Abstract
Light-dependent seed germination is a vital process for many seed plants. A decisive event in light-induced germination is degradation of the central repressor PHYTOCHROME INTERACTING FACTOR 1 (PIF1). The balance between gibberellic acid (GA) and abscisic acid (ABA) helps to control germination. However, the cellular mechanisms linking PIF1 turnover to hormonal balancing remain elusive. Here, employing far-red light-induced Arabidopsis thaliana seed germination as the experimental system, we identified PLANTACYANIN (PCY) as an inhibitor of germination. It is a blue copper protein associated with the vacuole that is both highly expressed in mature seeds and rapidly silenced during germination. Molecular analyses showed that PIF1 binds to the miR408 promoter and represses miR408 accumulation. This in turn posttranscriptionally modulates PCY abundance, forming the PIF1-miR408-PCY repression cascade for translating PIF1 turnover to PCY turnover during early germination. Genetic analysis, RNA-sequencing, and hormone quantification revealed that PCY is necessary and sufficient to maintain the PIF1-mediated seed transcriptome and the low-GA-high-ABA state. Furthermore, we found that PCY domain organization and regulation by miR408 are conserved features in seed plants. These results revealed a cellular mechanism whereby PIF1-relayed external light signals are converted through PCY turnover to internal hormonal profiles for controlling seed germination.
The conserved blue copper protein PLANTACYANIN is regulated by PIF1 and miR408 to balance the ratio of gibberellic acid and abscisic acid which regulates far-red light dependent seed germination.
Introduction
The seed is an embryonic plant enclosed in a protective capsule. After reaching full size, the embryo undergoes elaborate dehydration to establish a dormant state that helps the embryo withstand extreme environments and survive for long periods (Bewley 1997; Finch-Savage and Leubner-Metzger, 2006; Angelovici et al., 2010). Given the right environmental conditions, the desiccated seed germinates by taking up water and resuming embryo development, utilizing energy and nutrient stored in the seed (Bewley 1997; Finch-Savage and Leubner-Metzger, 2006; Finkelstein et al., 2008; Née et al., 2017). Gibberellic acid (GA) and abscisic acid (ABA) are the main plant hormones that control seed dormancy and germination. As the embryo matures, ABA is synthesized and signals the embryo to initiate the buildup of storage compounds and undergo desiccation (Nambara and Marion-Poll, 2005; Finkelstein et al., 2008; Angelovici et al., 2010; Shu et al., 2016). ABA is also important for maintaining seed dormancy and preventing precocious germination (Bewley 1997; Finch-Savage and Leubner-Metzger, 2006; Finkelstein et al., 2008). Conversely, GA is a crucial hormone necessary to break down dormancy and promote germination (Kallioo and Piiroinen 1959; Bewley 1997; Finch-Savage and Leubner-Metzger, 2006; Yamaguchi, 2008). It is well established that the GA/ABA balance critically determines the germination capacity (Nambara and Marion-Poll, 2005; Yamaguchi, 2008; Shu et al., 2016; Née et al., 2017).
Seeds must monitor a wide range of environmental factors, including ambient light, for optimal germination (Oh et al., 2004; Finch-Savage and Leubner-Metzger, 2006; Seo et al., 2009; Jiang et al., 2016). Molecular mechanisms of light perception and signaling during germination are well understood in the model plant Arabidopsis thaliana (Arabidopsis). PIF1 is a basic helix–loop–helix type transcription factor that is an essential negative regulator of light-dependent germination (Oh et al., 2004; Leivar and Quail, 2010; Shi et al., 2015). PIF1 is stabilized by DEETIOLATED 1 and other signaling molecules and thus it accumulates to high levels in seeds kept in darkness (Oh et al., 2004; 2006; Shi et al., 2015). Following light irradiation, phytochromes (Phys) are activated and enter the nucleus to interact with PIF1, inhibiting its activity and promoting its degradation via the 26S proteasome (Oh et al., 2006; Castillon et al., 2007; Shen et al., 2008; Leivar and Quail, 2010). Rapid removal of PIF1 is critical for maintaining the light-regulated transcriptome in imbibed seed (Oh et al., 2009; Shi et al., 2013; Pfeiffer et al., 2014) and ultimately it is also critical for establishing the high-GA-low-ABA state (Oh et al., 2006; 2007). However, an extensive search has not revealed a direct link between PIF1 and genes involved in GA and ABA metabolism (Oh et al., 2007; 2009; Cho et al., 2012). Consequently, the cellular events resulting from PIF1 turnover that led to hormonal balancing have not been elucidated and it is not known whether these events are conserved in seed plants.
A fundamental cellular process that is required during seed germination is the mobilization of mineral nutrients sequestered in the storage vacuoles. This sustains embryo growth before efficient uptake systems are established in the root (Lanquar et al., 2005; Kim et al., 2006; Roschzttardtz et al., 2009; Née et al., 2017; Paszkiewicz et al., 2017). Studies in Arabidopsis and other plants have shown that transition metals such as iron and manganese are released from vacuolar stores via tonoplast-localized transporters and are then transported to various cellular destinations (Lanquar et al., 2005; Kim et al., 2006; Eroglu et al., 2017). While the physiological consequences of insufficient or misregulated mineral mobilization have been abundantly documented (Lanquar et al., 2005; Kim et al., 2006), whether metal mobilization contributes to the establishment of hormonal profiles in light dependent germination is not yet well characterized.
Copper is an essential transition metal by serving as the cofactor for a number of cuproproteins with vital functions (Burkhead et al., 2009; Peñarrubia et al., 2015). Because free cellular copper is highly reactive and produces detrimental hydroxyl radicals, elaborate homeostasis and transport systems are required for the precise control of copper delivery to specific targets (Burkhead et al., 2009). The Arabidopsis genome encodes approximately 260 copper-dependent proteins (Schulten et al., 2019). Among them, small blue copper proteins, which contain a characteristic mononuclear type-I copper binding site, play important roles in redox reactions and copper homeostasis (Rydén and Hunt 1993; Guss et al., 1998; De Rienzo et al. 2000; Giri et al. 2004). Plastocyanin is the most abundant small blue copper protein in plants and an indispensable electron carrier in the Z-scheme of photosynthesis (Molina-Heredia et al., 2003; Weigel et al., 2003). Plants also have a specific family of plastocyanin-like blue copper proteins known as phytocyanins. They are divided into four subfamilies: PCYs, uclacyanins, stellacyanins, and early nodulin-like proteins, based on differences in the copper binding site and domain organization (Guss et al., 1998; Nersissian et al., 1998; Sun et al., 2019). Phytocyanins have been implicated in plant development processes such as pollen tube chemotropism and nodule development (Kim et al. 2003; Dong et al. 2005; Sun et al., 2019). They have also been implicated in stress responses such as pathogen resistance and drought and salinity tolerance (Jung and Hwang 2000; Ruan et al. 2011; Feng et al., 2013). However, their involvement in seed germination has not been investigated.
In this study, we focused on PCY in Arabidopsis that is highly expressed in the seed with contrasting expression patterns during seed development and germination. Through comprehensive molecular and genetic analyses, we delineated the PIF1-miR408-PCY repression cascade for regulating PCY turnover during far-red light induced germination. We showed that PCY locates to the storage vacuole and is necessary and sufficient to maintain PIF1-mediated seed transcriptome and the low-GA-high-ABA state. These results identified PCY-based copper mobilization as a potentially conserved cellular mechanism for converting PIF1-relayed light signals into hormonal profiles that control seed germination.
Results
PCY exhibits a distinctive expression pattern in seed development and germination
Using a gene expression atlas ien Arabidopsis (eFP Browser; Winter et al., 2007), we profiled expression pattern of the phytocyanin-encoding genes during seed development. We found that 31 of the 37 phytocyanin genes were expressed during seed formation and germination (Figure 1). Among these, the locus encoding PCY (At2g02850) exhibited the highest expression level in mature seed (Figure 1A). According to a temporal progression scheme that divides seed development into 10 stages (Schmid et al., 2005), we found that the PCY transcript level was low in early seed development, but was drastically induced at stage 7, progressively increased thereafter, and peaked at stage 10 (seed maturation; Figure 1B). Interestingly, PCY was drastically silenced during storage and imbibition (Figure 1B).
Figure 1.
PCY is induced during seed development and rapidly silenced during germination. (A) Global expression profile of 31 phytocyanin genes in the seed based on data in the Arabidopsis eFP Browser. Expression levels are indicated by the color scale at the bottom. (B) Comparison of PCY expression patterns in the seed and other organs. Data are mean ± SD of three individual experiments obtained from the eFP Browser. (C) Diagram illustrating the phyAOFF and phyAON regimes. Imbibed seeds were sequentially treated with the indicated light conditions. Arrows indicate when time points in various assays were analyzed. (D) Relative PCY transcript levels during the time course of phyAOFF and phyAON described in (C), as determined by RT-qPCR analysis. Data are mean ± SD from three independent qPCR reactions performed on the same pool of reverse-transcribed cDNA. Different letters at the same time point denote significant differences (one-way ANOVA, P < 0.001, Supplemental Data Set 3).
To elucidate the dynamics of PCY repression, we employed a previously described far-red light initiated, phytochrome A (phyA)-dependent germination assay (Oh et al., 2006; 2009; Cho et al., 2012). In the phyAOFF regime of this assay, imbibed seeds are briefly exposed to far-red light to inactivate phyB, then kept in darkness to allow inactive phyA to accumulate so that the seed does not break dormancy. In the phyAON regime, a second far-red irradiation with longer duration is used to activate phyA that induces germination (Figure 1C). Using reverse transcription coupled quantitative PCR (RT-qPCR), we observed that PCY transcript level remained steady during the time course of phyAOFF (Figure 1D). In contrast, after the second far-red irradiation in phyAON, PCY level was drastically reduced by the 4 h time point and remained low thereafter (Figure 1D). These results indicate that PCY was highly expressed in mature seed and was rapidly silenced during light-induced germination.
To validate the PCY expression pattern at the protein level during the germination phase, we generated the PCYpro:PCY-GFP Arabidopsis plants in which the PCY coding sequence was fused to the green fluorescence protein (GFP) and driven by the native PCY promoter (Figure 2). Immunoblotting with an antibody against GFP revealed that the PCY–GFP fusion protein was drastically suppressed within 24 h after vernalization (Figure 2A). By analyzing PCY–GFP transiently expressed in onion epidermal cells, we found that the GFP signals predominantly aligned the periphery of the central vacuole (Figure 2B). Moreover, PCY–GFP colocalized with mCherry-tagged COPT5 (Figure 2B), which is a member of the CTR-like high-affinity copper transporters residing on the tonoplast (Klaumann et al., 2011). This indicated that transiently expressed PCY is associated with the vacuole.
Figure 2.
PCY is vacuole associated and removed upon far-red irradiation. (A) Detection of PCY–GFP in PCYpro:PCY-GFP plants using an anti-GFP antibody. Size markers are indicated on the right. RPT5 was used as the loading control. (B) Subcellular localization of PCY. PCY–GFP and COPT5-mCherry were transiently expressed in the same onion epidermal cells and examined by confocal fluorescence microscopy. Bar, 10 μm. (C) Confirmation of vacuole association of PCY by immuno-gold labeling. Cotyledon cells of imbibed PCYpro:PCY-GFP seed in phyAOFF were analyzed by immuno-electron microscopy using an antibody for GFP. Arrows indicate the size-defined electron-dense immunogold particles labeling PCY-GFP. Bar, 0.2 μm. (D–E) Co-localization of GFP fluorescence with vacuole autofluorescence in cotyledon cells of imbibed PCYpro:PCY-GFP seed in phyAOFF (D) and phyAON (E). Bar, 10 μm.
