Abstract
Background
Lignin is one of the main components of the cell wall and is directly associated with plant development and defence mechanisms in plants, especially in response to Fusarium graminearum (Fg) infection. Caffeoyl-coenzyme A O-methyltransferase (CCoAOMT) is the main regulator determining the efficiency of lignin synthesis and composition. Although it has been characterized in many plants, to date, the importance of the CCoAOMT family in wheat is not well understood.
Results
Here, a total of 21 wheat CCoAOMT genes (TaCCoAOMT) were identified through an in silico genome search method and they were classified into four groups based on phylogenetic analysis, with the members of the same group sharing similar gene structures and conserved motif compositions. Furthermore, the expression patterns and co-expression network in which TaCCoAOMT is involved were comprehensively investigated using 48 RNA-seq samples from Fg infected and mock samples of 4 wheat genotypes. Combined with qRT-PCR validation of 11 Fg-responsive TaCCoAOMT genes, potential candidates involved in the FHB response and their regulation modules were preliminarily suggested. Additionally, we investigated the genetic diversity and main haplotypes of these CCoAOMT genes in bread wheat and its relative populations based on resequencing data.
Conclusions
This study identified and characterized the CCoAOMT family in wheat, which not only provided potential targets for further functional analysis, but also contributed to uncovering the mechanism of lignin biosynthesis and its role in FHB tolerance in wheat and beyond.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-021-07849-y.
Keywords: CCoAOMT gene family, Fusarium head blight (FHB), Expression pattern, Regulation module, Genetic variation, Wheat
Background
Wheat is considered as one of the most important staple crops worldwide, which accounts for approximately 30 % of the global cultivated area, and provides 20 % of the world’s food for consumption [1, 2]. Wheat is also an important source of human protein and mineral element intake [3, 4]. Continuously increasing and stable production of wheat holds the promise for ensuring global food security under the challenge of population boom and climate change as well as limited resource input in the future [5]. Fusarium head blight (FHB), which is also called scab and is caused mainly by Fusarium graminearum (Fg), is one of the most destructive diseases of wheat, resulting in huge wheat yield loss and the imposition of substantial health threats in both humans and livestock due to the DON toxin [6, 7]. More concerning, FHB has gradually become the major hazard and limitation of wheat production in recent years because of climate change and the expansion of conservation agriculture [8]. Thus, revealing the mechanism underlying FHB resistance and breeding FHB-tolerant wheat varieties are crucial to address these problems.
The cell wall is mainly composed of polysaccharides, phenolic compounds and proteins. In plant cells, the cell wall always has mechanical and regulatory functions [9]. Extensive studies have reported that the components of the cell wall endow plants with the ability to resist the invasion of pathogens [10, 11]. Giancaspro et al. found that the pectin methylesterase PME-1 and β, 1–3 glucanase (Glu-1) could be considered putative causal genes of FHB resistance in wheat, as they are involved in cell wall metabolism and regulated the non-specific lipid transfer protein (nsLTP-1) [10]. Lionetti et al. showed that PMEIs (pectin methylesterase inhibitors) could dynamically regulate the PME (pectin methylesterase) activity during Botrytis infection, and AtPMEI10, AtPMEI11 and AtPMEI12 were verified to play the significant roles in cell wall metabolism to enhance plant immunity [12].
Lignin is the second most abundant component of the plant cell wall, and is also involved in the basal disease resistance in plants [13]. It was reported that lignin accumulated rapidly in the ear cell wall of both resistant and susceptible wheat spikes during Fusarium culmorum infection, and the content of lignin in resistant spikes was significantly higher than that in susceptible spikes [14]. Soni et al. found that silencing of TaNAC032 could significantly decrease the lignin content in the rachis, resulting in the increased susceptibility to Fg infection in transgenic wheat [15]. Dhokane et al. integrated metabolo-transcriptomics to reveal the FHB candidate resistance gene in wheat QTL-Fhb2 and found that CAD (cinnamyl alcohol dehydrogenase) might be the putative resistance gene localized within the QTL-Fhb2 region, which is the crucial gene for lignin synthesis [16]. These studies suggested that lignin might also be involved in the FHB response and tolerance in wheat.
Lignin has been demonstrated to polymerize via dehydrogenation three hydroxycinnamyl alcohols, p-coumaryl, coniferyl, and sinapyl alcohols, which contribute to lignin biosynthesis, and these three hydroxycinnamyl alcohols give rise to the p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units of the lignin polymer, respectively [17]. And O-Methyltransferases (OMTs) play an important role in regulating these secondary metabolic processes involved in lignin biosynthesis. OMTs are generally classified into two types, including caffeic acid O-methyltransferase (COMT) and caffeoyl-coenzyme A O-methyltransferase (CCoAOMT), of which COMT controls the S unit pathway and CCoAOMT affects the S and G unit pathways[18]. COMT can catalyse the O-methylation at the 5 position of the aromatic ring, and CCoAOMT functions to form the 3 position of the aromatic ring [19, 20]. The hydroxylation and methylation steps are crucial to determine the lignin composition and the S/G ratio is a major determinant of lignin quality [18].
In light of its significance, the CCoAOMT gene family has been systematically investigated and analysed in many plants, such as Arabidopsis and rice [21], citrus [22], switchgrass [23], dove tree [24], tea plant [25] and sorghum [26]. In wheat, Nguyen et al. analysed the expression patterns and potential functions of some genes involved in lignin biosynthesis including several CCoAOMTs, and indicated that lodging resistance, tolerance against biotic and abiotic stresses and feedstock quality of wheat biomass were closely associated with its lignin content [27]. Ma and Luo verified that TaCCoAOMT1 was an important gene for regulating lignin biosynthesis, which is critical for stem development [28].
Although some CCoAOMT genes have been functionally characterized in wheat, the genomic organization, evolutionary relationship and regulatory module of the wheat CCoAOMT family are not well understood at present, especially in association with FHB. In this study, we performed an in silico genome-wide search method to identify and characterize the CCoAOMT family in wheat using updated reference genome information. Then, their phylogenetic relationships, conserved motifs and cis-elements were systematically analysed. Furthermore, the expression patterns and co-expression network of TaCCoAOMT genes under Fg treatment were studied and FHB-responsive TaCCoAOMTs as well as the regulatory modules were obtained. Finally, the genetic diversity and divergence of CCoAOMT genes in different Triticum species were also investigated based on resequencing data to reveal the evolutionary effect of this family during wheat formation.
Results
Genome-wide identification of CCoAOMT genes in wheat
Using the genome-wide search method described in the Methods section, a total of 21 CCoAOMT genes were detected in the wheat genome. Since there is no standard nomenclature, these identified CCoAOMT genes were named as TaCCoAOMT1 to TaCCoAOMT21 based on their chromosome location (Table 1). These TaCCoAOMT genes were mainly located on group 7 chromosomes (66.67 %), but not on groups 2, 3 and 6 chromosomes. The sizes of TaCCoAOMT genes ranged from 447 (TaCCoAOMT17) to 4567 (TaCCoAOMT12) bp in length. The average lengths of the CDS and amino acid sequences were 761 bp and 253 aa, respectively. The isoelectric point (pI) ranged from 4.89 (TaCCoAOMT15) to 11.11 (TaCCoAOMT17) and the molecular weight (Mw) ranged from 16284.2 (TaCCoAOMT17) to 34158.48 (TaCCoAOMT21), respectively. A search for orthologues of TaCCoAOMT genes revealed that 20 (95.24 %) of the TaCCoAOMTs had orthologues in Arabidopsis and rice, with the only exception being TaCCoAOMT6. Subcellular localization prediction showed that most of them were located in the cytoplasm and chloroplasts, and only one gene was located in the mitochondria and nucleus, respectively. In these 21 TaCCoAOMT genes, we also found four homoeologous gene groups with each containing A, B and D homoeologous copies, and all of them were localized on chromosome group 7, resulting in chromosome group 7 having the most abundant TaCCoAOMT genes.
Table 1.
