Abstract
Purpose:
Tumor-associated stroma is comprised of fibroblasts, tumor infiltrating lymphocytes (TILs), macrophages, endothelial, and other cells, that interactively influence tumor progression through inflammation and wound repair. Although gene expression signatures reflecting wound repair predict breast cancer survival, it is unclear whether combined density of tumor-associated stromal cells, a morphological proxy for inflammation and wound repair signatures on routine hematoxylin and eosin (H&E)-stained sections, is of prognostic relevance.
Methods:
By applying machine learning to digitized H&E-stained sections for 2,084 breast cancer patients from China (n=596; 24-55years), Poland (n=810; 31-75years), and the United States (n=678; 55-78years), we characterized tumor-associated stromal cellular density (SCD) as the percentage of tumor-stroma that is occupied by nucleated cells. Hazard ratios (HR) and 95% confidence intervals (CI) for associations between SCD and clinical outcomes (recurrence (China); mortality (Poland and United States)) were estimated using Cox proportional hazard regression, adjusted for clinical variables.
Results:
SCD was independently predictive of poor clinical outcomes in hormone receptor-positive (luminal) tumors from China (multivariable HR(95%CI)fourth(Q4) vs first(Q1) quartile=1.86(1.06-3.26);Ptrend=0.03), Poland (HR(95%CI)Q4 vs Q1=1.80(1.12-2.89);Ptrend=0.01), and United States (HR(95%CI)Q4 vs Q1=2.42(1.33-4.42);Ptrend=0.002). In general, SCD provided more prognostic information than most classical clinicopathologic factors, including grade, size, PR, HER2, IHC4, and TILs, predicting clinical outcomes irrespective of menopausal or lymph nodal status. SCD was not predictive of outcomes in hormone receptor-negative tumors.
Conclusions:
Our findings support the independent prognostic value of tumor-associated SCD among ethnically-diverse luminal breast cancer patients.
Impact:
Assessment of tumor-associated SCD on standard H&E could help refine prognostic assessment and therapeutic decision-making in luminal breast cancer.
Keywords: breast cancer, tumor-associated stromal cellular density, prognosis, machine learning, tumor microenvironment
Introduction
Emerging evidence indicates that components of the tumor microenvironment (TME), including inflammation and wound repair, may possess independent prognostic properties that can aid to further stratify breast cancer patients into clinically relevant subgroups (1-6). To date, however, many of the existing prognostic parameters do not incorporate biological and/or molecular characteristics of the TME in determining tumor aggressiveness or in predicting treatment response (7-11).
Encompassing cancer-associated fibroblasts (CAFs), inflammatory cells (including tumor infiltrating lymphocytes (TILs) and macrophages), endothelial and other nucleated cells, the composition of the tumor-stroma resembles that of wound healing process except that it is not self-limiting in cancers (12). The prognostic value of individual stromal cell populations has been well-documented (13-16). In addition, results from recent studies suggest that gene expression signatures reflective of inflammatory and wound repair processes were predictive of clinical outcomes in breast cancer (6, 17, 18). However, widespread adoption of these signatures is hampered by complexity of their evaluation, including requirements for fresh frozen tissue, RNA sequencing technology, and sophisticated immunohistochemical staining protocols.
Identification of prognostically relevant proxies for inflammation and wound repair processes on hematoxylin and eosin (H&E)-stained tissue sections, which are routinely performed as part of the diagnostic workup for breast cancer patients, could address challenges associated with gene or protein expression-based signatures, and be transformative in breast cancer management. Although visual assessment of the tumor-stroma to quantify features on H&E is challenging (19, 20), machine learning is an emerging alternative that enables systematic evaluation of tissue composition as well as cell detection on H&E images (20-22).
In the current study, we leveraged machine learning algorithms for spatial characterization of digitized H&E-stained breast cancer tissue sections into epithelial (tumor) and stromal regions and to detect and count the total number of nucleated cells (encompassing CAFs, TILs, macrophages, endothelial, and other stromal cells) per unit area of stroma i.e. stromal cellular density (SCD). Optimized machine learning algorithms were applied to H&E images from 2,084 breast cancer patients from study populations in China, Poland, and the United States (US). We assessed the prognostic significance of SCD in all patients and in stratified analyses by hormone receptor expression. We also compared the prognostic power of SCD to that of standard clinical factors and, for a subset of patients with relevant data, we compared SCD to TILs, as well as CD3+ and CD8+ T cell-densities.
Methods and Materials
Study population
This analysis included 2,084 patients with histologically or cytologically confirmed invasive breast cancer from study populations in China, Poland, and the United States (US). Chinese breast cancer patients (n=596) were premenopausal women, aged 24-55 years, diagnosed, and received treatment for hormone receptor-positive (luminal) breast cancer at the Cancer Hospital, Chinese Academy of Medical Sciences (CHCAMS) between 2008-2012. The patients received adjuvant endocrine therapy for at least five years after surgery, with or without systemic adjuvant chemotherapy or radiotherapy. Polish patients (n=810) were women participating in the Polish Breast Cancer Study (PBCS), which is a population-based case-control study that recruited women aged 20-74 years across participating hospitals in Warsaw and Lodz between 2000-2003 (23). PBCS participants were unselected for hormone receptor-expression status or menopause. US patients were postmenopausal women with incident invasive breast cancer, unselected for hormone receptor status, participating in the National Cancer Institute’s (NCI’s) Prostate, Lung, Colorectal & Ovarian (PLCO) cancer screening trial, a randomized controlled screening trial that recruited individuals aged 55-87 years between 1993-2001 (24, 25).
The primary endpoints of interest were 10-year Disease-free survival (DFS i.e. recurrence) for CHCAMS patients and 10-year Overall Survival (OS) for PBCS and PLCO patients. 10-year breast cancer-specific survival (BCSS) was considered as a secondary endpoint in all the cohorts due to fewer number of confirmed events. For CHCAMS and PBCS, data on treatment (surgery, endocrine therapy, chemotherapy, and/or trastuzumab), tumor size, lymph nodal involvement, histologic grade, and immunohistochemical (IHC) markers, including hormone (estrogen and progesterone) receptor (ER and PR) expression status, human epidermal growth factor receptor 2 (HER2) expression, and KI67, were obtained from clinical records. IHC scoring protocols for all markers have previously been described for participants from both studies (26, 27). For PLCO, data on tumor characteristics were obtained from supplemental questionnaires that were administered to participants with a diagnosis of breast cancer. Treatment and mortality data were collected by means of an annual study questionnaire as well as linkage to the National Death Index (NDI) (24).
Breast cancer subtypes were defined based on published criteria (28, 29) incorporating IHC markers and grade, as follows: Luminal A-like (ER+/PR+/HER2−/grade 1); luminal B-like (ER+/PR+ and: HER2+ and/or grade 2/3; ER+/PR−/HER2±/irrespective of grade; ER−/PR+/HER2±/irrespective of grade); HER2− enriched (ER−/PR−/HER2+); and triple-negative breast cancer (TNBC: ER−/PR−/HER2−). In general, ER+ and/or PR+ tumors were considered as luminal whereas ER− and PR− tumors were defined as non-luminal.
A subset of luminal breast cancer patients from CHCAMS (n=425) and PBCS (n=468) had complete quantitative data on the relevant IHC markers (ER, PR, HER2, KI67) to allow for the calculation of the IHC4 score according to published equation by Cuzick et al (10).
All studies were approved by local as well as US National Institutes of Health (NIH) institutional review boards. CHCAMS also received exemption by the Office of Human Research Protections at the NIH since it did not involve interactions with human subjects and/or use of personal identifying information. All patients in PBCS and PLCO provided written informed consent. For CHCAMS, informed consents were not required for the use of existing pathology materials with no reveal of identifiable patient information.
Retrieval, digitization, and analysis of archival hematoxylin & eosin (H&E)-stained sections
The collection and processing of formalin-fixed paraffin-embedded (FFPE) tumor blocks have been previously described for CHCAMS (26), PBCS (30), and PLCO (31) studies. For the current analysis, we utilized archival H&E-stained sections that were stored within the US National Cancer Institute’s Division of Cancer Epidemiology and Genetics’ digital image repository, managed by the Molecular and Digital Pathology Laboratory (MDPL; Center for Genomic Research, Leidos).
