Abstract
Purpose:
Enzalutamide is a second-generation androgen receptor (AR) inhibitor which has improved overall survival (OS) in metastatic castration resistant prostate cancer (CRPC). However, nearly all patients develop resistance. We designed a phase 2 multicenter study of enzalutamide in metastatic CRPC incorporating tissue and blood biomarkers to dissect mechanisms driving resistance.
Experimental Design:
Eligible patients with metastatic CRPC underwent a baseline metastasis biopsy and then initiated enzalutamide 160 mg daily. A repeat metastasis biopsy was obtained at radiographic progression from the same site when possible. Blood for circulating tumor cell (CTC) analysis was collected at baseline and progression. The primary objective was to analyze mechanisms of resistance in serial biopsies. Whole exome sequencing was performed on tissue biopsies. CTC samples underwent RNA sequencing.
Results:
65 patients initiated treatment, of whom 22 (33.8%) had received prior abiraterone. Baseline biopsies were enriched for alterations in AR (mutations, amplifications) and tumor suppression genes (PTEN, RB1, and TP53) which were observed in 73.1% and 92.3% of baseline biopsies, respectively. Progression biopsies revealed increased AR amplifications (64.7% at progression versus 53.9% at baseline) and BRCA2 alterations (64.7% at progression versus 38.5% at baseline). Genomic analysis of baseline and progression CTC samples demonstrated increased AR splice variants, AR regulated-genes, and neuroendocrine markers at progression.
Conclusions:
Our results demonstrate that a large proportion of enzalutamide-treated patients have baseline and progression alterations in the AR pathway and tumor suppressor genes. We demonstrate an increased number of BRCA2 alterations post-enzalutamide highlighting importance of serial tumor sampling in CRPC.
Keywords: Androgen receptor, Castration resistance, Circulating tumor cells, Enzalutamide, Metastases, Prostate cancer, Resistance
Precis:
We report on a phase 2, multicenter, open-label, single-arm study of enzalutamide in men with metastatic castration resistant prostate cancer incorporating baseline and progression metastasis tissue sampling and serial analyses of circulating tumor cells (CTCs) to dissect mechanisms driving clinical resistance to enzalutamide. Our results demonstrate that a substantial proportion of enzalutamide-treated metastatic CRPC patients harbor alterations in the AR pathway and tumor suppressor genes which contribute to the resistance phenotype.
Introduction:
Metastatic castration resistant prostate cancer (CRPC) is a lethal disease with a relative 5-year survival of 29%.[1] While novel treatments, including androgen receptor (AR) directed therapies, have improved overall survival for patients with CRPC, resistance is observed in nearly all patients. Enzalutamide is a rationally designed second generation AR inhibitor which competitively binds to AR with great potency and also inhibits active AR nuclear translation, DNA binding, and coactivator recruitment.[2] Two large phase 3 trials demonstrated the efficacy of enzalutamide over placebo resulting in routine clinical use in metastatic CRPC.[3, 4] However, 10–25% of patients receiving enzalutamide have primary resistance and at 18-months 50–80% of patients have developed radiographic progression.[3, 4] Therefore, strategies to understand determinants of primary and acquired resistance are essential to developing therapeutic approaches to prolong the activity and durability of treatment.
Several preclinical and clinical studies have examined mechanisms of resistance to AR targeting agents. AR dependent mechanisms hypothesized to cause resistance to enzalutamide include AR mutations[5], amplifications[6, 7], splice variant emergence[8, 9], and altered steroidogenesis[10, 11]. Additionally, resistance may also be mediated by activation of parallel AR-independent signaling pathways[12]. While these studies have been informative, they did not integrate paired baseline and progression tumor sampling with comprehensive molecular analysis. Prospective studies embedding tumor tissue and blood-based analyses which are placed in the context of patient outcomes are needed to understand mechanisms that drive resistance to treatment. We report the results of a phase 2, multicenter, open-label, single-arm study of enzalutamide in men with metastatic CRPC incorporating baseline and progression metastasis tumor sampling and serial analyses of circulating tumor cells (CTCs) to dissect mechanisms driving de novo and acquired clinical resistance to enzalutamide.
Materials and Methods:
Patients:
This is a phase 2, single arm, open-label study of enzalutamide in metastatic CRPC (NCT01942837). Eligible patients had CRPC defined as disease progression despite a serum total testosterone <50 ng/dL and: 1) PSA progression as defined by the Prostate Cancer Clinical Trials Working Group (PCWG) 2[13], 2) soft tissue disease progression as defined by Response Evaluation Criteria In Solid Tumors (RECIST) version 1.1[14], or 3) bone disease progression as defined by PCWG2[13]. Additionally, patients had evidence of metastases with at least one metastatic site amendable to biopsy. Patients were required to have prostatic adenocarcinoma. Variant histologies, including neuroendocrine differentiation, were not permitted.
Patients may have received prior hormonal therapies (including ketoconazole, abiraterone, first-generation anti-androgens) and up to two cytotoxic therapies. Prior enzalutamide, apalutamide or darolutamide was not allowed. Other eligibility criteria included an Eastern Cooperative Oncology Group performance status ≤2 and adequate organ and bone marrow function. The study was conducted after Institutional Review Board approval at each of the participating institutions in accordance with the principals outlined in the Declaration of Helsinki. All patients provided written informed consent.
Treatment:
Following enrollment, patients had a baseline biopsy of a metastasis. Biopsies were performed after informed consent in the interventional radiology department. Subsequently, patients received enzalutamide 160 mg by mouth daily. Patients continued enzalutamide until radiographic progression, significant toxicity or patient/physician requested withdrawal. A repeat metastasis biopsy, with ongoing enzalutamide, was obtained at radiographic progression in patients completing ≥3 cycles (1 cycle=28 days). The protocol specified that when possible the progression biopsy should target the same site as the baseline biopsy. Though baseline and progression biopsies were mandatory, not all patients underwent biopsy at progression given lack of feasible biopsy site, clinical disease progression, or patient study withdrawal. Imaging assessments occurred every twelve weeks. PSA was measured every four weeks.
