ABSTRACT
Axial elongation of the neural tube is crucial during mammalian embryogenesis for anterior-posterior body axis establishment and subsequent spinal cord development, but these processes cannot be interrogated directly in humans as they occur post-implantation. Here, we report an organoid model of neural tube extension derived from human pluripotent stem cell (hPSC) aggregates that have been caudalized with Wnt agonism, enabling them to recapitulate aspects of the morphological and temporal gene expression patterns of neural tube development. Elongating organoids consist largely of neuroepithelial compartments and contain TBXT+SOX2+ neuro-mesodermal progenitors in addition to PAX6+NES+ neural progenitors. A critical threshold of Wnt agonism stimulated singular axial extensions while maintaining multiple cell lineages, such that organoids displayed regionalized anterior-to-posterior HOX gene expression with hindbrain (HOXB1) regions spatially distinct from brachial (HOXC6) and thoracic (HOXB9) regions. CRISPR interference-mediated silencing of TBXT, a Wnt pathway target, increased neuroepithelial compartmentalization, abrogated HOX expression and disrupted uniaxial elongation. Together, these results demonstrate the potent capacity of caudalized hPSC organoids to undergo axial elongation in a manner that can be used to dissect the cellular organization and patterning decisions that dictate early human nervous system development.
KEY WORDS: Stem cell biology, Neural tube development, Organoid models
Summary: An organoid model of neural tube extension can be used to dissect the cellular organization and patterning decisions that dictate early human nervous system development.
INTRODUCTION
The development of multicellular organisms depends on the organized emergence of tissues and organs, which in turn relies on the specialization and regionalization of numerous cell types. In human spinal cord development, multiple neural subtypes that are crucial to motor control arise from distinct progenitor domains in the developing neural tube (Tanabe and Jessell, 1996). The neural tube itself arises in tandem with the initial establishment of the anterior-posterior embryonic axis in a process known as axial elongation (Steventon et al., 2016). Defects in early neural tube morphogenesis result in severe congenital abnormalities, such as spina bifida. Furthermore, defects in spinal interneuron development can ultimately impact motor control and locomotion (Azim et al., 2014; Zhong et al., 2010; Crone et al., 2008), highlighting the clinical importance of understanding human axial elongation and neural tube development. However, despite the advances gleaned from vertebrate models such as mouse and chicken, many of the molecular mechanisms and cellular behaviors that regulate spinal cord development in humans remain unknown due to the difficulty of studying these dynamic post-implantation processes.
Advances over the past decade have demonstrated that, unlike anterior neural populations, the posterior neural tube is partially generated from a unique pool of axial stem cells called neuromesodermal progenitors (NMPs) (Henrique et al., 2015). NMPs are bipotent progenitors that reside at the node-streak border within the caudal lateral epiblast and later in the chordoneural hinge of the tail bud (Wilson et al., 2009; Cambray and Wilson, 2002; Wymeersch et al., 2016; Beddington and Robertson, 1999). The lack of a model for human posterior neural tube development has hindered our understanding of how NMPs regulate and coordinate the emergence of the posterior spinal cord. However, studies in model systems and 2D differentiation models implicate Wnt and FGF as crucial posteriorizing factors that regulate this process (Briscoe and Ericson, 1999; Ericson et al., 1997; Liem et al., 1995; Diez del Corral and Storey, 2004)
Differentiation protocols based on embryonic development have enabled the robust production of neuronal subtypes in vitro (Butts et al., 2017; Wichterle et al., 2002; Li et al., 2005; Meinhardt et al., 2014). In parallel, high-throughput organoid platforms have been developed that are capable of recapitulating structures reminiscent of early embryonic tissues (Lancaster and Knoblich, 2014). More specifically, protocols have capitalized on NMP differentiation (Gouti et al., 2014) to generate neuromuscular junctions (Faustino Martins et al., 2020), mimic spinal cord dorsal-ventral patterning (Zheng et al., 2019), or recapitulate the complex spatiotemporal expression profiles of gastrulation (in structures termed ‘gastruloids’) (Beccari et al., 2018; van den Brink, 2020; Moris et al., 2020; Veenvliet et al., 2020). However, existing organoid models do not perfectly recapitulate the spatiotemporal dynamics of gene expression and differentiation that occur within embryos. Moreover, reproducibility within differentiations, between cell lines and experiments, and across labs remain a significant technical hurdle that has hindered widespread organoid adoption (Ortmann and Vallier, 2017). Despite these limitations, in vitro models offer the opportunity to deconvolve embryonic signaling pathways in ways that are not possible in a complex in vivo system, highlighting the complementary benefit of organoids relative to embryo-based research.
Expanding on the foundation of established ‘gastruloid’ models and our group's experience with hindbrain interneuron models (Butts et al., 2017), we have developed a human organoid model that recapitulates key morphological processes that generate axial elongation during human spinal cord development in a reproducible manner across multiple human induced and embryonic stem cell lines (i.e. hiPSCs and hESCs). These organoids demonstrate extensive self-driven unidirectional axial elongation, cell specification of NMP differentiation and neural tube morphogenesis, and regionalized gene expression profiles with distinct caudal identities. This robust model of neural tube morphogenesis and axial elongation enables direct examination of previously inaccessible aspects of early human spinal cord development and patterning.
RESULTS
Wnt agonism induces axial extension of neuronal organoids
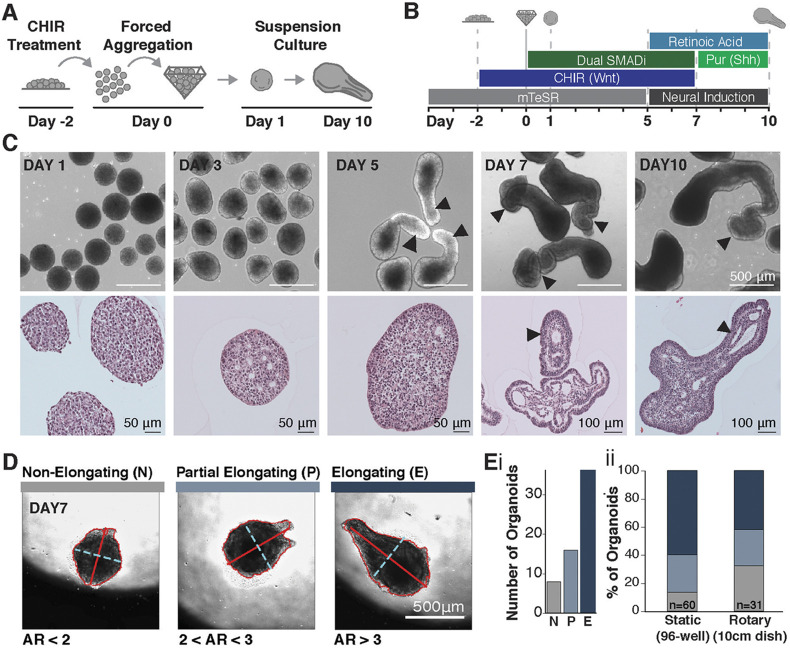
Exogenous canonical Wnt signaling induces morphogenesis resembling axial elongation in in vitro models of early gastrulation (Moris et al., 2020), where caudalization is crucially dependent on Wnt signaling (Wilson et al., 2009; Henrique et al., 2015; Yamaguchi, 2001). Therefore, to obtain a population of caudalized spinal neuron progenitors, we elaborated on a previously described 2D hindbrain interneuron differentiation protocol (Butts et al., 2017) that starts with promoting differentiation to the neuronal lineage by treating human PSCs with the small molecules SB43542 and LDN193189, which inhibit the TGFβ and BMP pathways, respectively (a process defined as dual SMAD inhibition). Small-molecule inhibition of these two pathways has been widely used to generate cranial (Shi et al., 2012; Chambers et al., 2009) and hindbrain (Butts et al., 2017) neuronal populations by mimicking the effects of morphogen gradients and their inhibitors within the embryo. In vivo, spinal progenitors emerge from regions of the epiblast exposed to high levels of Wnt/β-catenin signaling. Therefore, to generate a caudal neuronal population, we hypothesized that we would need to caudalize the progenitor population prior to neural commitment. We subsequently modified the original protocol by treating WTC-11 human induced pluripotent stem cells (hiPSCs) with 2 μM of the Wnt small-molecule agonist CHIR99021 (CHIR) starting 2 days prior to neuronal differentiation (Fig. 1A,B). CHIR treatment was initiated in mTeSR1 medium, which contains both FGF and activin (TGFβ) to keep hPSCs in a self-rrenewing pluripotent state (Ludwig et al., 2006). However, the introduction of CHIR to mTeSR1 medium results in morphogen milieu that mimics the signaling environment of the gastrulating embryo. The combination of Wnt agonism with FGF and activin allows for the emergence of neuromesodermal lineages and is crucial to the generation of spinal cord neurons and caudalization within the embryo anterior-posterior axis (Wilson et al., 2009; Henrique et al., 2015). Subsequently, in order to allow for 3D morphogenesis to occur, we generated 3D organoids by aggregating dissociated, single cells pretreated with CHIR in non-adherent pyramidal inverted wells (3000 cells/well) and culturing the organoids in bulk rotary suspension (Hookway et al., 2016) (at about 270 aggregates per well) or in static cultures of single aggregates. Aggregation was considered the start of differentiation (day 0), and CHIR treatment continued through day 7 mimicking the continued presence of Wnt signaling in the posterior of the embryo (Wilson et al., 2009; Yamaguchi, 2001) (Fig. 1A). At day 5, aggregates were transitioned from the mTeSR media+CHIR+dual SMAD inhibition to a neural maturation media (Butts et al., 2017; Amoroso et al., 2013) to facilitate the differentiation and maturation of caudal neural progenitors generated in the first 5 days of the differentiation. Finally, as retinoic acid and sonic hedgehog signaling are crucial for neuronal development and maturation (Briscoe and Ericson, 1999; Kim et al., 2009; Okada et al., 2004), both retinoic acid (RA) and the sonic hedgehog agonist purmorphamine (Pur) were added to the culture to facilitate the emergence of caudalized cell fates and limit the emergence of sensory dorsal neurons. After 5 days in suspension culture, some, but not all, of the CHIR-pretreated organoids displayed pronounced singular extensions (Fig. 1C). Notably, extension occurred both in bulk rotary cultures and in static cultures of single aggregates in non-adherent 96 wells, confirming elongated organoids formed through extension rather than fusion of multiple aggregates. To quantify the occurrence of elongations, we defined three categories of aggregates – non-elongating (N), partially elongating (P) and elongating (E) – based on the ratio of their long to short axes (Fig. 1D,E). Using this classification system, over 50% of aggregates were elongating at day 7 of culture in both bulk and single organoid culture formats. Elongated organoids were continuous, with single tissue protrusions extending outwardly from individual radially symmetric aggregates. Histological analysis revealed multiple internal, elongated epithelial compartments within the protrusions separated by regions devoid of cells (Fig. 1C). Interestingly, extensions occurred in organoids of various sizes (Fig. S1A) and organoids extended more robustly when cultured at low density (Fig. S1B), suggesting that density-mediated signaling parameters, such as paracrine effects or nutrient availability, modulate the induction and maintenance of axial extensions, similar to the documented role of Warburg glycolysis in vertebrate axial elongation (Oginuma et al., 2020, 2017).
Fig. 1.
CHIR treatment of neural PSC aggregates results in axial extension. (A,B) Schematic of experimental set-up and differentiation protocol using 2 μM CHIR. (C) Bright-field and histological images of elongating organoids (arrowheads indicate elongated structures). (D) Images from time-course of extension, displaying organoids that do not elongate (N), partially elongate (P) or fully elongate (E) with respective axis ratios (ARs) of AR<2, 2<AR<3 and AR>3 (minor axis dotted cyan line, major axis solid red line). (E,i) Elongation types detected during time lapse imaging (n=60 organoids). (E,ii) Quantification of elongating types across static (n=60 organoids) and rotary (n=31 organoids) cultures.
