Abstract
The insertion of organellar membrane proteins with the correct topology requires the following: First, the proteins must contain topogenic signals for translocation across and insertion into the membrane. Second, proteinaceous complexes in the cytoplasm, membrane, and lumen of organelles are required to drive this process. Many complexes required for the intracellular distribution of membrane proteins have been described, but the signals and components required for the insertion of plastidic β-barrel-type proteins into the outer membrane are largely unknown. The discovery of common principles is difficult, as only a few plastidic β-barrel proteins exist. Here, we provide evidence that the plastidic outer envelope β-barrel proteins OEP21, OEP24, and OEP37 from pea (Pisum sativum) and Arabidopsis thaliana contain information defining the topology of the protein. The information required for the translocation of pea proteins across the outer envelope membrane is present within the six N-terminal β-strands. This process requires the action of translocon of the outer chloroplast (TOC) membrane. After translocation into the intermembrane space, β-barrel proteins interact with TOC75-V, as exemplified by OEP37 and P39, and are integrated into the membrane. The membrane insertion of plastidic β-barrel proteins is affected by mutation of the last β-strand, suggesting that this strand contributes to the insertion signal. These findings shed light on the elements and complexes involved in plastidic β-barrel protein import.
Plastidic β-barrel proteins contain sequence-intrinsic signals for translocation and membrane insertion, the latter of which involves a complex formation with TOC75-V prior to the final membrane insertion.
Introduction
Most organellar proteins are synthesized at cytosolic ribosomes as precursor proteins. Consequently, the multiplicity of possible target destinations demands a high targeting accuracy. This specificity is generally guaranteed by the presence of a targeting signal that ensures the correct targeting and insertion of the proteins into the respective organelle and suborganellar compartments (Schatz and Dobberstein, 1996). This specificity appears to be high, as even plastids and mitochondria, which share many enzymatic steps due to their common Gram-negative bacterial origin, share only a small number of dual-targeted proteins (Carrie et al., 2009). Most precursor proteins of the mitochondrial matrix and the chloroplast stroma contain an N-terminal topogenic signal called the presequence or transit peptide, respectively, that is cleaved after successful translocation by intraorganellar processing peptidases (Schleiff and Becker, 2011; Jarvis and López-Juez, 2013). These topogenic signals have an overall positive charge and the tendency to form an amphiphilic α-helix (Bruce, 2001; Chacinska et al., 2009). In addition, chloroplast transit peptides are enriched in hydroxylated amino acids, which can be phosphorylated by cytosolic kinases (Martin et al., 2006). Presequences and transit peptides are divergent in length and primary structure, leading to the hypothesis that instead of a specific-sequence motif, a structural element or physiochemical pattern is important for recognition on the organellar receptors (Schleiff and Becker, 2011; Kunze and Berger, 2015).
The topogenic signal of some proteins, especially outer membrane proteins, appears to be distinct from this general scheme. One class with such distinct signals consists of the β-barrel proteins in the outer membranes of mitochondria and chloroplasts. In mitochondria of the yeast Saccharomyces cerevisiae, at least five β-barrel proteins were identified, namely, the 50-kDa component of the sorting and assembly machinery for β-barrel proteins (SAM50/TOB55), the 40-kDa subunit of the translocon of the outer mitochondrial membrane (TOM40), two homologs of the porin-like voltage-dependent anion channel (VDAC), and mitochondrial distribution and morphology protein 10 (MDM10). SAM50, a member of the OMP85 family, is the central component of the SAM complex (Kozjak et al., 2003; Paschen et al., 2003). TOM40 plays an essential role as the pore of the TOM complex (Hill et al., 1998; Künkele et al., 1998). MDM10 is required for mitochondrial biogenesis and organelle–organelle contact formation (Sogo and Yaffe, 1994; Boldogh et al., 2003; Youngman et al., 2004; Meisinger et al., 2007; Kornmann et al. 2009; Ellenrieder et al., 2016). However, MDM10 does not exist in plants (Flinner et al., 2013; Murcha et al., 2014). VDAC proteins belong to the porin family, although they consist of 19 β-strands, whereas bacterial porins contain an even number of β-strands (Cowan et al., 1992; Weiss and Schulz, 1992; Hiller et al., 2008; Ujwal et al., 2008). An additional plant-specific mitochondrial β-barrel protein of the porin family, the outer membrane protein of 47 kDa (OM47), was recently described (Li et al., 2016).
Six β-barrel proteins present in plastidic membranes are not involved in protein biogenesis. The outer envelope protein of 21 kDa (OEP21) constitutes an ATP-regulated anion-selective channel (Bölter et al., 1999). The outer envelope proteins of 24, 37, and 40 kDa (OEP24, OEP37, and OEP40, respectively) function as cation-selective solute channels with distinct substrate specificities (Pohlmeyer et al., 1998; Schleiff et al., 2003; Harsman et al., 2016). The predicted topology of OEP40 is atypical, as it consists of 10 β-strands in the center of the protein and a large, mainly unstructured C-terminal domain (Harsman et al., 2016). trigalactosyldiacylglycerol transport protein 4 and Lipopolysaccharide assembly protein D (LptD) are two additional 16 β-stranded plastidic β-barrel proteins involved in lipid trafficking (Xu et al., 2008; Haarmann et al. 2010; Hsueh et al., 2017a).
In addition, numerous plastidic OMP85 homologs were identified in Arabidopsis thaliana that are annotated according to their chromosomal locations: Toc75-I, Toc75-III, Toc75-IV, and Toc75-V/OEP80 (Jackson-Constan and Keegstra, 2001). Toc75-I is a pseudogene and Toc75-IV is thought to be expressed in specific organs or only under specific conditions (Baldwin et al., 2005). TOC75-III constitutes the pore-forming translocation channel of the translocon of the outer membrane of chloroplasts (TOC; Schleiff and Becker, 2011), while TOC75-V (Eckart et al., 2002; Patel et al., 2008) is thought to function in β-barrel protein insertion (Schleiff and Soll, 2005), which, however, still needs to be confirmed. As Toc75-V-knockdown plants show reduced levels of TOC75-III (Huang et al., 2011), the difference between the functions of TOC75-V in general β-barrel assembly versus the specific assembly of TOC75-III remains to be uncovered. However, both genes are essential, suggesting there is no functional overlap between TOC75-III and TOC75-V (Baldwin et al., 2005; Huang et al., 2011; Hsu et al., 2012). In addition, TOC75-V is not present in isolated TOC complexes (Schleiff et al., 2003b) and does not migrate with TOC75-III or other TOC components on native PAGE (Kikuchi et al., 2006; Ladig et al., 2011). P36 (Nicolaisen et al., 2015) and P39 (Hsueh et al., 2017b, 2018) are two additional nonessential β-barrel proteins that share evolutionary relationships with TOC75-V (Moslavac et al., 2005; Bredemeier et al., 2007).
The targeting signals, mode of import, and mode of assembly of β-barrel proteins into the outer membranes of mitochondria are well understood (Jores et al., 2016, 2018; Höhr et al., 2018). In contrast, little is known about these processes for plastidic β-barrel proteins. The existence of a cleavable, N-terminal targeting signal has only been described for TOC75-III and TOC75-V (Day et al., 2019; Gross et al., 2020). TOC75-III has a bipartite targeting signal (Tranel et al., 1995; Tranel and Keegstra, 1996). The N-terminal part of the targeting signal is responsible for its targeting to the chloroplast and translocation into the stroma, where it becomes cleaved by the stromal-processing peptidase, while the C-terminal part of the signal prevents full stromal translocation and is processed in the intermembrane space (IMS; Tranel and Keegstra, 1996). Moreover, the hydrophobicity profile in the penultimate β-strand of mitochondrial β-barrel proteins ensures the specificity of its targeting to this organelle; the manipulation of the hydrophobicity profile of OEP24 to VDAC-like resulted in its partial import into mitochondria (Klinger et al., 2019). Furthermore, the replacement of the C-terminal β-hairpin of the plastidic β-barrel protein OEP24 with the respective β-hairpin of the homolog VDAC-1 present in A. thaliana resulted in targeting of this chimeric protein to mitochondria in A. thaliana protoplasts (Jores et al., 2016). With respect to the translocation of plastidic β-barrel proteins, recent findings revealed a possible intermediate in the IMS (Klinger et al., 2019) derived by TOC-dependent translocation across the outer membrane (Day et al., 2019). All subsequent steps are currently unclear.
To gain insights into the targeting and insertion of β-barrel proteins in plants, we utilized a protoplast-based and an in vitro import assay to analyze the translocation of plastidic β-barrel proteins using OEP21, OEP24, OEP37, and P39 from pea (Pisum sativum), from A. thaliana, or both as examples. We dissected the importance of the different regions of plastidic β-barrel proteins for translocation and membrane insertion. Further, we examined the formation of intermediate complexes during the translocation of OEP37, as has been demonstrated numerous times for processes such as mitochondrial translocation (e.g. Model et al., 2001; Kutik et al., 2008) to shed light on the mode of β-barrel-type protein insertion into the outer envelope. Our findings provide additional evidence for the involvement of TOC in the translocation of plastidic β-barrel proteins and uncover a physical interaction of the substrate with TOC75-V in the IMS.
Results
OEP24 inserts into the outer membrane with its N- and C-termini exposed to the cytosol
OEP24 from pea was used as a model to establish a quantitative assay for β-barrel protein insertion because the targeting, translocation, and membrane insertion of OEP24 have been analyzed in vivo and in vitro (Machettira et al., 2011a, Ulrich et al., 2012; Jores et al., 2016; Klinger et al., 2019). Recent topology prediction suggested that PsOEP24 exhibits a 14 β-stranded topology (Supplemental Figure 1; Machettira et al., 2011a; Jores et al., 2016), whereas 12 β-strands were originally predicted (Schleiff et al., 2003).
The self-assembly GFP (saGFP) system was used for protein topology analysis in A. thaliana mesophyll protoplasts or onion (Allium cepa) epidermal cells (Figure 1). The two cellular systems were compared for the following reasons: (1) The two systems represent two different cell types. Thus, the mechanisms observed in both systems are likely to be universal and not cell type dependent. (2) The plastidic surface is larger in mesophyll cells than in epidermal cells. Thus, by using the two systems, dependence on the accessible organelle surface can be avoided. (3) Two different preparation and transfection methods are used, resulting in higher reliability of results. For example, protoplast purification results in a change in the cytoskeleton, whereas this structure is intact in onion cells (Scott et al., 1999; Jaedicke et al., 2011). Although the role of the cytoskeleton in targeting proteins to the surface of the chloroplast is currently unclear, a comparison between the two systems can avoid artifacts due to the cell purification method.
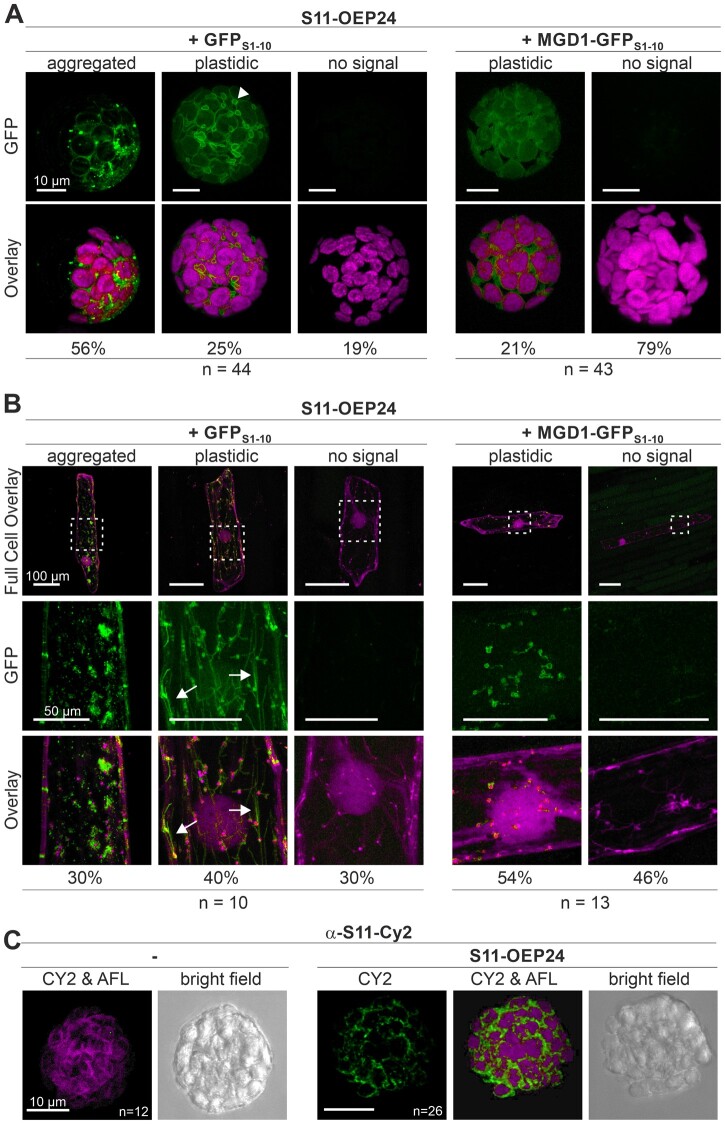
Figure 1.
Distribution of S11-OEP24 in A. thaliana protoplasts and A. cepa epidermal cells. A, Arabidopsis thaliana mesophyll protoplasts were co-transformed with S11-OEP24, mCherry, or mCherry-SKL (Supplemental Figure 2), GFPS1–10 (left) or MGD1-GFPS1–10 (right) and analyzed after 14±2 h using confocal laser scanning microscopy (CLSM). GFP signal (top) was overlaid with chlorophyll autofluorescence (bottom); scale bars represent 10 µm. A single stack is shown. B, A. cepa epidermal cells were co-transformed with S11-OEP24, SSUTP-mCherry (staining leucoplasts), GFPS1–10 (left), or MGD1-GFPS1–10 (right). Cells were analyzed after 14±2 h. GFP (middle) and mCherry-signals (bottom) were overlaid. The upper panel shows full cells (scale bar: 100 µm); the remaining panels show magnified views (scale bar: 50 µm). For A and B, a representative cell is shown for each indicated GFP signal category and its percentage is given below. The number of analyzed cells (n) in multiple biological replicates (br ≥3) is indicated. Arrows point to membrane extrusions. Additional signals are shown in Supplemental Figure 2. C, Arabidopsis thaliana mesophyll protoplasts were transformed with S11-OEP24 (right) and immune-stained with CY2-labeled α-S11 antibodies after 14±2 h, and fluorescence was recorded using CLSM. The C2 signal (CY2) was overlaid with chlorophyll autofluorescence (CY2 & AFL), and the bright field image is shown; scale bars represent 10 µm.
The saGFP system is based on the division of the GFP into a molecule containing the first 10 β-strands and a molecule consisting of only the 11th β-strand of the GFP barrel (Cabantous et al., 2005). In traditional BiFC assays using two rather equal parts of the fluorescent protein such as YFP, the two parts have a low affinity for each other and form a functional YFP only in case of high spatial proximity, which can be achieved, for example, by interactions of the fusion partners. In contrast, the two parts in the saGFP system have a higher affinity for each other, and functional GFPs can be formed when both components are in the same compartment (Cabantous et al., 2005; Sommer et al., 2011; Machettira et al., 2011b). This makes the split-YFP system optimal for the analysis of protein–protein interactions in vivo and the saGFP system for the analysis of intracellular localizations in vivo, which is why it was used in this work. For this, only two reporters were needed to verify the topology of the tagged proteins (Machettira et al., 2011b), and the resulting signals were characterized. The respective constructs were co-transformed with GFPS1–10 and the IMS reporter MGD1-GFPS1–10 to probe for cytosolic or IMS exposure of GFPS11, respectively. The co-transformation efficiency of multiple plasmids in A. thaliana protoplasts is in the range of 80% for the number of vectors used (Tripp et al., 2009; Sommer et al., 2013; Tillmann et al., 2015). General transformation was controlled by co-transformation with pML94-mCherry, pML94-mCherry-SKL, or pML94-SSUTP-mCherry; the latter was used to visualize the chlorophyll-free leucoplasts in A. cepa epidermal cells (Supplemental Figure 2). However, GFP assembly events are not as frequent as expected based on the co-transformation efficiency, because both fragments have to be present in the same compartment to yield fluorescence.
The different signal characteristics after GFP assembly and co-expression of GFPS11-OEP24 (S11-OEP24 hereafter) with reporter constructs were classified as “no GFP signal” (but mCherry signal), “(cytosolic) soluble,” “(cytosolic) aggregated,” “plastidic,” and “plastidic and aggregated” (Figures 1, A, B, and 2, A; Supplemental Figure 2). The latter category refers to cytosolic aggregation and plastidic localization in the same cell. Multiple cells from various independent experiments were analyzed, and the observed localizations were quantified to generate informative figures (Figure 1, A and B). The localization of the S11-OEP24 was further confirmed by immunodecorating protoplasts with antibodies against GFPS11 that had been labeled with the cyanine dye CY2. A specific, chloroplast surrounding fluorescence signal was detected after the expression of S11-OEP24 (Figure 1C), but not in untransformed protoplasts.
