Abstract
Nasal and nasopharyngeal swab specimens tested by the Cepheid Xpert Xpress SARS-CoV-2 were analyzed by whole-genome sequencing based on impaired detection of the N2 target. Each viral genome had at least one mutation in the N gene, which likely arose independently in the New York City and Pittsburgh study sites.
Keywords: SARS-CoV-2 diagnostic testing, Cepheid XpertXpress, COVID-19, SARS-CoV-2, single nucleotide polymorphisms
1. Research note
As the COVID-19 pandemic continues and cases increase, the on-going need to accurately detect SARS-CoV-2 RNA in clinical specimens relies significantly on the use of nucleic acid amplification test (NAAT)-based diagnostics. While NAATs using real-time RT-PCR, for example, are highly specific and sensitive, a single nucleotide polymorphism (SNP) within critical primer- or probe-binding regions could lead to false-negative results or reduced sensitivity. Mutations that impact detection are rare but have been reported in other viruses, such as influenza and HIV (Korn et al., 2009; Yang et al., 2014). For SARS-CoV-2, a report described the failure to detect a SARS-CoV-2 variant harboring a C26340T mutation using the Roche cobas® SARS-CoV-2 assay (Roche Diagnostics, Switzerland), which targets the viral E gene (Artesi et al., 2020). Other reports described mutations in the N gene that reduced assay sensitivity by the Xpert® Xpress SARS-CoV-2 (Xpert) test (Cepheid Inc.) (Hasan et al., 2021; Ziegler et al., 2020). We describe identification of SARS-CoV-2 variants harboring C29200T, C29197T and G29227T mutations in both New York City and Pennsylvania.
As part of routine clinical care, the New York City (NYC) Public Health Laboratory (PHL) and the University of Pittsburgh Medical Center (UPMC) performed SARS-CoV-2 testing of nasopharyngeal or nasal swabs collected in viral transport media using the Xpert test as previously described (Loeffelholz et al., 2020). The NYC PHL identified 2814 SARS-CoV-2-positive specimens from August 2020 to mid-January 2021 with the Xpert test. Twelve of these specimens representing 10 unique patients tested as presumptive positive, wherein the E target was detected but the N2 target was not detected. Six presumptive positive specimens from six unique patients were further analyzed (Table 1 ). Due to sequencing limitations when viral RNA levels are low, only specimens with Ct values below 33 were included. From October to December 2020, one presumptive positive specimen was identified among 599 positive specimens with the Xpert test at UPMC. A second UPMC specimen was identified that was strongly positive (Ct <20) for the E target but weakly positive for N2 (Ct >40) (Table 1).
Table 1.
Nucleic acid amplification test results from specimens with N2 mutations.
| Specimen code | SNP location | Pangolin lineage | Initial SARS-CoV-2 Test |
Follow-up SARS-CoV-2 Test |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Test name | Result | E Ct | N2 Ct | N1 Ct | Test name | Result | N1 Ct | N2 Ct | |||
| NYC1 | C29200T | B.1.1.434 | Cepheid Xpert Xpress SARS-CoV-2 | Presumptive Positive | 16.5 | ND | New York Real-Time RT-PCR | Positive | 19.9 | 21.0 | |
| NYC2 | C29200T | B.1 | Cepheid Xpert Xpress SARS-CoV-2 | Presumptive Positive | 17.8 | ND | New York Real-Time RT-PCR | Positive | 20.0 | 18.4 | |
| NYC3 | C29200T | B.1.298 | Cepheid Xpert Xpress SARS-CoV-2 | Presumptive Positive | 15.3 | ND | New York Real-Time RT-PCR | Positive | 19.7 | 19.4 | |
| NYC4 | C29197T, G29227T | B.1.1.519 | Cepheid Xpert Xpress SARS-CoV-2 | Presumptive Positive | 21.2 | ND | New York Real-Time RT-PCR | Positive | 25.8 | 25.8 | |
| NYC7 | C29197T, G29227T | B.1.1.519 | Cepheid Xpert Xpress SARS-CoV-2 | Presumptive Positive | 16.9 | ND | New York Real-Time RT-PCR | Positive | 20.3 | 19.4 | |
| NYC8 | C29197T | B.1.1.519 | Cepheid Xpert Xpress SARS-CoV-2 | Presumptive Positive | 24.5 | ND | New York Real-Time RT-PCR | Positive | 28.5 | 29.7 | |
| PGH1 | C29200T | B.1.2 | Cepheid Xpert Xpress SARS-CoV-2 | Presumptive Positive | 15.3 | ND | Hologic Aptima | Positive | |||
| PGH2 | C29200T | B.1.2 | Cepheid Xpert Xpress SARS-CoV-2 | Positive | 19.5 | 44.3 | Not performed | ||||
| PGH3 | C29200T | B.1.2 | CDC n2019 Real-Time RT-PCR | Positive | 23.8 | 22.9 | Not performed | ||||
| PGH4 | C29200T | B.1.2 | Cepheid Xpert Xpress SARS-CoV-2/Flu/RSV | Positive | 13.1 | Not performed | |||||
| PGH5 | C29200T | B.1.2 | Hologic Aptima | Positive | Not performed | ||||||
ND = not detected.