Elongating organoids consist of neural and mesodermal cells
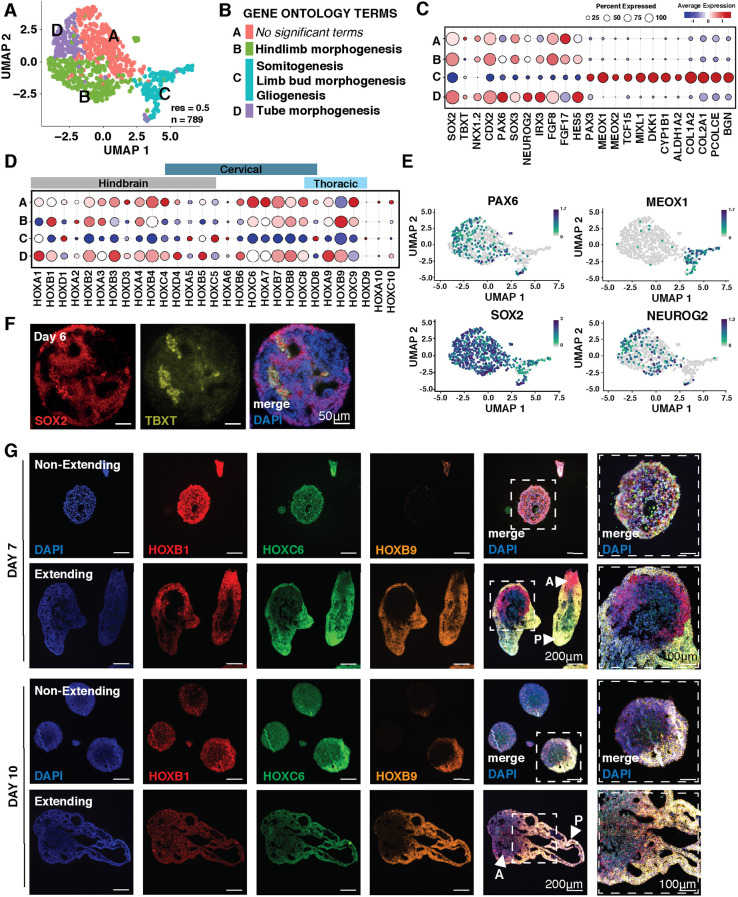
To examine cell diversity in extended organoids, we performed single-cell transcriptomics on elongated day 10 organoids induced by 2 μM CHIR. Shared nearest-neighbor computation revealed four clusters at resolution 0.5 (Fig. 2A,B; Fig. S2A-C). Neural markers, such as SOX2, were expressed in all clusters (Fig. 2C,E), but clusters A, B and D had higher expression levels of genes associated with the neural tube (SOX2, IRX3, PAX6 and NEUROG2) and the caudal lateral epiblast (CDX2, NKX1.2 and FGF8) (Fig. 2C,E; Fig. S2D). The highest differentially expressed genes (Fig. S2B,C) in cluster A were associated with regulation of apoptosis (BAX), neural homeostasis (NPY) and protein regulation (MLEC). Cluster B had multiple upregulated genes involved in tumor metastasis (FABP7, PTTG1 and NMP3), cell division (CDC20, NASP) and FGF signaling (FGF8 and FGFBP3), indicating replicating neural progenitors. Cluster D expressed genes associated with chromatin regulation (HIST1H4C and H1FX) and growth factor response (ZFP36L1), possibly indicating more mature neurons. In contrast, MIXL1, MEOX1, CYP1B1, MEOX2 and PAX3 were almost exclusively expressed in cluster C, indicating that this distinct cluster contained mesodermal cells (Fig. 2C,E; Fig. S2D). We did not observe expression of endoderm-specific markers such as SOX17, and only very low expression of FOXA2 and PAX9 in fewer than 1% of cells, indicating that, in contrast to previously reported gastruloid models (Beccari et al., 2018; van den Brink, 2020; Moris et al., 2020; Veenvliet et al., 2020), these elongated organoids consisted entirely of neural and mesodermal cell types. In addition, transcriptomic profiles revealed expression of HOX1-HOX9, demonstrating robust posteriorization (Fig. 2D). The expression of mesodermal genes in a caudalized spinal cord neuron differentiation led us to investigate whether an NMP population, which is known to give rise to both lineages, was present in our organoids. Through immunofluorescence staining of day 6 sectioned organoids, we identified a small population of SOX2+TBXT+ double-positive cells, a defining characteristic of NMP identity (Fig. 2F). Overall, these data indicate that elongating organoids contain cell populations that appear to be exclusively associated with mesoderm and neural tube, similar to the caudal lateral epiblast during axial extension and suggestive of an NMP-like origin of posterior neural identity.
Fig. 2.
Gene expression within elongating organoids. (A) UMAP of n=789 cells from elongating organoids exposed to CHIR (2 μM) at day 10 of differentiation from single-cell RNA-seq. (B) Significant (P<0.05) gene ontology classifications derived from the most differentially upregulated genes in elongating organoids from single-cell RNAseq. Gene ontology terms are assigned to identified clusters. (C) Dot plot displaying genes associated with neural and mesodermal lineages across clusters. (D) Dot-plot displaying HOX gene expression across clusters. (E) UMAPs showing cells expressing PAX6, MEOX1, SOX2 and NEUROG2. Color scale indicates a normalized increase in log2 fold change from min expression to max expression of the respective gene. (F) Immunofluorescence images of organoid paraffin sections stained for SOX2 and TBXT. (G) RNA-scope of sectioned elongating and non-elongating organoids cultured at low density and with 2 μM CHIR with probes for HOX genes marking different regions of the spine at day 7 and day 10 of differentiation [arrowheads mark anterior (A) and posterior (P) ends of organoids]. Scale bars: 200 μm; 100 μm (right-most column).
To obtain a more nuanced understanding of the caudalization of 2 μM CHIR-pretreated organoids, we examined HOX transcripts marking hindbrain (HOXB1), brachial (HOXC6) and thoracic (HOXB9) regions of the anterior-posterior axis via RNAscope at days 7 and 10 (Fig. 2G). Although non-elongating CHIR-pretreated organoids expressed both HOXB1 and HOXC6 HOX genes on day 7, their expression was equally distributed radially, and the expression of HOXB9 was not detected until day 10. By contrast, HOX expression was regionalized in the elongated organoids by day 7, with HOXB1 (hindbrain) enriched in the central aggregate mass and HOXC6 (brachial) and HOXB9 (thoracic) expressed in the elongated extension. Interestingly, HOXC6 expression was more pronounced at day 7 than at day 10 within elongating organoids, suggesting a transition to a more posterior fate over time (Fig. 2G). These data indicate that while organoid extension is not a requirement for posterior HOX gene expression, extension enables the stratification of distinct HOX domains.
Wnt signaling influences extension and differentiation across stem cell lines
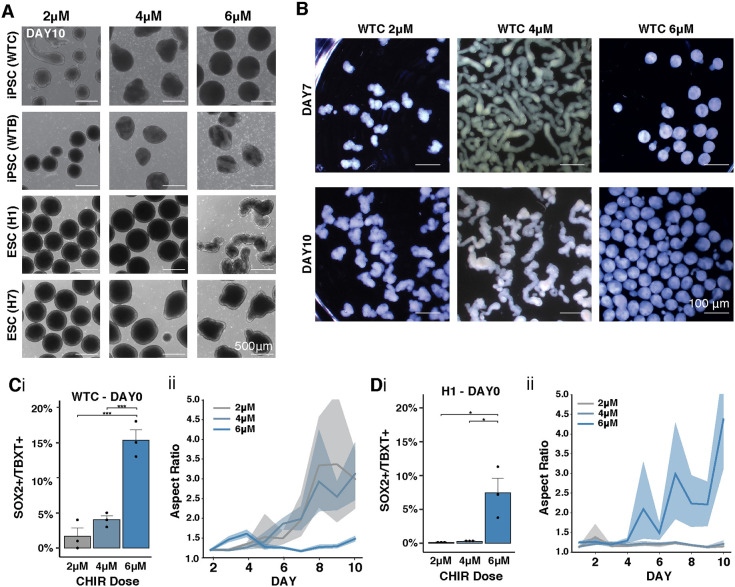
Because we observed heterogeneity in the length and frequency of organoid elongation, we examined the robustness of Wnt signaling effects on axial elongation in vitro across multiple human stem cell lines of both sexes. We differentiated two hiPSC lines (WTC and WTB) and two hESC lines (H1 and H7) into caudal neural organoids in the presence of varying amounts of CHIR (2 μM, 4 μM, 6 μM). Across a 10 day differentiation, the threshold of CHIR required to reliably generate extensions varied between lines (Fig. 3A). Whereas the WTC line produced elongations most robustly at a 4 μM dose of CHIR, all other lines tested only began to elongate when the CHIR dose was increased to 6 μM. For each cell line, the critical CHIR dose increased both the frequency of SOX2+TBXT+ cells at the start of the differentiation (Fig. 3Ci,Di) and the degree of extension (Fig. 3B,Cii,Dii, n≥3 experiments at each timepoint). Curiously, the WTC line demonstrated an intermediate maximum concentration for CHIR-induced extension, as 6 μM CHIR treatment actually reduced the number extensions (Fig. 3A,C). Importantly, all cell lines displayed internal epithelialization and cystic formation within extensions at the CHIR concentration that permitted elongation (Fig. S3A).
Fig. 3.
Wnt mediates organoid extension across human stem cell lines. (A) Bright-field images of organoids derived from iPSC and ESC lines at day 7 of differentiation with increased CHIR doses. (B) Stereoscope images of WTC-derived organoids at day 7 or day 10 of differentiation given 2 μM, 4 μM or 6 μM CHIR treatment. (C,D) (i) Quantification of SOX2+TBXT+ cells by flow cytometry in day 0 WTC hiPSCs organoids (C) or H1 ESC organoids (D) as a function of CHIR doses. (ii) Quantification of the length of extensions in WTC (C) and H1 (D) organoids measured as aspect ratio (i.e. major to minor). Solid line indicates mean; shading represents 95% confidence bounds. Student's t-test;*P<0.05 and ***P<0.0005.
To examine differences in cell populations induced by varying Wnt concentrations, we stained sections of WTC organoids treated with 2 μM, 4 μM or 6 μM of CHIR at day 10 of differentiation (Fig. S3B). Organoids exposed to 2 μM CHIR displayed SOX2 and PAX6 expression throughout the entire organoid, whereas organoids exposed to 4 μM or 6 μM CHIR showed regionalized expression of SOX2 and PAX6, suggesting that, as expected, non-neuronal cells (SOX2− PAX6−) emerge in the presence of high Wnt signaling. All organoids displayed N-cadherin and β III-tubulin expression, indicating the emergence of maturing populations of neurons. The H1 ESC line displayed regionalized expression of neural progenitor markers when exposed to 4 μM CHIR, similar to the WTC line (Fig. S3C). Altogether, the level of Wnt pathway activation that allows for robust emergence of an NMP progenitor pool and organoid extension varied across human iPSC and ESC lines – a commonly reported phenomenon in most differentiation protocols (Ortmann and Vallier, 2017). In all subsequent studies, unless otherwise stated, we used the critical elongation dose of 4 μM CHIR with the WTC cell line that elongated most consistently, and referred to it as a ‘high Wnt signaling’ condition.
Organoid extension is correlated with NMP presence
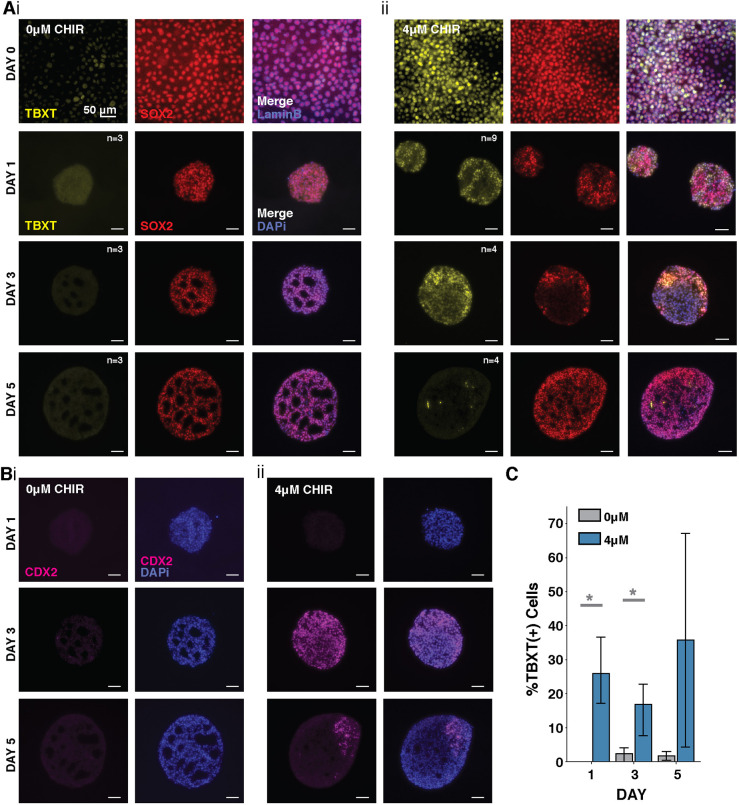
Because we hypothesized that the extensions observed in CHIR-treated organoids were due to the presence of a NMP population, we next quantified the correlation between organoid elongation, CHIR treatment and NMP presence. Therefore, cells were exposed to 0 μM or 4 μM CHIR from day −2 through day 7 of differentiation, and examined immediately before and after aggregation (day 0-1), and then through day 5 of differentiation (Fig. 4A,C). Consistent with previous observations, SOX2+TBXT+ cells, which are indicative of NMP-like cells, were present in 4μM CHIR organoids at the time of aggregation (after 2 days of treatment, day 0 of differentiation). In the absence of CHIR treatment, organoids lacked SOX2+TBXT+ cells and did not extend. Interestingly, the organoids with 0 μM CHIR treatment developed rosette structures of SOX2+ cells that were absent in the 4 μM CHIR-treated organoids (Fig. 4A). The 4 μM CHIR-treated organoids displayed high levels of TBXT expression at day 1 that then became progressively restricted to polarized crescents by day 3 and eventually to very small pockets of TBXT+ cells by day 5. Additionally, the 4 μM CHIR treatment resulted in the emergence of the caudal marker CDX2 throughout aggregates at day 3 but restricted to the extending region by day 5 (Fig. 4B), indicating both organoid polarization and posteriorization in the presence of high-Wnt signaling. These observations confirm that the presence of NMPs within CHIR-treated organoids precedes and correlates with axial elongation.