The cytosolic small heat-shock protein HSP18.5 (HSP18.5-S11; Siddique et al., 2008; Supplemental Figure 3) and the mitochondrial protein VDAC-1 (S11-VDAC-1; images in Klinger et al., 2019) were used to define thresholds for the reliability of the quantification of the saGFP system, because neither is thought to be targeted to or localized to plastids. Co-expression of HSP18.5-S11 with GFPS1–10 (cytosol), but not with MGD1-GFPS1–10 (IMS), yielded GFP fluorescence in most of the protoplasts and in many epidermal cells (Figure 2, A and B). Co-expression of S11-VDAC-1 with the GFPS1–10 yielded a GFP signal in most protoplasts and a co-localization of the GFP signal with plastids in 30% of the epidermal cells (Figure 2, A and B). In turn, co-expression of S11-VDAC-1 with MGD1-GFPS1–10 resulted in GFP fluorescence overlaying with the autofluorescence of plastids in 42% of all protoplasts, but did not result in GFP signal in epidermal cells (Figure 2, A and B). Hence, a reliable conclusion on protein localization requires a signal in one compartment of at least 40% of all analyzed protoplasts and in more than 30% of epidermal cells.
Figure 2.
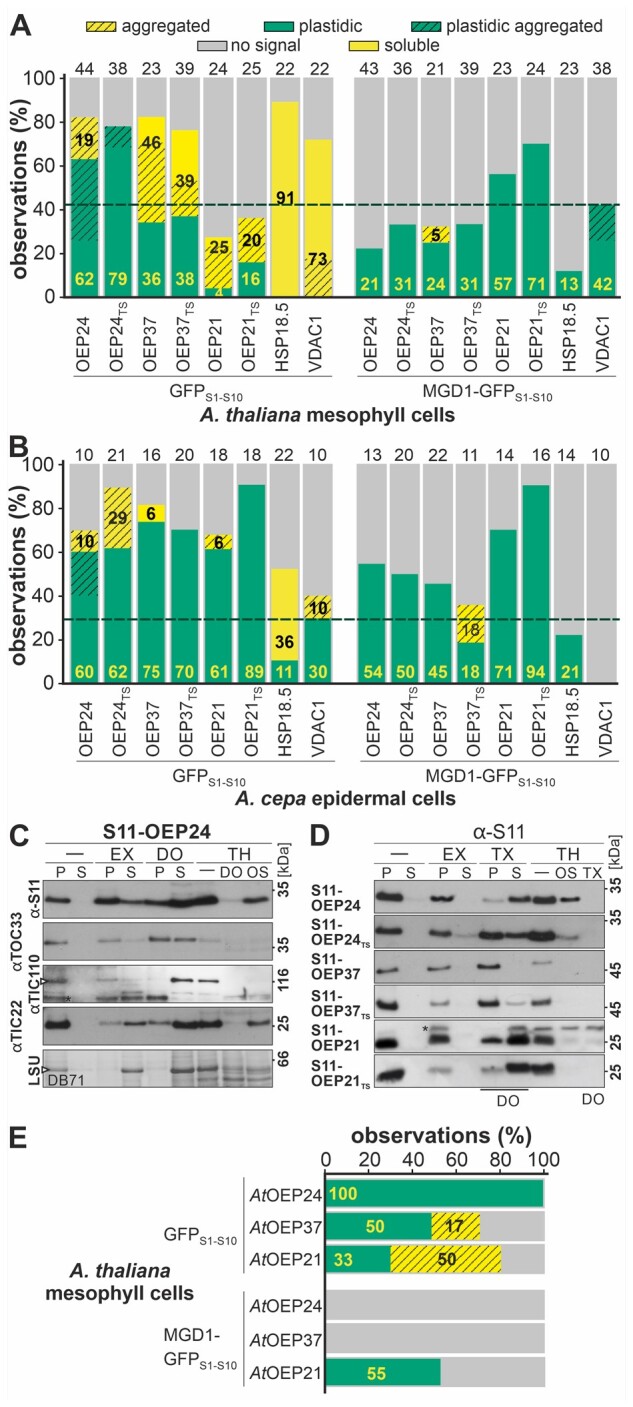
Quantitative analysis of the in vivo localizations of plastidic β-barrel proteins. A and B, Distribution of GFP fluorescence (% of all) in A. thaliana mesophyll protoplasts (A) and A. cepa epidermal cells (B), respectively, for the indicated S11-tagged proteins co-expressed with cytosolic (GFPS1–10) and intermembrane space reporter (MGD1-GFPS1–10 is shown [soluble (yellow), cytosolic-aggregated (yellow hashed), plastidic (green), plastidic aggregated (green hashed), no GFP signal (gray)]. Conditions and representative examples are provided (“Methods” section; Figure 1; Supplemental Figures 2–5). Numbers in boldface indicate the percentage of observations in the cytosol (black, soluble + cytosolic-aggregated) and plastids (yellow, plastids + plastidic aggregated). The number of analyzed cells is given above the bar (biological replicates br > 3). The dashed line indicates the reliability threshold for positive detection of a defined localization based on the nonplastidic controls. C, A. thaliana protoplasts were co-transformed with S11-OEP24 and GFPS1–10. After 14±2 h, the cells were harvested, lysed, and collected in isosmotic buffer (-) with 100 mM Na2CO3 (EX), 1.5% DOMA (DO), 200 µg/mL TH (TH/-), or DOMA and TH. One fraction was collected in osmolysis buffer (OS) containing TH. Samples from untreated, carbonate-, and DO-treated cells were separated into pellet (P) and supernatant (S), precipitated, and immunodecorated with αGFPS11, αTOC33, αTIC110, and αTIC22 antibodies. The lowest panel shows DB71 staining of RubisCO (LSU). D, A. thaliana protoplasts were co-transformed with S11-tagged proteins and GFPS1–10 and processed as in Figure 2C. Note, for all constructs except for S11-OEP21TS, 1.5% Triton X-100 was used instead of 1.5% DOMA. E, Distribution of GFP fluorescence (% of all observations) in A. thaliana mesophyll protoplasts for S11-tagged proteins co-expressed with GFPS1-10 (Supplemental Figure 4) or MGD1-GFPS1–10 as in A.
Co-transformation of S11-OEP24 with the cytosolic reporter resulted in chloroplast-localized GFP signal in more than 60% of the analyzed cells, irrespective of the cell type (GFPS1–10, Figures 1, 2, A and B). When S11-OEP24 was co-expressed with MGD1-GFPS1–10, only ∼20% of the analyzed protoplasts (Figures 1, A and 2, A) and ∼55% of transformed A. cepa epidermal cells showed chloroplast-localized GFP signals (Figures 1, B and 2, B).
The membrane insertion of S11-OEP24 after expression in protoplasts was confirmed by protoplast fractionation. To evaluate the fractionation efficiency, the presence of TOC33, a surface-exposed outer envelope protein (Seedorf et al., 1995), TIC110, an inner envelope protein with soluble domains (Jackson et al., 1998), TIC22, a chloroplast IMS component (Kouranov et al., 1998), and the stromal-localized large subunit of RubisCO (visualized by DB71 staining) were monitored. All control proteins were pelleted when the cell suspension was centrifuged (Figure 2C, -/P&S). Carbonate treatment to remove membrane-associated proteins (Fujiki et al., 1982) resulted in the solubilization of most of RubisCO and TIC22, while most of the three other proteins were pelleted (Figure 2C, EX/P&S). The addition of detergent (n-dodecyl β-d-maltoside, DO; Triton X-100, TX) to distinguish between aggregated and membrane-inserted proteins by solubilizing lipid bilayers resulted in a presence of all proteins in the supernatant, although a large fraction of TOC33 and a minor fraction of TIC22 remained insoluble (Figure 2C, DO/P&S). Treatment of the cell lysates with thermolysin (TH) resulted in the degradation of the surface-exposed TOC33, but not the proteins localized to plastids (Figure 2C, TH/-). The addition of detergent for organelle solubilization resulted in general sensitivity to protease (Figure 2C, TH/DO). In contrast, protease treatment during osmolysis resulted in the degradation of TOC33 and TIC110 (which contain large soluble domains), while TIC22 remained largely protease resistant (Figure 2C, TH/OS).
S11-OEP24 was pelleted before and after carbonate treatment (Figure 2C, D, -/P; EX/P). Detergent treatment resulted in the solubilization of most of the protein, irrespective of whether n-dodecyl β-d-maltoside (Figure 2C, DO/P versus S) or Triton X-100 was used for solubilization (Figure 2D, TX/P versus S). Treatment of the cell lysate with TH did not result in the degradation of S11-OEP24 (Figure 2C, D, TH/-). This result is rather expected, as OEP24 was previously shown to be resistant to TH (Schleiff et al., 2003, Ulrich et al., 2012). Furthermore, the first amino acid of OEP24 is considered to be integrated into the membrane (Supplemental Figure 1). The resistance of the S11 tag against TH might be attributed to the existence of charged amino acids, and its removal likely depends on the existence of available cleavage sites in the mature domain of the carrier. Osmolysis did not yield an efficient proteolysis of S11-OEP24 (TH/OS), because the membrane remains intact, while removing lipids by detergent treatment resulted in the accessibility of cleavage sites and degradation of the protein by TH (Figure 2C, TH/DO; Figure 2D, TH/TX).
Efficient membrane insertion of bacterial β-barrel proteins depends on the nature of the C-terminal amino acid (Struyvé et al., 1991). To probe for endogenous C-terminal amino acids that are important for targeting, translocation, or membrane insertion, a GFPS11-tagged OEP24 mutant protein was produced containing two additional hydroxylated amino acids, threonine (T) and serine (S) (GFPS11-OEP24TS). Utilizing the in vivo approach (Supplemental Figure 3), an efficiency of targeting and translocation of this protein into chloroplasts comparable to that of wild-type protein was observed, as judged by the detection of GFP fluorescence while co-expressing the respective reporters (Figure 2, A and B). Moreover, S11-OEP24TS remained in the pellet fraction after treating the cell suspension with sodium carbonate (Figure 2D, EX/P&S) and was largely solubilized by adding detergent (lane TX/P&S). Like the wild-type protein, S11-OEP24TS was protected against TH before but not after detergent treatment (lane TH/- & TX). Interestingly, the protein with two additional amino acids (S11-OEP24TS) was slightly more sensitive to protease treatment after osmolysis than the native protein (S11-OEP24; lane TH/OS).
The targeting and translocation behavior of OEP24 from pea was comparable to the behavior of the endogenous protein. The localization of S11-AtOEP24 (OEP24 from A. thaliana) after co-expression with GFPS1–10 or MGD1–GFPS1–10 in A. thaliana mesophyll protoplasts (Supplemental Figure 4) resulted in efficient GFP assembly during co-expression of the cytosolic marker, but not while using the IMS marker (Figure 2E).
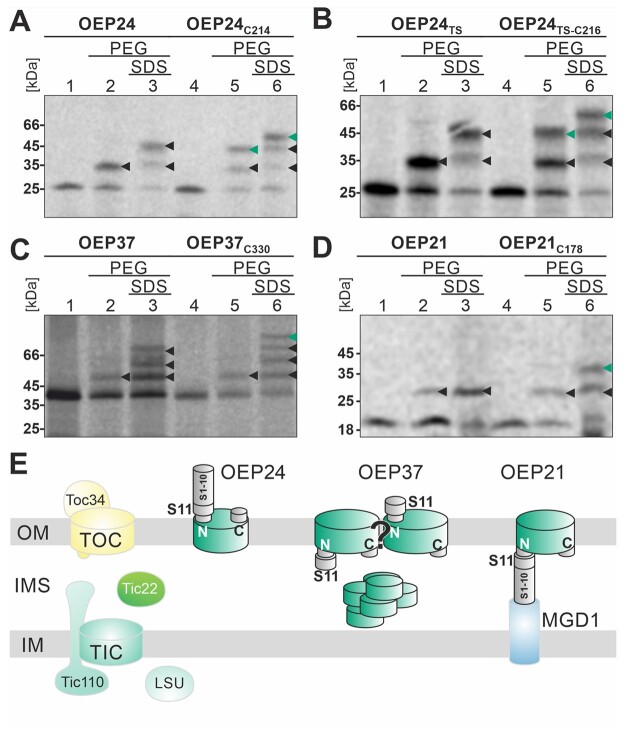
The topology of S11-OEP24 deduced from in vivo experiments (Figures 1 and 2) was confirmed by PEGylation of cysteine residues after import into chloroplasts in vitro. Adding PEG-Mal to chloroplasts after the import of OEP24 or OEP24TS resulted in the modification of a single cysteine, yielding a size shift of approximately 10 kDa (Figure 3, A and B, lanes 1, 2). Before solubilization, one cysteine was modified (lane 2). Solubilization of chloroplasts prior to the addition of PEG-Mal resulted in the modification of two cysteines (lane 3). Introduction of a cysteine behind the C-terminal amino acid (OEP24C214 or OEP24TS-C216) yielded an additional PEGylation before and after the solubilization of chloroplasts (lane 5 versus 6, green arrow), confirming the exposure of C-terminal residues to the cytoplasm. Furthermore, the PEGylation of the endogenous cysteines fits with their predicted positions in one cytosolic and one IMS-exposed loop.
Figure 3.
Topology of the plastidic β-barrel proteins based on PEGylation. A–D, OEP24 and OEP24C214 (A), OEP24TS and OEP24TSC216 (B), OEP37 and OEP37C330 (C), and OEP21 and OEP21C178 (D) were imported into chloroplasts (lanes 1 and 4), followed by the addition of PEG-Mal (lanes 2 and 5) in the presence of 1% SDS (lane 3 and 6). PEGylated endogenous cysteines are marked by black arrowheads, and PEGylated C-terminal-fused cysteine by green arrowheads. E, Schematic representation of the localization of the tested protein based on the observations presented.
In summary, fractionation confirmed that the majority of S11-OEP24 was inserted into the membrane. The GFP assembly experiments indicated that the N-terminus of OEP24 was exposed to the cytosol, while PEGylation revealed a cytosolic exposure of the C-terminus. Unfortunately, because OEP24 is embedded deep in the membrane and is known to be resistant to TH (Schleiff et al., 2003, Ulrich et al., 2012), the processing results could not be used to support or reject the topology prediction. Nevertheless, the combination of fluorescence, fractionation, and PEGylation results confirm the previously proposed topology of OEP24, including the cytosolic exposure of the N- and C-termini (Figure 2E; Jores et al., 2016; Klinger et al., 2019). The extension of the protein by one or two amino acids did not influence the efficiency of translocation across the membrane, and thus, it appears that a particular exposed C-terminal amino acid is not required for the biogenesis of OEP24.
OEP21 and OEP37 differ in their membrane topology
OEP37 and OEP21 were used as additional model proteins to generalize the information on β-barrel OMP topology. OEP37 is predicted to contain 18 β-strands (Supplemental Figure 1), with both termini exposed to the IMS (Ulrich et al., 2012), while the orientation of OEP21 is still under debate. The current model of OEP21 suggests that it contains 10 β-strands, while originally an 8 β-stranded topology was predicted (Supplemental Figure 1; Hemmler et al., 2006).
The in vivo approach (Supplemental Figure 5) revealed a low targeting and translocation efficiency of S11-OEP37 and S11-OEP37TS in protoplasts, which was below the threshold (Figure 2A). Nevertheless, the efficiency of GFP assembly on chloroplasts was slightly higher for the cytosolic reporter compared to the IMS reporter. Similar to OEP24, the results could be reproduced for the protein from A. thaliana (Figure 2E;Supplemental Figure 6). The targeting to plastids in epidermal cells is more efficient, as more plastidic GFP assembly during the expression of the cytosolic reporter was observed (Figure 2B). The fractionation of protoplasts transformed with S11-OEP37 or S11-OEP37TS yielded protein in the pellet fraction after isosmotic, carbonate, and Triton X-100 treatment (Figure 2D, -, EX, TX). The latter is consistent with the large amount of aggregated protein in protoplasts (Figure 2A). However, detergent treatment resulted in a soluble fraction of S11-OEP37TS. Treatment with TH resulted in a reduced amount of protein in isosmotic buffer and complete degradation after osmolysis or solubilization (Figure 2D, TH/- and OS and TX). Thus, it appears that a large fraction of S11-OEP37 or S11-OEP37TS is not inserted in the membrane but instead forms aggregates.
PEGylation of in vitro imported OEP37 pointed toward an IMS exposure of the C-terminus, as the addition of PEG-Mal after solubilizing the chloroplasts resulted in labeling of the additional C-terminally fused cysteine in OEP37C330 (Figure 3C, lane 3 versus 6). Before solubilization, just one cysteine was modified in OEP37 and OEP37C330, respectively (Figure 3C, lane 2 versus 5). This is consistent with the predicted topology of OEP37. Hence, our results point toward an IMS exposure of the C-terminus of OEP37. In contrast, the localization of the N-terminus could not be determined with high certainty (Figure 3E), especially since porins with even or uneven numbers of β-strands have been identified in different species (Cowan et al. 1992; Weiss and Schulz, 1992; Hiller et al., 2008; Ujwal et al., 2008).