To examine PCY localization in the seed, we performed immunoelectron microscopy on the PCYpro:PCY-GFP seed using a GFP antibody. The round, size-defined electron-dense immunogold particles labeling PCY–GFP were concentrated in the storage vacuoles and mainly surround the tonoplast (Figure 2C). Note that the dark particles outside the vacuole, which are less electron-dense and with no clearly defined sizes and shapes, are not immunogold-labeled PCY-GFP but are other substances stained by uranyl acetate. Using the PCYpro:PCY-GFP seed, we examined whether vacuole-associated PCY would be removed by far-red irradiation. In imbibed seed kept in darkness, GFP fluorescence was observed surrounding autofluorescence of storage vacuoles (Figure 2D). Consistent with the profile of the PCY transcript (Figure 1D), the GFP signals disappeared after far-red irradiation (Figure 2E). These observations indicated that storage vacuole-associated PCY was rapidly silenced following phyA activation.
PCY negatively regulates germination
The expression pattern of PCY inspired us to investigate its role in germination genetically. We used the previously characterized Arabidopsis mutant with an intronic T-DNA insertion and named this knockdown allele pcy-1 (Supplemental Figure 1) (Dong et al., 2005). We also generated a deletion mutant using the CRISPR-Cas9 system with paired guide RNAs. The resulting homozygous mutant, which contained a 506-bp deletion that spans the entire coding region, was named pcy-2 (Supplemental Figure 1A). Assays using RT-qPCR confirmed the absence of the PCY transcript in pcy-2 relative to the wild type (Supplemental Figure 1B). Consistent with previous characterization of pcy-1 (Dong et al., 2005), we found that both pcy mutants exhibited no apparent defects in the appearance of the mature seed (Supplemental Figure 1C). However, in contrast to the wild-type seed that failed to germinate in the phyAOFF regime, by 120 h in the phyAOFF regime, the germination frequency of pcy-1 and pcy-2 increased to 17% and 233%, respectively. This was significantly higher than the wild-type based on one-way analysis of variance (ANOVA) (Figure 3). In phyAON, the 48% and 63% germination rates for pcy-1 and pcy-2 were significantly higher than the approximately 40% germination rate of the wild type (Figure 3A and 3C). These results indicate that PCY is necessary for inhibiting germination in both phyAOFF and phyAON.
Figure 3.
PCY negatively regulates germination. (A) Representative plates showing the germination states of wild type and pcy seeds in phyAOFF (top) and phyAON (bottom). (B–C) Quantification of the germination rate over the time course of phyAOFF (B) and phyAON (C). Data are mean ± SD from three individual experiments. Different letters represent genotypes with significant differences at 120 h (one-way ANOVA, P < 0.01, Supplemental Data Set 3). (D) RT-qPCR analysis of relative PCY transcript level in the indicated genotypes without and with the application of β-estradiol. Data are mean ± SD from three replicates performed on the same cDNA. Different letters denote genotypes with significant differences (one-way ANOVA, P < 0.05, Supplemental Data Set 3). (E) Representative plates showing the germination states of the wild type and iPCY-OX seeds in phyAOFF and phyAON under the indicated treatments. (F) Quantification of germination rates of the wild type and iPCY-OX seeds. Data are mean ± SD from three independent experiments. Different letters denote genotypes with significant differences (one-way ANOVA, P < 0.05, Supplemental Data Set 3).
To test whether PCY was sufficient for repressing germination, we generated the iPCY-OX Arabidopsis plants in which PCY was controlled by a β-estradiol-inducible promoter (Zuo et al., 2000). The PCY transcript was induced to high levels in the iPCY-OX seed, which exhibited no morphological difference from the wild type (Supplemental Figure 1C) under both phyAOFF and phyAON after the application of β-estradiol (Figure 3D;Supplemental Figure 1D). This treatment significantly reduced the germination rate of the iPCY-OX seed, but not that of the wild type, in both phyAOFF and phyAON (Figure 3E and F), indicating that PCY accumulation is sufficient for inhibiting germination.
Young etiolated seedlings germinated in darkness switch to a different growth mode, termed greening, upon exposure to light (Zhong et al., 2009). Visual inspection revealed that the greening rate of dark-germinated pcy-2 seedlings (referred to as pcy hereafter) was significantly higher than that of the wild type after exposure to white light for 24 h (Figure 4). Fluorescence spectral analysis confirmed that the levels of both chlorophylls and their precursor protochlorophyllide in pcy seedlings were significantly higher than those in the wild type (Figure 4C and D). Moreover, we examined key pigment biosynthetic genes by RT-qPCR, including GLUTAMYL-TRNA REDUCTASE (HEMA1), GENOMES UNCOUPLED 4/5 (GUN4/5), and PROTOCHLOROPHYLLIDE OXIDOREDUCTASE A/B/C (PORA/B/C) (Tanaka et al., 2011). We found that, except for HEMA1, the expression of pigment genes was significantly upregulated in pcy in comparison to the wild type (Figure 4E). Collectively, these results established PCY as a negative regulator for germination and post-germinative growth in light.
Figure 4.
PCY negatively regulates seedling greening. (A) Representative wild type and pcy seedlings. Seedlings were grown in the dark for 96 h and then exposed to continuous white light for 24 h. Bar, 1 mm. (B) Quantified greening rate as described in Methods. Data are mean ± SD from three individual experiments. Different letters denote significant differences (P < 0.05 by Student’s t- test, Supplemental Data Set 3). (C–D) Comparison of pigment profiles in pcy versus wild-type seedlings. Protochlorophyllide and chlorophylls were assayed by spectral analysis in 4-day-old etiolated seedlings and etiolated seedlings exposed to white light for 24 h, respectively. Shown on the left are results from a representative experiment. Shown on the right is quantification of the protochlorophyllide fluorescence at 634 nm and chlorophyll fluorescence at 678 nm. Data are mean ± SD from three individual experiments. Different letters denote significant differences (Student’s t-test, P < 0.05 for C, P < 0.01 for D). (E) RT-qPCR analysis of the relative transcript levels of pigment biosynthetic genes in 4-day-old etiolated seedlings. Data are mean ± SD from three replicates performed on the same cDNA. For each gene, different letters denote significant differences (P < 0.05 by Student’s t-test, Supplemental Data Set 3).
PCY is silenced by miR408 during germination
PCY is a proven target of miR408 (Abdel-Ghany and Pilon, 2008; Zhang and Li, 2013). Using the standard assay of 5′ RNA ligation-based amplification of cDNA ends (RACE), we validated miR408-guided cleavage of the PCY mRNA in imbibed seed. This cleavage occurred between the 10th and 11th nucleotides in the miR408 recognition site (Figure 5). From degradome sequencing data on young seedlings, we retrieved reads that were mapped to the predicted miR408 recognition site in PCY (Figure 5B). This confirmed miR408 mediated cleavage of the PCY transcript. RT-qPCR analysis showed that the miR408 level in the seed was stable in the phyAOFF regime but drastically elevated 2 h after the second far-red irradiation and that it peaked at 8 h in phyAON (Figure 5C). Thus, miR408 and PCY exhibit reciprocal expression pattern following phyA activation (compare Figures 1D and 5C).
Figure 5.
miR408 represses PCY expression during germination. (A) Confirmation of miR408 targeting to PCY in the seed by RNA-ligation-based amplification of cDNA ends. The structure of PCY (top) and base pairing between miR408 and PCY (bottom) is shown. Arrows mark the detected transcript ends along with frequency of the corresponding clones. (B) Degradome sequencing data supporting miR408-guided cleavage of the PCY transcript. The frequencies of the sequenced 5′ ends are plotted against nucleotide position in the PCY transcript. The red dot indicates the position of the miR408 recognition site. (C) RT-qPCR analysis of relative miR408 levels over the time course of phyAOFF and phyAON. Data are means ± SD from three replicates performed on the same cDNA. (D) Relative PCY transcript levels in seeds of the indicated genotypes under phyAOFF and phyAON. Data are means ± SD from three replicates performed on the same cDNA. Different letters denote genotypes with significant differences (one-way ANOVA, P < 0.01, Supplemental Data Set 3).
To test the effect of miR408 on PCY expression during germination, we employed the miR408-overexpressing line (miR408-OX), in which the enhanced cauliflower mosaic virus 35S promoter (35Spro) was used to drive miR408 expression and the miR408-silencing line (amiR408) generated by an artificial miRNA that cleaves the miR408 precursor (Zhang and Li, 2013; Zhang et al., 2014). In the miR408-OX and amiR408 seeds, expression of PCY in phyAOFF was significantly decreased and increased in comparison to the wild type, respectively (Figure 5D). In phyAON, while PCY level was generally lowered compared to that in phyAOFF, the trend of relative PCY abundance in the wild type, miR408-OX, and amiR408 seeds maintained the same (Figure 5D). Taken together, these results indicate that miR408 negatively modulates PCY expression via the canonical transcript cleavage mechanism during early seed germination.
miR408 is a positive regulator of germination
To investigate the role of miR408 in germination, we examined phenotypes of the miR408-OX and amiR408 seeds (Figure 6). In phyAOFF, the wild type and amiR408 seeds both failed to germinate while the germination frequency of miR408-OX seeds increased to about 80% over the examined time course (Figure 6B). In phyAON, nearly 100% of the miR408-OX seed germinated after 120 h (Figure 6C). In contrast, the germination frequency of amiR408 reached only about 30%, significantly lower than that of the wild type (Figure 6C). We also generated another miR408-silencing mutant (STTM-miR408) using the short tandem target mimic approach that sequesters the mature miRNA (Yan et al., 2012). We found that STTM-miR408 seeds showed reduced germination in phyAON similar as amiR408 (Supplemental Figure 2). Additionally, we found that the greening rate and pigment contents of the miR408-OX and amiR408 seeds were significantly higher and lower than those of the wild type, respectively (Figure 6D–I). These results demonstrate that, contrary to PCY, miR408 is a positive regulator for germination and post-germinative growth.
Figure 6.
miR408 promotes seed germination and seedling greening. (A) Representative plates showing the germination states of the indicated genotypes under phyAOFF (left) and phyAON (right). (B–C) Quantification of germination rate of the indicated genotypes over the time course of phyAOFF (B) and phyAON (C). Data are mean ± SD from three individual experiments. Different letters denote genotypes with significant differences at 120 h (one-way ANOVA, P < 0.05 for B, P < 0.001 for C, Supplemental Data Set 3). (D) Representative etiolated seedlings of the indicated genotypes that were exposed to white light for 24 h. Bar, 1 mm. (E) Quantified greening rates of miR408-OX and amiR408 in comparison to the wild type. Data are mean ± SD from three independent experiments. Different letters represent significant differences (one-way ANOVA, P < 0.05, Supplemental Data Set 3). (F–I) Protochlorophyllide and chlorophylls were assayed in 4-day-old etiolated seedlings and etiolated seedlings exposed to white light for 24 h, respectively. Results from a representative spectral analysis are shown in F and H. The protochlorophyllide fluorescence at 634 nm (G) and chlorophyll fluorescence at 678 nm (I) were quantified. Data are mean ± SD from three individual experiments. Different letters represent significant differences (one-way ANOVA, P < 0.001, Supplemental Data Set 3).