Fig. 4.
Emergence of TBXT, SOX2 and CDX2 over time in 4 μM CHIR organoids. (A) Fluorescence images of histological sections of cultures exposed to (i) 0 μM CHIR and (ii) 4 μM CHIR before and after aggregation showing increases in TBXT+SOX2+ cells with CHIR treatment. (B) Fluorescence images of histological sections of cultures exposed to (i) 0 and (ii) 4 μM CHIR showing CDX2 emergence and localization over time. (C) Quantification of TBXT loss in fluorescence images (n>3 per day, *P=0.04; data are mean±s.e.m.).
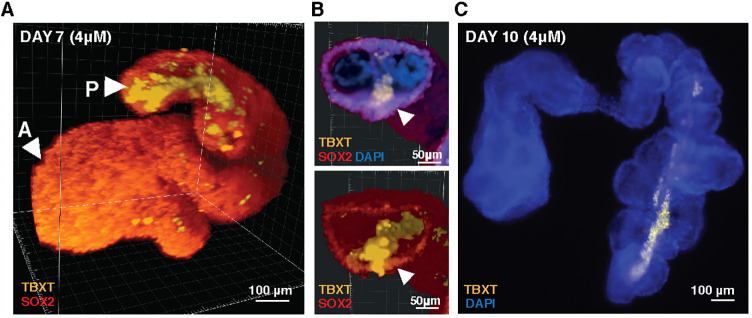
At day 7 of differentiation, in most imaged organoids grown in 4 μM CHIR, the TBXT+ population formed a streak along the protruding extension and accumulated at the end of the extension (Fig. 5A-C). The morphological arrangement of this population was reminiscent of primitive streak and notochord in vivo models and posterior TBXT+ cells in gastruloid models (Moris et al., 2020; Veenvliet et al., 2020). However, expression of the notochord marker homeobox protein notochord (NOTO) was not detectable by immunofluorescence at day 7 and 10 or in our single-cell RNA-sequencing dataset at day 10 of differentiation. The lack of NOTO expression is likely due to inhibition of TGFβ signaling (via the small molecule SB43542), which is known to influence NOTO expression during the initial neural-biased differentiation protocol (Zhang et al., 2020). In contrast to their distribution in 4 μM CHIR organoids, TBXT+ cells in 2 μM CHIR organoids were sparsely distributed across the surface of the spherical organoid with rare regions of SOX2+TBXT+ co-expression (Fig. S4A). This observation, combined with the histology at earlier timepoints, suggests that the TBXT+ population starts off stochastically distributed throughout the aggregate and then becomes spatially restricted, suggesting that NMPs specified early in differentiation cluster together post-aggregation in order to coordinate subsequent organoid elongation.
Fig. 5.
TBXT streak in CHIR-treated elongating organoids. (A) 3D reconstruction based on light sheet fluorescence imaging of a day 7 4 μM elongating organoid showing a streak of TBXT traveling down its length. (B) Cross-section through the 3D reconstruction shown in A displaying a TBXT+ streak and TBXT+SOX2+ cells adjacent to the streak (arrowheads). (C) Fluorescence image of a representative day 10 organoid with a TBXT streak [all aggregates shown cultured in high CHIR condition (4 μM)].
Elongating organoids display proliferating neuroepithelial compartments
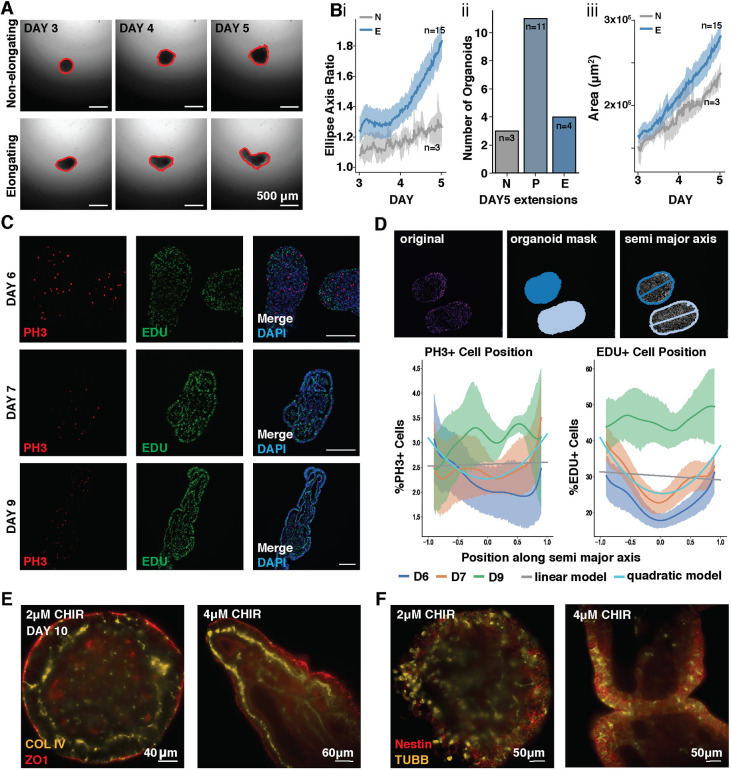
To examine the kinetics and diversity of elongating organoids, single organoids from the same starting population were imaged continuously over 48 h, revealing robust elongation from a single point in the presence of 4 μM CHIR. At day 5, over 50% of 4 μM CHIR organoids were partially elongating (P) or elongating (E) (Fig. 6A,B; Movies 1 and 2). Furthermore, the 4 μM CHIR-treated organoids had a higher total area and perimeter by day 5 of the time-course (Fig. 6Biii; Fig. S5).
Fig. 6.
Organoids contain proliferative neuroepithelium. (A) Frames from video time-course tracking the diversity of 4 μM CHIR-treated organoid extensions in static cultures. Red outline defines area measured. (Bi) Quantification of ellipse axis ratios from day 3 to day 5 in 4 μM CHIR organoids labeled as non-elongating (N; grey) or elongating (E; blue). (Bii) Quantification of non-elongating (N), partially elongating (P) and elongating organoids (E) with axis ratios (ARs) of AR<2, 2<AR<3 and AR>3, respectively. (iii) Quantification of the surface area of non-elongating (NE; gray) or elongating (E; blue) organoids treated 4 μM CHIR. Solid lines represent the mean ratio (dark color) with 95% confidence interval (light colored shading); n=18 organoids. (C) Immunofluorescence images of EdU incorporation and phospho-histone 3 (pH3) localization in 4 μM CHIR organoids taken from paraffin sections. (D) Top: segmentation of organoid sections showing pH3 staining (left), segmentation of nuclei by DAPI staining (middle) and a total least squares elliptical fit of the segmentation contour with the major axis superimposed (right). Bottom: line plots for pH3 or EdU expression along the normalized semi-major axis of 4 μM CHIR organoids at day 6, 7 and 9. Linear and quadratic least squares fits of pH3 and EdU expression along the normalized semi-major axis for all days; n>4 independent organoid sections at each timepoint. (E,F) Optical sections from light-sheet microscopy of low CHIR (2 μM) and high CHIR (4 μM) organoids stained for COL IV and ZO1 or Nestin and βIII-Tubulin (TUBB3).
Because of the uni-directional extension of our organoids, we examined whether extensions resulted from localized populations of proliferating cells, similar to what is seen in the developing embryo. To this end, we measured the percentage of actively dividing cells along the long axis of sectioned organoids by EdU incorporation and phospho-histone 3 (pH3) staining on days 6, 7 and 9 of differentiation in 4 μM CHIR (Fig. 6C,D, n≥4). A 2 h EdU pulse labeled over 50% of the cells in elongating organoids, and EdU+ cells were observed increasingly at the outer edges of day 6 and 7 elongating aggregates (quadratic model fit, P<0.001), with an ∼15% increase in EdU labeling from center to edge of the organoids. pH3 staining revealed a similar, though less stark, trend of increased proliferation at organoid edges (1% or less change, quadratic model fit, P=0.015). Overall, cell division increased at both ends of the organoid surface, and we did not observe a pole-specific proliferation bias, indicating that cell division alone could not explain uniaxial extension.
To examine epithelial morphology, we stained for collagen IV (COL IV), a marker of the basement membrane, and zona occludens-1 (ZO-1), a marker of tight junctions at cellular apical domains in low CHIR (2 μM) and high CHIR (4 μM) conditions (Fig. 6E; Fig. S6A). Basement membrane deposition and apical-basal cell polarization, determined by apical location of tight junctions, was observed within the extension of organoids between days 7 and 10 of differentiation in high CHIR organoids (Fig. S6B), paralleling the lumen formation by polarized epithelia documented in existing organoid models (Faustino Martins et al., 2020). After 10 days, ZO-1 was apparent at the apical domain of cells in the outer epithelial layers of the extensions, as well as in cells facing the internal lumens. Distinct layers of basement membrane marked by collagen IV were observed basally to ZO1, forming a bilayer between lumens and organoid exterior. This organization occurred in all organoids, although to a lesser extent in low CHIR conditions (Fig. 6E; Fig. S6A,Bi, Movies 3 and 4). Furthermore, at day 10 of differentiation, these epithelial layers contained cells expressing the neuronal progenitor markers Nestin and β-III-tubulin, indicating that the majority of cells at the surface layers differentiated to a neural fate (Fig. 6F, Fig. S6Bii, Movies 5 and 6). These patterns of polarized apical basal proteins suggest that elongating organoids undergo tissue polarization via axial extensions that consist of sheets of proliferating neuroepithelial cells with neural tube progenitor identity surrounding organized basement lamina and internal lumens (Martins-Green and Erickson, 1986; O'Shea (1987; Hay and Meier, 1974).
Elongating organoids progress display developmental ontogeny of gene expression
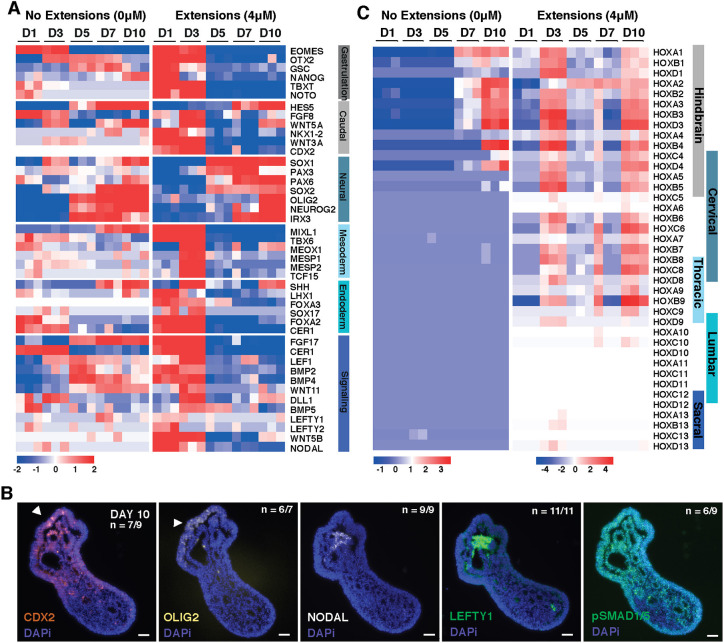
To investigate the transcriptional differences resulting from Wnt agonism, we compared organoids with 0 μM or 4 μM CHIR treatment via bulk RNA-sequencing at different stages of differentiation. Analysis of the top divergent genes between the 0 μM CHIR (no extensions) and the 4 μM CHIR conditions (Fig. 7A, Fig. S7) revealed unique components of pattern specification. At day 1, 4 μM CHIR organoids were enriched for genes involved in pattern specification and embryonic morphogenesis (WNT5B and CDH2). 4 μM CHIR organoids then progressed through distinct developmental stages, beginning with increased expression of genes involved in gastrulation, such as TBXT, eomesodermin (EOMES), NOTO and goosecoid (GSC), which was paired with reduced expression of pluripotency markers such as Nanog. Beginning at day 3, 4 μM CHIR organoids transitioned through a caudal epiblast phenotype characterized by increased expression of caudal genes CDX2, FGF8, WNT3A and WNT5A. At this stage, 4 μM CHIR organoids also displayed a reduction in the forebrain marker OTX2, whereas its expression was maintained in 0 μM CHIR organoids, further indicating that the 4 μM CHIR signaling condition promotes the emergence of caudal fates (Fig. 7A). Finally, starting at day 3 and continuing throughout the rest of the 10 day differentiation, 4 μM CHIR organoids displayed an increase in neuronal markers associated with the developing neural tube (IRX3, NEUROG2, OLIG2, PAX6, SOX1 and SOX2). Overall, this analysis indicates that early stimulation of Wnt signaling induces pattern specification in organoids that mirrors the successive in vivo phases of gastrulation, caudal epiblast formation and neural tube development associated with caudal neurogenesis.
