Abstract
Amphibians such as Xenopus tropicalis exhibit a remarkable capacity for tissue regeneration after traumatic injury. Although transforming growth factor-β (TGF-β) receptor signaling is known to be essential for tissue regeneration in fish and amphibians, the role of TGF-β ligands in this process is not well understood. Here, we show that inhibition of TGF-β1 function prevents tail regeneration in Xenopus tropicalis tadpoles. We found that expression of tgfb1 is present before tail amputation and is sustained throughout the regeneration process. CRISPR-mediated knock-out (KO) of tgfb1 retards tail regeneration; the phenotype of tgfb1 KO tadpoles can be rescued by injection of tgfb1 mRNA. Cell proliferation, a critical event for the success of tissue regeneration, is downregulated in tgfb1 KO tadpoles. In addition, tgfb1 KO reduces the expression of phosphorylated Smad2/3 (pSmad2/3) which is important for TGF-β signal-mediated cell proliferation. Collectively, our results show that TGF-β1 regulates cell proliferation through the activation of Smad2/3. We therefore propose that TGF-β1 plays a critical role in TGF-β receptor-dependent tadpole tail regeneration in Xenopus.
Keywords: TGF-β1, Xenopus tail regeneration, cell proliferation, Smad2/3
Introduction
Xenopus tropicalis tadpoles are able to regenerate appendages, including tails and limbs, through the activation of cell proliferation [1]. By contrast, mammals have a limited capacity for regeneration of damaged tissues due in part to the inability to reactivate cell proliferation after traumatic injury, indicating that cell proliferation is an important component of tissue regeneration [2, 3]. TGF-β signaling regulates several aspects of regeneration such as wound healing, cell proliferation, and tissue differentiation in the Xenopus tadpole tail and also in the zebrafish heart and axolotl limbs [4, 5, 6]. Thus, the function of TGF-β signaling is conserved among animal species that can accomplish tissue regeneration. However, it is not clear whether TGF-β ligands are required for tissue regeneration.
The TGF-β superfamily of ligands includes TGFβs, activins, bone morphogenetic proteins (BMPs), and growth and differentiation factors (GDFs) [7]. TGF-βs are produced in an inactive form that associates with the latency associated peptide (LAP) and are stored in the extracellular matrix (ECM). The latent TGF-βs are activated following the cleavage of LAP by protease digestion and are released from the ECM [7, 8]. TGF-β signaling is initiated upon ligand binding to the receptor, which phosphorylates downstream Smad2/3 transcription factors. Following phosphorylation, the pSmad2/3 form a complex with Smad4 and translocate into the nucleus to regulate the transcription of target genes [7]. During Xenopus tadpole tail regeneration, inhibition of TGF-β signaling causes a reduction of pSmad2 expression and of the number of mitotically dividing cells [4]. Several TGF-β superfamily ligands (tgfb1, tgfb2, inhba and gdf11) are expressed in the regenerating Xenopus tail. In zebrafish, it has been shown that morpholino-mediated knock-down of inhba impairs fin regeneration [9]. These studies suggest that multiple TGF-β superfamily ligands might be involved in the activation of Smad2/3 and cell proliferation for tissue regeneration. Therefore, determining the function of TGF-β superfamily ligands is essential for a better understanding of how TGF-β signaling regulates tissue regeneration.
In this study, we investigated X. tropicalis tadpole tail regeneration and show that tgfb1 is strongly expressed throughout the regeneration processes. Using the CRISPR/Cas9 technique, we found that TGF-β1 is required for tail regeneration and for activation of Smad2/3 that is crucial for cell proliferation. Furthermore, TGF-β1 was found to positively regulate cell proliferation and tissue differentiation during tail regeneration. These results suggest that TGF-β1 is a key regulator of Xenopus tadpole tail regeneration; our findings also contribute to understanding the regulatory mechanisms of tissue regeneration.
Materials and Methods
Animals, microinjection, and amputation
X. tropicalis tadpoles were obtained and maintained as described previously [10]. CRISPR/Cas9 mutagenesis was carried out with minor modifications of previously described protocol [10]. In brief, 1000 pg of sgRNAs were injected (for combinatorial injection, 333 pg each of three sgRNAs) with Cas9 protein (1 ng, Integrated DNA Technologies) into fertilized eggs in 6% Ficoll or 0.2% methylcellulose solution containing 0.1% BSA, 0.5X MMR and 50 μg/ml gentamycin. Tails were amputated from tadpoles at stage 41/42 and the tadpoles were maintained for 72 hours post amputation (hpa). To inhibit TGF-β signaling, tadpoles were treated with 12.5 μM of SB-505124 (Cayman Chemical) and DMSO (Nacalai) from 1 hour before tail amputation. Animal experiments followed the guidelines of the Animal Experimentation Ethics Committee of Hiroshima University and international regulations.
