SUMMARY
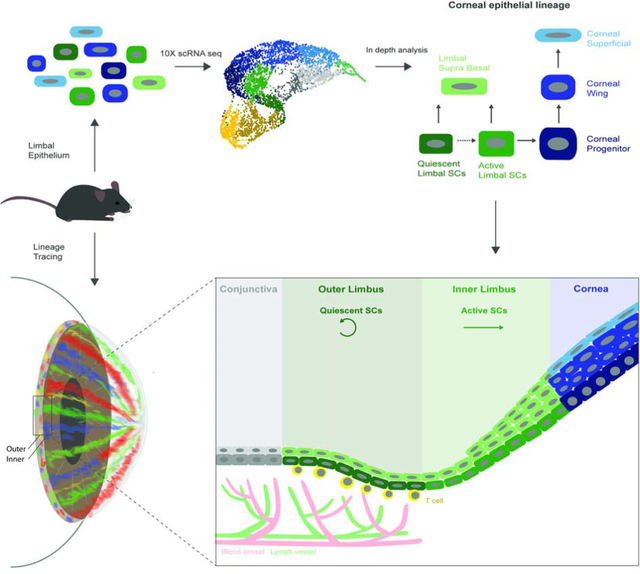
The accessibility and transparency of the cornea permit robust stem cell labeling and in vivo cell fate mapping. Limbal epithelial stem cells (LSCs) that renew the cornea are traditionally viewed as rare, slow-cycling cells that follow deterministic rules dictating their self-renewal or differentiation. Here, we combined single-cell RNA sequencing and advanced quantitative lineage tracing for in-depth analysis of the murine limbal epithelium. These analysis revealed co-existence of two LSC populations localized in separate and well-defined sub-compartments, coined the “outer” and the “inner” limbus. The primitive population of quiescent outer LSCs participates in wound healing and boundary formation, and are regulated by T cells which serve as a niche. In contrast, the inner peri-corneal limbus hosts active LSCs that maintain corneal epithelial homeostasis. Quantitative analyses suggest that LSC populations are abundant, following stochastic rules and neutral drift dynamics. Together these results demonstrate that discrete LSC populations mediate corneal homeostasis and regeneration.
eTOC
Single cell RNA-sequencing combined with quantitative lineage tracing revealed the existence, signature and dynamics of two distinct murine LSC populations, which reside in spatially defined sub-compartments, termed the “outer” and “inner” limbus. Each LSC populations exhibit distinct cycling properties, regulation by the niche and function under homeostasis and regeneration.
Graphical Abstract
INTRODUCTION
The traditional stem cell (SC) paradigm describes “true” SCs as rare, quiescent, and highly potent cells (Cotsarelis et al., 1989) (TILL and McCULLOCH, 1961)(Barrandon and Green, 1987). The proposed benefits of quiescence include reduced biochemical damage and minimalized accumulation of disease-causing mutations(Cho et al., 2019) (Tümpel and Rudolph, 2019). However, markers that reliably identify such scarce SCs are usually not available. The alternative equipotent SC model describes SCs as more frequently dividing abundant cells with dynamics that fit with the neutral drift model(Li and Clevers, 2010) (Clevers and Watt, 2018) (Colom and Jones, 2016) (Park et al., 2016) (Clayton et al., 2007) (Klein et al., 2007). This paradigm challenged the conventional SC dogma that considered quiescence as an integral feature of SCs.
The transparent cornea is unique in the sense that epithelial cells are exposed to harmful conditions that include sun irradiation, toxicants, injuries, and infections(Secker and Daniels, 2008). The clarity and high accessibility of the cornea, however, allows robust SC lineage tracing(Amitai-Lange et al., 2015a) (Amitai-Lange et al., 2015b) (Di Girolamo et al., 2015)(Dorà et al., 2015) and SC depletion using simple vital microscopy (Nasser et al., 2018) (Park et al., 2019) (Ishii et al., 2020). Corneal epithelial SCs are known as rare slow-cycling cells that reside in the opaque limbus, a ring-shaped zone at the corneal-conjunctival boundary (Cotsarelis et al., 1989) (Sartaj et al., 2017) (Ebato et al., 1988) (Pellegrini et al., 1999). However, previously identified limbal SC (LSC) markers typically label the broad limbal basal cell population, suggesting that the current model may be inaccurate. LSCs are believed to produce short-lived progenitors that undergo centripetal movement towards the central cornea, or delaminate, become post-mitotic, and eventually terminally differentiate and desquamate. The differentiation program and the key cell states of the corneal epithelial lineage are not well characterized.
Here we combined single-cell transcriptomics and quantitative lineage tracing to capture LSC populations and their signature. This work provides useful atlas of the entire murine corneal epithelial lineage, reveals the signature of two previously undescribed LSC states, and the role of T cells as niche cells.
RESULTS
Single-cell sequencing identifies discrete cell states in the corneal epithelial lineage
Careful inspection of the limbus of Krt15-GFP transgenic mice showed that GFP+ cells are confined to an “inner” limbal zone, suggesting that the “outer” limbal region (the space between Krt15-GFP+ cells and the Krt8+ conjunctiva) (Fig. 1A, S1A) contains a different LSC population. To identify LSC populations, we performed a single-cell transcriptomic analysis. To gain robust statistical data, we isolated epithelial cells from the limbus (with marginal conjunctiva and corneal periphery) of 5 individual adult mice (2.5 months old, n=10 eyes, Fig. 1A, see Methods) and performed 10x Chromium single-cell RNA sequencing (scRNA-seq). Initial raw data analysis and quality controls were performed with Cell Ranger software and 4,787 high quality cells passed rigorous quality tests exhibiting significant mean reads (98,763 per cell) and a substantial median number of 2,479 detected genes per cell.
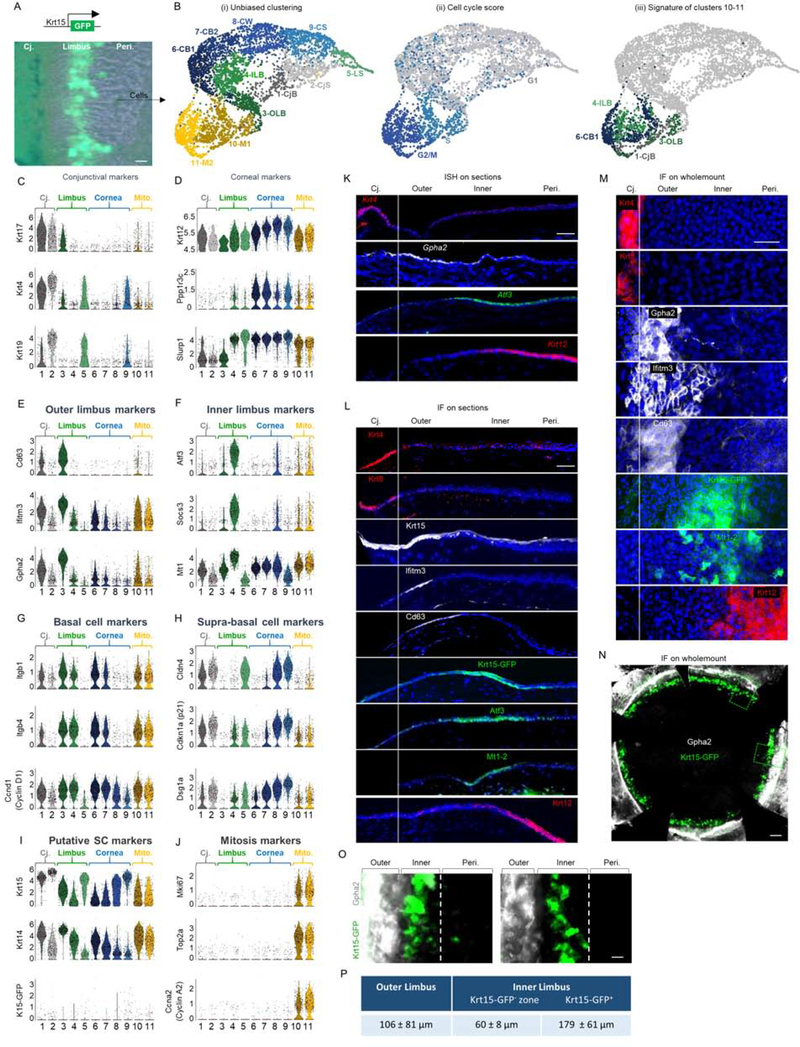
Figure 1: scRNA-seq reveals corneal epithelial cell states including 2 distinct basal limbal epithelial cell populations.
(A-B) The limbus (with marginal conjunctiva and cornea) of 10 corneas of 2.5-month old Krt15-GFP transgenic mice was dissected, pooled and epithelial cells were subjected to scRNA-seq analysis. (B) UMAP plots presentations of (i) Unbiased clustering revealed 11 distinct groups of cells. (ii) Cell cycle score analysis revealed that most cells in clusters 10–11 display the signature of cells captured in specific stages of mitosis. (iii) Cell type classification for clusters 10–11 shows they consist of a mixture of cells with a hallmark of different ocular basal cells. (C-J) Violin plots of differentially expressed markers in each cluster. Y-axis in C-J represents expression level. (K-O) In situ hybridization (ISH) (K) or immunofluorescent staining (IF) on tissue sections (L) or wholemount (M-O) of 2-month-old mice for the indicated markers. (P) Analysis of the width (mean ± standard deviation) of limbal sub-compartments. n=3 corneas. Scale bars were 50 μm (A-M, O-P) or 300 μm (N). Nuclei were counter stained by DAPI. Abbreviations: Cj., conjunctiva; Mito., mitosis; Peri., periphery; CjB, conjunctival basal; CjS, conjunctival suprabasal; OLB, outer limbal basal; ILB, inner limbal basal; LS, limbal superficial; CB1, corneal basal 1; CB2, corneal basal 2; CW, corneal wing; CS, corneal superficial, M1/M2, cells in mitosis.
See also Figures S1, S2, and S3.
Cell filtering and unbiased clustering (R package Seurat(Butler et al., 2018)) defined 11 cell populations (Fig. 1B(i), S1B). Over 99% of the cells were ocular epithelial cells (see Methods), while all clusters had a comparable contribution from all samples (Fig. S1C). Classification of clusters based on gene expression revealed clusters of conjunctival (1–2), limbal (3–5), corneal epithelial cells (6–9), and a mixture of cells from all types that were harvested during mitosis (10–11) (Fig. 1C–J, S1D–E, G–H). The estimated numbers of cells in each cluster/tissue (Fig. S1B) fits well with tissue dissection/harvesting (Fig. 1A) of marginal conjunctiva (~800 cells), limbus (~1400 cells), and a significant portion of the peripheral cornea (~2500 cells).
Conjunctival cells were identified based on the expression of Keratin 17 (Krt17), Krt4, Krt19 (Fig. 1C) and Krt6a, Krt13, Krt8 (Fig. S1D)(Kivelä and Uusitalo, 1998)), the lack or low expression of corneal markers Krt12, Slurp1(Kasper et al., 1988) (Swamynathan et al., 2015), and protein phosphatase 1 (Ppp1r3c) (Fig. 1D). Cluster 1 clearly expressed basal (Itgb1, Itgb4, Ccnd1 (Cyclin D1)(Stepp, 2006) (Joyce et al., 1996)) cell markers while cluster 2 expressed supra-basal cell markers (Cld4, Dsg1a, Cdkn1a (P21)(Yoshida et al., 2009) (King et al., 1997) (Brandner et al., 2002) (Fig. 1G–H). Krt15 and Krt14 marked conjunctival basal cells whereas Krt15 was even higher in conjunctival supra-basal cells (Fig. 1I).
Limbal cells were identified based on the lack or lower expression of both conjunctival (Fig. 1C, S1D) and corneal (Fig. 1D) markers. While cluster 5 displayed supra-basal cell phenotype, clusters 3–4 were basal cells (Fig. 1G–H) expressing a set of limbal specific markers (Fig. 1E–F). As compared to cluster 4, cluster 3 was hallmarked by much higher levels of Krt15 and Krt14 (Fig. 1I) and lower Krt12 (Fig. 1D), suggesting that it represents a more undifferentiated limbal cell population.
Although the corneal specific Krt12 mRNA was found at a certain level in all groups, clusters 6–9 expressed much higher levels (Fig. 1D). Clusters 6–7 were basal cells, although cluster 7 displayed attenuated basal cell phenotype (Fig. 1G–H). Cluster 8 displayed a partially differentiated phenotype that fits with corneal wing cells whereas cluster 9 represented terminally differentiated superficial cells (Fig. G-H). In line with previous reports(Nasser et al., 2018) (Yoshida et al., 2006), while Krt15 and Krt15-GFP labeled basal conjunctival/limbal epithelial cells, they also marked supra-basal cells (Fig. 1I).
