Abstract
The worldwide circulation of different viruses coupled with the increased frequency and diversity of new outbreaks, strongly highlight the need for new antiviral drugs to quickly react against potential pandemic pathogens. Broad-spectrum antiviral agents (BSAAs) represent the ideal option for a prompt response against multiple viruses, new and re-emerging. Starting from previously identified anti-flavivirus hits, we report herein the identification of promising BSAAs by submitting the multi-target 2,6-diaminopurine chemotype to a system-oriented optimization based on phenotypic screening on cell cultures infected with different viruses. Among the synthesized compounds, 6i showed low micromolar potency against Dengue, Zika, West Nile and Influenza A viruses (IC50 = 0.5–5.3 μM) with high selectivity index. Interestingly, 6i also inhibited SARS-CoV-2 replication in different cell lines, with higher potency on Calu-3 cells that better mimic the SARS-CoV-2 infection in vivo (IC50 = 0.5 μM, SI = 240). The multi-target effect of 6i on flavivirus replication was also analyzed in whole cell studies (in vitro selection and immunofluorescence) and against isolated host/viral targets.
Keywords: Diaminopurine, Broad-spectrum antivirals, Dengue, Zika, West nile virus, Influenza, SARS-CoV-2
Graphical abstract
Abbreviations
- BSAA
broad-spectrum antiviral agent
- DENV
Dengue virus
- ZIKV
Zika virus
- WNV
West Nile virus
- COVID-19
coronavirus disease 2019
- CCR5
C–C chemokine receptor type 5
- SAR
structure−activity relationship
- PDD
phenotypic drug discovery
- TDD
target-based drug discovery
- SYRA
secondary yield reduction assay
- DYRA
direct yield reduction assay
- PRA
plaque reduction assay
- IA
immunodetection assay
- RSA
radical scavenging activity
- CPE
cytopathic effect
- RdRp
RNA-dependent RNA-polymerase
- PR8 H1N1
influenza A Puerto Rico/8/34H1N1 virus
- p2009H1N1
pandemic A/California/04/09H1N1 virus
- Ulster H7N1
avian A/parrot/Ulster/73H7N1
- ICW
In Cell Western assay
- MTT
[3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay
- HA
Hemagglutinin
- FLU
influenza
- CC50
50% cytotoxic concentration
- IC50
50% inhibitory concentration
- SI
Selectivity Index
1. Introduction
Viruses make up roughly two-thirds of all newly discovered human pathogens — far more than either bacteria or fungi. Despite much social visibility and alertness to the potential impact of widespread epidemics of devastating viral infections, a significant number of viral pathogens remains without effective treatment or cure, whilst only a few major viral infections (except HIV) can be prevented with vaccines [1,2]. In 2018, the Global Virome Project estimated that our planet harbours around 1.6 million yet-to-be-discovered potentially zoonotic viruses, and less than 1% of these has been identified to date [3]. In addition, old viruses previously confined in specific and isolated areas are re-emerging and rapidly spreading as a consequence of globalization and climate changes, but also those viruses that were thought to be eradicated with vaccination campaigns are reappearing due to public misinformation, no-vax campaigns and geopolitical instability in specific territories. As human activity takes people into new environments or down to anti-scientific pathways, the risk of zoonotic virus spillover into humans increases and, along with that, the possibility of new epidemic outbreaks in the coming decades. The ongoing severe coronavirus disease 2019 (COVID-19) pandemic caused by the new severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is just one of the possible outcome, but emerging widespread viruses with moderate mortality rate (e.g. Flaviviruses) may also represent a serious epidemic threat [4]. In fact, viruses are subject to continuous evolution and new variants may become more virulent and/or acquire different tissue tropism, as recently documented for Zika virus (ZIKV) isolates causing microcephaly and other congenital anomalies in fetus as well as neurologic disorders in adults. As an example, a single casual mutation (S139 N) in ZIKV has been recently linked to an increased infectivity in vitro, to more severe microcephaly and higher mortality rates in neonatal mice [5]. The National Institute of Allergy and Infectious Diseases (NIAID) has therefore classified Dengue virus (DENV), West Nile virus (WNV), and ZIKV as categories A and B emerging infectious pathogens, accounting for 17% of infections worldwide and for which no drugs or universally protective vaccines are currently available. The epidemic potential of these flaviviruses is well represented by the increasing number of autochthonous vector-borne flavivirus infections in the last decade [6,7]. An additional problem with flavivirus infections is represented by coinfections, not only by different viruses in the same family (e.g. DENV/ZIKV [8,9]), but also by different virus families (e.g. DENV/SARS-CoV-2 [10,11] or DENV/influenza [12]), which creates immune response disorders, confusion in diagnosis and delays in treatment, leading to additive or synergistic morbidities [13,14]. These diagnostic and therapeutic issues are even more serious in the case of newly emerging potentially pandemic viruses or in case of emerging variants with higher mortality rate since the time needed to develop a specific vaccine or drug is not compatible with a prompt response. Therefore, development of pan-viral drugs (broad-spectrum antiviral agents; BSAAs) capable of inhibiting the replication of multiple viruses belonging to different families, is an ambitious but attractive option in preparedness for outbreaks of previously unknown viral diseases and also for the treatment of widespread viral infections where an effective drug is not available yet. However, to date, the rarely used and mildly effective Ribavirin (RBV) is the only BSAA approved for the treatment of infections caused by different virus families. Other nucleoside antivirals, despite acting on the highly conserved viral transcription process, are approved for the treatment of infections caused by a single virus family. On the other hand, a number of experimental non-nucleoside BSAAs with mechanism of action (MOA) that is often complementary to that of nucleoside inhibitors are also under clinical development for the treatment of multiple virus families, even if sometime their MOA is not fully understood due to their effect on multiple targets (e.g. Nitazoxanide, [71] Arbidol [72]) (Fig. 1 ).
Fig. 1.
Selection of approved broad-spectrum antiviral agents (BSAAs), experimental BSAAs, 2,6-diaminopurine-based antivirals 1–3 and target BSAAs reported in this work.
To come up with new antivirals needed to fight the growing circulation of different viruses, the antiviral drug discovery community has been gradually revising its original “one-drug, one virus” dogma, based on targeting specific viral proteins with a single drug, passing through different paradigm shifts: i) the “one-drug, multiple viruses” paradigm, based on the design of molecules targeting viral proteins highly conserved among different viruses (e.g. RNA/DNA polymerases); ii) the “host-targeting antivirals” paradigm, based on targeting host-proteins (e.g. CCR5 with Maraviroc) used by many viruses for their replication [15]; iii) the “hypothesis-free drug repositioning” approach, based on testing safe in man compounds as antiviral candidates, independently from their original indication, for an accelerated approval. The latter approach is being strongly pursued to quickly discover new antivirals for emerging diseases, despite no clinical success has been reported in the last two decades [16]. A recent addition to the field is the “one-drug, multiple-targets” paradigm (polypharmacology), which represents a more realistic approach that takes into account the high number of on- and off-targets interacting with a drug candidate and the presence of compensatory pathways that may limit the effectiveness of highly specific drugs [17,18]. In fact, it is not uncommon that looking back at old antivirals (e.g. Ribavirin; Fig. 1) with this new awareness, new pharmacological targets or mechanisms are found involved in determining the observed therapeutic effect and, possibly, the pan-antiviral activity [19]. Partial inhibition of multiple targets with a single multi-target drug seems more effective than full inhibition of a single target, reducing on- and off-target-related toxicity and attrition rate in clinical development [17]. In line with this paradigm shift in antiviral drug discovery, our research group has successfully reported the development of multi-target antivirals active on multiple viruses and related diseases in vitro [[20], [21], [22], [23]].
As a continuation of our previous work in this field, we describe herein a phenotypic-based study focused at the optimization of 2,6-diaminopurine multi-target anti-flavivirus agents (e.g. 1–3, Fig. 1), [20,22] to identify new easily accessible BSAAs as suitable candidates for the treatment of multiple viral infections. Compound 6i was identified as the most promising BSAA able to reduce the replication of viruses belonging to different families (DENV, ZIKV, WNV, Influenza A, SARS-CoV-2) at low micromolar concentration while the reference BSAA Ribavirin does not show significant activity against Influenza A virus and SARS-CoV-2 [34,36]. Despite the full characterization of the antiviral MOA of 6i against all viruses investigated is not the scope of the present work, we also conducted a series of biochemical studies (in whole cell and against isolated targets) to confirm the original multi-target hypothesis for the inhibition of Flavivirus replication.
2. Results and discussion
By exploiting the complementary advantages of target-based drug discovery (TDD) and phenotypic drug discovery (PDD), a first generation of purine-based multi-target antivirals (e.g. compounds 1–2, Fig. 1) was previously reported by us [20]. These compounds proved to inhibit DENV replication (the only affected virus), blocking the interaction between the recombinant viral proteins NS5/NS3 and inhibiting, at the same time, the activity of host c-Src/Fyn kinases. The following target-based optimization based on the allosteric cavity B of NS5 led to a second generation of derivatives (e.g. compound 3, Fig. 1) with a wider spectrum of activity, blocking the replication of all four DENV serotypes and ZIKV [22]. Unfortunately, this targeted optimization did not allow to improve the antiviral potency of the second generation derivatives as we expected from the modeling predictions. A summary of antiviral potency for compounds 1–3 is reported in Fig. 1. Here we reasoned that a more efficient approach to optimize multi-target antivirals could be based on phenotypic considerations resulting from functional screening of focused derivatives since: i) the antiviral effect in a cellular context may be the result of a synergic activity on multiple targets; ii) unexpected targets or novel modes of action can be responsible for the antiviral effect. To further explore the antiviral potential of the 2,6-diaminopurine chemotype we planned to build a new series of derivatives by combining the C2 substituents of the hit compounds 1–3 with different substituted anilines in C6 (Scheme 1 ). Compounds 6a-k were thus obtained by applying the fast and efficient two-step microwave-assisted protocol previously developed, starting from commercially available 2,6-dichloropurine (4) [20]. Compounds 6l,m were then obtained by hydrolysis of the corresponding methyl esters 6d,f. In addition, we also prepared the two “inverted” derivatives of compounds 6c and 6i where C2 and C6 substituents were exchanged: starting from 4, the phenoxyethylamino chain was initially introduced in C6 (compound 7) followed by C2 nucleophilic substitution with the opportune substituted aniline to give the desired “inverted” compounds 8a,b (Scheme 1).
Scheme 1.
aReagents and conditions: i.) amines, n-BuOH, Et3N, 70–100 °C, 10–50 min, μW; ii.) amines, n-BuOH, TFA, 150–170 °C, 40–70 min, μW; iii.) 6d or 6f, LiOH, THF/MeOH, r.t., 24 h (for 6l,m).
Compounds 6a-m and 8a,b where then analyzed for their anti-DENV and anti-ZIKV activity in a Huh7 hepatoma cell-based secondary yield reduction assay (SYRA) [24] which quantifies infectious culture supernatants by immunodetection (Table 1 ). All non-toxic compounds tested showed promising antiviral activity against both DENV and ZIKV. Within the 3′-F and 3′-CF3 series (compounds 6a-c and 6g-i, respectively) the selectivity index (SI) with both viruses increased with the length of the C2 side chain. The 3′-CO2Me series (compounds 6d-f) was characterized by a similar potency on ZIKV and, surprisingly, the 2-phenoxyethylamino derivative 6f did not show any activity on DENV. Compared to the 3′-CO2Me series, the 3′-CO2H series (compounds 6l-m) showed a reduced potency and opposite specificity since the 2-phenoxyethylamino derivative 6m was active against both DENV and ZIKV (compared to 6f) while the 2-methylamino derivative 6l was active only against ZIKV (compared to 6d). Also the 3′,4′-OH derivative 6j was active only against ZIKV while the trimethoxy derivative 6k was quite cytotoxic. It was also interesting to note that the two “inverted” compounds 8a,b, analogues of the active compounds 6c and 6i, showed high cytotoxicity and were not evaluated for antiviral potency. Out of these 15 tested compounds, 6i showed the most potent antiviral activity against both ZIKV (SI 182) and DENV (SI 77) with IC50 values in the low micromolar range, comparable to or slightly better than those of the reference drugs Sofosbuvir and Ribavirin. Based on these results and on those reported by us for the previous generations of 2,6-diaminopurine antivirals, a few rough SAR considerations can be drawn: i) substituted anilines in C2 lead to cytotoxic compounds (e.g. 8a,b and 12c-e ref. [22]); ii) cyclic secondary amines in C2 lead to inactive or poorly active antivirals (e.g. 12f ref. [22], 16a-c ref. [20]); iii) increasing the length of the C2 substituent (primary amines) increase the selectivity index and the antiviral spectrum; iv) different functional groups on the C6 aniline moiety are well tolerated for the antiviral activity (see Table 1), with a few exceptions (e.g. 6k).
Table 1.
DENV-2/ZIKV/WNV replication inhibitory effect.
| DENV-2 |
ZIKV |
WNV |
|||||
|---|---|---|---|---|---|---|---|
| Cpd. | CC50a (μM) | IC50b (μM) | SIc | IC50 (μM) | SI | IC50 (μM) | SI |
| 6a | 84.0 | 9.3 ± 4.2 | 9 | 14.7 ± 5.7 | 6 | ND | – |
| 6b | 136.0 | 29.8 ± 0.3 | 5 | 8.9 ± 0.9 | 15 | ND | – |
| 6c | 200.0 | 12.0 ± 0.2 | 17 | 2.9 ± 0.9 | 68 | ND | – |
| 6d | 200.0 | 18.0 ± 12.2 | 11 | 8.0 ± 1.5 | 25 | ND | – |
| 6e | 200.0 | 17.5 ± 7.3 | 11 | 6.2 ± 2.5 | 32 | ND | – |
| 6f | 100.0 | NAd | – | 8.2 ± 1.5 | 12 | ND | – |
| 6g | 200 | 3.5 ± 4.0 (4.9 ± 2.6)f |
57 41 |
ND | – | ND (NA)f |
– |
| 6h | 50.0 | 4.5 ± 0.2 | 11 | 10.6 ± 0.5 | 5 | ND (1.9 ± 2.9)f (1.5 ± 1.3)g |
– 26 33 |
| 6i | 200.0 | 2.6 ± 0.3 (NA)f (0.9 ± 0.04)g |
77 - 222 |
1.1 ± 0.4 (0.8 ± 0.3)f (0.5 ± 0.32)g |
182 250 400 |
ND (NA)f (3.9 ± 0.67)g |
– – 51 |
| 6j | 200.0 | NA | – | 25.0 ± 7.1 | 8 | ND | – |
| 6k | 7.5 | ND | - | ND | – | ND | – |
| 6l | 200.0 | NA | – | 33.5 ± 7.6 | 6 | ND | – |
| 6m | 200.0 | 28.2 ± 3.9 | 7 | 29.0 ± 4.8 | 7 | ND | – |
| 8a | 8.0 | ND | - | ND | – | ND | – |
| 8b | 6.0 | ND | - | ND | – | ND | – |
| Sofosbuvir | 200.0 | 4.7 ± 0.7 (4.6 ± 1.4)f (3.8 ± 1.1)g |
42 43 53 |
3.2 ± 0.7 (2.7 ± 0.5)f (2.0 ± 1.1)g |
62 74 100 |
ND (1.7 ± 0.5)f (1.2 ± 0.3)g |
– 117 166 |
| Ribavirin | 200.0 | 4.0 ± 0.6 (7.6 ± 1.2)f (4.1 ± 0.6)g |
50 26 49 |
4.4 ± 0.6 (3.8 ± 0.6)f (2.2 ± 0.6)g |
45 53 91 |
ND (9.5 ± 4.0)f (6.7 ± 0.6)g |
– 21 30 |
aCC50: half-maximal cytotoxic concentration (μM); bIC50: half-maximal inhibitory concentration calculated through SYRA; each value is the mean of three experiments ± standard deviation (SD); cSI: selectivity index (SI = CC50/IC50). dNA: not active; eND: not determined; fIC50: half-maximal inhibitory concentration calculated through DYRA; gIC50: half-maximal inhibitory concentration calculated through PRA. DMSO was used as control in all cytotoxicity and antiviral assays.
As previously reported, comparison between SYRA and direct yield reduction assay (DYRA) data can be used to obtain preliminary information about early versus late antiviral activity of drug candidates [[24], [25], [26]]. Compound 6i was thus analyzed through DYRA against ZIKV and DENV replication. As shown in Table 1, 6i confirmed the antiviral potency seen in SYRA against ZIKV but it did not show any antiviral activity against DENV in DYRA. These results suggest that 6i exerts the antiviral activity against DENV not at early stages of viral replication and probably at a later stage than Sofosbuvir, which showed the same potency in SYRA and DYRA, as expected.
The anti-DENV efficacy of 6i was finally confirmed by plaque reduction assay (PRA), giving an IC50 of 0.90 ± 0.04 μM (in agreement with SYRA) that is four-fold the IC50 and SI of both Sofosbuvir and Ribavirin. Curiously, 6i showed low micromolar potency in SYRA/PRA but also in DYRA against ZIKV, suggesting an effect on replication of this virus at an earlier stage compared to DENV. The antiviral activity of the most potent DENV inhibitors 6h and 6i was also assessed against WNV through DYRA and PRA in comparison with Sofosbuvir/Ribavirin. All compounds were active in the low micromolar range but, again, 6i proved to inhibit WNV replication in PRA and not in DYRA, matching the result obtained with DENV. Overall, this phenotypic optimization led to the identification of 6i as the most interesting broad-spectrum anti-flavivirus candidate, whose potency and SI was generally superior to that of Sofosbuvir and Ribavirin.