S11-OEP21 and S11-OEP21TS were targeted to (and translocated into) chloroplasts with high efficiency in both systems (Figure 2, A and B; Supplemental Figure 5). Co-expressing S11-OEP21 and S11-OEP21TS with GFPS1–10 resulted in GFP signal in a large number of epidermal cells, but not in protoplasts. In turn, co-transfection with MGD1-GFPS1–10 yielded a GFP signal in almost all protoplasts or epidermal cells, which suggests that the N-terminus was exposed to the IMS (Figure 2, A and B). The analysis of OEP21 from A. thaliana in protoplasts confirmed the species-independent behavior of this protein (Figure 2E;Supplemental Figure 6). The fractionation demonstrated that both proteins were resistant to isosmotic and carbonate treatment, but could be largely solubilized by treatment with detergent (Figure 2D, -, EX, TX). The proteins were unaffected by proteolytic digestion with TH in isosmotic buffer but were sensitive to this treatment after osmolysis or solubilization (Figure 2D, TH/- and OS and TX). Analysis of the localization of the C-terminus of OEP21C178 by PEGylation revealed no additional PEGylation before chloroplast solubilization but additional PEGylation after this process compared to OEP21 (Figure 3D, lane 2 versus 5 and lane 3 versus 6). Together, these results suggest a topology in which the N- and C termini of OEP21 are exposed to the IMS (Figure 3E).
The N-terminal domain of β-barrel proteins contains information for translocation in vivo
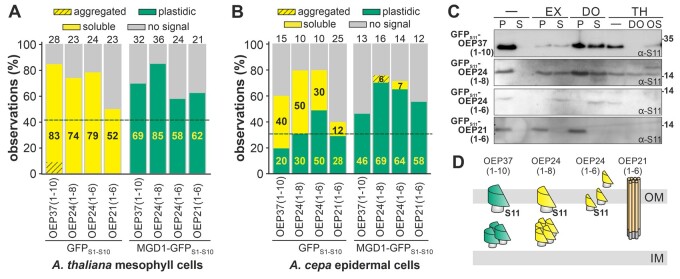
The capability of different regions of plastidic β-barrel proteins for targeting, translocation, and membrane insertion was probed since a defined signal for these processes is not yet known. C-terminal truncations were created to analyze the impact of the N-terminal domains. The C-terminal truncations of the three β-barrel proteins annotated as S11-OEP37(1-10) (representing the first ten predicted β-strands), S11-OEP24(1–8), S11-OEP24(1–6), and S11-OEP21(1–6) were expressed in protoplasts with the appropriate reporter and the localization quantified. The constructs were generated in this manner because eight strands are considered to be the minimal unit of a monomeric barrel (Mirus et al., 2010). Hence, the first two proteins could still form a monomeric β-barrel, while S11-OEP24(1–6) and S11-OEP21(1–6) either formed a multimeric β-barrel or did not form a β-barrel at all. When co-expressed with GFPS1-10, all proteins yielded cytosolic GFP fluorescence (Figure 4A, left; Supplemental Figure 7), while co-expression with MGD1-GFPS1–10 resulted in a chloroplast-localized GFP signal in the majority of the protoplasts (Figure 4A, right; Supplemental Figure 7).
Figure 4.
In vivo localization of the N-terminal fragments of plastidic β-barrel proteins. A, The distribution of the observed GFP fluorescence in protoplasts transformed with the constructs indicated is shown, as described in the legend of Figure 2A. B, The distribution of the GFP fluorescence in epidermal cells for the indicated constructs, as described in the legend of Figure 2A. C, The indicated proteins were expressed in A. thaliana protoplasts together with GFPS1–10 and processed as in Figure 2C. D, Schematic representation of the localization of the tested proteins based on the observations presented.
Membrane extrusions are indicative of the (massive) membrane insertion of proteins (Machettira et al., 2011a). Inspection of the underlying shape of the GFP fluorescence pattern after co-expressing the proteins with MGD1-GFPS1–10 yielded a significantly higher occurrence of extrusions for S11-OEP21 compared to S11-OEP21(1–6), and for S11-OEP24 compared to S11-OEP24(1–6) (Supplemental Figure 8). This observation suggests that only a minor fraction of the truncated proteins is inserted into the membrane. The frequency of extrusions observed while expressing S11-OEP37(1–10) or S11-OEP24(1–8) was comparable to that when full-length proteins were expressed (Supplemental Figure 8). In epidermal cells, GFP was efficiently assembled by co-expressing MGD1-GFPS1–10 with all four proteins (Figure 4B, right; Supplemental Figure 7). In some cases, co-expression with GFPS1-10 resulted in a rim-like GFP fluorescence surrounding the plastids, as visualized by co-expression of pML94-SSUTP-Cherry, but the percentage of cells was above the threshold only for S11-OEP24(1–6) (Figure 4B, left, Supplemental Figure 7).
Fractionation of protoplasts revealed that S11-OEP37(1–10) was partially extracted by sodium carbonate and partially solubilized by detergent treatment (Figure 4C, first panel: EX, DO). This finding, together with the observation that this protein was sensitive to protease following solubilization with detergent (Figure 4C, TH/DO), confirms the notion that parts of the protein are inserted into the membrane, while the detergent-resistant fraction likely represents IMS-localized aggregates (Figure 4D).
Similar results were obtained for S11-OEP24(1–8) as for S11-OEP37(1–10) (Figure 4C, second panel), although some of the proteins were not degraded by protease treatment even after the addition of detergent. This indicates that a portion of the protein is aggregated, and considering the high fraction of protoplasts with fluorescence via MGD1-GFPS1–10 expression, these aggregates are likely present in the IMS (Figure 4D). S11-OEP24(1–6) was largely extracted by both carbonate and detergent treatment (Figure 4C, third panel: EX, DO) and was fully degraded by protease treatment after, but not before, the disruption of the membrane (Figure 4C, TH). Thus, S11-OEP24(1–6) is likely trapped in the IMS in a rather soluble state (Figure 4D).
Finally, S11-OEP21(1–6) remained in the pellet fraction after carbonate or detergent treatment (Figure 4C, last panel: EX, DO) but was fully degraded by protease treatment even before lysis of the membranes (Figure 4C, last panel: TH/-). The homogeneous distribution of the GFP fluorescence from MGD1-GFPS1–10 surrounding the chloroplasts, and the formation of extrusions suggest that the majority of the protein are present in the OM, most likely as translocation intermediates (Figure 4D). We conclude that the N-terminal section of plastidic β-barrel proteins contains information required for the proper translocation across the membrane, or at least engagement with the translocon.
The C-terminal regions of β-barrel proteins affect membrane insertion
The signal for membrane insertion of mitochondrial β-barrel proteins (Kutik et al., 2008; Jores et al., 2016) and the discrimination signal for translocation into mitochondria or chloroplasts (Klinger et al., 2019) reside in the C-terminal β-hairpin of plant β-barrel proteins. Therefore, the impact of the C-terminal region of β-barrel proteins on translocation and membrane insertion was analyzed using proteins with N-terminal truncations: S11-OEP37(9–18) (representing the last 10 predicted β-strands of the protein), S11-OEP24(7–14), S11-OEP24(9–14), and S11-OEP21(5–10). As before, these proteins represent two forms, which contain a number of strands sufficient to form a monomeric β-barrel, while S11-OEP24(9–14) and S11-OEP21(5–10) proteins form either multimeric or no β-barrel structure.
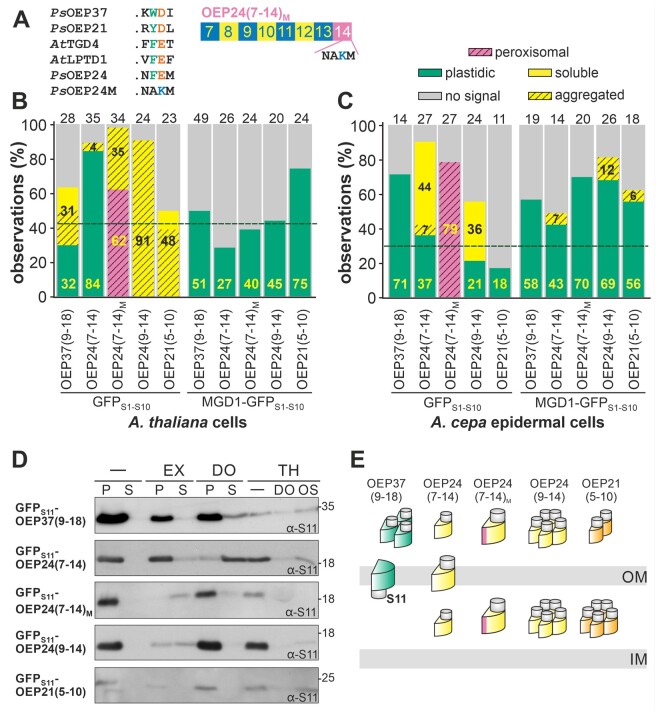
Furthermore, a comparison of known plastidic β-barrel proteins revealed that the penultimate (second from last) amino acid of the last β-strand is acidic and that the antepenultimate (third from last) amino acid is aromatic, while the most C-terminal amino acid is variable (Figure 5A). Therefore, a mutant of S11-OEP24(7–14) was engineered in which the PheGluMet (FEM) motif was replaced by AlaLysMet [AKM, GFPS11-OEP24(7–14)M; Figure 5A].
Figure 5.
In vivo localization of the C-terminal fragments of plastidic β-barrel proteins. A, The last four amino acids of the five plastidic β-barrel proteins as well as the mutant are shown for comparison. Bottom: schematic representation of OEP24(7-14)M, which contains a mutation in the last β-strand. B and C The distribution of the GFP fluorescence in protoplasts (B) or epidermal cells (C) for the constructs indicated is shown as described in the legend of Figure 2A and according to the legend given above. To analyze peroxisomal distribution, mCherry-SKL was co-expressed with these constructs. D, The indicated proteins were expressed in A. thaliana protoplasts together with GFPS1–10 and processed as in Figure 2B. E, Schematic representation of the localization of the tested proteins based on the observations presented.
Quantification of protein localization in protoplasts revealed a signal distribution of S11-OEP37(9–18), S11-OEP24(7–14), and S11-OEP21(5–10) comparable to that of full-length proteins (Figure 2A versus 5B; Supplemental Figure 9). In contrast, co-expression of S11-OEP24(9–14) with GFPS1–10 did not result in a chloroplast-localized GFP signal, and co-expression with MGD1-GFPS1–10 did not yield GFP fluorescence in general (Figure 5B). Furthermore, co-expression of S11-OEP24(7–14) with GFPS1–10 yielded a shape of the GFP fluorescence pattern within cells that was comparable to that of the full-length proteins. This was not observed when S11-OEP24(9–14), S11-OEP21(5–10), or S11-OEP37(9–18) were co-expressed with MGD1-GFPS1–10 (Supplemental Figure 8).
Co-expression of S11-OEP24(7–14)M with GFPS1–10 resulted in punctuated GFP fluorescence signals in the cytoplasm of most cells (Figure 5B;Supplemental Figure 10). Overlay with peroxisomal-targeted mCherry signals revealed that most of these structures represent a peroxisomal localization. In line with this notion, most protoplasts expressing S11-OEP24(7–14)M and MGD1-GFPS1–10 did not show GFP fluorescence (Figure 5B;Supplemental Figure 10).
In epidermal cells, the signal distribution of S11-OEP37(9–18) was comparable to that of the full-length protein (Figure 5C;Supplemental Figure 9). For S11-OEP24(7–14), a lower frequency of cells with chloroplast-localized GFP signal was generally found irrespective of the reporter (Figure 5C) compared to the full-length protein. In turn, for S11-OEP24(9–14) and S11-OEP21(5–10), the signal with the cytosolic reporter was largely absent, while pronounced fluorescence was obtained with the IMS reporter (Figure 5C). The distribution of S11-OEP24(7–14)M in epidermal cells was by and large comparable to that in protoplasts; only the fraction of cells with plastidic GFP fluorescence while co-expressing MGD1-GFPS1-10 increased (Figure 5, B and C).
Analysis of the membrane insertion of the proteins revealed that S11-OEP24(7–14)M was fully extracted by carbonate treatment, while the other four proteins were partially or fully resistant to this treatment (Figure 5D, EX). Moreover, S11-OEP24(7–14) was largely solubilized by detergent treatment, and S11-OEP37(9–18) was partially solubilized by this treatment, while the other three proteins remained insoluble. This is indicative of the strong aggregation of all proteins except S11-OEP24(7–14) and S11-OEP37(9–18). In line with an at least partial accumulation in the IMS, all proteins except S11-OEP37(9–18) were partially resistant against protease treatment, but all proteins became protease sensitive after the addition of detergent (Figure 5D, TH/- and DO).
In summary, the results suggest that S11-OEP37(9–18) and S11-OEP24(7–14) are at least partially inserted into the membrane, while S11-OEP24(9–14) and S11-OEP21(5–10) are likely aggregated in the IMS (Figure 5E). In contrast, S11-OEP24(7–14)M was largely imported into peroxisomes. A small portion of this protein accumulated in the IMS as well, while the mutation obviously abolished membrane insertion. Thus, the C-terminal fragments of the β-barrel proteins are translocated across the outer membrane, but with lower efficiency than the wild-type proteins. Moreover, this domain contains information in the last eight β-strands for membrane insertion, and the extreme C-terminus appears to be important for efficient membrane insertion.
The N-terminal domain of plastidic β-barrel proteins is required for efficient translocation in vitro
The efficient translocation across the membrane of truncated proteins representing the N-terminal β-strands (Figure 4) and the lower translocation efficiency of the truncated proteins representing the C-terminal β-strands of plastidic β-barrel proteins (Figure 5) in the protoplast system was challenged by in vitro import experiments. The four variants OEP24, OEP24(1–8), OEP24(1–6), and OEP24(7–14) were in vitro translated (Figure 6A, TP) and incubated with isolated chloroplasts. Following re-isolation of the organelles, treatment with iso-osmotic buffer, and subsequent centrifugation, the resulting pellet and supernatant fractions were precipitated and analyzed by SDS–PAGE (lanes -/P and S). All four proteins were present in the pellet. To evaluate the fraction of membrane-inserted or aggregated protein, the organelles were treated with sodium carbonate or Triton X-100 as described above. The majority of OEP24 was resistant to extraction but solubilized by Triton X-100 (Figure 6, EX, TX/P and S); these results are consistent with respect to the membrane insertion of this protein. In line with the efficient insertion of OEP24, the protein was protease resistant even after osmolysis (lanes TH/- and OS), but became protease sensitive after treatment of the membrane with detergent (lanes TH/TX). These results parallel the observations for S11-OEP24 (Figure 2C).
Figure 6.
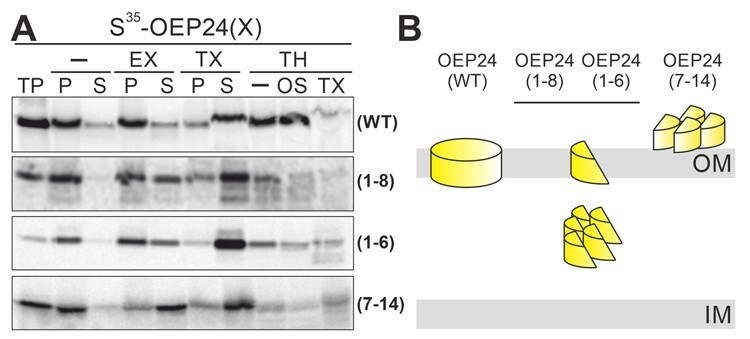
In vitro import of PsOEP24 variants in isolated chloroplasts from P. sativum. Isolated chloroplasts were incubated for 30 min at 25°C with 35S-labeled proteins that had been translated in rabbit reticulocyte lysate (proteins indicated on the right). Organelles were re-isolated and treated with different reagents to evaluate the suborganellar localization of imported proteins. Samples were loaded on SDS–PAGE and transferred to a PVDF membrane, and the autoradiogram visualized. TP: translated protein, —: isosmotic buffer, EX: extraction with 100 mM carbonate, TX: solubilization with 1.5% TX, TH: 200 µg/mL thermolysin, OS: osmolysis, P: pellet and S: supernatant after centrifugation. B, Schematic representation of the localization of the tested proteins based on the observations presented.
Only ∼50% of OEP24(1–8) and OEP24(1–6) were resistant to carbonate extraction, while OEP24(7–14) was mostly extricated by carbonate extraction (Figure 6A, EX/P and S). All three proteins became largely soluble by detergent treatment (lanes TX/P and S). In addition, OEP24(1–8) was partially sensitive to protease and was largely degraded after osmolysis (lanes TH/- and O S), while OEP24(7–14) was mainly protease sensitive before lysis (lanes TH/-). In contrast, OEP24(1–6) was barely sensitive to protease and became partially degraded after osmolysis or detergent treatment (lanes TH/- and OS and DO). These results suggest that the majority of OEP24(1–8) and OEP24 (1–6) are translocated across the membrane, while only a minor fraction is inserted into the membrane (Figure 6B). Moreover, the majority of OEP24(7–14) is not even translocated across the membrane (Figure 6B). This supports the notion that the N-terminal strands contain the translocation signal (Figure 4), while the C-terminal strands are required for membrane insertion (Figure 5), also in vitro. However, in vivo, the extreme C-terminus does not have to be exposed, and the conserved motif in the last strand is not essential for the insertion process of the full-length protein (Figures 1–3).
