Abstract
The plant phenylpropanoid pathway generates a major class of specialized metabolites and precursors of essential extracellular polymers that initially appeared upon plant terrestrialization. Despite its evolutionary significance, little is known about the complexity and function of this major metabolic pathway in extant bryophytes, which represent the non-vascular stage of embryophyte evolution. Here, we report that the HYDROXYCINNAMOYL-CoA:SHIKIMATE HYDROXYCINNAMOYL TRANSFERASE (HCT) gene, which plays a critical function in the phenylpropanoid pathway during seed plant development, is functionally conserved in Physcomitrium patens (Physcomitrella), in the moss lineage of bryophytes. Phylogenetic analysis indicates that bona fide HCT function emerged in the progenitor of embryophytes. In vitro enzyme assays, moss phenolic pathway reconstitution in yeast and in planta gene inactivation coupled to targeted metabolic profiling, collectively indicate that P. patens HCT (PpHCT), similar to tracheophyte HCT orthologs, uses shikimate as a native acyl acceptor to produce a p-coumaroyl-5-O-shikimate intermediate. Phenotypic and metabolic analyses of loss-of-function mutants show that PpHCT is necessary for the production of caffeate derivatives, including previously reported caffeoyl-threonate esters, and for the formation of an intact cuticle. Deep conservation of HCT function in embryophytes is further suggested by the ability of HCT genes from P. patens and the liverwort Marchantia polymorpha to complement an Arabidopsis thaliana CRISPR/Cas9 hct mutant, and by the presence of phenolic esters of shikimate in representative species of the three bryophyte lineages.
The catalytic properties and physiological function of a key enzyme in the phenylpropanoid pathway were conserved across the 500 million years of embryophyte evolution.
Introduction
Land colonization by plants, about 500 million years ago (Wickett et al., 2014; Puttick et al., 2018; Morris et al., 2018), was one of the most important evolutionary events associated with terraformation. Through photosynthetic activity and rock weathering, early land plants contributed to the rise of atmospheric oxygen, carbon sequestration, and the development of soils (Retallack, 1997; Lenton et al., 2016; Porada et al., 2016). Plant settlement on land therefore paved the way for the development of rich terrestrial ecosystems and the emergence of new life forms (Kenrick and Crane, 1997).
This transition from water to land exposed plants to challenging terrestrial conditions, such as drought, harmful levels of solar (UV) radiation, lack of buoyancy, extended temperature range, and novel pathogenic microorganisms (Rensing et al., 2008; de Vries and Archibald, 2018). Successful land colonization thus required specific developmental and metabolic adaptations (Reski, 2018). The formation of extracellular, or apoplastic, protective barriers was probably one of the most critical innovations of land plants, as they shield cells from damaging environmental conditions and allow the formation of specialized structures required for water and nutrient management (e.g. cuticles and vasculature). In angiosperms, such structures are essentially comprised of four canonical hydrophobic biopolymers—cutin, suberin, sporopollenin, and lignin—that reinforce and waterproof the polysaccharide-based cell wall (Nawrath et al., 2013).
Some precursors of these polymers are generated through the phenylpropanoid pathway, one of the most important branches of so-called plant specialized metabolism, which also allows the accumulation of powerful UV screens and antioxidants (Vogt, 2010; Weng and Chapple, 2010; Renault et al., 2019). The ability to synthesize phenylpropanoids evolved during the course of terrestrialization and is often regarded as a key adaptation by plants to life on land (Weng and Chapple, 2010; de Vries et al., 2017; Renault et al., 2019). The most common products generated by the phenylpropanoid pathway—flavonoids, soluble phenolic esters, and biopolymer precursors—all derive from p-coumaroyl-CoA (Figure 1, A). This hub molecule is produced through the activities of three essential enzymes in the initial steps of the phenylpropanoid pathway: phenylalanine ammonia-lyase; cinnamate 4-hydroxylase (C4H), which belongs to cytochrome P450 family 73 (CYP73); and 4-coumarate:CoA ligase (4CL; Figure 1, A). In flowering plants, further functionalization of the phenolic ring requires shikimate ester intermediates and a two-enzyme module involving hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase (HCT), which catalyzes transfer of the p-coumaroyl moiety from p-coumaroyl-CoA to shikimate (Hoffmann et al., 2003, 2004), and a second cytochrome P450, p-coumaroyl-shikimate 3′-hydroxylase (C3′H or CYP98), to generate caffeoyl-shikimate (Schoch et al., 2001; Franke et al., 2002; Alber et al., 2019; Figure 1, A). HCT was shown to transfer the caffeoyl moiety back to coenzyme A to form caffeoyl-CoA in vitro (Hoffmann et al., 2003; Vanholme et al., 2013). Later studies reported the existence of a second route toward caffeoyl-CoA in flowering plants, involving the combined action of caffeoyl shikimate esterase (CSE) and 4CL (Vanholme et al., 2013; Saleme et al., 2017; Ha et al., 2016; Figure 1, A). Recently, a bifunctional p-coumarate 3-hydroxylase/ascorbate peroxidase (C3H/APX) was also characterized, revealing a metabolic shunt for phenolic ring 3-hydroxylation, directly from free p-coumaric acid (Barros et al., 2019; Figure 1, A).
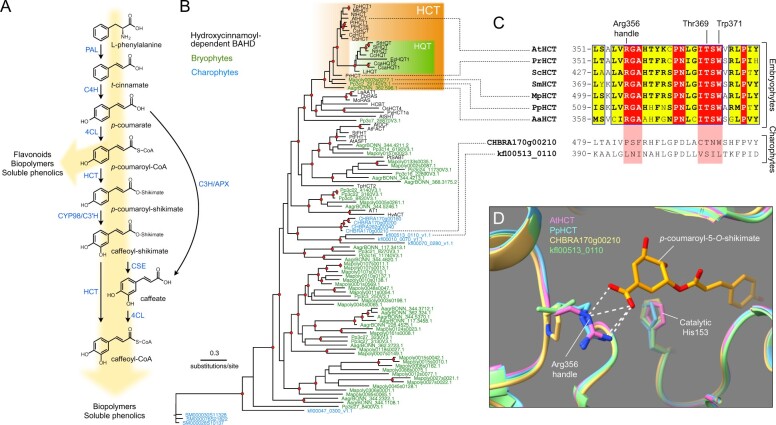
Figure 1.
Evolutionary history of the HCT gene family. A, Schematic representation of the phenylpropanoid pathway of angiosperms leading to caffeoyl-CoA. Enzyme names are indicated in blue. PAL, phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate:CoA ligase; HCT, hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase; C3′H, p-coumaroyl ester 3′hydroxylase; CSE, caffeoyl-shikimate esterase; C3H/APX, p-coumarate 3-hydroxylase/ascorbate peroxidase. B, Unrooted protein tree describing the phylogenetic relationships between 34 hydroxycinnamoyl-CoA-dependent BAHD acyltransferases of known biochemical function and all BAHDs from K. nitens, C. braunii, S. muscicola, P. patens, M. polymorpha, and A. agrestis. The tree is drawn to scale. Red dots indicate a maximum-likelihood ratio test branch support ≧ 0.80. Lists of characterized and uncharacterized BAHD proteins are available in Supplemental Tables S1, S2, respectively. C, Multiple sequence alignment highlighting the region comprising the three residues controlling shikimate acylation in selected HCT orthologs from the five major embryophyte groups. Corresponding regions from charophyte HCT homologs are displayed at the bottom for comparison. ScHCT, Salvinia cucullata Sacu_v1.1_s0010.g004618; SmHCT, Selaginella moellendorffii 152997. Red box/white character, strict identity; yellow box/black bold character, in-group similarity score >0.7; blue frame/yellow box, cross-group similarity score >0.7. D, Overlay of protein three-dimensional structures depicting the Arg356 handle interaction with shikimate. Models of P. patens (PpHCT, Pp3c2_29140, blue), C. braunii (CHBRA170g00210, yellow), and K. nitens (kfl00513_0110, green) HCT homologs were reconstructed using the crystal structure of AtHCT in complex with p-coumaroyl-5-O-shikimate (PDB entry: 5kju; pink). Residues are numbered according to AtHCT. White dashed lines represent predicted hydrogen bonds
HCT belongs to clade V of the BAHD acyltransferase superfamily, which features enzymes that use coenzyme A-activated acyl donors and chemically diverse acceptors, such as organic acids, amines, or fatty acids (D’Auria, 2006). Clade V also includes the closely related enzymes hydroxycinnamoyl-CoA:quinate hydroxycinnamoyl transferases (HQT), which use quinate as a preferred acceptor, rather than shikimate. HQT is involved in the production of chlorogenic acids (caffeoyl-quinates), which are widespread among angiosperms, though absent from Arabidopsis thaliana (Niggeweg et al., 2004; Guo et al., 2014). Unlike caffeoyl-shikimate, caffeoyl-quinate is not considered to be a key intermediate in lignin biosynthesis, but rather involved in responses to biotic and abiotic stressors, especially UV radiation (Niggeweg et al., 2004; Clé et al., 2008). An investigation of HCT catalytic properties revealed broad acceptor substrate permissiveness, extending beyond shikimate (Hoffmann et al., 2003, 2004; Sander and Petersen, 2011; Eudes et al., 2016). However, structural studies of HCT/HQT uncovered key amino acid residues that control shikimate and/or quinate acylation, thereby specifying the two types of enzymes (Lallemand et al., 2012; Levsh et al., 2016; Chiang et al., 2018). HCT represents a pivotal step in controlling lignin biosynthesis and composition, as demonstrated by HCT silencing studies in seed plants that consistently alter lignin content and/or composition, and often lead to adverse effects on growth (Hoffmann et al., 2004; Besseau et al., 2007; Wagner et al., 2007; Gallego-Giraldo et al., 2011).
Bryophytes and charophyte algae, the embryophyte sister group, are devoid of lignin, although some of them seem to possess parts of the genetic toolkit required to synthesize phenolic intermediates (de Vries et al., 2017; Renault et al., 2019; Jiao et al., 2020). The nature and role of such early phenylpropanoid derivatives are poorly documented. We recently showed through a molecular genetic approach, targeting the Physcomitrium patens (Physcomitrella) CYP98 gene, that a moss phenylpropanoid pathway is involved in the synthesis of caffeate precursors necessary to support cuticular biopolymer formation and erect (3D) growth (Renault et al., 2017). The major acylated products formed by the moss were shown to be threonate esters (p-coumaroyl-threonate and caffeoyl-threonate), while shikimate and quinate esters were not detected (Renault et al., 2017). However, a survey of embryophyte CYP98 substrate preference in vitro showed that the moss enzyme poorly converts p-coumaroyl-threonates, compared with p-coumaroyl-shikimate (Alber et al., 2019), leaving the nature of the native pathway intermediates in the moss unclear. Here, we sought to address this question by performing a functional analysis of a candidate P. patens HCT gene, which encodes the enzyme generating the CYP98 substrate. Combining in silico, in vitro, and in vivo analyses, we demonstrate conservation of HCT catalytic properties and physiological function across the 500 million years of embryophyte evolution.
Results
A bona fide HCT gene emerged in an embryophyte progenitor and was subsequently conserved
We performed a search for potential HCT genes in fully sequenced charophyte and bryophyte genomes. All BAHD acyltransferase protein sequences from the charophytes Klebsormidium nitens (Klebsormidiophyceae), Chara braunii (Characeae), and Spirogloea muscicola, (Zygnematophyceae) and from the bryophytes P. patens (moss), Marchantia polymorpha (liverwort), and Anthoceros agrestis (hornwort) were aligned with 34 already characterized hydroxycinnamoyl-CoA-dependent BAHD transferases prior to phylogeny reconstruction (complete lists in Supplemental Tables S1, S2). The resulting protein tree structure revealed a well-supported HCT clade with single members for each bryophyte species at its root (Figure 1, B). The angiosperm-specific HQT proteins clustered as a sister group to angiosperm HCTs, suggesting they originated from HCT duplication (Figure 1, B). BAHD enzymes from the charophytes C. braunii and K. nitens were not closely associated with HCTs, but rather clustered as a sister group to characterized hydroxycinnamoyl-CoA-dependent BAHD. Proteins from the Zygnematophyceae S. muscicola were found to be even more divergent from characterized HCT proteins (Figure 1, B andSupplemental Data Set S1). Multiple protein alignment revealed a strict conservation in representative embryophyte HCTs of the three residues (Arg356, Thr369, and Trp371) previously shown to be critical for HCT activity (Lallemand et al., 2012; Levsh et al., 2016; Chiang et al., 2018), whereas their conservation was only partial in charophyte homologs (Figure 1, C andSupplemental Figure S1). More particularly, the Arg356 handle was absent from charophyte BAHDs (Figure 1, C andSupplemental Figure S1). Finer details were gained through homology-modelling of HCT candidate proteins from P. patens (PpHCT, Pp3c2_29140), C. braunii (CHBRA170g00210), and K. nitens (kfl00513_0110) using the crystal structure of A. thaliana AtHCT in complex with p-coumaroyl-5-O-shikimate as a template. The predicted protein structures indicated that, similar to AtHCT, PpHCT binds the shikimate carboxyl group through an arginine handle (Figure 1, D). In charophyte proteins, the critical arginine residue was replaced by proline (CHBRA170g00210) or leucine (kfl00513_0110), neither of which is predicted to form hydrogen bonds with shikimate (Figure 1, D). Taken together, these data point to the emergence of bona fide HCT genes in the last common ancestor of embryophytes about 500 Ma, concurrently with the appearance of cuticles (Philippe et al., 2020) and prior to the capacity to produce the phenolic biopolymer lignin.
