ABSTRACT
Epithelial attachment to the basement membrane (BM) is essential for mammary gland development, yet the exact roles of specific BM components remain unclear. Here, we show that Laminin α5 (Lama5) expression specifically in the luminal epithelial cells is necessary for normal mammary gland growth during puberty, and for alveologenesis during pregnancy. Lama5 loss in the keratin 8-expressing cells results in reduced frequency and differentiation of hormone receptor expressing (HR+) luminal cells. Consequently, Wnt4-mediated crosstalk between HR+ luminal cells and basal epithelial cells is compromised during gland remodeling, and results in defective epithelial growth. The effects of Lama5 deletion on gland growth and branching can be rescued by Wnt4 supplementation in the in vitro model of branching morphogenesis. Our results reveal a surprising role for BM-protein expression in the luminal mammary epithelial cells, and highlight the function of Lama5 in mammary gland remodeling and luminal differentiation.
KEY WORDS: Laminin alpha 5, Extracellular matrix, Basement membrane, Mammary gland, Luminal epithelial cell, Wnt4
Summary: Luminal mammary epithelial cells produce basement membrane laminin α5 necessary for mammary epithelial growth and differentiation. Laminin α5 loss compromises hormone receptor-positive luminal cell function and Wnt4-mediated crosstalk between epithelial cells.
INTRODUCTION
Postnatal development of the mammary gland occurs by branching and elongation during puberty, and by extensive remodeling during pregnancy (Macias and Hinck, 2012; Paine and Lewis, 2017). The fully formed adult mammary epithelium consists of a bilayered duct, in which apicobasally polarized keratin 8/18+ luminal cells, which can be subdivided into hormone receptor-expressing and non-expressing luminal cells (HR+ and HR−, respectively), face the ductal lumen and are surrounded by keratin 5/14+ basal myoepithelial cells (Macias and Hinck, 2012; Adriance et al., 2005).
The mammary microenvironment contributes to various properties of mammary epithelial cells (MECs), including proliferation, survival and differentiation (Inman et al., 2015). In particular, attachment to the specialized layer of the extracellular matrix (ECM) called the basement membrane (BM) is crucial for cultured mammary epithelial cells and for mammary gland morphogenesis in vivo (Inman et al., 2015; Weaver et al., 1997; LaBarge et al., 2009). The BM acts as a physical barrier separating the epithelium from the stroma and as a scaffold supporting epithelial adhesion and tissue architecture (Yurchenco, 2011). Moreover, the BM regulates tissue homeostasis by supplying cells with growth factors and other signaling molecules, and by regulating their availability to the cells (Yurchenco, 2011).
Laminins are the main components of the BM that together with collagen IV form self-assembling networks, which provide epithelial cells an anchoring platform and various survival and differentiation signals (Hohenester and Yurchenco, 2013). Laminins are heterotrimers consisting of α, β and γ subunits, which are expressed in a tissue-specific and temporally controlled manner (Ahmed and Ffrench-Constant, 2016). Several laminin isoforms have been detected in the mammary gland, and earlier studies suggest that laminin-111 (containing α1, β1 and γ1 subunits), laminin-332 and laminin-511/521 are the most common forms in the adult glands (Goddard et al., 2016; Gudjonsson et al., 2002; Prince et al., 2002), yet laminin-211 and laminin-411/421 are also found. Microarray studies and later single-cell sequencing efforts have indicated high expression of Lama1 and Lama3 in the basal cells, with Lama5 being highest in luminal cells (Lim et al., 2010; Bach et al., 2017; Tabula Muris Consortium, 2018). However, what the spatial and temporal expression patterns of these laminins within intact tissue are and what their exact roles in mammary development and function are is unclear.
We set out to study the expression pattern and function of different laminin isoforms in the mouse mammary gland. We demonstrate that Lama5 produced by luminal cells is necessary for normal mammary gland growth and development during both puberty and pregnancy. Mechanistically, we show that Lama5 loss alters differentiation of HR+ luminal MECs, and consequently their Wnt4-mediated interactions with basal cells during gland remodeling. Our results reveal that Lama5 acts as a key microenvironmental effector of mammary gland function by orchestrating HR+ luminal cell specification during puberty and pregnancy.
RESULTS
Differential expression of laminin isoforms in distinct cell types of the mammary epithelium
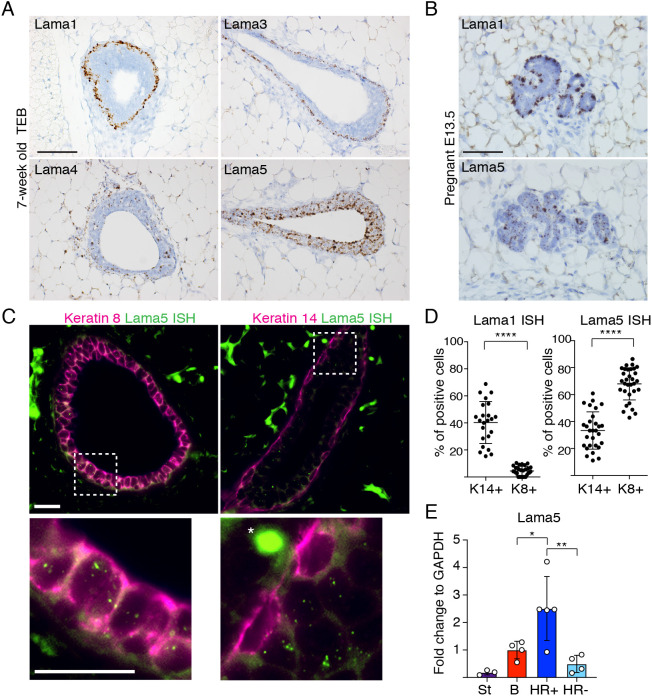
To explore the role of specific laminins in the mammary gland, we first studied their expression during the pubertal gland expansion, which is marked by terminal end bud (TEB) structures (Macias and Hinck, 2012). Using in situ hybridization (ISH) to detect the expression of Lama1, 3, 4 and 5 (encoding the laminin α1, α3, α4 and α5 subunits), we observed that both Lama1 and 3 were expressed by the basal cells of growing TEBs (Fig. 1A) and established ducts (Fig. S1A). Lama3 expression was also detected in some luminal cells of the mature ducts (Fig. S1A, red arrowheads). In striking contrast, Lama5 was strongly expressed by luminal epithelial cells, particularly in TEBs, and Lama4 showed widespread expression also in stroma. As Lama1 and Lama5 showed the most cell type-specific expression patterns, we next analyzed their expression during the pregnancy-induced gland remodeling. Although separation of the two cellular layers is less evident in the differentiated alveoli, Lama1 expression appeared to be restricted to basal cells and Lama5 was mostly restricted to luminal cells also during pregnancy (Fig. 1B). To quantitatively address expression of Lama1 and Lama5 in the luminal and basal MECs, we combined laminin ISH with keratin 8 (K8, also known as Krt8) and keratin 14 (K14, also known as Krt14) antibody staining in the pubertal glands (Fig. 1C; Fig. S1B,C). Whereas Lama1 was strictly expressed by basal (K8− and K14+) cells, Lama5 was significantly enriched in luminal (K8+ and K14−) epithelial cells with some expression in a subset of basal cells (Fig. 1D; Fig. S1C). Moreover, by further subdividing CD29low/CD24+ luminal cells into HR− and HR+ with fluorescence-activated cell sorting (FACS; Fig. S1D) by cell surface markers Sca1 (also known as Ly6a) and CD49b (Shehata et al., 2012), we found that HR-expressing luminal cells are the major source for Lama5 in the mammary epithelium (Fig. 1E).
Fig. 1.
Expression of laminin α chain isoforms in the mammary gland is cell type-specific. (A) Representative images of RNA ISH performed with probes for Lama1, 3, 4 and 5 on TEBs of 7-week-old pubertal mammary glands. (B) Representative images of RNA ISH of mammary glands from embryonic day 13.5 pregnant mice performed with probes for Lama1 and 5 on TEBs. (C) Representative images of Lama5 RNA ISH coupled with either K8 or K14 antibody staining. Asterisk denotes background staining. The dashed boxes indicate the magnified regions in the lower panels. (D) Quantification of the percentage of K14+ or K8+ cells showing either Lama1 or Lama5 expression. Lama1 quantification in K14+ and K8+ cells (K14, n=23 ducts; K8, n=28 ducts; from three animals). Lama5 quantification in K14+ (n=29) and K8+ cells (n=31) ducts from three animals. Data points correspond to individual ducts analyzed. (E) qRT-PCR analysis of Lama5 expression compared with GAPDH in populations of stromal (St), basal (B), luminal HR+ and luminal HR− cells FACS sorted from mammary glands (St, n=3; B, n=4; HR+, n=5; HR−, n=4 mice analyzed). Data are mean±s.d. An unpaired two-tailed Student's t-test was used to compare indicated groups (*P<0.05, **P<0.01, ****P<0.0001). Scale bars: 50 µm (A); 50 µm (B); 20 µm.
The expression of the BM component Lama5 in luminal cells was surprising as luminal cells are reported to have only very limited contacts with the BM (Smith and Medina, 1988; Lafkas et al., 2013). However, the Pan-laminin staining that we observed mostly in the periductal region suggested that the luminally expressed laminin proteins are also deposited on the BM surrounding the gland (Fig. S1E). Providing a potential route for luminal cells to secrete laminins into the BM, we detected contacts between luminal cells and the BM (Fig. S1F) in tamoxifen-induced Lgr6-CreERT2:Rosa26-tdTomato mice in which clones of exclusively luminal or basal cells can be visualized (Blaas et al., 2016). Taken together, these results indicate that laminin isoforms have distinct expression patterns within the mammary gland (summarized in Fig. S1G), and also infer that the luminal epithelial cells, in addition to the BM-adjacent basal cells, may contribute to the laminin pool of the BM.
