Abstract
Purpose:
Exome and whole-genome sequencing of muscle-invasive bladder cancer (BC) has revealed important insights into the molecular landscape; however, there are few studies of non-muscle invasive BC with detailed risk factor information.
Experimental Design:
We examined the relationship between smoking and other BC risk factors and somatic mutations and mutational signatures in bladder tumors. Targeted sequencing of frequently mutated genes in BC was conducted in 322 formalin-fixed paraffin-embedded bladder tumors from a population-based case-control study. Logistic regression was used to calculate odds ratios (ORs) and 95% confidence intervals (CIs), evaluating mutations and risk factors. We used SignatureEstimation to extract four known single base substitution mutational signatures and Poisson regression to calculate risk ratios (RRs) and 95%CIs, evaluating signatures and risk factors.
Results:
Non-silent KDM6A mutations were more common in females than males (OR=1.83,95%CI:1.05–3.19). There was striking heterogeneity in the relationship between smoking status and established single base substitution signatures: current smoking status was associated with greater ERCC2-Signature mutations compared to former (p-value=0.024) and never smoking (RR=1.40, 95% CI: 1.09–1.80, p-value=0.008); former smoking was associated with greater APOBEC-Signature13 mutations (p-value=0.05); and never smoking was associated with greater APOBEC-Signature2 mutations (RR=1.54, 95% CI: 1.17–2.01, p-value=0.002). There was evidence that smoking duration (the component most strongly associated with BC risk) was associated with ERCC2-Signature mutations and APOBEC-Signature13 mutations among current (p-trend=0.005) and former smokers (p=0.0004), respectively.
Conclusions:
These data quantify the contribution of BC risk factors to mutational burden and suggest different signature enrichments among never, former, and current smokers.
Keywords: Bladder Cancer, Risk Factors, Sequencing, Somatic Mutation, Epidemiology
INTRODUCTION
In Europe and North America, bladder cancer is the 9th most commonly diagnosed cancer among men and women of all ages.[1] Major established risk factors for bladder cancer include increasing age, male sex, a history of cigarette smoking, and occupational exposures.[2] The major subtypes of bladder cancer are non-muscle-invasive bladder cancer (NMIBC, the tumor is confined to the mucosa or the lamina propria) and the less common, but more aggressive, muscle-invasive bladder cancer (MIBC, tumor invades muscle). While exome and whole-genome sequencing of MIBC has revealed important molecular insights, the mutational profile of NMIBC remains poorly characterized, despite the fact that NMIBC represents 70% of incident diagnoses.[3–11] Studies combining mutational profiles with detailed risk factor information are needed to learn more about possible mechanisms by which the major risk factors for bladder cancer influence carcinogenesis.
Here, we used deep, targeted amplicon sequencing of the coding regions of 44 genes frequently mutated in bladder cancer to characterize somatic mutations in 307 formalin-fixed paraffin-embedded (FFPE) bladder tumors (260 NMIBC and 47 MIBC) from a population-based case-control study and evaluated the associations between somatic mutations and detailed information on smoking as well as other major risk factors for bladder cancer.
MATERIAL AND METHODS
Study Population and Tumor Tissue Collection
The New England Bladder Cancer Study (NEBCS) is a large population-based case-control study that includes 1,213 cases and 1,418 controls. Cases in the NEBCS were patients with histologically confirmed bladder cancer newly diagnosed between 2001 and 2004 among residents of Maine, New Hampshire, and Vermont, ages 30 to 79 years. A total of 1,213 patients were ascertained through hospital pathology departments and hospital and state cancer registries and interviewed (65% of eligible cases were interviewed). A standardized histopathology review to assign stage and grade was carried out by a study pathologist.[12] Tumors were staged according to the tumor, node, and metastases (TNM) criteria of the American Joint Commission on Cancer[13, 14] and graded according to both the 1973 World Health Organization (WHO) and 2004 WHO/International Society of Urologic Pathologists (ISUP) criteria.[15, 16] Participants were interviewed by trained interviewers to obtain detailed information on demographics, use of tobacco products[17], lifetime occupational histories[18], and other risk factors. All participants provided written consent. The study protocol was conducted in accordance with the ethical guidelines outlined in the Belmont report and was approved by all appropriate institutional review boards. Blinding and randomization are not relevant to this study.
Three hundred and twenty-two FFPE tumor tissue blocks from NEBCS patients with urothelial cell carcinoma enrolled in Maine and Vermont returned pathology consent forms, completed the interview, and had DNA available for the current analysis. There were no significant differences between these cases and those not sequenced with respect to age, gender, state of residence, education, history of work in a high-risk occupation for bladder cancer, cigarette smoking status, and tumor stage and grade (Supplemental Table 1). Tumor regions as annotated on digital images were hand macrodissected from three 5 μm sections and placed into 1.5 ml microcentrifuge tubes. DNA was isolated using the phenol-based AutoGenprep 245T Animal Tissue DNA Extraction Kit (Autogen) and DNA yield and purity were determined by NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
Targeted Sequencing, Variant Calling and Quality Control
Using previously published papers[3, 7, 9, 10, 19], we identified 44 genes that are frequently mutated in bladder cancer (Supplemental Table 2). The Ampliseq Designer was used to design a panel of amplicons to cover the coding region and splice site regions of each exon. The panel was tested on a pilot set of FFPE samples from the NEBCS that included 20 duplicate DNAs. From these data, we selected an quality score of 50 and an allele fraction of 10% as cutoff values for somatic variants. For each sample, 30 ng of tumor and germline DNA was used for amplification, and library construction, and sequencing that was performed on an Ion Torrent S5 sequencer. Variants were called in the Ion Torrent Variant Caller software using the paired sample option. All variants recurrent in more than three samples were manually reviewed in the Integrative Genomics Viewer and variants determined to be sequencing artifacts were removed. When comparing all variants in the TP53 gene identified in the same samples in the current study and, previously, by Sanger sequencing, we observed 99.3% concordance. Prior data, from Sanger sequencing of selected FGFR3 exons, were also incorporated.[20–23] The gene-wise mutation frequency plots for NMIBC and MIBC were generated using the R package maftools (oncoplot function).