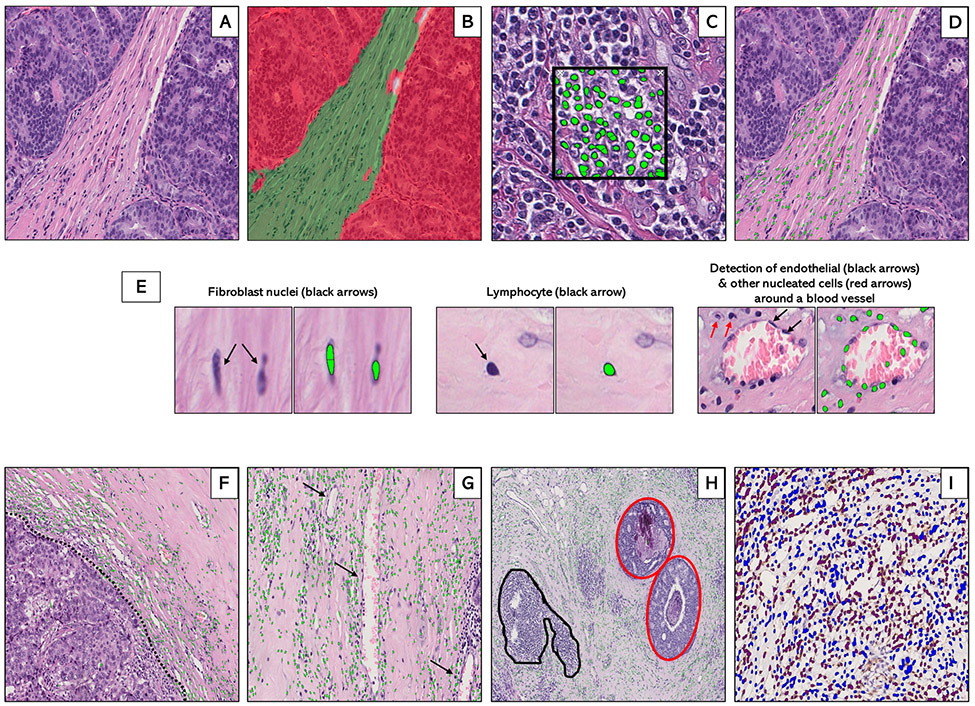
Image analysis was performed by using commercially available Halo software algorithm (Indica Labs, Albuquerque, NM). To account for pre-analytical variability in staining, pathologist-supervised machine learning scripts were developed for each study (Supplementary Table S1). Tissue classifier scripts were initially trained to segment regions on H&E images comprised of tumor (invasive and in-situ), stroma, and adipose tissue (Figure 1; A: H&E (unanalyzed); B: H&E (analyzed)). Next, cell detection scripts were trained to identify and count nucleated cells (green dots) in several regions of interest (Figure 1C). Training was based on nuclear morphology, including nuclear size (NS), minimum nuclear roundness (MNR), nuclear detection weight (NDW), and nuclear contrast threshold (NCT). These parameters were tuned to detect nucleated stromal cells, while excluding tumor-nuclei. Apart from NDW, which is a staining-based parameter, all other nuclear detection parameters were similar between the three study populations (Supplementary Table S1). The optimized cell detection script demonstrated excellent agreement (correlation=0.94; P-value<0.001) with manual cell counts across several regions of interest (Supplementary Figure S1). By embedding the previously optimized tissue classification script (Figure 1B) within the cell detection script (Figure 1C), cell detection was confined only to the stromal compartment (Figure 1D).
Figure 1.
Supervised machine learning for tissue segmentation and cell detection in breast cancer hematoxylin and eosin (H&E)-stained whole tissue sections. Machine-learning algorithms were applied to digitized hematoxylin and eosin (H&E)-stained sections (A) to generate data on stromal cellular density. First, an optimized tissue classification script was used to segment the tumor into epithelial (red) and stromal (green) areas (B). Next, a cell detection script was trained to segment and detect cells (green dots) based on color deconvolution as well as nuclear characteristics (C). Tissue classification and cell detection scripts were combined to enable cell detection to be limited to the stromal compartment (D). Examples of individual stromal cells, including fibroblasts, lymphocytes, and endothelial cells are also shown (E). In general, the machine counted stromal cells (green dots) at the tumor-stroma border (F: dotted black line), around blood vessels (G: black arrows) and in the stroma surrounding foci of ductal carcinoma in-situ (H: inset, red) while excluding tertiary lymphoid structures and lymphoid aggregates (H: inset, black). For a subset of patients with immunohistochemical staining on CD3+ and CD8+ T-cells, optimized scripts were used to quantify the densities of IHC-positive (brown) and IHC-negative (blue) cells within the stroma (I).
In addition to counting cells (shown as green dots) in the intervening stroma between infiltrating tumor nests or masses (Figure 1D), the machine counted cells along the tumor-stroma border (Figure 1F); and in stroma surrounding blood/lymphatic vessels (Figure 1G). For invasive tumors with an in-situ component, cell detection was limited to the surrounding stroma (red inset; Figure 1H). To capture only tumor-associated stromal cells, algorithms were trained to exclude tertiary lymphoid structures and lymphoid aggregates from the total stromal cell count (black inset; Figure 1H).
Based on our algorithms, we generated data on total tissue area (mm2) on H&E images, tumor area (mm2), stroma area (mm2), and total number of nucleated cells within the stromal compartment. Standard SCD was calculated by dividing total number of nucleated cells by stromal area. To convert this to a percentage, which is more intuitive and readily interpretable, we calculated total nuclei area (by multiplying the total number of nucleated cells by the average area (mm2) of a single nucleus (~2.0×10−4)), divided this by the combined stroma and nuclei area (i.e. stroma area + total nuclei area), and multiplied by 100. As would be expected, there was near-perfect positive correlation (r=0.99) between percent and standard SCD in all three study populations (Supplementary Figure S2).
In addition to SCD, we obtained data on stromal TILs for all CHCAMS patients (n=596). TIL scoring was performed by a pathologist (JZ) using recommended international guidelines (32). Subsets of these patients were also selected based on having high (n=100) and low (n=60) SCD and the corresponding FFPE blocks were retrieved and re-sectioned for CD3 and CD8 IHC staining. CD3+ and CD8+ T-cell densities were obtained by using optimized tissue segmentation and cell detection scripts as described for SCD. An additional color deconvolution step was used to separate IHC (3,3’-diaminobenzidine (DAB))-positive from DAB-negative cells (Figure 1H). The correlations between scripts that were independently trained by two digital pathology experts (MA and SML) for CD3+ and CD8+ T-cell detection were 0.93 (P-value <0.001) and 0.85 (P-value <0.001), respectively, demonstrating excellent reproducibility.
Statistical analysis
Frequency tables were used to assess the distributions of patients’ baseline characteristics. Histograms, box plots, and Kruskal-Wallis test were used to examine differences in the distributions of SCD by study and by relevant tumor characteristics. Associations between tumor characteristics at baseline and clinical outcomes were assessed in age-adjusted Cox proportional hazard regression models. Associations between SCD quartiles (Q1-Q4) and clinical outcomes (10-year DFS in CHCAMS and 10-year OS in PBCS and PLCO) were assessed in Kaplan-Meier survival curves and in Cox proportional hazard regression models. SCD was modelled both as quartiles (Q2, Q3, Q4 vs Q1) and continuous (0-100%) variables with adjustments for histologic grade (low(reference), intermediate, high), tumor size (≤2cm(reference), >2cm), lymph nodal involvement (negative(reference), positive), tumor subtype (luminal A-like(reference), luminal B-like, HER2-enriched, and TNBC), treatment (endocrine therapy (yes vs none), chemotherapy (yes vs none), and/or Herceptin (yes vs none)) and total tissue area (mm2). Violation of the Cox model proportionality assumption was assessed by modeling SCD as a time varying covariate – no violations were observed. Analyses were performed overall and by hormone receptor status. The contributions of SCD and standard clinical parameters to prognosis were assessed by determining the change in likelihood ratio chi-square (ΔLRχ2) following LR tests when each factor was removed from a fully adjusted model comprising of all other factors. The joint prognostic value of SCD and standard clinical parameters was assessed by creating a compositive variable combining binary categories of SCD and individual clinical parameters and modelling this in multivariable Cox models. Agreements between SCD, TILs, CD3+, and CD8+ T-cell densities were assessed using Pearson’s correlation and two-way scatter plots. In sensitivity analyses, we evaluated the association between SCD and 10-year BCSS in all cohorts. We also investigated SCD in relation to treatment (CHCAMS and PBCS) and DFS by surgery type (radical vs. breast-conserving). All tests were two-sided, and analyses were conducted using Stata statistical software version 16.1 (StataCorp, Lakeway Drive, TX, USA).