Characteristics of the CCoAOMT genes identified in wheat
| TaCCoAOMT | Transcript | Gene Length (bp) |
Protein Length (aa) | Exon number | Splice variant | pI | Mw (Da) | Subcellular localization | Orthologs |
|---|---|---|---|---|---|---|---|---|---|
| TaCCoAOMT1 | TraesCS1B02G023200.2 | 2088 | 238 | 5 | 2 | 5.03 | 26054.06 | Cytoplasmic | AtCCoAMT3 |
| TaCCoAOMT2 | TraesCS1B02G049800.1 | 1168 | 210 | 2 | 1 | 5.55 | 23350.86 | Cytoplasmic | AtCCoAMT3 |
| TaCCoAOMT3 | TraesCS1D02G019000.1 | 1245 | 248 | 2 | 1 | 5.02 | 27035.03 | Cytoplasmic | - |
| TaCCoAOMT4 | TraesCS4A02G442400.1 | 1355 | 246 | 2 | 1 | 4.98 | 27050.77 | Cytoplasmic | OsCCoAOMT3 |
| TaCCoAOMT5 | TraesCS4D02G362500.1 | 942 | 190 | 1 | 1 | 5.04 | 20387.36 | Chloroplast | OsCCoAOMT5 |
| TaCCoAOMT6 | TraesCS5A02G257600.1 | 1136 | 252 | 5 | 2 | 5.39 | 27304.38 | Chloroplast | - |
| TaCCoAOMT7 | TraesCS5D02G265900.1 | 1586 | 242 | 5 | 1 | 5.15 | 26189.03 | Chloroplast | OsCCoAOMT6 |
| TaCCoAOMT8 | TraesCS7A02G068600.1 | 1648 | 288 | 4 | 2 | 7.01 | 31886.32 | Mitochondrial | OsCCoAOMT5 |
| TaCCoAOMT9 | TraesCS7A02G127600.1 | 1093 | 284 | 3 | 1 | 5.04 | 31491.93 | Cytoplasmic | OsCCoAOMT1 |
| TaCCoAOMT10 | TraesCS7A02G240300.1 | 1753 | 247 | 2 | 1 | 4.99 | 27088.92 | Cytoplasmic | OsCCoAOMT3 |
| TaCCoAOMT11 | TraesCS7A02G240400.1 | 1219 | 245 | 3 | 1 | 4.95 | 27071.88 | Cytoplasmic | OsCCoAOMT4 |
| TaCCoAOMT12 | TraesCS7A02G302500.1 | 4567 | 300 | 10 | 2 | 8.59 | 32885.04 | Chloroplast | AtCCoAOMT5 |
| TaCCoAOMT13 | TraesCS7B02G027200.1 | 1729 | 267 | 4 | 1 | 5.11 | 29506.65 | Cytoplasmic | OsCCoAOMT1 |
| TaCCoAOMT14 | TraesCS7B02G135900.1 | 1854 | 302 | 2 | 1 | 5.54 | 33139.88 | Chloroplast | OsCCoAOMT3 |
| TaCCoAOMT15 | TraesCS7B02G136000.1 | 1734 | 245 | 3 | 1 | 4.89 | 27124.85 | Cytoplasmic | OsCCoAOMT4 |
| TaCCoAOMT16 | TraesCS7B02G202600.1 | 4218 | 294 | 9 | 1 | 7.57 | 32151.1 | Chloroplast | OsCCoAOMT2 |
| TaCCoAOMT17 | TraesCS7D02G063100.1 | 447 | 148 | 1 | 1 | 11.11 | 16284.2 | Nuclear | AtCCoAOMT3 |
| TaCCoAOMT18 | TraesCS7D02G126000.1 | 2465 | 260 | 4 | 1 | 5.11 | 28864.96 | Cytoplasmic | OsCCoAOMT1 |
| TaCCoAOMT19 | TraesCS7D02G239200.1 | 1548 | 247 | 2 | 1 | 4.98 | 27074.9 | Cytoplasmic | OsCCoAOMT3 |
| TaCCoAOMT20 | TraesCS7D02G239400.1 | 1185 | 245 | 3 | 1 | 4.94 | 27121.94 | Cytoplasmic | OsCCoAOMT3 |
| TaCCoAOMT21 | TraesCS7D02G297800.1 | 3828 | 311 | 9 | 2 | 8.38 | 34158.48 | Chloroplast | OsCCoAOMT2 |
Phylogenetic relationship, exon-intron structure and conserved motif analysis
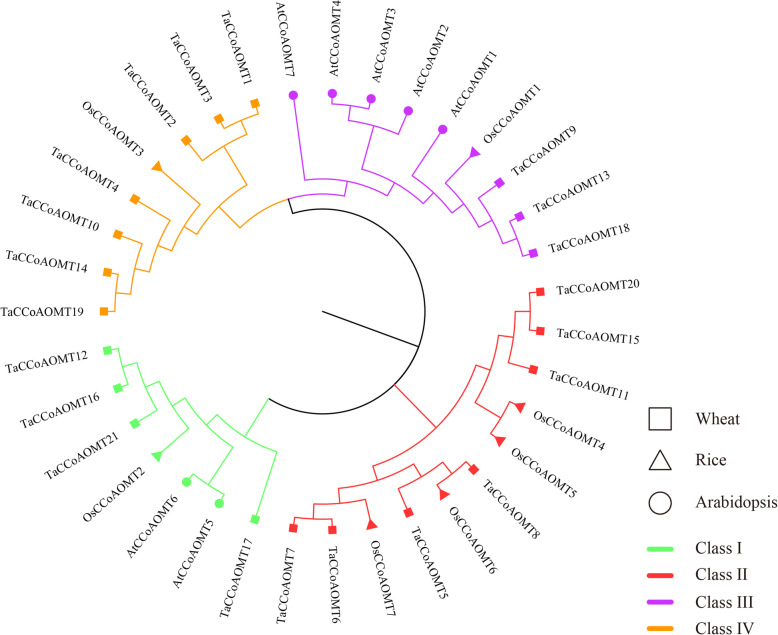
A phylogenetic tree was constructed using the full-length protein sequences of the CCoAOMT genes in wheat, Arabidopsis and rice (Fig. 1). The corresponding nomenclature information of CCoAOMT genes in Arabidopsis and rice is provided in Table S1. The results indicated 4 groups (designated classes I to IV) with 4, 7, 3 and 7 TaCCoAOMT genes, respectively. Meanwhile, TaCCoAOMT and OsCCoAOMT genes were distributed in all of groups, but AtCCoAOMT genes were mainly clustered in class III, and there were just two genes (AtCCoAOMT5 and AtCCoAOMT6) in class I.
Fig. 1.
Phylogenetic relationship of CCoAOMT genes in Arabidopsis, rice and wheat
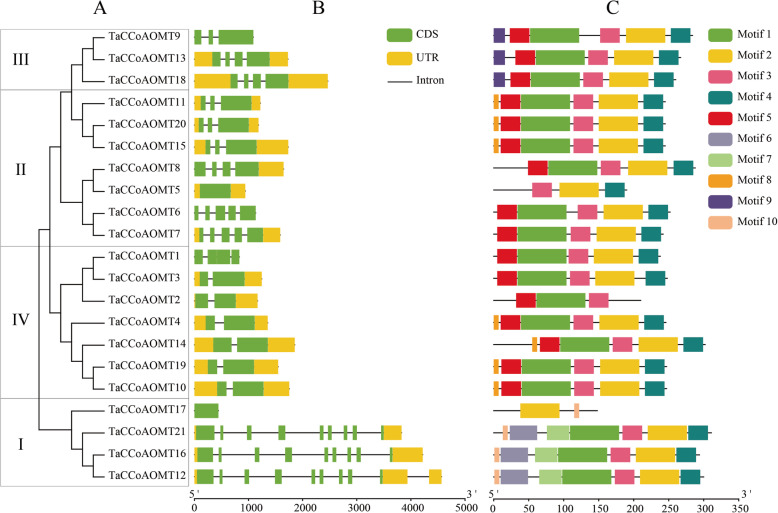
Then, the exon-intron and motif structures of TaCCoAOMTs were further analysed (Fig. 2). The exon number of TaCCoAOMTs ranged from 1 to 10, of which two genes contained only one exon, and 85.71 % of genes had 5 exons or less (Fig. 2B). Furthermore, 10 high confidence motifs were predicted (Fig. 2 C and Figure S1). Compared to other classes, motifs 6 and 9 were exclusively found in classes I and III, respectively. Although few differences in motifs were found between classes II and III, the exon number of TaCCoAOMT genes in class II was greater that in class III. Motif 2 was identified in 20 (95.24 %) TaCCoAOMTs, apart from TaCCoAOMT2 and motif 3 was also identified in 20 TaCCoAOMTs apart from TaCCoAOMT17. Motif 1, 2, 3, 4 were abundant in TaCCoAOMTs (Fig. 2), and all of them were found to be related to O-methyltransferase based on PFAM analysis, which further supported the prediction. Meanwhile, the members in the same groups shared the similar exon-intron structures and motif compositions (Fig. 2).
