Abstract
BRAF inhibitors are insufficient monotherapies for BRAF-mutated cancer; therefore, we investigated which inhibitory pathway would yield the most effective therapeutic approach when targeted in combination with BRAF inhibition. The oncogenic BRAF inhibitor, PLX4720, increased basal autophagic flux in BRAF-mutated cells compared to wild-type (WT) BRAF cells. Interestingly, early autophagy inhibition improved the effectiveness of PLX4720 regardless of BRAF mutation, whereas late autophagy inhibition did not. Although ATG5 knockout led to PLX4720 resistance in both WT and BRAF-mutated cells, the MEK inhibitor trametinib exhibited a synergistic effect on PLX4720 sensitivity in WT BRAF cells but not in BRAF-mutated cells. Conversely, the prolonged inhibition of endoplasmic reticulum (ER) stress reduced basal autophagy in BRAF-mutated cells, thereby increasing PLX4720 sensitivity. Taken together, our results suggest that the combined inhibition of ER stress and BRAF may simultaneously suppress both pro-survival ER stress and autophagy, and may therefore be suitable for treatment of BRAF-mutated tumors whose autophagy is increased by chronic ER stress. Similarly, for WT BRAF tumors, therapies targeting MEK signaling may be a more effective treatment strategy. Together, this study presents a rational combination treatment strategy to improve the efficacy of BRAF inhibitors depending on BRAF mutation status.
Keywords: BRAF, Autophagy, Mutation, Melanoma, TFEB, Cancer
INTRODUCTION
Activating BRAF mutations have been implicated in up to 70% of all melanomas, 10-15% of colorectal cancers, and several malignant gliomas (Davies et al., 2002). These mutations lead to the constitutive activation of the RAF-MEK-ERK signaling cascade, which drives cancer growth (Pisapia et al., 2020). The first B-Raf-targeting drug, sorafenib, is a broad-spectrum kinase inhibitor with higher potency against RAF-1 than BRAF (Wilhelm et al., 2004), but was ineffective in BRAF mutant melanomas (Eisen et al., 2006). Thus, second-generation inhibitors such as PLX4720 and dabrafenib, targeting mutant BRAF, have been developed (Tsai et al., 2008). However, these inhibitors caused a problem of MAPK paradoxical activation by RAF dimerization in wild type (WT) BRAF melanomas (Hatzivassiliou et al., 2010). The third generation BRAF inhibitors (referred to as pan-RAF inhibitors) such as LY3009120 was developed to address this problem by interfering with the monomeric and dimeric RAF complexes (Sullivan et al., 2020). Although the use of BRAF inhibitors initially results in effective treatment of cancer patients with oncogenic BRAF mutations, the effectiveness of the treatment is limited by the onset of acquired resistance to BRAF inhibitors (Luebker and Koepsell, 2019). The mechanisms of resistance to BRAF inhibitors are complex and multifaceted; however, recent studies have indicated that autophagy might play an important role in acquiring resistance to BRAF inhibitor (Xie et al., 2015).
Autophagy participates in the progression of various diseases by acting as an important catabolic process that maintains cellular homeostasis (Ichimiya et al., 2020). Since autophagy can both promote and inhibit tumor growth, there has been controversy over whether or how autophagy should be manipulated in cancer therapy. Previously, we reported that autophagy deficiency enhances susceptibility to cellular transformation (Hwang et al., 2018); however, induced autophagy may contribute toward tumor survival during cancer progression (Choi, 2012). The identification of autophagy as a resistance mechanism against anticancer drugs has reinforced the use of autophagy inhibitors as a novel cancer treatment strategy (Poklepovic and Gewirtz, 2014). However, the role of autophagy as a mechanism of acquiring resistance to anticancer drugs requires further confirmation.
Recent studies have reported that melanomas harboring mutant BRAF exhibit an increase in the basal autophagic rate for maintaining cellular homeostasis (Corazzari et al., 2013). In addition, autophagy plays a positive role in overcoming senescence and enhancing the growth of mutant BRAF-induced melanomas in mice (Xie et al., 2015) and has been shown to be consistently induced in melanoma patients receiving oncogenic BRAF inhibitors (Ma et al., 2014). Our previous study implicated autophagy in acquired resistance to BRAF inhibitors (Ahn and Lee, 2013); therefore, the combination of autophagy and BRAF inhibition may be particularly effective in BRAF mutant cancers, in which induced autophagy may serve as a survival mechanism during cancer progression (Zahedi et al., 2019).
In this study, we investigated the mechanisms via which autophagy is upregulated by BRAF inhibitors and how the sensitivity of tumor cells to BRAF inhibitors could be improved. We found that BRAF inhibition induced autophagy via the nuclear translocation of transcription factor EB (TFEB) in BRAF-mutated cells. In addition, our results suggest that the combined inhibition of endoplasmic reticulum (ER) stress and BRAF may be suitable for the treatment of BRAF mutant cancer cells, whereas therapeutic kinase inhibitors that target MEK may be a more effective treatment strategy for BRAF WT cells.
MATERIALS AND METHODS
Reagents and antibodies
Rabbit polyclonal anti-LC3 antibodies were purchased from Sigma-Aldrich (St. Louis, MO, USA), while anti-phospho-MEK, anti-phospho-ERK, ant-phospho-eIF2α, and anti-XBP-1s antibodies were obtained from Cell Signaling Technology (Danvers, MA, USA). SDS–polyacrylamide gel electrophoresis reagents were purchased from Bio-Rad (Hercules, CA, USA). Hydroxychloroquine (HCQ), chloroquine (CQ), and rapamycin were obtained from Sigma-Aldrich. SBI-0206965 was purchased from Cell Signaling Technology. PLX4720 and trametinib were obtained from Selleck Chemicals (Houston, TX, USA).
Cell lines and cell culture
Three melanoma cell lines (A375P, G-361, and SK-MEL-2) and one colon carcinoma cell line (HCT116) were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) or Korean Cell Line Bank (KCLB; Seoul, Korea). Among the two BRAF mutant cell lines, A375P is homozygous for V600E, whereas G-361 is heterozygous. Two RAS mutant cell lines, SK-MEL-2 (NRAS: Q61R/Q61R) and HCT116 (KRAS: G13D/+), are homozygous for WT BRAF. These cell lines were maintained at 37°C in Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher Scientific, Carlsbad, CA, USA) supplemented with 10% fetal calf serum (Thermo Fisher Scientific), and penicillin–streptomycin (Thermo Fisher Scientific). For the experiments, cells were cultured in T-75 tissue culture flasks until they reached 60-70% confluency.
Plasmid DNA and transient transfection
The ATG5 CRISPR/Cas9 construct was obtained from ToolGen (Seoul, Korea). EGFP-LC3 was a gift from Karla Kirkegaard (Addgene plasmid #11546). pCI-neo-mApg5 was a gift from Noboru Mizushima (Addgene plasmid #22956). ptfLC3 was a gift from Tamotsu Yoshimori (Addgene plasmid #21074). pEGF-TFEB (Addgene #38119) and pCIP-Flag-TfebAA (Addgene #79014) were gifts from Reuben Shaw (UCSD, San Diego, CA, USA). Cells were transiently transfected with expression plasmids encoding the indicated proteins using Lipofectamine 2000 (Thermo Fisher Scientific).
