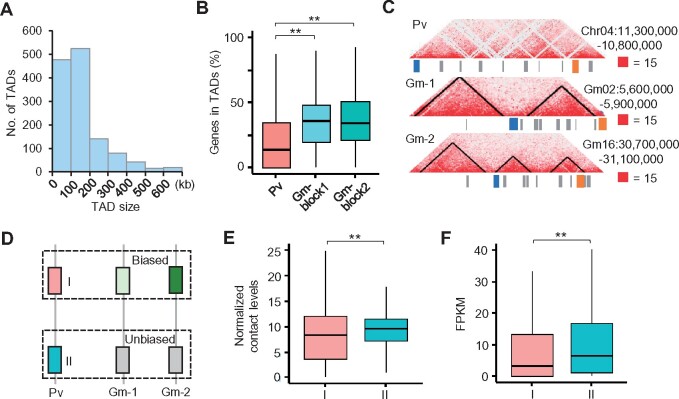
Since diverging from common bean, soybean underwent specific polyploidy and diploidization (Lavin et al., 2005). Through comparing chromatin architecture of soybean and common bean, we examined 3D genome structure rearrangement during polyploidization and diploidization. We first identified 1,304 TADs in the common bean genome at 5-kb resolution (Figure 5A;Supplemental Data Set S3), of which ∼13% (170/1304) are orthologous TADs in the soybean genome. In the collinearity blocks between one copy in common bean and two homologous copies in soybean, more genes were located in TADs in soybean than in common bean (P < 0.001, Wilcoxon rank-sum test) (Figure 5B). However, we cannot confidently exclude the possibility that TAD divergence between common bean and soybean is derived from quality discrepancy of different Hi-C libraries. Based on the differences in retention rates of genes (Zhao et al., 2017), soybean duplicated block pairs were divided into block1 (higher retention rate) and block2 (lower retention rate). The proportions of the genes located in TADs did not show significant difference between the two soybean homoeologous blocks (Figure 5B).
Figure 5.
Chromatin conformation changes between soybean and common bean. A, Size distributions of TADs in common bean (5-kb resolution). B, TAD divergence in syntenic blocks between P. vulgaris (Pv) and G. max (Gm) (5-kb resolution). **Indicates P < 2e−10 (Wilcoxon rank-sum test). C, An example showing TAD divergence of orthologous genes between P. vulgaris (one copy, Pv) and G. max (two copies, Gm-1 and Gm-2). Blue and orange boxes indicate two orthologous pairs (Phvul.004G071400-Glyma.02G064800-Glyma.16G145900; Phvul.004G069600-Glyma.02G066700-Glyma.16G147800). Hi-C contact matrixes were plotted at 2-kb resolution. Black lines indicate the TADs and boundaries. D, An schematic plot showing genes in P. vulgaris whose orthologous pairs in soybean have biased (group I) or unbiased (Group II) interactions. E and F, Normalized contact levels (E) and expression levels (F) of genes in Groups I and II. **Indicates P < 5e−20 (Wilcoxon rank-sum test).
We then identified 12,129 homologous genes with one copy in common bean and two copies in soybean. Remarkably, 5,711 (47%) genes in the common bean genome were void of TADs, but at least one of their homologs was present in TADs in soybean (Supplemental Figure S8A; Supplemental Data Set S7). On the other hand, only 1,527 (13%) genes were found in TADs in common beans and at least one copy of their homologs was apart from TADs in soybean (Supplemental Figure S8A; Supplemental Data Set S7). For example, Phvul.004G071400 and Phvul.004G069600 were apart from TADs in common bean, but in soybean one of their homologs maintained the ancestral state and another copy was located in TADs in soybean (Figure 5C). These results suggest that polyploidization may induce changes in TAD architecture. Notably, similar TAD divergences of the collinearity blocks and orthologous genes were also found from common bean to soybean at 10-kb resolution (Supplemental Figure S8, B and C).
To test if chromatin status of ancestral gene determines chromatin interaction divergence of homeologous genes after polyploidy, we classified genes into groups I and II in common bean, in which soybean homologs of genes in group I showed significant divergence of G–G and D–G chromatin interactions, and soybean homologs of genes in group II had similar intensity of chromatin interactions (Figure 5D). Interestingly, genes in group II had significantly more chromatin contacts and higher expression levels than genes in group I in common bean (Figure 5, E and F), indicating relatively inactive genes tend to exhibit chromosomal interaction differentiation after polyploidy.
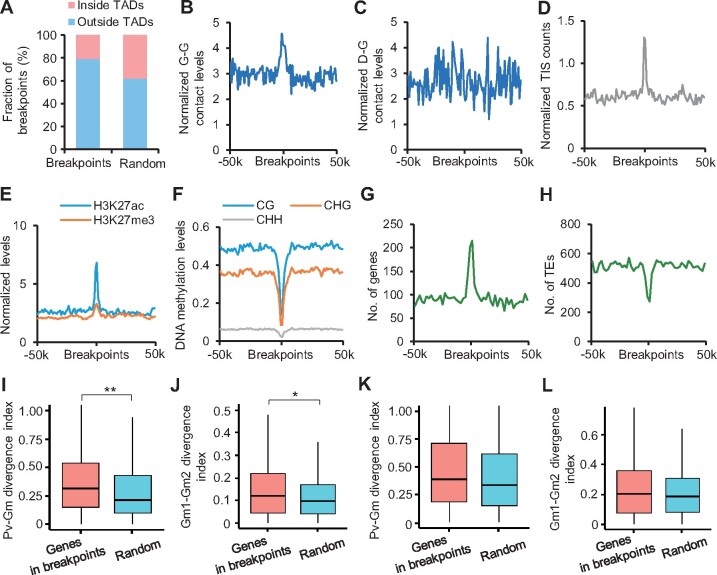
During post-polyploid genome diploidization, chromosomes are fractured and rearranged (Schmutz et al., 2014; Zhao et al., 2017). We identified 600 potential chromosomal breakpoints by comparing genome sequences of soybean and common bean (Supplemental Data Set S8). We found that chromosomal breakpoints were void of TADs (Figure 6A;Supplemental Figure S8D) and had higher G–G contacts (Figure 6B), but were not associated with D–G contacts (Figure 6C). The breakpoints also displayed higher levels of chromatin accessibility (Figure 6D), H3K27ac and H3K27me3 (Figure 6E), but lower CG, CHG, and CHH methylation levels (Figure 6F). Meanwhile, genes were highly enriched in breakpoints (Figure 6G), whereas transposable elements were excluded (Figure 6H).