Results
Patient population and description of baseline characteristics
As shown in Table 1, participants’ ages differed by study population, which may be reflective of differences in patient selection among studies. Whereas CHCAMS (median (range)= 44 (24-55) years) and PLCO (67 (55-87) years) enrolled pre- and post-menopausal patients, respectively, PBCS (55 (31-75) years) patients were unselected for menopausal status. The median (range) follow-up durations were 7 (0.2-11.2), 15 (0.2-18.5), and 11 (0.1-21) years in CHCAMS, PBCS, and PLCO. All CHCAMS patients, as well as the majority of PBCS (68.6%) and PLCO (75.8%) patients, had ER+ tumors. Although the frequency of HER2 overexpression was higher in CHCAMS (22%) than PBCS (16.7%) or PLCO (15.5%) patients, the majority of the tumors were low or intermediate grade, small (≤2cm), and node-negative in all the studies. The majority (>86%) of the patients had stage I or II disease at diagnosis, with 13%, 2.5%, and 5.2% having stage III/IV disease in CHCAMS, PBCS, and PLCO studies, respectively. Most tumor characteristics at baseline demonstrated associations with clinical outcomes that were in the expected directions.
Table 1:
Baseline patient characteristics and age-adjusted hazard ratios (HR) for tumor characteristics in relation to disease-free survival (CHCAMS) and overall survival (PBCS and PLCO)
| Study population | ||||||
|---|---|---|---|---|---|---|
| CHCAMS (n = 596) | PBCS (n = 810) | PLCO (n = 678) | ||||
| Freq (%) | HR (95% CI) | Freq (%) | HR (95% CI) | Freq (%) | HR (95% CI) | |
| Age, years | ||||||
| <35 | 60 (10.1) | 6 (0.7) | NA | |||
| 35-50 | 467 (78.3) | 215 (26.5) | NA | |||
| 50-65 | 69 (11.6) | 378 (46.7) | 227 (33.5) | |||
| >65 | NA | 211 (26.1) | 451 (66.5) | |||
| Median age (range) | 44 (24-55) | 55 (31-75) | 67 (55-87) | |||
| Median FU (range), years | 7 (0.2-11.2) | 15 (0.2-18.5) | 11 (0.1-21) | |||
| Events (DFS/OS) | ||||||
| Absent | 475 (81.9) | 443 (55.4) | 486 (71.7) | |||
| Present | 105 (18.1) | 356 (44.6) | 192 (28.3) | |||
| Endocrine | NA | |||||
| No | 97 (16.3) | 464 (57.3) | ||||
| Yes | 499 (83.7) | 346 (42.7) | ||||
| Trastuzumab | NA | |||||
| No | 567 (95.1) | 798 (98.5) | ||||
| Yes | 29 (4.9) | 12 (1.5) | ||||
| ER status | ||||||
| Negative | 0 () | 249 (31.4) | 1.00 (reference) | 153 (24.2) | 1.00 (reference) | |
| Positive | 596 (100) | 544 (68.6) | 0.60 (0.46, 0.78) | 480 (75.8) | 0.70 (0.48, 1.00) | |
| PR status | ||||||
| Negative | 31 (5.2) | 1.00 (reference) | 373 (47.1) | 1.00 (reference) | 273 (46.1) | 1.00 (reference) |
| Positive | 565 (94.8) | 0.70 (0.32, 1.50) | 419 (52.9) | 0.53 (0.41, 0.69) | 319 (53.9) | 0.76 (0.53, 1.09) |
| HER2 | ||||||
| Negative | 415 (77.4) | 1.00 (reference) | 590 (83.3) | 1.00 (reference) | 365 (84.5) | 1.00 (reference) |
| Positive | 121 (22.6) | 0.62 (0.36, 1.07) | 118 (16.7) | 1.73 (1.25, 2.39) | 67 (15.5) | 1.71 (1.08, 2.73) |
| Subtype | ||||||
| Lum A-like | 326 (60.a8) | 1.00 (reference) | 282 (40.0) | 1.00 (reference) | 129 (35.7) | 1.00 (reference) |
| Lum B-like | 210 (39.2) | 0.94 (0.62, 1.42) | 256 (36.3) | 1.55 (1.11, 2.18) | 132 (36.6) | 1.28 (0.74, 2.19) |
| HER2-enriched | NA | NA | 54 (7.6) | 2.79 (1.73, 4.49) | 28 (7.7) | 2.12 (0.98, 4.58) |
| TNBC | NA | NA | 114 (16.1) | 2.33 (1.56, 3.49) | 72 (20.0) | 1.86 (1.03, 3.37) |
| IHC4 score | NA | |||||
| Median | 48.9 | 67.5 | ||||
| Q1 | 107 (25.2) | 1.00 (reference) | 117 (25.0) | 1.00 (reference) | ||
| Q2 | 105 (24.7) | 2.08 (0.98, 4.43) | 117 (25.0) | 1.12 (0.66, 1.92) | ||
| Q3 | 105 (24.7) | 1.14 (0.51, 2.58) | 117 (25.0) | 1.49 (0.89, 2.49) | ||
| Q4 | 108 (25.4) | 1.82 (0.86, 3.84) | 117 (25.0) | 1.37 (0.81, 2.31) | ||
| Grade | ||||||
| Low | 71 (11.9) | 1.00 (reference) | 141 (17.4) | 1.00 (reference) | 158 (24.8) | 1.00 (reference) |
| Intermediate | 385 (64.6) | 1.42 (0.71, 2.86) | 455 (56.2) | 1.07 (0.73, 1.57) | 278 (43.6) | 1.64 (1.02, 2.62) |
| High | 140 (23.5) | 1.66 (0.79, 3.51) | 214 (26.4) | 1.93 (1.30, 2.87) | 201 (31.6) | 2.16 (1.33, 3.50) |
| Size (cm) | ||||||
| ≤2 | 220 (67.7) | 1.00 (reference) | 398 (51.8) | 1.00 (reference) | 475 (70.5) | 1.00 (reference) |
| >2 | 105 (32.3) | 1.62 (0.98, 2.66) | 371 (48.2) | 2.25 (1.71, 2.96) | 199 (29.5) | 2.08 (1.50, 2.88) |
| Node status | ||||||
| Negative | 351 (58.9) | 1.00 (reference) | 448 (59.0) | 1.00 (reference) | 463 (71.7) | 1.00 (reference) |
| Positive | 245 (41.1) | 2.52 (1.71, 3.73) | 311 (41.0) | 2.10 (1.61, 2.73) | 183 (28.3) | 2.07 (1.47, 2.90) |
| Stage | ||||||
| I | 265 (44.5) | 1.00 (reference) | 135 (25.8) | 1.00 (reference) | 383 (56.8) | 1.00 (reference) |
| II | 252 (42.3) | 1.69 (1.08, 2.66) | 375 (71.7) | 2.47 (1.52, 4.00) | 256 (38.0) | 1.75 (1.24, 2.46) |
| III/IV | 79 (13.3) | 3.26 (1.94, 5.48) | 13 (2.5) | 1.10 (0.26, 4.70) | 35 (5.2) | 4.62 (2.72, 7.84) |
CHCAMS: Cancer Hospital, Chinese Academy of Medical Sciences; PBCS: Polish Breast Cancer Study; PLCO: Prostate, Lung, Colorectal and Ovarian cancer screening trial in the United States. Breast cancer patients from CHCAMS were premenopausal women, aged 24-55 years, with hormone receptor-positive (HR+) breast cancer that were diagnosed between 2008-2012. Patients from PBCS were women aged 31-75 years, unselected for hormone receptor-status, diagnosed between 2000-2003. PLCO patients were postmenopausal women, aged 55-87 years, unselected for hormone receptor-status, that were participating in the PLCO trial (1993-2001). NA: Not available or applicable. FU: follow-up. HR: Hazard ratio; 95% CI: 95% Confidence interval. DFS: Disease-free survival. OS: Overall survival. ER: estrogen receptor. PR: progesterone receptor. HER2: human epiderma growth factor receptor 2.