Tumor Tissue Genomic Sequencing:
Biopsies were prioritized for pathologic assessment followed by whole exome sequencing. Whole exome sequencing was performed on tumor biopsies and collected blood normal using a customized version of a previously described protocol.[15] After DNA shearing, hybridization and exome capture were performed using either Illumina’s Rapid Capture Exome Kit or the Agilent SureSelect Human All Exon 44Mb v2.0 bait set (Supplementary Table 1S).[16] Libraries were sequenced with 76 bp paired-end reads on an Illumina instrument.
Reads were aligned using BWA v0.5.9 and somatic mutations called using a customized version of the Getz Lab CGA WES Characterization Pipeline (https://portal.firecloud.org/#methods/getzlab/CGA_WES_Characterization_Pipeline_v0.1_Dec2018/). [17] Briefly, we used ContEst to estimate contamination, MuTect and Strelka to call SNVs and indels, DeTiN to estimate tumor-in-normal contamination, and Orientation Bias Filter and MAFPoNFilter to filter sequencing artifacts.[18–23] For target intervals, we used an intersection of Illumina Rapid Capture Exome and Agilent SureSelect regions, created using bedtools.[24] Variants were annotated using VEP, Oncotator, and vcf2maf v.1.6.17 (https://github.com/mskcc/vcf2maf). [25, 26] Copy number alterations, purity, ploidy, and whole genome doubling status were called using FACETS v0.5.14.[27] In cases where FACETS fit an incorrect copy number profile, ABSOLUTE with manual review was used instead.[28] Copy number alterations were evaluated with respect to whole genome doubling status. Samples were included in the final cohort if they had contamination <4%, purity >20%, tumor coverage >50x, and normal coverage >30x. Presence of biallelic alterations was defined as 1) the presence of a loss of function mutation in addition to allelic full deletion, or 2) two allelic full deletions.
To compare mutations between distinct samples from the same patient, we used a previously described method designed to recover evidence for mutations called in one sample in all other samples derived from the same individual.[29] In brief, the ‘force-calling’ method uses the strong prior of the mutation being present in at least one sample in the patient to more sensitively detect and recover mutations that might otherwise be missed. Successfully sequenced tissue samples were part of larger cohort analyses.[30, 31]
Circulating Tumor Cell Analysis:
Blood for CTC gene expression analysis was collected in vacutainer tubes (BD Biosciences) with EDTA anticoagulant at baseline and off treatment. Mononuclear cells were isolated with a Ficoll-Pacque Plus (GE Healthcare) gradient before undergoing CD45 depletion (Miltenyi Biotec). The VERSA platform[32] was used for the live cell capture of CTCs using an anti-EpCAM antibody (R&D) conjugated to paramagnetic particles (Life Technolgies). Cells were lysed in the VERSA with a modified LIDs buffer (10 mM Tris-HCL, 500 mM lithium chloride, 1 % Igepal® CA-630 (Sigma-Aldrich, USA), 5 mM ethylenediaminetetraacetic acid (EDTA), 5 mM dithiothreitol, pH 7.5) and mRNA was extracted with olgio(dt)25 Dynabeads® (Life Technologies, USA)[33].
Extracted mRNA was reverse transcribed using a High Capacity cDNA Reverse Transcriptase kit (Life Tech, USA), according to manufacturer’s directions using Bio-Rad C1000 Thermo Cycler (Bio-Rad, USA). The RT reaction was amplified for 14 cycles using TaqMan® PreAmp (Life Tech) according to manufacturer’s directions and diluted 1:20 in 1x TE (10 mM Tris-HCL pH8, 1 mM EDTA). For TaqMan® assays, 5 μL of diluted cDNA template was mixed with 10 μL iTaq® master mix (Bio-Rad), 1 μL TaqMan® Gene Expression Assay (Life Technologies) and 4 μL nuclease free (NF) water. Each reaction was amplified for 45 cycles (denatured at 95 °C for 15 seconds followed by annealing at 60°C for 1 minute) using a CFX Connect® Real-Time PCR System (Bio-Rad). A table of genes of interest and primers used is available in Supplementary Table 2S. Samples are reported as 38-cycle threshold value with cycle threshold values less than 38 considered positive for expression.
Statistical Analysis:
The primary objective was to analyze mechanisms of de novo and acquired resistance to enzalutamide in serial CRPC tumor biopsies. This was assessed by tumor exome sequencing. The trial design assumed AR related resistance parameters to be measured as continuous variable. Sample size of 40 with serial biopsies was targeted to detect a standardized effect size of 0.454 for the changes in a set of resistance parameters at progression compared to baseline having 80% power with 1-sided alpha=0.025 using the paired t-test. The planned enrollment was 66 patients to obtain 40 evaluable patients with paired samples.
Secondary endpoints included toxicity, PSA and investigator-assessed radiographic response, time to PSA and investigator-assessed radiographic progression. Toxicity was summarized using Common Terminology Criteria for Adverse Events version 4.0. Radiographic response as defined by RECIST version 1.1 was summarized with 95% exact binomial confidence interval (CI)[14]. PSA response and progression were defined by PCWG2 criteria[13]. Time to PSA progression was defined from treatment initiation to PSA progression or censored at the date of last PSA evaluation. Radiographic progression was defined by RECIST version 1.1 for soft tissue and visceral disease and PCWG2 for bone disease[13, 14]. Progression-free survival (PFS) was defined as time from treatment initiation to radiographic progression or death from any cause, whichever came first, or censored at the date of last evaluation. Time to event endpoints were summarized using Kaplan-Meier method. We evaluated outcomes in the overall cohort, by type of prior therapy, and by CTC biomarker status. Patients with a positive CTC biomarker were defined as those positive for expression of AR variants, synaptophysin, and/or two or more AR-regulated genes (KLK2, LKL3, TMPRSS2, FOLH1, or NKX3.1). Comparisons between biomarker groups were conducted using the log-rank test.
Results:
Baseline Characteristics:
At data lock in April 2020, 67 patients were enrolled. The final analysis cohort for clinical outcomes consists of 65 men who received ≥1 dose of enzalutamide, and three patients remained on treatment; two men who never initiated treatment were excluded. Patient were enrolled between November 2013–May 2017: Dana-Farber Cancer Institute (n=39), University of Washington (n=19), Beth Israel Deaconess Medical Center (n=6), and South Shore Hospital (n=1) (Table 1). The median age was 70 years. Sixteen patients (24.6%) received prior chemotherapy, 22 (33.8%) prior abiraterone, and nine (13.8%) prior ketoconazole. All patients had metastases, of whom 32 (49.2%) had measurable disease.