Fig. 2.
Phylogenetic tree (A), gene structure (B) and conserved motif analysis (C) of the CCoAOMT gene family in wheat
Cis-element analysis of TaCCoAOMTs
A total of 44 types of cis-elements were identified in the 1.5-kb genomic sequences upstream from the transcription start sites (TSS) of TaCCoAOMT genes, with functions primarily associated with the stress response (Table S2 and Figure S2). The CAAT-box was identified in all TaCCoAOMTs (21), followed by the CGTCA-motif (19) and TGACG-motif (19). Together with the CGTCA motif and TGACG motif, the ABRE motif related to abscisic acid responsiveness was identified in 18 TaCCoAOMTs. In addition, CCAAT-box (MYBHv1 binding site), MBS (MYB binding site involved in drought-inducibility), TCA-element (cis-acting element involved in salicylic acid responsiveness), TC-rich repeats (cis-acting element involved in defence and stress responsiveness), MRE- (MYB binding site involved in light responsiveness), SARE- (cis-acting element involved in salicylic acid responsiveness) and WUN-motif (wound-responsive element) were also identified in 13, 7, 7, 2, 1, 1 and 1 TaCCoAOMTs, respectively.
To obtain some clues about the biological function of TaCCoAOMTs, we performed GO (gene ontology) enrichment analysis of them with all wheat proteins as background. The results showed that TaCCoAOMTs were significantly enriched in 10 terms in biological process, 10 terms in molecular function and one in cell component (Figure S3 and Table S3). Among them, lignin biosynthetic process (GO:0009809), O-methyltransferase activity (GO:0008171) and caffeoyl-CoA O-methyltransferase activity (GO:0042409) were significantly enriched, verifying the correlation between TaCCoAOMTs and lignin biosynthesis.
Expression patterns of TaCCoAOMT genes in different tissues and under abiotic stresses
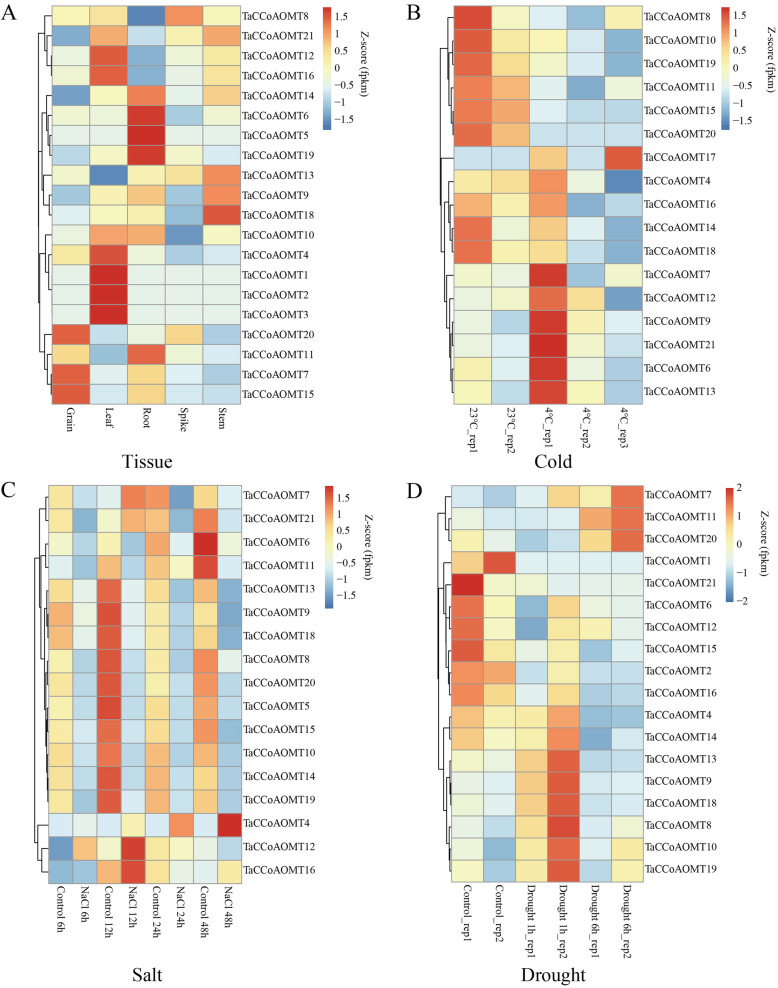
The spatio-temporal expression patterns of TaCCoAOMTs were investigated using RNA-seq samples of different tissues as well as under low temperature, salt, heat and drought stresses. A total of 20 TaCCoAOMTs were found to be expressed in different tissues and some genes exhibited tissue-specific expression. Totally, 3 genes (TaCCoAOMT7, 15, 20) were highly expressed in grain (Fig. 3 A). Meanwhile, 7 and 6 genes displayed high expression in leaves and roots, respectively. TaCCoAOMT8 was expressed only in the spike while 3 genes (TaCCoAOMT9, 13, 18) showed specifically high expression in stem tissue.
Fig. 3.
Expression patterns of TaCCoAOMT genes in different tissues (A) and under diverse abiotic stresses (B to D)
Under low temperature treatment (Fig. 3B and Table S4), we found 3 differentially expressed TaCCoAOMTs, of which TaCCoAOMT9 displayed up-regulated expression, while TaCCoAOMT20 and TaCCoAOMT15 were down-regulated. Under salt stress (Fig. 3 C and Table S4), 3 genes (TaCCoAOMT5, 8, 20) were significantly down-regulated and two (TaCCoAOMT4, 12) were up-regulated at 6 h, and 4 and 5 TaCCoAOMTs were significantly down-regulated and up-regulated at 12 h respectively. Only TaCCoAOMT4 and TaCCoAOMT5 showed differential expression at 24 and 48 h, respectively. Under drought stress (Fig. 3D and Table S4), 5 and 6 TaCCoAOMTs were differentially expressed at 1 and 6 h, respectively, of which 3 genes (TaCCoAOMT2, 13, 20) were differentially expressed at both time points.
Comprehensive analysis of the expression profiles of TaCCoAOMT genes under Fg infection
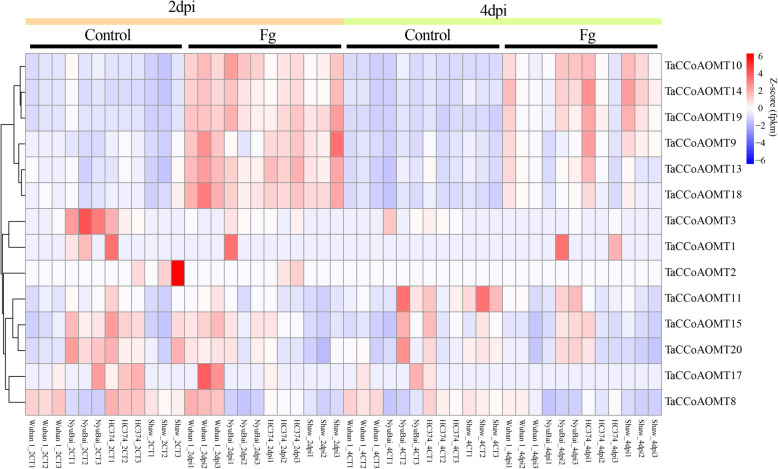
According to a previous study, lignin biosynthesis process-related genes played a significant role in the response to Fg infection [29]. To identify the TaCCoAOMTs involved in the Fg response, we further used 48 RNA-seq samples from Fg infection and mock samples of three FHB-tolerant varieties (HC374, NyuBai, Wuhan 1) and one FHB-susceptible variety (Shaw) at 2 days and 4 days post inoculation (dpi) [30]. Expression patterns of these TaCCoAOMTs showed obvious differentiation between 2 and 4 dpi. At 2 dpi, 13, 12, 12 and 11 of TaCCoAOMTs were expressed in genotypes HC374, NyuBai, Wuhan 1 and Shaw, respectively (Fig. 4 and Table S5), of which down-regulated genes accounted for 64.3 % and up-regulated genes accounted for 35.7 %. The number of DEG in Wuhan1 was the lowest, and that in Shaw was the highest. Furthermore, three homoeologues of TaCCoAOMT (8, 15, 20) showed differential expression among three varieties (HC374, NyuBai, Shaw), of which TaCCoAOMT 8 was differentially expressed among all four varieties. At 4 dpi, there were 12, 11, 10 and 10 TaCCoAOMT genes were expressed in genotypes HC374, NyuBai, Wuhan 1 and Shaw (down-regulated: 58.3 %; up-regulated: 41.7 %), respectively (Fig. 4 and Table S5), of which TaCCoAOMT 11 showed differential expression among all four varieties. Genotype Shaw also had the largest number of DEGs, while the number of DEGs in the other three varieties was basically the same. In general, the number of down-regulated genes was greater than the number of up-regulated genes after Fg injection, and the number of DEGs in Shaw was the hihgest. Compared to different dpi, the number of DEGs in Wuhan 1 increased from 2 dpi to 4 dpi, while the number of DEGs in the other three varieties decreased (HC374, NyuBai) or remained unchanged (Shaw). By comparing resistant and susceptible varieties, TaCCoAOMT3 and TaCCoAOMT17 showed significant down-regulated expression only in three resistant varieties; at the same time, TaCCoAOMT2 was significantly down-regulated only in the susceptible variety. It is interesting that TaCCoAOMT 10, 14 and 19 still showed up-regulated expression between Fg infection and mock samples in all four varieties, suggesting that they might play a vital role in the response to Fg infection in wheat.