Because the germination phenotype of miR408-OX was stronger than that of pcy, we examined other miR408 target genes. In Arabidopsis, miR408 has four validated targets that all encode cuproproteins, including PCY, LACCASE 3/12/13 (Abdel-Ghany and Pilon, 2008; Zhang and Li, 2013; Zhang et al., 2014). We found that the three LAC genes did not exhibit substantial expression in the seed (Supplemental Figure 3) (Winter et al., 2007; Zhuang et al., 2020). Phenotypic comparison of the lac3, lac12, lac13, and pcy single mutants revealed that only pcy displayed significantly elevated germination frequency than the wild type in both phyAOFF and phyAON (Supplemental Figure 3B). We next generated the lac12 lac13 double mutant and the pcy lac12 lac13 triple mutant. The pcy lac12 lac13 seed exhibited the same germination phenotype as pcy, while lac12 lac13 showed no difference from the wild type (Supplemental Figure 3C). These results indicated that other miR408 target genes were not involved in regulating germination. Whether the relatively weaker phenotype of pcy was due to the compensation by other phytocyanins warrants further investigation.
PIF1 directly suppresses miR408 to promote PCY expression
Our next goal was to find out how the miR408-PCY module is regulated by light signaling. Previously, we reported that ELONGATED HYPOCOTYL 5 (HY5), activates miR408 in response to increasing light irradiation by binding to the G box in the miR408 promoter (Zhang et al., 2014). Since PIFs and HY5 could antagonistically adjust the expression of common target genes (Chen et al., 2013; Toledo-Ortiz et al., 2014; Shi et al., 2018), we investigated the possibility that PIF1 transcriptionally represses miR408. Using PIF1 chromatin immunoprecipitation (ChIP) sequencing data (Pfeiffer et al., 2014), we identified a significantly enriched PIF1-binding region in the miR408 promoter encompassing the G box (Figure 7; Supplemental Figure 4). Using the PIF1-OX line expressing MYC-tagged PIF1 driven by 35Spro (Oh et al., 2004), we performed ChIP with an anti-MYC antibody followed with qPCR analysis. We found that PIF1 occupancy at the G box containing DNA fragment was enriched over fivefold in PIF1-OX relative to the wild type (Figure 7B), but was not enriched in the surrounding genomic regions (Supplemental Figure 4). These results demonstrate that PIF1 is a direct upstream regulator of miR408.
Figure 7.
PIF1 suppresses miR408 expression by binding to the miR408 promoter. (A) PIF1 occupancy profile at the miR408 locus. Significantly enriched PIF1 ChIP-sequencing reads were obtained from Pfeiffer et al. (2014) and mapped onto the Arabidopsis genome coordinates. Loci are represented by grey and black arrows. The blue circle marks the G box (CACGTG) in the miR408 promoter (horizontal line). (B) ChIP-qPCR confirming PIF1 binding to the miR408 promoter. An anti-MYC antibody was used to precipitate chromatin from PIF1-OX and wild-type seeds. Enrichment of PIF1 binding was determined by qPCR analysis. Data are mean ± SD from three qPCR performed on the same DNA. Different letters denote significant differences (P < 0.05 by Student’s t-test, Supplemental Data Set 3). (C) Transient dual luciferase assay showing PIF1 repression of miR408. The miR408pro:LUC reporter concatenated to 35Spro:REN was used to transform tobacco protoplasts with either the empty vector (−PIF1) or a PIF1-expressing construct (+ PIF1). (D) Quantification of the LUC/REN luminescence ratio. Data are mean ± SD from three independent transfections. Different letters denote significant differences (P < 0.05 by Student’s t-test, Supplemental Data Set 3). (E) Histochemical staining for GUS activity from miR408pro:GUS expressed in the wild type or pif1 seeds in phyAOFF and phyAON. Bar, 0.5 mm. (F) Quantification of GUS concentration by ELISA. Data are mean ± SD from three individual experiments. Statistical analysis was performed for data from phyAOFF and phyAON separately and different letters denote significant differences (P < 0.05 by Student’s t-test, Supplemental Data Set 3). DW, dry weight. (G–H) RT-qPCR analysis of relative miR408 (G) and PCY (H) transcript levels. Data are mean ± SD from three replicates performed on the same cDNA. Statistical analysis was performed for data from phyAOFF and phyAON separately and different letters denote genotypes with significant differences (one-way ANOVA, P < 0.05, Supplemental Data Set 3).
To ascertain the effect of PIF1 on miR408, we employed the firefly luciferase (LUC) and Renilla luciferase (REN) dual reporter system (Hellens et al., 2005). We generated the miR408pro:LUC reporter and the 35Spro:PIF1 effector constructs (Figure 7C). Following transfection of tobacco leaf protoplasts, we found that co-expression of PIF1 significantly reduced the LUC/REN ratio (Figure 7D). This indicated that PIF1 negatively modulates the miR408 promoter. To corroborate this relationship in Arabidopsis, we fused the β-glucuronidase (GUS) coding region with the miR408 promoter and expressed the same reporter gene in either the wild type (miR408pro:GUS/WT) or the pif1 (miR408pro:GUS/pif1) background (Figure 7E). We found that GUS activity, mainly detected in the cotyledons, was higher in the pif1 background in phyAOFF (Figure 7E). In phyAON where PIF1 is degraded, GUS activity in the pif1 and the wild-type backgrounds appeared to be comparable (Figure 7E). This pattern was corroborated by quantification of GUS concentration using an enzyme-linked immunosorbent assay (ELISA) (Figure 7F). These results confirmed PIF1-mediated suppression of miR408.
To monitor the influence of PIF1 on miR408 and PCY transcript accumulation, we performed RT-qPCR analysis of the pif1 and PIF1-OX seeds. This analysis revealed that miR408 abundance significantly increased in pif1 but decreased in PIF1-OX with reference to the wild type in phyAOFF (Figure 7G). Conversely, PCY abundance significantly increased in PIF1-OX but decreased in pif1 (Figure 7H). In phyAON, the trend remained while the miR408 level was not significantly altered in pif1 relative to the wild type (Figure 7G). Taken together, these results demonstrate that PIF1 binds to the miR408 promoter and represses miR408 accumulation in phyAOFF, which in turn posttranscriptionally silences PCY, thereby forming the PIF1-miR408-PCY repression cascade.
PIF1, miR408, and PCY act in the same pathway to regulate germination
Consistent with previously reports (Oh et al., 2004), we found that the pif1 seed completely germinated by 72 h in both phyAOFF and phyAON (Figure 8). This enhanced germination phenotype was observed for the miR408-OX and pcy seeds as well (Figures 3B–C and 6B–C). Conversely, the PIF1-OX seed failed to germinate even by 120 h in phyAON (Figure 8B). We showed that amiR408 and β-estradiol treated iPCY-OX seeds exhibited similar phenotypes as PIF1-OX (Figures 3F and 6C). These results indicate that the molecularly delineated PIF1-miR408-PCY repression cascade was in line with the germination phenotypes of the relevant mutants.
Figure 8.
Genetic analysis of the PIF1-miR408-PCY pathway. (A) The amiR408 line was crossed with pif1 to generate the pif1 amiR408 double mutant. Seeds from these lines and the wild type were assayed for germination rates over the time course of phyAOFF (left) and phyAON (right). (B) The miR408-OX line was crossed with PIF1-OX to generate the PIF1-OX miR408-OX double over-expression line. Seeds from these lines and wild type plants were assayed for germination rates in phyAOFF and phyAON. (C) Comparison of germination rates of the PIF1-OX, pcy, and PIF1-OX pcy seeds. All data are all means ± SD from three individual experiments. Different letters denote genotypes with significant differences at 120 h (one-way ANOVA, P < 0.05, Supplemental Data Set 3).
To further confirm that PIF1, miR408, and PCY act in the same genetic pathway, we generated and analyzed three double mutants involving pif1 and PIF1-OX. We found that the germination rate of the pif1 amiR408 seed was substantially reduced compared to that of pif1 (Figure 8A). By 120 h in phyAOFF and phyAON, the nearly 100% germination frequency of pif1 was lowered to about 40% and 80% by amiR408, respectively (Figure 8A). Regarding the PIF1-OX miR408-OX double overexpression seed, over 20% germinated by 120 h in phyAON (Figure 8B), indicating that miR408-OX was able to partially rescue the germination defect of PIF1-OX. These results demonstrate that miR408 acts downstream of PIF1 in the same pathway. We also tested the genetic relationship between PCY and PIF1 by generating the PIF1-OX pcy line. We found that the germination rate was substantially increased in this line compared to PIF1-OX in both phyAOFF and phyAON (Figure 8C), indicating that PCY also acts downstream of PIF1. Thus, PIF1, miR408, and PCY appear to act sequentially in the same pathway to regulate seed germination.
PIF1, miR408, and PCY regulate overlapping cohorts of genes
As a master transcriptional regulator, PIF1 programs the seed germination-related transcriptome (Oh et al., 2009; Shi et al., 2013; Pfeiffer et al., 2014). To test whether miR408-PCY participates in regulating that transcriptome, we performed RNA-sequencing analysis of the wild type, pif1, and miR408-OX seeds imbibed in darkness. The iPCY-OX imbibed seed with or without β-estradiol treatment was also analyzed. Consistent with previous reports (Oh et al., 2009; Shi et al., 2013), we identified 5,640 genes that were differentially expressed between pif1 and the wild type. These genes were defined as the PIF1-regulated set (Figure 9). Genes differentially expressed between miR408-OX and the wild type were defined as the miR408-regulated set, which included 4,294 genes (Figure 9A). Differentially expressed genes in iPCY-OX by β-estradiol treatment were defined as the PCY-regulated set, which included 14,646 genes (Figure 9A).
Figure 9.
Transcriptomic analysis of the PIF1-miR408-PCY pathway. (A) Venn diagram showing the relationships of PIF1-, miR408-, and PCY-regulated genes. Differentially expressed genes were identified from RNA-sequencing analyses of pif1, miR408-OX, and iPCY-OX seeds versus the respective controls. (B) Scatter plots showing pairwise correlation of the relative expression levels of the three sets of coregulated genes in pif1, miR408-OX, and iPCY-OX versus the respective controls. R, Pearson correlation coefficient. (C) Hierarchical clustering of the 2,290 genes that were differentially expressed in pif1, miR408-OX, and iPCY-OX vs. the respective controls. Colors represent the Log2-transformed fold changes. (D) Clustering analysis of the 218 genes associated with the GO term “seed germination” (GO:0009845). The genes were divided into three groups based on their relative expression levels in pif1 versus in the wild type. Group I, repressed in pif1; Group II, not differentially expressed; Group III, induced in pif1. (E) Expression pattern of representative Group I (SOM and RVE2) and Group III (JMJ22 and MAN7) genes in the indicated RNA-sequencing data sets.
Venn diagram analysis revealed that there were 2,651 genes in common between the PIF1-regulated and the miR408-regulated sets, 4,482 in common between the PIF1-regulated and the PCY-regulated sets, and 3,294 in common between the miR408-regulated and the PCY-regulated sets (Figure 9A). Based on Pearson correlation coefficients (R) of fold changes, we found that the common genes exhibited high pairwise correlations (Figure 9B). Venn diagram analysis further revealed 2,290 genes that were differentially expressed in pif1, miR408-OX, and iPCY-OX compared to the respective controls (Figure 9A). Clustering analysis showed that the vast majority of these genes were regulated in the same direction in both pif1 and miR408-OX, but in the opposite direction in iPCY-OX (Figure 9C). Together these results indicate that the PIF1-miR408-PCY pathway regulates large cohorts of common target genes in the seed.