Cell cycle analysis indicated that clusters 1–9 were in G1 state, while clusters 10–11 exhibited a signature of cells in S and G2/M (Fig. 1B(ii)) and expressed mitosis specific genes (Fig. 1J, S1G). Clusters 10–11 consisted of a mixture of cells with a hallmark (i.e. score of top 10 genes) of all cell types (Fig. 1B(iii)), in line with validation by immunostaining (Fig. S1H–I). Reduced (~45%) occurrence for cells with the signature of cluster 3 suggested less frequent cell division (see Figs 2–3).
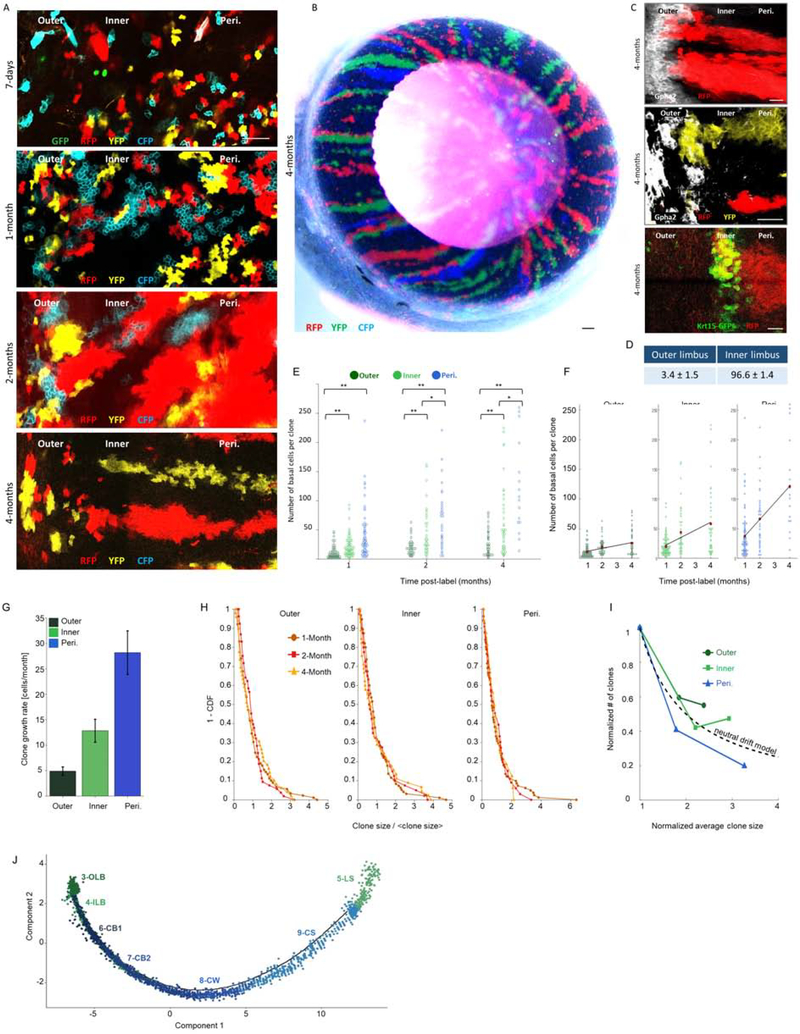
Fig 2: Quantitative “Confetti” lineage tracing unravels the dynamics of outer and inner LSCs.
Lineage tracing was induced by tamoxifen injection in 2-month-old UBC-CreERT2; Brainbow2.1“Confetti” mice (see Fig. S4B–C). Confetti-positive clones were analyzed at the indicated time points post-induction. Typical tile sean confocal images of the limbal zones are shown in (A, C) and the entire eye (cornea view) is shown in (B). The same pictures shown here in A, are also shown in Fig. S4D together with Gpha2 co-staining. (D) The percentage of stripes that emerged from the outer or inner limbus. (E-F) A Scatter plot of clone size distributions shows the indicated regions over time (E) or across the different time points for the different colonial regions (F). Red dots denote the average clone size and the black line indicates the linear model that fits the data. (G) The clonal effective growth rate, as estimated from the linear fit. Error bars are 95% confidence interval (CI). Note that the 95% CI do not overlap. (H) One minus the cumulative distribution function of the clone size divided by the mean clone size. The scaled distributions collapse onto the same curve (compare to Fig. S4G, which shows the non-scaled distributions). (I) The normalized number of clones as a function of normalized clone size. The dotted line denotes the inverse relation between clone size and clone number as expected from the neutral drift model. (J) Pseudotime plot predicts the differentiation process across clusters. Scale bars are 100 μm (A, C) and 50 μm for (B). n=15 areas from 3–5 corneas of 3 individuals. Statistical significance was calculated using the Kolmogorov-Smirnov test. (*, p-value < 0.05; ** p-value <0.005). Abbreviations: Peri., Periphery; OLB, outer limbal basal; ILB, inner limbal basal; LS, limbal superficial; CB1, corneal basal 1; CB2, corneal basal 2; CW, corneal wing; CS, corneal superficial.
See also Figure S4.
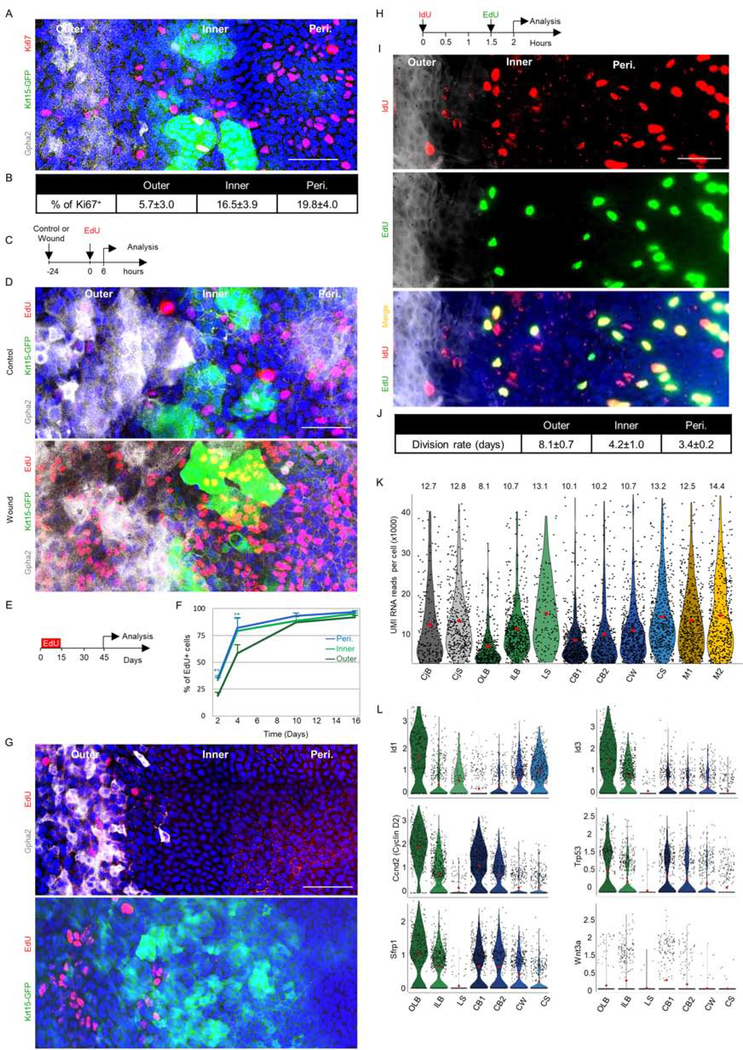
Fig 3: Proliferation analysis of slow-cycling and frequently dividing LSCs.
Adult 2–3- month- old Krt15-GFP mice were used for all experiments. The limbal region markers in wholemount corneas were defined by Gpha2/K15-GFP, nuclei were detected by DAPI counter staining (A, D, G, I) while Ki67+ cells were stained (A) and quantified (B). (C-D). Corneal epithelial debridement was followed by EdU injection and 6 hours later, cells in S-phase (EdU+) were identified in wounded or uninjured controls. A schematic illustration is shown in (C) and a typical tile scan confocal image of wholemount immunostaining is shown in (D). (E-G) Water-based EdU administration for 15-days (pulse) was followed by a 30-day chase. Schematic illustration (E), quantification of EdU accumulation in basal cells over time (F), and typical tile scan confocal image of wholemount staining with regional markers (G) is shown. (H-J) Double nucleotide (IdU/EdU) injection with an interval of 1.5 hours was followed (0.5 an hour later) by tissue harvesting and quantitative analysis (see methods) on wholemount staining. Schematic illustration (H), representative image (I), and an estimated cell cycle for each zone (J) is shown (n=15 areas from 3 corneas of 3 individuals). (K) Violin plot diagram of unique molecular identifiers (UMI) RNA reads per cell in each cluster. (L) Violin plot showing the expression of the indicated genes involved in SC quiescence or activation. Y-axis in L represents expression level. Statistical significance was calculated using the One-way ANOVA test followed by the Bonferroni test (*, p-value<0.05). Data represented as mean ± standard deviation, Abbreviations: Peri., periphery; CjB, conjunctival basal; CjS, conjunctival suprabasal; OLB, outer limbal basal; ILB, inner limbal basal; LS, limbal superficial; CB1, corneal basal 1; CB2, corneal basal 2; CW, corneal wing; CS, corneal superficial; M1/M2, mitosis. Scale bars, 50 μm.
Identification of two limbal sub-compartments
To certify the cluster identification in vivo, we performed in situ hybridization (ISH) on tissue sections of 2–3 months old C57BL/6 mice. Both basal and supra-basal conjunctival and corneal cells were labeled using ISH probes for Krt4 and Krt12, respectively. Interestingly, a well-demarcated signal of Gpha2 labeled the “outer limbus” while the “inner limbus” was marked by Atf3 (Fig. 1K). Immunostaining confirmed that outer limbal basal epithelial cells (cluster 3) were Krt15+/Ifitm3+/Cd63+, inner limbal epithelial cells were Krt15-GFP+/Atf3+/Mt1–2+ (Fig. 1L) and limbal supra-basal cells (cluster 5) were Prdm1 positive (Fig. S1E–F). Wholemount immunostainings were advantageous (Fig. 1M–O), avoiding detachments of outer/inner limbus in tissue sections. Notably, in line with previous reports (Morris et al., 2004), the endogenous Krt15 expression did not overlap with Krt15-GFP (Fig. 1I, 1L).
Quantitative analysis showed that the average width of the Gpha2+ outer limbus was ~100 μm, while the inner limbus was much wider (~240 μm, comprised of Krt15-GFP(-) and peri-corneal Krt15-GFP(+) subzones) (Figures 1O and 1P). The pattern of Krt15-GFP signal was often continuous, or nearly continuous with outer limbus markers like Gpha2, but never overlapped with the labeling of the outer limbus (Fig. 1N–O). Interestingly, flattened basal cells found in both sub-compartments were hallmarked by a unique pattern of nuclei staining that differed from that of the cuboidal basal cells that were detected in the marginal inner limbus and corneal periphery (Fig. 1L, S1J–K). Finally, analysis of selected keratin proteins confirmed that basal outer limbal epithelial cells were Krt15hi/Krt14hi/Krt17med/Krt12neg, basal inner limbal epithelial cells were Krt15neg/Krt14med/Krt17neg/Krt12low, while conjunctival cells were Krt19hi/Krt17hi/Krt6hi/Krt8med/Krt12neg (Fig. 1L, S1D, L–M).
Next, WebGestalt pathway enrichment analysis (Zhang et al., 2005) was applied to identify pathways that are enriched in clusters 3 or 4 in comparison to all other clusters. Significant enrichment in cancer pathways was evident, in line with the fact that SC and cancer pathways often overlap. Particularly, P53, Pi3K/AKT, and Rap1 signaling pathways were enriched in cluster 3 while cluster 4 was enriched with TNF pathways (Fig. S2). Additional analysis revealed a list of differentially expressed genes that were enriched by clusters 3 or 4 (Fig. S3A–B). Expectedly, genes related to hemi-desmosomes increased in clusters of basal cells (clusters 1, 3–4, 6–7) and desmosome-related and gap- junction associated genes increased in supra-basal clusters (clusters 2, 5, 8–9) (Fig. S3C). Taken together, these data illuminate the major cell states across the corneal epithelial lineage, and in particular, the co-existence of two discrete limbal sub-compartments.