Laminin α5 is required for normal pubertal mammary gland morphogenesis
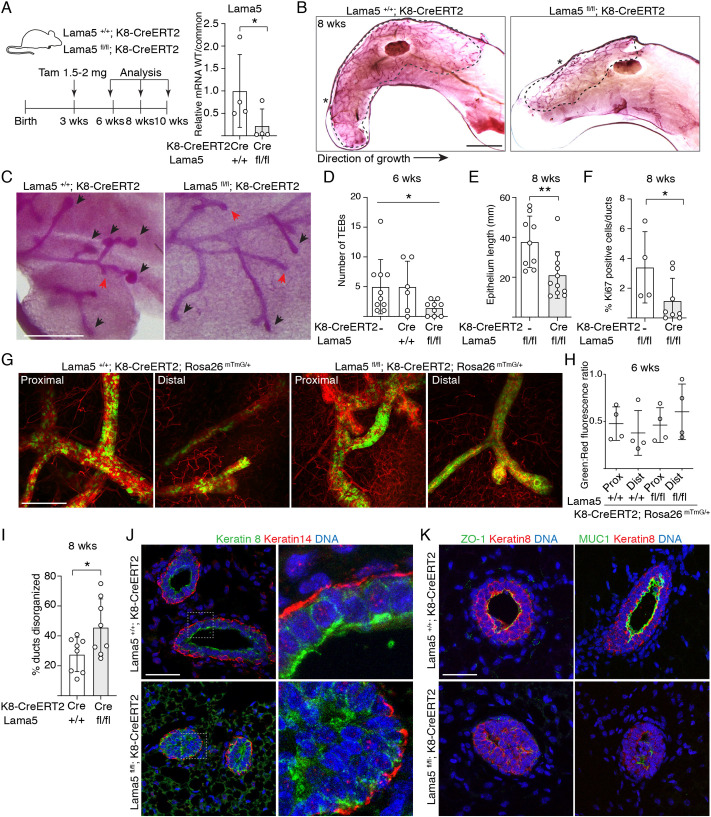
To address the functional role of the luminally expressed Lama5, we generated Lama5fl/fl;K8-CreERT2 mice (Nguyen et al., 2005; Van Keymeulen et al., 2011), which allow tamoxifen-inducible deletion of Lama5 in luminal epithelial cells. To determine the effects of Lama5 loss during puberty, we injected 3-week-old female mice with 1.5-2 mg of tamoxifen, and analyzed glands at 6, 8 or 10 weeks of age (Fig. 2A). Consistent with previous reports (Shehata et al., 2014; Scheele et al., 2017), we observed a significant decrease in epithelial growth and TEB number even with such a low tamoxifen dose, forbidding use of higher doses (Fig. S2A). To validate effective recombination, we first analyzed allelic frequencies in FACS-sorted luminal and basal epithelial cells 3 days after tamoxifen treatment (Fig. S2B), and in bulk epithelial cells 3 weeks after tamoxifen (Fig. S2C). Recombined Lama5 allele was observed only from the luminal epithelial cells, and a high level of recombination was evident 3 weeks after tamoxifen (Fig. S2B,C). Moreover, allele-specific analysis of mRNA levels (Fig. S2D) revealed an 88% reduction in the relative abundance of wild-type mRNA in luminal epithelial cells sorted from tamoxifen-treated 8-week-old Lama5fl/fl;K8-CreERT2 mice compared with Lama+/+;K8-CreERT2 controls (Fig. 2A; Fig. S2E). Thus, stable and cell type-specific deletion of Lama5 was achieved in the majority of the luminal cells, even with the lower tamoxifen dose used.
Fig. 2.
Luminal laminin α5 is required for pubertal growth of the mammary epithelium. (A) Schematic showing the outline of tamoxifen induction of K8-CreERT2 during puberty, and qPCR analysis of Lama5 mRNA expression in luminal MECs (+/+ and fl/fl, n=4 samples analyzed) by using wild-type (WT) allele-specific primers compared with primers common for both + and flox alleles (common). (B) Representative images of carmine alum-stained #4 mammary glands of 8-week-old Lama5+/+;K8-CreERT2 and Lamafl/fl;K8-CreERT2 treated with tamoxifen. The dashed line shows the outline of ductal growth and the asterisks mark the beginning of epithelium. (C) Representative images of TEBs and ductal ends (marked by black arrowheads and red arrows, respectively) in 6-week-old transgenic mice. (D) Quantification of the number of TEBs in 6-week-old transgenic mice analyzed from #4 mammary glands (K8-CreERT2, n=10; Lama5+/+;K8-CreERT2, n=6, Lama5fl/fl;K8-CreERT2, n=9 glands analyzed). (E) Quantification of epithelium length in 8-week-old transgenic mice analyzed from #4 mammary glands (Lama5fl/fl; n=9, Lama5fl/fl;K8-CreERT2, n=11 glands analyzed). (F) Quantification of Ki67 positivity in mammary glands of 8-week-old transgenic mice (Lama5fl/fl; n=4; Lama5fl/fl;K8-CreERT2, n=8 individuals analyzed). (G) Representative images of #4 mammary glands of 6-week-old Lama5+/+;K8-CreERT2;R26mTmG/+ and Lama5fl/fl;K8-CreERT2;R26mTmG/+ mice. Images show proximal (closer to the beginning of epithelium) and distal parts of the duct. (H) Quantification of green to red fluorescence ratio in proximal and distal parts of the duct in Lama5+/+;K8-CreERT2;R26mTmG/+ and Lama5fl/fl;K8-CreERT2;R26mTmG/+ mice (+/+ and fl/fl, both proximal and distal part, n=4 glands quantified). (I) Quantification of epithelial disorganization in ducts of 8-week-old Lama5+/+;K8-CreERT2 (n=8) and Lama5fl/fl;K8-CreERT2 (n=8) animals. (J) Representative immunofluorescence images of 8-week-old Lama5+/+;K8-CreERT2 and Lama5fl/fl;K8-CreERT2 glands immunostained with K8 and K14 antibodies. Boxes indicate the magnified regions in the right panels. (K) Representative immunofluorescence images of 8-week-old Lama5+/+;K8-CreERT2 and Lama5fl/fl;K8-CreERT2 glands immunostained with ZO-1 or MUC1, and K8 antibodies. Data points indicate analysis of individual mice. Data are mean±s.d. *P<0.05, **P<0.01 [unpaired two-tailed Student's t-test (A,E,I) or Welch's ANOVA test (D)]. Scale bars: 5 mm (B); 0.5 mm (C); 200 µm (G); 50 µm (J,K).
Lama5 deletion in luminal MECs led to delayed development of the mammary epithelium (Fig. 2B), which was accompanied by a reduction in TEB size and number (Fig. 2C,D). Consequently, the extent of the epithelial network was significantly reduced and contained fewer Ki-67+ cells at 8 weeks (Fig. 2E,F). However, when Lama5 was deleted in the post-pubertal mature glands of 8-week-old mice and analyzed at 12 weeks of age, no difference in the ductal end number was observed (Fig. S2F,G). These data suggest that although Lama5 is required for mammary epithelium growth and morphogenesis during pubertal remodeling, loss of Lama5 in adult mammary glands has little effect on adult homeostasis.
To investigate whether the defective gland growth upon Lama5 targeting was due to impaired function of the Lama5-deleted luminal cells, or due to their loss, we crossed Lama5fl/fl;K8-CreERT2 mice with the R26-mTmG reporter line (Muzumdar et al., 2007). Immunofluorescence analysis of the glands 3 weeks post tamoxifen showed comparable frequencies of recombined cells in Lama5fl/fl;K8-CreERT2;R26mT/mG and control mice (Fig. 2G,H; Fig. S2H). Although recombination frequency of the reporter and Lama5 loci may differ (Vooijs et al., 2001), these results are in line with our analysis of recombination efficiency (Fig. S2E), and indicate that Lama5-deficient cells are not lost and contribute to blunted epithelial growth.
Interestingly, although parts of the ducts appeared ultrastructurally normal, histological analysis of Lama5fl/fl;K8-CreERT2 glands at 8 weeks revealed a frequent loss of normal tissue architecture at certain gland regions. The regional variability may reflect the development of ducts before and after tamoxifen-induced Lama5 deletion, but even in blinded analysis of the full mammary gland, aberrant ducts were significantly more frequent in Lama5-deficient mice than in control mice (Fig. 2I). In the aberrant ducts, K8+ luminal cells were atypically organized into multiple layers (Fig. 2J). These observations suggested defects in cellular polarity in the Lama5-deficient epithelium, and we therefore inspected the localization and expression of apical polarity markers ZO-1 (also known as Tjp1) and MUC1 (Fig. 2K). We noticed that even in the aberrant Lama5fl/fl;K8-CreERT2 ducts localization of ZO-1 and MUC1 was mostly marking the apical surfaces facing the ductal lumens, similarly to controls (Fig. 2K). However, Lama5fl/fl;K8-CreERT2 ducts showed great variability in intensity and localization of the polarity markers, suggesting that loss of Lama5 from the luminal epithelium affects luminal cell polarity but the polarity is not completely lost. Interestingly, both ZO-1 and MUC1 also exhibited a dramatic reduction in general staining intensity in the Lama5-deficient ducts. MUC1 is a differentiation marker of luminal mammary epithelial cells (Shehata et al., 2012), raising the possibility of defective luminal differentiation in Lama5-deleted glands. In conclusion, Lama5 in the luminal epithelial cells is necessary for normal pubertal mammary gland growth and properly polarized duct architecture.