Figure 6.
Association between chromatin conformation and genome reconstructions during diploidization. A, Percentage of breakpoints inside or outside TADs (5-kb resolution). Randomly selected regions were used as control. B and C, Distributions of normalized G–G (B) and D–G (C) contacts around breakpoints. Y-axis indicates contacts normalized by ICE method and bin length. D–F, Levels of chromatin accessibility (D), histone modifications (E), and DNA methylation (F) around breakpoints. G and H, Distributions of genes (G) and TEs (H) around breakpoints. I and K P. vulgaris versus G. max (Pv–Gm) divergence index of G–G (I) and D–G (K) contacts in genes overlapping with breakpoints or random genes. Pv–Gm divergence index = abs(Gm1+Gm2–Pv)/Pv. Gm1 and Gm2 indicate duplicate pairs in G. max. *Indicates P < 0.05 and **indicates P < 4e−05 (Wilcoxon rank-sum test). J and L, Gm1–Gm2 divergence index of G–G (J) and D–G (L) contacts between WGD genes overlapping with breakpoints or random genes. Gm1–Gm2 divergence index = abs(Gm1−Gm2)/(Gm1+Gm2).
Next, we explored the impacts of chromosomal reconstructions on chromatin contacts of flanking regions. Interestingly, genes in breakpoints showed significant G–G contact changes from common bean to soybean (Figure 6I), and two homoeologous copies in soybean were also significantly divergent (Figure 6J). A similar trend was also found for D–G contacts (Figure 6, K and L), although it did not reach statistical significance. Taken together, these results indicated dramatic rearrangement of 3D chromatin architecture during polyploidization and diploidization.
Chromatin reorganization was involved in morphological changes during soybean domestication
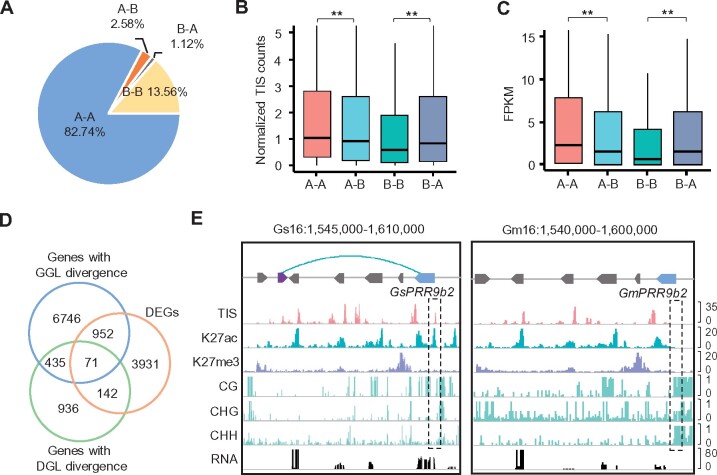
To gain insights into how 3D genome organization shapes gene expression during soybean domestication, we compared chromatin interaction maps between cultivated soybean (G. max) and its wild relative (G. soja). In total, 43,861 orthologous genes were identified between wild and cultivated soybean. The majority of them were conserved in the A–A (82.7%) and B–B (13.6%) compartments (Figure 7A), and only ∼3.7% showed A/B compartment shifts. The genes (2.6%) with A to B shift from wild species to cultivar showed lower chromatin accessibilities (Figure 7B) and expression levels (Figure 7C) than the genes conserved in the A compartments in wild soybean. Opposite results were observed for the genes with B to A shift compared with the genes conserved in B compartments (Figure 7, B and C).
Figure 7.
3D genome changes during domestication of soybean. A, A/B compartment conversions of orthologous genes between G. soja (Gs) and G. max (Gm). A–B or B–A indicates the compartment conversions from A to B or B to A for Gm compared with Gs. A–A or B–B indicate same compartment types between Gs and Gm. B and C, Chromatin accessibility (B) and expression levels (C) of genes in A–A, A–B, B–A, and B–B compartments in G. soja. **Indicates P < 7e−4 (Wilcoxon rank-sum test). D, Venn diagram showing the overlap among genes with GGL divergence, genes with DGL divergence and DEGs between Gs and Gm. DEGs were identified by P < 0.01 (ANOVA test) and log2(Fold change) ≥1. E, Chromatin contacts, epigenetic states, and expression levels of PRR9b2 in G. soja and G. max. Blue and gray arrowed blocks indicate orthologous genes. A purple arrowed block indicates a gene present in G. soja but absent in G. max.
We further identified 8,204 GGL and 1,584 DGL divergent genes, wherein loops only exclusively detected in wild or cultivated soybean. Differentially expressed orthologous genes between wild and cultivated soybean were significantly enriched in genes with divergent GGLs and DGLs (P < 0.05, Hypergeometric test) (Figure 7D;Supplemental Data Set S9). Interestingly, differentially expressed genes (DEGs) with GGL not DGL divergence showed significant changes of chromatin accessibility, H3K27ac, H3K27me3, CG, and CHG methylation between wild and cultivated soybean (Supplemental Figure S9), indicating epigenetic modifications also contribute to expression changes of genes with GGL divergence during domestication. For example, the pseudo-response regulator (PRR) gene PRR9b2 showed loop divergence between wild and cultivated soybean (Figure 7E) (Wang et al., 2020). GsPRR9b2 (Glysoja.16G042449) but not its homologous gene GmPRR9b2 (Glyma.16G018000) formed loops with a gene whose homolog was lost in the cultivated soybean, showing higher levels of chromatin accessibilities and H3k27ac but depleted of DNA methylation in the wild soybean (Figure 7E). As a result, PRR9b2 had higher levels of gene expression in the wild soybean (Figure 7E). These data suggest a potential role of chromatin contact divergence in expression variation of the genes during domestication.