Distribution of SCD by study and by tumor characteristics
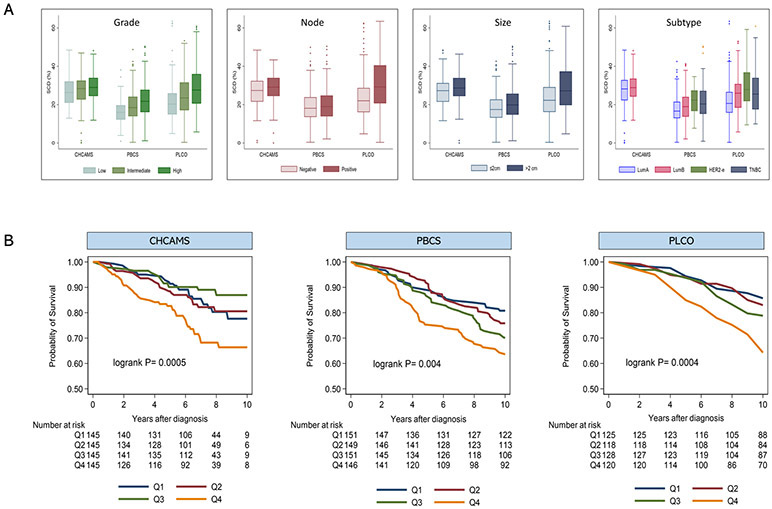
The median (range) SCD score was 28.4% (0.1-48.4%), 18.7% (0.4-50.4%), and 23.6% (0.3-63.5%) for patients in CHCAMS, PBCS, and PLCO studies, respectively (Supplementary Figure S3). The distribution of SCD varied by tumor characteristics, being higher in relation to more aggressive tumor characteristics. In general, SCD was highest in patients with grade 3, larger (>2cm) and node-positive tumors (Figure 2A). In addition, SCD was higher in patients with luminal B-like, HER2-enriched, and TNBC than in those with luminal A-like breast cancer. The distribution of SCD by tumor subtype was strikingly similar in both PBCS and PLCO studies, with SCD being highest in the HER2-enriched, followed by TNBC and luminal B-like tumors (Figure 2A).
Figure 2.
Tumor-associated stromal cellular density (SCD) in relation to clinicopathological characteristics and breast cancer clinical outcomes. (A) Distribution of SCD by grade, lymph nodal involvement, tumor size, and subtype according to study population. (B) Kaplan-Meier survival curves for the associations between strata (Q1-Q4) of SCD and clinical outcomes (10-year disease-free survival (DFS) and 10-year overall survival (OS)) among patients with luminal breast cancer from three independent study populations, including 580 Chinese patients from CHCAMS (DFS); 597 Polish patients from PBCS (OS); and 492 US patients from the PLCO study (OS). Patients from CHCAMS were premenopausal women, aged 24-55 years, with luminal (HR+) breast cancer that were diagnosed between 2008-2012. Patients from PBCS were women aged 31-75 years, unselected for hormone receptor-status, diagnosed between 2000-2003. PLCO patients were postmenopausal women, aged 55-87 years, unselected for hormone receptor-status, that were participating in the PLCO trial (1993-2001).
Associations between tumor-associated stromal cellular density (SCD) and clinical outcomes in breast cancer
Overall (combining luminal and non-luminal tumors), high SCD was statistically significantly associated with worse clinical outcomes in partially-adjusted models involving patients from both PBCS and PLCO. Following adjustment for standard clinical parameters, however, SCD remained statistically significantly predictive of 10-year OS only in PLCO (Table 2). In stratified analysis, increasing SCD was statistically significantly associated with worse clinical outcomes among luminal breast cancer patients from all three study populations both as a categorical (Figure 2B) and continuous measure (Table 2). After accounting for standard clinical parameters in multivariable models, increasing SCD remained statistically significantly associated with worse clinical outcomes among luminal breast cancer patients from all study populations: CHCAMS (HR(95% CI)Q4 vs Q1=1.86(1.06-3.26); P-trend=0.03); PBCS (HR(95% CI)Q4 vs Q1=1.80(1.12-2.89); P-trend=0.01); and PLCO (HR(95% CI)Q4 vs Q1=2.42(1.33-4.42); P-trend=0.002) studies (Table 2). In sensitivity analyses using 10-year BCSS as clinical outcome (Supplementary Table S2), the results were less precise but consistent with worse 10-year BCSS among patients with higher SCD in all three study populations: CHCAMS (HR(95%CI) Q4 vs Q1=3.21(0.87-11.90);P-trend=0.07), PBCS (HR(95%CI)Q4 vs Q1=1.06(0.41-2.76);P-trend=0.89), and PLCO (HR(95%CI)Q4 vs Q1=5.63(1.92-16.53);P-trend<0.001).