Because 4,482 out of 5,640 (79.5%) of the PIF1-regulated genes were differentially regulated in iPCY-OX (Figure 9A), which also exhibited the highest pairwise correlation among the genotypes (R = 0.884; Figure 9B), we selected the PIF1-PCY coregulated genes for further investigation (Supplemental Data Set 1). Gene Ontology (GO) analysis revealed that the PIF1-PCY coregulated genes were preferentially associated with terms in two categories: seed development and germination, and hormone metabolism and signaling (Supplemental Figure 5). For example, the GO term “seed germination” was associated with 218 genes in Arabidopsis (Supplemental Data Set 2). These genes could be divided into three groups based on their expression pattern in pif1 (Figure 9D). Genes in group I (40 out of 218, or 18.3%) and III (41 out of 218, or 18.8%) were substantially downregulated and upregulated in pif1 compared to the wild type, respectively (Figure 9D). These genes were reversely modulated in iPCY-OX (Figure 9D).
Further inspection revealed that individual genes that have been genetically or functionally implicated in germination-related processes were included in group I and III (Supplemental Figure 6). For example, SOMNUS (SOM) and REVEILLE 2 (RVE2) from group I were reported as inhibiting light-dependent germination (Kim et al., 2008; Jiang et al., 2016). On the contrary, JUMONJI DOMAIN-CONTAINING PROTEIN 22 (JMJ22) and ENDO-BETA-MANNASE 7 (MAN7) from group III were reported as promoting seed germination (Iglesias-Fernández et al., 2011; Cho et al., 2012). These two types of genes were regulated in an opposite pattern in pif1 and iPCY-OX (Figure 9E). Collectively, these results indicate not only that PCY is a key node downstream of PIF1 but also that it mediates the transcriptomic changes underlying germination.
The PIF1-miR408-PCY pathway differentially regulates GA and ABA metabolic genes
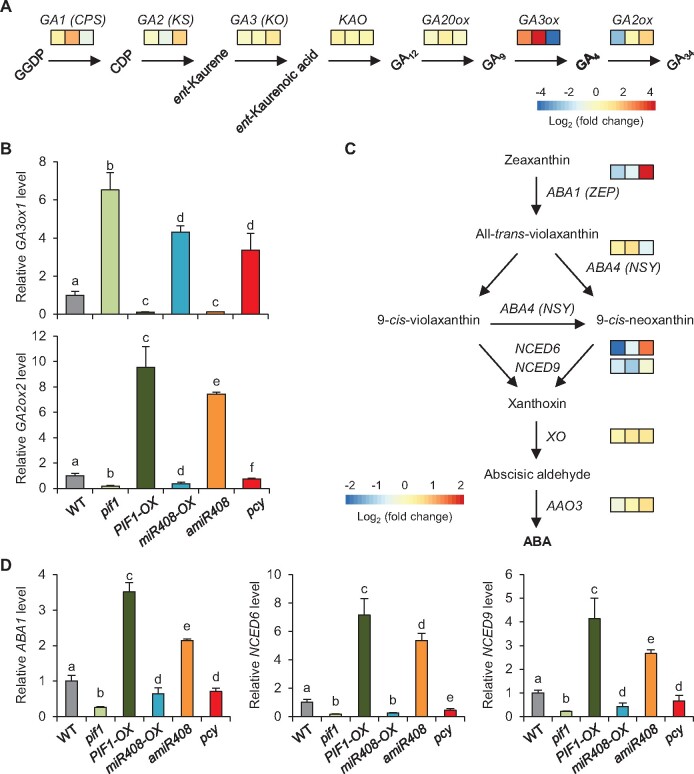
Because removal of PIF1 is critical for adjusting GA and ABA biosynthesis, we examined whether the PIF1-miR408-PCY pathway regulates GA and ABA metabolic genes. Inspection of the gene expression profile along the GA metabolic pathway using the RNA-sequencing data revealed that PIF1, miR408, and PCY consistently impacted the transcript levels of GA3ox1 and GA2ox2 (Figure 10), which encode the GA3-oxidase that catalyzes the terminal GA biosynthetic step and the GA2-oxidase that catabolizes bioactive GAs, respectively (Yamaguchi, 2008). This finding was corroborated by RT-qPCR analysis showing that GA3ox1 was upregulated in pif1, miR408-OX, and pcy, but it was downregulated in PIF1-OX and amiR408 compared to the wild type in both phyAOFF (Figure 10B) and phyAON (Supplemental Figure 7). In contrast, GA2ox2 was upregulated in PIF1-OX and amiR408 but down-regulated in pif1, miR408-OX, and pcy seeds relative to the wild type (Figure 10B;Supplemental Figure 7B). The expected expression pattern of GA3ox1 and GA2ox2 was confirmed in β-estradiol treated iPCY-OX seeds (Supplemental Figure 8). These results indicate that the PIF1-miR408-PCY pathway targets the GA3ox and GA2ox steps to repress accumulation of bioactive GA.
Figure 10.
The PIF1-miR408-PCY pathway differentially regulates GA and ABA metabolic genes. (A) Diagram of a simplified GA biosynthesis pathway illustrating genes influenced by the PIF1-miR408-PCY pathway. Genes associated with the individual biosynthesis steps are shown above the arrows. From left to right, colored boxes indicate relative expression levels of the corresponding gene in pif1, miR408-OX, and iPCY-OX against the respective controls. (B) RT-qPCR analysis of relative transcript levels of GA3ox1 and GA2ox2 in the indicated seeds under phyAOFF. Data are mean ± SD from three replicates performed on the same cDNA. Different letters denote genotypes with significant differences (one-way ANOVA, P < 0.05 for GA3ox1, P < 0.01 for GA2ox2, Supplemental Data Set 3). (C) Diagram of a simplified ABA biosynthesis pathway illustrating genes influenced by the PIF1-miR408-PCY pathway. Colored boxes indicate relative expression levels of the corresponding gene in pif1, miR408-OX, and iPCY-OX against the respective controls. (D) RT-qPCR analysis of the relative transcript levels of ABA metabolic genes ABA1, NCED6, and NCED9 in the indicated seeds. Data are mean ± SD from three replicates performed on the same cDNA. Different letters denote genotypes with significant differences (one-way ANOVA, p < 0.05, Supplemental Data Set 3).
Transcriptomic profiling also revealed that ABA DEFICIENT 1 (ABA1), NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 6 (NCED6) and NCED9 were among the most substantially influenced ABA biosynthetic genes by the PIF1-miR408-PCY pathway in phyAOFF (Figure 10C). According to the RNA-sequencing data, ABA1, NCED6, and NCED9 were downregulated in pif1 and miR408-OX but upregulated in β-estradiol treated iPCY-OX seeds compared to the controls (Figure 10C). RT-qPCR analysis confirmed that ABA1, NCED6, and NCED9 were downregulated in phyAON compared to phyAOFF in the wild type (Supplemental Figure 7A). In the series of mutants disrupting the PIF1-miR408-PCY pathway, we found that expression levels of ABA1, NCED6, and NCED9 were significantly altered compared to the wild type in both phyAOFF (Figure 10D) and phyAON (Supplemental Figure 7C). The expected expression pattern of these genes was further confirmed in β-estradiol treated iPCY-OX seeds (Supplemental Figure 8). These results collectively indicate that PIF1-miR408-PCY overall promotes expression of ABA biosynthetic genes in darkness.
PIF1-miR408-PCY modulates the GA/ABA ratio
To determine whether the PIF1-miR408-PCY pathway impacts the GA/ABA ratio, which is critical for germination, we directly quantified the amount of bioactive GA in the seed using the ultrahigh-performance liquid chromatography–triple quadrupole mass spectrometry (UPLC-MS/MS) method (Fu et al. 2012; Ma et al., 2015). Compared to the wild type, the level of GA4, the major bioactive GA in Arabidopsis seed (Oh et al., 2006), was significantly elevated in pif1, miR408-OX, and pcy seeds (Figure 11). In PIF1-OX and amiR408, GA4 level was significantly reduced compared to the wild type (Figure 11A). We further quantified the amount of ABA in the seed using the UPLC-MS/MS method. We found that endogenous ABA levels in pif1, miR408-OX, and pcy seeds were significantly reduced relative to the wild type, but PIF1-OX and amiR408 maintained ABA levels comparable to the wild type (Figure 11B). Calculation of the GA/ABA ratio revealed that the pif1, miR408-OX, and pcy seeds, while exhibiting elevated germination rates (Figure 8), displayed GA/ABA ratios significantly higher than wild-type seeds (Figure 11C). In contrast, PIF1-OX and amiR408 seeds, while exhibiting reduced germination (Figure 8), displayed GA/ABA ratios significantly lower than the wild type (Figure 11D). These results indicate that the PIF1-miR408-PCY pathway acts to adjust the GA/ABA ratio.
Figure 11.
PIF1-miR408-PCY controls germination by modulating the GA/ABA ratio. (A–B) Quantification of endogenous GA4 (A) and ABA (B) levels in imbibed seeds of the indicated genotypes. Data are mean ± SD from three individual experiments. Different letters denote genotypes with significant differences (one-way ANOVA, p < 0.001 for A, P < 0.05 for B, Supplemental Data Set 3). FW, fresh weight. (C–D) Calculated GA/ABA ratios in the indicated genotypes. Data are mean ± SD from three individual experiments. Different letters denote genotypes with significant differences (one-way ANOVA, P < 0.001 for C, P < 0.01 for D, Supplemental Data Set 3). (E) Germination rates of the indicated seeds in phyAOFF and phyAON with different chemical treatments. Mock, no chemical treatment; GA3, 10 μM GA3; Paclobutrazol, 100 μM paclobutrazol; ABA, 5 μM ABA. Data are mean ± SD from three individual experiments.
To test if the germination behaviors of mutants disrupting the PIF1-miR408-PCY pathway are dependent on the GA/ABA ratio, we performed pharmacological analyses that skew the GA–ABA balance. We found that application of exogenous bioactive GA3, which would elevate the endogenous GA/ABA ratio, promoted all seeds, including PIF1-OX and amiR408, to complete germination in phyAOFF (Figure 11E). Conversely, paclobutrazol, an inhibitor of GA biosynthesis, blocked germination of all seeds, including pif1, miR408-OX, and pcy, in phyAON in a dosage-dependent manner (Figure 11E;Supplemental Figure 9). Furthermore, application of exogenous ABA, which would decrease the GA/ABA ratio, inhibited germination of all seeds, including pif1, miR408-OX, and pcy in phyAON in a dosage-dependent manner (Figure 11E;Supplemental Figure 10). Thus, results from gene expression, hormone quantification, and pharmacological analyses coherently demonstrate that the PIF1-miR408-PCY cascade regulates germination by modulating the GA/ABA ratio.
