Abstract
Arbutin is a naturally occurring glycosylated product of hydroquinone. With the ability to interrupt melanin biosynthesis in epidermal cells, it is a promising cosmetic ingredient. In this study, a novel amylosucrase, Asmet, identified from a thermal spring metagenome, has been characterized for arbutin biosynthesis. Asmet was able to catalyze transglucosylation of hydroquinone to arbutin, taking sucrose as glycosyl donor, in the temperature range of 20 °C to 40 °C and pH 5.0 to 6.0, with the relative activity of 80% or more. The presence of chloride salts of Li, K, and Na at 1 mM concentration did not exhibit any notable effect on the enzyme's activity, unlike Cu, Ni, and Mn, which were observed to be detrimental. The hydroquinone (20 mM) to sucrose ratio of 1:1 to 1:10 was appropriate for the catalytic biosynthesis of arbutin. The maximum hydroquinone to arbutin conversion of 70% was obtained in 24 h of Asmet led catalysis, at 30 °C and pH 6.0. Arbutin production was also demonstrated using low-cost feedstock, table sugar, muscovado, and sweet sorghum stalk extract, as a replacement for sucrose. Whole-cell catalysis of hydroquinone to arbutin transglucosylation was also established.
Keywords: Arbutin, Hydroquinone, Amylosucrase, Transglucosylation, Skin-lightening, Cosmetic ingredient
Introduction
Arbutin (Hydroquinone β-D-glucopyranoside) is a glycosylated hydroquinone product naturally present in bearberry, mulberry, cranberry, pear, and wheat (Carmen et al. 2009). In cosmetics, it is used as an ingredient conferring mild anti-inflammatory and antioxidant properties (Bang et al. 2008; Ioku et al. 1992; Lee et al. 2012). Arbutin reduces melanin production in melanocytes (Funayama et al. 1995; Hori et al. 2004; Maeda and Fukuda 1996), hence reducing excessive skin pigmentation in patients with melasma or sunburn. Melanin is a skin pigment produced from tyrosine (via 3,4 dihydroxy-phenylalanine) by tyrosinase's catalytic action. Excessive melanin production causes hyperpigmentation. Hyperpigmentation is reduced by the inhibition of tyrosinase activity, which can be achieved by arbutin. Thus, arbutin, a natural skin-whitening agent, is a biomolecule of high demand in the pharmaceutical and cosmetic industries. Further, this natural compound is known to exert antimicrobial, antioxidative, and anti-inflammatory effects. It helps in wound healing (Polouliakh et al. 2020) and exerts radioprotective (Nadi et al. 2019), cryoprotective (Seyfizadeh et al. 2012), and estrogen-like effects (Zeng et al. 2018). Further, it can be used as an anti-ulcer agent (Taha et al. 2012). In the human body, arbutin is metabolized and excreted in its glucuronide and sulphate forms (Schindler et al. 2002).
The natural β-arbutin molecule can be extracted from plant sources, but the process is tedious, with stubby yield (Parejo et al. 2001). Transglucosylation of hydroquinone by the catalytic action of microbial glycosyltransferases results in the formation of α-arbutin (4-hydroquinone-α-D-glucopyranoside). It has about ten times greater tyrosinase inhibition activity than β-arbutin. Thus, α-arbutin is a promising biomolecule that significantly diminishes melanin synthesis without any noticeable effect on cell viability (Sugimoto et al. 2004). Chemical synthesis of arbutin is also possible but limited by substantial energy requirements, labour-intensive process, chemical wastes, and low yield (Zhu et al. 2019). In contrast, the enzymatic approach for α-arbutin production is environmentally friendly and offers a higher yield. Arbutin has been enzymatically produced from hydroquinone using different donor molecules such as sucrose, maltose, and maltopentaose. Some of the enzymes used for arbutin production are sucrose phosphorylase from Leuconostoc mesenteroides (Kitao et al. 1994), α-amylase from Bacillus subtilis (Nishimura et al. 1994), α-glucosidase from Saccharomyces cerevisiae and Xanthomonas campestris (Prodanovic et al. 2005a,b; Sato et al. 2012), dextransucrase from Leuconostoc mesenteroides (Seo et al. 2009), sucrose isomerase from Erwinia rhapontici (Zhou et al. 2011), cyclodextrin glycosyltransferase from Thermoanaerobacter sp. (Mathew et al. 2013), whole cells of X. campestris WU-9701 and X. maltophilia BT-112 (Sato et al. 2012; Liu et al. 2013), amylosucrase from Deinococcus geothermalis (Seo et al. 2012), Cellulomonas carboniz T26 (Yu et al. 2018), and X. campestris (Yang et al. 2019; Zhu et al. 2019) (Table 1).
Table 1.