Table 2:
Hazard ratios (HRs) and 95% confidence intervals (CIs) for the associations between tumor-associated stromal cellular density (SCD) and clinical outcomes in breast cancer patients from three independent study populations, overall and stratified by hormone receptor-expression
| Study population | ||||||
|---|---|---|---|---|---|---|
| CHCAMS | PBCS | PLCO | Meta-analysis | |||
| SCD categories | HR (95% CI)ϯ | HR (95% CI)ф | HR (95% CI) ф | HR (95% CI) | P het | |
| Univariable | ||||||
| All subjects | ||||||
| Q1 | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | |||
| Q2 | 1.17 (0.78, 1.74) | 1.21 (0.73, 2.00) | 1.18 (0.80, 1.57) | |||
| Q3 | 1.43 (0.98, 2.09) | 1.22 (0.74, 2.01) | 1.34 (0.92, 1.76) | |||
| Q4 | 1.60 (1.09, 2.32) | 2.52 (1.60, 3.95) | 1.93 (1.07, 2.80) | |||
| Trend | 1.17 (1.04, 1.32) | 1.36 (1.17, 1.57) | 1.25 (1.07, 1.44) | 0.13 | ||
| P trend | 0.007 | <0.001 | ||||
| Continuous, per 10% | 1.14 (0.98, 1.29) | 1.31 (1.16, 1.45) | 1.23 (1.06, 1.39) | |||
| P value | 0.08 | <0.001 | ||||
| HR+ (luminal) | ||||||
| Q1 | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | ||
| Q2 | 1.08 (0.61, 1.92) | 1.31 (0.80, 2.14) | 1.15 (0.60, 2.22) | 1.18 (0.78, 1.58) | ||
| Q3 | 0.66 (0.34, 1.24) | 1.64 (1.03, 2.62) | 1.53 (0.83, 2.83) | 1.19 (0.50, 1.87) | ||
| Q4 | 2.03 (1.22, 3.39) | 2.21 (1.40, 3.47) | 3.14 (1.77, 5.57) | 2.26 (1.56, 2.96) | ||
| Trend | 1.24 (1.04, 1.48) | 1.30 (1.13, 1.49) | 1.50 (1.24, 1.81) | 1.32 (1.19, 1.44) | 0.35 | |
| P trend | 0.02 | <0.001 | <0.001 | |||
| Continuous, per 10% | 1.32 (1.05, 1.59) | 1.26 (1.05, 1.47) | 1.40 (1.23, 1.57) | 1.34 (1.22, 1.46) | ||
| P value | 0.02 | 0.01 | <0.001 | |||
| HR− (non-luminal) | ||||||
| Q1 | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | |||
| Q2 | 1.70 (0.83, 3.46) | 0.42 (0.16, 1.10) | 0.91 (0.31, 2.12) | |||
| Q3 | 0.78 (0.36, 1.71) | 0.69 (0.30, 1.54) | 0.73 (0.27, 1.19) | |||
| Q4 | 1.03 (0.47, 2.26) | 0.74 (0.30, 1.81) | 0.86 (0.28, 1.44) | |||
| Trend | 0.92 (0.73, 1.17) | 0.92 (0.67, 1.24) | 0.92 (0.75, 1.09) | 1.00 | ||
| P trend | 0.52 | 0.56 | ||||
| Continuous, per 10% | 0.91 (0.60, 1.23) | 0.98 (0.69, 1.27) | 0.95 (0.73, 1.16 | |||
| P value | 0.58 | 0.91 | ||||
| Multivariable | ||||||
| All subjects | ||||||
| Q1 | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | |||
| Q2 | 1.19 (0.80, 1.78) | 1.11 (0.66, 1.85) | 1.16 (0.78, 1.54) | |||
| Q3 | 1.21 (0.82, 1.78) | 1.08 (0.65, 1.81) | 1.16 (0.78, 1.53) | |||
| Q4 | 1.26 (0.86, 1.85) | 1.84 (1.13, 3.00) | 1.41 (0.91, 1.91) | |||
| Trend | 1.07 (0.95, 1.20) | 1.22 (1.04, 1.43) | 1.13 (0.98, 1.27) | 0.21 | ||
| P trend | 0.27 | 0.01 | ||||
| Continuous, per 10% | 0.99 (0.83, 1.15) | 1.18 (1.02, 1.34) | 1.08 (0.90, 1.27) | |||
| P value | 0.88 | 0.02 | ||||
| HR+ (luminal) | ||||||
| Q1 | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | ||
| Q2 | 0.87 (0.46, 1.64) | 1.29 (0.79, 2.11) | 1.06 (0.55, 2.05) | 1.06 (0.55, 2.05) | ||
| Q3 | 0.54 (0.26, 1.10) | 1.38 (0.85, 2.21) | 1.35 (0.73, 2.52) | 1.01 (0.41, 2.25) | ||
| Q4 | 1.86 (1.06, 3.26) | 1.80 (1.12, 2.89) | 2.42 (1.33, 4.42) | 1.92 (1.29, 2.55) | ||
| Trend | 1.24 (1.02, 1.51) | 1.20 (1.04, 1.39) | 1.37 (1.13, 1.67) | 1.25 (1.10, 1.37) | 0.58 | |
| P trend | 0.03 | 0.01 | 0.002 | |||
| Continuous, per 10% | 1.31 (1.02, 1.62) | 1.14 (0.92, 1.36) | 1.31 (1.12, 1.51) | 1.25 (1.12, 1.38) | ||
| P value | 0.03 | 0.20 | 0.001 | |||
| HR− (non-luminal) | ||||||
| Q1 | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | |||
| Q2 | 2.21 (0.99, 4.92) | 0.34 (0.11, 1.00) | 1.02 (0.74, 2.78) | |||
| Q3 | 1.32 (0.58, 3.00) | 0.39 (0.15, 1.03) | 0.68 (0.16, 1.52) | |||
| Q4 | 1.06 (0.46, 2.45) | 0.42 (0.14, 1.29) | 0.61 (0.04, 1.18) | |||
| Trend | 0.95 (0.75, 1.20) | 0.70 (0.48, 1.02) | 0.84 (0.59, 1.08) | 0.16 | ||
| P trend | 0.65 | 0.07 | ||||
| Continuous, per 10% | 0.94 (0.64, 1.24) | 0.77 (0.43, 1.12) | 0.87 (0.64, 1.09) | |||
| P value | 0.69 | 0.20 | ||||
Hazard ratios (HRs) and 95% confidence intervals (CIs) were obtained from Cox proportional hazard regression models. Univariable HRs were adjusted for age and total tissue area while multivariable models had additional adjustments for histologic grade, tumor size, lymph nodal involvement, subtype, and systematic treatment (endocrine therapy, and/or trastuzumab, or adjuvant chemotherapy). Meta-analysis of HR estimates was performed using the random effects modeling approach. Breast cancer patients from the Cancer Hospital Chinese Academy of Medical Sciences (CHCAMS) were premenopausal women, aged 24-55 years, with hormone receptor-positive (HR+ (luminal)) breast cancer, while those from the Prostate, Lung, Colorectal and Ovarian cancer (PLCO) screening trial were postmenopausal women, aged 55-87 years, unselected for hormone receptor-status. Polish breast cancer patients from the Polish Breast Cancer Study (PLCO) were aged 31-75 years and unselected for menopausal status.
10-year disease-free survival (DFS).
10-year overall survival (OS).
Although increasing SCD was suggestively associated with favorable clinical outcomes in patients with non-luminal breast cancer from PBCS and PLCO, these findings did not attain statistical significance (Table 2).
Prognostic value of tumor-associated stromal cellular density (SCD) in relation to standard clinical factors among luminal breast cancer patients
In all studies, SCD provided prognostic information beyond standard clinical parameters (ΔLRχ2 = 14.7 (P-value=0.002), 4.2 (P-value=0.04), and 11.5 (P-value=0.009) in CHCAMS, PBCS, and PLCO, respectively). Furthermore, SCD provided more prognostic information than histologic grade in all studies and, with the exception of PBCS, than PR, HER2, subtype, and tumor size. In both CHCAMS and PBCS studies where we had data on IHC4 score, SCD (ΔLRχ2 =13.8 (P-value=0.003) and 9.1 (P-value=0.03) in CHCAMS and PBCS, respectively) provided more prognostic information than IHC4 score (ΔLRχ2 =2.0 (P-value=0.73) and 3.3 (P-value=0.51) in CHCAMS and PBCS, respectively).
In general, patients with SCD above the 75th percentile, corresponding to 33%, 24%, and 31% in CHCAMS, PBCS, and PLCO studies, respectively, had worse clinical outcomes than those with values below this threshold. In post-hoc analysis dichotomizing the data at this threshold, patients with high (>75th percentile) had statistically significantly worse clinical outcomes than those with low (≤75th percentile) SCD (HR (95% CI) >75th vs ≤75th percentile =2.02 (1.35-3.03); 1.46 (1.04-2.05); and 2.09 (1.36-3.21) in CHCAMS, PBCS, and PLCO, respectively).
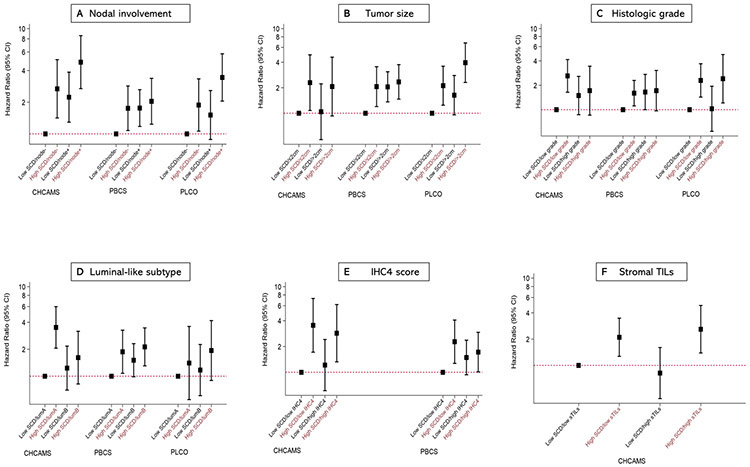
In stratified analysis by tumor characteristics (Figure 3), high SCD was associated with poor clinical outcomes in luminal breast cancer patients with node-positive or node-negative disease, small or large tumors, high or low grade tumors, luminal A-like or B-like disease, and irrespective of whether the patients had low (HR (95% CI) = 2.11 (1.27-3.48)) or high (HR (95% CI) = 2.60 (1.39-4.86)) TILs (Figure 3).