PSA and Radiographic Response:
Thirty-eight patients (58.5%) achieved a ≥50% PSA reduction and 20 patients (30.8%) had a ≥90% decline in PSA (Figure 1). The PSA response rates in patients having received prior abiraterone, ketoconazole, or chemotherapy were 22.7% (n=5/22), 33.3% (n=3/9), and 68.8% (n=11/16), respectively (Supplementary Table 3S). Of the 32 patients with measurable disease, 11 (34.4%) had an objective response (Supplementary Table 4S).
Figure 1. Waterfall plot of best PSA response to therapy with enzalutamide.
Each bar represents an individual patient. Best percent change of PSA was calculated using date of first cycle of PSA as reference. Each green, blue and red bar indicates those who received prior therapies of abiraterone acetate/ketoconazole only (n=20), chemotherapy only (n=10) and both (n=6), respectively. The gray bars represent patients without prior treatment with chemotherapy, abiraterone acetate, or ketoconazole.
PSA and Radiographic Progression:
Forty-three (66.1%) patients experienced PSA progression. Median time to PSA progression was 5.6 months (95% CI 3.7, 10.1) (Supplementary Figure 1S) in the overall cohort and 2.8 months (95% CI 1.9, 5.6; n=14 events/22 patients), 11.0 months (95% 1.8, not reached; n=5/9), and 6.4 months (95% CI 1.8, 13.8; n=10/16) in patients having received prior abiraterone, ketoconazole, and chemotherapy, respectively (Supplementary Table 5S).
Overall, 33 patients (50.8%) experienced radiographic progression. Median radiographic PFS was 11.0 months (95% CI 8.1, 19.6) (Supplementary Figure 1S): 5.3 months (95% CI 2.7, 8.1; n=13/22) for prior abiraterone (Supplementary Table 5S) and 19.1 months (95% CI 2.1, not reached) for prior chemotherapy.
Toxicity:
Patients received a median nine cycles of enzalutamide (range <1–56) with a median duration of 8.6 months (range 0.1–51.6). Five patients (7.7%) had a dose reduction to 120 mg daily and 8 patients (12.3%) experienced a treatment hold. Six patients (9.7% among 62 patients who discontinued therapy of any reason) discontinued treatment due to unacceptable toxicity.
Overall, 24.6% (n=16), 36.9% (n=24), 30.8% (n=20), and 4.6% (n=3) reported maximum grade 1, 2, 3, and 4 toxicity of any attribution. There were no grade 5 events. The most common treatment-associated adverse events of any grade included fatigue, pain, hypertension, and back pain. The most common grade 3 toxicity was hypertension. Falls occurred in six patients and all were grade 1 or 2.
Metastasis Biopsy Samples:
Overall, 66 patients underwent a baseline biopsy (including one patient who consented but did not initiate treatment) and 28 patients underwent a biopsy at progression (Supplementary Table 7S; Supplementary Figure 2S). The majority of biopsies were from bone (n=65, 68%) followed by lymph nodes (n=25, 26%). Following quality control including assessment of tumor purity, tumor and normal coverage, and contamination, successful sequencing analysis was performed on 26 (39%) baseline and 17 (61%) progression biopsies. Successful sequencing analysis was performed on 42% of bone biopsies (n=27/65) and 64% (n=16/25) of soft tissue biopsies.
Tumor Sequencing Analysis:
Androgen Receptor Alterations
Of patients with baseline biopsies (n=26), 14 (53.9%) had AR amplifications and seven (26.9%) had AR mutations of whom four (28.6%) and three patients (42.9%) had a PSA response (≥50% PSA reduction from baseline), respectively (Figure 2, Supplementary Table 8S). Of the seven AR mutations presents at baseline, patients with a T878S, T979A, L702H or W742L did not experience a PSA response (Supplementary Table 9S). The PSA response rate in patients without AR amplifications was 75% (n=9/12) and 52.6% (n=10/19) in patients without AR mutations. Of the nine patients who received prior abiraterone, four (44.4%) had AR mutations and six (66.7%) had AR amplifications at baseline biopsy. PSA responses to enzalutamide were low in abiraterone pretreated individuals with AR amplifications (n=2/6, 33.3%) or AR mutations (n=1/4, 25.0%).
Figure 2. Integrative landscape analysis of somatic alterations obtained through DNA sequencing from baseline metastasis biopsies prior to treatment with enzalutamide and progression metastasis biopsies following treatment with enzalutamide.
Columns represent an individual metastasis biopsy. The left panel of columns represents paired baseline and progression metastasis biopsies obtained from the same individual (total 10 patients). The middle panel represents baseline biopsies only (total 16 patients). The right panel represents progression biopsies only (total 7 patients). Mutations per Mb are shown in the upper histogram. Biopsy type, prior abiraterone exposure, and presence of PSA ≥50% response from baseline are delineated in the first three rows. The remaining rows represent specific genes of interest. Color legend of the alterations are displayed. Multiple mutations in a gene are represented by triangles. Copy number calls are allelic and relative to whole genome doubling status, with calls for the two alleles indicated by two triangles. Allelic deletions that are not complete deletions are possible in samples with whole genome doubling. Because AR is on the X chromosome and has only a single allele in men, its copy number is represented as a box. Complex indicates that a copy number breakpoint occurred within the body of the gene. Putative loss of function (LoF) missense mutations were annotated as LoF or likely LoF in OncoKB or mutated the same amino acid as a LoF mutation. This plot was created using the CoMut software.
With regards to individuals with progression biopsies (n=17), 11 patients had AR amplifications (64.7%) and three (17.6%) had AR mutations (Figure 2, Supplementary Table 8S, Supplementary Table 9S). Of the six patients who received prior abiraterone, five (83.3%) demonstrated AR amplifications and one (16.7%) demonstrated an AR mutation. PSA responses were observed in seven patients (63.6%) with AR amplifications and all patients with AR mutations at progression.
