Abstract
Pancreatic cancer is an increasingly common cause of cancer mortality with a tight correspondence between disease mortality and incidence. Furthermore, it is usually diagnosed at an advanced stage with a very dismal prognosis. Due to the high heterogeneity, metabolic reprogramming, and dense stromal environment associated with pancreatic cancer, patients benefit little from current conventional therapy. Recent insight into the biology and genetics of pancreatic cancer has supported its molecular classification, thus expanding clinical therapeutic options. In this review, we summarize how the biological features of pancreatic cancer and its metabolic reprogramming as well as the tumor microenvironment regulate its development and progression. We further discuss potential biomarkers for pancreatic cancer diagnosis, prediction, and surveillance based on novel liquid biopsies. We also outline recent advances in defining pancreatic cancer subtypes and subtype-specific therapeutic responses and current preclinical therapeutic models. Finally, we discuss prospects and challenges in the clinical development of pancreatic cancer therapeutics.
Subject terms: Cancer metabolism, Cancer microenvironment
Introduction
Pancreatic cancer, with one of the highest mortality rates of all malignancies, is the seventh leading cause of cancer-related death worldwide.1 With almost as many deaths (n = 466,000) as new cases (n = 496,000) according to GLOBOCAN 2020 estimates,1 pancreatic cancer has become the third leading cause of cancer-related death in the United States.2 Despite improvements in surgical techniques and therapy regimens, the 5-year survival rate for pancreatic cancer is still the lowest (9%) of any cancer type.3,4
Approximately 90% of pancreatic cancers are characterized as pancreatic ductal adenocarcinoma (PDAC). Risk factors associated with pancreatic cancer include familial risk due to susceptibility gene mutations, chronic pancreatitis, pancreatic cysts, and diabetes mellitus.5,6 Other risk factors include smoking, alcohol abuse, obesity or metabolic syndrome, aging, and occupational exposure.4,7 Approximately 80% of pancreatic cancer patients present with advanced disease or distant metastases and have no effective treatment options.8 Even early-stage patients eligible for resection have a 5-year survival rate of <31%.9 Advances in next-generation genome sequencing (NGS) have brought new excitement to the field by supporting the identification of molecular alterations that govern pancreatic cancer progression. Sequencing data have revealed that pancreatic cancer comprises highly heterogeneous tumors that develop resistance to traditional chemotherapy and radiation therapy.
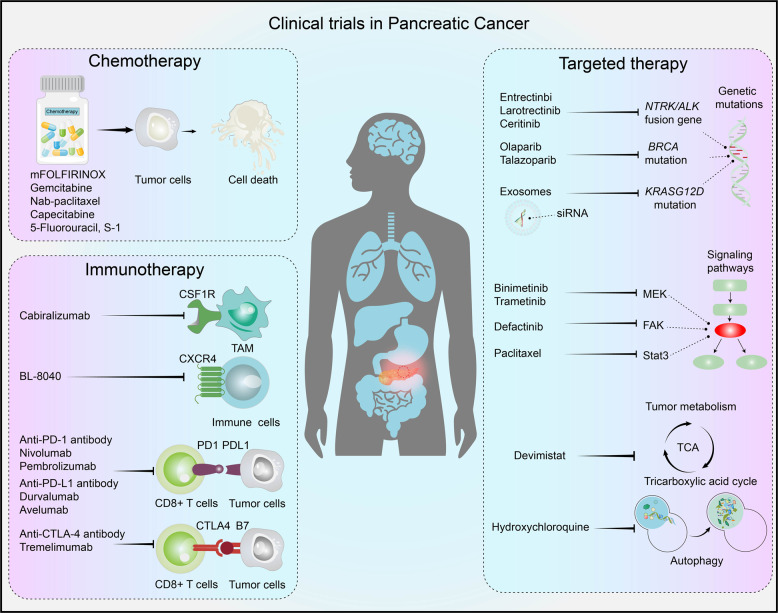
Gemcitabine has been a reference first-line therapeutic drug for pancreatic cancer patients since 1997.10 However, traditional chemotherapy and radiation therapy have not shown improved 5-year survival rates.11,12 Modified FOLFIRINOX (mFOLFIRINOX) (i.e., 5-fluorouracil (5-FU), leucovorin, oxaliplatin, and irinotecan) and nab-paclitaxel plus gemcitabine are generally considered as the best adjuvant chemotherapy regimens, although they have shown only modest survival benefits with considerable toxicity.13 Furthermore, multiple novel-targeted therapies (i.e., cetuximab, bevacizumab, axitinib, and aflibercept) have failed to effectively improve overall survival (OS), while erlotinib combined with gemcitabine showed a marginal clinical benefit.14,15 Therefore, new therapies, e.g., innovative immunotherapies and combination therapies with increased antitumor potency, are urgently needed.
In this review, we first provide a general discussion of pancreatic cancer. This is followed by a review of novel biomarkers for their diagnosis, treatment monitoring, and prognosis based on liquid biopsy. We conclude with a discussion of the clinical perspectives of current advances. We anticipate that novel therapeutic methods and strategies based on subtype-specific therapy and preclinical therapeutic models might have a great impact on pancreatic cancer treatment and management, thereby improving patient outcomes.
Characteristics of pancreatic cancer
Genetics and epigenetics of pancreatic cancer
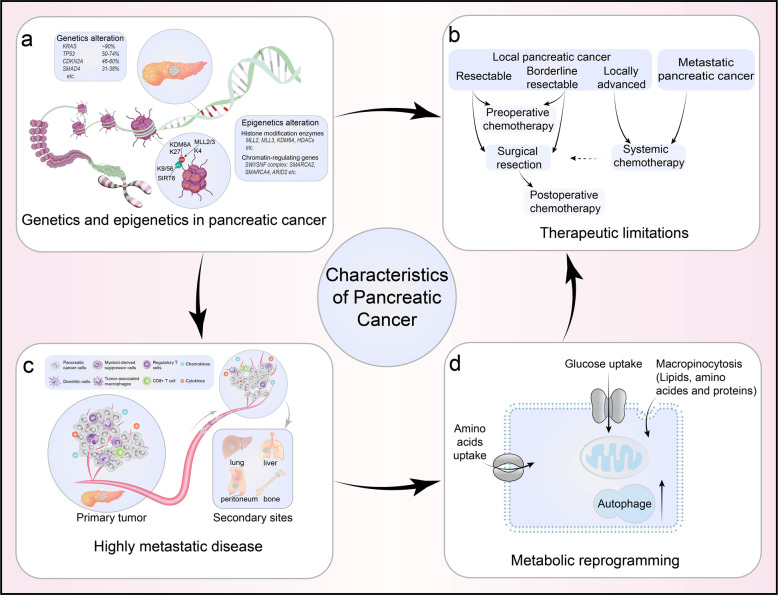
Via the use of NGS and computational biology, we have gained a deeper understanding of the genetic alterations underlying the genesis and progression of pancreatic cancer, including gene expression changes, copy number aberrations, chromosomal rearrangements, and epigenetic alterations (Fig. 1a). Pancreatic cancer typically starts as a precancerous lesion, i.e., a pancreatic intraepithelial neoplasia (PanIN), which accumulates gene mutations over time, ultimately progressing to a more dysplastic state.16–18 Approximately 90% of pancreatic cancers of all grades have activating oncogenic Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations. Among the oncogenic KRAS mutations associated with pancreatic cancer in humans, the GAT (aspartic acid; G12D), GTT (valine; G12V), and TGT (cysteine; G12C) mutations are the most common, while the CGT (arginine (Arg); G12R) and GCT (alanine; G12A) mutations as well as other point mutations at codons 11, 13, 61, or 146 appear to be less common.17,18 As the most common activating mutations in pancreatic cancer, KRAS mutations (which have also been investigated in depth in various other cancers, including metastatic colorectal cancer and non-small cell lung cancer) impair the intrinsic GTPase activity of KRAS and prevent GTPase-activating proteins (GAPs) from converting the active GTP-bound form to the GDP-bound inactive form. Clinical studies have also shown that KRAS mutations could be considered as a marker for the poor prognosis of pancreatic cancer.17 However, it has been reported that various KRAS mutations affect diverse signaling pathways, leading to distinct functional consequences. Pancreatic cancer patients with KRAS at codon 61 mutations exhibit lower extracellular signal-regulated kinase (ERK) activation compared with patients with other KRAS alleles, and the former harbor significantly better prognosis.19–21 It is evident that KRAS mutation seems to be necessary but not sufficient for pancreatic cancer development. Other genes, including tumor protein p53 (TP53), cyclin-dependent kinase inhibitor 2A (CDKN2A), and SMAD family member 4 (SMAD4), are also frequently involved in pancreatic cancer tumorigenesis and metastasis.22 Approximately 50–74% of pancreatic cancers have inactivating mutations in the tumor suppressor gene TP53.16,23 TP53 inactivation impairs DNA damage recognition and blocks cell cycle arrest, allowing cells to bypass cell cycle checkpoints and evade apoptotic signals.16,23 Mutations in CDKN2A, which can lead to loss of regulation of the cyclin-dependent kinase (CDK) 4 and CDK6 cell cycle checkpoints and, thus, dysregulation of the cell cycle and subsequent carcinogenesis, are detected in ~46–60% of pancreatic cancers.16,23,24 In addition, ~31–38% of pancreatic cancers have mutations in SMAD4, which can occur in the late stages of pancreatic carcinogenesis.23,25 In pancreatic cancer, the frequent loss of SMAD4 via homozygous deletion or mutation leads to decreasing SMAD4-dependent inhibition of transforming growth factor-β (TGF-β) and the promotion of noncanonical TGF-β signaling, thereby facilitating pro-tumorigenic responses.23,26,27 Recently, high-throughput sequencing studies have revealed genes with novel mutations/alterations with individual frequencies <20%, including lysine demethylase 6A (KDM6A) (18%), rac family small GTPase 1 (RAC1) (10%), ring-finger protein 43 (RNF43) (10%), AT-rich interaction domain 1A (ARID1A) (9%), B-Raf proto-oncogene, serine/threonine kinase (BRAF) (3%), TGF-β receptor 2 (TGFBR2) (3%), mitogen-activated protein kinase kinase kinase 21 (MAP3K21) (3%), switch/sucrose nonfermentable (SWI/SNF)-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 4 (SMARCA4) (3%), activin A receptor type 2A (ACVR2A) (2%), activin A receptor type 1B (ACVR1B) (2%), N-ras proto-oncogene, GTPase (NRAS) (1%), family with sequence similarity 133 member A (FAM133A) (<1%), and zinc-finger matrin-type 2 (ZMAT2) (<1%).24,28,29 It has been reported that 4–7% of pancreatic cancer patients have a germline DNA repair associated (BRCA) mutation.30 Furthermore, germline BRCA2 mutations have been observed in 5–17% of familial pancreatic cancer patients.31 Mutation of the GNAS complex locus (GNAS) at codon 201 has been observed exclusively in intraductal papillary mucinous neoplasms (IPMNs), a precursor to pancreatic cancer, at a rate of 41–75%.32 However, several studies have reported that ~4% of pancreatic cancer patients have GNAS mutations.24 Most of these low-frequency mutations are in genes related to cellular processes such as cell survival, cell fate determination, and genome maintenance, involving various signaling pathways.28,33 In addition, recurrent noncoding mutations, which are enriched in transcriptionally active regions of the genome, also play an essential role in pancreatic cancer.34
Fig. 1.