PpHCT is co-expressed with the p-coumaroyl ester 3′-hydroxylase PpCYP98
We then undertook a functional study of the P. patens HCT candidate gene (PpHCT, Pp3c2_29140) identified in the phylogenetic analysis. First, we used publicly available co-expression data derived from the P. patens gene atlas project (Perroud et al., 2018). This indicated that the expression profile of PpHCT is tightly correlated with those of two genes encoding enzymes flanking the HCT step, potentially forming a core enzymatic module in the moss phenolic pathway: 4-coumarate-CoA ligase 1 (Pp4CL1; Pp3c18_6360; Silber et al., 2008) and the functionally characterized PpCYP98 (Pp3c22_19010; Renault et al., 2017; Figures 1, A and 2, A). These data were supported by our RT-qPCR analysis, which showed higher (at least four-fold) expression levels of all three genes in gametophores than in protonema (Figure 2, B). To increase the spatial resolution of the PpHCT expression analyses, we generated knock-in PpHCT:uidA reporter lines by homologous recombination (Supplemental Figure S2). GUS staining was restricted to the apex of both young and old gametophores (Figure 2, C), which is very similar to the previously reported PpCYP98:uidA GUS staining profile (Renault et al., 2017).
Figure 2.
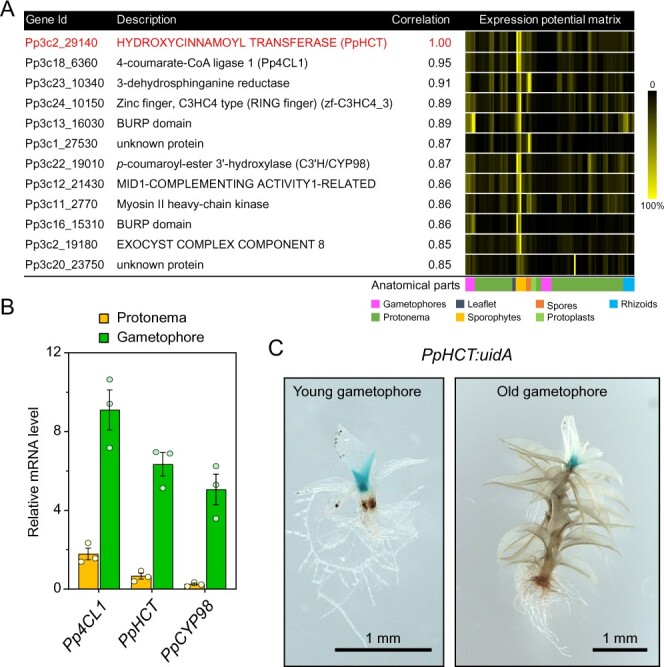
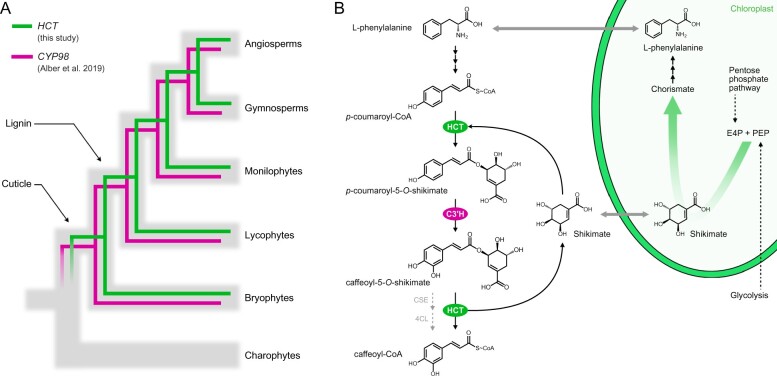
PpHCT co-expression network and expression pattern. A, List of genes co-expressed with PpHCT (Pp3c2_29140). Each element of the matrix represents the expression potential (% of maximum expression across the matrix) of a given gene (line) in a defined condition (column) derived from various anatomical parts. Data are retrieved from the P. patens gene atlas project (Perroud et al., 2018). B, RT-qPCR monitoring of Pp4CL1, PpHCT, and PpCYP98 expression in protonema and gametophore tissues. Data are the mean ± SEM of three biological replicates derived from three independent cultures. C, GUS staining pattern in PpHCT:uidA lines indicating prominent expression in the apex of gametophores.
PpHCT demonstrates substrate permissiveness in vitro
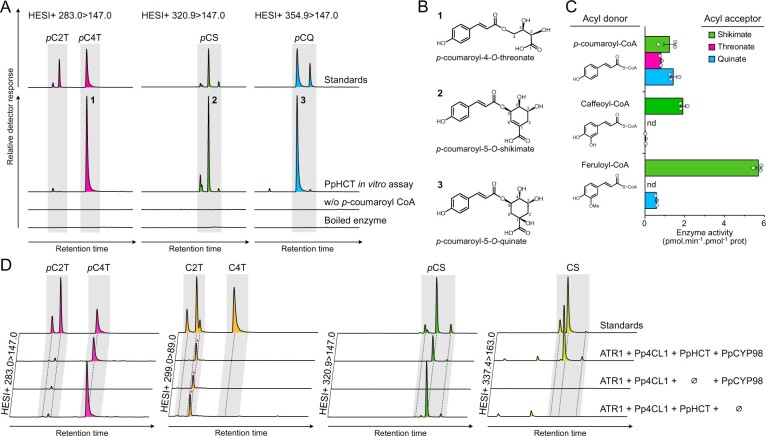
Previous data suggested that phenolic esters of threonic acid are the most likely intermediates in the P. patens phenylpropanoid pathway (Renault et al., 2017). Accordingly, we hypothesized that PpHCT generates p-coumaroyl-threonate from p-coumaroyl-CoA and l-threonic acid, and we tested this with in vitro assays using recombinant PpHCT expressed in Escherichia coli. We observed that PpHCT produced mainly p-coumaroyl-4-O-threonate and a minor amount of p-coumaroyl-2-O-threonate in vitro, indicative of substantial regiospecificity (Figure 3, A and B). We then tested shikimic acid and quinic acids as acyl acceptors, since they are native and accepted substrates of tracheophyte HCTs, respectively (Hoffmann et al., 2003; Chiang et al., 2018). PpHCT catalyzed the transfer of p-coumarate from p-coumaroyl-CoA to both of them (Figure 3, A). A strong regiospecificity favored the 5-position for shikimate and quinate acylation (Figure 3, A and B). We then investigated PpHCT acyl-CoA donor preference using end-point enzyme assays, testing all pairwise combinations of the donors p-coumaroyl-CoA, caffeoyl-CoA, and feruloyl-CoA with the acceptors threonate, shikimate, and quinate. As shown in Figure 3, C, PpHCT used all three acyl donors when shikimate was the acceptor, and the highest activity was observed with the combination of shikimate and feruloyl-CoA. PpHCT thus displayed significant donor and acceptor permissiveness. This was more pronounced with p-coumaroyl-CoA, the P. patens native acyl donor, which in addition was the only donor that coupled with threonate (Figure 3, C). A striking difference between PpHCT and orthologs from vascular plants is the presence of a 144-amino acid flexible loop joining the two main folded domains of the protein (Supplemental Figure S3). To test the effect of this loop on enzyme activity, a truncated version of PpHCT lacking the internal loop domain was produced in E. coli (Supplemental Figure S4). The truncation did not affect substrate preference; however, it caused a minor decrease in activity with threonate as the acceptor, without altering the shikimate and quinate acylation activity (Supplemental Figure S4).
Figure 3.
Investigation of recombinant PpHCT catalytic properties. A, Overlay of UHPLC–MS/MS chromatograms showing the production of p-coumaroyl esters by PpHCT in vitro. The recombinant protein was incubated with p-coumaroyl-CoA and one of the three different acyl acceptors: quinate, shikimate or threonate. Assays with boiled enzyme or without p-coumaroyl-CoA were used as controls. pC2T, p-coumaroyl-2-O-threonate; pC4T, p-coumaroyl-4-O-threonate; pCS, p-coumaroyl-shikimate; pCQ, p-coumaroyl-quinate. B, Structures of the main p-coumaroyl esters detected and numbered in (A). C, PpHCT activity for all pairwise combinations involving p-coumaroyl-CoA, caffeoyl-CoA and feruloyl-CoA as acyl donor, together with quinate, threonate and shikimate as an acyl acceptor. Enzyme activity was calculated based on end-point assays analyzed by HPLC-UV. Data are the mean ± SEM of three independent enzyme assays. nd, not detected. D, Overlay of UHPLC–MS/MS chromatograms showing the production of phenolic esters in whole-cell assays using engineered Saccharomyces cerevisiae strains expressing different combinations of Pp4CL1, PpHCT, PpCYP98, and ATR1 genes. p-coumaroyl-threonates (pC2T, pC4T), caffeoyl-threonates (C2T, C4T), p-coumaroyl-shikimates (pCS), and caffeoyl-shikimates (CS) esters were simultaneously analyzed from yeast culture supplemented with p-coumarate and l-threonate using HESI+ MRM methods. Y-axes of yeast extract chromatographs are linked to show each phenolic ester. For caffeoyl-threonate, a nonspecific signal was detected regardless of the gene set (red asterisks).
PpHCT kinetic parameters largely favor shikimate acylation
To gain deeper insights into PpHCT catalytic properties, enzyme kinetic parameters were determined from activity saturation curves and Michaelis–Menten nonlinear regression (Supplemental Figure S5). We focused the kinetic analysis on the three acyl acceptors, threonate, quinate, and shikimate, and on the native acyl donor p-coumaroyl-CoA. The results, summarized in Table 1, revealed an obvious preference of PpHCT for shikimate as an acyl acceptor, in terms of affinity (Km: 0.22 mM) and velocity (kcat: 5.1 s−1), compared with threonate (Km: 17.2 mM, kcat: 0.16 s−1). The calculated catalytic efficiency with shikimate (kcat/Km) was ∼2,500-fold higher than with threonate (Table 1). PpHCT activity with quinate was mixed, exhibiting low affinity (Km: 9.4 mM) but a rather high velocity (kcat: 3.5 s−1). PpHCT affinity for p-coumaroyl-CoA was 60 µM when shikimate was used as acceptor, a value seven times lower than when threonate was used as an acceptor (Table 1). PpHCT kinetic parameters with shikimate were found overall to be similar to those of angiosperm orthologs (Hoffmann et al., 2003; Levsh et al., 2016).
Table 1.
Summary of PpHCT kinetic parameters
| Substrates |
K m (mM) | k cat (s−1) | k cat/Km (s−1.M−1) | |
|---|---|---|---|---|
| Fixed | Variable | |||
| p-coumaroyl-CoA | Threonate | 17.2 (14.9–19.9) | 0.16 (0.15–0.17) | 9 |
| Shikimate | 0.22 (0.17–0.29) | 5.1 (4.8–5.4) | 23,005 | |
| Quinate | 9.4 (6.7–13.7) | 3.5 (3.0–4.2) | 368 | |
| Threonate | p-coumaroyl-CoA | 0.43 (0.27–0.76) | 0.18 (0.14–0.25) | 407 |
| Shikimate | p-coumaroyl-CoA | 0.06 (0.03–0.10) | 17.0 (14.1–20.7) | 283,333 |
Enzyme affinity (Km) and velocity (kcat) constants were determined from PpHCT activity saturation curves, based on nonlinear Michaelis–Menten regression (see the “Materials and methods” section; Supplemental Figure S5). Results are the means of three independent enzyme reactions; 95% confidence intervals (profile likelihood) are provided within brackets.
The PpHCT kinetic parameters suggest that threonate would need to be present in much higher concentrations than shikimate in P. patens to be used as a substrate by PpHCT. To investigate this hypothesis, we first determined the absolute levels of the three potential acyl acceptors, (−)-shikimate, d-quinate, and l-threonate, in the gametophyte of the three bryophytes P. patens, A. agrestis, and M. polymorpha, and in the angiosperm A. thaliana by ultra-high-performance liquid chromatography (UHPLC)– mass spectrometry (MS)/MS (see the “Materials and M=methods” section). Levels of l-phenylalanine, the amino acid that initiates the phenylpropanoid pathway, and l-malate, an intermediate of the Krebs cycle, were simultaneously measured and served as metabolic benchmarks. Metabolic profiling revealed a higher level of threonate in P. patens than in the other species, while phenylalanine and malate levels remained similar among them (Supplemental Table S3). At the level of the whole P. patens gametophore, threonate was found to be ∼250 times more abundant than shikimate; in the other species, the threonate/shikimate ratio ranged from 0.1 to 2 (Supplemental Table S3). Concentrations and ratios of these compounds might however differ in specific tissues, cell types or subcellular compartments. Quinate was not detected in any of the plant extracts, in accordance with the absence of a bona fide quinate dehydrogenase in the investigated species (Guo et al., 2014; Carrington et al., 2018). To test whether a 250-fold excess in threonate would shift the equilibrium from shikimate to threonate acylation, we incubated recombinant PpHCT with p-coumaroyl-CoA in the presence of 2.5-mM l-threonate and 10-µM (−)-shikimate, recapitulating a 250:1 molar ratio of competing substrates in vitro. Under these conditions, PpHCT activity was 37 times higher with shikimate than with threonate as an acyl acceptor (Supplemental Figure S6). These results confirm that the catalytic properties of PpHCT favor shikimate acylation, even in the presence of a far higher concentration of threonate.