In analyzing the paired baseline and progression samples (n=10), baseline AR alterations [n=2 mutations (W552C, T695A), n=5 allelic amplification] were present in seven patients (70%) of whom five (71.4%) experienced a PSA response (Figure 2). The two patients who did not experience a PSA response were abiraterone exposed. Acquired AR alterations present in progression metastases only were observed in four patients (40%) [n=1 mutation (S234C), n=3 allelic amplification] and all individuals experienced a PSA response to enzalutamide
Tumor Suppressor Genes Alterations
Of individuals with a baseline metastasis biopsy (n=26), alterations in tumor suppressor genes were present in 24 patients (92.3%) [TP53 n=18 (69.2%); RB1 n=18 (69.2%); PTEN n=17 (65.4%)] (Figure 2, Supplementary Table 8S). Biallelic alterations were observed in eight patients (30.7%) [TP53 n=4/26 (15.4%); RB1 n=1/26 (3.8%); PTEN n=5/26 (19.2%)]. The PSA response rate was 50.0% for patients with TP53 alterations (n=9/18), 50.0% for RB1 (n=9/18), and 47.1% for PTEN (n=8/17). The PSA response was 50% (n=4/8) in patients with biallelic alterations [TP53 n=2/4 (50%); RB1 n=0/1 (0%); PTEN n=3/5 (60%)]. PSA response rates were similar to those without tumor suppressor gene alterations [50.0% TP53 wildtype/neutral (n=4/8), 50.0% RB1 wildtype/neutral (n=4/8), and 55.6% PTEN wildtype/neutral (n=5/9)]. The frequency of tumor suppressor gene alterations was similar between patients with or without abiraterone exposure (Figure 2, Supplementary Table 8S). PSA response rates were lower in patients pretreated with abiraterone with tumor suppressor gene alterations; 25% of patients (n=2/8) pretreated with abiraterone with tumor suppressor gene alterations had a PSA response.
With regards to individuals with a progression metastasis biopsy (n=17), all patients (100%), including those with (n=6) and without (n=11) prior abiraterone exposure, had a tumor suppressor gene alteration [TP53 n=12/17 (70.6%); RB1 n=13/17 (76.5%); PTEN n=13/17 (76.5%)] (Figure 2, Supplementary Table 8S). Biallelic alterations at progression were observed in five patients (29.4%) [TP53 n=4/17 (23.5%); RB1 n=0/17 (0%); PTEN alterations n=1/17 (5.9%)]. The PSA response rate was 75.0% for patients with TP53 alterations (n=9/12), 61.5% for RB1 (n=8/13), and 61.5% for PTEN (n=8/13). The frequency of tumor suppressor gene alterations in progression biopsies was numerically higher in patients with prior abiraterone exposure compared to those naïve to abiraterone. Of patients pretreated with abiraterone with tumor suppressor gene alterations at progression (n=6), one patient (20%) experienced a PSA response.
From the paired biopsy samples (n=10), tumor suppressor gene alterations were present at baseline in nine patients (90.0%) [TP53 n=7/10 (70.0%); RB1 n=7/10 (70.0%); PTEN n=6/10 (60.0%)], including all three patients exposed to abiraterone (Figure 2). Acquired tumor suppressor gene alterations not present at baseline were observed in eight patients (80%) [(PTEN alteration n=4/10 (40.0%), RB1 alteration n=4/10 (40.0%), TP53 alteration n=2/10 (20.0%)].
DNA Repair Gene Alterations
Of individuals with a baseline biopsy (n=26), DNA repair genes alterations were present in 15 patients (57.7%) [BRCA2 n=10/26 (38.5%); CKD12 n=4/26 (15.4%); ATM n=1/26 (3.5%)] (Figure 2, Supplementary Table 8S). Biallelic alterations were observed in three patients (11.5%) [BRCA2 n=3/26 (11.5%)]. The PSA response rate was 50.0% (n=5/10), 25% (n=1/4), and 100% (n=1/1) for patients with BRCA2, CDK12, and ATM alterations, respectively. The PSA response was 0% in the three patients with biallelic alterations. In wildtype/neutral patients, PSA response rates were 50.0% (n=8/16), 54.5% (n=12/22), and 48.0% (n=12/25) for patients without BRCA2, CDK12, and ATM alterations. Of the patients pretreated with abiraterone with DNA repair alterations (n=7), only one patient (14.3%) with a CDK12 alteration had a PSA response.
Of those with a progression metastasis biopsy (n=17), 64.7% (n=11/17) had DNA repair alteration: 64.7% BRCA2 (n=11/17), 5.9% CDK12 (n=1/17), and 5.9% ATM (n=1/17) (Figure 2, Supplementary Table 8S). PSA response rates were observed in 54.5% of patients with BRCA2 alterations (n=6/11). One patient had a biallelic alteration in BRCA2 and did not experience a PSA response. Of the six patients with prior abiraterone exposure and a progression biopsy, five (83.3%) had DNA repair alterations at progression (n=5/6 with BRCA2 alterations, n=1/6 with a CDK12 alteration), of whom two experienced a PSA response (33.3%).
From the paired metastasis samples (n=10), BRCA2 gene alterations were observed in four patients (40.0%) at baseline (Figure 2). One patient each had a baseline CDK12 and ATM alteration. Acquired BRCA2 alterations not observed at baseline were seen in four patients (40%), in whom none were bilallelic and all co-occurred with RB1 alterations. No patient was previously exposed to a PARP inhibitor.
SPOP and CHD1 Alterations
In the baseline biopsy samples (n=26), SPOP alterations were observed in nine patients (34.6%) of whom four (44.4%) experienced a PSA response (Figure 2, Supplementary Table 8S). In patients having received prior abiraterone (n=9), SPOP alterations were present in four individuals (44.4%) of whom two (50.0%) had a PSA response to enzalutamide. CHD1 alterations were observed in 10 patients (38.5%) and seven (70.0%) experienced a PSA response to enzalutamide. Three patients (11.5%) had co-occurring SPOP and CHD1 alterations at baseline, two (66.7%) of whom had a PSA response.
In patients with evaluable progression biopsies (n=17), SPOP alterations were observed in five individuals (29.4%) (Figure 2, Supplementary Table 8S). Two patients (40.0%) with progression SPOP alterations experienced a PSA response. CHD1 alterations were present in nine (52.9%) patients at progression of whom seven (77.8%) had a PSA response to enzalutamide. Two patients (11.8%) had co-occurring SPOP and CHD1 alterations at progression and one of these individuals experienced a PSA response.