Comparative data of arbutin biosynthesis from hydroquinone by different biocatalyst systems
| Biocatalyst | Hydroquinone (mM) | Donor: HQ | Glycosyl donor | Optimum pH | Optimum temperature (℃) | Molar arbutin yield (%) | Ref |
|---|---|---|---|---|---|---|---|
| Sucrose phosphorylase (L. mesenteroides) | 18 | 8:1 | Sucrose | 7.5 | 42 | 46.5 | Kitao et al. 1994 |
| Dextransucrase (L. mesenteroides) | 450 | 1:2 | Sucrose | 5.2 | 28 | 0.4 | Seo et al. 2009 |
|
Amylosucrase (D. geothermalis) |
23.6 | 10:1 | Sucrose | 7 | 35 | 90 | Seo et al. 2012 |
|
Amylosucrase (C. carboniz) |
5 | 4:1 | Sucrose | 7 | 35 | 44.7 | Yu et al. 2018 |
|
Amylosucrase (X. campestris) |
10 | 80:1 | Sucrose | 7.5 | 35 | 95 | Zhu et al. 2019 |
| X. maltophilia BT-112 cells | 120 | 2:1 | Sucrose | 7 | 30 | 93.6 | Liu et al. 2013 |
|
α- amylase (B. subtilis) |
90 | 3.8:1 | Maltopentose | 5.5 | 40 | 24.8 | Nishimura et al. 1994 |
| α-glucosidase (S. cerevisiae) | 9 | 167:1 | Maltose | 5 | 30 | 14 | Prodanović et al. 2005a |
| α-glucosidase (S. cerevisiae) | 50 | 30:1 | Maltose | 5.5 | 30 | 4.6 | Prodanović et al. 2005b |
| α-glucosidase (X. campestris WU-9701) | 45 | 27:1 | Maltose | 7 | 40 | 55.6 | Sato et al. 2012 |
| X. campestris WU-9701 cells | 45 | 27:1 | Maltose | 7.5 | 40 | 93 | Kurosu et al. 2002 |
| Cyclodextrin glucanotransferase (Thermoanaerobacter sp.) | 9 | 11:1 | Maltodextrin | 5.5 | 40 | 21.2 | Mathew et al. 2013 |
|
Sucrose isomerase (E. rhapontici) |
50 | 1:50 | Sucrose | – | – | 33.2 | Zhou et al. 2011 |
|
Transglucosidase (E. coli) |
100 | 12:1 | Maltose | 7.2 | 40 | 76 | Wu et al. 2008 |
| Amylosucrase (Asmet) | 20 | 5:1 | Sucrose | 6 | 30 | 75 | This study |
Amylosucrase is a glycosyltransferase belonging to α-amylase superfamily of glycoside hydrolase (GH) 13 (Skov et al. 2001). Like GH13 family enzymes, the protein structure of amylosucrase comprises domains A, B, and C (Ramasubbu et al. 1996). Domain A adopts the characteristic (β/α)8-barrel structure, forming the catalytic core of the protein (Stam et al. 2006). The catalytic core of amylosucrase contains the conserved two catalytic acidic residues associated with α-amylases, aspartic acid and glutamic acid (Mirza et al. 2001; Moulis et al. 2016). Moreover, the mechanism of enzymatic activity of amylosucrase is similar to the α-amylase enzyme, i.e., the formation of a glucosyl-enzyme complex as an intermediate (Mirza et al. 2001). Apart from α-amylase like A, B, and C domains, amylosucrase contains B’ and N domains N. N-terminal domain plays a critical role in substrate preference of the enzyme (Lee et al. 2002). Domain B’ possibly attributes transferase activity to this enzyme (Mirza et al. 2001). Amylosucrase has been placed in polyspecific subfamily GH13_4 that exhibit more than one catalytic activity (Stam et al. 2006). Amylosucrase executes three main reactions- hydrolysis, transglycosylation, and polymerization (Choi et al. 2019). It catalyzes sucrose hydrolysis, followed by transfer of its glucose moiety to acceptors like water, fructose, and hydroquinone, resulting in diverse types of glycosylated products, e.g., turanose, trehalulose, α-1,4 glucans, arbutin, etc. (Tian et al. 2018). Amylosucrase has been experienced to display high catalytic efficiency for arbutin production compared to other transglucosidases (Tian et al. 2018). Therefore, it is desirable to develop amylosucrase based biocatalyst systems for arbutin production.
A novel amylosucrase (Asmet) was recently identified by exploring a thermal reservoir metagenome (Agarwal et al. 2019). In the present study, Asmet was characterized for the transformation of hydroquinone to arbutin in the presence of sucrose or other sucrose-containing low-cost feedstock as the donor of glycosyl moiety. Furthermore, Asmet expressing whole-recombinant cells were demonstrated to be a potential biocatalyst system for arbutin production.
Materials and methods
Materials
Sucrose, Luria–Bertani, Luria-Agar, Coomassie blue, Imidazole, and Tris base were procured from HiMedia (India). Terrific broth was purchased from Central Drug House (India). Ammonium per-sulphate (APS), α-arbutin, sodium acetate, sodium chloride, and isopropyl β-D-1-thiogalactopyranoside (IPTG) were purchased from Sigma Aldrich (USA). Muscovado (or Khandsari) and table sugar were purchased from the local market. The substrate hydroquinone was procured from Merck Millipore (USA). Sweet sorghum (Sorghum bicolor) cultivar, CSH-22ss, was grown in the field of CIAB campus, Mohali, India.
Enzyme preparation
The gene, Asmet (MK442921), cloned in the pET43a vector (Novagen, USA), was expressed in Escherichia coli BL21 strain, as described previously (Agarwal et al. 2019). The recombinant cells were grown in terrific broth media at 37 °C, with continuous shaking at 200 RPM. Protein expression was induced at 0.6 OD (600 nm) by introducing 0.3 mM IPTG in the cell culture, followed by incubation at 16 °C and 150 RPM for 16–18 h. Then, the recombinant cells were harvested at 4 °C. The cell pellet was suspended in 50 mM Tris–HCl buffer (pH 7.0). Cell disruption was performed by ultra-sonication for 3 min (3 s pulse on, 15 s pulse off). The cell lysate was clarified by centrifugation and filtration of the supernatant. Asmet was purified by nickel nitrilotriacetic acid (Ni- NTA) affinity column chromatography. The column was equilibrated with the binding buffer, comprised of 50 mM tris (pH 7.0), 10 mM imidazole, and 300 mM NaCl. For efficient binding of the His-tagged Asmet, the clarified cell-lysate was passed twice through the column. The unbound protein molecules were removed by washing the column using the wash buffer comprised of 50 mM tris (pH 7.0), 20 mM imidazole, and 300 mM NaCl. The elution buffer, comprising 50 mM sodium phosphate (pH 6.0), 200 mM imidazole, and 300 mM NaCl, was passed through the column to recover the purified Asmet fraction. All the steps of protein purification were performed in cold (4 °C) condition. The protein was concentrated using Amicon Ultra-15 Centrifugal filter of 30 kDa cutoff. The enzyme concentration was determined by Bradford's method using bovine serum albumin (BSA) as standard.
Enzyme assay
The standard enzymatic assays were done in 50 mM sodium phosphate buffer (pH 6.0) containing 20 mM hydroquinone and 40 mM sucrose, treated with 0.3 mg mL−1 Asmet at 30 °C for 1 h. The enzymatic reactions were stopped by heating at 100 °C for 10 min.