Mutational Signatures
Based on single nucleotide variants (SNVs), we define 96 trinucleotides as possible mutation types[24] to construct a mutational catalogue matrix M. Because of a small number of genes being sequenced, it was difficult to decompose M de novo to identify mutational signatures directly. Instead, we leveraged the current knowledge of major mutational signatures for bladder cancer and defined a 96×4 mutational signature matrix P consisting of COSMIC signatures 1 (age-related), APOBEC-mediated signatures 2 and 13 and 5*/ERCC2 signature [4, 25] (5*/ERCC2 signature cosine similarity of 0.90 to COSMIC 5). Given these matrices M and P, we extracted the exposure matrix E by minimizing Frobenius norm ∥M-PE∥ using the R package SignatureEstimation.[26]
Statistical Analysis
Multivariate logistic regression was used to calculate odds ratios (ORs) and 95% confidence intervals (95% CI) for the relationship between bladder cancer risk factors and presence or absence of somatic mutation (missense, nonsense, splice site, insertions, deletions) as well as the presence or absence of indels (any deletion, any insertion, one base pair deletions, one base pair insertions) in all 44 genes. Risk factors evaluated included age at diagnosis, sex, race (white/non-white), smoking characteristics (as described below), and ever employment in a high-risk occupation (yes/no). High-risk occupation was defined as a priori “suspect occupation” with an odds ratio of 1.1 or higher based on 15 or more exposed individuals in this study.[18] Smoking characteristics evaluated included cigarette smoking status (never, former, current), duration (in years), combined status and duration (never, former (and duration among former smokers), current (and duration among current smokers), intensity (cigarettes per day), and pack-years (all characteristics are defined in Baris et al.[17])). Risk factors, as well as tumor cellularity (defined as the percent of tumor cells by visual inspection of study pathologist), were included in multivariate models when they changed parameter estimates by more than 10%. Somatic mutations were also evaluated as a group assigned to the following major pathways altered in bladder cancer[4, 19]: cell-cycle regulation, histone modification/chromatin remodeling, transcription factors, RTK/Ras/PI(3)K, DNA damage response (DDR). Poisson regression was used to evaluate the relationship between risk factors and counts of mutations corrected for total mutational burden (TMB) (or, the proportion of mutations) contributing to each of the four major single base substitution (SBS) signatures in bladder tumors (Signature 1, C>T at CpG; APOBEC-Signature 2 C>T in TC[A/T] motifs; APOBEC-Signature 13 C>G in TC[A/T] motifs; ERCC2-Sig mutations). Genotypes of the germline single nucleotide polymorphism (SNP) rs1014971, previously associated with bladder cancer risk and APOBEC-signature mutation load [27], were available from a previous genome-wide association study.[28]
RESULTS
Of the 322 bladder tumors sequenced, 15 were excluded due to poor sequencing coverage (mean coverage <100x) leaving 307 samples for analysis (accession number phs002318.v1.p1). The resultant mean sequencing coverage across all targeted exons was 630x. The median number of mutations per sample was 5 with a range of 0–39. Two hundred and sixty patients had NMIBC (84.7%) and 47 had MIBC (15.3%).
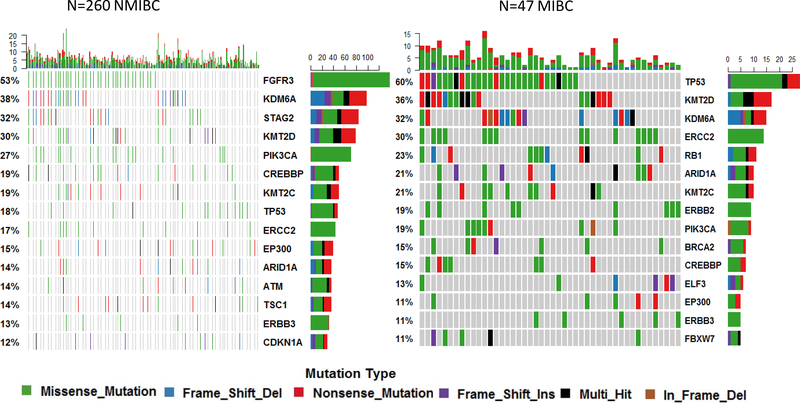
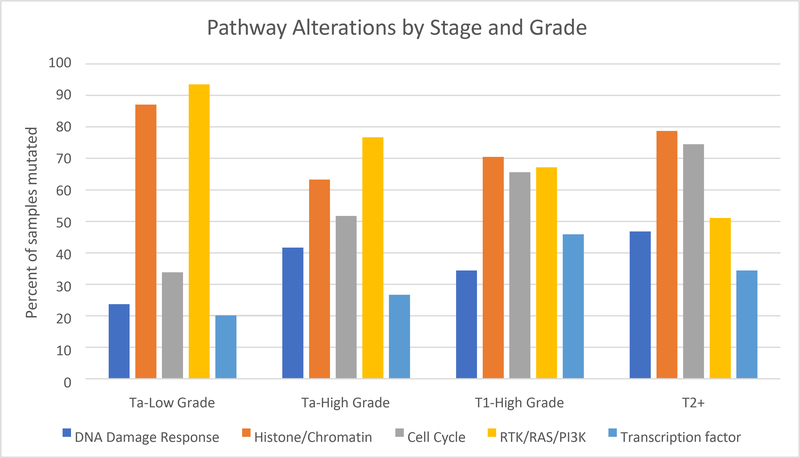
First, we analyzed individual mutations. The most frequently mutated gene in NMIBC was FGFR3 (53% mutated) and in MIBC tumors was TP53 (60% mutated, Figure 1). Mutations in histone/chromatin regulating genes (87% of samples mutated) and RTK/Ras/PI(3)K pathway alterations (93.5% samples mutated) were present at a high frequency in low grade Ta tumors. High grade, T1 tumors showed the highest frequency of mutation in genes in transcription factor pathways (45.9%) while MIBC had the highest frequency of cell-cycle alterations (74.5%) compared to other stage/grade subtypes (Figure 2).
Figure 1.
Top 15 genes with somatic mutations in non-muscle invasive (NMIBC) and muscle-invasive bladder cancer (MIBC) in the New England Bladder Cancer Study.
Figure 2.