To confirm the Xpert results, the six NYC specimens were tested with the New York SARS-CoV-2 Real-Time RT-PCR Diagnostic Panel which detects two SARS-CoV-2 N-gene targets: N1 and N2 (Wadsworth Center New York State Department of Health 2020). Both targets were detected in all six specimens, and their Ct values were within two cycles of each other (Table 1). One UPMC specimen (PGH1) was tested using the Hologic Aptima SARS-CoV-2 assay per the manufacturer's instructions and was positive (Aptima SARS-CoV-2 Assay Panther System, instructions for use, 2020). The Aptima assay utilizes transcription-mediated amplification, which is measured in relative light units instead of Ct values.
Whole genome sequencing (WGS) was used to identify SNPs in the N2 target region as defined by the CDC 2019-nCoV Real-time RT-PCR Diagnostic Panel primer sequences (2019-nCoV Real-time rRT-PCR panel primers and probes, 2020). The NYC PHL used two nucleic acid extraction platforms: NUCLISENS easyMag (bioMerieux) for specimens NYC1-4 and the KingFisher Flex system with the MagMAX Viral/Pathogen Nucleic Acid Isolation Kit (ThermoFisher) for specimens NYC7-8. The COVID-19 ARTIC protocol was followed with the v3 Illumina library construction on an Illumina MiSeq instrument (Farr et al., 2020). WGS for UPMC specimens was performed using Illumina viral RNA for enrichment. Briefly, RNA was extracted using the QIAamp Viral RNA Mini kit (Qiagen) according to the manufacturer's instructions. Following reverse transcription, the libraries were constructed using Nextera Flex. The normalized libraries were run on an Illumina NextSeq 550 platform. The resulting reads were processed using BreSeq and SNPs were identified by alignment to the Wuhan-1 reference genome. Full coverage of the N genes was obtained for all specimens. Genetic lineages were assigned using the Phylogenetic Assignment of Named Global Outbreak LINeages (Pangolin) (Rambaut et al., 2020). RAxML created the phylogenetic tree using a general time reversible model of evolution (GTRCAT) and 100 bootstrap replicates.