Figure 3.
Joint associations of tumor associated stromal cellular density (SCD) and tumor characteristics with clinical outcomes among luminal breast cancer patients. Hazards ratios (HR) and 95% confidence intervals for the joint associations between SCD and lymph nodal involvement (A), tumor size (B), histologic grade (C), luminal-like subtype (D), IHC4 score (E), and stromal tumor infiltration lymphocytes, sTILs (F) and clinical outcomes (10-year disease-free survival (DFS) and 10-year overall survival (OS)) among hormone receptor-positive breast cancer patients from three independent study populations including CHCAMS (China; DFS), PBCS (Poland; OS) and PLCO (United States; OS). Hazard ratios and corresponding estimates were obtained from multivariable Cox proportional hazard regression models accounting for standard clinical factors, including age, lymph nodal involvement, tumor size, histologic grade, subtype, and systemic therapy, as well as total tissue area. Each primary model exempted the variable in the joint SCD-clinical factor classification.
In sensitivity analyses investigating interactions between SCD and systemic treatment, we did not observe statistically significant differences in the prognostic value of SCD according to whether or not the patients received endocrine therapy (P-heterogeneity = 0.36 and 0.25 for CHCAMS and PBCS, respectively) or whether or not they received adjuvant chemotherapy (P-heterogeneity = 0.55 (PBCS only)). In addition, the associations between SCD and clinical outcomes did not differ by the amount of stroma or tumor on the H&E image upon which SCD was assessed (Supplementary Table S3) or, for 10-year DFS in CHCAMS, by surgery type (radical or breast-conserving surgery, P-heterogeneity=0.29).
Tumor-associated stromal cellular density (SCD) in relation to stromal tumor infiltrating lymphocytes (TILs), CD3+ and CD8+ T-cell densities among luminal breast cancer patients
In general, TILs (r =0.13), CD3+ (r =0.26) and CD8+ (r =0.17) T-cell densities correlated weakly but statistically significantly (P<0.05) with H&E-based SCD. However, the combination of all stromal cells on IHC-stained sections i.e. IHC-SCD showed considerably better correlation (r =0.53) with H&E-based SCD (Supplementary Figure S4), with ~96% of the measures being within the limits of agreement (Supplementary Figure S5). For each of the IHC markers, we observed the non-staining stromal cell population (i.e. CD3− cells, R2=57%; CD8− cells, R2=44%) to explain more of the variation in IHC-SCD than the corresponding positive staining cell populations (CD3+ cells, R2=21%; CD8+ cells, R2=29%). As shown in Supplementary Figure S6, we did not find TILs levels to be significantly associated with clinical outcomes among patients with luminal breast cancer (HR (95% CI) high vs low TILs = 1.06 (0.66-1.70)), which contrasts with the SCD finding for the same set of patients (HR (95% CI) high vs low SCD =2.02 (1.35-3.03)). High CD3+ (HR (95% CI) = 0.79 (0.39-1.58)) and high CD8+ (HR (95% CI) = 0.46 (0.23-0.94)) T-cell densities were associated with better clinical outcomes than low densities of these markers but estimates were statistically significant only in relation to CD8+ T-cell density. Conversely, high IHC-SCD was associated with worse clinical outcomes (HR (95% CI)= 2.51 (1.24-5.08)) in the same patients (Supplementary Figure S6).
Discussion
In this multi-ethnic analysis of 2,084 invasive breast cancer patients from study populations in China, Poland, and USA, we applied machine learning algorithms to archival H&E-stained sections and generated tumor-associated SCD as a morphological proxy for inflammation and wound repair signatures in breast tumors. The distribution of SCD varied between studies, which is likely reflective of the differences between the study populations in terms of ethnicity, age, menopausal status, and tumor characteristics. Nevertheless, the associations between SCD, tumor characteristics, and clinical outcomes were strikingly consistent across the studies. In general, SCD was associated with poor prognostic tumor characteristics, particularly histologic grade, tumor size, HER2-enriched and TNBC subtypes. In the analysis evaluating the prognostic value of SCD, we found high SCD to be independently and strongly predictive of poor 10-year DFS and OS among patients with luminal breast cancer from all studies. For these patients, SCD provided more prognostic information than many standard clinical parameters. In addition, our findings indicate that SCD is distinct from TILs scoring both in terms of cellular composition and prognostic property. Taken together, these findings suggest that assessment of tumor-associated SCD on routinely performed H&E-stained sections can provide additional prognostic information to patients with luminal breast cancer beyond what is obtained in many standard clinical factors.
While the inclusion of CAFs, TILs, macrophages, endothelial, and other stromal cells as part of SCD supports the idea of SCD as a proxy for the extent of inflammation and wound repair processes in tumors, SCD may also be reflective of other biological processes. For instance, SCD measures may encompass tumor microenvironment of metastasis (TMEM), a tripartite arrangement of endothelial cells, macrophages, and invasive carcinoma cells within the stroma that has been shown to predict clinical outcomes in luminal breast cancer (5, 33). SCD may also reflect a yet to be understood feature of TME characterized by spatial distributions of nucleated cells within the stroma. The latter idea is supported by results from a study by Beck and colleagues, demonstrating strong associations between stromal features, including presence of nucleated stromal regions on H&E-stained sections, and survival outcomes among breast cancer patients (3). Unlike the current study, however, the Beck study did not specifically quantify density of nucleated cells in tumor-stroma. In addition, this study was based on whole-slide images while the Beck study was based on tissue microarrays, which may not be representative of entire tumor volume.
The finding that SCD, and not TILs, was predictive of clinical outcomes in luminal breast cancer patients suggests that SCD is not the same as TILs and that non-TILs composition of SCD may be contributing to its prognostic property. This notion is supported by several of our observations. First, we observed weak correlations between TILs, CD3+, and CD8+ T-cell densities with SCD. Second, the correlation between SCD on IHC and H&E staining was substantially higher than that between individual immune cells and SCD. Third, whereas high CD8+ density was associated with favorable clinical outcomes in luminal breast cancer patients, which is in line with previous reports (34, 35), its combination with other stromal cells as part of IHC-SCD was associated with unfavorable prognosis. Our findings may also be reflective of a dominant pro-tumor immune response driven by FOXP3+ Tregs, which correlate with poor clinical outcomes in luminal breast cancer (32, 36). However, high SCD was strongly predictive of poor DFS even among patients with low TILs (who will, presumably, have low FOXP3+ cells) thereby making it highly unlikely that our SCD findings were driven by FOXP3+ Tregs alone.
It is unclear why high SCD leads to poor clinical outcomes among luminal breast cancer patients. A possible explanation may relate to differences in biologic behavior of the TME by tumor subtype. Also, since SCD is characterized by CAFs, inflammatory and endothelial cells, patients with high SCD may be at a more advanced phase along the cascade of biological processes, including extracellular matrix (ECM) remodeling and neovascularization, leading to metastasis. This is buttressed by our observation that among luminal breast cancer patients, SCD was higher among those with the more clinically aggressive luminal B-like than A-like subtype (27). High SCD may also be reflective of molecular changes that confer poor responsiveness to endocrine therapy, irrespective of luminal breast cancer subtype. This latter idea is supported by our observation that high SCD was predictive of unfavorable clinical outcomes among patients with both luminal A-like and B-like tumors.
Overall, our findings support the prognostic value of tumor-associated SCD and suggest that subject to additional validation SCD can be used in conjunction with other clinical parameters to guide treatment decision-making for luminal breast cancer, a clinically heterogeneous entity comprising >70% of all breast cancers (37-40). Although several molecular tests have been developed to aid clinical decision-making in luminal breast cancer (41, 42), many of these are limited by proprietary constraints, cost, or both (43). Moreover, some molecular tests may not provide prognostic information beyond what is contained in traditional IHC markers. The IHC4 score, for instance, has previously been shown to provide comparative prognostic information to Oncotype DX recurrence score (10). Our finding that SCD, which can be more easily assessed on H&E-stained sections compared to protein or gene expression-based assays, provided significantly more prognostic information than the IHC4 score, suggests that SCD could be used as an adjunct or alternative to molecular tests, particularly in low-resource settings.
