Abstract
The unprecedented COVID-19 pandemic has had major impact on human health worldwide. Whilst national and international COVID-19 lockdown and travel restriction measures have had widespread negative impact on economies and mental health, they may have beneficial effect on the environment, reducing air and water pollution. Mass bathing events (MBE) also known as Kumbh Mela are known to cause perturbations of the ecosystem affecting resilient bacterial populations within water of rivers in India. Lockdowns and travel restrictions provide a unique opportunity to evaluate the impact of minimum anthropogenic activity on the river water ecosystem and changes in bacterial populations including antibiotic-resistant strains. We performed a spatiotemporal meta-analysis of bacterial communities of the Godavari River, India. Targeted metagenomics revealed a 0.87-fold increase in the bacterial diversity during the restricted activity of lockdown. A significant increase in the resilient phyla, viz. Proteobacteria (70.6%), Bacteroidetes (22.5%), Verrucomicrobia (1.8%), Actinobacteria (1.2%) and Cyanobacteria (1.1%), was observed. There was minimal incorporation of allochthonous bacterial communities of human origin. Functional profiling using imputed metagenomics showed reduction in infection and drug resistance genes by − 0.71-fold and − 0.64-fold, respectively. These observations may collectively indicate the positive implications of COVID-19 lockdown measures which restrict MBE, allowing restoration of the river ecosystem and minimise the associated public health risk.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00248-021-01781-0.
Keywords: COVID-19, Lockdown measures, Kumbh Mela, Bacterial populations, Antibiotic resistance, Public health, Targeted metagenomics
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel beta-CoV group 2B zoonotic infection of humans first identified in December 2019 from a cluster of human coronavirus disease 2019 (COVID-19) cases in Wuhan, China [1, 2]. The ensuing COVID-19 global pandemic has severely impacted the healthcare systems worldwide, causing ~ 3.39 million deaths as of May 19, 2021 [2–4]. World leaders responded by imposing restriction on the movement of people, lockdowns and implementation of public health measures including social distancing, hand washing and wearing of face masks [5]. India was not an exception and imposed its nationwide lockdown on March 25, 2020. Considering India’s population and the population density, an event of community transmission might have led to a havoc [6]. Most countries have implemented lockdown measures with national and international travel restrictions, which have affected all facets of life globally with major negative social and economic consequences [7]. On the other hand, the reduced movement of humans may have played a role in cleansing and restoration of environment, biodiversity and ecosystems that were under the perpetual anthropogenic stress [8]. Hence, it became important to evaluate the implications and/or beneficial impact of COVID-19 pandemic on the environment [9].
A recent study by Somani et al. [6], detailing the comprehensive analysis of various pollutants, suggested positive implications of restricted anthropogenic activities during the COVID-19 lockdown, particularly on the quality of water, level of air pollution, noise pollution, waste generation, etc. Restricted movement and lower traffic were also found to aid in dropping the levels of nitrogen dioxide and noise pollution [10]. Similarly, a review by Rume and Islam [9] on environmental implications of COVID-19 underscores the improved quality of air with the reduction in emission of greenhouse gases and decrease in water pollution across the globe. Additionally, the review also highlights the plausibility of ecological restoration especially at the tourist spots. In Indian, the concentrations of pollutant such as PM2.5, PM10, O3, NO2, SO2 and CO were consistently noted to be higher than its permissible limits, but during the course of lockdown, it was found to be under the permissible limits (with some occasional spikes) in the various megacities [5, 11, 12]. Additionally, the stringent lockdown had its contribution in cleansing the rivers and other aquatic ecosystems which are otherwise under the continuous burden of discharges from untreated sewage and industries [13–15]. Surprisingly, in the matter of weeks of lockdown, the water quality of the longest river of India, i.e. Ganga, was more pronounce than the collective efforts of more than a decade [16–18]. Singhal and Matto [17] highlighted that the sudden decrease in the number of pilgrims and the 500% decrease in the introduction of domestic and industrial effluents have led the improvement of the water quality. Similarly, in case of rivers Yamuna, Cauvery and Krishna, Central Pollution Control Board [19] reported concentrations of various factors like dissolved oxygen (DO), pH and coliform count to be under the permissible limits during the lockdown. Alternatively, the COVID-19 was associated with the inevitable situation of generation of enormous biomedical waste. Ineffective disposal of the biomedical waste was related to the environmental hazard especially affecting the water bodies [9]. The latter was also supported by a case study from Bangladesh indicating responders suffering effective disposal of COVID-19 preventives, leading to contamination of the water bodies [20].
Apart from the factors known to influence the water pollution, in India, the recurring pilgrimages that are hosted on the banks of the river are a source of unique perturbations to the aquatic ecosystem [21]. One such pilgrimage is the Kumbh Mela, the world’s largest religious mass gathering event hosted along the banks of holy rivers in India, and it attracts millions of pilgrims from India and other countries [22–24]. The Kumbh Mela involves the ritual practice of bathing in the river which substantially increases the risk associated with the spread of water infectious diseases among the fellow pilgrims [23] and also causes perturbations of the ecosystems. Our spatiotemporal investigation of the river Godavari during the 2015 Kumbh Mela event by using bacterial communities as a proxy revealed the deleterious impact of the mass bathing event (MBE). It was observed to be associated with the loss of diversity, with invasive establishment of allochthonous bacterial communities, enormous introduction of human faecal microbiota and increased virulence and drug resistance genes [25].
The COVID-19 lockdown measures, travel restrictions and associated scaling down of mass gathering religious events over the past year provide a unique opportunity to evaluate the impact of minimum anthropogenic activity on the river water ecosystem. Recent studies on the river Ganga found a reduction in the biological oxygen demand (BOD), faecal coliform count and nitrate (NO3) concentration and an increase in dissolved oxygen during the lockdown [26, 27]. However, both the studies primarily focus on the quality of the river water and adopted a classical approach of assessment of physiochemical factors and microbial contaminants, and investigation was restricted to the faecal coliform and total coliform counts [26]. However, it fails to provide insight into the bacterial communities thriving in the environment at given time and space. As the bacterial communities are sensitive to subtle variations in their environments, it can serve as a proxy to delineate the extend of ecological disturbances or restoration, if any [28]. In the recent year, the next-generation sequencing has widely been used and has proven its competency in determining such anomalies associated with changing environment or in human health [29].