Fig. 7.
CHIR-treated organoids transition through multiple developmental stages. (A) Heat map of gene expression in 0 μM or 4 μM CHIR organoids over time displaying genes associated with gastrulation, caudal epiblast, neuroepithelium, mesoderm, endoderm and major signaling pathways, as determined by bulk RNAseq where the heat map displays the relative expression of a gene compared with the mean expression of all genes across all days. (B) Heat map of HOX gene relative expression compared with the mean expression across days in 0 μM or 4 μM CHIR organoids from bulk RNAseq. Scale bars: 50 μm. (C) Immunofluorescence images at day 10 of differentiation from paraffin sections in 4 μM CHIR organoids examining axis and developmental patterning markers (arrowheads indicate elongating region).
We next examined whether the spatial distribution of gene expression was analogous to anterior-posterior patterning in embryonic development. Polarized expression of the posterior marker CDX2 (Beck et al., 1995) was detected at day 5 and day 10 of differentiation in 4 μM CHIR organoids (Fig. 7B). By day 10, compartments of cells expressing the ventral spinal cord marker OLIG2 were observed lining the epithelial cysts of 4 μM CHIR organoids (Fig. 7C). Combined with the expression of Nestin and β-III-tubulin at day 10 (Fig. 6F), this expression confirms that the later stages of the 4 μM CHIR differentiation shift toward neural fates. Interestingly, at day 10 some 4 μM CHIR organoids displayed clusters of NODAL- and LEFTY-positive cells, and polarized pSMAD1/5 expression was detected along the length of the organoid (Fig. 7B; Fig. S8A,B), suggesting the presence of an organizer-like population. Interestingly, multiple clusters of NODAL-positive cells were occasionally detected in 4 μM CHIR organoids with multiple extensions (Fig. S8A). These observations suggest that the 4 μM CHIR organoids exhibit spatiotemporal gene expression and morphogenic organization analogous to those observed during the establishment of the anterior-posterior axis of the embryo.
In addition to the emergence of genes associated with the caudal epiblast, we also examined the timing and extent of HOX gene expression in 0 μM CHIR (non-elongating) and the 4 μM CHIR (elongating) organoids. 0 μM CHIR organoids began to express detectable anterior HOX transcripts at day 7 and adopted a hindbrain identity after 10 days of differentiation (Fig. 7C). In contrast, 4 μM CHIR organoids expressed both hindbrain and cervical HOX transcripts as early as day 3, with a notable upregulation in thoracic HOX genes by day 10. This temporal change in HOX gene expression suggests that the onset of HOX gene expression in 4 μM CHIR organoids mimics aspects of the progression of anterior-to-posterior elongation of in vivo embryonic development.
Brachyury is required for the formation of singular extensions
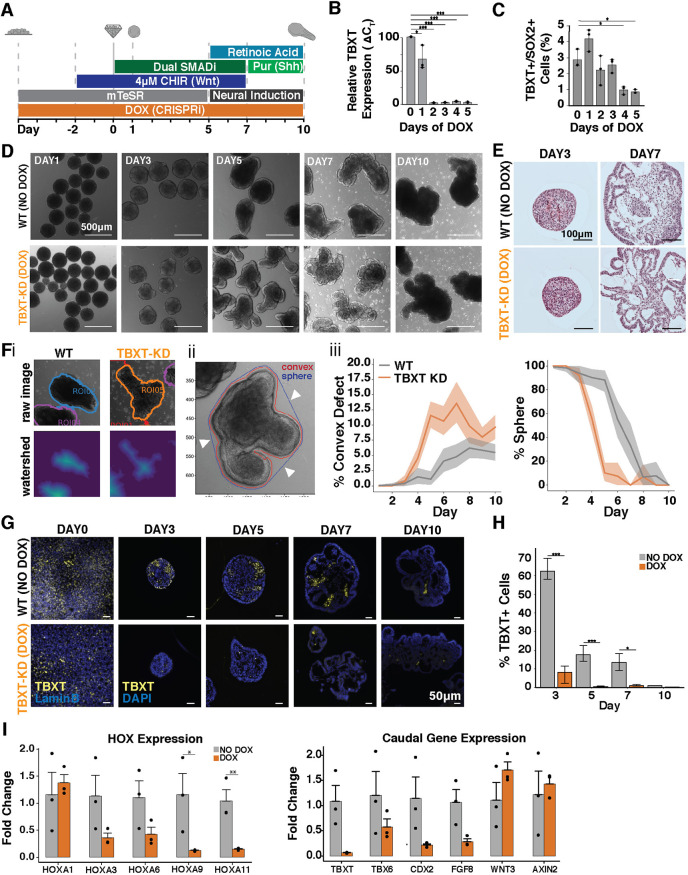
In vivo, Wnt signaling directly upregulates the expression of the transcription factor TBXT, which in turn mediates both Wnt and FGF posterior-to-anterior gradients (Amin et al., 2016). The importance of TBXT for proper axial elongation has been demonstrated in mouse knockout models, which have severely disrupted trunk morphogenesis and fail to generate the notochord and posterior somites (Amin et al., 2016; Pennimpede et al., 2012; Wilson and Beddington, 1997; Wilson et al., 1993; Chesley, 1935; Guibentif et al., 2021). Thus, we interrogated the role of TBXT in axial elongation in our organoids, using an inducible CRISPR interference (CRISPRi) system in human iPSCs to knock down TBXT expression (Larson et al., 2013).
First, we integrated RNA guides targeting TBXT into a Lamin-B::GFP WTC hiPSC line harboring a doxycycline (DOX)-inducible CRISPRi system (Libby et al., 2018; Mandegar et al., 2016). DOX induction for 5 days prior to cell aggregation (day 0) reduced the number of TBXT transcripts by 90% (Fig. 8A,B) and significantly reduced the frequency of TBXT+SOX2+ cells (Fig. 8C). Interestingly, by day 5 of differentiation under 4 μM CHIR conditions, the TBXT knockdown (TBXT KD) organoids displayed multiple protrusions rather than coordinated singular extensions (Fig. 8D). Histology revealed folded layers of epithelial sheets, indicating that increased epithelial tissue formation rather than coordinated morphogenesis was the likely cause of the multiple protrusions in the TBXT KD organoids (Fig. 8E). To quantify these folds, each organoid was segmented (Fig. 8Fi) and the number of convex indentations from the perimeter of each organoid was quantified (Fig. 8ii, arrows). This analysis showed that the TBXT KD organoids had an increased percentage of convex defects compared with the wild-type organoids, diverging as early as day 4 of the differentiation with the highest number of defects apparent at day 7 (n>14 per timepoint). The TBXT KD organoids also lost spherical morphology 2 days before wild-type organoids started extending (Fig. 7Fiii) and reduced levels of TBXT expression were maintained in KD organoids through day 10 (Fig. 8G,H). Interestingly, the KD organoids maintained a small proportion of TBXT+ cells that escaped knockdown, but not enough to trigger elongation, suggesting a threshold of TBXT+ cells is required for coordinated elongation. TBXT KD organoids and isogenic (non-silenced) controls expressed the neuronal marker Nestin, the caudal marker CDX2 and extracellular matrix protein fibronectin (Fb) (Fig. S9). However, there was a reduction in caudal HOX genes in the TBXT KD organoids at days 5 and 10 of differentiation, as well as a reduction in CDX2 and FGF8 expression (Fig. 8I), which are both highly expressed in the caudal lateral epiblast and reduced in TBXT knockout mice (Amin et al., 2016; Pennimpede et al., 2012). Interestingly, organoids maintained expression of Wnt signaling molecules, such as AXIN2 and WNT3, even after the termination of CHIR-induced Wnt agonism, suggesting that TBXT is necessary for unidirectional extension and caudal fate specification in organoids, but not essential for endogenous Wnt expression and signaling.
Fig. 8.
TBXT knockdown leads to increased epithelial folding. (A) Schematic of differentiation protocol timeline with TBXT knockdown (KD) induction via doxycycline (DOX). (B) Quantification of TBXT-KD efficiency by qPCR showing reduction in mRNA expression levels on day 0 of differentiation (day 5 of DOX treatment). (C) Quantification by flow cytometry of TBXT+SOX2+ cells with DOX treatment over time. (D) Bright-field images of the differentiation of TBXT-KD hiPSC line with and without DOX treatment (wild type; WT) over time. (E) Histological sections of organoids with and without DOX treatment at day 3 and 7 of differentiation. (F,i) Example images of organoid segmentation; (ii) example image of convex indentations on organoids (arrowheads indicate convex defects); (iii) quantification of the percent of organoids with convex defects and quantification of the loss of sphere morphology in organoids over time (n>14 organoids at each timepoint). (G) TBXT immunofluorescence staining of fixed cells (day 0) or organoid paraffin sections (days 3-10) with and without DOX treatment. (H) Quantification of the immunofluorescence signal indicating loss of TBXT (Student's t-test;*P<0.05 and ***P<0.0005). (I) qPCR quantification of HOX genes and genes related to axial pattern specification from day 10 organoids with and without DOX treatment (Student's t-test; *P<0.05 and **P<0.005).
DISCUSSION
Here, we have generated an organoid model of neural tube formation that mimics several aspects of early spinal cord development, including axial elongation, the maintenance of a population of SOX2+TBXT+ NMPs, and the generation of neural tube-like neuroepithelial cells. This model is based on aggregates of hPSCs that undergo robust unidirectional axial extension when exposed to a critical dose of Wnt signaling. In organoids that were exposed to the Wnt agonist, CHIR, the number of TBXT+SOX2+ NMP-like cells increased proportionally to CHIR dose as well as the presence of both mesoderm and neurectoderm during elongation of the organoid, similar to the embryonic body axis (Wymeersch et al., 2021). Single-cell RNA sequencing of elongating organoids confirmed the presence of caudal cell phenotypes, in addition to neural progenitors and paraxial mesoderm populations. Elongating organoids displayed anterior-posterior patterning and a more posterior HOX gene profile than non-elongating organoids in equivalent culture conditions. Finally, NMP generation, coordinated axial elongation and adoption of a caudal identity were all abrogated in organoids with TBXT silenced (Pennimpede et al., 2012; Guibentif et al., 2021). Overall, these elongating organoids provide a significant step towards reliable in vitro models that can be used to further interrogate the gene regulatory networks that contribute to morphogenetic elongation, the multicellular morphogenesis involved in extension organization, and the stratification of anterior and posterior populations that contribute to the development of the central nervous system in humans.
In this model, elongating organoids often developed multiple internal epithelial compartments rather than a single cohesive neural tube. We hypothesize that this multi-tube-like morphology is due to the absence of surrounding tissue(s) that are present in vivo and likely regulate proper neural tube formation via a combination of mechanical and biochemical cues. In support of this hypothesis, mis-regulated Wnt signaling has been shown to generate excessive neural tubes at the expense of mesoderm (Chapman and Papaioannou, 1998; Takemoto et al., 2011; Shum et al., 1999). The exogenous addition of the ventralizing sonic hedgehog agonist, purmorphamine, to the culture was carried out intentionally to limit the generation of sensory dorsal neural progenitors, but purmorphamine might also disrupt the ability of the organoid to form a single neural tube (Murdoch and Copp, 2010). Furthermore, the elongating spinal cord is physically situated between multiple tissues (gut, lateral plate mesoderm, etc.) that present different local tissue mechanics (Zhou et al., 2009), which may also contribute to the morphogenesis of a single neural tube. Finally, recent reports in chick have described the fusion of multiple lumens into a single neural tube during secondary neurulation, when the most caudal region of the embryo is formed (Gonzalez-Gobartt et al., 2021). Thus, it is possible that human PSC-derived elongating organoids emulate early stages of a similar lumen fusion process to form a single neural tube.