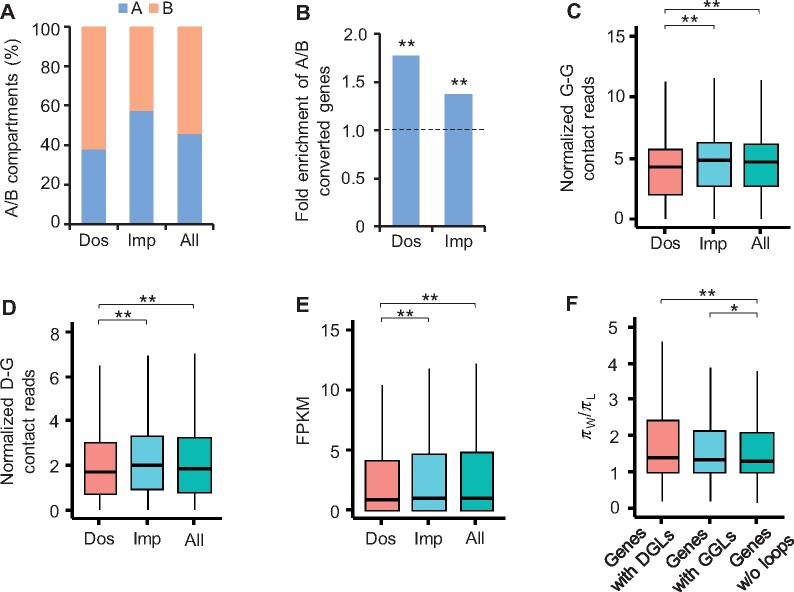
Based on published resequencing data for wild soybean and cultivated soybean including landraces and cultivars (Zhou et al., 2015), we found that domestication (from wild to landrace) selective sweeps tended to occur in the B compartments, whereas improvement (from landrace to cultivar) selective sweeps tended to coincide in the A compartments (P < 2.2e−16, Fisher’s exact test) (Figure 8A). The genes with A/B compartment shifts were significantly enriched in both domestication and improvement selective sweeps (Figure 8B). We further found that the genes coincident in domestication sweeps contained significantly fewer G–G (Figure 8C) and D–G (Figure 8D) contacts compared with genes in improvement sweeps and whole genome (P < 6e−6, Wilcoxon rank-sum test). Meanwhile, those genes in domestication selective sweeps showed lower expression (Figure 8E). It is worth noting that the genes with GGLs and DGLs showed stronger selective signals in the processes of domestication than the genes without loops (Figure 8F), indicating a selection preference for loop-coupled genes during domestication.
Figure 8.
Genes with chromatin loops were under stronger selection. A, Distributions of A/B compartments in domestication (Dos) and improvement (Imp) selective sweeps. Whole genome (All) was used as control. B, Fold enrichment of genes with A/B compartment conversions between wild and cultivated soybeans in domestication and improvement selective sweeps relative to all genes. **Indicates P < 8e−6 (hypergeometric test). C and D, Normalized G–G (C) and D–G (D) contacts of genes in domestication and improvement selective sweeps compared with all genes in soybean. Y-axis indicates contacts normalized by ICE method and gene length. **Indicates P < 6e−6 (Wilcoxon rank-sum test). E, FPKM of genes in domestication and improvement selective sweeps compared with all genes. **Indicates P < 5e−3 (Wilcoxon rank-sum test). F, πW/πL values of genes with DGLs, GGLs, or without (w/o) loops. πW,π values in wild soybean; πL,π values in landrace. *Indicates P < 0.05 and **Indicates P < 1e−3 (Wilcoxon rank-sum test).
Discussion
Polyploidy is a pervasive evolutionary feature of all angiosperms (Song and Chen, 2015). Although rapid genomic changes can occur in some plant polyploids such as resynthesized Brassica napus and Tragopogon miscellus (Xiong et al., 2011; Chester et al., 2012), many polyploids such as Arabidopsis and cotton are genetically stable and show conserved gene content and synteny (Wang et al., 2006; Chen et al., 2020). This suggests a mechanism to maintain genomic stability during polyploid evolution. Using high-resolution (∼2 kb) in situ Hi-C maps for G. max, G. soja, and P. vulgaris, we established comprehensive chromatin architecture maps at domain- and gene levels during polyploid formation and diversification (G. max versus P. vulgaris) and soybean domestication (G. max versus G. soja). Although the soybean genome experienced dramatic chromosomal rearrangements during diploidization (Schmutz et al., 2010), the majority (88.5%) of WGD genes have two homoeologous genes located in the same type of A/B compartments (Figure 4A), indicating that local A/B compartment partitions are relatively stable once established. The A/B compartments and TADs are highly associated with epigenetic modifications and chromatin accessibility, which may help to maintain chromatin architecture during polyploidization and subsequent diploidization. Moreover, these retained WGD genes are associated with more long-range chromosomal interactions and higher levels of active histone marks and gene expression but void of DNA methylation, compared with single-copy and small-scale duplicated genes (Figure 3). These data suggest a coordinated role of chromatin interactions and epigenetic modifications in maintaining transcriptional stability of WGD genes and likely the overall subgenomic stability in polyploids. It has been reported that expression divergence between duplicate genes is related to biased levels of H3K27me3 and gene-body methylation (Berke et al., 2012; Wang et al., 2015b). We found that chromatin contacts including GGLs and DGLs are also implicated in the expression divergence of WGD genes (Figure 4, D–F), although the cause and consequence of these changes need to be further investigated.
Chromatin architecture at the TAD level is often conserved in mammals (Dixon et al., 2012). However, TADs are not so conserved in various plant species, including maize, tomato (Solanum lycopersicum), sorghum (Sorghum bicolor), foxtail millet (Setaria italica), and rice (Dong et al., 2017). We found obvious TAD variation in the collinearity blocks between soybean and common bean (Figure 5), and more orthologous genes are located in TADs in soybean than in common bean. Soybean underwent a lineage-specific WGD after divergence with common bean (Lavin et al., 2005). These data suggest that polyploidy and subsequent diploidization may facilitate formation and distribution of TAD structure.