Therefore, the current study attempts to elucidate the implication of COVID-19 lockdown on the environment by analysing change in the river bacterial communities. Using targeted metagenomics, we performed a spatiotemporal and meta-analysis of bacterial community structure and composition of the Godavari River, India, during the COVID-19 lockdown period (1) to understand the impact of COVID-19–mediated restrictions on the river ecosystem and (2) to examine the extent of ecosystem restoration, if any.
Material and Methods
Sampling Location
The current study is an extension of the spatiotemporal investigation of the river bacterial communities under the influence of the mass bathing event at Nashik Kumbh Mela 2015 [25], and it involved sample collection from same sites to reduce experimental heterogeneity. Five different sampling sites were utilised during the 2015 event [25] representing upstream sites (low human interference) and mass bathing sites (higher human activity). Water samples were collected in triplicate from the mass bathing sites located in Nashik City (Anandvalli, Gharpure Ghat and Tapovan) during the lockdown period (June 2020) implemented as a measure of containment of COVID-19 spread. However, due to travel restrictions imposed due to COVID-19 pandemic, samples were not collected from the upstream sites located in ‘Trimbakeshwar’ near the ‘Anjaneri’ forest. The samples collected during the lockdown period were termed as LS3, LS4 and LS5 corresponding to sampling site Anandvalli, Gharpure Ghat and Tapovan, respectively (Fig. 1).
Fig. 1.
The map depicts the location of the sampling sites along the bank of the Godavari River, Nashik, India. Samples collected before the event are named as BS3, BS4 and BS5; samples collected during the event are DS3, DS4 and DS5; and samples collected during the COVID lockdown were termed as LS3, LS4 and LS5 that correspond to locations Anandvalli, Gharpure Ghat and Tapovan, respectively
Sample Collection and Processing
Surface water samples were collected in triplicates from the defined bathing areas located at the river bank. The samples were stored in the sterile containers and transported to the laboratory by maintaining the cold chain. Each sample aliquot was subjected to physicochemical analysis at the Department of Microbiology, KTHM College, Nashik. Environmental parameters such as turbidity (OD), total dissolved solid (TDS), total solids (TS), total suspended solids (TSS), DO, BOD, faecal coliforms and most probable number (MPN) of total coliforms were assessed as per the procedures of APHA (1998) whilst pH (Pocket digital pH meter, Milwaukee pH600) and temperature (Vintage USA E8000) were recorded onsite. The remaining water sample was filtered through 0.22-µ Millipore filters (Merck Millipore, USA) and stored at − 20 °C until further processing.
High-Throughput Sequencing and Data Analysis
Community DNA extraction was carried out from the stored 0.22-µ Millipore filters using DNeasy Power Water Kit (Qiagen, The Netherlands). The qualitative and quantitative assessment of the DNA was performed in accordance with Sharma et al. [30]. The DNA was processed for the targeted amplicon sequencing by amplification of the V4-hyper variable region of the 16S rRNA gene using primers 515F-806R [31]. Library preparation was performed by following the protocols of Illumina, Inc. (USA). The obtained libraries were sequenced on Illumina MiSeq using 2 × 250 bp v2 chemistry. The sequences are available on the NCBI SRA portal under BioProject Id PRJNA698474.
In order to maintain the uniformity, the amplicon sequences generated during the Kumbh Mela 2015 [25] study were retrieved prior to analysing the current sequencing data (BioProject Id PRJNA383664). The bioinformatics analysis was in accordance with Jani et al. [25] which involved analysis using QIIME (Quantitative Insights Into Microbial Ecology, v1.9) [32]. Assembly of the paired-end reads using FLASH (Fast Length Adjustment of Short reads) tool, followed by adaptor and quality-filtering using Cutadapt (v3.2) and mothur (v1.32), respectively [33–35]. The operational taxonomic unit (OTU)-picking was performed using a closed reference approach against the Greengenes database with the UCLUST algorithm with sequence similarity of 97% [36, 37]. Taxonomic assignment of the OTUs was performed by adopting the RDP naïve Bayesian classifier against the Greengenes database [38]. Further, downstream analysis precludes sequence rarefaction to the lowest number of reads per sample. Estimates of the alpha diversity indices, viz. OTU richness, Shannon, Chao1, and Good’s coverage, were carried out using QIIME (v1.9). Statistical analysis and data visualisation were performed in R using the packages cowplot, corrplot, Hmisc, PerformanceAnalytics, phyloseq, vegan and ggplot2 [39–45].
Tracking the Source of Bacterial Communities
The source and establishment of the human microbiota was assessed by employing an approach of Bayesian mixing model [46]. The model aids in the identification and quantification of potential microbial communities of human origin. Microbiome data (examining various body sites, i.e. skin, oral and stool) of Indian sub-population representing both healthy and disease states was collected and compiled. The human microbiome data was used as a potential source (a reference dataset) of microbial communities whereas the mass bathing data (generated in this study and BioProject ID PRJNA383664) was used as a sink (target dataset) to deduce the origin of microbiota observed during the mass bathing event [47, 48].
Metagenomic Imputations
Functional profiling of these bacterial communities was performed by an approach of imputed metagenomics using PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) [49]. Specifically, the computational argument of “metagenomic contributions” was applied to decipher the functional potential of bacterial communities observed before and during the mass bathing event as well as during the minimal state of perturbation to the river, i.e. COVID-19 lockdown [50].
Results and Discussion
Estimation of Environmental Parameters
Assessment of the environmental parameters along mass bathing sites across three points, viz. before MBE, during MBE and lockdown, indicates substantial variability, especially when compared between the groups, i.e. before vs during MBE and during MBE vs lockdown (Table 1). The estimates of the TS, TDS and TSS were found to increase during the course of MBE (by 2.08-, 1.72- and 2.45-fold, respectively) and decrease during the minimal anthropogenic activity, i.e. the period of lockdown (− 0.74-, − 0.65- and − 0.81-fold, respectively). The significant increase in the TS (p ≤ 0.01), TDS (p = 0.055) and TSS (p ≤ 0.005) leads to a surge in the turbidity of water. A significant variation in turbidity was noted under the temporal gradient (p ≤ 0.02). The turbidity increased by 2.26-fold during the course of mass bathing event. In contrast, the turbidity was found to degrease by − 0.79-fold during the lockdown period. Increase in the turbidity and solute concentration during MBE corroborated the fact that the event attracts participation by millions of pilgrims at the event. Additionally, the untreated human solid and sewer waste along with the industrial and household waste being routinely discharged into the river lead to deterioration of water quality [22, 23]. Contamination of freshwater ecosystem consequently has a deleterious impact on the aquatic biota, including microorganisms [15, 51]. In contrast, the imposed restriction due the COVID-19 pandemic has been found to improvise the water quality and ecosystem, which was also noted in the case of river Damodar [27]. The observation was further supported by the study on river Ganga, suggesting positive implications of the lockdown towards ecosystem restoration [26]. Further, a study by Cooke et al. [7] highlighted both the benefits and risk due to pandemic on the freshwater fishes. In contrast, pH and temperature of the water were not found to differ significantly (p = 0.65 and p = 0.62, respectively).