An important element of axial elongation is the progressive activation of HOX gene clusters both spatially and temporally. Although our elongating system recapitulates some of this pattern and initiates expression of relatively posterior HOX profiles, we did observe day-to-day fluctuations in HOX gene expression via bulk sequencing. Wnt and FGF signaling have been shown to specify brachial and thoracic HOX identity by upregulating expression of the transcription factor CDX2, which allows for HOX1-HOX9 expression, forming a posterior axial identity (Mazzoni et al., 2013; Metzis et al., 2018; van den Akker et al., 2002) In our system, high expression of FGF17, WNT5B and CDX2 were observed starting at day 1 through day 3 in the 4uM CHIR treated organoids, consistent with early expression of HOX genes (Fig. 6). However, HOX expression was then generally reduced at days 5 and 7 before increasing again at day 10. There could be several explanations for this phenomenon. As the gene expression is averaged across the entire tissue, bulk sequencing may not provide sufficient resolution to identify clear trends in expression, particularly as organoids grow and generate a greater diversity of more complex cell types. Additionally, recent studies have shown that the temporal regulation of HOX genes can be influenced by tissue culture conditions (Mouilleau et al., 2021). In particular, the regulation of FGF, RA and Wnt signaling influence HOX gene emergence, which may in part be the reason that fluctuations in HOX gene expression are observed as the organoids elongate over the course of differentiation. For example, high FGF signaling and Wnt agonism in stem cell media with CHIR early on (days −2 to 5) is transitioned to neural induction media that lacks those factors and is supplemented with RA from days 7 to 10. Even so, the induction of posterior HOX genes demonstrates caudalization of an in vitro model of neural development and is an important bridge for studying human neural tube formation.
Various in vitro techniques, from micropatterns to Matrigel embedding, have been shown to impact TBXT expression and tissue organization (Sagy et al., 2019; Blin et al., 2018; Muncie et al., 2020). In our organoids, reduction in TBXT expression resulted in loss of posterior HOX gene expression and uncoordinated folding of tissue, reminiscent of the loss of posterior somite formation in murine models. The appearance of multiple protrusions in the TBXT knockdown organoids likely reflects an excess of anterior neural tissue at the expense of NMPs and mesoderm. The confined spherical expansion of a proliferating neuroepithelial sheet would result in epithelial sheet buckling (Nelson et al., 2011; Trushko et al., 2020; Murisic et al., 2015) and the appearance of multiple protrusions. In particular, the proliferation analysis in the wild-type organoids showed actively dividing epithelial sheets in extending organoids (Fig. 6C,D); therefore, it is possible that the reduction of TBXT+ cells in the TBXT KD organoids results in a loss of an organizer to drive polarized extension, leaving only epithelial proliferation that mechanically would result in tissue buckling. Another hypothesis is that TBXT expression is necessary for the coalescence of an organizer-like domain from the initial Wnt-stimulated population. Reduced levels of TBXT in the KD organoids would therefore cause tissue polarity to be lost, resulting in multiple asynchronous protrusions.
Overall, this work contributes to the growing body of knowledge showing that stem cell-derived organoid models that reflect multiple organ systems (Lancaster and Knoblich, 2014; Murisic et al., 2015; Lindborg et al., 2016) and stages of development (Faustino Martins et al., 2020; Zheng et al., 2019; Beccari et al., 2018; Eiraku et al., 2011; Broutier et al., 2016) can be used to investigate aspects of human embryogenesis that were previously elusive due to poor tissue quality, technical difficulties isolating and imaging embryonic tissues, and ethical concerns regarding tissue procurement. Although all organoids are imperfect models of embryogenesis, they are highly amenable to variations in culture conditions (Lindborg et al., 2016; Eiraku et al., 2011; Lancaster et al., 2013), to genetic modification (Broutier et al., 2016; Fujii et al., 2015) or to small-molecule drug screens (Zhou et al., 2017; Marikawa et al., 2020), enabling the systematic manipulation of experimental conditions on a larger scale and at a higher throughput than is possible in model organisms. As such, the organoid model described in this report enables future scientific inquiry that can address its current limitations. For example, this system can be used to interrogate the multicellular organization events underlying tissue extension decoupled from the influences of surrounding tissues that would be present in an embryo. The system can be adapted further to identify conditions that allow for the generation of a single continuous neural tube, which has yet to be shown in a human organoid model(Moris et al., 2020; Marikawa et al., 2020) but has been in murine gastruloids (Veenvliet et al., 2020) in vitro. Furthermore, although an anterior-posterior axis is clearly developed in our human elongated neural organoids, the emergence of a well-defined dorsal-ventral axis remains elusive, allowing for future elucidation of the independent and interdependent mechanisms that regulate these two axes. Overall, this system advances models of neural tube self-assembly beyond cell signaling toward coordinated multicellular organization and is a facultative stepping point to directly interrogate mechanisms of human morphogenesis and developmental paradigms.
MATERIALS AND METHODS
Human induced pluripotent stem cell line generation and culture
All work with human induced pluripotent stem cells (iPSCs) or human embryonic stem cells (ESCs) was approved by the University of California, San Francisco Human Gamete, Embryo, and Stem Cell Research (GESCR) Committee. Human iPSC lines were derived from the WTC11 line (Coriell, #GM25256), the WTB line (Conklin Lab) (Miyaoka et al., 2014) and the Allen Institute WTC11-LaminB cell line (AICS-0013 cl.210). The human ESCs were the H7 and H1 lines (WiCell). All cell lines were karyotyped by Cell Line Genetics and reported to be karyotypically normal. Additionally, all cell lines tested negative for mycoplasma using a MycoAlert Mycoplasma Detection Kit (Lonza).
Human iPSCs were cultured on growth factor reduced Matrigel (Corning Life Sciences) and fed daily with mTeSRTM-1 medium (STEMCELL Technologies) (Ludwig et al., 2006). Cells were passaged by dissociation with Accutase (STEM CELL Technologies) and re-seeded in mTeSRTM-1 medium supplemented with the small molecule Rho-associated coiled-coil kinase (ROCK) inhibitor Y276932 (10 μM; Selleckchem) (Park et al., 2015) at a seeding density of 12,000 cells per cm2.
The generation of the TBXT CRISPRi line first involved TALEN-mediated insertion of the CRIPSRi cassette pAAVS1-NDi-CRISPRi (Gen2) (Addgene) to the AAVS1 locus of the Allen Institute WTC11-LaminB cell line. Following antibiotic selection of clones that received the CRIPSRi cassette, CRISPRi gRNAs were generated targeting TBXT (Table S2) using the Broad Institute GPP Web Portal and cloned into the gRNA-CKB (Addgene) following the previously described protocol (Libby et al., 2018). Guide RNA vectors were nucleofected into the LaminB CRISPRi iPSC line using a P3 Primary Cell 96-well Nucleofector Kit (Lonza) and the 4D Nucleofector×Unit (Lonza) following the manufacturer's instructions. Nucleofected cells were allowed to recover in mTeSR-1 medium supplemented with Y276932 (10 μM) and then underwent antibiotic selection with blasticidin (ThermoFisher Scientific; 10 μg/ml) following the previously published protocol (Beck et al., 1995; Amin et al., 2016). Knockdown efficiency was evaluated by addition of doxycycline to the daily feeding media over the course of 5 days, collection of mRNA and subsequent quantification of gene expression by qPCR.
Organoid differentiation
Organoid differentiation was performed according to a modified version of a previously published spinal cord interneuron differentiation protocol (Butts et al., 2017). Human iPSCs or ESCs were seeded at 125 000 cells/cm2 in mTeSR-1 medium supplemented with the small-molecule Rho-associated coiled-coil kinase (ROCK) inhibitor Y276932 (10 μM; Selleckchem) and small-molecule GSK inhibitor CHIR99021 (2 μM, 4 μM or 6 μM; Selleckchem). Two days later, cells were singularized with Accutase (STEMCELL Technologies), counted using a Countess II FL (Life Technologies) and seeded into 800 μm×800 μm PDMS microwell inserts in a 24-well plate (∼270 wells/insert) (Hookway et al., 2016). After ∼18 h, condensed organoids were transferred to rotary culture in six-well plates in mTeSR-1 medium supplemented with Y276932 (10 μM; Selleckchem), CHIR99021 (2 μM, 4 μM, or 6 μM; Selleckchem), ALK5 small-molecule inhibitor SB431542 (10 μM, Selleckchem) and small-molecule BMP inhibitor LDN193189 (0.2 μM, Selleckchem) at an approximate density of 270 aggregates per well unless otherwise mentioned in the figure legend. Organoids were fed every other day for up to 17 days. Y276932 was removed from the media on day 3. At day 5, organoids were transferred to Neural Induction Media [DMEM F:12 (Corning), N2 supplement (Life Technologies), L-Glutamine (VWR), 2 μg/ml heparin (Sigma Aldrich), non-essential amino acids (Mediatech), penicillin-streptomycin (VWR), supplemented with fresh 0.4 μg/ml ascorbic acid (Sigma Aldrich) and 10 ng/ml brain derived neurotrophin factor (BDNF, R&D Systems)] supplemented with CHIR99021 (2 μM, 4 μM, or 6 μM; Selleckchem), SB431542 (10 μM, Selleckchem) and LDN193189 (0.2 μM, Selleckchem). From day 7 onwards, organoids were fed with Neural Induction Media supplemented with retinoic acid (10 nM, Sigma Aldrich), purmorphamine (300 nM, EMD Millipore) and N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT D5942, 1 μM, Sigma-Aldrich).
Organoid elongation imaging and quantification
Day 5 organoids from a single 10 cm dish were individually transferred using wide-bore pipette tips into the center 60 wells of an uncoated ultra-low attachment 96-well plate (Corning), seeding exactly one organoid per well, with the remaining organoids maintained in rotary culture through day 7. Using an inverted Axio Observer Z1 (Zeiss) microscope with incubation (Zeiss Heating Unit XL S, maintained at 37°C, 5% CO2), all 60 wells were imaged using an AxioCam MRm (Zeiss) digital CMOS camera at 5× magnification (NA 0.16, 2.6 μm×2.6 μm per pixel). Each well was imaged in TL Brightfield every 20 min for 48 h giving a total of 145 frames. At the end of imaging (day 7), 31 organoids from the parallel rotary culture were imaged at 5× magnification to generate a comparison image set.
To segment the organoids, all well images were first aligned by fitting a truncated quadratic curve to the average image intensity, then solving for the peak of maximum intensity, which was assumed to be the well center. Next, the average lighting inhomogeneity was calculated as the pixel-wise median of all 60 aligned well images, which was then subtracted from the individual aligned frames. After background correction, individual organoids were isolated by finding objects brighter than 0.83% of maximum intensity, but less than 3.0% of maximum intensity, with object size greater than 2000 pixels, eccentricity greater than 0.1 and solidity greater than 40%. A bounding box 2 mm×2 mm around the center of each of these objects was calculated and all frames of the time series cropped to this bounding box to reduce memory usage. To detect the region of maximum motion in the time series, the difference between each pair of sequential images was calculated and then the pixel wise standard deviation was calculated over all these differences in a given region. This standard deviation image was then thresholded at between 0.01 and 0.03 (AU) depending on the remaining lighting inhomogeneity in the image, producing a ring-shaped mask around the periphery of each organoid. Finally, using the interior of the mask as the organoid seed and the exterior as the background seed for the first frame, organoids were segmented using anisotropic diffusion (Grady, 2006), evolving the foreground and background seeds using the contour calculated from the previous frame for subsequent segmentations. Segmentation, labeling and metrology were all performed using the python package sckit-image (van den Walt et al., 2014).
Segmentations were manually inspected for accuracy, with 45 out of 60 determined as having no or only minor flaws, with the remaining 15 excluded from automated analysis. Using the high-quality segmentations only, each organoid time series was then analyzed to examine geometry change over time. For each contour at each time point, we calculated contour area, contour perimeter, minimum, maximum and mean distance from contour center of mass to the perimeter. As additional non-dimensional measures of shape, we calculated the ratio of maximum to minimum radius and organoid circularity. Organoids were also manually classified as ‘elongating’, ‘partially elongating’ or ‘non-elongating’ by examining each video. Organoids assigned to ‘elongating’ exhibited at least one, and at most two large protuberances that extended at least 100 μm from the main body. Partially elongating organoids exhibited at least one, but often many, protuberances, all of which failed to extend robustly past the 100 μm demarcation. Non-elongating organoids were any organoids that failed to generate any extensions over the observation period.
To estimate the number of extensions, TBXT-KD organoids were segmented in bright field as described above. The number of organoid extensions was assessed by analyzing deviation from convex planform area. Specifically, the convex hull of each organoid contour was calculated from organoid perimeter contour, then the organoid planform area was subtracted from the organoid convex area. Resulting regions of at least 30 μm2 were classified as convexity ‘defects’ and further analyzed. Organoids with zero defects were classified as ‘round’. Organoids with one or two defects were classified as ‘single extensions’. Organoids with more than three defects were classified as containing multiple extensions. Furthermore, a ratio of defect area to convex area for each organoid was calculated, to estimate how perpendicular each extension was from the main organoid body. Finally, organoid perimeter tortuosity was assessed as the ratio of convex perimeter to segmented perimeter, with ratios of 1.0 approaching perfect smoothness, and ratios of zero approaching levels of convolution.