Quantitative lineage tracing suggests distinct clonal dynamics for outer and inner limbal epithelial cells
Comparative analysis (WebGestalt) between clusters 3 and 4 suggested that a central difference between these clusters lies in their cell cycle and motility characteristics (Fig. S4A). To gain insights on proliferation dynamics, centripetal movement, and discover the potential hierarchy between all clusters, we designed lineage tracing experiments. We used the efficient UBC-CreERT2 driver (recombinase under the Ubiquitin C promoter) for tracking potentially rare LSC populations. Upon transient exposure to tamoxifen, Cre recombinase translocated into the nucleus, inducing stochastic and irreversible expression of one out of four “confetti” (Brainbow2.1) fluorescent proteins (nuclear green (GFP), cytoplasmic red (RFP) or yellow (YFP), or membrane cyan (CFP), Fig. S4B–E) (Amitai-Lange et al., 2015a)(Nasser et al., 2018). Compatible induction in all regions was found 7 days post tamoxifen injection (Figure S4E). We defined three types of clones depending on the location of their base (circumferential margin) in the inner, outer, or peripheral cornea. For example, outer limbal clones were those that emerged from Gpha2+ outer limbus, even if they extended to the inner limbus or peripheral cornea.
Interestingly, at all time-points, the average clone size (i.e. number of basal cells) in the outer limbus was much smaller compared to other zones while corneal peripheral clones were largest (Fig. 2A, E–F). Surprisingly, 97% of radial stripes possessed a base in the inner limbus while only 3% in the outer limbus (Fig. 2A–D). Moreover, outer limbal clones often displayed a centrifugal (circumferential) pattern (Fig. 2A, 2–4-months). The detection of stripes with a “foot” in the outer and inner limbal zones after very long-term tracing, suggests that both limbal zones contain bona fide LSCs.
In tightly packed multi-layered epithelium, cells do not simply “migrate” between compartments. The primary driving force of cell movement is proliferation that is coupled with delamination of neighboring cells(Strinkovsky et al., 2021) (Mesa et al., 2018) (Miroshnikova et al., 2018). Accordingly, the rarity of the centripetal pattern of the outer limbal clones, and their predominant association with the inner limbus, suggest that the inner limbus maintains homeostasis while the outer limbus serves as boundary and SC reservoir.
The relatively uniform pattern of marker expression and clonal growth dynamics in the outer and inner limbal zones implied that limbal epithelial cells might follow stochastic rules and neutral drift(Klein and Simons, 2011). The main prediction of this model is that the clone size would grow linearly with time(Jones et al., 2019). In this case, the slope of this linear trend is proportional to the product of the doubling rate and the probability of symmetric division in the epithelial plane (Methods). These results suggest that clone size dynamics in all regimes can be captured by a linear growth and that the slopes are significantly different and ordered centripetally – each regime has a slope that is about twice higher than its neighboring zone (Fig. 2F–G).
To further analyze the clonal dynamics, we aimed to characterize not only the changes of the average over time (Fig. S4F) but also the dynamics of the entire clonal probability distribution. Another prediction of the neutral drift model is that the clone size distributions scale with their averages. That is, the size distribution of the clone size normalized by their mean should be similar. Fig. 2H (and Fig. S4G) shows the probability of having a clone that is larger than some value (one minus the cumulative probability distribution) for the normalized size distributions. All regimes show clear scaling of their clone size for all time points.
It is insightful to consider the changes in the number of clones and average size on the same graph. As clones are growing and competing with each other, we expect an inverse relationship between the average clone size and their number. Under the assumption of the neutral drift model, the number of clones should decrease inversely with the clone size (Fig. 2I). IIn our case, there is a clear difference among the three different zones tested. While there is an inverse relationship between size and number, in the outer and inner regimes, after 4 months, the reduction is smaller than expected by the neutral drift model. The fact that the number of clones for a given area in the peripheral cornea is less than expected by the neutral drift model is consistent with biased centripetal pattern dynamics (Strinkovsky et al., 2021). Taken together, this data suggests that the outer and the inner limbus contain LSC populations that exhibit similar stochastic dynamics but different doubling times.
To further assess the hierarchical relations between clusters in the corneal epithelial lineage, we plotted the cells based on their progression through biological process dynamics using the Pseudotime algorithm for limbal/corneal epithelial clusters (clusters 3–9). This algorithm infers the sequential changes of gene expression between clusters and thereby provides insights into the dynamics of biological processes, in this case cell differentiation. This analysis suggested that cluster 3 (outer limbal basal cells) can give rise to cluster 4 (inner limbal basal cells) that derives clusters 6–7 (corneal basal cells) and clusters 8–9 (corneal wing and superficial cells), while cluster 5 (limbal superficial) that represents post-mitotic supra-basal cells (hence very different from limbal basal clusters 3–4), was positioned next to supra-basal corneal clusters (Fig. 2J). Further analysis should be done to better understand the identity of cluster 5. Taken together, the combined quantitative lineage tracing and in silico analysis illuminates the dynamics and hierarchy between corneal epithelial cell populations.
Outer and inner limbus represent segregated sub-compartments for quiescent and active LSCs
Interestingly, Ki67+ staining (Fig. 3A–B) and 5-Ethynyl-2′-deoxyuridine (EdU) nucleotide analogue incorporation assay (Fig. 3C–D) showed much less frequent labeling of dividing cells in the outer limbus, as compared to other zones. However, in response to corneal injury, numerous outer (and inner) limbal cells entered the cell cycle (Fig. 3D). To examine the presence of slow-cycling cells, a water-based EdU “pulse” of 15-days was followed by a “chase” period of 30-days in the absence of EdU. The outer limbus accumulated signal more slowly than other zones, and eventually, nearly all cells were comparably labeled in all compartments (Fig. 3E–F). During the chase period, the label declines by 50% within every cell division and becomes undetectable after ~4 successive division rounds(West et al., 2018) (Sartaj et al., 2017).In agreement, EdU+ label-retaining cells were predominantly found in the outer limbus, whereas rare cells were occasionally found in the inner limbus (Fig. 3G), suggesting that during the 30-day chase period most outer LSCs exceeded sufficient division rounds and lost labeling and only a few which underwent less replications could be detected. Finally, we validated that label-retaining cells were epithelial cells, by co-staining of EdU+ label-retaining cells with Krt17 antibody (Fig. S1M).
The relative homogeneity of Gpha2/Ifitm3/Cd63 labeling by the outer LSCs, together with the absence of Ki67 associated with the broad slow growth of clones in the outer limbus, implied that the vast majority, perhaps entire, basal cell population in the outer limbal zone divides less frequently. To gain direct evidence on the cell cycle of each population and evaluate the rate of cell division in each zone we performed a double nucleotide-analogue injection. As depicted in Fig. 3H, 1.5 hours following injection of iododeoxyuridine (IdU, red), EdU (green) was injected, and 30 minutes later tissues were harvested, stained, and analyzed (Fig. 3I–J). As detailed in Methods, S-phase length was first calculated and the division frequency was estimated as previously reported(Martynoga et al., 2005). This analysis suggests that outer limbal basal cells divide on average every ~8 days, inner limbal basal cells divide every ~4 days while corneal peripheral basal cells divide every ~3.5 days (Fig. 3J). Taken together, since the outer limbus contains cells that divide at a significantly lower rate, they were termed quiescent LSCs (qLSCs), and the fast-cycling inner LSCs renewed the cornea, they were referred to as active LSCs (aLSCs).
Cellular quiescence is linked with reduced transcriptional activity (Cho et al., 2019) (Tümpel and Rudolph, 2019). Indeed, cluster 3 (qLSCs) displayed the lowest value of RNA reads per cell (Fig. 3K). Previously, BMP was linked with epithelial SC quiescence while WNT repressed the BMP pathway and induced SC activation(Mira et al., 2010) (Genander et al., 2014) (Manrique et al., 2015) (Kishimoto et al., 2000). In agreement, cells of cluster 3 (qLSCs) preferentially expressed the BMP-regulated transcription factors Id1/Id3 and the Wnt inhibitor, Sfrp1(Lim et al., 2016), while Wnt3a was higher in cluster 4 of aLSCs (Fig. 3L). Interestingly, cluster 3 expressed Cyclin D2(Meyyappan et al., 1998) (Salpeter et al., 2011) and the tumor suppressor Trp53(Liu et al., 2009), both of which were linked with growth arrest induced by various stimuli.
IFITM3 and GPHA2 marked KRT15+ human LSCs and IFITM3 supported undifferentiated state
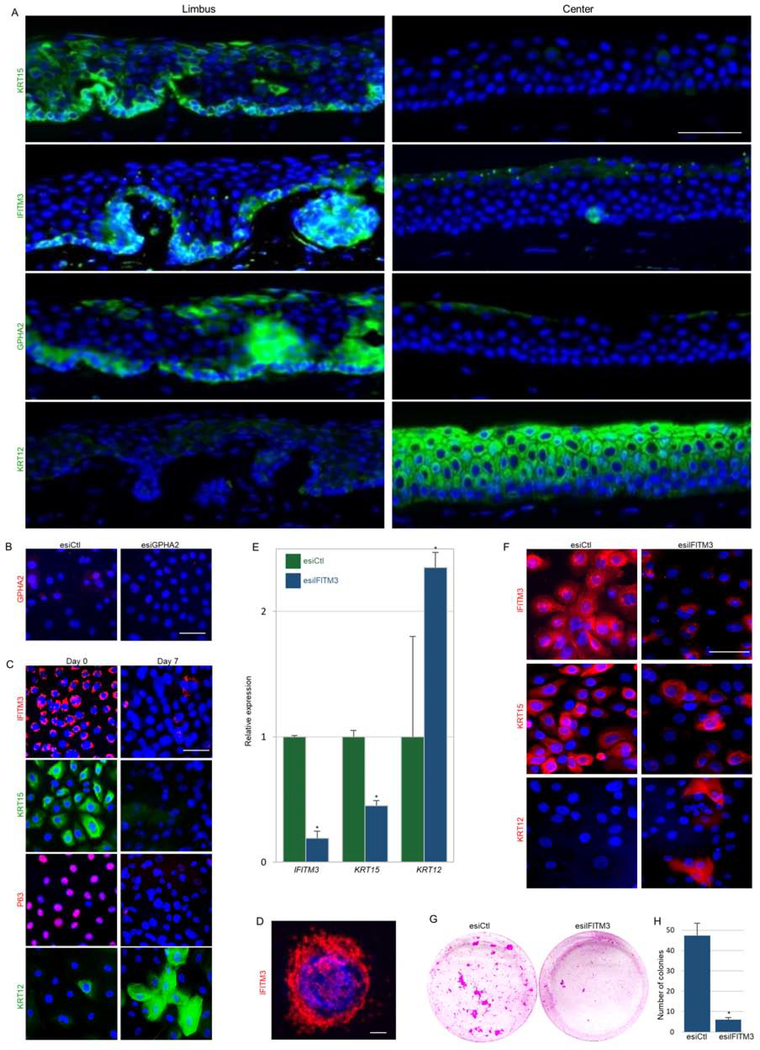
Next, we explored the relevance of the newly identified markers of murine LSCs to human. Immunofluorescent staining revealed that KRT15, IFITM3, and GPHA2 are expressed by human basal limbal epithelial cells in vivo (Fig. 4A). Interestingly, however, only GPHA2 expression was drastically downregulated upon cultivation in vitro (Fig. 4B–C). Calcium induced LSC differentiation enhanced KRT12 and reduced KRT15, IFITM3, and P63 (Fig. 4C). In line with the previous report that linked IFITM3 with endo-membrane localization and anti-viral activity(Wu et al., 2018), the IFITM3 signal was confined to cellular vesicles in the cytoplasm of undifferentiated limbal cells (Fig. 4D).
Figure 4: GPHA2 and IFITM3 marked qLSCs while IFITM3 supported the undifferentiated state in vitro.