OEP37 inserts into the membrane by forming intermediate complexes
Having established constraints for import and insertion, we aimed to obtain some insights into the translocation and insertion process. The complex formation of rabbit reticulocyte lysate-translated S35-labeled β-barrel proteins in chloroplasts was monitored under in vitro import conditions by histidine-deoxycholate-native (HDN) –PAGE (Ladig et al., 2011). Loading was controlled by Coomassie staining of the HDN–PAGE (Figure 7A, top-right), analysis of the import by SDS–PAGE (Figure 7a, bottom-left), and if required, immunodecoration of the fraction with antibodies to confirm the organelle purity (Figure 7a, bottom-right).
Figure 7.
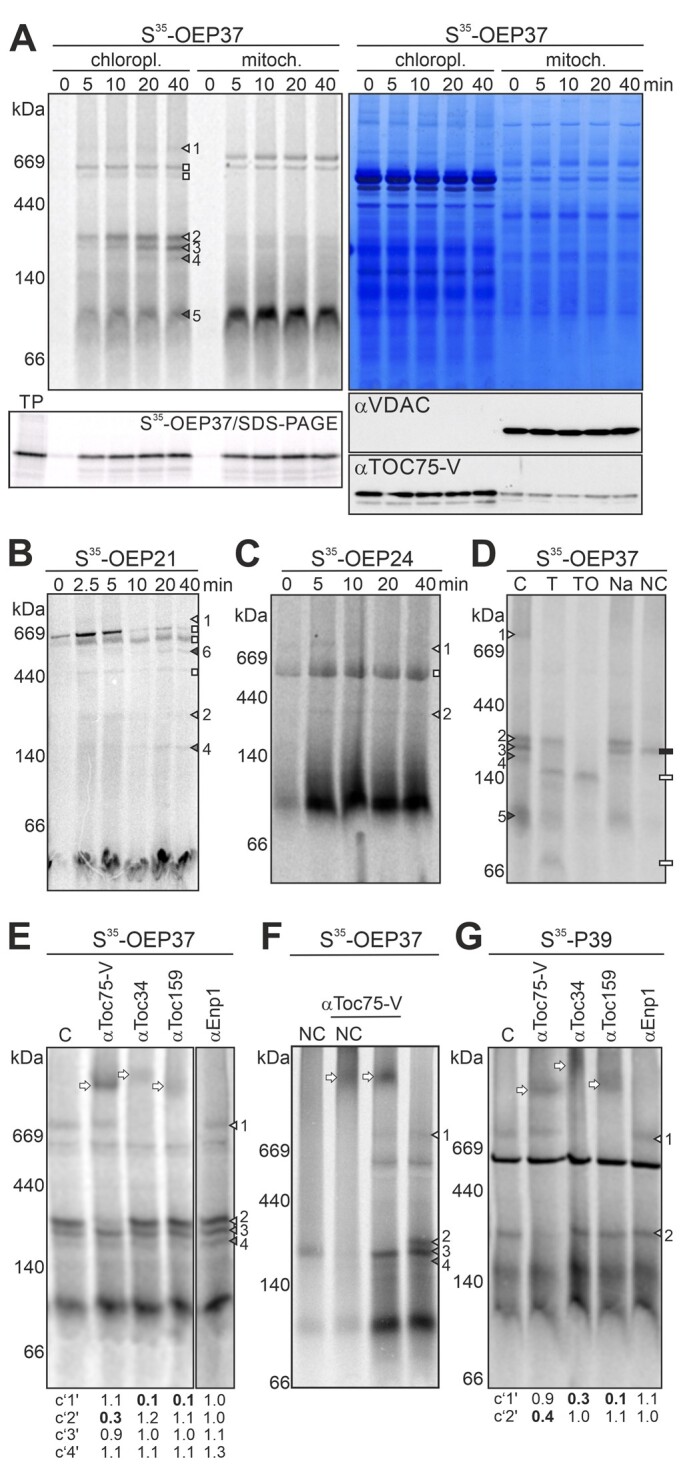
Translocation intermediates during the membrane insertion of OEP37. A, S35-OEP37 was incubated with chloroplasts or mitochondria under in vitro import conditions. Organelles (20 µg chlorophyll; 50 µg mitochondrial protein) were re-purified after the indicated times, solubilized, and subjected to HDN–PAGE. Radioactive signals were visualized by phosphor-imaging (left) and proteins by Coomassie Blue staining (right). The complexes of OEP37 in chloroplasts are indicated. 15% of the samples were subjected to SDS–PAGE, and radioactive signals were visualized (bottom, left; TP = 1.5% of used translation product). 15% of the samples were used for immunoblotting and immunodecoration with antibodies against mitochondrial VDAC1 and chloroplast TOC75-V (bottom right). B, C, Chloroplasts were incubated with S35-OEP21 (B) or S35-OEP24 (C) at 25°C for the indicated times, re-purified, solubilized, and subjected to HDN–PAGE. Complexes formed are marked according to the complexes of OEP37. Complex “6” is unique for OEP21. D, S35-OEP37 was incubated with chloroplasts (20 min, 25°C). Re-purified chloroplasts were either untreated (C) or treated with TH under isosmotic (T) or osmotic conditions (TO), with 1 M NaCl (Na) or 100 mM Na2CO3 (NC), followed by harvesting, solubilization, and HDN–PAGE analysis (Supplemental Figure 14) as in A. White rectangles: degradation products of OEP37. Black rectangles: carbonate-resistant band of OEP37. E Chloroplasts incubated with S35-OEP37 (15 min, 25°C) were solubilized and remained untreated (C) or were incubated with the indicated antibodies. Samples were subjected to HDN–PAGE, and radioactivity was visualized. Bands resulting from antibody addition are labeled by arrows. F, S35-OEP37 was incubated with chloroplasts (20 min, 25°C). After re-purification, organelles remained untreated (right) or were treated with 100 mM Na2CO3 (NC, left). After harvesting, organelles were solubilized (left and right lanes) and incubated with αToc75-V antibodies (middle lanes). The samples were subjected to HDN–PAGE analysis. G Chloroplasts incubated with S35-P39 (20 min, 25°C) were solubilized. Samples were loaded directly (C) or incubated with the indicated antibodies. Protein complexes were separated by HDN–PAGE analysis. Bands occurring at ∼700 kDa and at ∼230 kDa are marked as in A, arrows: bands occurring by antibody addition. In E and G, the intensity of the bands was quantified by ImageJ, and the values were normalized to the control. Drastic changes are highlighted in boldface.
The time- and temperature-dependent formation of at least five distinct complexes was observed for OEP37 (Figure 7A, triangles; Supplemental Figures 11 and 12; 4°C versus 25°C). Most of the observed bands are specific for chloroplasts, as they did not appear when OEP37 was incubated with mitochondria (Figure 7A). One of the five assigned complexes migrated at ∼700 kDa (Figure 7A, complex 1). Three complexes migrated between 180 and 230 kDa: These complexes are annotated as complexes 2, 3, and 4 from the largest to the smallest molecular weight. In addition, time-dependent chloroplast association of OEP37 was observed in the lower molecular weight region at both temperatures as well as in mitochondria (Figure 7A, complex 5; Supplemental Figure 11). Thus, at least the major amount of OEP37 in complex 5 is considered to be a membrane-bound translation product and is not discussed further. The formation of all complexes (1–4) is ATP dependent, as their occurrence was drastically reduced when the import assays were performed in the absence of ATP. The intermediate formation could be rescued by the addition of 3 mM ATP to the import reaction (Supplemental Figure 13).
S35-OEP21 (Figure 7B) and S35-OEP24 (Figure 7C) formed at least complexes migrating at similar molecular weights to complexes 1 and 2 observed for OEP37 (Figure 7A), although at lower intensity than found for OEP37. In addition, two (OEP21) or one (OEP24) radioactively labeled band already occurred after 0 min of incubation, which is thus likely unspecific. Moreover, while importing OEP21, an additional band at 500 kDa was observed, which was annotated as complex 6 (Figure 7B;Supplemental Figure 12).
To further analyze the nature of these complexes, we concentrated on OEP37 as an example. The localization and membrane insertion of the observed S35-OEP37 import intermediates was probed by treating chloroplasts with TH, sodium carbonate, and high levels of salt. In parallel, the same treatments were performed following import of the precursor of the small subunit of RubisCO (pSSU), which assembles into soluble complexes. Treating chloroplasts with externally active TH did not influence RubisCO (Supplemental Figure 14), whereas OEP37 in complexes 1, 3, and 4 was degraded by this treatment (Figure 7D, T). Additionally, TH treatment of chloroplasts after incubation with OEP37 resulted in the appearance of two novel bands that migrated slightly above 140 and 66 kDa, respectively (Figure 7D, T, squares). The nature of OEP37 occurring after protease treatment was confirmed by 2D-gel analysis, showing that both newly occurring bands represent the degradation products of OEP37 (Supplemental Figure 14). Thus, OEP37 in complexes 1, 3, and 4 is at least partially exposed to the cytosol. This is also the case for a large portion of OEP37 in complex 5, which is consistent with the above-mentioned hypothesis that this mainly represents the surface-bound translation product. Protease treatment of lysed chloroplasts resulted in the degradation of RubisCO, confirming the lysis and protease activity (Supplemental Figure 14), as well as the degradation of full-length OEP37 in all complexes (Figure 7D, TO; Supplemental Figure 14).
The membrane integration of the different OEP37 intermediates was probed by treating chloroplasts with 1 M NaCl (Figure 7D, Na). The addition of NaCl did not affect the abundance of RubisCO or OEP37-containing complexes, except complex 1 (Figure 7D, Na; Supplemental Figure 14). Treatment with sodium carbonate drastically reduced the content of RubisCO, confirming the removal of soluble or membrane-associated proteins (Supplemental Figure 14). This treatment resulted in the removal of OEP37 from all complexes (1–4), except for one migrating between complexes 2 and 3 (Figure 7D, NC, black square).
Plastidic β-barrel proteins interact with the TOC complex and with TOC75-V
To probe for the possible presence of TOC components or TOC75-V in the complexes identified while importing OEP37, we performed an antibody shift assay (e.g. Truscott et al., 2002). Chloroplasts were incubated with S35-OEP37 under import conditions; solubilized; and purified antibodies against TOC components or TOC75-V were added (Figure 7E;Supplemental Figure 13). After the addition of antibodies against TOC34 or TOC159, the high molecular weight band assigned as complex 1 was shifted (Figure 7E, complex 1, quantification). Moreover, antibody-induced shifts were not observed after incubating translated OEP37 with any of the antibodies in the absence of chloroplasts (Supplemental Figure 15). The addition of antibodies against TOC75-V resulted in a shift of complex 2 formed by OEP37 (Figure 7E, complex 2, quantification). The specificity of the shift was confirmed using antibodies against the nuclear protein ENP1 (Figure 7E; αENP1), which is not related to protein translocation into chloroplasts (Missbach et al., 2013). Competition by pre-incubating the antibodies with overexpressed TOC75-V POTRA or solubilized outer envelope vesicles (OEVs) confirmed the specificity of the antibody shift (Supplemental Figure 15). These results suggest that OEP37 engages the TOC complex (complex 1) as well as TOC75-V (complex 2) during import.
To explore whether the complex shifted by antibodies against TOC75-V is the membrane integral complex, the antibody shift of untreated and carbonate-treated organelles was compared (Figure 7F). Indeed, a clear shift of the carbonate-resistant band was detectable after the addition of αTOC75-V (Figure 7F, NC, αTOC75-V). This suggests that OEP37 in the TOC75-V-containing complex is at least partially inserted in the membrane. The kinetics of the occurrence of the carbonate-resistant band further supports its assignment as complex 2, although it migrates at a somewhat smaller molecular weight (Figure 6F). Similar to complex 2, the resistant band already reached its maximum after 10–20 min (Supplemental Figure 16).
To generalize the observation obtained for the porin-type β-barrel proteins OEP21, OEP24, and OEP37, the β-barrel outer membrane protein P39 of the OMP85 family protein without a transit peptide (Hsueh et al., 2017b, 2018) was incubated with chloroplasts and the resulting intermediates were analyzed by HDN–PAGE. P39 formed one intermediate complex at ∼700 kDa (Figure 7G, white triangle) and one at ∼230 kDa (Figure 7G, dark gray triangle). The 700-kDa complex was shifted by adding antibodies against the TOC components TOC34 and TOC159 (Figure 7G; αTOC34; αTOC159). As observed for OEP37, P39 in this complex was protease sensitive and extractable by salt treatment (Supplemental Figure 15). These observations indicate that P39 is TOC dependent as well and the intermediate assigned as complex 1.
Furthermore, incubation with αTOC75-V (Figure 7G; αTOC75-V), but not with αENP1 (Figure 7G; αEnp1), resulted in a size-shift of imported P39 migrating at 230 kDa. Similar to OEP37, P39 of this complex was protected from protease and resistant to salt or carbonate treatment, but became protease sensitive after osmolysis (Supplemental Figure 15). The addition of αTOC75-V antibodies to chloroplasts after import of the radioactively labeled α-helical outer membrane protein TOC34 did not result in a change in the migration behavior of the observed bands (Supplemental Figure 15). Therefore, our results suggest that β-barrel proteins are substrates for TOC75-V.
OEP37 is translocated across the outer membrane by TOC
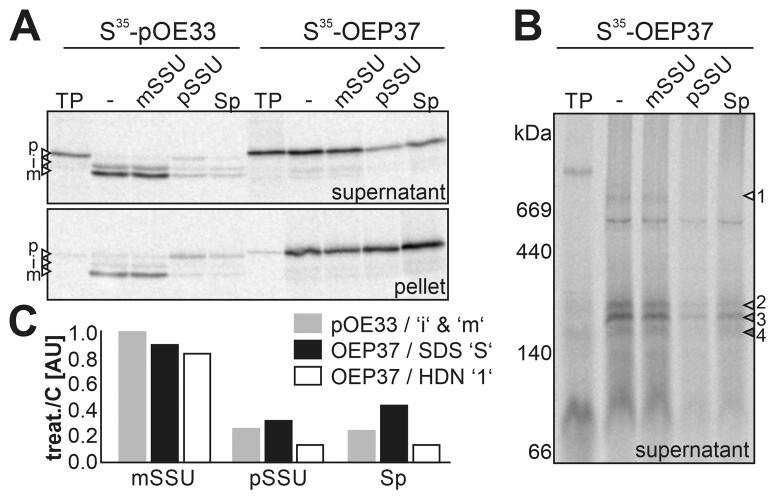
To confirm the existence of the suggested IMS-localized translocation intermediate and its dependence on TOC activity, we conducted in vitro import of OEP37 into isolated chloroplasts in the presence of inhibitors. Excess amounts of substrates of the TOC complex compete for translocation of in vitro translated precursor proteins (Qbadou et al., 2003). The presence of excess amounts of the precursor (pSSU), but not the mature form (mSSU) of the small subunit of RubisCO (Supplemental Figure 17), considerably reduced the import of the precursor of the thylakoid-localized oxygen evolving a complex subunit of 33 kDa (pOE33) and OEP37 (Figure 8A, “- “versus pSSU or mSSU). Furthermore, the addition of spermine (Qbadou et al., 2003) led to a reduction in the import of both proteins (Figure 8A, “- “versus Sp). Analysis of the complexes formed by OEP37 during translocation by HDN–PAGE revealed a drastic reduction of the high molecular weight band (Figure 8B, white triangle, complex “1”). Quantification of the import efficiency revealed a similar sensitivity of pOE33 and OEP37 import to the inhibitors (Figure 8C). These results confirm the notion that the translocation of β-barrel proteins is dependent on the TOC complex.
Figure 8.
OEP37 translocation is affected by inhibitors of the TOC complex. A, S35-pOE33 or S35-OEP37 (TP) was incubated with untreated chloroplasts (-) or chloroplasts pre-treated with 2 µM mSSU, pSSU (Supplemental Figure 17) or 10 mM spermine (Sp) for 30 min. After import, the chloroplasts were washed, solubilized by 1% digitonin, and fractionated into the soluble fraction (top) and pellet (bottom). 15% of the samples were loaded onto SDS–PAGE, and the autoradiogram is shown. The migration of the precursor (p), intermediate (i), and mature (m) form of OE33 is indicated by arrowheads. B, 85% of the samples of solubilized proteins prepared as in A were subjected to HDN–PAGE. The radioactivity was visualized by phosphor imaging. The labeling of the bands by arrows is the same as in Figure 7A. C, Quantification of S35-pOE33 (gray) and S35-OEP37 signal (black) in the soluble fraction after import (in A: SDS gel, soluble fraction) and of the band of S35-OEP37 marked as complex “1” (in B; white; HDN–PAGE, complex 1) in the presence of the indicated additive (treatment: treat) normalized to the sample without treatment (control: C) is shown.