Establishment of the ATG5 knockout (KO) cell line
ATG5 KO cells were established using the ATG5 CRISPR/Cas9 construct, as described previously (Hwang et al., 2018), with the following target single guide (sg) RNA: 5’-AAG ATG TGC TTC GAG ATG TGT GG-3′. The following primer sets were used to evaluate ATG5 KO: forward, 5’-GCT TCG AGA TGT GTG GTT TG-3’ and reverse, 5’-CAG TGG TGT GCC TTC ATA TT-3′. PCR products were verified through electrophoresis using 2% agarose gel.
Cell survival assay
The effect of each treatment on cell viability was determined through WST-1 assay. Briefly, cells were seeded at 5×103 cells per well into 96-well plates and were treated with the indicated reagents for the indicated days. Next, WST-1 (10 μL/well) was added into each well and cultured for additional 4 h. Absorbance was measured at 450 nm using a SpectraMax 190 microplate reader (Molecular Devices, Sunnyvale, CA, USA). Assays were performed in quadruplicate for each treatment and the average absorbance was normalized to that of the control (100%) to determine relative cell viability.
Analysis of autophagic flux using a tandem mRFP-GFP-LC3 reporter
Cells were seeded in a 60-mm dish and transiently transfected with ptfLC3 (tandem mRFP-GFP-LC3) plasmids at 70-80% confluency. After 24 h, cells were re-plated in 4-well chamber slides and treated with the indicated reagents before being fixed in 10% neutral-buffered formalin for 10 min, rinsed three times with DPBS for 5 min, and mounted on slides using VECTASHIELD Antifade mounting media (#H-100, Vector Laboratories, Burlingame, CA, USA). Slides were imaged using an Axio Imager Z1 upright trinocular fluorescence microscope equipped with an Axiocam 506 color (Carl Zeiss, Thornwood, NY, USA). Approximately 20 transfected cells were quantitatively analyzed per treatment. The ptfLC3 plasmids simultaneously detect LC3-positive immature autophagosomes (yellow) and mature autolysosomes (red) (Kimura et al., 2007). The numbers of LC3 puncta per cell were calculated using ImageJ software (NIH, Bethesda, MD, USA).
Preparation of cell lysates and immunoblot analysis
To prepare whole-cell lysates, cells were scraped into lysis buffer containing 1% Triton X-100 and then were centrifuged at 15,000×g and 4°C for 10 min. Protein concentration in the lysates was quantified using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Immunoblot analysis was performed as previously described (Hwang et al., 2018). Chemiluminescent signals were captured using a Bio-Rad ChemiDoc XRS+ instrument and quantified using Bio-Rad Image Lab software, version 5.2.1 (Bio-Rad).
Synergy measurements
The combination index (CI) was calculated according to the method of Chou and Talalay (Chou, 2010) using CompuSyn software (downloaded from http://www.combosyn.com/), with values of <1, =1, and >1 indicating synergism, additive effects, and antagonism, respectively.
RESULTS
Effects of PLX4720 on cell lines with different BRAF mutational status
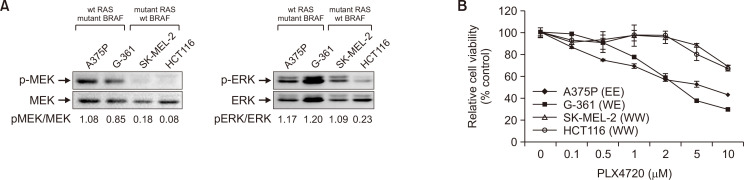
BRAF activating mutations drive tumorigenesis by constitutively activating the MAPK pathway (Cantwell-Dorris et al., 2011). As expected, the two BRAF mutant cell lines (A375P and G-361) had high basal levels of MEK/ERK phosphorylation (Fig. 1A), whereas much lower levels were observed in the two wild-type (WT) BRAF cell lines bearing activating RAS mutations (SK-MEL-2 and HCT116), despite the MAPK signaling cascade being a downstream regulator of RAS activation. The cell survival assay revealed that sensitivity to the BRAF inhibitor, PLX4720, correlated well with the BRAF mutational status of the cell lines (Fig. 1B). In particular, the BRAF mutant cells were much more sensitive to PLX4720 than the WT BRAF cells. MEK is a common downstream effector of RAF; therefore, MEK inhibitors have the potential to target all tumors dependent on MAPK signaling. We observed that the MEK inhibitor trametinib and ERK inhibitor SCH772984 showed efficacy against all cell lines tested, irrespective of BRAF mutation (Supplementary Fig. 1). Interestingly, A375P cells displayed stronger resistance to both MEK and ERK inhibitors than the other three cell lines.
Fig. 1.
Effects of PLX4720 on cell growth and autophagy in cells with different BRAF mutational status. (A) Whole-cell lysates were immunoblotted with antibodies against phospho-ERK, ERK, phospho-MEK and MEK. Cells were BRAF homozygous or heterozygous for wild-type (WT), or V600E (E) mutation. (B) Cell viability, measured through WST-1 assay, relative to vehicle controls in four cell lines exposed to PLX4720 (0.1-10 μM) for 72 h. Values represent the mean ± SD of quadruplicate determinants from one of three representative experiments.
Association of autophagy with mutant BRAF
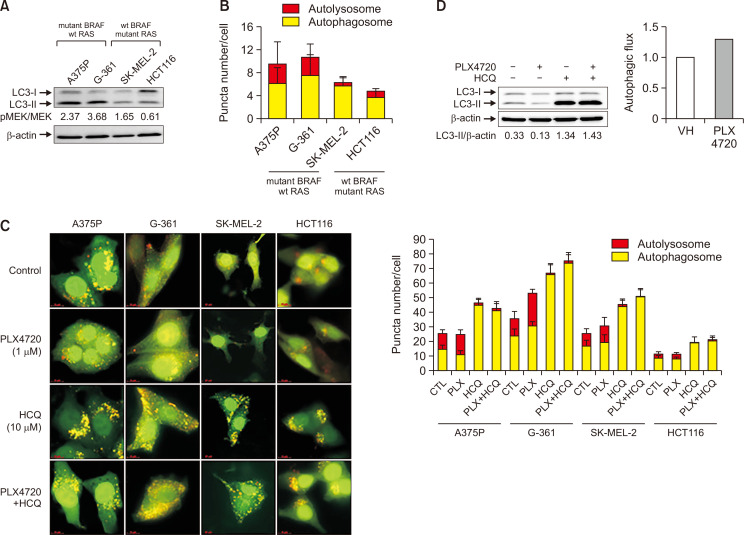
Tumor cells harboring mutant BRAF have been shown to exhibit an increased basal rate of autophagy (Rather et al., 2020). In this study, we investigated the link between sensitivity to BRAF inhibitors and basal autophagy by comparing the basal autophagic flux in four cell lines with different BRAF mutational status using LC3 conversion and tfLC3 imaging assays. As shown in Fig. 2A, levels of the autophagic marker LC3-II were elevated in two BRAF-mutated cell lines (A375P and G-361) but were lower in two WT BRAF cell lines (SK-MEL-2 and HCT116). To confirm these observations, we measured autophagic flux using ptfLC3 plasmids. Overall, a higher number of autolysosomes were observed in the BRAF mutant cells compared to WT BRAF cells (Fig. 2B). In addition, we assessed the effect of PLX4720 on autophagic flux by visualizing tandem mRFP-GFP-tagged LC3. Increased autophagosome formation was evident in all cell lines, but particularly BRAF-mutated cells, after PLX4720 treatment in the presence of the autophagy inhibitor HCQ (Fig. 2C). PLX4720-induced autophagy was also confirmed in A375P cells in the presence of HCQ by LC3 western blotting, in which autophagic flux was calculated based on the LC3-II band normalized to β-actin (Fig. 2D). Importantly, the LC3-I band is not an index of autophagic flux, and therefore, the LC3-II band should not be normalized to the LC3-I band for the evaluation of autophagic flux (Zhang et al., 2016).