Real time quantitative polymerase chain reaction
Total RNA was isolated from organoid samples using an RNAeasy Mini Kit (QIAGEN) according to manufacturer's instructions. Subsequently, cDNA was generated using an iScript cDNA Synthesis kit (BIORAD) and the reaction was run on a SimpliAmp thermal cycler (Life Technologies). A quantitative PCR reaction using Fast SYBR Green Master Mix (ThermoFisher Scientific) was run on a StepOnePlus Real-Time PCR system (Applied Biosciences). Relative gene expression was determined by normalizing to the housekeeping gene 18S rRNA, using the comparative threshold (CT) method. Gene expression was displayed as fold change of each sample versus control. The primer sequences were obtained from the Harvard Primer bank or designed using the NCBI Primer-BLAST website (Table S1).
Owing to low RNA abundance in the dissected organoids, PreAmp Master Mix (FLUIDIGM) was used to amplify cDNA. Briefly, 20 ng of cDNA were amplified per 5 μl reaction volume for 15 cycles on a SimpliAmp thermal cycler (Life Technologies). Amplified cDNA was diluted fivefold using nuclease-free water. 1 μl of amplified diluted cDNA was used for each 20 μl quantitative PCR reaction using Fast SYBR Green Master Mix (ThermoFisher Scientific) and run on a StepOnePlus Real-Time PCR system (Applied Biosciences), following normal qPCR methods described above.
Histology, immunocytochemistry and imaging
Organoids were fixed with 4% paraformaldehyde (VWR) for 40 min and washed three times with PBS. Organoids to be used for histology were embedded in HistoGel Specimen Processing Gel (Thermo Fisher) prior to paraffin processing. Paraffin-embedded samples were sectioned at 5 μm, and subsequently stained using Hematoxylin and Eosin. For immunofluorescent staining, slides were deparaffinized as for Hematoxylin and Eosin staining. Epitope retrieval was performed by submerging slides in citrate buffer (pH 6.0; Vector Laboratories) in a 95°C water bath for 35 min. Samples were permeabilized in 0.2% Triton X-100 (Sigma-Aldrich) for 5 min, blocked in 1.5% normal donkey serum (Jackson Immunoresearch) for 1 h, and probed with primary antibodies against SOX2, PAX6, TBXT, NES, TUBB3 and CDH2 (Table S3) overnight at 4°C and with secondary antibodies (Alexa Fluor-488/555/647, ThermoFisher Scientific) for 1 h at room temperature. Nuclei were stained with a 1:10,000 dilution of Hoechst 33342 (Thermo Fisher) included with secondary antibodies. Coverslips were mounted with anti-fade mounting medium (ProlongGold, Life Technologies) and samples were imaged on a Zeiss Axio Observer Z1 inverted microscope equipped with a Hamamatsu ORCA-Flash 4.0 camera.
Whole-mount light sheet imaging
4% paraformaldehyde-fixed paraffin-embedded samples (see ‘Histology, immunocytochemistry and imaging’ section) were permeabilized with 1.5% Triton X-100 (Sigma Aldrich) for 1 h, blocked in 5% normal donkey serum (Jackson Immunoresearch) for 1 h, and probed with primary and secondary antibodies (Table S3) overnight. Nuclei were stained with a 1:10,000 dilution of Hoechst 33342 (Thermo Fisher) included with secondary antibodies. Samples were then embedded in 1.5% low melt agarose (BioReagent) and drawn up into ∼1 mm imaging capillaries and subsequently imaged on the Zeiss Z.1 light sheet microscope equipped with two PCO.edge SCMOS cameras at 5× and 20× (NA 1.34, aqueous objective).
Flow cytometry
WTC and H1 cells were pretreated with either 2, 4 or 6 μM CHIR for 2 days, then dissociated from tissue culture plates with Accutase (STEMCELL Technologies) and washed with PBS. Similarly, the LBC-TBXT knockdown cells were pretreated with between 0 and 5 days of doxycycline concurrent with a final 2 days of 4 μM CHIR treatment, then dissociated and washed as described above. Cells were fixed for 20 min with 4% paraformaldehyde and washed three times for 3 min with PBS. Samples were permeabilized in 0.5% Triton-X-100 (Sigma Aldrich) for 30 min, then blocked in 1% normal donkey serum (Jackson Immunoresearch) for 1 h, and probed with primary and secondary antibodies (Table S3) overnight. Samples were run on a LSR-II analyzer (BD Biosciences). Singlets were first identified by gating on forward-scatter to side-scatter ratio, then samples were gated into SOX2+ and TBXT+ using single-stained controls for each cell line, then finally samples were assessed for percent SOX2+/TBXT+ cells. Analysis was conducted with a minimum of 20,000 events per sample.
Bulk RNA-sequencing sample and library preparation
Whole organoids differentiated in either 0 μM CHIR or 4 μM CHIR at days 1, 3, 5, 7 and 10 of the differentiation protocol (n=3 per condition per day) were lysed with RPE buffer with 5 mM 2-mercaptoethanol, and RNA was extracted using the RNeasy Mini Kit (Qiagen) and quantified using the NanoDrop 2000c (ThermoFisher Scientific). RNA-seq libraries were generated using the SMARTer Stranded Total RNA Sample Prep Kit (Takara Bio) and sequenced using NextSeq500/550 High Output v2.5 kit to a minimum depth of 25 million reads per sample. The sequences were aligned to GRCh37 using HiSat2 (Kim, et al., 2015), reads were quantified using the featureCounts tool in the subread package (Liao et al., 2014), and differential expression between 0 μM and 4 μM CHIR-treated organoids was assessed at each day using the edgeR differential expression pipeline with limma/voom normalization (Law et al., 2014). Longitudinal differential expression was assessed for each CHIR condition using the ‘l’ normalization algorithm (Zhang et al., 2019). Raw data have been deposited in GEO under the accession number GSE155382.
Single cell RNA-sequencing sample and library preparation
Multiple organoid samples were combined and processed together using the MULTI-Seq technology (McGinnis et al., 2019). Organoids were singularized using Accutase (STEMCELL Technologies) and washed with ice-cold PBS. Cells were resuspended in PBS with lipid-modified Anchor and Barcode oligonucleotides (gifts from Zev Gartner, UCSF, USA) and incubated on ice for 5 min. A co-Anchor oligo was then added in order to stabilize membrane retention of the barcodes incubated for an additional 5 min on ice. Excess lipid-modified oligos were quenched with 1% BSA in PBS, washed with ice-cold 1% BSA solution, and counted using a Countess II FL (Life Technologies). Single cell GEMs and subsequent libraries were then prepared using the 10× Genomics Single Cell V2 protocol with an additional anchor-specific primer during cDNA amplification to enrich barcode sequences. Short barcode sequences (∼65-100 bp determined by Bioanalyzer) were purified from cDNA libraries with two sequential SPRI bead cleanups. Barcode library preparation was performed according to the KAPA HiFi Hotstart (Kapa Biosystems) protocol to functionalize with the P5 sequencing adapter and library-specific RPIX barcode. Purified ∼173 bp barcode fragments were isolated with another SPRI bead cleanup and validation by Bioanalyzer. Raw data are have been deposited in GEO under accession number GSE155383.
The sample library was sequenced on an Illumina NovaSeq yielding an average of 41,112 reads per cell and 6444 cells. The MULTI-Seq barcode library was sequenced on an Illumina NextSeq yielding an average of 9882 reads per barcode and enabling sample assignment for 4681 of 6124 unique UMIs detected (76.4% recovery), using the demultiplexing code provided by the MULTI-Seq protocol (McGinnis et al., 2019).
Genome annotation, RNA-seq read mapping, and estimation of gene and isoform expression
The sample library was aligned to the human GRCh38 reference genome using Cell Ranger v1.2.0 (10× Genomics). Gene expression levels were assessed using the Seurat v3.0.0 analysis pipeline (Butler et al., 2018). First cells were removed with fewer than 200 detected genes, with fewer than 1000 total detected transcripts or with more than 10% mitochondrial gene expression. Next, expression levels were log normalized, and the top 2000 variable genes calculated using the VST algorithm. The top 20 principal components were used to group cells into two clusters using a resolution of 0.3. Finally, top markers were detected for each cluster by detecting the top differentially expressed genes between both clusters, where at least 25% of cells in the cluster expressed the gene and expression of the gene was at least a 0.25 log2 fold-change different from the remaining population. Clusters and gene expression were visualized on a two-dimensional UMAP projection of the first 15 principal components.
Cluster analysis
To assign cluster identity, the top markers for each cluster were tested for GO term enrichment using the biological process ‘enrichGO’ function in the R package ‘clusterProfiler’ v3.12 (Yu et al., 2012). In addition, differentiation maturity in each cluster was assessed by examining expression level of panels of early neuroectoderm markers, proliferation markers, markers of neuron fate commitment, and markers of cell types present in neural tube formation and axial extension (Tanabe and Jessell, 1996). Finally, to assess anterior-posterior position of each, panels of HOX genes were examined to assign rough position of each cluster along the head-tail axis (Bel-Vialar et al., 2002; Carpenter, 2002).
Quantification of EdU and PH3 localization
Sections stained for DAPI, EdU and pH3 were segmented by detecting the peaks of DAPI staining for each section using non-maximum suppression. The exterior of each section was then segmented by grouping DAPI+ cells into spatially contiguous clusters consisting of at least 5000 pixels that touch the image border in less than 1% of their total area. The exterior contour was then calculated for each region and fit to an elliptical model by total least squares (Halir and Flusser, 2020). All detected cells were projected onto the ellipse major axis, which was then normalized by total length. Where more detections fell on the left half of the major axis, the coordinates were reversed such that the right half of each projected section always contained the majority of detections. Detected cells that corresponded to pH3 or EdU fluorescence at 20% or more above the background level outside of any section were counted as positive detections. A 15-bin histogram was then used to calculate the percentage of projected cells for each day at each point along the semi-major axis, and then gaussian kernel density estimation was used to produce the empirical distribution. Linear and quadratic models were fit using ordinary least squares over the total population.
RNAScope
In situ hybridization for HOXB1, HOXC6 and HOXB9 (probe information in Table S4) was performed on sections of 4% paraformaldehyde-fixed paraffin-embedded samples (see ‘Histology, immunocytochemistry and imaging’ section) using the RNAscope Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics) and following the protocol outlined in User Manual 323100-USM. Sections were imaged on a Zeiss Axio Observer Z1 inverted microscope equipped with a Hamamatsu ORCA-Flash 4.0 camera.
Statistical analysis
Each experiment was performed with at least three biological replicates. Multiple comparisons were used to compare multiple groups followed by unpaired t-tests (two tailed) between two groups subject to a post-hoc Bonferroni correction. In gene expression analysis, three replicates were used for each condition, and all gene expression was normalized to control wild-type populations followed by unpaired t-tests (two-tailed). Significance was specified as P<0.05, unless otherwise specified in figure legends. All error bars represent standard error of the mean (s.e.m.) unless otherwise noted in the figure legend.
Supplementary Material
Acknowledgements
We thank the Gladstone Light Microscopy and Histology Core, the Gladstone Stem Cell Core, the Gladstone Flow Cytometry Core and the Gladstone Graphics Team for their support and experimental expertise. Additionally, we specifically thank Dr Vaishaali Natarajan for her expertise and assistance with flow cytometry.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.R.G.L., D.A.J., T.C.M.; Methodology: A.R.G.L., D.A.J., E.A.B., F.M.-C., J.C.B.; Software: D.A.J., M.Z.K.; Validation: A.R.G.L., D.A.J., N.H.E., E.A.B., M.Z.K., E.A.G., F.M.-C.; Formal analysis: A.R.G.L., D.A.J., N.H.E., E.A.B., M.Z.K.; Investigation: A.R.G.L., D.A.J., N.H.E., E.A.B., E.A.G., F.M.-C., T.C.M.; Data curation: A.R.G.L., D.A.J., N.H.E., E.A.B., T.C.M.; Writing - original draft: A.R.G.L., D.A.J.; Writing - review & editing: A.R.G.L., D.A.J., N.H.E., E.A.B., M.Z.K., E.A.G., J.C.B., T.C.M.; Visualization: A.R.G.L., D.A.J., E.A.B.; Supervision: A.R.G.L., D.A.J., T.C.M.; Project administration: T.C.M.; Funding acquisition: A.R.G.L., T.C.M.
Funding
This research was supported by funding from the California Institute of Regenerative Medicine (LA1_C14-08015) and the National Science Foundation (CBET 0939511). A.R.G.L. was supported by a fellowship from the National Heart Lung and Blood Institute (1F31HL140907-01). Deposited in PMC for release after 12 months.