In the attempt to further investigate and expand the multi-target propensity of the 2,6-diaminopurine class of molecules (generally referred to as privileged structure), we aimed at introducing small structural modifications that may confer additional antioxidant properties to 6i and closely related analogues. The induction of oxidative stress is in fact recognized as a common trigger of exacerbation in diseases caused by different viruses, including flavivirus, coronavirus and, particularly, Influenza virus [27]. A few literature reports show that opportune functionalization of the purine scaffold can lead to derivatives also endowed with antioxidant properties by scavenging reactive oxygen species (ROS) [[28], [29], [30]]. Among the many possible functionalization options, we were intrigued by a recent work from the Feringa group, describing the development of photoswitchable purine derivatives bearing an azobenzene moiety in C6 [31]. The synthesis of these compounds pass through a 6-phenylhydrazino intermediate that is easily converted into the desired azobenzene derivative by O2-mediated oxidation: although the hydrazine motif is generally considered a toxicophore moiety, we thought that opportunely substituted C6-phenylhydrazino derivatives may counteract the virus-induced oxidative stress converting themselves into stable azo-derivatives. This choice was also supported by the low cytotoxicity of 6-phenylhydrazino purines previously reported as anti-malaria agents [32]. The synthesis of our target compounds 10a-e followed the same synthetic sequence used for the synthesis of 6a-m: microwave-assisted C6 nucleophilic substitution on 4 with substituted phenylhydrazines afforded intermediates 9a-e that were finally converted into 10a-e by reaction with phenoxyethylamine at high temperature for 90 min (Scheme 2 ). The latter compounds were slowly converted into stable aza-derivatives 11a-d by simply stirring their methanolic solution at room temperature under air for 96 h. It was interesting to note that, under the same oxidative conditions, the 3-CF3 derivative 11e tend to decompose and the corresponding azo-derivative 11e was not isolated.
Scheme 2.
aReagents and conditions: i.) amines, n-BuOH, Et3N, 70 °C, 20–40 min, μW; ii.) phenoxyethylamine, n-BuOH, TFA, 170 °C, 120 min, μW; iii.) MeOH, air, r.t., stirring, 96 h.
Compounds 10a-e and 11a-d were then submitted for biological evaluation against flaviviruses. Unfortunately, most of the compounds displayed high cytotoxicity in Huh7 cells and were not tested for inhibition of virus replication. Only 3′-F derivatives 10a and 11a, which showed acceptable cytotoxic effect, were analyzed for their anti-flavivirus potency (Table 2 ): both compounds displayed low micromolar activity against DENV, ZIKV and WNV with the highest SI against the latter virus. Interestingly, the target compound 10a and the corresponding oxidized derivative 11a showed the same antiviral activity, supporting the idea that 10a may exert a synergic antiviral/antioxidant activity in infected cells and then being converted into a stable metabolite (11a) that still displays antiviral activity (dual action). The potential antioxidant effect of compound 10a was then experimentally evaluated in terms of ‘radical scavenging activity’ (RSA) using a ABTS radical decolorization method in comparison with 11a, 6i and with a potent antioxidant molecule (ascorbic acid) used as reference. [33].
Table 2.
DENV-2/ZIKV/WNV replication inhibitory effect.
| DENV-2 |
ZIKV |
WNV |
|||||
|---|---|---|---|---|---|---|---|
| Cpd. |
Huh7 CC50a (μM) |
IC50b (μM) | SIc | IC50 (μM) | SI | IC50 (μM) | SI |
| 10a | 20.0 | 3.4 ± 0.9 | 6 | 2.4 ± 0.3 | 8 | (1.1 ± 1.5)d | 18 |
| 10b | 8.5 | NDe | - | ND | – | ND | – |
| 10c | 5.0 | ND | - | ND | – | ND | – |
| 10d | 8.0 | ND | - | ND | – | ND | – |
| 10e | 7.5 | ND | - | ND | – | ND | – |
| 11a | 18.5 | 4.0 ± 1.02 | 5 | 2.8 ± 0.8 | 7 | (0.6 ± 0.2)d (0.6 ± 0.4)f |
28 30 |
| 11b | 12 | ND | - | ND | – | ND | – |
| 11c | 7.5 | ND | - | ND | – | ND | – |
| 11d | 8.0 | ND | – | ND | – | ND | – |
| Sofosbuvir | 200.0 | 4.7 ± 0.7 | 42 | 3.2 ± 0.7 | 62 | (1.7 ± 0.5)d (1.2 ± 0.3)f |
117 166 |
CC50: half-maximal cytotoxic concentration (μM).
IC50: half-maximal inhibitory concentration calculated through SYRA, each value is the mean of three experiments ± standard deviation (SD).
SI: selectivity index (SI = CC50/IC50).
IC50: half-maximal inhibitory concentration calculated through DYRA.
ND: not determined.
IC50: half-maximal inhibitory concentration calculated through PRA. DMSO was used as control in all cytotoxicity and antiviral assays.
As reported in Fig. 2 , compound 10a showed a clear concentration-dependent radical scavenging activity that was not observed for 11a and 6i. In addition, the stability of 11a towards oxidative stress was further assessed by reaction with H2O2 in argon atmosphere: while compound 10a was quickly converted into 11a, the latter compound and 6i were stable for days in the presence of 40 mM H2O2 in ethanol (data not shown).
Fig. 2.
Radical scavenging activity of compounds 10a, 11a and 6i expressed as % inhibition of the ABTS absorbance at 734 nm. The potent antioxidant ascorbic acid was used as a positive control.
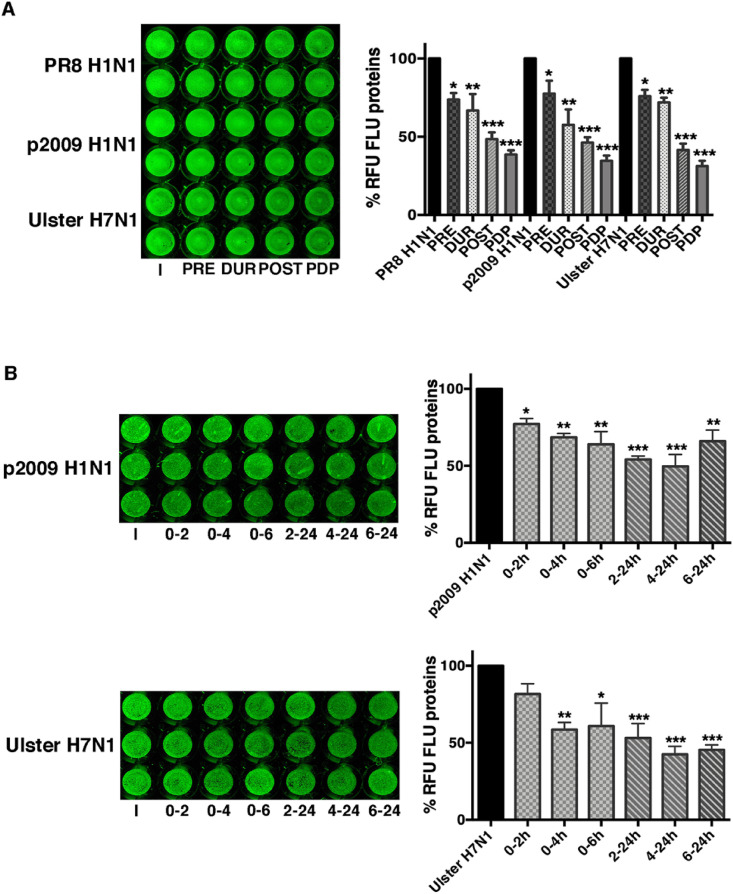
To further explore the spectrum of antiviral activity of the most promising inhibitors herein reported, compounds 6c, 6i, 10a were also evaluated against a segmented negative RNA virus (Influenza A Puerto Rico/8/34H1N1, PR8). MDCK and A549 cell lines were used to evaluate compounds’ cytotoxicity and antiviral potency. The BSAA Ribavirin (RBV) is reported in Table 3 as reference. [34]. The selected compounds did not display cytotoxicity on both cell lines and, by comparing the CC50 of Ribavirin, the three compounds showed higher CC50 values. Regarding the antiviral effect, 6c, 6i and 10a inhibited the replication of Influenza A virus at low micromolar concentration. In particular, 6i and 10a showed higher SI and IC50 with respect to 6c, and a stronger anti-influenza activity in comparison with RBV. Since compound 10a inhibited replication when added after the infection and due to its antioxidants activity, we hypothesize its role in maintaining reducing conditions into the cells. In fact it has been demonstrated that influenza virus induces redox changes in terms of ROS production and glutathione depletion, to activate redox-sensitive pathways useful for viral replication [35]. Further studies are in progress to evaluate this hypothesis. To get further insights on the effect of compound 6i on Influenza virus replication, it was added at different times: i) 2 h before infection (PRE); ii) for 1 h during viral adsorption (DUR); iii) after viral infection for 24 h (POST) or iv) before, during and after infection for the following 24 h (PDP). Supernatants from infected cells were recovered at 24 h from infection and used to infect a fresh cell monolayer. The expression of viral protein (hemagglutinin; HA) was quantified as percentage of relative fluorescence units (RFU) analyzed by ICW, as described in Methods Section. As shown in Fig. 3 A, the percentage of HA was similarly reduced (50% inhibition) when 6i was added during (DUR) or post infection (POST). On the contrary, pretreatment alone (PRE) was not quite effective. Next, specific steps of infection were evaluated by using a time-of-addition assay. In particular, compound 6i was added: i) immediately after infection for 2, 4 and 6 h on A549 cell monolayers and then maintaining cells in fresh medium without the compound until 24 h from infection; ii) after 2, 4 and 6 h from infection and maintaining the compound until 24 h from infection. Supernatants were recovered and used to infect fresh monolayers of MDCK cells. As shown in Fig. 3B, the percentage of HA expression was mostly reduced when 6i was added within the first 6 h from infection. Indeed, if the compound was added after 6 h, we did not find a significant inhibition, suggesting that the early phases of infection were impaired by 6i addition.
Table 3.
Influenza A virus and SARS-CoV-2 replication inhibitory effect.
| Influenza A Puerto Rico 8/34/H1N1 |
SARS-CoV-2d |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cpd | MDCK |
A549 |
Huh7 |
Caco-2 |
Vero E6 |
Calu-3 |
||||||||||||
| CC50a (μM) | IC50b (μM) | SIc | CC50 (μM) | IC50 (μM) | SI | CC50 (μM) | IC50 (μM) | SI | CC50 (μM) | IC50 (μM) | SI | CC50 (μM) | IC50 (μM) | SI | CC50 (μM) | IC50 (μM) | SI | |
| 6c | 77.0 | 14.0 | 5.5 | 58.0 | 20.8 | 2.8 | NDe | ND | – | ND | ND | – | ND | ND | – | ND | ND | – |
| 6i | 77.0 | 5.3 | 14.5 | 106.0 | 9.9 | 11 | 200 | 2.7 ± 0.1 | 74 | 81.3 | 5.9 ± 1.8 | 14 | 200.0 | 53.4 ± 1.1 | 4 | 120.0 | 0.5 ± 0.1 (1.9 ± 0.4)f | 240 |
| 10a | 136.0 | 6.3 | 21.6 | 52.4 | 14.9 | 3.5 | ND | ND | – | ND | ND | – | ND | ND | – | ND | ND | – |
| RBV | (>50) | (9.5)34 | (>5.3) | (>50) | (37.4)34 | (>1.3) | ND | ND | – | ND | ND | – | (>400) | (109.5)36 | – | ND | ND | – |
| RMD | ND | ND | – | ND | ND | – | 77 | 0.004 ± 0.005 (0.002) [37] | 19250 | 80.0 | 0.01 ± 0.006 (0.38) [39] | 8000 | 166.0 | 6.0 ± 1.1 (26.9) [38] | 28 | 97.0 | 0.11 ± 0.04 (0.28) [37] | 882 |
| CMT | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 200.0 | 0.82 ± 0.32 (0.04 ± 0.01)f | 243 |
CC50: half-maximal cytotoxic concentration (μM).
IC50: half-maximal inhibitory concentration, each value is the mean of three experiments ± standard deviation (SD) at 0.01 MOI.
SI: selectivity index (SI = CC50/IC50).
IC50: half-maximal inhibitory concentration calculated through DYRA.
ND: Not determined.
IC50 calculated incubating with the ENTRY-DYRA protocol. Literature antiviral data for Remdesivir (RMD) and Ribavirin (RBV) are reported in parenthesis. CMT: Camostat. DMSO was used as control in all cytotoxicity and antiviral assays.
Fig. 3.
Compound 6i interferes with early steps of influenza virus replication. A549 cells were infected with PR8 and treated or not with 6i (20 μM) at different phases of the virus life-cycle: the compound was added for 2 h before (PRE); during viral adsorption for 1 h (DUR); immediately after viral adsorption for 24 h (POST); before, during and for the following 24 h (PDP). The supernatants were recovered and used to infect fresh A549 monolayers for the following 24 h (A); compound 6i was immediately added after viral adsorption for 2 (0–2 h), 4 (0–4 h) or 6 (0–6 h) hours. Then cells were maintained up to 24 h without 6i in fresh medium plus 2% FBS; compound 6i was added after 2 (2–24 h), 4 (4–24 h) or 6 (6–24 h) hours infection and maintained until 24 h from infection. The supernatants were recovered and used to infect fresh MDCK monolayers for 24 h (B).
The expression of hemagglutinin (HA) was analyzed by ICW, using LI-COR Image Studio Software, as described in Methods. The percentage of relative fluorescence units (RFU) was calculated in comparison to untreated-infected cells (PR8 H1N1) (considered 100%). Data represent the mean ± S.D. of 3 replicates (A) or 4 replicates (B) obtained from two experiments performed. Statistical significance of the data vs PR8 H1N1 was defined as ∗P < 0.05 and ∗∗P < 0.01.
To evaluate whether 6i was effective against other influenza strains, we performed similar treatments on A549 cells infected with three different strains: the human PR8 H1N1, the human pandemic 2009H1N1 or the avian Ulster H7N1 as described in Methods. In the first set of experiments, cell monolayers were infected with the three strains and the expression of viral proteins, stained with an anti-FLU antibody capable of recognizing more viral proteins belonging to different strains, was observed directly on the infected monolayers at 24 h p.i. by ICW assay. As shown in Fig. 4 A, the addition of 6i inhibited viral proteins expression at all the conditions performed, even though the treatment with the compound after infection (POST) and before, during and after infection (PDP) was the most effective. Then, A549 cells were infected with the three strains and treated with 6i at different times from infection, as described before. Supernatants of treated cells were recovered after 24 h infection and used to infect fresh MDCK monolayers. As shown in Fig. 4B, the treatment with 6i was effective when added in the first 6 h from infection, thus suggesting an impairment in the early phases of virus life-cycle. Nevertheless, we cannot exclude that the compound partly affects host cell membrane [as indicated by about 20–26% inhibition during the pretreatment (PRE) of cell monolayers] or the viral attach/entry [28–45% inhibition during the viral adsorption phase (DUR)] (Fig. 3, Fig. 4A). Further studies are needed to ascertain those steps of viral infection.
Fig. 4.
Compound 6i is effective against different influenza A virus strains. A549 cells were infected with influenza A virus strains as described in Methods and treated or not with 6i (20 μM) at different phases of the virus life-cycle: A) compound 6i was added for 2 h before (PRE); during viral adsorption for 1 h (DUR); immediately after viral adsorption for 24 h (POST); before, during and for the following 24 h (PDP). Infected treated cells were fixed and stained with anti-FLU antibodies as already described; B) compound 6i was immediately added after viral adsorption for 2 (0–2 h), 4 (0–4 h) or 6 (0–6 h) hours. Then cells were maintained without the compound up to 24 h in fresh medium plus 2% FBS; the compound was added after 2 (2–24 h), 4 (4–24 h) or 6 (6–24 h) hours infection and maintained until 24 h from infection. The supernatants were recovered and used to infect fresh MDCK monolayers for 24 h.
The expression of FLU proteins was analyzed by ICW, using LI-COR Image Studio Software. The percentage of relative fluorescence units (RFU) was calculated in comparison to untreated-infected cells (considered 100%). Values are the mean ± S.D. of 2 replicates (A) or 3 replicates (B) obtained from one of the two experiments performed. Statistical significance of the data vs untreated-infected cells was defined as ∗P < 0.05, ∗∗ P < 0.01 and ∗∗∗P < 0.001.
Finally, the BSAA 6i was selected as the most interesting candidate to be evaluated against SARS-CoV-2: it was noteworthy to find that 6i inhibited the replication of this new coronavirus in the low micromolar range in all human cell lines evaluated (Table 3). Compound 6i showed submicromolar potency in SARS-CoV-2-infected Calu-3 lung cell line and a substantial reduction of efficacy in the monkey kidney VERO E6 cell line as observed for Remdesivir. [[37], [38]] Compound 6i also showed low micromolar anti-SARS-CoV-2 potency in liver Huh7 and intestinal Caco-2 cells, while Remdesivir showed low nanomolar potency on both cell lines in agreement with literature data [37,39]. The activity of 6i against SARS-CoV-2 was examined in depth in the pulmonary Calu-3 cell line, which better mimics the SARS-CoV-2 infection in vivo. Further experiments were performed incubating the cells and the virus with scalar concentration of 6i at two different conditions: 6i was (i) incubated with cells and removed after virus-cell adsorption (ENTRY-DYRA) or simply added after viral adsorption as previously performed (DYRA). The IC50 obtained in ENTRY-DYRA (1.9 ± 0.4 μM) and DYRA (0.5 ± 0.1 μM) were quite similar and the difference observed (3.8-fold) excludes a possible mechanism of action as entry inhibitor. On the other hands, using the entry inhibitor Camostat (CMT) as reference compound, [73] its antiviral potency differs of more than 20-fold when tested with ENTRY DYRA (0.04 ± 0.01 μM) or DYRA (0.82 ± 0.32 μM) protocol. Probably, the 6i DYRA IC50 is slightly lower with respect to that observed in ENTRY-DYRA, because the compound is never removed from the cells and it's available for 72 h of incubation. Considering that DYRA measures only the SARS-CoV-2 N protein reduction, we also investigated the role of 6i in the inhibition of viral replication. To assess the viral infectivity, we collected the supernatants derived from DYRA and we titred them infecting in quadruplicate VERO E6 cell line. The titer of each supernatant expressed as 50% tissue infectious culture dose (TCID50) was determined by Reed and Muench method and compared to the control virus titers. At the concentration of 20 μM and 0.8 μM, 6i was able to reduce the viral replication of 91.9 ± 5.1% and 83.5 ± 5.1% respectively.