Table 1.
Environmental parameters and alpha diversity across the temporal variation of mass bathing event and COVID lockdown
| Sample ID | Event | pH | Temperature (°C) | OD | TS (mg/l) | TDS (mg/l) | TSS (mg/l) | DO (mg/l) | BOD (mg/l) | MPN (per 100 ml) | Faecal coliform (per 100 ml) | Chao1 | Observed OTUs | Shannon |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BS3 | Before | 6.8 | 25.2 | 0.04 | 811 | 436 | 375 | 3.86 | 19.8 | 1.40E + 04 | 320 | 4181.95 | 2919 | 6.45 |
| BS4 | Before | 6.5 | 23.3 | 0.07 | 682 | 430 | 252 | 2.89 | 15.1 | 1.10E + 04 | 298 | 3975.94 | 2521 | 6.83 |
| BS5 | Before | 6.8 | 23.5 | 0.08 | 835 | 323 | 512 | 2.07 | 17.8 | 1.40E + 04 | 1007 | 5528.73 | 3702 | 7.19 |
| DS3 | During | 7.2 | 23.1 | 0.16 | 1856 | 694 | 1162 | 1.82 | 31.1 | 1.10E + 05 | 1007 | 3570.94 | 2181 | 3.57 |
| DS4 | During | 6.8 | 25.4 | 0.16 | 2068 | 1021 | 1047 | 0.77 | 29.7 | 1.70E + 05 | 400 | 1686.41 | 1062 | 2.75 |
| DS5 | During | 6.5 | 27.7 | 0.3 | 3250 | 1529 | 1721 | 0.88 | 51.5 | 2.40E + 06 | 1820 | 1265.5 | 841 | 3.13 |
| LS3 | Lockdown | 7 | 25.1 | 0.03 | 700 | 395 | 305 | 2.8 | 2.24 | 1.60E + 03 | 47 | 1836.44 | 1337 | 5.22 |
| LS4 | Lockdown | 6.7 | 24.3 | 0.06 | 650 | 415 | 235 | 6.16 | 2.16 | 1.60E + 03 | 80 | 2082.19 | 1609 | 6.18 |
| LS5 | Lockdown | 6.8 | 24.9 | 0.04 | 510 | 322 | 188 | 3.2 | 1.6 | 1.60E + 03 | 286 | 2201.25 | 1651 | 6.38 |
Further, evaluation of the oxygen relationship depicted a significant variation in the BOD (p ≤ 0.009) under temporal variation across mass bathing sites. The BOD was increased by 1.13-fold during the mass bathing event but was found to decrease by − 0.94-fold during the lockdown. The variations in BOD was the consequence of substantial variation in the concentration of the DO (p = 0.11). The concentration of the DO was decreased by − 0.6-fold but raised by 2.5-fold during the lockdown. Similarly, the microbiological assessment of the river water indicated a 67.7-fold increase in the MPN count (p = 0.34) which was found to reduce to − 0.99-fold during the lockdown. Similarly, the faecal coliform count (p = 0.052) raised by 0.98-fold and lowered to − 0.87 during the lockdown. The increase in the BOD is suggestive of the enormous influx of solutes due to bathing in the river, sediment resuspension, discharge from industries and leaching from unattended solid wastes during the MBE [51–53]. These changes in the river physiochemistry collectively have deleterious impact on the river microbial communities. Perturbation to the river bacterial communities and augmentation of specific bacterial groups like faecal coliforms pose a risk of spread of gastrointestinal diseases during the event [54, 55]. However, the lockdown was also found to be beneficial concerning the oxygen relationship of the river water which also coincides with observations of Dutta et al. [26] depicting the increased level of DO and reduction in BOD and faecal coliform counts in the river Ganga.
Structure and Composition of Bacterial Community
Analysis of high-quality paired-end reads using QIIME yielded 7469 OTUs. The reads were normalised to 105,246 prior to performing further downstream analysis. The estimates of alpha diversity index revealed substantial variability under the spatiotemporal realm; however, it was not be supported by the statistical significance (Table 1). The estimates of the non-parametric Shannon index depicted variance with respect to the time; i.e. it was found to decrease by − 0.53-fold during the mass bathing event whilst it was found to recover during the lockdown by 0.87-fold. This observation suggested potential re-establishment of diversity as minimal divergence of − 0.13-fold was noted between before MBE and lockdown samples. It was further supported by the assessment of Chao1 index and observed OTUs (Table 1). In summary, the species richness was found to follow an order of during MBE < lockdown < before MBE samples (Fig. S1). Loss of bacterial diversity found during the MBE might be supported by deteriorated quality of the river water and enormous influx of organic and inorganic waste having adverse impact on the microbial communities and river ecosystem [52, 53, 56]. Alternatively, a 0.87-fold increase in the Shannon index implied the events of ecosystem restoration under the improvised water quality during the imposed lockdown period [9, 26].