Loss of laminin α5 compromises HR+ cell differentiation
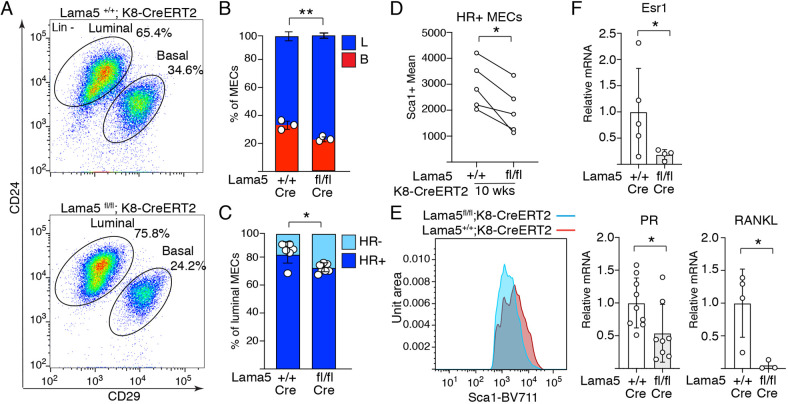
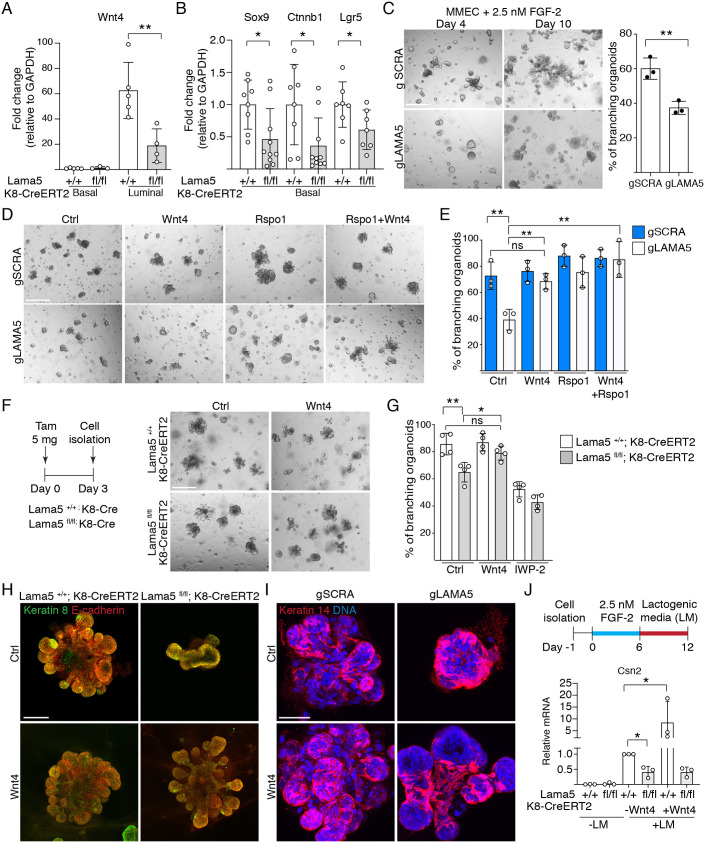
The aberrant organization and reduction in luminal cell differentiation marker MUC1 prompted us to investigate cellular ratios of the Lama5-deficient mammary epithelium. We noticed a significant increase in luminal (CD29low/CD24+) to basal (CD29hi/CD24+) cell ratio in Lama5fl/fl;K8-CreERT2 glands in comparison with controls (Fig. 3A,B; Fig. S3A,B). Interestingly, we also noted a 1.7-fold relative increase in the Sca1−/CD49b+ luminal cell subpopulation comprising HR− cells (Fig. 3C) (Shehata et al., 2012; Sleeman et al., 2007) in Lama5-lacking animals. Moreover, the remaining HR+ cells expressed Sca1 on a lower level (Fig. 3D,E). This indicated that prepubertal loss of Lama5 compromises specifically the pool of HR+ cells during pubertal gland growth. To further investigate whether the expression of HR+ luminal epithelial differentiation markers are altered in Lama5-deficient glands, we performed qPCR on estrogen receptor alpha (Esr1), progesterone receptor (PR, also known as Pgr) and Receptor Activator of Nuclear factor Kappa B ligand (RANKL, also known as Tnfsf11). Expression of all three markers of HR+ differentiation in the Lama5-targeted luminal cells was reduced significantly and beyond the level expected based on the observed reduction in HR+ cell frequency (Fig. 3F, compare with Fig. 3C). These data indicate that Lama5 is indeed necessary for HR+ luminal cell differentiation and for maintaining a normal ratio of epithelial cells.
Fig. 3.
Laminin α5 is needed for HR+ luminal epithelial cell identity. (A) Representative CD24/CD29 FACS plot showing relative amounts of lineage marker negative luminal and basal MECs from 10-week-old Lama5fl/fl;K8-CreERT2 and control animals. Gating strategy for the same sample is shown in Fig. S3A. (B,C) Quantification of the percentages of basal and luminal MECs (+/+, n=3; fl/fl, n=3 individuals analyzed) and (C) luminal HR+ and HR− subpopulations from 10-week-old Lama5fl/fl;K8-CreERT2 and control mice (+/+, n=6; fl/fl, n=6 individuals analyzed). Data points indicate samples analyzed from individual mice. (D) Mean intensity of Sca1+ luminal population from Lama5fl/fl;K8-CreERT2 and control mice. Data show mean from n=5 separate experiments, in which 3-4 individual animals were analyzed per group. Data points indicate mean values from five separate experiments. (E) Representative Sca1-BV711 histograms of individual Lama5+/+;K8-CreERT2 and Lama5fl/fl;K8-CreERT2 mice. (F) qRT-PCR analysis of Esr1, PR and RANKL expression compared with GAPDH in luminal MECs from 10-week-old Lama5fl/fl;K8-CreERT2 and control mice (Esr1 +/+, n=5; fl/fl, n=4; PR +/+, n=9; fl/fl, n=8; RANKL +/+, n=4; fl/fl, n=3). Data points indicate samples analyzed from individual mice. Data are mean±s.d. *P<0.05, **P<0.01 [unpaired two-tailed Student's t-test (B,C,F); paired t-test (D)].
Laminin α5 is necessary for normal alveologenesis
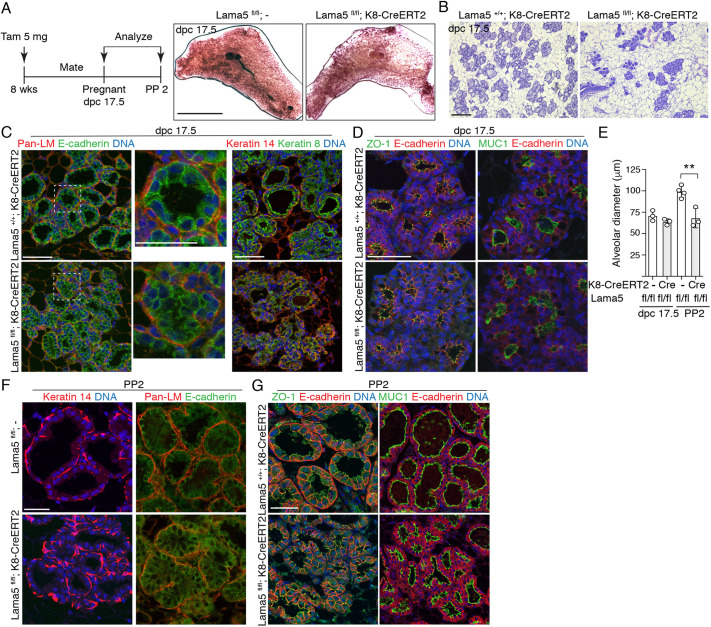
We next assessed the role of Lama5 in the pregnancy-induced growth and functional differentiation of the mammary gland. Lama5 was deleted from the luminal cells of 8-week-old female mice 1-2 weeks before pregnancy, and glands were analyzed 17.5 days post coitum (dpc; Fig. 4A; Fig. S4A). Alveolar development was dramatically reduced in Lama5-deficient glands, as evidenced by fewer and less densely packed alveoli compared with control glands (Fig. 4A,B; Fig. S4B). Moreover, Lama5-deficient alveoli often failed to form round structures, and were instead devoid of distinct lumen and exhibited aberrant organization of both luminal and basal cells (Fig. 4C). However, the apicobasal polarity of Lama5-deficient glands appeared only modestly affected at this stage of pregnancy (Fig. 4D) yet, intriguingly, also at this stage, MUC1 staining intensity was reduced. Analysis at postpartum day 2 (PP2) revealed that alveoli in Lama5-deleted glands were significantly smaller and exhibited disorganized cellular organization, even though Pan-laminin staining indicated that the BM remained intact (Fig. 4E,F). Again, cell polarity was largely unaffected, as demonstrated by staining of ZO-1 and MUC1 (Fig. 4G). However, the Lama5-deficient females often had less pups, which at this stage can also affect gland morphology. Taken together, our characterization of the mouse model with luminal-specific Lama5 deletion demonstrates an important in vivo role for Lama5 in luminal MECs during the key stages of mammary gland development and function.
Fig. 4.
Laminin α5 is essential for pregnancy-induced growth and differentiation of the mammary gland. (A) Schematic showing the experimental outline of tamoxifen induction of K8-CreERT2 before pregnancy. Representative images of carmine alum-stained #4 mammary glands of 17.5 dpc pregnant Lamafl/fl;- and Lamafl/fl;K8-CreERT2 animals. (B) Representative H&E images of 17.5 dpc pregnant Lama5+/+;K8-CreERT2 and Lama5fl/fl;K8-CreERT2 mice. (C) Representative immunofluorescence images of 17.5 dpc pregnant Lama5+/+;K8-CreERT2 and Lama5fl/fl;K8-CreERT2 glands immunostained with Pan-laminin and E-cadherin, or K8 and K14 antibodies. Square shows area of inset. (D) Representative immunofluorescence images of 17.5 dpc pregnant Lama5+/+;K8-CreERT2 and Lama5fl/fl;K8-CreERT2 glands immunostained with ZO-1 or MUC1, and E-cadherin antibodies. (E) Quantification of the alveolar diameter of 17.5 dpc (+/+, n=3; fl/fl, n=3 individuals analyzed) and PP2 mice (+/+, n=4; fl/fl, n=3 individuals analyzed). N=3-4 individual animals. Data points represent mean alveolar diameter quantified from individual animals. (F) Representative immunofluorescence images of PP2 mammary glands immunostained with K14 or Pan-laminin and E-cadherin antibodies. (G) Representative immunofluorescence images of PP2 mammary glands immunostained with ZO-1 or MUC1, and E-cadherin antibodies. Data are mean±s.d. **P<0.01 (unpaired two-tailed Student's t-test). Scale bars: 10 mm (A); 200 µm (B); 50 µm (C,D,F,G).