Overall, phenotypic screening of the synthesized purine derivatives against different RNA viruses and different cell lines, clearly identified compound 6i as the most promising BSAA that can be quickly produced by two simple synthetic steps in only 60 min. Although 6i is just a promising lead compound, its cheap and easy synthetic accessibility is particularly relevant in comparison with the many synthetic steps that are needed to synthesize the reference nucleoside antivirals Sofosbuvir [40] and Remdesivir [41].
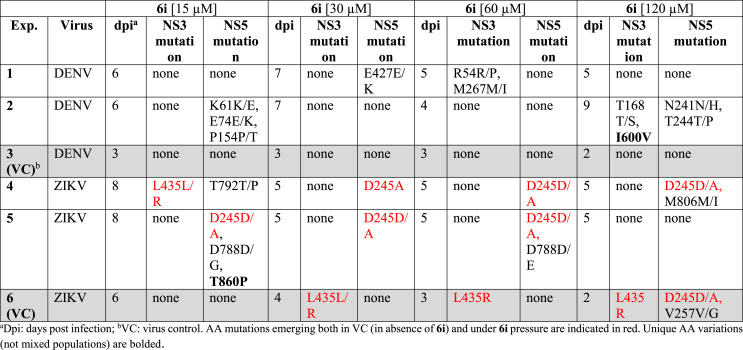
Considering the high anti-DENV and anti-ZIKV activity of 6i in Huh7 (SI 222 and 400 respectively in PRA), in vitro selection experiments were performed to investigate the 6i genetic barrier. ZIKV and DENV inputs (0.05 MOI), each in duplicate, were used to infect Huh-7 cells in the presence of increasing concentration of 6i, starting from 15 μM of 6i up to 120 μM. Uninfected cells treated with 6i were used as negative control to discriminate between cytopathic effect (CPE) derived from 6i cytotoxicity and physiological cell mortality. 6i pressure delayed viral growth with respect to the no-drug virus control (VC). Indeed, around 80% CPE was reached at 5–9 days post infection (dpi) with 15–120 μM 6i while the VC was consistently collected at 2–4 dpi (Table4 ). In vitro selection experiments were stopped after a mean of 23.8 ± 1.5 dpi when the drug pressure was 133-fold and 240-fold higher than 6i IC50 against DENV and ZIKV wild type, respectively. Aminoacidic (aa) variations in NS5 and NS3 regions selected under 6i pressure are indicated in Table 4. It must be noted that some mutations were transiently or permanently observed in the absence of drug pressure, suggesting that changes in the viral genome driven by virus adaptation to the cell line used or stochastically also occur.
Table 4.
In vitro ZIKV and DENV resistance selection to 6i.
Even if at different timing, both ZIKV and DENV escaped from 6i drug pressure, selecting mutation mixtures which were then lost at subsequent drug increment steps. When considering mutation profiles including pure mutations, the two viral strains emerging at 120 μM (I600V in DENV NS3) and at 15 μM (T860P in ZIKV NS5) were not quantifiable by IA. The same occurred with the virus carrying the mixed aa profile selected in ZIKV NS5 at 60 μM 6i (D245D/A combined with D788D/E). On the basis of these observations, we can hypothesize that these profiles selected by 6i impair the viral fitness resulting in lack of CPE at subsequent infection rounds. In this context, CPE is probably caused not only by replicating virus but also by the accumulation of viral transcripts and non-replication competent viruses. The mixed and pure mutations selected under 6i drug pressure were distributed in different NS5 domains for ZIKV and DENV.
With the exception of DENV E427 E/K and ZIKV D788D/G/E, all mutations selected in NS5 occurred at conserved sites among different flaviviruses [42]. The majority of NS5 aa variations selected (D788D/G, D788D/E, T792 T/P, M806 M/I, T860P for ZIKV and E427 E/K for DENV) were located in the polymerase region (RdRp); none of them were located in the catalytic site.43 The E427 is located between the G and F motifs of DENV RdRp: these two motifs interact with the nascent RNA template. D788, T792, M806 and T860 are located in the ZIKV priming loop which is involved in the correct positioning of nucleotides during polymerization. Interestingly, the T860P mutation emerged in ZIKV is located in the allosteric RdRp cavity B: it has been observed that mutated residues located in this cavity near T860 (W859A, I863A in DENV) significantly reduced viral replication and this could explain the fitness impairment of strains carrying this mutational profile [44]. All the remaining mixed mutations (K61 K/E, E74 E/K, P154 P/T, N241 N/H and T244 T/P) emerged in DENV were located in the N terminal region of NS5 in the methyltransferase domain (MTase) which interacts with the C-terminal domain of NS3 involved in the unwinding of double stranded RNA intermediate (ATPase/helicase activity) and in viral RNA 5′-capping reaction (triphosphatase) [45]. Under 6i drug pressure, no pure or mixed mutations were selected in ZIKV NS3. The mixed R54 R/P substitution selected in DENV NS3 is located in the N terminal domain near the catalytic site (His53, Asp77 and Ser138) of the trypsin-like serine protease [43]. The mixed substitution M267 M/I is located in the C terminal subdomain I of NS3; the conserved motifs in subdomains I and II are key regions for binding to RNA to generate the ATP binding cleft [46]. It's important to underline that, the only pure aa variation I600V acquired in the presence of 120 μM 6i by DENV is located in the subdomain 3 of the helicase region of the NS3 protein and between two α-helices (559–607 residues) α9 and α10 [47]. Interestingly, the subdomain III (NS3 483–618 aa) contains the interaction site with NS5 (320–368 aa) of the RdRP domain [48,49]. In comparison to our previously published work [24] investigating the genetic barrier of sofosbuvir against WNV with the same methodology, 6i seems not to have a high genetic barrier. Indeed the selected mutations were lost in the subsequent culture passages with increasing drug concentration. In addition, the time of viral breakthrough in the presence of 6i was not significantly increased with respect to VC. However, the I600V substitution for DENV NS3 and the D788D/G/E alone or coupled with the T860P substitution for ZIKV NS5 reduced dramatically the viral fitness. Indeed, mutated profiles were not able to replicate in subsequent cultures in the presence of 6i and reverted to wild type when grown in the absence of 6i (data not shown). While the mutations emerging from this study cannot explain the mechanism of action of 6i in detail, their location supports the targeting of the replication machinery and the interaction between NS3 and NS5 as per our original design [20]. To further corroborate the interaction between 6i and DENV/ZIKV NS5, we evaluated the inhibition of both MTase and polymerase enzymatic activity in cell free experiments. Compounds 6c, 10a and 11a were tested as comparison together with the lead compound 6i (Table 5 ). Increasing concentrations of inhibitor were incubated with the purified MTase or polymerase and the respective enzymatic activity was determined as previously described [50,51]. The IC50 of compounds, deduced from Hill slope curve-fitting, are presented in Table 5. The results show that the compounds barely inhibit the MTase activity. Conversely we observed that 6c and 6i inhibit the polymerase of DENV and ZIKV at a micromolar concentrations slightly higher than the corresponding IC50s in cell, further suggesting a multi-target mechanism for the inhibition of DENV/ZIKV replication.
Table 5.
IC50 values of compounds 6c, 6i, 10a and 11a against DENV/ZIKV polymerases and MTases.
| Cpds. | IC50 (μM)a |
IC50 (μM)a |
|||
|---|---|---|---|---|---|
| D2polb | D2 NS5c | ZIKV NS5d | DENV NS5-MTase | ZIKV NS5-MTase | |
| 6c | 31.2 ± 7.8 | >100 | 18.9 ± 3.1 | NA | NA |
| 6i | 20.8 ± 5.7 | 43.7 ± 12.0 | 12.8 ± 2.2 | 112.0 ± 51.5 | NA |
| 10a | >100 | 153.2 ± 38.2 | 105 ± 24.3 | NA | NA |
| 11a | >100 | >100 | >100 | 226.1 ± 107.4 | NA |
| 3'dATP control | 0.27 ± 0.03 | 0.32 ± 0.05 | 1 ± 0.2 | – | – |
| Sinefungin | – | – | – | (0,63 ± 0,04)52 | (1,18 ± 0,05)53 |
inhibition concentration 50%.
D2pol: polymerase domain of dengue 2 virus.
D2 NS5: full length DENV protein.
ZIKV NS5: full length ZIKV protein. Literature data for Sinefungin are reported in parenthesis.
In vitro resistance selection experiments showed that DENV and ZIKV escape 6i pressure by selecting a number of mutations in the NS5 and NS3 proteins that may impair their activity and/or the formation of the replication complex. Cell free experiment on isolated NS5 confirmed a moderate inhibitory effect of 6i, mostly on the polymerase activity. To further investigate the effect of 6i on the formation of the DENV replication complex in whole-cell context, we conducted an immunofluorescence analysis to follow the contemporary intracellular localization of NS5 and NS3 proteins at 8, 12, 16 and 24 h post infection with and without the presence of 6i or Sofosbuvir using non-toxic concentration (50 and 100 μM, respectively). Preliminary confocal microscopy analysis revealed that compound 6i significantly reduced the expression of NS3 (p ≤ 0.05 at 12 h) and NS5 (p ≤ 0.0001 at 16 h) proteins (Fig. 5 , panels B and C). On the other hands, treatment with Sofosbuvir significantly reduced NS3 and NS5 proteins expression starting from 12 h (p ≤ 0.05 and p ≤ 0.01, respectively) (Fig. 5, panels B and C). Next, the localization of both NS5 and NS3 in untreated control (VC) was analyzed at different time points: although flavivirus RNA replication occurs in the membrane-bound multi-protein assembly (the replication complex) localized in the cytoplasm, NS5 nuclear accumulation is a key process in flavivirus replication [[54], [55], [56]] while the contemporary NS3 localization has been less investigated [57]. Results from our experiments showed that initially (8 h) the NS3 protein had predominantly nuclear localization and moved to the cytoplasm at 24 h.
Fig. 5.
Immunofluorescence analysis. DENV-2 infected cells were treated with Sofosbuvir (100 μM) or 6i compound (50 μM) and stained at different time points with DENV NS3 antibody (detected using AlexaFluor® 488 labeled secondary antibody) and DENV NS5 antibody (detected using AlexaFluor® 568 labeled secondary antibody), and examined by confocal microscopy. A) Cells stained with DAPI and counted at 40× magnification in 5 random fields per well (∗ p ≤ 0.05). B) Expression of the viral NS3 protein in cells counted at 40× magnification in 5 random fields per well (∗ p ≤ 0.05; ∗∗ p ≤ 0.01; ∗∗∗∗ p ≤ 0.0001). C) Expression of the viral NS5 protein in cells counted at 40× magnification in 5 random fields per well (∗∗ p ≤ 0.01; ∗∗∗ p ≤ 0.001; ∗∗∗∗ p ≤ 0.0001). D) Individual antibody stained as well as merged images in untreated control, cells treated with Sofosbuvir, and cells treated with 6i compound. Each experiment was repeated at least two times.
On the other hand, NS5 protein initially (8 h) had predominantly cytoplasmic localization and translocation to the nucleus began at 16 h to be completed within 24 h. This analysis on the untreated control showed a sort of concerted nucleocytoplasmic shuttling of NS5 and NS3 proteins in opposite directions during infection: the NS3:NS5 colocalization peak, which could be ideally associated with the formation of the replication complex, occurred at 16 h, mainly at nuclear and perinuclear level (Fig. 5, panel D, merge line). Looking at the localization of NS5:NS3 in treated cells, sofosbuvir seems to abolish the formation of the replication complex: NS5 translocation into the nucleus was reduced or absent while, at the same time, NS3 protein did not translocate into the cytoplasm (Fig. 5, panel D, Sofosbuvir column). In cells treated with 6i, NS3:NS5 colocalization was significantly reduced in treated cells (Fig. 5, panel D, 6i column). The NS3:NS5 colocalization reduction observed in 6i treated cells was confirmed and quantified by colocalization analysis, with the greatest reduction observed at 16 h (MCC NS3/NS5 = 0.404 ± 0.106 and MCC NS5/NS3 = 0.397 ± 0.101; Costes P = 100%) compared to the untreated control (MCC NS3/NS5 = 0.726 ± 0.135 and MCC NS5/NS3 = 0.752 ± 0.125; Costes P = 100%) (p ≤ 0.0001) (Fig. 6 ).
Fig. 6.
Immunofluorescence analysis. DENV-2 infected cells were treated with 6i (50 μM) and stained at different time points with DENV NS3 antibody (detected using AlexaFluor® 488 labeled secondary antibody) and DENV NS5 antibody (detected using AlexaFluor® 568 labeled secondary antibody), and examined by confocal microscopy. NS3:NS5 stained cells merged images and white dots colocalization (WDC) images in untreated control (above) and treated with 6i compound (below) are shown as indicated. Each experiment was repeated at least two times.
It was interesting to observe that, although 6i and Sofosbuvir showed a similar effect on the reduction of NS5/NS3 expression over time, they induced a completely different effect on the localization of the same proteins, which reflects the different mechanism of action of the two compounds and indirectly support the difference seen in the DYRA/SYRA assays and the mutational profile that is observed under 6i pressure (not compatible with that of a nucleoside inhibitor).
Finally, to verify the multi-target effect of this class of compounds, the most interesting 2,6-diaminopurine derivatives synthesized in this work (6c, 6i, 10a and 11a) were also evaluated against selected host kinases that are known from the literature to be involved in the replication of flaviviruses, Influenza viruses and Coronaviruses (Table 6 ) [[58], [59], [60]]. The above compounds were tested at fixed concentration corresponding to their median antiviral IC50 (5 μM) in comparison with the hit 3. Despite potent nanomolar kinase inhibitors with no antiviral activity have been reported among 2,6-diaminopurine derivatives, [61] our compounds showed a moderate inhibitory effect on the selected kinases at micromolar concentration: in particular, compound 6i reduced the activity of Src, Abl and PI3Kα by roughly 50% at 5 μM. The get a wider picture on the kinase specificity of 6i, it was also tested against an additional panel of 15 host kinases selected among those known from the literature to be involved in the replication of different viruses (Table S3, Supporting Information) [74]. However, no significant inhibition of these additional targets was found testing 6i at 5 μM concentration, making Src, Abl and PI3Kα the only kinases (among the few selected) that are mildly inhibited by our lead compound. This moderate host kinases inhibition together with the observed effect on NS5/NS3 may therefore account for the anti-flaviviral effect of these multi-target inhibitors. As expected from the literature, [[58], [59], [60]] the effect of 6i on the above kinases could also play a partial role in inhibiting the replication of Influenza A and SARS-CoV-2 but the full understanding of its MOA is a complex endeavour and major efforts will be required to solve this challenge in the future. It is also important to remember that the precise knowledge of a drug's target is not an essential requirement in drug discovery and around 10–20% of approved drugs have unknown target or MOA [62,63]. Especially in the discovery of new BSAAs for emerging diseases, the right compromise between target identification and efficacy in multiple functional assays should probably drive the rapid development of new and efficient candidates for preclinical studies.
Table 6.
Inhibitory effect of BSAAs 6c, 6i, 10a and 11a against selected kinases.
| Residual enzymatic activity (%)b |
|||||
|---|---|---|---|---|---|
| Cpd.a | Src | Abl | Fyn | PI3Kα | CDK9/cT1 |
| 6c | 45.0 ± 20.0 | 54.0 ± 4.0 | NAc | 69.0 ± 1.0 | 87.0 ± 14.0 |
| 6i | 47.0 ± 20.0 | 70.0 ± 10.0 | NA | 66.0 ± 0.2 | NA |
| 10a | 84.0 ± 1.0 | 67.0 ± 2.0 | NA | 77.0 ± 3.2 | 88.0 ± 10.0 |
| 11a | NA | NA | 89.0 ± 2.0 | 78.0% ± 0.3 | NA |
| 3 | 59.0 ± 9.4 | 65.0 ± 5.6 | 45.0 ± 6.9 | ND | NA |
Each compound was tested at fixed 5 μM concentration. Values for reference compound 3 reported from the literature.
Values are the mean ± SD of two independent experiments.
NA: residual enzymatic activity >98%.
3. Conclusion
The development of new BSAAs represents the ideal option to counteract the spreading of different viruses (new and re-emerging), to deal with viral co-infections and in preparedness for new pandemics. Considering also the fact that the majority of the worlds’ population lives in underprivileged regions, efforts should be placed on the development of cheap and easy to be produced drugs. In our previous study on the identification of multi-target anti-flavivirus agents, we reported the design and identification of the hit compounds 1 and 2, as the most interesting multi-target compound endowed with anti-Src/Fyn activity, micromolar activity against DENV-2 (the only affected virus) and inhibition of the interaction between recombinant NS3 and NS5 proteins in an AlphaScreen assay at high micromolar concentration [20]. The following target-based optimization study led to the identification of compound 3 as a promising derivative active against all four DENV serotypes and ZIKV but possessing a potency (IC50 DENV-2 = 10 μM; IC50 ZIKV = 16.7 μM) comparable to that of compounds 1 and 2 [22]. Considering the sub-optimal results of this target-based optimization and the advantages of a system-oriented investigation in deciphering the functional efficacy of multi-target antivirals, we have reported here a phenotypic hit-to-lead optimization that has allowed us to identify compound 6i as a promising BSAA active against all tested viruses in the submicromolar to low micromolar range (IC50 = 0.5–5.3 μM).
