I hate it when a recipe calls for 2 ounces of heavy cream and the smallest container I can find to purchase is 16 ounces; I have to figure out what to do with all that excess cream (it being not the healthiest option to use it liberally in coffee). What for meis a minor inconvenience in the kitchen can be a big problem for multimeric protein complexes in cells. Cells typically maintain tight control of the production of protein subunits in the amounts needed to meet the stoichiometric requirements of the final complex, as amounts in excess of these requirements can lead to detrimental aggregation and the buildup of toxic intermediates or breakdown products (as if the extra cream had spilled and spoiled all the fresh produce in the refrigerator).
The photosynthetic carbon-fixing enzyme Rubisco makes an excellent case study in this regard: chloroplasts are packed with Rubisco, which makes up about 3% of the total mass of leaves (Bar-On and Milo, 2019), and the most abundant Form I Rubisco (present in all land plants, green algae, and cyanobacteria) is composed of eight chloroplast-encoded large subunits (LSUs) and eight nucleus-encoded small subunits (SSU) arranged in a stoichiometric L8S8 structure. Since the subunits are encoded in separate cellular compartments, all the steps involved in biosynthesis: transcription and translation of these components, transport of the SSU to the chloroplast, and assembly of the holoenzyme, must be highly coordinated. Cells can handle a certain amount of “excess” functional holoenzyme through regulation of activity, and, in the case of Rubisco, this may be a regulatory mechanism (Quick et al., 1991). However, the uncoordinated production of subunits of multimeric complexes appears to be a matter of cellular concern, as cells have evolved complex mechanisms to avoid it (Taggart et al. 2020).
New work by Wietrzynski et al. (2021) provides a detailed picture of feedback control of Rubisco LSU translation in Chlamydomonas reinhardtii (Chlamydomonas). Wietrzynski et al. conducted an extensive characterization of LSU intermediates formed during Rubisco biogenesis using a series of site-directed mutants of Chlamydomonas combined with biochemical and molecular genetic analysis to show that LSU translation is controlled by its ability to assemble with the SSU, via the autoregulatory feedback mechanism Control by Epistasy of Synthesis (CES). Epistasy refers to interactions between genes or gene products; CES is a mechanism by which the translation rate of one or more subunits of a protein complex is regulated by the presence or absence of (and thus the ability to interact with) assembly partners. CES has been shown to occur for a number of photosynthetic complexes, especially when subunits are encoded in distinct intracellular compartments, as for PSI, PSII, cyt b6f, and ATP synthase (Choquet and Wollman, 2009), and the current work confirms that Rubisco can be added to this list. But how does it work?
Wietrzynski et al. focused on LSU biogenesis and previous observations in maize and tobacco chloroplasts that unassembled LSU somehow exerts negative feedback control on its own translation (Wostrikoff and Stern, 2007; Wostrikoff et al., 2012). But the “somehow” of the effect on LSU translation, the nature of assembly intermediates, and their relationship to the SSU, were largely unknown. In this study, the author identifies LSU assembly intermediates associated with the RAF1 chaperone and show that, in the absence of SSU, an LSU8-RAF1 complex likely turns into an inhibitor that exerts negative autoregulatory feedback on further LSU translation (see Figure).
The authors use a series of cleverly designed site-directed mutants to show that inhibition of LSU translation (from the rbcL gene) in the absence of SSU is dependent on regulatory sequence present in the rbcL 5′ UTR, and it is unassembled LSU itself that functions as the negative regulator of its own translation. They further sought to determine the nature of the LSU intermediate and how it acquires this autoinhibitory function, a challenging task as Rubisco assembly is fast and does not permit significant accumulation of assembly intermediates. Analyzing native PAGE gels using extracts from mutant strains under a variety of experimental conditions led the authors to focus on RAF1 as a potentially important component of an LSU assembly intermediate complex. A key set of experiments using epitope-tagged RAF1 in a ΔRBCS strain and structure-guided mutagenesis experiments (based on mutations altering the formation or stability of LSU dimers or oligomerization in octamers), provided evidence that an LSU-RAF1 complex accumulates in vivo and RAF1 plays a role in LSU stabilization, and that an LSU8-RAF18 complex may constitute the last assembly intermediate prior to SSU binding. The authors propose a model whereby the LSU8-RAF1 intermediate provides a platform for SSU binding to form the holoenzyme, and when SSU is not available, this complex exerts negative regulation on LSU translation initiation (Figure).