Phenotyping of tgfb1 KO tadpoles
We carried out CRISPR/Cas9-mediated mutagenesis of tgfb1 in X. tropicalis embryos following a published F0 mutagenesis strategy [11]. To ensure phenotypic consistency among tgfb1 KO tadpoles, we generated three sgRNAs targeting different sites of the tgfb1 gene. We monitored tadpoles daily following injection of the sgRNAs until 72 hpa to evaluate the effect of tgfb1 KO on tail regeneration. Tadpoles were graded for the extent of tail regeneration based on tail length at 72 hpa as follows: normal tail regeneration, weakly delayed tail regeneration, and severely delayed tail regeneration. The efficiency of tgfb1 KO was determined using a T7E1 assay and by TA cloning-based genotyping. A rescue experiment using tgfb1 mRNA was performed to examine the specificity of tgfb1 KO.
Cloning of the tgfb1 gene
Full-length tgfb1 was amplified from X. tropicalis embryo (stage 29/30) cDNAs using the following primers: forward 5’- AAG GCC TCA ACC AGG ATC TCC CAC ACT −3’ and reverse 5’- GCT CTA GAT GTG GGT TGC GTT GTT TCT A −3’. The amplified product was digested with StuI and XbaI and subcloned into pDH105 (pDH105-tgfb1).
Whole-mount immunostaining and in situ hybridization
Immunostaining of phosphorylated Histone H3 (pH3) and pSmad2/3 was performed as previously described with a minor modification [10]. For pSmad2/3 staining, tadpoles were treated with a permeabilization solution (1% NP-40, 1X PBST) for 30 min after bleaching. The following primary and secondary antibodies were used: anti-pH3 antibody at 1:500 dilution (Upstate Biotechnology); anti-pSmad2/3 antibody at 1:500 dilution (Cell Signaling Technology); Alexa Fluor 488 goat anti-rabbit antibody at 1:500 dilution (Molecular Probe); Alexa Fluor 488 goat anti-mouse antibody at 1:500 dilution (Molecular Probe). Whole-mount in situ hybridization (WISH) and probe synthesis were carried out as described previously [12, 13]. The tgfb1 antisense and sense probes were generated from pDH105-tgfb1.
Preparation of sgRNAs and mRNA synthesis
sgRNAs were designed and produced as previously described [10, 14]. The following forward primers for sgRNAs targeting tgfb1 locus were used: sg 1, 5’- ATT TAG GTG ACA CTA TAG GTG TCT ACC TGT AAG ACT GGT TTT AGA GCT AGA AAT AGC AAG −3’; sg 2, 5’- ATT TAG GTG ACA CTA TAG GAG AAT TGA AGC CAT CAG GGT TTT AGA GCT AGA AAT AGC AAG −3’; sg 3, 5’- ATT TAG GTG ACA CTA TAG GTT TAC AAT AGC ACC TTG GGT TTT AGA GCT AGA AAT AGC AAG −3’. Capped tgfb1 mRNA was generated by in vitro transcription of pDH105-tgfb1 using an SP6 transcription kit (Invitrogen).
Genotyping
Lysis, PCR, T7E1 assay, and TA cloning were performed as described previously [10]. To confirm the genotype of tgfb1 KO tadpoles, the following primers were used for PCR amplification of the tgfb1 locus: forward 5’- AAG ACG GGA CAG CAA CTT TC −3’ and reverse 5’- TGG CAC ACA TGC AGA ACT ATC −3’.
Quantification and statistical analysis
Fluorescent images were captured with a Zeiss Axio Zoom V-16 system. CellSens standard software (Olympus) was used to measure tail lengths. Statistical comparisons were performed using Student’s t-tests (*P < 0.05, ***P < 0.001). The fluorescent intensity of pSmad2/3 immunostaining was quantified using Zen Blue software (Zeiss). A previous report [4] and the present analysis observed a non-specific signal of pSmad2/3 staining in regenerating tails that was not reduced after TGF-β receptor inhibition. To measure the specific fluorescent intensity of pSmad2/3, the mean value of the fluorescence signal in tyrosinase KO and tgfb1 KO tadpoles was subtracted by that of the non-specific signal in SB-505124-treated tadpoles.