Starting from preliminary SAR consideration on previously published 2,6-diaminopurine antivirals, we developed a first set of molecules (6a-m; 8a,b) that showed improved antiviral potency against selected flaviviruses (DENV-2, ZIKV and WNV). We then explored the possibility to functionalize the 2,6-diaminopurine scaffold with a ROS scavenging moiety to relieve the cellular oxidative stress generated by viral infections. Within this second set of molecules (10a-e; 11a-d), only two derivatives (10a and 11a) showed interesting antiviral results against selected flaviviruses.
The three most promising compounds identified from the phenotypic screening against flaviviruses (6c, 6i and 10a) were then evaluated against Influenza A (human and avian viruses) thus identifying 6i as the most potent and promising candidate, which retained activity on different Influenza strains (the human PR8 H1N1 and the pandemic 2009H1N1 or the avian Ulster H7N1). Compound 6i was also tested against the novel coronavirus SARS-CoV-2 proving to inhibit its replication at low micromolar and submicromolar concentrations in all human cell lines evaluated.
We also conducted a series of biochemical studies in the attempt to confirm the multi-target effect of this class of compounds on viral and host targets previously employed for the design of the antiviral hits 1 and 2. The original rationale that led us to the development of the anti-Flaviviral 2,6-diaminopurines was based on the contemporary inhibition of Src/Fyn host kinases and blockage of viral NS3:NS5 interaction. In vitro selection experiments were thus performed on DENV and ZIKV under 6i pressure: even if compound 6i seems not to have a high genetic barrier to develop resistance it selected few mutations in important functional sites of both NS5 and NS3 which dramatically reduced the viral fitness. Interestingly, mutations on the NS5 allosteric site involved in NS3:NS5 interaction were selected both on DENV (I600V) and ZIKV (T860P), in line with the original rationale. Immunofluorescence experiments on DENV-infected cells also showed a significant reduction of NS5/NS3 colocalization after treatment with 6i. These results are in line with the in vitro selection experiments and further indicate, in a whole cell context, that the anti-DENV effect of 6i is directly connected with a negative modulation of the replication complex as originally suggested by AlphaScreen experiments on recombinant NS3 and NS5 proteins [20]. In addition, it was also possible to highlight that 6i and Sofosbuvir act on the replication complex with two different MAO, thus suggesting a possible synergy between nucleoside antivirals and our multi-target molecules.
Finally, the multi-target effect of this class of 2,6-diaminopurine antivirals was also supported by the moderate inhibitory activity of 6i against ZIKV/DENV polymerase and against Src, Abl and PI3Kα out of a panel of 20 selected kinases. While a direct correlation between the inhibition of these host kinases and the broad-spectrum antiviral activity of 6i has not been demonstrated at the moment, Src, Abl and PI3Kα are validated and well recognized targets for the development or repurposing of antivirals against flavivirus, Influenza A virus and SARS-CoV-2.[74] In summary, the moderate inhibition of multiple isolated targets (and possibly others still unknown) herein observed, well describes the advantages of combining multi-target TBDD with the efficacy of a PDD approach for the identification and optimization of new promising BSAAs.
Overall, the identified lead compound 6i represents a very interesting BSAA capable of inhibiting the replication of DENV-2, ZIKV, WNV, Influenza A virus and SARS-CoV-2 with high selectivity index. By comparison with the reference antiviral drugs used in this work (Sofosbuvir, Ribavirin, Remdesivir and Camostat), compound 6i is very easy/cheap to be produced (just two synthetic steps from commercial materials) and displays at low micromolar and submicromolar antiviral efficacy in all infected cell lines while presenting a unique broad-spectrum profile that is not shown by any of these drugs. Further studies on this class of 2,6-diaminopurine antivirals are currently ongoing to evaluate the ADME profile and suitable formulations for in vivo studies, while additional functionalization in N9 are being introduced to fully explore the biologically relevant chemical space around this chemotype. Additional results will be published in due course.
4. Experimental section
4.1. Chemistry
General. All commercially available chemicals were purchased from Merck or Fluorochem and, unless otherwise noted, used without any previous purification. Solvents used for work-up and purification procedures were of technical grade. TLC was carried out using Merck TLC plates (silica gel on Al foils, SUPELCO Analytical). Where indicated, products were purified by silica gel flash chromatography on columns packed with Merck Geduran Si 60 (40–63 μm). 1H and 13C NMR spectra were recorded on BRUKER AVANCE 300 MHz and BRUKER AVANCE 400 MHz spectrometers. Chemical shifts (δ scale) are reported in parts per million relative to TMS. 1H NMR spectra are reported in this order: multiplicity and number of protons; signals were characterized as: s (singlet), d (doublet), dd (doublet of doublets), ddd (doublet of doublet of doublets), t (triplet), m (multiplet), bs (broad signal). Low resolution mass spectrometry measurements were performed on quattromicro API tandem mass spectrometer (Waters, Milford, MA, USA) equipped with an external APCI or ESI ion source. ESI-mass spectra are reported in the form of (m/z). Melting points were taken using a Gallenkamp melting point apparatus and were uncorrected. The FDA-approved anti-HCV drugs sofosbuvir (MCE® cat. HY-15005), remdesivir (MCE® cat. HY-104077) and ribavirin (MCE® cat. HY-B0434) used as reference compounds, were supplied as powder and dissolved in 100% dimethyl sulfoxide (DMSO). Camostat mesylate (MCE® cat. HY-13512), an inhibitor of TMPRSS2 showing antiviral activity against SARS-CoV-2 was dissolved in water and used as reference compound in ENTRY-DYRA assay. All final compounds were screened for PAINS using the Free ADME-Tox Filtering Tool (FAF-Drugs4) program (http://fafdrugs4.mti.univ-paris-diderot.fr/) [70]: as expected, the azo-derivatives 11a-d were flagged as potential PAINS. Elemental analyses were performed on a ThermoQuest (Italia) FlashEA 1112 Elemental Analyzer. All final compounds were >95% pure as determined by elemental analysis. data for C, H, and N (within 0.4% of the theoretical values; Table S4).
Microwave Irradiation Experiments. Microwave reactions were conducted using a CEM Discover Synthesis Unit (CEM Corp., Matthews, NC). The machine consists of a continuous focused microwave power delivery system with an operator-selectable power output from 0 to 300 W. The temperature inside the reaction vessel was monitored using a calibrated infrared temperature control mounted under the reaction vessel. All experiments were performed using a stirring option whereby the reaction mixtures were stirred by means of a rotating magnetic plate located below the floor of the microwave cavity and a Teflon-coated magnetic stir bar in the vessel.
General Procedure for the Synthesis of intermediate 5a-e: In a microwave tube 2,6-dichloro-9H-purine 4 (100 mg; 0.53 mmol) and the proper amine (2.645 mmol) were suspended in n-BuOH (3 mL). NEt3 (258 μL; 1.852 mmol) was added, except for 5e, and the tube was heated at 70–120 °C until the starting material was consumed (max μW power input: 100 W; ramp time: 1 min; power max: off; maximum pressure: 260 psi). At the end of irradiation, the solid obtained was isolated by filtration over a Buchner funnel after the addiction of cold n-hexane and ethyl acetate. In a few cases (5b, 5d), at the end of irradiation, the reaction mixture was concentrated under vacuum, dissolved with ethyl acetate, and washed with NaHCO3. The combined organic were washed with brine, dried over anhydrous Na2SO4, filtered and concentrated. The crude material was purified by silica gel chromatography, using the proper eluent: 5b: CH2Cl2/MeOH 99:1–96:4; 5d: CH2Cl2/Formic Acid/MeOH 93:3:4.
2-chloro-N-(3-fluorophenyl)-9H-purin-6-amine (5a): 1st Irradiation cycle: 20 min, 100 °C; 2nd Irradiation cycle: 35 min, 120 °C. Yield: 67%. MS (ESI) [M+H]+: 264.14 m/z. 1H NMR (DMSO‑d 6, 400 MHz): δ 5.37 (s, 1H), 6.29 (m, 1H), 6.99 (dd, 1H, J = 15.4, 7.9 Hz), 7.39 (d, 1H, J = 7.4 Hz), 7.88 (d, 1H, J = 12.1 Hz), 8.34 (s, 1H), 10.44 (s, 1H).
Methyl 3-((2-chloro-9H-purin-6-yl)amino)benzoate (5b): 1st Irradiation cycle: 20 min, 70 °C; 2nd Irradiation cycle: 15 min, 100 °C. Yield: 25%. MS (ESI) [M+H]+: 304.12 m/z. 1H NMR (DMSO‑d 6, 400 MHz): δ 3.87 (s, 3H), 7.51 (m, 1H), 7.66 (m, 1H), 8.14 (m, 1H), 8.33 (s, 1H), 8.54 (s, 1H), 10.42 (s, 1H), 13.36 (s, 1H).
2-chloro-N-(3-(trifluoromethyl)phenyl)-9H-purin-6-amine (5c): 1st Irradiation cycle: 15 min, 100 °C. Yield: 96%. MS (ESI) [M+H]+: 314.23 m/z. 1H NMR (DMSO‑d 6, 400 MHz): δ 7.42 (d, 1H, J = 8 Hz), 7.60 (t, 1H, J = 8 Hz), 8.15 (d, 1H, J = 8 Hz), 8.36 (s, 1H), 8.39 (s, 1H), 10.54 (s, 1H), 13.34 (s, 1H).
4-((2-chloro-9H-purin-6-yl)amino) benzene-1,2-diol (5d): 1st Irradiation cycle: 20 min, 70 °C; 2nd Irradiation cycle: 15 min, 120 °C. Yield: 29%. 1H NMR (DMSO‑d 6, 400 MHz): δ 6.71 (d, 1H, J = 8 Hz), 7.02 (d, 1H, J = 8 Hz), 7.22 (s, 1H), 8.33 (s, 1H), 9.90 (s, 1H).
2-chloro-N-(3,4,5-trimethoxyphenyl) -9H-purin-6-amine (5e): 1st Irradiation cycle: 20 min, 70 °C. Yield: 70%. 1H NMR (DMSO‑d 6, 400 MHz): δ 3.65 (s, 3H), 3.78 (s, 3H), 7.40 (s, 2H), 8.30 (s, 1H), 10.12 (s, 1H), 13.33 (s, 1H).
General Procedure for the Synthesis of Compound 6a-m: In a microwave tube 5a-e (0.19 mmol) and the proper amine (0.570 mmol) were suspended in n-BuOH (3 mL). TFA (0.19 mmol) was added and the tube was heated under microwave irradiation at 150–170 °C for 40–70 min (max μW power input: 300 W; ramp time: 1 min; power max: off; maximum pressure: 260 psi). The reaction mixture was concentrated under vacuum, dissolved with EtOAc and washed with NaHCO3. The combined organic phases were washed with brine, dried over anhydrous Na2SO4, and evaporated to dryness. The crude material was purified by silica gel flash chromatography, using the proper eluent: 6a, 6c, 6d, 6f, 6i: CH2Cl2/Formic Acid/MeOH 96:3:1; 6b, 6e: CH2Cl2/Formic Acid/MeOH 92:3:5; 6g: CH2Cl2/MeOH 96:4; 6h CH2Cl2/MeOH 9:1 plus 1% of formic acid; 6j CH2Cl2/Formic Acid/MeOH 95:3:2–96:1:3; 6k CH2Cl2/MeOH/Formic Acid 96:3:1.
N6-(3-fluorophenyl)-N2-methyl-9H-purine-2,6-diamine (6a): 1st Irradiation cycle: 10 min, 170 °C; 2nd Irradiation cycle: 30 min, 150 °C. Yield: 20%. Mp: 266–267 °C. MS (ESI) [M+H]+: 259.17 m/z. 1H NMR (DMSO‑d 6, 400 MHz): δ 2.83 (d, 3H, J = 2 Hz), 6.74 (s, 1H), 6.77 (t, 1H, J = 4 Hz), 7.29 (q, 1H, J = 15.2, 8 Hz), 7.76 (d, 1H, J = 8.4 Hz), 7.88 (s, 1H), 8.17 (m, 1H), 9.70 (s, 1H), 12.64 (bs, 1H). 13C NMR (DMSO‑d 6, 100 MHz): δ 28.89, 107.03, 108.44, 115.94, 130.24, 137.25, 142.79, 151.85, 153.34, 159.84, 161.43, 163.48. Anal. (C12H11FN6) C, H, N.
2-((6-((3-fluorophenyl)amino)-9H-purin-2-yl)amino)ethan-1-ol (6b): 1st Irradiation cycle: 10 min, 170 °C; 2nd Irradiation cycle: 30 min, 150 °C. Yield: 27%. MS (ESI) [M+H]+: 289.12 m/z. 1H NMR (DMSO‑d 6, 400 MHz): δ 3.36 (m, 2H), 3.56 (m, 2H), 6.59 (s, 1H), 6.75 (m, 1H), 7.28 (q, 1H J = 8 Hz), 7.78 (d, 1H, J = 4 Hz), 7.84 (s, 1H), 8.10 (s, 1H), 9.62 (s, 1H), 12.50 (s, 1H). 13C NMR (DMSO‑d 6, 100 MHz): δ 44.49, 60.55, 106.65, 106.92, 108.13, 115.86, 130.15, 130.25, 142.71, 142.83, 159.51, 161.43, 163.81. Anal. (C13H13FN6O) C, H, N.
N6-(3-fluorophenyl)-N2-(2-phenoxyethyl)-9H-purine-2,6-diamine (6c): 1st Irradiation cycle: 10 min, 170 °C; 2nd Irradiation cycle: 30 min, 150 °C. Yield: 28%. Mp: 217–219 °C. MS (ESI) [M+H]+: 365.20 m/z. 1H NMR (DMSO‑d 6, 400 MHz): δ 3.67 (q, 2H, J = 6.32 Hz), 4.14 (t, 2H, J = 6.32 Hz), 6.75 (t, 1H, J = 10.84 Hz), 6.94 (m, 4H), 7.27 (m, 3H), 7.77 (d, 1H, J = 10.12 Hz), 7.87 (s, 1H), 8.13 (s, 1H), 9.64 (s, 1H), 12.54 (s, 1H). 13C NMR (DMSO‑d 6, 100 MHz): δ 41.11, 66.50, 106.77, 107.03, 108.48, 114.86, 115.96, 120.98, 129.97, 137.23, 142.61, 142.72, 152.02, 158.97, 159.22, 161.43, 163.51, 163.81. Anal. (C19H17FN6O) C, H, N.
Methyl 3-((2-(methylamino)-9H-purin-6 yl)amino)benzoate (6d): 1st Irradiation cycle: 10 min, 170 °C; 2nd Irradiation cycle: 40 min, 150 °C. Yield: 55%. Mp: 255–257 °C. MS (ESI) [M+H]+: 299.29 m/z. 1H NMR (DMSO‑d 6, 400 MHz): δ 2.86 (d, 3H, J = 4.4 Hz), 3.84 (s, 3H), 6.49 (q, 1H, J = 8 Hz), 7.41 (m, 1H), 7.55 (d, 1H, J = 8 Hz), 7.84 (s, 1H), 8.13 (s, 1H), 8.22 (s, 1H), 9.66 (s, 1H), 12.42 (bs, 1H). 13C NMR (DMSO‑d 6, 100 MHz): δ 28.99, 52.51, 120.97, 122.61, 124.75, 129.09, 130.28, 141.40, 152.01, 153.63, 160.06, 160.15, 163.52, 166.94. Anal. (C14H14N6O2) C, H, N.
Methyl 3-((2-((2-hydroxyethyl)amino)-9H-purin-6-yl)amino)benzoate (6e): 1st Irradiation cycle: 10 min, 170 °C; 2nd Irradiation cycle: 30 min, 150 °C. Yield: 45%. Mp: 250–252 °C. MS (ESI) [M+H]+: 329.36 m/z. 1H NMR (DMSO‑d 6, 300 MHz): δ 3.41 (m, 2H), 3.56 (d, 2H, J = 3 Hz), 3.86 (s, 3H), 6.35 (t, 1H, J = 3 Hz), 7.42 (t, 1H, J = 9 Hz), 7.56 (d, 1H, J = 6 Hz), 7.84 (s, 1H), 8.25 (s, 1H), 8.71 (s, 1H), 9.65 (s, 1H), 12.45 (bs, 1H). 13C NMR (DMSO‑d 6, 75 MHz): δ 44.59, 52.53, 60.61, 120.97, 122.65, 124.87, 129.12, 130.27, 137.10, 141.33, 152.10, 159.51, 163.57, 166.91. Anal. (C15H16N6O3) C, H, N.
Methyl 3-((2-((2-phenoxyethyl)amino)-9H-purin-6-yl)amino)benzoate (6f): 1st Irradiation cycle: 10 min, 170 °C; 2nd Irradiation cycle: 30 min, 150 °C. Yield: 32%. Mp: 209–211 °C. MS (ESI) [M+H]+: 405.36 m/z. 1H NMR (DMSO‑d 6, 400 MHz): δ 3.71 (d, 2H, J = 5.6 Hz), 3.84 (s, 3H), 4.14 (t, 2H, J = 4 Hz), 6.71 (t, 1H, J = 4 Hz), 6.92 (m, 3H), 7.26 (t, 2H, J = 8 Hz), 7.39 (t, 1H, J = 8 Hz), 7.55 (d, 1H, J = 7.6 Hz), 7.85 (s, 1H), 8.26 (s, 1H), 8.73 (s, 1H), 9.70 (s, 1H), 12.44 (s, 1H). 13C NMR (DMSO‑d 6, 100 MHz): δ 41.21, 52.52, 66.72, 114.87, 120.95, 121.06, 122.73, 124.94, 129.12, 129.95, 130.30, 137.09, 141.27, 152.30, 159.03, 159.34, 163.51, 166.89. Anal. (C21H20N6O3) C, H, N.