Data availability
The bulk and single cell RNA-seq data have been deposited in GEO under the accession numbers GSE155382 and GSE155383, respectively. The data analysis code generated in the live imaging analysis studies is available in GitHub https://github.com/david-a-joy/organoid-shape-tools.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.198275
References
- Amin, S., Neijts, R., Simmini, S., Van Rooijen, C., Tan, S. C., Kester, L., Van Oudenaarden, A., Creyghton, M. P. and Deschamps, J. (2016). Cdx and T Brachyury Co-activate growth signaling in the embryonic axial progenitor niche. Cell Rep. 17, 3165-3177. 10.1016/j.celrep.2016.11.069 [DOI] [PubMed] [Google Scholar]
- Amoroso, M. W., Croft, G. F., Williams, D. J., O'keeffe, S., Carrasco, M. A., Davis, A. R., Roybon, L., Oakley, D. H., Maniatis, T., Henderson, C. E.et al. (2013). Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J. Neurosci. 33, 574-586. 10.1523/JNEUROSCI.0906-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azim, E., Jiang, J., Alstermark, B. and Jessell, T. M. (2014). Skilled reaching relies on a V2a propriospinal internal copy circuit. Nature 508, 357-363. 10.1038/nature13021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beccari, L., Moris, N., Girgin, M., Turner, D. A., Baillie-Johnson, P., Cossy, A.-C., Lutolf, M. P., Duboule, D. and Arias, A. M. (2018). Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature 562, 272-276. 10.1038/s41586-018-0578-0 [DOI] [PubMed] [Google Scholar]
- Beck, F., Erler, T., Russell, A. and James, R. (1995). Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev. Dyn. 204, 219-227. 10.1002/aja.1002040302 [DOI] [PubMed] [Google Scholar]
- Beddington, R. S. P. and Robertson, E. J. (1999). Axis development and early asymmetry in mammals. Cell 96, 195-209. 10.1016/S0092-8674(00)80560-7 [DOI] [PubMed] [Google Scholar]
- Bel-Vialar, S., Itasaki, N. and Krumlauf, R. (2002). Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxBgenes in two distinct groups. Development 129, 5103-5115. 10.1242/dev.129.22.5103 [DOI] [PubMed] [Google Scholar]
- Blin, G., Wisniewski, D., Picart, C., Thery, M., Puceat, M. and Lowell, S. (2018). Geometrical confinement controls the asymmetric patterning of brachyury in cultures of pluripotent cells. Development 145, dev166025. 10.1242/dev.166025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe, J. and Ericson, J. (1999). The specification of neuronal identity by graded sonic hedgehog signalling. Semin. Cell Dev. Biol. 10, 353-362. 10.1006/scdb.1999.0295 [DOI] [PubMed] [Google Scholar]
- Broutier, L., Andersson-Rolf, A., Hindley, C. J., Boj, S. F., Clevers, H., Koo, B.-K. and Huch, M. (2016). Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat. Protoc. 11, 1724-1743. 10.1038/nprot.2016.097 [DOI] [PubMed] [Google Scholar]
- Butler, A., Hoffman, P., Smibert, P., Papalexi, E. and Satija, R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411-420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butts, J. C., Mccreedy, D. A., Martinez-Vargas, J. A., Mendoza-Camacho, F. N., Hookway, T. A., Gifford, C. A., Taneja, P., Noble-Haeusslein, L. and Mcdevitt, T. C. (2017). Differentiation of V2a interneurons from human pluripotent stem cells. Proc. Natl. Acad. Sci. USA 114, 4969-4974. 10.1073/pnas.1608254114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambray, N. and Wilson, V. (2002). Axial progenitors with extensive potency are localised to the mouse chordoneural hinge. 129, Development 4855-4866. 10.1242/dev.129.20.4855 [DOI] [PubMed] [Google Scholar]
- Carpenter, E. M. (2002). Hox genes and spinal cord development. Dev. Neurosci. 24, 24-34. 10.1159/000064943 [DOI] [PubMed] [Google Scholar]
- Chambers, S. M., Fasano, C. A., Papapetrou, E. P., Tomishima, M., Sadelain, M. and Studer, L. (2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275-280. 10.1038/nbt.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, D. L. and Papaioannou, V. E. (1998). Three neural tubes in mouse embryos with mutations in the T-box gene Tbx6. Nature 391, 695-697. 10.1038/35624 [DOI] [PubMed] [Google Scholar]
- Chesley, P. (1935). Development of the short-tailed mutant in the house mouse. J. Exp. Zool. 70, 429-459. 10.1002/jez.1400700306 [DOI] [Google Scholar]
- Crone, S. A., Quinlan, K. A., Zagoraiou, L., Droho, S., Restrepo, C. E., Lundfald, L., Endo, T., Setlak, J., Jessell, T. M., Kiehn, O.et al. (2008). Genetic ablation of V2a ipsilateral interneurons disrupts left-right locomotor coordination in mammalian spinal cord. Neuron 60, 70-83. 10.1016/j.neuron.2008.08.009 [DOI] [PubMed] [Google Scholar]
- Diez Del Corral, R. and Storey, K. G. (2004). Opposing FGF and retinoid pathways: a signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. BioEssays 26, 857-869. 10.1002/bies.20080 [DOI] [PubMed] [Google Scholar]
- Eiraku, M., Takata, N., Ishibashi, H., Kawada, M., Sakakura, E., Okuda, S., Sekiguchi, K., Adachi, T. and Sasai, Y. (2011). Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51-56. 10.1038/nature09941 [DOI] [PubMed] [Google Scholar]
- Ericson, J. Fischer, C., Urzi, A., Vidal, R., Kunz, S., Ruffault, P.-L., Kabuss, L., Hube, I., Gazzerro, E. and Birchmeier, C.et al. (1997). Graded sonic hedgehog signaling and the specification of cell fate in the ventral neural tube. Cold Spring Harbor Symp. Quant. Biol. 62, 451-466. 10.1101/SQB.1997.062.01.053 [DOI] [PubMed] [Google Scholar]
- Faustino Martins, J.-M., Fischer, C., Urzi, A., Vidal, R., Kunz, S., Ruffault, P.-L., Kabuss, L., Hube, I., Gazzerro, E., Birchmeier, C.et al. (2020). Self-organizing 3D human trunk neuromuscular organoids. Cell Stem Cell 26, 172-186.e6. 10.1016/j.stem.2019.12.007 [DOI] [PubMed] [Google Scholar]
- Fujii, M., Matano, M., Nanki, K. and Sato, T. (2015). Efficient genetic engineering of human intestinal organoids using electroporation. Nat. Protoc. 10, 1474-1485. 10.1038/nprot.2015.088 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Gobartt, E., Blanco-Ameijeiras, J., Usieto, S., Allio, G., Benazeraf, B. and Martí, E. (2021). Cell intercalation driven by SMAD3 underlies secondary neural tube formation. Dev. Cell 56, 1147-1163. 10.1016/j.devcel.2021.03.023 [DOI] [PubMed] [Google Scholar]
- Gouti, M., Tsakiridis, A., Wymeersch, F. J., Huang, Y., Kleinjung, J., Wilson, V. and Briscoe, J. (2014). In vitro generation of neuromesodermal progenitors reveals distinct roles for wnt signalling in the specification of spinal cord and paraxial mesoderm identity. PLoS Biol. 12, e1001937. 10.1371/journal.pbio.1001937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady, L. (2006). Random walks for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 28, 1768-1783. [DOI] [PubMed] [Google Scholar]
- Guibentif, C., Griffiths, J. A., Imaz-Rosshandler, I., Ghazanfar, S., Nichols, J., Wilson, V., Göttgens, B. and Marioni, J. C. (2021). Diverse routes toward early somites in the mouse embryo. Dev. Cell 56, 141-153.e6. 10.1016/j.devcel.2020.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halir, R. and Flusser, J. (2020). Numerically stable direct least squares fitting of ellipses. In Proc. 6th International Conference in Central Europe on Computer Graphics and Visualization. WSCG 98, 125-132. [Google Scholar]
- Hay, E. D. and Meier, S. (1974). Glycosaminoglycan synthesis by embryonic inductors: neural tube, notochord, and lens. J. Cell Biol. 62, 889-898. 10.1083/jcb.62.3.889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrique, D., Abranches, E., Verrier, L. and Storey, K. G. (2015). Neuromesodermal progenitors and the making of the spinal cord. Development 142, 2864-2875. 10.1242/dev.119768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hookway, T. A., Butts, J. C., Lee, E., Tang, H. and Mcdevitt, T. C. (2016). Aggregate formation and suspension culture of human pluripotent stem cells and differentiated progeny. Methods 101, 11-20. 10.1016/j.ymeth.2015.11.027 [DOI] [PubMed] [Google Scholar]
- Kim, M., Habiba, A., Doherty, J. M., Mills, J. C., Mercer, R. W. and Huettner, J. E. (2009). Regulation of mouse embryonic stem cell neural differentiation by retinoic acid. Dev. Biol. 328, 456-471. 10.1016/j.ydbio.2009.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D., Langmead, B. and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357-360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster, M. A. and Knoblich, J. A. (2014). Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 345, 1247125. 10.1126/science.1247125 [DOI] [PubMed] [Google Scholar]
- Lancaster, M. A., Renner, M., Martin, C.-A., Wenzel, D., Bicknell, L. S., Hurles, M. E., Homfray, T., Penninger, J. M., Jackson, A. P. and Knoblich, J. A. (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373-379. 10.1038/nature12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson, M. H., Gilbert, L. A., Wang, X., Lim, W. A., Weissman, J. S. and Qi, L. S. (2013). CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8, 2180-2196. 10.1038/nprot.2013.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law, C. W., Chen, Y., Shi, W. and Smyth, G. K. (2014). voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29. 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X.-J., Du, Z.-W., Zarnowska, E. D., Pankratz, M., Hansen, L. O., Pearce, R. A. and Zhang, S.-C. (2005). Specification of motoneurons from human embryonic stem cells Nature Biotechnology. Nat. Biotechnol. 23, 215-221. 10.1038/nbt1063 [DOI] [PubMed] [Google Scholar]
- Liao, Y., Smyth, G. K. and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923-930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Libby, A. R., Joy, D. A., So, P.-L., Mandegar, M. A., Muncie, J. M., Mendoza-Camacho, F. N., Weaver, V. M., Conklin, B. R. and Mcdevitt, T. C. (2018). Spatiotemporal mosaic self-patterning of pluripotent stem cells using CRISPR interference. eLife 7, e36045. 10.7554/eLife.36045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem, K. F., Tremml, G., Roelink, H. and Jessell, T. M. (1995).Dorsal differentiation of neural plate cells induced by BMP-mediated signals from epidermal ectoderm. Cell 82, 969-979. 10.1016/0092-8674(95)90276-7 [DOI] [PubMed] [Google Scholar]
- Lindborg, B. A., Brekke, J. H., Vegoe, A. L., Ulrich, C. B., Haider, K. T., Subramaniam, S., Venhuizen, S. L., Eide, C. R., Orchard, P. J., Chen, W.et al. (2016). Rapid induction of cerebral organoids from human induced pluripotent stem cells using a chemically defined hydrogel and defined cell culture medium. Stem Cells Transl. Med. 5, 970-979. 10.5966/sctm.2015-0305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, T. E., Bergendahl, V., Levenstein, M. E., Yu, J., Probasco, M. D. and Thomson, J. A. (2006). Feeder-independent culture of human embryonic stem cells. Nat. Methods 3, 637-646. 10.1038/nmeth902 [DOI] [PubMed] [Google Scholar]
- Mandegar, M. A., Huebsch, N., Frolov, E. B., Shin, E., Truong, A., Olvera, M. P., Chan, A. H., Miyaoka, Y., Holmes, K., Spencer, C. I.et al. (2016). CRISPR interference efficiently induces specific and reversible gene silencing in human iPSCs. Cell Stem Cell 18, 541-553. 10.1016/j.stem.2016.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marikawa, Y., Chen, H.-R., Menor, M., Deng, Y. and Alarcon, V. B. (2020). Exposure-based assessment of chemical teratogenicity using morphogenetic aggregates of human embryonic stem cells. Reprod. Toxicol. 91, 74-91. 10.1016/j.reprotox.2019.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-Green, M. and Erickson, C. A. (1986). Development of neural tube basal lamina during neurulation and neural crest cell emigration in the trunk of the mouse embryo. Development 98, 219-236. 10.1242/dev.98.1.219 [DOI] [PubMed] [Google Scholar]