As WGD is more common in plants than in animals (Van de Peer et al., 2017), the higher divergence of TADs across plants may be partly due to polyploidy. We found that chromosomal rearrangements are overrepresented in the regions with higher levels of chromatin contacts, DNA accessibility, and active histone modifications (Figure 6). Consistent with this notion, chromosomal rearrangements tend to take place outside of TADs (Figure 6A), as lower levels of DNA accessibility and less active histone modifications were observed in the interior of TADs compared with TAD boundaries (Figure 1, D and E). These results suggest that the chromatin status of TADs may contribute to maintaining TAD integrity during diploidization.
Cultivated soybean is derived from wild species, accompanied by important agronomic traits changes, like reduced seed dormancy, loss of seed shattering (Sedivy et al., 2017). We observed stronger artificial selection for chromatin looped genes than the genes without loops (Figure 8F). In addition, we demonstrated that chromatin loops including GGLs and DGLs were involved in expression changes during soybean domestication (Figure 7). One PRR gene, PRR9b2, showed decreased expression levels in cultivated soybean, concurrent with reduction or removal of long-range chromatin contacts with another gene (Figure 7E). Functional analysis of other genes with loop divergence between wild and cultivated soybeans could help us to understand the roles of chromatin conformation remodeling in morphological changes during domestication.
In general, euchromatic regions (A compartments) have much higher recombination rates than heterochromatic regions (B compartments) (Li et al., 2014). Genes in recombination-rich regions tend to generate novel allelic combinations that could be exploited by breeding programs. Interestingly, we found that domestication selective sweeps tended to occur in the B compartments, whereas improvement selective sweeps tended to coincide in the A compartments (Figure 8A). This result suggests genes in A compartments are widely utilized during modern improvement and genes in B compartments were prone to be exploited during early domestication. Further utilization of genes in B compartments by genomic technologies or by increasing recombination rates would enhance modern soybean breeding. Collectively, we provided comprehensive 3D chromatin conformation maps of cultivated soybean, wild soybean, and common bean, and revealed the role of genome topology in the evolution and domestication of paleopolyploid soybean.
Materials and methods
Plant materials and growth conditions
Seeds of P. vulgaris (G18842), G. soja (W05), and G. max (Williams 82) were planted in growth room with 28°C/25°C (day/night) and 12-h light/12-h dark (while fluorescent light; 200 μmol m−2 s−1 light intensity) cycles. Two independent pools of leaves at 28 days after sowing as two biological replicates were collected for library construction of Hi-C, ATAC-seq, ChIP-seq, MethylC-seq, and RNA-seq.
Hi-C library construction
Hi-C libraries were constructed according to the published protocol with modifications (Liu, 2017). About 0.5 g leaves for each replicate were harvested and immediately crosslinked in 1% formaldehyde buffer (10 mM potassium phosphate, 50 mM NaCl, 0.1 M sucrose, and 1% formaldehyde) for 30 min. Glycine buffer (10 mM potassium phosphate, 50 mM NaCl, 0.1 M sucrose, and 0.15 M glycine) was added to quench the reaction. Crosslinked leaves were rinsed thrice by deionized water and immediately frozen in liquid nitrogen. Leaves were ground into fine powder and transferred to nuclei isolation buffer [40% glycerol, 0.25 M sucrose, 20 mM HEPES, 1 mM MgCl2, 5 mM KCl, 0.25% TritonX-100, 0.1 mM PMSF, 1× Protease Inhibitor Cocktail (Roche Basel, Switzerland), and 0.1% 2-mercaptoethanol]. After mixing thoroughly, the slurry was kept on ice for 30 min and filtered by a 70 μm strainer. After centrifugation (3,000g for 5 min), the nuclei pellet was resuspended in 200 μL of 0.5% sodium dodecyl sulfate (SDS). Then the suspension was split into four tubes and incubated at 62°C for 5 min. After adding 145 μL water and 25 μL 10% Triton X-100, each tube was incubated at 37°C for 15 min to quench the SDS. The chromatin was digested by adding 25 μL 10× NEBuffer 3 and 50 U DpnII (NEB, Ipswich, MA, USA; R0543) at 37°C overnight. On the following day, after incubating at 62°C for 20 min to inactivate DpnII, the restriction fragments were filled by adding tagging buffer (10 μL 1 mM biotin-14-dCTP, 1 μL 10 mM dATP, 1 μL 10 mM dGTP, 1 μL 10 mM dTTP, 40 U Klenow (NEB, Ipswich, MA, USA; M0210), and 25 μL ddH2O) and incubated at 22°C for 4 h. Next, filled fragments were proximally ligated by adding ligation buffer (663 μL water, 120 μL 10× T4 DNA ligase buffer, 100 μL 10% Triton X-100 and 2000 U T4 DNA Ligase (NEB, Ipswich, MA, USA; M0202)) and kept at 22°C for 4 h. The mixture was centrifuged at 1,000 g for 3 min, and the pellet was resuspended with 500 μL nuclei lysis buffer (50 mM Tris–HCl, 1% SDS, 10 mM EDTA). Proteinase K (10 μL) was added and the tubes were kept at 55°C for 30 min. Crosslinking was reversed at 65°C overnight after adding 50 μL of 5 M NaCl. DNA was purified using QIAquick PCR Purification Kit (Qiagen, Hilden, Germany ) and sheared to 300–500 bp by Covaris M220 (Covaris, Woburn, MA, USA). Ligated fragments were pulled down by Dynabeads MyOne Streptavidin T1 beads (Invitrogen, Carlsbad, CA, USA; Cat No. 65601). Libraries were constructed using NEBNext Ultra II DNA Library Prep Kit (NEB, Ipswich, MA, USA; E7645L), and sequenced on NovaSeq platform (Illumina, San Diego, CA, USA) for 150-bp paired-end reads.