TOC75-V is involved in the membrane insertion path of OEP37
To further confirm the intermediate complex formation between TOC75-V and OEP37, we performed pull-down experiments using untagged and Twin-Strep-tagged OEP37 (Figure 9A). The protein was imported and assembled the same complexes as OEP37, although an N-terminal tag was present (Supplemental Figure 17). After solubilizing the chloroplasts, PsTOC75-V was co-purified with Twin-Strep-OEP37 (Figure 9A, last lane, bottom panel, gray arrowhead). In contrast, PsTOC75-V was not precipitated when the experiment was conducted with untagged OEP37 (Figure 9A, fourth lane, bottom panel). These results are consistent with the involvement of TOC75-V in β-barrel protein insertion into the outer membrane of chloroplasts.
Figure 9.
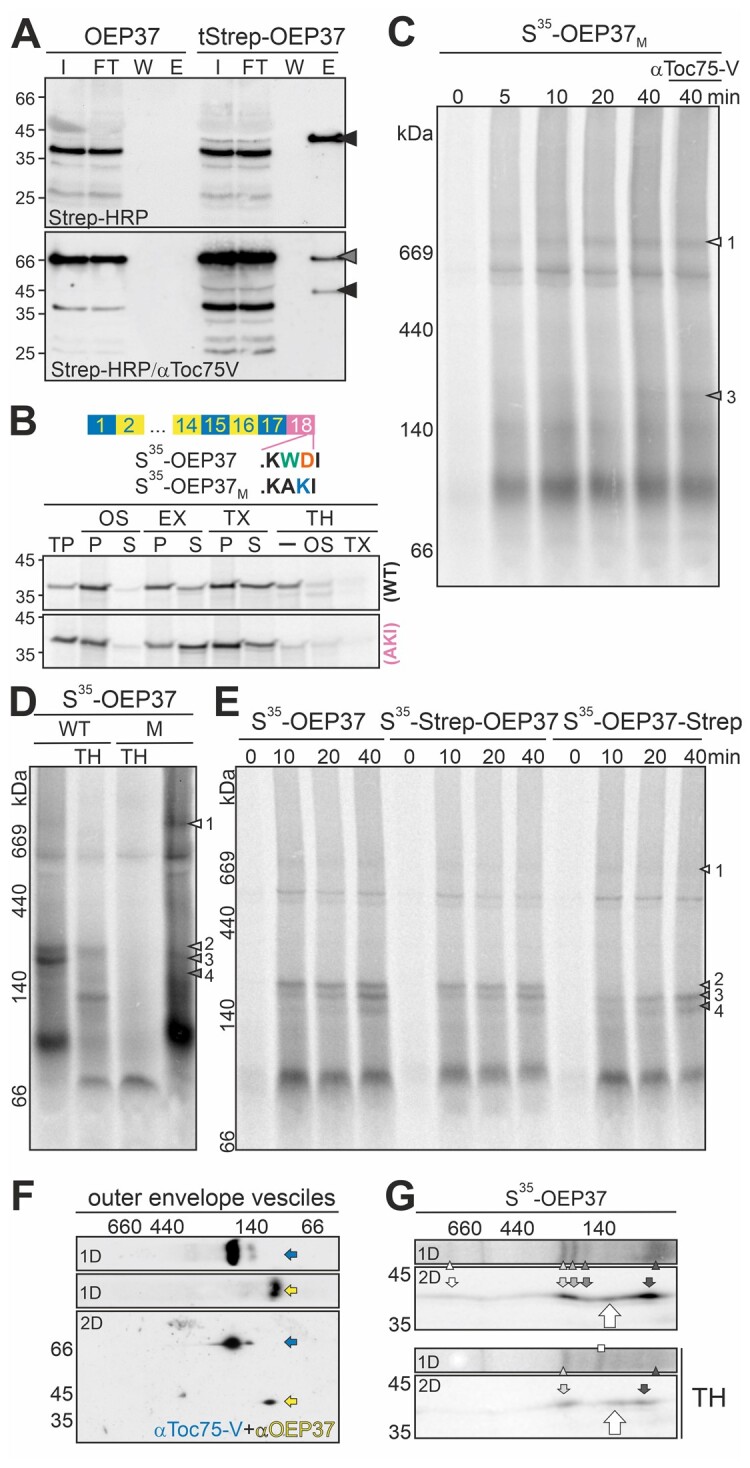
OEP37 interacts with TOC75-V prior to insertion into the membrane. A, OEP37 and Twin-Strep-OEP37 were incubated with chloroplasts. The chloroplasts were solubilized and bound to MagStrep XT beads. Input prior to binding (I: 1%), flow through (FT: 1%), last wash (W: 10%), and elution (E: 100%) were subjected to SDS-PAGE followed by immunoblotting. The membrane was decorated with Strep-Tactin-conjugated HRP (Strep-HRP, top), and subsequently with αToc75-V antibodies (bottom). Signals of Twin-Strep-OEP37 and TOC75-V are indicated by arrows. B, Chloroplasts were incubated with S35-OEP37 or 35S-OEP37M (AKI; indicated on the right; 30 min, 25°C). Organelles were isolated, treated as indicated, loaded on SDS–PAGE, and the autoradiogram was visualized. TP: translated protein, EX: extraction with 100 mM carbonate, TX: solubilization with 1.5% TX, TH: 200 µg/mL thermolysin, OS: osmolysis, P: pellet and S: supernatant after centrifugation. C, S35-OEP37M was incubated with chloroplasts. After the indicated times, the organelles were re-purified and solubilized. In the last lane, αToc75-V antibodies were added. Samples were subjected to HDN–PAGE, and the radioactive signal was visualized by phosphor-imaging. D, S35-OEP37 (WT, left) or S35-OEP37M (M, right) was incubated with chloroplasts. After harvesting, the chloroplasts remained untreated (lanes left and right) or were incubated with TH under iso-osmotic conditions (TH). The samples were solubilized and subjected to HDN–PAGE analysis. The main complexes of OEP37M are indicated as in Figure 7A. E, S35-OEP37, S35-Strep-OEP37, and S35-OEP37-Strep were incubated for the indicated times with chloroplasts (25°C), and HDN-PAGE was performed as in Figure 7A. F, HDN–PAGE-resolved envelope complexes (1D) were separated by SDS–PAGE (2D). First (1D) and second dimension (2D) were immunodecorated with αToc75-V (upper and lower panel, blue) and subsequently with αOEP37 antibodies (middle and lower panel, yellow). G, Chloroplasts were incubated with S35-OEP37, re-purified, and remained untreated (top) or were treated with TH under iso-osmotic conditions (bottom; TH). After solubilization, the complexes were separated by HDN–PAGE, followed by 2D–SDS–PAGE. The radioactivity was visualized by phosphor imaging. The full gels are shown in Supplemental Figure 14.
Remarkably, the insertion of OEP24 into the membrane was drastically reduced by the mutation of the antepenultimate and penultimate amino acids in the last β-strand in the C-terminal construct (Figure 5). The low membrane insertion efficiency was due to both reduced translocation efficiency and the tendency of the translocated protein to aggregate in the IMS. Thus, a mutant of OEP37 was created that served as a probe for the complex formation of β-barrel proteins during translocation and membrane insertion containing the semi-conserved motif at its C-terminus similar to that of OEP24. In the mutant, the amino acids TrpAsp (Figure 5A) were substituted by AlaLys (Figure 9B). Following incubation with chloroplasts under in vitro import conditions, both S35-OEP37 and S35-OEP37M were co-purified with organelles (Figure 9B; -/P). However, compared to the 10% input, the wild-type OEP37 was more efficiently associated. Extraction of proteins not embedded in the membrane by the addition of sodium carbonate yielded a large fraction of wild-type protein in the pellet, while the majority of the mutant protein was found to be soluble (Figure 9B; EX/P and S). The addition of detergent yielded an equal distribution of OEP37 between the pellet and supernatant (Figure 9B; TX/P and S, upper panel), while the majority of the mutant was found in the pellet (Figure 9B; TX/P and S, lower panel). These results suggest that the mutant shows a larger tendency for aggregation than the wild-type protein. The wild-type OEP37 was largely resistant to TH treatment of intact chloroplasts (Figure 9B; TH/-, upper panel). In contrast, only a small portion of the mutant protein was resistant to protease treatment (Figure 9B; TH/-, lower panel versus TP). After detergent treatment, both proteins were largely degraded. Therefore, the mutation of the last putative β-strand of OEP37 strongly reduced the translocation as well as the membrane insertion of this protein, similar to the C-terminal mutation of OEP24 (Figure 5).
Thus, we searched for the import intermediates formed by the OEP37 mutant. Complex 1 accumulated in a time-dependent manner (Figure 9C;Supplemental Figures 12 and 17), while the formation of complexes 2, 3, and 4 was either abolished or drastically reduced, with only complex 3 being faintly detectable after 40 min of import (Figure 9C). The assignment of the observed band as complex 3 was based on its migration behavior compared to the wild-type OEP37 (Supplemental Figure 17), as well as the resistance against the addition of antibodies against TOC75-V (Figure 9C, the last lane). Moreover, in contrast to the TOC75-V and OEP37-containing complex, the complexes formed by the mutant of OEP37 were sensitive to protease treatment (Figure 9D). Thus, the mutation of the last β-strand inhibited a formation of complex 2, and although the association with the TOC complex was not compromised (Figure 9, C and D), the transfer to TOC75-V was inhibited. Remarkably, extension by two amino acids did not compromise the targeting or membrane insertion of the β-barrel proteins. Thus, we fused ten additional amino acids in the form of the Strep-tag and two amino acids as a spacer to both the N- and C-termini of OEP37. While S35-Strep-OEP37 formed the same intermediates as S35-OEP37 during import (Figure 9E;Supplemental Figure 12), the presence of the ten amino acids at the C-terminus inhibited the formation of an HDN–PAGE-detectable complex 2, but not complex 1 (Figure 9E). This further strengthens the notion that the semi-conserved motif must be present in proximity of the C-terminus.
Finally, to determine whether OEP37 is assembled into a native-like state while importing the protein into chloroplasts and after forming a membrane-inserted complex with TOC75-V, the migration of the endogenous protein was analyzed by 2D-HDN–PAGE. TOC75-V migrated at ∼200 kDa, while OEP37 migrated at ∼100 kDa (Figure 9F). Thus, none of the identified intermediates (1–4) represent the final inserted protein, nor does the mature OEP37 co-migrate with the membrane-associated translation product assigned as complex 5. However, analysis of the migration of OEP37 after import by 2D PAGE revealed a band migrating at a higher molecular weight than complex 5 and a lower molecular weight than complex 4 (Figure 9G, large arrow). Remarkably, this band was detectable after TH treatment as well (Figure 9G). Thus, it is tempting to speculate that OEP37 assembles into native complexes, although with low efficiency.
Discussion
The signals of plastidic β-barrel proteins for translocation and membrane insertion
How plastidic β-barrel proteins are translocated and inserted into the membrane is a puzzle. TOC75-III and TOC75-V contain a cleavable N-terminal signal (Tranel et al., 1995; Tranel and Keegstra, 1996; Day et al., 2019; Gross et al., 2020), which stands in contrast to the other OEPs and the other OMP85 family proteins. Thus, a distinct signal must exist for the latter. Moreover, OEP40 is an atypical β-barrel protein (based on topology prediction) with large N- and C-terminal soluble domains (Harsman et al., 2016). Furthermore, the proteins of the LptD family contain an N-terminal soluble domain of approximately 70 amino acids that is thought to be important for substrate binding (Hsueh et al., 2017a). A similar but short soluble N-terminal region is predicted for OEP37, while the plastidic β-barrel proteins OEP21 and OEP24 are thought to start with a β-strand (Supplemental Figure 1).
Using the porin-type OEP37, OEP24, and OEP21 as examples, we observed that the N-terminal β-strands contain information for translocation across the membrane. Based on the analysis of mutants of OEP24 and OEP21, we propose that the first six β-strands form a unit sufficient for translocation across the outer envelope membrane (Figures 4 and 6). The N-terminal signal is required for translocation across the membrane in vitro, as exemplified for OEP24 mutants without N-terminal β-strands (Figure 6). This requirement can be partially overruled in vivo, as a reduction but not a loss of translocation across the membrane was obtained using the C-terminal β-strands (Figures 5 and 6). This finding hints at the engagement of cytosolic targeting factors. Whether the cytosolic proteins involved in targeting are comparable to those found in mitochondria (Jores et al., 2018) remains to be investigated.
The N-terminal β-strands do not have to be positioned at the extreme N-terminus. Short N-terminal polypeptide strands are tolerated as naturally occurring for LptD or OEP37, or as enforced by the N-terminal fusion with GFPS11 or the Strep-tag (Figures 1–4 and 9). GFPS11 on its own does not serve as a targeting signal, because mitochondrial β-barrel proteins with a disturbed last β-sheet aggregated in the cytoplasm instead of being redirected to chloroplasts (Klinger et al., 2019). This is consistent with the existence of cleavable signals in the case of TOC75-III and TOC75-V (Tranel et al., 1995; Tranel and Keegstra, 1996; Day et al., 2019; Gross et al., 2020), which both contain large POTRA domains, while TOC75-IV and P39 do not contain POTRA domains and do not possess a cleavable signal (Baldwin et al., 2005; Hsueh et al., 2017b). Thus, we conclude that a signal for translocation exists in the six β-strand-containing N-terminal regions of proteins without a large N-terminal soluble domain (Figure 10, step a).
Figure 10.
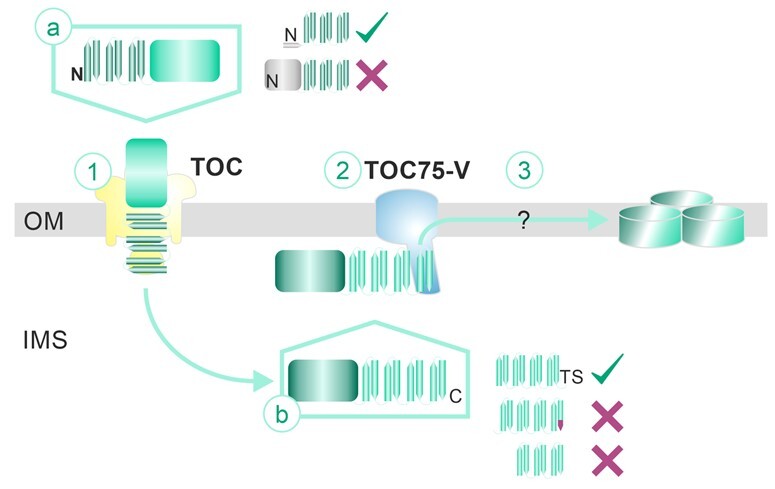
Model for the insertion of plastidic β-barrel proteins. The signal for translocation across the membrane is present in the five to six N-terminal β-strands. Short N-terminal extensions are tolerated, but large soluble domains inhibit translocation. Translocation across the membrane requires the action of the TOC complex. Within the intermembrane space, the plastidic β-barrel protein is transported to TOC75-V. The chaperones involved are yet unknown. The most C-terminal strand is required for insertion into the membrane and likely for the interaction with TOC75-V. Again, few additional amino acids are tolerated, while larger segments inhibit membrane insertion. Furthermore, mutation of a semi-conserved motif in the C-terminal region of the last strand leads to the disruption of this complex (indicated as orange tip). Through interaction with TOC75-V, the plastidic β-barrel protein becomes inserted into the membrane and assembles into complexes by a yet unknown mechanism. For further details, see the Discussion. (TS: Thr/Ser; N: N-terminus; C: C-terminus)
The signal for membrane insertion appears to be present in the C-terminal domain of plastidic β-barrel proteins (Figure 10, step b), which would be comparable to the mode of translocation and membrane insertion found for mitochondrial β-barrel proteins (Kutik et al., 2008; Jores et al., 2016). Consistent with the notion that the C-terminal motif functions in membrane insertion, the N-terminal fragments largely aggregated in the IMS [Figure 4; OEP37(1–10) and OEP24(1–8)]. Mutating the semi-conserved antepenultimate and penultimate amino acids in the last β-strand largely omitted the membrane insertion of this protein (Figure 5; OEP24(7–14)M; Figure 9; OEP37M). Moreover, it appears that this signal is required for the interaction with TOC75-V, as a mutation of the motif in the proposed last β-strand of OEP37 inhibited this interaction (Figure 9). Nevertheless, the motif does not have to end exactly at the penultimate amino acid of the last β-strand, as the addition of two hydroxylated amino acids (ThrSer) did not disturb the membrane insertion (Figures 1–3). In contrast, the addition of a C-terminal 10-amino acid Strep-tag inhibited the association with TOC75-V (Figure 9). Remarkably, this indicates that the amino acid motif required for the interaction is not restricted to the mutated antepenultimate and penultimate amino acids. The Strep-tag (WSHPQFEK) contains an aromatic amino acid at the antepenultimate position and an acidic amino acid at the penultimate position, but this sequence does not form a β-strand (Schmidt et al., 1996). Thus, while the motif is important in the context of the last β-strand of the analyzed proteins OEP37 and OEP24, additional structural properties are important as well, which could be the ability to form a β-strand. This notion is consistent with the finding that proteins of the TOC75 family do not contain the conserved motif at their C-termini (TOC75-III/TOC75-IV: GERY; TOC75-V: GLRN; P36/P39: ASST).