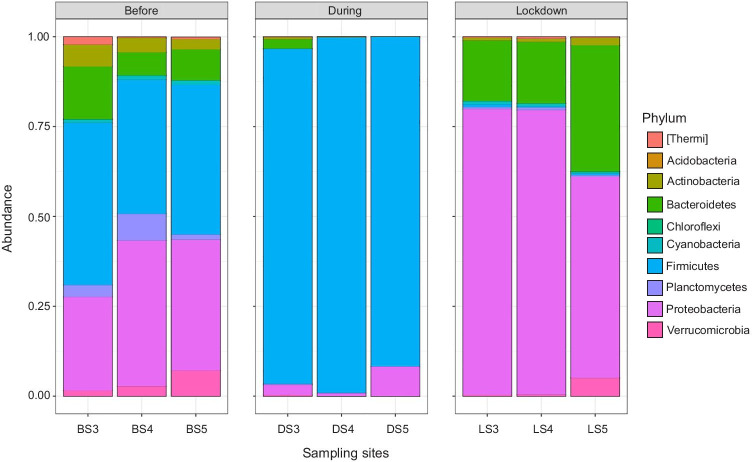
The most abundant bacterial phyla found under the spatiotemporal constraint consist of Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, Verrucomicrobia, Planctomycetes, Cyanobacteria, [Thermi], Chlorobi and Chloroflexi contributing for 99.9% of bacterial community (Fig. 2). The river bacterial community composition before the MBE was comprised of varied bacterial taxa having relative abundance ≥ 1%, viz. Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, Planctomycetes, Verrucomicrobia and Cyanobacteria. These phyla are known as the major constituents of river autochthonous communities driving key functions of primary production in river ecosystem [57, 58]. Conversely, members of bacterial phylum Actinobacteria are effective players in the degradation of a variety of compounds [59]. However, these phyla were found to be replaced by the selective augmentation of phylum Firmicutes (94.6%) along the mass bathing sites during the course of mass bathing event. It indicated the potential of members of phylum Firmicutes to withstand and flourish in the oxygen-limiting and high-solute conditions [60]. Minimal human activity during the period of lockdown sites featured re-establishment of various bacterial phyla Proteobacteria (70.6%), Bacteroidetes (22.5%), Verrucomicrobia, Actinobacteria and Cyanobacteria having relative abundance of ≥ 1%. It further supported the observed increase in the Shannon index by 0.87-fold during the lockdown period.
Fig. 2.
Distribution of most abundant bacterial phyla across the temporal variation of mass bathing event and lockdown period
Similarly, localisation of bacterial communities with respect to the time depicted the predominance of Planococcaceae, followed by Comamonadaceae, Bacillaceae, Flavobacteriaceae, ACK-M1, Moraxellaceae, Verrucomicrobiaceae, Rhodocyclaceae and Oxalobacteraceae before the MBE (relative abundance ≥ 1%). In contrast, enrichment of Planococcaceae (53.1%) (Fig. S2), Bacillaceae (41.1%) and Sphingomonadaceae (2%) was noted during the course of mass bathing due to its virtue of sustaining the extreme conditions of high-solute concentration and low-oxygen conditions [60, 61]. Furthermore, due to minimal human interference during the lockdown period, augmentation of diverse families, viz. Moraxellaceae, Flavobacteriaceae, [Chromatiaceae], Comamonadaceae, Sphingomonadaceae, Rhodobacteraceae, Pseudomonadaceae, Verrucomicrobiaceae, Oxalobacteraceae and Rhodocyclaceae (relative abundance ≥ 1%), was observed.
At the genus level (relative abundance ≥ 1%), predominance of Bacillus, Lysinibacillus, Flavobacterium, Acinetobacter, Limnohabitans and Planomicrobium was observed as a constituent of autochthonous river microbiota before MBE (Fig. S3). Conversely, Lysinibacillus (43.8%), Bacillus (41%) and Planomicrobium (4.9%) were found to flourish during the MBE. Moreover, the correlation between the changing environmental factors and bacterial community at the level of genus was carried which revealed a distinct influence of environmental factors on the bacterial communities (Fig. S4). The variations in the optical density of the water along with the TS, TDS, TSS and BOD were found to favour the bacterial genera Lysinibacillus, Bacillus and Planomicrobium (p ≤ 0.05). However, the dissolved oxygen was found to cast negative impact on the Lysinibacillus, Bacillus and Planomicrobium (p ≤ 0.05). Additionally, the substantial variation in the dissolved oxygen was found to be positively correlated with the Rhodobacter and Novosphingobium (p ≤ 0.05), whereas changes in the estimates of faecal coliforms were found to favour the Lysinibacillus (p ≤ 0.05). Interestingly, the Lysinibacillus, Bacillus and Planomicrobium showed a positive correlation between themselves (p ≤ 0.001); in contrast, they cast negative impact on the bacterial genera, viz. Flavobacterium (p ≤ 0.05), Acinetobacter (p ≤ 0.05), Rheinheimera, Rhodobacter (p ≤ 0.05), Novosphingobium and Limnohabitans. Conversely, Acinetobacter shares a positive correlation between Rhodobacter (p ≤ 0.001) and Novosphingobium (p ≤ 0.01). Similarly, Rhodobacter and Novosphingobium depict a significant positive correlation (p ≤ 0.01). Enumeration of members of Lysinibacillus and Bacillus indicates the potential shedding of human skin–associated microbiota during the MBE. Enrichment of Planomicrobium corroborated the facts of the ability of various Planomicrobium species to thrive in a variety of halophilic environments and their association with recreational waters [28, 60]. Although species of Lysinibacillus represents common members of human skin microbiota, some species of Lysinibacillus have also been associated with the sepsis in immunocompromised patients [61, 62]. Thus, an increase in abundance of Lysinibacillus may pose a serious health risk for the fellow pilgrims bathing in the river. Interestingly, during the lockdown period, re-establishment of few bacterial genera that were observed before the MBE, viz. Acinetobacter, Flavobacterium and Limnohabitans (relative abundance ≥ 1%), was observed.
Impact of Lockdown on the Bacterial Communities
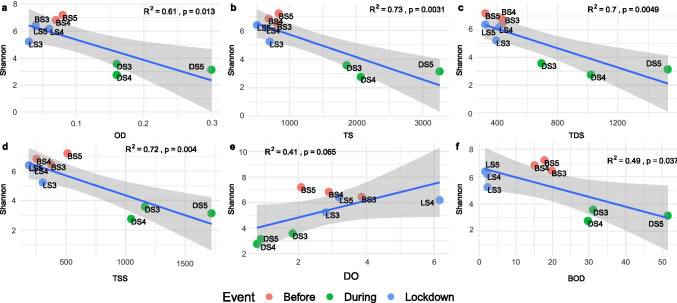
The relationship between the bacterial communities and the changing physiochemical conditions over the period investigation was derived by applying the linear regression model. The analysis revealed a significant influence of varying environmental factors such as optical density of water, TS, TDS, TSS followed by dissolved oxygen and BOD (R2 = 0.61 [p = 0.013], R2 = 0.73 [p = 0.0031], R2 = 0.7 [p = 0.0049], R2 = 0.72 [p = 0.004], R2 = 0.41 [p = 0.065] and R2 = 0.49 [p = 0.037], respectively). The environmental factors were found to cast negative impact on the bacterial communities during the course of mass bathing [25]. However, the current analysis depicts the positive correlation between above-mentioned factors and the bacterial communities which support composite increase (based on the Shannon index) in river bacterial diversity during the period of lockdown (Fig. 3, Fig. S1).