Results
tgfb1 is expressed throughout Xenopus tail regeneration
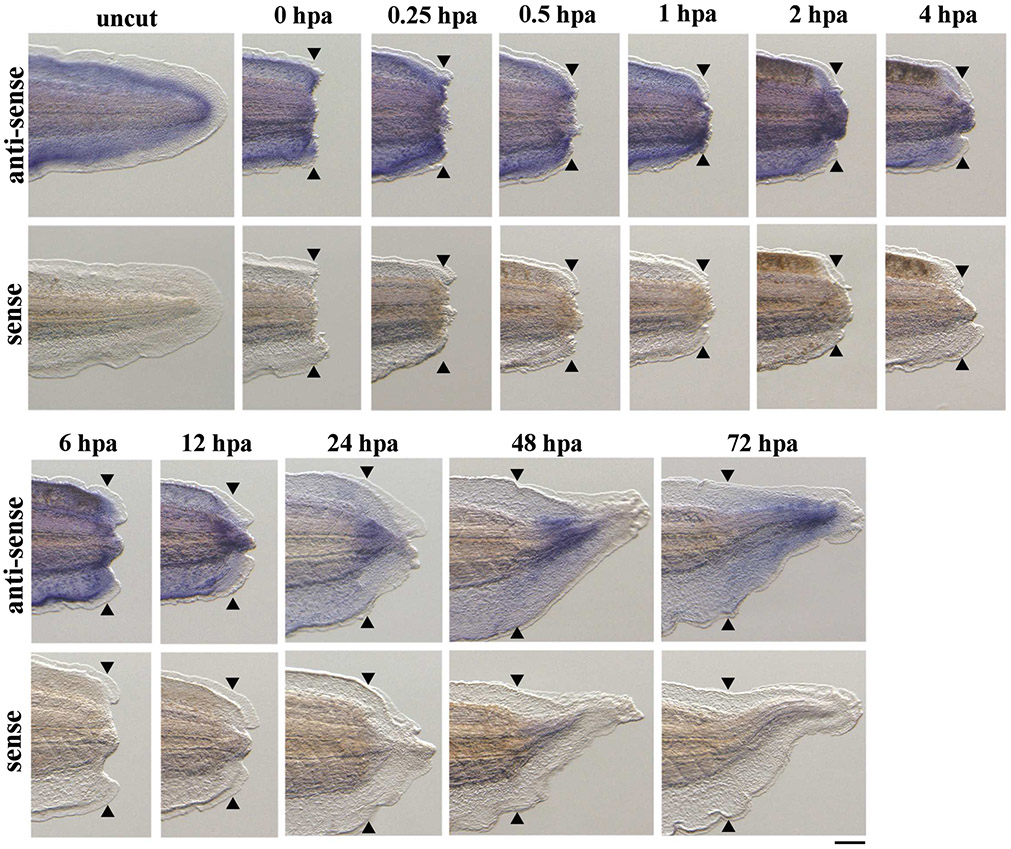
Expression of tgfb1 has previously been described during X. laevis tadpole tail regeneration [4]. Consistently, RNAseq analysis of regenerating X. tropicalis tails has indicated that tgfb1 is strongly expressed compared to other TGF-β superfamily ligands [15]. Therefore, we performed a detailed examination of tgfb1 expression in regenerating X. tropicalis tails using WISH (Fig. 1). Before amputation, tgfb1 expression was detectable throughout the entire tail, especially in the inner fin region. Following amputation, tgfb1 expression was observed in the amputation plane at 0–1 hpa. At 2–12 hpa, tgfb1 expression commenced in the regenerating tail tip. Subsequently, at 24–72 hpa, tgfb1 transcripts were widely present in the regenerating tissues. No signals were detected in regenerating tail tissues using a sense probe as a negative control. These results suggest that tgfb1 was expressed before and after tail amputation, and that TGF-β1 might be involved in Xenopus tadpole tail regeneration.
Figure 1. Expression of tgfb1 in X. tropicalis tadpoles before and after tail amputation.
Lateral views of uncut tadpole tails and amputated tails at 0, 0.25, 0.5, 1, 2, 4, 6, 12, 24, 48 and 72 hours post amputation (hpa) after whole-mount in situ hybridization using tgfb1 antisense and sense RNA probes. Black arrowheads show amputation sites. Scale bar, 200 μm.
TGF-β1 is required for Xenopus tail regeneration
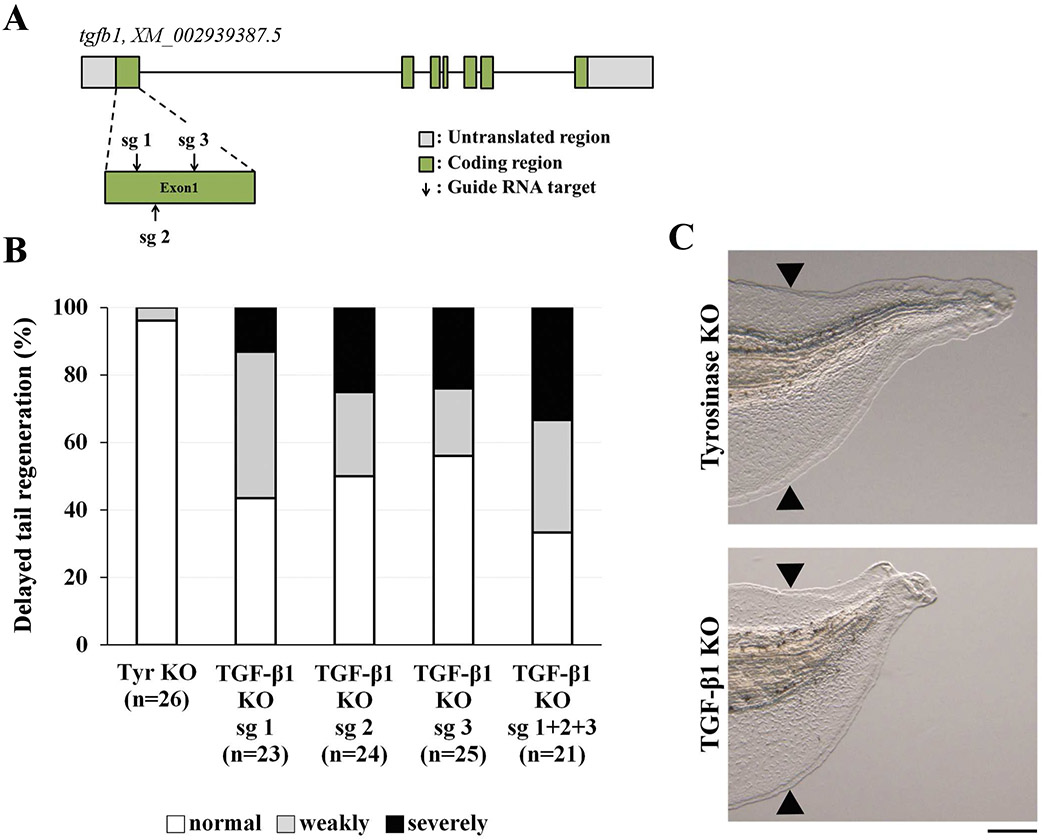
To determine whether TGF-β1 is required for Xenopus tail regeneration, we conducted a CRISPR-mediated loss-of-function experiment. First, we generated tgfb1 KO tadpoles using single sgRNAs (sg 1, sg 2, or sg 3) that target the latency associated peptide of TGF-β1; the sgRNA was injected into fertilized eggs with Cas9 protein (Fig. 2A). tgfb1 KO tadpoles injected with single sgRNAs showed a delay in tail regeneration (sg 1, 56.5%; sg 2, 50%; sg 3, 44%; when weakly and severely delayed phenotypes were combined), while tyrosinase KO tadpoles did not show this delay (tyrosinase KO, 3.8%; Fig. 2B). This phenotypic consistency suggested that the regeneration defect of tgfb1 KO tadpoles was caused by loss of TGF-β1 function. As single sgRNAs induced a moderate rate of tgfb1 mutations as determined by a T7E1 assay (data not shown), we injected a combination of all three sgRNAs (sg 1 + sg 2 + sg 3) into fertilized eggs to increase the