Fig. 4.
Expression patterns of TaCCoAOMT genes in four different wheat genotypes inoculated with water (Mock) or Fg at 2 dpi and 4 dpi. Wuhan 1, HC374, Nyubai and Shaw are the names of the four wheat varieties. dpi: days post inoculation
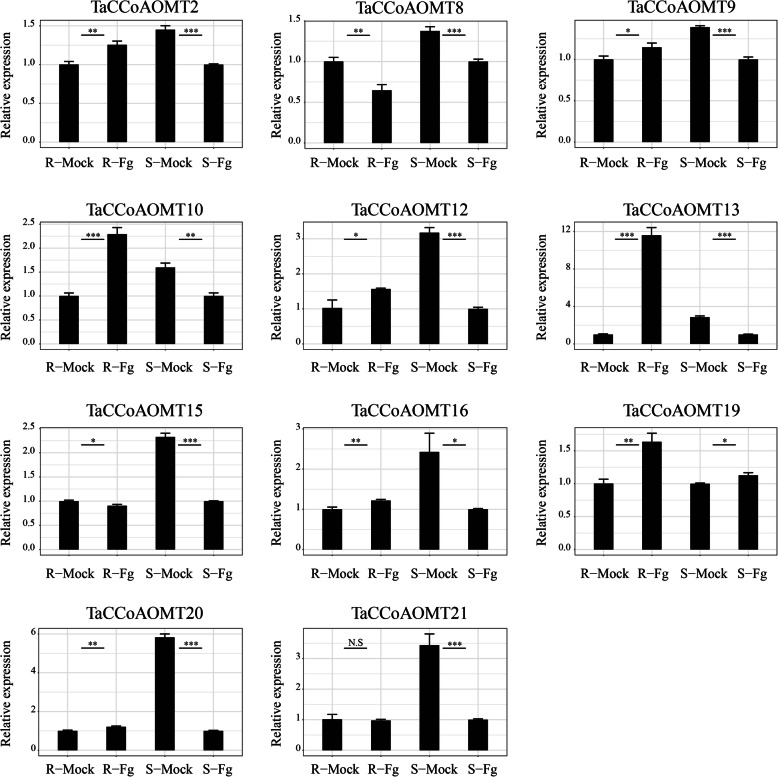
To further verify the expression of TaCCoAOMTs under Fg infection, 11 genes were randomly selected to validate their expression between the resistant and susceptible recombinant inbred lines by qRT-PCR analysis (Fig. 5). The results demonstrated that the expression trend of these genes was consistent with that of RNA-seq analysis. Compared to mock, almost all of the tested genes displayed differential expression under Fg infection in both resistant and susceptible genotypes, of which TaCCoAOMT19 were up-regulated after Fg infection in both resistant and susceptible genotypes, while TaCCoAOMT8, 15, 21 showed down-regulated expression in both genotypes. It is obvious that the expression levels of 7 genes (TaCCoAOMT2, 9, 10, 12, 13, 16, 20) were up-regulated in the resistant genotype but down-regulated in the susceptible genotype, suggesting that they might be involved in Fg resistance.
Fig. 5.
qRT-PCR analysis of 11 FHB-responsive TaCCoAOMT genes. R-Mock: resistant genotype R75 under control condition; R-Fg: resistant genotype R75 under Fg treatment; S-Mock: susceptible genotype S98 under control condition; S-Fg: susceptible genotype under Fg treatment. Significance between mock and Fg infection samples in resistant genotype R75 and susceptible genotype S98 were analyzed using student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.001, N.S: Not significant). The exact sample size for each experimental condition of each TaCCoAOMT gene was 15 spikes, classified into three biological replicates. The bars display the means of TaCCoAOMT gene expression in the mock or control. The error bars represent standard error of mean (SEM) of the three separate technical replicates of qRT-PCR experiments
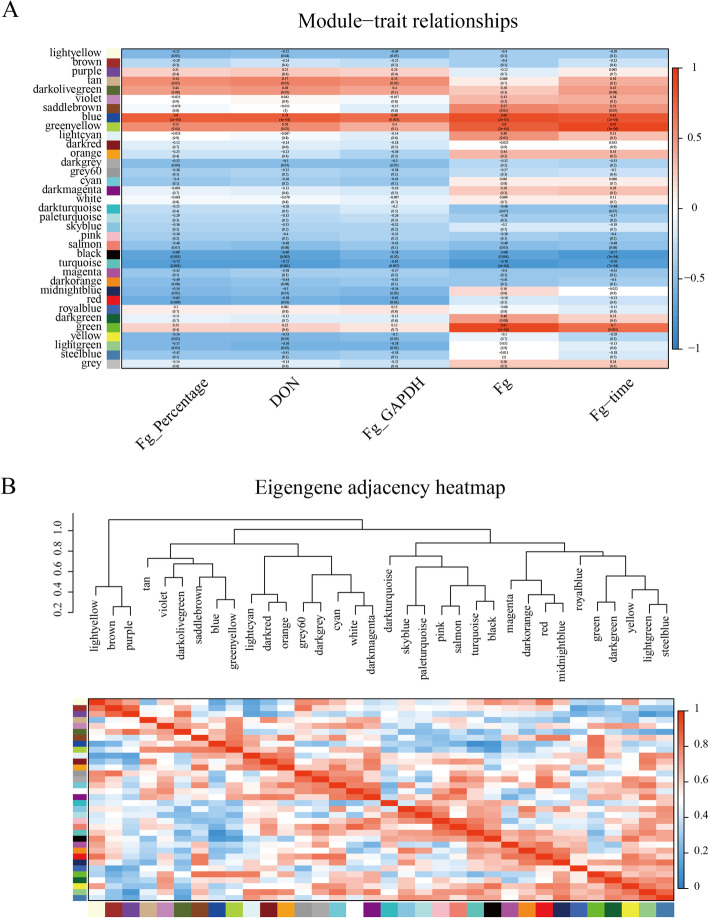
Co-expression network and regulation module of FHB-responsive TaCCoAOMTs
To better understand the function and regulatory network of the identified FHB-responsive TaCCoAOMTs, we further constructed a WGCNA co-expression network based on these RNA-seq data. By constructing a weighted correlation network, 34 co-expression modules were obtained (Fig. 6). Then, we linked the co-expression modules with the available phenotypic data of the Fg inoculation, including percentage (Fusarium oxysporum inoculum), DON (deoxynivalenol), GAPDH (glyceraldehyde-3-phosphate dehydrogenase content) and inoculation time [30]. The results showed that tan, darkolivegreen, blue and greenyellow modules were highly correlated with Fg percentage, DON, GAPDH and inoculation time, respectively (Fig. 6 A). Interestingly, 2, 5, 1, 1 and 2 FHB-responsive TaCCoAOMTs were found in the blue, green, light yellow, turquoise and yellow modules, respectively. Furthermore, these modules were also associated with Fg inoculation (Fig. 6B). A total of 93 genes in these modules showed similar expression patterns. GO enrichment analysis found that they were mainly enriched in terms related to defence response, such as GO:0052544 (defense response by callose deposition in cell wall), GO:0010294 (abscisic acid glucosyltransferase activity) and GO:0000165 (MAPK cascade). It is obvious that the TaCCOAOMTs are located at the hubs of these modules. Among them, TaCCoAOMT19, which is associated with FHB tolerance, could interact with TaCCoAOMT13 to regulate 19 other genes to form a regulatory module (Fig. 7), which might play a potentially important role in FHB tolerance.