Moreover, only proteins consisting of eight β-strands are inserted into the membrane, as (for example) OEP24(9–14) or OEP21(5–10) remain largely soluble (Figure 5). It is tempting to speculate that this represents a structural constraint, as eight strands are considered to be the minimal unit for monomeric β-barrel proteins (e.g. Wimley, 2003; Mirus et al., 2010). Whether a proof-reading mechanism exists or whether the shorter proteins that are not properly assembled in the membrane are degraded remains to be further investigated.
We conclude that plastidic β-barrel proteins contain the translocation signal in their N-terminal regions, while the mitochondrial targeting of β-barrel proteins requires information in the C-terminal β-hairpin (Jores et al., 2016; Klinger et al., 2019). In turn, the signal for membrane insertion of plastidic β-barrel proteins is likely present in the C-terminal β-hairpin, which would be comparable to the mode of membrane insertion found for mitochondrial β-barrel proteins (Kutik et al., 2008; Jores et al., 2016).
The emerging import path of plastidic β-barrel proteins
Both the TOC complex and TOC75-V are involved in the insertion of β-barrel-type proteins into the outer envelope membrane of plastids, as exemplified here for the outer envelope proteins OEP37 and P39. This finding confirms the recently observed engagement of the TOC complex in this process (Day et al., 2019). The initial step involves the formation of a complex that migrates at ∼700 kDa in HDN–PAGE annotated as complex 1, which is shifted by antibodies against the TOC components TOC34 and TOC159 (Figure 7) and can be inhibited by the addition of TOC-specific inhibitors (Figure 8). A similar complex is formed by OEP24 and OEP21, although it is less pronounced. This lower abundance might be due to the reduced stability of the complex during isolation or a faster translocation of the substrate (Figure 7). At this stage, the protein is still exposed to the cytoplasm and has not yet integrated into the membrane (Figures 7 and 9). We conclude that the first step (Figure 10, step 1) of β-barrel insertion involves the translocation of proteins synthesized in the cytosol across the outer envelope membrane by the TOC complex in an ATP-dependent manner (Supplemental Figure 13, Day et al., 2019).
Subsequently, a complex annotated as complex 2 that migrates at ∼230 kDa in HDN–PAGE is formed by OEP37, OEP24, OEP21, or P39 (Figure 7). As exemplified for OEP37 or P39, the β-barrel protein in this complex is protease-protected, and the complex contains TOC75-V (Figure 7). The physical interaction between TOC75-V and OEP37 was confirmed by affinity-tag co-precipitation and was found to be dependent on the composition of the last β-strand (Figure 9). Thus, the observed dependence of OEP membrane insertion on the composition of the last β-strand likely represents the existence of a signal for the interaction with TOC75-V.
Within complex 2, OEP37 is already (partially) inserted into the membrane, as this protein is resistant to sodium carbonate treatment (Figure 7). However, the complex migrated after carbonate treatment at a somewhat smaller molecular weight, which suggests that an additional associated protein might be involved in this complex formation, a notion that needs to be addressed in the future. Nevertheless, we propose that as the second step of the translocation path, TOC75-V recognizes the IMS-localized form of the β-barrel protein, as exemplified for OEP37 and P39 (Figure 10, step 2). The participation of TOC75-V in the insertion path of β-barrel proteins is consistent with the previously proposed function of TOC75-V (Schleiff and Soll, 2005), as well as its close phylogenetic relationship with the bacterial OMP85 proteins (Moslavac et al., 2005; Bredemeier et al., 2007). It is further consistent with the essential function of TOC75-V for plant viability (Huang et al., 2011; Hsu et al., 2012).
After interacting with TOC75-V, OEP37 engages in a final membrane insertion stage (Figure 10, step 3). After import, a very small fraction of OEP37 migrated at a comparable molecular weight to the endogenous protein (Figure 9). However, whether this is the endogenous oligomeric state (which is not yet known) needs to be further confirmed. Nevertheless, even this complex is of low abundance and only becomes detectable by 2D analysis (Figures 7–9). Thus, the endogenous state is formed at low efficiency, perhaps due to the absence of additional proteins associated with the assembly machinery at the cytosolic surface, as observed for mitochondria (Schleiff and Becker, 2011). These proteins might have been lost during the isolation of chloroplasts. An alternative explanation might exist as well, and thus, the assembly of OEP37 into native complexes needs to be addressed in subsequent studies.
During the insertion of OEP37 and OEP21, but not OEP24, P39, or OEP37M, additional complexes were observed (Figures 7 and 9; complexes 3, 4, 6). However, for OEP37, we demonstrated that these complexes were protease- and carbonate-sensitive and thus likely exposed to the surface of the chloroplasts (Figure 7). These complexes did not contain TOC75-V or typical TOC subunits (Figure 7). However, the nature of these complexes remains unknown, and thus, these complexes need to be further analyzed in subsequent studies.
Taken together, although the formation of additional intermediate assemblies is still possible, the current and previous results suggest that the membrane insertion of plastidic β-barrel proteins without cleavable signal involves at least three events: (1) translocation across the outer envelope membrane by TOC; (2) recognition by TOC75-V in the IMS; and (3) the formation of a membrane embedded final assembly. Thus, our results allow us to propose a mechanistic model for plastid β-barrel protein insertion into the outer envelope membrane (Figure 10).
Materials and methods
Bioinformatics analysis
The annotation of the secondary structure elements, specifically the β-strands, was performed as described (Jores et al., 2016). In brief, for each sequence of the corresponding protein from A. thaliana (OEP21, OEP24, OEP37) and Pisum sativum (OEP21, OEP24, OEP37), two β-barrel predictions were carried out using the BOCTOPUS 2 (Hayat et al., 2012), and PredTMBB (Bagos et al., 2004) servers. In addition, a consensus secondary structure and disorder prediction was carried out using the GeneSilico Metaserver (Kurowski and Bujnicki, 2003). For each sequence, these predictions were combined into one meta-prediction. A position was considered to be within the membrane when it was predicted to be a transmembrane β-strand by both prediction servers or predicted to be located in a transmembrane β-strand by one server and its secondary structure was predicted not to be disordered and to be a β-strand. The predictions for one protein family were then combined. A multiple alignment by MAFFT (Katoh and Sandley, 2013) was used to determine homologous amino acid positions within each family. A position was considered to be in a transmembrane β-strand if (1) at least half of the meta-predictions assigned the amino acid to be within a β-strand according to the above stated criteria and (2) the multiple alignment showed the characteristic alternating hydrophobic pattern of β-strands.
Molecular cloning
Standard protocols for PCR, DNA restriction reactions, DNA ligation, transformation, bacterial growth, and plasmid mini- and midi-preparation via alkaline lysis were used (Green and Sambrook, 2012). cDNA was generated by reverse transcription using RevertAid Reverse Transcriptase (Thermo Scientific, Germany) according to the manufacturer’s recommendations. Gibson cloning (Gibson et al., 2009) was performed omitting the ligase step. Primers were designed using the NEB assembly online tool (nebuilder.neb.com). Oligonucleotides (Supplemental Table 1) were ordered from Sigma Aldrich (Munich, Germany). DNA sequencing was performed by GATC Biotech (Konstanz, Germany). Clone Manager 9 software (Sci-Ed software) was used for in silico cloning.
For cell transformation, pAVA derivatives (von Arnim et al., 1998) were used. Inserts were cloned via KpnI/SpeI restriction sites (Gross et al., 2011). Genes for plastidic β-barrel proteins originated from P. sativum cDNA (Supplemental Table 2). For in vitro transcription, pGem4, pSP65 (Promega, Madison, WI, USA), or pBat (Annweiler et al., 1991) derivatives were used (Supplemental Table 2). All constructs with shortened β-barrels ended directly at the end of the last included β-strand. Only OEP247–14 (pAVA-GFPS11-OEP247–14) and OEP249–14 (pAVA-GFPS11-OEP249–14) contained half the loop in front of β-strands 7 and 9, respectively. OEP37_AKI was generated by PCR using pGem4_OEP37 (primers: Supplemental Table 1, Ulrich et al., 2012). The product was cloned into pGem4 via SacI/PstI. AtOEP21, AtOEP24, and AtOEP37 were amplified from cDNA from 7-day-old wild-type Col-0 seedlings grown in a climate chamber (8-h light provided by LED (spectra provided upon request), 120 µmol m−2s−1, 22°C; 16-h dark, 20°C) and cloned with SpeI/XbaI (AtOEP21) and KpnI/XbaI into pAVA, which included an N-terminal GFPS11 tag.
OEP37 N- and C-Strep constructs were generated by amplifying OEP37 from pGem_OEP37 (Ulrich et al., 2012). The tag was introduced with the primer and the product cloned into pSP65 (NcoI/XbaI). The N-terminal Twin-Strep tag was introduced by primer annealing to pSP65 via NcoI/BamHI. OEP37 was inserted into the pSP65_N-Twin Strep vector (BamHI/XbaI). mSSU was amplified by PCR from pET21d_pSSU_his (Qbadou et al., 2006) and cloned into pET21d (NcoI/XhoI). The DNA encoding the POTRA domains of AtToc75-V was amplified and cloned into pET24c (pET24c_Toc75-V_P1-3, NdeI/NotI). The construct encoding the Toc75-V POTRA domains with N-terminal MBP-tag and C-terminal 6xHIS-tag (pMal_Toc75-V_P1-3_his) was generated by amplifying the coding region from pET24c_Toc75-V_P1-3 and cloning into pMal (NEB, G; BamHI/NotI). A construct of Toc75-V POTRA1-3 with an N-terminal 6xHIS- and MBP tag was generated using primers with BamHI and SalI restriction sites. The PCR product was inserted into a pMal vector. The MBP_75VP1-3 expression cassette was excised by digestion with NdeI/SalI and cloned into pCold1, resulting in an N-terminal His-tag followed by MBP.
C-terminal cysteine constructs (OEP21C178, OEP24C214, OEP24TS-C216, and OEP37C330) were generated by site-directed mutagenesis PCR (QuickChange mutagenesis kit; Stratagene, La Jolla, CA) on pSP65_OEP21, pGEM4Z_OEP24, pGEM4Z_OEP24TS, and pGEM4_OEP37. The cysteines were introduced as the last C-terminal amino acid before the stop codon. For further identification of mutants, a restriction site (OEP21 and OEP37: XhoI; OEP24 and OEP24TS: XbaI) was introduced.
Arabidopsis mesophyll protoplast isolation and transformation
Arabidopsis thaliana Col-0 wild-type plants were grown in a climate chamber (8-h light provided by LED, 120 µmol m−2s−1, 22°C; 16-h dark, 20°C) on Hawitta sowing or cutting soil for 7 weeks. Mesophyll protoplasts were isolated from leaves and transformed as in Gross et al., (2011) with the following modifications: 5–10 µg DNA per plasmid was used for transformation. To avoid disintegration of the vitamins, the K3 solution was prepared freshly every time and filter-sterilized through a 0.2-µm Supor filter membrane (PALL Life Sciences, Dreieich, Germany). To prevent the bacterial growth, 0.1 mg/mL ampicillin was added to the K3 solution. The plasmids were described in Gross et al., (2011), Machettira et al., (2011a), Sommer et al., (2011), Ulrich et al., (2012), Klinger et al., (2019) or listed in Supplemental Table 2. Immunochemical staining was performed as previously described (Mishra et al., 2002).
Particle bombardment of onion epidermal cells
The protocol for onion epidermal cell bombardment was adopted from Schmidt von Braun et al., (2007). Organic onions (A. cepa) were purchased at a local market. Biolistic transformation of A. cepa epidermal cell tissue was conducted with a PDS-1000/He™ Biolistic Particle Delivery System according to the manufacturer’s instructions (Bio-Rad, Hercules, CA, USA) with a particle traveling distance of 6 cm and a launch pressure of 75 bar (Bio-Rad, Germany). Gold particles 1 µm in diameter were coated with a total of 5-µg plasmid DNA under constant vortexing in low-binding Eppendorf tubes (Sarstedt, Nümbrecht, Germany). After bombardment, the onion epidermis was kept on MS plates in the dark for at least 14 ± 2 h.
Confocal laser scanning microscopy and quantification
Confocal laser scanning microscopy was performed on a Leica TCS SP5 or Zeiss LSM 780 microscope as described (Klinger et al., 2019). To analyze protoplasts, an HCX PL APO Lambda Blue 63 × 1.4 oil objective (SP5) or a Plan Apochromat 63 × 1.4 oil DIC M27 objective (LSM780) was used. For A. cepa epidermal cells, an HCX PL APO CS 20×/0.7 DRY objective (SP5) or a Plan Apochromat 20×/0.8 M27 objective (LSM 780) was used. To stain the mitochondria, MitoTracker™ Orange CMTMRos or MitoTracker™ Deep Red FM (Thermo Scientific™, Waltham, MA, USA) was added to the cells. Fluorescence was excited and detected as stated in Supplemental Table 3.
Images were processed with Leica LAS AF Lite (SP5) or Zeiss Zen 2.3 blue and black lite software (LSM 780). Transformed A. thaliana protoplasts were identified by exciting mCherry-fluorescence to detect the co-transformed pML94-mCherry-SKL, followed by GFP fluorescence detection. The same procedure was applied to A. cepa epidermal cells co-transformed with pML94-SSUTP-mCherry. For each combination of GFPS11-tagged protein and saGFP reporter, the observed GFP fluorescence signals were classified as “soluble,” “aggregated,” “plastidic,” and “no signal,” and their percentile distribution was calculated (Klinger et al., 2019).
Localization assay in A. thaliana mesophyll protoplasts
106A. thaliana protoplasts were isolated and co-transformed with plasmids encoding the GFPS11-tagged protein of interest and the cytosolic GFPS1–10 reporter protein (50 µg each). After 14 ± 2 h, the protoplasts were collected (100g, 5 min) and dissolved in 1 mL K3 buffer (20 mM MES/KOH pH 6, 0.4 M sorbitol, 1 mM CaCl2, 1× MS salts).
Four aliquots with 8×104 cells were retained, centrifuged (100g, 5 min), and dissolved in the respective treatment buffer. The aliquot was dissolved in 100 µL K3 buffer for the control, 100 µL 100 mM Na2CO3 for carbonate treatment, and 100 µL K3 buffer with 1.5% TX or 1.5% DOMA for Triton X-100 (TX) or dodecyl-maltoside treatment, respectively. The fourth aliquot was used for TH/osmolysis in 100 µL buffer containing 20 mM MES/KOH pH 6, 1 mM CaCl2, and 200 µg/mL TH.
TH treatment was performed using K3 buffer with 200 µg/mL TH, and TH/detergent treatment was performed using K3 buffer with 200 µg/mL TH and 1.5% detergent. The residual protoplasts were lysed on ice by pressing them three times through a cannula (0.45 × 25 mm) and gauze to rupture the protoplast membrane. In each case, 80 μL of the cell lysate was combined with TH solution to a total volume of 100 µL.
All treatments were performed for 30 min on ice, and TH activity was inhibited by the addition of 10 mM ethylenediaminetetraacetic acid (EDTA). Untreated, carbonate- and detergent-treated samples were centrifuged (100,000g, 30 min) to separate the pellet and supernatant. Supernatants were collected and pellets dissolved in 500 µL K3 buffer. All samples were precipitated by the addition of 15% (v/v) trichloroacetic acid followed by incubation for 1 hour on ice. Precipitated proteins were collected by centrifugation at 25,000g for 15 min. The pellets were washed with 1 mL 80% (v/v) acetone. After centrifugation at 25,000g for 15 min, the pellets were dried at 45°C for 10 min, dissolved in 20 µL 4× Schägger loading buffer (30% glycerol, 0.05% Coomassie Brilliant Blue, 12% SDS, 6% β-mercaptoethanol, 150 mM Tris–HCl pH 7), and dissolved under a constant shaking (1200 rpm) at 46°C for 10 min.
Organelle and envelope vesicle isolation
Pisum sativum “Arvica” seedlings were grown in the greenhouse on vermiculite for seven to eight days under native light shifts at 22°C during the day and 18°C at night. Chloroplasts and chloroplast OEVs were isolated as described (Schleiff et al., 2003b). Mitochondria were isolated from seedlings grown under the same conditions (Rudhe et al., 2002; Rödiger et al., 2010).
In vitro import into isolated Pisum sativum organelles
In vitro transcription (omitting the capping of the mRNA) and translation in reticulocyte lysate were performed according to Bionda and Schleiff (2010). In brief, plasmid DNA (Hemmler et al., 2006; Qbadou et al., 2006; Ulrich et al., 2012; Lumme et al. 2014; Hsueh et al., 2017b; Supplemental Table 2) was linearized by digestion with restriction enzymes cutting downstream of the 3′-end of the coding sequence. In vitro transcription was performed using SP6 polymerase (Thermo Scientific, Germany) according to the supplier’s recommendations. In vitro translations and radiolabeling were performed using the Flexi Rabbit Reticulocyte Lysate system (Promega, Germany) and an amino acid mixture lacking methionine (Promega, Germany) in the presence of S35 EasyTag™ EXPRESS35S Protein Labeling Mix (Perkin Elmer, Germany). To remove aggregates, translation products were centrifuged (300,000g, 10 min, 4°C) prior to use in experiments. For ATP depletion experiments, the translation products were treated with 0.05 U apyrase (10 min, 30°C).
