Abstract
The coordinated development of sporophytic and gametophytic tissues is essential for proper ovule patterning and fertility. However, the mechanisms regulating their integrated development remain poorly understood. Here, we report that the Swi2/Snf2-Related1 (SWR1) chromatin-remodeling complex acts with the ERECTA receptor kinase-signaling pathway to control female gametophyte and integument growth in Arabidopsis thaliana by inhibiting transcription of the microRNA gene MIR398c in early-stage megagametogenesis. Moreover, pri-miR398c is transcribed in the female gametophyte but is then translocated to and processed in the ovule sporophytic tissues. Together, SWR1 and ERECTA also activate ARGONAUTE10 (AGO10) expression in the chalaza; AGO10 sequesters miR398, thereby ensuring the expression of three AGAMOUS-LIKE (AGL) genes (AGL51, AGL52, and AGL78) in the female gametophyte. In the context of sexual organ morphogenesis, these findings suggest that the spatiotemporal control of miRNA biogenesis, resulting from coordination between chromatin remodeling and cell signaling, is essential for proper ovule development in Arabidopsis.
SWR1-mediated nucleosome structure and ERECTA signaling play a critical role in repressing the transcription of MIR398c during ovule development.
Introduction
Development in multicellular organisms requires coordinated growth and morphogenesis between tissues. In Arabidopsis thaliana, ovules comprise the haploid female gametophyte (also called the embryo sac) and the surrounding diploid sporophytic tissues. Development of these distinct tissues must be integrated for proper ovule morphogenesis and successful fertilization (Bencivenga et al., 2011; Chevalier et al., 2011). The Arabidopsis ovule primordium originates from the placenta, elongates along the proximal/distal axis, and develops into three regions; the nucellus, chalaza, and funiculus. The female germline initiates in the distal nucellus, followed by development of the female gametophyte. The proximal chalaza and funiculus differentiate into the integuments and stalk, respectively (Figure 1A). Arabidopsis ovules contain inner and outer integuments, each composed of two cell layers (Coen et al., 2017). The integuments envelop the female gametophyte while leaving a small opening known as a micropyle, through which pollen tubes enter to deliver sperm to the female gametophyte.
Figure 1.
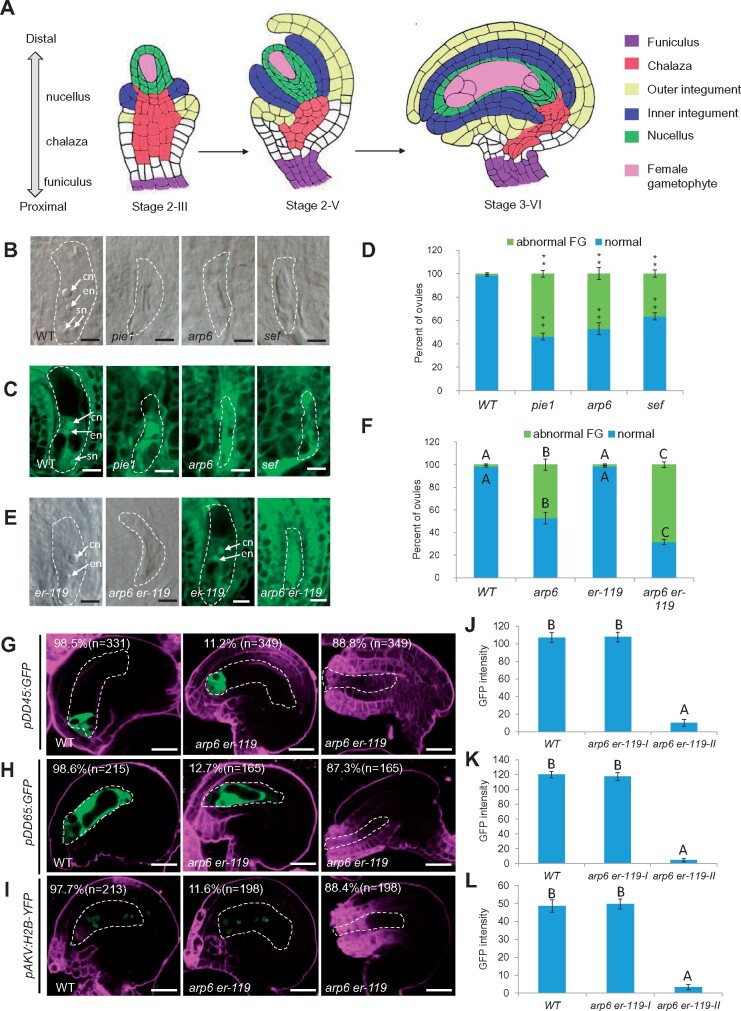
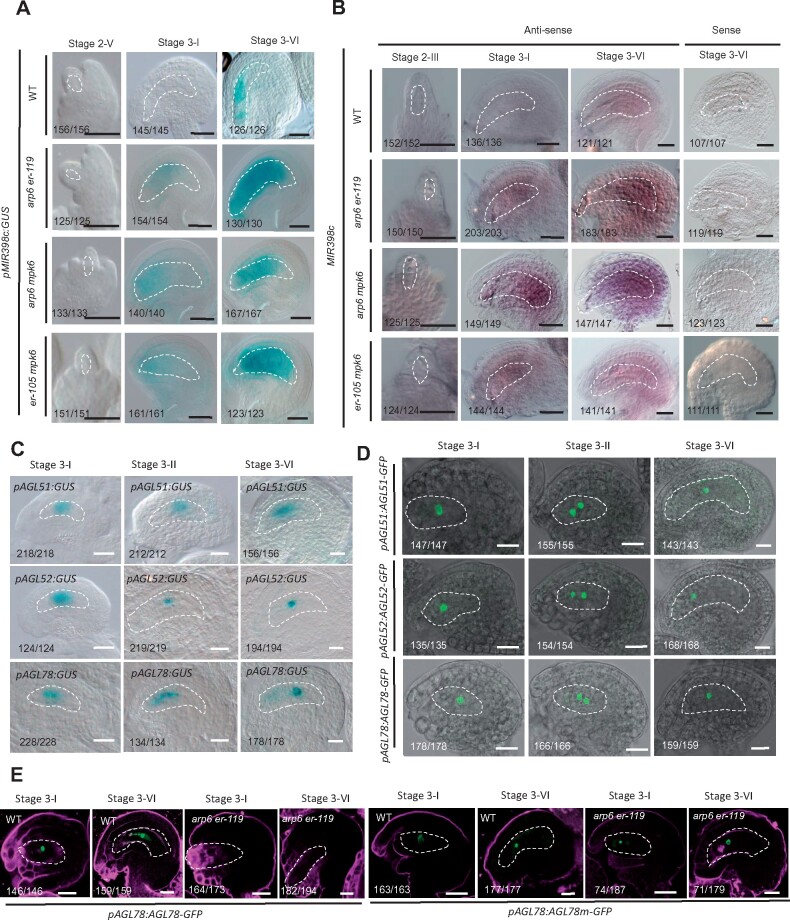
The er-119 mutation enhanced the female gametophyte developmental defects in SWR1 complex mutant. A, Diagram illustrating the ovule structure and developmental process in A. thaliana. B, DIC observation of ovules at Stage 3-VI. C, Confocal observation of ovules at Stage 3-VI. D, Quantification of normal and abnormal female gametophytes (FGs) at Stage 3-VI for each sample. The number of ovules observed is shown in Supplemental Data Set 1. Asterisks above the columns indicate significant differences compared to WT (**P < 0.01 by t-test). E, DIC (left panel) and confocal (right panel) observation of ovules at Stage 3-VI. F, Quantification of normal and abnormal FGs at Stage 3-VI for each sample. The number of ovules observed is shown in Supplemental Data Set 1. Different letters above the columns indicate significant differences at P < 0.01, as determined by one-way ANOVA. G–I, Signal corresponding to egg cell marker pDD45:GFP (G), central cell marker pDD65:GFP (H), and female gametophyte marker pAKV:H2B-YFP (I) in ovules at Stage 3-VI. J–L, The quantification of GFP/YFP signal intensity corresponding to egg cell marker pDD45:GFP (J), central cell marker pDD65:GFP (K), and female gametophyte marker pAKV:H2B-YFP (L) in WT and arp6 er-119 mutant ovules at Stage 3-VI. Error bars indicate ± SD (n = 10 biological replicates). Different letters above the columns indicate significant differences at P < 0.01, as determined by one-way ANOVA. cn, central cell nucleus; en, egg cell nucleus; sn, synergid cell nucleus. Bars in (B)–(E), 5 μm. Bars in (G)–(I), 10 μm. The female gametophyte is outlined by white dot lines in (B) and (C), (E), and (G)–(I).
The development of the haploid female gametophyte is regulated by genes expressed in the female gametophyte and/or the surrounding ovule integuments (Yang and Sundaresan, 2000; Yadegari and Drews, 2004). Integument-expressed proteins that are important for the development of the female gametophyte include transcription factors (TFs) such as BELL1 (a homeodomain TF; Reiser et al., 1995), INNER NO OUTER (INO, a YABBY TF; Villanueva et al., 1999), and AINTEGUMENTA (ANT, an AP2 TF; Elliott et al., 1996; Klucher et al., 1996), and kinases such as TOUSLED (a nuclear Ser/Thr protein kinase; Roe et al., 1997), ERECTA (ER), ERECTA-LIKE 1 (ERL1), and ERL2 (leucine-rich repeat receptor-like kinases; Pillitteri et al., 2007), and MPK3 and MPK6 (mitogen-activated protein kinases, MPKs; Wang et al., 2008). Plant hormone signaling in the integuments also regulates female gametophyte development, including auxin (Pagnussat et al., 2009; Lituiev et al., 2013) and cytokinin (Kinoshita-Tsujimura and Kakimoto, 2011; Bencivenga et al., 2012; Cheng et al., 2013). Mutations in integument-expressed genes can cause defective integument elongation along with arrested female gametophyte development (Baker et al., 1997). Although many mutations in female gametophyte-expressed genes do not lead to obvious sporophytic integument growth defects (Colombo et al., 2008; Rabiger and Drews, 2013), altered integument gene expression has been detected in ovule mutants that lack a female gametophyte (Johnston et al., 2007; Armenta-Medina et al., 2013; Zhao et al., 2014). These reports suggest that the gametophyte and integument communicate and mutually influence each other’s development.
Epigenetic regulatory pathways likely contribute to the coordinated development of the integument and female gametophytic tissues in the ovule. For example, a mutation in the SET DOMAIN GROUP2 (SDG2) gene, which mediates histone H3 lysine 4 trimethylation (H3K4me3), causes defective female gametophyte formation and inhibits integument growth (Berr et al., 2010). The ATP-dependent CHD3 chromatin remodeler PICKLE (PKL), which can be either positively or negatively associated with H3K27me3 in genes important for seed germination, was also reported to control female gametophyte development and integument growth (Carter et al., 2016). Several microRNAs (miRNAs), including miR167 and miR165/6, regulate ovule morphogenesis and fertility by restricting the expression of their target genes AUXIN RESPONSE FACTORS ARF6 and ARF8 and the Class III homeodomain leucine zipper (HD-Zip III) gene PHABULOSA (PHB), respectively (Wu et al., 2006; Hashimoto et al., 2018). However, how these factors mediate the integrated development of the ovule integument and the female gametophyte is not well understood.
The ATP-dependent chromatin-remodeling complex SWR1 (SWI2/SNF2-Related 1) plays a crucial role in regulating gene expression by exchanging histone H2A-H2B dimers with H2A.Z-H2B dimers (Aslam et al., 2019). The H2A.Z histone variant is highly conserved throughout eukaryotes (van Daal et al., 1990). H2A.Z deposition into nucleosomes plays roles in fine-tuning gene expression, both positively and negatively, by changing chromatin architecture and transcription factor accessibility (Marques et al., 2010; Dai et al., 2017). In Arabidopsis, various SWR1 complex subunits such as PHOTOPERIOD INDEPENDENT EARLY FLOWERING1 (PIE1), ACTIN-RELATED PROTEIN6 (ARP6), SERRATED LEAVES AND EARLY FLOWERING (SEF), SWR1 COMPLEX SUBUNIT2 (SWC2), SWC4, and METHYL-CpG-BINDING DOMAIN9 (MBD9) have been identified (Lazaro et al., 2008; March-Diaz and Reyes, 2009; Potok et al., 2019; Sijacic et al., 2019; Luo et al., 2020). SWR1 orchestrates numerous aspects of growth and development, including leaf shape, organ size, flowering time, meiosis, and germline specification (Choi et al., 2005; Deal et al., 2005; Choi et al., 2007; Deal et al., 2007; Lazaro et al., 2008; March-Diaz and Reyes, 2009; Choi et al., 2013; Rosa et al., 2013; Zhao et al., 2018). Mutations in SWR1 complex subunit genes cause similar pleiotropic developmental phenotypes, although pie1 mutants exhibit more severe phenotypes than arp6 and sef (Mizuguchi et al., 2004).
Using genetic enhancer screening, we found that SWR1 genetically interacts with the ER-MPK-signaling pathway in the control of Arabidopsis ovule development. Specifically, they regulate the coordinated growth and development of the female gametophyte and integument by repressing the expression of MIR398c during early-stage megagametogenesis. We further show that SWR1 and ER-MPK signaling are critical for spatially inhibiting miR398 accumulation in the developing female gametophyte. miR398 would otherwise inhibit three target AGAMOUS-LIKE (AGL) genes (AGL51, AGL52, and AGL78) and disturb their function in female gametophyte development and integument growth. We also identified two mechanisms that prevent miR398 accumulation in the female gametophyte: 1) pri-miR398c is generated in the female gametophyte but is translocated to and processed in the ovule sporophytic tissues and 2) AG010 plays a key role in sequestering miR398 in the chalaza. Our results indicate that chromatin remodeling is coordinated with a kinase-signaling pathway to ensure integrated development between sporophytic and gametophytic tissues, via precise spatial and temporal control of miR398 biogenesis. These findings provide new insights into sexual organ morphogenesis in higher plants.
Results
The SWR1 chromatin-remodeling complex genetically interacts with the ER-MPK-signaling pathway to control female gametophyte development
The SWR1 chromatin-remodeling complex was shown to regulate female meiosis during female gametophyte development (Rosa et al., 2013; Qin et al., 2014). Consistent with this regulatory function, in a subset of mature Stage 3-VI ovules (Schneitz et al., 1995), the SWR1 subunit single mutants showed abnormal female gametophytes at frequencies (57.6% in pie1, 51.2% in arp6, and 37.1% in sef) significantly higher than that in wild-type (WT; 0.5%, P < 0.01, see Supplemental Data Set 1 for statistical data). The mutants either lacked female gametophyte cell nuclei or contained smaller female gametophytes compared to wild-type ovules, which typically contained a kidney-shaped female gametophyte with a central cell, an egg cell, two synergid cells, and three antipodal cells nuclei (Figure 1A–D).