The characteristics of pancreatic adenocarcinoma. Pancreatic cancer is a common cause of cancer mortality characterized by high heterogeneity, a dense stromal tumor microenvironment (TME), highly metastatic propensity, metabolic reprogramming, and limited benefits from current conventional therapies. a Genetic and epigenetic changes in pancreatic cancer. KRAS (~90%), TP53 (50–74%), CDKN2A (46–60%), and SMAD4 (31–38%) are the most frequently mutated genes known to modulate the initiation and progression of pancreatic cancer. Epigenetic regulatory genes, including MLL2/3, KDM6A, and multiple HDACs encoding genes, regulate histone modification. SMARCA2/4 and ARID2 modulate chromatin remodeling. b Therapeutic limitations in pancreatic cancer. Surgical resection is the only potentially curative choice for pancreatic cancer patients. Adjuvant chemotherapy can only partially improve the overall survival of pancreatic cancer patients c Pancreatic cancer is an extremely aggressive tumor with high metastatic propensity. The immunosuppressive TME plays an important role in modulating the metastasis of pancreatic cancer cells to the liver, lungs, peritoneum, and bone. d Metabolic reprogramming of pancreatic cancer. Pancreatic cancer cells can survive and proliferate in stressful microenvironments by reprogramming energy metabolism to increase glucose and glutamine uptake, macropinocytosis, and autophagy
Epigenetic alterations, which can regulate histone or DNA modification, play significant roles in molecular aspects of pancreatic tumorigenesis.35 In pancreatic cancer, epigenetic regulatory genes are frequently mutated, including those encoding SWI/SNF-mediated chromatin remodeling complexes and histone modification enzymes.36 Commonly mutated histone modification enzymes include the mixed-lineage leukemia (MLL) histone methylases (MLL2 and MLL3), histone methyltransferases, and the KDM6A histone demethylase.25,37 Furthermore, MLL2/3 and KDM6A exist in the same complex, which drives transcriptional activation through coordinated regulation of histone 3 lysine 4 (H3K4) methylation and histone 3 lysine 27 (H3K27) demethylation.38 During pancreatic cancer development, tumors with genetic defects in MLLs are more likely to induce expression of chromatin-regulating genes and cell proliferation-associated genes (including members of the SWI/SNF chromatin remodeling complex) as well as genes involved in cell cycle progression and cellular proliferation.37 The SWI/SNF complexes comprise multiple components encoded by different genes, including SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 2 (SMARCA2), SMARCA4, SMARCE1, SMARCB1, AT-rich interaction domain 2 (ARID2), ARID1A, AT-rich interaction domain 1B (ARID1B), and double PHD fingers 2 (DPF2). The overall prevalence of gene mutations or copy number variations in SWI/SNF complex components is >10%; thus, such alterations have important effects on chromatin remolding in pancreatic cancer pathogenesis.20,28,36 Histone deacetylases (HDACs), which catalyze histone deacetylation, are classified into two enzyme subtypes: the NAD+-dependent sirtuins (SIRT1–7) and the zinc-dependent HDAC1–11 proteins.39 Interestingly, HDAC1 and HDAC2 are highly expressed in pancreatic cancer, and they can be recruited to the epithelial-cadherin (CDH1) promoter (which is involved in histone deacetylation) by zinc-finger E-box binding homeobox 1 (ZEB1) or Snail, thereby promoting epithelial-to-mesenchymal transition (EMT) and tumor metastasis.40,41 4SC-202, a specific inhibitor of class I-HDACs, can inhibit TGF-β-induced EMT and p21 cyclin-dependent kinase inhibitor 1A (CDKN1A)-mediated cell proliferation.42 SIRT6 ablation can potently cooperate with activated KRAS to promote pancreatic cancer metastasis via the hyperacetylation of histone 3 lysine 9 (H3K9) and histone 3 lysine 56 (H3K56) at the promoter of the lin-28 homolog B (LIN28b) gene, resulting in Myc recruitment and Lin28b upregulation. As a result, the expression levels of high-mobility group AT-hook 2 (HMGA2), insulin-like growth factor 2 mRNA-binding protein (IGF2BP) 1, and IGF2BP3, which are downstream of target genes let-7, are increased.43
Subtyping of pancreatic cancer
Pancreatic cancer is a deadly malignancy that lacks effective therapeutics (Fig. 1b). This is in part due to the inter- and intratumor heterogeneity.44–46 Pancreatic cancer molecular subtype identification has the potential to improve clinical outcomes by allowing the development of individualized treatments. Recently, various molecular subtypes and subtype-specific treatment responses in pancreatic cancer have been recognized.36,44,47 The two predominant transcriptomic-based subtypes, which have been validated across multiple studies, are the classical/pancreatic progenitor subtype and the basal-like/squamous/quasi-mesenchymal (QM-PDA) subtype.44,46,47 On the other hand, the existence of the exocrine-like/aberrantly differentiated endocrine exocrine (ADEX) subtype and the immunogenic subtype, which were proposed by Collisson et al. and Bailey and co-workers, have been controversial.44,47,48 Due to the desmoplastic nature of pancreatic cancer, low tumor cellularity remains a major concern in sampling and defining the molecular subtypes of this disease. Collisson et al. identified classical and QM-PDA subtypes within a large panel of pancreatic cancer cell lines, but did not detect an exocrine-like subtype. The Cancer Genome Atlas (TCGA) project found that the ADEX subtype and immunogenic subtype were associated with low cellularity, likely representing non-transformed cells.49 Later, Puleo et al. confirmed that the ADEX tumor subtype resulted from contamination of their pancreatic exocrine and endocrine components with adjacent normal pancreatic acinar cells.48 However, Noll et al. and Knudsen et al. identified the exocrine subtype in their patient-derived pancreatic cancer models.50,51 In addition, the exocrine subtype showed resistance to tyrosine kinase inhibitors and paclitaxel treatment via the expression of cytochrome P450 family 3 subfamily A member 5 (CYP3A5).50 A recent study reported significant variation in the tissue composition of pancreatic tumors from the Moffitt, Bailey, and TCGA cohorts. Up to 45% of the tumor samples from various public cohorts were reclassified after removal of the non-epithelial signal from the tumor bulk expression profiles.52
Signaling pathways regulating pancreatic cancer tumorigenesis and metastasis
Signaling pathways (e.g., RAS, phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT), nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), janus kinase (JAK)/signal transducer and activator of transcription (STAT), Hippo/yes-kinase-associated-protein (YAP), Wingless/int1 (WNT), etc.) have been linked to a variety of cancer-related cellular processes, including cell proliferation, differentiation, apoptosis, migration, angiogenesis, metabolism, and immune regulation. They have been implicated in pancreatic cancer tumorigenesis, prognosis, and resistance to therapy, it is likely that a deep understanding of these pathways will support the development of molecularly targeted pancreatic cancer therapies.
RAS pathway
RAS signaling is centered on the activated small GTPase RAS, which drives the activation of three major effector pathways, RAS-like proto-oncogene A/B, RAF/MAP kinase (MEK)/ERK, and PI3K. Oncogenic activation of RAS due to missense mutations is frequently detected in several types of cancer, including pancreatic cancer.53 Given that mutations in one RAS protein isoform, KRAS, are found in nearly 90% of pancreatic cancers, RAS signaling appears to play a critical role in both pancreatic cancer initiation and maintenance. Activated RAS can activate effector signaling pathways and transcription factors involved in cell transformation, proliferation, and metastasis. Activated RAS also promotes pro-inflammatory signaling via activation of NF-κB, signal transducer and STAT3, and glycogen synthase kinase-3/nuclear factor of activated T cell signaling.54–56 Pancreatic cancers without KRAS mutations show RAS activation via upstream signaling through receptor tyrosine kinases (RTKs), e.g., epidermal growth factor receptor (EGFR), and oncogenic activation of the downstream B-Raf proto-oncogene (B-RAF) molecule is detected in a small number of patients.57 Despite extensive insight into the molecular mechanisms of KRAS in multiple cancer-promoting effects, it has been challenging to obtain clinically effective KRAS inhibitors, with the exception of a KRAS G12C (carried by ~1.5% of pancreatic cancer patients)-selective inhibitor AMG 510.58 In mouse models, KRAS inhibition can cause the activation of AKT, erb-b2 RTK2 (HER2), platelet-derived growth factor receptor alpha, and EGFR, which might explain the inefficacy of these inhibitors.28
PI3K/AKT pathway
Since PI3K can be activated through an initial phosphorylation in response to oncogenic RAS, the PI3K/AKT pathway is frequently activated in human pancreatic cancer and mouse models of KRAS-driven pancreatic cancer.28,59 Abnormal overexpression or activation of AKT is associated with >40% of pancreatic cancer patients. Abnormal AKT overexpression or activation was also reported to be associated with short survival and the pathologic grade of pancreatic cancer.60,61 It was demonstrated that PI3K/AKT activation appears to occur at the earliest stages of tumor evolution and that it controls pancreatic cell plasticity and carcinogenesis, as strong activation of PI3K signaling was observed in human acinar–ductal metaplasia (ADM) and in both low- and high-grade PanIN.62 Interestingly, PI3K p110α is required for pancreatic cell plasticity and cancer initiation induced by oncogenic KRAS,63 and constitutively active AKT1 cooperates with active KRAS (G12D) in the AKT pathway to accelerate pancreatic tumor onset and progression.64 Insulin-like growth factor (IGF) has been demonstrated to enhance the invasion and proliferation of pancreatic cancer cells via activation of the PI3K/AKT signaling pathway.65 Our recent study demonstrated that the PI3K/AKT/mTOR pathway regulates enolase 2 (ENO2) K394 deacetylation upon IGF-1 stimulation, thereby promoting liver metastasis in pancreatic cancer.66 Abnormal overexpression of IGF2BP2 caused by genomic amplification or posttranscriptional regulation promotes pancreatic cancer proliferation by activating the PI3K/AKT signaling pathway.67 In addition, multiple noncoding RNAs (ncRNAs), including miR-107, lncRNA ABHD11-AS1, lncRNA SNHG1, lncRNA AB209630, and transmembrane protein 158 have been reported to govern tumorigenesis, progression, metastasis, apoptosis, or drug resistance in pancreatic cancer cells via the PI3K/AKT pathway.68,69 Currently, several PI3K/AKT inhibitors are under investigation, but none are in routine clinical use. Some clinical trials of PI3K/AKT inhibitors (alone or in combination with chemotherapy drugs) in pancreatic cancer patients might reveal promising therapeutic effects.
NF-κB pathway
Constitutive activation of NF-κB, a transcription factor that is essential for inflammatory responses, has been frequently observed in pancreatic cancer. Increasing evidence suggests that both canonical and noncanonical NF-κB pathways can affect pancreatic cancer progression, metastasis, and drug resistance.70,71 KRAS mutation as well as oncogenic mutations in other genes, e.g., EGFR, PI3K, and TP53, also contribute to NF-κB activation in pancreatic cancer.72 Elevated levels of inflammatory cytokines and chemokines are observed in pancreatic cancer cells, and these increased levels are generally associated with the enhanced NF-κB signaling.73 For example, activated NF-κB translocates into the nucleus to enhance transcription of downstream inflammatory target genes, including interleukin-6 (IL-6), IL-8, and IL-18, and increased levels of these cytokines lead to the activation of NF-κB signaling, thereby forming a positive feedback loop.74,75 Several studies have revealed that ncRNAs regulate the NF-κB signaling pathway by directly interacting with the functional domain or its transcripts. Overexpression of lncRNA-PLACT1 promotes pancreatic cancer progression via constitutive activation of NF-κB signaling. MiR-628-5p functions as a tumor-suppressive microRNA (miRNA) in pancreatic cancer by negatively regulating phospholipid scramblase 1 and insulin receptor substrate 1 expression, which inhibits NF-κB signaling.76 Furthermore, NF-κB is reported to be involved in antitumor immunity. NF-κB in pancreatic stromal cells (PSCs) contributes to tumor growth by increasing the expression of C–X–C motif chemokine ligand 12 (CXCL12), which prevents cytotoxic T cells from infiltrating the tumor and killing cancer cells.77 NF-κB is required for pancreatic cancer cells to evade macrophage surveillance, and growth differentiation factor 15-mediated inhibition of NF-κB signaling in infiltrating macrophages blocks the antitumor immune response during the early stages of tumorigenesis.78 A variety of NF-κB pathway inhibitors, including small molecules, peptides, small DNA/RNAs, viral proteins, and natural compounds, despite not being tested specifically in a pancreatic cancer model, have shown great promise.79
JAK/STAT pathway
The JAK/STAT pathway is clearly involved in many types of human cancer, including pancreatic cancer. High JAK2 expression predicts a poor prognosis in patients with PDAC.80 Several studies have shown that the JAK/STAT pathway is involved in inflammatory processes in pancreatic cancer. Both type 1 (i.e., IFNα and IFNβ) and type 2 (i.e., IFNγ) interferons can upregulate programmed cell death 1-ligand 1 (PD-L1) expression via the JAK-STAT signaling pathway in pancreatic cancer.81,82 Sustained JAK/STAT pathway activation mediates chronic inflammation and impairs cytotoxic T lymphocyte (CTL) activation in pancreatic cancer, and the JAK-STAT inhibitor ruxolitinib can increase CTL infiltration to induce a Tc1/Th1 immune response in the tumor microenvironment of pancreatic cancer.83 In the PDAC microenvironment, it was demonstrated that tumor-derived IL-1 induces LIF expression and downstream JAK/STAT activation to generate inflammatory cancer-associated fibroblasts (iCAFs), whereas tumor-derived TGF-β antagonizes this process by downregulating IL-1 receptor type 1 expression and promoting differentiation into myofibroblasts (myCAFs).84
Hippo/YAP pathway
YAP and its transcriptional coactivator with PDZ-binding motif (TAZ) are the two major downstream effectors of the Hippo pathway.85 Several studies have revealed that YAP is upregulated in pancreatic cancer patients, and YAP overexpression is correlated with liver metastasis and poor prognosis of pancreatic cancer.86 YAP has been reported to be insufficient to drive an initial step in the progression to pancreatic cancer with ADM. However, YAP is required for the induction of ADM progression to PanIN, and PanIN progression to pancreatic cancer.87,88 Accumulating studies have demonstrated that YAP is a critical player in pancreatic cancer progression in KRAS mutant mice.88 Active YAP promotes pancreatic tumor tumorigenesis, development, metastasis, stromal response, drug resistance, and metabolic homeostasis in KRAS-driven pancreatic cancer.89,90 However, it has been demonstrated that YAP is sufficient to drive PDAC recurrence in the absence of KRAS via bypass mechanisms involving YAP.91 Recent studies have also demonstrated that YAP is a major driver of the squamous subtype of pancreatic cancer, which is notably less dependent on oncogenic KRAS.92 These findings indicate that YAP not only acts as a pancreatic cancer driver downstream of KRAS but that it also substitutes for loss of oncogenic KRAS.93 Furthermore, YAP was identified as a critical regulator of the immunosuppressive microenvironment. YAP and TAZ can modulate the behavior of pancreatic stellate cells (PSCs) and influence the recruitment of tumor-associated macrophages (TAMs) and myeloid-derived suppressor cells. For example, YAP has been shown to promote the expression and secretion of multiple cytokines and chemokines, which in turn promote the differentiation and accumulation of myeloid-derived suppressor cells (MDSCs) in pancreatic cancer.94–96