N2-methyl-N6-(3-(trifluoromethyl)phenyl)-9H-purine-2,6-diamine (6g): 1st Irradiation cycle: 10 min, 170 °C; 2nd Irradiation cycle: 20 min, 170 °C. Yield: 40%. 1H NMR (DMSO‑d 6, 400 MHz): δ 2.83 (d, 3H, J = 4.8), 6.59 (d, 1H, J = 4.4), 7.28 (d, 1H, J = 7.6 Hz), 7.50 (t, 1H, J = 8 Hz), 7.86 (s, 1H), 8.26 (s, 1H); 8.67 (s,1H), 9.79 (s, 1H), 12.44 (s, 1H). 13C NMR (DMSO‑d 6, 100 MHz): δ 28.90, 113.91, 116.32, 117.96, 123.53, 123.62, 126.23, 129.78, 136.98, 141.86, 152.07, 153.45, 160.08. Anal. (C13H11F3N6) C, H, N.
2-((6-((3-(trifluoromethyl)phenyl)amino)-9H-purin-2-yl)amino)ethan-1-ol (6h): 1st Irradiation cycle: 10 min, 170 °C; 2nd Irradiation cycle: 20 min, 150 °C. Yield: 35%. 1H NMR (DMSO‑d 6, 400 MHz): δ 3.39 (t, 2H, J = 8 Hz), 3.57 (t, 2H, J = 4 Hz), 6.41 (t, 1H, J = 5.2 Hz), 7.28 (d, 1H, J = 8 Hz), 7.50 (t, 1H, J = 8 Hz), 7.87 (s, 1H), 8.15 (s, 1H), 8.32 (s, 1H), 8.49 (s, 1H), 12.44 (bs, 1H). 13C NMR (DMSO‑d 6, 100 MHz): δ 31.16, 60.48, 116.27, 118.05, 118.07, 123.46, 123.64, 126.17, 129.44, 129.76, 129.84, 137.13, 141.75, 152.01, 159.44. Anal. (C14H13F3N6O) C, H, N.
N [2]-(2-phenoxyethyl)-N [6]-(3-(trifluoromethyl)phenyl)-9H-purine-2,6-diamine (6i): 1st Irradiation cycle: 10 min, 170 °C; 2nd Irradiation cycle: 50 min, 150 °C;. Yield: 56%. Mp: 180–181 °C. MS (ESI) [M+H]+: 415.21 m/z. 1H NMR (DMSO‑d 6, 400 MHz): δ 3.68 (q, 2H, J = 6 Hz), 4.12 (t, 2H, J = 6 Hz), 6.78 (t, 1H, J = 6 Hz), 6.90 (m, 3H), 7.26 (t, 3H, J = 7.6 Hz), 7.48 (t, 1H, J = 8 Hz), 7.87 (s, 1H), 8.29 (s, 1H), 8.48 (s, 1H), 9.81 (s, 1H), 12.48 (bs, 1H). 13C NMR (DMSO‑d 6, 100 MHz): δ 41.16, 66.59, 114.83, 116.35, 118.19, 120.97, 123.73, 126.17, 129.50, 129.81, 129.86, 129.97, 141.71, 159.00, 159.30. Anal. (C20H17F3N6O) C, H, N.
4-((2-((2-phenoxyethyl)amino)-9H-purin-6-yl)amino)benzene-1,2-diol (6j): 1st Irradiation cycle: 10 min, 170 °C; 2nd Irradiation cycle: 60 min, 150 °C. Yield: 39%. Mp: 170–172 °C. MS (ESI) [M+H]+: 379.39 m/z. 1H NMR (DMSO‑d 6, 300 MHz): δ 3.63 (q, 2H, J = 6 Hz), 4.11 (t, 2H, J = 6 Hz), 6.44 (t, 1H, J = 6 Hz), 6.61 (d, 1H, J = 9 Hz), 6.95 (m, 3H), 7.26 (m, 3H), 7.77 (s, 1H), 8.20 (s, 1H), 8.93 (s, 1H), 12.42 (bs, 1H). 13C NMR (DMSO‑d 6, 100 MHz): δ 41.10, 66.67, 109.87, 112.38, 114.88, 115.48, 120.94, 129.97, 132.68, 141.06, 145.18, 159.02, 159.47, 163.91. Anal. (C19H18N6O3) C, H, N.
N2-(2-phenoxyethyl)-N6-(3,4,5-trimethoxyphenyl)-9H-purine-2,6-diamine (6k): 1st Irradiation cycle: 30 min, 170 °C; 2nd Irradiation cycle: 30 min, 170 °C. Yield: 52%. MS (ESI) [M+H]+: 437,47 m/z. 1H NMR (DMSO‑d 6, 400 MHz): δ 3.62 (s, 3H), 3.72 (m, 2H), 3.78 (s, 6H), 4.12 (t, 2H, J = 6.4 Hz), 6.93 (m, 3H), 7.27 (m, 2H), 7.43 (s, 1H), 7.89 (s, 1H), 8.14 (s, 1H), 9.27 (s, 1H), 12.63 (m, 1H). 13C NMR (DMSO‑d 6, 100 MHz): δ 41.08, 56.20, 60.55, 66.45, 98.22, 114.02, 115.06, 121.01, 121.73, 129.98, 133.01, 136.69, 151.90, 152.97, 158.21, 158.88, 163.49. Anal. (C22H24N6O4) C, H, N.
General Procedures for the Synthesis of compounds 6l-6m: Compound 6d (45 mg; 0.151 mmol) or 6f (33 mg; 0.082 mmol) were dissolved in a 1:1 mixture of THF/MeOH (1.53 mL), treated with 2 N aqueous solution of LiOH (0.30 mL) was added and the resulting mixtures were stirred for 24 h at room temperature. At the end of reaction, the solvent was evaporated under vacuum. The crude material was purified by silica gel flash chromatography using the proper eluent: 6l: CH2Cl2/NEt3/MeOH 82:3:15; 6m: CH2Cl2/Formic Acid/MeOH 95:3:2.
3-((2-(methylamino)-9H-purin-6-yl)amino)benzoic acid (6l): Yield: 16%. Mp: 211–213 °C. MS (ESI) [M+H]+: 285.26 m/z. 1H NMR (DMSO‑d 6, 300 MHz): δ 2.85 (d, 3H, J = 4.5 Hz), 6.42 (d, 1H, J = 3 Hz), 7.38 (t, 1H, J = 8.1 Hz), 7.53 (d, 1H, J = 7.2 Hz), 7.82 (s, 1H), 8.24 (m, 1H), 8.65 (s, 1H), 9.58 (s, 1H), 12.38 (bs, 1H). 13C NMR (DMSO‑d 6, 100 MHz): δ 28.70, 113.88, 121.41, 122.89, 124.52, 128.85, 131.61, 136.80, 141.18, 152.24, 158.44, 160.18, 168.06. Anal. (C13H12N6O2) C, H, N.
3-((2-((2-phenoxyethyl)amino)-9H-purin-6-yl)amino)benzoic acid (6m): Yield: 91%. Mp: 209–211 °C. MS (ESI) [M+H]+: 391.46 m/z. 1H NMR (DMSO‑d 6, 300 MHz): δ 3.70 (q, 2H, J = 4.2, 8.7 Hz), 4.14 (t, 2H, J = 4.2 Hz), 6.68 (t, 1H, J = 3.6 Hz), 6.92 (m, 2H); 7.27 (t, 1H, J = 7.2 Hz), 7.38 (t, 1H, J = 8 Hz), 7.55 (d, 1H, J = 8 Hz), 7.85 (s, 1H), 8.27 (s, 1H), 8.62 (s, 1H), 9.63 (s, 1H) 12.43 (s, 1H). 13C NMR (DMSO‑d 6, 100 MHz): δ 41.20, 66.78, 114.88, 120.96, 123.08, 124.65, 128.99, 129.97, 131.48, 136.98, 141.00, 159.02, 159.35, 168.00. Anal. (C20H18N6O) C, H, N.
Synthesis of 2-chloro-N-(2-phenoxyethyl)-9H-purin-6-amine (7): In a microwave tube 2,6-dichloro-9H-purine 4 (100 mg; 0.53 mmol) and 2-phenoxyethylamine (1.06 mmol) were suspended in n-BuOH (3 mL). NEt3 (258 μL; 1.852 mmol) was added and the tube was heated at 70 °C for 30 min (max μW power input: 100 W; ramp time: 1 min; power max: off; maximum pressure: 260 psi). At the end of irradiation, the solid obtained was isolated by filtration over a Buchner funnel and washed with MeOH and EtOAc. Yield: 85%. 1H NMR (DMSO‑d 6, 300 MHz): δ 3.21 (t, 2H, J = 3 Hz), 4.17 (t, 2H, J = 6 Hz), 6.96 (m, 3H), 7.30 (m, 2H), 8.15 (s, 1H), 8.30 (s, 1H), 9.00 (bs, 1H).
General Procedures for the Synthesis of compounds 8a,b: In a microwave tube 7 (50 mg; 0.173 mmol) and 3-fluoroaniline or 3-trifluoromethylaniline (0.865 mmol) were suspended in n-BuOH (2 mL). TFA (0.259 mmol) was added and the tube was heated under microwave irradiation for two consecutive cycles: 1st Irradiation cycle: 10 min, 170 °C; 2nd Irradiation cycle: 40 min, 150 °C (max μW power input: 300 W; ramp time: 1 min; power max: off; maximum pressure: 260 psi). The reaction mixture was concentrated under vacuum, diluted with ethyl acetate, and washed with NaHCO3. The combined organic were washed with NH4Cl, brine, dried over anhydrous Na2SO4, filtered and concentrated. The crude material was purified by silica gel flash chromatography using the proper eluent: 8a: CH2Cl2/Formic Acid/MeOH 95:3:2; 8b: CH2Cl2/Formic Acid/MeOH 96.5:3:0.5.
N2-(3-fluorophenyl)-N6-(2-phenoxyethyl)-9H-purine-2,6-diamine (8a): Yield: 33%. Mp: 208–210 °C. MS (ESI) [M+H]+: 365.38 m/z. 1H NMR (DMSO‑d 6, 400 MHz): δ 3.89 (s, 2H), 4.20 (m, 2H), 6.25 (m, 1H), 6.93 (m, 3H), 7.20 (m, 1H), 7.30 (m, 2H), 7.48 (d, 1H, J = 1.2 Hz), 7.63 (s, 1H), 7.87 (s, 1H), 7.95 (d, 1H, J = 13.2 Hz), 9.15 (s, 1H), 12.54 (s, 1H). 13C NMR (DMSO‑d 6, 100 MHz): δ 40.66, 66.31, 104.65, 104.91, 106.34, 106.55, 114.20, 114.87, 121.06, 129.98, 130.07, 137.47, 144.10, 144.22, 156.26, 158.89, 161.71, 163.49, 164.08. Anal. (C19H17FN6O) C, H, N.
N [6]-(2-phenoxyethyl)-N [2]-(3 (trifluoromethyl) phenyl)-9H-purine-2,6-diamine (8b): Yield: 42%. Mp: 211–213 °C. MS (ESI) [M+H]+: 415.39 m/z. 1H NMR (DMSO‑d 6, 400 MHz): δ 3.89 (s, 2H), 4.20 (m, 2H), 6.91 (m, 3H), 7.15 (d, 1H, J = 8 Hz), 7.27 (m, 2H), 7.41 (t, 1H, J = 8 Hz), 7.68 (s, 1H), 7.88 (s, 1H), 7.97 (s, 1H), 8.42 (s, 1H), 9.30 (s, 1H), 12.53 (bs, 1H). 13C NMR (DMSO‑d 6, 100 MHz): δ 40.66, 66.39, 114.23, 114.85, 116.41, 121.06, 121.81, 123.64, 126.35, 129.52, 129.69, 129.83, 129.98, 143.06, 156.21, 158.91, 163.50. Anal. (C20H17F3N6O) C, H, N.
General Procedures for the Synthesis of intermediates 9a-e: In a microwave tube 2,6-dichloropurine (100 mg; 0,529 mmol) and the proper phenylhydrazine (2645 mmol) were suspended in n-BuOH (3 mL). NEt3 (221 μL; 1587 mmol) was added and the tube was heated under microwave irradiation at 70 °C for 20–40 min (max μW power input: 100 W; ramp time: 1 min; power max: off; maximum pressure: 260 psi). When the reaction ended, the mixture was concentrated under vacuum, diluted with ethyl acetate, and washed with NH4Cl. The combined organic were washed with NH4Cl, brine, dried over anhydrous Na2SO4, filtered and concentrated under vacuum. The crude material was purified by silica gel flash chromatography using the proper eluent: 9a: CH2Cl2/MeOH 96:4; 9b-d: hexane/acetone 1:1; 9e: CH2Cl2/MeOH 97:3.
(E)-2-chloro-6-(2-(3-fluorophenyl)hydrazinyl)-9H-purine (9a): 1st Irradiation cycle: 10 min, 70 °C; 2nd Irradiation cycle: 10 min, 70 °C. Yield: 60%. 1H NMR (DMSO‑d 6, 400 MHz): δ 6.58 (m, 2H), 6.30 (m, 1H), 6.18 (m, 1H), 7.83 (s, 1H), 8.14 (s, 1H), 9.56 (s, 1H), 12,41 (m, 1H).
(E)-2-chloro-6-(2-(3,5-difluorophenyl) hydrazinyl)-9H-purine (9b): 1st Irradiation cycle: 10 min, 70 °C; 2nd Irradiation cycle: 10 min, 70 °C; 3rd Irradiation cycle: 20 min, 70 °C. Yield: 37%. MS (ESI) [M+H]+: 297.13 m/z. 1H NMR (DMSO‑d 6, 400 MHz): δ 6.47 (m, 3H), 8.23 (s, 1H), 8.72 (s, 1H), 9.99 (s, 1H), 12.88 (m, 1H).
(E)-6-(2-(3,5-bis(trifluoromethyl)phenyl) hydrazinyl)-2-chloro-9H-purine (9c): 1st Irradiation cycle: 10 min, 70 °C; 2nd Irradiation cycle: 10 min, 70 °C. Yield: 73%. MS (ESI) [M+H]+: 397.06 m/z. 1H NMR (DMSO‑d 6, 400 MHz): δ 7.28 (m, 2H), 7.42 (m, 2H), 8.28 (s, 1H); 9.11 (m, 1H), 10.02 (m, 1H), 10.25 (m, 1H), 12.99 (m, 1H).
(E)-2-chloro-6-(2-(4-(trifluoromethyl)phenyl) hydrazinyl)-9H-purine (9d): 1st Irradiation cycle: 10 min, 70 °C; 2nd Irradiation cycle: 10 min, 70 °C. Yield: 68%. MS (ESI) [M+H]+ = 331.20 m/z. 1H NMR (DMSO‑d 6, 400 MHz): δ 6.86 (d, 2H, J = 8 Hz), 7.48 (d, 2H, J = 7.6 Hz), 8.23 (s, 1H), 8.79 (s, 1H), 10.09 (s, 1H), 12.9 (m, 1H).
2-chloro-6-(2-(3-(trifluoromethyl)phenyl) hydrazinyl)-9H-purine (9e): 1st Irradiation cycle: 10 min, 70 °C; 2nd Irradiation cycle: 10 min, 70 °C. Yield: 54%. MS (ESI) [M+H]+: 328.69 m/z. 1H NMR (DMSO‑d 6, 400 MHz): δ 7.14 (m, 4H), 8.24 (m, 1H), 8.63 (m, 1H), 10.00 (m, 1H), 12.97 (m, 1H).
General procedures for the Synthesis of compounds 10a-e: In a microwave tube 9a-e and the proper amine (2.5 equivalent) were suspended in n-BuOH (2 mL). TFA (1 equivalent) was added and the tube was heated at 170 °C for 4 consecutive cycles of 30 min each (max μW power input: 300 W; ramp time: 1 min; power max: off; maximum pressure: 300 psi). The reaction mixture was concentrated under vacuum. Products were purified by silica gel flash chromatography, using the proper eluent: 10a: hexane/acetone 3:7 plus 1% of formic acid; 10b: hexane/acetone 3:7; 10c: hexane/acetone 4:6 plus 1% of formic acid; 10d: hexane/acetone 2:8 plus 1% of formic acid; 10e: CHCl3/MeOH 94:4 plus 1% of formic acid.
6-(2-(3-fluorophenyl) hydrazinyl)-N-(2-phenoxyethyl)-9H-purin-2-amine (10a): Yield: 46%. MS (ESI) [M+H]+ = 380.29 m/z. 1H NMR (DMSO‑d 6, 300 MHz): δ 3.50 (s, 2H), 3.91 (s, 2H), 6.43 (m, 3H), 6.58 (d, 1H, J = 9 Hz), 6.90 (m, 3H), 7.13 (q, 1H, J = 9 Hz), 7.25 (t, 2H, J = 9 Hz), 7.77 (s, 1H), 8.06 (s, 1H), 9.17 (s, 1H). 13C NMR (DMSO‑d 6, 100 MHz): δ 40.97, 66.42, 99.15, 104.41, 108.69, 114.76, 114.86, 120.84, 120.93, 121.23, 129.88, 130.56, 130.70, 153.10, 155.45, 158.98, 159.47, 162.42, 164.84. Anal. (C19H18FN7O) C, H, N.
6-(2-(3,5-difluorophenyl) hydrazinyl)-N-(2-phenoxyethyl)-9H-purin-2-amine (10b): Yield: 42%. MS (ESI) [M+H]+ = 398,30 m/z. 1H NMR (DMSO‑d 6, 300 MHz): δ 3.50 (d, 2H, J = 6), 3.92 (m, 2H), 6.31 (m, 3H), 6.51 (m, 1H), 6.87 (m, 3H), 7.24 (q, 2H, J = 6, 11.2 Hz), 7.76 (s, 1H), 8.35 (s, 1H), 9.30 (s, 1H), 12.36 (s, 1H). 13C NMR (CDCl3+CD3OD, 75 MHz): δ 41.24, 66.42, 92.89, 94.99, 95.25, 114.75, 114.94, 120.87, 129.91, 153.97, 155.27, 158.98, 159.28, 162.44, 162.60, 163.50, 164.84, 165.00. Anal. (C19H17F2N7O) C, H, N.