To identify additional genes involved in SWR1 control of female gametophyte development, we performed an EMS mutagenesis screen for enhancers of the arp6 mutation. In this screen, we identified an ERECTA (ER) point mutation allele er-119, a C-to-T mutation resulting in the conversion of 397th amino acid in the LRR domain from Arg to a premature stop codon. er-119 is a knock-out allele that significantly enhanced the fertility defects in arp6 (Cai et al., 2017; Supplemental Figure S1A). The phenotype of er-119 is similar to that of er-105.
To examine if the reduced fertility in arp6 er-119 double mutants results from male or female tissues, we conducted reciprocal crosses and found that the low fertility of the arp6 er-119 double mutant was specifically due to defects in the female tissues (Supplemental Figure S2A and B). An in vivo pollen tube guidance assay showed that many arp6 er-119 ovules did not receive pollen tubes (Supplemental Figure S2C). Consistent with these findings, arp6 er-119 showed a significantly increased percentage of developmentally stunted ovules with an abnormal female gametophyte, compared to the arp6 and er-119 single mutants (Figure 1E and F; P < 0.01, Supplemental Data Set 1).
To test if gamete cell differentiation is also affected in the arp6 er-119 female gametophyte, we separately introduced egg cell, central cell, and female gametophyte cell markers into arp6 er-119. The pDD45:GFP egg cell marker, the pDD65:GFP central cell marker, and the pAKV:H2B-YFP female gametophyte marker were expressed in only 11.2%, 12.7%, and 11.6% of arp6 er-119 ovules, respectively, significantly lower than that (98.5%, 98.6%, and 97.7%) in wild-type ovules (Figure 1G–I; P < 0.01, Supplemental Data Set 1). The GFP/YFP signal was significantly decreased in arp6 er-119 compared to WT (Figure 1J–L; P < 0.01, Supplemental Data Set 1), suggesting that ARP6 and ER are required for the differentiation of the female gametophyte.
A complementation construct containing the genomic ER sequence restored the reduced seed set and female gametophyte defects of arp6 er-119 to levels comparable to those in arp6 (Supplemental Figure S1A and B). This analysis confirmed that the enhanced female gametophyte defects of arp6 er-119 were caused by the er-119 mutation. A second er allele, er-105, similarly enhanced the gametophytic defects of arp6, further supporting the involvement of both ARP6 and ER in female gametophyte development. Specifically, the arp6 er-105 double mutant had reduced seed set and an increased percentage of abnormal female gametophytes compared to arp6 (Supplemental Figure S1A–D). To investigate whether ER functions specifically with the ARP6 subunit of the SWR1-remodeling complex in female gametophyte regulation, we generated the sef er-105 double mutant. Compared to the sef single mutant, the double mutant had enhanced defects in seed set and female gametophyte development (Supplemental Figure S1A–D).
MPK3 and MPK6 act redundantly downstream of ER in multiple developmental processes, including ovule development (Pillitteri et al., 2007; Wang et al., 2008). To test if the MPK-signaling pathway also functions with the SWR1 complex in female gametophyte regulation, we generated the arp6 mpk6 and sef mpk6 double mutants. Similar to the arp6 er, sef er, and er mpk6 double mutants, arp6 mpk6 and sef mpk6 double mutants had reduced fertility and enhanced female gametophyte defects compared to the arp6 and sef single mutants (Supplemental Figure S1A–D), as indicated by abnormal expression of the female gametophyte marker and the central cell and egg cell markers (Supplemental Figure S3A–C). The GFP/YFP signal was significantly decreased in arp6 mpk6 and er-105 mpk6 compared to WT (Supplemental Figure S3D–F; P < 0.01, Supplemental Data Set 1). Collectively, these results link SWR1 and the ER-MPK-signaling pathway in the regulation of female gametophyte development.
SWR1 and ER-MPK coordinately repress MIR398c expression in ovules
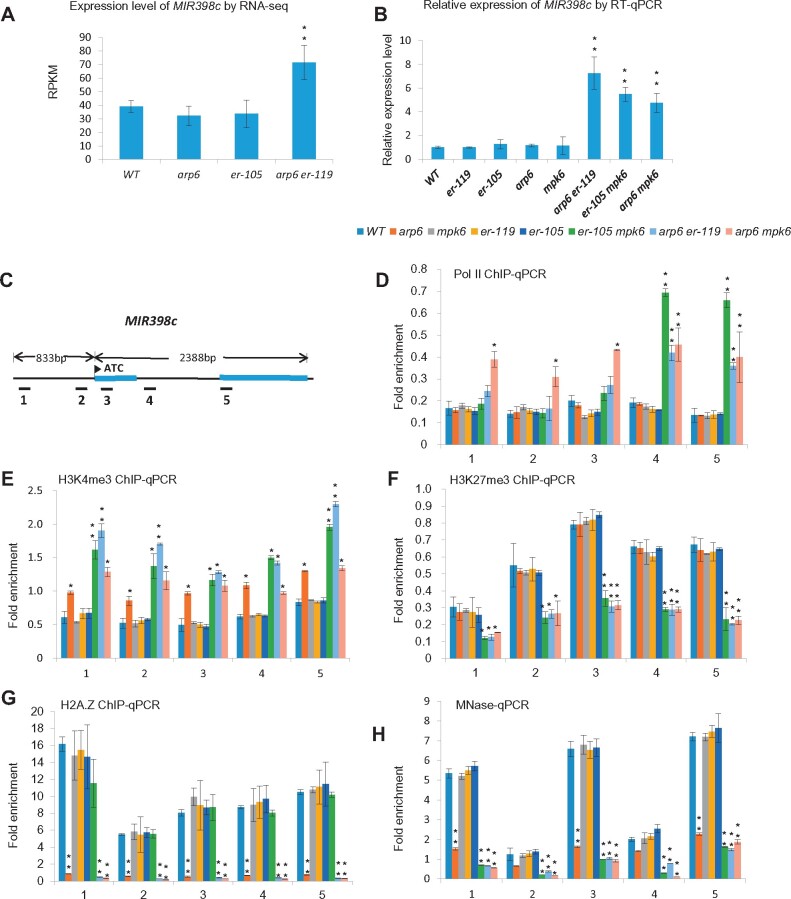
Next, we focused on the identification of components downstream of both the SWR1 complex and the ER-MPK-signaling pathway in the regulation of female gametophyte development. As the SWR1 complex controls activation or repression of gene expression (Ojolo et al., 2018), we profiled changes in gene expression in arp6 er-119 double mutant ovules. Specifically, we performed comparative RNA-seq analysis of wild type, arp6, er-105, and arp6 er-119 ovules from Stage 2-III to Stage 3-VI (Cai et al., 2017). Because the female gametophyte defects in arp6 er-119 ovules were significantly greater than in arp6, we focused only on genes with significantly altered expression (fold change ≥ 2, FDR ≤ 0.05; Supplemental Data Set 2) in the arp6 er-119 double mutant compared to arp6. Among the 55 differentially expressed genes, MIR398c was the only miRNA-encoding gene and ranked second in the list of genes with increased expression (Supplemental Data Set 2 and Figure 2A). Reverse transcription-quantitative PCR (RT-qPCR) confirmed the increased MIR398c transcript levels in arp6 er-119 ovules compared to WT, arp6, and er-105 ovules (Figure 2B). Similar increases in MIR398c expression were also detected in arp6 mpk6 and er-105 mpk6 double mutant ovules compared to WT, arp6, er-105, er-119, and mpk6 ovules (Figure 2B), indicating that ARP6, ER, and MPK6 cooperatively repress MIR398c expression in ovules.
Figure 2.
MIR398c transcription is repressed by SWR1 and the ER-MPK-signaling pathway. A, reads per kilobase per million mapped reads (RPKM) of MIR398c in ovules detected by RNA-seq. B, Relative MIR398c (pri-miR398c) levels in ovules tested by RT-qPCR analysis. C, Diagram of the MIR398c gene. Exons are indicated as blue boxes, and the promoter and introns are indicated as black lines. The flag indicates the transcription start site. Regions amplified by PCR primer sets are indicated with black bars below the diagram. Primer set numbers correspond to the numbers on the x-axis of the graphs in (D)–(H). D–G, Chromatin immunoprecipitation–quantitative polymerase chain reaction (ChIP-qPCR) analysis for the enrichment of RNA Pol II (D), H3K4me3 (E), H3K27me3 (F), and H2A.Z (G) at MIR398c in WT and the indicated mutants. H, Nucleosome occupancy at MIR398c as measured by MNase treatment followed by qPCR. Values are means ± SD from three biological replicates. Each biological replicate corresponds to three technical replicates. Asterisks above the columns indicate significant differences compared to WT (**P < 0.01, *P < 0.05 by t test).
Because RNA polymerase II (Pol II) occupancy is a reliable indicator of active gene transcription, we compared Pol II occupancy at the MIR398c locus in WT, arp6, er-119, er-105, mpk6, arp6 er-119, arp6 mpk6, and er-105 mpk6. For this analysis, we performed chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) using floral bud tissues and a Pol II antibody. Pol II occupancy at the MIR398c gene body increased in arp6 er-119, arp6 mpk6, and er-105 mpk6 compared to WT and each of the single mutants (Figure 2C and D). We also found that the abundance of H3K4me3, a marker of active transcription, at MIR398c was significantly increased in arp6 er-119, arp6 mpk6, and er-119 mpk6 (Figure 2C and E). Moreover, the H3K27me3 repressive mark was significantly reduced at the MIR398c locus in the double mutants compared to wild type and the single mutants (Figure 2C and F). Together, these results demonstrate that MIR398c is actively transcribed in arp6 er-119, arp6 mpk6, and er-105 mpk6.
As a chromatin-remodeling complex, SWR1 regulates gene transcription by facilitating the deposition of the H2A.Z variant into nucleosomes and modifying histone-DNA interactions (Marques et al., 2010). To test if H2A.Z deposition at the MIR398c gene is mediated by ARP6, we performed ChIP-qPCR using an H2A.Z antibody. In wild-type floral buds, we detected a high level of H2A.Z deposition at the MIR398c promoter (Figure 2C and G). In contrast, H2A.Z occupancy at the MIR398c promoter was depleted in arp6 floral buds, as well as arp6 er-119 and arp6 mpk6 floral buds (Figure 2C and G).
H2A.Z-containing nucleosomes have altered DNA interactions compared to H2A-containing nucleosomes, thereby affecting nucleosomal stability (Kumar and Wigge, 2010). We therefore evaluated nucleosome occupancy at MIR398c in the presence or absence of ARP6 using micrococcal nuclease (MNase) digestion followed by qPCR (Petesch and Lis, 2008). The results showed that nucleosome occupancy at regions occupied by H2A.Z was greatly decreased in the arp6 single mutant and the arp6 er-119 and arp6 mpk6 double mutants, compared to wild type (Figure 2C and H). However, the activation of MIR398c transcription was only observed in the arp6 er-119, arp6 mpk6, and er-105 mpk6 double mutants compared to wild type and not in the arp6 single mutant (Figure 2A and B). The present findings therefore suggest that while SWR1 contributes to the transcriptional inhibition of MIR398c, a gene regulated by ER-MPK, changes in nucleosome status controlled by the SWR1 complex alone are not sufficient to alter MIR398c transcription.
MIR398c overexpression disrupted female gametophyte development
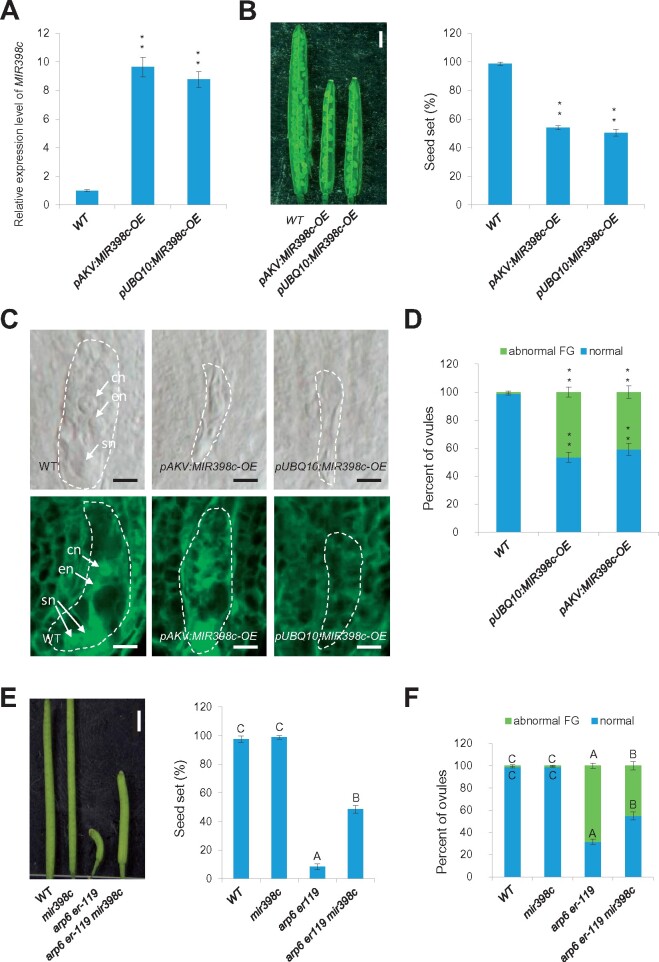
We next tested whether increased MIR398c expression leads to abnormal female gametophyte development. If MIR398c levels are increased in the double mutant ovules and if MIR398c expression is normally suppressed by SWR1 and ER, then MIR398c overexpression in wild type should recapitulate the female gametophyte defects of the arp6 er double mutants. To obtain MIR398c overexpression lines, we generated the constructs using the genomic MIR398c sequence driven by the constitutive UBIQUITIN 10 (UBQ10) promoter (pUBQ10:MIR398c-OE) and the female gametophyte cell-specific AKV promoter (pAKV:MIR398c-OE) and transformed the constructs into wild-type plants, separately. RT-qPCR confirmed that pUBQ10:MIR398c-OE and pAKV:MIR398c-OE had increased MIR398c transcript levels (Figure 3A). Moreover, pUBQ10:MIR398c-OE and pAKV:MIR398c-OE plants had reduced fertility (Figure 3B) and abnormal female gametophytes (Figure 3C and D; significantly different from wild type, P < 0.01, Supplemental Data Set 1), which resembled the defects of the arp6 er-119, er-105 mpk6, and arp6 mpk6 double mutants.
Figure 3.