Figure.
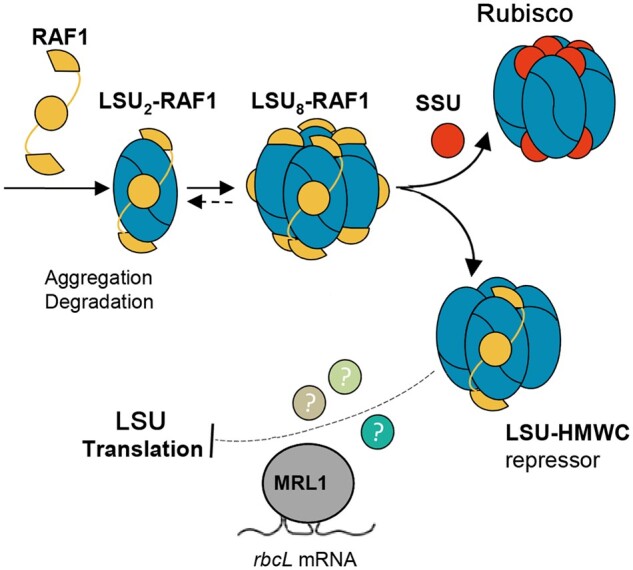
Model of CES regulation of Rubisco LSU (rbcL) translation. The figure depicts key steps of later stages of LSU assembly. LSU associates with RAF1 and then further oligomerizes to form the Rubisco core LSU8-RAF1, which provides a scaffold for SSU binding. RAF1 is substituted by SSU to form the complete holoenzyme. When SSU is absent or limiting, the LSU8-RAF1 high molecular weight complex (LSU-HMWC) becomes a repressor of LSU translation (CES process), thereby preventing wasteful production of LSU. Adapted from Wietrzynski et al. (2021), Figure 12.
Future experiments will need to determine the exact composition of the LSU8-RAF1 complex, the function of RAF1 itself (whether it participates in translation repression or only stabilizes the complex), and whether there are additional interacting partners. Genetic evidence of function could provide critical information, and a raf1 mutant (unavailable to date) could provide the cream on top of the pudding in this respect.
References
- Bar-On YM, Milo R (2019) The global mass and average rate of rubisco. Proc Natl Acad Sci USA 116: 4738–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y, Wollman F-A (2009) The CES process. InHarris EH, Stern DB, Witman GB, eds, The Chlamydomonas Sourcebook, Chapter 29. Academic Press, London, pp. 1027–1063 [Google Scholar]
- Quick WP, Schurr U, Fichtner K, Schulze ED, Rodermel SR, Bogorad L, Stitt M (1991) The impact of decreased Rubisco on photosynthesis, growth, allocation and storage in tobacco plants which have been transformed with antisense rbcS. Plant J 1: 51–58 [Google Scholar]
- Taggart JC, Zauber H, Selbach M, Li GW, Mcshane E (2020) Keeping the proportions of protein complex components in check. Cell Syst 10.1016/j.cels.2020.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wietrzynski W, Traverso E, Wollman F-A, Wostrikoff K (2021) The state of oligomerization of Rubisco controls the rate of synthesis of the Rubisco large subunit in Chlamydomonas reinhardtii. Plant Cell 33: 1706–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K, Stern D (2007) Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proc Natl Acad Sci USA 104: 6466–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K, Clark A, Sato S, Clemente T, Stern D (2012) Ectopic expression of Rubisco subunits in maize mesophyll cells does not overcome barriers to cell type-specific accumulation. Plant Physiol 160: 419–432 [DOI] [PMC free article] [PubMed] [Google Scholar]