(A) Human cornea frozen sections were subjected to immunofluorescence staining of indicated markers. (B) Human limbal epithelial cells were transfected with endoribonuclease prepared small interfering RNA against GPHA2 (esiGPHA2) or control (esiCtl) and cells were immunostained for GPHA2. (C) Human limbal epithelial cells were cultured and maintained undifferentiated at low calcium (Day 0) or induced to differentiate (Day 7) and stained for the indicated markers. (D) High magnification of IFITM3 staining (day 0). (E-H) Undifferentiated cells were transfected with esiIFITM3 or esiCtl. (E-F) The expression of the indicated markers was examined 4–5 days post-transfection by quantitative real-time polymerase chain reaction (E) and immunofluorescent staining (F). (G-H) Transfectants were seeded at clonal density and allowed to grow for 2–3 weeks. Rhodamine-stained colonies are shown in (G) and quantification in (H). Nuclei were detected by DAPI counter staining (A-D, F). Scale bars are 50 μm or 10 μm (D). Statistical significance was calculated using t-test (*, p-value<0.05). Data are represented as mean ± SD.
To test whether IFITM3 influences stemness and differentiation, we performed a knock-down experiment. To achieve efficient and specific knock-down, we used endoribonuclease-prepared silencing RNA (esiRNA). Knock-down of IFITM3 resulted in differentiation phenotype (Fig. 4E–F) and reduction of colony formation capacity (Fig. 4G–H). Taken together, this set of experiments suggests that IFITM3 and GPHA2 can be used to identify LSCs and that IFITM3 controls the undifferentiated state.
T cells serve as niche cells for qLSC regulating cell proliferation and wound closure
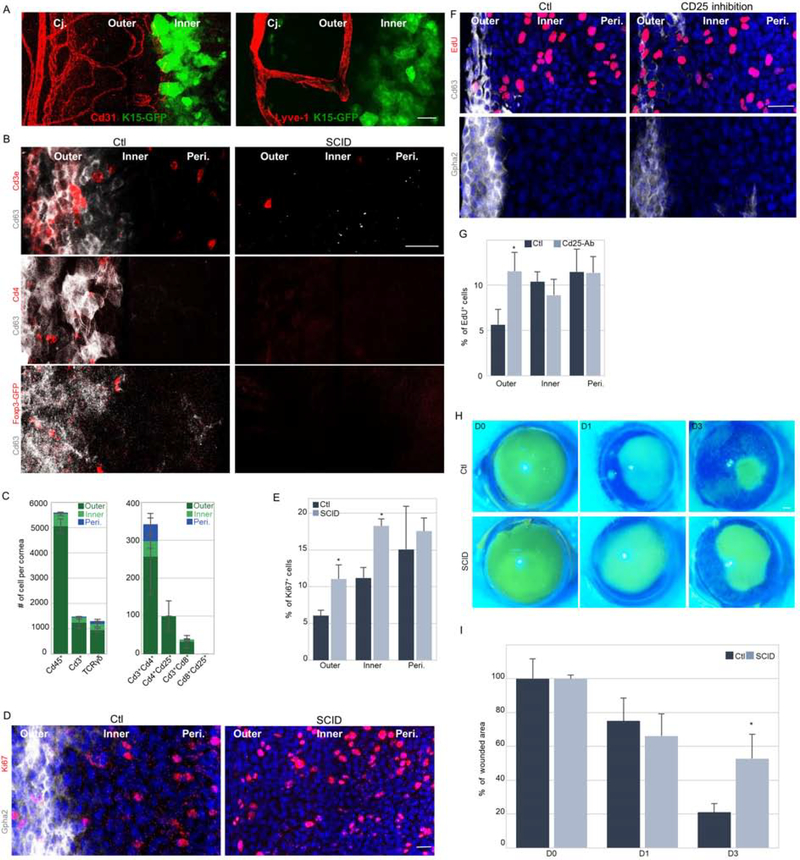
The striking segregation of the two populations into demarcated sub-compartments suggests a role for the microenvironment (niche). Indeed, the outer limbus was demarcated by blood/lymph vasculature (Fig 5A) and the presence of immune cells. Staining of surface markers of immune cell sub-populations on wholemount cornea and confocal Z-stack images of anterior stroma were collected covering ≤20 microns distance from the epithelial basal layer. Numerous Cd45+ cells were specifically located in the outer limbus, among them, Cd3+ T cells, including Cd3+Cd4+ and Cd4+Cd25+ (or Foxp3+) regulatory T cells and fewer Cd3+/Cd8+ T cells (Fig. 5B–C and S5A–C). No Cd8+/Cd25+ T cells and neither Cd19+ B cells (Fig. 5C, S5B) were identified.
Figure 5: T cells regúlate qLSC proliferation and response to wound Stimulus.
(A) Wholemount immunostaining of corneas was performed to label blood (Cd31) and lymph (Lyve-1) vessels of 2–3 months old Krt15-GFP mice. (B,D) Wholemount immunostaining of corneas of immunodeficient mice (SCID) or controls (Ctl) to identify qLSCs (Cd63+ or Gpha2+) or T cell populations (Cde3+ and Cd4+, or FoxP3-GFP+ transgene) or dividing cells (Ki67+). (C) Quantification of the different immune cell populations in a cornea (n=3). (E) Quantification of Ki67 expression in different limbal zones. (F) Six-days post subconjunctival injection of anti-Cd25 antibody (inhibits regulatory T cells) or vehicle (Ctl) to Krt15-GFP mice, EdU was injected and 6-hours later wholemount immunostaining of the indicated qLSC markers and EdU (mitotic cells) was performed. (G) Quantification of EdU labeling in different limbal zones. (H-I) Corneal epithelial debridement was performed in 2-month old mice of the indicated genotype. Fluorescein dye stain was performed to follow wound closure. Typical images (H) and quantification (I) are shown. Nuclei were detected by DAPI counter staining (D,F). Scale bars are 50 μm. Data represented as mean ± standard error. Statistical significance was calculated using t-test (*, p-value<0.05). Abbreviations: Cj., conjunctiva; Peri., periphery.
See also Figures S5.
Next, we explored the limbus of two laboratory mouse strains that fail to maturate T and B lymphocytes, namely, severe combined immunodeficiency (SCID) and non-obese diabetic SCID (NOD/SCID), and BALB/c mice served as a control group. Similar to the C57BL/6 genetic background (Fig 1–3), the outer limbus of BALB/c mice was hallmarked by Gpha2+/Cd63+/Ifitm3+ that less frequently proliferate (Fig. 5B, D–G, S5A–C, S5F). Curiously, an extensive reduction of Gpha2 and Cd63 proteins to levels that became barely detectable was found in SCID mice (Fig. 5B,D) as well as in NOD/SCID mice (Fig. S5C), while Ifitm3 was unaffected (Fig. S5B). Moreover, a higher index of Ki67 labeling was found in the outer limbus (as well as inner limbus) of SCID mice (Fig. 5D–E) that displayed mild epithelial thickening (Fig. S5D). Similar defects were found in NOD/SCID mice (Fig. S5C). Since no B cells (Cd19+) were detected in the limbus (Fig. S5B), we assumed that this phenotype is caused by the absence of T cells in SCID mice. Interestingly, thymus deficient Foxn1-deficient (“Nude”) mice displayed similar but more severe corneal defects including loss of qLSC markers, corneal vascularization, and opacification (Fig. S5E–F). However, since SCID (Prkdc-null) and NOD/SCID (Prkdc-null/II2rg-null) mice that also lack T lymphocytes displayed a milder phenotype, it is likely that the severe phenotype of Nude mice is partly associated with a cell-autonomous role of Foxn1 in LSCs (independent of T cell absence) (Niederkorn et al., 1990) (Kaminska and Niederkorn, 1993).
To substantiate the crosstalk of T cells – qLSC in adulthood and exclude the possibility of developmental failure, we repressed the ocular immune system by topically applying the corticosteroid Dexamethasone (Fig. S5G), or alternatively and more specifically, performed inhibition of regulatory T cells by sub-conjunctival injection of anti-Cd25 antibody (PC61.5). Intriguingly, 6-days post antibody injection, Cd63 and Gpha2 drastically decreased whereas cell proliferation increased (Fig. 5F–G), strongly suggesting that T cells directly regulate quiescence in the outer limbus. Similar results were obtained following Dexamethasone treatment (Fig. S5G). Finally, to link between T cell regulation and qLSC functionality, we performed corneal epithelial debridement and followed epithelial closure by fluorescein dye penetration. As shown in Fig. 5H–I, immunodeficient mice displayed delayed wound closure. Taken together, this set of experiments suggests that T cells, serve as a niche for qLSCs and play a critical role in quiescence maintenance, control of epithelial thickness, and wound healing.
DISCUSSION
In 1989, the seminal study by Cotsarelis and Lavker reported the identification of SCs in the limbus (Cotsarelis et al., 1989). More recent studies evaluated different SC hypotheses (Mort et al., 2012) and estimated a higher frequency of slow-cycling LSCs (Sagga et al., 2018). Here we captured the signature of primitive qLSCs and propose that these cells are more prevalent than previously estimated. Based on (i) relatively uniform marker expression, (ii) quantitative clonal growth dynamics and (iii) proliferation analysis, we propose that the outer limbus hosts equipotent qLSCs which divide every ~8 days. This frequency of cell division of outer qLSCs is much higher than that of other tissue-specific slow-cycling SCs(Wilson et al., 2008) (Waghmare et al., 2008). The interval of 30-days of chase in the absence of EdU (Fig. 3E–G) should theoretically allow ~4 division rounds for outer qLSCs, and it is therefore not surprising that most outer limbal basal cells lost labeling while only few retained low EdU signal. One-month post Confetti tracing, all outer limbus clones (n=89) divided at least once (≥2 cells), 93% divided at least twice, and on average they all divided 3.5 times. Hence, one could not exclude the possibility that an even slower cell population co-exists in the outer limbus. However, if so, such cells must be quite rare and probably not too slow.
In contrast to the previous assumption that LSCs are islands of rare cells surrounded by their progeny, this model posits an abundance of qLSCs. This hypothesis fits well with the equipotent SC model proposed to describe epithelial SC dynamics in other tissues(Clayton et al., 2007) (Snippert et al., 2010) (Giroux et al., 2017). In these tissues, abundant equipotent SCs were shown to be extremely fast-cycling. Epidermal and esophageal SCs divide every 2–3 days, similar to aLSCs, and faster than qLSCs. Collectively, it suggests that under distinct physiological conditions, tissues may display tailored regenerative strategies for reasons of cost and benefit tradeoff.
The lineage tracing experiment implies that aLSCs play a key role in corneal homeostasis, as most stripes emerged from this region. By contrast, a key feature of qLSCs is their ability to rapidly exit the dormant state and enter the cell cycle in response to injury(Gadye et al., 2017) (Fig. 3). This suggests that qLSC serve as a reservoir for tissue regeneration while their persistent localization at the boundary and circumferentially elongated clonal growth pattern strongly implies that these cells play a key role in boundary maintenance. In agreement with the present study, in vivo imaging of limbal cells performed by two-photon microscopy ((Farrelly et al., 2020), published back-to-back with this study) demonstrated the clear bi-compartmentalized organization of the limbus.
The analysis of the clone size distributions showed that they scale with the average number of clones in all the regions. This suggests that the underlying stochastic dynamics of all the regions are similar. The clone size dynamics are consistent with the neutral drift dynamics model and suggest that the doubling time in the inner limbus is twice the doubling time in the outer limbal regime, which is consistent with the cell cycle length measurements. In all regimes, there is a reduction in the number of clones with time, as expected. However, analyzing the quantitative relation between clone size and clone number reveals a striking difference between the outer and inner limbal regimes and the periphery. While all zones exhibit clone size distributions that are consistent with the neutral drift model, the decrease in the inner and outer regimes is slower than expected by this model. This saturation in clone number decrease rate might indicate that the clonal neutral competition is limited in these areas. This could be due to the tight spatial boundary conditions of the limbal regimes.
Our working model is in agreement with the hematopoietic, hair follicle(Li and Clevers, 2010), neural (Wang et al., 2011), and muscle (Brack and Rando, 2012) lineages, all of which incorporate qSCs that can produce aSCs(Li and Clevers, 2010). What are the costs and benefits of having two distinct SC states? What is the mechanism that controls the transition between SC states? Which physiological constraints influence SC division rates? The location of LSCs in an extremely hazardous environment may require unique mechanisms to prevent neoplasm. This must entail fundamental considerations for SC well-being such as localizing the SCs at the most well-protected zone (opaque limbus) and accrediting SCs with special metabolism and DNA repair mechanisms.