Fig. 3.
a–f Correlation between the bacterial diversity and the changing environmental parameters across the studied time points. Colour code defines the temporal variations in the samples, i.e. before (red) and during (green) the event and COVID-19 lockdown (blue)
The beta diversity analysis was carried out to determine the divergence in the bacterial communities, under the differential anthropogenic impact across the studied time points. Principal component analysis (PCA) analysis demonstrated three distinct groups based on the temporal variations (i.e. before MGE, during MGE and lockdown) indicating a significant impact of mass bathing on the river bacterial communities. The samples belonging to the before MGE and lockdown period showed lower intra-group variability (Fig. S5). However, the during MGE samples showed higher intra-group variability which corroborates the fact of samples DS4 and DS5 being sink to other mass bathing sites. Although the Shannon index demonstrates the increase in bacterial diversity during the lockdown period whilst, the distinctive clustering observed during the beta diversity analysis indicates the variation in the bacterial community structure and composition across the studied time points. The latter is supported by the observed changes in bacterial community structure and composition at the level of family and genera (Figs. S2 and S3). The composite observation of distinct clustering, lower intra-group variability during the lockdown period, significant increase in bacteria diversity (0.87-fold increase in Shannon index) and enumeration of resilient bacterial communities imply a positive impact of lockdown on the river ecosystem and, consequently, on the river bacterial communities [9, 26].
Microbial Source Tracking
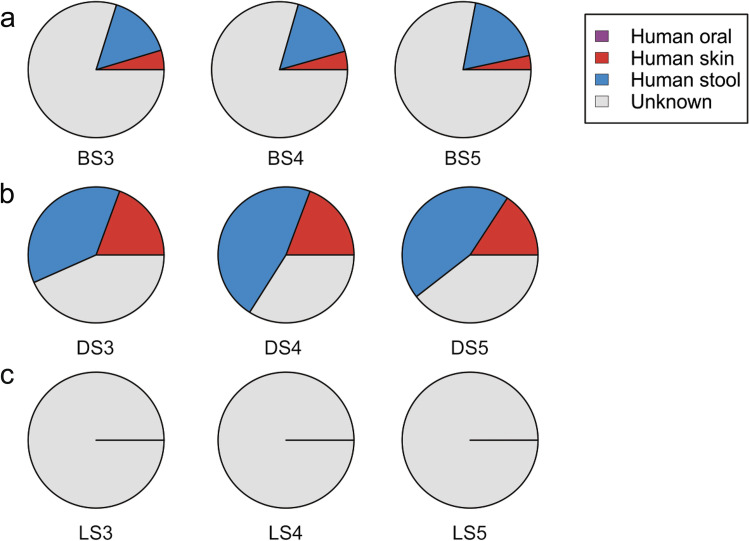
The attempt to identify the source of diverse bacterial communities using SourceTracker (a Bayesian mixing model) with microbiome profile of healthy Indian sub-population as a reference dataset revealed a significant increase in skin and stool–associated microbiota in the river during the course of mass bathing event (p ≤ 0.01). A 3.37-fold (p = 0.003) increase in skin associated bacterial communities was recorded across the mass bathing sites during the course of Kumbh Mela 2015. Similarly, the proportion of the human stool–associated microbiota also found to increase by 1.56-fold (p = 0.009) during the MBE, which was supported by the elevated levels of MPN and faecal coliform counts across mass bathing sites. Interestingly, during the lockdown period, a − 0.99-fold decrease in both human skin and stool–associated microbiota was observed, when compared with the mass bathing period (p = 0.004) (Fig. 4a–c). It is suggestive of the fact that the lockdown had a positive impact, as it indirectly led to cleaning of the river ecosystem. Although health risk associated with such alien communities remains largely unknown however, assessing the source of these microbial communities might aid to devise the mitigation strategies for potential infections related to the eye, skin and bowel [63, 64]. As observed, earlier prevalence of Lysinibacillus during the MBE might have an unfavourable outcome of sepsis [61]. It is in conjunction with the observation from the previous study on sea bathing where a bather itself acts a source of non-point pollution [65].
Fig. 4.