WNT pathway
The WNT signaling pathway includes the canonical and noncanonical pathways and controls the maintenance of somatic stem cells in many tissues. Both the canonical (β-catenin-dependent) and noncanonical (β-catenin-independent) pathways have been implicated in pancreatic carcinogenesis, tumor progression, and therapeutic resistance.97,98 WNT signaling is required for KRAS-induced PanIN lesions and pancreatic cancer formation.99 KRAS activation can promote the invasion and migration of pancreatic cancer cells by regulating the WNT/β-catenin signaling pathway or by increasing the interaction between β-catenin and cyclic AMP-response element-binding protein-binding protein.100 Increased WNT/β-catenin signaling activation results in an enhanced stem cell-like phenotype of pancreatic cancer.101 Activation of the canonical pathway can prevent β-catenin degradation and promote its nuclear translocation, which enhances the transcription of targeted genes such as cyclin D1 and c-Myc.102,103 Aberrant nuclear accumulation of β-catenin is frequently found in PanIN and pancreatic cancer and is associated with their development. Canonical WNT ligands, such as Wnt family member 2, Wnt family member 5A, and Wnt family member 7A, have been observed to be increased in pancreatic cancer tissues, and activated the WNT pathway, leading to the progression of pancreatic cancer.104,105 In addition, noncanonical ligands, such as mucin (MUC) family members (MUC1 and MCU4) and R-spondin, activate the WNT pathway, leading to pancreatic cancer progression.106–108 Pancreatic cancer is characterized by hypoxic conditions. Hypoxia-inducible factor-2α (HIF-2α) modulates WNT signaling by maintaining the levels of both SMAD4 and β-catenin during PanIN progression.109 Moreover, hypoxic conditions in pancreatic tumors stabilize HIF-2α, which interacts with β-catenin, leading to elevated canonical WNT/β-catenin activity while favoring tumor progression.110
High invasion and metastasis of pancreatic cancer
Pancreatic cancer is generally considered an extremely aggressive tumor with high metastatic propensity. Most patients are diagnosed with advanced metastatic disease with a dismal prognosis. A better understanding of the mechanisms underlying metastatic progression is required to develop improved therapeutic interventions.111
It has been demonstrated that metastasis can occur during the early stages of pancreatic cancer, even before large mass formation by the primary tumor112,113 (Fig. 1c). It has been conceptualized that a stepwise accumulation of genetic and epigenetic alterations is the driving force for this process.114 Previous efforts to identify a signature of prometastatic genes or a consistent pattern of metastasis-driving mutations via genomic sequencing of human primary pancreatic tumors and their metastatic sites have failed.115 Nevertheless, distant metastatic lesions are hypothesized to evolve from the original tumor cells by acquiring distinct gene mutations.116
To metastasize, neoplastic cells must detach from the primary tumor and travel through the blood vessels or lymphatic system. A number of ncRNAs,117,118 transcription factors (e.g., Kruppel-like factor 4 (KLF4),119 KLF5,120 KLF7121), growth factors (e.g., vascular endothelial growth factor (VEGF)122), and oxygen conditions123 have been reported to drive pancreatic cancer metastasis. Using an organoid culture system, Roe et al. studied how transcription and the enhancer landscape evolves during the metastatic transition in a pancreatic cancer mouse model. They found that forkhead box A1-dependent enhancer reprogramming can promote the acquisition of metastatic traits.124 In pancreatic cancer, the most common metastatic site is the liver, followed by the lungs, peritoneum, and bones.125–127 Recently, Lee et al. reported that activation of STAT3 in hepatocytes and the subsequent production of serum amyloid A1 and A2 in the bloodstream direct pancreatic cancer liver metastasis.128 Interestingly, lineage-tracing analysis in a mouse model of pancreatic cancer revealed that metastases in the lung and liver are monoclonal, whereas those in the peritoneum and diaphragm tend to be polyclonal. These findings indicate that clonal diversity depends on the metastatic site.129 Global gene expression and molecular profiling studies showed that cytokines (e.g., IL-8), growth factors (e.g., hepatocyte growth factor and VEGF), and matrix metalloproteinase (e.g., extracellular matrix (ECM) protease)122,130,131 as well as membrane receptors were key differentiating factors underlying pancreatic cancer-derived hematogenous metastasis vs. peritoneal dissemination.127 In pancreatic cancer, lymph node metastasis is considered a critical risk factor in patients with high-risk features.132 By using data and tumor samples from three independent cohorts, a recent study identified a miRNA signature associated with pancreatic cancer patients at risk for lymph node metastasis.133 Additional evidence has demonstrated that extracellular vesicles (EVs), such as exosomes, are essential for pancreatic cancer initiation and metastasis.134 Pancreatic cancer cell-derived exosomes can be taken up by Kupffer cells, resulting in TGF-β secretion and upregulation of fibronectin production by hematopoietic stem progenitor cells, which then induce liver pre-metastatic niche formation.135 Moreover, exosome-derived integrin (α6β4, α6β1, and αvβ5) uptake by resident cells (liver Kupffer cells, lung fibroblasts, or epithelial cells) contributes to organ-specific pre-metastatic niche formation via activation of SRC proto-oncogene (Src) phosphorylation and pro-inflammatory S100 gene expression.136 The extensively studied metastatic process of EMT, which represents the transition of epithelial cells into mesenchymal cells, has been shown to be pivotal during pancreatic cancer cell invasion and metastasis.137–139 TGF-β was the first identified cytokine that induced EMT in pancreatic cancer cells via the RAS-MEK-ERK signaling pathway.140 Subsequent studies have demonstrated that EMT in pancreatic cancer cells can be triggered by various factors, including growth factors, cytokines, transcription factors, and miRNAs.141,142 Our group found that IGF-1 induced ENO2 deacetylation by HDAC3-enhanced EMT, thus promoting liver metastasis of pancreatic cancer.66 In addition, pancreatic cancer cells undergoing EMT can induce a cancer stem cell (CSC) phenotype as well as drug resistance.143,144
The pancreatic tumor microenvironment is indispensable for pancreatic cancer progression. Reciprocal communication between cancer cells and stromal cells induces changes in cellular components of the pancreatic tumor microenvironment, which can prime the primary tumor for metastasis and cell migration.145 Primary pancreatic tumors can modulate the local microenvironment in metastatic sites by secreting exosomes and soluble factors to promote colonization.146,147 Various stromal cells, e.g., TAMs, also participate in angiogenesis and metastasis of pancreatic cancer via the secretion of VEGF, CXCL1, and CXCL8.148,149
Metabolism dysregulation in pancreatic cancer
Metabolic reprogramming is a hallmark of cancer, and it influences the survival and growth of cancer cells by providing energy and macromolecular precursors.150 Pancreatic cancer is characterized by hypovascularization, and pancreatic cancer cells are surrounded by a tight desmoplasia, thus creating a highly hypoxic and nutrient-limited microenvironment.151,152 Malignant pancreatic cancer cells proliferate uncontrollably and have a proclivity for distant metastasis, which results in increased demand for both energy and biosynthetic precursors (Fig. 1d).153 Metabolic reprogramming is central to the pathogenesis of pancreatic cancer driven by KRAS (Fig. 2). A recent study reported that pancreatic cancer tumors can be classified into four metabolic subgroups, i.e., quiescent, glycolytic, cholesterogenic, and mixed metabolic profiles, based on the expression levels of glycolytic and cholesterogenic genes. Glycolytic tumors correlate with poor outcomes in pancreatic cancer patients, whereas patients with cholesterogenic tumors tend to have better survival, possibly due to their higher energy expenditure.154
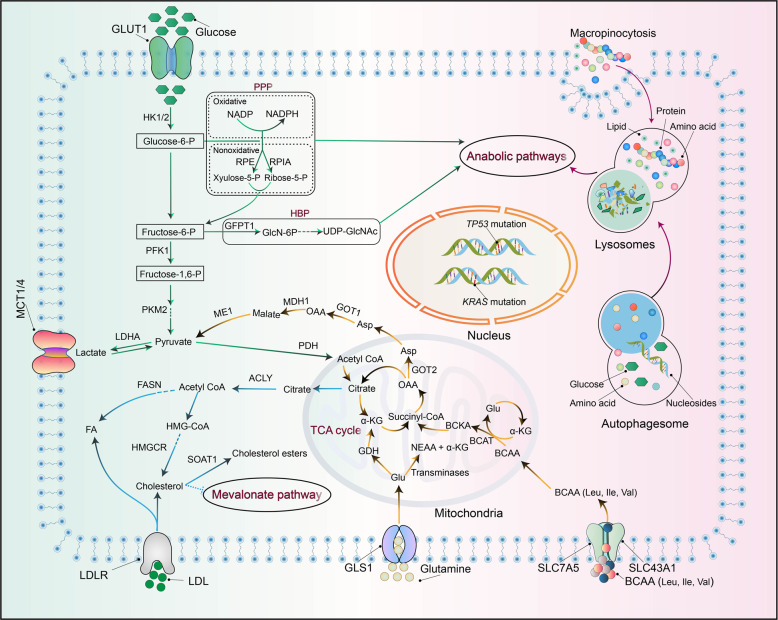
Fig. 2.
Metabolic reprogramming in pancreatic cancer cells. KRAS activation and mutant TP53 enhance glucose metabolism to provide biosynthetic precursors for anabolic pathways, including the non-oxidative arm of the pentose phosphate pathway (PPP) and the hexosamine biosynthesis pathway (HBP). KRAS activation reprograms glutamine metabolism to sustain cellular redox homeostasis by increasing the NADPH/NADP+ ratio and recycling glutathione (GSH) via reduction of oxidized GSH. The BCAT2-mediated BCAA catabolism driven by KRAS plays a critical role during pancreatic cancer development. Enhanced nutrient salvaging, via the induction of macropinocytosis and autophagy, provides energy and regenerative nutrients, including glucose, amino acids, lipids, and nucleosides
Reprogrammed glucose metabolism in pancreatic cancer
Cancer cells can survive and proliferate in stressful microenvironments by reprogramming their energy metabolism to increase glucose uptake and enhance glycolysis and lactate production despite the presence of oxygen (known as the Warburg effect).155 Notably, pancreatic cancer cells exhibit extensive glucose metabolic reprogramming, including glycolytic enzyme overexpression and increased lactate production.152,156 It has been demonstrated that oncogenic KRAS promotes glycolysis by enhancing glucose uptake via upregulation of the glucose transporter (GLUT1) and several rate-limiting glycolytic enzymes, including hexokinase 1 and 2, phosphofructokinase-1, and lactate dehydrogenase A.156,157 Furthermore, in pancreatic cancer cells, mutant KRAS signaling promotes mitochondrial translocation of phosphoglycerate kinase-1, resulting in the production of phosphorylated pyruvate dehydrogenase kinase-1 and restricted oxidative phosphorylation (OXPHOS).158 Enhanced glycolysis induced by oncogenic KRAS provides biosynthetic precursors for anabolic pathways, including the hexosamine biosynthesis pathway (HBP) and the non-oxidative arm of the pentose phosphate pathway (PPP).156 KRAS mutations drive the HBP, a side path of glycolysis, to increase the generation of biosynthetic precursors required for protein glycosylation and the synthesis of proteoglycans, glycolipids, and glycosyl phosphatidylinositol anchors.156,159 This process depends on both increased glycolysis and transcriptional upregulation of glutamine (Gln) fructose 6-phosphate transamidase 1 in the HBP.156 Similarly, oncogenic KRAS enhances the flux of glucose-derived carbon into the non-oxidative arm of the PPP to generate ribose-5-phosphate for de novo nucleotide biosynthesis, thus fueling proliferation.160 In addition, p53 also plays a vital role in metabolic reprogramming in pancreatic cancer cells. It has been demonstrated that KRAS and p53 can promote metabolic changes even before the occurrence of malignant transformation.161 In pancreatic cancer cells, mutant TP53 can activate GLUT1-mediated glucose transport by upregulating paraoxonase 2 expression and impairing the expression of TP53-induced glycolysis regulatory phosphatase to promote glycolysis.162 A gain-of-function TP53 allele was demonstrated to enhance the Warburg effect by inducing GLUT1 translocation to the plasma membrane, which is regulated by the RAS homolog family member A/rho-associated coiled-coil containing protein kinase/GLUT1 signaling pathway.163 In addition, TP53 mutations can decrease mitochondrial activity to suppress pancreatic cancer progression.164 Highly hypoxic microenvironments commonly form during pancreatic cancer progression, thus increasing the expression and stability of HIF-1. The hypoxic condition in the tumor microenvironment and HIF-1α upregulation contributes to elevated glycolytic activity and these factors cooperate with mutant KRAS to sustain cytosolic ATP generation even though knockdown of its expression has limited influence on metabolism-related enzyme expression.156,165 HIF-1α upregulation has been shown to upregulate GLUT1 as well as the expression levels of glycolysis-related genes to sustain cytosolic ATP levels in pancreatic cancer cells.166,167 HIF-1α also upregulates the expression of GFPT2 and inhibits the expression of pyruvate dehydrogenase to restrict mitochondrial oxidation.168
Reprogrammed amino acid metabolism in pancreatic cancer
It has been demonstrated that amino acid metabolism plays a crucial role in pancreatic cancer progression.169,170 Among the amino acids, Gln is the most abundant in circulation and is a major source of carbon and nitrogen for cancer cells.171 Notably, Gln is essential to support pancreatic cancer growth via redox homeostasis in a KRAS-driven metabolic pathway.156,172,173 Inhibition of downstream components of Gln metabolism leads to decreased tumor growth.151,172 Modification of the activity of glutaminase (GLS), which catalyzes the first step in glutaminolysis, i.e., the conversion of Gln to glutamate (Glu) in mitochondria, represents a viable therapeutic strategy. Interestingly, pancreatic cancer cells exhibit compensatory metabolic networks that sustain progression after GLS inhibition.174 When GLS1 was inhibited in pancreatic tumors, the GLS2 pathway was upregulated for Glu production.175
Arg can be directly catalyzed by oxide synthases (NOS1–3) into citrulline and nitric oxide (NO). NO promotes pancreatic cancer cell proliferation, and this effect can be dampened by knocking down NOS3 activity in KRAS mutant tumors.176 Arg deprivation is currently being evaluated for safety and efficacy in clinical trials. A recent study reported that Arg deprivation inhibits pancreatic cancer cell migration, invasion, and EMT.177 The mammalian target of rapamycin complex 1 (mTORC1) kinase is a master growth regulator that senses many environmental cues, including amino acid levels.178,179 Via solute carrier family 38 member 9, Arg serves as a lysosomal messenger that couples mTORC1 activation to the lysosomal release of essential amino acids needed to drive pancreatic cancer cell growth.179,180
In addition, proline dehydrogenase (PRODH)/proline oxidase, which catalyzes the first step of proline degradation, has been extensively linked to the progression of cancer, including pancreatic cancer.181 PRODH1 expression is elevated in pancreatic cancer cells to sustain cellular survival and proliferation under low-glucose or Gln-limited conditions.182 Via experiments with genetically engineered mouse models (GEMMs) and primary pancreatic epithelial cells, a recent study demonstrated that induction of the serine–glycine one-carbon pathway plays a vital role in tumorigenesis.183 In recent years, branched-chain amino acid (BCAA) metabolism has been tentatively linked to pancreatic cancer development.184 As essential amino acids, BCAA must be derived from the extracellular milieu, and specific transporters responsible for BCAA (isoleucine, leucine, and valine) absorbance include members of the solute carrier family 7 (SLC7) family (SLC7A5A and SLC7A8) and the solute carrier family 43 (SLC43) family (SLC43A1 and SLC43A2).185,186 It has been reported that plasma BCAAs are elevated in patients with increased pancreatic cancer risk and in mice with early-stage pancreatic cancer driven by mutant KRAS.187 BCAA transaminase 2 (BCAT2), a BCAA transaminase, is upregulated in advanced pancreatic cancer.188 A recent study demonstrated that BCAT2 plays a central role in the development of ductal-derived PanIN lesions by sustaining BCAA catabolism, and BCAT2-mediated BCAA catabolism driven by KRAS is critical for pancreatic cancer development
Reprogrammed lipid metabolism in pancreatic cancer