This study has several strengths and some limitations. An important strength of the study is the inclusion of population-based studies comprising of patients with different ethnic backgrounds, enrolment periods, and duration of follow-up after diagnosis. The treatments received are also likely to differ between the different studies. Despite these differences, our results were consistent across studies, demonstrating robustness as well as external generalizability of SCD as a prognostic marker in luminal breast cancer. Further, the availability of several clinicopathologic factors allowed us to account for many standard clinical parameters in our analysis and to assess the prognostic value of SCD in different clinical scenarios. The innovative application of computational pathology to generate SCD on H&E- and, for a subset of patients, IHC-stained sections is another important strength of this study. Despite modest correlation between SCD on H&E and IHC-staining, ~96% of the measures were within the limits of agreement and SCD was predictive of clinical outcome irrespective of whether assessment was performed on H&E- or IHC-stained images. Nonetheless, further work is required to validate our findings and to standardize the assessment of SCD on H&E- or IHC-stained images.
Our assessment of SCD as a summary score of stromal cellular composition by using H&E did not allow assessment of the contributions of different stromal cell populations to SCD’s prognostic property, which is necessary to unravel biologically and therapeutically relevant details. Addressing this limitation will require the use of special stains such as multiplex immunofluorescence staining to identify several cell phenotypes on the same tissue sections as SCD. Such an approach can also facilitate the discovery of parsimonious cellular combinations, explaining different biologic pathways, that are most predictive of clinical outcomes in breast cancer. Notwithstanding these limitations, our observation that SCD on H&E-stained sections was independently predictive of clinical outcomes among luminal breast cancer patients suggests that the dominant biological processes underpinning SCD are pro-tumorigenic. The retrospective-prospective design and lack of comprehensive treatment data, particularly for chemotherapy and anti-HER2 therapy, from all of the studies meant that we could not assess the predictive value of SCD for any particular treatment strategy. However, for those studies in which treatment data were available, we did not observe evidence for interaction between SCD and treatment. Further studies evaluating the predictive value of SCD for treatment response will be required.
In conclusion, our finding that high tumor-associated SCD portended worse clinical outcomes in patients with luminal breast cancer may be indicative of its value in identifying a subset of patients with higher risk of metastasis or poor endocrine therapy responsiveness. Subject to further validation, these findings could have important implications for refining prognostic assessment and therapeutic decision-making for luminal breast cancer patients.
Supplementary Material
Table 3:
Change in likelihood ratio chi-square (ΔLRχ2) and corresponding p values comparing the contribution of tumor-associated stromal cellular density (SCD) and other clinicopathological parameters to a fully adjusted prognostic model for hormone receptor-positive (luminal) breast cancer patients from three independent study populations.
| CHCAMS ϯ |
PBCS ф |
PLCO ф |
||||
|---|---|---|---|---|---|---|
| Marker | ΔLRχ2 | P value | ΔLRχ2 | P value | ΔLRχ2 | P value |
| SCD | 14.7 | 0.002 | 4.2 | 0.04 | 11.5 | 0.009 |
| PR | 1.0 | 0.31 | 6.4 | 0.01 | 1.2 | 0.55 |
| HER2 | 3.6 | 0.17 | 5.3 | 0.07 | 2.3 | 0.32 |
| Subtype | 0.4 | 0.53 | 7.0 | 0.03 | 0.5 | 0.79 |
| IHC4 score | 2.0 | 0.73 | 3.3 | 0.51 | NA | |
| Grade | 0.6 | 0.75 | 3.9 | 0.14 | 0.8 | 0.85 |
| Size | 1.3 | 0.53 | 10.6 | 0.01 | 5.6 | 0.02 |
| Nodes | 16.6 | <0.001 | 8.9 | 0.01 | 4.9 | 0.08 |
The primary model comprised of SCD, PR, HER2, histologic grade, tumor size, and lymph nodal involvement. In separate models, PR and HER2 were substituted for subtype (luminal B-like vs A-like) or IHC4 score. All models were constructed separately for each study, with further adjustments for age at diagnosis and tissue area. The contribution of each marker to model fit was assessed by removing that marker from the full model containing all other prognostic parameters and assessing the change in likelihood ratio chi-squared (ΔLRχ2) and P value following a likelihood ratio test. NA: Not available. .
10-year disease-free survival (DFS).
10-year overall survival (OS).
Acknowledgements
We would like to thank all the participants from the Cancer Hospital, Chinese Academy of Medical Sciences (CHCAMS), Beijing, China; the Polish Breast Cancer Study (PBCS), Poland; as well as from the Prostate, Lung, Colorectal, and Ovarian (PLCO), cancer screening trial, USA. We are also thankful to the staff members who contributed to the study management and provided data support for these studies. This work was supported by intramural research funds from the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, USA. M. Abubakar, T.U. Ahearn, H. Koka, M. Garcia-Closas, and X.R. Yang were supported by intramural research funds from the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health.
Financial Support:
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics, USA.
Abbreviations:
- CAFs
cancer-associated fibroblasts
- CHCAMS
Cancer Hospital Chinese Academy of Medical Sciences
- CI
confidence interval
- DFS
disease-free survival
- ECM
extracellular matrix remodeling
- ER
estrogen receptor
- H&E
hematoxylin & eosin
- FFPE
formalin-fixed paraffin-embedded
- HER2
human epidermal growth factor receptor 2
- HR
hazard ratio
- IHC
immunohistochemistry
- LR
likelihood ratio
- NDI
National Death Index
- OS
overall survival
- PBCS
Polish Breast Cancer Study
- PLCO
Prostate, Lung, Colorectal, Ovarian cancer screening trial
- SCD
stromal cellular density
- TILs
tumor infiltrating lymphocytes
- TME
tumor microenvironment
- TMEM
tumor microenvironment of metastasis
- WRS
wound response signature
Footnotes
Disclosures of Potential Conflicts of Interest:
The authors declare no conflicts of interest
References
- 1.Natrajan R, Sailem H, Mardakheh FK, Arias Garcia M, Tape CJ, Dowsett M, et al. Microenvironmental Heterogeneity Parallels Breast Cancer Progression: A Histology–Genomic Integration Analysis. PLOS Medicine. 2016;13(2):e1001961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heindl A, Sestak I, Naidoo K, Cuzick J, Dowsett M, Yuan Y. Relevance of Spatial Heterogeneity of Immune Infiltration for Predicting Risk of Recurrence After Endocrine Therapy of ER+ Breast Cancer. JNCI: Journal of the National Cancer Institute. 2017;110(2):166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, van de Vijver MJ, et al. Systematic Analysis of Breast Cancer Morphology Uncovers Stromal Features Associated with Survival. Science Translational Medicine. 2011;3(108):108ra13–ra13. [DOI] [PubMed] [Google Scholar]
- 4.Yuan Y, Failmezger H, Rueda OM, Ali HR, Gräf S, Chin S-F, et al. Quantitative Image Analysis of Cellular Heterogeneity in Breast Tumors Complements Genomic Profiling. Science Translational Medicine. 2012;4(157):157ra43–ra43. [DOI] [PubMed] [Google Scholar]
- 5.Rohan TE, Xue X, Lin H-M, D’Alfonso TM, Ginter PS, Oktay MH, et al. Tumor Microenvironment of Metastasis and Risk of Distant Metastasis of Breast Cancer. JNCI Journal of the National Cancer Institute. 2014;106(8):dju136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, et al. Gene Expression Signature of Fibroblast Serum Response Predicts Human Cancer Progression: Similarities between Tumors and Wounds. PLOS Biology. 2004;2(2):e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A Multigene Assay to Predict Recurrence of Tamoxifen-Treated, Node-Negative Breast Cancer. New England Journal of Medicine. 2004;351(27):2817–26. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, et al. A Comparison of PAM50 Intrinsic Subtyping with Immunohistochemistry and Clinical Prognostic Factors in Tamoxifen-Treated Estrogen Receptor–Positive Breast Cancer. Clinical Cancer Research. 2010;16(21):5222–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jerevall PL, Ma XJ, Li H, Salunga R, Kesty NC, Erlander MG, et al. Prognostic utility of HOXB13 : IL17BR and molecular grade index in early-stage breast cancer patients from the Stockholm trial. British Journal of Cancer. 2011;104(11):1762–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuzick J, Dowsett M, Pineda S, Wale C, Salter J, Quinn E, et al. Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health recurrence score in early breast cancer. Journal of Clinical Oncology. 2011;29(32):4273–8. [DOI] [PubMed] [Google Scholar]