Fig. 2.
Effects of PLX4720 on autophagy in cells with different BRAF mutational status. (A) Protein levels of autophagic markers were evaluated in four cell lines with different BRAF mutational status by monitoring LC3 conversion using western blotting. LC3 quantification reflects the LC3II/LC3I ratio. Protein levels were normalized to β-actin. (B) Representative histograms and quantification of basal autophagy. Autophagic flux was measured using the ptfLC3 plasmid that simultaneously expresses mRFP- and GFP-tagged LC3 protein. Quantification of the ratio of autophagosomal to autolysosomal LC3 puncta (n=20). (C) Cells were transfected with the ptfLC3 plasmid and treated with PLX4720 (1 μM) or HCQ (10 μM) for 24 h alone or in combination. Autophagosomes (yellow) show GFP-mRFP colocalization. Autolysosomes appear red. Right panel: quantification of the ratio of autophagosomal to autolysosomal LC3 puncta (n=20). (D) Cells were treated with PLX4720 (1 μM) or HCQ (10 μM) for 24 h alone or in combination. LC3 conversion was detected by immunoblot analysis. Autophagic flux was calculated using the LC3-II band normalized to β-actin. Lower panel: autophagic flux based on the LC3-II band calculated as the difference between the HCQ-treated and untreated groups.
TFEB nuclear translocation and autophagy induction in BRAF-mutated cells treated with PLX4720
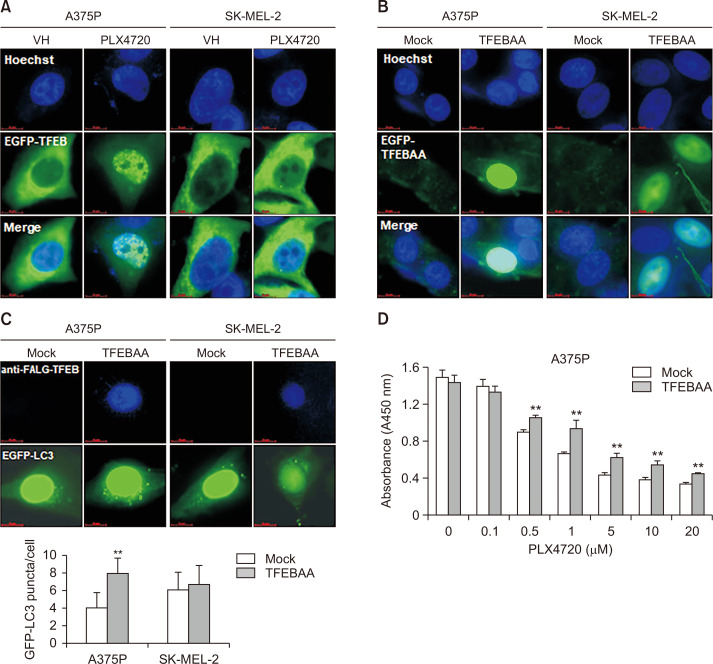
FEB regulates the expression of most ATG genes, along with genes involved in lysosomal biogenesis (Settembre et al., 2012). Unphosphorylated TFEB has been reported to translocate from the cytoplasm to the nucleus, and act as a transcription factor that coordinates expression of lysosomal hydrolases, membrane proteins and genes involved in autophagy (Settembre et al., 2011). In BRAF mutant cancer, constitutive TFEB phosphorylation by the BRAF downstream effector ERK has been shown to cause its cytoplasmic retention, leading to the down-regulation of autophagy–lysosome target genes (Li et al., 2019). Consistently, BRAF inhibition with PLX4720 treatment strongly triggered the nuclear translocation of TFEB in BRAF-mutated A375P cells in this study, but only weakly triggered translocation in WT BRAF SK-MEL-2 cells (Fig. 3A). To further explore the role of TFEB in autophagy induction, we investigated the effect of the forced overexpression of constitutively active S142A/S211A-TFEB mutant (TFEBAA) in A375P and SK-MEL-2 cells. We found that TFEBAA was mainly localized in the nucleus of both A375P and SK-MEL-2 cells (Fig. 3B). Moreover, we observed an increase in GFP-LC3 puncta in A375P cells co-transfected with TFEBAA and ptf-LC3 (Fig. 3C). In addition, The TFEBAA overexpression caused an increase in short-term proliferation of A375P cells after PLX4720 treatment as compared with mock-transfected cells (Fig. 3D). Together, these findings suggest that BRAF inhibitors induce autophagy via the nuclear translocation of TFEB, resulting in resistance to BRAF inhibition in BRAF-mutated cells.
Fig. 3.
TFEB nuclear translocation and autophagy induction in PLX4720-treated A375P cells. (A) Cells were transiently transfected with pEGFP-TFEB, exposed to PLX4720 (1 μM) for 24 h, and then fixed and counter-stained with Hoechst 33342 to identify the nucleus. (B) Cells were transiently transfected with FLAG-tagged TFEBAA (constitutively active S142A/S211A-TFEB mutant), and then fixed and counter-stained with Hoechst 33342 to identify the nucleus. (C) A375P and SK-MEL-2 cells were co-transfected with FLAG-tagged TFEBAA and pEGFP-LC3 before being stained with anti-FLAG antibodies. TFEBAA localization and LC3 puncta formation were detected by fluorescence microscopy. Lower panel: quantification of GFP-LC3 puncta per cell. Data are expressed as mean ± SD. **p<0.01 as compared with mock-transfected cells, as determined by unpaired t-test. (D) A375P cells were transfected with mock or FLAG-tagged TFEBAA for 24 h. The cells were washed, treated with increasing concentrations of paclitaxel ranged from 0.1 to 0.5 μM for 72 h. Cell viability was determined by the WST-1 assay. All values are relative to the vehicle control and are presented as mean ± SD (n=4). **p<0.01 as determined by the Dunnett’s t-test compared to control cells. Values represent the mean ± SD of quadruplicate determinants from one of three representative experiments.
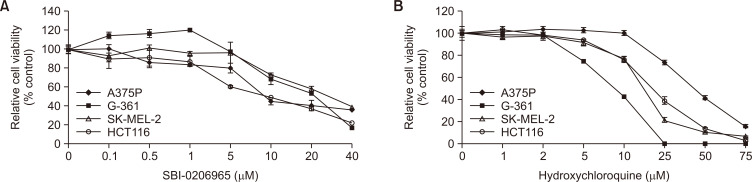
Pharmacologic inhibition of early-stage autophagy enhances the efficacy of BRAF inhibition
Since our results showed that basal autophagy was enhanced in BRAF-mutated cells, we assessed the sensitivity of BRAF mutant cells to BRAF and autophagy inhibition by treating BRAF-mutated and WT cells with early-(SBI-0206965) or late-(HCQ) stage autophagy inhibitors in the presence of a BRAF inhibitor. SBI-0206965 is an ATP-competitive ULK1 kinase inhibitor that has been shown to effectively inhibit early-stage autophagy (Egan et al., 2015), whereas HCQ is thought to inhibit autophagosome fusion with lysosomes at the late stage of autophagy (Piao and Amaravadi, 2016). The exposure of cells to these autophagy inhibitors reduced cell viability in a dose-dependent manner in all cell lines (Fig. 4). Combined treatment with PLX4720 and SBI-0206965 resulted in significantly greater cytotoxicity than either treatment alone (Supplementary Fig. 2A); however, this combination treatment was not effective in colon carcinoma cell line, HCT116, suggesting that the efficacy can be variable depending on the cell types. On the other hand, PLX4720 sensitivity was not significantly improved by the addition of HCQ (Supplementary Fig. 2B). To assess whether the combined efficacy is due to a synergistic effect, we calculated CI values using the Chou–Talalay method (Table 1) (Chou, 2010). Our data indicate that early autophagy inhibition improves the effectiveness of BRAF inhibitors, irrespective of BRAF inhibitor sensitivity.