6-(2-(3,5-bis(trifluoromethyl)phenyl) hydrazinyl)-N-(2- phenoxyethyl)-9H-purin-2-amine (10c): Yield: 38%. MS (ESI) [M+H]+ = 498.29 m/z. 1H NMR (DMSO‑d 6, 400 MHz): δ 3.58 (q, 2H, J = 8 Hz), 3.97 (t, 2H, J = 8 Hz), 6.18 (t, 1H, J = 4 Hz); 6.89 (m, 3H), 7.19 (s, 1H), 7.24 (t, 2H, J = 4 Hz), 7.32 (s, 1H), 7.73 (s, 1H), 8.14 (s, 1H), 8.54 (s, 1H). 13C NMR (DMSO‑d 6, 100 MHz): δ 40.91, 66.23, 106.69, 111.65, 114.62, 114.64, 120.85, 122.64, 125.36, 128.07, 129.87, 131.11, 131.43, 137.14, 137.76, 158.88, 163.55. Anal. (C21H17F6N7O) C, H, N.
N-(2-phenoxyethyl)-6-(2-(4-(trifluoromethyl)phenyl) hydrazinyl)-9H-purin-2- amine (10d): Yield: 41%. MS (ESI) [M+H]+ = 430.36 m/z. 1H NMR (DMSO‑d 6, 300 MHz): δ 3.76 (q, 2H, J = 6 Hz), 4.15 (t, 2H, J = 6 Hz), 6.89 (t, 2H, J = 9 Hz), 6.96 (d, 2H, J = 9 Hz), 7.26 (m, 2H), 7.37 (bs, 1H), 7.68 (s, 1H), 8.03 (s, 1H), 8.05 (s, 1H), 8.25 (s, 1H), 8.28 (s, 1H), 8.37 (s, 1H). 13C NMR (DMSO‑d 6, 75 MHz): δ 49.12, 66.38, 111.87, 114.73, 114.92, 120.83, 121.00, 124.33, 126.54, 127.37, 129.87, 129.96, 154.56, 158.95, 159.47, 159.88. Anal. (C20H18F3N7O) C, H, N.
N-(2-phenoxyethyl)-6-(2-(3- (trifluoromethyl)phenyl) hydrazinyl)-9H-purin-2- amine (10e): MS (ESI) [M+H]+ = 430.15 m/z. 1H NMR (DMSO, 400 MHz) δ 3.50 (m, 2H); 3.89 (m, 2H), 6.46 (bs, 1H), 6.90 (t, 1H, J = 8 Hz), 6.99 (m, 4H), 7.28 (m, 4H), 7.76 (s, 1H), 8.21 (s, 1H), 9.28 (bs, 1H), 12.34 (bs, 1H). 13C NMR (DMSO‑d 6, 100 M Hz) δ 41.10, 66.30, 114.81, 114.93, 120.94, 121.00, 122.88, 125.56, 129.97, 130.62, 130.73, 131.05, 131.54, 131.55, 143.09, 158.97, 163.55. Anal. (C20H18F3N7O) C, H, N.
General procedures for synthesis of compounds 11a-d: Compounds 10b-e (20 mg each) were dissolved in MeOH and stirred at room temperature under air atmosphere for 96 h. The corresponding oxidized compounds 11a-d were obtained as pure products in quantitative yields.
6-((3-fluorophenyl)diazenyl)-N-(2-phenoxyethyl)-9H-purin- 2-amine (11a): MS (ESI) [M+H]+: 378.28 m/z. 1H NMR (CDCl3, 400 MHz): δ 4.00 (m, 2H), 4.21 (m, 2H), 6.11 (bs, 1H), 6.94 (m, 3H), 7.27 (m, 3H), 7.50 (q, 1H, J = 8 Hz), 7.73 (s, 1H), 7.89 (d, 1H, J = 4 Hz), 8.24 (s, 1H). 13C NMR (CDCl3 +CD3OD, 100 MHz): δ 41.22, 66.45, 108.88, 109.18, 114.46, 119.95, 120.93, 121.34, 129.45, 130.47, 130.58, 153.87, 158.58, 159.78, 161.50, 164.81. Anal. (C19H16FN7O) C, H, N.
6-((3,5-difluorophenyl)diazenyl)-N-(2-phenoxyethyl)-9H-purin-2-amine (11b): [M+H]+: 396.22 m/z. 1H NMR (CDCl3+CD3OD, 400 MHz): δ 3.93 (t, 2H, J = 4 Hz), 4.19 (t, 2H, J = 4 Hz), 6.91 (m, 3H), 7.02 (q, 1H, J = 4 Hz), 7.25 (t, 2H, J = 8 Hz), 7.28 (s, 1H), 7.70 (d, 2H, J = 4 Hz), 8.12 (s, 1H), 13.65 (s, 1H). 13C NMR (CDCl3+CD3OD, 100 MHz): δ 41.23, 66.41, 106.91, 107.18, 107.72, 107.98, 108.21, 114.45, 120.96, 129.46, 154.23, 158.58, 159.71, 162.02, 164.38. Anal. (C19H15F2N7O) C, H, N.
6-((3,5-bis(trifluoromethyl)phenyl)diazenyl)-N-(2-phenoxyethyl)-9H-purin-2-amine (11c): MS (ESI) [M+H]+ : 496.34 m/z. 1H NMR (CDCl3, 400 MHz): δ 4.02 (m, 2H), 4.23 (m, 2H), 6.13 (bs, 1H), 6.95 (m, 3H), 7.28 (m, 2H), 8.08 (s, 1H), 8.26 (s, 1H), 8.56 (s, 1H). 13C NMR (CDCl3+CD3OD, 100 MHz): δ 41.51, 66.42, 114.45, 121.09, 121.38, 123.76, 124.09, 125.98, 129.53, 132.58, 132.92, 133.26, 133.60, 152.48, 158.50, 159.93. Anal. (C21H15F6N7O) C, H, N.
N-(2-phenoxyethyl)-6-((4- (trifluoromethyl)phenyl)diazenyl)-9H-purin-2-amine (11d): MS (ESI) [M+H]+: 428.15 m/z. 1H NMR (CDCl3, 400 MHz): δ 4.01 (t, 2H, J = 4 Hz), 4.22 (t, 2H, J = 4 Hz), 6.09 (s, 1H), 6.94 (m, 3H), 7.27 (m, 2H), 7.81 (d, 2H, J = 8 Hz), 8.15 (d, 2H, J = 8 Hz), 8.24 (s, 1H), 13.65 (bs, 1H) . 13C NMR (CDCl3+CD3OD, 100 MHz): δ 45.12, 70.29, 118.35, 118.39, 124.84, 126.19, 127.89, 128.90, 130.42, 133.35, 137.92, 138.24, 158.23, 162.54, 162.59, 163.67. Anal. (C20H16F3N7O) C, H, N.
4.2. ABTS radical assay
ABTS assay was performed according to the protocol reported on reference 33. The stock solution was prepared by mixing ABTS (2, 2 azobis-(3-ethylbenzothiozoline-6-sulphonic acid), dissolved in water to a 7 mM concentration, with 2.45 mM potassium persulfate and allowing the mixture to stand in the dark at room temperature for 12–16 h before use. The latter solution, opportunely diluted in ethanol (1 : 100, v/v), gave an absorbance of 0.70 (±0.02) at 734 nm and was used as control solution. 3.0 mL of the latter control solution were added to 30 μL of the reference compound (Ascorbic Acid) or selected compounds dissolved in DMSO, at different concentrations (10 μM, 50 μM, 0.1 mM, 0.2 mM, 0.5 mM, 1 mM). The absorbance at 734 nm was read exactly after 10 min of incubation time. The scavenging activity was estimated based on the percentage of ABTS radicals scavenged by the following formula:
| %scavenging = [(A0 − As)/A0] × 100 |
where A0 is absorption of control solution (run in each assay), AS is absorption in the presence of reference compound or sample.
4.3. Biology
4.3.1. Cells and viruses
The H/PF/2013 ZIKV strain belonging to the Asian lineage, the New Guinea C DENV serotype 2 strain and WNV lineage 1 (Italy/2009) strain were kindly provided by the Istituto Superiore di Sanità (Rome, Italy) while the SARS-CoV-2 strain, belonging to lineage B1 was kindly provided by the Department of Biomedical and Clinical Sciences Luigi Sacco, University of Milan. [64] Vero E6 (ATCC catalogue no. CRL-1586), Huh7 (kindly provided by Istituto Toscano Tumori, Core Research Laboratory, Siena, Italy), A549 (ATCC catalogue no. CCL-185), Caco-2 (ATCC catalogue HTB-37) and Calu-3 (ATCC catalogue no. HTB-55), MDCK (ATCC catalogue no. CCL-34) were used to determine the antiviral activity of candidate compounds. Cell lines were maintained in high glucose Dulbecco's Modified Eagle's Medium with sodium pyruvate and l-glutamine (DMEM; Euroclone), Minimum Essential Medium Eagle (EMEM; Euroclone) or Roswell Park Memorial Institute medium (RPMI-1640, Sigma) supplemented with 10% Fetal Bovine Serum (FBS; Euroclone) and 1% Penicillin/Streptomycin (Pen/Strep, Euroclone). The same medium was used but with a lower FBS concentration for the viral propagation and drug susceptibility testing (1–2%) and for in vitro selection experiments (3%). Cells were incubated at 37 °C in a humidified incubator supplemented with 5% CO2. All the viral stocks were titrated by plaque reduction assay (PRA) as previously described [65]. Briefly, confluent Vero E6 (WNV, DENV, SARS-CoV-2) and A549 (ZIKV) in 6-well format were infected with three tenfold dilutions of viral stock and after 1 h adsorption at 37 °C, the cells were washed with PBS and 0.75% Sea Plaque Agarose (Lonza) was added to each well. After 3 (WNV), 5 (ZIKV, SARS-COV-2) and 10 (DENV) days incubation at 37 °C, the monolayers were fixed with 10% formaldehyde (Carlo Erba Chemicals) and stained with 0.1% crystal violet (Carlo Erba Chemicals). After 3 h incubation, the agar overlay was removed by water washing and PFU were counted. Allantoic cavities of 11-day-old embryonated chicken eggs were used to grow influenza viruses A/Puerto Rico/8/34H1N1 (PR8 H1N1), pandemic A/California/04/09H1N1 (p2009H1N1) and avian A/parrot/Ulster/73H7N1 (Ulster H7N1). Viral suspension was inoculated in the allantoic cavity and incubated for 48 h at 37 °C, then infected eggs were maintained overnight at 4 °C. Subsequently, the allantoic fluid was collected and clarified by centrifugation (2500×g for 30 min). The recovered virus was used for the infection of MDCK and A549 cells. Allantoic fluid from uninfected eggs was used as reference for mock infection [68].
4.3.2. Drugs and cytotoxicity assay
Cytotoxicity of investigational compounds was determined by CellTiter-Glo 2.0 Luminescent Cell Viability Assay (Promega) according to the manufacturer's protocol in the cells used to subsequently test flaviviruses and SARS-COV-2 (Vero E6, Huh7 A549 Caco-2 and Calu-3). The luminescence values obtained from cells treated with investigational compounds or DMSO control were measured through the GloMax® Discover Multimode Microplate Reader (Promega) and elaborated with the GraphPad PRISM software version 6.01 (La Jolla) to calculate the half-maximal cytotoxic concentration (CC50). The cytotoxicity assay of candidate compounds was evaluated on cell lines used for influenza virus infection by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay [69]. Briefly, cells were seeded in 96-well plates at a density of 2 × 104 cells/well in 100 μL of complete RPMI without phenol red for 24 h at 37 °C. Subsequently, cell monolayers were treated or not with increasing concentrations (0.5, 2.5, 5, 10 and 20 μM) of compounds for 24 h at 37 °C. After 24 h, 10 μL of MTT solution (5 mg/mL) were added to each well for 3–4 h at 37 °C. Afterwards, each sample was acidified by adding 0.1 N HCl in isopropanol (100 μL/well) for 30 min in slow agitation to ensure that all formazan crystals were dissolved. Absorbance of samples was read at 570 nm, using an automatic plate reader (Multiskan EX, Ascent Software, Thermo Fisher Scientific). Untreated cells were used as control. The 50% cytotoxic concentration (CC50), defined as the compound's concentration required to reduce cell viability by 50%, was calculated using the GraphPad, Prism 6 Software.
4.3.3. Antiviral assays
To determine the antiviral activity of candidate compounds against WNV, DENV and ZIKV, a DYRA based on infection of cells in presence of serial drug dilutions was performed as previously described [24,66]. Briefly, 7000 Huh7 cells per well were infected with 50 TCID50 of DENV or ZIKV and 25 TCID50 of WNV. Viral adsorption was performed in 96-well plates for 1 h at 37 °C with 5% CO2 and after virus removal, serial dilutions of each tested compound were added to the cells and incubated at 37 °C with 5% CO2. After 48 h (WNV) and 72 h (DENV and ZIKV) of incubation, the antiviral activity was measured on cell monolayer by immunodetection assay (IA). ZIKV DENV and WNV harvested supernatants, were used to infect pre-seeded Huh7 cells in SYRA or PRA as previously described [24,67]. Yield reduction protocol was adapted to evaluate the antiviral activity of candidate compounds against SARS-COV-2 in Vero E6, Caco-2, Calu-3 and Huh7 using the MOI of 0.01. Viral adsorption was performed in 96-well plates for 2 h at 37 °C with 5% CO2 and after virus removal, serial dilutions of each tested compound were added to the cells and incubated 48 h at 37 °C with 5% CO2 (DYRA). To discriminate the activity of drug as entry inhibitor, serial dilutions of reference compound and 6i were added onto the Calu-3 cell and removed after cell-virus adsorption (ENTRY-DYRA). The CPE was used as read out in Vero E6 and measured by CellTiter-Glo 2.0 Luminescent Cell Viability Assay (Promega) according to the manufacturer's protocol. The IA protocol was adapted for the detection of SARS nucleocapsid protein in infected Caco-2 cells. Briefly, fixation and permeabilization were performed as previously described and cell lines were incubated for 1 h with a monoclonal SARS Nucleocapsid Protein Antibody (Novus, cat. AP201054) diluted 1:1000 in blocking buffer (PBS containing 1% bovine serum albumine (BSA) and 0.1% Tween 20). After washing, monolayers were incubated for 1 h with a polyclonal HRP-coupled anti-mouse IgG secondary antibody (Novus Bio NB7570) diluted 1: 5000 in blocking buffer. After cell washing, the 3,3′,5,5′-Tetramethylbenzidine substrate (Sigma Aldrich) was added to each well and the reaction was stopped with one volume of 0.5 M sulfuric acid. Absorbance was measured at 450 nm optical density (OD450) using the Absorbance Module of the GloMax® Discover Multimode Microplate Reader (Promega). The half-maximal inhibitory concentration (IC50) was calculated through a non-linear regression analysis of the dose-response curves generated with GraphPad PRISM software version 6.01. In each test, sofosbuvir and remdesivir were used as reference compounds against flaviviruses and SARS-CoV-2 respectively. Infected and uninfected cells without drugs were used to calculate the 100% and 0% of viral replication, respectively. Selectivity Index (SI) was calculated as ratio between CC50 and IC50.
The anti-influenza activity of candidate compounds was evaluated by Hemagglutination assay (HAU) in the supernatants of infected cells recovered after 24 h from infection [69]. Briefly, MDCK and A549 cells seeded in 12-well plates at a density of 2.5 × 105 cells/well, were infected with influenza virus A/Puerto Rico/8/34H1N1 and incubated for 1 h at 37 °C. After viral adsorption, cells monolayers were washed with PBS and then, fresh medium supplemented with 2% FBS with different concentrations (0.5, 2.5, 5, 10 and 20 μM) of selected compounds was added for 24 h at 37 °C. Each compound solved in DMSO was diluted in RPMI (the highest concentration of DMSO added in the culture medium was equal to 0.2% of final volume). As reference, infected cells were treated with DMSO alone. The IC50 of each compound was calculated using the GraphPad, Prism 6 Software. The SI was calculated as described above.
4.3.4. In cell western assay
This is an innovative cell-based technique that allows a sensitive quantification of influenza virus proteins expressed on cell monolayers [69]. Briefly, permissive cells were seeded in 96-well plates at a density of 2 × 104 cells/well for 24 h at 37 °C in an atmosphere of 5% CO2. Confluent cell monolayers were infected with different influenza A virus strains or with supernatants recovered from infected cells for 1 h with PBS and then fresh medium supplemented with 2% FBS was added for 24 h at 37 °C. Subsequently, cell monolayers were fixed with 3.7% formaldehyde for 20 min, permeabilized with 0.1% Triton X-100 and incubated with Odyssey blocking buffer (LI-COR Bioscience, Lincoln, NE) for 60 min at room temperature. The cells were stained at 4 °C overnight with primary antibodies against influenza Hemagglutinin (HA) or anti-FLU. After incubation, three washes with Phosphate Buffered Saline (PBS) plus 0.1% Tween 20 were performed and then the cells were stained with a mixture of fluorochrome-conjugated secondary antibodies (fluorescence emission at 800 nm), properly diluted in Odyssey blocking buffer and fluorochrome-conjugated Cell Tag (fluorescence emission at 700 nm), for 1 h at room temperature. Cell Tag was used as control of the integrity of cell monolayer. Subsequently, three washes with PBS plus 0.1% Tween 20 were performed and plates were analyzed by the Odyssey infrared imaging system (LI-COR). Integrated intensities of fluorescence were determined by the LI-COR Image Studio software and the relative fluorescence units (RFU) were expressed as percentage compared to untreated infected cells (100%).