PCY is conserved in seed plants
Finally, to provide phylogenetic evidence supporting PCY as a key node in germination, we examined PCY conservation in seed plants. By searching the sequenced genomes, we identified 276 putative PCY orthologs from 52 seed plants but none in nonseed plants (Figure 12; Supplemental Figure 11). Comparison of PCY sequences and domain organization to the most homologous blue copper proteins in three representative nonseed plants revealed two salient features of PCY. First, all PCYs were found to contain only a signal peptide at the N-terminus and a type-I copper binding motif at the C-terminus (Figure 12; Supplemental Figure 11). These two domains exhibited an extremely compact organization. For example, the Arabidopsis PCY possessed 129 amino acid residues of which 33 (26%) were devoted to the signal peptide and 95 (74%) to the copper-binding motif (Figure 12). Second, a miR408 recognition site was found near the 5′ end of the coding region of all examined PCY transcripts (Figure 12), which has been experimentally validated in several species (Abdel-Ghany and Pilon, 2008; Zhou et al., 2010; Feng et al., 2013; Zhang and Li, 2013). Moreover, length and domain organization suggest that PCY in Ginkgo biloba may resemble the prototype of this protein, which was further evolved in angiosperm by trimming the C-terminus to a bare-bones copper binding motif (Figure 12). Taken together, our results indicate that PCY has specifically evolved in seed plants as a miR408-targeted, storage vacuole-associated compact cuproprotein to balance GA and ABA levels for controlling germination (Figure 13).
Figure 12.
PCY exhibits features conserved in seed plants. Comparison of PCY and related blue copper proteins in representative land plants. Shown on the left is a species tree showing the reconstructed order of divergence of the indicated plant species using information obtained from the TimeTree database at timetree.org. The branch length reflects evolutionary divergence time in millions of years. Species with identified PCYs are shaded in green. PCY domains are shown with different colors on the right. Scale represents accumulative number of amino acid residues. The alignment of the PCY related proteins is provided as a text file in Supplemental File 1.
Figure 13.
Working model of far-red light-dependent seed germination mediated by PCY. The PIF1-miR408 module is critical for regulating PCY abundance in far-red light-induced seed germination. In darkness, due to the absence of active phyA, PIF1 is stabilized and binds to the G box of the miR408 promoter. This leads to transcriptional repression of miR408 and allows PCY to accumulate. Upon irradiation with far-red light, phyA-mediated PIF1 degradation leads to transcriptional derepression of miR408, which in turn silences PCY. Removal of PCY from storage vacuoles facilitates an increase in the GA/ABA ratio, and this ultimately sets germination in motion.
DISCUSSION
Seeds have elaborate molecular mechanisms to monitor and transduce the light signals for proper germination, which is a process vital for survival of many seed plants (Oh et al., 2004; Finch-Savage and Leubner-Metzger, 2006; Seo et al., 2009; Shi et al., 2015). Decades of research have shown that a decisive event downstream of light signaling is the establishment of the high-GA-low-ABA hormonal state (Nambara and Marion-Poll, 2005; Oh et al., 2006; 2007; Yamaguchi, 2008; Seo et al., 2009; Shu et al., 2016). Elucidating how the light signals are converted into the hormonal profiles is critical to our understanding of seed biology, and this has immediate relevance to agriculture and human nutrition.
A long repression cascade regulating light-dependent germination
In this study, we used far-red light triggered, phyA-dependent germination as our experiment model (Oh et al., 2004; 2006; Cho et al., 2012). Incorporation of findings from our molecular and genetic analyses to established light signaling framework (Castillon et al., 2007; Leivar and Quail, 2010) delineated the phyA-PIF1-miR408-PCY repression cascade as a key regulatory mechanism in germination (Figure 13). Through transcriptome profiling and quantification of endogenous GA and ABA levels, we demonstrated that this signal relay chain is required for the conversion of light signals into hormonal profiles (Figures 9–11). In darkness, absence of active phyA leads to PIF1 accumulation (Shen et al., 2008), which suppresses transcription of miR408 via binding to the G box (Figure 7). A low level of miR408 in turn allows PCY to accumulate in the storage vacuole (Figures 1, 2, and 5), and this correlates with the low-GA-high-ABA state (Figures 9–11). Upon far-red irradiation, phyA is rapidly activated to destabilize PIF1 (Oh et al., 2007; Shen et al., 2008), releasing miR408 from transcriptional inhibition (Figure 7). Accumulation of miR408 then silences PCY (Figures 1 and 5), correlating with the high-GA-low-ABA state (Figures 9–11). The multistep repressor cascades are typical for developmental transcription networks, enabling them to generate robust temporal delay (Rosenfeld and Alon, 2003; Shoval and Alon, 2010). The phyA-PIF1-miR408-PCY repression cascade may therefore help to specify the time from light perception to PCY turnover.
Based on our phylogenetic findings, the phyA-PIF1-miR408-PCY cascade appears to have formed by sequentially adding downstream components to the chain during evolution of the seed plants. The phytochrome signaling pathway and the PIF family were shown to originate in the ancestors of charophytes (Han et al., 2019). Inhibition of PIFs by phytochromes to regulate light responses is conserved in vascular plants and at least the liverworts (Lee and Choi, 2017; Han et al., 2019). On the other hand, miR408 is present in all examined land plants including moss (Pan et al., 2018; Guo et al., 2020) while PCY has acquired the miR408 recognition site after the emergence of seed plants (Figure 12). These observations suggest that the repression cascade has specifically evolved in seed plants, taking advantage of extant regulatory modules, for controlling seed germination.
PCY links copper mobilization to hormone metabolism
PCY belongs to the phytocyanin family of small blue copper proteins, which are ancient copper-containing redox proteins widely distributed in microorganisms and plants (Rydén and Hunt 1993; Guss et al., 1998; De Rienzo et al. 2000; Giri et al. 2004). Different from other phytocyanins, PCYs are extremely compact, with all 120–130 amino acid residues of the proteins devoted to three conserved motifs. Besides the characteristic type-I copper binding motif on the C-terminus, the signal peptide and the miR408 recognition site superimpose on the N-terminus (Figure 12; Supplemental Figure 11). Our results provided two important clues to PCY function. First, through quantification of endogenous hormones, genetic analysis, gene expression profiling, and pharmacological analyses (Figures 8–11), we demonstrated that PCY acts as a switch downstream of PIF1 and is both necessary and sufficient to modulate the GA/ABA ratio in the seed. Previously, high expression of PCY in the transmitting tract of the pistil was reported (Dong et al. 2005). Overexpression of PCY was found to disrupt pollen tube guidance into the style and to reduce seed set (Dong et al. 2005). The latter was corroborated by observations that overexpression of miR408 resulted in larger seed size and higher grain yield (Pan et al., 2018). Thus, besides seed germination, PCY may participate in other aspects of seed biology and reproductive development. It will be interesting to test if balancing endogenous hormone levels is the unifying function of PCY in these processes.
Second, through RNA-sequencing, we found that PCY is both necessary and sufficient to regulate an overwhelming portion of the PIF1-dependent transcriptome underlying germination (Figure 9; Supplemental Figure 5; Supplemental Data Set 1). This finding suggests that PCY turnover is associated with major changes to cellular state. Because PCY is a vacuole-located cuproprotein that has a basic type-I copper motif and is highly expressed in mature seed (Figures 2 and 12), we contemplate that PCY is a key carrier for copper mobilization. In the seed, mineral nutrients are sequestered in the storage vacuoles (Lanquar et al., 2005; Kim et al., 2006; Roschzttardtz et al., 2009; Eroglu et al., 2017). Right after imbibition, there likely is no new assimilation of transition metals before extensive translation and protein synthesis is taking place (Née et al., 2017; Paszkiewicz et al., 2017). Thus, mineral elements need to be mobilized from vacuolar stores and transported to the cytoplasm and other organelles for reconstituting biochemical activity. Disrupting these processes has been shown to lead to severe germination defects (Lanquar et al., 2005; Kim et al., 2006). Previously, we have shown that miR408 promotes copper allocation to the plastid and enhances photosynthesis via elevating plastocyanin abundance (Zhang et al., 2014; Pan et al., 2018). Taken together, we speculate that the miR408-PCY module controls copper redistribution between the vacuole and the plastid.
How does PCY regulate the GA/ABA ratio?
PCY turnover as a means for copper mobilization and delivery to the plastid is consistent with previous studies on the effects of copper on plastid physiology and biochemistry. The plastid is known to be the major cellular copper sink in plants (Burkhead et al., 2009), whereby the transition metal acts as a cofactor for plastocyanin in the thylakoid lumen, which is an indispensable electron carrier in the Z-scheme of photosynthesis (Molina-Heredia et al., 2003; Weigel et al., 2003). Copper is also a critical cofactor for the copper- and zinc-containing superoxide dismutase in the stroma, which neutralizes reactive oxygen species in order to maintain proper redox state in the plastid (Gupta et al., 1993). Copper allocation to the plastid is critical for plastocyanin abundance and activity (Weigel et al., 2003; Zhang et al., 2014; Pan et al., 2018). The copper level was also reported to impact the number of chloroplasts per cell, thylakoid stacking, and grana size in soybean (Glycine max) cell suspensions (Bernala et al., 2006). It is intriguing to note that GA biosynthesis initiates in the plastid (Sun and Kamiya 1997; Yamaguchi, 2008). Thus, proper regulation of PCY degradation may promote copper translocation or allocation to the plastid. Copper-induced plastid development may in turn provide the structural and biochemical basis for initiating GA biosynthesis in the seed.
Alternatively, the effect of PCY turnover on hormone rebalancing may be explained by a direct impact on ABA synthesis taking place in the cytosol. In Arabidopsis, ABSCISIC ALDEHYDE OXIDASE 3 (AAO3) encodes an aldehyde oxidase that catalyzes the last step of ABA biosynthesis, the conversion of abscisic aldehyde to ABA (Seo et al., 2000). AAO3 is a cytosolic molybdoenzyme that requires the molybdenum cofactor for catalytic activity (Seo et al., 2000). Structural and biochemical analyses have shown that the final step of molybdenum cofactor biosynthesis is dependent on a copper–dithiolate complex that protects the reactive dithiolate before molybdenum insertion (Kuper et al., 2004). Perhaps PCY is one of the copper donors and, through unidentified cytoplasmic chaperones, it passes on copper to the dithiolate group for synthesizing the molybdenum cofactor (Peñarrubia et al., 2015). Induction of PCY expression during late seed development (Figure 1) is consistent with this scenario, whereby elevated PCY helps to maintain AAO3 activity and hence ABA accumulation during seed maturation. Upon light irradiation, rapid PCY turnover would deplete copper supply for AAO3 and impede ABA synthesis. This is consistent with the fourfold decline of ABA content in the pcy seed relative to the wild type (Figure 11B). The finding in rice (Oryza sativa) that exogenous copper increases ABA accumulation and inhibits germination (Ye et al., 2014) provides another line of evidence for this model.
As PCY in the seed is associated with the storage vacuole (Figure 2), it is expected that copper released from PCY upon its degradation would be transported from the vacuole to other cellular destinations. The plausible candidate for this function in Arabidopsis is COPT5, the tonoplast-located high-affinity copper transporter (Klaumann et al., 2011). Further characterization of COPT5 and the related cellular copper homeostasis and allocation machineries could shed more light on hormone synthesis and balancing during seed development and germination.