Fig. 4.
Growth response to autophagy inhibitors in cell lines with different BRAF mutational status. Four cell lines were also treated with increasing concentrations of either early autophagy inhibitor SBI-0206965 (A) or late autophagy inhibitor HCQ (B) for 72 h. Cell growth was evaluated using a WST-1 assay. The relative viability of cells treated with the vehicle alone was regarded as 100%. Values represent the mean ± SD of quadruplicate determinants from one of three representative experiments.
Table 1.
Combination index (CI) for early or late-stage autophagy or MEK inhibitors combined with PLX4720 in BRAF WT and mutant cells
| Cell line | PLX4720 (μM) | ||||
|---|---|---|---|---|---|
| 0.1 | 0.5 | 1 | 5 | ||
| SBI (μM) | |||||
| A375P | 10 | 0.76 | 0.50 | 0.50 | 0.68 |
| G-361 | 10 | 0.92 | 0.78 | 0.73 | 0.87 |
| SK-MEL-2 | 10 | 0.46 | 0.47 | 0.40 | 0.41 |
| HCT116 | 10 | 1.52 | 1.56 | 1.43 | 1.19 |
| HCQ (μM) | |||||
| A375P | 10 | 3,287.86 | 1.91 | 1.45 | 7.51 |
| G-361 | 10 | 2.51 | 2.36 | 2.33 | 2.09 |
| SK-MEL-2 | 10 | 2.97 | 5.16 | 1.75 | 2.52 |
| HCT116 | 10 | 1.09 | 0.97 | 0.86 | 1.08 |
| Trametinib (μM) | |||||
| A375P | 0.1 | 0.03 | 0.12 | 0.20 | 0.66 |
| G-361 | 0.1 | 0.15 | 0.14 | 0.14 | 0.15 |
| SK-MEL-2 | 0.1 | 0.02 | 0.02 | 0.02 | 0.05 |
| HCT116 | 0.1 | 0.26 | 0.27 | 0.26 | 0.25 |
Four cell lines were treated with increasing PLX4720 doses in combination with SBI-0206965 (10 μM), HCQ (10 μM) or trametinib (0.1 μM) for 3 days. Cell viability was assessed using WST-1 assays. The Chou–Talalay equation was used to calculate the IC50 and determine synergism. CI values of <1, =1, and >1 indicate synergy, additives, and antagonism, respectively.
Synergistic effects of combined treatment with PLX4720 and MEK inhibitor on cell survival
The constitutive re-activation of downstream MEK/ERK signaling is thought to be the primary mechanism underlying intrinsic resistance to BRAF inhibitors (Lito et al., 2013). Therefore, a popular approach for treating cancer patients with BRAF mutations involves simultaneous targeting of BRAF and MEK (Cheng et al., 2018), although resistance to combined therapy is still observed in these patients (Holderfield et al., 2014). We found that the MEK inhibitor trametinib increased the efficacy of PLX4720 more effectively than autophagy inhibitors in both BRAF-mutated and WT cells (Table 1, Supplementary Fig. 3).
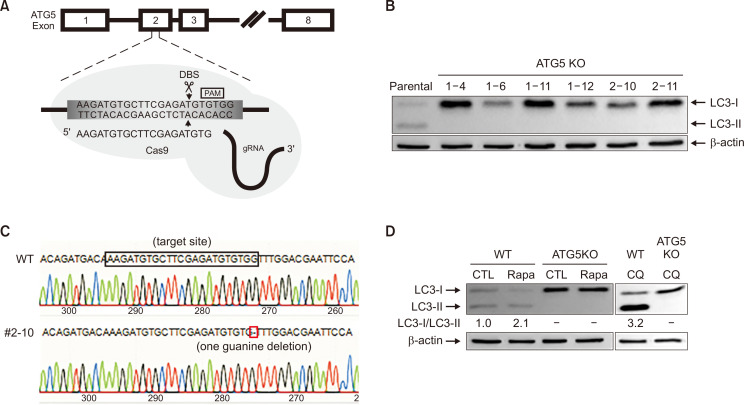
Establishment and validation of ATG5 KO cell lines
To further elucidate the role of autophagy in PLX4720-induced cell death, we established a cell line knocked-out for ATG5, which is necessary for autophagosome elongation. ATG5 KO in the A375P cell line was established as described previously (Kim et al., 2016) and CRISPR/Cas9 genome editing was employed to establish ATG5 KO in SK-MEL-2 cells. ATG5 sgRNA and target site were shown in Fig. 5A. Six clones were probed for ATG5 protein expression using immunoblot assays (Supplementary Fig. 4) and the lack of autophagy in ATG5 KO SK-MEL-2 cells was confirmed by LC3 conversion (Fig. 5B). LC3-II was not observed in ATG5 KO cells. From the initial screening, ATG5 gene disruption was confirmed in one clone (#2-10) by genomic DNA sequencing (Fig. 5C) and the lack of functional autophagy in this clone was further verified by the conversion of non-autophagic LC3-I into autophagic LC3-II in response to 250 nM rapamycin, an autophagy inducer (Fig. 5D). As expected, autophagic LC3-II was not detectable in the ATG5 KO cells and there was pronounced non-autophagic LC3-I accumulation. Treatment with CQ, an established inducer of autophagosome accumulation, led to the prominent accumulation of LC3-II in SK-MEL-2 cells but not its ATG5 KO cells.
Fig. 5.
Generation and validation of ATG5 knockout (KO) in SK-MEL-2 cells. (A) ATG5 sgRNA and target site. The 20-nt guide sequence comprising the 5′-end of the chimeric sgRNA is shown. DSB, double-strand break. (B) ATG5 knockout was confirmed by monitoring LC3 conversion using immunoblot analysis. β-actin was used as an internal control. (C) ATG5 KO was confirmed by sequencing for genomic editing at the target site. (D) Autophagy deficiency in the ATG5 KO cell line was confirmed by treating cells with rapamycin (250 nM) or CQ (20 μM) for 24 h. LC3 conversion was identified using immunoblotting. β-actin expression was used as a protein loading control.