Qualitative and quantitative analysis
The qualitative detection of arbutin was done by thin layer chromatography (TLC). The reaction samples (2 µL) were spotted on the TLC plate (aluminium TLC plate, silica gel coated with fluorescent indicator F254 (MERCK Millipore, Germany). The plate was kept in a TLC chamber containing a mobile phase comprising of ethyl acetate, acetic acid, and water in the ratio 3:1:1. After completing the run, the TLC plate was air-dried, followed by the spray of a methanol-based solution containing 0.5% (w/v) 1-naphthyl ethylenediamine dihydrochloride and 5% sulfuric acid. Then, the TLC plate was heated at 120 °C for 10 to 15 min until the spots developed. Qualitative analysis of the reaction product was done by high-performance liquid chromatography (HPLC) (Water ACQUITY) system, equipped with C-18 column and UV detector (222 nm). A degassed solution of methanol, water, and 0.1 M HCl in the ratio of 89:10:1 was used as the mobile phase, keeping the column temperature 25 °C. Sample elution was done at 1 mL min−1.
Temperature and pH profiling
To study the effect of temperature on arbutin biosynthesis, the enzyme assays were conducted in the temperature range of 20 °C to 70 °C. The optimum pH for arbutin biosynthesis was determined by performing assays in the buffers of a wide pH range of 4.0 to 8.0. The buffer systems used were 50 mM sodium acetate buffer (pH 4.0–5.5), 50 mM sodium phosphate buffer (pH 6.0) and 50 mM Tris–HCl buffer (7.0–8.0).
Effect of metal ions and ascorbic acid
The effect of different metals on the transglucosylation activity of Asmet for arbutin biosynthesis was examined by introducing 1 mM of different metal salts (CaCl2, CoCl2, CuSO4, MgCl2, MnCl2, LiCl, FeCl3, KCl, NaCl, NiCl2, and ZnCl2) in the reaction assays. Reactions were also performed in the presence of different hydroquinone (20 mM) to ascorbic acid ratios viz. 1: 0.05, 1:0.1, 1:0.2, 1:0.3, 1:0.5, 1:0.8, and 1:1.
Substrate concentration
The ratio of glycosyl donor and acceptor molecule was optimized for the maximum arbutin yield. The enzymatic reactions were performed taking different hydroquinone (20 to 80 mM) to sucrose ratio (1:1, 1:2, 1:5 1:8, 1:10, and 1:20).
Low-cost feedstock in place of pure sucrose
Different low-cost feedstock, e.g., table sugar, muscovado, and sweet sorghum juice (SSJ), were examined for their potential to be used in the place of sucrose for arbutin production. Sweet sorghum juice was concentrated in a rotary evaporator to achieve a high concentration of sucrose. The catalytic reactions were performed, taking hydroquinone (20 mM) to sucrose feedstock ratio of 1:5 under the optimum reaction conditions. The reaction was stopped by boiling the reaction mixture for 10 min. Arbutin yield was analyzed at different time points using HPLC.
Whole-cell catalytic system for arbutin production
Asmet expressed recombinant E. coli cells were used to perform whole-cell catalysis reaction in 50 mM sodium phosphate buffer (pH 6.0). The hydroquinone (20 mM) to sucrose ratio of 1:1, 1:3, and 1:5 were treated with recombinant E. coli cells corresponding to OD 3.2 (OD600) of the cells. One unit of Asmet was defined as 1 μmol of substrate utilized min−1 g−1 (wet weight) of the cell (Zhang et al. 2013). The reaction was performed under the optimal pH and temperature conditions. Then, the cells were pelleted, and the reaction product was micro-filtered, followed by boiling for 10 min.
Statistical analysis
The enzymatic reactions were performed in triplicates, and the mean values of three replications ± standard deviation were presented in tables and figures. All the graphs were prepared using SigmaPlot 10.0. The significant difference between treatments was determined by Analysis of Variance (ANOVA) test at 5% probability or 95% confidence (p ≤ 0.05), followed by Tukey’s post hoc test. The statistical tests were performed employing Minitab® 20 statistical software.
Results and discussion
Hydroquinone glycosylation by Asmet
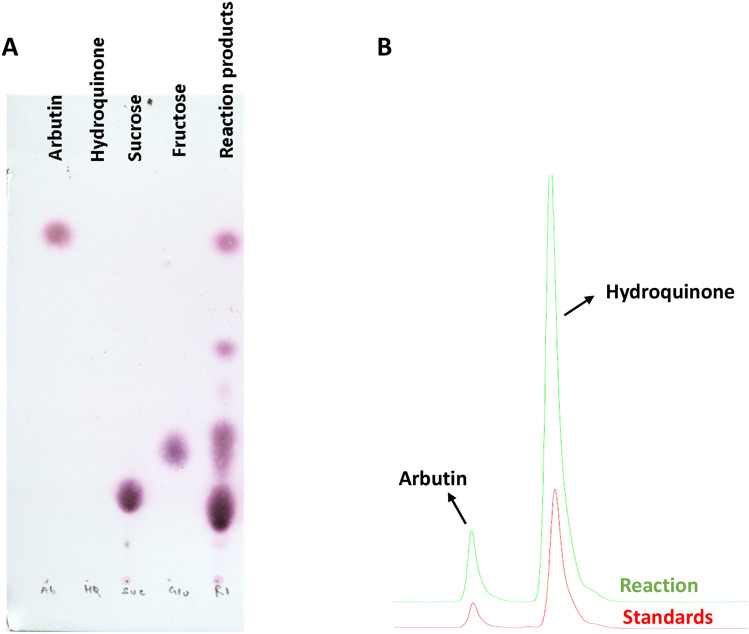
Amylosucrase had been characterized from different microbial sources, e.g., Bifidobacterium thermophilum, B. dentium, B. longum, B. pseudocatenulatum, Neisseria polysaccharea, N. subflava, Truepera radiovictrix, Cellulomonas carbonis, Methylomicrobium alcaliphilum, Synechococcus sp., Methylobacillus flagellates, Arthrobacter chlorophenolicus, Deinococcus geothermalis, D. radiodurans, D. radiopugnans, D. wulumuqiensis, and Alteromonas macleodii (Kim et al. 2014, 2019, 2020; Tian et al 2018). However, amylosucrase from a limited source, e.g., D. geothermalis, Cellulomonas carboniz, and X. campestris, had been exploited to catalyze transglycosylation of hydroquinone (acceptor) in the presence of donor (sucrose) molecules, generating α-1,4-linked glucose-transferred product, α-arbutin (Table 1). In this study, the catalytic potential of a novel amylosucrase (Asmet), identified from a thermal aquatic habitat metagenome, was examined for hydroquinone glycosylation. The enzymatic assay, containing sucrose as a glucosyl donor and hydroquinone as acceptor, resulted in hydroquinone glycosylation. TLC and HPLC analysis confirmed the catalytic product of Asmet as α-arbutin (Fig. 1a, b). Thus, similar to the amylosucrases from D. geothermalis and Cellulomonas carboniz, Asmet was capable of transferring the glucosyl unit onto both the sugar (D-fructose) and non-sugar (hydroquinone) molecules, resulting biosynthesis of turanose and arbutin, respectively (Wang et al. 2017; Guérin et al. 2012; Seo et al. 2012; Yu et al. 2018; Agarwal et al. 2019).