ATAC-seq library construction
Construction of ATAC-seq libraries was performed as previously described with modifications (Bajic et al., 2018). Approximately 0.1 g leaves per replicate were crosslinked in 35 mL cross-linking buffer (0.4 M sucrose, 10 mM Tris–HCl, 1 mM PMSF, 1 mM EDTA, and 1% formaldehyde) for 20 min, which were stopped by adding 2.2 mL of 2 M glycine. After washed thrice, crosslinked leaves were immediately frozen in liquid nitrogen and ground into fine powder. Nuclei was isolated by adding the powder into 10 mL nuclei extraction buffer 1 (20 mM MOPS, 40 mM NaCl, 9 0mM KCl, 2 mM EDTA, 0.5 mM EGTA, 0.5 mM spermidine, 0.2 mM spermine, and 1× Cocktail). The mixture was filtered by 70 μm strainer and washed by 1 mL nuclei extraction buffer 2 (0.25 M Sucrose, 10 mM Tris–HCl, 10 mM MgCl2, 1% Triton X-100, and 1× Cocktail). Following resuspended by 300 μL nuclei extraction buffer 3 (1.7 M Sucrose, 10 mM Tris–HCl, 2 mM MgCl2, and 0.15% Triton X-100 and 1× Cocktail), the nuclei suspension was loaded on the surface of 600 μL nuclei extraction buffer 3 and centrifuged at 16,000g at 4°C for 10 min. The purified nuclei were resuspended by 1 mL nuclei extraction buffer 1, stained with 4,6-diamidino-2-phenylindole and loaded into a hemocytometer to calculate nuclei counts. About 50,000 nuclei were incubated with Tn5 transposase (Vazyme, Nanjing, China; TD501) at 37°C for 30 min. Next, crosslink reversal was performed at 65°C overnight using solution (50 mM Tris–HCl, 1 mM EDTA, 1% SDS, 0.2 M NaCl and 5 ng/mL proteinase K). DNA was recovered by MinElute PCR Purification Kit (Qiagen, Hilden, Germany) and amplified for 11–13 cycles. In addition, genomic DNA was used for library construction as input control. Libraries were sequenced on NovaSeq platform (Illumina, San Diego, CA, USA) for 150-bp paired-end reads.
ChIP-seq library construction
Chromatin immunoprecipitation was performed following a general protocol (Yamaguchi et al., 2014). Briefly, ∼0.5 g leaves for each replicate were crosslinked as described in the ATAC-seq protocol. After nuclei were isolated and lysed, chromatin was sonicated to 300–500 bp. Pre-equilibrated Protein A/G MagBeads (GenScript, Piscataway, New Jersey, USA) were added and rotated for 2 h at 4°C to remove background noise. Nearly 2.5 μg antibodies against H3K14ac (Abcam, Cambridge, UK; ab52946), H3K27ac (Abcam; ab4729), H3K27me3 (Abcam; ab6002), H3K4me3 (Abcam; ab8580), and H4K12ac (Cell signaling, Danvers, MA, USA; 13944) were added to the solution and rotated at 4°C for overnight. Immunoprecipitated chromatin was pull down by Protein A/G MagBeads and crosslink was reversed by adding 5 M NaCl at 65°C for overnight. Libraries were constructed by NEBNext Ultra II DNA Library Prep Kit (NEB, Ipswich, MA, USA; E7645L) and sequenced on NovaSeq platform (Illumina, San Diego, CA, USA) for 150-bp paired-end reads.
MethylC-seq library construction
MethylC-seq was performed as described previously (Song et al., 2015). Genomic DNA was isolated by CTAB method (Allen et al., 2006) and sonicated into 300–500 bp. End repair, dA-tailing, and methylated adapter ligation were performed using NEBNext Ultra II DNA Library Prep Kit (NEB, Ipswich, MA, USA), followed by bisulfite conversion using EZ DNA Methylation-Gold kit (ZYMO research, Irvine, CA, USA). The bisulfite DNA was amplified by 15 cycles, and purified by VAHTSTM DNA Clean Beads (Vazyme, Nanjing, China). Libraries were sequenced on NovaSeq platform (Illumina, San Diego, CA, USA) for 150-bp paired-end reads.
mRNA-seq library construction
Total RNA was extracted by TRIzol reagent (Invitrogen, Carlsbad, California, USA). Strand-specific mRNA-seq libraries were constructed using NEBNext Ultra Directional RNA Library Prep Kit (NEB, Ipswich, MA, USA) and sequenced on NovaSeq platform (Illumina, San Diego, CA, USA) for 150-bp paired-end reads.
Hi-C data processing
After removing adaptors and low-quality reads by TrimGalore (version 0.6.4), clean reads were aligned to reference genome (William82 genome version 4 for cultivated soybean, W05 genome for wild soybean, and G19983 genome version 2 for common bean) using HiC-Pro (version 2.11.1) with default parameters (Servant et al., 2015). Low-quality mapped reads (mapping quality (MAPQ) < 20) and duplication were discarded. After removing self-circle, dangling-end, religation, and dumped reads, raw contacts were normalized by iterative correction and eigenvector decomposition (ICE) method to minimize the biases due to PCR amplification, mappability, and distance-related interaction decay at 2-kb resolution. The normalized contact matrices were used for contact comparison between different groups. To visualize Hi-C map, valid pairs between two highly correlated replicates were merged to build contact matrix by Juicer tools (version 1.13.02) (Durand et al., 2016a) with KR balancing method. Hi-C resolution was calculated as previous definition (Rao et al., 2014). Contact maps were visualized by Juicebox (version 1.11.08) (Durand et al., 2016b).
A/B compartments were identified by “Eigenvector” of Juicer tools with 100-kb resolution. TADs were called by “Arrowhead” of Juicer tools with 5- and 10-kb resolutions. FitHiC2 (version 2.0.7) (Kaul et al., 2020) was used to identify intra (q-value <0.05, contact reads ≥6) and interchromosomal (q-value <0.05, contact reads ≥ 10) loops with 2-kb resolution with parameters “-L 4000 -U 2000000.” Loops were plotted by Sushi (version 1.24.0) (Phanstiel et al., 2014), an R package for visualizing genome data.
To compare loops and TADs divergence between different species, reads were down-sampled to same level of sequencing depth to reduce the effects of sequencing depth variation.
ATAC- and ChIP-seq processing
Adaptors and low-quality reads were removed by TrimGalore (version 0.6.4). Clean data were mapped onto reference genome by Bowtie2 (version 2.2.9) (Langmead and Salzberg, 2012) with default parameters. Duplications were removed by Picard (version 2.17.6) and concordantly mapped reads with MAPQ ≥ 20 were used for further analysis. Bam files were converted to bed formats using BEDTools (version 2.26.0) to calculate reads and Tn5 insertion site distributions around specific regions, including TSSs of genes. BigWig files were generated by BamCoverage (version 3.3.0) with 1× average coverage in deepTools (Ramírez et al., 2016) and visualized by IGV (version 2.3.32).