4.3.5. Time-of-addition assay
A549 cells were seeded in 12-well plates at a density of 2.5 × 105 cells/ml for 24 h at 37 °C, infected with different influenza A virus strains as described before and treated or not with 6i at different phases of virus life-cycle: the compound was added for 2 h before (PRE); during viral adsorption for 1 h (DUR); immediately after viral adsorption for 24 h (POST); before, during and for the following 24 h (PDP). After the conditions of pretreatment and during, cells were washed with PBS, and successively maintained in fresh medium for the following 24 h. In another set of experiments, i) the compound was immediately added after viral adsorption for 2 (0–2 h), 4 (0–4 h) or 6 (0–6 h) hours. Then cells were washed with PBS and maintained up to 24 h without 6i in fresh medium plus 2% FBS; ii) the compound was added after 2 (2–24 h), 4 (4–24 h) or 6 (6–24 h) hours infection and maintained until 24 h from infection. The supernatants were recovered and used to infect fresh MDCK monolayers for 24 h. Untreated infected cells were used as control.
The expression of viral proteins was analyzed directly on the infected cell monolayers or on other monolayers infected for 24 h with the recovered supernatants. Statistical significance of the data was analyzed by means of a one-way ANOVA Multiple comparisons test and ∗P values of <0.05 were considered significant. Data were expressed as mean ± the standard deviation (SD).
4.3.6. In vitro selection experiments
The in vitro selection experiments were performed as previously described [67]. Huh7 cells at 70% confluence were infected in duplicate with ZIKV and DENV at 0.005 multiplicity of infection (MOI). After 1 h adsorption at 37 °C with 5% CO2, the virus inoculum was removed and cells were treated with an initial concentration of 15 μM 6i, corresponding to 17-fold and 30-fold IC50 (as observed by PRA) against DENV and ZIKV, respectively. Cell cultures were monitored every 24 h and when 80% viral CPE was observed, cells and supernatant were freeze-thawed, cleared by centrifugation and viral stocks were used to re-infect fresh pre-seeded Huh7 cells with a twofold increased 6i concentration (30, 60 and 120 μM). All viral stocks collected during in vitro selection experiments were analyzed by population sequencing to detect emergent mutations in the NS5 and NS3 regions. Wild type and mutant viruses were titrated by IA.
4.3.7. Viral RNA amplification and sequencing
Total RNA was extracted in duplicate from 150 μL of viral stocks derived from in vitro selection experiments, using the ZR Viral RNA Kit (Zymo Research) according to the manufacturer's protocol. Ten microliters of extracted ZIKV RNA were denatured at 70 °C for 5 min and then added to a mixture including 664 μM dNTPs, 6 μL of 5X ImProm-II TM Reaction Buffer, 50 ng Hexanucleotides, 1.5 mM MgCl2, 20U RNasin® Plus RNase Inhibitor and 1U of ImProm-II™ Reverse Transcriptase (Promega) in a final volume of 30 μL. Reverse transcription reactions were run in Eppendorf Mastercycler Nexus (Eppendorf) with an initial step at 25 °C for 5 min, followed by a reverse transcription step at 37 °C for 30 min and a final step at 80 °C for 5 min cDNA was used as the template for PCR amplification of the whole NS5 and NS3 genes, using the Q5 Hot Start High-Fidelity DNA Polymerase (NEB) protocol.
To design primers with a high degree of conservation, the ZIKV and DENV alignment available at the NCBI website was used (https://www.ncbi.nlm.nih.gov/genomes/VirusVariation); the primer sequences and coordinates, as referred to the H/PF/2013 ZIKV strain (Gen Bank Accession Number KJ776791) and DENV DENV-2 New Guinea C (Gen Bank Accession Number KM204118.1) are indicated in Table S1 (Supporting information). Bidirectional DNA sequencing was performed using the BrilliantDye TM Terminator Kit v1.1 (Nimagen) with 8 and 6 different primers spanning the whole NS5 and NS3 regions of ZIKV and DENV, respectively (Table S1; Supporting Information). The sequencing reactions were treated with the X-Terminator®Purification kit (AppliedBiosystems) in a 96-well plate as suggested by the manufacturer, resolved by capillary electrophoresis on the 3130 XL Genetic Analyzer (Applied Biosystems) and analyzed with the DNAStar Lasergene 7.1.0 SeqMan Pro module.
4.3.8. Immunofluorescence
DENV NS3 and NS5 protein expression and colocalization were visualized by confocal microscopy analysis. Huh-7 cells were seeded 1 × 104 cells/well onto 1 cm round glass coverslips, infected with DENV-2 (MOI 0.05), treated with Sofosbuvir (100 μM) or compound 6i (50 μM), and fixed in 4% paraformaldehyde/PBS at different time-points (8-12-16-24 h). After permeabilization (0,5% Triton-X 100, 7 min), non-specific binding sites were blocked in 3% BSA for 1 h and cells were incubated overnight with primary antibody (anti-NS3 and anti-NS5, diluted in PBS containing 0.5% BSA + 0.5% Triton-X 100). After washing (PBS containing 0.5% BSA + 0.5% Triton-X 100 + 0.05% Tween 20), cells were incubated for 1 h with AlexaFluor® 488- and 568-labeled secondary antibody diluted in PBS with 0.5% BSA + 0.5% Triton-X 100. Cells were stained with 4′,6-diamidino-2-phenylindole (DAPI, 10 min). Coverslips were mounted in Fluoromount Aqueous Mounting Medium and pictures of stained cells were taken through a confocal microscope (Zeiss LSM500; Carl Zeiss, Germany). Reagents for immunofluorescence (PBS, Triton-X 100, Tween 20, anti-NS3 and anti-NS5 antibodies, and Fluoromount) were purchased from Sigma-Aldrich (St. Louis, MO, USA). BSA was purchased from Gibco by Life Technologies (Carlsbad, CA, USA) and secondary antibodies were from Invitrogen (Carlsbad, CA, USA). The number of cells was determined by the count of cells stained with DAPI, and randomly counted (5 fields) at 40× original magnification. NS3 and NS5 expressing cells were counted and data were reported as percentage of positive cells/well. Images were pre-processed using ImageJ software (U. S. National Institutes of Health, Bethesda, MD, USA). The colocalization of NS3 and NS5 proteins was represented as white dots, using the Colocalization plug-in. The quantitative colocalization analyses were performed using the JACoP plug-in to determine Manders' colocalization coefficients (MCCs) and Costes' randomization (Costes P) values. Costes P values ≥ 95% were considered as a colocalization signal. Statistical analyses were performed using Prism 6 software (Graphpad Software, La Jolla, CA, USA). Differences between treated and untreated control were estimated using 2-way ANOVA followed by Dunnett's multiple comparisons test. Values were expressed as the mean ± SD and p < 0.05 was considered statistically significant.
4.3.9. Statistical analysis
Statistical analyses were performed using Prism 6 software (Graphpad Software, La Jolla, CA, USA). Differences between treated and untreated control were estimated using 2-way ANOVA followed by Dunnett's multiple comparisons test. Values were expressed as the mean ± SD and p < 0.05 was considered statistically significant.
4.3.10. In vitro kinase inhibition assay
PIK3CA substrate. 1.1 mg of phosphatidylinositol (PI) (Sigma, Darmstadt, Germany) and 3.3 mg of 2-oleoyl-1-palmitoyl-sn-glycero-3-phospho-l-serine (PS) (Sigma, Darmstadt, Germany) were dissolved in 9:1 chloroform/methanol solution in TPX Polymethylpentene microtubes (Diagenode – Liege, Belgium). The lipid mixture was dried, resuspended in distilled water and sonicated for 13 cycles (1 min ON and 30 s OFF) by the Bioruptor® Plus sonication device (Diagenode – Liege, Belgium). The PI:3 PS lipid solution was finally transferred into low-binding tubes and stored at −20 °C. PI:3 PS stock solution was than prepared to a final concentration of 750 μM in TrisHCl 50 mM, EGTA 1.25 mM.
Kinase reactions. All kinase reactions were performed according to manufacturer's instructions, using 10–50 ng of enzyme. The nature of the substrates and their concentration and the presence of NP-40 or BSA are reported in Table S2 (Supporting information). All reactions were performed in 10 μL at 30 °C for 10 min using protein low-binding tubes. Reactions were stopped by adding 5 μL of phosphoric acid 0.8%. For protein kinases, aliquots (10 μL) were then transfer into a P30 Filtermat (PerkinElmer), washed five times with 75 mM phosphoric acid and once with acetone for 5 min. For PIK3CA, aliquots (10 μL) were then transfer into a GF/C Filtermat A (PerkinElmer), washed five times with 75 mM phosphoric acid and once with distilled water for 5 min. The filter was dried and transferred to a sealable plastic bag and 4 mL scintillation cocktail was added. Spotted reactions were read in a scintillation counter (Trilux, PerkinElmer).
4.3.11. In vitro polymerase and MTase assays
The polymerase assays were performed as previously described [51]. Briefly, the compounds were added in wells (2 μL, 5% DMSO final concentration), and the enzyme was distributed in wells. Reactions were started by the addition of the nucleotide mix (100 mM ATP) and were incubated at 30 C for 10 min before stopping by EDTA (100 mM) addition. Reactions were revealed by addition of Picogreen® fluorescent reagent (Molecular probes), diluted to 1/800 in TE buffer and the fluorescence signal was then read at 480 nm (excitation) and 530 nm (emission) using a TecanSafire2. IC50 was determined using the equation: % of active enzyme ¼ 100/(1þ(I)2/IC50), where I is the concentration of inhibitor and 100% of activity is the fluorescence intensity without inhibitor. The MTase activity was determined by filter binding assay as previously described [50]. The MTase activity was assayed by incubating the MTase in presence of increasing inhibitor concentration together with 7MeGpppAC4 and [3H] AdoMet. The assay was performed in 20 μL samples containing 40 mM Tris–HCl pH 7.5, 5 mM DTT, 10 μM AdoMet (0.2–2 μCi [3H] AdoMet), 1 μM of MTase and 1 μM 7MeGpppAC4 [52]. Reactions were incubated at 30 °C for 30 min and stopped by 20-fold dilution in ice-cold 100 μM AdoHcy solution. Samples were then transferred onto DEAE membrane (DEAE Filtermat; Wallac) by a Filtermat Harvester (Packard Instruments) washed with 0.01 M ammonium formate (pH 8.0), water and ethanol, and the radioactivity transferred onto RNA was measured using a Wallac 1450 MicroBeta Trilux Liquid Scintillation Counter. DENV3 N7-MTase activity was performed as described above but using of 0.5 μM GpppA2′OMe-RNA74 as substrate. The different protein used for polymerase and MTase assays were produces and purified as preciously describes [51,53].
Author contributions
The manuscript was written by M.R. through contributions of all authors, who have given approval to the final version of the manuscript.
Notes
M. Z. reports consultancy for ViiV Healthcare, Gilead Sciences and Janssen-Cilag, and grants for his institution from ViiV Healthcare and Gilead outside the submitted work. All other authors declare no competing financial interest.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the Ministero dell'Istruzione, dell'Università della Ricerca Italiano (MIUR), PRIN 2017 project (cod. 2017BMK8JR) “ORIGINALE CHEMIAE in Antiviral Strategy - Origin and Modernization of Multi-Component Chemistry as a Source of Innovative Broad Spectrum Antiviral Strategy” to M.R., M.Z., L.N., by Tuscany region, project Tuscany Antiviral Research Network -TUSCAVIR.NET (Bando Ricerca Salute 435, 2018) (to M.Z.) and by Sapienza University of Rome grants (L.N.). E.C. was supported by AIRC: MFAG Id.18811. G.M. was supported by Fondazione Banca del Monte di Lombardia. Francesca Preziuso is kindly acknowledged for the support on the ABTS assay. We also thank Priscila Sutto-Ortiz for excellent technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejmech.2021.113683.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Table S1-S4, 1H and 13C NMR spectra can be found in the supporting information.
References
- 1.De Clercq E., Li G. Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 2016;29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Symons J.A., Deval Innovation and trends in the development and approval of antiviral medicines: 1987–2017 and beyond. J. Antiviral Res. 2018;155:76–88. doi: 10.1016/j.antiviral.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll D., Daszak P., Wolfe N.D., Gao G.F., Morel C.M., Morzaria S., Pablos-Méndez A., Tomori O., Mazet J.A.K. The global Virome project. Science. 2018;359:872–874. doi: 10.1126/science.aap7463. [DOI] [PubMed] [Google Scholar]
- 4.Pierson T.C., Diamond M.S. The continued threat of emerging flaviviruses. Nat. Microbiol. 2020;5:796–812. doi: 10.1038/s41564-020-0714-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan L., et al. A single mutation in the prM protein of Zika virus contributes to fetal microcephaly. Science. 2017;358:933–936. doi: 10.1126/science.aam7120. [DOI] [PubMed] [Google Scholar]
- 6.Tomasello D., Schlagenhauf P. Chikungunya and dengue autochthonous cases in Europe, 2007-2012. Travel. Med. Infect. Dis. 2013;11:274–284. doi: 10.1016/j.tmaid.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Gossner C.M., Ducheyne E., Schaffner F. Increased risk for autochthonous vector-borne infections transmitted by Aedes albopictus in continental Europe. Euro Surveill. 2018;23(24):1800268. doi: 10.2807/1560-7917.ES.2018.23.24.1800268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez-Morales A.J., Villamil-Gómez W.E., Franco-Paredes C. The arboviral burden of disease caused by co-circulation and co-infection of dengue, chikungunya and Zika in the Americas. Trav. Med. Infect. Dis. 2016;14:177–179. doi: 10.1016/j.tmaid.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Olawoyin O., Kribs C. Coinfection. Altered vector infectivity, and antibody-dependent enhancement: the dengue–zika interplay. Bull. Math. Biol. 2020;82:1–13. doi: 10.1007/s11538-019-00681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorenz C., Azevedo T.S., Chiaravalloti-Neto F. COVID-19 and dengue fever: a dangerous combination for the health system in Brazil. Trav. Med. Infect. Dis. 2020;35:101659. doi: 10.1016/j.tmaid.2020.101659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harapan H., Ryan M., Yohan B., Abidin R.S., Nainu F., Rakib A., Jahan I., Emran T.B., Ullah I., Panta K., Dhama K., Sasmono R.T. Covid-19 and dengue: double punches for dengue-endemic countries in Asia. Rev. Med. Virol. 2021;31(2) doi: 10.1002/rmv.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmid M.A., Gonzalez K.N., Shah S., Pena J., Mack M., Talarico L.B., Polack F.P., Harris E. Influenza and dengue virus co-infection impairs monocyte recruitment to the lung, increases dengue virus titers, and exacerbates pneumonia. Eur. J. Immunol. 2017;47:527–539. doi: 10.1002/eji.201646675. [DOI] [PubMed] [Google Scholar]
- 13.Hansen S., Hotop S.-K., Faye O., Ndiaye O., Böhlken-Fascher S., Pesso^a R., Hufert F., Stahl-Hennig C., Frank R., Czerny C.-P., Schmidt-Chanasit J., Sanabani S.S., Sall A.A., Niedrig M., Brönstrup M., Fritz H.-J., El Wahed A.A. Diagnosing Zika virus infection against a background of other flaviviruses: studies in high resolution serological analysis. Sci. Rep. 2019;9:3648. doi: 10.1038/s41598-019-40224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henrina, J.; Santosa Putra, I. C.; Lawrensia, S.; Handoyono, Q. F.; Cahyadi, A. Coronavirus disease of 2019: a mimicker of dengue infection? Sn Compr. Clin. Med. 10.1007/s42399-020-00364-3. [DOI] [PMC free article] [PubMed]
- 15.Kumar N., Sharma S., Kumar R., Tripathi B.N., Barua S., Ly H., Roused B.T. Host-directed antiviral therapy. Clin. Microl. Rev. 2020;33(3) doi: 10.1128/CMR.00168-19. e00168-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards A. What are the odds of finding a COVID-19 drug from a lab repurposing screen? J. Chem. Inf. Model. 2020;60(12):5727–5729. doi: 10.1021/acs.jcim.0c00861. in press. [DOI] [PubMed] [Google Scholar]
- 17.Morphy J.R., editor. RSC Drug Discovery Series. RSC Publ; Cambridge: 2012. Designing multi-target drugs. [Google Scholar]
- 18.Bizzarri M., Giuliani A., Monti N., Verna R., Pensotti A., Cucina A. Rediscovery of natural compounds acting via multitarget recognition and noncanonical pharmacodynamical actions. Drug Discov. Today. 2020;25:920–927. doi: 10.1016/j.drudis.2020.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Nystrom K., Waldenstrom J., Tang K.-W., Lagging M. Ribavirin: pharmacology, multiple modes of action and possible future perspectives. Future Virol. 2019;14:153–160. [Google Scholar]