rate of mutations. Sequencing analysis of tgfb1 KO tadpoles produced by the combination of the three sgRNAs showed that all had mutations in the tgfb1 locus: in-frame mutations, 13.7%; out-of-frame mutations, 86.3% (Supplementary Fig. 1). Furthermore, tgfb1 KO tadpoles (sg 1 + sg 2 + sg 3) showed a clear delay in tail regeneration (66.6%; Fig. 2B and C) compared to tadpoles injected with a single sgRNA. Therefore, we used tgfb1 KO tadpoles (sg 1 + sg 2 + sg 3) in the following experiments. We performed a rescue experiment of tgfb1 KO tadpoles to explore the specificity of the knockout. Overexpression of tgfb1 partially but significantly rescued the knockout phenotype, indicating that the delay in tail regeneration in tgfb1 KO tadpoles resulted from inactivation of tgfb1 (Supplementary Fig. 2).
Figure 2. TGF-β1 is required for Xenopus tail regeneration.
(A) Schematic drawing of sgRNA target sites (sg 1, sg 2 and sg 3) in the tgfb1 locus. Grey boxes, untranslated region; green boxes, coding region; arrows, sgRNA target sites; bars, intron regions. (B) Delayed tail regeneration in tgfb1 KO tadpoles. The extent of tail regeneration was classified at 72 hpa as normal tail regeneration, weakly delayed tail regeneration, and severely delayed tail regeneration. (C) The phenotypes of tyrosinase KO (control) and tgfb1 KO tadpoles (sg 1 + 2 + 3) at 72 hpa. Black arrowheads show amputation sites. Scale bar, 200 μm.
TGF-β1 is important for tissue differentiation and cell proliferation
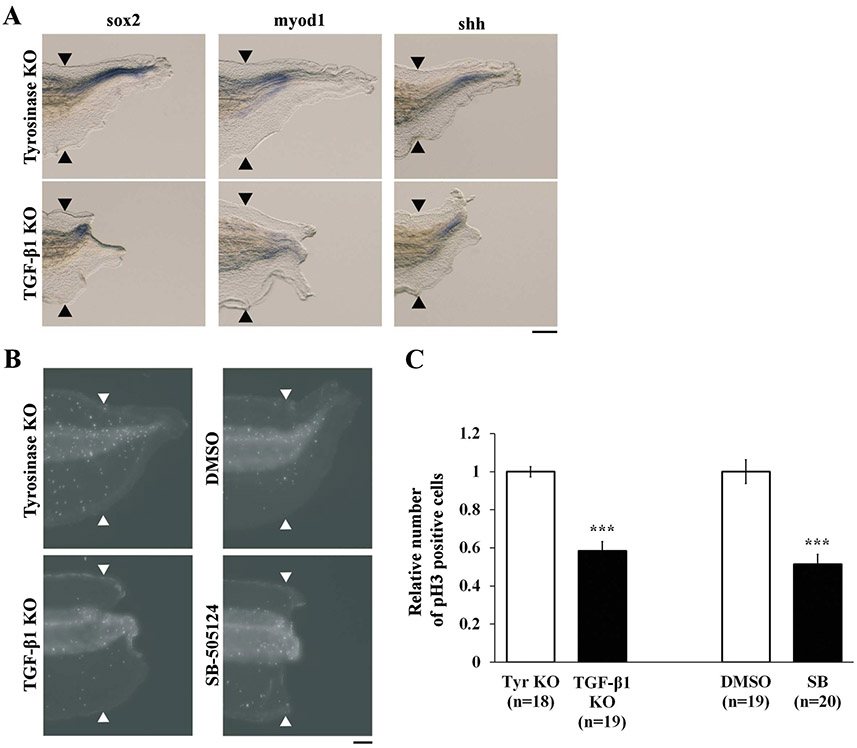
Next, we analyzed the effects of tgfb1 KO on tissue differentiation and cell proliferation in Xenopus tail regeneration. The tadpole tail is composed of several types of tissue including spinal cord, muscle, and notochord. We evaluated tissue differentiation at 72 hpa using WISH with sox2, myod1, and shh probes [10, 16, 17]. Differentiation of tail tissues was greatly reduced in tgfb1 KO tadpoles but was present in tyrosinase KO tadpoles, demonstrating that TGF-β1 is necessary for proper tissue differentiation (Fig. 3A). Since blastema cell proliferation precedes tissue differentiation [18], we performed whole-mount immunostaining for pH3 in regenerating tails at 48 hpa to examine cell proliferation in tgfb1 KO tadpoles. It has been shown that the numbers of mitotic cells in regenerating tail in Xenopus tadpoles are reduced by treatment with a TGF-β receptor inhibitor [4]. In agreement with this report, we found that cell proliferation in the regenerating