Lipid metabolism is an essential cellular process that converts nutrients into metabolic intermediates for the maintenance of cellular structures, energy storage, and the generation of signaling molecules production. It is increasingly reported that dysregulation of lipid metabolism is associated with the progression of various cancers, including pancreatic cancer.189,190
In mice, a high-fat diet can increase oncogenic KRAS activity, leading to fibrosis, inflammation, and enhanced PDAC development.191 It has been demonstrated that multiple enzymes that catalyze de novo fatty acid (FAs) synthesis, including ATP citrate lyase, FA synthase (FASN), and stearoyl-CoA desaturase, are obviously upregulated in pancreatic cancer.192,193 In addition, increased FASN expression was correlated with poor survival and poor gemcitabine responsiveness through enhanced estrogen receptor stress and cancer stemness. Inhibition of FA biosynthesis could be a promising strategy to overcome gemcitabine resistance in pancreatic cancer.194 Interestingly, different FAs might play different roles in pancreatic cancer. Saturated FAs promote tumor progression by increasing cyclooxygenase-2, VEGF, and caveolin 1 expression and the production of lipid droplets (LDs). High-fat diets enriched in monounsaturated FAs and ω6 polyunsaturated FAs (PUFAs) can increase tumor size in pancreatic cancer mouse models, and ω6 PUFA-rich diets frequently lead to liver metastasis by increasing hepatic ω6 PUFAs levels.195 ω3 PUFAs decrease pancreatic cancer cell proliferation by reducing AKT phosphorylation, while ω6 PUFAs can promote tumor growth by increasing AKT phosphorylation.196 FA β-oxidation (FAO) is essential for energy maintenance to support the proliferation and survival of cancer cells.197,198 A recent study demonstrated that the main source of ATP production in pancreatic cancer depends on FAO rather than glycolysis.199 Excess lipids stored as LDs can be converted into FAs by adipose triglyceride lipase and hormone-sensitive lipase (HSL) to provide ATP for pancreatic cancer metastasis. A recent study demonstrated that oncogenic mutant KRAS in pancreatic cancer facilitated LD utilization via suppressing the expression and phosphorylation of HSL, thereby fueling cancer cell invasion.200,201
In an oncogenic KRAS mouse model, the comparative transcriptomic analysis identified lipid-related metabolic pathways, in particular cholesterol uptake, as being the most highly enriched in pancreatic cancer (compared with a normal pancreas).192 Pancreatic cancer patients with increased expression of cholesterol synthesis genes showed a survival benefit in resectable and metastatic cases.154 Furthermore, cholesterol metabolism plays an important role in controlling pancreatic cancer development and differentiation. By using a GEMM driven by KRASG12D mutation and homozygous TP53 loss, a recent study demonstrated that inhibition of cholesterol biosynthesis with statins, a widely used inhibitor of the mevalonate pathway, or NAD(P)-dependent steroid dehydrogenase-like enzyme inhibitor can switch glandular pancreatic carcinomas to a basal phenotype.202 Loss of function of TP53 can drive tumorigenesis via downregulation of cholesterol transporter gene ATP-binding cassette subfamily A member 1 and activation of PI3K/sterol regulatory element-binding protein 2 (SREBP2) maturation. Consequently, the mevalonate pathway, which is responsible for cholesterol and sterol biosynthesis, is upregulated.203,204 Statins can modestly affect cholesterol homeostasis and significantly reduce the synthesis of electron carrier coenzyme Q, leading to severe oxidative stress and apoptosis in pancreatic cancer.205 By using organoid and mouse models, a recent study demonstrated that the loss of sterol O-acyltransferase 1 (SOAT1), a key enzyme catalyzing cholesterol to inert cholesterol esters to sustain mevalonate pathway flux, significantly impairs pancreatic cancer progression. Mechanistically, mutant TP53 and TP53 LOH can promote the expression of SOAT1, which upholds SREBP2-driven cholesterol biosynthesis to sustain the levels of multiple nonsterol isoprenoids, e.g., farnesyl pyrophosphate and geranylgeranyl pyrophosphate, ultimately resulting in RAS and Rho activation. These findings indicate that SOAT1 inhibition might be a potential therapeutic modality for pancreatic cancer patients with mutant TP53 and TP53 LOH.206
Autophagy and nutrient salvage regulation in pancreatic cancer
Autophagy, a cellular “self-eating” process, allows cells to degrade and recycle their own cellular components, thereby enhancing their survival under stress conditions. Notably, it seems that autophagy has conflicting functions during cancer progression, including both tumor-suppressive and tumor-promoting effects, which are likely context-dependent. The role of autophagy in pancreatic cancer is also complex. During early stages, autophagy is antitumorigenic because of its function in cellular quality control, while in established cancer, autophagy can support cancer cell survival and progression by providing resources for macromolecule biosynthesis and bioenergetics.207 A growing body of evidence suggests that autophagy is required for pancreatic cancer cell survival and metabolism. Genetic depletion of autophagic components or pharmacological inhibition of autophagy with chloroquine can decrease tumorigenicity in vivo.208,209 Oncogenic KRAS can activate autophagy, and knockdown of KRAS expression or inhibition of its downstream MEK/ERK signaling cascade further induces autophagy and decreases both glycolytic and mitochondrial functions. Combined autophagy inhibition by hydroxychloroquine (HCQ) and MEK/ERK inhibitors showed enhanced antitumor activity in vivo.209,210 Although autophagy is usually regulated by nutrient and oxygen availability, pancreatic cancer cells often display high basal levels of autophagic flux even under nutrient-rich conditions.211 Pancreatic cancer cells use autophagy to obtain glucose and amino acids from catabolized substrates to fuel the citric acid cycle (TCA) cycle, OXPHOS, and ATP biosynthesis. It has been shown that autophagy induction in pancreatic cancer cells occurs as part of a broader transcriptional program regulated by the microphthalmia/transcription factor E (MiT/TFE) family of transcription factors.212 MiT/TFE-dependent autophagy–lysosome activation is required to maintain intracellular amino acid pools, and knockdown of the encoding genes strongly suppresses tumor progression.212 In vitro, autophagy inhibition leads to disrupted redox state, elevated DNA damage, and decreased levels of metabolic substrates, resulting in suppression of pancreatic cancer cell proliferation.209,213 Notably, the disrupted redox state and defective mitochondrial respiration caused by autophagy inhibition can be rescued by supplying antioxidants and TCA cycle intermediates.213,214 Autophagy is also a key regulator of immunogenicity in pancreatic cancer cells by virtue of its selective targeting of major histocompatibility complex class I (MHC-I) for degradation. Elevated autophagy is strongly correlated with increased immune evasion and reduced CD8+ T cell infiltration in pancreatic cancer.215,216
The tumor microenvironment of pancreatic cancer
The tumor microenvironment in pancreatic cancer is highly immunosuppressive and is characterized by an abundant stromal response as a desmoplastic reaction. Accumulating research has highlighted the critical roles of the tumor microenvironment in maintaining pancreatic cancer development; hence, the tumor microenvironment acts as a key determinant by which this malignancy acquires therapeutic resistance to currently available treatment.
The pancreatic stroma in pancreatic cancer
Pancreatic cancer is characterized by an extensive and dense fibrous stroma, and the stromal composition can account for up to 90% of the total tumor volume. The crosstalk between tumor cells and the stromal microenvironment is complex, and stromal elements regulate pancreatic cancer progression in a more complex manner. The paradoxical effects of the pancreatic cancer-associated stroma on pancreatic cancer cells, which include both tumor-promoting and tumor-suppressive regulations, might represent a target for new therapeutic strategies based on context-dependent stromal alterations.217,218
The composition of the pancreatic cancer stroma
Pancreatic cancer is known for its stroma/desmoplastic reaction, comprising a heterogeneous mass of cells, including PSCs, fibroblasts, immune cells, ECM, and soluble proteins such as cytokines and growth factors.219 The main types of stromal cells that contribute to tumor progression are PSCs, CAFs, TAMs, regulatory T cells (Tregs), and MDSCs.219 PSCs can be classified as quiescent PSCs (qPSCs) or activated PSCs (aPSCs) depending on their activation state. As lipid storage cells in the pancreas, qPSCs are located in surrounding perivascular regions or basolateral aspects of acinar cells.220 qPSCs can be activated under some environmental stress conditions, including inflammation, hypoperfusion, and oxidative stress, etc. aPSCs can transdifferentiate into myofibroblast-like phenotype, constituting a growth-permissive microenvironment in the tumor tissue.221 However, aPSCs are not permanently sustained because they can revert to their inactivate state under the influence of several factors, e.g., apoptosis, senescence, tissue regression, or recovery.220 Macrophages, a key class of innate immune cells, are derived from the mononuclear phagocyte system in the bone marrow, and they contribute to desmoplasia and immunosuppression of pancreatic cancer. TAMs can respond to environmental signals (e.g., inflammatory cytokines IL-8, IL-6, IL-1β, IL-10),222 and it is thought that a high number of infiltrating TAMs may correlate with tumor size, prognosis, and patient survival in pancreatic cancer.223 Forkhead box P3-positive (Foxp3+) Tregs are a subtype of T cells with significant roles in immunological self-tolerance maintenance and immunosuppression modulation during tumor progression. Foxp3+ Tregs have been shown to interact with tumor-associated CD11c+ dendritic cells and to restrain the immunogenic activation of CD8+ T cells.224 In pancreatic cancer, Foxp3+ Tregs are an essential source of TGF-β ligands, promoting the differentiation of SMA+ fibroblasts (myCAF) for tumor progression. Furthermore, Foxp3+ Treg depletion results in the differentiation of inflammatory fibroblast subsets, which in turn drive increased myeloid cells recruitment and impair the alleviation of immunosuppression.225
MDSCs are a heterogeneous population of immature myeloid cells that can be found in both tumors and the spleen. MDSCs usually participate in various pathological processes, including tissue inflammation and tumor progression. MDSCs can build crosstalk with TAMs, Tregs, and other immune cells to suppress effector T cells in the tumor microenvironment.226 Pancreatic tumors can create a hypoxic environment that recruits the MDSCs to the tumor microenvironment where they exert their immunosuppressive effect.227
The origin and functions of CAFs in pancreatic cancer
CAFs are one of the most prominent and active components of the desmoplastic stroma of pancreatic cancer. They possess diverse functions, including a robust fibro-inflammatory stromal effect and extensive reciprocal crosstalk with pancreatic cancer cells.228 The exact origin of CAFs is unclear at this time because they are derived from various types of cells, e.g., adipocytes, epithelial cells, resident fibroblasts, and bone marrow-derived mesenchymal stem cells.221 The general consensus is that aPSCs are one of the predominant origins of CAFs in the pancreatic cancer stroma.229 Moreover, CAFs activation involves multiple complex pathways and cytokines, including EMT, the sonic hedgehog (SHH) pathway, and various cytokines, including IL-1, IL-6, IL-10, TGF-β, and tumor necrosis factor (TNF-α).228,230
As major players in pancreatic tumorigenesis, activated CAFs from PSCs have numerous anti- and pro-tumorigenic influences on cancer cells in the tumor microenvironment. CAFs establish signal communication with tumor cells through a variety of mechanisms, e.g., EVs and classical paracrine.221 CAFs can increase the pancreatic cancer aggressiveness via EVs crosstalk that carries the annexin A6 (ANXA6)/LDL receptor-related protein 1/thrombospondin 1 complex under physiopathologic conditions (macrophages invasion, hypoxia, and lipid starvation).231 The presence of ANXA6+ EVs in the peripheral blood is restricted to pancreatic cancer patients, establishing these EVs as a potential biomarker of pancreatic cancer aggressiveness.231 Moreover, CAF-derived EVs can inhibit the metabolic reaction of mitochondrial OXPHOS and increase glycolysis and Gln-dependent reductive carboxylation in pancreatic cancer cells.232 Under chemotherapy, exposure of pancreatic cancer to gemcitabine might increase the release of CAF-derived EVs, which might further enhance chemoresistance and tumor cell survival.233 High expression of fibroblast activation protein (FAP) in CAFs has been established as a key enhancer of PDAC progression. Genetic deletion of FAP in the KPC mouse model results in delayed primary tumor formation in mice bearing pancreatic cancer tumors, leading to improved survival. Currently, researchers are attempting to establish FAP as a precise molecular target for guiding pancreatic cancer therapy.234 CAFs can enhance aerobic glycolysis and secrete high-energy metabolites, including lactate and pyruvate.235 Moreover, it has been demonstrated that enhanced autophagy in CAFs contributes to the release of amino acids, which tumor cells can use as anabolic substrates.236 CAFs also supply amino acids, TCA cycle intermediates, and lipids to tumor cells via exosomes, thus promoting pancreatic tumor growth, especially under nutrient-limited conditions.232 Lysophosphatidylcholine released from CAFs can be converted into lysophosphatidic acid (LPA) by autotaxin within the tumor microenvironment, subsequently exerting mitogenic signals via binding with the LPA-receptor on the surface of tumor cells, to support tumor growth.237
CAF heterogeneity in pancreatic cancer
Due to differences in their sources of origination and activation mechanisms, CAFs display heterogeneity that is characterized by several subgroups with different phenotypes and functions in pancreatic cancer tissue. For a long time, CAFs were only considered tumor promoters in the tumor microenvironment of the pancreatic cancer cells based on their involvement in tumor cell proliferation and survival via facilitating tumor invasion, metastasis, and immunosuppression.238,239 For example, as one of the traditional CAF activation pathways, the SHH pathway has been targeted by genetic deletion or chemical inhibition for tumor suppression. However, some recent studies have found that tumors can become more aggressive and undifferentiated upon Shh deletion in a pancreatic cancer mouse model.218,240 This evidence suggests that CAFs might comprise several subsets that have different or even opposing effects on tumor progression. Biffi et al. have identified two CAFs subtypes with either myofibroblastic characteristics or inflammatory phenotypes, which they have named “myofibroblastic CAFs” and “inflammatory CAFs,” respectively. Despite these observations, the exact origins of CAFs and their roles in pancreatic cancer progression have not yet been fully explored.84 It has been reported that genetically regulated phenotypes of pancreatic cancer affect pancreatic cancer progression by increasing matricellular fibrosis and tissue tension.241 Tape et al. found that oncogenic KRASG12D engages heterotypic fibroblasts, which subsequently instigate reciprocal signaling in tumor cells.242
Classical therapeutic approaches that focus on the depletion of the stromal desmoplasia usually lead to a disappointing result. In light of the complexity of the pancreatic cancer microenvironment,243 future therapeutic approaches that target both the tumor and the stroma may be able to achieve a better outcome.