- 20.Vincetti P., Caporuscio F., Kaptein S., Gioiello A., Mancino V., Suzuki Y., Yamamoto N., Crespan E., Lossani A., Maga G., Rastelli G., Castagnolo D., Neyts J., Leyssen P., Costantino G., Radi M. Discovery of multi-target antivirals acting on both the dengue virus NS5-NS3 interaction and the host Src/Fyn kinases. J. Med. Chem. 2015;58:4964–4975. doi: 10.1021/acs.jmedchem.5b00108. [DOI] [PubMed] [Google Scholar]
- 21.Tassini S., Sun L., Lanko K., Crespan E., Langron E., Falchi F., Kissova M., Armijos-Rivera J.I., Delang L., Mirabelli C., Neyts J., Pieroni M., Cavalli A., Costantino G., Maga G., Vergani P., Leyssen P., Radi M. Discovery of multi-target agents active as broad-spectrum antivirals and correctors of cystic fibrosis transmembrane conductance regulator (CFTR) for associated pulmonary diseases. J. Med. Chem. 2017;60:1400–1416. doi: 10.1021/acs.jmedchem.6b01521. [DOI] [PubMed] [Google Scholar]
- 22.Kaptein S.J.F., Vincetti P., Crespan E., Armijos-Rivera J.I., Costantino G., Maga G., Neyts J., Radi M. Identification of broad-spectrum dengue/Zika virus replication inhibitors by functionalization of quinoline and 2,6-diaminopurine scaffolds. ChemMedChem. 2018;13:1371–1376. doi: 10.1002/cmdc.201800178. [DOI] [PubMed] [Google Scholar]
- 23.Tassini S., Langron E., Delang L., Mirabelli C., Lanko K., Crespan E., Kissova M., Tagliavini G., Fontò G., Bertoni S., Palese S., Giorgio C., Ravanetti F., Ragionieri L., Zamperini C., Mancini A., Dreassi E., Maga G., Vergani P., Neyts J., Radi M. Multi-target CFTR modulators endowed with multiple beneficial side effects for cystic fibrosis patients: toward a simplified therapeutic approach. J. Med. Chem. 2019;62:10833–10847. doi: 10.1021/acs.jmedchem.9b01416. [DOI] [PubMed] [Google Scholar]
- 24.Vicenti I., Dragoni F., Giannini A., Giammarino F., Spinicci M., Saladini F., Boccuto A., Zazzi M. Development of a cell-based immunodetection assay for simultaneous screening of antiviral compounds inhibiting Zika and dengue virus replication. SLAS Discov. 2020;25:506–514. doi: 10.1177/2472555220911456. [DOI] [PubMed] [Google Scholar]
- 25.Puertas M.C., Buzón M.J., Ballestero M., Van Den Eede P., Clotet B., Prado J.G., Martinez-Picado J. Novel two- round phenotypic assay for protease inhibitor susceptibility testing of recombinant and primary HIV-1 isolates. J. Clin. Microbiol. 2012;50:3909–3916. doi: 10.1128/JCM.01636-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saladini F., Giannini A., Boccuto A., Vicenti I., Zazzi M. Agreement between an in-house replication competent and a reference replication defective recombinant virus assay for measuring phenotypic resistance to HIV-1 protease, reverse Transcriptase, and integrase inhibitors. J. Clin. Lab. Anal. 2017;32 doi: 10.1002/jcla.22206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Checconi P., De Angelis M., Marcocci M.E., Fraternale A., Magnani M., Palamara A.T., Nencioni L. Redox-modulating agents in the treatment of viral infections. Int. J. Mol. Sci. 2020;21:4084. doi: 10.3390/ijms21114084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duarte L.F.B., Oliveira R.L., Rodrigues K.C., Voss G.T., Godoi B., Schumacher R.F., Perin G., Wilhelm E.A., Luchese C., Alves D. Organoselenium compounds from purines: synthesis of 6-arylselanylpurines with antioxidant and anticholinesterase activities and memory improvement effect. Bioorg. Med. Chem. 2017;25:6718–6723. doi: 10.1016/j.bmc.2017.11.019. [DOI] [PubMed] [Google Scholar]
- 29.Brathe A., Andresen G., Gundersen L.-L., Malterud K.E., Rise F. Antioxidant activity of synthetic cytokinin analogues: 6-alkynyl- and 6-alkenylpurines as novel 15-lipoxygenase inhibitors. Bioorg. Med. Chem. 2002;10:1581–1586. doi: 10.1016/s0968-0896(01)00427-8. [DOI] [PubMed] [Google Scholar]
- 30.Djuidje E.N., Dissette V., Bino B., Benetti S., Balzarini J., Liekens S., Manfredini S., Vertuani S., Baldisserotto A. A multitarget approach toward the development of 8- substituted purines for photoprotection and prevention of UV-related damage. ChemMedChem. 2017;12:760–769. doi: 10.1002/cmdc.201700137. [DOI] [PubMed] [Google Scholar]
- 31.Kolarski D., Szymanski W., Feringa B.L. Two-step, one-pot synthesis of visible-light-responsive 6-azopurines. Org. Lett. 2017;19:5090–5093. doi: 10.1021/acs.orglett.7b02361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Too K., Brown D.M., Bongard E., Yardley V., Vivas L., Loakesa D. Anti-malarial activity of N6-modified purine analogues. Bioorg. Med. Chem. 2007;15:5551–5562. doi: 10.1016/j.bmc.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 33.Re R., Pellegrini N., Proteggente A., Pannala A., Yang M., Rice-Evans C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999;26:1231–1237. doi: 10.1016/s0891-5849(98)00315-3. [DOI] [PubMed] [Google Scholar]
- 34.Park J., Ávila-Pérez G., Nogales A., Blanco-Lobo P., de la Torre J., Martínez-Sobrido L. Identification and characterization of novel compounds with broad-spectrum antiviral activity against influenza A and B viruses. J. Virol. 2020;94 doi: 10.1128/JVI.02149-19. e02149-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amatore D., Sgarbanti R., Aquilano K., Baldelli S., Limongi D., Civitelli L., Nencioni L., Garaci E., Ciriolo M.R., Palamara A.T. Influenza virus replication in lung epithelial cells depends on redox-sensitive pathways activated by NOX4-derived ROS. Cell Microbiol. 2015;17:131–145. doi: 10.1111/cmi.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-NCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pruijssers A., George A., Schäfer A., Leist S., Gralinksi L., Dinnon K., Yount B., Agostini M., Stevens L., Chappell J., Lu X., Hughes T., Gully K., Martinez D., Brown A., Graham R., Perry J., Du Pont V., Pitts J., Ma B., Babusis D., Murakami E., Feng J., Bilello J., Porter D., Cihlar T., Baric R., Denison M., Sheahan T. Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep. 2020;32:107940. doi: 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choy K., Wong A., Kaewpreedee P., Sia S., Chen D., Hui K., Chu D., Chan M., Cheung P., Huang X., Peiris M., Yen H. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Meyer S., Bojkova D., Cinatl J., Van Damme E., Buyck C., Van Loock M., Woodfall B., Ciesek S. Lack of antiviral activity of darunavir against SARS-CoV-2. Int. J. Infect. Dis. 2020;97:7–10. doi: 10.1016/j.ijid.2020.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sofia M.J., Bao D., Chang W., Du J., Nagarathnam D., Rachakonda S., Reddy P.G., Ross B.S., Wang P., Zhang H.-R., Bansal S., Espiritu C., Keilman M., Lam A.M., Micolochick Steuer H.M., Niu C., Otto M.J., Furman P.A. Discovery of a β-D-20-Deoxy-2’-α-fluoro-20-β-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J. Med. Chem. 2010;53:7202–7218. doi: 10.1021/jm100863x. [DOI] [PubMed] [Google Scholar]
- 41.Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., Perry J., Perron M., Stray K.M., Barauskas O., Feng J.Y., Xu Y., Lee G., Rheingold A.L., Ray A.S., Bannister R., Strickley R., Swaminathan S., Lee W.A., Bavari S., Cihlar T., Lo M.K., Warren T.K., Mackman R.L. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of ebola and emerging viruses. J. Med. Chem. 2017;60:1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 42.Dubankova A., Boura E. Structure of the yellow fever NS5 protein reveals conserved drug targets shared among flaviviruses. Antivir. Res. 2019;169:104536. doi: 10.1016/j.antiviral.2019.104536. [DOI] [PubMed] [Google Scholar]
- 43.Wu J., Liu W., Gong P. A structural overview of RNA-dependent RNA polymerases from the flaviviridae family. Int. J. Mol. Sci. 2015;16:12943–12957. doi: 10.3390/ijms160612943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou G., Chen Y.-L., Dong H., Lim C.C., Yap L.J., Yau Y.H., Shochat S.G., Lescar J., Shi P.-Y. Functional analysis of two cavities in flavivirus NS5 polymerase. J. Biol. Chem. 2011;286:14362–14372. doi: 10.1074/jbc.M110.214189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tay M.Y.F., Vasudevan S.G. The transactions of NS3 and NS5 in flaviviral RNA replication. Adv. Exp. Med. Biol. 2018;1062:147–163. doi: 10.1007/978-981-10-8727-1_11. [DOI] [PubMed] [Google Scholar]
- 46.Sarto C., Kaufman S.B., Estrin D.A., Arrar M. Nucleotide-dependent dynamics of the dengue NS3 helicase. Biochim. Biophys. Acta Protein Proteonomics. 2020;1868:140441. doi: 10.1016/j.bbapap.2020.140441. [DOI] [PubMed] [Google Scholar]
- 47.Wu J., Bera A.K., Kuhn R.J., Smith J.L. Structure of the flavivirus helicase: implications for catalytic activity, protein interactions, and proteolytic processing. J. Virol. 2005;79:10268–10277. doi: 10.1128/JVI.79.16.10268-10277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saw W.G., Pan A., Subramanian Manimekalai M.S., Grüber A., Grüber G. Structure and flexibility of non-structural proteins 3 and -5 of dengue- and Zika viruses in solution. Prog. Biophys. Mol. Biol. 2019;143:67–77. doi: 10.1016/j.pbiomolbio.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 49.Tay M.Y.F., Saw W.G., Zhao Y., Chan K.W.K., Singh D., Chong Y., Forwood J.K., Ooi E.E., Grüber G., Lescar J., Luo D., Vasudevan S.G. The C-terminal 50 amino acid residues of dengue NS3 protein are important for NS3-NS5 interaction and viral replication. J. Biol. Chem. 2015;290:2379–2394. doi: 10.1074/jbc.M114.607341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barral K., Sallamand C., Petzold C., Coutard B., Collet A., Thillier Y., Zimmermann J., Vasseur J.-J., Canard B., Rohayem J., Debart F., Decroly E. Development of specific dengue virus 2′-O- and N7-methyltransferase assays for antiviral drug screening. Antivir. Res. 2013;99:292–300. doi: 10.1016/j.antiviral.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Benmansour F., Eydoux C., Querat G., de Lamballerie X., Canard B., Alvarez K., Guillemot J.-C., Barral K. Novel 2-phenyl-5-[(E)-2-(Thiophen-2-Yl)Ethenyl]-1,3,4-Oxadiazole and 3-phenyl-5-[(E)-2-(Thiophen-2-Yl)Ethenyl]-1,2,4-Oxadiazole derivatives as dengue virus inhibitors targeting NS5 polymerase. Eur. J. Med. Chem. 2016;109:146–156. doi: 10.1016/j.ejmech.2015.12.046. [DOI] [PubMed] [Google Scholar]
- 52.Selisko B., Peyrane F.F., Canard B., Alvarez K., Decroly E. Biochemical characterization of the (Nucleoside-2’O)-Methyltransferase activity of dengue virus protein NS5 using purified capped RNA oligonucleotides 7MeGpppACn and GpppACn. J. Gen. Virol. 2010;91(1) doi: 10.1099/vir.0.015511-0. 112–12.1. [DOI] [PubMed] [Google Scholar]
- 53.Coutard B., Barral K., Lichière J., Selisko B., Martin B., Aouadi W., Lombardia M.O., Debart F., Vasseur J.-J., Guillemot J.C., Canard B., Decroly E. Zika virus methyltransferase: structure and functions for drug design perspectives. J. Virol. 2017;91(5) doi: 10.1128/JVI.02202-16. e02202-16, e02202-e02216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hannemann H., Sung P.-Y., Chiu H.-C., Yousuf A., Bird J., Lim S.P., Davidson A.D. Serotype-specific differences in dengue virus non-structural protein 5 nuclear localization. J. Biol. Chem. 2013;288:22621–22635. doi: 10.1074/jbc.M113.481382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ji W., Luo G. Zika virus NS5 nuclear accumulation is protective of protein degradation and is required for viral RNA replication. Virology. 2020;541:124–135. doi: 10.1016/j.virol.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 56.Lopez-Denman A.J., Russo A., Wagstaff K.M., White P.A., Jans D.A., Mackenzie J.M. Nucleocytoplasmic shuttling of the West Nile virus RNA -dependent RNA polymerase NS5 is critical to infection. Cell Microbiol. 2018;20 doi: 10.1111/cmi.12848. [DOI] [PubMed] [Google Scholar]
- 57.García Cordero J., León Juárez M., González-Y-Merchand J.A., Cedillo Barrón L., Gutiérrez Castañeda B. Caveolin-1 in lipid rafts interacts with dengue virus NS3 during polyprotein processing and replication in HMEC-1 cells. PloS One. 2014;9 doi: 10.1371/journal.pone.0090704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar N., Sharma S., Kumar R., Tripathi B.N., Barua S., Ly H., Rouse B.T. Host-directed antiviral therapy. Clin. Microbiol. Rev. 2020;33(3) doi: 10.1128/CMR.00168-19. e00168-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meineke R., Rimmelzwaan G., Elbahesh H. Influenza virus infections and cellular kinases. Viruses. 2019;11(2):171. doi: 10.3390/v11020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weisberg E., Parent A., Yang P.L., Sattler M., Liu Q., Liu Q., Wang J., Meng C., Buhrlage S.J., Gray N., Griffin J.D. Repurposing of kinase inhibitors for treatment of COVID-19. Pharm. Res. (N. Y.) 2020;37(9):167. doi: 10.1007/s11095-020-02851-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang H., Ma J., Shi J., Meng L., Jiang H., Ding J., Liu H. Discovery of novel purine derivatives with potent and selective inhibitory activity against c-src tyrosine kinase. Bioorg. Med. Chem. 2010;18:4615–4624. doi: 10.1016/j.bmc.2010.05.032. [DOI] [PubMed] [Google Scholar]
- 62.Davis R.L. Mechanism of action and target identification: a matter of timing in drug discovery. iScience. 2020;23(9):101487. doi: 10.1016/j.isci.2020.101487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moffat J.G., Vincent F., Lee J.A., Eder J., Prunotto M. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug Discov. 2017;16(8):531–543. doi: 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- 64.Lai A., Bergna A., Caucci S., Clementi N., Vicenti I., Dragoni F., Cattelan A.M., Menzo S., Pan A., Callegaro A., Tagliabracci A., Caruso A., Caccuri F., Ronchiadin S., Balotta C., Zazzi M., Vaccher E., Clementi M., Galli M., Zehender G. Molecular tracing of SARS-CoV-2 in Italy in the first three months of the epidemic. Viruses. 2020;12(8):798. doi: 10.3390/v12080798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vicenti I., Boccuto A., Giannini A., Dragoni F., Saladini F., Zazzi M. Comparative analysis of different cell systems for Zika virus (ZIKV) propagation and evaluation of anti-ZIKV compounds in vitro. Virus Res. 2018;244:64–70. doi: 10.1016/j.virusres.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 66.Dragoni F., Boccuto A., Picarazzi F., Giannini A., Giammarino F., Saladini F., Mori M., Mastrangelo E., Zazzi M., Vicenti I. Evaluation of sofosbuvir activity and resistance profile against West Nile virus in vitro. Antivir. Res. 2020;175:104708. doi: 10.1016/j.antiviral.2020.104708. [DOI] [PubMed] [Google Scholar]
- 67.Di Sotto A., Checconi P., Celestino I., Locatelli M., Carissimi S., De Angelis M., Rossi V., Limongi D., Toniolo C., Martinoli L., Di Giacomo S., Palamara A.T., Nencioni L. Antiviral and antioxidant activity of a hydroalcoholic extract from humulus lupulus L. Oxid. Med. Cell. Longev. 2018;2018:1–14. doi: 10.1155/2018/5919237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Checconi P., Sgarbanti R., Celestino I., Limongi D., Amatore D., Iuvara A., Alimonti A., Garaci E., Palamara A.T., Nencioni L. The environmental pollutant cadmium promotes influenza virus replication in MDCK cells by altering their redox state. Int. J. Mol. Sci. 2013;14:4148–4162. doi: 10.3390/ijms14024148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bizzarri B.M., Fanelli A., Piccinino D., De Angelis M., Dolfa C., Palamara A.T., Nencioni L., Zippilli C., Crucianelli M., Saladino R. Synthesis of stilbene and chalcone inhibitors of influenza A virus by sba -15 supported hoveyda-grubbs metathesis. Catalysts. 2019;9:983. [Google Scholar]
- 70.Lagorce D., Bouslama L., Becot J., Miteva M.A., Villoutreix B.O. FAF-Drugs4: free ADME-tox filtering computations for chemical biology and early stages drug discovery. Bioinformatics. 2017;33:3658–3660. doi: 10.1093/bioinformatics/btx491. [DOI] [PubMed] [Google Scholar]
- 71.Stachulski A.V., Taujanskas J., Pate S.L., Rajoli R.K.R., Aljayyoussi G., Pennington S.H., Ward S.A., Hong W.D., Biagini G.A., Owen A., Nixon G.L., Leung S.C., O’Neill P.M. Therapeutic potential of nitazoxanide: an appropriate choice for repurposing versus SARS-CoV-2? ACS Infect. Dis. 2021;7(6):1317–1331. doi: 10.1021/acsinfecdis.0c00478. [DOI] [PubMed] [Google Scholar]
- 72.Blaising J., Polyak S.J., Pécheur E.-I. Arbidol as a broad-spectrum antiviral: an update. Antivir. Res. 2014;107:84–94. doi: 10.1016/j.antiviral.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xiu S., Dick A., Ju H., Mirzaie S., Abdi F., Cocklin S., Zhan P., Liu X. Inhibitors of SARS-CoV-2 entry: current and future opportunities. J. Med. Chem. 2020;63:12256–12274. doi: 10.1021/acs.jmedchem.0c00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raghuvanshi R., Bharate S.B. Recent developments in the use of kinase inhibitors for management of viral infections. J. Med. Chem. 2021 doi: 10.1021/acs.jmedchem.0c01467. (acs.jmedchem.0c01467). In press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.