Pathway alterations by stage and grade of bladder cancer
The presence of non-silent mutations was not associated with age at diagnosis or race group. Non-silent KDM6A mutations were significantly more common in females compared to males, OR=1.83, 95% CI: 1.05–3.19; in females these mutations largely occurred in Ta tumors (86%, OR=2.17, 95% CI: 1.17–4.22) (Supplemental Table 3). A history of employment in a high-risk occupation was significantly associated with mutations in cell cycle pathway genes OR=1.89, 95% CI: 1.01–3.55 (Supplemental Table 4). This was predominantly driven by the strong positive association between TP53 mutations in those with a history of employment in a high-risk occupation, OR=5.73, 95% CI: 2.07–15.83, p-value=0.0008 (Supplemental Table 4). There were no significant associations between any metrics of smoking and the presence of any somatic mutation in any of the 44 genes frequently mutated in bladder tumors.
Next, we analyzed four major mutational signatures, which were constructed using data on 1,472 SNVs in the 44 sequenced genes. These SBS signatures included Signature 1, C>T at CpG, overall mean contribution: 0.115; Signature 2, APOBEC C>T in TC[A/T] motifs, overall mean contribution: 0.316; Signature 13 APOBEC C>G in TC[A/T] motifs, overall mean contribution: 0.121; ERCC2-Signature mutations, overall mean contribution: 0.448. Signature 1 mutations were significantly associated with age at bladder cancer diagnosis (p=0.007), ERCC2-Signature mutations were significantly associated with non-silent ERCC2 mutations (p=3.24 ×10−11), and the bladder cancer risk allele of a germline SNP rs1014971 was associated with APOBEC-signature 13 mutations (β=0.29, p=0.044) in multivariate Poisson regression models.
The incorporation of detailed cigarette smoking history showed several notable associations with all mutational signatures tested except Signature 1. Current smoking was associated with greater ERCC2-Signature mutations compared to both never smokers (RR=1.40, 95% CI: 1.09–1.80, Table 2) and former smokers (p-value=0.0238, Supplemental Table 5); among cases with wild-type ERCC2, the relative proportion of ERCC2-Signature mutations was even stronger among current smokers (compared to never smokers, RR=1.52, 95% CI: 1.13–2.05). APOBEC-Signature 13 mutations were highest among former smokers (RR=1.58, 95% CI: 1.00–2.50, p-value=0.050), and APOBEC-Signature 2 mutations were highest among never smokers (RR=1.54, 95% CI: 1.17–2.01, p-value=0.0019, Table 2). Further, there was significant evidence that smoking duration was associated with ERCC2-Signature mutations among current smokers (p-trend=0.005) and with APOBEC-Signature 13 mutations among former smokers (p=0.0004, Supplemental Table 5). There were no significant trends between intensity or pack-years smoked and any of the signatures. Among former smokers, APOBEC-Signature 13 mutations were also increased with a shorter time since quitting (RR=2.62, 95% CI: 1.59– 4.30, p-value = 0.0002, Table 2).
Table 2.
Association between smoking characteristics and mutational signaturesa
| Smoking Characteristic | APOBEC-Sig 2 Mutations (C>T in TC[A/T] motifs) | APOBEC-Sig 13 Mutations (C>G in TC[A/T] motifs) | ERCC2-Sig Mutations | ||||
|---|---|---|---|---|---|---|---|
| N Cases | Risk Ratiob (95% CI) | p-val | Risk Ratiob,c (95% CI) | p-val | Risk Ratiob,c,d (95% CI) | p-val | |
| Smoking Status | |||||||
| Never smoked | 46 | 1.54 (1.17, 2.01) | 0.0019 | Ref | Ref | ||
| Former Smokers | 154 | 1.13 (0.90, 1.42) | 0.2803 | 1.58 (1.00, 2.50) | 0.050 | 1.15 (0.90, 1.47) | 0.2581 |
| Current Smokers | 103 | Ref | 0.86 (0.51, 1.43) | 0.5625 | 1.40 (1.09, 1.80) | 0.0077 | |
| Duration Smoked, years | |||||||
| Never smoked | 46 | 1.46 (1.09, 1.95) | 0.0102 | Ref | Ref | ||
| ≤25 | 61 | 0.93 (0.68, 1.26) | 0.6259 | 0.94 (0.54, 1.63) | 0.8148 | 1.31 (1.00, 1.72) | 0.0537 |
| >25–35 | 66 | 1.09 (0.82, 1.44) | 0.5493 | 1.84 (1.13, 3.02) | 0.0147 | 1.03 (0.78, 1.36) | 0.8433 |
| >35–45 | 56 | 1.13 (0.83, 1.56) | 0.4373 | 1.18 (0.68, 2.07) | 0.5581 | 1.33 (1.00, 1.78) | 0.0519 |
| >45 | 73 | Ref | 1.17 (0.70, 1.95) | 0.5516 | 1.37 (1.05, 1.79) | 0.0206 | |
| p-trend | 0.0980 | 0.5149 | 0.0068 | ||||
| Status and Duration | |||||||
| Never smoked | 46 | 1.55 (1.12, 2.13) | 0.0075 | Ref | Ref | ||
| Former Smoker | |||||||
| <30 years | 76 | 1.15 (0.85, 1.55) | 0.3748 | 1.45 (0.88, 2.39) | 0.1423 | 1.21 (0.93, 1.58) | 0.1571 |
| ≥30 years | 77 | 1.15 (0.86, 1.55) | 0.3454 | 2.00 (1.21, 3.31) | 0.0072 | 1.08 (0.82, 1.42) | 0.5993 |
| Current Smoker | |||||||
| <45 years | 51 | 1.02 (0.68, 1.52) | 0.9421 | 0.49 (0.23, 1.04) | 0.063 | 1.33 (0.98, 1.79) | 0.0631 |
| ≥45 years | 52 | Ref | 1.18 (0.67, 2.06) | 0.5623 | 1.46 (1.11, 1.93) | 0.0069 | |
| p-trend | 0.0078 | 0.3316 | 0.0052 | ||||
| Intensity smoked (packs per day) | |||||||
| Never Smoked | 46 | 1.32 (1.01, 1.72) | 0.0399 | Ref | Ref | ||
| <1 pack | 52 | 0.66 (0.47, 0.93) | 0.0189 | 1.59 (0.93, 2.71) | 0.0901 | 1.31 (0.98, 1.74) | 0.0709 |
| 2 packs | 110 | 0.96 (0.77, 1.20) | 0.7178 | 1.03 (0.63, 1.69) | 0.8963 | 1.31 (1.02, 1.68) | 0.0327 |
| 3+ packs | 95 | Ref | 1.39 (0.86, 2.24) | 0.1767 | 1.17 (0.90, 1.51) | 0.2473 | |
| p-trend | 0.2964 | 0.4507 | 0.4533 | ||||
| Pack-years | |||||||
| Never Smoked | 46 | 1.35 (1.05, 1.74) | 0.0213 | Ref | Ref | ||
| <40 | 133 | 0.97 (0.78, 1.19) | 0.744 | 1.19 (0.75, 1.91) | 0.4608 | 1.23 (0.96, 1.56) | 0.102 |
| ≥40 | 128 | Ref | 1.34 (0.84, 2.13) | 0.2246 | 1.22 (0.95, 1.56) | 0.1235 | |
| p-trend | 0.0657 | 0.2069 | 0.2217 | ||||
| Time since quitting (among Former Smokers) | |||||||
| Never Smoked | 46 | 1.41 (1.03, 1.95) | 0.0348 | Ref | Ref | ||
| ≥26 years | 49 | 1.00 (0.71, 1.40) | 0.9957 | 1.45 (0.84, 2.50) | 0.187 | 1.23 (0.91, 1.65) | 0.1777 |
| 13–25 | 54 | 1.16 (0.85, 1.59) | 0.3557 | 1.05 (0.59, 1.85) | 0.876 | 1.12 (0.84, 1.50) | 0.4273 |
| ≤ 13 | 51 | Ref | 2.62 (1.59, 4.30) | 0.0002 | 1.07 (0.79, 1.44) | 0.6788 | |
| p-trend | 0.0863 | 0.0002 | 0.9201 | ||||
Number of cases may not add up to total (n=307) due to exclusion of occasional smokers (n=4) and/or missing information for smoking duration or intensity.