Future studies should address the mechanism of LSC self-renewal and differentiation. The bioinformatic analysis of limbal clusters suggests that qLSC engage the Pi3K pathway, the BMP-regulated transcription factors Id1/3(Mira et al., 2010) (Genander et al., 2014) (Manrique et al., 2015), the tumor suppressor gene Trp53(Cho et al., 2019), Cyclin D2(Meyyappan et al., 1998) (Salpeter et al., 2011) and the Wnt inhibitor Srf1(Tumbar et al., 2004), all of which were linked with growth arrest. Of interest, aLSCs were enriched for TNF, WNT, and Ap-1 pathways (Fig. S2–3) that were linked with proliferation or SC activation. Atf3 which is a structural protein of the Ap-1 complex was shown to repress Id1 and enhance epithelial cell proliferation(Kang et al., 2003).
The discovery of new LSC self-renewal factors may facilitate the generation of high-quality organoids, corneal bioengineering, and better LSC-based therapies. GPHA2 expression was lost ex vivo, in line with the fact that culture conditions are sub-optimal and lacking essential components of the niche. T cells may secret cytokines to control SC function (Naik et al., 2018) and play a dual role, not only for immune surveillance but also to regulate stemness and regeneration. Based on the literature and our atlas of gene expression, IL-10, IL-35, TGF-ß, Amphiregulin (AREG) which are suppressor cytokines for effector T cells, may act directly or via antigen-presenting cells, engaging key accessory molecules to which other cells express counter receptors, among them GITR, CTLA-4, TIGIT (Shevach, 2018).
In the clinics, dexamethasone is used to prevent uncontrolled corneal inflammation and neovascularisation and is considered a highly effective drug for nonspecific suppression of inflammation. Anti-inflammatory steroids must be used for a short time as they may have adverse effects including corneal wound healing delay (Petroutsos et al., 1982). The awareness and recognition of such an adverse effect are of particular importance given the observed interference of T cells – LSC crosstalk. Future studies will be needed to illuminate these interactions and translate this knowledge into better preventive and curative strategies for treating blindness in patients that suffer from LSC deficiency(Pellegrini and De Luca, 2014).
We identified here a set of new markers that allow identifying and perhaps purifying LSCs. Their function and regulation are largely unknown. Transgenic overexpression of Gpha2 showed no gross phenotype(Okada et al., 2006). In the present study, GPHA2 levels were reduced in cases where the niche was disturbed, for example, when LSCs were disconnected from the niche and grown in vitro, or following immune cell repression or absence in vivo. In both cases, outer LSCs were affected, suggesting that GPHA2 may be regulated by T cells and controls this cell state. By contrast, IFITM3 expression was not dependent on the presence of immune cells, suggesting that it is regulated by other niche means. Interestingly, both IFITM3 and CD63 reside in cellular endomembranes (Muñoz-Moreno et al., 2016). Moreover, IFITM3 is known for its antiviral specific functions (Diamond and Farzan, 2013), protecting against entry and replication of several types of viruses including the severe acute respiratory syndrome coronavirus 2 (Wu et al., 2018). Therefore, IFITM3 may protect SCs and control their functions. No major developmental disorder was reported in IFITM3 knockout mice (Lange et al., 2008), however, GPHA2 and IFITM3 are expressed by epidermal SCs in the G0 state(Dekoninck et al., 2020). Future studies will be needed to illuminate the role of these genes in vivo.
In conclusion, this report provides a useful atlas that uncovers the main corneal epithelial cell populations, capturing the signature and the niche of quiescent and active LSC states. These data open new research avenues for studying the mechanisms of LSC proliferation and differentiation as well as their applications in regenerative medicine.
Limitations of the Study
This study demonstrates the existence of two LSC populations that reside in separate and well-defined sub-compartments. The interchange between the two compartments should be studied, possibly by lineage tracing following a unique promoter for each population. While our results show that T cells regulate qLSC proliferation and response to wound stimulus, further experiments are needed to unravel the specific mechanism and secreted factors involved. Moreover, the specific niche cell types involved in the regulation of the active SCs are still to be discovered.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ruby Shalom-Feuerstein (shalomfe@technion.ac.il).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The single cell mRNA-seq data generated during this study are available at GEO (Gene Expression Omnibus) GEO: GSE167992.
This study did not generate code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Animal care and use conformed to the Association for Research in Vision and Ophthalmology (ARVO) statement for the use of animals in ophthalmic and vision research. The mouse strains B6.Cg-Tg(Krt1–15-EGFP)2Cot/J (referred to as Krt15-GFP) (Fuchs et al., 2013), Gt(ROSA)26Sortm1(CAG-Brainbow2.1)Cle/Jand B6.Cg-Ndor1Tg(UBC-CreERT2)1Ejb/2J (referred to as UBC-CreERT2) were from The Jackson Laboratory (Bar Harbor, ME). The mouse strains: C57BL/6JOlaHsd (referred to as C57BL/6), NOD.CB17-Prkdcscid/NCrHsd (referred to as Nod-Scid), C.B-17/IcrHsd-Prkdcscid (referred to as Scid) and Hsd:Athymic Nude-Foxn1nu (referred to as Nude) and their controls BALB/cOlaHsd (referred to as BALB) were from Envigo RMS Ltd (Israel). UBC-CreERT2; Brainbow2.1 homozygous mice were generated by crossing UBC-CreERT2 with R26R-Brainbow2.1. Both male and female age-matched mice from 7 to 14 weeks of age were used for all experiments in this study. Littermates of the same genotype, sex, and age were randomly assigned to experimental groups. All mice were housed under specific-pathogen-free SPF conditions at the Technion animal facilities.
In vitro culture
Human limbal rings from cadaveric corneas were obtained under the approval of the local ethical committee and declaration of Helsinki from at least 3 corneas from 3 different donors. The epithelium was separated from the underlying stroma following incubation with dispase II (Gibco, Life Technologies, USA). Cells were cultured at 37°C, 5% CO2, and 20% O2. For clonogenicity assay cells were grown in co-culture with mitomycinized growth-arrested J2-NIH3T3 cells in Green medium (60% DMEM (Gibco), 30% DMED F12 (Gibco), 10% FCII serum (Hyclone), 1mM L-Glutamine (Biological Industries), 1mM Sodium Pyruvate (Biological Industries), 0.2mM Adenine (Sigma), 5μg/ml Insulin (Sigma), 0.5μg/ml Hydrocortisone (Sigma), 10mM Choleratoxin (Sigma), 10ng/ml EGF (Peprotec)) (Rheinwald and Green, 1977) and split in 80% confluence. For efficient transfection and for controlled calcium-induced differentiation, cells were switched to defined EPIGRO medium with supplements (SCMK001, Millipore, USA) containing 1% penicillin/streptomycin and low calcium (150μM).
METHOD DETAILS
Single cell isolation from limbal epithelium
Ten eyes of 2.5-month-old Krt1–15-EGFP mice were enucleated, the limbus (with marginal conjunctiva and peripheral cornea) was dissected (~0.5mm), tissues were pooled and incubated in 300μl of trypsin (X10 Biological Industries) for 10 minutes (37°C). Supernatant was collected into 10ml RPMI (Biological Industries) containing 10% chelated fetal calf serum. Trypsinization was repeated for 10 cycles adding fresh trypsin in each cycle. Cell suspension was centrifuged (8 minutes at 300g), re-suspended, and filtered using a cell strainer (VWR) to achieve 1200cells/μL
Immunostaining
Paraffin sections:
Eyes were fixed (overnight, 4% formaldehyde), washed briefly with phosphate buffered saline (PBS), incubated in increasing ethanol concentrations (1-hour incubation in 70%, 80%, twice 95%, twice 100%), 45 minutes in a 1:1 mixture of 100% Ethanol and Xylene, two 1-hour incubations in Xylene, and three cycles of 90 minutes in paraffin (60°C). Tissues were then embedded in paraffin blocksand 5μm sections were cut. Sections were incubated overnight (ON) at 37°C and then rehydrated (two 15 minutes incubation in Xylene, 5 minutes incubation in 100%, 95%, 90%, 80%, 70%, 50% Ethanol, and 5 minutes incubation in distilled water). Antigen retrieval was performed with an unmasking solution (Vector Laboratories, H3300), blocked (0.2% Tween20 and 0.2% gelatin, 2 hours), incubated with primary antibodies (overnight, 4°C), secondary antibodies (Invitrogen, 1:500, 2 hours) followed by 4′,6-diamidino-2-phenylindole (DAPI) staining, and mounting (Thermo Scientific).
Frozen sections:
Eyes were fixed (4 hours, 4% formaldehyde), washed briefly (PBS), incubated in 30% sucrose (overnight) and embedded in optimal cutting temperature compound (OCT). Sections (8μm) were fixed (4% paraformaldehyde), permeabilized (0.1% TritonX-100, 20 minutes), blocked (1% BSA, 1% gelatin, 2.5% Normal goat serum, 2.5% Normal donkey serum, 0.3% TritonX-100, 1 hour), incubated with primary antibodies (overnight, 4°C), secondary antibodies (1:500, 1 hour), DAPI and mounting (Thermo Scientific).
Wholemount:
Corneas were isolated, fixed (2% formaldehyde, 2 hours, room temperature), permeabilized (0.5% Triton, 5 hours), blocked (0.1% TritonX-100, 2% Normal donkey serum, 2.5% BSA, 1 hour) incubated with primary antibodies (overnight, 4°C on a shaker), secondary antibodies (1:500, 1 hour), followed by DAPI, tissue flattening under a dissecting binocular and mounting (Thermo Scientific).
Cell culture staining:
Cells grown on cover-slips were fixed (4% formaldehyde, 15 minutes), then permeabilized (0.1% Triton X-100, 10 minutes), blocked (2.5% BSA, 30 minutes), incubated with primary antibodies overnight at 4°C, washed and incubated with secondary antibodies (1:500, 1h at room temperature). Nuclei were stained with DAPI (Sigma) and mounted (Thermo).
Immunohistochemistry:
Paraffin sections were deparaffinized by heating for 1 hour at 60°C then rehydrated twice in Xylene for 5 minutes followed by two incubations in 100% Ethanol for 5 minutes. Suppression of endogenous peroxidase activity was achieved by incubation in Methanol with 1% H2O2, incubation in 70% Ethanol for 2 minutes followed by a rinse in Distilled water. Antigen retrieval was performed with an unmasking solution (Vector Laboratories, H3300), followed by blocking (10% goat serum), and incubation with a primary antibody (overnight, 4°C), secondary antibodies (Universal Immuno peroxidase Polymer anti-rabbit/mouse, a ready-made solution, 1 hour) followed by substrate addition (AEC solution), Hematoxylin staining and mounting (Thermo Scientific).
H&E (Hematoxylin and eosin) staining:
Paraffin sections were deparaffinized in Xylene for 10 minutes, rehydrated (2 incubations of 3 minutes in 100% and 95% Ethanol), incubated with Hematoxylin (1 minute), washed (tap water, 15 minutes), stained with Eosin (5 minutes), dehydrated (2 incubations of 3 minutes in 95% and 100% Ethanol), incubated with Xylene (15 minutes), and mounted (DPX Mountant for histology).
In situ hybridization (ISH)
ISH was performed according to the manufacturer’s instructions for frozen tissues (LGC Biosearch Technologies). All probe sets contained ≥ 28 oligonucleotides each (20-mer). Hybridization mixture contained 125 nM of each probe set: Krt4 (48 probes), Gpha2 (28 probes), Atf3 (48 probes), or Krt12 (47 probes) (Sequences provided in Table S1).
Edu labelling
Single intraperitoneal injection of 200μl (7.5mg/ml) EdU (Sigma) was performed. Six hours later tissues were processed. For pulse-chase experiments, EdU (0.5mg/ml) solution was administered via drinking water for 15 consecutive days and tissues processed after additional 30-days. The water was changed every 3 days. Isolated corneas were fixed (2% PFA, 1 hour) and stained (Click-iT, Invitrogen) according to the manufacturer’s instructions followed by wholemount staining protocol (described above).
Short double pulse-chase experiment
For the calculation of cell cycle length (Tc) 200 ul of 1.7mg/ml IdU (Abcam) was injected and 1.5 hours later 200 ul of 7.5mg/ml EdU was injected. Tissues were harvested 30 minutes later. Corneas were isolated and fixed in cold Methanol (4°C, 10 minutes), washed twice with a PBS solution containing 0.5% triton for 5 minutes and Edu labelled as described above. Next, DNA hydrolysis was done by incubation in 2N HCl solution (37°C, 15 minutes) followed by neutralization with 0.1M Borate Buffer pH 8.5 (room temperature, 20 minutes). Tissues were then washed with PBS, blocked (0.1% TritonX-100, 4% Normal donkey serum, 0.1% BSA, 2 hour), and incubated with an IdU primary antibody (overnight, 4°C), extensively washed with PBS (three times, 20 minutes), incubated with a secondary antibody (1:500, 3h at room temperature), and washed again as described above. Nuclei were stained with TOPRO (30 minutes, room temperature, Thermo Fisher Scientific) and mounted (Thermo Scientific).