In assessing the paired metastasis biopsies (n=10), SPOP alterations were present at baseline in four patients (40.0%) of whom two (50.0%) had been previously exposed to abiraterone and two (50.0%) had a PSA response to enzalutamide (Figure 2). Acquired SPOP alterations, not present at baseline but present at progression, were observed in two individuals (20.0%), both of whom developed emergent co-occurring RB1 alterations and one developed emergent AR amplification.
Association of Tumor Gene Status with Outcomes
In evaluating baseline and progression biopsy samples, AR amplification was more prevalent in tumors samples at progression compared to baseline (Figure 3); no other gene was associated with presence in the progression biopsy in this analysis.
Figure 3. Association of AR amplification in progression samples (n=17) compared to baseline samples (n=26).
To assess significant changes in copy number before and after enzalutamide exposure, we combined p-values from a paired, two-sided Mann-Whitney U test for paired biopsies (n=10 pairs) and an unpaired, two-sided Mann-Whitney U test using unpaired biopsies (n=16 baseline, n=7 progression). The p-values were combined using a partially-mixed pooling approach designed to enable robust analysis of combined paired and unpaired data. AR=Androgen receptor, WGD=Whole genome duplication.
Circulating Tumor Cell Analysis:
Of the 65 patients who received at least one dose of enzalutamide, 52 had blood samples collected at baseline of whom 21 patients (40%) had adequate CTCs for gene expression analysis (Supplementary Figure 2S). Progression samples were collected from 37 patients of whom 23 (62%) had adequate samples. Reasons for the inability to perform the CTC assay for the remainder of patients included shipping delays/shipping protocol deviations (17% baseline, 5% progression) and inadequate blood volume or reagent issues (43% baseline, 22% progression). Collection methods were revised mid study that resolved these issues. Paired baseline and progression samples were available from 11 patients. Matched biopsy and CTC samples were available for six patients at baseline and 10 at progression (Supplementary Figure 3S).
We measured gene expression of splice variant AR, AR-regulated genes, and neuroendocrine markers. In swimmer plots, we observe shorter survival in patients who had detectable expression, either at baseline or progression, of genes in these pathways related to enzalutamide resistance (Figure 4). Overall survival was shorter in patients positive for enzalutamide resistant gene expression at baseline (median overall survival 17.7 months versus not reached in patients positive or negative for enzalutamide resistant gene expression respectively, HR 6.29 95% CI 1.22 to 32.5, p= 0.01) (Supplementary Figure 4S). When comparing the paired baseline and progression CTC samples (n=11), there was increased frequency of AR splice variants, AR regulated genes, and neuroendocrine markers in progression samples compared to baseline (Figure 5).
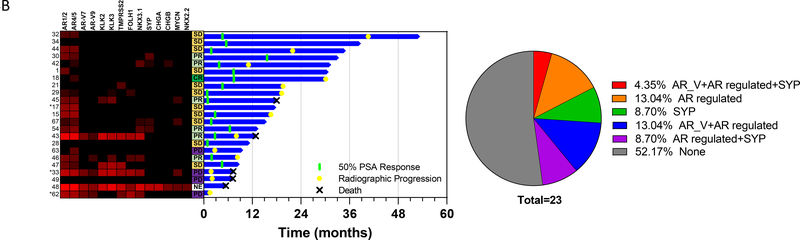
Figure 4. CTC gene expression analysis at baseline (A) and at progression (B).
Left panel: Heatmap showing expression of genes of interest. Each row represents an individual patient. Each column represents an individual gene. Red denotes increased expression. Middle panel: Swimmer plot of patient outcomes. Each row represents an individual patient. Column to the left of the Swimmer plot denotes best objective response on radiographic imaging (PR=partial response; SD=stable disease; PD=progressive disease). Right panel: Pie chart demonstrating percent expression of AR splice variants, AR regulated genes, and synaptophysin.
A) Baseline CTC expression analysis correlated with patient outcomes.
B) Progression CTC expression analysis correlated with patient outcomes.
Figure 5. CTC gene expression analysis of paired baseline and progression samples.
A: Heatmaps showing expression of genes of interest. Each row represents an individual patient. Each column represents an individual gene. Red denotes increased expression. Left panel denotes baseline pre-treatment CTC samples. Right panel denotes progression CTC samples. B: Pie chart demonstrating percent expression of AR splice variants, synaptophysin, and AR regulated genes. Left pie chart denotes baseline pre-treatment CTC samples. Right pie chart denotes progression CTC samples. C: Swimmer plot of patient outcomes. Each row represents an individual patient. Column to the left of the Swimmer plot denotes best objective response on radiographic imaging (PR=partial response; SD=stable disease; PD=progressive disease).
Discussion:
In this phase 2 study, embedding tissue and CTC-based genomic analyses, we investigate mechanisms of resistance to enzalutamide in metastatic CRPC. This analysis is important to understanding therapy selection and developing strategies to overcome resistance. Our analysis of tumor genomics with tissue and blood-based assays, confirms the landscape of CRPC alterations and reveals several insights about mechanisms of resistance to enzalutamide.[30]
A critical initial step to the molecular characterization of metastatic CRPC is the procurement of tumor tissue for genomic profiling. Successful sequencing of a metastasis biopsy requires sufficient tumor for isolation of high-quality nucleic acid. The majority of prostate cancer patients have bone metastases and bone-predominant disease. Bone metastases are frequently associated with a dense sclerotic reaction making biopsy itself and DNA preparation technically challenging; decalcification procedures may have a negative impact on nucleic acid quality and quantity. While use of archival primary prostate tumor tissue could overcome some of these challenges, treatment-naïve tumors will not capture alterations that emerge as a consequence of systemic therapy.[34]
In our study, of the 94 biopsies performed, 46% underwent successful whole exome sequencing. Larger efforts profiling the genomic landscape of metastatic CRPC, either do not report on successful sequencing yield from patients who underwent metastasis biopsy or report slightly higher yields than documented in our series.[30, 31, 35] Ongoing refinement of tissue biopsy and processing procedures will maximize the success of future genomic analyses in CRPC; these efforts are especially important in the current era of PARP inhibitor therapy. Given the challenges associated with metastasis biopsy and limitations in capturing the scope of tumor heterogeneity from an isolated metastatic site, minimally invasive blood-based “liquid” biopsies have emerged as an alternative to tissue sampling. Liquid biopsies enable frequent and sequential monitoring of tumor molecular dynamics; however, the concordance of tissue and blood-based methods for genomic assessment has varied.[36] In our study, we utilize an integrated molecular CTC assay to complement tissue analyses. Of the 89 patients with blood samples collected for CTC analysis, 49% underwent successful gene expression analysis. Because of an initial low success rate, we refined our methods for sample collection, shipping, and processing which resulted in higher yields in collected progression samples.