MIR398c acts downstream of SWR1 and ER in female gametophyte regulation. A, Relative MIR398c levels in ovules of WT and MIR398c overexpression (OE) lines by RT-qPCR analysis. Data are means ± SD (n = 3 biological replicates; **P < 0.01 by t test). B, Siliques (left panel) and quantification of seed-set percentage (right panel). Data are means ± SD (n = 10 siliques from five independent plants, two independent siliques from each plant; **P < 0.01 by t test). Seed-set percentage was calculated corresponding to the percentage of aborted seeds/ovules. Bars = 1 mm. C, DIC (top panel) and confocal (bottom panel) observation of MIR398c-OE ovules at Stage 3-VI. cn, central cell nucleus; en, egg cell nucleus; sn, synergid cell nucleus. Bars = 5 μm. The female gametophyte (embryo sac) is outlined by white dot lines. D, Quantification of female gametophyte phenotype at Stage 3-VI for each sample. The number of ovules observed is shown in Supplemental Data Set 1. **Significant differences at P < 0.01 by t test. E, Siliques (left panel) and quantification of seed-set percentage (right panel). Data are means ± SD (n = 10 biological replicates, 10 siliques from five independent plants, two independent siliques from each plant; different letters above the columns indicate statistically significant differences at P < 0.01, as determined by one-way ANOVA). Seed-set percentage was calculated corresponding to the percentage of aborted seeds/ovules. Bars = 1 mm. F, Quantification of female gametophyte phenotype at Stage 3-VI for each sample. The number of ovules observed is shown in Supplemental Data Set 1. Different letters above the columns indicate statistically significant differences at P < 0.01, as determined by one-way ANOVA.
We also investigated whether knockout/knockdown of MIR398c could suppress the female gametophyte defects seen in arp6 er-119 double mutants. A mutant line with a T-DNA insertion in the MIR398c locus (Dugas and Bartel, 2008) with reduced MIR398c transcripts (Supplemental Figure S4C), but without ovule development defects (Figure 3E and F), was crossed with the arp6 er-119 double mutant to obtain arp6 er-119 mir398c plants. These plants had partially restored plant fertility and an increased percentage of normal female gametophytes compared to arp6 er-119 (Figure 3E and F; P < 0.01, Supplemental Data Set 1). Together, these analyses show that elevated expression of MIR398c in arp6 er double mutants is sufficient to disrupt female gametophyte development.
miR398 target genes AGL51/52/78 act downstream of SWR1 and ER-MPK signaling in female gametophyte development
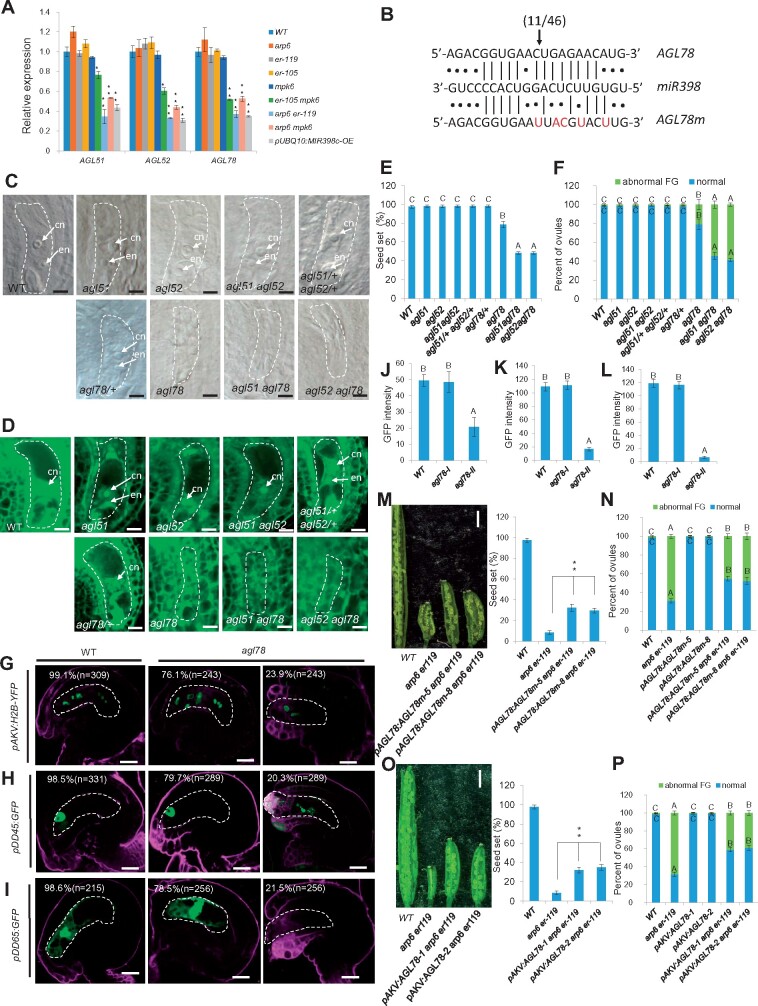
To investigate the molecular function of miR398 in female gametophyte development, we used the online tool psRNATarget (http://plantgrn.noble.org/psRNATarget/; Dai and Zhao, 2011) to identify putative miR398 targets. The identified candidate targets included three closely related AGAMOUS-LIKE (AGL) genes, AGL51, AGL52, and AGL78, which belong to the Mβ subclade of the MADS-box gene family (Parenicova et al., 2003). RT-qPCR analysis of AGL51, AGL52, and AGL78 in wild type, pUBQ10:MIR398c-OE, arp6, and the ER-MPK pathway mutants showed that all three genes had reduced transcript levels in pUBQ10:MIR398c-OE, arp6 er-119, arp6 mpk6, and er-105 mpk6 ovules compared to wild type and the single mutants (Figure 4A). We chose AGL78 as a representative gene to confirm the bioinformatic miRNA target prediction using a modified 5’-RACE PCR technique that enables precise determination of the cleavage sites of miRNAs on their mRNA targets (Llave et al., 2002). The results showed cleavage of AGL78 mRNA within the predicted miR398 target site, between positions 10 and 11 of the miR398 nucleotides (Figure 4B).
Figure 4.
AGL51/52/78, targets of miR398, regulate female gametophyte development. A, Relative mRNA levels of AGL51/52/78 in ovules by RT-qPCR analysis. Data represent means ± SD (n = 3 biological replicates; **P < 0.01, *P < 0.05 by t test). B, Alignment of the miR398 sequence with the corresponding complementary site of AGL78 and a miR398-resistant form of AGL78 (AGL78m). The arrow indicates the 5′ terminus cleavage product for the number of clones indicated. Cleavage was experimentally validated using 5′-RACE PCR. Red letters indicate the mutation sites of AGL78m. The lines represent base pairing; the dots represent nonpairing. C, DIC observation of WT and mutant ovules at Stage 3-VI. Bars = 5 μm. cn, central cell nucleus; en, egg cell nucleus; sn, synergid cell nucleus. D, Confocal observation of WT and mutant ovules at Stage 3-VI. Bars = 5 μm. cn, central cell nucleus; en, egg cell nucleus. E, Quantification of seed-set percentage for each sample. Data are means ± SD (n = 10 siliques from five independent plants, two independent siliques from each plant). Different letters above the columns indicate statistically significant differences at P < 0.01, as determined by one-way ANOVA. Seed-set percentage was calculated corresponding to the percentage of aborted seeds/ovules. F, Quantification of female gametophyte phenotype at Stage 3-VI for each sample. The number of ovules observed is shown in Supplemental Data Set 1. Different letters above the columns indicate statistically significant differences at P < 0.01, as determined by one-way ANOVA. G–I, Signal corresponding to the female gametophyte marker pAKV:H2B-YFP (G), egg cell marker pDD45:GFP (H), and central cell marker pDD65:GFP (I) in Stage 3-VI ovules. Bars = 10 μm. J–L, The quantification of GFP/YFP signal intensity corresponding to egg cell marker pDD45:GFP (J), central cell marker pDD65:GFP (K), and female gametophyte marker pAKV:H2B-YFP (L) in WT and agl78 mutant ovules at Stage 3-VI. Error bars indicate ± SD (n = 10 biological replicates). Different letters above the columns indicate statistically significant differences at P < 0.01, as determined by one-way ANOVA. M, Siliques (left panel) and quantification of seed-set percentage (right panel). Data represent means ± SD (n = 10 siliques from five independent plants, two independent siliques from each plant; **P < 0.01 by t test). Seed-set percentage was calculated corresponding to the percentage of aborted seeds/ovules. Bars = 1 mm. N, Quantification of female gametophyte phenotype at Stage 3-VI for each sample. The number of ovules observed is shown in Supplemental Data Set 1. Different letters above the columns indicate statistically significant differences at P < 0.01, as determined by one-way ANOVA. O, Siliques (left panel) and quantification of seed-set percentage (right panel). Data represent means ± SD (n = 10 siliques from five independent plants, two independent siliques from each plant; **P < 0.01 by t test). Seed-set percentage was calculated corresponding to the percentage of aborted seeds/ovules. Bars = 1 mm. P, Quantification of female gametophyte phenotype at Stage 3-VI for each sample. The number of ovules observed is shown in Supplemental Data Set 1. Different letters above the columns indicate statistically significant differences at P < 0.01, as determined by one-way ANOVA. The female gametophyte is outlined by white dot lines in (C), (D), and (G)–(I).
We then tested whether loss of function of any of these three AGL genes would lead to compromised female gametophyte development. agl78 had abnormal female gametophyte development and low seed set (Figure 4C–F), similar to the phenotypes of MIR398c-OE, arp6 er-119, er-105 mpk6, and arp6 mpk6. We also analyzed several gametophytic cell-specific GFP markers in agl78. The results showed that 23.9%, 20.3%, and 21.5% of agl78 ovules did not properly express the pAKV:H2B-YFP (female gametophyte cells), pDD45:GFP (egg cell), and pDD65:GFP (central cell) markers, respectively (Figure 4G–I; significantly different from wild type, P < 0.01, Supplemental Data Set 1); also, the GFP/YFP signal was significantly decreased in agl78 compared to WT (Figure 4J–L; P < 0.01, Supplemental Data Set 1). agl78/+ heterozygous mutants, agl51 and agl52 single mutants, agl51 agl52 double mutants, and agl51/+ agl52/+ double heterozygous mutants did not exhibit female gametophyte defects (Figure 4C–F), but the agl51 and agl52 single mutations could enhance the female gametophyte and fertility defects of agl78 in agl51 agl78 and agl52 agl78 double mutants (Figure 4C–F; P < 0.01, Supplemental Data Set 1). Taken together, these results suggest that increased miR398 accumulation in ARP6 and ER-MPK pathway mutants disrupts normal female gametophyte development by inhibiting the expression of three redundant genes, AGL51/52/78, among which AGL78 plays a dominant role in gametophyte development.
We conducted two experiments to test whether AGL51/52/78 act downstream of ARP6 and the ER-MPK pathway. First, we introduced a miR398-resistant form of AGL78 driven by the native promoter and that contained five mutation sites (pAGL78:AGL78m; Figure 4B) into arp6 er-119. This line had significantly increased seed set and a reduced percentage of ovules with an abnormal female gametophyte compared to the arp6 er-119 double mutant (Figure 4M and N; P < 0.01, Supplemental Data Set 1). Second, we overexpressed AGL78 driven by the female gametophyte cell-specific AKV promoter in the arp6 er-119 double mutant and found that it partially rescued the low seed set and female gametophyte defects of arp6 er-119 (Figure 4O and P; P < 0.01, Supplemental Data Set 1). These data collectively indicate that AGL51/52/78 function redundantly downstream of ARP6 and the ER-MPK pathway to regulate female gametophyte development.
Expression pattern of AGL51/52/78 and MIR398c in the developing ovule
Our results identified AGL51/52/78 as positive regulators of female gametophyte development and MIR398c as a direct negative regulator of AGL51/52/78 expression in ovules (Figure 4). However, the ovule is composed of both the diploid integument and haploid female gametophyte. We therefore addressed the cell-specific spatial and temporal expression patterns of AGL51/52/78 and MIR398c in ovules during female gametophyte development.
For MIR398c, we constructed a MIR398c promoter-GUS fusion (pMIR398c:GUS) and examined its expression in wild type. pMIR398c:GUS was expressed in the mature female gametophyte of ovules at Stage 3-VI, but not at earlier ovule stages such as 2-V and 3-I (Figure 5A). Consistent with the qPCR results (Figure 2B), pMIR398c:GUS expression in the female gametophyte of arp6 er-119, arp6 mpk6, and er-105 mpk6 ovules showed two abnormalities. First, pMIR398c:GUS expression was significantly increased in the female gametophyte compared to the level in wild type, and pMIR398c:GUS was expressed at earlier stages of female gametophyte development such as 3-I (Figure 5A). The expression of pMIR398c:GUS in arp6+/− er-119+/− ovules is comparable to that in wild type (Supplemental Figure S5), suggesting that the regulation of MIR398c expression by SWR1 and ER-signaling pathway likely act sporophytically. To investigate the expression pattern of SWR1 and ER pathway genes in the ovule, we performed pARP6:GUS (Qin et al., 2014), ER in situ hybridization, and pMPK6:MPK6-GFP (Wang et al., 2008) expression analysis and found that ARP6, ER, and MPK6 are expressed broadly in the ovule and present in both the female gametophyte and the ovule sporophytic tissue (Supplemental Figure S6).
Figure 5.
The expression pattern of MIR398c and AGL51/52/78 in ovules. A, pMIR398c:GUS expression pattern in WT, arp6 er-119, arp6 mpk6, and er-105 mpk6 ovules. Bars = 10 μm. B, Whole-mount in situ hybridization of antisense and sense MIR398c probe in WT, arp6 er-119, arp6 mpk6, and er-105 mpk6 ovules. Bars = 10 μm. C, pAGL51:GUS (top row), pAGL52:GUS (middle row), and pAGL78:GUS (bottom row) expression pattern in ovules. Bars = 10 μm. D, pAGL51:AGL51-GFP (top row), pAGL52:AGL512-GFP (middle row), and pAGL78:AGL78-GFP (bottom row) expression pattern in ovules. Bars = 10 μm. E, Signal corresponding to pAGL78:AGL78-GFP and pAGL78:AGL78m-GFP expression in WT and arp6 er-119 ovules. Bars = 10 μm. The female gametophyte is outlined by white dot lines in (A)–(E).
To confirm the results of the pMIR398c:GUS reporter analysis, we performed ovule whole-mount in situ hybridization using an antisense MIR398c probe. The hybridization results confirmed the preferential expression of MIR398c in the mature wild-type female gametophyte and the increased expression of MIR398c in arp6 er-119, arp6 mpk6, and er-105 mpk6 premature female gametophytes compared to wild type (Figure 5B). These results indicated that the combined activities of SWR1 and ER-MPK are required to fully repress MIR398c expression in the early-stage megagametogenesis (Figure 5A and B). The mutations in arp6 er-119, arp6 mpk6, and er-105 mpk6 also led to increased levels of MIR398c expression in the mature female gametophyte (Figure 5A and B), suggesting that SWR1 and ER-MPK also regulate MIR398c expression levels at the mature female gametophyte stage.