Immunomodulation in pancreatic cancer
Pancreatic cancer is often considered to be immunosuppressive based on a massive infiltration of immunosuppressive leukocytes and minimal antitumor T cell infiltration.244 Pancreatic cancer cells have been shown to cooperate with other regulatory immune cells to evade immune surveillance and to resist the cytotoxic effect of T lymphocytes (Fig. 3).245 Previous studies have shown that there is a correlation between the clinical significance and the composition and quantity of tumor-infiltrating immune cells and tumor-associated myofibroblasts.246 Mahajan et al. described a novel prognostic signature comprising distinct immune cell and stromal components for risk assessment of pancreatic cancer patients and prediction of their progression-free survival (PFS).247 Research has also shown that patients with higher levels of CD4+ and/or CD8+ T cells have significantly longer survival.248
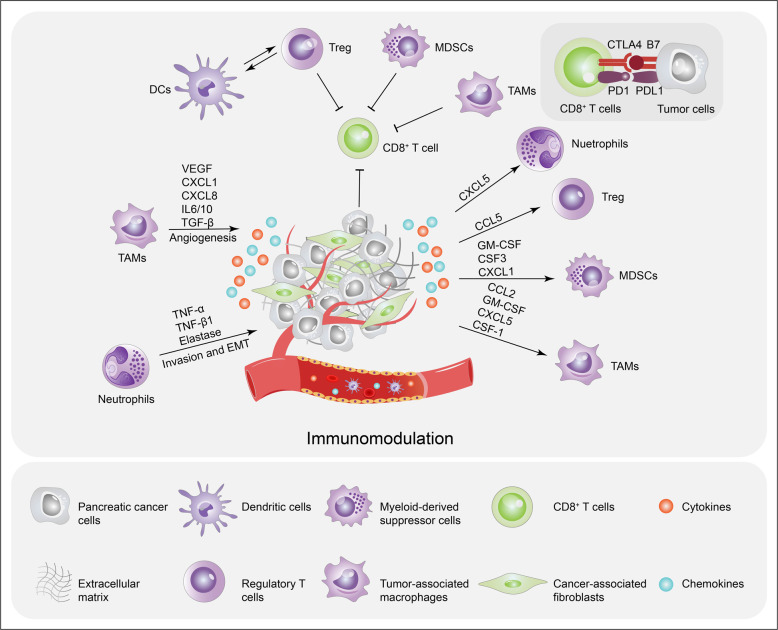
Fig. 3.
Immune evasion orchestrated by pancreatic cancer cells and stromal cells. The secretion and immunomodulation of pro-tumorigenic cytokines by pancreatic cancer cells and stromal cells are tightly regulated by oncogenic KRAS- or mutant TP53-dependent pathways. Pancreatic cancer cells secrete cytokines and, chemokines and recruit immunosuppressive cells, including MDSCs, TAMs, Treg cells, and neutrophils, which suppress the activity and functions of CD8+ cytotoxic T cells. Pancreatic cancer cells also evade the immune system by expressing PDL-1 to promote CD8+ T cell exhaustion. Immune cell infiltration also releases cytokines and growth factors that directly stimulate tumor growth by promoting angiogenesis and increasing the invasive ability of pancreatic cancer cells
The immunosuppressive tumor microenvironment of pancreatic cancer can be largely attributed to tumor cell-intrinsic pathways. Oncogenic KRAS mutations and KRAS-induced colony-stimulating factor (CSF) 2 (GM-CSF) production, promote pancreatic neoplasia progression via an influx of CD11b+Gr1+ immunosuppressive cells and suppressed antitumor T cell function.249 PI3K is a critical downstream effector of KRAS. One of its catalytic subunits, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha, has been shown to reduce the expression of MHC class I and CD80 in pancreatic cancer cells, leading to limited T cell recognition and pancreatic cancer cell clearance.250 Moreover, p53 and KRAS can cooperatively target the ADP ribosylation factor 6-ArfGAP with SH3 domain, and ankyrin repeat and PH domain 1 pathway to promote pancreatic cancer immune evasion, PD-L1 dynamics, and malignancy.251 Loss of p53 function activates JAK2-STAT3 signaling, which induces macrophage and neutrophil infiltration, reduces CD8+ T cell levels, and promotes tumor growth in pancreatic tumors in mice.252 In neoplastic pancreatic cancer cells, focal adhesion kinase (FAK) activity is hyperactivated and is correlated with high levels of TAMs, MDSCs, and Treg cells and low CD8+ cytotoxic T cell infiltration.253,254 In addition, FAK inhibition increases immune surveillance by mitigating the immunosuppressive pancreatic cancer tumor microenvironment, thereby rendering tumors responsive to immunotherapy.254 Similarly, inhibition of endogenous Myc triggers ubiquitous regression of pancreatic tumors by decreasing the levels of neutrophil and macrophage infiltration.255 Studies in a pancreatic cancer mouse model have demonstrated that Myc activation alone is sufficient to trigger the release of instructive signals that cooperatively coordinate changes in multiple immune and stromal cell types.256 Moreover, Myc and KRASG12D cooperatively regulate gene expression resulting in natural killer cell-mediated immune surveillance.257
Aside from genetic alterations that regulate the tumor microenvironment, pancreatic cancer can adopt other strategies of immune evasion, including the production of metabolites and immunosuppressing chemokines and cytokines. Lactate is robustly exported from pancreatic cancer cells via elevated monocarboxylate transporter 4 (MCT4)4 expression, and neighboring tumor cells in normoxic regions overexpress the lactate importer MCT1 to fuel OXPHOS and promote tumor growth.168,258 Furthermore, lactate secreted from pancreatic cancer cells remodels the tumor microenvironment and contributes to the M2-like polarization of TAMs, which leads to suppressed immunity.259,260 TAMs participate in metabolic crosstalk and elevate the glycolytic signature, which in turn promotes pancreatic cancer vascularization and metastasis.261 TAM-secreted C–C motif chemokine ligand 18 (CCL18) interacts with ITPNM family member 3 to induce vascular cell adhesion molecule-1 overexpression in pancreatic cancer cells, thereby promoting lactate production accompanied by elevated aerobic glycolysis. This effect activates macrophages into a TAM-like phenotype, forming a positive feedback loop.259 Pancreatic cancer cells can secrete CCL5 to promote the recruitment of Treg cells into the tumor site.262,263 The levels of tumor-derived GM-CSF,249 M-CSF,249 CSF1,264 CCL2, and CXCL5265,266 are often highly elevated by mutant KRAS expression and are associated with TAM infiltration. The chemokine CCL2, which can be produced by pancreatic cancer cells, facilitates the recruitment of immunosuppressive C–C motif chemokine receptor 2-positive TAMs into the tumor microenvironment, thereby limiting T cell infiltration.267,268 CXCL5 is an important attractant for immune cell accumulation. Pancreatic cancer patients with high CXCL5 expression have more intratumoral M2-polarized macrophages, neutrophils, and IgG+ plasma cells than those with low CXCL5 expression.269 Studies have demonstrated that tumor-derived CXCL1,270 GM-CSF,271 and CSF-3272 mediate the recruitment of MDSCs to the tumor microenvironment. CXCL1 has been shown to facilitate the recruitment of myeloid cells and inhibition of the T cell infiltration into tumors.273 Moreover, CSF1 serum levels are upregulated in pancreatic cancer, and higher levels correlate with increased macrophage infiltration and advanced tumor stage.274,275 Pancreatic cancer cells also secrete various immunosuppressive cytokines, including TGF-β and IL-10, which (1) coordinate immunosuppressive tumor microenvironment formation and (2) recruit cells involved in immune evasion, e.g., TAMs and Treg cells, to evade antitumor immunity.276,277 Foxp3 expressed by pancreatic cancer cells can activate CCL5 to promote the recruitment of Treg cells from peripheral blood to the tumor site, which negatively correlates with poor prognosis in pancreatic cancer.262 It has been demonstrated that neutrophils can increase pancreatic cancer metastasis by releasing a collection of cytokines, including TNF-α, TGF-β1, and elastase.278,279 Moreover, increased PD-L1 expression on pancreatic cancer cells can restrain T cell function by promoting CD8+ T cell exhaustion. Pancreatic cancer can even induce CD8+ T cell apoptosis via a Fas/FasL counterattack.280,281
Clinical perspectives of current advances in pancreatic cancer
Novel biomarkers of pancreatic cancer based on liquid biopsy
Currently, pancreatic cancer detection and diagnosis primarily rely on imaging modalities, including transabdominal ultrasonography, computed tomography (CT), magnetic resonance imaging), positron emission tomography, endoscopic retrograde cholangio-pancreatography, and endoscopic ultrasonography.282 These screening technologies have limitations in their detection capability. For instance, they are ineffective at detecting early-stage pancreatic cancer and small metastases or peritoneal lesions, etc.283,284 Serum tumor markers such as carcinoembryonic antigen and carbohydrate antigen have been widely used for pancreatic cancer diagnosis in clinical settings. However, they lack sensitivity and specificity as pancreatic cancer biomarkers.285,286 To date, novel markers such as ncRNAs (miRNA and lncRNA),287,288 genetic markers (e.g., KRAS, TP53, SMAD4, and CDKN2A),17,289 circulating tumor DNA (ctDNA),290 circulating tumor cells (CTCs),291,292 and exosomes293,294 have been explored. They show great potential for pancreatic cancer early detection, and might provide significant improvement in the future management and treatment outcome of pancreatic cancer. A liquid biopsy allows for serial, real-time monitoring of the dynamics and complex nature of cancer. In this section, we focus on the most promising liquid biopsy markers: CTCs, ctDNA, and exosomes, and their clinical applications in pancreatic cancer.