Adjusted for age
Additionally adjusted for non-silent ERCC2 mutations
Additionally adjusted for state
A strong positive association was also observed between presence of any 1-bp deletions and both status and duration of smoking (RRFormer,<30yrs= 3.09, 95% CI: 0.80–11.89; RRFormer,≥30yrs= 3.57, 95% CI: 0.93–13.80; RRCurrent,<45yrs= 4.54, 95% CI: 1.13–18.27; RRCurrent,≥45yrs= 6.89, 95% CI: 1.77–26.82; p-trend=0.0027, Table 3). These deletions were most common in genes influencing histone modification/chromatin remodeling (67%). There were no positive associations between any smoking characteristic and the presence of 1-bp insertions.
Table 3.
Association between smoking characteristics and 1-bp indelsa
| 1-bp deletion | 1-bp insertion | |||||||
|---|---|---|---|---|---|---|---|---|
| (+) N Cases | (−) N Cases | Odds Ratiob (95% CI) | p-val | (+) N Cases | (−) N Cases | Odds Ratiob (95% CI) | p-val | |
| Smoking Status | ||||||||
| Never smoked | 3 | 43 | Ref | 10 | 36 | Ref | ||
| Former Smokers | 27 | 127 | 3.42 (0.96, 12.13) | 0.0574 | 34 | 120 | 0.83 (0.35, 1.93) | 0.6584 |
| Current Smokers | 27 | 76 | 5.77 (1.61, 20.67) | 0.0071 | 14 | 89 | 0.46 (0.18, 1.18) | 0.1056 |
| Duration Smoked, years | ||||||||
| Never smoked | 3 | 43 | Ref | 10 | 36 | Ref | ||
| ≤25 | 12 | 49 | 4.10 (1.06, 15.91) | 0.0416 | 13 | 48 | 0.90 (0.34, 2.38) | 0.8235 |
| >25–35 | 13 | 53 | 3.56 (0.93, 13.57) | 0.0629 | 8 | 58 | 0.41 (0.14, 1.17) | 0.0957 |
| >35–45 | 10 | 46 | 3.38 (0.83, 13.66) | 0.0881 | 15 | 41 | 1.09 (0.41, 2.91) | 0.8653 |
| >45 | 18 | 55 | 6.00 (1.57, 22.93) | 0.0087 | 12 | 61 | 0.49 (0.18, 1.35) | 0.1698 |
| p-trend | 0.0269 | 0.3057 | ||||||
| Status and Duration | ||||||||
| Never smoked | 3 | 43 | Ref | 10 | 36 | Ref | ||
| Former Smoker | ||||||||
| <30 years | 12 | 64 | 3.09 (0.80, 11.89) | 0.1007 | 15 | 61 | 0.72 (0.28, 1.85) | 0.4960 |
| ≥30 years | 14 | 63 | 3.57 (0.93, 13.80) | 0.0647 | 19 | 58 | 0.91 (0.35, 2.33) | 0.8382 |
| Current Smoker | ||||||||
| <45 years | 12 | 39 | 4.54 (1.13, 18.27) | 0.0332 | 8 | 43 | 0.63 (0.21, 1.89) | 0.4107 |
| ≥45 years | 15 | 37 | 6.89 (1.77, 26.82) | 0.0054 | 6 | 46 | 0.33 (0.11, 1.06) | 0.0631 |
| p-trend | 0.0027 | 0.0769 | ||||||
| Intensity smoked (packs per day) | ||||||||
| Never Smoked | 3 | 43 | Ref | 10 | 36 | Ref | ||
| <1 pack | 11 | 41 | 4.47 (1.13, 17.76) | 0.0334 | 13 | 39 | 0.93 (0.35, 2.48) | 0.8800 |
| 2 packs | 28 | 82 | 5.61 (1.58, 19.94) | 0.0078 | 15 | 95 | 0.45 (0.18, 1.14) | 0.0918 |
| 3+ packs | 15 | 80 | 2.93 (0.78, 10.95) | 0.1110 | 20 | 75 | 0.80 (0.32, 1.98) | 0.6292 |
| p-trend | 0.2659 | 0.4531 | ||||||
| Pack-years | ||||||||
| Never Smoked | 3 | 43 | Ref | 10 | 36 | Ref | ||
| <40 | 27 | 106 | 4.06 (1.15, 14.35) | 0.0295 | 26 | 107 | 0.76 (0.32, 1.78) | 0.5293 |
| ≥40 | 27 | 101 | 4.50 (1.25, 16.17) | 0.0214 | 23 | 105 | 0.59 (0.24, 1.44) | 0.2496 |
| p-trend | 0.0404 | 0.2357 | ||||||
| Time since quitting (among Former Smokers) | ||||||||
| Never Smoked | 3 | 43 | Ref | 10 | 36 | Ref | ||
| ≥26 years | 8 | 41 | 4.32 (0.97, 19.14) | 0.0544 | 11 | 38 | 0.68 (0.23, 1.99) | 0.4825 |
| 13–25 | 9 | 45 | 3.87 (0.94, 16.06) | 0.0619 | 13 | 41 | 0.79 (0.29, 2.17) | 0.6517 |
| ≤ 13 | 10 | 41 | 4.87 (1.15, 20.69) | 0.0318 | 10 | 41 | 0.72 (0.25, 2.10) | 0.5504 |
| p-trend | 0.0556 | 0.6504 | ||||||
Number of cases may not add up to total (n=307) due to exclusion of occasional smokers (n=4) and/or missing information for smoking duration or intensity.