For dOCR identification, reads of ATAC-seq and input library were down-sampled to same level by picard (version 2.17.6). Peaks were called by MACS2 (version 2.1.2) (Zhang et al., 2008) with parameters “–nomodel –shift -100 –extsize 200 –broad –keep-dup all -q 0.01.” Peaks which were excluded from 2-kb upstream of TSSs, gene body, and 500-bp downstream of transcriptional terminated sites were considered as dOCRs.
MethylC-seq and mRNA-seq data processing
After trimming low-quality reads, MethylC-seq reads were mapped to reference genome by Bismark (version 0.15.1) (Krueger and Andrews, 2011) with default parameters. Only reads mapped to the unique sites were retained, and the reads aligned to the same site were collapsed into a single consensus read to reduce clonal bias. Weighted methylation levels were used for further analysis.
Clean mRNA-seq data were mapped by HISAT2 (version 2.1.0) (Kim et al., 2015a) with the parameter of “–dta –rna-strandness RF.” Uniquely mapped reads were maintained. StringTie (Pertea et al., 2015) was used to assemble the potential transcripts and calculate FPKM of genes with parameters “-B -A –rf.”
Identification of WGD genes in soybean
Protein sequences in Williams82 genome (version 4) were all-by-all blasted against themselves with the E-value <1e−10. Hits with identity >50 and gene coverage >50% were retained. MCScanX (Wang et al., 2012) was used to detect collinear blocks and syntenic gene pairs with default parameters. Whole-genome duplicate, tandem, proximal, dispersed, and singleton genes were identified by the “duplicate_ gene_ classifier” tools in MCScanX.
Identification of collinear blocks/syntenic gene pairs between common bean and soybean
Protein sequences of G19983 genome were all-by-all blasted against protein sequences of Williams82 genome and collinear blocks or syntenic gene pairs were detected using MCScanX. Due to the specific WGD in soybean, synteny blocks with only one copy in common bean and two copies in soybean were retained for further analysis. Then 1:2 orthologous genes (one copy in common bean and two copies in soybean) were identified by following three criteria: (1) the genes between common bean and soybean were orthologous; (2) two copies in soybean were WGD genes; and (3) these genes located in colinear blocks.
Identification of orthologous TADs between common bean and soybean
A pair of orthologous TADs was defined by following two criteria as described (Dong et al., 2017). (1) The TAD in common bean contains at least five P. vulgaris–G.max orthologous genes. (2) More than 50% of orthologous genes in the TAD of common beans are also located in a TAD of soybean.
Identification of breakpoints during diploidization
First, collinear blocks between common beans and soybean were identified as described above. Second, transcriptional start or terminated sites of genes in boundaries of blocks were extracted as endpoints of blocks. Third, 5-kb upstream and downstream sequences of endpoints were considered as potential breakpoints during diploidization.
Motif analysis
OCRs overlapping 10-kb flanking sequences of TAD boundaries were used to detect motifs by MEME software (version 5.0.3) (Bailey et al., 2009) with the parameters “-meme-minw 6 -meme-maxw 10 -dreme-e 0.05.” An equal amount of OCRs was randomly selected as control.
Construction of chromatin interaction network
Chromatin interaction networks were constructed as previously described (Li et al., 2019). Interchromosomal interaction networks were constructed by igraph package in R software using all interchromosomal loops in cultivated soybean. Chromatin interaction networks with at least three chromatin interactions were kept. The nodes in chromatin interaction networks present loop anchors in chromatin interactions. Each edge in chromatin interaction networks presents corresponding chromatin interaction. Cytoscape software was used to visualize networks (Shannon et al., 2003).
Identifications of domestication/improvement selective sweeps
For identification of domestication and improvement sweeps, π values were calculated by vcftools (version 0.1.16) (Danecek et al., 2011) with 100-kb windows and 10-kb step using published resequencing data (Zhou et al., 2015). The regions with top 5% of π ratios (πwild/πLandrace for domestication and πLandrace/πCultivar for improvement) were considered as threshold of domestication or improvement sweeps (Wang et al., 2017).
Accession numbers
The data reported in this article have been deposited in NCBI Sequence Read Archive (SRA) under Accession Number PRJNA657728.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genome-wide Hi-C interaction maps of cultivated soybean, wild soybean, and common bean.
Supplemental Figure S2. Chromatin interaction maps, A/B compartments, and epigenetic marks throughout chromosome 02 of wild soybean, and chromosome 03 of common bean.
Supplemental Figure S3. Epigenetic features of TAD boundaries, distal OCRs, and genes with or without loops in soybean.
Supplemental Figure S4. Characters of interchromosomal interactions.
Supplemental Figure S5. Chromatin states around TSSs and TTSs of WGD, small-scale duplicate and single-copy genes.
Supplemental Figure S6. Distributions of chromatin accessibility, histone modifications, and DNA methylation around TSSs of differentially expressed WGD genes.
Supplemental Figure S7. Chromatin interactions and epigenetic states of WGD genes that are not differentially expressed.
Supplemental Figure S8. TAD divergence between soybean and common bean.
Supplemental Figure S9. Epigenetic changes of DEGs between G. soja (Gs) and G. max (Gm) with loop divergence during domestication.
Supplemental Data Set S1. Summary of sequence data.
Supplemental Data Set S2. A and B compartments in cultivated soybean, wild soybean, and common bean.
Supplemental Data Set S3. TADs in cultivated soybean, wild soybean, and common bean.
Supplemental Data Set S4. Intrachromosomal loops in wild and cultivated soybeans.
Supplemental Data Set S5. Distal OCRs in wild soybean, cultivated soybean, and common bean.
Supplemental Data Set S6. Interchromosomal loops in cultivated soybean.
Supplemental Data Set S7. TAD divergence of orthologous genes between soybean and common bean.
Supplemental Data Set S8. Breakpoints in common beans during diploidization.