Combination of PLX4720 and trametinib exerts a synergistic effect on reducing the survival of ATG5 KO SK-MEL-2 cells
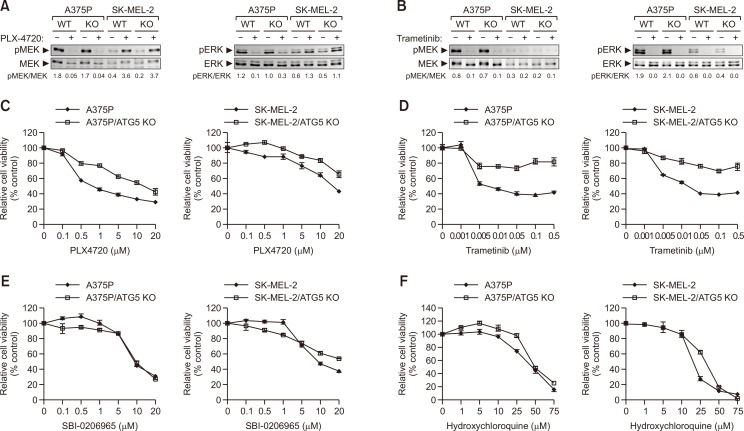
Next, we investigated the effect of PLX4720 on MEK/ERK activation status in both ATG5 KO A375P and SK-MEL-2 cells (Fig. 6A). Consistent with a previous study (Poulikakos et al., 2010), PLX4720 completely inhibited MEK/ERK phosphorylation in BRAF-mutated A375P cells but induced MEK/ERK phosphorylation in WT BRAF SK-MEL-2 cells. However, the changes in MEK/ERK phosphorylation caused by PLX4720 did not differ between ATG5 KO and parental cells, irrespective of BRAF mutation. The MEK inhibitor trametinib also drastically reduced MEK/ERK phosphorylation in both A375P and SK-MEL-2 cells regardless of ATG5 KO (Fig. 6B). Although MEK and ERK activation were inhibited by PLX4720 or trametinib in ATG5 KO cells, these cells showed stronger resistance to both inhibitors than their parental cells (Fig. 6C, 6D), suggesting that functional autophagy is required for the cytotoxic action of PLX4720 and trametinib against tumor cells. Neither autophagy inhibitor (SBI-0206965 and HCQ) altered cell survival between autophagy-proficient (parental) and deficient (ATG5 KO) cells (Fig. 6E, 6F).
Fig. 6.
Combined effect of PLX4720 and autophagy inhibitors on cell survival. In (A) and (B), A375P, SK-MEL-2, and their ATG5 KO cells were treated with 1 μM of PLX4720 (A) or trametinib (B). Whole-cell lysates were immunoblotted with antibodies to phospho-ERK, ERK, phospho-MEK and MEK. In (C-F), A375P, SK-MEL-2, and their ATG5 KO cells were treated with increasing concentrations of PLX4720 (C), trametinib (D), SBI-0206965 (E), or HCQ (F). Cell viability was determined using the WST-1 assay. All values are relative to the vehicle control and are presented as mean ± SD (n=4).
We also examined the effect of early or late-stage autophagy or MEK inhibitors on ATG5 KO cells in the presence of BRAF inhibition (Table 2, Supplementary Fig. 5). As expected, ATG5 KO abolished the synergistic effects of combined treatment with PLX4720 and SBI-0206965. In addition, HCQ had no synergistic effect with PLX4720 in ATG5 KO A375P or SK-MEL-2 cells, consistent with its effects in parental cells. Interestingly, trametinib exerted a synergistic effect on sensitivity to BRAF inhibitors in cells bearing WT BRAF but not activating BRAF mutants when ATG5 was knocked out.
Table 2.
Combination index (CI) for early or late-stage autophagy or MEK inhibitors combined with PLX4720 in ATG5 KO cells
| PLX4720 (μM) | A375P/ATG5 KO | SK-MEL-2/ATG5KO | |||||
|---|---|---|---|---|---|---|---|
| SBI (10 μM) |
HCQ (10 μM) |
Trametinib (0.1 μM) |
SBI (10 μM) |
HCQ (10 μM) |
Trametinib (0.1 μM) |
||
| 0.1 | 3.71 | 9.34 | 3.71 | 1.13 | 134.43 | 0.09 | |
| 0.5 | 0.64 | 0.93 | 3.33 | 1.37 | 570.61 | 0.10 | |
| 1 | 0.61 | 0.43 | 2.20 | 1.24 | 1115.84 | 0.09 | |
| 5 | 1.03 | 1.55 | 1.87 | 1.65 | 80.63 | 0.14 | |
ATG5 KO cells were treated with increasing PLX4720 doses in combination with SBI-0206965 (10 μM), HCQ (10 μM) or trametinib (0.1 μM) for 3 days. Cell viability was assessed using WST-1 assays. The Chou–Talalay equation was used to calculate the IC50 and determine synergism. CI values of <1, =1, and >1 indicate synergy, additives, and antagonism, respectively.
ER stress inhibition enhances the cytotoxic effect of PLX4720 in A375P cells
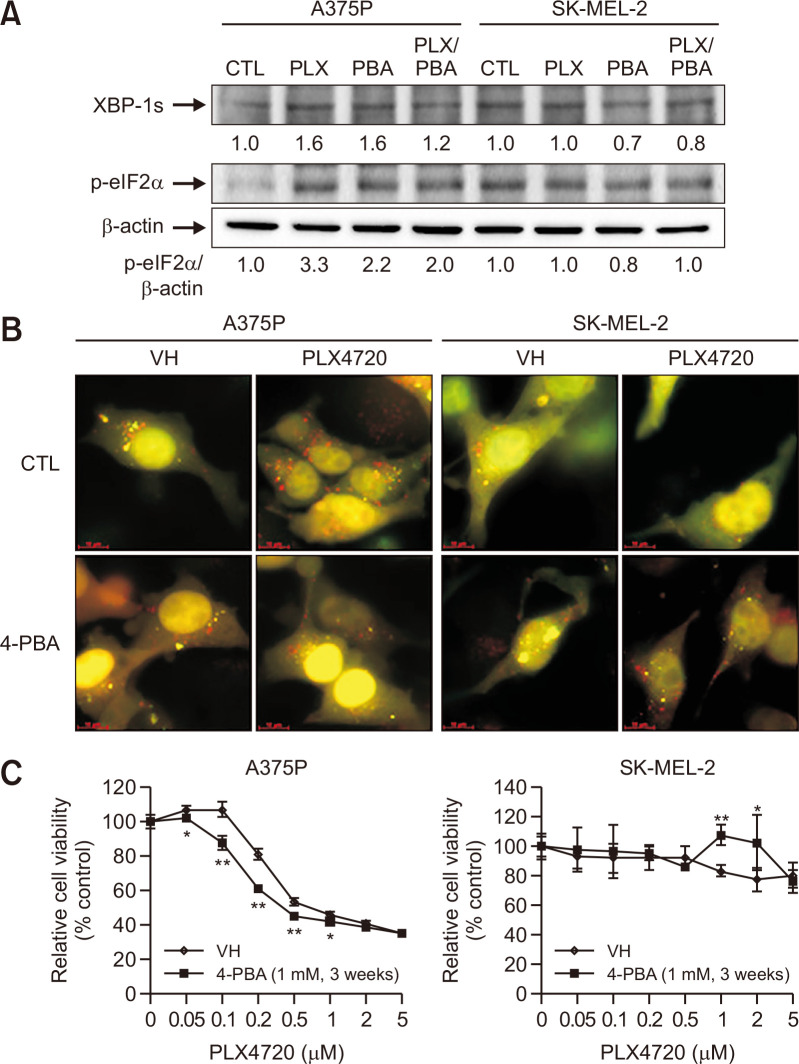
Oncogenic BRAF has been reported to induce chronic ER stress, thereby enhancing basal autophagy (Rather et al., 2020). Also, ER stress has been shown to provoke and precede BRAF inhibitor-induced autophagy (Ma et al., 2014). In this study, PLX4720 treatment significantly upregulated several critical ER stress markers, including XBP-1s and p-eIF2α (normalized to β-actin expression), in A375P cells compared to the untreated control (Fig. 7A). Relatively high basal p-eiF2α levels were observed in SK-MEL-2 cells but there was no change in the expression of ER stress markers after PLX4720 treatment. Three weeks of 4-phenylbutyric acid (4-PBA; ER stress inhibitor) treatment attenuated the upregulation of ER stress markers in response to PLX4720 in A375P cells, thus relieving the ER stress induced by BRAF inhibition.