Adjusted for age, race, state, sex and tumor cellularity.
1-bp indels: insertion or deletion of one base pair.
All analyses conducted separately for non-muscle invasive and muscle-invasive disease can be found in Supplemental Tables 7-10.
DISCUSSION
In the current analysis, we observed several associations between major bladder cancer risk factors and cancer gene mutations, as well as mutational signatures derived from 44 genes frequently mutated in bladder cancer. Notably, there was striking heterogeneity in the relationship between cigarette smoking and the established bladder cancer SBS and indel signatures that directly track with the components of smoking (status and duration) that drive the risk for bladder cancer. These results show a clear imprint of cigarette smoking in bladder tumors and provide new insights into the mechanisms by which cigarettes influence carcinogenesis.
The mutational profile of bladder tumors has been described to include four major SBS signatures[4, 25] and, more recently, doublet base substitution and indel signatures.[29] Interestingly, the SBS mutational signature associated with tobacco smoking (COSMIC 4) has not been identified in bladder cancer[30], but rather a nucleotide-excision repair (NER) pathway-related mutational signature, associated with recurrent somatic ERCC2 mutations, has been associated with ever (but not former/current) smoking status.[4, 25] Here, we also show a link between cigarette smoking and ERCC2-Signature mutations and, by using detailed lifetime cigarette smoking history, we further clarify that current smoking is associated with the greatest ERCC2-Signature mutational burden compared to both former and never smokers. Our results are consistent with a recent reanalysis of TCGA data that showed a significantly higher burden of COSMIC signature 5 (cosine similarity of 0.90 to ERCC2-signature) among current smokers.[31] The activity of this signature has also been shown to be positively correlated with pack-years smoked.[25, 30] Here, we demonstrate a dose-dependent increase in the proportion of ERCC2-signaure mutations with duration smoked (among current smokers), but not with intensity or pack-years of smoking. The above observations are directly supported by epidemiologic evidence that status and duration of cigarette smoking are the components of smoking that drive increasing risk for bladder cancer.[17]
NER is the major pathway for the repair of bulky DNA adducts induced by carcinogens in cigarette smoke,[32] thus, it is not surprising that ERCC2 (a major NER gene) is frequently altered in bladder cancer and that ERCC2-Signature mutational burden is highest among current smokers. Consistent with other reports, we also observed a higher proportion of ERCC2-Signature mutations in smokers compared to never smokers among those with wild-type ERCC2 [4, 25], but found that this was significantly higher in current (and not former) smokers. It has been suggested that the observed enrichment of ERCC2-Signature mutations in smokers with wild-type ERCC2 may be due to the influence of ERCC2 germline variants or other unidentified DNA repair genes/pathways contributing to ERCC2-Signature mutational burden.[25] Future integration of detailed smoking history with both somatic and germline NER pathway variant characterization may yield additional insights into the contribution of ERCC2-Signature mutations, particularly among lifelong smokers, and the mechanisms by which cigarette smoking influences bladder carcinogenesis.
APOBEC-signature mutations accounted for 43% of SNVs in our dataset of largely NMIBC, confirming that APOBEC-signature mutation load is likely generated early in the carcinogenic process. Here, we demonstrate a strong link between certain smoking characteristics that differ between the two APOBEC mutational signatures and show novel additional evidence of the imprint of cigarette smoking in bladder tumors. Loads of APOBEC-Signature 13 mutations (C>G in TC[A/T] motifs) were found to be highest among former smokers, compared to never or current smokers. These mutations were significantly enriched for mutations in histone-modifying and chromatin remodeling genes (Supplemental Table 11). Although this link has not been reported before in bladder cancer, pack-years of smoking has been linked to elevated APOBEC-Signature 13 mutational burden in smokers with lung adenocarcinoma.[25, 30] In our study, we also observed a significant increase in APOBEC-Signature 13 mutations with smoking duration and with shorter time since quitting among former smokers, which provides additional evidence that increasing exposure increases APOBEC-Signature 13 mutational load in former smokers. We also note, that APOBEC-Sig 13 mutational burden was not associated with intensity of smoking or with pack-years (the primary metric of cumulative cigarette smoking used to explore this relationship in prior analyses).[25, 30] APOBECs introduce mutations by editing single-stranded DNA, which is abundant under conditions associated with DNA damage, repair, and replication.[33, 34] Furthermore, expression of APOBEC3A and APOBEC3B genes that encode main mutagenic enzymes, is elevated in bladder tumors [4, 27]) and was found to be induced directly by DNA damage in bladder cancer cell lines.[27] In the process of bioactivation/detoxification, tobacco-derived carcinogens accumulate in urine where they form adducts with DNA at the bladder epithelium; resolution of these adducts through DNA breaks generates single-stranded DNA that activates the APOBEC system. Thus, the induction of APOBEC-signature mutations in bladder cancer due to cigarette smoking is plausible.
APOBEC-Sig 2 mutations (C>T in TC[A/T] motifs) were found to be highest among never smokers. This contrasts with reports based on TCGA data that never smokers had higher C>G mutations [19] and APOBEC-Signature 13 mutations (C>G in TC[A/T] motifs).[31] Despite the highly statistically significant finding for never smokers in our study, we only had 46 never-smoking bladder cancer cases; thus, our findings would need to be replicated. We note, however, the possibility of misclassification of the smoking data in TCGA, particularly among never smokers, who comprise an unusually high proportion (27.8%) of MIBC cases in that study[4] compared to only 11.5% of MIBC cases in the population-based NEBCS.[17] Alternatively, TCGA may not be representative of all MIBC. Additional exploration of the role of environmental tobacco smoke (ETS) among never smokers in our study showed no association between ETS and APOBEC-Sig 2 mutations. Thus, more work is needed to fully understand if there is a divergent etiology associated with the two APOBEC signatures in bladder tumors.