Lama5-deficient HR+ luminal MECs are unable to support basal cells
Ductal elongation during puberty and alveologenesis during pregnancy both require accurate interplay between the luminal and basal MECs (Macias and Hinck, 2012; Inman et al., 2015). HR+ luminal MECs are considered to orchestrate mammary growth and development by responding to systemic hormonal cues and transforming them into paracrine signals (Tanos et al., 2012). Accordingly, prepubertal loss of Esr1 from luminal cells impedes the development of the mammary epithelium during puberty (Mallepell et al., 2006; Feng et al., 2007), and Esr1 loss during pregnancy results in defective alveologenesis and milk production (Feng et al., 2007). As Lama5 expression was highest in the luminal HR+ cells (Fig. 1E), and luminal Lama5 loss reduced HR+ cell frequency and differentiation, as well as Esr1, PR, and RANKL expression (Fig. 3C-E), we hypothesized that defects in the growth and differentiation of Lama5 lacking mammary epithelium are due to compromised function of HR+ cells.
Wnt4 is an important paracrine mediator that is expressed in the HR+ MECs in response to progesterone (Tanos et al., 2012). Together with R-spondin 1 (Rspo1) from HR− luminal MECs, Wnt4 activates Wnt signaling in the basal cells to regulate growth of the mammary epithelium (Rajaram et al., 2015; Cai et al., 2014; Brisken et al., 2000). Interestingly, expression of Wnt4 was significantly reduced in Lama5-deficient luminal cells, whereas Rspo1 showed only a trend of modest reduction (Fig. 5A; Fig. S5A). Moreover, the expression of Wnt-responsive genes Sox9 and Lgr5 regulated directly via transcriptional control (Blache et al., 2004; Schuijers and Clevers, 2012), or Ctnnb1 regulated via mRNA stability (Bikkavilli and Malbon, 2010), was significantly reduced in the basal cells (Fig. 5B). Jointly these data indicate that defects in the HR+ cells after luminal Lama5 loss does indeed compromise the luminal-basal interactions.
Fig. 5.
Lama5-deficient luminal MECs are unable to support basal MECs due to defective Wnt signaling. (A) qRT-PCR analysis of Wnt4 expression compared with GAPDH in basal (CD29hi/CD24+) and luminal (CD29low/CD24+) MECs FACS sorted from 8-10-week-old Lama5+/+;K8-CreERT2 and Lama5fl/fl;K8-CreERT2 glands treated with tamoxifen starting from 3 weeks of age (+/+ luminal and basal, fl/fl basal, n=3; fl/fl luminal, n=4 samples analyzed). (B) qRT-PCR analysis of Sox9, Ctnnb1 and Lgr5 expression compared with GAPDH in basal MECs from 10-week-old Lama5fl/fl;K8-CreERT2 and control mice (Sox9 and Ctnnb1 +/+, n=8; fl/fl, n=10; Lgr5 +/+, n=7; fl/fl, n=7 samples analyzed). In A and B, data points indicate RNA samples analyzed from individual mice. (C) Primary MMECs carrying either LAMA5 or SCRA CRISPR guides (gLAMA5 and gSCRA) grown in Matrigel in the presence of 2.5 nM FGF-2 for 4 or 10 days, and quantification of the percentage of branching organoids at day 10 (gSCRA, n=3; gLAMA, n=3). (D) gLAMA5 or gSCRA expressing primary MMECs grown in Matrigel in the presence of 2.5 nM FGF-2 and either Wnt4, Rspo1 or both ligands for 7 days. (E) Quantification of the percentage of branching organoids at day 7 grown as in D (all treatments n=3). (F) Schematic showing the outline of tamoxifen induction and representative images. MECs were isolated from 10-14-week-old Lama5+/+;K8-CreERT2 and Lama5fl/fl;K8-CreERT2 mice treated with tamoxifen 3 days earlier, and grown in Matrigel in the presence of 2.5 nM FGF-2 and either Wnt4 or IWP-2 for 7 days. (G) Quantification of the percentage of branching organoids at day 7 grown as in F (all treatments n=4). In C, E and G, data points indicate RNA samples analysed from individual mice. (H) Representative keratin 8 and E-cadherin immunofluorescence images of primary MMECs organoids treated as in F and grown for 7 days. (I) Representative keratin 14 immunofluorescence images of primary MMECs organoids expressing either gLAMA5 or gSCRA treated as in D and grown for 7 days. (J) Schematic showing the outline of organoid treatment with lactogenic medium. Quantitative PCR analysis of Csn2 gene expression in MECs isolated from 10-14-week-old Lama5+/+;K8-CreERT2 and Lama5fl/fl;K8-CreERT2 mice treated with tamoxifen 3 days earlier, and grown as described in the schematic with or without Wnt4 (all treatments n=3). Data are normalized to lactogenic medium-treated Lama5+/+;K8-CreERT2 for each experiment. Data points indicate RNA samples analysed from independent organoid experiments. Data are mean±s.d. *P<0.05; **P<0.01; ns, not significant [unpaired one-tailed (J) or two-tailed (A,B,C,E,G) Student's t-test]. Scale bars: 100 µm (C,D,H); 50 µm (I).
To directly test whether the altered paracrine Wnt signaling contributes to the defective growth of the mammary gland after luminal Lama5 loss, we modeled gland growth in vitro. Ductal growth by the TEB structures can be modeled in vitro with FGF-2 supplemented three-dimensional cultures, in which MEC organoids form branching structures (Ewald et al., 2008). Under these conditions, lentiviral CRISPR/Cas9-mediated Lama5 editing resulted in Lama5 mRNA downregulation and a significant reduction in organoid branch formation and elongation (Fig. 5C; Fig. S5B,C), thereby resembling the effects of luminal Lama5 deficiency in vivo. Strikingly, exogenous addition of Wnt4 and Rspo1 completely rescued the effects that Lama5 deletion had on branching frequency (Fig. 5D,E). These effects are likely to reflect the general activation of the canonical Wnt-pathway as Wnt3a supplementation recapitulated the results seen with Wnt4 (Fig. S5D). Highlighting the role of Wnt signaling, inhibition of Wnt production by porcupine inhibitor IWP-2 reduced the branching of control organoids but had no further effects on Lama5-edited organoids (Fig. S5D). In order to model the phenotypes we observed in the mouse model more closely, and to assess the cell type-specific effect of Lama5 in organoid growth, we next isolated MECs from Lama5fl/fl;K8-CreERT2 and control mice 3 days after tamoxifen treatment (Fig. 5F). Such preparations contain both luminal and basal cells, but Lama5 loss is exclusive to luminal cells (Fig. S2B). Recapitulating results with lentiviral targeting, branching frequency was decreased in organoids with luminal-specific Lama5 deletion, and exogenous Wnt4 rescued the branching defect (Fig. 5F,G). Moreover, branching was inhibited in control and Lama5-deficient organoids with the addition of IWP-2. These data further demonstrate that Lama5, specifically in luminal cells, is necessary for branching morphogenesis via paracrine signaling.
Targeting Lama5 or the rescue of branching with exogenous Wnt ligands did not significantly change the frequency of Ki67+ cells (Fig. S5E), suggesting mechanisms other than proliferation. When stained for K8 and E-cadherin to visualize the luminal organoid structure, both lentivirally Lama5-edited and Lama5fl/fl;K8-CreERT2 organoids showed similarly altered morphology with blunted branching (Fig. 5H; Fig. S5F). However, upon Wnt4 treatment both formed elongated branches that contained K14-expressing basal cells, and were structurally similar to branches in control organoids (Fig. 5H,I; Fig. S5F,G). Further supporting the role of Wnt signaling in promoting basal cell function during branching, exogenous Wnt3a and Rspo1 also resolved the structure of Lama5-deficient organoids (Fig. S5G).
Finally, to probe the possible interdependence of the in vivo observed phenotypes on gland growth and differentiation during pregnancy, we tested the lactogenic differentiation of organoids lacking luminal Lama5. MECs were isolated from Lama5fl/fl;K8-CreERT2 and control mice 3 days after tamoxifen, and organoids were first grown in conditions inducing branching for 6 days (Fig. 5J). Next, differentiation was induced with lactogenic medium (LM) for an additional 6 days (Sumbal et al., 2020), leading to a robust increase in the milk protein genes in control organoids (Fig. S6A). Consistent with our in vivo results, loss of Lama5 led to a decreased induction of Csn2 expression following the use of lactogenic medium (Fig. 5J; Fig. S6B). However, although Wnt4 further increased Csn2 expression in controls, and it rescued the branching defect of Lama5-deficient organoids as before, it did not rescue the defective lactogenic induction in Lama5-deficient organoids. Thus, although Wnt4 can resolve the defective organoid morphology in Lama5-deficient organoids, it is not sufficient to support the full functional differentiation of luminal epithelium lacking Lama5.
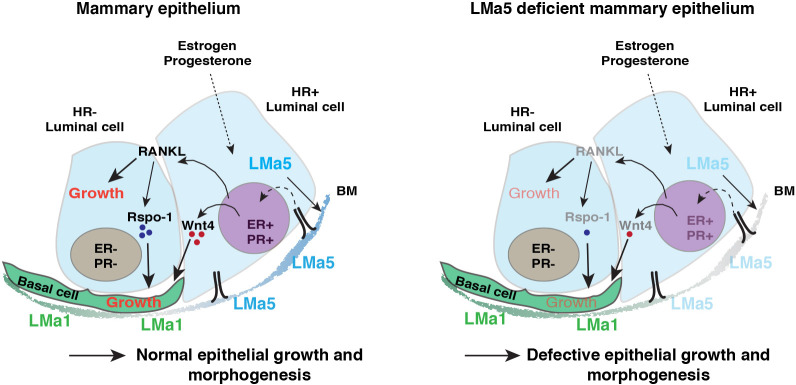
In conclusion, our data suggest that downstream of attenuated HR+ cell differentiation, loss of Lama5 in the luminal epithelium results in defective gland growth due to altered interplay between basal and luminal epithelial cells mediated by Wnt, and in defective functionalization upon pregnancy (Fig. 6).
Fig. 6.