Supplemental Data Set S9. Loop divergent genes and DEGs between wild and cultivated soybeans.
Supplementary Material
Acknowledgments
We thank Dr Hon-Ming Lam for providing W05 seeds and Bioinformatics Center at Nanjing Agricultural University for data analysis.
Funding
This work was supported by Jiangsu Specially-Appointed Professor Program to Q.S. Z.J.C. is the D. J. Sibley Centennial Professor of Plant Molecular Genetics.
Conflict of interest statement. The authors declare that they have no competing interest.
Q.S. conceived and designed the research. L.W., G.J., and S.C. performed the experiments. L.W., X.J., and Q.S. analyzed the data. L.W., Z.J.C., and Q.S. wrote the manuscript. All authors read and approved the final manuscript.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Qingxin Song (qxsong@njau.edu.cn).
REFERENCES
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF (2006) A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protocols 1: 2320–2325 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME Suite: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic M, Maher KA, Deal RB (2018) Identification of open chromatin regions in plant genomes using ATAC-Seq. Methods Mol Biol 1675: 183–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagan JA, Phillips-Cremins JE (2020) On the existence and functionality of topologically associating domains. Nat Genet 52: 8–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke L, Sanchez-Perez GF, Snel B (2012) Contribution of the epigenetic mark H3K27me3 to functional divergence after whole genome duplication in Arabidopsis. Genome Biol 13: R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Sreedasyam A, Ando A, Song Q, De Santiago LM, Hulse-Kemp AM, Ding M, Ye W, Kirkbride RC, Jenkins J. et al. (2020) Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat Genet 52: 525–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester M, Gallagher JP, Symonds VV, Cruz da Silva AV, Mavrodiev EV, Leitch AR, Soltis PS, Soltis DE (2012) Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc Natl Acad Sci USA 109: 1176–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. (2011) The variant call format and VCFtools. Bioinformatics 27: 2156–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P, Tu X, Chu PY, Lu P, Zhu N, Grierson D, Du B, Li P, Zhong S (2017) 3D chromatin architecture of large plant genomes determined by local A/B compartments. Mol Plant 10: 1497–1509 [DOI] [PubMed] [Google Scholar]
- Dong Q, Li N, Li X, Yuan Z, Xie D, Wang X, Li J, Yu Y, Wang J, Ding B, et al. (2018) Genome-wide Hi-C analysis reveals extensive hierarchical chromatin interactions in rice. Plant J 94: 1141–1156 [DOI] [PubMed] [Google Scholar]
- Dong Y, Yang X, Liu J, Wang BH, Liu BL, Wang YZ (2014) Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat Commun 5: 3352. [DOI] [PubMed] [Google Scholar]
- Du J, Tian Z, Sui Y, Zhao M, Song Q, Cannon SB, Cregan P, Ma J (2012) Pericentromeric effects shape the patterns of divergence, retention, and expression of duplicated genes in the paleopolyploid soybean. Plant Cell 24: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Liu L, Peng Y, Li M, Li Y, Liu D, Li X, Zhang Z (2020) UNBRANCHED3 expression and inflorescence development is mediated by UNBRANCHED2 and the distal enhancer, KRN4, in maize. PLoS Genet 16: e1008764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SS, Huntley MH, Lander ES, Aiden EL (2016a) Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst 3: 95–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, Aiden EL (2016b) Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst 3: 99–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Schubert V, Zhai J, Pellegrini M, Jacobsen SE (2014) Genome-wide Hi-C analyses in wild-type and mutants reveal high-resolution chromatin interactions in Arabidopsis. Mol Cell 55: 694–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH. et al. (2009) An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462: 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob S, Grossniklaus U (2019) Invasive DNA elements modify the nuclear architecture of their insertion site by KNOT-linked silencing in Arabidopsis thaliana. Genome Biol 20: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaaslan ES, Wang N, Faiss N, Liang Y, Montgomery SA, Laubinger S, Berendzen KW, Berger F, Breuninger H, Liu C (2020) Marchantia TCP transcription factor activity correlates with three-dimensional chromatin structure. Nat Plants 6: 1250–1261 [DOI] [PubMed] [Google Scholar]
- Kaul A, Bhattacharyya S, Ay F (2020) Identifying statistically significant chromatin contacts from Hi-C data with FitHiC2. Nat Protocol 15: 991–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015a) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KD, Shin JH, Van K, Kim DH, Lee SH (2009) Dynamic rearrangements determine genome organization and useful traits in soybean. Plant Physiol 151: 1066–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KD, El Baidouri M, Abernathy B, Iwata-Otsubo A, Chavarro C, Gonzales M, Libault M, Grimwood J, Jackson SA (2015b) A comparative epigenomic analysis of polyploidy-derived genes in soybean and common bean. Plant Physiol 168: 1433–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Van K, Kang YJ, Kim KH, Lee SH (2012) Tracing soybean domestication history: from nucleotide to genome. Breed Sci 61: 445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF (2005) Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol 54: 575–594 [DOI] [PubMed] [Google Scholar]
- Li E, Liu H, Huang L, Zhang X, Dong X, Song W, Zhao H, Lai J (2019) Long-range interactions between proximal and distal regulatory regions in maize. Nat Commun 10: 2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Liu YL, Reif JC, Liu ZX, Liu B, Mette MF, Chang RZ, Qiu LJ (2014) Biparental resequencing coupled with SNP genotyping of a segregating population offers insights into the landscape of recombination and fixed genomic regions in elite soybean. G3 (Bethesda) 4: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Watanabe S, Uchiyama T, Kong F, Kanazawa A, Xia Z, Nagamatsu A, Arai M, Yamada T, Kitamura K, et al. (2010) The soybean stem growth habit gene Dt1 is an ortholog of Arabidopsis TERMINAL FLOWER1. Plant Physiol 153: 198–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C (2017) In situ hi-C library preparation for plants to study their three-dimensional chromatin interactions on a genome-wide scale. Methods Mol Biol 1629: 155–166 [DOI] [PubMed] [Google Scholar]
- Liu C, Cheng YJ, Wang JW, Weigel D (2017) Prominent topologically associated domains differentiate global chromatin packing in rice from Arabidopsis. Nat Plants 3: 742–748 [DOI] [PubMed] [Google Scholar]