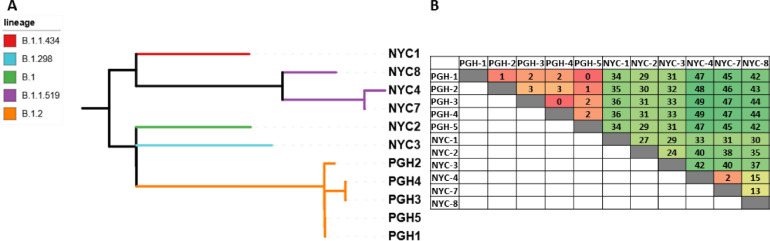
WGS analysis of the eight SARS-CoV-2 genomes revealed that the two UPMC genomes and three of the six NYC PHL genomes had the previously described C29200T SNP (Table 1) (Ziegler et al., 2020). The other three NYC specimens (NYC4, 7 and 8) had a C>T change at position 29197. Additionally, viral genomes from specimens NYC4 and NYC7 possessed a second SNP in the N2 region: a G>T change at position 29227. The G29227T SNP was only observed in conjunction with C29197G, and the two genomic sequences in which the SNPs were found together are closely related (Fig 1 A; B.1.1.519). However, for one sequence (NYC8), C29197T was present without the G29227T mutation, indicating that the C29197T mutation alone is sufficient for impaired N2 detection by the Xpert test. The Pittsburgh WGS database of 347 SARS-CoV-2 sequences was reviewed to determine whether additional specimens tested with other assays carried either the C29197T or C29200T N2 mutations. Three additional specimens carrying the C29200T mutation were identified (Table 1, PGH 3-5), resulting in a prevalence of 1.4% in the Pittsburgh database of 347 sequences. No Pittsburgh genomes with the C29197T mutation were identified. A phylogeny based upon pair-wise SNP differences among all eleven genomes demonstrates that the C29200T mutation arose independently in NYC (Fig. 1A). While the Pittsburgh B.1.2 lineage bearing the C29200T mutation is distinct from NYC lineages, the Pittsburgh lineage shows evidence of local transmission with 0to 3 SNP differences (Fig. 1B). In contrast, the NYC lineages bearing the C29197T mutation show evidence of genetic relatedness and potential transmission with 2-15 SNP differences (Fig. 1B).
Fig. 1.
Relatedness of the N2-mutant genomes (A) SNP- based phylogenetic tree with each lineage represented by colored branches. (B) Heat map table of the pairwise SNP differences.
The three SNPs are located within the N2 primer/probe-binding regions of both the New York and CDC SARS-CoV-2 RT-PCR Panels: C29299T and C29197T are in the probe-binding site and G29227T is in the reverse primer-binding site. However, these mutations do not interfere with detection with the New York RT-PCR assay (Table 1). Without knowledge of the specific sequences targeted in the Xpert test, it is not possible to determine if the identified SNPs are responsible for the failure to detect N2. Presumably, a molecular beacon analogous to the ones described for the Xpert® MTB/RIF assay is used for the SARS-CoV-2 Xpert test, and its binding may be similarly abrogated by the presence of a SNP within that region, thereby impeding N2 detection (Lawn and Nicol, 2011).
Most clinical diagnostic tests for SARS-CoV-2 target at least two independent genes/genetic regions providing detection redundancy. However, some laboratory-based tests in widespread use (such as SalivaDirect) and some available for use in CLIA-waived settings (like Abbott ID NOW COVID-19) detect only a single target and are therefore more vulnerable to false-negative results due to genetic variants. Cepheid has released a newer SARS-CoV-2 test, which also incorporates FluA/B and RSV targets in a multiplex assay. Although the same two SARS-CoV-2 targets are amplified, thus maintaining detection redundancy, the assay does not differentiate between them. Instead, one SARS-CoV-2 Ct value is reported, as demonstrated by specimen PGH-4 (Table 1). Clinical and public health laboratories should remain vigilant for evidence of genetic changes in the targets of NAAT diagnostic assays and may consider incorporating on-going genomic surveillance to identify and characterize variants.
Author contributions
Mindy Leelawong: Data Curation, Methodology, Writing—original draft; Stephanie L. Mitchell: Conceptualization, Supervision, Writing—original draft; Randal C. Fowler: Conceptualization, Visualization, Writing—review and editing; Edimarlyn Gonzalez: Formal analysis; Scott Hughes: Project administration, Writing—review and editing; Marissa P. Griffith: Software, Formal analysis, Writing—review and editing; Jane W. Marsh: Software, Writing—review and editing; Lee H. Harrison: Software, Supervision, Writing—review and editing; Jennifer L. Rakeman: Conceptualization, Supervision, Writing—review and editing;
Funding
This work was supported by a Centers for Disease Control and Prevention (CDC) ELC Enhanced Detection grant (ML, RCF, EG, SH, JLR) and the University of Pittsburgh COVID Medical Response Office and the University of Pittsburgh Medical Center Infection Prevention (MPG, JWM, LHH).