Working model of how luminal Lama5 regulates mammary epithelial growth and crosstalk. Schematic showing how the production of LMα5 by luminal MECs supports specifically the HR+ luminal cells, in which it enhances gene expression of ER, PR, Wnt4 and RANKL. The products of these genes then act on neighboring basal and HR− epithelial cells to orchestrate epithelial growth and morphogenesis during puberty and pregnancy. Deficiency of LMα5 in the BM environment in turn leads to reduced gene expression in these genes, and defective intercellular communication and epithelial growth.
DISCUSSION
The mammary ECM is essential for normal physiology and pathology of the mammary gland (LaBarge et al., 2009; Maller et al., 2010; Goddard et al., 2016). However, the exact functions of many specific BM constituents in mammary gland physiology have remained unclear. Here, we show that BM laminin subunits α1, α3, α4 and α5 exhibit distinct gene expression patterns in the luminal, basal and stromal compartments of the mammary gland. We discovered that Lama5 is predominantly expressed in the HR+ luminal epithelial cells, and when Lama5 was deleted in the luminal cells at various stages, we found that it has a critical role during pubertal growth and pregnancy-induced remodeling and growth. Our results are in concert with previous studies showing that the downregulation of laminin-binding adhesion receptors, including β1- and β4-integrins, results in the inhibition of mammary epithelial growth and alveologenesis (Li et al., 2005; Naylor et al., 2005; Walker et al., 2020; Romagnoli et al., 2020). Together, these data underline the importance of laminin and BM microenvironment in the regulation of mammary epithelial growth and differentiation.
The growth of the mammary epithelium occurs in a coordinated manner, requiring interplay between both luminal and basal cell types (Macias and Hinck, 2012; Inman et al., 2015). HR+ cells have been indicated as essential mediators of this interplay by sensing systemic hormones and producing paracrine signals for the neighboring HR− luminal cells and the basal cells (Tanos et al., 2012). We observed a moderate reduction in the number of HR+ luminal epithelial cells in the Lama5-deficient mammary epithelium, but a more marked reduction in the HR+-specific expression of Esr1, PR and RANKL. Additionally, Sca1 marking both HR+ cells and their progenitors (Sleeman et al., 2007; Shehata et al., 2012), showed a decrease after Lama5 loss. These data suggest qualitative changes in the Lama5-deficient HR+ luminal cells, which we postulate impairs their capability to sense and respond to systemic and paracrine signals. Previous studies have established Wnt signaling as an important mediator of the interactions between HR+ and basal epithelial cells to induce mammary epithelial growth (Zeng and Nusse, 2010; Rajaram et al., 2015; Cai et al., 2014). We show that Lama5-deficient luminal cells produce less Wnt4, which is expressed specifically by HR+ cells, and functions as a downstream effector of Esr1 and PR (Cai et al., 2014; Rajaram et al., 2015). We also demonstrate decreased expression of Wnt target genes Lgr5, Ctnnb1 and Sox9 in the basal epithelial cells of Lama5-deficient epithelia. Moreover, in our in vitro studies, exogenous administration of Wnt4 rescued organoid branching defects caused by Lama5 loss. Thus, we conclude that the altered luminal-basal interactions, which are mediated by Wnt signaling, can in part explain the defective growth and remodeling of Lama5-deficient glands.
Loss of Lama5 from the adult mammary epithelium prior to pregnancy led to reduced growth and defective differentiation of alveoli during pregnancy-induced functionalization. Correspondingly, downregulation of Lama5 in vitro led to defective lactogenic differentiation in organoid cultures. Intriguingly, in these settings, the addition of Wnt4 was unable to rescue the differentiation defect, suggesting a lack of other essential factors. Expansion of luminal epithelial cells during pregnancy is largely attributable to HR− epithelial cells (Van Keymeulen et al., 2017), presumably mostly in response to progesterone-mediated signals from HR+ cells (Brisken et al., 1998). Therefore, it is interesting to hypothesize that the paracrine signaling between HR− and HR+ cells is also impaired in Lama5 lacking epithelium. Indeed, we observed that RANKL, which is expressed by the HR+ cells downstream of progesterone (Fata et al., 2000; Asselin-Labat et al., 2010), was significantly downregulated in the Lama5-deficient epithelial cells. Furthermore, loss of RANKL has been shown to inhibit alveologenesis and lactogenic differentiation (Fata et al., 2000), which could underlie the Lama5-dependent deficiency in lactogenic differentiation observed in vitro. However, the small number of HR+ cells shown to be present in the pregnant mammary gland (Van Keymeulen et al., 2017) could in part explain the moderate phenotype observed in pregnant glands.
Lama5 deficiency also affected the ductal epithelial organization in puberty and pregnancy. Our results imply that the Lama5-deficient luminal cells proliferate, albeit less than cells in control glands, but fail to maintain normal contacts with the BM. This may result in the observed accumulation of cells filling the gland lumen. Luminal-ECM interactions are necessary for alveolar differentiation and, thus, functionalization and milk secretion (Inman et al., 2015; Streuli et al., 1995; Roskelley et al., 1994). Indeed, mammary epithelia lacking the adhesion receptor β1-integrin exhibit defective apicobasal polarization, surplus luminal cells within the ducts and impaired alveolar cell function (Naylor et al., 2005; Akhtar and Streuli, 2013). We noted that apicobasal polarity is affected also in the Lama5-deficient luminal epithelial cells, suggesting it may contribute to the surplus of luminal cells during puberty. Cell polarity was better conserved in the Lama5-deficient glands during pregnancy, suggesting the most aberrant glands may not be able to contribute to pregnancy-induced gland remodeling. However, addressing the exact order of events downstream of Lama5 deletion and loss of tissue architecture during puberty requires further experiments.
Finally, how adhesion to Lama5-encoded Lama5 regulates Wnt4 signaling, and why particularly HR+ cells are influenced by Lama5 loss remain as open questions. As HR+ cells express more Lama5 than HR− luminal cells (Tabula Muris Consortium, 2018), and HR+ are responsive to systemic cues, it is attractive to postulate that they require Lama5-mediated contacts with the BM in order to sense the systemic hormonal signals. HR+ luminal cells also express, for example, Integrin b4 (Itgb4) capable of adhering to Lama5 (Yurchenco, 2011), whereas Itgb4 expression in HR− cells is lower (Tabula Muris Consortium, 2018). The slender projections of luminal cells that we noted reaching past basal cells may therefore represent an HR+ cell-specific feature of the luminal layer.
Taken together, our data suggest that Lama5 loss in the mammary epithelium results in adhesion and organization defects of luminal MECs, leads to a deficient HR+ luminal MEC pool with insufficient capability to sustain luminal-basal interactions, and gland remodeling in puberty and pregnancy. Further experiments are required to address whether additional functions of Lama5 partake mammary gland biology in vivo and whether the mechanisms described here contribute to pathologies with distorted tissue organization, such as cancer.
MATERIALS AND METHODS
Animals
Animal studies were approved by the National Animal Ethics Committee of Finland and conducted in the Laboratory Animal Center of the University of Helsinki under institutional guidelines or approved by the ethics committee of the Board of Agriculture, Experimental Animal Authority, Stockholm, Sweden. The Lama5 fl mouse was a kind gift from Dr Jeffrey Miner (Nguyen et al., 2005). Lgr6-EGFP-IRES-CreERT2, Rosa26-tdTomato, K8-CreERT2 and R26-mTmG mouse lines have been described previously (Muzumdar et al., 2007; Van Keymeulen et al., 2011; Nguyen et al., 2005; Blaas et al., 2016). Genotyping of the animals was performed using primers described previously. Excised Lama5 allele was detected using the following primers: Fwd, 5′-ACCTGGCTTTGACGGTCCT-3′; Rev, 5′-GTTGAAGCCAAAGCGTACAGCG-3′. All animals used were females between 6 and 20 weeks of age at the time of sacrifice. For Lgr6-Cre induction, a single dose of 1 mg tamoxifen (Sigma-Aldrich) in 50 µl sunflower oil (Sigma-Aldrich) was injected intraperitoneally in 2-week-old mice. For K8-CreERT2 induction, a single dose of 1.5-2 mg tamoxifen in corn oil (Sigma-Aldrich) was injected intraperitoneally in 3-week-old mice, and 5 mg of tamoxifen in 8-week-old mice.
RNA in situ hybridization
Gene expression analysis by RNA ISH was performed using an RNAscope 2.5 HD Reagent Kit-Brown or an RNAscope Multiplex Kit (Advanced Cell Diagnostics Srl) according to manufacturer's instructions. Paraffin-embedded sections (5 µm) were deparaffinized, rehydrated and pretreated with RNAscope Target Retrieval Reagents. A barrier was created around each tissue section on the slides, dried overnight and used the following day. Pretreated samples were hybridized with either laminin α1, α3, α4 or α5 probes (custom made by Advanced Cell Diagnostics) for 2 h at 40°C. Thereafter, signal amplification hybridization was performed, followed by detection with DAB (3,3′-diaminobenzidine) and counterstained with Hematoxylin. Alternatively, when a Multiplex Fluorescent assay was used, after signal amplification hybridization, the slides were immunostained with primary antibodies (K8, 1:1000, TROMA-1, Developmental Studies Hybridoma Bank; K14, 1:2000, BioLegend, PRB 155P) for 1 h. Thereafter, samples were washed three times with PBS, 5 min each, and incubated with secondary antibodies in 10% normal goat serum in immunofluorescence buffer for 1-2 h. Slides were next washed three times, followed by counterstaining of nuclei with DAPI and mounting. Samples were imaged using a Zeiss Axio Imager.M2 and AxioCam HRc camera using Plan Neofluar 20× (NA 0.5) or Plan Apochrom 63× (NA 1.4, oil) objectives and Zen software, or using a Nikon Inverted widefield system with a 60× Plan apochromat objective (NA 1.2, water) and Andor Xyla 4.2+ camera, and NIS elements software (Nikon).
Whole-mount stainings
Mammary gland tissue samples for whole-mount staining were fixed in 4% paraformaldehyde (PFA) overnight. For whole-mount staining, #4 inguinal mammary glands were stained for several hours in carmine-alumn staining solution (2% w/v Carmine, Sigma-Aldrich; 5% w/v aluminum potassium sulfate, Sigma-Aldrich). After the desired color had developed, glands were mounted to glass coverslips and imaged.