CTCs, which can be found in pancreatic cancer patients’ circulation, provide useful information for diagnosis, staging, and prognosis, and they even can act as novel personalized treatment targets.295,296 Some studies have reported that CTCs can be detected and used as an early-stage diagnostic marker using a number of methods, including negative enrichment, immunofluorescence and in situ hybridization (FISH) of chromosome (NE-iFISH) system, and subtraction enrichment and immunostaining-fluorescence in situ hybridization (SE-iFISH). NE-iFISH exhibits a very high CTC detection rate (90%) in pancreatic cancer patients. SE-iFISH has both high sensitivity (88%) and high specificity (90%) in pancreatic cancer with a cutoff value of 2 cells/7.5 ml.297,298 CTCs in venous blood samples from pancreatic cancer patients were evaluated using a microfluidic NanoVelcro CTC chip and CTCs were detected in 54 out of 72 patients with a sensitivity and specificity of 75.0% and 96.4%, respectively.299 Despite differences in CTCs enrichment and detection methods, studies have consistently shown that a higher number of CTCs could be predictive of reduced survival.297,300–302 In addition, a higher number of CTCs in the portal vein correlated well with a higher risk of cancer recurrence and liver metastases after surgery. For example, at a 3-year follow-up with patients who underwent surgical resection, portal vein CTC-positive patients presented a higher rate of liver metastases than CTC-negative patients.303 In another study, 84.6% of patients with a high portal CTCs count (defined as >112 CTCs/2 ml blood) developed liver metastases within 6 months after surgery.304 CTCs can also be a promising biomarker or useful for monitoring the response to chemotherapy of pancreatic cancer patients. It has been reported that triploid CTCs (which show three hybridization signals for a given chromosome) <3 groups displayed significantly increased 1-year survival and OS compared with the triploid CTCs ≥3 group.298 Moreover, advanced pancreatic cancer patients can be tested for the presence of circulating tumor microemboli, whose presence predicts a low response to chemotherapy and poor survival.298,305
ctDNA, which represents a small fraction of the total cell-free DNAs (cfDNA), is derived from either the CTCs or the tumor cells.306 The sensitivity of ctDNA detection is generally lower than that of CA19-9 detection for early pancreatic cancer diagnosis.307 However, ctDNA proved to be a more suitable biomarker than CA19-9 and CTCs for the prognosis for pancreatic cancer patients. The higher burden of KRAS mutation on cfDNA levels has been shown to be negatively correlated with OS.308–310 Furthermore, the ctDNA level is a good indicator of pancreatic cancer progression during or after treatment (chemotherapy and/or surgery). Pancreatic cancer patients with detectable ctDNA showed poorer OS and a higher risk of cancer recurrence compared with those with undetectable ctDNA.37,311–313
Tumor-derived exosomes are enriched with DNA, mRNA, miRNA, and protein both inside and on their surface, establishing them as potential biomarkers for early-stage diagnosis of pancreatic cancer.314–316 Melo et al. reported that the biomarker glypican-1 (GPC1) was specifically enriched on pancreatic cancer exosomes.317 GPC1(+) GPC1-circulating exosomes were detected in the serum of pancreatic cancer patients with 100% specificity and sensitivity, thereby distinguishing pancreatic cancer patients from patients with nonmalignant pancreatic disease and healthy subjects.317–319 In addition, certain exosomal miRNAs (miR-10b, -21, -30c, -181a, and -let7a) had 100% sensitivity and 100% specificity, and ephrin receptor A2-EV had 94% sensitivity and 85% specificity in distinguishing pancreatic cancer from healthy controls.320,321 With such high specificity and sensitivity, pancreatic tumor-derived exosomes might be one of the most promising markers for both early-stage pancreatic cancer diagnosis and monitoring of the therapeutic response during treatment.321–324
While liquid biopsies are increasingly being adopted for potential biomarkers for cancer diagnosis, prediction, and surveillance, there is a need for developments in assay technology for the isolation, quantification, and analysis of biomarkers based on liquid biopsy. For example, it is difficult to isolate tumor-specific DNA because of the limitations of the currently available techniques.325 Despite these shortcomings, significant research is still dedicated to liquid biopsies because of their potential advantages. One well-validated, Food and Drug Administration (FDA)-approved liquid biopsy test panel, MSK-IMPACT, was developed at the Memorial Sloan Kettering Cancer Center. They have also developed another liquid biopsy assay called the Analysis of Circulating Cell-free DNA to Evaluate Somatic Status.326 By using the latter panel, Razavi et al. identified cancer-derived somatic variants in plasma-circulating cfDNA and highlighted the importance of matched cfDNA white blood cell sequencing for accurate variant interpretation.327 This panel was also used to identify clinically relevant mutations and mutation signatures as well as novel noncoding alterations.328 This panel was subsequently expanded via Clinical Laboratory Improvement Amendment-certified testing for the analysis of tumor-derived and matched germline DNA samples, which has also been approved by the FDA.329 However, the majority of liquid biopsy assays still lack sufficient evidence of clinical validity and utility. Only after the demonstration of clinical validity and clinical utility, liquid biopsies may further reach their full potential and impact on the clinical management of pancreatic cancer patients.
Subtype-specific therapeutic responses in pancreatic cancer
Pancreatic cancer is extensive heterogeneity amongst patients with respect to treatment response. Basal-like tumors have a higher frequency of TP53 mutations,36,46,48 while the classical subtype is characterized by GATA binding protein 6 (GATA6) expression and KRAS dependency.44,46 Importantly, patients with basal-like tumors have higher pathologic grades and worse OS compared with those with classical tumors.44,46,48,50,330 Nevertheless, there is no consensus on subtype-specific treatment regimens. Although Collisson et al. reported that QM-PDA subtype patients had significantly worse outcomes compared with patients with other subtypes, they found that in pancreatic cancer cells, the QM-PDA subtype was more sensitive to gemcitabine. By contrast, classical subtype cell lines were more sensitive to erlotinib.44 One possible explanation for these findings is that in vitro 2D-cultured cell lines might not manifest expected bulk RNA-sequencing (RNA-seq(-based molecular signatures, as Moffitt et al. reported that traditional 2D-cultured cell lines lack the classical subtype.47 Moffitt et al. also demonstrated that patients with basal-like tumors showed a tendency to benefit more from adjuvant therapy, although the effect was not significant.47 In vivo patient-derived xenograft (PDX) models or 3D organoid-based approaches might be more effective tools for investigating subtype-specific treatment responses.45 Interestingly, by using an organoid-based model, Tiriac et al. found that the basal-like subtype was enriched in an oxaliplatin-nonsensitive group, while it was present at similar frequencies in gemcitabine-sensitive and nonsensitive groups.45
Recently, Aung et al. reported the results of the COMPASS trial, showing that basal-like subtype tumors showed a poorer response to first-line chemotherapy (1/12 vs. 13/38 had a partial response in the basal-like vs. classical subtype, respectively). Patients with classical subtype tumors treated with mFOLFIRINOX had the best PFS.331 More recently, Chan et al. devised a more comprehensive method for molecular subtyping and for developing subtype-specific treatment regimens in pancreatic cancer.46 They reclassified pancreatic cancer samples into five subtypes: basal-like A, basal-like B, hybrid, classical A, and classical B. Distinguishing the basal-like A, basal-like B, and hybrid subtypes from the former basal-like/squamous subtype allowed the detection of more subtle subtype-specific chemotherapy responses; furthermore, Chan et al. reported that the basal-like A subtype showed the poorest responses to gemcitabine-based and mFOLFIRINOX chemotherapies.46 Remarkably, single-cell RNA-seq data provided evidence of the coexistence of basal-like and classical expression signatures within a single cancer sample; furthermore, subtype switching, a rare event due to the outgrowth of minor clones driven by therapeutic selection and genetic instability, was observed.46 Additional evidence of intratumoral transcriptional heterogeneity in pancreatic cancer was found by Hayashi et al. via multiregional sampling.332 In addition, Er et al. demonstrated a squamous subtype-specific synergistic effect of Src and MEK1/2 inhibitors with gemcitabine in pancreatic cancer cell lines.333
Limited access to freshly frozen samples and cost-prohibitive issues have limited the use of transcriptomic-based subtyping. Puleo et al. evaluated the transcriptome of formalin-fixed paraffin-embedded pancreatic cancer samples and successfully identified basal-like and classical subtypes.48 Law et al. reported proteomic-based subtyping of pancreatic cancer liver metastases and identified four distinct pancreatic cancer subtypes, i.e., metabolic, progenitor-like, proliferative, and inflammatory.330 Pancreatic cancer patients with metabolic and progenitor-like subtypes showed significant benefit from FOLFIRINOX treatment.
Hayashi et al. and Kalimuthu et al. provided paradigms for integrating clinical morphological and histological features with transcriptomic expression profiles such that pancreatic cancer molecular subtypes can be predicted based on morphological classification.332,334 Hayashi et al. found that histological squamous features and glandular patterns were consistent with the RNA-seq-defined basal-like and classical subtypes, respectively.332 They also identified intratumoral heterogeneity in transcriptional subtypes and suggested a subclonal feature of pancreatic cancer subtypes, which was inconsistent with Chan et al.46 Kalimuthu et al. identified four morphological patterns that segregated into two components, i.e., gland-forming and non-gland-forming, that corresponded to the classical and basal-like subtypes.334 They even found that morphological pattern-based subgroups provide better prediction of clinical outcomes than transcriptional subtypes. Efforts have also been made to simplify the classification of pancreatic cancer samples using surrogate markers, e.g., TP63,335 YAP1,92 HNF1 homeobox A, keratin 81 (KRT81),50 GATA6,44,46 and KRT17.336 Interestingly, Kaissis et al. reported that a machine learning-based analysis of preoperative CT images could predict molecular pancreatic cancer subtypes.337 These findings might facilitate clinical patient stratification and provide guidance for precision medicine.
Although some studies have demonstrated the feasibility of prospective genomic profiling of advanced pancreatic cancer, this approach is time-consuming and costly.331,338 In addition, clustering algorithms are threshold sensitive, and molecular classifiers are heavily confounded by limitations such as sampling methods, tumor cellularity, RNA quality, and tumor heterogeneity.338,339 Nevertheless, molecular subtyping now guides the optimization of therapeutic strategies and provides valuable opportunities for improving pancreatic cancer patient outcomes.
Current preclinical and clinical developments in pancreatic cancer
Current preclinical therapeutic models in pancreatic cancer
Identification of clinically applicable approaches for pancreatic cancer treatment relies heavily on the availability of preclinical models. In recent decades, novel preclinical models, e.g., PDX systems and patient-derived organoids (PDO), have been developed and are becoming increasingly used in drug screening, biomarker development, and evaluation of personalized therapeutic strategies. PDX and PDO models have positive predictive values of 80 and 88% for patient-specific sensitivities to therapeutic agents, respectively.340–342 However, PDX and PDO models are primarily suitable for basic research and have severe limitations for clinical use. For example, it normally takes several months for a PDX model to yield preclinical results. Moreover, it is infeasible to study early carcinogenesis, progression, and tumor immunology based on PDX or PDO models.
Recently, advances in genetic engineering techniques and an increased understanding of the mechanisms of pancreatic oncogenesis have supported the development of GEMMs useful as pancreatic cancer preclinical models (Table 1). Most of the GEMMs currently used in translational oncological studies are Cre/loxP-based models. Endogenous expression of oncogenic KRASG12D induces PanIN in mice, and a subset of these mice develop pancreatic cancer tumors at an advanced age, revealing that additional events are necessary for tumor formation onset.343
Table 1.
An overview of common genetically engineered mouse models of pancreatic cancer and their key characteristics
| Genotype | Preneoplastic lesion | Cancer phenotype | Metastasis | Features | References |
|---|---|---|---|---|---|
| Pdx1; KRASG12D/+ | PanIN | PDAC | Yes | Long latency | 343 |
| Ptf1a+/Cre; KRASG12D/+ | PanIN | PDAC | Yes | Long latency | 343 |
| KRASG12D/+; Ela-TGFa | PanIN | IPMN | Yes (50%) | Moderate latency | 350 |
| Pdx1-Cre; LSL-KRASG12D/+; Ink4a/Arflox/lox | PanIN | PDAC | Yes (21%) | Short latency and high penetrance | 346 |
| Pdx1-Cre; LSL-KRASG12D/+; LSL-TP53R172H/+ | PanIN | PDAC | Yes (63%) | Accelerated development of metastatic PDAC | 345 |
| LSL-KRASG12D; TP53fl/fl (in situ electroporation of Cre recombinase and myrAkt2) | PanIN | PDAC | Yes (>60%) | High efficiency, short latency | 384 |
| Co-electroporation of SB13 transposase with a KRASG12V-expressing transposon, Cre recombinase and myrAkt2 in TP53fl/fl mice | PanIN | PDAC | Yes (>70%) | High efficiency, short latency | 384 |
| Pdx1-Cre; KRASG12D/+; Ink4a−/− TP53lox/lox | PanIN | PDAC | Yes (20%) | Short latency and high penetrance | 347 |
| Pdx1-Cre; KRASG12D/+; Smadlox/lox | IPMN | PDAC | Yes (38%) | Model of IPMN to PDAC progression | 351,352 |
| Ptf1a+/Cre; KRASG12D/+; Smadlox/lox | MCN | PDAC | Yes (18%) | MCNs resembling human disease | 385 |
| Ptf1a+/Cre; KRASG12D/+; ATMlox/lox | PanIN | PDAC | Yes (78%) | High metastasis tendency | 348 |
IPMN intraductal papillary mucinous neoplasm, MCN mucinous cystic neoplasm, PanIN pancreatic intraepithelial neoplasia, PDAC pancreatic ductal adenocarcinomas
Strains for targeting pancreatic progenitor cells, such as Pdx1-Cre transgenic or Ptf1a+/Cre knock-in strains in the context of endogenous mutant KRASG12D (generally referred to as KC mice) have been successfully used to recapitulate the development and progression of PanIN lesions to pancreatic cancer.343,344 Furthermore, the KPC PDAC mouse model, first described in 2005, allows conditional activation of the KRASG12D mutation and a TP53 mutation (Pdx1-Cre; LSL-KRASG12D/+; LSL-TP53R172H/+) via Cre-Lox technology.345 In addition, combined with the KRASG12D mutation, Ink4a/Arf deficiency (Pdx1-Cre; LSL-KRASG12D/+; Ink4a/Arflox/lox), ATM deficiency (Ptf1a+/Cre; KRASG12D/+; ATMlox/lox), or a TP53lox/lox background plus P16Ink4a deficiency (Pdx1-Cre; KRASG12D/+; Ink4a−/− TP53lox/lox) result in accelerated progression from PanIN to invasive PDAC.345–349 Concomitant expression of TGFα and KRASG12D leads to the development of cystic papillary lesions that resemble human IPMN (KRASG12D/+; Ela-TGFa).350 Concomitant presence of KRASG12D and SMAD deficiency (Pdx1-Cre; KRASG12D/+; Smadlox/lox or Ptf1a+/Cre; KRASG12D/+; Smadlox/lox) leads to the development of IPMN or mucinous cystic neoplasm, respectively.351,352 Compared with traditional xenograft models, these GEMMs more faithfully recapitulate key morphological and molecular PDAC features. They can be used to study early carcinogenesis, progression, and tumor immunology of traditional xenograft models. GEMM models also offer higher predictive value for clinical diagnostic and therapeutic interventions. However, despite the increasing repertoire of animal models that emulate key genetic driver mutations found in pancreatic cancer, it is important to emphasize that many GEMMs do not fully recapitulate all aspects of the clinical population by fully covering the full range of genetic diversity and metastatic spread, as demonstrated by multiple failed clinical trials.353,354
Clinical strategies in pancreatic cancer
For pancreatic cancer patients, the only potentially curative choice is surgical resection of the pancreas. This approach is restricted to 20% of cases with anatomically resectable disease. Moreover, up to 50% of patients have incomplete resection accompanied by positive surgical margins. For those patients, the overall 5-year survival sharply decreases to 7%.16,355,356 It has been demonstrated that adjuvant chemotherapy can improve OS, but postoperative complications limit the intended treatment in 50% of patients.357 For patients with borderline or locally advanced pancreatic cancer, neoadjuvant therapy can convert unresectable disease into a potentially resectable state, which benefits OS.358
Current guidelines recommend FOLFIRINOX, mFOLFIRINOX, gemcitabine, or gemcitabine plus nab-paclitaxel for preoperative pancreatic cancer treatment. From 1997 to 2011, the first-line chemotherapy for patients with advanced pancreatic cancer was gemcitabine monotherapy.10 The treatment method changed in 2011 when FOLFIRINOX demonstrated a better survival benefit compared with that of gemcitabine monotherapy.11 Thereafter, gemcitabine plus nab-paclitaxel was adopted as another first-line treatment option. This treatment method had a better survival benefit (median OS 8.5 vs. 6.7 months, hazard ratio 0.72, 95% confidence interval 0.62–0.83, P < 0.001) compared with gemcitabine monotherapy.12 According to a large-scale retrospective study, FOLFIRINOX and nab-paclitaxel plus gemcitabine have similar outcomes.359 However, these therapeutic methods are reserved for otherwise healthy patients with good performance status. For the elderly and patients with a poor performance status, gemcitabine monotherapy is still considered a more tolerable treatment.360 The updated pancreatic cancer treatment guidelines recommend pembrolizumab immunotherapy for patients that carry mismatch repair (MMR) deficiencies or microsatellite instability after the failure of first-line therapies.361 As a second-line treatment option for pancreatic cancer, a combination of 5-FU and liposomal irinotecan is the only approved therapeutic method.362 In addition to these cytotoxic chemotherapies, novel agents are also under active investigation. The addition of leucovorin to S-1 improved PFS in patients with gemcitabine-refractory advanced pancreatic cancer.363 For patients with pancreatic cancer, novel therapeutic trials targeting DNA repair, gene mutations, tumor metabolism, the tumor microenvironment, or immune checkpoints might improve their prognosis (Fig. 4 and Table 2).