In vitro import experiments were performed according to Rudhe et al., (2002), Bionda and Schleiff (2010), and Ruprecht et al., (2010) for single chloroplast imports. In short, chloroplasts (20 µg chlorophyll per import reaction) were isolated and mixed with 5 or 8% (v/v) 35S radioactively labeled proteins in a 100-µL import reaction containing 3 mM ATP. If not otherwise indicated, import reactions were performed at 25°C. For SDS-PAGE analyses of post-import treated OEP24, its variants, OEP37 and OEP37M, the reactions were performed for 30 min. For HDN-PAGE analyses, import times are indicated in the figure legends. Chloroplasts were recovered by centrifugation over a 40% Percoll cushion (5,200g, 5 min, pellet). After washing the isolated organelles thrice, post-import treatments were performed as described below or the organelles were solubilized for HDN–PAGE.
For import experiments under ATP depletion, chloroplasts were incubated with 5 µM nigericin and 5 µM CCCP (30 min, ice, dark) prior to import. For import competition, pSSU_his and mSSU_his were subjected to buffer exchange via Amicon (3 kDa cutoff, Merck) to 50 mM HEPES/KOH pH 7.6 and 6 M urea. Import samples were supplemented with 2 µM mSSU, 2 µM pSSU, or 10 mM spermine. The concentration of urea in the import reaction was adjusted to 400 mM (Ruprecht et al., 2010). For comparability, all other samples of the competition experiment were supplemented with 400 mM urea (final).
Post-import treatments
Intact chloroplasts after import were osmotically lysed by the addition of 50 mM HEPES/KOH pH 7.6, while sodium chloride, sodium carbonate, and TX treatments were performed by the addition of 1M NaCl, 0.1 M Na2CO3, or 50 mM HEPES/KOH pH 7.6 containing the indicated amounts of Triton X-100, respectively. Samples were incubated in the dark (30 min, ice) and subsequently centrifuged (100,000g, 30 min, 4°C). For TH treatment, chloroplasts equal to 20 µg chlorophyll were resuspended in 100 µl of 50 mM HEPES/KOH pH 7.6, 330 mM sorbitol, 1 mM CaCl2, and TH (Calbiochem, Germany) at a final concentration of 120 or 200 µg/mL (30 min, ice, dark), followed by the addition of 10 mM EDTA. Organelles were either harvested and washed with 50 mM HEPES/KOH pH 7.6 and 330 mM sorbitol or re-purified by centrifuging through a 40% Percoll (50 mM HEPES/KOH pH 7.6, 330 mM sorbitol) cushion and subsequently washed. TH treatment under hypo-osmotic conditions was performed as above in 50 mM HEPES/KOH pH 7.6 and collected by centrifugation (100,000g, 30 min, 4°C) or directly supplemented with SDS loading buffer and 10 mM EDTA (final).
Post-import PEGylation assay
Intact chloroplasts were PEGylated after import as described (Gross et al., 2020) with the following modifications: Prior to PEGylation, the third wash step after import was performed with 1 M NaCl followed by a fourth washing step with 100 mM Tris/HCl pH 7, 330 mM sorbitol, and 1 mM EDTA. For PEGylation, the buffer was supplemented with 10 mM PEG-Mal (20 min, ice, dark). PEGylation during SDS-based solubilization was performed in the buffer with PEG-Mal and additional 1% SDS (20 min, ice, dark). PEGylation was quenched by adding 100 mM DTT (final) and incubating for 5 min on ice (dark). Intact chloroplasts were harvested (1,500g, 1 min) and resuspended in SDS loading buffer. SDS loading buffer was directly added to SDS-solubilized samples. SDS-solubilized OEP21 samples were resuspended in loading buffer without SDS. For OEP24, OEP24ts, and OEP37, acetone precipitation was used to remove the excess noncrosslinked PEG-Mal (Fic et al., 2010). Samples (corresponding to 10 µg chlorophyll) were subjected to SDS–PAGE.
Purification of recombinant proteins and generation of antibodies
6xHIS-tagged pSSU (Qbadou et al., 2006), mSSU, and AtToc75-V amino acids 154–396 (pET24c_Toc75-V_P1-3; Supplemental Table 2) were produced in E. coli following the addition of 1 mM IPTG and incubation for 3 h at 37°C. For AtToc75-V_P1-3, cells were resuspended in 50 mM Tris–HCl pH 8. The cells were lysed in a French press and centrifuged, and the pellets were washed once with wash buffer (50 mM Tris–HCl pH 8, 1 M urea) containing 1% (v/v) Triton X-100 and once with wash buffer lacking detergent. The inclusion bodies were solubilized in 50 mM Tris–HCl, 300 mM NaCl, and 8 M urea (overnight, room temperature). For immunization, the proteins were subjected to SDS gel electrophoresis, reversibly stained by 100 mM KCl, and directly excised. Two rabbits were immunized (Pineda Antikörper-Service, Germany). Other antibodies were previously described (Supplemental Table 4; Schleiff et al., 2003b; Sommer et al., 2011; Missbach et al., 2013; Rudolf et al., 2013; Jores et al., 2016). Purification of αToc75-V antibodies with MBP_Toc75-V_P1-3_his after DnaK depletion (Rial and Ceccarelli, 2002) is described in Supplemental Table 4.
For mSSU_his and pSSU_his, cell pellets were resuspended in 50 mM HEPES/KOH pH 7.6, 150 mM NaCl, 1 mM EDTA, and 2 mg/mL lysozyme (20 min, room temperature). Cell lysis was performed by sonication, and after centrifugation, the pellets were washed with 50 mM HEPES/KOH pH 7.6, 100 mM NaCl, 1 M urea, 1% (v/v) Triton X-100, and 10 mM β-mercaptoethanol. Pellets were solubilized in 50 mM HEPES/KOH, 100 mM NaCl, 10 mM β-mercaptoethanol, and 8 M urea (overnight, RT). mSSU_his and pSSU_his were purified through a Ni-NTA gravity flow column using 50 mM HEPES/KOH, 300 mM NaCl, 8 M urea, and 15 mM imidazole as wash buffer. Elution was performed with 50 mM HEPES/KOH pH 7.6, 6 M urea, and 500 mM imidazole. The imidazole was removed using Amicon Ultra-0.5 centrifugal filters.
Native and 2D PAGE analyses
All samples were solubilized in phosphate-buffered saline containing 1% (w/v) digitonin, 10% (v/v) glycerol, and 1% (v/v) Protease Inhibitor Cocktail—DMSO solution (Sigma-Aldrich) if not otherwise specified. Prior to loading, the samples were subjected to a clearing centrifugation (50,000g, 30 min, 4°C). HDN gels were prepared as described (Ladig et al., 2011) and run overnight at 95 V. For 2D electrophoresis, gels were incubated in denaturing buffer (100 mM Tris–HCl pH 6.8, 66 mM sodium carbonate, 0.67% [v/v] β-mercaptoethanol, 2% [w/v] SDS; 20 min, room temperature) prior to SDS–PAGE (Reisinger and Eichacker, 2007). For antibody-based mobility shift assays, solubilized samples were incubated with 10 µg affinity-purified antibody (1h, ice, dark) if not otherwise indicated. Prior to HDN–PAGE, the samples were subjected to a clearing centrifugation (25,000g, 15 min).
Pull-down of OEP37-N-Twin strep import intermediates
OEP37 and OEP37-N-Twin-Strep were in vitro translated in rabbit reticulocyte lysate using unlabeled amino acids (Promega). The proteins were incubated with isolated chloroplasts (800 µg chlorophyll per protein; 30 min, 25°C). Organelles were placed on the top of a 40% Percoll cushion and re-purified by centrifugation. The collected organelles were washed twice with HMS buffer (50 mM HEPES/KOH pH 7.6, 330 mM sorbitol, 3 mM MgCl2). Chloroplasts were solubilized in pull-down buffer (100 mM Tris–HCl pH 8, 150 mM NaCl, 10% glycerol, 1 mM EDTA) containing 1% (v/v) protease inhibitor cocktail, 1% (w/v) DIG, and 200 µg/ml avidin (30 min, ice). After centrifugation (50,000g, 30 min, 4°C), the supernatants were incubated with pre-equilibrated 1-µL net resin volume of MagStrep “type3” XT beads (IBA, Germany; 4°C, 2.5 h). The beads were washed three times with 500 µL and once with 100 µL pull-down buffer containing 0.5% DIG. Bound proteins were eluted with 25 µL SDS loading buffer.
Protein analysis
Proteins or protein complexes were separated by Tris–glycine-based SDS–PAGE (Laemmli, 1970), Tris–tricine-based SDS–PAGE (Schägger, 2006), or HDN–PAGE (Ladig et al., 2011). For proteins with a very low-molecular weight (e.g. in vitro translated PsOEP241–6), a precast NuPAGE™ gradient (4%–12%) Bis–Tris gel from Invitrogen by Thermo Scientific™ (Waltham, NA, USA) was used. The gels were stained with Coomassie Brilliant Blue (Green and Sambrook, 2012), radioactivity was analyzed by phosphorimager (Typhoon 9400, GE Healthcare, Chicago, USA), or incubated with primary antibody (Supplemental Table 4) after transfer to PVDF or nitrocellulose-blotting membrane (Merck, Darmstadt, Germany) via semi-dry electro-blotting (Green and Sambrook, 2012).
Accession numbers
The following genes were used for biological experiments or bioinformatics analysis in this study: from N. tabacum NtSSU: RBS_TOBAC, P69249; A. thaliana AtENP1: At1g31660; AtHSP18.5: AT2G19310; AtLPTD1: AT2G44640; AtOEP21: AT1G76405 and AT1G20816; AtOEP24: AT1G45170 and AT5G42960; AtOEP37: AT2G43950; AtP39: AT3G44160; AtTGD4: AT3G06830; AtTOC75-V: AT5G19620; AtVDAC1: AT3G01280; and Pisum sativum PsOE33; PSBO_PEA, P14226; PsOEP21:OEP21_PEA, Q9SM57; PsOEP24: OEP24_PEA, O49929; PsOEP37: OEP37_PEA, Q4LDF9; PsToc34: TOC34_PEA, Q41009; PsTOC159: Q9LKR1_PEA, Q9LKR1.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure 1. Topology of the plastidic β-barrel proteins.
Supplemental Figure 2. GFPS11-OEP24 distribution in A. thaliana protoplasts and A. cepa epidermal cells.
Supplemental Figure 3. Analysis of the GFP fluorescence of Hsp18.5-GFPS11 and GFPS11-OEP24TS.
Supplemental Figure 4. Analysis of the GFP fluorescence of GFPS11-AtOEP24.
Supplemental Figure 5. Analysis of the GFP fluorescence of GFPS11-OEP37, GFPS11-OEP37TS GFPS11-OEP21, and GFPS11-OEP21TS.
Supplemental Figure 6. Analysis of the GFP fluorescence of GFPS11-AtOEP37 and GFPS11-AtOEP21.
Supplemental Figure 7. Analysis of GFP fluorescence of the N-terminal regions of plastidic β-barrel proteins.
Supplemental Figure 8. Analysis of the shape of intracellular GFP fluorescence of the N- and C-terminal regions of plastidic β-barrel proteins.
Supplemental Figure 9. Analysis of GFP fluorescence of the C-terminal regions of plastidic β-barrel proteins.
Supplemental Figure 10. Analysis of GFP fluorescence of GFPS11-OEP24 (7–14)M.
Supplemental Figure 11. The temperature- and time-dependent formation of OEP37 import intermediates.
Supplemental Figure 12. Quantification of the time-dependent formation of import intermediates.
Supplemental Figure 13. ATP dependence of OEP37 translocation into chloroplast membranes.
Supplemental Figure 14. Surface exposure and membrane insertion of OEP37 import intermediates.
Supplemental Figure 15. Specificity of the antibody shift of chloroplast-associated OEP37.
Supplemental Figure 16. Time dependence of the formation of membrane-inserted import intermediates of OEP37.
Supplemental Figure 17. The mode of OEP37 import.
Supplemental Table 1. Oligonucleotides used in this study.
Supplemental Table 2. Constructs and plasmids used in this study.
Supplemental Table 3. Excitation and emission parameters of fluorophores.
Supplemental Table 4. Antibodies used in this study.
Supplementary Material
Acknowledgments
We thank Daniela Bublak (Goethe University) and Maike Ruprecht (Goethe University) for constant technical support and Doron Rapaport (University Tübingen) for critical discussion.
Funding
This study was supported by the Deutsche Forschungsgemeinschaft (EXC 115: Macromolecular Complexes; DFG SCHL 585-7; DFG SCHL 585-9).
Conflicts of interest. All authors declare no actual or potential conflict of interest or competing financial interests.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is Enrico Schleiff (schleiff@bio.uni-frankfurt.de).
Conceptualization: E.S.; proposal and design of experiments: L.E.G., A.K., R.L., E.S.; in vivo import: A.K., T.E., U.B.; in vitro import: A.K., L.E.G., N.S.; bioinformatics analysis: N.F., S.S.; data analysis: L.E.G., A.K., E.S.; draft writing: L.E.G., A.K., U.B., E.S.; draft editing and approval: all authors.