For whole-mount imaging and immunostaining of Lgr6-CreERT2;Rosa26-tdTomato or Lama5 fl;K8-CreERT2;Rosa26-mTmG mammary glands, the dissected mammary glands were pre-fixed in 4% PFA for 30–60 min at room temperature to preserve native fluorescence. For whole-mount immunostaining, the tissue was permeabilized with two washes of PBS0.5T (PBS plus 0.5% v/v Triton X-100) for 1 h, and the samples were incubated overnight at 4°C with primary antibody against keratin 14 (1:500, BioLegend PRB 155P) diluted in PBD0.2T [PBS, 1% bovine serum albumin (BSA), 1% v/v DMSO and 0.2% v/v Triton X-100]. The next day, they were washed four times with PBD0.2T for 1 h. Tissues were incubated with secondary antibodies plus DAPI (Roche) at 4°C overnight. After four washes with PBD0.2T for 1 h, tissues were optically cleared in 80% glycerol in PBS at room temperature. The stained mammary glands were mounted between two cover slips and sealed with silicone. Imaging of the whole mounts was performed using a Zeiss LSM710 confocal microscope. A dry lens (5×, EC Plan Neofluar, NA 0.16) or a water immersion lens (20× W Plan Apochromat NA 1.0 or 40× C-Apochromat, NA 1.2) was used to record optical sections at 512×512, 1024×1024 or 2048×2048 pixels, and data were processed using ZEN software (Zeiss). Images were converted to RGB images with ImageJ software.
Morphometric analysis of the mammary gland
For quantification of the number of terminal end buds (TEB) and ductal ends in the mammary glands and the length of the epithelium, #4 mammary glands stained with carmine-alumn whole mount were imaged using a Leica S9i stereomicroscope equipped with an integrated 10 MP camera. The amount of TEBs and ductal ends was quantified from the images based on morphology. The length of the epithelium was measured from the same images using ImageJ with its line tool. The length of the whole epithelial network was recorded.
For quantification of ductal density, images of Hematoxylin and Eosin (H&E)-stained pregnant mammary glands were acquired using a Leica Microscope DM2000 with a Leica MC 190 HD camera with an N Plan 10× objective. The percentage of ductal density was quantified from 10× images by applying a series of Gaussian filtering and thresholding for brightness to separate stained tissue area from the lumen and surrounding adipose tissue using a custom Java script that can be obtained from the authors. The ratio of the red channel to the other channels, balanced with the brightness, was used to further separate Hematoxylin-stained nucleus-rich ductal tissue from Eosin-stained connective tissue, and produce a binary image. A size-based filter was applied to remove single nuclei and debris. The remaining area representing the ducts was calculated for each tissue slide.
Tissue immunostainings
Mammary gland tissue samples for immunostaining were fixed in 4% PFA overnight and embedded in paraffin for sections. For H&E stainings, 5 µm sections were used. Stainings were performed in the Finnish Center for Laboratory Animal Pathology (FCLAP) at the University of Helsinki. Imaging for representative images was performed using a Pannoramic 250 Flash II high-throughput brightfield scanner (3DHISTECH) with a 20× (NA 0.8) objective and Pannoramic Viewer software.
For immunofluorescence stainings, 6-8 µm sections were deparaffinized and rehydrated, and antigen retrieval was performed with 1 mM EDTA for 5-8 min in a microwave oven, followed by 20 min incubation at room temperature. Thereafter, non-specific binding sites were blocked with 10% normal goat serum (NGS, Gibco) in PBS for 30 min at room temperature. Next, samples were incubated with primary antibodies (pan-laminin 1:500, Abcam, ab11575; K14, 1:500, BioLegend, PRB 155P; K8, 1:1000, TROMA-1, Developmental Studies Hybridoma Bank; Ki-67, 1:500, Abcam, ab15580; E-cadherin 36/E, 1:500, BD 610181; ZO-1, 1:500, Thermo Fisher Scientific, 61-7300; and MUC, 1:500, Abcam, ab15481) overnight at 4°C. The next day, samples were washed three times with PBS, 5 min each, and incubated with secondary antibodies in 10% NGS in PBS for 1-2 h. Thereafter, samples were washed three times with PBS, 5 min each, followed by counterstaining of nuclei with Hoechst 33342 (Sigma-Aldrich). All slides were mounted using Immu-Mount (Thermo Scientific) mounting reagent. Images were acquired using a Leica TCS SP8 STED 3X CW 3D confocal with an HC PL APO 63× water (NA 1.20) motCORR CS2 objective and LAS AF software, or a Leica SP8 upright confocal microscope with an HC PL APO 63× glycerol (NA 1.3) objective and LAS AF software. The same settings for laser power and detector gain were maintained within one experiment.
Quantification of fluorescence in mammary gland whole mounts
Mammary gland whole mounts of Lama5+/+;K8-CreERT2;Rosa26-mTmG and Lama5fl/fl;K8-CreERT2;Rosa26-mTmG were imaged for 3-5 proximal and distal areas of the mammary epithelium. Z-stacks were converted to Z projections using the sum slice method. Regions of interest (ROI) were drawn to encompass the epithelium and three spots were drawn to quantify the background fluorescence. The integrated density of green and red channels of ROIs was measured, and mean background fluorescence was subtracted from the epithelial ROIs. From these values, the green to red fluorescent ratio was calculated for both proximal and distal areas. Data are shown as mean per mammary gland.
FACS analysis and sorting of primary MMECs
Single-cell suspension of isolated primary mouse mammary epithelial cells (MMECs) was resuspended in 0.2% BSA in Dulbecco's PBS, and the cells were incubated with the following primary antibodies for analysis: CD29-FITC (Miltenyi Biotech, 130-102-975); CD24-APC (M1/69, BioLegend, 101814); CD31-BV421 (MEC13.3, BD Biosciences, 562939); CD45-BV421 (30 F11, BD Biosciences, 563890); Ter119-BV421 (BD Biosciences, 563998); Sca1-BV711 (D7, BioLegend, 108131); and CD49b-PE (HMα2, BioLegend, 103506). All antibodies were diluted 1:500 at 4°C for 30 min. Cells were washed with PBS and resuspended in 0.2% BSA in Dulbecco's PBS with 1:500 Sytox Blue (Life Technologies, Thermo Fisher Scientific) to exclude dead cells. Sorting was performed using a FACSAria Fusion cell sorter (Becton Dickinson). FlowJo V10 was used for post-analysis of sorted cells.
RNA isolation and qPCR
RNA isolation was performed using an RNeasy isolation kit (Qiagen) combined with On-Column DNase digestion (Qiagen), or using TRIzol (Life Technologies, Thermo Fisher Scientific; from FACS-sorted cells) according to manufacturer's instructions. TRIzol-isolated RNA was treated with DNase I (Thermo Fisher Scientific) according to manufacturer's protocol. cDNA synthesis was performed using a Revert Aid cDNA synthesis kit (Thermo Fisher Scientific) starting with 250 or 500 ng of RNA. Quantitative PCR reaction was performed using Power SYBR green master mix (Applied Biosystems) and a Bio-Rad CFX384 Touch Real-Time PCR detection system. Data were analyzed using a Bio-Rad CFX Manager program. Relative mRNA amounts were assayed by comparing PCR cycles to GAPDH using the ddCT method and normalizing to control samples. The following primers were used: mGAPDH fwd, 5′-AAGGTCGGAGTCAACGGATT-3′; mGAPDH rev, 5′-TTGATGACAAGCTTCCCGTT-3′; mLama5 WT fwd, 5′-GGGTGGAGTTACTGAGTGCC-3′; mLama5 WT rev, 5′-AGCTACGGCAGCCAAAGTAG-3′; mLama5 Common fwd, 5′-GCCAGCAAGGCGATCCAAG-3′; mLama5 Common rev, 5′-CCTGGTCTACGCTGAACACA-3′; mESR1 fwd, 5′-CCTCCCGCCTTCTACAGGT-3′; mESR1 rev, 5′-CACACGGCACAGTAGCGAG-3′ (Harvard Primer Bank ID, 6679695a1); mPR fwd, 5′-CTCGGACGTGTCGTCTGTAG-3′; mPR rev, 5′-CCTGTCTTTCCGTCTGGGAG-3′ (Harvard Primer Bank ID, 112363097c1); mRANKL fwd, 5′-CGCTCTGTTCCTGTACTTTCG-3′; mRANKL rev, 5′-GAGTCCTGCAAATCTGCGTT-3′ (Harvard Primer Bank ID, 114842414c2); mWnt4 fwd, 5′-GTACCTGGCCAAGCTGTCAT-3′; mWnt4 rev, 5′-CTTGTCACTGCAAAGGCCAC-3′; mRspo1 fwd, 5-TTCTGCTGGAGAGGAACGAC-3′; mRspo1 rev, 5′-GCCTCACAGTGCTCGATCTT-3′; mLgr5 fwd, 5′-ACCCGCCAGTCTCCTACATC-3′; mLgr5 rev, 5′-GCATCTAGGCGCAGGGATTG-3′; mSox9 fwd, 5′-GAGCCGGATCTGAAGAGGGA-3′; mSox9 rev, 5′-GCTTGACGTGTGGCTTGTTC-3′; mCtnnb1 fwd, 5′-TGACTAGGGCTCAGAGGGTC-3′; and mCtnnb1 rev, 5′-TCAGCTCAGGAATTGCACGT-3′.