Materials and methods
Plant materials and growth conditions
The wild-type Arabidopsis thaliana used in this study was Col-0. The pif1, PIF1-OX, miRpro408:GUS, miR408-OX, and amiR408 plants were as previously described (Oh et al., 2004; Zhang and Li, 2013; Zhang et al., 2014). The template of the STTM-miR408 module (5′-GCTCTAGAGCCAGGGAAGACTAGGCAGTGCATGTTGTTGTTGTTATGGTCTAATTTAAATATGGTCTAAAGAAGAAGAATGCCAGGGAAGACTAGGCAGTGCATCTCGAGCGG) (Yan et al., 2012) was chemically synthesized, PCR amplified, and cloned into the modified pJim19-35S vector (Zhang et al., 2008). This construct was used to transform wild-type plants, and homozygous STTM-miR408 lines were selected for BASTA resistance at the T2 generation. To delete PCY, a CRISPR-Cas9 system employing the modified pCAMBIA1300 vector was used (Mao et al., 2013) in which the 35S and the AtU6-26 promoters, respectively drive Cas9 and a pair of sgRNAs that were designed to target both ends of the PCY coding region. The resulting construct was used to transform the wild type, and T1 plants were individually genotyped by PCR and sequencing to identify deletion events. Approximately 100 individual T2 plants were genotyped to identify Cas9-free homozygous pcy-2 lines. The iPCY-OX transgenic plants were obtained by cloning the PCY-coding sequence into the pER8 vector (Zuo et al., 2000) and transforming the wild-type plants. Homozygotes were selected for hygromycin resistance in the T2 population. The PCYpro:PCY-GFP construct was obtained by cloning the PCY coding sequence and its native promoter into the modified pJim19-GFP vector, replacing the 35S promoter in the vector. Following transformation of the wild-type plants with the PCYpro:PCY-GFP construct, homozygotes were selected by Kanamycin resistance in the T2 population. Sequences of the relevant primers are listed in Supplemental Table 1. The miR408pro:GUS/pif1, PIF1-OX miR408-OX, pif1 amiR408, PIF1-OX pcy, lac12 lac13, and pcy lac12 lac13 lines were generated by crossing and selection for homozygotes at the F2 generation.
Adult Arabidopsis plants were grown in commercial soil at 22°C, ∼60% relative humidity, and under long day (16 h light/8 h dark) condition in a growth chamber. Light was provided by white fluorescent bulbs with an intensity of ∼120 μmol m−2 s−1. Tobacco plants (Nicotiana benthamiana) used for transient expression experiments were maintained in a growth chamber with 16 h light/8 h dark a light at intensity of ∼200 μmol m−2 s−1 that was provided by white fluorescent bulbs, a relative humidity of ∼60%, and a temperature setting of 25°C/21°C.
Germination assays
Care was taken to maintain parent plants side by side under the same growth conditions. For each experiment, the seeds were harvested from the same batch of plants at approximately the same time. After harvesting, the seeds were dried at room temperature and stored under the same conditions for 6–8 weeks prior to germination. The far-red light induced germination assay was performed as described with minor modifications (Oh et al., 2004). Briefly, a triplicate set of 50–75 seeds for each sample was surface sterilized with 30% liquid bleach and plated on half-strength MS aqueous agar medium (0.6% agar, 1% sucrose, pH 5.7) under ambient light. One h after the start of sterilization, the plated seeds were irradiated with 3.2 μM m−2 s−1 far-red light for 5 min and then incubated in the dark at 22°C for 48 h. For phyAOFF, the imbibed seeds were continuously placed in darkness for 4 h. For phyAON, the seeds were treated with a second far-red irradiation for 4 h and then in darkness at 22°C (Figure 1C). For pharmacological treatments, paclobutrazol, GA3, or ABA (Sigma-Aldrich) was added to the medium at the indicated final concentrations. Germination rates were determined by examining radicle formation at the indicated time points.
Statistical analyses
Student’s t-test was used for comparing means between two samples. For all multiple comparisons, one-way ANOVA was first conducted to test the significance of the difference among different group means using Excel. Dunnett’s test was used post hoc for multiple comparisons between two group means at the indicated significance levels. Detailed statistical reports are shown in Supplemental Data Set 3.
Analysis of seedling greening
Vernalized seeds were germinated and grown in the dark for 4 days and then transferred to continuous white light (100 μM m−2 s−1) for 24 h. The greening rate, calculated as the ratio of green seedlings to the total germinated seedlings as previously described (Zhong et al., 2009), was determined by three individual experiments each containing 50 seedlings. Pigments were extracted from etiolated seedlings in the dark at room temperature using 90% acetone containing 0.1% NH3 as previously described (Zhong et al., 2014). Supernatants containing the pigments were subject to fluorescence spectral analysis using an Infinite M200 microplate reader (Tecan). The excitation wavelength was 443 nm and emission spectra were recorded from 610 to 740 nm with 1 nm bandwidth. The protochlorophyllide and chlorophyll fluorescence intensity was further quantified at 634 nm and 678 nm, respectively. Measurements were performed on three independent biological samples each including 20 seedlings.
Transcript quantification
Total RNA from imbibed seeds was isolated using the Quick RNA Isolation Kit (Huayueyang). For each experiment, mRNA and miRNA from at least three independent biological samples were reverse transcribed into cDNA using the PrimeScript II 1st Strand cDNA Synthesis Kit (TaKaRa) and the miRcute Plus miRNA First-Stand cDNA Synthesis Kit (Tiangen), respectively. qPCR was performed using SYBR Green Mix (TaKaRa) on the 7500 Fast Real-Time PCR System (Applied Biosystems). ACTIN7 (At5g09810) and 5S RNA were used for mRNA and miRNA normalization, respectively. Quantification was derived from three independent qPCR reactions performed on the same pool of cDNA. All RT-qPCR experiments were repeated at least three times and data from one representative experiment were shown. Sequences of the primers are listed in Supplemental Table 1.
RNA-sequencing analyses
The miR408-OX, pif1, and the control wild-type seeds were grown on half-strength MS medium (Murashige and Skoog, 1962). The iPCY-OX and the control wild-type seeds were grown on medium supplemented with or without 5 μM β-estradiol (Sigma-Aldrich). All seeds were treated with the phyAOFF condition for 12 h before sample collection. Total RNA was isolated using the Quick RNA Isolation Kit (Huayueyang). Library preparation and RNA-sequencing were performed on the Illumina HiSeq 2000 platform. For each genotype, three paired-end libraries from independent biological samples were prepared. At least 16 million raw paired-end reads were generated from each library. Quality control was conducted using fastQC. Cutadapt and a custom Perl script were used to trim adaptors with the parameter Q30 and the first nine bases following the adaptors with low fastQC score. After trimming, reads longer than 100 bases were retained and the R1 and R2 files were paired simultaneously. The clean reads were mapped to the TAIR10 Arabidopsis genome build using STAR with an average mapping rate of ∼90% and unique mapping rate above 80%. Transcript quantification by Stringtie was processed by Cuffdiff to identify differentially expressed genes using a fold change cutoff of 1.5 against the respective controls. The false discovery rate (FDR) method was used for correcting multiple comparisons and a FDR cutoff of 0.05 was applied. Clustering and correlation analyses were performed and visualized using R scripts. GO analysis was carried out using AgriGO (http://bioinfo.cau.edu.cn/agriGO/).
ChIP-qPCR
ChIP was carried out on 4-day-old dark-grown PIF1-OX and wild-type seedlings using a monoclonal anti-MYC antibody (Sigma-Aldrich, catalog # C3956, lot # 022M4795V, 1:1,000 (v/v) dilution) as previously described (Pfeiffer et al., 2014). After ChIP, equal amount of input DNA was subjected to qPCR analysis of the target DNA fragments. Fold of enrichment was calculated between PIF1-OX and the wild-type input. Fold of enrichment in PIF1-OX against the wild type was calculated from three independent qPCR reactions performed on the same DNA.
Immunoblotting
The PCYpro:PCY-GFP line was used for protein analysis. Dry seed was incubated in water at room temperature for 0.5 h or at 4°C for 24 h before sample collection. Total protein was isolated with an extraction buffer containing 10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% (v/v) glycerol, 1 mM phenylmethylsulfonyl fluoride, and 1× protease inhibitor mixture (Roche). Following electrophoresis, standard immunoblotting analyses (Zhang et al., 2008) were performed. The blots were probed with antibodies against GFP (Abmart, catalog # 200004S, lot # 313769, 1:1,000 (v/v) dilution) and RPT5 (Abcam, catalog # ab22676, laboratory storage, 1:5,000 (v/v) dilution). Relative protein abundance was detected using the ChemiDoc XRS+ imaging system (Bio-Rad). Replicates for immunoblotting experiments were individually prepared proteins isolated from pooled seeds.
Immuno-electron microscopy
Imbibed seeds were fixed with 4% (w/v) paraformaldehyde in 0.1 M phosphate buffer (pH 7.4) for 2 h at room temperature and overnight at 4°C, washed four times in 0.1 M phosphate buffer, dehydrated through an alcohol series (30%, 50%, 70%, 85%, 95%, and 100%), and embedded in LR White resin (Sigma-Aldrich). The resin blocks were polymerized for 48 h at −10°C by exposing to UV light, ultrathin-sectioned (75 nm) using an UC7 ultramicrotome, and mounted on 100-mesh nickel grids. The mounted sections were floated on TBS (137 mM NaCl, 2.7 mM KCl, 50 mM Tris–HCl, pH 7.4) drops, incubated twice for 5 min, blocked with TBG (TBS containing 0.1% BSA-c) for 10 min, and then incubated with anti-GFP antibody (Abcam, catalog # ab6556, laboratory storage, 1:5 (v/v) dilution) at 4°C overnight in TBG in an enclosed humidity chamber. After five washes in TBG, the sections were incubated with goat anti-rabbit IgG conjugated to 10 nm colloidal gold (1:50, Sigma-Aldrich) for 1 h at room temperature. Following washes with TBS, sections were postfixed in 0.1% (v/v) glutaraldehyde in TBS for 10 min, washed with distilled water, counterstained with 2% uranyl acetate, and observed with a Tecnai G2 Spirit electron microscope (FEI) at 120 kV.
Histochemical staining for GUS activity
Dry seeds (50 mg per sample) were grown for 12 h in phyAOFF and phyAON. After stripping away the seed coat in green safe light, seeds were incubated in a standard GUS staining solution for 3 h at 37°C. Following removal of the staining solution, seeds were washed with several changes of 75% ethanol until pigments were no longer visible. Images of the GUS staining pattern were taken with a digital camera. To quantify GUS concentration, the stained seeds were ground into fine powder in liquid nitrogen and analyzed by ELISA using the Plant β-GUS ELISA Kit (3A Chemicals). Absorbance at 450 nm was measured by a Cytation 5 Imaging Reader (BioTek) to infer GUS concentration according to the manufacturer’s instruction. Statistical analysis was performed on three experiments using individual batches of seeds.
Live-cell imaging
Cotyledons of the PCYpro:PCY-GFP seed were dissected away from the testa and endosperm by the application of gentle pressure to seed held between a microscope slide and a cover slip. Fluorescence images were obtained using a Nikon A1R si+ laser scanning confocal microscope equipped with an APO 40 × 1.25 NA water immersion objective. Excitation and emission wavelengths were 488/500 to 550 nm for GFP and 405/425 to 475 nm for vacuole autofluorescence. Autofluorescence spectra were obtained using a 32-PMT spectral detector. Spectral unmixing and image analysis were performed using the NIS Elements AR software (Nikon Instruments).