Reconstitution of the moss phenolic pathway in yeast
The in vivo functionality of PpHCT and its ability to operate with potential partner enzymes were investigated in engineered Saccharomyces cerevisiae co-expressing Pp4CL1, PpHCT, and PpCYP98, as well as A. thaliana ATR1 (At4g24520) encoding a P450 reductase to ensure sufficient electron supply to PpCYP98 (Urban et al., 1997). Since S. cerevisiae does not naturally synthesize phenylpropanoids or threonate, we supplemented the yeast culture media with p-coumarate and l-threonate 6 h after the onset of galactose-induced recombinant protein production. UHPLC–MS/MS analysis of yeast culture extracts revealed the production of p-coumaroyl-4-O-threonate but not p-coumaroyl-2-O-threonate (Figure 3, D), consistent with PpHCT having a strong regiospecificity. Notably, caffeoyl-threonate was not detected in the yeast culture extracts (Figure 3, D). Saccharomyces cerevisiae synthesizes shikimate as an intermediate of aromatic amino acid biosynthesis and, accordingly, we detected p-coumaroyl-shikimate in extracts of all PpHCT-expressing yeast strains (Figure 3, D), which confirmed PpHCT promiscuity in vivo. Caffeoyl-shikimate was readily detected in the yeast extracts, indicating that shikimate esters were intermediates, allowing an effective coupling of PpHCT and PpCYP98 activities (Figure 3, D). The major p-coumaroyl ester isomers produced in yeast were similar to those predominantly generated by PpHCT in vitro (Figure 3, B). In parallel, we used the yeast platform to assess the catalytic activity of the K. nitens HCT homolog kfl00513_0110 (see Figure 1), and found that it did not lead to the production of detectable p-coumaroyl-shikimate when co-expressed with Pp4CL1, PpCYP98, and ATR1 (Supplemental Figure S7). This supports the idea that charophyte HCT homologous proteins do not act as canonical HCT enzymes.
PpHCT efficiently converts caffeoyl-5-O-shikimate into caffeoyl-CoA in vitro
Earlier work on recombinant proteins from Nicotiana tabacum and Arabidopsis showed that, in vitro, HCT catalyzes the conversion of caffeoyl-5-O-quinate or caffeoyl-5-O-shikimate into caffeoyl-CoA, using coenzyme A as acyl acceptor (Hoffmann et al., 2003; Vanholme et al., 2013)—a reaction hereafter referred to as the HCT reverse reaction (Figure 4, A). However, there has subsequently been little additional experimental support for the existence of this reaction, in particular in plant lineages other than angiosperms. We therefore tested in vitro the ability of recombinant PpHCT to catalyze the production of caffeoyl-CoA from caffeoyl esters. We first developed a sensitive HPLC-UV method that provides a linear response to caffeoyl-CoA (Figure 4, B; see the “Materials and Methods” section). We then performed in vitro assays with recombinant PpHCT, using coenzyme A as acyl acceptor and either caffeoyl-5-O-shikimate or caffeoyl-5-O-quinate as acyl donors. Caffeoyl-threonates, which are not commercially available and are not synthesized efficiently by CYP98 enzymes in vitro (Alber et al., 2019), could not be tested as substrates. HPLC-UV analysis of enzyme reaction products showed a sharp decrease of caffeoyl-5-O-shikimate in the PpHCT assay with a concomitant appearance of a peak that was identified as caffeoyl-CoA, based on retention time and absorbance spectrum comparison with an authentic standard (Figure 4, C and D). Minor amounts of additional caffeoyl-shikimate isomers were present in the reaction, but were not effectively metabolized by PpHCT (Figure 4, C). Based on caffeoyl-CoA production, PpHCT reverse activity with caffeoyl-5-O-shikimate as an acyl donor was comparable to that of the conjugating activity producing p-coumaroyl-5-O-shikimate. However, PpHCT reverse activity using caffeoyl-5-O-quinate as an acyl donor was seven times lower (Figure 4, E), and this result was confirmed by the amount of caffeoyl esters remaining at the end of the enzyme assay (Figure 4, F).
Figure 4.
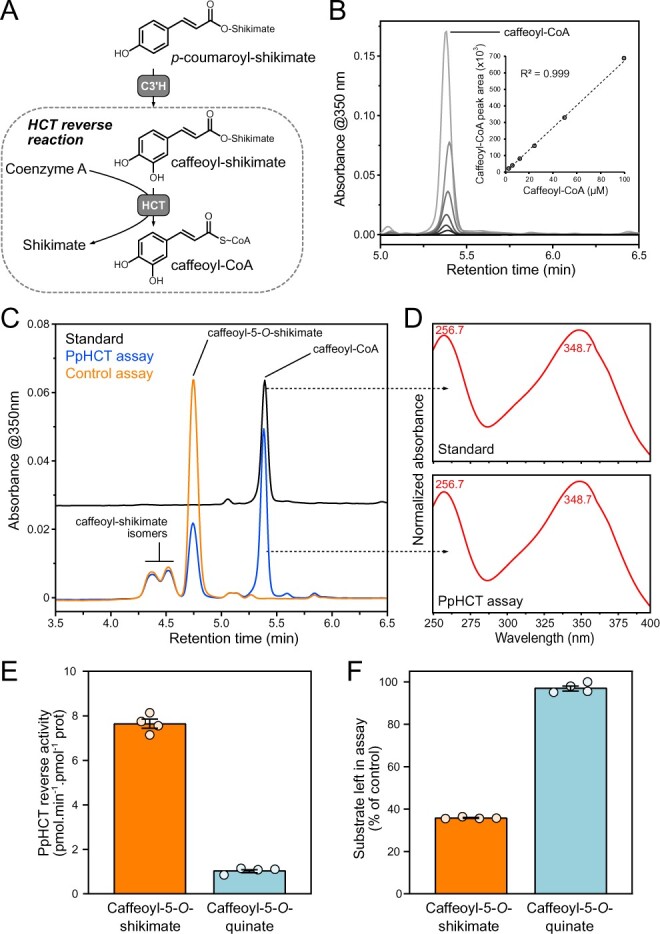
Investigation of the HCT reverse reaction in vitro. A, Scheme depicting the HCT reverse reaction that leads from caffeoyl-shikimate to caffeoyl-CoA thioester. B, HPLC–UV detection of authentic caffeoyl-CoA molecule at 350 nm and associated calibration curve exhibiting linearity over at least three orders of concentration. C, Representative HPLC–UV chromatograms showing the conversion of caffeoyl-5-O-shikimate into caffeoyl-CoA by PpHCT in vitro. Control assay was performed with boiled PpHCT enzyme. D, Absorbance spectrum from 240 to 400 nm of PpHCT assay and caffeoyl-CoA standard peaks. Wavelengths corresponding to maximum absorbance are indicated. E, PpHCT reverse activity with caffeoyl-5-O-shikimate or caffeoyl-5-O-quinate as acyl donor. F, Relative level of caffeoyl-5-O-shikimate and caffeoyl-5-O-quinate left in PpHCT assay (% of control assay). Data are the mean ± SEM of four independent enzyme assays.
PpHCT produces p-coumaroyl-shikimate in planta as a precursor of caffeate derivatives
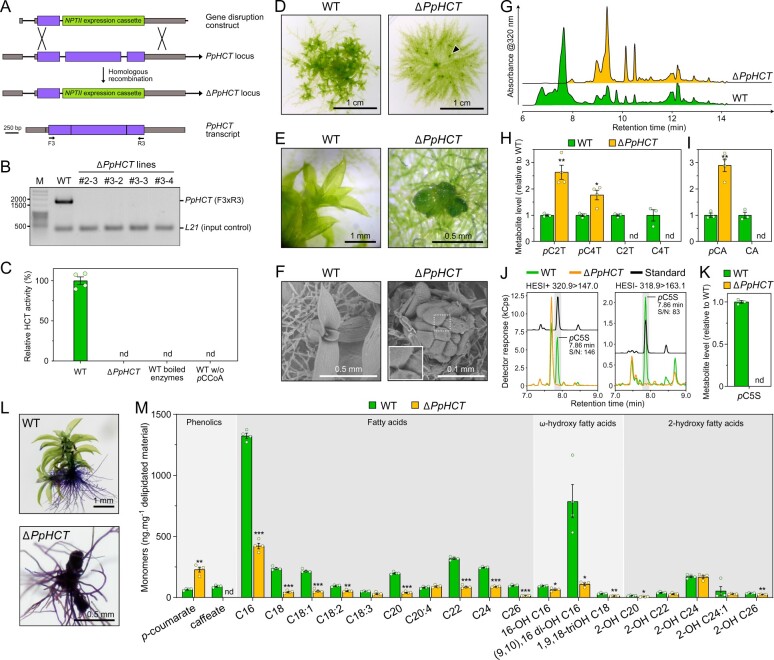
Next, we generated four independent PpHCT deletion mutants (ΔPpHCT) via homologous recombination (Figure 5, A andSupplemental Figure S8) in order to address the in planta function of PpHCT. The four ΔPpHCT lines lacked full-length PpHCT transcripts (Figure 5, B), leading to a complete abolishment of HCT activity in gametophore protein extracts (Figure 5, C). The ΔPpHCT lines phenocopied ΔPpCYP98 mutants (Renault et al., 2017) and newly reported PpHCT loss-of-function mutant Ppnog2-R (Moody et al., 2020), characterized by defective gametophore development (Figure 5, D–F and Supplemental Figure S8). UV-fingerprinting of gametophore metabolite extracts revealed the absence of major peaks in the ΔPpHCT mutant chromatogram (Figure 5, G). This low-resolution UV analysis was refined by targeted UHPLC–MS/MS analysis, which revealed both qualitative and quantitative differences in threonate esters. As expected, if PpHCT generates the substrate(s) of PpCYP98, caffeoyl-threonates were absent from ΔPpHCT (Figure 5, H). Unexpectedly, however, levels of p-coumaroyl-threonate esters were higher in the ΔPpHCT lines (Figure 5, H). Taken together, these data suggest that p-coumaroyl-threonate esters are not (i) derived from PpHCT activity, implying the existence of another dedicated enzyme in P. patens or (ii) the native substrates of PpCYP98, although they could be metabolized in vitro (Renault et al., 2017). We addressed the identity of this putative enzyme by testing the ability of each of the twelve full-length, expressed BAHD proteins from P. patens to catalyze the formation of p-coumaroyl-threonate in yeast. Only PpHCT was found to catalyze threonate acylation (Supplemental Figure S9, A). This result was corroborated by in vitro assays with protein extracts from P. patens gametophores, which did not yield detectable p-coumaroyl-4-O-threonate (Supplemental Figure S9, B). Next, to investigate the existence of potentially overlooked hydroxycinnamoyl intermediates, and in particular caffeoyl conjugates, gametophore extracts were submitted to acid hydrolysis to release hydroxycinnamate moieties prior to UHPLC–MS/MS analysis. Caffeate was not detected in mutant gametophore hydrolyzed extracts (Figure 5, I). This result indicates that PpHCT provides the only route toward caffeate derivatives in P. patens, in accordance with the absence of a bona fide C3H/APX gene in P. patens (Barros et al., 2019). A large increase in the amount of p-coumarate in hydrolyzed extracts (Figure 5, I) was consistent with the previously reported accumulation of p-coumaroyl-threonates in ΔPpHCT mutant lines (Figure 5, H). Taking advantage of the increased sensitivity and resolution provided by a new UHPLC–MS/MS analytical platform, we searched for shikimate esters in gametophore extracts in which we had not detected these compounds previously (Renault et al., 2017). To improve the detection threshold, extracts were also concentrated five-fold, and under these conditions we detected p-coumaroyl-5-O-shikimate in gametophore extracts from wild-type P. patens, but not in those from ΔPpHCT (Figure 5, J and K). The results were orthogonally validated by both retention time comparison with molecular standards and two different mass transitions in positive and negative modes (signal-to-noise ratio > 80). Taken together, the metabolic analysis of the ΔPpHCT mutants confirmed that HCT has a key function in the production of caffeate derivatives in P. patens via the formation of a p-coumaroyl-5-O-shikimate intermediate and did not support the existence of alternative pathways.
Figure 5.
Investigation of PpHCT function in planta. A, Homologous recombination-mediated strategy for PpHCT gene disruption. A genomic fragment encompassing the second and third PpHCT exons was excised with simultaneous insertion of the NPTII selection cassette conferring resistance to G418. Binding sites of oligonucleotides used for characterization of the transgenic lines are shown. Gray box, UTR; black line, intron; purple box, exon. B, Agarose gel photograph produced from RT-PCR analysis reporting the absence of PpHCT transcripts in the four ΔPpHCT KO lines. M, DNA size marker. C, HCT activity in protein extracts from wild-type (WT) and ΔPpHCT gametophores. HCT activity was measured in vitro using shikimate and p-coumaroyl-CoA as substrates. Negative WT control assays involved boiled protein extracts or omission of p-coumaroyl-CoA (pCCoA). Results are the mean ± SEM of four independent enzyme assays, performed with protein extracts from each of the four independent ΔPpHCT mutant lines. nd, not detected. D, Phenotype of 4-week-old P. patens WT and ΔPpHCT colonies. Arrowhead points to a ΔPpHCT gametophore. E, Magnified image of gametophores visible in (D). F, SEM micrographs of 4-week-old gametophores. For ΔPpHCT, inset shows intercellular adhesive structures (enlargement of boxed region). G, Representative HPLC-UV chromatograms of WT and ΔPpHCT gametophore extracts. H, Relative levels of phenolic threonate esters in gametophore extracts. pC2T, p-coumaroyl-2-O-threonate; pC4T, p-coumaroyl-4-O-threonate; C2T, caffeoyl-2-O-threonate; C4T, caffeoyl-4-O-threonate. I, Relative levels of free hydroxycinnamic acids in gametophore extracts after acid hydrolysis. pCA, p-coumaric acid; CA, caffeic acid. J, Overlay of representative UHPLC-MS/MS chromatograms showing the absence of p-coumaroyl-5-O-shikimate (pC5S) in ΔPpHCT gametophore extracts. Gray shaded regions highlight relevant elution time windows. K, Relative levels of p-coumaroyl-5-O-shikimate (pC5S) in gametophore extracts. Results are the mean ± SEM of three independent WT biological replicates and four independent ΔPpHCT mutant lines. L, Toluidine blue staining of WT and a ΔPpHCT mutant. Protonema and rhizoids do not have a cuticle, and so are readily stained. M, Compositional analysis of WT and ΔPpHCT gametophore cuticular biopolymers. Data are the mean ± SEM of four WT biological replicates and four independent ΔPpHCT mutant lines. WT versus mutant t test adjusted P-value: *P<0.05; **P<0.01; ***P<0.001.