Fig. 6.
WGCNA based on 48 RNA-seq samples in wheat. (A). Correlation between co-expression modules and traits. A negative value represents the negative correlation, and a positive value represents the positive correlation. (B). Heatmap of correlation between co-expression modules
Fig. 7.
miRNA-mRNA interaction network of TaCCoAOMT genes under Fg stress
Furthermore, we predicted the miRNAs that could target on TaCCoAOMTs to regulate their expression. Totally, 15 TaCCoAOMT genes were found to be targeted by 25 miRNAs (Table S6) and 50 miRNA-TaCCoAOMT interactions were constructed. Combined with the miRNA-TaCCoAOMT relationship and co-regulation modules of TaCCoAOMTs, we further obtained the miRNA-mediated networks associated with the FHB response and resistance involving TaCCoAOMTs in wheat (Fig. 7), which provided some insights into the regulation of TaCCoAOMTs expression to control lignin biosynthesis and enhanced FHB tolerance through a post-transcriptional approach.
Genetic diversity and haplotype analysis of the CCoAOMT family in wheat and its relatives
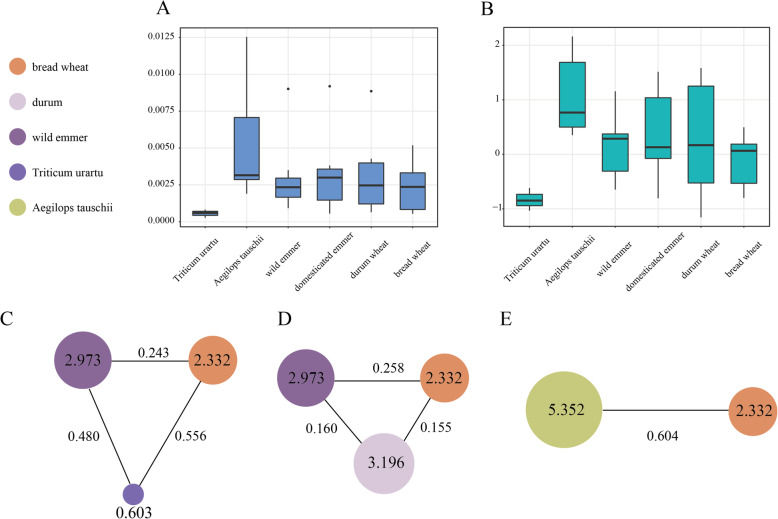
Based on the resequencing data of Triticum species [31], the genetic variations of CCoAOMT genes in wheat and its diploid and tetraploid relatives were investigated, including the nucleotide diversity (π), population divergence (Fst) and Tajima’s D index. The average values of π in Triticum urartu, Aegilops tauschii, wild emmer, domesticated emmer, durum wheat and bread wheat were 0.000565, 0.00535, 0.00297, 0.00321, 0.00320 and 0.00233, respectively (Fig. 8 A), and the average values of Tajima’s D was − 0.834, 1.094, 0.145,0.370, 0.292 and − 0.137, respectively (Fig. 8B). Due to few SNPs identified from the re-sequencing data, T. urartu showed the abnormally lower value of nucleotide diversity. Ae. tauschii displayed the highest nucleotide diversity in CCoAOMT genes, while bread wheat had the lowest nucleotide diversity apart from T. urartu, with a value that decreased by 2 times, suggesting that a significant genetic bottleneck occurred in the CCoAOMT gene family during wheat evolution. Then, the gene flow and genetic divergence between wheat subgenome and its relatives were also detected. In the A subgenome, the Fst value between bread wheat and T. urartu was 0.556, ranking as the largest, followed by that of wild emmer and T. urartu with the value of 0.480, and bread wheat and wild emmer with a value of 0.248, indicating the high divergence between bread wheat and T. urartu, compared to that of wild emmer at the A subgenome level from the perspective of CCoAOMT gene family (Fig. 8 C). In the B subgenome, the divergence between bread wheat and wild emmer was larger than that of durum and wild emmer, and bread wheat was closer to durum wheat than wild emmer wheat (Fig. 8D). In the D subgenome, the Fst value of between bread wheat and Ae. tauschii was 0.604 (Fig. 8E). Overall, the genetic divergence at the D subgenome was highest, followed by the A and B subgenomes.
Fig. 8.
Genetic diversity, population divergence and Tajima’s D across the TaCCoAOMT genes in bread wheat and its relative populations. (A). Nucleotide diversity across five Triticum species among the whole genome. (B). Tajima’s D across five Triticum species in the whole genome. (C). Nucleotide diversity (π × 10− 3) and population divergence (Fst) across the Triticum urartu, wild emmer and bread wheat among the A genome. (D). Nucleotide diversity and population divergence across the wild emmer, durum and bread wheat among the B genome. (E). Nucleotide diversity and population divergence across the Aegilops tauschii and bread wheat among the D genome
Finally, we identified the haplotype organization and frequency of each TaCCoAOMT gene in these populations based on the resequencing data (Table S7). A total of 13 TaCCoAOMT genes were found to have the genetic variations among these populations, of which 4, 2 and 7 genes were located in the A, B, D subgenomes, respectively. Then, the main haplotype and its frequencies were investigated (Figure S4, S5 and S6). It is obvious that the percentage of the main haplotype in cultivated wheat was significantly larger than that of wild species at all subgenome levels, indicating that the artificial selection exerted on these CCoAOMT genes resulted in the decline of genetic diversity and genetic bottlenecks during wheat domestication and improvement processes.
Discussion
Lignin is the main component of the cell wall and is involved in the response to abiotic and biotic stresses [19]. The characteristics of the CCoAOMT gene involved in the lignin synthesis pathway has been analysed in Arabidopsis, sorghum, and other plants. In this study, we identified 21 CCoAOMTs in wheat at the genome level. Based on the phylogenetic relationship, these TaCCoAOMTs were classified into four groups, and the TaCCoAOMTs belonging to the same group shared similar gene structures and motif organizations. In sorghum, the CCoAOMT proteins were classified into clade 1a, clade 1b, clade 1c and clade 2, of which clade 1a was the orthologous gene with AtCCoAOMT1 and OsCCoAOMT1, which was considered as the true CCoAOMT gene, while the other classes were considered as CCoAOMT-like genes [26]. In our results, AtCCoAOMT1 and OsCCoAOMT1 were clustered into the same group (class III), which was consistent with that of sorghum. Meanwhile, CCoAOMT genes in rice and wheat can be found in each group, but AtCCoAOMTs just were found in classes I and III, indicating that there was some divergence between the monocot and dicot CCoAOMT families. Additionally, classes I and III have the specific motifs 6 and 9, respectively. Although no specific motif was found in classes II and IV, their gene structures displayed differences with the obvious variations in exon number, suggesting that more types of splice variants or binding-sites might be present in these two classes.
Based on RNA-seq samples, the expression levels of these wheat CCoAOMT genes in different tissues were comprehensively analysed, and the tissue-specific TaCCoAOMTs were obtained, which provided useful targets for further functional study. Simultaneously, 7 TaCCoAOMTs showed significantly up-regulated expression after Fg treatment compared to mock treatment, of which 3 genes (TaCCoAOMT10, 14 and 19) were shared by three resistant varieties and one susceptible variety, suggesting that they might play an important role in the response to FHB. Interestingly, TaCCoAOMT10, 14 and 19 were the A, B and D homoeologous copies of the same homoeologous group respectively, of which the expression level of TaCCoAOMT19 was the highest, indicating its dominant role in the response to FHB and asymmetric expression between homoeologous copies. Otherwise, the difference between the four varieties showed the potential FHB tolerance candidates. For example, TaCCoAOMT3 and 17 were specifically down-regulated in three resistant varieties, indicating the repression effect of them on Fg tolerance. TaCCoAOMT2 displayed the specifical down-regulation in susceptible variety Shaw, which further demonstrates the uniqueness of the sensitive variety.