tail excluding the fin was significantly downregulated by treatment with the TGF-β receptor inhibitor SB-505124. Moreover, the numbers of mitotic cells in tgfb1 KO tadpoles were significantly decreased at 48 hpa compared to tyrosinase KO tadpoles (Fig. 3B and C). These results suggest that TGF-β1 is required for cell proliferation and subsequent tissue differentiation in Xenopus tail regeneration.
Figure 3. TGF-β1 regulates tissue differentiation and cell proliferation.
(A) Lateral views of WISH performed with sox2 (spinal cord), myod1 (muscle) and shh (notochord) antisense RNA probes at 72 hpa. Scale bar, 200 μm. (B) Whole-mount immunostaining of phosphorylated Histone H3 (pH3) at 48 hpa. Scale bar, 100 μm. (C) Quantification of mitotic cells in the regenerating tail. The number of pH3 positive cells in tgfb1 KO and SB-505124-treated tadpoles was normalized against tyrosinase KO and DMSO-treated control tadpoles, respectively. Black and white arrowheads indicate amputation sites. ***P < 0.001.
TGF-β1 activates Smad2/3, which is important for cell proliferation
After tail amputation, several mediators of the early responses to injury (reactive oxygen species, mitogen-activated protein kinase, and Smad2/3) are activated in the amputation plane [4, 19, 20]. Among these mediators, pSmad2/3, a downstream signal transducer of the TGF-β receptor, is known to be essential for TGF-β-mediated wound healing and cell proliferation [4]. To investigate whether TGF-β1 is involved in the activation of Smad2/3 in Xenopus tail regeneration, we performed a whole-mount immunostaining analysis of pSmad2/3 in the regenerating tails of tgfb1 KO tadpoles. We found that pSmad2/3 expression was reduced in tgfb1 KO tadpoles at 6 hpa (Fig. 4). Collectively, the results of these analyses demonstrate that TGF-β1 controls cell proliferation through the activation of Smad2/3 in Xenopus tail regeneration.
Figure 4. TGF-β1 regulates the activation of Smad2/3.
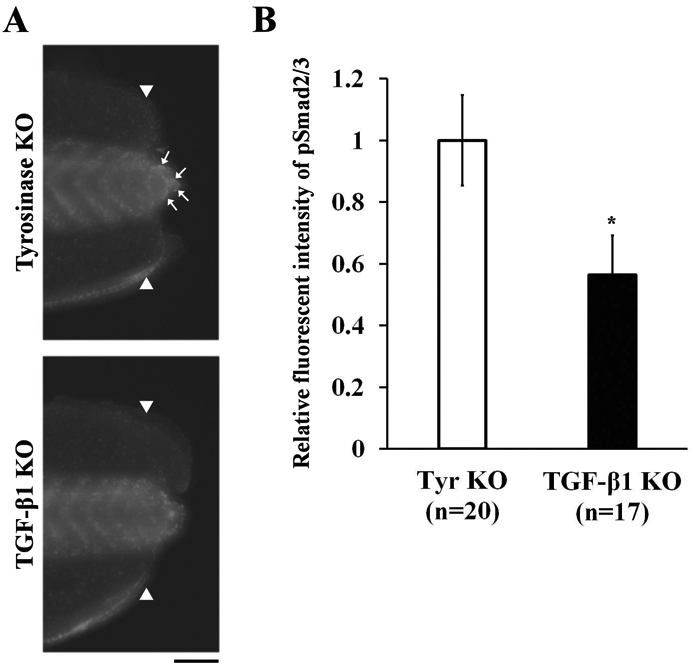
(A) Whole-mount immunostaining of phosphorylated Smad2/3 (pSmad2/3) at 6 hpa. Scale bar, 100 μm. (B) Quantitative fluorescence intensities of pSmad2/3 immunostaining in regenerating tails. The vertical axis indicates the average fluorescence intensity in regenerating tails of tgfb1 KO tadpoles normalized against tyrosinase KO control tadpoles. White arrowheads indicate amputation sites. White arrows indicate the localization of fluorescence signals in the regenerating tail tip. *P < 0.05.