The Arabidopsis genome contains three MIR398 genes. In order to know whether MIR398a and MIR398b were also expressed in ovule, we constructed pMIR398a:GUS and pMIR398b:GFP transgenes and examined their expression patterns in ovules. pMIR398a:GUS was expressed in early-stage floral buds, but not in ovules (Supplemental Figure S7A). Whereas, pMIR398b:GFP was only expressed in the inner integument primordia or distal epidermal nucellus cells of ovules at meiosis stage (Stage 2-III; Supplemental Figure S7B). No signal was detected prior to Stage 2-II or post-meiosis (Supplemental Figure S7B). These data indicated that the expression patterns of MIR398a and MIR398b are different from that of MIR398c. We also found that either mir398a or mir398b mutation failed to restore fertility to arp6 er-119, although the sequences of mature miR398a and miR398b are near identical or identical to miR398c (Supplemental Figure S4A–B, D–F).
To characterize the cell-specific expression of AGL51/52/78 in ovules, we generated transgenic plants for each of the promoter-GUS fusion constructs, pAGL51:GUS, pAGL52:GUS, and pAGL78:GUS. The promoter activities of AGL51/52/78 were detected in the developing female gametophyte from post-meiotic Stages 3-I to mature Stage 3-VI (Figure 5C). Consistent with these GUS reporter results and as expected from their predicted transcription factor activity (Parenicova et al., 2003), GFP translationally fused to each of the three AGL proteins (pAGL51:AGL51-GFP, pAGL52:AGL52-GFP, and pAGL78:AGL78-GFP) localized to the nuclei of female gametophyte cells at post-meiotic Stage 3-I to mature Stage 3-VI ovules (Figure 5D). To further confirm the female gametophyte-specific expression of AGL51/52/78 in ovules, we performed laser capture microdissection (LCM) of female gametophyte and surrounding sporophytic ovule tissue at the mature stage (Stage 3-VI) followed by real-time RT-PCR and found that the transcripts of AGL51/52/78 in the mature stage ovule were mainly detected in the female gametophyte and barely detected in the sporophytic tissue (Supplemental Figure S8A). Consistently, whole-mount ovule in situ hybridization assay also showed AGL78 signal only in the female gametophyte of wild-type ovule from post-meiotic Stage 3-I to mature Stage 3-VI, but not in the surrounding sporophytic ovule tissue (Supplemental Figure S8B).
To confirm that AGL51/52/78 expression is controlled by ARP6 and the ER-MPK pathway, we introduced pAGL78:AGL78-GFP (as a representative of the three AGLs analyzed) into arp6 er-119. The pAGL78:AGL78-GFP construct was sufficient to rescue the mutant phenotype in agl78 (Supplemental Figure S9), suggesting that this construct plays a role similar to that of endogenous AGL78. Compared to wild type, we detected a reduced expression of pAGL78:AGL78-GFP in arp6 er-119 ovules from post-meiotic Stage 3-I to mature Stage 3-VI (Figure 5E). This finding is consistent with our results in arp6 er-119 and arp6 mpk6 ovules, in which both pMIR398c:GUS activity and MIR398c transcript levels were increased from Stage 3-I to mature Stage 3-VI (Figures 2B and 5A and B). Introduction of the miR398-resistant form of AGL78 (pAGL78:AGL78m-GFP) into the arp6 er-119 double mutant revealed that its expression was comparable in arp6 er-119 and WT ovules (Figure 5E). Based on these results, we concluded that ARP6 and the ER-MPK-signaling pathway activate AGL51/52/78 expression in the developing female gametophyte by repressing MIR398c transcription.
SWR1 and the ER-MPK-signaling pathway are required to prevent miR398 accumulation in the developing female gametophyte
As described above, pMIR398c:GUS was expressed in the mature female gametophyte of ovules at Stage 3-VI (Figure 5A), which coincided with the expression of its targets AGL51/52/78, according to promoter activity and protein-GFP fusion analyses (Figure 5C and D). One possible explanation for this unexpected expression overlap is that precursor or mature miR398 may migrate away from the mature female gametophyte, where MIR398c is transcribed, and localize elsewhere. This putative migration would allow AGL51/52/78 expression in the female gametophyte without inhibition by miR398.
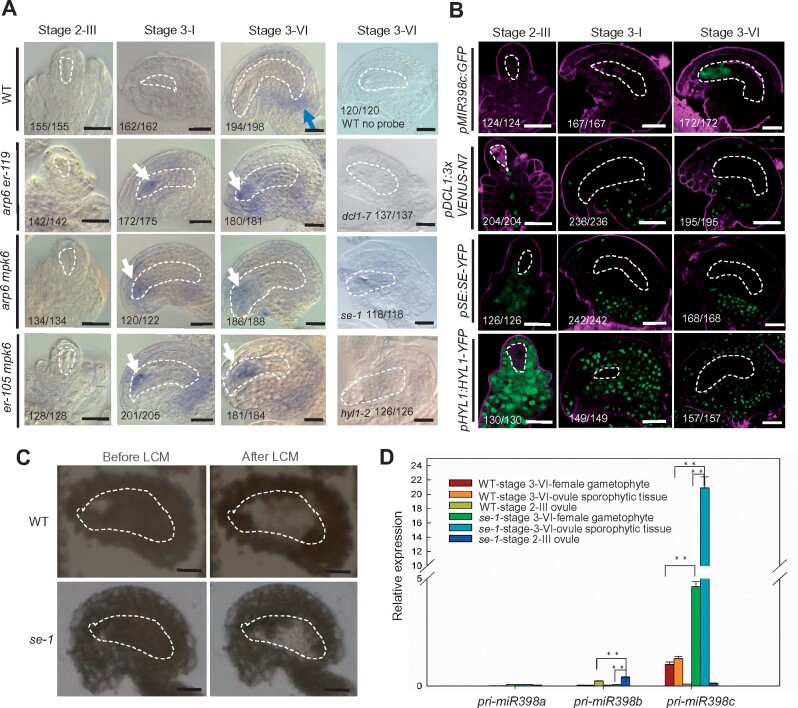
To test this hypothesis, we examined the localization of mature miR398 in ovules by performing whole-mount ovule in situ hybridization assay with miR398 locked nucleic acid (LNA) oligonucleotide probes. In wild type, miR398 signal was not detected in ovules until Stage 3-VI (Figure 6A). miR398 signal was detected in the chalaza of mature wild-type ovule, but not in the female gametophyte (Figure 6A), where MIR398c promoter activity was detected (Figure 5A). However, increased and ectopic miR398 signal was detected in the developing female gametophyte of arp6 er-119, arp6 mpk6, and er-105 mpk6 double mutant ovules from the post-meiotic (Stage 3-I) to mature stages (Figure 6A). No miR398 signal was detected in wild-type ovules without probe, or in ovules of the miRNA-processing mutants dcl1-7, se-1, and hyl1-2 with the miR398 LNA probe (Figure 6A).
Figure 6.
pri-miR398c is produced in the female gametophyte but is translocated to and processed in the surrounding sporophytic ovule tissues. A, Whole-mount ovule in situ localization of miR398 using LNA oligonucleotide probes. Blue arrow points to the chalaza-localized miR398 in WT. White arrows point to the ectopic localization of miR398 in the female gametophyte in the indicated mutants. B, Signal corresponding to pMIR398c:GFP, pDCL1:3xVENUS-N7, pSE:SE-YFP, and pHYL1:HYL1-YFP expression in ovules. C, Paraffin-embedded WT and se-1 ovule at mature Stage 3-VI before and after LCM. D, Relative levels of pri-miR398a/b/c in LCM isolated female gametophyte and the surrounding sporohytic tissues of WT and se-1 ovule at mature Stage 3-VI and the whole WT and se-1 ovule at Stage 2–III undergoing meiosis tested by RT-qPCR analysis. Bars in (A)–(C), 10 μm. Data are means ± SD (n = 3 biological replicates; **P < 0.01 by t test). The female gametophyte is outlined by white dot lines in (A)–(C).
The promoter-GUS activity and in situ hybridization analyses showed that the MIR398c gene was mainly transcribed in the female gametophyte, but that mature miR398 was present in the chalaza and not in the female gametophyte of wild-type ovules. Analysis using the MIR398c promoter-GFP fusion construct confirmed that MIR398c promoter activity was restricted to the female gametophyte at mature stage (Figure 6B), prompting us to investigate where miR398 is processed. To this end, we determined the expression patterns of genes that encode the microprocessor, which processes primary miRNAs (pri-miRNAs). These included DICER-LIKE1 (DCL1, an RNase III enzyme), SERRATE (SE, a zinc finger protein), and HYPONASTIC LEAVES1 (HYL1, a dsRNA-binding protein). DCL1-promoter GFP fusion (pDCL1:3×VENUS-N7) is only expressed in the ovule-sporophytic tissue (Figure 6B). Similarly, for SE and HYL1, YFP fusions driven by native promoters (Fang and Spector, 2007) were detected exclusively in ovule sporophytic tissue and not in the female gametophyte (Figure 6B). SE-YFP was enriched in the chalaza (Figure 6B). Thus, the expression patterns of MIR398c and the microprocessor genes are mutually exclusive. To further determine whether the biogenesis of miR398 occurs in ovule sporophytic or gametophytic tissues, we performed in situ hybridization assay to detect mature miR398 in ovules from dcl1-7 +/− plants. Although half of the female gametophyte is dcl mutant, 93.9% of ovules from a dcl1-7 +/− plant (n = 214) exhibited miR398 signal in the chalaza (Supplemental Figure S10A), similar to that in WT (94.9%, n = 178), indicating that the processing of pri-miR398 does not occur in the female gametophyte. In line with this conclusion, phenotypes of dcl1 mutants show maternal effects while homozygous dcl1 mutants have reduced fertility, dcl1 gametophytes do not show defects when the sporophyte is dcl1/+ (Golden et al., 2002; Supplemental Figure S10B–D).
We therefore hypothesize that pri-miR398c needs to be translocated from the female gametophyte to the surrounding sporophytic ovule tissues to be processed. To test this hypothesis, we performed LCM of the female gametophyte and surrounding sporophytic ovule tissue at the mature stage (Stage 3-VI) followed by real-time RT-PCR in wild type (Figure 6C). Although both pMIR398c:GUS and pMIR398c:GFP showed that pri-miR398c was mainly produced in the female gametophyte, we found that the level of pri-miR398c in the ovule sporophytic tissue at the mature stage was comparable to that in the female gametophyte (Figure 6D), suggesting that pri-miR398 was translocated from the female gametophyte to the surrounding ovule sporophytic tissues. No or very low levels of pri-miR398c were detected in the laser capture dissected whole ovule undergoing meiosis (Stage 2-III; Figure 6D), consistent with the results from pMIR398c:GUS and pMIR398c:GFP analysis. If pri-miR398c was produced in female gametophyte but translocated into the ovule sporophytic tissue for processing, then it is expected that in a miRNA-processing mutant, pri-miR398c would accumulate more in the ovule sporophytic tissue than in the female gametophyte. Indeed, we detected increased pri-miR398c in both se-1 female gametophyte and se-1 surrounding ovule sporophytic tissues at the ovule mature stage compared to wild type (Figure 6D). More importantly, the level of pri-miR398c in se-1 ovule sporophytic tissue was significantly higher than that in se-1 female gametophytes (Figure 6D). No or very low levels of pri-miR398a and pri-miR398b were detected in the laser capture dissected ovules (Figure 6D), consistent with previous pMIR398a:GUS and pMIR398b:GFP expression pattern analysis. Together these results suggest that pri-miR398c was produced in the female gametophyte, but was translocated to and processed in the ovule sporophytic tissue. The movement of pri-miRNA and the sporophyte-specific expression pattern of the miRNA-processing proteins may serve as an efficient strategy to prevent the accumulation of miRNAs in the female gametophyte. The expression pattern of HYL1-YFP in wild type and arp6 er-119 ovule is comparable (Supplemental Figure S11A and B), suggesting that SWR1 and the ER-MPK pathway may not affect pri-miRNA398c processing.
SWR1 and the ER-MPK-signaling pathway also activate AGO10 expression in the chalaza to sequester miR398
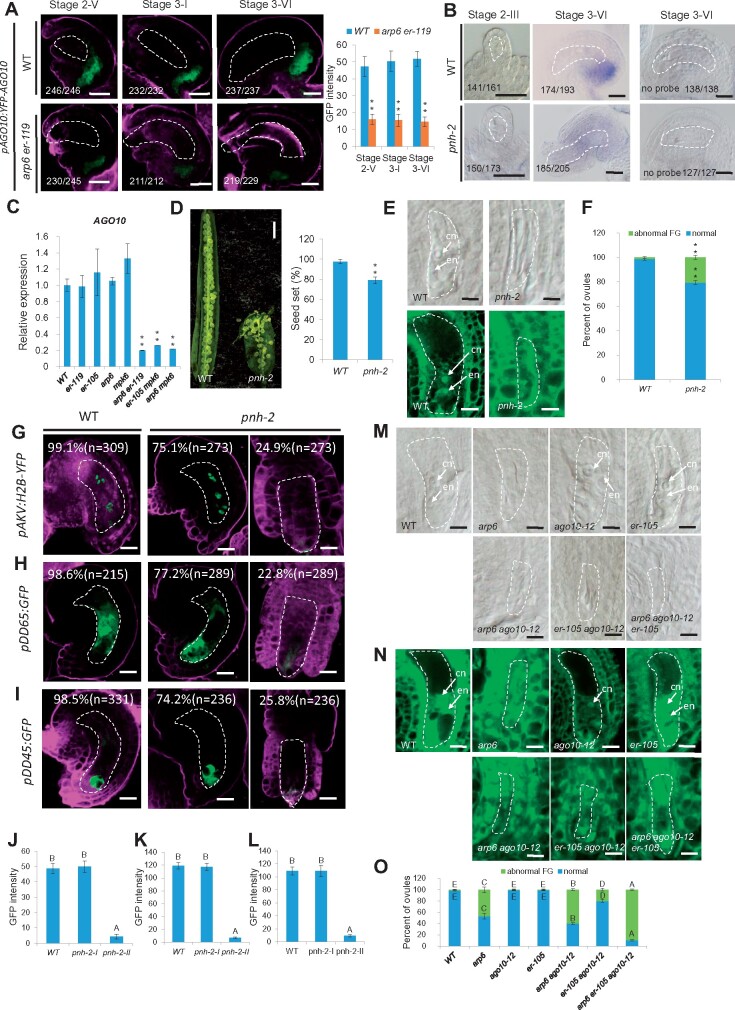
MiRNAs can move from cell to cell by passive diffusion (Carlsbecker et al., 2010) but an unknown gating mechanism can provide directionality in miRNA trafficking (Skopelitis et al., 2018). As mature miR398 accumulated in the chalaza, but not in the female gametophyte in WT ovule, we hypothesized that there may be a barrier that prevents mature miR398 from moving back to the female gametophyte. AGO10 has been shown to attenuate miR165/6 activity by sequestering the miRNA (Zhu et al., 2011), and to promote its degradation (Yu et al., 2017). Intriguingly, previous data showed that miR398 was the second most abundant miRNA species found in AGO10 immunoprecipitation (Yu et al., 2017). We thus examined the expression pattern of pAGO10:YFP-AGO10 (Yu et al., 2017) in ovules and found that AGO10 was present in the chalaza of ovules from Stage 2-V to Stage 3-VI (Figure 7A), the tissue where miR398 is located.