Fig. 4.
Current clinical strategies in pancreatic cancer. For patients with pancreatic cancer, the primary clinical strategies rely on chemotherapy, whereas novel therapeutic agents targeting DNA repair, gene mutations, tumor metabolism, tumor microenvironments, or immune checkpoints might improve their prognosis. Currently, increasing interest has emerged in combined chemotherapy and immunotherapy or targeted therapy
Table 2.
Ongoing clinical trials for pancreatic cancer treatment
| Condition or disease | Intervention/treatment | Target | ClinicalTrials.gov identifier | Phase | Status and results | Reference |
|---|---|---|---|---|---|---|
| Metastatic PDAC | Mesenchymal stromal cell-derived exosomes with KRASG12D siRNA | KRAS | NCT03608631 | 1 | Not yet recruiting | 365 |
| NTRK fusion-positive solid tumors | Entrectinbi | NTRK inhibitor | NCT02568267 | 2 | Recruiting | 366 |
| NTRK fusion-positive solid tumors | Larotrectinib | NTRK inhibitor | NCT02576431 | 2 | Recruiting | 367 |
| Advanced solid tumors | Ceritinib | ALK inhibitor | NCT02227940 | 1 | Completed | |
| BRCA-mutated metastatic pancreatic cancer | Olaparib maintenance | PARP inhibitor | NCT02184195 | 3 | Completed, ORR and PFS improved | 14 |
| Advanced PDAC | Nivolumab with cabiralizumab with or without chemotherapy | Anti-PD-1 antibodies and CSF-1R antibodies | NCT03336216 | 2 | Active, not recruiting | |
| Metastatic pancreatic cancer | Pembrolizumab and BL-8040 | Anti-PD-1 antibodies and CXCR4 antagonist | NCT02907099 | 2 | Active, not recruiting | |
| Non-colorectal MSI-H/dMMR-deficient cancer | Pembrolizumab | anti-PD-1 antibodies | NCT02628067 | 2 | Completed, ORR 18.2%, CR 4.5%, and PR 13.6% for PDAC | 373 |
| Metastatic PDAC | Durvalumab with or without tremelimumab | Anti-PD-L1 antibodies, anti-CTLA-4 antibodies | NCT02558894 | 2 | Completed, tolerated, ORR 3.1% for combination therapy | |
| Locally advanced or metastatic RAS-mutant solid tumors | Avelumab, binimetinib, and talazoparib, second line | Anti-PD-L1 antibodies, MEK inhibitor, PARP inhibitor | NCT03637491 | 2 | Recruiting | |
| Metastatic PDAC | Devimistat with mFOLFIRINOX | Tricarboxylic acid cycle modulation | NCT03504423 | 3 | Recruiting | 378 |
| Advanced or metastatic PDAC | Hydroxychloroquine with gemcitabine/nab-paclitaxel | Autophagy inhibitor | NCT01506973 | 1/2 | Active, not recruiting | |
| Advanced or metastatic PDAC | Hydroxychloroquine and trametinib | Autophagy inhibitor, MEK inhibitor | NCT03825289 | 1 | Recruiting | |
| Advanced or metastatic PDAC | Paclitaxel liposome and S-1 | – | NCT04217096 | Active, not recruiting | ||
| Stage IV untreated PDAC | PEGPH20 based on nab-paclitaxel and gemcitabine | Recombinant human hyaluronidase | NCT01839487 | 2 | Completed, improved PFS and ORR | 375 |
|
Previously treated Hyaluronan high metastatic PDAC |
PEGPH20 with pembrolizumab | Recombinant human hyaluronidase, anti-PD-1 antibodies | NCT03634332 | 2 | Recruiting | |
| Advanced solid tumors | Defactinib with pembrolizumab | FAK inhibitor, PD-1 antagonist | NCT02758587 | 1/2 | Recruiting | |
| PDAC | GSK2256098 with trametinib | FAK inhibitor, MEK inhibitor | NCT02428270 | 1 | Active, not recruiting | |
| Unresectable pancreatic cancer | Pamrevlumab based on gemcitabine and nab-paclitaxel | CTGF antagonism | NCT02210559 | 1/2 | Active, not recruiting | |
| Pancreatic cancer | CAR T cells, second line | NCT03323944 | 1 | Active, not recruiting | ||
| Metastatic pancreatic cancer | Paclitaxel and gemcitabine plus or without napabucasin | STAT3 and cancer cell stemness inhibitor | NCT03721744 | 3 | Recruiting | |
| Metastatic PDAC | Napabucasin plus nab-paclitaxel with gemcitabine | Cancer stemness inhibitor | NCT02993731 | 3 | Active, not recruiting |
CR complete response, ORR objective response rate, PARP poly-(ADP-ribose) polymerase, PDAC pancreatic ductal adenocarcinomas, PFS progression-free survival, PR partial response
The high prevalence of KRAS mutations (carried by >90% of pancreatic cancer patients) has led to considerable interest in KRAS-targeted therapies. Unfortunately, current approaches for direct targeting of mutant KRAS protein are ineffective due to their high affinity for GTP and/or GDP.36,364 An alternative strategy was developed to target KRAS by using exosomes or small EVs loaded with small interfering RNAs that target KRASG12D. This study has entered a phase 1 clinical trial for patients with metastatic pancreatic cancer (NCT03608631).365 Furthermore, the National Cancer Institute in the United States also established the RAS Initiative in 2013 to explore effective therapies for RAS-related cancers (https://www.cancer.gov/research/key-initiatives/ras).
Increasing interest has also emerged in the targeting of low-prevalence, actionable aberrations, including BRCA1/2, NTRK1/2/3, or MMR deficiencies. In three phase 1 and 2 clinical trials evaluating the treatment of patients with advanced or metastatic NTRK fusion-positive solid tumors with a potent TRK inhibitor, entrectinib, a complete response (CR) of 7% and a partial response (PR) of 50% were achieved (NCT02097810, NCT02568267, EudraCT 2012-000148-88).366 In other clinical trials evaluating the efficacy of the TRK inhibitor larotrecitinib, the objective response rate (ORR) reached 75% among 55 patients with NTRK fusion-positive tumors, including one pancreatic cancer patient who had a PR (NCT02122913, NCT02637687, NCT02576431).367 Larotrectinib and entrectinib received FDA approval for use as tissue-agnostic indicators in patients with solid tumors with NTRK fusions in November 2018 and August 2019, respectively. Furthermore, the use of ALK inhibitors in pancreatic cancer patients is currently under investigation. ALK rearrangements have also been established as a promising molecular target in other malignancies, e.g., non-small cell lung cancer.368 In a 2017 study, five cases harboring an ALK fusion gene were identified via comprehensive genomic profiling of 3170 pancreatic cancer patients. Among them, four patients were treated with an ALK inhibitor, three of which showed a stable radiographic response and/or normalization of serum CA19-9.369 A phase 1 study to combine ceritinib, a novel ALK inhibitor, with chemotherapy for the treatment of advanced pancreatic cancer has just completed (NCT02227940).
Therapeutic strategies based on BRCA-poly (ADP-ribose) polymerase (PARP) synthetic lethality have shown to be effective for the treatment of patients with BRCA1/2 mutations. A multicenter phase 2 trial of olaparib (a PARP inhibitor) for the treatment of patients with a germline BRCA1/2 mutation and recurrent cancer achieved an ORR of 26.2% overall and an ORR of 21.7% for pancreatic cancer (NCT01078662).370 In the POLO trial and a randomized phase 3 trial for patients with metastatic pancreatic cancer with a germline BRCA1/2 mutation after at least 16 weeks of platinum-based chemotherapy, olaparib as a maintenance therapy achieved an ORR of 20% with a PFS median of 7.4 months, compared with 10% and 3.8 months in the placebo group (NCT02184195).14
The tumor microenvironment is another area of interest in pancreatic cancer therapeutic exploration. Immune checkpoint inhibitors (ICIs) have emerged as a new treatment paradigm for patients with certain types of tumors. However, the results of early clinical trials investigating the efficacy of the anti-CTLA-4 antibody ipilimumab or the anti-PD-L1 antibody BMS-936559 in patients with advanced pancreatic cancer were disappointing.371,372 In the phase 2 KEYNOTE-158 study of pembrolizumab (an anti-PD-1 monoclonal antibody), an ORR of 34.3% was achieved in patients with previously treated, advanced non-colorectal high microsatellite instability (MSI-H)/DNA MMR (dMMR)-deficient cancer, and in pancreatic cancer patients, an ORR of 18.2%, a CR of 4.5%, and a PR of 13.6% were achieved (NCT02628067).373 Based on these results, the FDA granted accelerated approval to pembrolizumab for adult and pediatric patients with unresectable MSI-H or dMMR solid tumors in May 2017. Clinical trials of PD-1 blockade or PD-L1 combined with other ICIs or blockade are currently ongoing (NCT03336216, NCT02907099, NCT02558894, NCT03637491). Hyaluronic acid is a hydrophilic glycosaminoglycan whose overproduction in the stroma of patients with pancreatic cancer leads to increased interstitial tumor pressure, reducing tumor perfusion and access of anticancer drugs to the tumor.374 A phase 2 trial showed that the addition of PEGPH20 (pegvorhyaluronidase-α, recombinant human hyaluronidase) to nab-paclitaxel and gemcitabine resulted in significant improvements in ORR (45 vs. 31%) and PFS (9.2 vs. 5.2 months) in pancreatic cancer patients with high hyaluronic acid expression levels (NCT01839487).375 However, a later phase 3 study failed to show improvements in OS or PFS, and the results did not support additional development of PEGPH20 as a treatment for metastatic PDAC, as a high rate of dose holds and reductions in the PEGPH20 arm might have led to lower chemotherapy drug exposure, which could have contributed to the inferior survival outcomes with PEGPH20.376
Metabolic reprogramming can also be a therapeutic target in pancreatic cancer. Devimistat, which inhibits enzymes in the mitochondrial tricarboxylic acid cycle, is hypothesized to act synergistically with cytotoxic agents to induce decreased production of anabolic intermediates required for DNA damage repair. A phase 1 study of devimistat combined with mFOFIRINOX for the treatment of metastatic pancreatic cancer patients showed an ORR of 61%, including a CRR of 17% (NCT01835041).377 Additional clinical trials evaluating the safety and efficacy of devimistat combined with gemcitabine and nab-paclitaxel or mFOLFIRINOX are ongoing (NCT03435289, NCT03504423).378 Inhibition of autophagy in pancreatic cancer primary tumors contributes to a metabolic defect that leads to decreased mitochondrial OXPHOS and significant growth suppression.213 In a phase 2 study, autophagy inhibition using HCQ in patients with metastatic pancreatic cancer failed to achieve therapeutic efficacy.379 Further clinical trials to test the effectiveness of HCQ combined with cytotoxic agents or MEK inhibitors are undergoing (NCT01506973, NCT03825289).
A number of combined therapeutic approaches have already been introduced. Preclinical research has shown that FAK inhibition can lead to decreased levels of fibrosis, synergy with chemotherapy and ICIs, and improved survival outcomes in the KPC mouse model of pancreatic cancer.254 Currently, a number of clinical trials to investigate the efficacy of combinations of FAK inhibitors and ICIs and/or cytotoxic agents or MEK inhibitors are ongoing (NCT02546531, NCT02758587, NCT02428270). Connective tissue growth factor (CTGF), a member of the CCN family of secreted proteins, is involved in ECM production, desmoplasia, and tumor progression.380 Preclinical data indicated that CTGF antagonism with the therapeutic monoclonal human antibody pamrevlumab enhanced the response to gemcitabine in pancreatic cancer mouse models.381 The safety and efficacy of the agent in combination with gemcitabine and nab-paclitaxel for the treatment of patients with unresectable pancreatic cancer were evaluated in a phase 1/2 trial (NCT02210559). The results indicated that the addition of pamrevlumab to neoadjuvant therapy in locally advanced pancreatic cancer patients might lead to higher resectability and resection rate. Currently, two phase 3 studies to investigate the efficacy of the addition of napabucasin to gemcitabine and nab-paclitaxel for treating metastatic pancreatic cancer patients are ongoing (NCT03721744, NCT02993731).382
Preclinical investigations in pancreatic cancer PDX models identified a small subset of tumor cells (termed CSCs) that determines self-renewal and metastatic phenotypes of pancreatic cancer.383 A phase 1b/2 study of the cancer stemness inhibitor napabucasin combined with gemcitabine and nab-paclitaxel for treating metastatic pancreatic cancer achieved a CR of 3% and a PR of 42%.