Discussion
We demonstrate here that TGF-β1 is required for tissue regeneration in the X. tropicalis tadpole tail. Previous studies investigated the role of TGF-β signaling in tissue regeneration using TGF-β receptor inhibitors in regenerating tissues of several animal species [4, 5, 6, 21, 22]. However, this approach did not enable a detailed analysis of the role of TGF-β ligands in TGF-β receptor-dependent tissue regeneration. As multiple TGF-βs are expressed during tissue regeneration [4, 5, 22], it has been difficult to identify which TGF-βs are critical for regeneration. Here, we show that knockout of tgfb1 prevents cell proliferation and Smad2/3 activation, and impairs tail regeneration. Our observations clearly show that TGF-β1 is an essential TGF-β ligand for TGF-β receptor-dependent tissue regeneration in Xenopus tadpoles.
Interestingly, treatment of tadpoles with a TGF-β receptor inhibitor caused a more severe delay in tail regeneration than the tgfb1 KO (data not shown) [4]. The TGF-β receptor inhibitors SB-505124 and SB-431542 are potent inhibitors of ALK4, 5 and 7 that interact with multiple TGF-β superfamily ligands (TGF-βs, activins, and GDFs) [23, 24, 25]. It has also been reported that inhba and gdf11 are upregulated at 4 and 48 hpa, respectively, during X. laevis tail regeneration [4]. This suggests that in addition to TGF-β1, other TGF-β superfamily ligands might function in Xenopus tail regeneration.
Similar functional redundancy may also occur among TGF-βs. Both tgfb1 and tgfb2 are expressed during X. tropicalis tail regeneration (Fig. 1; data not shown) [15]. In zebrafish, expression of tgfb2 during Müller glia-mediated retinal regeneration is activated at a similar time-course as tgfb1 expression. Additionally, although administration of TGF-β1 increases the numbers of mitotic cells after retinal injury, knock-down of tgfb1 did not significantly affect glial proliferation [21, 22]. These studies and our results suggest that both TGF-β1 and TGF-β2 might contribute to tissue regeneration. Further detailed analyses will be necessary to resolve the likely redundancy of TGF-βs in tissue regeneration.
Expression of tgfb1 has been shown to gradually increase after tail amputation in X. laevis tadpoles [4]. However, the pattern of tgfb1 expression in undamaged tadpole tails is not well documented. In the present study, we showed that tgfb1 expression was present in undamaged tails (prior to tail amputation at stage 41/42) of X. tropicalis tadpoles (Fig. 1). This suggests that in addition to injury-induced expression of tgfb1, TGF-β1 protein might be stored in the ECM of undamaged tails and might contribute to the activation of Smad2/3 immediately after tail amputation. The rapid mobilization of TGF-β1 protein from the ECM may be crucial to the restoration of damaged tissues and for protection against infection, as TGF-β signaling is important not only for cell proliferation but also for the formation of wound epithelium that occurs at an early stage of tail regeneration processes [4]. Consistently, after amputation of X. laevis tails or limbs, tgfb1 is expressed in the apical epithelial cap (AEC) which is located at the distal part of the wound epithelium [26]. As described above, tgfb1 is the first of the TGF-β superfamily ligands to be expressed during Xenopus tadpole tail regeneration and thus it may function as a master regulator to orchestrate the initial responses to injury that eventually result in cell proliferation and differentiation. In addition to Xenopus tails and limbs, tgfb1 expression has been observed during regeneration of other tissues and organs (e.g., fin, heart, retina, and spinal cord) in zebrafish [5, 9, 22, 27]. Therefore, TGF-β1 may be widely involved in the regeneration of lost appendages and of damaged tissues/organs.
Until now, the contribution of TGF-β1 to tissue regeneration had not been completely revealed. Our results clearly demonstrate the essential role of TGF-β1 in the promotion of tissue regeneration through the regulation of cell proliferation activated by pSmad2/3. Thus, this study provides new insights into the molecular mechanisms of TGF-β signal-dependent tissue regeneration upon injury in animals.
Supplementary Material
Supplementary Figure 1. Genotyping of tgfb1 KO (sg 1 + sg 2 + sg 3) tadpoles.
Sequencing analysis was performed using tyrosinase KO (n = 6) and tgfb1 KO (n = 6) tadpoles. At the top, three sgRNA target sites and the PAM sequence are highlighted in blue and green, respectively. Deleted sequences are shown as black dashes, and insertions and substitutions are shown in blue and red font, respectively.
Supplementary Figure 2. Rescue experiment of the tgfb1 KO phenotype by injection of tgfb1 mRNA (A) Representative phenotypes observed in this experiment. Tadpoles were categorized into three types: normal tail regeneration, weakly delayed tail regeneration, and severely delayed tail regeneration. (B) Rescue of delayed tail regeneration in the tgfb1 KO tadpoles by injection of tgfb1 mRNA. The extent of tail regeneration was classified at 72 hpa. Black arrowheads show amputation sites. Scale bar, 200 μm.