References
- Annweiler A, Hipskind RA, Wirth T (1991) A strategy for efficient in vitro translation of cDNAs using the rabbit beta-globin leader sequence. Nucleic Acids Res 19:3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ (2004) PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res 32:W400–W404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin AJ, Wardle A, Patel R, Dudley P, Park SK, Twell D, Inoue K, Jarvis P (2005) A molecular-genetic study of the arabidopsis Toc75 gene family. Plant Physiol 138:715–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bionda T, Schleiff E (2010) Chloroplast isolation and in vitro protein import. J Endocytobiosis Cell Res 20:17–26 [Google Scholar]
- Boldogh IR, Nowakowski DW, Yang H, Chung H, Karmon S, Royes P, Pon LA (2003) A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol Biol Cell 14:4618–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölter B, Soll J, Hill K, Hemmler R, Wagner R (1999) A rectifying ATP-regulated solute channel in the chloroplastic outer envelope from pea. EMBO J 18:5505–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeier R, Schlegel T, Ertel F, Vojta A, Borissenko L, Bohnsack MT, Groll M, von Haeseler A, Schleiff E (2007) Functional and phylogenetic properties of the pore-forming beta-barrel transporters of the Omp85 family. J Biol Chem 282:1882–1890 [DOI] [PubMed] [Google Scholar]
- Bruce BD (2001) The paradox of plastid transit peptides: conservation of function despite divergence in primary structure. Biochim Biophys Acta 1541:2–21 [DOI] [PubMed] [Google Scholar]
- Carrie C, Giraud E, Whelan J (2009) Protein transport in organelles: dual targeting of proteins to mitochondria and chloroplasts. FEBS J 276:1187–1195 [DOI] [PubMed] [Google Scholar]
- Cabantous S, Terwilliger TC,, Walso GS (2005) Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat Biotechnol 23:102–107 [DOI] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N (2009) Importing mitochondrial proteins: machineries and mechanisms. Cell 138:628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan SW, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit RA, Jansonius JN, Rosenbusch JP (1992) Crystal structures explain functional properties of two E. coli porins. Nature 358:727–733 [DOI] [PubMed] [Google Scholar]
- Day PM, Inoue K, Theg SM (2019) Chloroplast outer membrane β-barrel proteins use components of the general import apparatus. Plant Cell 31:1845–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckart K, Eichacker LA, Sohrt K, Schleiff E, Heins L, Soll J (2002) A Toc75-like protein import channel is abundant in chloroplasts. EMBO Rep 3:557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenrieder L, Opaliński Ł, Becker L, Krüger V, Mirus O, Straub SP, Ebell K, Flinner N, Stiller SB, Guiard B,. et al. (2016) Separating mitochondrial protein assembly and endoplasmic reticulum tethering by selective coupling of Mdm10. Nat Commun 7:13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fic E, , Kedracka-Krok S, , Jankowska U, , Pirog A, , Dziedzicka-Wasylewska M ( 2010) Comparison of protein precipitation methods for various rat brain structures prior to proteomic analysis. Electrophoresis 31: 3573–3579 [DOI] [PubMed] [Google Scholar]
- Flinner N, Ellenrieder L, Stiller SB, Becker T, Schleiff E, Mirus O (2013) Mdm10 is an ancient eukaryotic porin co-occurring with the ERMES complex. Biochim Biophys Acta 1833:3314–3325 [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB (1982) Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol 93:97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–5 [DOI] [PubMed] [Google Scholar]
- Green MR, Sambrook J (2012). Molecular Cloning: A Laboratory Manual,4th edn,Cold Spring Harbor Laboratory Press [Google Scholar]
- Gross LE, Machettira AB, Schleiff E, Sommer MS (2011). GFP-based in vivo protein topology determination in plant protoplasts. J Endocytobiosis Cell Res 21:89–97 [Google Scholar]
- Gross LE, Spies N, Simm S,, Schleiff E (2020) Toc75-V/OEP80 is processed during translocation into chloroplasts and this leads to an exposure of the POTRA domains to the intermembrane space. FEBS Open Bio 10: 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarmann R, Ibrahim M, Stevanovic M, Bredemeier R, Schleiff E (2010) The properties of the outer membrane localized Lipid A transporter LptD. J Phys Condens Matter 22:454124. [DOI] [PubMed] [Google Scholar]
- Harsman A, Schock A, Hemmis B, Wahl V, Jeshen I, Bartsch P, Schlereth A, Pertl-Obermeyer H, Goetze TA, Soll J, Philippar K, Wagner R (2016) OEP40, a regulated glucose-permeable β-barrel solute channel in the chloroplast outer envelope membrane. J Biol Chem 291:17848–17860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayat S, Elofsson A (2012) BOCTOPUS: improved topology prediction of transmembrane β-barrel proteins. Bioinformatics 28:516–522 [DOI] [PubMed] [Google Scholar]
- Hemmler R, Becker T, Schleiff E, Bölter B, Stahl T, Soll J, Götze TA, Braams S, Wagner R (2006) Molecular properties of OEP21, an ATP-regulated anion-selective solute channel from the outer chloroplast membrane. J Biol Chem 281:12020–12029 [DOI] [PubMed] [Google Scholar]
- Hill K, Model K, Ryan MT, Dietmeier K, Martin F, Wagner R, Pfanner N (1998) Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature 395:516–521 [DOI] [PubMed] [Google Scholar]
- Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G (2008) Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 321:1206–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhr AIC, Lindau C, Wirth C, Qiu J, Stroud DA, Kutik S, Guiard B, Hunte C, Becker T, Pfanner N,. et al. (2018) Membrane protein insertion through a mitochondrial β-barrel gate. Science 359: pii: eaah6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SC, Nafati M, Inoue K (2012) OEP80, an essential protein paralogous to the chloroplast protein translocation channel Toc75, exists as a 70-kD protein in the Arabidopsis thaliana chloroplast outer envelope. Plant Mol Biol 78:147–158 [DOI] [PubMed] [Google Scholar]
- Hsueh Y-C, Ehmann C, Flinner N, Ladig R, Schleiff E (2017a) The plastid outer membrane localized LPTD1 is important for glycerolipid remodelling under phosphate starvation. Plant Cell Environ 40:1643–1657 [DOI] [PubMed] [Google Scholar]
- Hsueh YC, Flinner N, Gross LE, Haarmann R, Mirus O, Sommer MS, Schleiff E (2017b) Chloroplast outer envelope protein P39 in Arabidopsis thaliana belongs to the Omp85 protein family. Proteins Struct Funct Bioinforma 85:1391–1401 [DOI] [PubMed] [Google Scholar]
- Hsueh YC, Nicolaisen K, Gross LE, Nöthen J, Schauer N, Vojta L, Ertel F, Koch I, Ladig R, Fulgosi H, et al. (2018) The outer membrane Omp85-like protein P39 influences metabolic homeostasis in mature Arabidopsis thaliana. Plant Biol 20:825–833 [DOI] [PubMed] [Google Scholar]
- Huang W, Ling Q, Bédard J, Lilley K, Jarvis P (2011) In vivo analyses of the roles of essential Omp85-related proteins in the chloroplast outer envelope membrane. Plant Physiol 157:147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Constan D, Keegstra K (2001) Arabidopsis genes encoding components of the chloroplastic protein import apparatus. Plant Physiol 125:1567–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DT, Froehlich JE, Keegstra K (1998) The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J Biol Chem 273:16583–16588 [DOI] [PubMed] [Google Scholar]
- Jaedicke K, Rösler J, Gans T, Hughes J (2011) Bellis perennis: a useful tool for protein localization studies. Planta 234:759–768 [DOI] [PubMed] [Google Scholar]
- Jarvis P, López-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nature Rev Mol Cell Biol 14:787–802 [DOI] [PubMed] [Google Scholar]
- Jores T, Klinger A, Groß LE, Kawano S, Flinner N, Duchardt-Ferner E, Wöhnert J, Kalbacher H, Endo T, Schleiff E, et al. (2016). Characterization of the targeting signal in mitochondrial β-barrel proteins. Nat Commun 7:12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jores T, Lawatscheck J, Beke V, Franz-Wachtel M, Yunoki K, Fitzgerald JC, Macek B, Endo T, Kalbacher H, Buchner J, et al. (2018) Cytosolic Hsp70 and Hsp40 chaperones enable the biogenesis of mitochondrial β-barrel proteins. J Cell Biol 217:3091–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Hirohashi T, Nakai M (2006) Characterization of the preprotein translocon at the outer envelope membrane of chloroplasts by blue native PAGE. Plant Cell Phys 47:363–371 [DOI] [PubMed] [Google Scholar]
- Klinger A, Gosch V, Bodensohn U, Ladig R, Schleiff E (2019) The signal distinguishing between targeting of outer membrane β-barrel protein to plastids and mitochondria in plants. Biochim Biophys Acta Mol Cell Res 1866:663–672 [DOI] [PubMed] [Google Scholar]
- Kouranov A, Chen X, Fuks B, Schnell DJ (1998) Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J Cell Biol 143:991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P (2009) An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325:477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozjak V, Wiedemann N, Milenkovic D, Lohaus C, Meyer HE, Guiard B, Meisinger C, Pfanner N (2003) An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J Biol Chem 278:48520–48523 [DOI] [PubMed] [Google Scholar]
- Künkele K-P, Heins S, Dembowski M, Nargang FE, Benz R, Thieffry M, Walz J, Lill R, Nussberger S, Neupert W (1998) The preprotein translocation channel of the outer membrane of mitochondria. Cell 93:1009–1019 [DOI] [PubMed] [Google Scholar]
- Kunze M, Berger J (2015) The similarity between N-terminal targeting signals for protein import into different organelles and its evolutionary relevance. Front Physiol 6:259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutik S, Stojanovski D, Becker L, Becker T, Meinecke M, Krüger V, Prinz C, Meisinger C, Guiard B, Wagner R. et al. (2008) Dissecting membrane insertion of mitochondrial beta-barrel proteins. Cell 132:1011–1024 [DOI] [PubMed] [Google Scholar]
- Kurowski MA, Bujnicki JM (2003) GeneSilico protein structure prediction meta-server. Nucleic Acids Res 31:3305–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladig R, Sommer MS, Hahn A, Leisegang MS, Papasotiriou DG, Ibrahim M, Elkehal R, Karas M, Zickermann V, Gutensohn M, et al. (2011) A high-definition native polyacrylamide gel electrophoresis system for the analysis of membrane complexes. Plant J 67:181–194 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- Li L, Kubiszewski-Jakubiak S, Radomiljac J, Wang Y, Law SR, Keech O, Narsai R, Berkowitz O, Duncan O, Murcha MW. , et al. (2016) Characterization of a novel β-barrel protein (AtOM47) from the mitochondrial outer membrane of Arabidopsis thaliana. J Exp Bot 67:6061–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumme C, Altan-Martin H, Dastvan R, Sommer MS, Oreb M, Schuetz D, Hellenkamp B, Mirus O, Kretschmer J, Lyubenova S, et al. (2014) Nucleotides and substrates trigger the dynamics of the Toc34 GTPase homodimer involved in chloroplast preprotein translocation. Structure 22:526–538 [DOI] [PubMed] [Google Scholar]
- Machettira AB, Groß LE, Tillmann B, Weis BL, Englich G, Sommer MS, Königer M, Schleiff E (2011a) Protein-induced modulation of chloroplast membrane morphology. Front Plant Sci 2:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machettira AB, Gross LE, Sommer MS, Weis BL, Englich G, Tripp J, Schleiff E (2011b) The localization of Tic20 proteins in Arabidopsis thaliana is not restricted to the inner envelope membrane of chloroplasts. Plant Mol Biol 77:381–390 [DOI] [PubMed] [Google Scholar]
- Martin T, Sharma R, Sippel C, Waegemann K, Soll J, Vothknecht UC (2006) A protein kinase family in Arabidopsis phosphorylates chloroplast precursor proteins. J Biol Chem 281:40216–40223 [DOI] [PubMed] [Google Scholar]
- Meisinger C, Pfannschmidt S, Rissler M, Milenkovic D, Becker T, Stojanovski D, Youngman MJ, Jensen RE, Chacinska A, Guiard B, et al. (2007) The morphology proteins Mdm12/Mmm1 function in the major beta-barrel assembly pathway of mitochondria. EMBO J 26:2229–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirus O, Hahn A, Schleiff E (2010) Outer membrane proteins. InKönig H., Claus H., Varma, eds A.. Prokaryotic Cell Wall Compounds. Structure and Biochemistry, Springer-Verlag, pp. 175–230 [Google Scholar]
- Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf KD (2002) In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev 16:1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missbach S, Weis BL, Martin R, Simm S, Bohnsack MT, Schleiff E (2013) 40S ribosome biogenesis co-factors are essential for gametophyte and embryo development. PLoS One 8:e54084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Model K, Meisinger C, Prinz T, Wiedemann N, Truscott KN, Pfanner N, Ryan MT (2001) Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat Struct Biol 8:361–370 [DOI] [PubMed] [Google Scholar]
- Moslavac S, Mirus O, Bredemeier R, Soll J, von Haeseler A, Schleiff E. ( 2005) Conserved pore-forming regions in polypeptide-transporting proteins. FEBS J 272:1367–1378 [DOI] [PubMed] [Google Scholar]
- Murcha MW, Kmiec B, Kubiszewski-Jakubiak S, Teixeira PF, Glaser E, Whelan J (2014) Protein import into plant mitochondria: signals, machinery, processing, and regulation. J Exp Bot 65:6301–6335 [DOI] [PubMed] [Google Scholar]
- Nicolaisen K, Missbach S, Hsueh Y-C, Ertel F, Fulgosi H, Sommer MS, Schleiff E (2015) The Omp85-type outer membrane protein p36 of Arabidopsis thaliana evolved by recent gene duplication. J Plant Res 128:317–325 [DOI] [PubMed] [Google Scholar]
- Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W (2003) Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature 426:862–866 [DOI] [PubMed] [Google Scholar]
- Patel R, Hsu S-C, Bédard J, Inoue K, Jarvis P (2008) The Omp85-related chloroplast outer envelope protein OEP80 is essential for viability in Arabidopsis. Plant Physiol 148:235–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmeyer K, Soll J, Grimm R, Hill K, Wagner R (1998) A high-conductance solute channel in the chloroplastic outer envelope from pea. Plant Cell 10:1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qbadou S, Tien R, Soll J, Schleiff E (2003). Membrane insertion of the chloroplast outer envelope protein, Toc34: constrains for insertion and topology. J Cell Sci 116:837–846 [DOI] [PubMed] [Google Scholar]
- Qbadou S,, Becker T, Mirus O, Tews I, Soll J, Schleiff E (2006) The molecular chaperone Hsp90 delivers precursor proteins to the chloroplast import receptor Toc64. EMBO J 25:1836–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisinger V, Eichacker L.A (2007) Analysis of membrane protein complexes by Blue Native PAGE. Proteomics 6:6–15 [DOI] [PubMed] [Google Scholar]
- Rial DV, Ceccarelli EA (2002) Removal of DnaK contamination during fusion protein purifications. Protein Expr Purif 25:503–507 [DOI] [PubMed] [Google Scholar]
- Rödiger A, Baudisch B, Klösgen RB (2010) Simultaneous isolation of intact mitochondria and chloroplasts from a single pulping of plant tissue. J Plant Physiol 167:620–624 [DOI] [PubMed] [Google Scholar]
- Rudhe C, Chew O, Whelan J, Glaser E (2002) A novel in vitro system for simultaneous import of precursor proteins into mitochondria and chloroplasts. Plant J 30:213–220 [DOI] [PubMed] [Google Scholar]
- Rudolf M, Machettira AB, Groß LE, Weber KL, Bolte K, Bionda T, Sommer MS, Maier UG, Weber APM, Schleiff E,. et al. (2013) In vivo function of Tic22, a protein import component of the intermembrane space of chloroplasts. Mol Plant 6:817–829 [DOI] [PubMed] [Google Scholar]
- Ruprecht M, Bionda T, Sato T, Sommer MS, Endo T, Schleiff E (2010) On the impact of precursor unfolding during protein import into chloroplasts. Mol Plant 3:499–508 [DOI] [PubMed] [Google Scholar]
- Schägger H (2006) Tricine–SDS-PAGE. Nature Protoc 1:16–22 [DOI] [PubMed] [Google Scholar]
- Schatz G, Dobberstein B (1996) Common principles of protein translocation across membranes. Science 271:1519–1526 [DOI] [PubMed] [Google Scholar]
- Schleiff E, Soll J (2005) Membrane protein insertion: mixing eukaryotic and prokaryotic concepts. EMBO Rep 6:1023–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E,, Becker T (2011) Common ground for protein translocation: access control for mitochondria and chloroplasts. Nat Rev Mol Cell Biol 12:48–59 [DOI] [PubMed] [Google Scholar]
- Schleiff E, Eichacker LA, Eckart K, Becker T, Mirus O, Stahl T, Soll J (2003) Prediction of the plant beta-barrel proteome: a case study of the chloroplast outer envelope. Protein Sci 12:748–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E,, Soll J, Küchler M, Kühlbrandt W, Harrer R (2003b) Characterization of the translocon of the outer envelope of chloroplasts. J Cell Biol 160:541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TG, Koepke J, Frank R, Skerra A (1996) Molecular interaction between the Strep-tag affinity peptide and its cognate target, streptavidin. J Mol Biol 255:753–766 [DOI] [PubMed] [Google Scholar]
- Schmidt von Braun S, Sabetti A, Hanic-Joyce PJ, Gu J, Schleiff E, Joyce PB (2007) Dual targeting of the tRNA nucleotidyltransferase in plants: not just the signal. J Exp Bot 58:4083–4093 [DOI] [PubMed] [Google Scholar]
- Scott A, Wyatt S, Tsou P-L, Robertson D, Strömgren-Allen N (1999) Model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques 26:7. [DOI] [PubMed] [Google Scholar]
- Seedorf M, Waegemann K, Soll J (1995) A constituent of the chloroplast import complex represents a new type of GTP-binding protein. Plant J 7:401–11 [DOI] [PubMed] [Google Scholar]
- Siddique M, Gernhard S, Von Koskull-Döring P, Vierling E, Scharf KD (2008) The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones 13:183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo LF,, Yaffe MP. ( 1994) Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol 126:1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer MS, Daum B, Gross LE, Weis BL, Mirus O, Abram L, Maier UG, Kühlbrandt W, Schleiff E (2011) Chloroplast Omp85 proteins change orientation during evolution. Proc Natl Acad Sci USA 108:13841–13846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer M, Rudolf M, Tillmann B, Tripp J, Sommer MS, Schleiff E. ( 2013) Toc33 and Toc64-III cooperate in precursor protein import into the chloroplasts of Arabidopsis thaliana. Plant Cell Environ 36:970–983 [DOI] [PubMed] [Google Scholar]
- Struyvé M, Moons M, Tommassen J (1991). Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol 218:141–148 [DOI] [PubMed] [Google Scholar]
- Tillmann B, Röth S, Bublak D, Sommer M, Stelzer EHKK, Scharf KD, Schleiff E (2015) Hsp90 is involved in the regulation of cytosolic precursor protein abundance in tomato. Mol Plant 8:228–241 [DOI] [PubMed] [Google Scholar]
- Tranel PJ, Froehlich JE, Goyal A, Keegstra K (1995) A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J 14:2436–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranel PJ,, Keegstra K (1996) A novel, bipartite transit peptide targets OEP75 to the outer membrane of the chloroplastic envelope. Plant Cell 8:2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp J, Mishra SK, Scharf KD (2009) Functional dissection of the cytosolic chaperone network in tomato mesophyll protoplasts. Plant Cell Environ 32:123–133 [DOI] [PubMed] [Google Scholar]
- Truscott KN, Wiedemann N, Rehling P, Müller H, Meisinger C, Pfanner N, Guiard B (2002) Mitochondrial import of the ADP/ATP carrier: the essential TIM complex of the intermembrane space is required for precursor release from the TOM complex. Mol Cell Biol 22:7780–7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujwal R, Cascio D, Colletier J-P, Faham S, Zhang J, Toro L, Ping P, Abramson J (2008) The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating. Proc Natl Acad Sci 105:17742–17747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich T, Gross LE, Sommer MS, Schleiff E, Rapaport D (2012) Chloroplast beta-barrel proteins are assembled into the mitochondrial outer membrane in a process that depends on the TOM and TOB complexes. J Biol Chem 287:27467–27479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW, Stacey MG (1998) Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221:35–43 [DOI] [PubMed] [Google Scholar]
- Weiss MS, Schulz GE (1992) Structure of porin refined at 1.8 A resolution. J Mol Biol 227:493–509 [DOI] [PubMed] [Google Scholar]
- Wimley WC (2003) The versatile β-barrel membrane protein. Curr Opin Struct Biol 13:404–411 [DOI] [PubMed] [Google Scholar]
- Xu C, Fan JF, Cornish AJ, Benning C (2008) Lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis requires the extraplastidic TGD4 protein. Plant Cell 20:2190–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman MJ, Hobbs AEA, Burgess SM, Srinivasan M, Jensen RE (2004) Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J Cell Biol 164:677–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.