Primary cell isolation and culture
Primary MMECs were isolated from 10-16-week-old virgin female mice, unless otherwise stated. Mammary glands #3-#5 were dissected, and the lymph node in #4 glands was removed and the glands were finely chopped. Tissue was incubated with 0.01 mg of Collagenase A (Sigma-Aldrich) per 1 g of tissue in Dulbecco's modified Eagle medium (DMEM)/F12 growth medium (Life Technologies) containing 2.5% fetal calf serum (FCS), 5 µg/ml insulin, 50 µg/ml gentamicin and 2 mM glutamine with gentle shaking (120 rpm in an environmental shaker) at 37°C for 2 h. The resulting cell suspension was then first centrifuged at 400 g for 10 min and consecutively pulse-centrifuged 3-5 times 400 g to yield preparation free of cells other than MMEC organoids. Next, organoids were trypsinized with 0.05% Trypsin-EDTA (Difco, J.T. Baker) for 5-10 min to obtain smaller organoid units and drained through a 70 µm cell strainer (Becton Dickinson), and resuspended in MMEC growth medium [DMEM/F12 medium containing 5 µg/ml insulin, 1 µg/ml hydrocortisone, 10 ng/ml mouse EGF, 2 mM glutamine, 50 µg/ml gentamycin and penicillin and streptomycin (all from Sigma-Aldrich) supplemented with 10% FCS (Gibco)].
Lentiviral virus production and infection
Lentiviruses were produced in 293fT cells (Thermo Fisher Scientific; regularly tested for mycoplasma contamination) grown in DMEM (Sigma-Aldrich) supplemented with 10% FCS (Gibco), Penicillin/Streptomycin (Orion/Sigma-Aldrich) and 2 mM glutamine (Sigma-Aldrich). Transfections of transfer vector (pLentiCRISPRv2 gSCRA, pLentiCRISPRv2 gLAMA5) and packaging plasmids (CMV-VSVg and Delta8.9) were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. To concentrate lentiviral particles, medium from transfected cells was harvested 72 h post-transfection and concentrated by ultracentrifugation at 87,000 g for 120 min (Sorvall Discovery 90SE Ultracentrifuge). The viral pellet was then resuspended in PBS at a ∼300-fold concentration and left to dissolve at 4°C for 18 h. Viral titer was determined using a p24 ELISA test measuring viral capsid protein p24 (Perkin Elmer, performed in the Biomedicum Functional Genomics Unit). Primary MMECs were infected on 24-well low adhesion plates overnight using multiplicity of infection 5 in an 800 µl volume and washed with growth medium the next day.
Plasmid construction for CRISPR guides
CRISPR guides were designed to target LAMA5 using the CRISPR Design tool (http://crispr.mit.edu). The following target sequences were used: gLAMA5, 5′-CCATCGATGGCACGGAGCGC-3′, and gSCRA, 5′-CTAAAACTGCGGATACAATC-3′. Oligos with target sequences were designed according to instructions published previously (Shalem et al., 2014), annealed and cloned into the pLentiCRISPRv2 vector.
Three-dimensional organotypic cell culture assays
Three-dimensional organotypic culture was performed in growth factor-reduced BM from Engelbreth–Holm–Swarm mouse sarcoma (Matrigel, Becton Dickinson), which was prepared according to the manufacturer's instructions. Isolated primary MMECs grown on low adhesion plates were trypsinized with 0.05% Trypsin-EDTA for 5-10 min, centrifuged and suspended with liquid Matrigel, and plated onto 8-well chamber slides at ∼1500 cells/well. Organoids were grown in DMEM/F12 supplemented with ITS medium supplement (Sigma-Aldrich), penicillin and streptomycin and 2.5 nM FGF-2 (Sigma-Aldrich). Wnt3a and Wnt4 (both R&D Biosystems) were used in 100 ng/ml, Rspo1 (R&D Biosystems) in 500 ng/ml, and porcupine inhibitor IWP-2 (Sigma-Aldrich) in 2 µM, and were added on the starting day of the cultures. Medium was refreshed every 3-4 days. For lactogenic differentiation, organoids were first grown for 6 days in the DMEM/F12 medium supplemented with 2.5 nM FGF-2, ITS and penicillin and streptomycin, and thereafter, 6 days in the same culture medium additionally supplemented with 1 μg/ml mouse prolactin (Sigma-Aldrich) and 1 μg/ml hydrocortisone (Sigma-Aldrich).
Three-dimensional organoid immunofluorescence staining and imaging
Three-dimensional organoids were fixed with 2% PFA for 20 min at room temperature and thereafter washed with PBS. Organoids were permeabilized with 0.25% Triton X-100 in PBS for 10 min at 4°C, and thereafter washed with PBS. The non-specific binding sites were blocked with 10% normal goat serum (Gibco) for 1-2 h. The primary antibodies (keratin 14, Covance, 1:300; Keratin 8, TROMA-1, 1:300; Ki-67, Abcam, 1:500) were incubated in the blocking solution overnight at 4°C. Following the incubation, structures were washed three times with PBS, 15 min each wash, and then incubated with appropriate Alexa Fluor secondary antibody diluted in blocking solution. After 40-50 min incubation at room temperature, the structures were washed with IF buffer as before and the nuclei were counterstained with Hoechst 33342. All slides were mounted using Immu-Mount (Thermo Scientific) mounting reagent. Images were acquired using a Leica TCS SP8 STED 3X CW 3D confocal microscope with an HC PL APO 63× water (NA 1.20) motCORR CS2 objective and LAS AF software, or a Leica SP8 Upright confocal microscope with an HC PL APO 63× glycerol (NA 1.3) objective.
Quantification and statistical analysis
Data are presented as mean±s.d. from at least three independent experiments or three individual animals quantified, unless otherwise stated in the figure legend. An unpaired two-tailed Student's t-test was used to compare two groups, unless otherwise stated in the figure legend. Statistical tests were performed using GraphPad Prism 8.
Supplementary Material
Acknowledgements
We thank J. Bärlund and M. Simula for excellent technical assistance, and members of the Katajisto laboratory for comments and discussion. The Light Microscopy Unit at the Institute of Biotechnology, University of Helsinki and Live Cell Imaging Unit/Nikon Center of Excellence at the Department of Biosciences and Nutrition, Karolinska Institutet, are acknowledged for assistance with microscopy.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.I.E., L.B., P.K.; Methodology: J.I.E., A.R., L.B., H.C., N.P., J.D.; Investigation: J.I.E., A.R., L.B., H.C., N.P.; Resources: A.R., S.I., M.P.; Writing - original draft: J.I.E., P.K.; Writing - review & editing: J.I.E., P.K.; Visualization: J.I.E.; Funding acquisition: P.K.
Funding
The study was funded by grants from the European Research Council (677809 to P.K.), the Suomen Akatemia (266869 and 304591 to P.K.), the Knut och Alice Wallenbergs Stiftelse (KAW 2014.0207), the Center for Innovative Medicine, Cancerfonden, the Sigrid Juséliuksen Säätiö, and the Finnish Cancer Society. Deposited in PMC for immediate release.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.199281
References
- Adriance, M. C., Inman, J. L., Petersen, O. W. and Bissell, M. J. (2005). Myoepithelial cells: good fences make good neighbors. Breast Cancer Res. 7, 190-197. 10.1186/bcr1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, M. and Ffrench-Constant, C. (2016). Extracellular matrix regulation of stem cell behavior. Curr Stem Cell Rep 2, 197-206. 10.1007/s40778-016-0056-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar, N. and Streuli, C. H. (2013). An integrin-ILK-microtubule network orients cell polarity and lumen formation in glandular epithelium. Nat. Cell Biol. 15, 17-27. 10.1038/ncb2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin-Labat, M.-L., Vaillant, F., Sheridan, J. M., Pal, B., Wu, D., Simpson, E. R., Yasuda, H., Smyth, G. K., Martin, T. J., Lindeman, G. J.et al. (2010). Control of mammary stem cell function by steroid hormone signalling. Nature 465, 798-802. 10.1038/nature09027 [DOI] [PubMed] [Google Scholar]
- Bach, K., Pensa, S., Grzelak, M., Hadfield, J., Adams, D. J., Marioni, J. C. and Khaled, W. T. (2017). Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing. Nat. Commun. 8, 2128. 10.1038/s41467-017-02001-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikkavilli, R. K. and Malbon, C. C. (2010). Dishevelled-KSRP complex regulates Wnt signaling through post-transcriptional stabilization of β-catenin mRNA. J. Cell Sci. 123, 1352-1362. 10.1242/jcs.056176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaas, L., Pucci, F., Messal, H. A., Andersson, A. B., Josue Ruiz, E., Gerling, M., Douagi, I., Spencer-Dene, B., Musch, A., Mitter, R.et al. (2016). Lgr6 labels a rare population of mammary gland progenitor cells that are able to originate luminal mammary tumours. Nat. Cell Biol. 18, 1346-1356. 10.1038/ncb3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blache, P., van de Wetering, M., Duluc, I., Domon, C., Berta, P., Freund, J.-N., Clevers, H. and Jay, P. (2004). SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J. Cell Biol. 166, 37-47. 10.1083/jcb.200311021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisken, C., Park, S., Vass, T., Lydon, J. P., O'malley, B. W. and Weinberg, R. A. (1998). A paracrine role for the epithelial progesterone receptor in mammary gland development. Proc. Natl. Acad. Sci. USA 95, 5076-5081. 10.1073/pnas.95.9.5076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisken, C., Heineman, A., Chavarria, T., Elenbaas, B., Tan, J., Dey, S. K., Mcmahon, J. A., Mcmahon, A. P. and Weinberg, R. A. (2000). Essential function of Wnt-4 in mammary gland development downstream of progesterone signaling. Genes Dev. 14, 650-654. [PMC free article] [PubMed] [Google Scholar]
- Cai, C., Yu, Q. C., Jiang, W., Liu, W., Song, W., Yu, H., Zhang, L., Yang, Y. and Zeng, Y. A. (2014). R-spondin1 is a novel hormone mediator for mammary stem cell self-renewal. Genes Dev. 28, 2205-2218. 10.1101/gad.245142.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald, A. J., Brenot, A., Duong, M., Chan, B. S. and Werb, Z. (2008). Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev. Cell 14, 570-581. 10.1016/j.devcel.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fata, J. E., Kong, Y.-Y., Li, J., Sasaki, T., Irie-Sasaki, J., Moorehead, R. A., Elliott, R., Scully, S., Voura, E. B., Lacey, D. L.et al. (2000). The osteoclast differentiation factor osteoprotegerin-ligand is essential for mammary gland development. Cell 103, 41-50. 10.1016/S0092-8674(00)00103-3 [DOI] [PubMed] [Google Scholar]