Highlights.
tgfb1 is expressed in Xenopus tadpole tail before and after tail amputation
TGF-β1 is essential for tail regeneration
TGF-β1 regulates cell proliferation during regeneration by activation of Smad2/3
Acknowledgments
We thank the National Xenopus Resource (RRID: SCR_013731) at the Marine Biological Laboratory (MBL) for technical support with the CRISPR/Cas9-mediated mutagenesis and genotyping, the National Bio-Resource Project (NBRP) of Japan Agency for Medical Research and Development (AMED) under Grant Number JP18km0210085 for providing the X. tropicalis frogs, and Dr. Kikuchi Yutaka and his laboratory members at Hiroshima University for helpful discussions. A. S. and K. T.-S. were supported by Grant-in-Aids for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan: Japan Society for the Promotion of Science (JSPS) KAKENHI, 19K07247 and 20K07225, Japan. This work was also supported by Hiroshima University Natural Science Center for Basic Research and Development and by National Institutes of Health (NIH) P40 OD010997 and R24 OD 030008, United States, to M.E.H.
Footnotes
To the best of our knowledge, the named authors have no conflict of interest, financial or otherwise.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- [1].Phipps LS, Marshall L, Dorey K, Amaya E, Model systems for regeneration: Xenopus, Development 147 (2020) devl80844, http://doi.org/10/1242/dev.180844. [DOI] [PubMed] [Google Scholar]
- [2].Ali SR, Hippenmeyer S, Saadat LV, Luo L, Weissman IL, Ardehadi R, Existing cardiomyocytes generate cardiomyocytes at a low rate after birth in mice, Proc. Natl. Acad. Sci. U. S. A 111 (2014) 8850–8855, 10.1073/pnas.1408233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mohammadi MM, Abouissa A, Azizali I, Xie Y, Cordero J, Shirvani A, Gigina A, Engelhardt M, Trogisch FA, Geffers R, Dobreva G, Bauersachs J, Heineke J, Induction of cardiomyocyte proliferation and angiogenesis protects neonatal mice from pressure overload-associated maladaptation, JCI Insight 4 (2019) e128336, 10.1172/jci.insight.128336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ho DM, Whitman M, TGF-β signaling is required for multiple processes during Xenopus tail regeneration, Dev. Biol 315 (2008) 203–216. 10.1016/j.ydbio.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chablais F, Jazwinska A, The regenerative capacity of the zebrafish heart is dependent on TGFβ signaling, Development 139 (2012) 1921–1930, 10.1242/dev.078543. [DOI] [PubMed] [Google Scholar]
- [6].Levesque M, Gatien S, Finnson K, Desmeules S, Villiard E, Pilote M, Philip A, Roy S, Transforming growth factor: β signaling is essential for limb regeneration in axolotls, PLOS ONE 11 (2007) e1227, 10.1371/journal.pone.0001227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Weiss A, Attisano L, The TGFbeta superfamily signaling pathway, Wiley Interdiscip. Rev. Dev. Biol 2 (2013) 47–63, 10.1002/wdev.86. [DOI] [PubMed] [Google Scholar]
- [8].Santibanez JF, Obradovic H, Kukolj T, Krstic J, Transforming growth factor-β, matrix metalloproteinases, and urokinase-type plasminogen activator interaction in the cancer epithelial to mesenchymal transition, Dev. Dyn 247 (2018) 382–395, 10.1002/dvdy.2455. [DOI] [PubMed] [Google Scholar]
- [9].Jazwinska A, Badakov R, Keating MT, Activin-βA signaling is required for zebrafish fin regeneration, Curr. Biol 17 (2007) 1390–1395, 10.1016/j.cub.2007.07.019. [DOI] [PubMed] [Google Scholar]
- [10].Nakamura M, Yoshida H, Takahashi E, Wlizla M, Takebayashi-Suzuki K, Horb ME, Suzuki A, The AP-1 transcription factor JunB functions in Xenopus tail regeneration by positively regulating cell proliferation, Biochem. Biophys. Res. Commun 522 (2020) 990–995, 10.1016/j.bbrc.2019.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Square TA, Jandzik D, Massey JL, Romasek M, Stein HP, Hansen AW, Purkayastha A, Cattell MV, Medeiros DM, Evolution of the endothelin pathway drove neural crest cell diversification, Nature 585 (2020) 563–568, 10.1038/s41586-020-2720-z. [DOI] [PubMed] [Google Scholar]
- [12].Harland RM, In situ hybridization: an improved whole-mount method for Xenopus embryos, Methods Cell Biol. 36 (1991) 685–695, 10.1016/S0091-679X(08)60307-6. [DOI] [PubMed] [Google Scholar]
- [13].Takebayashi-Suzuki K, Kitayama A, Terasaka-Iioka C, Ueno N, Suzuki A, The forkhead transcription factor FoxB1 regulates the dorsal-ventral and anterior-posterior patterning of the ectoderm during early Xenopus embryogenesis, Dev. Biol. 360 (2011) 11–29, 10.1016/j.ydbio.2011.09.005. [DOI] [PubMed] [Google Scholar]