Fig. 1.
a Thin layer chromatography demonstrating the production of arbutin from hydroquinone treated with Asmet in the presence of sucrose. b HPLC chromatograms of standards and the reaction product, arbutin
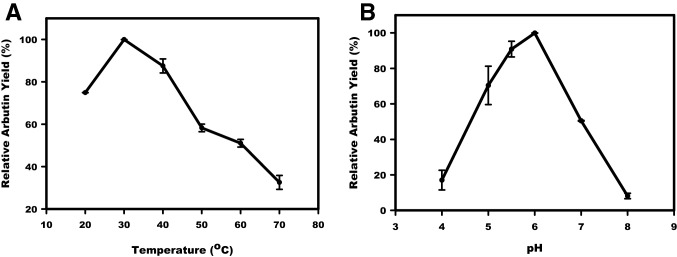
Effects of temperature and pH on hydroquinone glycosylation
The molar production of arbutin over the hydroquinone was measured in Asmet led catalytic reactions performed in the temperature range of 20 °C to 70 °C (Fig. 2a). The maximum arbutin biosynthesis was obtained at 30 °C. However, Asmet was able to catalyze arbutin biosynthesis in the high-temperature range of 50–60 °C, with about 50% relative activity and the low temperature, 20 °C, with 75% relative activity. Thus, Asmet can be considered as a cold-active biocatalyst for hydroquinone glycosylation, in contrast to the optimum sugar glycosylation at the high temperature (50 ℃) (Agarwal et al. 2019). The cold-active property of Asmet is an essential aspect for industrial production of arbutin, since it offers a relatively less investment of energy for executing catalytic reaction (Kuddus 2018). The variable pH and temperature activity profiles for transglucosylation reaction could be ascribed to the changes in the enzyme's shape after binding with different acceptor molecule types, i.e., sugar or non-sugar (Robinson 2015). A similar trend of differential optimum pH and temperature for sugar and non-sugar glycosylation have been reported in the case of amylosucrases from D. geothermalis and Cellulomonas carboniz. The poor hydroquinone glycosylation at the higher temperatures (50 ℃ or higher) is in accordance with amylosucrases from D. geothermalis and Cellulomonas carboniz, and X. campestris (Seo et al. 2012; Yu et al. 2018; Sato et al. 2012).
Fig. 2.
a Temperature and b pH optima of Asmet for arbutin biosynthesis
Similar to the amylosucrases from D. geothermalis and X. campestris, the pH activity profiling of Asmet recorded 6.0 as the optimum pH for arbutin biosynthesis. However, unlike other amylosucrases, alteration in pH affected arbutin biosynthesis with about 30% and 50% loss in the activity at pH 5.0 and 7.0, respectively (Fig. 2b). The results indicated a slightly acidic medium as the favorable environment for Asmet to glycosylate hydroquinone. In contrast, the alkaline medium (pH 8.0) was appreciative for optimal glycosylation of sugar molecule by Asmet (Agarwal et al. 2019).
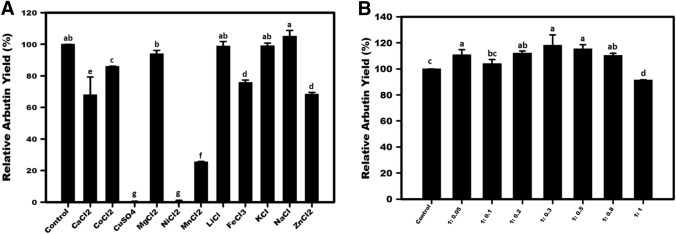
Effect of metal ions and ascorbic acid on hydroquinone glycosylation
The hydroquinone glycosylation efficiency was unaffected by the presence of metal ions such as Mg, Li, K, and Na. The reactions containing Ca, Co, Zn, and Fe, experienced 14% to 32% loss in the enzyme’s activity, whereas Mn lessened the glycosylation by about 75% (Fig. 3a). Interestingly, a nil amount of arbutin was produced in the presence of Cu and Ni in the reaction assay. The decrease in the relative activity (%) of Asmet for arbutin synthesis, caused by Ca, Co, Cu, Ni, Fe, Zn, and Mn, was statistically significant. This metal activity profile of Asmet was more or less similar to that of X. campestris Amy-1 (Zhu et al. 2019). The results suggested that the maximum possible exclusion of these activity-inhibiting metals should be assured in Asmet catalytic reaction system for arbutin production.
Fig. 3.