Acknowledgements
The authors like to thank the NYC PHL whole-genome sequencing team for sequencing and analyzing specimens, as well as the staff at the NYC COVID Express Quickie Laboratories for their dedication during the COVID-19 pandemic. In addition, The authors thank the members of the Microbial Genomic Epidemiology Laboratory and the Microbial Genome Sequencing Center for their expert technical performance.
Declaration of competing interest
No conflicts of interest.
References
- Artesi M, Bontems S, Gobbels P, Franckh M, Maes P, Boreux R, et al. A recurrent mutation at position 26340 of SARS-CoV-2 is associated with failure of the E gene quantitative reverse transcription-PCR utilized in a commercial dual-target diagnostic assay. J Clin Microbiol. 2020;58:e01598–20. doi: 10.1128/JCM.01598-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr B, Rajan D, Betteridge E, Shirley L, Quail M, Park N, et al. protocols.io; 2020. COVID-19 ARTIC v3 illumina library construction and sequencing protocol V.5. Available at: dx.doi.org/10.17504/protocols.io.bibtkann. Accessed January 28, 2020. [Google Scholar]
- Hasan MR, Sundararaju S, Manickam C, Mirza F, Al-Hail H, Lorenz S, et al. A novel point mutation in the N gene of SARS-CoV-2 may affect the detection of the virus by Reverse Transcription-Quantitative PCR. J Clin Microbiol. 2021;59:e03278–20. doi: 10.1128/JCM.03278-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control . U.S. Department of Health and Human Services; 2020. 2019-nCoV Real-time rRT-PCR panel primers and probes.https://www.cdc.gov/coronavirus/2019-ncov/downloads/rt-pcr-panel-primer-probes.pdf Available at: Accessed January 29, 2021. [Google Scholar]
- Korn K, Weissbrich B, Henke-Gendo C, Heim A, Jauer CM, Taylor N, et al. Single-point mutations causing more than 100-fold underestimation of human immunodeficiency virus type 1 (HIV-1) load with the Cobas TaqMan HIV-1 real-time PCR assay. J Clin Microbiol. 2009;47:1238–1240. doi: 10.1128/JCM.02204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawn SD, Nicol MP. Xpert(R) MTB/RIF assay: development, evaluation and implementation of a new rapid molecular diagnostic for tuberculosis and rifampicin resistance. Future Microbiol. 2011;6:1067–1082. doi: 10.2217/fmb.11.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffelholz MJ, Alland D, Butler-Wu SM, Pandey U, Perno CF, Nava A, et al. Multicenter evaluation of the cepheid Xpert Xpress SARS-CoV-2 test. J Clin Microbiol. 2020;58:e00926–20. doi: 10.1128/JCM.00926-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Holmes EC, O'Toole A, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hologic . Hologic Inc; San Diego, CA: 2020. Aptima SARS-CoV-2 Assay (Panther System), instructions for use. AW-21492-001 Rev. 005. [Google Scholar]
- Wadsworth Center New York State Department of Health . New York State Department of Health; Albany, NY: 2020. New York SARS-CoV-2 Real-time Reverse Transcriptase (RT)-PCR diagnostic panel, instructions for use. [Google Scholar]
- Yang JR, Kuo CY, Huang HY, Wu FT, Huang YL, Cheng CY, et al. Newly emerging mutations in the matrix genes of the human influenza A(H1N1)pdm09 and A(H3N2) viruses reduce the detection sensitivity of real-time reverse transcription-PCR. J Clin Microbiol. 2014;52:76–82. doi: 10.1128/JCM.02467-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler K, Steininger P, Ziegler R, Steinmann J, Korn K, Ensser A. SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Euro Surveill. 2020;25:2001650. doi: 10.2807/1560-7917.ES.2020.25.39.2001650. [DOI] [PMC free article] [PubMed] [Google Scholar]