- [14].Nakayama T, Blitz IL, Fish MB, Odeleye AO, Manohar S, Cho KWY, Grainger RM, Cas9-based genome editing in Xenopus tropicalis, Methods Enzymol. 546 (2014) 355–375, 10.1016/B978-0-12-801185-0.00017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chang J, Baker J, Wills A, Transcriptional dynamics of tail regeneration in Xenopus tropicalis, Genesis 55 (2017) e23015, 10.1002/dvg.23015. [DOI] [PubMed] [Google Scholar]
- [16].Sugiura T, Taniguchi Y, Tazaki A, Ueno N, Watanabe K, Mochii M, Differential gene expression between the embryonic tail bud and regenerating larval tail in Xenopus laevis, Devlop. Growth Differ. 46 (2004) 97–105, 10.1111/j.1440-169X.2004.00727.x. [DOI] [PubMed] [Google Scholar]
- [17].Taniguchi Y, Watanabe K, Mochii M, Notochord-derived hedgehog is essential for tail regeneration in Xenopus tadpole, BMC Dev. Biol 14 (2014) 27, 10.1186/1471-213X-14-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Love NR, Chen Y, Bonev B, Gilchrist MJ, Fairclough L, Lea R, Mohun TJ, Paredes R, Zeef LA, Amaya E, Genome-wide analysis of gene expression during Xenopus tropicalis tadpole tail regeneration, BMC Dev. Biol 11 (2011) 70, 10.1186/1471-213X-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Love NR, Chen Y, Ishibashi S, Kritsiligkou P, Lea R, Koh Y, Gallop JL, Dorey K, Amaya E, Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration, Nat. Cell Biol 15 (2013) 222–229, 10.1038/ncb2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sato K, Umesono Y, Mochii M, A transgenic reporter under control of an es1 promoter/enhancer marks wound epidermis and apical epithelial cap during tail regeneration in Xenopus laevis tadpole, Dev. Biol 433 (2018) 404–415, 10.1016/j.ydbio.2017.08.012. [DOI] [PubMed] [Google Scholar]
- [21].Sharma P, Gupta S, Chaudhary M, Mitra S, Chawla B, Khursheed MA, Saran NK, Ramachandran R, Biphasic role of Tgf-β signaling during müller glia reprogramming and retinal regeneration in zebrafish, iScience 23 (2020) 100817, 10.1016/j.isci.2019.100817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lee MS, Wan J, Goldman D, Tgfb3 collaborates with PP2A and notch signaling pathways to inhibits retina regeneration, eLife 9 (2020) e55137, 10.7554/eLife.55137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Byfield SD, Major C, Laping NJ, Roberts AB, SB-505124 is a selective inhibitor of transforming growth factor-β type I receptors ALK4, ALK5, and ALK7, Mol. Pharmacol 65 (2004) 744–752, 10.1124/mol.65.3.744. [DOI] [PubMed] [Google Scholar]
- [24].Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS, SB-431542 is a potent and specific inhibitor of transforming growth factor-β superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7, Mol. Pharmacol 62 (2002) 65–74, 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- [25].Laping NJ, Grygielko E, Mathur A, Butter S, Bomberger J, Tweed C, Martin W, Fornwald J, Lehr R, Harling J, Gaster L, Callahan JF, Olson BA, Inhibition of transforming growth factor (TGF)-β1–induced extracellular matrix with a novel inhibitor of the TGF-β type I receptor kinase activity: SB-431542, Mol. Pharmacol 62 (2002) 58–64, 10.1124/mol.62.1.58. [DOI] [PubMed] [Google Scholar]
- [26].Okumura A, Hayashi T, Ebisawa M, Yoshimura M, Sasagawa Y, Nikaido I, Umesono Y, Mochii M, Cell type-specific transcriptome analysis unveils secreted signaling molecule genes expressed in apical epithelial cap during appendage regeneration, Develop. Growth Differ. 61 (2019) 447–456, 10.1111/dgd.12635. [DOI] [PubMed] [Google Scholar]
- [27].Hui SP, Sengupta D, Lee SGP, Sen T, Kundu S, Mathavan S, Ghosh S, Genome wide expression profiling during spinal cord regeneration identifies comprehensive cellular responses in zebrafish, PLOS ONE 9 (2014) e84212, 10.1371/journal.pone.0084212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Genotyping of tgfb1 KO (sg 1 + sg 2 + sg 3) tadpoles.
Sequencing analysis was performed using tyrosinase KO (n = 6) and tgfb1 KO (n = 6) tadpoles. At the top, three sgRNA target sites and the PAM sequence are highlighted in blue and green, respectively. Deleted sequences are shown as black dashes, and insertions and substitutions are shown in blue and red font, respectively.
Supplementary Figure 2. Rescue experiment of the tgfb1 KO phenotype by injection of tgfb1 mRNA (A) Representative phenotypes observed in this experiment. Tadpoles were categorized into three types: normal tail regeneration, weakly delayed tail regeneration, and severely delayed tail regeneration. (B) Rescue of delayed tail regeneration in the tgfb1 KO tadpoles by injection of tgfb1 mRNA. The extent of tail regeneration was classified at 72 hpa. Black arrowheads show amputation sites. Scale bar, 200 μm.