Recent data have characterized indel signatures, classified as deletions or insertions of a single base or longer fragments in many tumor types. In our dataset, the most common pattern was for 1-bp indels. When we evaluated the relationship between the presence of these indels and the various smoking characteristics, we observed a strong positive association between smoking status and duration smoked (p=0.0027) and 1-bp deletions. Nearly all the cases harboring these 1-bp deletions were smokers (54/57 tumors), further suggesting that these are driven by tobacco exposure. A more detailed analysis of specific types of 1-bp deletions was not possible due to the limited sample size, however we observed that 67% percent of these deletions occurred in genes influencing histone modification/chromatin remodeling. The mechanism by which observed mutations in genes involved in these processes impact bladder cancer development is not well understood, but histone methylation is generally described to influence gene expression and numerous cellular processes.[35] Future studies that aim to evaluate cigarette-induced histone and other epigenomic changes in experimental studies of bladder cancer, like those done for lung cancer[36, 37], are warranted given our findings.
There are sex differences in the incidence of bladder cancer with a male-to-female ratio of about 4.0, which cannot be entirely attributed to cigarette smoking.[2] Still, the molecular basis of this sex disparity is not well understood. Cross-cancer analysis of sex-based differences suggested a higher proportion of gene expression and DNA methylation differences in bladder cancer were significantly enriched in the sex chromosomes.[38] Recently, case-only findings by Hurst et al.[39] and by Nassar et al.[40] revealed that mutations in the X-linked KDM6A gene were more frequent in tumors from women compared to those from men. Here, we confirm this observation in 199 Ta tumors and find that 53.6% of Ta tumors from women and 35.6% of Ta tumors from men had at least one KDM6A mutation. This difference was statistically significant with multivariable adjustment for other bladder cancer risk factors that differ by sex, most notably, cigarette smoking. And, case-control analyses suggest a true increase in the risk for KDM6A positive tumors in women (p-heterogeneity=0.02, see Supplemental Table 3a). Among the 35 women with mutated KDM6A in our study, 9 had more than one KDM6A mutation, and two others have a likely loss of heterozygosity (allele fraction ≥70%). Several pieces of evidence support the tumor suppressor function of KDM6A in bladder cancer.[7, 41] Coupled with KDM6A escape from X-chromosome inactivation [42], this may partially explain the lower incidence rates for bladder cancer among women compared to men. Despite this protection, when women do develop bladder cancer, KDM6A mutations and reduced expression were shown to be predictive of reduced disease-free survival in MIBC (not observed in men).[41] Papillary (Ta) NMIBC, is frequently characterized by recurrent FGFR3 mutations (and now also KDM6A mutations) and recent data suggest that the co-occurrence of these alterations cooperate in bladder cells to reduce the expression of luminal genes, promoting a more basal (aggressive) cell state.[43] These findings provide some insights into the progression of some papillary tumors to more consequential subtypes; unfortunately, we do not have disease recurrence, progression, or survival information in this study to explore this question.
Over 40 high-risk occupations have been identified for bladder cancer, most notably from historic exposure to aromatic amine chemicals.[2] Lifetime assessment of occupational histories (all jobs held for at least 6 months since age 16) from patients in this study show the highest frequency of ever being employed in transportation occupations, as mechanics and repairers, in military occupations, as handlers, equipment cleaners and laborers, or in service occupations (except private households and protective) (Supplemental Table 5). The mutational profile of subjects ever employed in a high-risk occupation suggests an excess of mutation in genes regulating the cell-cycle, particularly for mutations in TP53. Our data also show a higher burden of ERCC2-Signature mutations among those employed in high-risk occupations compared to those who were never employed in these jobs (Table 4). Our results suggest that exogenous exposures (other than smoking) contribute to the mutational burden in bladder tumors and are consistent with the observed excess risk for bladder cancer due to occupational carcinogens. One other independent study also identified an excess of TP53 mutations among men who held high-risk occupations [44] but we are not aware of similar reports linking high-risk occupations to mutational profiles in bladder tumors, thus these results require replication. Since the exposures in the noted high-risk occupations are highly heterogenous, studies that also include detailed characterization of specific exposures and putative mechanisms that are driving these associations are needed.
Table 4.
Association between high-risk occupation and mutational signatures
| N | APOBEC-Sig 2 Mutations (C>T in TC[A/T] motifs) | APOBEC-Sig 13 Mutations (C>G in TC[A/T] motifs) | ERCC2-Sig Mutations | Sig 1 Mutations C>T at CpG dinucleotides | |||||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Risk Ratioa (95% CI) | p-val | Risk Ratioa (95% CI) | p-val | Risk Ratioa (95% CI) | p-val | Risk Ratioa (95% CI) | p-val | |
| High-risk occupation | |||||||||
| No | 63 | Ref | Ref | Ref | Ref | ||||
| Yes | 241 | 0.67 (0.52, 0.84) | 0.0008 | 1.34 (0.87, 2.05) | 0.1781 | 1.40 (1.09, 1.79) | 0.0380 | 1.43 (0.88, 2.34) | 0.1501 |
Adjusted for age, race, state, sex, smoking status and duration, tumor cellularity, and non-silent ERCC2 mutations.
Our study has several strengths, most notably, a detailed characterization of all bladder cancer risk factors from an extensive interview with study participants. This allowed for a comprehensive analysis of the relationship between smoking characteristics (status, duration, intensity, pack-years, and ETS) and somatic mutations in bladder cancer. We were also able to assess the impact of lifelong occupational exposures, which are not routinely collected in the clinical setting. The weaknesses of the study include limitations in the sample size to look at individual gene mutations and the size of the targeted sequencing panel. Despite the small number of SNVs detected, we were able to reliably characterize the four main SBS signatures previously reported in bladder tumors. This is supported by the observed relationship in our data to established characteristics of these signatures, i.e., age at diagnosis and COSMIC SBS signature 1[30], non-silent ERCC2 mutation and ERCC2-Signature mutations[25], and the germline bladder cancer risk variant rs1014971[28] associated with increased APOBEC mutational load.[27] Still, some misclassification of the contribution to each signature is possible. As an added check of the signature derivation from SignatureEstimation, we manually annotated all mutations according to stringent APOBEC context [45] (to compare observed results between APOBEC mutagenesis and smoking characteristics) and found the same association between former smoking duration and APOBEC-Signature 13 C>G in TC[A/T] mutations and APOBEC-Signature 2 C>T in TC[A/T] mutations and never smoking (data not shown). In addition, when we extracted the four signatures from the bladder cancer samples in the publicly available TCGA dataset (which includes only MIBC) using only the 44 genes included in our panel, the relative contributions of the signatures was observed to be comparable to those from muscle-invasive cancers in our dataset (Supplemental Table 12). Other limitations in the study include our inability to characterize mutations in the TERT promoter using our panel. The TERT promoter, although commonly mutated in bladder cancer, is very GC-rich and effective analysis requires a specific PCR-based approach or whole genome sequencing which was not possible in our study on FFPE tumors.