While only 25% of patients in our cohort (n=16/65) had biopsies with matched CTC analysis and the methodologies of analysis differed by specimen source, there were notable similarities in tissue and CTC molecular profiles. Specimens with AR alterations in tissue had increased expression of AR and AR regulated genes in CTCs. Our work aligns with other studies demonstrating conservation of AR alterations between CTCs and biopsies suggesting that CTCs can serve as a non-invasive surrogate for characterizing tumor molecular alterations.[37–39]
We demonstrate that BRCA2 alterations were acquired in 40% (n=4/10) of patients with paired metastasis biopsies following treatment with enzalutamide. Whether these alterations are true driver events remains to be determined given that most were monoallelic losses co-occurring with RB1 alterations. Prior reports have demonstrated the presence of alterations in homologous recombination repair (HRR) genes in tumors post AR signaling inhibitors (ARSIs), however these reports were without analysis of tumor samples prior to ARSI exposure.[31] BRCA2 and RB1 are both located on chromosome 13q, 16 megabases apart, thus there is a tendency for co-occurrence of alterations in these genes.[30] Moreover, while our numbers are small, these data underscore the value of serial tumor sampling in patients with CRPC to identify potential molecular targets with vulnerabilities to systemic treatment and also to evaluate for the emergence of neuroendocrine prostate cancer. Patients with HRR alterations, particularly BRCA2 mutations, can be responsive to PARP inhibitors or platinum chemotherapy, therefore testing for the emergence of such alterations is critical to therapy selection for patients. Our RNA-based CTC assay did not integrate HRR gene status, though continued assay refinement to integrate assessment of DNA and RNA is currently in process.
We confirm that AR pathway alterations, namely amplifications and mutations, are drivers of resistance to enzalutamide. Point mutations in the AR ligand-binding domain have been associated with resistance to AR-targeted therapy, including F877L and T878A, which have been associated with resistance to ARSIs.[5, 40–42] Other mutations, including T878S, have been associated with receptor promiscuity with increased sensitivity to steroids or AR antagonists.[42] L702H has been observed to emerge following glucocorticoid exposure and confers resistance to enzalutamide.[43] In our study, patients harboring baseline T878A, T878S, W742L, and L702H did not have a PSA response to enzalutamide. This raises the question of whether alternate AR antagonists, such as darolutamide, which have demonstrated in vitro activity against mutant AR, would be more effective in metastatic CPRC harboring these mutations or in the post abiraterone setting.[44] Our work highlights the difficulty of individual real-time tumor analysis, however targeted patient/tumor specific therapy remains a laudable goal.
We demonstrate that CTC transcriptomic interrogation is feasible and results in meaningful information that can elucidate both primary and secondary resistance mechanisms to enzalutamide. The CTC gene set analyzed in this cohort included splice variant AR, AR regulated genes, and neuroendocrine markers. While the well-studied AR variant-7 has clinical relevance given that detection in CTCs is predictive of resistance to enzalutamide and abiraterone, additional AR variants have been discovered that confer resistance to ARSIs[45]. In our CTC analysis, expression of AR variants and AR-regulated genes was seen in a higher proportion of progression CTC samples, consistent with findings observed from tumor genomic profiling. Given that CTCs are shed from tumor into circulation, it is expected that some concordance between tissue and CTC profiling would exist. Prior studies have demonstrated conservation between CTC and tissue AR pathway alterations, including AR variants and amplifications.[38] We confirm that possible AR dependency remains a persistent mechanism of resistance in CPRC and can be recapitulated in CTC analysis.
It is recognized that a proportion of CRPC tumors develop histologic neuroendocrine transformation as an AR-independent mechanism of treatment resistance. These tumors often have low or absent AR and/or AR-regulated genes and increased expression of classic neuroendocrine markers, including chromogranin and synaptophysin[46]. Frequently, these tumors harbor loss of RB1 and TP53, however these alterations are not specific to neuroendocrine CRPC[47]. In our CTC analysis, we demonstrate that progression samples exhibit increased expression of markers of resistance including neuroendocrine markers, AR variants but also increase in AR-regulated genes. This result likely reflects the heterogeneity of both AR-dependent and independent resistance mechanisms observed in advanced CRPC and lineage plasticity occurring in a subset of resistance clones. ARSI exposed tumors have a higher percentage of histologic neuroendocrine features and have higher neuroendocrine expression scores[30]. A recent study demonstrated that a targeted genomic and epigenomic gene set applied to cfDNA was capable of identifying patients with neuroendocrine CRPC with high concordance between cfDNA and tissue[48]. Patients with neuroendocrine CPRC are candidates for platinum chemotherapy, although subsequent effective treatments are limited for this poor risk population.
We evaluate the relevance of SPOP alterations in baseline and progression samples and demonstrate a PSA response rate of 55% (n=6/11) in patients with baseline SPOP alterations. Furthermore, two of the three abiraterone pretreated patients with SPOP alterations demonstrated a response to enzalutamide. Prior studies have demonstrated that SPOP-mutated prostate cancer is associated with more favorable prognosis, enrichment in earlier stage disease relative to CRPC, and improved responses to ARSI[30]. While our numbers are low, our data corroborate these findings.
Lastly, we confirm that prior exposure to CYP-17 inhibition, including abiraterone or ketoconazole, results in blunted efficacy to enzalutamide. In patients without abiraterone and/or ketoconazole exposure, PSA ≥50% responses were observed in 79.5% (n=31/39), while 26.9% of abiraterone and/or ketoconazole exposed patients (n=7/26) and 13.2% of abiraterone exposed patient (n=5/22) experienced a PSA response. This is consistent with prior studies which have demonstrated that sequential use of ARSIs results in cross-resistance and decreased efficacy[49, 50]. In a randomized, phase 2 crossover trial evaluating abiraterone followed by enzalutamide, PSA responses to second-line enzalutamide were seen in 36% of patients compared to 68% in patients receiving first-line abiraterone[49]. Additionally, a meta-analysis of eight studies including 643 patients demonstrated decreased PSA responses to second-line ARSIs[50].