Transient expression in onion epidermal cells was performed as previously described (Wang and Frame, 2009). The gold particles were coated with plasmid DNA containing the expression cassette for PCY-GFP or COPT5-mCherry. The Biolistic PDS-1000/He Particle Delivery System (Bio-Rad) was used for bombarded with the following settings: 1,100 psi rupture disc, 25–26-inch Hg vacuum, and target distance of 10 cm. After bombardment, the explants were kept in dark at 25°C for 16–18 h and observed with a Nikon A1R si+ microscope. Excitation and emission wavelengths were 488/500 to 550 nm for GFP and 561/570 to 620 nm for mCherry, respectively.
Quantification of endogenous GA and ABA
For each genotype, 500 mg of seed grown in phyAOFF for 24 h was collected and ground into fine powder in liquid nitrogen. Endogenous ABA was purified and measured as previously described (Fu et al. 2012) with minor modifications to the detection procedure. Briefly, UPLC-MS/MS analysis was performed on a UPLC system (Waters) coupled to the 5500 Qtrap system (AB SCIEX). Chromatography separation was achieved with a BEH C18 column (Waters) with mobile phase 0.05% HAc (A) and 0.05% HAc in acetonitrile (B). The gradient was set initially with 20% B and increased to 70% B within 6 min. ABA was detected in the MRM mode with transition 263/153 and the isotope dilution method was used for quantification. Statistical analysis was performed on three experiments using individual batches of seeds (Supplemental Data Set 3).
Quantitative GA measurements were performed as previously described using UPLC-MS/MS (Ma et al., 2015). Chromatographic separation was achieved using a BEH C18 column (Waters) with mobile phase 0.05% HAc (A) and 0.05% HAc in acetonitrile (B). The gradient was set as: 0–17 min, 3% B to 65% B; 17–18.5 min, 65% B to 90% B; 18.5–19.5 min, 90% B; 19.5–21 min, 90% B to 3% B; and 21–22.5 min, 3% B. GA4 was detected in the negative MRM mode and quantified with an MRM transition. The source parameters were: IS voltage −4500 V, TEM 600°C, GS1 45, GS2 55, and curtain gas 28. Statistical analysis was performed on three experiments using individual batches of seeds (Supplemental Data Set 3).
Transient expression in tobacco protoplasts
The promoter sequence of miR408 was cloned from Arabidopsis genomic DNA, inserted into the pGreen II 0800-LUC vector (Hellens et al., 2005), and used as the reporter. The PIF1 coding sequence was cloned from Arabidopsis cDNA, inserted into pGreen II 62-SK (Hellens et al., 2005), and used as the effector. Tobacco protoplasts were freshly prepared as described previously (Yoo et al., 2007). The effector or the empty vector was combined with the report construct and used to transiently transform the protoplasts using the Dual-Luciferase Reporter System (Promega), following the manufacturer’s instruction. Transfected protoplasts were incubated under low light for 16 h and chemiluminescence was determined using a LB942 Multimode Reader (Berthold Technologies).
PCY domain and phylogenetic analyses
PCY in Arabidopsis was used as query to perform a BLASTP search against all proteins in 52 plant species covering all main clades of land plants. A total of 276 PCY sequences were identified based on two criteria: E-value ≤ e−10 and a “plantacyanin” annotation term assigned by InterProScan. N-terminal signal peptides were predicted using SignalP (Almagro Armenteros et al., 2019). The binding sites for miR408 were predicted using psRNATarget (Dai et al., 2018). To show the evolutionary trajectory, PCYs from 14 representative plant species and the most homologous genes encoding small blue copper proteins from Physcomitrella patens, Salvinia cucullate, and Azolla filiculoides were selected and mapped to a species tree obtained from TimeTree (http://www.timetree.org/). The alignment of the PCY related proteins is provided as a text file in Supplemental File 1.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: miR408 (At2g47015), PIF1 (At2g20180), PHYA (At1g09570), PCY (At2g02850), GUN4 (At3g59400), GUN5 (At5g13630), HEMA1 (At1g58290), PORA (At5g54190), PORB (At4g27440), PORC (At1g03630), LAC3 (At2g30210), LAC12 (At5g05390), LAC13 (At5g07130), GA3ox1 (At1g15550), GA2ox2 (At1g30040), ABA1 (At5g67030), NCED6 (At3g24220), NCED9 (At1g78390), SOM (At1g03790), RVE2 (At5g37260), JMJ22 (At5g06550) and MAN7 (At5g66460). T-DNA insertion mutants used are pif1 (SALK_072677), pcy-1 (SALK_091945), lac3 (SALK_031901C), lac12 (SALK_087122), and lac13 (SALK_023935). RNA sequencing data can be found at the National Center for Biotechnology Information Sequence Read Archive under the accession number PRJNA633227. Accession numbers for the PCY-related proteins are Cucumis melo basic blue protein-like, XP_008465594.1; Lactuca sativa basic blue protein-like, XP_023755703.1; Nelumbo nucifera basic blue protein-like, XP_010270378.1; which can be found in GenBank (https://www.ncbi.nlm.nih.gov/genbank/); Prunus persica basic blue protein-like, Prupe.8G016000.1.p; Glycine max basic blue protein-like, Glyma06g12680.1; Populus trichocarpa basic blue protein-like, Potri.001G209300.1; Vitis vinifera basic blue protein-like, GSVIVT01023002001; Oryza sativa basic blue protein-like, LOC_Os02g49850.1; Musa acuminata basic blue protein-like, GSMUA_Achr8P23110_001; Amborella trichopoda basic blue protein-like, evm_27.model.AmTr_v1.0_scaffold00010.92; Physcomitrella patens basic blue protein-like, Pp3c6_3530V3.1.p; which can be found in Phytozome (v12) (https://phytozome.jgi.doe.gov/pz/portal.html); Pinus tabuliformis basic blue protein-like, A0A0K0M6V7; Picea sitchensis basic blue protein-like, A9NT06; which can be found in InterPro (https://www.ebi.ac.uk/interpro/); Ginkgo biloba basic blue protein-like, Gb_11841, which can be found in GigaDB (http://gigadb.org/); Azolla filiculoides basic blue protein-like, Azfi_s0019.g015312; Salvinia cucullata basic blue protein-like, Sacu_v1.1_s0032.g010905; which can be found in Fernbase (https://www.fernbase.org).
Supplemental Data
Supplemental Figure 1. Generation and characterization of PCY-related mutants.
Supplemental Figure 2. Both amiR408 and STTM-miR408 seeds exhibit reduced germination.
Supplemental Figure 3 . The miR408-PCY module specifically regulates germination.
Supplemental Figure 4. Analysis of PIF1 binding at the miR408 locus by ChIP-qPCR.
Supplemental Figure 5. Enriched GO terms associated with PIF1 and PCY coregulated genes.
Supplemental Figure 6. Exemplar genes regulated by the PIF1-miR408-PCY pathway.
Supplemental Figure 7. PIF1-miR408-PCY regulates GA and ABA metabolic genes in phyAON.
Supplemental Figure 8. Induction of PCY Mis-regulates GA and ABA metabolic genes.
Supplemental Figure 9. Blocking GA biosynthesis suppresses germination of the examined mutants.
Supplemental Figure 10. Exogenous ABA suppresses germination of the examined mutants.
Supplemental Figure 11. PCY has a compact domain organization.
Supplemental Table 1. Oligonucleotide sequences of the primers used in this study.
Supplemental Data Set 1 . List of PIF1 and PCY coregulated genes.
Supplemental Data Set 2. Expression profile of 218 germination-related genes.
Supplemental Data Set 3. Results of statistical analyses.
Supplemental File 1. Sequence alignment of the PCY-related proteins in Figure 12.
Supplementary Material
Acknowledgements
We thank Drs Xiangdong Fu, Xing-Wang Deng, and Hongwei Guo for providing some of the plasmids and seeds used in this study. Dr Yingchun Hu is acknowledged for technical assistance with TEM analysis. Part of the bioinformatic analysis was performed on the High-Performance Computing Platform of the Center for Life Science at Peking University.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2017YFA0503800) and the National Natural Science Foundation of China (31621001).
Conflict of interest statement. None declared.
L.L. designed and supervised the research. A.J., J.P., Y.Y., Y.Z., D.Z., and C.H. performed the experiments. Z.G. (Guo), Z.G. (Gao), and S.Z. analyzed the data. P.X. and J.C quantified hormone levels. A.J. and L.L. wrote the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Author (https://academic.oup.com/plcell) is: Lei Li (lei.li@pku.edu.cn).