PpHCT deficiency impairs cuticle development
A previous analysis of a ΔPpCYP98 mutant led us to conclude that the availability of caffeate, or a derivative, is required for normal P. patens gametophore development and cuticle formation (Renault et al., 2017). Since ΔPpHCT lines essentially phenocopied ΔPpCYP98 at macroscopic and metabolic levels, we tested tissue surface permeability of mutant and WT gametophores using a toluidine blue staining assay, to assess for similar cuticle defects. The strong blue staining of the ΔPpHCT lines confirmed their increased surface permeability compared to WT (Figure 5, L andSupplemental Figure S8), consistent with reduced cuticle barrier properties associated with the PpHCT deletion. We also characterized the monomeric composition of the cuticular biopolymer from the ΔPpHCT gametophore and found differences in aliphatic or phenolic components compared with WT (Figure 5, M). The ΔPpHCT cuticle appeared to be devoid of caffeate residues, but showed a three-fold increase in p-coumarate units compared with WT, consistent with the analysis of soluble phenolic compounds (Figure 5, H and I). This change in phenolic composition was accompanied by a substantial decrease in long-chain fatty acids (LCFAs) and ω-hydroxylated LCFA, especially in the two most abundant monomers, palmitic acid (C16) and (9,10),16 di-hydroxypalmitic acid. A minor decrease in the total amounts of 2-OH-VLCFA (very-LCFAs), derived from membrane sphingolipids (Molina et al., 2006) indicated that plasma membranes were only slightly affected, in contrast to the cuticular biopolymer (Figure 5, M).
Conservation of HCT properties between bryophytes and angiosperms
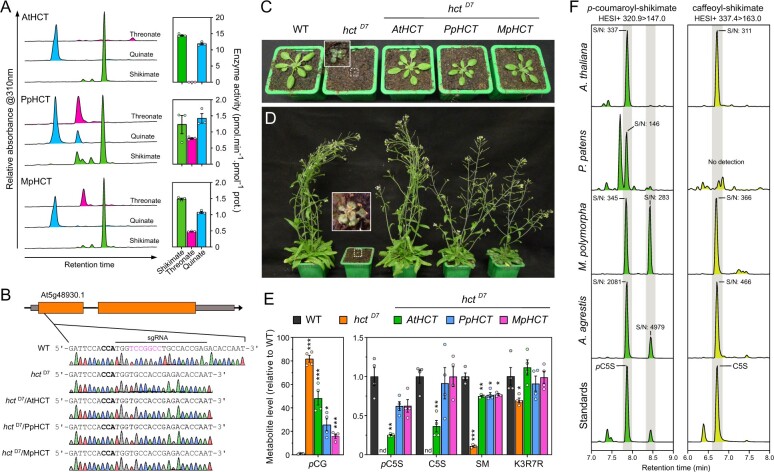
Functional analysis of PpHCT suggested that HCT function was conserved over the ∼500 million years that span embryophyte evolution. To test this hypothesis, we first investigated in vitro the acyl acceptor permissiveness of recombinant HCT from M. polymorpha (MpHCT), which belongs to another major bryophyte group, and A. thaliana (AtHCT; Figure 6, A). In contrast to PpHCT and MpHCT, AtHCT activity using threonate as an acyl acceptor was barely detectable (Figure 6, A). However, all three proteins had in common a preference for shikimate or quinate as an acceptor, indicative of conservation of HCT enzyme properties in embryophytes (Figure 6, A). To further assess the functional conservation of HCT genes across embryophyte evolution, we conducted transcomplementation experiments. The first step was to generate an A. thaliana hct null mutant since only RNA-interference lines, with residual HCT expression, were available. Following a CRISPR/Cas9-mediated strategy, we isolated a mutant allele characterized by a deletion of seven nucleotides in the AtHCT first exon, hereafter termed the hctD7 mutant (Figure 6, B), which introduces a frameshift leading to a premature stop codon.
Figure 6.
Evolutionary conservation of HCT function in embryophytes. A, AtHCT, PpHCT, and MpHCT acyl acceptor permissiveness was investigated using threonate, quinate, or shikimate, and p-coumaroyl-CoA as an acyl donor in in vitro end-point assays. Representative HPLC-UV chromatograms (left) and corresponding HCT activity (right) are shown for the different acyl acceptors. Results are the mean ± SEM of three independent enzyme assays. Note that the results for PpHCT are the same as those reported in Figure 3, C. B, Schematic representation of the AtHCT locus and sequence of the CRISPR/Cas9 target site. Gray box, UTR; black line, intron; orange box, exon. The protospacer adjacent motif (NGG) is highlighted in bold. Sanger sequencing chromatograms of wild-type (WT) and the homozygous hctD7 mutant transformed, or not, with AtHCT, PpHCT, or MpHCT show the seven-nucleotide (in pink) deletion in the HCT gene of hctD7 plants. C, D, Phenotypes of 3-week-old (C) and 60-day-old (D) A. thaliana WT and the hctD7 null mutant transformed, or not, with AtHCT, PpHCT, or MpHCT genes. E, Relative levels of the phenolic esters p-coumaroyl-glucose (pCG), p-coumaroyl-5-O-shikimate (pC5S), caffeoyl-5-O-shikimate (C5S) and sinapoyl-malate (SM), and of the flavonol kaempferol 3-O-rhamnoside 7-O-rhamnoside (K3R7R) in the rosettes of 3-week-old plants. Results are the means ± SEM of four independent biological replicates. WT versus mutant t test adjusted P-value: *P<0.05; **P<0.01; ***P<0.001. F, Representative UHPLC-MS/MS chromatograms reporting the search for p-coumaroyl-5-O-shikimate and caffeoyl-5-O-shikimate in the bryophytes P. patens, M. polymorpha and A. agrestis, and the angiosperm A. thaliana. Additional positional isomers of p-coumaroyl-shikimate may occur in M. polymorpha and A. agrestis. S/N, signal-to-noise ratio. Gray shaded regions highlight relevant elution time windows.
We then transformed heterozygous hctD7+/− plants with AtHCT, PpHCT, and MpHCT coding sequences under control of the A. thaliana C4H promoter, which efficiently drives gene expression in phenylpropanoid-accumulating tissues (Weng et al., 2008, 2011; Sibout et al., 2016; Alber et al., 2019), and selected plants homozygous for both hctD7 allele and complementation constructs (Figure 6, B). The AtHCT null mutation led to reduced growth (Figure 6, C and D), similar to previous observations of HCT-RNAi lines (Besseau et al., 2007; Li et al., 2010a), but this abnormal phenotype was entirely abolished by introducing an HCT coding sequence from A. thaliana, and almost completely in the case of PpHCT or MpHCT (Figure 6, C and D). Similar to P. patens, disruption of HCT function in Arabidopsis led to an increased permeability of aerial tissues to toluidine blue (Supplemental Figure S10). The HCT null mutation resulted in obvious changes in UV chromatograms (Supplemental Figure S11), which was confirmed by targeted analysis of diagnostic phenylpropanoid molecules. The targeted profiling revealed an 80-fold accumulation of p-coumaroyl-glucose in the hctD7 mutant compared with WT, while p-coumaroyl and caffeoyl esters of shikimate were absent from the mutant (Figure 6, E). Residual levels of sinapoyl-malate, the main soluble phenolic ester in A. thaliana leaves, were detected in hctD7 (∼10% of wild-type level), possibly due to the alternative C3H/APX pathway using free p-coumarate, or promiscuous activities of C3’H on accumulating p-coumaroyl esters (e.g. p-coumaroyl-glucose; Figure 6, F). The level of the main A. thaliana leaf flavonoid, the flavonol kaempferol 3-O-rhamnoside 7-O-rhamnoside (Li et al., 2010a; Yin et al., 2014), was slightly reduced in hctD7 compared with WT (Figure 6, E). This result does not match data from previous analyses of RNAi-HCT lines (Besseau et al., 2007; Li et al., 2010a), but is corroborated by HPLC-UV analysis indicating that no other phenylpropanoids, including flavonoids, overaccumulated in the hctD7 null mutant to levels similar to wild-type sinapoyl-malate under our growth conditions (Supplemental Figure S11). All hctD7 plant metabolic defects were, at least partially, complemented by transformation with AtHCT, PpHCT, or MpHCT under the control of the AtC4H promoter (Figure 6, E). In particular, the ability to synthesize p-coumaroyl-5-O-shikimate and caffeoyl-5-O-shikimate was restored in all the HCT-complemented lines (Figure 6, E), consistent with functional conservation of bryophyte and angiosperm HCT genes. Threonate esters were not detected in any of the investigated Arabidopsis genotypes. To further establish phenolic shikimate esters as conserved metabolic intermediates during embryophyte evolution, we checked for their presence in representative species of the three major bryophyte lineages. In addition to P. patens and A. thaliana (Figure 6, E and F and Supplemental Figure S12), targeted analysis revealed the presence of p-coumaroyl-5-O-shikimate in the liverwort M. polymorpha and the hornwort A. agrestis (Figure 6, F andSupplemental Figure S12). With the exception of P. patens, the 3′-hydroxylated form of p-coumaroyl-5-O-shikimate, caffeoyl-5-O-shikimate, was detected in all plant samples. The results were consistently confirmed by both retention time comparison with molecular standards and simultaneous MS/MS analysis in positive and negative modes (Figure 6, F andSupplemental Figure S12). Parallel profiling of threonate esters in the same plant extracts suggested a lineage-specific pattern, since they were detected only in P. patens extracts (Supplemental Figure S13).
Discussion
The silencing of HYDROXYCINNAMOYL-CoA:SHIKIMATE HYDROXYCINNAMOYL TRANSFERASE in seed plants typically results in a strong reduction in the abundance, and/or compositional modification, of the biopolymer lignin, and is usually associated with stunted growth (Hoffmann et al., 2004; Chen et al., 2006; Wagner et al., 2007; Besseau et al., 2007; Gallego-Giraldo et al., 2011). Parallel in vitro and structural studies showed that tracheophyte HCTs consistently use shikimate as a preferred acyl acceptor to form p-coumaroyl-shikimate esters (Hoffmann et al., 2003; Lallemand et al., 2012; Levsh et al., 2016; Chiang et al., 2018), which in turn serve as substrates for C3′H enzymes (Schoch et al., 2001; Alber et al., 2019). Taken together, these data suggested a deep evolutionary conservation of HCT function in vascular plants. Here, through a multidisciplinary study of the bryophyte model P. patens, we are able to extend HCT functional conservation throughout the entirety of embryophyte evolution, pointing to an emergence in the last common ancestor of land plants, approximately 500 Ma (Figure 7, A). New methodologies have allowed us to refine our previous studies (Renault et al., 2017) and we conclude, based on new evidence, that shikimate esters are the native intermediates for phenolic ring 3-hydroxylation in P. patens (Figure 7, B). The absence of a bona fide C3H/APX gene and the presence of only distantly-related CSE homologs in P. patens (Ha et al., 2016; Barros et al., 2019) further supports a pivotal function of HCT in the moss phenylpropanoid pathway, for both producing and deconjugating shikimate esters (Figure 7, B).
Figure 7.
Evolutionary and metabolic models of HCT function. A, Evolutionary model for the conservation of the HCT/CYP98 pair in embryophytes. Gray branches represent organismal evolution of species. Green and cyan branches represent HCT (this study) and CYP98 (Alber et al., 2019) gene evolutions, respectively. CYP98 encodes p-coumaroyl 3′hydroxylase (C3′H). B, Metabolic model of P. patens phenylpropanoid pathway. HCT plays a pivotal role in controlling the formation and deconjugation of shikimate esters, the evolutionarily conserved intermediates for phenolic ring functionalization by C3′H enzyme. Shikimate partly derives from the pentose phosphate pathway and is a precursor of l-phenylalanine, potentially establishing a regulatory control of photosynthetic carbon allocation to the phenylpropanoid pathway through two critical steps. The occurrence of a CSE/4CL alternative route toward caffeoyl-CoA in P. patens remains an open question. E4P, D-erythrose 4-phosphate; PEP, phosphoenolpyruvate.