Despite this being a prospective, multicenter phase 2 study interrogating mechanisms of resistance to enzalutamide, several limitations exist. The study required blood and tissue collection at baseline and progression; however, samples passing quality control were limited, resulting in a smaller sample size than projected. Additionally, the small sample size limited our ability to make inferences regarding less common genomic events.
Despite limitations our results confirm previously published data that resistance to enzalutamide is driven by alterations in the AR pathway and tumor suppressor genes. Our work was performed within the framework of a prospective, multicenter phase 2 trial leveraging paired tissue sampling and minimally invasive liquid biopsies in a patient population representative of standard enzalutamide treatment. Larger prospective studies, with integrated tissue analyses, are ongoing to validate the CTC gene expression panel utilized in this study. This panel has the potential to guide therapy selection between AR targeting agents, PARP inhibitors, chemotherapy, and clinical trials. Our data underscore the need for novel treatments and combinations to enhance efficacy to AR targeting agents and overcome or prolong resistance.
Supplementary Material
Table 1.
Baseline patient and disease characteristics.
| Characteristic | N | Median (q1-q3) or % |
|---|---|---|
| Institution | ||
| Dana-Farber Cancer Institute | 39 | 60.0% |
| University of Washington | 19 | 29.2% |
| Beth Israel Deaconess Medical Center | 6 | 9.2% |
| South Shore Hospital | 1 | 1.5% |
| Age at baseline (years) | 65 | 70 (66–75) |
| ECOG performance status | ||
| 0 | 44 | 67.7% |
| 1 | 21 | 32.3% |
| Gleason score at diagnosis | ||
| 6 | 6 | 9.2% |
| 7 | 22 | 33.8% |
| 8 | 9 | 13.8% |
| 9 | 22 | 33.8% |
| 10 | 3 | 4.6% |
| Missing | 3 | 4.6% |
| Metastases at diagnosis | 18 | 27.7% |
| Prior chemotherapy | 16 | 24.6% |
| Prior docetaxel | 15 | 23.1% |
| Prior abiraterone* | 22 | 33.8% |
| Prior ketoconazole | 9 | 13.8% |
| Prior sipuleucel-T | 15 | 23.1% |
| Prior first-generation anti-androgens** | 53 | 81.5% |
| Measureable disease at baseline | 32 | 49.2% |
| Non-measurable disease at baseline | 65 | 100% |
| Sites of metastasis at baseline*** | ||
| Bone | 58 | 89.2% |
| Lymph nodes | 32 | 49.2% |
| Lung | 8 | 12.3% |
| Liver | 1 | 1.5% |
| Other**** | 9 | 13.8% |
| Laboratory data at baseline | ||
| PSA (ng/mL) | 65 | 14.2 (6.9–140.6) |
| Albumin (g/dL) | 65 | 4.2 (4.0–4.4) |
| Alkaline phosphatase (U/L) | 65 | 80 (66–116) |
| Calcium (mg/dL) | 65 | 9.6 (9.2–9.8) |
| Hemoglobin (g/dL) | 65 | 12.8 (11.7–13.6) |
| Platelets (K/UL) | 65 | 211 (183–268) |
| White blood cells (K/uL) | 65 | 6.4 (5.5–7.6) |
ECOG=Eastern Oncology Cooperative Group, PSA=prostate specific antigen.
17 patients received prior abiraterone without ketoconazole; 5 patients received prior abiraterone and ketoconazole.
First-generation anti-androgens include bicalutamide or nilutamide,
Percentage for each category is calculated based on N=65, regardless of measurable/non-measurable disease.
Others include bladder, pelvis, paraspinal lesion, peritoneum/omentum or prostate.
Acknowledgements:
We would like to thank the patients and family members who participated in this clinical trial. This study was funded by Medivation/Astellas (M. Taplin). It was also supported by the Fairweather Family Fund (P. Taplin) and Fat Boys Slim Sisters Fund (M. Taplin) at the Lank Center for Genitourinary Oncology DFCI, PCF Challenge Award (Award Number 16CHAL03 – M. Taplin), and the National Cancer Institute (Award Numbers P01CA163227, R01CA234715 and the DH/HCC Prostate Cancer SPORE NCI P50 CA090381 – M. Taplin; PNW Prostate Cancer Spore NCI P50 CA097186 – P. Nelson).
Conflict of Interest:
RRM received research funding from Bayer, Pfizer, Tempus; serves on Advisory Board for AstraZeneca, Bayer, Bristol Myers Squibb, Calithera, Exelixis, Janssen, Merck, Novartis, Pfizer, Sanofi, Tempus; is a consultant for Dendreon, Vividion. PSN served received renumeration for consultant/advisory services to Bristol Myers Squibb, Astellas, and Janssen. XW received research funding from Bristol Myers Squibb. EMV received renumeration for consultant/advisory services to Tango Therapeutics, Genome Medical, Invitae, Enara Bio, Janssen, Manifold Bio, Monte Rosa, and received research funding from Novartis, Bristol Myers Squibb. MET served on the Advisory Board for Bayer, AstraZeneca, Janssen, Abbvie, and Astellas. The remaining authors have no disclosures.