- 11.Candido dos Reis FJ, Wishart GC, Dicks EM, Greenberg D, Rashbass J, Schmidt MK, et al. An updated PREDICT breast cancer prognostication and treatment benefit prediction model with independent validation. Breast Cancer Research. 2017;19(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schäfer M, Werner S. Cancer as an overhealing wound: an old hypothesis revisited. Nature Reviews Molecular Cell Biology. 2008;9(8):628–38. [DOI] [PubMed] [Google Scholar]
- 13.Place AE, Jin Huh S, Polyak K. The microenvironment in breast cancer progression: biology and implications for treatment. Breast Cancer Research. 2011;13(6):227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eming SA, Krieg T, Davidson JM. Inflammation in Wound Repair: Molecular and Cellular Mechanisms. Journal of Investigative Dermatology. 2007;127(3):514–25. [DOI] [PubMed] [Google Scholar]
- 15.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nature Reviews Cancer. 2006;6(5):392–401. [DOI] [PubMed] [Google Scholar]
- 16.Tonnesen MG, Feng X, Clark RAF. Angiogenesis in Wound Healing. Journal of Investigative Dermatology Symposium Proceedings. 2000;5(1):40–6. [DOI] [PubMed] [Google Scholar]
- 17.Chang HY, Nuyten DSA, Sneddon JB, Hastie T, Tibshirani R, Sørlie T, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(10):3738–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Troester MA, Lee MH, Carter M, Fan C, Cowan DW, Perez ER, et al. Activation of Host Wound Responses in Breast Cancer Microenvironment. Clinical Cancer Research. 2009;15(22):7020–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kos Z, Roblin E, Kim RS, Michiels S, Gallas BD, Chen W, et al. Pitfalls in assessing stromal tumor infiltrating lymphocytes (sTILs) in breast cancer. npj Breast Cancer. 2020;6(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amgad M, Stovgaard ES, Balslev E, Thagaard J, Chen W, Dudgeon S, et al. Report on computational assessment of Tumor Infiltrating Lymphocytes from the International Immuno-Oncology Biomarker Working Group. npj Breast Cancer. 2020;6(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B. Histopathological image analysis: a review. IEEE Rev Biomed Eng. 2009;2:147–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xing F, Yang L. Robust Nucleus/Cell Detection and Segmentation in Digital Pathology and Microscopy Images: A Comprehensive Review. IEEE Rev Biomed Eng. 2016;9:234–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang XR, Sherman ME, Rimm DL, Lissowska J, Brinton LA, Peplonska B, et al. Differences in Risk Factors for Breast Cancer Molecular Subtypes in a Population-Based Study. Cancer Epidemiology Biomarkers & Prevention. 2007;16(3):439–43. [DOI] [PubMed] [Google Scholar]
- 24.Prorok PC, Andriole GL, Bresalier RS, Buys SS, Chia D, David Crawford E, et al. Design of the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Controlled Clinical Trials. 2000;21(6, Supplement 1):273S–309S. [DOI] [PubMed] [Google Scholar]
- 25.Gohagan JK, Prorok PC, Hayes RB, Kramer B-S. The Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial of the National Cancer Institute: History, organization, and status. Controlled Clinical Trials. 2000;21(6, Supplement 1):251S–72S. [DOI] [PubMed] [Google Scholar]
- 26.Abubakar M, Guo C, Koka H, Sung H, Shao N, Guida J, et al. Clinicopathological and epidemiological significance of breast cancer subtype reclassification based on p53 immunohistochemical expression. npj Breast Cancer. 2019;5(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abubakar M, Figueroa J, Ali HR, Blows F, Lissowska J, Caldas C, et al. Combined quantitative measures of ER, PR, HER2, and KI67 provide more prognostic information than categorical combinations in luminal breast cancer. Modern Pathology. 2019;32(9):1244–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thürlimann B, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Annals of Oncology. 2013;24(9):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coates AS, Members P, Winer EP, Members P, Goldhirsch A, Members P, et al. Tailoring therapies—improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Annals of Oncology. 2015;26(8):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Closas M, Brinton L, Lissowska J, Chatterjee N, Peplonska B, WF Anderson N. Established breast cancer risk factors by clinically important tumour characteristics. British journal of cancer. 2006;95(1):123–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu CS, Huang W-Y, Pinsky PF, Berg CD, Sherman M, Yu KJ, et al. The Prostate, Lung, Colorectal and Ovarian Cancer (PLCO) Screening Trial Pathology Tissue Resource. Cancer Epidemiology Biomarkers & Prevention. 2016;25(12):1635–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hendry S, Salgado R, Gevaert T, Russell PA, John T, Thapa B, et al. Assessing Tumor-infiltrating Lymphocytes in Solid Tumors: A Practical Review for Pathologists and Proposal for a Standardized Method From the International Immunooncology Biomarkers Working Group Part 1 Assessing the Host Immune Response, TILs in Invasive Breast Carcinoma and Ductal Carcinoma In Situ, Metastatic Tumor Deposits and Areas for Further Research. Advances in Anatomic Pathology. 2017;24(5):235–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson BD, Sica GL, Liu Y-F, Rohan TE, Gertler FB, Condeelis JS, et al. Tumor Microenvironment of Metastasis in Human Breast Carcinoma: A Potential Prognostic Marker Linked to Hematogenous Dissemination. Clinical Cancer Research. 2009;15(7):2433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahmoud S, Lee A, Ellis I, Green A. CD8+ T lymphocytes infiltrating breast cancer. OncoImmunology. 2012;1(3):364–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ali HR, Provenzano E, Dawson S-J, Blows FM, Liu B, Shah M, et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann Oncol. 2014;25 8:1536–43. [DOI] [PubMed] [Google Scholar]
- 36.Shou J, Zhang Z, Lai Y, Chen Z, Huang J. Worse outcome in breast cancer with higher tumor-infiltrating FOXP3+ Tregs : a systematic review and meta-analysis. BMC Cancer. 2016;16(1):687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howlader N, Altekruse SF, Li CI, Chen VW, Clarke CA, Ries LAG, et al. US Incidence of Breast Cancer Subtypes Defined by Joint Hormone Receptor and HER2 Status. Journal of the National Cancer Institute. 2014;106(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciriello G, Sinha R, Hoadley KA, Jacobsen AS, Reva B, Perou CM, et al. The molecular diversity of Luminal A breast tumors. Breast Cancer Res Treat. 2013;141(3):409–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howell SJ. Advances in the treatment of luminal breast cancer. Current Opinion in Obstetrics & Gynecology. 2013;25(1):49–54. [DOI] [PubMed] [Google Scholar]
- 40.Ignatiadis M, Sotiriou C. Luminal breast cancer: from biology to treatment Nature Reviews Clinical Oncology 2013;10:13. [DOI] [PubMed] [Google Scholar]
- 41.Sotiriou C, Pusztai L. Gene-Expression Signatures in Breast Cancer. New England Journal of Medicine. 2009;360(8):790–800. [DOI] [PubMed] [Google Scholar]
- 42.Lal S, McCart Reed AE, de Luca XM, Simpson PT. Molecular signatures in breast cancer. Methods. 2017;131:135–46. [DOI] [PubMed] [Google Scholar]
- 43.Arranz EE, Vara JÁF, Gámez-Pozo A, Zamora P. Gene Signatures in Breast Cancer: Current and Future Uses. Translational Oncology. 2012;5(6):398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.