It is well known that cis-regulatory elements can regulate gene expression levels by binding to corresponding transcription factors, and then might determine the specific expression patterns in different tissues and stresses [32]. The CGTCA-motif and TGACG-motif, which are related to MeJA-responsiveness, were identified in the promoter regions of all up-regulated TaCCoAOMTs. A previous study proved that MeJA could not only help to delay the necrosis process of susceptible varieties in wheat but also increase the activities of enzymes related to pathogen defence [33]. Therefore, these up-regulated TaCCoAOMTs containing CGTCA-motif and TGACG-motif might play a crucial role in regulating FHB tolerance through MeJA mediation. We further reconstructed the co-expression network by the WGCNA method based on 48 RNA-seq samples of four wheat varieties [30]. Results showed that seven modules were significantly associated with Fg infection, of which 5 modules harboured the TaCCoAOMT genes as the hub factors, and two modules (blue and green) were positively correlated with Fg infection and three modules (lightyellow, turquoise and yellow) were negative. These modules provided some insight into the genetic basis and regulatory network of TaCCoAOMT, which might contribute to the molecular mechanism underlying FHB resistance. Furthermore, eight microRNAs were found to target five TaCCoAOMT genes involved in the Fg infection-related modules, including tae-miR167a and tae-miR1119. Previous studies have been reported that miR167a could meditate auxin signalling to respond to biotic stresses in tomato [34], and miR1119 was proven to regulate the expression of actin under stress conditions and activate the plant defence signalling pathway in barley [35]. We postulated that these two miRNAs also play the regulatory role in the response to Fg infection by controlling the expression of CCoAOMT genes in wheat.
Lignin is mainly involved in the basal disease resistance in plants [36, 37]. Furthermore, we validated the expression patterns of 11 selected FHB-responsive TaCCoAOMT genes between mock and Fg infection samples in the resistant RIL genotype R75 and susceptible RIL genotype S98 using the spray inoculation method. Almost all 11 selected TaCCoAOMTs displayed differential expression during Fg infection although different change trends were also found between them. At the same time, the expressioin patterns of them detected by qRT-PCR were also consistent with those of RNA-seq analysis, which not only demonstrated the roles of TaCCoAOMTs involved in the response to Fg infection, but also provided some candidates for further functional studies.
Finally, we used resequencing data to investigate the genetic variations and divergence of the CCoAOMT family in wheat and its relative populations. Results indicated that wild species showed high genetic diversity and rich haplotype composition in this family compared to cultivated species, suggesting selection effect was exerted on this family and obvious genetic bottleneck has occurred at it during wheat domestication and improvement processes [38]. Wild populations possessed specific haplotypes of the CCoAOMT genes that were lost in cultivated populations, which holds promising for enriching the genetic diversity and also improving the traits controlled by the CCoAOMT genes in cultivated wheat, such as FHB resistance.
Conclusions
This is the first study to identify the CCoAOMT family in wheat at the genome level. The genomic organization, phylogenetic relationship, exon-intron structure and cis-elements as well as the expression profiles of this family were systematically investigated and characterized. Furthermore, the expression patterns and co-expression network of these genes involved in Fg infection were also investigated. providing some useful insights on the roles of TaCCoAOMTs in the FHB response and tolerance. Additionally, the genetic diversity and divergence of these CCoAOMT genes in wheat and its relative populations were analysed based on resequencing data. This study not only shed light on the potential function of the CCoAOMT family in regulating wheat lignan biosynthesis and the FHB response but also provided some clues for the evolution of this family in wheat and other plants.
Methods
Identification of CCoAOMT genes in wheat
For the identification of the CCoAOMT family in wheat, the protein sequences of the wheat genome were retrieved from the Ensembl plant database to use as the local protein database (ftp://ftp.ensemblgenomes.org/pub/plants/release-50/gff3/triticum_aestivum). CCoAOMT genes in Arabidopsis and rice [25] were used to perform a BLASTP search against the local protein database with the threshold of E-value < 1e− 5 [39]. The PFAM profile (PF01596) was downloaded from the PFAM database (https://pfam.xfam.org/) and used as the query to search against the local protein database using HMMER 3.0 with the threshold of E-value < 1e− 5. The results of HMMER and BLASTP were integrated together, and the redundant were manually removed. Then, the putative wheat CCoAOMT genes were submitted to SMART (http://smart.embl-heidelberg.de/) and PFAM database (PF01596) (http://pfam.xfam.org/search) to predict the conserved protein domain, and those containing a complete CCoAOMT domain were remained as candidates. The candidate TaCCoAOMT proteins were submitted to the ExPASy database (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) to compute the theoretical isoelectric point (pI) and molecular weight (Mw). The cello tool (http://cello.life.nctu.edu.tw/) was used to predict the subcellular localization. The orthologues of each putative TaCCoAOMT gene with Arabidopsis or rice were determined by BLASTP results and further validated by retrieval in the Ensembl plant database.
Phylogenetic, gene structure, conserved motif and cis-element analysis of TaCCoAOMTs
Multiple sequence alignment was performed using ClustalX v2.0 [40]. The neighbor-joining method embedded in the MEGA-X program was used to construct the phylogenetic tree, and bootstrapping was set to 1000 [41]. Additionally, conserved motifs of TaCCoAOMT proteins were predicted using MEME v5.2.0 with the default parameters. The gene and motif structures were displayed based on GTF annotation files using TBtools [42]. The upstream 1500 bp region of each TaCCoAOMT gene was extracted and submitted to the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to predict cis-elements.
Expression analysis of TaCCoAOMT genes using RNA-seq
A total of 106 RNA-seq samples in different tissues (grain, leaf, root, spike, stem) and under diverse stress treatments (heat, salt, low temperature, drought, Fg inoculation) were downloaded from the URGI (http://wheat-urgi.versailles.inra.fr/) and the NCBI database (https://www.ncbi.nlm.nih.gov/) (accession nos. SRP045409, SRP062745, SRP043554 and SRP045409). All of these RNA-seq data were mapped to the wheat reference genome IWSGCv1.1 by STAR v2.7.6a [43] and the fragments per kilobase per million (FPKM) were calculated by StringTie v2.1.2 [44]. The expression patterns were displayed using R featureCounts v2.0.1 [45]and the edgeR package was used for reads normalization, while |log2-fold change| > = 1 and P values < = 0.05 were used as the parameters to identify differential expressed gene. Gene ontology (GO) enrichment was performed using AgriGO v2 (http://systemsbiology.cau.edu.cn/agriGOv2/index.php) and T. aestivum was set as the background. The GO enrichment results were plotted by ggplot2 and divided into three classes.
FHB-related coexpression network construction and miRNA analysis
A co-expression network was constructed using the WGCNA tools based on the 48 RNA-seq samples of four wheat varieties under Fg treatment. To obtain the related module and clarify gene interactions, we set the restricted minimum gene number to 30 for each module and used a threshold of 0.25 to merge the similar modules. Genes had higher weights in important modules were chosen to construct a co-expression network. Publicly available trait data, including Fg treatment, Fg time, Fg percent, Fg GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and DON(deoxynivalenol), were used for trait-module correlation analysis [30]. miRNA binding sites were predicted using psRNATarget (http://plantgrn.noble.org/psRNATarget/analysis) with default parameters and all of the wheat miRNAs were used. The regulatory network of the TaCCoAOMT gene and miRNA were visualized using Cytoscape v3.8.0 [46].
Haplotype and population genetics analysis of TaCCoAOMT
VCF files of wheat resequencing were downloaded from Genome Variation Map (https://bigd.big.ac.cn/gvm) (accession no. GVM000082) [31], which contained the genome variations of a total of 163 bread wheat accessions, 13 durum wheat accessions, 29 domesticated emmer wheat accessions, 28 wild emmer wheat accessions, 29 T. urartu accessions and 30 Aegilops tauschii accessions. SNPs in the coding region of TaCCoAOMT genes were extracted based on the chromosome location using TBtools. Furthermore, the haplotype organization and frequency were investigated by an in-house Python script.