- Mazzoni, E. O., Mahony, S., Peljto, M., Patel, T., Thornton, S. R., Mccuine, S., Reeder, C., Boyer, L. A., Young, R. A., Gifford, D. K.et al. (2013). Saltatory remodeling of Hox chromatin in response to rostrocaudal patterning signals. Nat. Neurosci. 16, 1191-1198. 10.1038/nn.3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcginnis, C. S., Patterson, D. M., Winkler, J., Conrad, D. N., Hein, M. Y., Srivastava, V., Hu, J. L., Murrow, L. M., Weissman, J. S., Werb, Z.et al. (2019). MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat. Methods 16, 619-626. 10.1038/s41592-019-0433-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt, A., Eberle, D., Tazaki, A., Ranga, A., Niesche, M., Wilsch-Bräuninger, M., Stec, A., Schackert, G., Lutolf, M. and Tanaka, E. M. (2014). 3D reconstitution of the patterned neural tube from embryonic stem cells. Stem Cell Rep. 3, 987-999. 10.1016/j.stemcr.2014.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzis, V., Steinhauser, S., Pakanavicius, E., Gouti, M., Stamataki, D., Ivanovitch, K., Watson, T., Rayon, T., Mousavy Gharavy, S. N. and Lovell-Badge, R.et al. (2018). Nervous system regionalization entails axial allocation before neural differentiation. Cell 175, 1105-1118.e17. 10.1016/j.cell.2018.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka, Y., Chan, A. H., Judge, L. M., Yoo, J., Huang, M., Nguyen, T. D., Lizarraga, P. P., So, P.-L. and Conklin, B. R. (2014). Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat. Methods 11, 291-293. 10.1038/nmeth.2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moris, N., Anlas, K., Van Den Brink, S. C., Alemany, A., Schröder, J., Ghimire, S., Balayo, T., Van Oudenaarden, A. and Martinez Arias, A. (2020). An in vitro model of early anteroposterior organization during human development. Nature 582, 410-415. 10.1038/s41586-020-2383-9 [DOI] [PubMed] [Google Scholar]
- Mouilleau, V., Vaslin, C., Gribaudo, S., Robert, R., Nicolas, N., Jarrige, M., Terray, A., Lesueur, L., Mathis, M. W., Croft, G.et al. (2021). Dynamic extrinsic pacing of the HOX clock in human axial progenitors controls motor neuron subtype specification. Development 148, dev194514. 10.1242/dev.194514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muncie, J. M., Ayad, N. M. E., Lakins, J. N., Xue, X., Fu, J. and Weaver, V. M. (2020). Mechanical tension promotes formation of gastrulation-like nodes and patterns mesoderm specification in human embryonic stem cells. Dev. Cell 55, 679-694.e11. 10.1016/j.devcel.2020.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch, J. N. and Copp, A. J. (2010). The relationship between Sonic hedgehog signaling, cilia and neural tube defects. Birth Defects Res. A Clin. Mol. Teratol. 88, 633-652. 10.1002/bdra.20686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murisic, N., Hakim, V., Kevrekidis, I. G., Shvartsman, S. Y. and Audoly, B. (2015). From discrete to continuum models of three-dimensional deformations in epithelial sheets. Biophys. J. 109, 154-163. 10.1016/j.bpj.2015.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, M. R., Howard, D., Jensen, O. E., King, J. R., Rose, F. R. A. J. and Waters, S. L. (2011). Growth-induced buckling of an epithelial layer. Biomech. Model. Mechanobiol. 10, 883-900. 10.1007/s10237-010-0280-0 [DOI] [PubMed] [Google Scholar]
- Oginuma, M., Moncuquet, P., Xiong, F., Karoly, E., Chal, J., Guevorkian, K. and Pourquié, O. (2017). A gradient of glycolytic activity coordinates FGF and Wnt signaling during elongation of the body axis in amniote embryos. Dev. Cell 40, 342-353.e10. 10.1016/j.devcel.2017.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oginuma, M., Harima, Y., Tarazona, O. A., Diaz-Cuadros, M., Michaut, A., Ishitani, T., Xiong, F. and Pourquié, O. (2020). Intracellular pH controls WNT downstream of glycolysis in amniote embryos. Nature 584, 98-101. 10.1038/s41586-020-2428-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, Y., Shimazaki, T., Sobue, G. and Okano, H. (2004). Retinoic-acid-concentration-dependent acquisition of neural cell identity during in vitro differentiation of mouse embryonic stem cells. Dev. Biol. 275, 124-142. 10.1016/j.ydbio.2004.07.038 [DOI] [PubMed] [Google Scholar]
- Ortmann, D. and Vallier, L. (2017). Variability of human pluripotent stem cell lines. Curr. Opin. Genet. Dev. 46, 179-185. 10.1016/j.gde.2017.07.004 [DOI] [PubMed] [Google Scholar]
- O'Shea, K. S. (1987). Differential deposition of basement membrane components during formation of the caudal neural tube in the mouse embryo. Development 99, 509-519. 10.1242/dev.99.4.509 [DOI] [PubMed] [Google Scholar]
- Park, S., Kim, D., Jung, Y.-G. and Roh, S. (2015). Thiazovivin, a Rho kinase inhibitor, improves stemness maintenance of embryo-derived stem-like cells under chemically defined culture conditions in cattle. Anim. Reprod. Sci 161, 47-57. 10.1016/j.anireprosci.2015.08.003 [DOI] [PubMed] [Google Scholar]
- Pennimpede, T., Proske, J., König, A., Vidigal, J. A., Morkel, M., Bramsen, J. B., Herrmann, B. G. and Wittler, L. (2012). In vivo knockdown of Brachyury results in skeletal defects and urorectal malformations resembling caudal regression syndrome. Dev. Biol. 372, 55-67. 10.1016/j.ydbio.2012.09.003 [DOI] [PubMed] [Google Scholar]
- Sagy, N., Slovin, S., Allalouf, M., Pour, M., Savyon, G., Boxman, J. and Nachman, I. (2019). Prediction and control of symmetry breaking in embryoid bodies by environment and signal integration. Development 146, dev181917. 10.1242/dev.181917 [DOI] [PubMed] [Google Scholar]
- Schiffmann, Y. (2006). Symmetry breaking and convergent extension in early chordate development. Prog. Biophys. Mol. Biol. 92, 209-231. 10.1016/j.pbiomolbio.2005.10.002 [DOI] [PubMed] [Google Scholar]
- Shi, Y., Kirwan, P., Smith, J., Robinson, H. P. C. and Livesey, F. J. (2012). Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat. Neurosci. 15, 477-486. 10.1038/nn.3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum, A. S. W., Poon, L. L. M., Tang, W. W. T., Koide, T., Chan, B. W. H., Leung, Y.-C. G., Shiroishi, T. and Copp, A. J. (1999). Retinoic acid induces down-regulation of Wnt-3a, apoptosis and diversion of tail bud cells to a neural fate in the mouse embryo. Mech. Dev. 84, 17-30. 10.1016/S0925-4773(99)00059-3 [DOI] [PubMed] [Google Scholar]
- Steventon, B., Duarte, F., Lagadec, R., Mazan, S., Nicolas, J.-F. and Hirsinger, E. (2016). Species-specific contribution of volumetric growth and tissue convergence to posterior body elongation in vertebrates. Development 143, 1732-1741. 10.1242/dev.126375 [DOI] [PubMed] [Google Scholar]
- Takemoto, T., Uchikawa, M., Yoshida, M., Bell, D. M., Lovell-Badge, R., Papaioannou, V. E. and Kondoh, H. (2011). Tbx6-dependent Sox2 regulation determines neural or mesodermal fate in axial stem cells. Nature 470, 394-398. 10.1038/nature09729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe, Y. and Jessell, T. M. (1996). Diversity and Pattern in the Developing Spinal Cord. Science 274, 1115-1123. 10.1126/science.274.5290.1115 [DOI] [PubMed] [Google Scholar]
- Trushko, A., Di Meglio, I., Merzouki, A., Blanch-Mercader, C., Abuhattum, S., Guck, J., Alessandri, K., Nassoy, P., Kruse, K., Chopard, B.et al. (2020). Buckling of an epithelium growing under spherical confinement. Dev. Cell 54, 655-668.e6. 10.1016/j.devcel.2020.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Akker, E., Forlani, S., Chawengsaksophak, K., De Graaff, W., Beck, F., Meyer, B. I. and Deschamps, J. (2002). Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development 129, 2181-2193. 10.1242/dev.129.9.2181 [DOI] [PubMed] [Google Scholar]
- van den Brink, S. C. . ., Alemany, A., van Batenburg, V., Moris, N., Blotenburg, M., Vivié, J., Baillie-Johnson, P., Nichols, J., Sonnen, K. F., Martinez Arias, A.et al. (2020). Single-cell and spatial transcriptomics reveal somitogenesis in gastruloids. Nature 582, 405-409. 10.1038/s41586-020-2024-3 [DOI] [PubMed] [Google Scholar]
- van der Walt, S., Schönberger, J. L., Nunez-Iglesias, J., Boulogne, F., Warner, J. D., Yager, N., Gouillart, E. and Yu, T. and the scikit-image contributors (2014). scikit-image: image processing in Python. PeerJ 2, e453. 10.7717/peerj.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenvliet, J. V., Bolondi, A., Kretzmer, H., Haut, L., Scholze-Wittler, M., Schifferl, D., Koch, F., Guignard, L., Kumar, A. S., Pustet, M.et al. (2020). Mouse embryonic stem cells self-organize into trunk-like structures with neural tube and somites. Science 370, eaba4937. 10.1126/science.aba4937 [DOI] [PubMed] [Google Scholar]
- Wichterle, H., Lieberam, I., Porter, J. A. and Jessell, T. M. (2002). directed differentiation of embryonic stem cells into motor neurons. Cell 110, 385-397. 10.1016/S0092-8674(02)00835-8 [DOI] [PubMed] [Google Scholar]
- Wilson, V. and Beddington, R. (1997). Expression of T protein in the primitive streak is necessary and sufficient for posterior mesoderm movement and somite differentiation. Dev. Biol. 192, 45-58. 10.1006/dbio.1997.8701 [DOI] [PubMed] [Google Scholar]
- Wilson, V., Rashbass, P. and Beddington, R. S. (1993). Chimeric analysis of T (Brachyury) gene function. Development 117, 1321-1331. 10.1242/dev.117.4.1321 [DOI] [PubMed] [Google Scholar]
- Wilson, V., Olivera-Martinez, I. and Storey, K. G. (2009). Stem cells, signals and vertebrate body axis extension. Development 136, 1591-1604. 10.1242/dev.021246 [DOI] [PubMed] [Google Scholar]
- Wymeersch, F. J., Huang, Y., Blin, G., Cambray, N., Wilkie, R., Wong, F. C. K., Wilson, V. (2016). Position-dependent plasticity of distinct progenitor types in the primitive streak. eLife 5, e10042. 10.7554/eLife.10042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymeersch, F. J., Wilson, V. and Tsakiridis, A. (2021). Understanding axial progenitor biology in vivo and in vitro. Development 148, dev180612. 10.1242/dev.180612 [DOI] [PubMed] [Google Scholar]
- Yamaguchi, T. P. (2001). Heads or tails: Wnts and anterior–posterior patterning. Curr. Biol. 11, R713-R724. 10.1016/S0960-9822(01)00417-1 [DOI] [PubMed] [Google Scholar]
- Yu, G., Wang, L.-G., Han, Y. and He, Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A J. Integr. Biol. 16, 284-287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Yu, D., Seo, M., Hersh, C. P., Weiss, S. T. and Qiu, W. (2019). Novel Data Transformations for RNA-seq differential expression analysis. Sci. Rep. 9, 4820. 10.1038/s41598-019-41315-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Zhang, Z., Chen, P., Ma, C. Y., Li, C., Au, T. Y. K., Tam, V., Peng, Y., Wu, R., Cheung, K. M. C.et al. (2020). Directed differentiation of notochord-like and nucleus pulposus-like cells using human pluripotent stem cells. Cell Rep. 30, 2791-2806.e5. 10.1016/j.celrep.2020.01.100 [DOI] [PubMed] [Google Scholar]
- Zheng, Y., Xue, X., Resto-Irizarry, A. M., Li, Z., Shao, Y., Zheng, Y., Zhao, G. and Fu, J. (2019). Dorsal-ventral patterned neural cyst from human pluripotent stem cells in a neurogenic niche. Sci. Adv. 5, eaax5933. 10.1126/sciadv.aax5933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, G., Droho, S., Crone, S. A., Dietz, S., Kwan, A. C., Webb, W. W., Sharma, K. and Harris-Warrick, R. M. (2010). Electrophysiological Characterization of V2a Interneurons and Their Locomotor-Related Activity in the Neonatal Mouse Spinal Cord. J. Neurosci. 30, 170-182. 10.1523/JNEUROSCI.4849-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Kim, H. Y., Davidson, L. A. (2009). Actomyosin stiffens the vertebrate embryo during crucial stages of elongation and neural tube closure. Development 136, 677-688. 10.1242/dev.026211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, T., Tan, L., Cederquist, G. Y., Fan, Y., Hartley, B. J., Mukherjee, S., Tomishima, M., Brennand, K. J., Zhang, Q., Schwartz, R. E.et al. (2017). High-content screening in hPSC-neural progenitors identifies drug candidates that inhibit Zika virus infection in fetal-like organoids and adult brain. Cell Stem Cell 21, 274-283.e5. 10.1016/j.stem.2017.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.