Figure 7.
AGO10 expression activated by SWR1 and the ER-signaling pathway is required for the sequestration of miR398 in the chalaza. A, Signal corresponding to pAGO10:YFP-AGO10 expression in WT and arp6 er-119 ovules (left panel) and quantification of YFP signal intensity corresponding to pAGO10:YFP-AGO10 in WT and arp6 er-119 ovules (right panel). Error bars indicate ± SD (n = 10 biological replicates; **P < 0.01 by t test). B, In situ localization of miR398 in WT and pnh-2 ovules using miR398 LNA oligonucleotide probes. C, Relative mRNA levels of AGO10 in ovules by RT-qPCR analysis. Data are means ± SD (n = 3 biological replicates; **P < 0.01 by t test). D, Siliques (left panel) and quantification of seed-set percentage (right panel) of WT and pnh-2. Data are means ± SD (n = 10 siliques from five independent plants, two independent siliques from each plant; **P < 0.01 by t test). Seed-set percentage was calculated corresponding to the percentage of aborted seeds/ovules. E, DIC (top panel) and confocal (bottom panel) observation of WT and pnh-2 ovules at Stage 3-VI. F, Quantification of female gametophyte phenotype in WT and mutant at Stage 3-VI for each sample. The number of ovules observed is shown in Supplemental Data Set 1. (**P < 0.01 by t test). (G–I) Signal corresponding to the female gametophyte marker pAKV:H2B-YFP (G), the central cell marker pDD65:GFP (H), and the egg cell marker pDD45:GFP (I) in ovules at Stage 3-VI. J–L, The quantification of GFP/YFP signal intensity corresponding to female gametophyte marker pAKV:H2B-YFP (J) central cell marker pDD65:GFP (K) and egg cell marker pDD45:GFP (L) in WT and pnh-2 mutant ovules at Stage 3-VI. Error bars indicate ±SD (n = 10 biological replicates). Different letters above the columns indicate statistically significant differences at P < 0.01, as determined by one-way ANOVA. M, DIC observation of WT and mutant ovules at Stage 3-VI. N, Confocal observation of WT and mutant ovules at Stage 3-VI. O, Quantification of female gametophyte phenotype in WT and mutant at Stage 3-VI for each sample. The number of ovules observed was showed in Supplemental Data Set 1. Different letters above the columns indicate statistically significant differences at P < 0.01, as determined by one-way ANOVA. cn, central cell nucleus; en, egg cell nucleus. Bars in (A), and (B), 10 μm; bar in (D), 1 mm; bars in (E), (M), and (N), 5 μm; bars in (G)–(I), 10 μm. The female gametophyte is outlined by white dot lines in (A), (B), (E), (G)–(I), (M), and (N).
This led to the hypothesis that AGO10 sequesters miR398 in the chalazal region. To test this hypothesis, we performed ovule whole-mount in situ hybridization with the miR398 LNA probe using the ovules of wild type and pnh-2 (a loss-of-function ago10 mutant; Liu et al., 2009). We observed expanded localization of miR398 in pnh-2 ovules, in contrast to the chalaza-specific localization of miR398 in wild-type ovules (Figure 7B). These results demonstrate that AGO10 prevents the movement of miR398 to the female gametophyte by sequestering miR398 in the chalaza.
MiR398 was not detected in the wild-type female gametophyte but ectopically accumulated in the female gametophyte of arp6 er-119, arp6 mpk6, and er-105 mpk6 double mutant ovules (Figure 6A). These findings prompted us to investigate how SWR1 and the ER-MPK pathway might repress the movement of mature miR398 to the female gametophyte. We therefore quantified the transcript levels of AGO10 in the ovules of wild type and the SWR1 and ER-MPK pathway mutants. RT-qPCR analysis showed that AGO10 transcripts were significantly decreased in the arp6 er-119, arp6 mpk6, and er-105 mpk6 double mutants compared to wild type, arp6, er-119, er-105, and mpk6 (Figure 7C). We also crossed pAGO10:YFP-AGO10 into arp6 er-119 and found that the expression of pAGO10:YFP-AGO10 in arp6 er-119 ovules was significantly reduced compared to wild type (Figure 7A), consistent with the RT-qPCR results.
The next question we addressed was whether AGO10 is required for ovule development. Analysis of the ovule morphology of pnh-2 showed reduced fertility (Figure 7D), an aberrant female gametophyte (Figure 7E and F), as indicated by abnormal expression of the female gametophyte marker and the central cell and egg cell markers (Figure 7G–I; significantly different from wild type, P < 0.01, Supplemental Data Set 1) and the GFP/YFP signal was significantly decreased in pnh-2 compared to WT (Figure 7J–L; P < 0.01, Supplemental Data Set 1), suggesting the involvement of AGO10 in female gametophyte development. We also introduced the weak point mutation allele ago10-12 (Ji et al., 2011), which does not show any obvious female gametophyte developmental defects, into arp6, er-105, and arp6 er-105 and observed severe ovule developmental defects in arp6 ago10-12, er-105 ago10-12, and arp6 ago10-12 er-105 ovules (Figure 7M–O). These data indicate that SWR1 and the ER-MPK-signaling pathway also activate AGO10 expression to sequester miR398 in the chalaza to ensure proper female gametophyte development.
Female gametophyte-expressed AGL51/52/78 control sporophytic integument growth
In addition to the female gametophyte defects, we noted that integument growth was also compromised in the agl78, agl51 agl78, and agl52 agl78 mutants. Considering that AGL51/52/78 were specifically expressed in the female gametophyte and not in sporophytic tissues in both developing and mature ovules (Figure 5C and D), these sporophyte integument defects were surprising. To test the possibility that female gametophyte-expressed AGL51/52/78 affect integument development, we characterized integument development in wild type and agl mutant ovules. In wild type, the female gametophyte was fully enveloped by integument tissue, which curled around the gametophyte to form a micropyle close to the funiculus (Supplemental Figure S12A and B). However, in a subset of agl78, agl51 agl78, and agl52 agl78 mutant ovules, the integument did not elongate sufficiently to cover the female gametophyte; as a result, the micropyle did not bend as far as in wild type, leading to an atypical protruding female gametophyte (Supplemental Figure S12A and B; P < 0.01, Supplemental Data Set 1). To determine whether the integument growth defect in the mutants was due to reduced cell proliferation, we stained mature Stage 3-VI ovules with Calcofluor White and quantified the integument cell numbers in the outermost layer. The results revealed significantly fewer cells in the outer integuments of agl78, agl51 agl78, and agl52 agl78 compared to wild type (Supplemental Figure S12C and D; P < 0.01, Supplemental Data Set 1).
We performed two experiments to test whether miR398 and AGL51/52/78 are important for ovule sporophytic tissue development. First, we examined if pUBQ10:MIR398c-OE and pAKV:MIR398c-OE ovules exhibit integument growth defects. Indeed, we detected ovules with reduced integument growth and the protruding embryo sac phenotype (Supplemental Figure S12B), as well as reduced outer integument cell numbers in pUBQ10:MIR398c-OE and pAKV:MIR398c-OE ovules (Supplemental Figure S12B and D; significantly different to wild type, P < 0.01, Supplemental Data Set 1). Second, we knocked down MIR398c and induced AGL78 overexpression driven by native promoter or the female gametophyte cell-specific AKV promoter, and both approaches could complement the protruding female gametophyte and integument defects in arp6 er-119 ovules, to varying degrees (Supplemental Figure S12C and D; Supplemental Data Set 1). These data indicate that in addition to regulating female gametophyte development, AGL51/52/78 also affects sporophytic integument growth.
Since MIR398c acts downstream of SWR1 and the ER-MPK pathway and since the movement of miR398 is dependent on SWR1 and ER-MPK, we investigated whether single and double ARP6 and ER-MPK pathway mutants exhibit integument growth defects. The protruding female gametophyte phenotype was detected in the pie1 and pnh-2 single mutants, but the phenotype was very weak or not detected in the arp6, sef, er-119, and er-105 single mutants (Supplemental Figures S12C and D, S13–S15). However, the arp6 er-119, arp6 er-105, arp6 mpk6, er-105 mpk6, sef er-105, sef mpk6, arp6 ago10-12, and er-105 ago10-12 double mutants and arp6 ago10-12 er-105 triple mutants all exhibited the protruding female gametophyte phenotype, and the outer integument cell numbers were also significantly reduced in these double mutants compared to the single mutants and wild type (Supplemental Figure S12C and D and S13–S15; Supplemental Data Set 1). Taken together, these results indicate that SWR1 and the ER-MPK pathway control female gametophyte-specific AGL51/52/78 expression, which in turn at least partially affects sporophytic integument growth in ovules.
Discussion
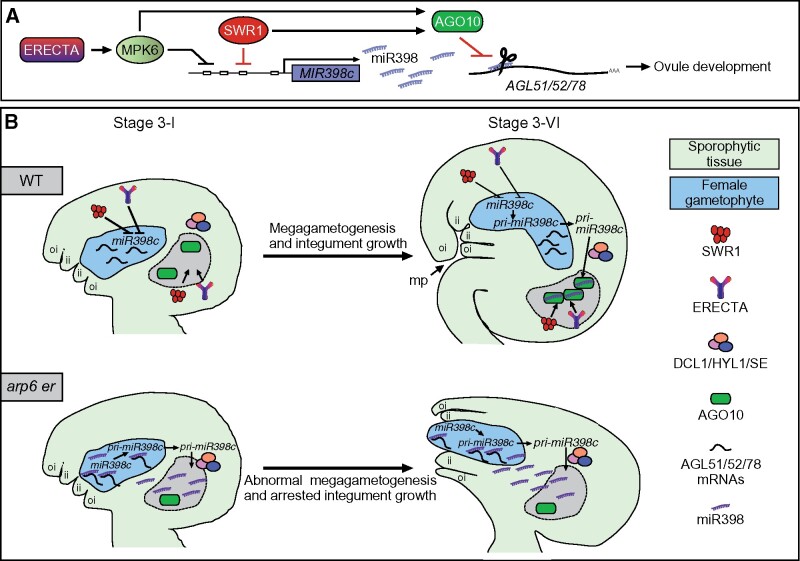
Coordinated cell expansion and cell proliferation are critical for plant growth and establishing organ shape. During ovule development, the coordinated growth of the sporophytic integuments and the gametophytic embryo sac required for ovule morphogenesis has long been believed to be regulated by active signaling events. Here, we provide developmental genetic evidence that ER-MPK signaling, in coordination with the ATP-dependent chromatin-remodeling complex SWR1, regulates ovule development by inhibiting the transcription of pri-miR398 in early stage of megagametogenesis and preventing the accumulation of miR398 inside the female gametophyte (Figure 8). This work highlights the importance of coordination between signal transduction pathways and chromatin-remodeling factors in the temporal control of miRNA biogenesis and spatial control of miRNA distribution during development.
Figure 8.
Proposed model for SWR1 coordination with ER signaling in regulating ovule development through the miR398-AGLs module. A, A diagram showing that SWR1 and ER–MPK6 repress the transcription of MIR398c and the accumulation of miR398. miR398 regulation of its target genes AGL51/52/78 is attenuated by AGO10. The expression of AGO10 is promoted by SWR1 and ER–MPK6. B, A diagram showing the temporal and spatial control of miR398 biogenesis by SWR1 and ER to ensure female gametophyte development and proper ovule morphogenesis. In WT ovules, SWR1 and ER repress the transcription of MIR398c at the early stage of megagametogenesis. In the mature stage of ovule, pri-miR398c is transcribed in the female gametophyte. But, it is translocated from the female gametophyte to surrounding sporophytic tissues, where it is processed by DCL1, HYL1, and SE. SWR1 and ER–MPK6 also activate AGO10 expression in the chalaza, which is essential for sequestering miR398 and preventing its movement into the female gametophyte to access its targets AGL51/52/78. These AGLs are expressed in the female gametophyte and are essential for female gametophyte development as well as integument growth. In arp6 er ovules, increased MIR398c expression results in the over-accumulation of miR398. At the same time, the reduced AGO10 fails to sequester miR398, leading to ectopic accumulation of miR398 in the female gametophyte and inhibition of AGL51/52/78 expression. As a result, these mutant ovules exhibit compromised female gametophyte development and a protruding embryo sac. The chalaza is labeled and outlined by black dot lines in B.
Coordinated activity of SWR1 and the ER-MAPK-signaling pathway controls ovule integument development by repressing MIR398c transcription
Although SWR1 has been showed to be involved in multiple growth and developmental processes (Aslam et al., 2019), it remains unclear how SWR1 modulation of histone–DNA interactions controls gene transcription in specific developmental contexts. Here, we show that MIR398c expression was significantly increased in arp6 er and arp6 mpk6 ovules but not in the arp6 single mutant. Previous studies have shown that ARP6 plays a key role in female meiosis and ∼45% of arp6 ovules have meiotic defects, leading to abnormal female gametophyte development (Qin et al., 2014), which is associated with reduced recombinase DISRUPTED MEIOTIC cDNA1 (DMC1) expression, but not increased MIR398c expression. We revealed that the increased MIR398c expression and reduced AGL51/52/78 expression cause the enhanced female gametophyte defects in arp6 er and arp6 mpk6 double mutants. Therefore, the pAGL78:AGL78m-GFP and pAGL78:AGL78m-GFP constructs could only partially rescue the female gametophyte defects in the arp6 er-119 double mutant. We also showed that overexpression of AGL78 driven by the female gametophyte cell-specific AKV promoter also partially complements the arp6 er-119 double mutant phenotype, indicating that miR398 target genes AGL51/52/78 act downstream of ARP6 and the ER-MPK pathway in the control of ovule development.
We also found that the arp6 mutation caused reduced H2A.Z and nucleosome occupancy at the MIR398c promoter. These observations indicate that through nucleosome dynamics, SWR1 contributes to the transcriptional inhibition of MIR398c, a gene regulated by ER-MPK. However, changes in nucleosome structure alone were not sufficient to alter MIR398c transcription. A recent study revealed that H2A.Z activates the expression of MIR156a/MIR156c in early shoot development (Xu et al., 2018), further indicating that SWR1 contributes to the fine control of plant development by maintaining a balance between miRNAs and their target mRNAs. In addition to altering nucleosome structure, H2A.Z also regulates gene transcription by affecting histone-modification status (Dai et al., 2017; Cai et al., 2019). Consistent with this role, we observed enhanced H3K4me3 and reduced H3K27me3 levels at MIR398c, alongside the reduced H2A.Z level in the arp6 mutant. H2A.Z deposition at MIR398c was independent of ER-MPK signaling (Figure 2). A question for future study is how SWR1 and H2A.Z are specifically recruited at MIR398c.