Cell cycle length (Tc) was calculated according to the following formula previously described (Martynoga et al., 2005):
Tamoxifen induction
To induce Cre recombinase activity, 20mg/ml Tamoxifen (T5648, Sigma) dissolved in corn oil was intraperitoneally injected (200μl) for 3 consecutive days, as previously reported (Amitai-Lange et al., 2015a) (Amitai-Lange et al., 2015b). The induction efficiency of RFP/YFP/CFP was high and comparable (~40%, ~30%, and ~25% respectively) while the GFP labeling was low (~5%).
Human Epithelial Limbal Cell Differentiation
Purified human limbal epithelial cells were seeded and grown in EPIGRO medium to 80–100% confluence and then switched to high (1.2mM) calcium for up to 1 week.
Human Epithelial Limbal Transfection
Cells were transfected using Lipofectamine 3000 or RNAimax with 50nM esiRNA targeting EGFP (EHUEGFP, Sigma) or IFITM3 (EHU222801, Sigma) according to the manufacturers’ instructions. Cells were collected 72 hours after transfection.
Real-time PCR analysis
Cells were washed with cold PBS and RNA was isolated using TRI-Reagent (Sigma) according to the manufacturers’ instructions. cDNA was prepared by reverse transcription polymerase chain reaction (RT-PCR) using the Qscript cDNA synthesis kit (Quantabio) according to the manufacturer’s instructions. Quantitative real-time polymerase chain reaction (qPCR) was performed with Fast SYBR green master mix (Thermo). Samples were cycled using StepOnePlus (Applied Biosystems) qPCR system. Relative gene expression was normalized to GAPDH and calculated according to the ΔΔCT method for qPCR.
Depletion of T regulatory cells
Animals were anaesthetized (2% Isoflurane) and a subcutaneous injection of the analgesic Buprenorphine (0.03mg/ml,50 μl) was performed (day 0). For sub-conjunctival injection, the bulbar conjunctiva was pulled using forceps and 30 μl (250 μg) anti-Cd25 antibody (Clone:PC61, antiCd25 IL-2Rα) (Fujisaki et al., 2011) or rat IgG as control (BioCell InVivoMAb) was injected under the binocular using a 30-gauge needle connected to 1 ml syringe. Slow injection into the space between the conjunctiva and the sclera was performed to create a ballooning effect in the peri-limbal conjunctival zone. A second sub-conjunctival anti-Cd25 antibody (or control) injection was performed two-days later. Mice were sacrificed three days later, corneas were dissected and prepared for wholemount staining and analysis of proliferating cells, immune cell populations or LSC markers was performed as described above.
Corneal wounding
Mice were anesthetized (2% Isoflurane) and injected subcutaneously with the analgesic Buprenorphine (0.03mg/ml, 50 μl). Central corneal wounding (2mm diameter) was performed using an ophthalmic rotating burr (Algerbrush) debridement under a fluorescent binocular. The epithelial layer was scraped off by gentle strokes of the Algerbrush over the cornea taking care not to penetrate the stroma. The wounded corneas were stained by the application of 1% fluorescein solution followed by an extensive PBS wash. The wounded area was imaged and quantified (NIS-Elements analysis D).
Dexamethasone application
Dexamethasone (Sigma) was dissolved in PBS (0.5%) and administered as drops (15 μl) topically over the ocular surface of anesthetized (2% Isoflurane) mice, for 10 minutes every 12 hours for 4 consecutive days.
QUANTIFICATION AND STATISTICAL ANALYSIS
Library prep and data generation
An RNA library was produced according to the 10x Genomics protocol (Chromium Single Cell 3’ Library & Gel Bead Kit v2, PN-120237) using 18000 input cells. Single cell separation was performed using the Chromium Single Cell A Chip Kit (PN-120236). The RNAseq data was generated on Illumina NextSeq500, 150bp paired-end reads, high-output mode (Illumina, FC-404–2005) according to 10X recommendations read 1– 26bp and read 2– 98bp.
Processing of 10X Genomics data
Cell ranger (version 2.2.0) was used to analyze the single cell 3’ RNA-seq data that was produced by the 10x Chromium Platform. Alignment, filtering, barcode counting, and UMI counting were performed using the count function. Transcripts were mapped to the mm10 reference genome (refdata-cellranger-mm10–2.1.0/GRCm38.84), with the addition of the eGFP gene sequence.
Downstream analyses
Downstream analyses including clustering, classifying single cells and differential expression analyses were performed using R package – Seurat (version 2.3.4) (Butler et al., 2018). Low-quality cells (>5% mitochondrial UMI counts, or <200 or >8000 expressed genes(Ilicic et al., 2016)) and genes detected in <3 cells were excluded, and eventually, 14,941 genes across 4,738 cells were analyzed. To identify significant PCs, we used the Jackstraw function. The top 10 PCs were used for clustering with the FindClusters function to generate cell clusters, 11 clusters were detected using a resolution of 0.8. In-silico analysis of non-epithelial cell markers confirmed the absence (<0.1%) of corneal endothelial cells (Clrn1, Pvrl3, Gpc4, Slc9a7, Slc4a4, Grip1, Hrt1d, Irx2), corneal stromal keratocytes (Vim, Thy1, S1004a4), goblet cells (Muc2, Muc5ac, Muc5b), melanocytes (Dct, Mitf, Ptgs, Tyrp1, Melana, Tyr), T cells (Cd3d, Cd3e, Cd4, G2mb), B cells (Cd19, Cd79a, Cd79b) confirming that most (>99%) cells were ocular epithelial cells.
Pathway enrichment analysis
A list of differentially expressed genes between chosen clusters was analyzed using WebGestalt (Zhang et al., 2005). Over-Representation Enrichment Analysis (ORA) was used for pathway analysis with KEGG functional database.
Sample identity distribution
Sample identity and the distribution of pooled epithelial cells from 5 mice were performed using the Souporcell algorithm as previously described (Heaton et al., 2020).
Cell cycle scoring
Cell cycle scores were assigned using Seurat’s CellCycleScoring function. Scores were assigned to cells based on the expression of genes that are associated with G1, S, or G2/M phase (Kowalczyk et al., 2015). For each cell type (conjunctiva basal, outer limbus basal, inner-limbus basal, and corneal basal cells) we calculated a signature score based on the top 10 enriched genes in the relevant cluster (1, 3, 4, and 5+6 respectively) using Seurat’s AddModuleScore function (Tirosh et al., 2016). Based on these scores, dividing cells in clusters 10 and 11 were classified into the four basal cell types. To analyze the percentage of mitotic cells, values were normalized to the number of cells in the relevant cluster and to the S-phase length.
Imaging
Images were acquired using 20X/0.8 M27 Plan-Apochromat objective on LSM 880 airyscan laser scanning confocal system (Zeiss, Oberkochen, Germany) or by Nikon Eclipse NI-E upright Microscope using Plan Apo λ 20x objective.
Quantification analyses
Lineage tracing analysis, Edu accumulation, clonogenicity, ki67 positive cells quantification, and immune cells quantification were done by count function using Nis-elements Analysis D software. The outer limbus was marked by Gpha2 or Cd63 staining, the adjacent 100 μm was defined as the inner limbus, while the peripheral cornea was defined as the 100 μm zone adjacent to the K15-GFP+ zone (or regime that begins 200 μm radially to the outer limbus, in case that none K15-GFP mice were used). At least 5 different fields were calculated from each cornea and at least 3 corneas were quantified from at least 3 different mice.
Clone size and number analysis
Potential clonal merging was calculated as the fraction of clones that were 2t/td, where “t” is the time post induction and “td” is the doubling time. Clones that are smaller than this value originated from a single cell. Given t=10-days (3-consecutive days of injection following by chase after a week) and td of 3-days, ~94 of the RFP clones and 98% of the other colored clones are not merged.
As described previously (Klein et al., 2007) (Clayton et al., 2007) (Jones et al., 2019), one of the main predictions of the neutral drift dynamics model is that the average number of cells in a given clone as a function of time is given by, where ρ is the fraction of basal cells that divide, 2r is the probability for symmetric division and λ is the division rate. The number of clones is expected to decrease with time as, . Thus, the relation between the clone size and clone number is given by .
Statistical analysis
Data are presented as mean ± standard deviation (SD). T-test, analysis of variance (ANOVA) followed by Bonferroni test, or Kolmogorov-Smirnov test were performed using GraphPad Prism software to calculate p-values as indicated in legends. Differences were considered to be statistically significant from a p-value < 0.05.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-Krt4 | Abcam | Cat#ab183329 |
| Rabbit monoclonal anti-Krt8 | Abcam | Cat#ab53280; RRID: AB_869901 |
| Rabbit Polyclonal IgG anti-Krt6a | BioLegend | Cat#905702; RRID: AB_2734680 |
| Mouse monoclonal anti-Gpha2 | Santa Cruz | Cat#sc-390194 |
| Mouse monoclonal anti-Cd63 | Santa Cruz | Cat#sc-5275; RRID: AB_627877 |
| Rabbit monoclonal anti-Ifitm3(Fragilis) | Abcam | Cat#ab15592; RRID: AB_2122095 |
| Rabbit Polyclonal anti-Atf3 | Santa Cruz | Cat#AB_225851 |
| Mouse monoclonal anti-Mt1-2(Methallothionein) | Abcam | Cat#ab12228; AB_298949 |
| Rabbit monoclonal anti-Krt12 | Abcam | Cat#ab185627 |
| Mouse monoclonal anti-Krt15 | Santa Cruz | Cat#sc-47697; RRID:AB_627847 |
| Rabbit monoclonal anti-Krt17 | Cell Signaling | Cat#4543S; RRID: AB_2133014 |
| Rabbit monoclonal anti-Krt19 | Abcam | Cat#ab52625; RRID:AB_228102 |
| Mouse monoclonal anti-Krt14 | Millipore | Cat#CBL197; RRID:AB_2132747 |
| Mouse monoclonal anti-P63 (4A4) | Santa Cruz | Cat#sc-8431; RRID: AB_628091 |
| Rabbit monoclonal anti-CD31 | Abcam | Cat#ab-231436 |
| Rabbit Polyclonal anti-Lyve1 | Abcam | Cat#ab-14917; RRID:AB_301509 |
| Armenian Hamster IgG anti- CD3ε | Biolegend | Cat#100312; RRID:AB_312677 |
| Rat monoclonal FITC conjugated anti-CD8a | Biolegend | Cat#100706; RRID:AB_312745 |
| Rat monoclonal FITC conjugated anti-CD4 | Biolegend | Cat#100405; RRID:AB_312690 |
| Rabbit monoclonal anti-IL-2 Receptor alpha | Abcam | Cat#ab-227834 |
| Armenian Hamster monoclonal FITC conjugated anti-TCR β chain | Biolegend | Cat#109205; RRID:AB_313428 |
| Rat monoclonal anti-CD45 | Biolegend | Cat#103102; RRID:312967 |
| Rat monoclonal anti-F4/80 | Biolegend | Cat#123102; RRID:AB_893506 |
| Rat monoclonal PE conjugated anti-CD19 | BD Biosciences | Cat#553786; RRID:AB_395050 |
| Rat monoclonal anti-IdU | Abcam | Cat#ab-187742 |
| Chicken polyclonal anti-GFP | Abcam | Cat#ab-13970; RRID:AB_300798 |
| Rabbit monoclonal anti-Ki67 | Abcam | Cat#ab-16667; RRID:AB_302459 |
| Rabbit monoclonal anti-Gapdh | Cell signaling | Cat#2118 RRID:AB_561053 |
| Mouse monoclonal anti-Krt13 | Abcam | Cat#ab-16112 |
| Rat monoclonal anti-Blimp-1 | Invitrogen | Cat#14-5963-82; RRID:AB_1907437 |
| InVivoMAb Rat IgG1 Isotype control; anti Horseradish Peroxidase Clone: HRPN | BioXCell | BE0088 |
| InVivoMAb anti-mouse CD25 (IL-2Rα) PC-61.5.3 | BioXCell | BE0012 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Molecular Probes | A21203; RRID:AB_141633 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Molecular Probes | A21202; RRID:AB_141607 |
| Donkey anti-Rat IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Molecular Probes | A21209; RRID:AB_2535795 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Molecular Probes | A21206; RRID:AB_2535792 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Molecular Probes | A21207; RRID:AB_141637 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Molecular Probes | A21208; RRID:AB_141709 |