- Feng, Y., Manka, D., Wagner, K.-U. and Khan, S. A. (2007). Estrogen receptor-alpha expression in the mammary epithelium is required for ductal and alveolar morphogenesis in mice. Proc. Natl. Acad. Sci. USA 104, 14718-14723. 10.1073/pnas.0706933104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard, E. T., Hill, R. C., Barrett, A., Betts, C., Guo, Q., Maller, O., Borges, V. F., Hansen, K. C. and Schedin, P. (2016). Quantitative extracellular matrix proteomics to study mammary and liver tissue microenvironments. Int. J. Biochem. Cell Biol. 81, 223-232. 10.1016/j.biocel.2016.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson, T., Rønnov-Jessen, L., Villadsen, R., Rank, F., Bissell, M. J. and Petersen, O. W. (2002). Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement membrane deposition. J. Cell Sci. 115, 39-50. 10.1242/jcs.115.1.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenester, E. and Yurchenco, P. D. (2013). Laminins in basement membrane assembly. Cell Adh. Migr. 7, 56-63. 10.4161/cam.21831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman, J. L., Robertson, C., Mott, J. D. and Bissell, M. J. (2015). Mammary gland development: cell fate specification, stem cells and the microenvironment. Development 142, 1028-1042. 10.1242/dev.087643 [DOI] [PubMed] [Google Scholar]
- Labarge, M. A., Nelson, C. M., Villadsen, R., Fridriksdottir, A., Ruth, J. R., Stampfer, M. R., Petersen, O. W. and Bissell, M. J. (2009). Human mammary progenitor cell fate decisions are products of interactions with combinatorial microenvironments. Integr. Biol. 1, 70-79. 10.1039/B816472J [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafkas, D., Rodilla, V., Huyghe, M., Mourao, L., Kiaris, H. and Fre, S. (2013). Notch3 marks clonogenic mammary luminal progenitor cells in vivo. J. Cell Biol. 203, 47-56. 10.1083/jcb.201307046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N., Zhang, Y., Naylor, M. J., Schatzmann, F., Maurer, F., Wintermantel, T., Schuetz, G., Mueller, U., Streuli, C. H. and Hynes, N. E. (2005). Beta1 integrins regulate mammary gland proliferation and maintain the integrity of mammary alveoli. EMBO J. 24, 1942-1953. 10.1038/sj.emboj.7600674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, E., Wu, D., Pal, B., Bouras, T., Asselin-Labat, M.-L., Vaillant, F., Yagita, H., Lindeman, G. J., Smyth, G. K. and Visvader, J. E. (2010). Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 12, R21. 10.1186/bcr2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias, H. and Hinck, L. (2012). Mammary gland development. Wiley Interdiscip. Rev. Dev. Biol. 1, 533-557. 10.1002/wdev.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallepell, S., Krust, A., Chambon, P. and Brisken, C. (2006). Paracrine signaling through the epithelial estrogen receptor α is required for proliferation and morphogenesis in the mammary gland. Proc. Natl. Acad. Sci. USA 103, 2196-2201. 10.1073/pnas.0510974103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller, O., Martinson, H. and Schedin, P. (2010). Extracellular matrix composition reveals complex and dynamic stromal-epithelial interactions in the mammary gland. J. Mammary Gland Biol. Neoplasia 15, 301-318. 10.1007/s10911-010-9189-6 [DOI] [PubMed] [Google Scholar]
- Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. and Luo, L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593-605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Naylor, M. J., Li, N., Cheung, J., Lowe, E. T., Lambert, E., Marlow, R., Wang, P., Schatzmann, F., Wintermantel, T., Schüetz, G.et al. (2005). Ablation of beta1 integrin in mammary epithelium reveals a key role for integrin in glandular morphogenesis and differentiation. J. Cell Biol. 171, 717-728. 10.1083/jcb.200503144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, N. M., Kelley, D. G., Schlueter, J. A., Meyer, M. J., Senior, R. M. and Miner, J. H. (2005). Epithelial laminin alpha5 is necessary for distal epithelial cell maturation, VEGF production, and alveolization in the developing murine lung. Dev. Biol. 282, 111-125. 10.1016/j.ydbio.2005.02.031 [DOI] [PubMed] [Google Scholar]
- Paine, I. S. and Lewis, M. T. (2017). The terminal end bud: the little engine that could. J. Mammary Gland Biol. Neoplasia 22, 93-108. 10.1007/s10911-017-9372-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince, J. M., Klinowska, T. C. M., Marshman, E., Lowe, E. T., Mayer, U., Miner, J., Aberdam, D., Vestweber, D., Gusterson, B. and Streuli, C. H. (2002). Cell-matrix interactions during development and apoptosis of the mouse mammary gland in vivo. Dev. Dyn. 223, 497-516. 10.1002/dvdy.10070 [DOI] [PubMed] [Google Scholar]
- Rajaram, R. D., Buric, D., Caikovski, M., Ayyanan, A., Rougemont, J., Shan, J., Vainio, S. J., Yalcin-Ozuysal, O. and Brisken, C. (2015). Progesterone and Wnt4 control mammary stem cells via myoepithelial crosstalk. EMBO J. 34, 641-652. 10.15252/embj.201490434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagnoli, M., Bresson, L., Di-Cicco, A., Perez-Lanzon, M., Legoix, P., Baulande, S., de la Grange, P., de Arcangelis, A., Georges-Labouesse, E., Sonnenberg, A.et al. (2020). Laminin-binding integrins are essential for the maintenance of functional mammary secretory epithelium in lactation. Development 147: dev181552. 10.1242/dev.181552 [DOI] [PubMed] [Google Scholar]
- Roskelley, C. D., Desprez, P. Y. and Bissell, M. J. (1994). Extracellular matrix-dependent tissue-specific gene expression in mammary epithelial cells requires both physical and biochemical signal transduction. Proc. Natl. Acad. Sci. USA 91, 12378-12382. 10.1073/pnas.91.26.12378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheele, C. L. G. J., Hannezo, E., Muraro, M. J., Zomer, A., Langedijk, N. S. M., van Oudenaarden, A., Simons, B. D. and VAN Rheenen, J. (2017). Identity and dynamics of mammary stem cells during branching morphogenesis. Nature 542, 313-317. 10.1038/nature21046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuijers, J. and Clevers, H. (2012). Adult mammalian stem cells: the role of Wnt, Lgr5 and R-spondins. EMBO J. 31, 2685-2696. 10.1038/emboj.2012.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem, O., Sanjana, N. E., Hartenian, E., Shi, X., Scott, D. A., Mikkelson, T. S., Heckl, D., Ebert, B. L., Root, D. E., Doench, J. G.et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84-87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata, M., Teschendorff, A., Sharp, G., Novcic, N., Russell, I. A., Avril, S., Prater, M., Eirew, P., Caldas, C., Watson, C. J.et al. (2012). Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 14, R134. 10.1186/bcr3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata, M., VAN Amerongen, R., Zeeman, A. L., Giraddi, R. R. and Stingl, J. (2014). The influence of tamoxifen on normal mouse mammary gland homeostasis. Breast Cancer Res. 16, 411. 10.1186/s13058-014-0411-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman, K. E., Kendrick, H., Robertson, D., Isacke, C. M., Ashworth, A. and Smalley, M. J. (2007). Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J. Cell Biol. 176, 19-26. 10.1083/jcb.200604065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, G. H. and Medina, D. (1988). A morphologically distinct candidate for an epithelial stem cell in mouse mammary gland. J. Cell Sci. 90, 173-183. 10.1242/jcs.90.1.173 [DOI] [PubMed] [Google Scholar]
- Streuli, C. H., Edwards, G. M., Delcommenne, M., Whitelaw, C. B. A., Burdon, T. G., Schindler, C. and Watson, C. J. (1995). Stat5 as a target for regulation by extracellular matrix. J. Biol. Chem. 270, 21639-21644. 10.1074/jbc.270.37.21639 [DOI] [PubMed] [Google Scholar]
- Sumbal, J., Chiche, A., Charifou, E., Koledova, Z. and Li, H. (2020). Primary Mammary Organoid Model of Lactation and Involution. Front. Cell Dev. Biol. 8, 68. 10.3389/fcell.2020.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabula Muris Consortium, Overall coordination, Logistical coordination, Organ collection and processing, Library preparation and sequencing, Computational data analysis, Cell type annotation, Writing group, Supplemental text writing group, Principal investigators. (2018). Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367-372. 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos, T., Rojo, L. J., Echeverria, P. and Brisken, C. (2012). ER and PR signaling nodes during mammary gland development. Breast Cancer Res. 14, 210. 10.1186/bcr3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Keymeulen, A., Rocha, A. S., Ousset, M., Beck, B., Bouvencourt, G., Rock, J., Sharma, N., Dekoninck, S. and Blanpain, C. (2011). Distinct stem cells contribute to mammary gland development and maintenance. Nature 479, 189-193. 10.1038/nature10573 [DOI] [PubMed] [Google Scholar]
- van Keymeulen, A., Fioramonti, M., Centonze, A., Bouvencourt, G., Achouri, Y. and Blanpain, C. (2017). Lineage-restricted mammary stem cells sustain the development, homeostasis, and regeneration of the estrogen receptor positive lineage. Cell Rep. 20, 1525-1532. 10.1016/j.celrep.2017.07.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vooijs, M., Jonkers, J. and Berns, A. (2001). A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2, 292-297. 10.1093/embo-reports/kve064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, M. R., Amante, J. J., Li, J., Liu, H., Zhu, L. J., Feltri, M. L., Goel, H. L. and Mercurio, A. M. (2020). Alveolar progenitor cells in the mammary gland are dependent on the beta4 integrin. Dev. Biol. 457, 13-19. 10.1016/j.ydbio.2019.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, V. M., Petersen, O. W., Wang, F., Larabell, C. A., Briand, P., Damsky, C. and Bissell, M. J. (1997). Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J. Cell Biol. 137, 231-245. 10.1083/jcb.137.1.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenco, P. D. (2011). Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb Perspect Biol 3, a004911. 10.1101/cshperspect.a004911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Y. A. and Nusse, R. (2010). Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell 6, 568-577. 10.1016/j.stem.2010.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.