Fig. 7.
The role of ER stress in PLX4720-induced cell death. (A) Control cells and cells pretreated with 4-PBA (1 mM) for 3 weeks were treated with PLX4720 (1 μM) for 24 h. Immunoblots against the indicated ER stress markers are shown. p-eIF2α expression was normalized to β-actin. Data represent three independent experiments. (B) Control cells and cells pretreated with 4-PBA (1 mM) for 3 weeks were transfected with the ptfLC3 plasmid and treated with PLX4720 (1 μM) for 24 h. Autophagosomes (yellow) show GFP-mRFP colocalization. Autolysosomes appear red. (C) Control cells and cells pretreated with 4-PBA (1 mM) for 3 weeks were treated with increasing concentrations of PLX4720 for 72 h. Cell viability was determined by the WST-1 assay. All values are relative to the vehicle control and are presented as mean ± SD (n=4). *p<0.05 and **p<0.01 as determined by the Dunnett’s t-test compared to control cells.
Next, we determined whether ER stress inhibition with chemical chaperones would decrease basal autophagy and increase PLX4720 susceptibility. The tfLC3 imaging assay revealed yellow and red fluorescent LC3 puncta, indicative of autophagosome and autolysosome accumulation, respectively (Fig. 7B). Three weeks of pre-exposure to 4-PBA significantly reduced PLX4720-induced autolysosome accumulation in A375P cells but not SK-MEL-2 cells. Therefore, 4-PBA appears to alleviate chronic ER stress in BRAF mutant cells and consequently reduce autophagic activity. In addition, we found that 4-PBA enhanced the sensitivity of BRAF mutant A375P cells to low concentration PLX4720 (0.05-1 μM; Fig. 7C), indicating a cytoprotective role for autophagy induced by chronic ER stress.
DISCUSSION
Tumor cells harboring oncogenic BRAF mutations have been shown to display an increased basal rate of autophagy (Corazzari et al., 2013). Consistently, this study confirmed an overall increase in the number of autolysosomes in BRAF mutant cells. Although RAS-driven cancer cells are also known to upregulate basal autophagy (Kinsey et al., 2019), the two RAS mutant cell lines exhibited a lower rate of basal autophagy than the two BRAF mutant cell lines, indicating that RAS and BRAF oncogenes differentially regulate the autophagic properties of the examined cancer cells. Many studies have revealed that chemotherapies such as BRAF inhibitors can induce autophagy to confer a significant growth advantage to cancer cells (Ma et al., 2014). Here, we found that PLX4720 generally increased the number of autolysosomes in BRAF mutant cells compared to BRAF WT cells and also triggered the nuclear translocation of TFEB much more strongly in BRAF mutant cells than in WT BRAF cells. TFEB phosphorylation by mTORC1 or ERK, the oncogenic downstream effector of BRAF, has been shown to lead to its cytoplasmic retention and impair autophagy-lysosomal function (Settembre et al., 2012; Li et al., 2019). Therefore, our results suggest that PLX4720 suppresses ERK by inhibiting oncogenic BRAF, in turn allowing the nuclear localization of TFEB and increasing the production of autophagy-lysosomal factors in BRAF mutant cells. The differences in autophagy induction by PLX4720 observed between BRAF mutant and WT cells may be due to the cytoplasmic retention of TFEB via paradoxical ERK activation in BRAF WT cells.
There have been conflicting reports regarding the role of autophagy in carcinogenesis; for instance, autophagy has been shown to promote growth of oncogenic BRAF-driven tumorigenesis (Xie et al., 2015), whereas reduced autophagy-related gene expression was observed in metastatic melanoma patients with oncogenic BRAF (Liu et al., 2014). Our previous study reported that autophagy deficiency enhanced susceptibility to cellular transformation (Hwang et al., 2018), suggesting a cytotoxic role for autophagy; however, various recent studies have suggested that autophagy may suppress tumor initiation but also promote tumor progression (Yang et al., 2014). Despite finding that PLX4720 and trametinib induced equivalent levels of MEK/ERK inhibition in ATG5 KO and parental cells, this study showed that ATG5 KO promoted resistance to both PLX4720 and trametinib in A375P and SK-MEL-2 cells, regardless of BRAF mutation. These results suggest that basal autophagy is required for the cytotoxic action of BRAF and MEK inhibitors against tumor cells. Conversely, a previous study indicated that BRAF inhibitor-induced autophagy is a pro-survival mechanism that limits BRAF inhibitor-induced cell death (Ma et al., 2014). While induced autophagy may contribute toward tumor development, basal autophagy may play a role in tumor suppression (Choi, 2012); however, the underlying molecular mechanisms remain unknown.
Due to the apparent pro-survival role of autophagy in PLX4720-induced cell death, it has been suggested that autophagy inhibition may improve the response of tumor cells to BRAF inhibition (Levy et al., 2014). In fact, autophagy inhibition was shown to be effective when combined with an oncogenic BRAF inhibitor in patients who express the BRAF V600E mutant (Levy et al., 2014); however, we found that early or late autophagy inhibition can exert different effects on cancer cell survival. The early autophagy inhibitor SBI-0206965 showed synergistic effects with PLX4720 in all cell lines, irrespective of their BRAF inhibitor sensitivity. Conversely, PLX4720 sensitivity was not significantly improved by the addition of HCQ, although it has been reported that late-stage autophagy inhibition improves the efficacy of BRAF inhibitors in BRAF mutant cancer (Mulcahy Levy et al., 2017). Our results are consistent with a previous study which showed that HCQ failed to augment PLX4720-induced growth impairment in BRAF inhibitor-sensitive A375P cells (Ma et al., 2014). Although CQ derivatives such as HCQ inhibit autophagy by increasing the intralysosomal pH, it has also been reported that CQ can activate TFEB by inhibiting mTORC1 activity (Zhitomirsky et al., 2018), thereby increasing the mRNA expression of autophagy-related genes. Thus, two contrasting processes appear to occur after HCQ treatment: 1) autophagic flux inhibition, and 2) TFEB activation, meaning that no synergistic effects between HCQ and PLX4720 are likely to be associated with its activating effects on TFEB.
Chronic ER stress due to oncogenic BRAF may play a key role in enhanced basal autophagy in BRAF-mutated cells (Rather et al., 2020). Moreover, BRAF inhibitors have been reported to increase the ER stress response soon after treatment, indicating that they may induce cytoprotective autophagy by activating an ER stress response (Ma et al., 2014). In this study, we found that PLX4720 induced ER stress in BRAF mutant cells but not WT BRAF cells, suggesting that the former may be more reliant on autophagy as a survival mechanism. In addition, the inhibition of ER stress with 4-PBA significantly increased the PLX4720 sensitivity of BRAF mutant cells; therefore, targeting ER stress may be a potential strategy for inhibiting both pro-survival ER stress and autophagy in BRAF mutant melanomas (Rather et al., 2020). Besides the molecular chaperone activity, 4-PBA functions as a histone deacetylase (HDAC) inhibitor (Kouraklis and Theocharis, 2006). Moreover, recent finding indicates that HDAC inhibitor reduces RTKs and PI3K signaling to synergise with BRAF inhibitor (Lai et al., 2013). Thus, it cannot be excluded the possibility that histone deacetylase activity may be partially responsible for the cytotoxic effect of PLX4720.