Effect of a different metals, and b ratio of hydroquinone (HQ): ascorbic acid (VC) on Asmet led biosynthesis of arbutin. The control reaction was performed with nil metal or nil ascorbic acid. The activity of the control reaction was taken as 100%. The relative activity (%) of the enzyme in the presence of metal or ascorbic acid was presented as compared to the control. Mean values not sharing common alphabets are statistically different at p ≤ 0.05
Oxidation of hydroquinone during the enzymatic assay is reported to cause benzoquinone production, turning the reaction sample brown and negatively affecting the arbutin yield (Seo et al. 2012). The semiquinone radicals produced could inhibit the function of amylosucrase. Therefore, it is advisable to add L-ascorbic acid in the reaction to avoid the oxidation of hydroquinone. In the present study, the hydroquinone to ascorbic acid ratio of 1:0.05 to 1:0.8 exerted a slightly positive effect on arbutin's catalytic production (Fig. 3b). In a previous study, ascorbic acid had been demonstrated to be a promising antioxidant to obtain a better bioconversion yield of arbutin in the reaction catalyzed by D. geothermalis amylosucrase (Seo et al. 2012). Nevertheless, in the present study, the addition of ascorbic acid in Asmet reaction did not lead to any remarkable increase in the bioconversion yield of arbutin (Fig. 3b). The results suggested that ascorbic acid may not be an essential component for the glycosylation of hydroquinone by Asmet. Similar results were demonstrated in the case of amylosucrase from C. carboniz and X. campestris (Yu et al. 2018; Yang et al. 2019).
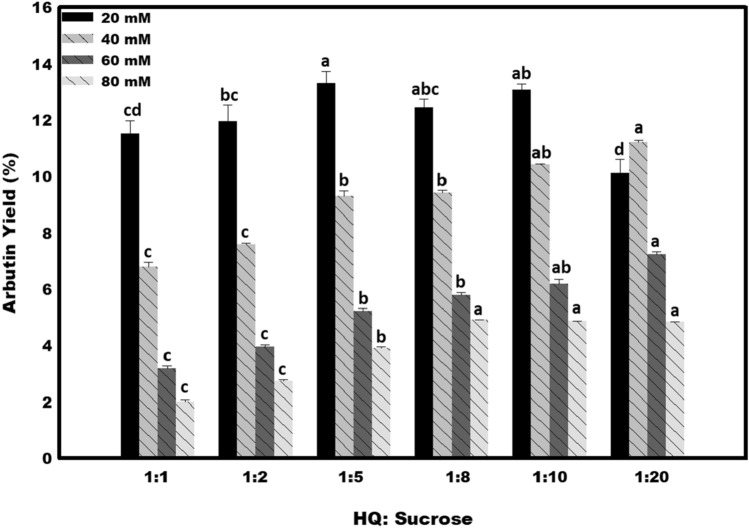
Optimum acceptor and donor ratio for arbutin biosynthesis
The acceptor and donor ratio has been discussed as an essential factor that can affect arbutin production. In the previous studies, the hydroquinone and sucrose molar ratio of 1:4 has been demonstrated to be appropriate for the maximum arbutin biosynthesis by amylosucrases from C. carboniz, whereas, in case of D. geothermalis amylosucrase, the best molar ratio was noted to be 1:10 (Seo et al. 2012; Yu et al. 2018). In a recent study, a high concentration of sucrose (acceptor: donor ratio of 1:80) was found essential to achieve the optimum hydroquinone conversion yield (Zhu et al. 2019). The optimum acceptor and donor ratio for arbutin production in the Asmet enzymatic assays was examined. Different concentrations of hydroquinone (20 to 80 mM) were used under variable acceptor: donor ratios (1:1 to 1:20), and the net arbutin yield was calculated. The hydroquinone concentration of 20 mM was noted to be the best concentration, with any amount of sucrose taken for catalytic glycosylation. In standard enzyme assay, the maximum arbutin yield was obtained in the reaction samples containing the donor to acceptor ratio of 1:5 in 1 h (Fig. 4). Therefore, this acceptor: donor ratio was taken forward for arbutin production. The reaction performed for 6 h resulted in hydroquinone to arbutin conversion yield of about 50%, whereas, in 24 h the yield of about 70% was obtained (Fig. 5a). After this, a small increase in the arbutin yield was noticed, with about 75% hydroquinone bioconversion in 48 h of reaction.
Fig. 4.
Effect of different hydroquinone (HQ): sucrose ratios on Asmet led biosynthesis of arbutin. Mean values not sharing common alphabets in the bars of same pattern are statistically different at p ≤ 0.05
Fig. 5.
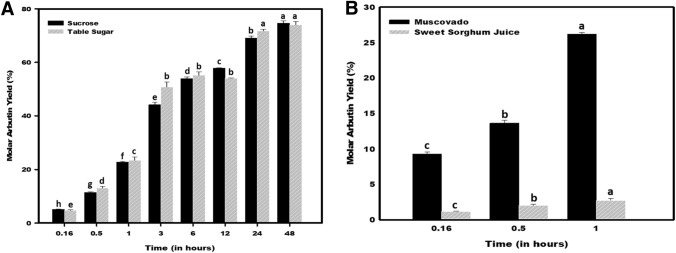
a Arbutin biosynthesis yield using hydroquinone (HQ): sucrose/table sugar (1:5). b Arbutin biosynthesis using low-cost feedstock (muscovado, and sweet sorghum stalk extract) in place of sucrose. Mean values not sharing common alphabets in the bars of same pattern are statistically different at p ≤ 0.05
In our previous study (Agarwal et al. 2019), Asmet was demonstrated to execute glycosylation of D-fructose in the presence of low-cost feedstock as the donor molecules. In the present study, the hydroquinone glycosylation was examined in the presence of the inexpensive feedstock- table sugar, muscovado, and sweet sorghum stalk-juice. Arbutin production was found comparable in the case of enzymatic reactions performed using pure sucrose or table sugar. In the case of muscovado, used as a replacement for sucrose, arbutin synthesis was estimated to be about 25% in 1 h. The sweet sorghum stalk-juice based reactions faced significantly reduced biosynthesis of arbutin (2.5% in 1 h) (Fig. 5b). This might be due to the combined effects of metals like Mn, Cu, and Ni, present in the juice (Sharma et al. 2020). The results indicated that contaminations of these metals should be avoided in Asmet based biocatalysis reaction for arbutin production.