| Donkey anti-Mouse IgG (H+L) AffinitPure Secondary Antibody, Alexa Fluor 647 | Jackson Immuno Research | 715-605-151; RRID:AB_2340863 |
| Biological Samples | ||
| Human Limbal rings | Department of Ophthalmology, Hillel Yaffe Medical Center, Hadera, Israel | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Trypsin 0.5% (×10) | Gibco | 15400054 |
| RPMI medium | Biological Industries | 01-100-1A |
| Tamoxifen | Sigma | T5648 |
| Dexamethasone | Sigma | D2915 |
| PBS | Biological Industries | 02-023-1A |
| Isoflurane | Piramal | NDC 66794 |
| EdU(5-ethynyl-2’-deoxyuridine) | Invitrogen | E10187 |
| Formaldehyde 35% | Bio-Lab | 6756523F1 |
| IDU(Idoxuridine, Antiviral thymidine analog) | Abcam | ab142581 |
| Ethanol absolute (Dehydrated) | Bio-Lab | 525050300 |
| Xylene | Gadot | 830119371 |
| Paraplast for tissue embedding | Sigma | P3558 |
| Antigen Unmasking Solution Citrate Based | Vector | H3300 |
| Tween 20 | Sigma | P2287 |
| Gelatin | Sigma | G1890 |
| DAPI | Sigma | MBD0015 |
| TO-PRO™-3 Iodide (642/661) | Thermo Fisher Scientific | T3605 |
| Immu-Mount | Thermo Scientific | 9990402 |
| DPX Mountant for histology | Sigma | 06522 |
| Eosin | Sigma | HT110116-500ML |
| Harris Hematoxylin | Bar Naor | bn1513 |
| OCT Tissue Freezing Medium | Leica Biosystems | LE-14020108926 |
| BSA | Sigma | A2153 |
| Normal Goat Serum | Jackson Immuno Research | 005-000-121 |
| Dispase | Gibco | 17105041 |
| Pen/Strep | Biological Industries | 03-031-1B |
| N-Histone Simpke Stain AEC solution | Nichirei Biosciences INC. | 415182F |
| N-Histone Simple Stain Max po | Nichirei Biosciences INC. | 414151F |
| Collagen I | Sigma | C8919 |
| Lipofectamine 3000 | Invitrogen | L3000-008 |
| RNAimax | Invitrogen | 13778075 |
| TRI-Reagent | Sigma | T9424 |
| Fluorescein | Haag-Streit International | N/A |
| Critical Commercial Assays | ||
| Chromium Single Cell 3’ Library & Gel Bead Kit v2 | Chromium | PN-120237 |
| Single Cell A Chip Kit | Chromium | PN-120236 |
| Click-iT™ Plus Alexa Fluor™ 488 Picolyl Azide Toolkit | Invitrogen | C10641 |
| Qscript CDNA synthesis kit | Quantabio | 95047 |
| Fast SYBR Green Master Mix | Applied Biosystems (Thermo Fisher Scientific) | AB-4385612 |
| Deposited Data | ||
| Single cell RNA Seq | This paper | GEO: GSE167992 |
| Experimental Models: Cell Lines | ||
| J2-NIH3T3 | A kind gift from Prof. Daniel Aberdam | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: B6.Cg-Tg(Krt1-15-EGFP)2Cot/J | Jackson Laboratory | JAX 005244 |
| Mouse: Gt(ROSA)26Sortm1(CAG-Brainbow2.1)Cle/J | Jackson Laboratory | JAX 013731 |
| Mouse: B6.Cg-Ndor1Tg(UBC-cre/ERT2)1Ejb/2J | Jackson Laboratory | JAX 008085 |
| Mouse: B6.Cg-Ndor1Tg(UBC-cre/ERT2)1Ejb/2J; Gt(ROSA)26Sortm1(CAG-Brainbow2.1)Cle/J | This paper | N/A |
| Mouse: C57BL/6JOlaHsd | Envigo | Cat#057 |
| Mouse: NOD.CB17-Prkdcscid/NCrHsd | Envigo | Cat#170 |
| Mouse: C.B-17/IcrHsd-Prkdcscid | Envigo | Cat#182 |
| Mouse: Hsd:Athymic Nude-Foxn1nu | Envigo | Cat#069 |
| Mouse: BALB/cOlaHsd | Envigo | Cat#162 |
| Oligonucleotides | ||
| esiRNA targeting EGFP | Sigma | Cat#EHUEGFP |
| esiRNA targeting IFITM3 | Sigma | Cat#EHU222801 |
| esiRNA targeting Gpha2 | Sigma | Cat#EHU036941 |
| Forward primer for GAPDH: GCCAAGGTCATCCATGACAAC | This paper | N/A |
| Backward primer for GAPDH: CTCCACCACCCTGTTGCTGTA | This paper | N/A |
| Forward primer for IFITM3: CTGGGCTTCATAGCATTCGCCT | This paper | N/A |
| Backward primer for IFITM3: AGATGTTCAGGCACTTGGCGGT | This paper | N/A |
| Forward primer for KRT15: GACGGAGATCACAGACCTGAG | This paper | N/A |
| Backward primer for KRT15: CTCCAGCCGTGTCTTTATGTC | This paper | N/A |
| Forward primer for KRT12: TGAATGGTGAGGTGGTCTCA | This paper | N/A |
| Backward primer for KRT15: TTTCAGAAGGGCAAAAAGGA | This paper | N/A |
| Stellaris FISH probes custom assay with Quasar 570 for: Krt12, Gpha2, Krt4 and Atf-3. See table S1 | Bio Search technologies | SMF-1063-5 |
| Software and Algorithms | ||
| Cell ranger (version 2.2.0) | 10X | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| R package – Seurat (version 2.3.4) | Butler et al., 2018 | https://satijalab.org/seurat/articles/install.html |
| Souporcell algorithm | Heaton et al., 2020 | https://github.com/wheaton5/souporcell |
| NIS-Elements analysis D | Nikon | https://www.microscope.healthcare.nikon.com/products/software/nis-elements/nis-elements-documentation |
| WebGestalt | Zhang et al., 2005 | http://www.webgestalt.org/ |
| Matlab 2019b | MathWorks, Inc | https://www.mathworks.com/ |
| Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| Cell strainer 70 μm | VWR | 10199-656 |
| Illumina NextSeq500, 150 bp paired-end reads, high-output mode | Illumina | FC-404-2005 |
| Algerbrush | Albert Heiss | H-7151 |
| LSM 880 airyscan laser scanning confocal system | Zeiss | https://www.zeiss.com/microscopy/us/dynamic-content/news/2014/news-lsm-880.html |
| Eclipse NI-E upright Microscope | Nikon | https://www.microscope.healthcare.nikon.com/products/upright-microscopes/eclipse-ni-e |
Highlights.
Discovery of two limbal stem cell (LSC) populations, their signature and niches
The outer limbus hosts quiescent LSCs while inner LSCs actively renew the cornea
LSCs are abundant in their niche, follow stochastic rules and neutral drift dynamics
T cells regulate outer LSC quiescence, marker expression and wound healing response
ACKNOWLEDGMENTS:
We thank L. Linde, S. Galili and their team for the scRNA-seq experiment, D. Aberdam and Y. Bar Onn for critical reading. RSF received funding from European Union’s Horizon 2020 research & innovation program (828931), the ISRAEL SCIENCE FOUNDATION (1308/19 and 2830/20), Rappaport Family Institute for Research in Medical Sciences, NIH-exploratory R21 (800040).
Footnotes
DECLARATION OF INTERESTS: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Amitai-Lange A, Altshuler A, Bubley J, Dbayat N, Tiosano B, and Shalom-Feuerstein R (2015a). Lineage tracing of stem and progenitor cells of the murine corneal epithelium. Stem Cells 33,230–239. [DOI] [PubMed] [Google Scholar]
- Amitai-Lange A, Berkowitz E, Altshuler A, Dbayat N, Nasser W, Suss-Toby E, Tiosano B, and Shalom-Feuerstein R (2015b). A Method for Lineage Tracing of Corneal Cells Using Multi-color Fluorescent Reporter Mice. J. Vis. Exp. e53370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrandon Y, and Green H (1987). Three clonal types of keratinocyte with different capacities for multiplication. Proc. Natl. Acad. Sci. U. S. A. 84,2302–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack AS, and Rando TA (2012). Tissue-specific stem cells: lessons from the skeletal muscle satellite cell. Cell Stem Cell 10,504–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandner JM, Kief S, Grund C, Rendl M, Houdek P, Kuhn C, Tschachler E, Franke WW, and Moll I (2002). Organization and formation of the tight junction system in human epidermis and cultured keratinocytes. Eur. J. Cell Biol. 81,253–263. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36,411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho IJ, Lui PP, Obajdin J, Riccio F, Stroukov W, Willis TL, Spagnoli F, and Watt FM (2019). Mechanisms, Hallmarks, and Implications of Stem Cell Quiescence. Stem Cell Reports 12,1190–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton E, Doupé DP, Klein AM, Winton DJ, Simons BD, and Jones PH (2007). A single type of progenitor cell maintains normal epidermis. Nature 446,185–189. [DOI] [PubMed] [Google Scholar]
- Clevers H, and Watt FM (2018). Defining Adult Stem Cells by Function, not by Phenotype. Annu. Rev. Biochem. 87,1015–1027. [DOI] [PubMed] [Google Scholar]
- Colom B, and Jones PH (2016). Clonal analysis of stem cells in differentiation and disease. Curr. Opin. Cell Biol. 43,14–21. [DOI] [PubMed] [Google Scholar]
- Cotsarelis G, Cheng SZ, Dong G, Sun TT, and Lavker RM (1989). Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: Implications on epithelial stem cells. Cell 57,201–209. [DOI] [PubMed] [Google Scholar]
- Dekoninck S, Hannezo E, Sifrim A, Miroshnikova YA, Aragona M, Malfait M, Gargouri S, de Neunheuser C, Dubois C, Voet T, et al. (2020). Defining the Design Principles of Skin Epidermis Postnatal Growth. Cell 181,604–620.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, and Farzan M (2013). The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 13,46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorà NJ, Hill RE, Collinson JM, and West JD (2015). Lineage tracing in the adult mouse corneal epithelium supports the limbal epithelial stem cell hypothesis with intermittent periods of stem cell quiescence. Stem Cell Res. 15,665–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebato B, Friend J, and Thoft RA (1988). Comparison of limbal and peripheral human corneal epithelium in tissue culture. Invest. Ophthalmol. Vis. Sci. 29,1533–1537. [PubMed] [Google Scholar]
- Farrelly O, Suzuki-horiuchi Y, Brewster M, Kuri P, Huang S, Rice G, Xu J, Dentchev T, Lee V, Rompolas P, et al. (2020). Bi-compartmentalized stem cell organization of the corneal limbal niche. BioRxiv 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Y, Brown S, Gorenc T, Rodriguez J, Fuchs E, and Steller H (2013). Sept4/ARTS regulates stem cell apoptosis and skin regeneration. Science 341,286–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisaki J, Wu J, Carlson AL, Silberstein L, Putheti P, Larocca R, Gao W, Saito TI, Lo Celso C, Tsuyuzaki H, et al. (2011). In vivo imaging of Treg cells providing immune privilege to the haematopoietic stem-cell niche. Nature 474,216–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadye L, Das D, Sanchez MA, Street K, Baudhuin A, Wagner A, Cole MB, Choi YG, Yosef N, Purdom E, et al. (2017). Injury Activates Transient Olfactory Stem Cell States with Diverse Lineage Capacities. Cell Stem Cell 21, 775–790.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genander M, Cook PJ, Ramsköld D, Keyes BE, Mertz AF, Sandberg R, and Fuchs E (2014). BMP signaling and its pSMAD1/5 target genes differentially regulate hair follicle stem cell lineages. Cell Stem Cell 15,619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Girolamo N, Bobba S, Raviraj V, Delic NC, Slapetova I, Nicovich PR, Halliday GM, Wakefield D, Whan R, and Lyons JG (2015). Tracing the fate of limbal epithelial progenitor cells in the murine cornea. Stem Cells 33, 157–169. [DOI] [PubMed] [Google Scholar]
- Giroux V, Lento AA, Islam M, Pitarresi JR, Kharbanda A, Hamilton KE, Whelan KA, Long A, Rhoades B, Tang Q, et al. (2017). Long-lived keratin 15+ esophageal progenitor cells contribute to homeostasis and regeneration. J. Clin. Invest. 127, 2378–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton H, Talman AM, Knights A, Imaz M, Gaffney DJ, Durbin R, Hemberg M, and Lawniczak MKN (2020). Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat. Methods 17,615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilicic T, Kim JK, Kolodziejczyk AA, Bagger FO, McCarthy DJ, Marioni JC, and Teichmann SA (2016). Classification of low quality cells from single-cell RNA-seq data. Genome Biol. 17,29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii R, Yanagisawa H, and Sada A (2020). Defining compartmentalized stem cell populations with distinct cell division dynamics in the ocular surface epithelium. Development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KB, Furukawa S, Marangoni P, Ma H, Pinkard H, D’Urso R, Zilionis R, Klein AM, and Klein OD (2019). Quantitative Clonal Analysis and Single-Cell Transcriptomics Reveal Division Kinetics, Hierarchy, and Fate of Oral Epithelial Progenitor Cells. Cell Stem Cell 24,183–192.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce NC, Meklir B, Joyce SJ, and Zieske JD (1996). Cell cycle protein expression and proliferative status in human corneal cells. Invest. Ophthalmol. Vis. Sci. 37, 645–655. [PubMed] [Google Scholar]