In conclusion, our study provides additional insight into the impact of cigarette smoking on the mutational burden in bladder cancer. In addition, we found that other major risk factors, including occupational exposures, also contribute to known bladder cancer mutations and mutational signatures. This work highlights the value of integrated analyses of risk factor information with somatic mutation characterization to add insight into the mechanism for bladder carcinogenesis.
Supplementary Material
Table 1.
Select characteristics among 307 patients with targeted sequencing data in the New England Bladder Cancer Study
| Characteristic | N | Percent |
|---|---|---|
| Age | ||
| <55 | 52 | 16.9 |
| 55–64 | 87 | 28.3 |
| 65–74 | 109 | 35.5 |
| 75+ | 59 | 19.2 |
| Sex | ||
| Female | 74 | 24.1 |
| Male | 233 | 75.9 |
| Race | ||
| White | 279 | 90.9 |
| Non-White | 28 | 9.1 |
| Smoking | ||
| Occasional smokers | 4 | 1.3 |
| Never smokers | 46 | 15.0 |
| Former smokers | 154 | 50.2 |
| Current smokers | 103 | 33.6 |
| High-Risk Occupation | ||
| No | 63 | 20.5 |
| Yes | 241 | 78.5 |
| Missing | 3 | 1.0 |
| Stage/Grade | ||
| Ta, Low Grade | 139 | 45.3 |
| Ta, High Grade | 60 | 19.5 |
| T1, High Grade | 61 | 19.9 |
| T2+ (MIBC) | 47 | 15.3 |
Translational Relevance:
Few studies have examined detailed risk factor information with somatic mutational profiling and mutational signatures in population-based studies. By analyzing 322 representative urothelial bladder carcinomas, we confirm an enrichment of KDM6A somatic mutations in women and show etiologic heterogeneity in risk. Mutational signature profiles demonstrate a clear imprint of cigarette smoking in bladder tumors and show that these track with the components of smoking (status and duration) that drive the risk for bladder observed in population-based epidemiologic studies. TP53 mutations were enriched in patients with a history of ever holding a high-risk occupation. These observations underscore the connection between established bladder cancer risk factors and the mutational burden in tumors and highlight the need for detailed clinical characterization of these factors.
Acknowledgments
Funding: This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics (ZIA CP010125-24).
Funding: This work was funded by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics (ZIA CP010125-24).
Footnotes
Author disclosures: The authors declare no potential conflicts of interest.
References
- 1.Ferlay J, Ervik M, Lam F, Colombet M, Mery L, Piñeros M, et al. Global Cancer Observatory: Cancer Today. https://gco.iarc.fr/today.
- 2.Silverman DT, Koutros S, Figueroa JD, Prokunina-Olsson L, Rothman N. Bladder Cancer. In: Thun MJ, Linet MS, Cerhan C, et al. , (eds). Schottenfeld and Fraumeni Cancer Epidemiology and Prevention. 4th ed. New York: Oxford University Press; 2018, 977–96. [Google Scholar]
- 3.Solomon DA, Kim JS, Bondaruk J, Shariat SF, Wang ZF, Elkahloun AG, et al. Frequent truncating mutations of STAG2 in bladder cancer. Nat Genet. 2013;45(12):1428–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell. 2017;171(3):540–56 e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pietzak EJ, Bagrodia A, Cha EK, Drill EN, Iyer G, Isharwal S, et al. Next-generation Sequencing of Nonmuscle Invasive Bladder Cancer Reveals Potential Biomarkers and Rational Therapeutic Targets. Eur Urol. 2017;72(6):952–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nordentoft I, Lamy P, Birkenkamp-Demtroder K, Shumansky K, Vang S, Hornshoj H, et al. Mutational context and diverse clonal development in early and late bladder cancer. Cell Rep. 2014;7(5):1649–63. [DOI] [PubMed] [Google Scholar]
- 7.Nickerson ML, Dancik GM, Im KM, Edwards MG, Turan S, Brown J, et al. Concurrent alterations in TERT, KDM6A, and the BRCA pathway in bladder cancer. Clin Cancer Res. 2014;20(18):4935–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isharwal S, Audenet F, Drill EN, Ostrovnaya I, Pietzak EJ, Al-Ahmadie H, et al. Next generation sequencing of urothelial bladder cancer: Memorial Sloan Kettering Cancer Center experience in 454 patients. Journal of Clinical Oncology. 2018;36(6_suppl):469. [Google Scholar]
- 9.Guo G, Sun X, Chen C, Wu S, Huang P, Li Z, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. 2013;45(12):1459–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balbas-Martinez C, Sagrera A, Carrillo-de-Santa-Pau E, Earl J, Marquez M, Vazquez M, et al. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat Genet. 2013;45(12):1464–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Audenet F, Isharwal S, Cha EK, Donoghue MTA, Drill EN, Ostrovnaya I, et al. Clonal Relatedness and Mutational Differences between Upper Tract and Bladder Urothelial Carcinoma. Clin Cancer Res. 2019;25(3):967–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schned AR, Andrew AS, Marsit CJ, Kelsey KT, Zens MS, Karagas MR. Histological classification and stage of newly diagnosed bladder cancer in a population-based study from the Northeastern United States. Scand J Urol Nephrol. 2008;42(3):237–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sobin LH, Gospodarowicz MK, Wittekind C. UICC TNM Classification of Malignant Tumours. Seventh ed. New York; 2009. [Google Scholar]
- 14.Sobin LH, Wittekind C. UICC TNM Classification of Malignant Tumours. Sixth ed. New York; 2002. [Google Scholar]
- 15.Eble JN, Sauter G, Epstein JI, Sesterhenn IA. Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs. In: World Health Organization Classification of Tumours, (ed). Lyon: IARC Press; 2004. [Google Scholar]
- 16.Mostfi FK, Sobin LH, Torloni H. Histological typing of urinary bladder tumours. Geneva: World Health Organization; 1973. [Google Scholar]