Proportion of human-associated microbiota (oral [purple], skin [red] and stool [blue]) determined using the Bayesian mixing model across the time points understudy. a Before the mass bathing event; b during the mass bathing event; c lockdown period
Infectious Diseases and Drug Resistance Genes
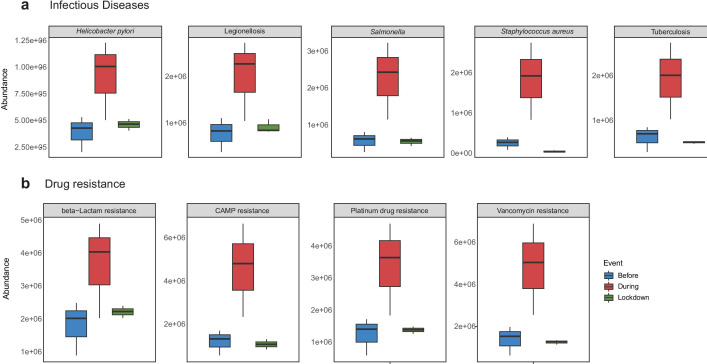
Assessment of the functional potential of the river bacterial communities across temporal divergence using an approach of metagenomics imputation depicted substantial variations in the abundance of the gene families involved in the metabolism and genetic information processing. However, gene families related to the infectious diseases and drug resistance were targeted to evaluate the impact of changes in the bacterial communities on the public health. Elevated levels (represented as fold change) in the abundance of gene families belonging to infectious diseases, particularly Helicobacter pylori infection (1.34-fold), Salmonella infection (2.87-fold), legionellosis (1.59-fold), Staphylococcus aureus infection (5.96-fold) and Tuberculosis (2.1-fold), were noted during the mass bathing event. However, during the lockdown, substantial reduction in the levels of gene families, viz. Helicobacter pylori infection (− 0.49-fold), Salmonella infection (− 0.74-fold), legionellosis (− 0.54-fold), Staphylococcus aureus infection (− 0.96-fold) and Tuberculosis (− 0.73-fold), was recorded, when compared to during MBE (Fig. 5a). Likewise, gene families driving the drug resistance in the bacterial communities were also found to rise with the MBE. Specifically, an increase in beta-lactam resistance (1.04-fold), vancomycin resistance (2.49-fold), cationic antimicrobial peptide (CAMP) resistance (2.86-fold) and platinum drug resistance (1.75-fold) was observed during the MBE. The plausible re-establishment of the bacterial communities during the lockdown appeared to have lower prevalence of the gene families associated with drug resistance. A reduction in the levels of beta-lactam resistance (− 0.39-fold), vancomycin resistance (− 0.73-fold), CAMP resistance (− 0.76-fold) and platinum drug resistance (− 0.59-fold) was noted during the lockdown period (Fig. 5b). Such elevated levels of gene families involved in the infectious diseases and drug resistance during the mass bathing event might pose a serious threat to the attendees which may have catastrophic outcomes like epidemics. Many recent studies on the river ecosystem and/or the religious pilgrimage support the observed increase in the abundance of gene families involved in infectious diseases and drug resistance. Specifically, the study on Ganga targeting the blaNDM-1 gene revealed an increase in antibiotic resistance during the period of pilgrimage [21]. Similarly, in our previous study, we also noted an increase in antibiotic resistance in bacteria isolated across the MBE and described a novel cefotaxime-resistant strain of Corynebacterium godavarianum from the Godavari River, India [66, 67]. The recent study by Yadav et al. [68] involving shotgun metagenomic analysis based on the MinION sequencing supports the observed elevated levels of antibiotic resistance genes during mass bathing.
Fig. 5.
Differential abundance of gene families associated with the a. infectious diseases and b. drug resistance across the various time points
Conclusion
In summary, a meta-analysis of the river bacterial communities under the extensive anthropogenic impact of mass bathing to minimal impact during the COVID-19 lockdown revealed a significant impact of MBE driving reduction in the bacterial diversity and an increase in potential risk to the human health. Alternatively, the lockdown provides a plausibility of natural restoration of ecosystem as noted by re-establishment of resilient bacterial communities. However, the extent of re-establishment of resilient bacterial communities from the end of previous mass bathing event (year 2015) and before the lockdown period could not be evaluated and could not be ignored. Our study represents a unique set of baseline data during the lockdown period which will serve as reference for comparative analysis in future studies on the Godavari River.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are grateful to Ms. Vaishali Tile for her help in collecting the samples during the lockdown.
Author Contribution
K.J., Y.S. and A.S. were involved in the study design. J.B. was involved in the sample collection and analysis of environmental parameters. K.J. and A.S. were involved in the experimental work, bioinformatics and statistical analysis. K.J., S.S., E.I.A., S.A., Y.S. and A.S. were involved in the interpretation of the data. All the authors were involved in the drafting of the manuscript.
Funding
This work was supported by the ‘Department of Biotechnology (DBT), Government of India’ (by Grant No. BT/Coord.II/01/03/2016), under the project Establishment of Centre for Excellence, National Centre for Microbial Resource (NCMR). Avinash Sharma acknowledges the support by the DBT/Wellcome Trust India Alliance under the project grant (IA/E/17/1/503700). Professor Sir Ali Zumla acknowledges the support from the European and Developing Countries Clinical Trials Partnership (EDCTP2) Programme, Horizon 2020, the European Union’s Framework Programme for Research and Innovation, grants PANDORA-ID-NET, TESA-2 and CANTAM-2.
Declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
All authors have read and approved the publication of the current manuscript.
Conflict of Interest
All authors have an interest in mass gathering medicine. All authors declare that there are no conflicts of interest.
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P (2019) A novel coronavirus from patients with pneumonia in China. NEJM. 2020 Jan 24.
- 2.Coronavirus disease (COVID-19) pandemic. World Health Organization, Geneva (2020) https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed 19 May 2021
- 3.Wang C, Pan R, Wan X, Tan Y, Xu L, Ho CS, Ho RC. Immediate psychological responses and associated factors during the initial stage of the 2019 coronavirus disease (COVID-19) epidemic among the general population in China. Int J Environ Res Public Health. 2020;17:1729. doi: 10.3390/ijerph17051729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO Coronavirus disease (COVID-19) dashboard. https://covid19.who.int/. Accessed 19 May 2021
- 5.Amanat F, Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Somani M, Srivastava AN, Gummadivalli SK, Sharma A. Indirect implications of COVID-19 towards sustainable environment: an investigation in Indian context. Bioresource Technology Reports. 2020;11:100491. doi: 10.1016/j.biteb.2020.100491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooke SJ, Twardek WM, Lynch AJ, Cowx IG, Olden JD, Funge-Smith S, Lorenzen K, Arlinghaus R, Chen Y, Weyl OL, Nyboer EA. A global perspective on the influence of the COVID-19 pandemic on freshwater fish biodiversity. Biol Conserv. 2020;9:108932. [Google Scholar]
- 8.Bates AE, Primack RB, Moraga P, Duarte CM. COVID-19 pandemic and associated lockdown as a “global human confinement experiment” to investigate biodiversity conservation. Biol Conserv. 2020;248:108665. doi: 10.1016/j.biocon.2020.108665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rume T, Islam SD (2020) Environmental effects of COVID-19 pandemic and potential strategies of sustainability. Heliyon e04965. [DOI] [PMC free article] [PubMed]
- 10.National Aeronautics and Space Administration (NASA) (2020). Airborne particle levels plummet in Northern India. Retrieved on April 14, 2021 from https://earthobservatory.nasa.gov/images/146596/airborne-particle-levels-plummet-in-northern-india.