- Kaminska GM, and Niederkorn JY (1993). Spontaneous corneal neovascularization in nude mice. Local imbalance between angiogenic and anti-angiogenic factors. Invest. Ophthalmol. Vis. Sci. 34,222–230. [PubMed] [Google Scholar]
- Kang Y, Chen C-R, and Massagué J (2003). A self-enabling TGFbeta response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol. Cell 11,915–926. [DOI] [PubMed] [Google Scholar]
- Kasper M, Moll R, Stosiek P, and Karsten U (1988). Patterns of cytokeratin and vimentin expression in the human eye. Histochemistry 89,369–377. [DOI] [PubMed] [Google Scholar]
- King IA, Angst BD, Hunt DM, Kruger M, Arnemann J, and Buxton RS (1997). Hierarchical expression of desmosomal cadherins during stratified epithelial morphogenesis in the mouse. Differentiation. 62,83–96. [DOI] [PubMed] [Google Scholar]
- Kishimoto J, Burgeson RE, and Morgan BA (2000). Wnt signaling maintains the hair-inducing activity of the dermal papilla. Genes Dev. 14,1181–1185. [PMC free article] [PubMed] [Google Scholar]
- Kivelä T, and Uusitalo M (1998). Structure, development and function of cytoskeletal elements in non-neuronal cells of the human eye. Prog. Retin. Eye Res. 17,385–428. [DOI] [PubMed] [Google Scholar]
- Klein AM, and Simons BD (2011). Universal patterns of stem cell fate in cycling adult tissues. Development 138,3103–3111. [DOI] [PubMed] [Google Scholar]
- Klein AM, Doupé DP, Jones PH, and Simons BD (2007). Kinetics of cell division in epidermal maintenance. Phys. Rev. E. Stat. Nonlin. Soft Matter Phys. 76, 21910. [DOI] [PubMed] [Google Scholar]
- Kowalczyk MS, Tirosh I, Heckl D, Rao TN, Dixit A, Haas BJ, Schneider RK, Wagers AJ, Ebert BL, and Regev A (2015). Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 25,1860–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange UC, Adams DJ, Lee C, Barton S, Schneider R, Bradley A, and Surani MA (2008). Normal germ line establishment in mice carrying a deletion of the Ifitm/Fragilis gene family cluster. Mol. Cell. Biol. 28,4688–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, and Clevers H (2010). Coexistence of quiescent and active adult stem cells in mammals. Science 327, 542–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim X, Tan SH, Yu K. Lou, Lim SBH, and Nusse R (2016). Axin2 marks quiescent hair follicle bulge stem cells that are maintained by autocrine Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. U. S. A. 113,E1498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Elf SE, Miyata Y, Sashida G, Liu Y, Huang G, Di Giandomenico S, Lee JM, Deblasio A, Menendez S, et al. (2009). p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell 4, 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrique I, Nguewa P, Bleau A-M, Nistal-Villan E, Lopez I, Villalba M, Gil-Bazo I, and Calvo A (2015). The inhibitor of differentiation isoform Id1b, generated by alternative splicing, maintains cell quiescence and confers self-renewal and cancer stem cell-like properties. Cancer Lett. 356,899–909. [DOI] [PubMed] [Google Scholar]
- Martynoga B, Morrison H, Price DJ, and Mason JO (2005). Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev. Biol. 283,113–127. [DOI] [PubMed] [Google Scholar]
- Mesa KR, Kawaguchi K, Cockburn K, Gonzalez D, Boucher J, Xin T, Klein AM, and Greco V (2018). Homeostatic Epidermal Stem Cell Self-Renewal Is Driven by Local Differentiation. Cell Stem Cell 23,677–686.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyyappan M, Wong H, Hull C, and Riabowol KT (1998). Increased expression of cyclin D2 during multiple states of growth arrest in primary and established cells. Mol. Cell. Biol. 18,3163–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira H, Andreu Z, Suh H, Lie DC, Jessberger S, Consiglio A, San Emeterio J, Hortigüela R, Marqués-Torrejón MA, Nakashima K, et al. (2010). Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell 7, 78–89. [DOI] [PubMed] [Google Scholar]
- Miroshnikova YA, Le HQ, Schneider D, Thalheim T, Rübsam M, Bremicker N, Polleux J, Kamprad N, Tarantola M, Wang I, et al. (2018). Adhesion forces and cortical tension couple cell proliferation and differentiation to drive epidermal stratification. Nat. Cell Biol. 20,69–80. [DOI] [PubMed] [Google Scholar]
- Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, Lin JS, Sawicki JA, and Cotsarelis G (2004). Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 22, 411–417. [DOI] [PubMed] [Google Scholar]
- Mort RL, Douvaras P, Morley SD, Dorà N, Hill RE, Collinson JM, and West JD (2012). Stem cells and corneal epithelial maintenance: insights from the mouse and other animal models. Results Probl. Cell Differ. 55, 357–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Moreno R, Cuesta-Geijo MÁ, Martínez-Romero C, Barrado-Gil L, Galindo I, García-Sastre A, and Alonso C (2016). Antiviral Role of IFITM Proteins in African Swine Fever Virus Infection. PLoS One 11, e0154366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Larsen SB, Cowley CJ, and Fuchs E (2018). Two to Tango: Dialog between Immunity and Stem Cells in Health and Disease. Cell 175,908–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasser W, Amitai-Lange A, Soteriou D, Hanna R, Tiosano B, Fuchs Y, and Shalom-Feuerstein R (2018). Corneal-Committed Cells Restore the Stem Cell Pool and Tissue Boundary following Injury. Cell Rep. 22,323–331. [DOI] [PubMed] [Google Scholar]
- Niederkorn JY, Ubelaker JE, and Martin JM (1990). Vascularization of corneas of hairless mutant mice. Invest. Ophthalmol. Vis. Sci. 31,948–953. [PubMed] [Google Scholar]
- Okada SL, Ellsworth JL, Durnam DM, Haugen HS, Holloway JL, Kelley ML, Lewis KE, Ren H, Sheppard PO, Storey HM, et al. (2006). A glycoprotein hormone expressed in corticotrophs exhibits unique binding properties on thyroid-stimulating hormone receptor. Mol. Endocrinol. 20,414–425. [DOI] [PubMed] [Google Scholar]
- Park M, Richardson A, Pandzic E, Lobo EP, Lyons JG, and Di Girolamo N (2019). Peripheral (not central) corneal epithelia contribute to the closure of an annular debridement injury. Proc. Natl. Acad. Sci. U. S. A. 116,26633–26643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Greco V, and Cockburn K (2016). Live imaging of stem cells: answering old questions and raising new ones. Curr. Opin. Cell Biol. 43,30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini G, and De Luca M (2014). Eyes on the prize: limbal stem cells and corneal restoration. Cell Stem Cell 15,121–122. [DOI] [PubMed] [Google Scholar]
- Pellegrini G, Golisano O, Paterna P, Lambiase A, Bonini S, Rama P, and De Luca M (1999). Location and clonal analysis of stem cells and their differentiated progeny in the human ocular surface. J. Cell Biol. 145, 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroutsos G, Guimaraes R, Giraud JP, and Pouliquen Y (1982). Corticosteroids and corneal epithelial wound healing. Br. J. Ophthalmol. 66, 705–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinwald JG, Green H. Epidermal growth factor and the multiplication of cultured human epidermal keratinocytes Nature, 265 (1977), pp. 421–424, 10.1038/265421a0. [DOI] [PubMed] [Google Scholar]
- Sagga N, Kuffová L, Vargesson N, Erskine L, and Collinson JM (2018). Limbal epithelial stem cell activity and corneal epithelial cell cycle parameters in adult and aging mice. Stem Cell Res. 33, 185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salpeter SJ, Klochendler A, Weinberg-Corem N, Porat S, Granot Z, Shapiro AMJ, Magnuson MA, Eden A, Grimsby J, Glaser B, et al. (2011). Glucose regulates cyclin D2 expression in quiescent and replicating pancreatic β-cells through glycolysis and calcium channels. Endocrinology 152,2589–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartaj R, Zhang C, Wan P, Pasha Z, Guaiquil V, Liu A, Liu J, Luo Y, Fuchs E, and Rosenblatt MI (2017). Characterization of slow cycling corneal limbal epithelial cells identifies putative stem cell markers. Sci. Rep. 7, 3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secker GA, and Daniels JT (2008). Corneal epithelial stem cells: deficiency and regulation. Stem Cell Rev. 4, 159–168. [DOI] [PubMed] [Google Scholar]
- Shevach EM (2018). Foxp3(+) T Regulatory Cells: Still Many Unanswered Questions-A Perspective After 20 Years of Study. Front. Immunol. 9,1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. (2010). Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143,134–144. [DOI] [PubMed] [Google Scholar]
- Stepp MA (2006). Corneal integrins and their functions. Exp. Eye Res. 83,3–15. [DOI] [PubMed] [Google Scholar]
- Strinkovsky L, Havkin E, Shalom-Feuerstein R, and Savir Y The role of replication-removal spatial correlations and cellular replicative lifespan in corneal epithelium homeostasis eLife, 10 (2021), p. e56404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamynathan S, Delp EE, Harvey SAK, Loughner CL, Raju L, and Swamynathan SK (2015). Corneal expression of SLURP-1 by age, sex, genetic strain, and ocular surface health. Investig. Ophthalmol. Vis. Sci. 56,7888–7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TILL JE, and McCULLOCH EA (1961). A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat. Res. 14,213–222. [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. (2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352,189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, and Fuchs E (2004). Defining the epithelial stem cell niche in skin. Science 303,359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tümpel S, and Rudolph KL (2019). Quiescence: Good and Bad of Stem Cell Aging. Trends Cell Biol. 29,672–685. [DOI] [PubMed] [Google Scholar]
- Waghmare SK, Bansal R, Lee J, Zhang YV, McDermitt DJ, and Tumbar T (2008). Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells. EMBO J. 27, 1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-Z, Plane JM, Jiang P, Zhou CJ, and Deng W (2011). Concise review: Quiescent and active states of endogenous adult neural stem cells: identification and characterization. Stem Cells 29,907–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JD, Mort RL, Hill RE, Morley SD, and Collinson JM (2018). Computer simulation of neutral drift among limbal epithelial stem cells of mosaic mice. Stem Cell Res. 30,1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, et al. (2008). Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135,1118–1129. [DOI] [PubMed] [Google Scholar]
- Wu X, Dao Thi VL, Huang Y, Billerbeck E, Saha D, Hoffmann H-H, Wang Y, Silva LAV, Sarbanes S, Sun T, et al. (2018). Intrinsic Immunity Shapes Viral Resistance of Stem Cells. Cell 172,423–438.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Shimmura S, Kawakita T, Miyashita H, Den S, Shimazaki J, and Tsubota K (2006). Cytokeratin 15 can be used to identify the limbal phenotype in normal and diseased ocular surfaces. Invest. Ophthalmol. Vis. Sci. 47,4780–4786. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Ban Y, and Kinoshita S (2009). Tight junction transmembrane protein claudin subtype expression and distribution in human corneal and conjunctival epithelium. Invest. Ophthalmol. Vis. Sci. 50,2103–2108. [DOI] [PubMed] [Google Scholar]
- Zhang B, Kirov S, and Snoddy J (2005). WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33, W741–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single cell mRNA-seq data generated during this study are available at GEO (Gene Expression Omnibus) GEO: GSE167992.
This study did not generate code.