The ER signal transduction pathway regulates ovule integument and female gametophyte development
MPK3/MPK6 and ER/ERL1/ERL2 regulate similar developmental processes in Arabidopsis. It was previously shown that the ovule integument defects of mpk3+/− mpk6−/− and er erl1 erl2+/− mutants are due to disrupted cell proliferation, suggesting that the MPK3/MPK6 cascade might function downstream of ER family receptors in ovule patterning regulation (Pillitteri et al., 2007; Wang et al., 2008). In this study, we demonstrated that the MPK6 cascade functions in the same pathway as ER in the regulation of ovule morphology and promotes female gametophyte development and integument cell proliferation. The YDA-MKK4/5-MPK3/6 cascade functions downstream of ER/ERLs in regulating inflorescence architecture by promoting cell division in pedicels, as demonstrated by epistasis analysis and gain- and loss-of-function analysis (Meng et al., 2012; Bemis et al., 2013). Correlative evidence also suggests the YDA-MKK4/5-MPK3/6 cascade is required to transduce the signals perceived by the ER receptor family in stomatal development and patterning (Wang et al., 2007; Kim et al., 2012; Pillitteri and Torii, 2012; Lampard et al., 2014). The YDA-MKK4/5-MPK3/6 module may similarly function downstream of ER in ovule development regulation.
Secreted EPF/EPFL peptides have been identified as ligands of ER receptors in multiple developmental processes (Abrash et al., 2011; Lee et al., 2012; Uchida et al., 2012). Given the large number of putative secreted proteins expressed in the developing ovule and the pivotal role of cell-cell communication in ovule development (Jones-Rhoades et al., 2007; Chevalier et al., 2011; Qu et al., 2015), it will be of interest to identify the ligands of the ER family in ovule development regulation. Components downstream of the ER-MPK cascade in stomatal development and substrates of MPK3/6 in stress responses and pollen development have been identified (Lampard et al., 2008; Bethke et al., 2009; Wang et al., 2010; Guan et al., 2014; Zhang et al., 2015). In contrast, the downstream components of the ER-MPK cascade in ovule integument development are largely unknown. Here, we show that the ER-MPK cascade is required to ensure the function of three redundant Mβ subclade MADS box genes AGL51/52/78 in the female gametophyte. Specifically, it represses MIR398c transcription and inhibits miR398 accumulation in the female gametophyte via a gate-keeping mechanism (Figure 8).
MiR398 target genes AGL51/52/78 regulate communication between the female gametophyte and surrounding sporophytic tissue to ensure proper ovule morphogenesis
The development of the haploid female gametophyte is tightly linked to the surrounding sporophyte (Yang and Sundaresan, 2000; Yadegari and Drews, 2004). Many sporophytic mutants with aberrant integument initiation and outgrowth, such as bel1 (BELL1, a homeodomain TF; Reiser et al., 1995), ino (INNER NO OUTER, a YABBY TF; Villanueva et al., 1999), ant (an AP2 TF; Elliott et al., 1996; Klucher et al., 1996), tsl (TOUSLED, a nuclear Ser/Thr protein kinase; Roe et al., 1997), hll (HUELLENOS, a mitochondrial ribosomal protein; Schneitz et al., 1998), and sin1/dcl1 (SHORT INTEGUMENTS1/DICER LIKE1, a ribonuclease; Robinson-Beers et al., 1992; Schauer et al., 2002) also exhibit arrested female gametophyte development, suggesting that integument formation promotes female gametophyte development. However, the effects of female gametophyte on the integument growth are less clear.
Here, we showed that AGL51/52/78, three Mβ subclade MADS-box genes (Parenicova et al., 2003) targeted by miR398, are expressed in the developing female gametophyte. Loss-of-function mutations in AGL51/52/78 disrupt female gametophyte development and inhibit integument cell proliferation, resulting in a protruding embryo sac and compromised ovule morphogenesis and fertility. The coordination of growth between the integuments and the female gametophyte is important for ovule morphogenesis and micropyle formation, which is essential for efficiently sending the attracting signal from the female gametophyte to pollen tube. The protruding female gametophyte phenotype in the double mutants including arp6 er-119, arp6 er-105, arp6 mpk6, er-105 mpk6, sef er-105, and sef mpk6 could cause less successful targeting by pollen tubes. Therefore, greater reductions in seed set are found in double mutants, compared to the frequency of abnormal female gametophytes. Our findings underscore the importance of crosstalk between the female gametophyte and surrounding sporophytic integument tissues during ovule development.
Spatial specificity of pri-miRNA processing and sequestration of miRNAs in sporophytic tissue affect the regulation of target mRNAs
Small RNAs are key signals in plant development, growth, and stress responses (Liu and Chen, 2018). Recently, mobile small RNAs have been characterized as a positional signal to drive developmental patterning (Benkovics and Timmermans, 2014). For example, the specification of leaf adaxial-abaxial polarity, the specification of the root central stele, and the maintenance of shoot stem cell competency all rely on the movement of small RNAs (Juarez et al., 2004; Carlsbecker et al., 2010; Knauer et al., 2013). The movement of small RNAs, such as AGO9-associated 24-nucleotide small RNAs and TAS3-derived transacting small interfering RNAs (ta-siRNAs), has also been shown to be involved in the specification of the megaspore mother cell during the early stages of ovule development (Olmedo-Monfil et al., 2010; Su et al., 2017; Su et al., 2020). Despite the fundamental roles of mobile small RNAs in plant growth and survival, little is known about how their movement is precisely controlled. The proposed processes for the control of small RNA movement include the plasmodesmata, phloem-dependent mechanisms, and a gate-keeping mechanism (Han and Kim, 2016; Skopelitis et al., 2018; Tsikou et al., 2018).
By examining the expression pattern of the MIR398c and miRNA-processing proteins, we found that pri-miR398 is mainly generated in the female gametophyte, but is then translocated to and processed in the surrounding sporophytic tissue, likely in the proximal chalaza. In addition to the reported mature miRNA mobility during a developmental process and rhizobial infection (Skopelitis et al., 2018; Tsikou et al., 2018), here we showed that the pri-miRNA is also mobile. The female gametophyte-to-sporophytic translocation of pri-miRNA and the sporophyte-specific expression pattern of the miRNA-processing proteins may serve as an efficient strategy to prevent the accumulation of miRNAs in the female gametophyte. Although the MIR398c promoter-GUS and -GFP fusions displayed female gametophyte-specific expression of pri-miR398c, some level of pri-miR398c signal in the ovule sporophytic tissues was detected by an in situ hybridization assay. The detection of pri-miR398c in the ovule sporophytic tissues could be due to the translocation of pri-miR398c from the female gametophyte to the ovule sporophytic tissues. The discrepancy could also be due to the absence of additional regulatory elements in the GUS and GFP fusions. Thus, the possibility of pri-miR398c production in the sporophyte is not completely ruled out. Nevertheless, based on the results from LCM followed by RT-qPCR analysis, gametophyte-to-sporophyte translocation of pri-miR398c is also not excluded.
AGO10 can lock or sequester miRNAs by preventing their normal regulatory roles, thereby helping to produce positional signals for multiple developmental processes (Manavella et al., 2011). The spatiotemporal sequestration of miR165/166 by AGO10 from their target HD-ZIP III genes in regulating shoot apical meristem maintenance was reported (Zhu et al., 2011; Zhou et al., 2015). In addition to association with miR165/166, AGO10 can also directly bind other miRNAs, including miR398 (Yu et al., 2017). Here, we further revealed that during ovule development, AGO10 sequesters miR398 in the chalaza to maintain the function of its target AGL genes in the female gametophyte. A recent study reported that miR165/166 also accumulates in the chalaza (Hashimoto et al., 2018), indicating that chalaza-localized AGO10 may serve as a gatekeeper that controls miRNA movement between the female gametophyte and sporophytic tissues. Notably, ectopic miR398 in the female gametophyte was detected in the arp6 er-119, arp6 mpk6, and er-105 mpk6 double mutants. We demonstrate that AGO10 expression in the ovule chalaza is activated by SWR1 and the ER-MPK pathway, suggesting that AGO10 mediated sequestration of miR398 in chalaza is dependent on ARP6 and the ER-MPK-signaling pathway, providing new insights into the mechanisms underlying small RNA movement during plant development and the reciprocal communication between these tissues.
Materials and methods
Plant material and growth conditions
The Arabidopsis thaliana ecotype Columbia (Col) was used as wild type. All mutants are in the Col background except for ago10-12 (hen6), which is in the Landsberg (Ler) background. The mutants and alleles used in this study were arp6 (Qin et al., 2014), sef (CS822749), pie1 (Salk_096434), agl51 (Salk_011841C), agl52 (Salk_030570C), agl78 (Salk_020476C), mir398a (CS433641), mir398b (SALK_122007), mir398c (Salk_038698C), dcl1-7 (CS3089), se-1 (CS3257), hyl1-2 (SALK_064863; from the Arabidopsis Biological Resource Center; ABRC), er-105 (Shpak et al., 2005), mpk6 (Liu and Zhang, 2004; Wang et al., 2007), pnh-2 (Liu et al., 2009), and ago10-12 (Ji et al., 2011). Plants were grown under 16 h light/8 h dark at 22°C.
Differential interference contrast observation of ovule structure
Ovules from WT, mutant, and transgenic plants were dissected from the pistils of Stages 8–13 flowers in a drop of chloral hydrate solution (chloral hydrate:H2O:glycerol = 8:2:1; Zhao et al., 2014). Cleared ovules were observed under a differential interference contrast (DIC) microscope and imaged using a ZEISS (Imager.A2) microscope with DIC optics.
Preparation of ovules for confocal laser scanning microscopy
For confocal fluorescence microscopy, ovules were prepared according to a previously reported method (Christensen et al., 1997) and mounted in 30% glycerol with 5 μM FM4-64 dye. The ovules were stained for 5 min and analyzed using a Leica TCS SP8 microscope.
Integument cell number counts
Ovules were analyzed after staining with a 0.1% solution of Calcofluor White for cell wall examination (Adamski et al., 2009) and imaged using a Leica SP8 confocal microscope. Cell number in the outmost layer of the outer integument was counted from the distal end (micropylar ends) to the proximal end (chalazal end). Significant differences were calculated by one-way ANOVA (P < 0.01; n = 6).
Plasmid construction
The pMIR398c:GFP/GUS constructs were generated by amplifying a 2-kb sequence upstream of the MIR398c gene from WT genomic DNA using the primers listed in Supplemental Table S1. The PCR product was cloned into the pENTR/D-TOPO vector (Invitrogen). pENTR/D-TOPO clones were recombined into the destination vectors pGWB604 (Nakagawa et al., 2007) and pGWB533 (Nakagawa et al., 2007) using LR Clonase II (Invitrogen). The PCR fragments were cloned into the pENTR/D-TOPO vector (Invitrogen), and the pENTR/D-TOPO clones were recombined into the destination vector pGWB502 (Nakagawa et al., 2007) using LR Clonase II (Invitrogen). pMIR398a: GUS and pMIR398b:GFP were generated by amplifying 681-bp and 608-bp upstream of the MIR398a and MIR398b gene sequences, respectively, from WT genomic DNA using the primers listed in Supplemental Table S1.
The PCR fragments were cloned into the pENTR/D-TOPO vector (Invitrogen), and the pENTR/D-TOPO clones were recombined into the destination vector pGWB533 and pGWB604 using LR Clonase II (Invitrogen). pAGL51:GUS, pAGL52:GUS, and pAGL78:GUS were generated by amplifying 1452-bp, 1594-bp, and 1490-bp upstream of the AGL51/52/78 genes, respectively, from WT genomic DNA using the primers listed in Supplemental Table S1. The PCR products were cloned into the pENTR/D-TOPO vector (Invitrogen). pENTR/D-TOPO clones were recombined into the destination vector pGWB533 using LR Clonase II (Invitrogen). pAGL51:AGL51-GFP, pAGL52:AGL52-GFP, pAGL78:AGL78-GFP, and pAGL78:AGL78m:GFP were generated by amplifying 2040 bp, 2581-bp, 2513-bp, and 2513-bp sequences, respectively, from WT genomic DNA and cDNA using the primers listed in Supplemental Table S1. The PCR products were cloned into the pENTR/D-TOPO vector (Invitrogen). pENTR/D-TOPO clones were recombined into the destination vector pGWB604 using LR Clonase II (Invitrogen). pAGL78:AGL78 was generated by amplifying a 2516-bp sequence from WT genomic DNA using the primers listed in Supplemental Table S1. The PCR product was cloned into the pENTR/D-TOPO vector (Invitrogen). pENTR/D-TOPO clones were recombined into the destination vector pGWB502 using LR Clonase II (Invitrogen). The PCR product was cloned into the pENTR/D-TOPO vector (Invitrogen). pENTR/D-TOPO clones were recombined into the destination vector pGWB518 (Nakagawa et al., 2007) using LR Clonase II (Invitrogen). pUBQ10:MIR398c was generated by amplifying a 638-bp sequence of UBQ10 promoter from WT genomic DNA and a 2375 bp of the MIR398c gene sequences using the primers listed in Supplemental Table S1. The PCR product was cloned into the pENTR/D-TOPO vector (Invitrogen). pENTR/D-TOPO clones were recombined into the destination vector pGWB501(Nakagawa et al., 2007) using LR Clonase II (Invitrogen). pAKV:AGL78 was generated by amplifying a 1854-bp sequence of AKV promoter from WT genomic DNA and a 1026-bp sequence of AGL78 CDS from cDNA using the primers listed in Supplemental Table S1. The PCR product was cloned into the pENTR/D-TOPO vector (Invitrogen). pENTR/D-TOPO clones were recombined into the destination vector pGWB501 using LR Clonase II (Invitrogen). The pER:ER construct was generated by amplifying a 7634-bp segment of ER genomic DNA using the primers listed in Supplemental Table S1. The PCR product was then cloned into pENTR/D-TOPO vector (Invitrogen) and recombined into the destination vectors pGWB404 using LR Clonase II (Invitrogen). pAKV:MIR398c was generated by amplifying an 1854-bp sequence of AKV promoter from WT genomic DNA and a 2375 bp of the MIR398c gene sequences using the primers listed in Supplemental Table S1. The PCR product was cloned into the pENTR/D-TOPO vector (Invitrogen). pENTR/D-TOPO clones were recombined into the destination vector pGWB501 using LR Clonase II (Invitrogen). pDCL1:3×VENUS-N7 were generated by amplifying 2000-bp upstream of the DCL1 gene sequence from WT genomic DNA using the primers listed in Supplemental Table S1. The PCR fragments were cloned into the pSPL4 3xVENUS-N7 vector with BamHI and XbaI restriction sites (Heisler et al., 2005).