REFERENCES
- Abdel-Ghany SE, Pilon M (2008) MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J Biol Chem 283: 15932–15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H (2019) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37: 420–423. [DOI] [PubMed] [Google Scholar]
- Angelovici R, Galili G, Fernie AR, Fait A. (2010). Seed desiccation: a bridge between maturation and germination. Trends Plant Sci 15: 211–218. [DOI] [PubMed] [Google Scholar]
- Bernala M, Ramiroa MV, Casesb R, Picorela R, Yruela I (2006) Excess copper effect on growth, chloroplast ultrastructure, oxygen-evolution activity and chlorophyll fluorescence in Glycine max cell suspensions. Physiologia Plantarum 127: 312–25. [Google Scholar]
- Bewley JD (1997) Seed germination and dormancy. Plant Cell 9: 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhead JL, Reynolds KA, Abdel-Ghany SE, Cohu CM, Pilon M (2009) Copper homeostasis. New Phytol 182: 799–816. [DOI] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E (2007) Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci 12: 514–521. [DOI] [PubMed] [Google Scholar]
- Chen D, Xu G, Tang W, Jing Y, Ji Q, Fei Z, Lin R (2013) Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25: 1657–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Ryu J, Jeong Y, Park J, Song J, Amasino RM, Noh B, Noh Y (2012) Control of seed germination by light-induced histone arginine demethylation activity. Dev Cell 22: 736–748. [DOI] [PubMed] [Google Scholar]
- Dai X, Zhuang Z, Zhao PX (2018) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 46: W49–W54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rienzo F, Gabdoulline RR, Menziani MC, Wade RC (2000) Blue copper proteins: a comparative analysis of their molecular interaction properties. Protein Sci 9: 1439–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Kim ST, Lord EM (2005) Plantacyanin plays a role in reproduction in Arabidopsis. Plant Physiol 138: 778–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu S, Giehl RFH, Meier B, Takahashi M, Terada Y, Ignatyev K, Andresen E, Küpper H, Peiter E, von Wirén N (2017) Metal tolerance protein 8 mediates manganese homeostasis and iron reallocation during seed development and germination. Plant Physiol 174: 1633–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Zhang Q, Wang Q, Wang X, Liu J, Li M, Huang L, Kang Z (2013) Target of tae-miR408, a chemocyanin-like protein gene (TaCLP1), plays positive roles in wheat response to high-salinity, heavy cupric stress and stripe rust. Plant Mol Biol 83: 433–443. [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G (2006) Seed dormancy and the control of germination. New Phytol 171: 501–523. [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59: 387–415. [DOI] [PubMed] [Google Scholar]
- Fu J, Chu J, Sun X, Wang J, Yan C (2012) Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC-MS/MS using single SPE purification and isotope dilution. Anal Sci 28: 1081–1087. [DOI] [PubMed] [Google Scholar]
- Giri AV, Anishetty S, Gautam P (2004) Functionally specified protein signatures distinctive for each of the different blue copper proteins. BMC Bioinformatics 9: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Kuang Z, Wang Y, Zhao Y, Tao Y, Cheng C, Yang J, Lu X, Hao C, Wang T, Cao X, Wei J, Li L, Yang X (2020) PmiREN: A comprehensive encyclopedia of plant miRNAs. Nucleic Acids Res 48: D1114–D1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta AS, Heinen JL, Holaday AS, Burke JJ, Allen RD (1993) Increased resistance to oxidative stress in transgenic plants that overexpress chloroplastic Cu/Zn superoxide dismutase. Proc Natl Acad Sci USA 90: 1629–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guss JM, Merritt EA, Phizackerley RP, Hedman B, Murata M, Hodgson KO, Freeman HC (1998) Phase determination by multiple-wavelength x-ray diffraction: crystal structure of a basic blue copper protein from cucumbers. Science 241: 806–811. [DOI] [PubMed] [Google Scholar]
- Han X, Chang X, Zhang Z, Chen H, He H, Zhong B, Deng XW (2019) Origin and evolution of core components responsible for monitoring light environment changes during plant terrestrialization. Mol Plant 12: 847–862. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias‐Fernández R, Rodríguez‐Gacio MC, Barrero‐Sicilia C, Carbonero P, Matilla AJ (2011) Three endo‐β‐mannanase genes expressed in the micropilar endosperm and in the radicle influence germination of Arabidopsis thaliana seeds. Planta 233: 25–36. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Xu G, Jing Y, Tang W, Lin R (2016) Phytochrome B and REVEILLE1/2-mediated signaling controls seed dormancy and germination in Arabidopsis. Nat Commun 7: 12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HW, Hwang BK (2000) Isolation, partial sequencing, and expression of pathogenesis-related cDNA genes from pepper leaves infected by Xanthomonas Campestris Pv. Vesicatoria. Mol Plant Microbe Interact 13: 136–142. [DOI] [PubMed] [Google Scholar]
- Kallioo P, Piiroinen P (1959) Effect of gibberellin on the termination of dormancy in some seeds. Nature 183: 1830–1831. [DOI] [PubMed] [Google Scholar]
- Kim DH, Yamaguchi S, Lim S, Oh E, Park J, Hanada A, Kamiya Y, Choi G (2008) SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20: 1260–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Mollet JC, Dong J, Zhang K, Park SY, Lord EM (2003) Chemocyanin, a small basic protein from the lily stigma, induces pollen tube chemotropism. Proc Natl Acad Sci USA 100: 16125–16130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, Punshon T, Lanzirotti A, Li L, Alonso JM, Ecker JR, Kaplan J, Guerinot ML (2006) Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314: 1295–1298. [DOI] [PubMed] [Google Scholar]
- Klaumann S, Nickolaus SD, Fürst SH, Starck S, Schneider S, Ekkehard Neuhaus H, Trentmann O (2011) The tonoplast copper transporter COPT5 acts as an exporter and is required for interorgan allocation of copper in Arabidopsis thaliana. New Phytol 192: 393–404. [DOI] [PubMed] [Google Scholar]
- Kuper J, Llamas A, Hecht HJ, Mendel RR, Schwarz G (2004) Structure of the molybdopterin-bound Cnx1G domain links molybdenum and copper metabolism. Nature 430: 803–806. [DOI] [PubMed] [Google Scholar]
- Lanquar V, Lelièvre F, Bolte S, Hamès C, Alcon C, Neumann D, Vansuyt G, Curie C, Schröder A, Krämer U, Barbier-Brygoo H, Thomine S (2005) Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. The EMBO J 24: 4041–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Choi G (2017) Phytochrome-interacting factor from Arabidopsis to liverwort. Curr Opin Plant Biol 35: 54–60. [DOI] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2010) PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Ma J, Zhai H, Xin P, Chu J, Qiao Y, Han L (2015) CHR729 is a CHD3 protein that controls seedling development in rice. PLoS One 10: e0138934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu JK (2013) Application of the CRISPR-Cas System for efficient genome engineering in plants. Mol Plant 6: 2008–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Heredia FP, Wastl J, Navarro JA, Bendall DS, Hervás M, Howe CJ, De La Rosa MA (2003) Photosynthesis: a new function for an old cytochrome? Nature 424: 33–34. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol Plant 15: 473–497. [Google Scholar]
- Nambara E, Marion-Poll A (2005). Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185. [DOI] [PubMed] [Google Scholar]
- Née G, Xiang Y, Soppe WJ (2017) The release of dormancy, a wake-up call for seeds to germinate. Curr Opin Plant Biol 35: 8–14. [DOI] [PubMed] [Google Scholar]
- Nersissian AM, Immoos C, Hill MG, Hart PJ, Williams G, Herrmann RG, Valentine JS (1998) Uclacyanins, stellacyanins, and plantacyanins are distinct subfamilies of phytocyanins: plant-specific mononuclear blue copper proteins. Protein Sci 7: 1915–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Kang H, Yamaguchi S, Park J, Lee D, Kamiya Y, Choi G (2009) Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Kim J, Park E, Kim JI, Kang C, Choi G (2004) PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16: 3045–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Hu JH, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Kamiya Y, Bae G, Chung WI, Choi G (2006) Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J 47: 124–139. [DOI] [PubMed] [Google Scholar]
- Pan J, Huang D, Guo Z, Kuang Z, Zhang H, Xie X, Ma Z, Gao S, Lerdau MT, Chu C, Li L (2018) Overexpression of microRNA408 enhances photosynthesis, growth, and seed yield in diverse plants. J Integr Plant Biol 60: 323–340. [DOI] [PubMed] [Google Scholar]
- Paszkiewicz G, Gualberto JM, Benamar A, Macherel D, Logan DC (2017) Arabidopsis seed mitochondria are bioenergetically active immediately upon imbibition and specialize via biogenesis in preparation for autotrophic growth. Plant Cell 29: 109–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñarrubia L, Romero P, Carrió-Seguí A, Andrés-Bordería A, Moreno J, Sanz A (2015) Temporal aspects of copper homeostasis and its crosstalk with hormones. Front Plant Sci 6: 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A, Shi H, Tepperman JM, Zhang Y, Quail PH (2014). Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol Plant 7: 1598–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschzttardtz H, Conéjéro G, Curie C, Mari S (2009) Identification of the endodermal vacuole as the iron storage compartment in the Arabidopsis embryo. Plant Physiol 151: 1329–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld N, Alon U (2003) Response delays and the structure of transcription networks. J Mol Biol 329: 645–654. [DOI] [PubMed] [Google Scholar]
- Ruan X, Luo F, Li D, Zhang J, Liu Z, Xu W, Huang G, Li X (2011) Cotton BCP genes encoding putative blue copper-binding proteins are functionally expressed in fiber development and involved in response to high-salinity and heavy metal stresses. Physiol Plant 141: 71–83. [DOI] [PubMed] [Google Scholar]
- Rydén LG, Hunt LT (1993). Evolution of protein complexity: the blue copper-containing oxidases and related proteins. J Mol Evol 36: 41–66. [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506. [DOI] [PubMed] [Google Scholar]
- Schulten A, Bytomski L, Quintana J, Bernal M, Krämer U (2019) Do Arabidopsis squamosa promoter binding protein-Like genes act together in plant acclimation to copper or zinc deficiency? Plant Direct 3: e00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Nambara E, Choi G, Yamaguchi S (2009) Interaction of light and hormone signals in germinating seeds. Plant Mol Biol 69: 463–472. [DOI] [PubMed] [Google Scholar]
- Seo M, Peeters AJ, Koiwai H, Oritani T, Marion-Poll A, Zeevaart JA, Koornneef M, Kamiya Y, Koshiba T (2000) The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc Natl Acad Sci USA 97: 12908–12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Zhu L, Castillon A, Majee M, Downie B, Huq E (2008) Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Lyu M, Luo Y, Liu S, Li Y, He H, Wei N, Deng XW, Zhong S (2018) Genome-wide regulation of light-controlled seedling morphogenesis by three families of transcription factors. Proc Natl Acad Sci USA 115: 6482–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang X, Mo X, Tang C, Zhong S, Deng XW (2015) Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination. Proc Natl Acad Sci USA 112: 3817–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Zhong S, Mo X, Liu N, Nezames CD, Deng XW (2013) HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 25: 3770–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoval O, Alon U (2010) SnapShot: network motifs. Cell 143: 326–e1. [DOI] [PubMed] [Google Scholar]
- Shu K, Liu XD, Xie Q, He ZH (2016) Two faces of one seed: hormonal regulation of dormancy and germination. Mol Plant 9: 34–45. [DOI] [PubMed] [Google Scholar]
- Sun TP, Kamiya Y (1997) Regulation and cellular localization of ent-kaurene synthesis. Physiol Plant 101: 701–708. [Google Scholar]
- Sun Y, Wu Z, Wang Y, Yang J, Wei G, Chou M (2019) Identification of phytocyanin gene family in legume plants and their involvement in nodulation of Medicago truncatula. Plant Cell Physiol 60: 900–915. [DOI] [PubMed] [Google Scholar]
- Tanaka R, Kobayashi K, Masuda T (2011) Tetrapyrrole metabolism in Arabidopsis thaliana. Arabidopsis Book 9: e0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Johansson H, Lee KP, Bou-Torrent J, Stewart K, Steel G, Rodríguez-Concepción M, Halliday KJ (2014) The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet 10: e1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Frame B (2009). Biolistic gun-mediated maize genetic transformation. Methods Mol Biol 526: 29–45. [DOI] [PubMed] [Google Scholar]
- Weigel M, Varotto C, Pesaresi P, Finazzi G, Rappaport F, Salamini F, Leister D (2003) Plastocyanin is indispensable for photosynthetic electron flow in Arabidopsis thaliana. J Biol Chem 278: 31286–31289. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 8: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251. [DOI] [PubMed] [Google Scholar]
- Yan J, Gu Y, Jia X, Kang W, Pan S, Tang X, Chen X, Tang G (2012) Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24: 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye N, Li H, Zhu G, Liu Y, Liu R, Xu W, Jing Y, Peng X, Zhang J (2014) Copper suppresses abscisic acid catabolism and catalase activity, and inhibits seed germination of rice. Plant Cell Physiol 55: 2008–2016. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang H, Li L (2013) SQUAMOSA promoter binding protein-like7 regulated microRNA408 is required for vegetative development in Arabidopsis. Plant J 74: 98–109. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhao X, Li J, Cai H, Deng XW, Li L (2014) MicroRNA408 is critical for the HY5-SPL7 gene network that mediates the coordinated response to light and copper. Plant Cell 26: 4933–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng S, Chen F, Chen H, Wang J, McCall C, Xiong Y, Deng XW (2008) Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell 20: 1437–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Shi H, Xue C, Wei N, Guo H, Deng XW (2014) Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc Natl Acad Sci USA 111: 3913–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Zhao M, Shi T, Shi H, An F, Zhao Q, Guo H (2009) EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci USA 106: 21431–21436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou MG, Gu LF, Li PC, Song XW, Wei LY, Chen ZY, Cao XF (2010) Degradome sequencing reveals endogenous small RNA targets in rice (Oryza sativa L. ssp. indica). Front Biol 5: 67–90. [Google Scholar]
- Zhuang Y, Zuo D, Tao Y, Cai H, Li L (2020) Laccase3-based extracellular domain provides possible positional information for directing Casparian strip formation in Arabidopsis. Proc Natl Acad Sci USA 117: 15400–15402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH (2000) An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.