Validation of the expression of TaCCoAOMTs through qRT-PCR analysis
For experimental verification, a FHB resistant RIL line (R75) and a FHB susceptible line (S98) from the wheat RIL population developed by single-seed descent from a cross between the susceptible US wheat variety Wheaton and the Chinese resistant wheat landrace HYZ, which were generously provided by Prof. Tao Li, Yangzhou University, China, were used [47]. The plant material was grown in a greenhouse to heading and then a spray inoculation method was used for Fg inoculation, following the method as described by Buerstmayr et al. with some modification [48]. A total of 5 inoculated spikes were collected to pool into one sample at 2 days post inoculation (dpi) and three biological replications were adopted. Meanwhile, the counterpart mock samples were collected by the same method. RNA Easy Fast Plant Tissue Kit (Tiangen, Beijing, China) was used to extract total RNA from all samples and RT Master Mix Perfect Real-Time kit (Takara, Dalian, China) was used to synthesize cDNA according to the manufacturer’s instruction. qRT-PCR reaction was performed on a QuantStudioTM 7 Flex System (Thermo Fisher Scientific, USA) using SYBR® Green Premix Pro Taq HS qPCR Kit (Accurate Biology, Hunan, China) with the following thermal cycling conditions: 95 ℃ for 30 s followed by 40 cycles of 95 ℃ for 3 s, 60 ℃ for 30 s. All reactions were performed in three separate technological replicates. The expression levels of these 11 randomly selected TaCCoAOMTs were calculated using the 2−ΔΔCT method with TaActin2 as the internal reference gene. The primers used in this study are listed in the supplementary file Table S8.
Supplementary Information
Additional file 1: Figure S1. Motifs found in TaCCoAOMT genes.Figure S2. Prediction of the cis-element in promoter regions of TaCCoAOMT genes.Figure S3. GO enrichment for TaCCoAOMT genes.Figure S4. Main haplotype and frequency of TaCCoAOMT genes in the A subgenome of Triticum.Figure S5. Main haplotype and frequency of TaCCoAOMT genes in the B subgenome of Triticum.Figure S6. Main haplotype and frequency of TaCCoAOMTgenes in the D subgenome of Triticum.
Additional file 2: Table S1. Corresponding ID of CCoAOMT genes in Arabidopsis and rice.Table S2. Characteristics of cis-acting regulatory elements in the promoter region of CCoAOMT genes in wheat.Table S3. GO enrichment analysis of the identified TaCCoAOMTs.Table S4. Differential expression analysis of TaCCoAOMT genes under abiotic stress.Table S5. Differential expression analysis of TaCCoAOMT genes under Fgtreatment.Table S6. Identification of miRNA bind sites in TaCCoAOMT genes.Table S7. Haplotype of CCoAOMT genes in five Triticum species.Table S8. The list of primers used for qRT-PCR in this study.
Acknowledgements
We thank the two anonymous reviewers so much for their insightful and constructive comments and also appreciate Prof. Tao Li’s help for providing the plant materials for qRT-PCR validation. We are grateful to all the labs whose data were used in our analyses and also grateful to High-Performance Computing center of Northwest A&F University for providing computational resources.
Research involving plants
All methods were performed in accordance with the relevant Chinese national guidelines.
Abbreviations
- FHB
Fusarium head blight
- Fg
Fusarium graminearum
- CCoAOMT
caffeoyl-coenzyme A O-methyltransferase
- TaCCoAOMT
Wheat CCoAOMT
- HMM
Hidden Markov Model
- CDS
Coding sequence
- GO
Gene ontology
- qRT-PCR
Quantitative real-time polymerase chain reaction
Authors' contributions
YG performed analysis and drafted the manuscript. PWQ collected data and performed the qRT-PCR experiments. ZRY and PY contributed to plant material collection. GQF completed the visualization of the data. SWN revised the manuscript. ZWJ and NXJ conceived this study and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was mainly supported supported by the National Science Foundation of China (Grant No.31561143005) and the Shaanxi Province Key Research and Development Program, China (Grant No. 2019NY-014).
Availability of data and materials
All of the datasets supporting the results of this article are included within the article and its Additional files. And the datasets generated for phylogenetic tree analysis during the current study are available in the Treebase repository, http://purl.org/phylo/treebase/phylows/study/TB2:S28407?x-access-code=96af71ab9297e6ae630774617ab5c9d&format=html.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
All authors have declared that there no competent interest existed.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Guang Yang and Wenqiu Pan contributed equally to this work
Contributor Information
Weijun Zheng, Email: zhengweijun@nwafu.edu.cn.
Xiaojun Nie, Email: small@nwsuaf.edu.cn.
References
- 1.Shewry PR: Wheat. J Exp Bot 2009, 60(6):1537–1553. [DOI] [PubMed]
- 2.Statistics FJR: Food and Agriculture Organization of the United Nations. 2010, 3(13):2012.
- 3.Gill BS, Appels R, Botha-Oberholster A-M, Buell CR, Bennetzen JL, Chalhoub B, Chumley F, Dvořák J, Iwanaga M, Keller BJG. A workshop report on wheat genome sequencing: International Genome Research on Wheat Consortium. Genetics. 2004;168(2):1087–1096. doi: 10.1534/genetics.104.034769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appels R, Eversole K, Stein N, Feuillet C, Keller B, Rogers J, Pozniak CJ, Choulet F, Distelfeld A, Poland JJS: Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361(6403). [DOI] [PubMed]
- 5.Miransari M, Smith DJCrib. Sustainable wheat (Triticum aestivum L.) production in saline fields: a review. Critical Reviews in Biotechnology. 2019;39(8):999–1014. doi: 10.1080/07388551.2019.1654973. [DOI] [PubMed] [Google Scholar]
- 6.Bai G, Shaner GJPd. Scab of wheat: prospects for control. Plant Disease. 1994;78(8):760–766. doi: 10.1094/PD-78-0760. [DOI] [Google Scholar]
- 7.Dexter J, Clear R, Preston KJCC. Fusarium head blight: effect on the milling and baking of some Canadian wheats. Cereal Chemistry. 1996;73(6):695–701. [Google Scholar]
- 8.Zhu ZW, Hao YF, Mergoum M, Bai GH, Humphreyse G, Cloutier S, Xia XC, He ZH. Breeding wheat for resistance to Fusarium head blight in the Global North: China, USA, and Canada. The Crop Journal. 2019;7(6):730–738. doi: 10.1016/j.cj.2019.06.003. [DOI] [Google Scholar]
- 9.Yokoyama R, Nishitani KJP, Physiology C. Genomic basis for cell-wall diversity in plants. A comparative approach to gene families in rice and Arabidopsis. Plant & Cell Physiology. 2004;45(9):1111–1121. doi: 10.1093/pcp/pch151. [DOI] [PubMed] [Google Scholar]
- 10.Giancaspro A, Lionetti V, Giove SL, Zito D, Fabri E, Reem N, Zabotina OA, De Angelis E, Monaci L, Bellincampi D, et al. Cell wall features transferred from common into durum wheat to improve Fusarium Head Blight resistance. Plant Science. 2018;274:121–128. doi: 10.1016/j.plantsci.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 11.Pogorelko G, Lionetti V, Bellincampi D, Zabotina O: Cell wall integrity: targeted post-synthetic modifications to reveal its role in plant growth and defense against pathogens. Plant Signal Behav 2013, 8(9). [DOI] [PMC free article] [PubMed]
- 12.Lionetti V. PECTOPLATE: the simultaneous phenotyping of pectin methylesterases, pectinases, and oligogalacturonides in plants during biotic stresses. Front Plant Sci. 2015;6:331. doi: 10.3389/fpls.2015.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sattler SE, Funnell-Harris DL. Modifying lignin to improve bioenergy feedstocks: strengthening the barrier against pathogens? Front Plant Sci. 2013;4:70. doi: 10.3389/fpls.2013.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang Z, Buchenauer H. Ultrastructural and immunocytochemical investigation of pathogen development and host responses in resistant and susceptible wheat spikes infected by Fusarium culmorum. Physiological and Molecular Plant Pathology. 2000;57(6):255–268. doi: 10.1006/pmpp.2000.0305. [DOI] [Google Scholar]
- 15.Soni N, Altartouri B, Hegde N, Duggavathi R, Nazarian-Firouzabadi F, Kushalappa AC: TaNAC032 transcription factor regulates lignin-biosynthetic genes to combat Fusarium head blight in wheat. Plant Science 2021, 304. [DOI] [PubMed]