References:
- 1.Sartor O, de Bono JS. Metastatic Prostate Cancer. N Engl J Med 2018; 378: 1653–1654. [DOI] [PubMed] [Google Scholar]
- 2.Tran C, Ouk S, Clegg NJ et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 2009; 324: 787–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beer TM, Armstrong AJ, Rathkopf DE et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med 2014; 371: 424–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scher HI, Fizazi K, Saad F et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012; 367: 1187–1197. [DOI] [PubMed] [Google Scholar]
- 5.Joseph JD, Lu N, Qian J et al. A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509. Cancer Discov 2013; 3: 1020–1029. [DOI] [PubMed] [Google Scholar]
- 6.Conteduca V, Wetterskog D, Sharabiani MTA et al. Androgen receptor gene status in plasma DNA associates with worse outcome on enzalutamide or abiraterone for castration-resistant prostate cancer: a multi-institution correlative biomarker study. Ann Oncol 2017; 28: 1508–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards J, Krishna NS, Grigor KM, Bartlett JM. Androgen receptor gene amplification and protein expression in hormone refractory prostate cancer. Br J Cancer 2003; 89: 552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antonarakis ES, Lu C, Wang H et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014; 371: 1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armstrong AJ, Halabi S, Luo J et al. Prospective Multicenter Validation of Androgen Receptor Splice Variant 7 and Hormone Therapy Resistance in High-Risk Castration-Resistant Prostate Cancer: The PROPHECY Study. J Clin Oncol 2019; 37: 1120–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C, Lou W, Zhu Y et al. Intracrine Androgens and AKR1C3 Activation Confer Resistance to Enzalutamide in Prostate Cancer. Cancer Res 2015; 75: 1413–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stanbrough M, Bubley GJ, Ross K et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res 2006; 66: 2815–2825. [DOI] [PubMed] [Google Scholar]
- 12.Crona DJ, Whang YE. Androgen Receptor-Dependent and -Independent Mechanisms Involved in Prostate Cancer Therapy Resistance. Cancers (Basel) 2017; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scher HI, Halabi S, Tannock I et al. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: recommendations of the Prostate Cancer Clinical Trials Working Group. J Clin Oncol 2008; 26: 1148–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenhauer EA, Therasse P, Bogaerts J et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009; 45: 228–247. [DOI] [PubMed] [Google Scholar]
- 15.Fisher S, Barry A, Abreu J et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol 2011; 12: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gnirke A, Melnikov A, Maguire J et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 2009; 27: 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cibulskis K, Lawrence MS, Carter SL et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013; 31: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cibulskis K, McKenna A, Fennell T et al. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics 2011; 27: 2601–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costello M, Pugh TJ, Fennell TJ et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res 2013; 41: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence MS, Stojanov P, Mermel CH et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014; 505: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saunders CT, Wong WS, Swamy S et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012; 28: 1811–1817. [DOI] [PubMed] [Google Scholar]
- 23.Taylor-Weiner A, Stewart C, Giordano T et al. DeTiN: overcoming tumor-in-normal contamination. Nat Methods 2018; 15: 531–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010; 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLaren W, Gil L, Hunt SE et al. The Ensembl Variant Effect Predictor. Genome Biol 2016; 17: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramos AH, Lichtenstein L, Gupta M et al. Oncotator: cancer variant annotation tool. Hum Mutat 2015; 36: E2423–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016; 44: e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carter SL, Cibulskis K, Helman E et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012; 30: 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stachler MD, Taylor-Weiner A, Peng S et al. Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat Genet 2015; 47: 1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abida W, Cyrta J, Heller G et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 2019; 116: 11428–11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson D, Van Allen EM, Wu YM et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015; 161: 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sperger JM, Strotman LN, Welsh A et al. Integrated Analysis of Multiple Biomarkers from Circulating Tumor Cells Enabled by Exclusion-Based Analyte Isolation. Clin Cancer Res 2017; 23: 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strotman L, O’Connell R, Casavant BP et al. Selective nucleic acid removal via exclusion (SNARE): capturing mRNA and DNA from a single sample. Anal Chem 2013; 85: 9764–9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Dessel LF, van Riet J, Smits M et al. The genomic landscape of metastatic castration-resistant prostate cancers reveals multiple distinct genotypes with potential clinical impact. Nat Commun 2019; 10: 5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sailer V, Schiffman MH, Kossai M et al. Bone biopsy protocol for advanced prostate cancer in the era of precision medicine. Cancer 2018; 124: 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wyatt AW, Annala M, Aggarwal R et al. Concordance of Circulating Tumor DNA and Matched Metastatic Tissue Biopsy in Prostate Cancer. J Natl Cancer Inst 2017; 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Josefsson A, Larsson K, Mansson M et al. Circulating tumor cells mirror bone metastatic phenotype in prostate cancer. Oncotarget 2018; 9: 29403–29413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Podolak J, Eilers K, Newby T et al. Androgen receptor amplification is concordant between circulating tumor cells and biopsies from men undergoing treatment for metastatic castration resistant prostate cancer. Oncotarget 2017; 8: 71447–71455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Punnoose EA, Ferraldeschi R, Szafer-Glusman E et al. PTEN loss in circulating tumour cells correlates with PTEN loss in fresh tumour tissue from castration-resistant prostate cancer patients. Br J Cancer 2015; 113: 1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen EJ, Sowalsky AG, Gao S et al. Abiraterone treatment in castration-resistant prostate cancer selects for progesterone responsive mutant androgen receptors. Clin Cancer Res 2015; 21: 1273–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rathkopf DE, Smith MR, Ryan CJ et al. Androgen receptor mutations in patients with castration-resistant prostate cancer treated with apalutamide. Ann Oncol 2017; 28: 2264–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taplin ME, Bubley GJ, Ko YJ et al. Selection for androgen receptor mutations in prostate cancers treated with androgen antagonist. Cancer Res 1999; 59: 2511–2515. [PubMed] [Google Scholar]
- 43.Carreira S, Romanel A, Goodall J et al. Tumor clone dynamics in lethal prostate cancer. Sci Transl Med 2014; 6: 254ra125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu J, Zhou P, Hu M et al. Discovery and biological evaluation of darolutamide derivatives as inhibitors and down-regulators of wild-type AR and the mutants. Eur J Med Chem 2019; 182: 111608. [DOI] [PubMed] [Google Scholar]
- 45.Tietz KT, Dehm SM. Androgen receptor variants: RNA-based mechanisms and therapeutic targets. Hum Mol Genet 2020; 29: R19–R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Epstein JI, Amin MB, Beltran H et al. Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. Am J Surg Pathol 2014; 38: 756–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beltran H, Rickman DS, Park K et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov 2011; 1: 487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beltran H, Romanel A, Conteduca V et al. Circulating tumor DNA profile recognizes transformation to castration-resistant neuroendocrine prostate cancer. J Clin Invest 2020; 130: 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khalaf DJ, Annala M, Taavitsainen S et al. Optimal sequencing of enzalutamide and abiraterone acetate plus prednisone in metastatic castration-resistant prostate cancer: a multicentre, randomised, open-label, phase 2, crossover trial. Lancet Oncol 2019; 20: 1730–1739. [DOI] [PubMed] [Google Scholar]
- 50.Mori K, Miura N, Mostafaei H et al. Sequential therapy of abiraterone and enzalutamide in castration-resistant prostate cancer: a systematic review and meta-analysis. Prostate Cancer Prostatic Dis 2020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.