Our data also highlight a previously unappreciated complexity in the bryophyte phenylpropanoid pathway, which in P. patens produces both soluble esters and precursors of a hydrophobic apoplastic biopolymer. This metabolic typology is akin to that of flowering plants, which often produce lineage-specific soluble phenolic esters (e.g. sinapoyl-malate or chlorogenic acids), as well as essential precursors of biopolymers, such as monolignols. Soluble esters act as UV screens and antioxidants (Lehfeldt et al., 2000; Clé et al., 2008) and, as such, may be advantageous in particular ecological niches (Li et al., 2010b). We propose that threonate esters, which we found only in P. patens, are specialized stress-mitigating molecules, while the shikimate esters are evolutionarily conserved intermediates involved in phenolic ring functionalization. Threonate originates from the degradation of the plant-specific antioxidant ascorbate, possibly in the cell wall (Green and Fry, 2005), suggesting a connection between P. patens threonate ester biosynthesis and stress acclimation. Despite our efforts, we failed to identify the enzyme(s) responsible for threonate ester production in P. patens. Our preliminary data suggest that this neither involves a BAHD acyl transferase, nor a coenzyme A thioester acyl donor. In this respect, serine carboxypeptidase-like enzymes would be good candidates for catalyzing such a reaction using glucose esters as acyl donors, as is the case for sinapoyl-malate biosynthesis (Lehfeldt et al., 2000). On the other hand, evolutionary selection has led to the coupling of phenol-containing biopolymer biosynthesis with shikimate, a widespread molecule found in plants, bacteria, and fungi. In plants, shikimate partly derives from the pentose phosphate pathway and is a precursor of aromatic amino acids, including l-phenylalanine (Maeda and Dudareva, 2012; Figure 7, B). Cellular concentrations of shikimate are assumed to be lower than the Km of HCT enzymes for shikimate. It was therefore proposed that shikimate availability might serve as a biochemical mechanism to regulate photosynthetic carbon allocation to phenylpropanoid production, with HCT playing a key role in such a mechanism (Schoch et al., 2006; Adams et al., 2019; Figure 7, B).
Here, we provide evidence that the HCT gene, and the associated shikimate-mediated regulation of the plant phenolic metabolism, appeared during plant terrestrialization in the last common ancestor of embryophytes, concomitant with the occurrence of a cuticle, but prior to lignin evolution (Figure 7, A). This evolutionary pattern matches that of CYP98, which encodes the downstream C3′H enzyme (Alber et al., 2019). A complex evolutionary interplay therefore likely shaped HCT and CYP98 macro-evolutions and established the HCT/CYP98 pairing as a core metabolic module within the phenylpropanoid pathway, deeply rooted in land plant evolution (Figure 7, A). The tight relationships between the two enzymes is further evidenced by their ability to physically interact and to form a supramolecular complex in A. thaliana (Bassard et al., 2012), a mechanism by which intermediate channeling could occur. Whether the phenylpropanoid pathway in P. patens is also organized at a supramolecular level however remains an open question. The CYP98/HCT pair also features lower scale evolutionary patterns, as illustrated by recurrent, independent duplications of the core HCT and CYP98 genes, which led to the emergence of specialized phenolic compounds, such as rosmarinic acid and phenolamides (Matsuno et al., 2009; Liu et al., 2016; Levsh et al., 2019).
Both PpCYP98 (Renault et al., 2017) and PpHCT knock-out mutants show stunted gametophore growth and organ fusion phenotypes, associated with a complete loss of cuticular caffeate units. Cuticles are essential to control water permeability, and provide plant protection against drought (Kosma et al., 2009; Lü et al., 2012) and other environmental stresses, including UV-B radiation (Krauss et al., 1997; Yeats and Rose, 2013). Thus, emergence of a cuticle with properties that enabled plant terrestrialization may have been dependent on the presence of a primordial phenylpropanoid pathway. The severe developmental defects of the ΔPpHCT and ΔPpCYP98 mutants unfortunately prevent meaningful evaluation of their stress tolerance. Although found in substantial amounts in the cuticle of some tracheophytes, such as the leaf cuticle of Solanum lycopersicum (Bolger et al., 2014), hydroxycinnamic acids usually represent small proportions of the cuticle of vascular plants (Fich et al., 2016). The presence of large amounts of hydroxycinnamic acids might therefore be a typical, and possibly essential, feature of bryophyte lineages (Caldicott and Eglinton, 1976; Buda et al., 2013; Kong et al., 2020). Hydroxycinnamic acids might play an important role, since they are covalently attached to fatty acid monomers (Riley and Kolattukudy, 1975). We show here that the absence of caffeate in P. patens prevents the formation of the cuticle and cuticular biopolymer polymerization, as evidenced by the large decreases in the major cutin monomers C16 FA and (9,10),16 di-OH C16 FA in the ΔPpHCT lines, as was previously shown in the PpCYP98 deletion mutants (Renault et al., 2017). A straightforward interpretation is that caffeate anchors, or shapes, the cuticle lipidic scaffold of P. patens. Such a function is apparently not fulfilled by p-coumarate, which accumulates in the ΔPpHCT cuticle. This might indicate an important role of phenolic ring functionalization for biopolymer formation, as is the case in natural plant lignins, which are predominantly derived from di- or tri-substituted phenolic precursors (Ralph et al., 2019). Whether the structural function of phenolic compounds in the cuticle is specific to bryophytes, or even P. patens, remains to be clarified. It was indeed shown that the absence of ferulate from A. thaliana cuticles did not noticeably reduce cuticle integrity (Rautengarten et al., 2012), while we report here an increased permeability to toluidine blue of the aerial tissues of the Arabidopsis hctD7 mutant. The enrichment of the P. patens cuticle with phenolic compounds potentially contributes various functional attributes, including UV protection, water/gas management, tissue scaffolding for erect growth and organ determination (i.e. organ fusion avoidance). We hypothesize that reduction of this bryophyte property was linked to the emergence of specialized biopolymers in tracheophytes, such as canonical lignin and suberin, which assumed some of the functions mediated by the phenol-enriched cuticle of bryophytes.
Materials and methods
Phylogenetic analysis
All BAHD sequences from P. patens (moss, bryophyte), M. polymorpha (liverwort, bryophyte), A. agrestis (hornwort, bryophyte), S. muscicola (Zygnematophyceae, charophyte), C. braunii (Charophyceae, charophyte), and K. nitens (Klebsormidiophyceae, charophyte) were retrieved by BLASTp search using AtHCT (At5g48930.1) as query (E-value < 0.01). Truncated proteins with less than 420 residues were discarded. Obtained bryophyte and charophyte BAHDs were aligned with 34 functionally characterized BAHD protein (full list in Supplemental Tables S1, S2) using the MUSCLE algorithm (Edgar, 2004; alignment file available as Supplemental Data Set S1). Ambiguous sites of the alignment were masked applying the Gblocks method (Castresana, 2000). Phylogenetic relationships were reconstructed with a maximum-likelihood approach using PhyML3.0 (Guindon et al., 2010). Selection of an evolution model that best fits the dataset was guided by the SMS software; the tree was ultimately inferred from the LG +G + I+F model (Le and Gascuel, 2008). Initial tree(s) for the heuristic search were obtained automatically by applying the BioNJ algorithm, and by selecting the topology with superior log likelihood value. Best of nearest neighbor interchange and subtree pruning and regrafting methods were used for improving the tree. Branch tree supports were calculated with the approximate likelihood ratio test (Anisimova and Gascuel, 2006). The tree file is available as supplemental material (Supplemental Data Set S2). Sequence manipulation was performed with Seaview 4 software (http://pbil.univ-lyon1.fr/) and phylogenetic analysis on the PhyML server (http://www.atgc-montpellier.fr/phyml/).
Homology modeling of proteins
3D models of P. patens (Pp3c2_29140), C. braunii (CHBRA170g00210), and K. nitens (kfl00513_0110) proteins were generated using the Modeler comparative module (Sali and Blundell, 1993) embedded in ChimeraX v1.0 software (Goddard et al., 2018) using A. thaliana HCT in complex with p-coumaroyl-5-O-shikimate (pdb entry: 5kju) as template. Prior to modeling, target proteins were aligned with embryophyte representative HCTs visible in Figure 1, B with the MUSCLE algorithm (alignment file available as Supplemental Data Set S3). Five models were automatically generated for each target protein; 3D models with the best GA341 and zDOPE scores were kept for subsequent analyses. Potential hydrogen bonds linking protein residues and p-coumaroyl-5-O-shikimate were predicted with the ChimeraX FindHBond tool. Overlay and visualization of 3D protein models were performed with ChimeraX.
Plant growth conditions
Physcomitrium patens (Physcomitrella; Hedw.) Bruch & Schimp., strain Gransden (IMSC acc. no. 40001; Lang et al., 2018) was cultured in liquid or on solid Knop medium (Reski and Abel, 1985) supplemented with 50-µmol⋅L−1 H3BO3, 50-µmol⋅L−1 MnSO4, 15-µmol⋅L−1 ZnSO4, 2.5-µmol⋅L−1 KI, 0.5-µmol⋅L−1 Na2MoO4, 0.05-µmol⋅L−1 CuSO4, and 0.05-µmol⋅L−1 CoCl2. Medium was solidified with 12-g⋅L−1 purified agar. Physcomitrium patens gametophores were propagated on agar plates or in liquid cultures established by soft tissue disruption (∼15 s). Liquid cultures were subcultured weekly and kept under constant agitation (130 rpm) for proper aeration. Marchantia polymorpha Tak-1 accession and A. agrestis Oxford accession were grown on half-strength Gamborg B5 medium solidified with 12-g⋅L−1 agar. Bryophytes were kept at 23°C under 16/8 h day/night cycle, with light intensity set to 70 µmol⋅m−2⋅s−1 (Osram BIOLUX L 36W/965). Arabidopsis thaliana plants (Col-0 background) were grown on soil, kept under a 22/18°C, 16-h/8-h light/dark regime (100 µmol⋅m−2⋅s−1 light intensity; Philips LEDtube 20W/840 white cool) and watered from the bottom every 2 days with tap water.
Determination of gene expression by RT-qPCR
Total RNA was isolated from 10 mg of lyophilized tissue with 1 ml of TriReagent (Sigma–Aldrich). Samples were agitated for 5 min at room temperature prior to centrifugation for 15 min at 13,000 g, 4°C. After transfer of the supernatant to a new microtube, an equal volume of chloroform was added and samples were thoroughly vortexed and centrifuged for 15 min at 13,000 g, 4°C to induce phase separation. The clear upper phase was recovered and transferred to a new microtube. Total RNA was precipitated by adding 0.1 volume of sodium acetate (NaOAc, 3 M, pH 5.2) and 2.5 volumes of absolute ethanol. After incubation at −20°C for 2 h, RNA was spun down by centrifugation for 30 min at 13,000 g, 4°C. Supernatant were discarded, the RNA pellet was washed with 1 ml of 70% ethanol, and then dried at room temperature for 10 min. Total RNA was finally resuspended in DEPC-treated water. Twenty micrograms of RNA was treated with 5 U of RQ1 DNaseI (Promega) and subsequently purified using phenol–chloroform (50/50, v/v) and precipitation by NaOAc/EtOH. One microgram of DNaseI-treated RNA was reverse-transcribed with oligo(dT) and the Superscript III enzyme (Thermo Scientific) in 20 μL reaction. RT-qPCR reactions consisted of 10 ng cDNA, 500 nM of each primer, and 5 µL of 2× LightCycler 480 SYBR Green I Master mix (Roche) in a 10-µL final volume. Reactions were run in triplicate on a LightCycler 480 II device (Roche). The amplification program was 95°C for 10 min and 40 cycles (95°C denaturation for 10 s, annealing at 60°C for 15 s, extension at 72°C for 15 s), followed by a melting curve analysis from 55°C to 95°C to check for transcript specificity. Crossing points (Cp) were determined using the manufacturer’s software. Cp values were corrected according to primer pair PCR efficiency computed with the LinReg PCR method (Ruijter et al., 2009). Pp3c19_1800 and Pp3c27_3270 genes were used as internal reference for expression normalization. List of qPCR primers is available in Supplemental Table S4.
GUS staining
Plant tissues were vacuum infiltrated for 10 min with X-Gluc solution (50-mM potassium phosphate buffer pH 7.0, 0.5-mM ferrocyanide, 0.5-mM ferricyanide, 0.1% Triton X-100, 0.5-mg/mL X-Gluc) and incubated at 37°C for 4.5 h. Chlorophyll was removed by washing tissues three times in 70% ethanol.
Recombinant protein production
Cloning of the AtHCT (At5g48930) coding sequence into the pGEX-KG vector and purification of the corresponding recombinant protein were performed as previously described (Hoffmann et al., 2003; Besseau et al., 2007). Coding sequences of PpHCT (Pp3c2_29140) and MpHCT (Mapoly0003s0277) were PCR-amplified from P. patens Gransden and M. polymorpha Tak-1 cDNA, respectively, using Gateway-compatible primers (Supplemental Table S4). The truncated PpHCT coding sequence, shown in Supplemental Figure S4, A, was ordered as double-stranded gBlock (Integrated DNA Technologies) with Gateway compatible extensions. CDS were cloned into the pDONR207 vector by the BP Clonase reaction, and then shuttled to the pHGWA expression vector by the LR clonase reaction, allowing N-terminal fusion of the protein with hexahistidine tag. Escherichia coli Rosetta2pLyS strain was transformed with recombined pHGWA plasmids and cultivated in ZYP-5052 autoinducible medium. Recombinant proteins were purified by immobilized metal affinity chromatography using an AKTA Pure 25 system equipped with HisTrap HP 1-mL column and submitted to gel filtration using a Superdex 200 increase 10/300 GL column (GE Healthcare). Purified recombinant proteins were conserved at −80°C in 1× PBS solution containing 10% glycerol.