- 16.Dhokane D, Karre S, Kushalappa AC, McCartney C. Integrated Metabolo-Transcriptomics Reveals Fusarium Head Blight Candidate Resistance Genes in Wheat QTL-Fhb2. PLoS One. 2016;11(5):e0155851. doi: 10.1371/journal.pone.0155851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Do CT, Pollet B, Thevenin J, Sibout R, Denoue D, Barriere Y, Lapierre C, Jouanin L. Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta. 2007;226(5):1117–1129. doi: 10.1007/s00425-007-0558-3. [DOI] [PubMed] [Google Scholar]
- 18.Fellenberg C, van Ohlen M, Handrick V, Vogt T. The role of CCoAOMT1 and COMT1 in Arabidopsis anthers. Planta. 2012;236(1):51–61. doi: 10.1007/s00425-011-1586-6. [DOI] [PubMed] [Google Scholar]
- 19.Whetten R, Sederoff R. Lignin Biosynthesis. The Plant Cell. 1995;7(7):1001–1013. doi: 10.2307/3870053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W. Lignin Biosynthesis and Structure. Plant Physiology. 2010;153(3):895–905. doi: 10.1104/pp.110.155119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamberger B, Ellis M, Friedmann M, de Azevedo Souza C, Barbazuk B, Douglas CJ. Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: the Populus lignin toolbox and conservation and diversification of angiosperm gene familiesThis article is one of a selection of papers published in the Special Issue on Poplar Research in Canada. Canadian Journal of Botany. 2007;85(12):1182–1201. doi: 10.1139/B07-098. [DOI] [Google Scholar]
- 22.Liu X, Luo Y, Wu H, Xi W, Yu J, Zhang Q, Zhou Z. Systematic analysis of O-methyltransferase gene family and identification of potential members involved in the formation of O-methylated flavonoids in Citrus. Gene. 2016;575(2 Pt 2):458–472. doi: 10.1016/j.gene.2015.09.048. [DOI] [PubMed] [Google Scholar]
- 23.Liu SJ, Huang YH, He CJ, Fang C, Zhang YW. Cloning, bioinformatics and transcriptional analysis of caffeoyl-coenzyme A 3-O-methyltransferase in switchgrass under abiotic stress. Journal of Integrative Agriculture. 2016;15(3):636–649. doi: 10.1016/S2095-3119(16)61363-1. [DOI] [Google Scholar]
- 24.Wu X, Yan Z, Dong X, Cao F, Peng J, Li M. Cloning and characterization of a CCoAOMT gene involved in rapid lignification of endocarp in dove tree (Davidia involucrata Baill.) Biotechnology & Biotechnological Equipment. 2019;32(6):1398–1406. doi: 10.1080/13102818.2018.1525324. [DOI] [Google Scholar]
- 25.Lin SJ, Yang YZ, Teng RM, Liu H, Li H, Zhuang J. Identification and expression analysis of caffeoyl-coenzyme A O-methyltransferase family genes related to lignin biosynthesis in tea plant (Camellia sinensis) Protoplasma. 2021;258(1):115–127. doi: 10.1007/s00709-020-01555-4. [DOI] [PubMed] [Google Scholar]
- 26.Rakoczy M, Femiak I, Alejska M, Figlerowicz M, Podkowinski J. Sorghum CCoAOMT and CCoAOMT-like gene evolution, structure, expression and the role of conserved amino acids in protein activity. Mol Genet Genomics. 2018;293(5):1077–1089. doi: 10.1007/s00438-018-1441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen TN, Son S, Jordan MC, Levin DB, Ayele BT. Lignin biosynthesis in wheat (Triticum aestivum L.): its response to waterlogging and association with hormonal levels. BMC Plant Biol. 2016;16:28. doi: 10.1186/s12870-016-0717-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma QH, Luo HR. Biochemical characterization of caffeoyl coenzyme A 3-O-methyltransferase from wheat. Planta. 2015;242(1):113–122. doi: 10.1007/s00425-015-2295-3. [DOI] [PubMed] [Google Scholar]
- 29.Bi C, Chen F, Jackson L, Gill BS, Li W. Expression of Lignin Biosynthetic Genes in Wheat during Development and upon Infection by Fungal Pathogens. Plant Molecular Biology Reporter. 2010;29(1):149–161. doi: 10.1007/s11105-010-0219-8. [DOI] [Google Scholar]
- 30.Pan Y, Liu Z, Rocheleau H, Fauteux F, Wang Y, McCartney C, Ouellet T. Transcriptome dynamics associated with resistance and susceptibility against fusarium head blight in four wheat genotypes. BMC Genomics. 2018;19(1):642. doi: 10.1186/s12864-018-5012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y, Zhao X, Li Y, Xu J, Bi A, Kang L, Xu D, Chen H, Wang Y, Wang YG, et al. Triticum population sequencing provides insights into wheat adaptation. Nat Genet. 2020;52:1412–1422. doi: 10.1038/s41588-020-00722-w. [DOI] [PubMed] [Google Scholar]
- 32.Hernandez-Garcia CM, Finer JJJPS. Identification and validation of promoters and cis-acting regulatory elements. Plant Science. 2014;217–218:109–119. doi: 10.1016/j.plantsci.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 33.Motallebi P, Niknam V, Ebrahimzadeh H, Enferadi ST, Hashemi M. The effect of methyl jasmonate on enzyme activities in wheat genotypes infected by the crown and root rot pathogen Fusarium culmorum. Acta Physiologiae Plantarum. 2015;37:237. doi: 10.1007/s11738-015-1988-3. [DOI] [Google Scholar]
- 34.Jodder J, Basak S, Das R, Kundu P. Coherent regulation of miR167a biogenesis and expression of auxin signaling pathway genes during bacterial stress in tomato. Physiological and Molecular Plant Pathology. 2017;100:97–105. doi: 10.1016/j.pmpp.2017.08.001. [DOI] [Google Scholar]
- 35.Colaiacovo M, Subacchi A, Bagnaresi P, Lamontanara A, Cattivelli L, Faccioli PJBg. A computational-based update on microRNAs and their targets in barley (Hordeum vulgare L.) BMC Genomics. 2010;11(1):595. doi: 10.1186/1471-2164-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tronchet M, Balague C, Kroj T, Jouanin L, Roby DJMpp. Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Molecular Plant Pathology. 2010;11(1):83–92. doi: 10.1111/j.1364-3703.2009.00578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee MH, Jeon HS, Kim SH, Chung JH, Roppolo D, Lee HJ, Cho HJ, Tobimatsu Y, Ralph J, Park OKJTEj. Lignin-based barrier restricts pathogens to the infection site and confers resistance in plants. The EMBO Journal. 2019;38(23):e101948. doi: 10.15252/embj.2019101948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127(7):1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 39.Cui L, Yang G, Yan J, Pan Y, Nie X. Genome-wide identification, expression profiles and regulatory network of MAPK cascade gene family in barley. BMC Genomics. 2019;20(1):750. doi: 10.1186/s12864-019-6144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 41.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C, Chen H, He Y, Xia RJB: TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018:289660.
- 43.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao Y, Smyth GK, Shi WJB. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 46.Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker TJB. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li T, Bai G, Wu S, Gu SJT, Genetics A. Quantitative trait loci for resistance to fusarium head blight in a Chinese wheat landrace Haiyanzhong. Theor Appl Genet. 2011;122(8):1497–1502. doi: 10.1007/s00122-011-1549-0. [DOI] [PubMed] [Google Scholar]
- 48.Buerstmayr H, Steiner B, Lemmens M, Ruckenbauer P: Resistance to fusarium head blight in winter wheat: heritability and trait associations. <bi>Crop Science</bi>, 2000, 40(4): 1012–1018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Motifs found in TaCCoAOMT genes.Figure S2. Prediction of the cis-element in promoter regions of TaCCoAOMT genes.Figure S3. GO enrichment for TaCCoAOMT genes.Figure S4. Main haplotype and frequency of TaCCoAOMT genes in the A subgenome of Triticum.Figure S5. Main haplotype and frequency of TaCCoAOMT genes in the B subgenome of Triticum.Figure S6. Main haplotype and frequency of TaCCoAOMTgenes in the D subgenome of Triticum.
Additional file 2: Table S1. Corresponding ID of CCoAOMT genes in Arabidopsis and rice.Table S2. Characteristics of cis-acting regulatory elements in the promoter region of CCoAOMT genes in wheat.Table S3. GO enrichment analysis of the identified TaCCoAOMTs.Table S4. Differential expression analysis of TaCCoAOMT genes under abiotic stress.Table S5. Differential expression analysis of TaCCoAOMT genes under Fgtreatment.Table S6. Identification of miRNA bind sites in TaCCoAOMT genes.Table S7. Haplotype of CCoAOMT genes in five Triticum species.Table S8. The list of primers used for qRT-PCR in this study.
Data Availability Statement
All of the datasets supporting the results of this article are included within the article and its Additional files. And the datasets generated for phylogenetic tree analysis during the current study are available in the Treebase repository, http://purl.org/phylo/treebase/phylows/study/TB2:S28407?x-access-code=96af71ab9297e6ae630774617ab5c9d&format=html.