The simultaneous targeting of BRAF and MEK is a major approach for treatment of BRAF-mutated cancer (Algazi et al., 2020); however, this strategy of ‘vertically’ inhibiting the MAP kinase pathway via combined BRAF and MEK inhibition has not proven satisfactory (Holderfield et al., 2014). In this study, combined treatment with PLX4720 and the MEK inhibitor trametinib reduced the viability of both BRAF-mutated and WT cells more efficiently than an autophagy inhibitor, irrespective of their BRAF inhibitor sensitivity. In particular, the MEK inhibitor exerted a synergistic effect on sensitivity to BRAF inhibitors in cells bearing WT BRAF but not activating BRAF mutants with ATG5 KO. These contrasting effects of PLX4720 were associated with its different effects on ERK activation, which was inhibited in BRAF-mutated cells but enhanced in WR BRAF cells. Therefore, WT BRAF cells are more reliably sensitive to MEK inhibition than BRAF mutant cells. Consistently, a previous study reported that BRAF and MEK inhibitors exert powerful synergistic effects in WT BRAF and NRAS-mutated melanoma cells (Dinter et al., 2019).
Taken together, the findings of this study demonstrate that PLX4720 exerts a synergistic effect with early autophagy inhibitors but not late autophagy inhibitors, regardless of BRAF mutation. Therefore, future studies should determine how specific autophagy pathways are activated or inhibited by various anticancer therapies. In addition, our results suggest that simultaneously targeting BRAF and MEK could be a therapeutic avenue for WT BRAF melanomas, whereas combined treatment with BRAF and ER stress inhibitors (which inhibit pro-survival ER stress and autophagy) may be a potential strategy for BRAF-mutated tumors. Therefore, it is important to elucidate and exploit the mechanisms that contribute toward resistance to BRAF inhibitors in order to design the most effective therapeutic strategies.
SUPPLEMENTARY MATERIALS
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT (MIST) (NRF-2019R1F1A1048733) and by the Incheon National University Research Grant in 2019.
Footnotes
CONFLICT OF INTEREST
The authors declare no potential conflicts of interest.
REFERENCES
- Ahn J. H., Lee M. Autophagy-dependent survival of mutant B-Raf melanoma cells selected for resistance to apoptosis induced by inhibitors against oncogenic B-Raf. Biomol. Ther. (Seoul) 2013;21:114–120. doi: 10.4062/biomolther.2013.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algazi A. P., Othus M., Daud A. I., Lo R. S., Mehnert J. M., Truong T. G., Conry R., Kendra K., Doolittle G. C., Clark J. I., Messino M. J., Moore D. F., Jr., Lao C., Faller B. A., Govindarajan R., Harker-Murray A. H., Dreisbach L., Moon J., Grossmann K. F., Ribas A. Continuous versus intermittent BRAF and MEK inhibition in patients with BRAF-mutated melanoma: a randomized phase 2 trial. Nat. Med. 2020;26:1564–1568. doi: 10.1038/s41591-020-1060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantwell-Dorris E. R., O'Leary J. J., Sheils O. M. BRAFV600E: implications for carcinogenesis and molecular therapy. Mol. Cancer Ther. 2011;10:385–394. doi: 10.1158/1535-7163.MCT-10-0799. [DOI] [PubMed] [Google Scholar]
- Cheng L., Lopez-Beltran A., Massari F., MacLennan G. T., Montironi R. Molecular testing for BRAF mutations to inform melanoma treatment decisions: a move toward precision medicine. Mod. Pathol. 2018;31:24–38. doi: 10.1038/modpathol.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K. S. Autophagy and cancer. Exp. Mol. Med. 2012;44:109–120. doi: 10.3858/emm.2012.44.2.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T. C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- Corazzari M., Fimia G. M., Lovat P., Piacentini M. Why is autophagy important for melanoma? Molecular mechanisms and therapeutic implications. Semin. Cancer Biol. 2013;23:337–343. doi: 10.1016/j.semcancer.2013.07.001. [DOI] [PubMed] [Google Scholar]
- Davies H., Bignell G., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M. J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B. A., Cooper C., Shipley J., Hargrave D., Pritchard-Jones K., Maitland N., Chenevix-Trench G., Riggins G. J., Bigner D. D., Palmieri G., Cossu A., Flanagan A., Nicholson A., Ho J. W., Leung S. Y., Yuen S. T., Weber B. L., Seigler H. F., Darrow T. L., Paterson H., Marais R., Marshall C. J., Wooster R., Stratton M. R., Futreal P. A. Mutations of the B-Raf gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Dinter L., Karitzky P., Kaeubler T., Niessner H., Wanke I., Beissert S., Meier F., Westphal D. BRAF/MEK inhibitors induce potent ER stress - Enforced apoptosis in BRAFwt/NRASmut melanoma cells - Insights into mode of action and resistance mechanisms. Proceedings of the 16th International Congress of the Society for Melanoma Research; 2019 Nov 20-21; Salt Lake City, UT, USA: The Society for Melanoma Research; 2019. p. 102. [Google Scholar]
- Egan D. F., Chun M. G., Vamos M., Zou H., Rong J., Miller C. J., Lou H. J., Raveendra-Panickar D., Yang C. C., Sheffler D. J., Teriete P., Asara J. M., Turk B. E., Cosford N. D., Shaw R. J. Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol. Cell. 2015;59:285–297. doi: 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen T., Ahmad T., Flaherty K. T., Gore M., Kaye S., Marais R., Gibbens I., Hackett S., James M., Schuchter L. M., Nathanson K. L., Xia C., Simantov R., Schwartz B., Poulin-Costello M., O'Dwyer P. J., Ratain M. J. Sorafenib in advanced melanoma: a Phase II randomised discontinuation trial analysis. Br. J. Cancer. 2006;95:581–586. doi: 10.1038/sj.bjc.6603291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzivassiliou G., Song K., Yen I., Brandhuber B. J., Anderson D. J., Alvarado R., Ludlam M. J., Stokoe D., Gloor S. L., Vigers G., Morales T., Aliagas I., Liu B., Sideris S., Hoeflich K. P., Jaiswal B. S., Seshagiri S., Koeppen H., Belvin M., Friedman L. S., Malek S. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- Holderfield M., Deuker M. M., McCormick F., McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat. Rev. Cancer. 2014;14:455–467. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S. H., Han B. I., Lee M. Knockout of ATG5 leads to malignant cell transformation and resistance to Src family kinase inhibitor PP2. J. Cell. Physiol. 2018;233:506–515. doi: 10.1002/jcp.25912. [DOI] [PubMed] [Google Scholar]
- Ichimiya T., Yamakawa T., Hirano T., Yokoyama Y., Hayashi Y., Hirayama D., Wagatsuma K., Itoi T., Nakase H. Autophagy and autophagy-related diseases: a review. Int. J. Mol. Sci. 2020;21:8974. doi: 10.3390/ijms21238974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N. Y., Han B. I., Lee M. Cytoprotective role of autophagy against BH3 mimetic gossypol in ATG5 knockout cells generated by CRISPR-Cas9 endonuclease. Cancer Lett. 2016;370:19–26. doi: 10.1016/j.canlet.2015.10.008. [DOI] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Kinsey C. G., Camolotto S. A., Boespflug A. M., Guillen K. P., Foth M., Truong A., Schuman S. S., Shea J. E., Seipp M. T., Yap J. T., Burrell L. D., Lum D. H., Whisenant J. R., Gilcrease G. W. r., Cavalieri C. C., Rehbein K. M., Cutler S. L., Affolter K. E., Welm A. L., Welm B. E., Scaife C. L., Snyder E. L., McMahon M. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat. Med. 2019;25:620–627. doi: 10.1038/s41591-019-0367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouraklis G., Theocharis S. Histone deacetylase inhibitors: a novel target of anticancer therapy (review) Oncol. Rep. 2006;15:489–494. doi: 10.3892/or.15.2.489. [DOI] [PubMed] [Google Scholar]