- 11.Sharma S, Zhang M, Gao J, Zhang H, Kota SH. Effect of restricted emissions during COVID-19 on air quality in India. Sci Total Environ. 2020;728:138878. doi: 10.1016/j.scitotenv.2020.138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venter ZS, Aunan K, Chowdhury S, Lelieveld J. COVID-19 lockdowns cause global air pollution declines. PNAS. 2020;117:18984–18990. doi: 10.1073/pnas.2006853117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaseem H, Banerjee TK. Evaluation of pollution of Ganga River water using fish as bioindicator. Environ Monit Assess. 2016;188:1–9. doi: 10.1007/s10661-016-5433-x. [DOI] [PubMed] [Google Scholar]
- 14.Yadav A, Pandey J. Water quality interaction with alkaline phosphatase in the Ganga River: implications for river health. B Environ Contam Tox. 2017;99:75–82. doi: 10.1007/s00128-017-2108-4. [DOI] [PubMed] [Google Scholar]
- 15.Jani K, Ghattargi V, Pawar S, Inamdar M, Shouche Y, Sharma A. Anthropogenic activities induce depletion in microbial communities at urban sites of the river Ganges. Curr Microbiol. 2018;75:79–83. doi: 10.1007/s00284-017-1352-5. [DOI] [PubMed] [Google Scholar]
- 16.Central Pollution Control Board (CPCB) (2020a). Impact of lockdown on water quality of river Ganga. Retrieved from: https://cpcb.nic.in/upload/Assessment-of-Impact-Lockdown-WQ-MajorRivers.pdf. Accessed 19 May 2021
- 17.Singhal S, Matto M (2020) COVID-19 lockdown: a ventilator for rivers. DownToEarth, Retrieved from https://www.downtoearth.org.in/blog/covid-19-lockdown-a-ventilator-for-rivers-70771. Accessed 14 April 2021
- 18.Katariy M (2020) 10 things that have happened for the first time in years during the coronavirus lockdown. Scoopwhoop, Retrieved from https://www.scoopwhoop.com/news/things-that-have-happened-for-the-first-time-in-years-during-coronavirus-lockdown/. Accessed 14 April 2021
- 19.Central Pollution Control Board (CPCBb) (2020) Report on assessment of impact of lockdown on water quality of river Yamuna-Delhi stretch Retrieved from https://www.cpcb.nic.in/latest-cpcb.php
- 20.Islam SD, Mondal PK, Ojong N, Bodrud-Doza M, Siddique MA, Hossain M, Mamun MA (2021) Water, sanitation, hygiene and waste disposal practices as COVID-19 response strategy: insights from Bangladesh. Environ Develop Sustain 1–22 [DOI] [PMC free article] [PubMed]
- 21.Ahammad ZS, Sreekrishnan TR, Hands CL, Knapp CW, Graham DW. Increased waterborne bla NDM-1 resistance gene abundances associated with seasonal human pilgrimages to the Upper Ganges River. Environ Sci Technol. 2014;48:3014–3020. doi: 10.1021/es405348h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vortmann M, Balsari S, Holman SR, Greenough PG. Water, sanitation, and hygiene at the world’s largest mass gathering. Curr Infect Dis Rep. 2015;17(2):461. doi: 10.1007/s11908-015-0461-1. [DOI] [PubMed] [Google Scholar]
- 23.David S, Roy N. Public health perspectives from the biggest human mass gathering on earth: KumbhMela, India. Int J Infect Dis. 2016;47:42–45. doi: 10.1016/j.ijid.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Memish ZA, Steffen R, White P, Dar O, Azhar EI, Sharma A, Zumla A. Mass gatherings medicine: public health issues arising from mass gathering religious and sporting events. Lancet. 2019;393:2073–2084. doi: 10.1016/S0140-6736(19)30501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jani K, Dhotre D, Bandal J, Shouche Y, Suryavanshi M, Rale V, Sharma A. World’s largest mass bathing event influences the bacterial communities of Godavari, a holy river of India. Microb Ecol. 2018;76:706–718. doi: 10.1007/s00248-018-1169-1. [DOI] [PubMed] [Google Scholar]
- 26.Dutta V, Dubey D, Kumar S. Cleaning the river Ganga: impact of lockdown on water quality and future implications on river rejuvenation strategies. Sci Total Environ. 2020;743:140756. doi: 10.1016/j.scitotenv.2020.140756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakraborty B, Roy S, Bera A, Adhikary PP, Bera B, Sengupta D, Bhunia GS, Shit PK. Cleaning the river Damodar (India): impact of COVID-19 lockdown on water quality and future rejuvenation strategies. Environ Dev Sustain. 2021;3:1–5. doi: 10.1007/s10668-020-01152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee CS, Kim M, Lee C, Yu Z, Lee J. The microbiota of recreational freshwaters and the implications for environmental and public health. Front Microbiol. 2016;7:1826. doi: 10.3389/fmicb.2016.01826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kajale S, Jani K, Sharma A. Contribution of archaea and bacteria in sustaining climate change by oxidizing ammonia and sulfur in an Arctic Fjord. Genomics. 2020 doi: 10.1016/j.ygeno.2020.11.005. [DOI] [PubMed] [Google Scholar]
- 30.Sharma A, Jani K, Thite V, Dhar SK, Shouche Y. Geochemistry shapes bacterial communities and their metabolic potentials in tertiary coalbed. Geomicrobiol J. 2019;36:179–187. doi: 10.1080/01490451.2018.1526987. [DOI] [Google Scholar]
- 31.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 35.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 38.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H (2020). vegan: community ecology package. R package version 2.5–7. https://CRAN.R-project.org/package=vegan. Accessed 19 May 2021
- 41.Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer-Verlag, New York. ISBN 978–3–319–24277–4. https://ggplot2.tidyverse.org. Accessed 19 May 2021
- 42.Wilke CO (2020). cowplot: streamlined plot theme and plot annotations for ‘ggplot2’. R package version 1.1.1. https://CRAN.R-project.org/package=cowplot. Accessed 19 May 2021
- 43.Wei T, Simko V (2017). R package “corrplot”: visualization of a correlation matrix (version 0.84). Available from https://github.com/taiyun/corrplot. Accessed 19 May 2021
- 44.Harrell FE Jr, with contributions from Charles Dupont and many others (2021). Hmisc: Harrell Miscellaneous. R package version 4.5–0. https://CRAN.R-project.org/package=Hmisc. Accessed 19 May 2021