In vitro enzyme assays
Five millimolar stock solutions of p-coumaroyl-CoA, caffeoyl-CoA, and feruloyl-CoA (PlantMetaChem or MicroCombiChem) were prepared in H2O. Eighty millimolar stock solution of l-threonic acid was prepared from its hemicalcium salt (Sigma–Aldrich) in H2O containing 40-mM EDTA to chelate calcium and improve solubility. Forty millimolar stock solutions of shikimate and d-quinate (Sigma–Aldrich) were prepared in H2O. Standard in vitro HCT assays were performed in 100 µL containing 50-mM potassium phosphate buffer pH 7.4, 1 mM dithiothreitol (DTT), 5 µg recombinant HCT protein, 5 mM acyl acceptor [(−)-shikimate, d-quinate, or l-threonate; Sigma–Aldrich] and 200 µM acyl-CoA. Reactions were initiated by the addition of acyl-CoA, incubated at 30°C for 25 min and stopped by addition of 100 µL acetonitrile. To determine PpHCT kinetic parameters, substrate and enzyme concentrations were optimized for each tested substrate. For shikimate, 50 ng protein, 200 µM p-coumaroyl-CoA, and 0.125–8 mM (−)-shikimate were used. For quinate, 100 ng protein, 200 µM p-coumaroyl-CoA, and 0.312–20 mM d-quinate were used. For threonate, 2 µg protein, 200 µM p-coumaroyl-CoA, and 4–32 mM l-threonate were used. For p-coumaroyl-CoA, 50 ng protein, 8 mM shikimate, and 12.5–400 µM p-coumaroyl-CoA, or 2 µg protein, 32 mM l-threonate, and 12.5–600 µM p-coumaroyl-CoA were used. Reactions were initiated by addition of the saturating substrate, incubated at 30°C for 10 min and stopped by the addition of 100 µL acetonitrile. Relative quantification of reaction products was performed by UHPLC–MS/MS. Absolute quantification of phenolic esters was performed on HPLC-UV with external calibration curves of corresponding free hydroxycinnamic acid (i.e. p-coumarate, caffeate, and ferulate; Sigma–Aldrich). Kinetic parameters were calculated with nonlinear Michealis–Menten regression using the GraphPad Prism v4.8 software (Supplemental Figure S5). Threonate/shikimate competition assays were performed in 50 µL containing 50-mM potassium phosphate buffer pH 7.4, 1-mM DTT, 100-ng recombinant PpHCT, 2.5-mM l-threonic acid, 10 µM (−)-shikimic acid, and 200-µM p-coumaroyl-CoA. Reactions were initiated by addition of p-coumaroyl-CoA from 5 mM stock solution, incubated for 15 min at 30°C and stopped with 50 µL methanol. Under these conditions, shikimate consumption was kept below 30%. Because of the complex nature of the competition assay, absolute quantification of reaction products p-coumaroyl-4-O-threonate and p-coumaroyl-5-O-shikimate was performed by UHPLC–MS/MS with external calibration curves of standard molecules. To test the PpHCT reverse reaction, in vitro assays were performed in 50 µL containing 50-mM potassium phosphate buffer pH 7.4, 1-mM DTT, 1-µg recombinant PpHCT, 250-µM coenzyme A (Sigma–Aldrich), and 100-µM caffeoyl ester. Reactions were initiated by addition of the coenzyme A from 5-mM stock solution, incubated for 30 min at 30°C and stopped with 50-µL methanol. Absolute quantification of the reaction product caffeoyl-CoA was performed by HPLC-UV using an external calibration curve of the authentic molecule.
Yeast metabolic engineering
For P. patens phenolic pathway reconstitution, Pp4CL1, PpHCT, and PpCYP98 coding sequences were PCR-amplified from Gransden cDNA using Gateway-compatible primers (Supplemental Table S4) and shuttled by LR recombination to yeast galactose-inducible expression vectors pAG424GAL, pAG423GAL and pAG425GAL (Alberti et al., 2007), respectively; A. thaliana ATR1 coding sequence was PCR-amplified from Col-0 cDNA and transferred to pAG426GAL yeast expression vector. Recombined vectors were introduced in INVSc1 S. cerevisiae yeast strain (ThermoFisher Scientific) following the lithium acetate/polyethylene glycol method. Yeast transformants were selected on SC- media lacking relevant molecule(s) (6.7 g⋅L-1 yeast nitrogen base without amino acids, 20 g⋅L-1 glucose, appropriate concentration of relevant Yeast Synthetic Drop-out Medium; Sigma–Aldrich) and incubated for 3 days at 30°C. For the whole-cell metabolic assay, a 2.5-mL SC-liquid culture was inoculated with a yeast colony and incubated overnight at 180 rpm and 30°C. Cultures were centrifuged for 5 min at 3,000 g, room temperature, and cell pellets were washed in 25-mL sterile ultra-pure water and centrifuged again for 5 min at 3,000 g. Cells were resuspended in 2.5 mL of liquid SC-medium supplemented with galactose instead of glucose to induce gene expression and incubated at 30°C, 180 rpm. Six hours after induction, yeast cultures were supplemented with 25 µL of 100-mM sterile p-coumarate solution in DMSO and 50 µL of 50-mM sterile l-threonic acid solution in water (5-mM final concentration each). Following substrate addition, cultures were incubated for 24 h at 30°C, 180 rpm. Metabolites were extracted from whole yeast cultures by adding one volume of methanol followed by thorough vortexing. Extracts were centrifuged at 16,000 g for 10 min to spin down yeasts. Supernatants were recovered, dried in vacuo, and resuspended in 50% methanol in one-fifth of the initial volume. Concentrated extracts were analyzed by UHPLC–MS/MS.
Generation of P. patens transgenic lines
ΔPpHCT knock-out mutants were generated by protoplast transfection with a genetic disruption construct allowing introduction of the NPTII expression cassette into the PpHCT locus by homologous recombination. The genetic construct was made by assembling two 750-bp genomic regions PCR-amplified from P. patens genomic DNA with the NPTII selection cassette by GIBSON cloning. The assembled fragment was then PCR-amplified and blunt-end cloned into the pTA2 vector using the pJET1.2 cloning kit (ThermoFisher Scientific). The PpHCT disruption construct was excised from the vector backbone by EcoRI digestion, using restriction sites introduced by PCR. Final sterile DNA solution used for PEG-mediated protoplast transfection contained 45 µg of excised fragment in 0.1 M Ca(NO3)2. Protoplast isolation, transfection and regeneration were performed according to (Hohe et al., 2004). Transformants were selected on Knop plates supplemented with 25-mg⋅L−1 geneticin (G418). For PpHCT:uidA reporter lines, two genomic regions for homologous recombination framing the PpHCT STOP codon were PCR-amplified from genomic DNA and assembled with the uidA reporter gene following the same procedures as described above. A linker sequence was introduced by PCR to limit GUS protein hindrance on PpHCT activity. The PpHCT:uidA construct was excised from vector backbone by NheI digestion. Fifty micrograms of excised fragment was used for protoplast transfection. Since PpHCT:uidA does not contain a selection marker, it was co-transfected with the pRT101 plasmid (Girke et al., 1998) containing the NPTII selection cassette. Transformants were selected on Knop plates supplemented with 25-mg L−1 geneticin (G418).
Molecular characterization of P. patens transgenic lines
Proper genomic integration of DNA construct was assessed using a tailored PCR strategy (Supplemental Figures S2, S8) with primers listed in Supplemental Table S4. Genomic DNA was extracted with DNA extraction buffer (75-mM Tris pH 8.8, 20-mM (NH4)2SO4, and 0.01% Tween 20) during 15 min incubation at 45°C under agitation (1,400 rpm). Two microliters was used for direct PCR using Phire II DNA polymerase (Thermo Scientific) in a final volume of 20 µL. ΔPpHCT mutant lines with seamless 5′ and 3′ integration of the genetic construct at the desired locus were checked for the absence of full-length transcript. Total RNA was isolated and retrotranscribed as described above. PpHCT transcripts were amplified from two microliters of cDNA using the Phire II DNA polymerase (Thermo Scientific). The constitutively expressed L21 gene (Pp3c13_2360), encoding a 60S ribosomal protein, was used as reference. Primers used for RT-PCR are listed in Supplemental Table S4. Four transgenic lines with complete absence of HCT transcripts were selected for subsequent investigations. The MassRuler DNA Ladder Mix (ThermoFisher Scientific) was used as DNA size marker.
Determination of HCT activity in P. patens protein extracts
Proteins were extracted from 3-month-old WT and ΔPpHCT gametophores in 2-mL microtubes containing five volumes of extraction buffer (100-mM Tris–HCl pH 7.4, 10% glycerol, 2-mM DTT, cOmplete EDTA-free Protease Inhibitor Cocktail). Samples were homogenized using 5 mm steel beads and a Tissuelyser II (Qiagen) operated at 30 Hz for 5 min. Following a centrifugation step (20,000 g, 4°C, 40 min), supernatants were recovered and transferred to a 50-mL conical tube. Proteins were precipitated by slow addition to samples of ammonium sulfate up to 0.5 g⋅mL-1 under constant agitation. Once ammonium sulfate was fully solubilized, samples were centrifuged for 20 min at 16,000 g and 4°C, supernatants discarded and protein pellets resuspended in 5 mL of extraction buffer. A second round of precipitation and centrifugation was performed to fully remove plant endogenous metabolites. Protein pellets were resuspended in 500 µL of extraction buffer. Next, samples were centrifuged (5 min, 18,000 g, 4°C) to pellet non-protein material, and supernatants were transferred to new microtubes. Protein concentration was assessed with the Qubit Protein Assay Kit (ThermoFisher Scientific) and adjusted to 200 ng⋅µL-1 with extraction buffer. All steps were performed at 4°C and samples were kept on ice. HCT activity in total protein preparations was evaluated from 50-µL end-point enzyme assays containing 50 mM potassium phosphate buffer (pH 7.4), 2.5 µg total proteins, 1 mM DTT, 200 µM p-coumaroyl-CoA, and 5 mM shikimate or threonate. Reactions were initiated by the addition of p-coumaroyl-CoA, incubated at 30°C and stopped after 1 h by the addition of 50 µL acetonitrile. Production of p-coumaroyl-shikimate was monitored by UHPLC–MS/MS. Relative HCT activity was computed from p-coumaroyl-shikimate peak area and expressed as a percentage of WT.
Plant tissue collection and metabolite extraction
Liquid cultured gametophores were harvested 5 weeks after the last tissue disruption, and 1 week after nutrient medium change. Plant material was collected by filtration on a 100-µm pore size sieve, quickly blotted on paper towel and snap-frozen in liquid nitrogen. For M. polymorpha and A. agrestis, 1-month-old thalli were harvested from Petri plates and snap-frozen in liquid nitrogen. For A. thaliana, whole 3-week-old rosettes were harvested from soil-grown plants and snap-frozen in liquid nitrogen. Samples were lyophilized for 2 days; dry material was homogenized using 5-mm steel beads and a Tissuelyser II (Qiagen) for 1 min at 30 Hz. Metabolites were extracted from 8-mg dry plant powder following a methanol:chloroform:water protocol as described previously (Renault et al., 2017), except that 500 µL methanol, 250 µL chloroform, and 500 µL water were used. For shikimate ester detection in P. patens, 250 µL of metabolic extracts was dried in vacuo, and dry residues were resuspended in 50 µL 50% methanol before analysis. Acid hydrolysis of metabolic extract was conducted as reported before (Renault et al., 2017).
HPLC–UV chromatography
Metabolite separation and detection were carried out on a high-performance liquid chromatography system (Alliance 2695; Waters) coupled to a photodiode array detector (PDA 2996; Waters). Ten to twenty microliters of metabolite extract was injected onto Kinetex Core-Shell C18 column (100 × 4.6 mm, 2.6-µm particle size, or 150 × 4.6 mm, 5-µm particle size; Phenomenex). Needle and injection loops were successively washed with weak (95% water/5% acetonitrile) and strong (20% water/80% acetonitrile) solvents. For phenolics, the mobile phase consisted of a mix of HPLC grade water (A) and acetonitrile (B), both containing 0.1% formic acid. The elution program was as follows: 0.0 min, 95% A; 15.0 min, 5% A (curve 8); 17.0 min, 5% A (curve 6); 18.0 min, 95% A (curve 6); 20.0 min, 95% A. The flow was set to 1 mL⋅min-1 and column temperature to 35°C. For acyl-CoA analysis, the mobile phase consisted of a mix of 20 mM sodium phosphate pH 5.3 prepared in HPLC grade water (A) and acetonitrile (B). The elution program was as follows: 0.0 min, 95% A; 1.0 min, 95% A; 9.0 min, 50% A; 10 min, 40% A; 10.5 min, 5% A; 12.0 min, 5% A. The flow was set to 1 mL⋅min-1 and column temperature to 35°C, and the absorbance was recorded between 250 and 400 nm. Data were processed with the Empower 3 Software (Waters).