Whole-cell catalysis for arbutin production
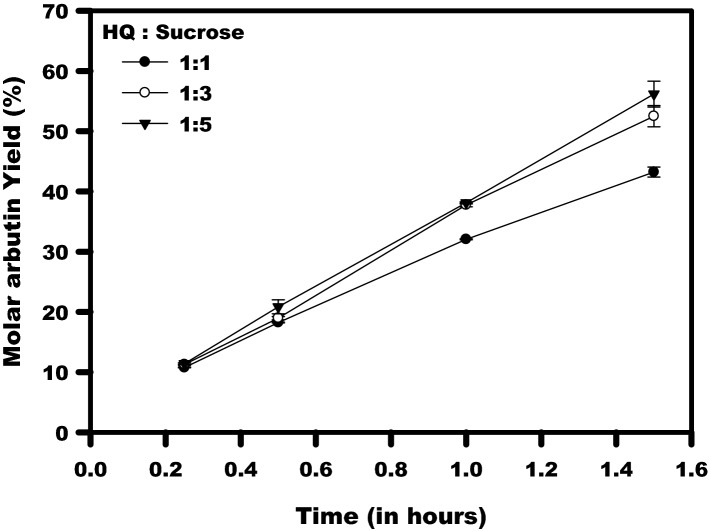
To further validate the industrial importance of the metagenomic amylosucrase, Asmet, arbutin production was demonstrated to be produced using whole recombinant cells as a biocatalyst in the reaction. The use of whole cells avoids the process of protein extraction and purification steps in the bioprocess. Asmet expressing E. coli cells were directly used to execute glycosylation of hydroquinone by whole-cell catalysis reaction. The catalytic performance of cells of different OD600 (1.5, 2, 2.5, 3, and 3.2) for arbutin biosynthesis were examined. TLC analysis confirmed the synthesis of arbutin in the catalysis reaction performed using the whole recombinant cells of 2.5, 3, and 3.2 OD600 (Fig. S1). The arbutin yield of about 50% was achieved using the cells of OD 3.2 (equivalent to about 0.7 U amylosucrase g−1 cells) in 1.5 h (Fig. 6). A higher rate of hydroquinone transglucosylation in the case of whole-cell catalysis reaction could be due to the elimination of the possible inhibition of enzyme molecules by the oxidized hydroquinone (Zhu et al. 2019).
Fig. 6.
Molar arbutin yield (%) by whole-cell catalysis reaction performed with 20 mM hydroquinone, taking variable concentration of sucrose in the ratio (HQ: sucrose) 1:1, 1:3, and 1:5
Conclusions
This study presents the biosynthesis of the skin-whitening cosmetic ingredient, arbutin, from the organic compound hydroquinone, using sucrose as the glycosyl donor and the metagenomic amylosucrase, Asmet, as a cold-active biocatalyst. For the first time, inexpensive feedstock, e.g., muscovado, and table sugar, in the place of refined sucrose, has been demonstrated as a suitable glycosyl donor for the transformation of hydroquinone into arbutin. Furthermore, Asmet expressing whole-cells, without involving protein extraction and purification steps, were established to produce arbutin with a reasonably high conversion rate. The results discerned Asmet as a potential biocatalyst for the industrial production of arbutin, the biomolecule of cosmetic and pharmaceutical significance.
Acknowledgements
Authors acknowledge the Department of Biotechnology (DBT), Government of India for facilitating the present work at Center of Innovative and Applied Bioprocessing (CIAB), Mohali. SPS acknowledges the DBT project-grant, BT/PR17586/PFN/20/1195. NA acknowledges the fellowship obtained from the DBT project, and the Department of Biotechnology, Panjab University, Chandigarh, for Ph.D. registration.
Authors contribution
SPS conceived and supervised the study. NA performed all the experiments. AKR helped in data analysis. SPS, NA, and AKR prepared the manuscript.
Declarations
Conflicts of interest
The authors declare that there is no conflict of interest.
Footnotes
The gene sequence is available under the NCBI accession number, MK442921.
References
- Agarwal N, Narnoliya LK, Singh SP. Characterization of a novel amylosucrase gene from the metagenome of a thermal aquatic habitat, and its use in turanose production from sucrose biomass. Enzyme Microb Technol. 2019;131:109372. doi: 10.1016/j.enzmictec.2019.109372. [DOI] [PubMed] [Google Scholar]
- Bang SH, Han SJ, Kim DH. Hydrolysis of arbutin to hydroquinone by human skin bacteria and its effect on antioxidant activity. J Cosmet Dermatol. 2008;7:189–193. doi: 10.1111/j.1473-2165.2008.00387.x. [DOI] [PubMed] [Google Scholar]
- Carmen POP, Vlase L, Tamas M. Natural resources containing arbutin. Determination of arbutin in the leaves of Bergenia crassifolia (L.) Fritsch. acclimated in Romania. Not Bot Horti Agrobo. 2009;37:129–132. [Google Scholar]
- Choi SW, Lee JA, Yoo SH. Sucrose-based biosynthetic process for chain-length defined α-glucan and functional sweetener by Bifidobacterium amylosucrase. Carbohydr Polym. 2019;205:581–588. doi: 10.1016/j.carbpol.2018.10.064. [DOI] [PubMed] [Google Scholar]
- Funayama M, Arakawa H, Yamamoto R, Nishino T, Shin T, Murao S. Effects of α-and β-arbutin on activity of tyrosinases from mushroom and mouse melanoma. Biosci Biotech Bioch. 1995;59:143–144. doi: 10.1271/bbb.59.143. [DOI] [PubMed] [Google Scholar]
- Guérin F, Barbe S, et al. Structural investigation of the thermostability and product specificity of amylosucrase from the bacterium Deinococcus geothermalis. J Biol Chem. 2012;287:6642–6654. doi: 10.1074/jbc.M111.322917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori I, Nihei KI, Kubo I. Structural criteria for depigmenting mechanism of arbutin. Phytother Res. 2004;18:475–479. doi: 10.1002/ptr.1456. [DOI] [PubMed] [Google Scholar]
- Ioku IK, Terao J, Nakatani N. Antioxidative activity of arbutin in a solution and liposomal suspension. Biosci Biotech Bioch. 1992;56:1658–1659. doi: 10.1271/bbb.56.1658. [DOI] [Google Scholar]
- Kim MD, Seo DH, Jung JH, Jung DH, Joe MH, Lim S, Lee JH, Park CS. Molecular cloning and expression of amylosucrase from highly radiation-resistant Deinococcus radiopugnans. Food Sci Biotechnol. 2014;23(6):2007 . doi: 10.1007/s10068-014-0273-3. [DOI] [Google Scholar]