Generation of transgenic plants and marker lines in WT or mutants background
The GV3101-Agrobacterium with the pER:ER or AGL78-relative complementation vectors was used to transform the arp6+/− er-119 +/− double heterozygote, and the transformants were selected by respective antibiotics. The arp6 er-119 double homozygote plants with the pER:ER or AGL78-relative complementation vectors were genotyped and selected. At least 10 independent lines were obtained for every complementation construct. The GV3101-Agrobacterium with the pUBQ10:MIR398c or pAKV:MIR398c vectors was used to transform the WT, and the transformants were identified by selecting seedlings on Murashige and Skoog (MS) plates containing hygromycin at 50 µgml−1. At least 10 independent T3 homozygous lines were used for phenotypic characterization. The overexpression lines were selected and established with similar methods, and T3 generation plants were used for phenotypic analysis. Various female gametophyte-specific marker lines were introgressed into the arp6 er-119, arp6 mpk6, er-105 mpk6, agl78, and pnh-2 background by crossing homozygous mutant plants to the marker lines and allowing the F1 to self-fertilize. PCR genotyping of the segregating F2 population was performed, and the marker gene was identified under a fluorescence microscope. F3 homozygous plants (mutant and marker lines both homozygous) were used in this study. The homozygous status of the plants was confirmed by the corresponding antibiotic selection marker in the transgene and Mendelian inheritance segregation ratios of the selection markers. At least 10 independent F3 generation lines crossed with markers were obtained for phenotypic analysis.
RNA-seq and analysis of differentially expressed genes
Ovaries with ovules from Stage 2-III to Stage 3-VI were collected from Stage 8 to Stage 12 WT, arp6, er-105, and arp6 er-119 flower buds. RNA isolation, sequencing, and data processing were conducted as previously described (Zhao et al., 2014). We used the TAIR10 Arabidopsis thaliana genome as the reference. Clean reads were aligned to the reference genome using STAR v2.5.0. The alignment results were processed using the SourceForge Subread package featureCount v1.5.0 for gene quantification. Finally, edgeR v3.12.0 was used to identify the differentially expressed genes (fold change ≥ 2; a value of FDR ≤ 0.05 was considered to be statistically significant) between samples.
RT-qPCR
The RNA samples were extracted from WT and mutants’ ovaries (with ovules from Stage 2-III to Stage 3-VI). In order to determine the relative transcript levels of selected genes, real-time qPCR was performed with specific primers (Supplemental Table S1) according to manufacturer’ instructions using the BIO-RAD real-time PCR system and the SYBR Premix Ex Taq II system (TaKaRa), and the details were described (Cai et al., 2017). The expression levels were analyzed by the Livak method (Livak and Schmittgen, 2001). Data are represented as the normalized relative expression level (2−△△CT) of the respective genes in various samples. The relative transcript levels of the analyzed genes were normalized to the transcript levels of HK2 (AT4G26410; Cai et al., 2017). Three biological replicates and two technical replicates for each sample were performed in the qPCR experiments.
In situ hybridization of whole-mount ovules
Ovules with placenta were dissected from Stage 8 to 12 pistils, fixed, and processed as previously described (Hejatko et al., 2006; Javelle and Timmermans, 2012). For the MIR398c and AGL78 probe, 299-bp and 263-bp fragments were cloned into the pTA2 vector (Toyobo), respectively. The PCR primers used are listed in Supplemental Table S1. PCR products were amplified using ExTaq DNA polymerase, and then cloned into the linearized pTA2 vector. For in situ localization of small RNAs, LNA oligonucleotide probes were used. miR398 DIG-labeled LNA probe was ordered directly from Exiqon (http://www.exiqon.com/microrna-in-situ-Hybridization).
ChIP-qPCR
ChIP was performed as previously described (Qin et al., 2014) using inflorescences with floral buds younger than 12c stage. Polyclonal H2A.Z antibodies (Deal et al., 2007), Pol II antibodies (Abcam, ab817), anti-H3K27me3 (Millipore, 07-449), and anti-H3K4me3 (Millipore, 07-473) were used. The relative enrichment of H2A.Z, H3K27me3, H3K4me3, and Pol II-associated DNA fragments was analyzed by qPCR as previously described (Qin et al., 2014). Our previous genome-wide analysis of the distribution patterns of H2A.Z, H3K4me3, H3K27me3 mark, and nucleosome occupancy had shown that H2A.Z regulates gene expression by modulating nucleosome structure at +1 nucleosome and -1 nucleosome in association with H3K4me3 and H3K27me3 (Dai et al., 2017). Therefore, the five regions (1 to 5) of the MIR398c locus across the promoter, -1 nucleosome, +1 nucleosome, and gene body were selected for ChIP-qPCR analysis. ChIP experiments results were calculated by the Fold Enrichment Method. Fold Enrichment value was obtained as follows: ChIP fraction Ct value for the normalized background (NIS/mock IP) fraction Ct value (first △△Ct), △△Ct [CHIP/NIS]= △Ct [normalized CHIP] - △Ct [normalized NIS/mock]. Fold Enrichment = 2(−△△Ct [CHIP/NIS]; Cai et al., 2019). Fold enrichment of ChIP experiments was calculated using HK2 (AT4G26410) gene as a reference. Primers used in ChIP-qPCR are listed in Supplemental Table S1.
Nucleosome occupancy
MNase digestion of enriched chromatin followed by qPCR was performed as previously described (Kumar and Wigge, 2010) with minor modifications. Oligos were designed to have 95–110-bp amplicons every 35–45 bp in MIR398c. Chromatin from inflorescences with floral buds younger than 12c stage was digested with MNase, and mononucleosome-sized fragments were gel purified and used in qPCR. Relative nucleosome occupancy was represented as the fraction of uncut chromatin DNA and was plotted against the MIR398c gene positions relative to the transcription start sites (TSS) for each primer pair; the position denotes the center of each amplicon. Oligonucleotide sequences used are provided in Supplemental Table S1.
5′-RACE PCR
To experimentally validate computationally predicted miRNA targets, 5′-RACE PCR was performed (Nadiya et al., 2018). The ligated product was reverse transcribed using 5′-RACE outer primer complementary to the linker and a gene-specific outer primer, followed by PCR amplification using 5′RACE inner primer and gene-specific inner primer. RACE products were purified using the Agarose Gel DNA Purification Kit (TaKaRa Bio), ligated into the pMD18-T vector (TaKaRa Bio), and sequenced. Gene-specific primers used are listed in Supplemental Table S1.
LCM
Ovaries from WT and se-1 mutant with ovules at mature Stage 3-VI and meiosis Stage 2-III fixed, embedded, and processed as previously described (Tang et al., 2012). TargetAmpTM 2-Round aminoally-aRNA amplification kit 1.0 (Epicentre, TAA2R4924) and Qiagen RNeasy MinElute Cleanup Kit (Qiagen, 74204), SuperScript II ReverseTranscriptase (Invitrogen, 18064-014), and SuperScript III ReverseTranscriptase (Invitrogen, 18064-044) were used for RNA amplification.
GUS staining
Inflorescence samples or pistils pollinated with pLAT52:GUS pollen and pMIR398c:GUS transgenic plant inflorescence and ovules samples were fixed, stained, and processed as previously described (Cai et al., 2017). After the staining, the ovules were observed under a ZEISS (Imager.A2) microscope.
Statistical analysis
All t-test analysis was conducted using Excel and the ANOVA analysis was conducted using SPSS software. To determine statistical significance, we employed independent t-tests with two-tail distribution between two groups and one-way ANOVA Turkey’s test among various genotypes. A value of P < 0.05 was considered to be statistically significant. The results of statistical analyses are shown in Supplemental Data Set 1.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ARP6 (AT3G33520), ERECTA (AT2G26330), PIE1 (AT3G12810), SEF (AT5G37055), MPK6 (AT2G43790), MIR398a (AT2G03445), MIR398b (AT5G14545), MIR398c (AT5G14565), AGL78 (AT5G65330), AGL51 (AT4G02235), AGL52 (AT4G11250), AGO10 (AT5G43810), DCL1 (AT1G01040), HYL1 (AT1G09700), and SE (AT2G27100). The RNA-seq data are deposited in the European Nucleotide Archive under accession number PRJEB42278.
Supplemental data
Supplemental Figure S1. SWR1 and ER-MPK pathway double mutants exhibited low seed set and compromised female gametophyte development.
Supplemental Figure S2. The reduced seed set of arp6 er-119 is due to female defects.
Supplemental Figure S3. The expression of the female gametophyte marker (pAKV:H2B-YFP), central cell marker pDD65:GFP, and egg cell marker pDD45:GFP is altered in arp6 mpk6 and er-105 mpk6 ovules.
Supplemental Figure S4. The functions of MIR398a and MIR398b are different from MIR398c.
Supplemental Figure S5. The expression of pMIR398c:GUS in arp6+/− er-119+/− ovule at Stage 3-VI is comparable to that in wild type.
Supplemental Figure S6. The expression of ARP6, ER, and MPK6 in the ovules.
Supplemental Figure S7. The expression patterns of MIR398a and MIR398b in flower buds and ovules.
Supplemental Figure S8. The expression pattern of AGL51/52/78 in female gametophyte.
Supplemental Figure S9. The pAGL78:AGL78-GFP construct is sufficient to complement the fertility defects in agl78.
Supplemental Figure S10. dcl1-7+/− heterozygote exhibited normal phenotype in ovule structure and plant fertility.
Supplemental Figure S11. The expression of pHYL1:HYL1-YFP is not affected in arp6 er-119.
Supplemental Figure S12. AGL51/52/78 and MIR398c also regulate sporophytic integument growth.
Supplemental Figure S13. The er-119 mutation enhanced the integument growth defects in SWR1 complex mutants.
Supplemental Figure S14. AGO10 regulates sporophytic integument growth.
Supplemental Figure S15. The ago10 mutation enhanced the integument growth defects of the arp6 er mutant.
Supplemental Table S1. Primers used in this study.
Supplemental Data Set 1. The comparison and statistical results.
Supplemental Data Set 2. RNA-seq expression values of the differentially expressed genes (DEGs) in arp6 er-119 double mutant ovules compared to arp6.
Supplementary Material
Acknowledgments
We thank Dr. K. Torii for sharing the er-105 seeds and Dr. R. Deal for sharing the H2A.Z antibody. We thank Dr. W. Tang for providing the LCM platform and technical guidance.
Funding
This work was supported by NSFC (grant nos. 31970333, U1605212 to Y.Q.; 31700279 to H.C.), Guangxi Distinguished Experts Fellowship and Science and Technology Major Project of Guangxi (Gui Ke 2018-266-Z01) to Y.Q., and NIH GM129373 to X.C.
Conflict of interest statement: The authors declare no competing interests.
H.C. cloned the genes; generated the transgenic lines; and performed phenotypic, expression, and genetic analyses. L.L. performed expression and genetic analyses. M.Z., M.C., F.C., M.Y., and Z.S. were involved in the phenotypic and genetic analyses. X.C., R.P., and I.H. interpreted the data and revised the manuscript. Y.Q. and H.C. designed the research and wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Yuan Qin (yuanqin@fafu.edu.cn).