Targeted metabolic profiling by UHPLC–MS/MS
Metabolites were separated and detected on a Dionex UltiMate 3000 UHPLC (ThermoFisher Scientific) system coupled to an EvoQ Elite LC‐TQ (MS/MS) mass spectrometer equipped with a heated electrospray ionization source (HESI; Bruker). Nitrogen was used as the drying (30-L⋅h-1 flow rate) and nebulizing (35 L⋅h-1 flow rate) gas. The interface temperature was set to 350°C and the source temperature to 300°C. The capillary voltage was set to 3.5 kV both for positive and negative ionization modes. MS data acquisition and LC piloting were performed with the Bruker MS Workstation 8 and Compass Hystar 4.1 SR1 softwares, respectively. Metabolites were ionized in either positive or negative modes and detected using specific MRM methods (Supplemental Table S5). Bruker MS Data Review software was used to integrate peaks and report corresponding areas. For phenolic molecules, 3 μL of sample was injected onto a C18 Cortecs UPLC T3 column (150 × 2.1 mm, 1.6 μm; Waters) and eluted with a mix of LC–MS grade water (A) and acetonitrile (B), both containing 0.1% formic acid to keep molecules in the protonated form. After each injection, the needle and injection loop were washed with 25% acetonitrile solution. The elution program was as follows: 0.0 min, 5% B; 1.0 min, 5% B; 11.5 min, 100% B (curve 8); 13.0 min, 100% B; 14.0 min, 5% B; 15.0 min, 5% B; total run time: 15 min. Flow was set to 0.400 mL⋅min-1 and column temperature to 35°C. Metabolite peak area was normalized to plant dry weight; metabolite level was expressed relative to WT. For central polar metabolites, 2 μL of the same plant extracts used for phenolics analysis was injected onto an Acquity UPLC HSS PFP column (150 × 2.1 mm, 1.8 μm; Waters) and eluted with a mix of LC–MS grade water (A) and acetonitrile (B), both containing 0.1% formic acid. After each injection, the needle and injection loop were washed with 25% acetonitrile solution. The elution program was as follows: 0.0 min, % B; 2.0 min, 0% B; 5.0 min, 25% B; 10.0 min, 35% B; 10.5 min, 95% B; 12.5 min, 95% B; 12.6 min, 0% B; 15.0 min, 0% B; total run time: 15 min. Flow was set to 0.250 mL⋅min-1 and column temperature to 40°C. The absolute level of polar central metabolites was calculated according to plant dry weight and external calibration curves of authentic molecules and expressed as µmoles of compound per g of plant dry weight (µmol⋅g−1 DW).
Cuticular biopolymer compositional analysis
Cutin monomer analysis was performed on the same gametophore samples used for metabolic analysis, following a previously published protocol (Renault et al., 2017). Briefly, tissues were delipidated by extensive washing with a series of solvents. The delipidated tissues, including cuticle, were dried, weighed and chemically treated (12:3:5 methanol: methyl acetate: 25% sodium methoxide, 60°C, o/n) to depolymerize cutin. Released monomers were then derivatized with pyridine and BSTFA (N,Obis(trimethylsilyl)trifluoroacetamide), dried again by heating under a stream of nitrogen, and resuspended in 100 µL of chloroform. The samples were analyzed by gas chromatography (GC) using an Agilent GC 6850 with a Flame Ionization Detector. Compounds were identified based on a comparison of retention times with standards, and by performing GC–MS using an Agilent GC 6890 coupled to a JEOL GC MATE II mass spectrometer. Monomer levels were normalized to internal standards and dry delipidated tissue weights.
Permeability assay
The permeability test was performed by immersing gametophores in a 0.05% toluidine blue solution for two minutes, and then rinsed with water until the washing solution was clear.
Production and complementation of an Arabidopsis hct null mutant
We generated an hct null mutant by CRISPR/Cas9-mediated gene inactivation as described earlier (DiGennaro et al., 2018). Briefly, BbsI restriction enzyme was used to introduce a double strand fragment resulting from 5′-GATTGCTCGGTGGCAGGCCGGACCA and 5′-AAACTGGTCCGGCCTGCCACCGAGC oligonucleotide annealing, which targets the HCT region CTCGGTGGCAGGCCGGACCATGG, into the At-psgR/GW plasmid. At-psgR/GW with HCT genomic target was transferred into the pEarleyGate 301 vector by the LR Clonase reaction (ThermoFisher Scientific). The recombined pEarleyGate 301 vector was introduced into Agrobacterium tumefaciens GV3101 and used to transform Arabidopsis Col-0 by the floral dip method (Clough and Bent, 1998). Genotyping of T1 and T2 plants was performed by PCR amplification of the genomic sequence spanning the HCT target site using 5′-CCTTCTGAGAGAGTTGGTCGAC and 5′-CTAGCTCGGAGGAGTGTTCG oligonucleotides, followed by AvaII restriction digestion and run on an agarose gel to assess restriction site loss. The loss of the At-psgR/GW cassette at T2 or subsequent generation was assessed by sensitivity to selective agent (glyphosate). A line, free of the AtpsgR/GW cassette and harboring a 7-bp deletion 28 bp after the initiation codon, was isolated for this study and named hctD7. Mutation at the desired locus was confirmed by Sanger sequencing using the PCR fragment generated with 5′-CCTTCTGAGAGAGTTGGTCGAC and 5′-CTAGCTCGGAGGAGTGTTCG oligonucleotides. hctD7 was subsequently used for transcomplementation assays with AtHCT, PpHCT, and MpHCT coding sequences. To this end, Gateway pENTRY vectors harboring coding sequences were recombined with the binary pCC0996 vector that contains a 2,977-bp promoter fragment from the A. thaliana C4H gene (Weng et al., 2011). Resulting plant expression vectors were introduced into Agrobacterium tumefaciens GV3101 and used to transform heterozygous hctD7 plants by the floral dip method. Transformants were selected with BASTA and the hctD7 allele was monitored along the selection process as described above. Experiments were performed with T3 plants homozygous for both the mutant allele and the transcomplementation construct.
Replication and statistical analyses
Unless otherwise noted, independent biological replicates correspond to pooled plants from an independent container. For P. patens, one replicate was derived from plants grown in one flask; for M. polymorpha and A. agrestis, one replicate was derived from plants grown in one Petri plate; for A. thaliana, one replicate was derived from plants grown in one pot. All statistical analyses were performed with GraphPad v8 software. For enzyme catalytic parameters, 95% confidence intervals were computed from nonlinear regression curves based on three independent enzyme assays. For metabolic profiling data, multiple two-tailed unpaired Student’s t test were performed to compare wild-type and mutant means; P-values were corrected using the Holm–Šídák method. Results from statistical analyses are shown in Supplemental Tables S6–S9.
Accession numbers
DNA and derived protein sequence data from this article are available in the Phytozome database (phytozome.jgi.doe.gov) under the following accession numbers: PpHCT (Pp3c2_29140); AtHCT (At5g48930); MpHCT (Mapoly0003s0277); Pp4CL1 (Pp3c18_6360); PpCYP98 (Pp3c22_19010); ATR1 (At4g24520).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Multiple sequence alignment of protein region containing residues critical for HCT activity.
Supplemental Figure S2. Molecular characterization of PpHCT:uidA reporter lines.
Supplemental Figure S3 . Multiple sequence alignment of representative embryophyte HCTs.
Supplemental Figure S4. Sequence, multiple sequence alignment and catalytic properties of a truncated PpHCT protein.
Supplemental Figure S5. Saturation curves of PpHCT activity used to infer kinetic parameters.
Supplemental Figure S6 . In vitro threonate/shikimate competition assay.
Supplemental Figure S7. Assessment of the catalytic function K. nitens HCT homologous protein kfl00513_0110 in yeast.
Supplemental Figure S8. Molecular and phenotypic characterization of ΔPpHCT mutant lines.
Supplemental Figure S9. Search for P. patens enzymes able to produce p-coumaroyl-threonate from p-coumaroyl-CoA.
Supplemental Figure S10. Toluidine blue staining assay of Arabidopsis hctD7 mutant.
Supplemental Figure S11. UV fingerprinting of metabolic extracts from hctD7 lines.
Supplemental Figure S12. Shikimate ester occurrence in bryophytes as evidenced by HESI UHPLC–MS/MS analysis in negative mode.
Supplemental Figure S13. Threonate esters occur in P. patens only as evidenced by HESI+ UHPLC–MS/MS analysis.
Supplemental Table S1. List of functionally characterized hydroxycinnamoyl-CoA-dependent BAHD acyltransferases used for the phylogenetic analysis.
Supplemental Table S2. List of uncharacterized BAHD acyltransferases used for the phylogenetic analysis.
Supplemental Table S3. Absolute level of l-phenylalanine, l-malate, d-quinate, l-threonate, and (−)-shikimate in plant tissues.
Supplemental Table S4. List of oligonucleotides used in the study.
Supplemental Table S5. List of multiple reaction monitoring methods used for metabolite analysis.
Supplemental Table S6. Results of t test from Figure 5, H.
Supplemental Table S7. Results of t test from Figure 5, I.
Supplemental Table S8. Results of t test from Figure 5, M.
Supplemental Table S9. Results of t test from Figure 6, E
Supplemental Data Set S1. Multiple protein alignment of BAHD protein sequences used for phylogeny reconstruction.
Supplemental Data Set S2. BAHD phylogenetic tree file.
Supplemental Data Set S3. Multiple protein alignment used to reconstruct HCT tridimensional models.
Supplementary Material
Acknowledgments
The authors would like to thank Pr. Takayuki Kohchi (Kyoto University) and Dr. Isabel Monte (University of Zürich) for providing M. polymorpha and A. agrestis plants, respectively. They are grateful to Annette Alber who performed initial in vitro PpHCT enzyme assays.
L.K. and H.R. designed the research. L.K., S.K., E.G., K.T., D.G., I.S., L.H., J.Z., and H.R. performed the research. L.K. and H.R. analyzed the data. L.K. and H.R. wrote the manuscript with critical input of J.K.C.R., R.R., and D.W.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Hugues Renault (renault@unistra.fr).
Funding
This work received support from the initiative of excellence IDEX-Unistra (ANR-10-IDEX-0002-02, H.R.), the Agence Nationale de la Recherche (ANR-19-CE20-0017, H.R.), the grant-in-aid “Diversity of Biological Mechanisms” from the Institut des Sciences Biologiques—CNRS (DBM2020, H.R.), the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy (EXC-2189—Project ID: 390939984, R.R.), the National Science Foundation (NSF-1517546, J.K.C.R.), and the Agriculture and Food Research Initiative of the United States Department of Agriculture (2016-67013-24732, J.K.C.R.).
Conflict of interest statement. None declared.