- 45.Peterson BG, Carl P (2020) PerformanceAnalytics: econometric tools for performance and risk analysis. R package version 2.0.4. https://CRAN.R-project.org/package=PerformanceAnalytics. Accessed 19 May 2021
- 46.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST. Bayesian community-wide culture-independent microbial source tracking. Nat Methods. 2011;8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaudhari DS, Dhotre DP, Agarwal DM, Gaike AH, Bhalerao D, Jadhav P, Mongad D, Lubree H, Sinkar VP, Patil UK, Salvi S. Gut, oral and skin microbiome of Indian patrilineal families reveal perceptible association with age. Sci Rept. 2020;10:1–3. doi: 10.1038/s41598-020-62195-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gaike AH, Paul D, Bhute S, Dhotre DP, Pande P, Upadhyaya S, Reddy Y, Sampath R, Ghosh D, Chandraprabha D, Acharya J (2020) The gut microbial diversity of newly diagnosed diabetics but not of prediabetics is significantly different from that of healthy nondiabetics. Msystems 5. [DOI] [PMC free article] [PubMed]
- 49.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RL, Knight R, Beiko RG. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotech. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:1–8. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev. 2011;75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spietz RL, Williams CM, Rocap G, Horner-Devine MC. A dissolved oxygen threshold for shifts in bacterial community structure in a seasonally hypoxic estuary. PLoS ONE. 2015;10:e0135731. doi: 10.1371/journal.pone.0135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amalfitano S, Corno G, Eckert E, Fazi S, Ninio S, Callieri C, Grossart HP, Eckert W. Tracing particulate matter and associated microorganisms in freshwaters. Hydrobiologia. 2017;800:145–154. doi: 10.1007/s10750-017-3260-x. [DOI] [Google Scholar]
- 54.Fries JS, Characklis GW, Noble RT. Attachment of fecal indicator bacteria to particles in the Neuse River Estuary, NC. J Environ Eng. 2006;132:1338–1345. doi: 10.1061/(ASCE)0733-9372(2006)132:10(1338). [DOI] [Google Scholar]
- 55.Fisher JC, Levican A, Figueras MJ, McLellan SL. Population dynamics and ecology of Arcobacter in sewage. Front Microbiol. 2014;5:525. doi: 10.3389/fmicb.2014.00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gibbons SM, Jones E, Bearquiver A, Blackwolf F, Roundstone W, Scott N, Hooker J, Madsen R, Coleman ML, Gilbert JA. Human and environmental impacts on river sediment microbial communities. PLoS ONE. 2014;9:e97435. doi: 10.1371/journal.pone.0097435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warnecke F, Amann R, Pernthaler J. Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ Microbiol. 2004;6:242–253. doi: 10.1111/j.1462-2920.2004.00561.x. [DOI] [PubMed] [Google Scholar]
- 58.Newton RJ, McLellan SL. A unique assemblage of cosmopolitan freshwater bacteria and higher community diversity differentiate an urbanized estuary from oligotrophic Lake Michigan. Front Microbiol. 2015;6:1028. doi: 10.3389/fmicb.2015.01028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van der Gucht K, Vandekerckhove T, Vloemans N, Cousin S, Muylaert K, Sabbe K, Gillis M, Declerk S, De Meester L, Vyverman W. Characterization of bacterial communities in four freshwater lakes differing in nutrient load and food web structure. FEMS Microbiol Ecol. 2005;53:205–220. doi: 10.1016/j.femsec.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 60.Jung YT, Kang SJ, Oh TK, Yoon JH, Kim BHPlanomicrobium flavidum sp. nov., isolated from a marine solar saltern, and transfer of Planococcus stackebrandtii Mayilraj, , et al. 2005 to the genus Planomicrobium as Planomicrobium stackebrandtii comb. nov. Int J Syst Evol Microbiol. 2009;59:2929–2933. doi: 10.1099/ijs.0.009191-0. [DOI] [PubMed] [Google Scholar]
- 61.Wenzler E, Kamboj K, Balada-Llasat JM. Severe sepsis secondary to persistent Lysinibacillus sphaericus, Lysinibacillus fusiformis and Paenibacillus amylolyticus bacteremia. Int J Infectious Diseases. 2015;35:93–95. doi: 10.1016/j.ijid.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 62.Ndiaye C, Lo CI, Bassene H, Raoult D, Lagier JC, Sokhna C (2019) Lysinibacillus timonensis sp. nov., Microbacterium timonense sp. nov., and Erwinia mediterraneensis sp. nov., three new species isolated from the human skin. New Microbes New Infect 31:100579. [DOI] [PMC free article] [PubMed]
- 63.Guidelines for safe recreational water environments. Volume 1: Coastal and Fresh Waters. WHO 2003. http://www.who.int/water_sanitation_health/publications/srwe1/en/. Accessed on April 14 2021. [PubMed]
- 64.Papastergiou P, Mouchtouri V, Pinaka O, Katsiaflaka A, Rachiotis G, Hadjichristodoulou C. Elevated bathing-associated disease risks despite certified water quality: a cohort study. Int J Environ Res Public Health. 2012;9:1548–1565. doi: 10.3390/ijerph9051548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fewtrell L, Kay D. Recreational water and infection: a review of recent findings. Curr Environ Health Rep. 2015;2:85–94. doi: 10.1007/s40572-014-0036-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jani K, Khare K, Senik S, Karodi P, Vemuluri VR, Bandal J, Shouche Y, Rale V, Sharma A. Corynebacterium godavarianum sp. nov., isolated from the Godavari river. India Int J Syst Evol Microbiol. 2018;68:241–247. doi: 10.1099/ijsem.0.002491. [DOI] [PubMed] [Google Scholar]
- 67.Jani K, Bandal J, Rale V, Shouche Y, Sharma A. Antimicrobial resistance pattern of microorganisms isolated and identified from Godavari River across the mass gathering event. J Biosci. 2019;44:1–6. doi: 10.1007/s12038-019-9941-z. [DOI] [PubMed] [Google Scholar]
- 68.Yadav R, Rajput V, Gohil K, Khairnar K, Dharne M. Comprehensive metagenomic insights into a unique mass gathering and bathing event reveals transient influence on a riverine ecosystem. Ecotoxicol Environ Saf. 2020;202:110938. doi: 10.1016/j.ecoenv.2020.110938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.