- Lai F., Guo S. T., Jin L., Jiang C. C., Wang C. Y., Croft A., Chi M. N., Tseng H. Y., Farrelly M., Atmadibrata B., Norman J., Liu T., Hersey P., Zhang X. D. Cotargeting histone deacetylases and oncogenic BRAF synergistically kills human melanoma cells by necrosis independently of RIPK1 and RIPK3. Cell Death Dis. 2013;4:e655. doi: 10.1038/cddis.2013.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy J. M., Thompson J. C., Griesinger A. M., Amani V., Donson A. M., Birks D. K., Morgan M. J., Mirsky D. M., Handler M. H., Foreman N. K., Thorburn A. Autophagy inhibition improves chemosensitivity in BRAF(V600E) brain tumors. Cancer Discov. 2014;4:773–780. doi: 10.1158/2159-8290.CD-14-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Song Y., Quach C., Guo H., Jang G. B., Maazi H., Zhao S., Sands N. A., Liu Q., In G. K., Peng D., Yuan W., Machida K., Yu M., Akbari O., Hagiya A., Yang Y., Punj V., Tang L., Liang C. Transcriptional regulation of autophagy-lysosomal function in BRAF-driven melanoma progression and chemoresistance. Nat. Commun. 2019;10:1693. doi: 10.1038/s41467-019-09634-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lito P., Rosen N., Solit D. B. Tumor adaptation and resistance to RAF inhibitors. Nat. Med. 2013;19:1401–1409. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- Liu H., He Z., Simon H. U. Autophagy suppresses melanoma tumorigenesis by inducing senescence. Autophagy. 2014;10:372–373. doi: 10.4161/auto.27163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luebker S. A., Koepsell S. A. Diverse mechanisms of BRAF inhibitor resistance in melanoma identified in clinical and preclinical studies. Front. Oncol. 2019;9:268. doi: 10.3389/fonc.2019.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. H., Piao S. F., Dey S., McAfee Q., Karakousis G., Villanueva J., Hart L. S., Levi S., Hu J., Zhang G., Lazova R., Klump V., Pawelek J. M., Xu X., Xu W., Schuchter L. M., Davies M. A., Herlyn M., Winkler J., Koumenis C., Amaravadi R. K. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J. Clin. Invest. 2014;124:1406–1417. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy Levy J. M., Zahedi S., Griesinger A. M., Morin A., Davies K. D., Aisner D. L., Kleinschmidt-DeMasters B. K., Fitzwalter B. E., Goodall M. L., Thorburn J., Amani V., Donson A. M., Birks D. K., Mirsky D. M., Hankinson T. C., Handler M. H., Green A. L., Vibhakar R., Foreman N. K., Thorburn A. Autophagy inhibition overcomes multiple mechanisms of resistance to BRAF inhibition in brain tumors. eLife. 2017;6:e19671. doi: 10.7554/eLife.19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao S., Amaravadi R. K. Targeting the lysosome in cancer. Ann. N. Y. Acad. Sci. 2016;1371:45–54. doi: 10.1111/nyas.12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisapia P., Pepe F., Iaccarino A., Sgariglia R., Nacchio M., Russo G., Gragnano G., Malapelle U., Troncone G. BRAF: a two-faced Janus. Cells. 2020;9:2549. doi: 10.3390/cells9122549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poklepovic A., Gewirtz D. A. Outcome of early clinical trials of the combination of hydroxychloroquine with chemotherapy in cancer. Autophagy. 2014;10:1478–1480. doi: 10.4161/auto.29428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos P. I., Zhang C., Bollag G., Shokat K. M., Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rather R. A., Bhagat M., Singh S. K. Oncogenic BRAF, endoplasmic reticulum stress, and autophagy: crosstalk and therapeutic targets in cutaneous melanoma. Mutat. Res. 2020;785:108321. doi: 10.1016/j.mrrev.2020.108321. [DOI] [PubMed] [Google Scholar]
- Settembre C., Di Malta C., Polito V. A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S. U., Huynh T., Medina D., Colella P., Sardiello M., Rubinsztein D. C., Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Zoncu R., Medina D. L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M. C., Facchinetti V., Sabatini D. M., Ballabio A. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan R. J., Hollebecque A., Flaherty K. T., Shapiro G. I., Ahnert J. R., Millward M. J., Zhang W., Gao L., Sykes A., Willard M. D., Yu D., Schade A. E., Crowe K., Flynn D. L., Kaufman M. D., Henry J. R., Peng S. B., Benhadji K. A., Conti I., Gordon M. S., Tiu R. V., Hong D. S. A phase I study of LY3009120, a pan-RAF inhibitor, in patients with advanced or metastatic cancer. Mol. Cancer Ther. 2020;19:460–467. doi: 10.1158/1535-7163.MCT-19-0681. [DOI] [PubMed] [Google Scholar]
- Tsai J., Lee J. T., Wang W., Zhang J., Cho H., Mamo S., Bremer R., Gillette S., Kong J., Haass N. K., Sproesser K., Li L., Smalley K. S., Fong D., Zhu Y. L., Marimuthu A., Nguyen H., Lam B., Liu J., Cheung I., Rice J., Suzuki Y., Luu C., Settachatgul C., Shellooe R., Cantwell J., Kim S. H., Schlessinger J., Zhang K. Y., West B. L., Powell B., Habets G., Zhang C., Ibrahim P. N., Hirth P., Artis D. R., Herlyn M., Bollag G. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm S. M., Carter C., Tang L., Wilkie D., McNabola A., Rong H., Chen C., Zhang X., Vincent P., McHugh M., Cao Y., Shujath J., Gawlak S., Eveleigh D., Rowley B., Liu L., Adnane L., Lynch M., Auclair D., Taylor I., Gedrich R., Voznesensky A., Riedl B., Post L. E., Bollag G., Trail P. A. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64:7099–7109. doi: 10.1158/0008-5472.CAN-04-1443. [DOI] [PubMed] [Google Scholar]
- Xie X., Koh J. Y., Price S., White E., Mehnert J. M. Atg7 overcomes senescence and promotes growth of BrafV600E-driven melanoma. Cancer Discov. 2015;5:410–423. doi: 10.1158/2159-8290.CD-14-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A., Rajeshkumar N. V., Wang X., Yabuuchi S., Alexander B. M., Chu G. C., Von Hoff D. D., Maitra A., Kimmelman A. C. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014;4:905–913. doi: 10.1158/2159-8290.CD-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahedi S., Fitzwalter B. E., Morin A., Grob S., Desmarais M., Nellan A., Green A. L., Vibhakar R., Hankinson T. C., Foreman N. K., Mulcahy Levy J. M. Effect of early-stage autophagy inhibition in BRAFV600E autophagy-dependent brain tumor cells. Cell Death Dis. 2019;10:679. doi: 10.1038/s41419-019-1880-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Singh R., Aschner M. Methods for the detection of autophagy in mammalian cells. Curr. Protoc. Toxicol. 2016;69:20.12.1–20.12.26. doi: 10.1002/cptx.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhitomirsky B., Yunaev A., Kreiserman R., Kaplan A., Stark M., Assaraf Y. G. Lysosomotropic drugs activate TFEB via lysosomal membrane fluidization and consequent inhibition of mTORC1 activity. Cell Death Dis. 2018;9:1191. doi: 10.1038/s41419-018-1227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.