References
- Adams ZP, Ehlting J, Edwards R (2019) The regulatory role of shikimate in plant phenylalanine metabolism. J Theor Biol 462: 158–170 [DOI] [PubMed] [Google Scholar]
- Alber AV, Renault H, Basilio-Lopes A, Bassard J-E, Liu Z, Ullmann P, Lesot A, Bihel F, Schmitt M, Werck-Reichhart D, et al. (2019) Evolution of coumaroyl conjugate 3-hydroxylases in land plants: lignin biosynthesis and defense. Plant J 99: 924–936 [DOI] [PubMed] [Google Scholar]
- Alberti S, Gitler AD, Lindquist S (2007) A suite of Gateway® cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24: 913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O (2006) Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55: 539–552 [DOI] [PubMed] [Google Scholar]
- Barros J, Escamilla-Trevino L, Song L, Rao X, Serrani-Yarce JC, Palacios MD, Engle N, Choudhury FK, Tschaplinski TJ, Venables BJ, et al. (2019) 4-Coumarate 3-hydroxylase in the lignin biosynthesis pathway is a cytosolic ascorbate peroxidase. Nat Commun 10: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassard J-E, Richert L, Geerinck J, Renault H, Duval F, Ullmann P, Schmitt M, Meyer E, Mutterer J, Boerjan W, et al. (2012) Protein-protein and protein-membrane associations in the lignin pathway. Plant Cell 24: 4465–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M (2007) Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19: 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A, Scossa F, Bolger ME, Lanz C, Maumus F, Tohge T, Quesneville H, Alseekh S, Sorensen I, Lichtenstein G, et al. (2014) The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat Genet 46: 1034–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buda GJ, Barnes WJ, Fich EA, Park S, Yeats TH, Zhao L, Domozych DS, Rose JK (2013) An ATP binding cassette transporter is required for cuticular wax deposition and desiccation tolerance in the moss Physcomitrella patens. Plant Cell 25: 4000–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldicott AB,, Eglinton G (1976) Cutin acids from bryophytes: an ω-1 hydroxy alkanoic acid in two liverwort species. Phytochemistry 15: 1139–1143 [Google Scholar]
- Carrington Y, Guo J, Le CH, Fillo A, Kwon J, Tran LT, Ehlting J (2018) Evolution of a secondary metabolic pathway from primary metabolism: shikimate and quinate biosynthesis in plants. Plant J 95: 823–833 [DOI] [PubMed] [Google Scholar]
- Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552 [DOI] [PubMed] [Google Scholar]
- Chen F, Srinivasa Reddy MS, Temple S, Jackson L, Shadle G, Dixon RA (2006) Multi-site genetic modulation of monolignol biosynthesis suggests new routes for formation of syringyl lignin and wall-bound ferulic acid in alfalfa (Medicago sativa L.). Plant J 48: 113–124 [DOI] [PubMed] [Google Scholar]
- Chiang Y-C, Levsh O, Lam CK, Weng J-K, Wang Y (2018) Structural and dynamic basis of substrate permissiveness in hydroxycinnamoyltransferase (HCT). PLoS Comput Biol 14: e1006511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clé C, Hill LM, Niggeweg R, Martin CR, Guisez Y, Prinsen E, Jansen MAK (2008) Modulation of chlorogenic acid biosynthesis in Solanum lycopersicum; consequences for phenolic accumulation and UV-tolerance. Phytochemistry 69: 2149–2156 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de Vries J, Archibald JM (2018) Plant evolution: landmarks on the path to terrestrial life. New Phytol 217: 1428–1434 [DOI] [PubMed] [Google Scholar]
- de Vries J, de Vries S, Slamovits CH, Rose LE, Archibald JM (2017) How embryophytic is the biosynthesis of phenylpropanoids and their derivatives in streptophyte algae? Plant Cell Physiol 58: 934–945 [DOI] [PubMed] [Google Scholar]
- D’Auria JC (2006) Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol 9: 331–340 [DOI] [PubMed] [Google Scholar]
- DiGennaro P, Grienenberger E, Dao TQ, Jun JH, Fletcher JC (2018) Peptide signaling molecules CLE5 and CLE6 affect Arabidopsis leaf shape downstream of leaf patterning transcription factors and auxin. Plant Direct 2: e00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eudes A, Pereira JH, Yogiswara S, Wang G, Teixeira Benites V, Baidoo EEK, Lee TS, Adams PD, Keasling JD, Loqué D (2016) Exploiting the substrate promiscuity of Hydroxycinnamoyl-CoA:Shikimate Hydroxycinnamoyl Transferase to reduce lignin. Plant Cell Physiol 57: 568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fich EA, Segerson NA, Rose JK (2016) The plant polyester cutin: biosynthesis, structure, and biological roles. Annu Rev Plant Biol 67: 207–233 [DOI] [PubMed] [Google Scholar]
- Franke R, Humphreys JM, Hemm MR, Denault JW, Ruegger MO, Cusumano JC, Chapple C (2002) The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J 30: 33–45 [DOI] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Jikumaru Y, Kamiya Y, Tang Y, Dixon RA (2011) Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol 190: 627–639 [DOI] [PubMed] [Google Scholar]
- Girke T, Schmidt H, Zahringer U, Reski R, Heinz E (1998) Identification of a novel delta 6-acyl-group desaturase by targeted gene disruption in Physcomitrella patens. Plant J 15: 39–48 [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE (2018) UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci 27: 14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MA, Fry SC (2005) Vitamin C degradation in plant cells via enzymatic hydrolysis of 4-O-oxalyl-L-threonate. Nature 433: 83–87 [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Guo J, Carrington Y, Alber A, Ehlting J (2014) Molecular characterization of quinate and shikimate metabolism in populus trichocarpa. J Biol Chem 289: 23846–23858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CM, Escamilla-Trevino L, Yarce JCS, Kim H, Ralph J, Chen F, Dixon RA (2016) An essential role of caffeoyl shikimate esterase in monolignol biosynthesis in Medicago truncatula. Plant J 86: 363–375 [DOI] [PubMed] [Google Scholar]
- Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D, Lapierre C, Pollet B, Legrand M (2004) Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16: 1446–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L, Maury S, Martz F, Geoffroy P, Legrand M (2003) Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J Biol Chem 278: 95–103 [DOI] [PubMed] [Google Scholar]
- Hohe A, Egener T, Lucht JM, Holtorf H, Reinhard C, Schween G, Reski R (2004) An improved and highly standardised transformation procedure allows efficient production of single and multiple targeted gene-knockouts in a moss, Physcomitrella patens. Curr Genet 44: 339–347 [DOI] [PubMed] [Google Scholar]
- Jiao C, Sorensen I, Sun X, Behar H, Alseekh S, Philippe G, Lopez KP, Sun L, Reed R, Jeon S, et al. (2020) The Penium margaritaceum genome: hallmarks of the origins of land plants. Cell 181: 1097–1111 [DOI] [PubMed] [Google Scholar]
- Kenrick P, Crane PR (1997) The origin and early evolution of plants on land. Nature 389: 33–39 [Google Scholar]
- Kong L, Liu Y, Zhi P, Wang X, Xu B, Gong Z, Chang C (2020) Origins and evolution of cuticle biosynthetic machinery in land plants. Plant Physiol 184: 1998–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma DK, Bourdenx B, Bernard A, Parsons EP, Lü S, Joubès J, Jenks MA (2009) The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol 151: 1918–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss P, Markstadter C, Riederer M (1997) Attenuation of UV radiation by plant cuticles from woody species. Plant Cell Environ 20: 1079–1085 [Google Scholar]
- Lallemand LA, Zubieta C, Lee SG, Wang Y, Acajjaoui S, Timmins J, McSweeney S, Jez JM, McCarthy JG, McCarthy AA (2012) A structural basis for the biosynthesis of the major chlorogenic acids found in coffee. Plant Physiol 160: 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Ullrich KK, Murat F, Fuchs J, Jenkins JW, Haas FB, Piednoel M, Gundlach H, Van Bel M, Meyberg R, et al. (2018) The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J 93: 515–533 [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25: 1307–1320 [DOI] [PubMed] [Google Scholar]
- Lehfeldt C, Shirley AM, Meyer K, Ruegger MO, Cusumano JC, Viitanen PV, Strack D, Chapple C (2000) Cloning of the SNG1 gene of Arabidopsis reveals a role for a serine carboxypeptidase-like protein as an acyltransferase in secondary metabolism. Plant Cell 12: 1295–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenton TM, Dahl TW, Daines SJ, Mills BJ, Ozaki K, Saltzman MR, Porada P (2016) Earliest land plants created modern levels of atmospheric oxygen. Proc Natl Acad Sci U S A 113: 9704–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levsh O, Chiang YC, Tung CF, Noel JP, Wang Y, Weng JK (2016) Dynamic conformational states dictate selectivity toward the native substrate in a substrate-permissive acyltransferase. Biochemistry 55: 6314–6326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levsh O, Pluskal T, Carballo V, Mitchell AJ, Weng JK (2019) Independent evolution of rosmarinic acid biosynthesis in two sister families under the Lamiids clade of flowering plants. J Biol Chem 294: 15193–15205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Bonawitz ND, Weng JK, Chapple C (2010a) The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22: 1620–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Bergelson J, Chapple C (2010b) The Arabidopsis accession Pna-10 is a naturally occurring sng1 deletion mutant. Mol Plant 3: 91–100 [DOI] [PubMed] [Google Scholar]
- Liu Z, Tavares R, Forsythe ES, Andre F, Lugan R, Jonasson G, Boutet-Mercey S, Tohge T, Beilstein MA, Werck-Reichhart D, et al. (2016) Evolutionary interplay between sister cytochrome P450 genes shapes plasticity in plant metabolism. Nat Commun 7: 13026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü S, Zhao H, Des Marais DL, Parsons EP, Wen X, Xu X, Bangarusamy DK, Wang G, Rowland O, Juenger T, et al. (2012) Arabidopsis ECERIFERUM9 involvement in cuticle formation and maintenance of plant water status. Plant Physiol 159: 930–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Dudareva N (2012) The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu Rev Plant Biol 63: 73–105 [DOI] [PubMed] [Google Scholar]
- Matsuno M, Compagnon V, Schoch GA, Schmitt M, Debayle D, Bassard J-E, Pollet B, Hehn A, Heintz D, Pascaline U, et al. (2009) Evolution of a novel phenolic pathway for pollen development. Science 325: 1688–1692 [DOI] [PubMed] [Google Scholar]
- Molina I, Bonaventure G, Ohlrogge J, Pollard M (2006) The lipid polyester composition of Arabidopsis thaliana and Brassica napus seeds. Phytochemistry 67: 2597–2610 [DOI] [PubMed] [Google Scholar]
- Moody LA, Kelly S, Clayton R, Weeks Z, Emms DM, Langdale JA (2020) NO GAMETOPHORES 2 is a novel regulator of the 2D to 3D growth transition in the moss Physcomitrella patens. Curr Biol 31: 1–9 [DOI] [PubMed] [Google Scholar]
- Morris JL, Puttick MN, Clark JW, Edwards D, Kenrick P, Pressel S, Wellman CH, Yang Z, Schneider H, Donoghue PCJ (2018) The timescale of early land plant evolution. Proc Natl Acad Sci U S A 115: 201719588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Schreiber L, Franke RB, Geldner N, Reina-Pinto JJ, Kunst L (2013) Apoplastic diffusion barriers in Arabidopsis. Arab B 11: e0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggeweg R, Michael AJ, Martin C (2004) Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat Biotechnol 22: 746–754 [DOI] [PubMed] [Google Scholar]
- Perroud P-F, Haas FB, Hiss M, Ullrich KK, Alboresi A, Amirebrahimi M, Barry K, Bassi R, Bonhomme S, Chen H, et al. (2018) The Physcomitrella patens gene atlas project: large-scale RNA-seq based expression data. Plant J 95: 168–182 [DOI] [PubMed] [Google Scholar]
- Philippe G, Sørensen I, Jiao C, Sun X, Fei Z, Domozych DS, Rose JK (2020) Cutin and suberin: assembly and origins of specialized lipidic cell wall scaffolds. Curr Opin Plant Biol 55: 11–20 [DOI] [PubMed] [Google Scholar]
- Porada P, Lenton TM, Pohl A, Weber B, Mander L, Donnadieu Y, Beer C, Poschl U, Kleidon A (2016) High potential for weathering and climate effects of non-vascular vegetation in the Late Ordovician. Nat Commun 7: 12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttick MN, Morris JL, Williams TA, Cox CJ, Edwards D, Kenrick P, Pressel S, Wellman CH, Schneider H, Pisani D, et al. (2018) The interrelationships of land plants and the nature of the ancestral embryophyte. Curr Biol 28: 733–745 [DOI] [PubMed] [Google Scholar]
- Ralph J, Lapierre C, Boerjan W (2019) Lignin structure and its engineering. Curr Opin Biotechnol 56: 240–249 [DOI] [PubMed] [Google Scholar]
- Rautengarten C, Ebert B, Ouellet M, Nafisi M, Baidoo EE, Benke P, Stranne M, Mukhopadhyay A, Keasling JD, Sakuragi Y, et al. (2012) Arabidopsis deficient in cutin ferulate encodes a transferase required for feruloylation of ω-hydroxy fatty acids in cutin polyester. Plant Physiol 158: 654–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault H, Alber A, Horst NA, Basilio Lopes A, Fich EA, Kriegshauser L, Wiedemann G, Ullmann P, Herrgott L, Erhardt M, et al. (2017) A phenol-enriched cuticle is ancestral to lignin evolution in land plants. Nat Commun 8: 14713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault H, Werck-Reichhart D, Weng J-K (2019) Harnessing lignin evolution for biotechnological applications. Curr Opin Biotechnol 56: 105–111 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud P-F, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Reski R (2018) Enabling the water-to-land transition. Nat Plants 4: 67–68 [DOI] [PubMed] [Google Scholar]
- Reski R, Abel WO (1985) Induction of budding on chloronemata and caulonemata of the moss, Physcomitrella patens, using isopentenyladenine. Planta 165: 354–358 [DOI] [PubMed] [Google Scholar]
- Retallack GJ (1997) Early forest soils and their role in devonian global change. Science 276: 583–585 [DOI] [PubMed] [Google Scholar]
- Riley RG, Kolattukudy PE (1975) Evidence for covalently attached p-coumaric acid and ferulic acid in cutins and suberins. Plant Physiol 56: 650–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleme MdLS, Cesarino I, Vargas L, Kim H, Vanholme R, Goeminne G, Van Acker R, Fonseca FCdA, Pallidis A, Voorend W, et al. (2017) Silencing CAFFEOYL SHIKIMATE ESTERASE affects lignification and improves saccharification in poplar. Plant Physiol 175: 1040–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234: 779–815 [DOI] [PubMed] [Google Scholar]
- Sander M, Petersen M (2011) Distinct substrate specificities and unusual substrate flexibilities of two hydroxycinnamoyltransferases, rosmarinic acid synthase and hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl-transferase, from Coleus blumei Benth. Planta 233: 1157–1171 [DOI] [PubMed] [Google Scholar]
- Schoch G, Goepfert S, Morant M, Hehn A, Meyer D, Ullmann P, Werck-Reichhart D (2001) CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J Biol Chem 276: 36566–36574 [DOI] [PubMed] [Google Scholar]
- Schoch GA, Morant M, Abdulrazzak N, Asnaghi C, Goepfert S, Petersen M, Ullmann P, Werck-Reichhart D (2006) The meta-hydroxylation step in the phenylpropanoid pathway: a new level of complexity in the pathway and its regulation. Environ Chem Lett 4: 127–136 [Google Scholar]
- Sibout R, Le Bris P, Legee F, Cezard L, Renault H, Lapierre C (2016) Structural redesigning Arabidopsis lignins into alkali-soluble lignins through the expression of p-coumaroyl-CoA:Monolignol Transferase PMT. Plant Physiol 170: 1358–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber MV, Meimberg H, Ebel J (2008) Identification of a 4-coumarate:CoA ligase gene family in the moss, Physcomitrella patens. Phytochemistry 69: 2449–2456 [DOI] [PubMed] [Google Scholar]
- Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D (1997) Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem 272: 19176–19186 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P, et al. (2013) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Vogt T (2010). Phenylpropanoid biosynthesis. Mol Plant 3: 2–20 [DOI] [PubMed] [Google Scholar]
- Wagner A, Ralph J, Akiyama T, Flint H, Phillips L, Torr K, Nanayakkara B, Te Kiri L(2007) Exploring lignification in conifers by silencing hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase in Pinus radiata. Proc Natl Acad Sci U S A 104: 11856–11861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng JK, Akiyama T, Ralph J, Chapple C (2011) Independent recruitment of an O-methyltransferase for syringyl lignin biosynthesis in Selaginella moellendorffii. Plant Cell 23: 2708–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng JK, Chapple C (2010) The origin and evolution of lignin biosynthesis. New Phytol 187: 273–285 [DOI] [PubMed] [Google Scholar]
- Weng JK, Li X, Stout J, Chapple C (2008) Independent origins of syringyl lignin in vascular plants. Proc Natl Acad Sci U S A 105: 7887–7892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA, et al. (2014) Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci U S A 111: E4859–E4868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Rose JKC (2013) The formation and function of plant cuticles. Plant Physiol 163: 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R, Han K, Heller W, Albert A, Dobrev PI, Zažímalová E, Schäffner AR (2014) Kaempferol 3-O-rhamnoside-7-O-rhamnoside is an endogenous flavonol inhibitor of polar auxin transport in Arabidopsis shoots. New Phytol 201: 466–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.