- 17.Baris D, Karagas MR, Verrill C, Johnson A, Andrew AS, Marsit CJ, et al. A case-control study of smoking and bladder cancer risk: emergent patterns over time. J Natl Cancer Inst. 2009;101(22):1553–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colt JS, Karagas MR, Schwenn M, Baris D, Johnson A, Stewart P, et al. Occupation and bladder cancer in a population-based case-control study in Northern New England. Occup Environ Med. 2011;68(4):239–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balbas-Martinez C, Rodriguez-Pinilla M, Casanova A, Dominguez O, Pisano DG, Gomez G, et al. ARID1A alterations are associated with FGFR3-wild type, poor-prognosis, urothelial bladder tumors. PLoS One. 2013;8(5):e62483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hafner C, van Oers JM, Vogt T, Landthaler M, Stoehr R, Blaszyk H, et al. Mosaicism of activating FGFR3 mutations in human skin causes epidermal nevi. J Clin Invest. 2006;116(8):2201–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernandez S, Lopez-Knowles E, Lloreta J, Kogevinas M, Jaramillo R, Amoros A, et al. FGFR3 and Tp53 mutations in T1G3 transitional bladder carcinomas: independent distribution and lack of association with prognosis. Clin Cancer Res. 2005;11(15):5444–50. [DOI] [PubMed] [Google Scholar]
- 23.Koutros S, Kogevinas M, Friesen MC, Stewart PA, Baris D, Karagas MR, et al. Diesel exhaust and bladder cancer risk by pathologic stage and grade subtypes. Environ Int. 2020;135:105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3(1):246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, Mouw KW, Polak P, Braunstein LZ, Kamburov A, Kwiatkowski DJ, et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat Genet. 2016;48(6):600–06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang X, Wojtowicz D, Przytycka TM. Detecting presence of mutational signatures in cancer with confidence. Bioinformatics. 2017; 10.1093/bioinformatics/btx604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Middlebrooks CD, Banday AR, Matsuda K, Udquim KI, Onabajo OO, Paquin A, et al. Association of germline variants in the APOBEC3 region with cancer risk and enrichment with APOBEC-signature mutations in tumors. Nat Genet. 2016;48(11):1330–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rothman N, Garcia-Closas M, Chatterjee N, Malats N, Wu X, Figueroa JD, et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet. 2010;42(11):978–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexandrov L, Kim J, Haradhvala NJ, Huang MN, Ng AWT, Boot A, et al. The Repertoire of Mutational Signatures in Human Cancer. bioRxiv. 2018; 10.1101/322859:322859. [DOI] [Google Scholar]
- 30.Alexandrov LB, Ju YS, Haase K, Van Loo P, Martincorena I, Nik-Zainal S, et al. Mutational signatures associated with tobacco smoking in human cancer. Science. 2016;354(6312):618–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fantini D, Seiler R, Meeks JJ. Molecular footprints of muscle-invasive bladder cancer in smoking and nonsmoking patients. Urol Oncol. 2018; 10.1016/j.urolonc.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 32.Formation Hang B. and repair of tobacco carcinogen-derived bulky DNA adducts. J Nucleic Acids. 2010;2010:709521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landry S, Narvaiza I, Linfesty DC, Weitzman MD. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 2011;12(5):444–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nowarski R, Kotler M. APOBEC3 cytidine deaminases in double-strand DNA break repair and cancer promotion. Cancer Res. 2013;73(12):3494–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alam H, Gu B, Lee MG. Histone methylation modifiers in cellular signaling pathways. Cell Mol Life Sci. 2015;72(23):4577–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaz M, Hwang SY, Kagiampakis I, Phallen J, Patil A, O’Hagan HM, et al. Chronic Cigarette Smoke-Induced Epigenomic Changes Precede Sensitization of Bronchial Epithelial Cells to Single-Step Transformation by KRAS Mutations. Cancer Cell. 2017;32(3):360–76 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sundar IK, Nevid MZ, Friedman AE, Rahman I. Cigarette smoke induces distinct histone modifications in lung cells: implications for the pathogenesis of COPD and lung cancer. J Proteome Res. 2014;13(2):982–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan Y, Liu L, Chen H, Wang Y, Xu Y, Mao H, et al. Comprehensive Characterization of Molecular Differences in Cancer between Male and Female Patients. Cancer Cell. 2016;29(5):711–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hurst CD, Alder O, Platt FM, Droop A, Stead LF, Burns JE, et al. Genomic Subtypes of Non-invasive Bladder Cancer with Distinct Metabolic Profile and Female Gender Bias in KDM6A Mutation Frequency. Cancer Cell. 2017;32(5):701–15 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nassar AH, Umeton R, Kim J, Lundgren K, Harshman L, Van Allen EM, et al. Mutational Analysis of 472 Urothelial Carcinoma Across Grades and Anatomic Sites. Clin Cancer Res. 2019;25(8):2458–2470. [DOI] [PubMed] [Google Scholar]
- 41.Kaneko S, Li X. X chromosome protects against bladder cancer in females via a KDM6A-dependent epigenetic mechanism. Sci Adv. 2018;4(6):eaar5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenfield A, Carrel L, Pennisi D, Philippe C, Quaderi N, Siggers P, et al. The UTX gene escapes X inactivation in mice and humans. Hum Mol Genet. 1998;7(4):737–42. [DOI] [PubMed] [Google Scholar]
- 43.Barrows D, Feng L, Carroll TS, and Allis D. Loss of UTX/KDM6A and the activation of FGFR3 converge to regulate differentiation gene-expression programs in bladder cancer. PNAS. 2020;117(41) 25732–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelsey KT, Hirao T, Hirao S, Devi-Ashok T, Nelson HH, Andrew A, Colt J, Baris D, Morris JS, Schned A, Karagas M. TP53 alterations and patterns of carcinogen exposure in a U.S. population-based study of bladder cancer. Int J Cancer. 2005;117(3):370–5. [DOI] [PubMed] [Google Scholar]
- 45.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45(9):970–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.