- Kim KT, Rha CS, Jung YS, Kim YJ, Jung DH, Seo DH, Park CS. Comparative study on amylosucrases derived from Deinococcus species and catalytic characterization and use of amylosucrase derived from Deinococcus wulumuqiensis. Amylase. 2019;3(1):19–31. doi: 10.1515/amylase-2019-0002. [DOI] [Google Scholar]
- Kim SY, Seo DH, Kim SH, Hong YS, Lee JH, Kim YJ, Jung DH, Yoo SH, Park CS. Comparative study on four amylosucrases from Bifidobacterium species. Int J Biol Macromol. 2020;155:535–542. doi: 10.1016/j.ijbiomac.2020.03.176. [DOI] [PubMed] [Google Scholar]
- Kitao S, Sekine H. a-D-Glucosyl transfer to phenolic compounds by sucrose phosphorylase from Leuconostoc mesenteroides and production of a-arbutin. Biosci Biotechnol Biochem. 1994;58:38–42. doi: 10.1271/bbb.58.38. [DOI] [PubMed] [Google Scholar]
- Kuddus M. Cold-active enzymes in food biotechnology: An updated mini review. J Appl Biol Biotechnol. 2018;6:58–63. doi: 10.7324/JABB.2018.60609. [DOI] [Google Scholar]
- Kurosu J, Sato T, Yoshida K, Tsugane T, Shimura S, Kirimura K, Kino K, Usami S. Enzymatic synthesis of α-arbutin by α-anomer-selective glucosylation of hydroquinone using lyophilized cells of Xanthomonas campestris WU-9701. J Biosci Bioeng. 2002;93:328–330. doi: 10.1016/S1389-1723(02)80037-8. [DOI] [PubMed] [Google Scholar]
- Lee HS, Kim MS, Cho HS, Kim JI, Kim TJ, Choi JH, Park C, Lee HS, Oh BH, Park KH. Cyclomaltodextrinase, neopullulanase, and maltogenic amylase are nearly indistinguishable from each other. J Biol Chem. 2002;277:21891–21897. doi: 10.1074/jbc.M201623200. [DOI] [PubMed] [Google Scholar]
- Liu CQ, Deng L, Zhang P, Zhang SR, Liu L, Xu T, Tan TW. Screening of high α-arbutin producing strains and production of α-arbutin by fermentation. World J Microbiol Biotechnol. 2013;29:1391–1398. doi: 10.1007/s11274-013-1302-8. [DOI] [PubMed] [Google Scholar]
- Maeda K, Fukuda M. Arbutin: mechanism of its depigmenting action in human melanocyte culture. J Pharmacol Exp Ther. 1996;276:765–769. [PubMed] [Google Scholar]
- Mathew S, Adlercreutz P. Regioselective glycosylation of hydroquinone to α-arbutin by cyclodextrin glucanotransferase from Thermoanaerobacter sp. Biochem Eng J. 2013;79:187–193. doi: 10.1016/j.bej.2013.08.001. [DOI] [Google Scholar]
- Mirza O, Skov LK, Remaud-Simeon M, Potocki de Montalk G, Albenne C, Monsan P, Gajhede M. Crystal structures of amylosucrase from Neisseria polysaccharea in complex with D-glucose and the active site mutant Glu328Gln in complex with the natural substrate sucrose. Biochemistry. 2001;40:9032–9039. doi: 10.1021/bi010706l. [DOI] [PubMed] [Google Scholar]
- Moulis C, André I, Remaud-Simeon M. GH13 amylosucrases and GH70 branching sucrases, atypical enzymes in their respective families. Cell Mol Life Sci. 2016;73:2661–2679. doi: 10.1007/s00018-016-2244-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadi S, Elahi M, Moradi S, Banaei A. Radioprotective Effect of Arbutin in Megavoltage Therapeutic X-irradiated Mice using Liver Enzymes Assessment. J Biomed Phys Eng. 2019;9:533. doi: 10.31661/jbpe.v0i0.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Kometani T, Takii H, Terada Y, Okada S. Purification and some properties of α-amylase from Bacillus subtilis X-23 that glucosylates phenolic compounds such as hydroquinone. J Ferment Bioeng. 1994;78:31–36. doi: 10.1016/0922-338X(94)90174-0. [DOI] [Google Scholar]
- Parejo I, Viladomat F, Bastida J, Codina C. A single extraction step in the quantitative analysis of arbutin in bearberry (Arctostaphylos uvae-ursi) leaves by high-performance liquid chromatography. Phytochem Analysis. 2001;12:336–339. doi: 10.1002/pca.602. [DOI] [PubMed] [Google Scholar]
- Polouliakh N, Ludwig V, Meguro A, Kawagoe T, Heeb O, Mizuki N. Alpha-arbutin promotes wound healing by lowering ROS and upregulating insulin/IGF-1 pathway in human dermal fibroblast. Front Physiol. 2020 doi: 10.3389/fphys.2020.586843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodanović R, Milosavić N, Sladić D, Zlatović M, Božić B, Veličković TĆ, Vujčić Z. Transglucosylation of hydroquinone catalysed by α-glucosidase from baker's yeast. J Mol Catal B Enzym. 2005;35:142–146. doi: 10.1016/j.molcatb.2005.06.011. [DOI] [Google Scholar]
- Prodanović RM, Milosavić NB, Sladić D, Veličković TĆ, Vujčić Z. Synthesis of hydroquinone-α-glucoside by α-glucosidase from baker’s yeast. Biotechnol Lett. 2005;27:551–554. doi: 10.1007/s10529-005-2880-9. [DOI] [PubMed] [Google Scholar]
- Ramasubbu N, Paloth V, Luo Y, Brayer GD, Levine MJ. Acta Crystallogr D Biol Crystallogr. 1996;52:435–446. doi: 10.1107/S0907444995014119. [DOI] [PubMed] [Google Scholar]