- Liu C, Wang C, Wang G, Becker C, Zaidem M, Weigel D (2016) Genome-wide analysis of chromatin packing in Arabidopsis thaliana at single-gene resolution. Genome Res 26: 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R. et al. (2015) Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161: 1012–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J. et al. (2017) A chromosome conformation capture ordered sequence of the barley genome. Nature 544: 427–433 [DOI] [PubMed] [Google Scholar]
- Peng Y, Xiong D, Zhao L, Ouyang W, Wang S, Sun J, Zhang Q, Guan P, Xie L, Li W, et al. (2019) Chromatin interaction maps reveal genetic regulation for quantitative traits in maize. Nat Commun 10: 2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phanstiel DH, Boyle AP, Araya CL, Snyder MP (2014) Sushi.R: flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics 30: 2808–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, et al. (2013) Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153: 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T (2016) deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, McClean PE, Mamidi S, Wu GA, Cannon SB, Grimwood J, Jenkins J, Shu S, Song Q, Chavarro C. et al. (2014) A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet 46: 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J. et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Sedivy EJ, Wu F, Hanzawa Y (2017) Soybean domestication: the origin, genetic architecture and molecular bases. New Phytol 214: 539–553 [DOI] [PubMed] [Google Scholar]
- Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, Heard E, Dekker J, Barillot E (2015) HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 16: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Zhang J, Liu Y, Liu S, Liu Z, Duan Z, Wang Z, Zhu B, Guo YL, Tian Z (2018) DNA methylation footprints during soybean domestication and improvement. Genome Biol 19: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofueva S, Yaffe E, Chan WC, Georgopoulou D, Vietri Rudan M, Mira-Bontenbal H, Pollard SM, Schroth GP, Tanay A, Hadjur S (2013) Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J 32: 3119–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Chen ZJ (2015) Epigenetic and developmental regulation in plant polyploids. Curr Opin Plant Biol 24: 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Guan X, Chen ZJ (2015) Dynamic roles for small RNAs and DNA methylation during ovule and fiber development in allotetraploid cotton. PLoS Genet 11: e1005724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotelo-Silveira M, Chavez Montes RA, Sotelo-Silveira JR, Marsch-Martinez N, de Folter S (2018) Entering the next dimension: plant genomes in 3D. Trends Plant Sci 23: 598–612 [DOI] [PubMed] [Google Scholar]
- Sun L, Jing Y, Liu X, Li Q, Xue Z, Cheng Z, Wang D, He H, Qian W (2020a) Heat stress-induced transposon activation correlates with 3D chromatin organization rearrangement in Arabidopsis. Nat Commun 11: 1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Dong L, Zhang Y, Lin D, Xu W, Ke C, Han L, Deng L, Li G, Jackson D. et al. (2020b) 3D genome architecture coordinates trans and cis regulation of differentially expressed ear and tassel genes in maize. Genome Biol 21: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z, Wang X, Lee R, Li Y, Specht JE, Nelson RL, McClean PE, Qiu L, Ma J (2010) Artificial selection for determinate growth habit in soybean. Proc Natl Acad Sci USA 107: 8563–8568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Mizrachi E, Marchal K (2017) The evolutionary significance of polyploidy. Nat Rev Genet 18: 411–424 [DOI] [PubMed] [Google Scholar]
- Wang C, Liu C, Roqueiro D, Grimm D, Schwab R, Becker C, Lanz C, Weigel D (2015a) Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res 25: 246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Beyene G, Zhai J, Feng S, Fahlgren N, Taylor NJ, Bart R, Carrington JC, Jacobsen SE, Ausin I (2015b) CG gene body DNA methylation changes and evolution of duplicated genes in cassava. Proc Natl Acad Sci USA 112: 13729–13734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tian L, Lee HS, Wei NE, Jiang H, Watson B, Madlung A, Osborn TC, Doerge RW, Comai L, et al. (2006) Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Tu L, Lin M, Lin Z, Wang P, Yang Q, Ye Z, Shen C, Li J, Zhang L, et al. (2017) Asymmetric subgenome selection and cis-regulatory divergence during cotton domestication. Nat Genet 49: 579–587 [DOI] [PubMed] [Google Scholar]
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, et al. (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yuan L, Su T, Wang Q, Gao Y, Zhang S, Jia Q, Yu G, Fu Y, Cheng Q, et al. (2020) Light- and temperature-entrainable circadian clock in soybean development. Plant Cell Environ 43: 637–648 [DOI] [PubMed] [Google Scholar]
- Xiong Z, Gaeta RT, Pires JC (2011) Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc Natl Acad Sci USA 108: 7908–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Winter CM, Wu MF, Kwon CS, William DA, Wagner D (2014) PROTOCOLS: chromatin immunoprecipitation from Arabidopsis tissues. Arabidopsis Book 12: e0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zheng R, Wang Y, Zhang Y, Hong P, Fang Y, Li G, Fang Y (2019) The effects of Arabidopsis genome duplication on the chromatin organization and transcriptional regulation. Nucleic Acids Res 47: 7857–7869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Wang S, Cao Z, Ouyang W, Zhang Q, Xie L, Zheng R, Guo M, Ma M, Hu Z, et al. (2019) Chromatin loops associated with active genes and heterochromatin shape rice genome architecture for transcriptional regulation. Nat Commun 10: 3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Zhang B, Lisch D, Ma J (2017) Patterns and consequences of subgenome differentiation provide insights into the nature of paleopolyploidy in plants. Plant Cell 29: 2974–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Xie W (2019) The role of 3D genome organization in development and cell differentiation. Nat Rev Mol Cell Biol 20: 535–550 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Jiang Y, Wang Z, Gou Z, Lyu J, Li W, Yu Y, Shu L, Zhao Y, Ma Y,. et al. (2015) Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol 33: 408–414 [DOI] [PubMed] [Google Scholar]
- Zhu W, Hu B, Becker C, Dogan ES, Berendzen KW, Weigel D, Liu C (2017) Altered chromatin compaction and histone methylation drive non-additive gene expression in an interspecific Arabidopsis hybrid. Genome Biol 18: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.