References
- Abrash EB, Davies KA, Bergmann DC (2011) Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand-receptor interactions. Plant cell 23: 2864–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamski NM, Anastasiou E, Eriksson S, O'Neill CM, Lenhard M (2009) Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc Natl Acad Sci USA 106: 20115–20120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armenta-Medina A, Huanca-Mamani W, Sanchez-Leon N, Rodriguez-Arevalo I, Vielle-Calzada JP (2013) Functional analysis of sporophytic transcripts repressed by the female gametophyte in the ovule of Arabidopsis thaliana. PLoS One 8: e76977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam M, Fakher B, Jakada BH, Cao S, Qin Y (2019) SWR1 chromatin remodeling complex: a key transcriptional regulator in plants. Cells 8:1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SC, Robinson-Beers K, Villanueva JM, Gaiser JC, Gasser CS (1997) Interactions among genes regulating ovule development in Arabidopsis thaliana. Genetics 145: 1109–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis SM, Lee JS, Shpak ED, Torii KU (2013) Regulation of floral patterning and organ identity by Arabidopsis ERECTA-family receptor kinase genes. J Exp Bot 64:5323–5333. [DOI] [PubMed] [Google Scholar]
- Bencivenga S, Colombo L, Masiero S (2011) Cross talk between the sporophyte and the megagametophyte during ovule development. Sex Plant Reprod 24:113–121. [DOI] [PubMed] [Google Scholar]
- Bencivenga S, Simonini S, Benkova E, Colombo L (2012) The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. Plant Cell 24: 2886–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkovics AH, Timmermans MC (2014) Developmental patterning by gradients of mobile small RNAs. Curr Opin Genet Dev 27: 83–91. [DOI] [PubMed] [Google Scholar]
- Berr A, McCallum EJ, Menard R, Meyer D, Fuchs J, Dong A, Shen WH (2010) Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell 22: 3232–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke G, Unthan T, Uhrig JF, Poschl Y, Gust AA, Scheel D, Lee J (2009) Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc Natl Acad Sci USA 106: 8067–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Zhang M, Chai M, He Q, Huang X, Zhao L, Qin Y (2019) Epigenetic regulation of anthocyanin biosynthesis by an antagonistic interaction between H2A.Z and H3K4me3. New Phytol 221:295–308. [DOI] [PubMed] [Google Scholar]
- Cai H, Zhao L, Wang L, Zhang M, Su Z, Cheng Y, Zhao H, Qin Y (2017) ERECTA signaling controls Arabidopsis inflorescence architecture through chromatin-mediated activation of PRE1 expression. New Phytol 214:1579–1596. [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vaten A, Thitamadee S, et al. (2010) Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter B, Henderson JT, Svedin E, Fiers M, McCarthy K, Smith A, Guo C, Bishop B, Zhang H, Riksen T, et al. (2016) Cross-talk between sporophyte and gametophyte generations is promoted by CHD3 chromatin remodelers in Arabidopsis thaliana. Genetics 203: 817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Mathews DE, Schaller GE, Kieber JJ (2013) Cytokinin-dependent specification of the functional megaspore in the Arabidopsis female gametophyte. Plant J 73: 929–940. [DOI] [PubMed] [Google Scholar]
- Chevalier E, Loubert-Hudon A, Zimmerman EL, Matton DP (2011) Cell-cell communication and signalling pathways within the ovule: from its inception to fertilization. New Phytol 192:13–28. [DOI] [PubMed] [Google Scholar]
- Choi K, Park C, Lee J, Oh M, Noh B, Lee I (2007) Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134: 1931–1941. [DOI] [PubMed] [Google Scholar]
- Choi K, Kim S, Kim SY, Kim M, Hyun Y, Lee H, Choe S, Kim SG, Michaels S, Lee I (2005) SUPPRESSOR OF FRIGIDA3 encodes a nuclear ACTIN-RELATED PROTEIN6 required for floral repression in Arabidopsis. Plant Cell 17: 2647–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Zhao X, Kelly KA, Venn O, Higgins JD, Yelina NE, Hardcastle TJ, Ziolkowski PA, Copenhaver GP, Franklin FC, . et al. (2013) Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat Genet 45: 1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CA, King EJ, Jordan JR, Drews GNJSPR (1997) Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sexual Plant Reproduct 10: 49–64 [Google Scholar]
- Coen O, Fiume E, Xu W, De Vos D, Lu J, Pechoux C, Lepiniec L, Magnani E (2017) Developmental patterning of the sub-epidermal integument cell layer in Arabidopsis seeds. Development 144: 1490–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Masiero S, Vanzulli S, Lardelli P, Kater MM, Colombo L (2008) AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. Plant J 54: 1037–1048. [DOI] [PubMed] [Google Scholar]
- Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39:W155–W–159.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Bai Y, Zhao L, Dou X, Liu Y, Wang L, Li Y, Li W, Hui Y, Huang X, et al. (2017) H2A.Z represses gene expression by modulating promoter nucleosome structure and enhancer histone modifications in Arabidopsis. Mol Plant 10:1274–1292. [DOI] [PubMed] [Google Scholar]
- Deal RB, Kandasamy MK, McKinney EC, Meagher RB (2005) The nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant Cell 17: 2633–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Topp CN, McKinney EC, Meagher RB (2007) Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19: 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas DV, Bartel B (2008) Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant Mol Biol 67:403–417. [DOI] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR (1996) AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8:155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Spector DL (2007) Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol 17: 818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden TA, Schauer SE, Lang JD, Pien S, Mushegian AR, Grossniklaus U, Meinke DW, Ray A (2002) SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol 130: 808–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Meng X, Khanna R, LaMontagne E, Liu Y, Zhang S (2014) Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genet 10: e1004384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Kim JY (2016) Integrating hormone- and micromolecule-mediated signaling with plasmodesmal communication. Mol Plant 9: 46–56. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Miyashima S, Sato-Nara K, Yamada T, Nakajima K (2018) Functionally diversified members of the mir165/6 gene family regulate ovule morphogenesis in Arabidopsis thaliana. Plant Cell Physiol 59: 1017–1026. [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911. [DOI] [PubMed] [Google Scholar]
- Hejatko J, Blilou I, Brewer PB, Friml J, Scheres B, Benkova E (2006) In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat Protoc 1: 1939–1946. [DOI] [PubMed] [Google Scholar]
- Javelle M, Timmermans MC (2012) In situ localization of small RNAs in plants by using LNA probes. Nat Protoc 7: 533–541. [DOI] [PubMed] [Google Scholar]
- Ji L, Liu X, Yan J, Wang W, Yumul RE, Kim YJ, Dinh TT, Liu J, Cui X, Zheng B. et al. (2011) ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet 7: e1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AJ, Meier P, Gheyselinck J, Wuest SE, Federer M, Schlagenhauf E, Becker JD, Grossniklaus U (2007) Genetic subtraction profiling identifies genes essential for Arabidopsis reproduction and reveals interaction between the female gametophyte and the maternal sporophyte. Genome Biol 8: R204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Borevitz JO, Preuss D (2007) Genome-wide expression profiling of the Arabidopsis female gametophyte identifies families of small, secreted proteins. PLoS Genet 3: 1848–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MC (2004) microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428: 84–88. [DOI] [PubMed] [Google Scholar]
- Kim TW, Michniewicz M, Bergmann DC, Wang ZY (2012) Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482: 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita-Tsujimura K, Kakimoto T (2011) Cytokinin receptors in sporophytes are essential for male and female functions in Arabidopsis thaliana. Plant Signal Behav 6: 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL (1996) The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8:137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer S, Holt AL, Rubio-Somoza I, Tucker EJ, Hinze A, Pisch M, Javelle M, Timmermans MC, Tucker MR, Laux T (2013) A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev Cell 24:125–132. [DOI] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA (2010) H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147. [DOI] [PubMed] [Google Scholar]
- Lampard GR, Macalister CA, Bergmann DC (2008) Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116. [DOI] [PubMed] [Google Scholar]
- Lampard GR, Wengier DL, Bergmann DC (2014) Manipulation of mitogen-activated protein kinase kinase signaling in the Arabidopsis stomatal lineage reveals motifs that contribute to protein localization and signaling specificity. Plant Cell 26: 3358–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A, Gomez-Zambrano A, Lopez-Gonzalez L, Pineiro M, Jarillo JA (2008) Mutations in the Arabidopsis SWC6 gene, encoding a component of the SWR1 chromatin remodelling complex, accelerate flowering time and alter leaf and flower development. J Exp Bot 59: 653–666. [DOI] [PubMed] [Google Scholar]
- Lee JS, Kuroha T, Hnilova M, Khatayevich D, Kanaoka MM, McAbee JM, Sarikaya M, Tamerler C, Torii KU (2012) Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev 26: 126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lituiev DS, Krohn NG, Muller B, Jackson D, Hellriegel B, Dresselhaus T, Grossniklaus U (2013) Theoretical and experimental evidence indicates that there is no detectable auxin gradient in the angiosperm female gametophyte. Development 140: 4544–4553. [DOI] [PubMed] [Google Scholar]
- Liu L, Chen X (2018) Intercellular and systemic trafficking of RNAs in plants. Nat Plants 4: 869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Yao X, Pi L, Wang H, Cui X, Huang H (2009) The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J 58: 27–40. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056. [DOI] [PubMed] [Google Scholar]
- Luo YX, Hou XM, Zhang CJ, Tan LM, Shao CR, Lin RN, Su YN, Cai XW, Li L, Chen S, et al. (2020) A plant-specific SWR1 chromatin-remodeling complex couples histone H2A.Z deposition with nucleosome sliding. EMBO J 39: e102008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella PA, Weigel D, Wu L (2011) Argonaute10 as a miRNA locker. Cell 145: 173–174. [DOI] [PubMed] [Google Scholar]
- March-Diaz R, Reyes JC (2009) The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol Plant 2: 565–577. [DOI] [PubMed] [Google Scholar]
- Marques M, Laflamme L, Gervais AL, Gaudreau L (2010) Reconciling the positive and negative roles of histone H2A.Z in gene transcription. Epigenetics 5: 267–272. [DOI] [PubMed] [Google Scholar]
- Meng X, Wang H, He Y, Liu Y, Walker JC, Torii KU, Zhang S (2012) A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24: 4948–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C (2004) ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348. [DOI] [PubMed] [Google Scholar]
- Nadiya F, Anjali N, Thomas J, Gangaprasad A, Sabu KK (2019) Deep sequencing identified potential miRNAs involved in defense response, stress and plant growth characteristics of wild genotypes of cardamom. Plant Biol 21: 3–14 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y. et al. (2007) Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100. [DOI] [PubMed] [Google Scholar]
- Ojolo SP, Cao S, Priyadarshani S, Li W, Yan M, Aslam M, Zhao H, Qin Y (2018) Regulation of plant growth and development: A review from a chromatin remodeling perspective. Front Plant Sci 9: 1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo-Monfil V, Duran-Figueroa N, Arteaga-Vazquez M, Demesa-Arevalo E, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Vielle-Calzada JP (2010) Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464: 628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Alandete-Saez M, Bowman JL, Sundaresan V (2009) Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science 324: 1684–1689. [DOI] [PubMed] [Google Scholar]
- Parenicova L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B. et al. (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15: 1538–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT (2008) Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134: 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri LJ, Torii KU (2012) Mechanisms of stomatal development. Annu Rev Plant Biol 63: 591–614. [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Bemis SM, Shpak ED, Torii KU (2007) Haploinsufficiency after successive loss of signaling reveals a role for ERECTA-family genes in Arabidopsis ovule development. Development 134: 3099–3109. [DOI] [PubMed] [Google Scholar]
- Potok ME, Wang Y, Xu L, Zhong Z, Liu W, Feng S, Naranbaatar B, Rayatpisheh S, Wang Z, Wohlschlegel JA. et al. (2019) Arabidopsis SWR1-associated protein methyl-CpG-binding domain 9 is required for histone H2A.Z deposition. Nat Commun 10: 3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Zhao L, Skaggs MI, Andreuzza S, Tsukamoto T, Panoli A, Wallace KN, Smith S, Siddiqi I, Yang Z. et al. (2014) ACTIN-RELATED PROTEIN6 regulates female meiosis by modulating meiotic gene expression in Arabidopsis. Plant Cell 26: 1612–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu LJ, Li L, Lan Z, Dresselhaus T (2015) Peptide signalling during the pollen tube journey and double fertilization. J Exp Bot 66: 5139–5150. [DOI] [PubMed] [Google Scholar]
- Rabiger DS, Drews GN (2013) MYB64 and MYB119 are required for cellularization and differentiation during female gametogenesis in Arabidopsis thaliana. PLoS Genet 9: e1003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser L, Modrusan Z, Margossian L, Samach A, Ohad N, Haughn GW, Fischer RL (1995) The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell 83: 735–742. [DOI] [PubMed] [Google Scholar]
- Robinson-Beers K, Pruitt RE, Gasser CS (1992) Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell 4: 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe JL, Nemhauser JL, Zambryski PC (1997) TOUSLED participates in apical tissue formation during gynoecium development in Arabidopsis. Plant Cell 9: 335–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa M, Von Harder M, Cigliano RA, Schlogelhofer P, Mittelsten Scheid O (2013) The Arabidopsis SWR1 chromatin-remodeling complex is important for DNA repair, somatic recombination, and meiosis. Plant Cell 25: 1990–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer SE, Jacobsen SE, Meinke DW, Ray A (2002) DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci 7: 487–491. [DOI] [PubMed] [Google Scholar]
- Schneitz K, Hulskamp M, Pruitt RE (1995) Wild‐type ovule development in Arabidopsis thaliana: a light microscope study of cleared whole‐mount tissue. Plant J 7: 731–749. [Google Scholar]
- Schneitz K, Baker SC, Gasser CS, Redweik A (1998) Pattern formation and growth during floral organogenesis: HUELLENLOS and AINTEGUMENTA are required for the formation of the proximal region of the ovule primordium in Arabidopsis thaliana. Development 125: 2555–2563. [DOI] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU (2005) Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293. [DOI] [PubMed] [Google Scholar]
- Sijacic P, Holder DH, Bajic M, Deal RB (2019) Methyl-CpG-binding domain 9 (MBD9) is required for H2A.Z incorporation into chromatin at a subset of H2A.Z-enriched regions in the Arabidopsis genome. PLoS Genet 15: e1008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopelitis DS, Hill K, Klesen S, Marco CF, von Born P, Chitwood DH, Timmermans MCP (2018) Gating of miRNA movement at defined cell-cell interfaces governs their impact as positional signals. Nat Commun 9: 3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Wang N, Hou Z, Li B, Li D, Liu Y, Cai H, Qin Y, Chen X (2020) Regulation of female germline specification via small RNA mobility in Arabidopsis. Plant Cell 32: 2842–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Zhao L, Zhao Y, Li S, Won S, Cai H, Wang L, Li Z, Chen P, Qin Y, Chen X (2017) The THO complex non-cell-autonomously represses female germline specification through the TAS3-ARF3 module. Curr Biol 27: 1597–1609 e1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WH, Zhang Y, Duvick J (2012) The application of laser microdissection to profiling fungal pathogen gene expression in planta. Methods Mol Biol 835: 219–236. [DOI] [PubMed] [Google Scholar]
- Tsikou D, Yan Z, Holt DB, Abel NB, Reid DE, Madsen LH, Bhasin H, Sexauer M, Stougaard J, Markmann K (2018) Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 362: 233–236. [DOI] [PubMed] [Google Scholar]
- Uchida N, Lee JS, Horst RJ, Lai HH, Kajita R, Kakimoto T, Tasaka M, Torii KU (2012) Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc Natl Acad Sci USA 109: 6337–6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Daal A, White EM, Elgin SC, Gorovsky MA (1990) Conservation of intron position indicates separation of major and variant H2As is an early event in the evolution of eukaryotes. J Mol Evol 30: 449–455. [DOI] [PubMed] [Google Scholar]
- Villanueva JM, Broadhvest J, Hauser BA, Meister RJ, Schneitz K, Gasser CS (1999) INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev 13: 3160–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu Y, Bruffett K, Lee J, Hause G, Walker JC, Zhang S (2008) Haplo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. Plant Cell 20: 602–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Du Y, Li Y, Ren D, Song CP (2010) Hydrogen peroxide-mediated activation of MAP kinase 6 modulates nitric oxide biosynthesis and signal transduction in Arabidopsis. Plant Cell 22: 2981–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Tian Q, Reed JW (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133: 4211–4218. [DOI] [PubMed] [Google Scholar]
- Xu M, Leichty AR, Hu T, Poethig RS (2018) H2A.Z promotes the transcription of MIR156A and MIR156C in Arabidopsis by facilitating the deposition of H3K4me3. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari R, Drews GN (2004) Female gametophyte development. Plant Cell 16 Suppl: S133–S–141.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, Sundaresan V (2000) Genetics of gametophyte biogenesis in Arabidopsis. Curr Opin Plant Biol 3: 53–57. [DOI] [PubMed] [Google Scholar]
- Yu Y, Ji L, Le BH, Zhai J, Chen J, Luscher E, Gao L, Liu C, Cao X, Mo B. et al. (2017) ARGONAUTE10 promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLoS Biol 15: e2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang P, Shao W, Zhu JK, Dong J (2015) The BASL polarity protein controls a MAPK signaling feedback loop in asymmetric cell division. Dev Cell 33: 136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, He J, Cai H, Lin H, Li Y, Liu R, Yang Z, Qin Y (2014) Comparative expression profiling reveals gene functions in female meiosis and gametophyte development in Arabidopsis. Plant J 80: 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Cai H, Su Z, Wang L, Huang X, Zhang M, Chen P, Dai X, Zhao H, Palanivelu R. et al. (2018) KLU suppresses megasporocyte cell fate through SWR1-mediated activation of WRKY28 expression in Arabidopsis. Proc Natl Acad Sci USA 115: E526–E535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Honda M, Zhu H, Zhang Z, Guo X, Li T, Li Z, Peng X, Nakajima K, Duan L, et al. (2015) Spatiotemporal sequestration of miR165/166 by Arabidopsis Argonaute10 promotes shoot apical meristem maintenance. Cell Rep 10: 1819–1827. [DOI] [PubMed] [Google Scholar]
- Zhu H, Hu F, Wang R, Zhou X, Sze SH, Liou LW, Barefoot A, Dickman M, Zhang X (2011) Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 145: 242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.