- Robinson PK. Enzymes: principles and biotechnological applications. Essays Biochem. 2015;59:1–41. doi: 10.1042/bse0590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Hasegawa N, Saito J, Umezawa S, Honda Y, Kino K, Kirimura K. Purification, characterization, and gene identification of an α-glucosyl transfer enzyme, a novel type α-glucosidase from Xanthomonas campestris WU-9701. J Mol Catal B Enzym. 2012;80:20–27. doi: 10.1016/j.molcatb.2012.04.014. [DOI] [Google Scholar]
- Schindler G, Patzak U, et al. Urinary excretion and metabolism of arbutin after oral administration of Arctostaphylos uvae ursi extract as film-coated tablets and aqueous solution in healthy humans. J Clin Pharmacol. 2002;42:920–927. doi: 10.1177/009127002401102740. [DOI] [PubMed] [Google Scholar]
- Seo ES, Kang J, Lee JH, Kim GE, Kim GJ, Kim D. Synthesis and characterization of hydroquinone glucoside using Leuconostoc mesenteroides dextransucrase. Enzym Microb Technol. 2009;45:355–360. doi: 10.1016/j.enzmictec.2009.07.011. [DOI] [Google Scholar]
- Seo DH, Jung JH, et al. High-yield enzymatic bioconversion of hydroquinone to α-arbutin, a powerful skin lightening agent, by amylosucrase. Appl Microbiol Biotechnol. 2012;94:1189–1197. doi: 10.1007/s00253-012-3905-7. [DOI] [PubMed] [Google Scholar]
- Seyfizadeh N, Mahjoub S, Zabihi E, Moghadamnia A, Pouramir M, Mir H, Elahimanesh F. Cytoprotective effects of arbutin against tert-butyl hydroperoxid induced toxicity in Hep-G2 cell line. World Appl Sci J. 2012;19:163–167. [Google Scholar]
- Sharma M, Sangwan RS, Khatkar BS, Singh SP. Development of a prebiotic oligosaccharide rich functional beverage from sweet sorghum stalk biomass. Waste Biomass Valorization. 2020;12(4):1–12. [Google Scholar]
- Skov LK, Mirza O, Henriksen A, De Montalk GP, Remaud-Simeon M, Sarçabal P, Gajhede M. Amylosucrase, a glucan-synthesizing enzyme from the α-amylase family. J Biol Chem. 2001;276(27):25273–25278. doi: 10.1074/jbc.M010998200. [DOI] [PubMed] [Google Scholar]
- Stam MR, Danchin EG, Rancure C, Coutinho PM, Henrissat B. Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of α-amylase-related proteins. Protein Eng Des Sel. 2006;19:555–562. doi: 10.1093/protein/gzl044. [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Nishimura T, Nomura K, Sugimoto K, Kuriki T. Inhibitory effects of α-arbutin on melanin synthesis in cultured human melanoma cells and a three-dimensional human skin model. Biol Pharm Bull. 2004;27:510–514. doi: 10.1248/bpb.27.510. [DOI] [PubMed] [Google Scholar]
- Taha MME, Salga MS, Ali HM, Abdulla MA, Abdelwahab SI, Hadi AHA. Gastroprotective activities of Turnera diffusa Willd. ex Schult. revisited: Role of arbutin. J Ethnopharmacol. 2012;141:273–281. doi: 10.1016/j.jep.2012.02.030. [DOI] [PubMed] [Google Scholar]
- Tian Y, Xu W, Zhang W, Zhang T, Guang C, Mu W. Amylosucrase as a transglucosylation tool: from molecular features to bioengineering applications. Biotechnol Adv. 2018;36:1540–1552. doi: 10.1016/j.biotechadv.2018.06.010. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xu W, Bai Y, Zhang T, Jiang B, Mu W. Identification of an α-(1, 4)-glucan-synthesizing amylosucrase from Cellulomonas carboniz T26. J Agric Food Chem. 2017;65:2110–2119. doi: 10.1021/acs.jafc.6b05667. [DOI] [PubMed] [Google Scholar]
- Wu PH, Nair GR, Chu IM, Wu WT. High cell density cultivation of Escherichia coli with surface anchored transglucosidase for use as whole-cell biocatalyst for α-arbutin synthesis. J Ind Microbiol Biotechnol. 2008;35(2):95. doi: 10.1007/s10295-007-0270-0. [DOI] [PubMed] [Google Scholar]
- Yang C, Fan W, Zhang R, Shi J, Knežević-Jugović Z, Zhang B. Study on transglucosylation properties of amylosucrase from Xanthomonas campestris pv. Campestris and its application in the production of α-arbutin. Catalysts. 2019;9(1):5. doi: 10.3390/catal9010005. [DOI] [Google Scholar]
- Yu S, Wang Y, Tian Y, Xu W, Bai Y, Zhang T, Mu W. Highly efficient biosynthesis of α-arbutin from hydroquinone by an amylosucrase from Cellulomonas carboniz. Process Biochem. 2018;68:93–99. doi: 10.1016/j.procbio.2018.02.012. [DOI] [Google Scholar]
- Zeng M, Zhang L, Li M, Zhang B, Zhou KY, Feng W, Zheng X. Estrogenic effects of the extracts from the Chinese yam (dioscorea opposite thumb.) and its effective compounds in vitro and in vivo. Molecules. 2018;23(2):11. doi: 10.3390/molecules23020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li Q, Ye T, Zhang Z, Li L. Optimization of the whole-cell catalytic activity of recombinant Escherichia coli cells with surface-immobilized organophosphorus hydrolase. J Environ Biol. 2013;34:315–319. [PubMed] [Google Scholar]
- Zhou X, Zheng YT, Wei XM, Yang KD, Yang XK, Wang YT, Xu LM, Du LQ, Huang RB. Sucrose isomerase and its mutants from Erwinia rhapontici can synthesise alpha-Arbutin. Protein Pept Lett. 2011;18:1028–1034. doi: 10.2174/092986611796378774. [DOI] [PubMed] [Google Scholar]
- Zhu L, Jiang D, Zhou Y, Lu Y, Fan Y, Chen X. Batch-feeding whole-cell catalytic synthesis of α-arbutin by amylosucrase from Xanthomonas campestris. J Ind Microbiol Biot. 2019;46:759–767. doi: 10.1007/s10295-019-02143-z. [DOI] [PubMed] [Google Scholar]