In summary, for patients with resectable pancreatic cancer, mFOLFIRINOX and nab-paclitaxel plus gemcitabine regimens were shown to achieve the longest median OS. For patients with advanced or metastatic pancreatic cancer, treatment regimens based on the identification of potentially actionable alterations in small subsets of patients via comprehensive genomic profiling and monitoring of the efficacy of therapeutic strategies are currently being explored. The POLO trial, which involved the identification of germline BRCA mutations and targeted therapy, proved to be effective, and such approaches are worth further exploration.
Conclusions and future perspectives
Pancreatic cancer remains one of the most common and deadly cancers with limited options for effective therapy. Meaningful clinical progress in diagnostic investigations, surgical techniques, and systemic therapies are certain to improve pancreatic cancer patient survival. A deeper understanding of the biology and genetics of pancreatic cancer, including new insight into driver gene mutations, tumor metabolism, and the tumor microenvironment, might lead to promising and innovative therapeutic strategies. It has been widely proposed that targeting a single molecule or pathway is unlikely to yield more pancreatic cancer therapies. Both subtype-specific therapy and combined therapy might represent more promising strategies to control tumor progression. Recent advances showing that pancreatic cancer patients with germline BRCA mutations benefit from PARP inhibitors might inspire novel strategies that further increase the clinical efficacy of subtype-specific therapy. While clinical progress has never been clearer in the improvement of outcomes in pancreatic cancer patients, the path to clinical translation of novel therapeutic approaches will be greatly enhanced by the use of more sophisticated animal models and multidisciplinary clinical collaborations.
Acknowledgements
This work was supported by the following: the National Key Research and Development Program of China (No. 2017YFC1308604); National Natural Science Foundation of China (Nos. 81802903, 81672820, 81940074, and 81872356); the Program of Shanghai Academic Research Leader (No. 20XD1400900); the Shanghai International Science and Technology Collaboration Program.
Author contributions
Q.Z.-D., L.-X.Q., and Y.Z. designed the review. S.W., Y.Z., and F.Y. drafted the manuscript. L.Z., X.-Q.Z., Z.-F.W., X.-L.W., C.-H.Z., J.-Y.Y., B.-Y.H., B.K., and D.-L.F. prepared the figures. C.B., L.-X.Q., and Q.-Z.D. participated in its coordination and modification. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Shun Wang, Yan Zheng
Contributor Information
Yue Zhao, Email: yue.zhao@uk-koeln.de.
Lun-Xiu Qin, Email: qinlx@fudan.edu.cn.
Qiong-Zhu Dong, Email: qzhdong@fudan.edu.cn.
References
- 1.Sung H, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 3.Miller KD, et al. Cancer statistics for adolescents and young adults, 2020. CA Cancer J. Clin. 2020;70:443–459. doi: 10.3322/caac.21637. [DOI] [PubMed] [Google Scholar]
- 4.Rawla P, Sunkara T, Gaduputi V. Epidemiology of pancreatic cancer: global trends, etiology and risk factors. World J. Oncol. 2019;10:10–27. doi: 10.14740/wjon1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abe T, et al. Deleterious germline mutations are a risk factor for neoplastic progression among high-risk individuals undergoing pancreatic surveillance. J. Clin. Oncol. 2019;37:1070–1080. doi: 10.1200/JCO.18.01512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kromrey ML, et al. Prospective study on the incidence, prevalence and 5-year pancreatic-related mortality of pancreatic cysts in a population-based study. Gut. 2018;67:138–145. doi: 10.1136/gutjnl-2016-313127. [DOI] [PubMed] [Google Scholar]
- 7.Kleeff J, et al. Pancreatic cancer. Nat. Rev. Dis. Prim. 2016;2:16022. doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 8.Kuehn BM. Looking to long-term survivors for improved pancreatic cancer treatment. JAMA. 2020;324:2242–2244. doi: 10.1001/jama.2020.21717. [DOI] [PubMed] [Google Scholar]
- 9.Picozzi VJ, et al. Five-year actual overall survival in resected pancreatic cancer: a contemporary single-institution experience from a multidisciplinary perspective. Ann. Surg. Oncol. 2017;24:1722–1730. doi: 10.1245/s10434-016-5716-z. [DOI] [PubMed] [Google Scholar]
- 10.Burris HA, 3rd, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J. Clin. Oncol. 1997;15:2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 11.Conroy T, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 12.Von Hoff DD, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 2013;369:1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conroy T, et al. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N. Engl. J. Med. 2018;379:2395–2406. doi: 10.1056/NEJMoa1809775. [DOI] [PubMed] [Google Scholar]
- 14.Golan T, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med. 2019;381:317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Assenat E, et al. Phase II study evaluating the association of gemcitabine, trastuzumab and erlotinib as first-line treatment in patients with metastatic pancreatic adenocarcinoma (GATE 1) Int. J. Cancer. 2021;148:682–691. doi: 10.1002/ijc.33225. [DOI] [PubMed] [Google Scholar]
- 16.Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet. 2020;395:2008–2020. doi: 10.1016/S0140-6736(20)30974-0. [DOI] [PubMed] [Google Scholar]
- 17.Buscail L, Bournet B, Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2020;17:153–168. doi: 10.1038/s41575-019-0245-4. [DOI] [PubMed] [Google Scholar]
- 18.Singh K, et al. Kras mutation rate precisely orchestrates ductal derived pancreatic intraepithelial neoplasia and pancreatic cancer. Lab. Invest. 2021;101:177–192. doi: 10.1038/s41374-020-00490-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Witkiewicz AK, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mann KM, Ying H, Juan J, Jenkins NA, Copeland NG. KRAS-related proteins in pancreatic cancer. Pharm. Ther. 2016;168:29–42. doi: 10.1016/j.pharmthera.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Qian Y, et al. Molecular alterations and targeted therapy in pancreatic ductal adenocarcinoma. J. Hematol. Oncol. 2020;13:130. doi: 10.1186/s13045-020-00958-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christenson ES, Jaffee E, Azad NS. Current and emerging therapies for patients with advanced pancreatic ductal adenocarcinoma: a bright future. Lancet Oncol. 2020;21:e135–e145. doi: 10.1016/S1470-2045(19)30795-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Connor AA, et al. Integration of genomic and transcriptional features in pancreatic cancer reveals increased cell cycle progression in metastases. Cancer Cell. 2019;35:267–282.e267. doi: 10.1016/j.ccell.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waddell N, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dardare, J., Witz, A., Merlin, J. L., Gilson, P. & Harlé, A. SMAD4 and the TGFβ pathway in patients with pancreatic ductal adenocarcinoma. Int. J. Mol. Sci.21, 3534 (2020). [DOI] [PMC free article] [PubMed]
- 27.Yachida S, et al. Clinical significance of the genetic landscape of pancreatic cancer and implications for identification of potential long-term survivors. Clin. Cancer Res. 2012;18:6339–6347. doi: 10.1158/1078-0432.CCR-12-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ying H, et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016;30:355–385. doi: 10.1101/gad.275776.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen GQ, Aleassa EM, Walsh RM, Morris-Stiff G. Next-generation sequencing in pancreatic cancer. Pancreas. 2019;48:739–748. doi: 10.1097/MPA.0000000000001324. [DOI] [PubMed] [Google Scholar]
- 30.Holter S, et al. Germline BRCA mutations in a large clinic-based cohort of patients with pancreatic adenocarcinoma. J. Clin. Oncol. 2015;33:3124–3129. doi: 10.1200/JCO.2014.59.7401. [DOI] [PubMed] [Google Scholar]
- 31.Murphy KM, et al. Evaluation of candidate genes MAP2K4, MADH4, ACVR1B, and BRCA2 in familial pancreatic cancer: deleterious BRCA2 mutations in 17% Cancer Res. 2002;62:3789–3793. [PubMed] [Google Scholar]
- 32.Tanaka M, et al. Revisions of international consensus Fukuoka guidelines for the management of IPMN of the pancreas. Pancreatology. 2017;17:738–753. doi: 10.1016/j.pan.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Chandana S, Babiker HM, Mahadevan D. Therapeutic trends in pancreatic ductal adenocarcinoma (PDAC) Expert Opin. Investig. Drugs. 2019;28:161–177. doi: 10.1080/13543784.2019.1557145. [DOI] [PubMed] [Google Scholar]
- 34.Feigin ME, et al. Recurrent noncoding regulatory mutations in pancreatic ductal adenocarcinoma. Nat. Genet. 2017;49:825–833. doi: 10.1038/ng.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hung, Y. H., Hsu, M. C., Chen, L. T., Hung, W. C. & Pan, M. R. Alteration of epigenetic modifiers in pancreatic cancer and its clinical implication. J. Clin. Med.8, 903 (2019). [DOI] [PMC free article] [PubMed]
- 36.Bailey P, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 37.Sausen M, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat. Commun. 2015;6:7686. doi: 10.1038/ncomms8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 39.Wang, M. & Lin, H. Understanding the function of mammalian sirtuins and protein lysine acylation. Annu. Rev. Biochem. 10.1146/annurev-biochem-082520-125411 (2021). [DOI] [PubMed]
- 40.Fritsche P, et al. HDAC2 mediates therapeutic resistance of pancreatic cancer cells via the BH3-only protein NOXA. Gut. 2009;58:1399–1409. doi: 10.1136/gut.2009.180711. [DOI] [PubMed] [Google Scholar]
- 41.Aghdassi A, et al. Recruitment of histone deacetylases HDAC1 and HDAC2 by the transcriptional repressor ZEB1 downregulates E-cadherin expression in pancreatic cancer. Gut. 2012;61:439–448. doi: 10.1136/gutjnl-2011-300060. [DOI] [PubMed] [Google Scholar]
- 42.Mishra VK, et al. Histone deacetylase class-I inhibition promotes epithelial gene expression in pancreatic cancer cells in a BRD4- and MYC-dependent manner. Nucleic Acids Res. 2017;45:6334–6349. doi: 10.1093/nar/gkx212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kugel S, et al. SIRT6 suppresses pancreatic cancer through control of Lin28b. Cell. 2016;165:1401–1415. doi: 10.1016/j.cell.2016.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collisson EA, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tiriac H, et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 2018;8:1112–1129. doi: 10.1158/2159-8290.CD-18-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan-Seng-Yue M, et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat. Genet. 2020;52:231–240. doi: 10.1038/s41588-019-0566-9. [DOI] [PubMed] [Google Scholar]
- 47.Moffitt RA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puleo F, et al. Stratification of pancreatic ductal adenocarcinomas based on tumor and microenvironment features. Gastroenterology. 2018;155:1999–2013.e1993. doi: 10.1053/j.gastro.2018.08.033. [DOI] [PubMed] [Google Scholar]
- 49.Cancer Genome Atlas Research Network. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell32, 185–203.e113 (2017). [DOI] [PMC free article] [PubMed]
- 50.Noll EM, et al. CYP3A5 mediates basal and acquired therapy resistance in different subtypes of pancreatic ductal adenocarcinoma. Nat. Med. 2016;22:278–287. doi: 10.1038/nm.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knudsen ES, et al. Pancreatic cancer cell lines as patient-derived avatars: genetic characterisation and functional utility. Gut. 2018;67:508–520. doi: 10.1136/gutjnl-2016-313133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maurer C, et al. Experimental microdissection enables functional harmonisation of pancreatic cancer subtypes. Gut. 2019;68:1034–1043. doi: 10.1136/gutjnl-2018-317706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tago K, et al. Oncogenic Ras mutant causes the hyperactivation of NF-κB via acceleration of its transcriptional activation. Mol. Oncol. 2019;13:2493–2510. doi: 10.1002/1878-0261.12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Loncle C, et al. IL17 functions through the novel REG3β-JAK2-STAT3 inflammatory pathway to promote the transition from chronic pancreatitis to pancreatic cancer. Cancer Res. 2015;75:4852–4862. doi: 10.1158/0008-5472.CAN-15-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baumgart S, et al. GSK-3β Governs Inflammation-Induced NFATc2 Signaling Hubs to Promote Pancreatic Cancer Progression. Mol. Cancer Ther. 2016;15:491–502. doi: 10.1158/1535-7163.MCT-15-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eibl G, Rozengurt E. KRAS, YAP, and obesity in pancreatic cancer: a signaling network with multiple loops. Semin. Cancer Biol. 2019;54:50–62. doi: 10.1016/j.semcancer.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hong DS, et al. KRAS(G12C) inhibition with sotorasib in advanced solid tumors. N. Engl. J. Med. 2020;383:1207–1217. doi: 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Igbinigie E, Guo F, Jiang SW, Kelley C, Li J. Dkk1 involvement and its potential as a biomarker in pancreatic ductal adenocarcinoma. Clin. Chim. Acta. 2019;488:226–234. doi: 10.1016/j.cca.2018.11.023. [DOI] [PubMed] [Google Scholar]
- 60.Ebrahimi S, et al. Targeting the Akt/PI3K signaling pathway as a potential therapeutic strategy for the treatment of pancreatic cancer. Curr. Med. Chem. 2017;24:1321–1331. doi: 10.2174/0929867324666170206142658. [DOI] [PubMed] [Google Scholar]
- 61.Song M, Bode AM, Dong Z, Lee MH. AKT as a therapeutic target for cancer. Cancer Res. 2019;79:1019–1031. doi: 10.1158/0008-5472.CAN-18-2738. [DOI] [PubMed] [Google Scholar]
- 62.Eser S, et al. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell. 2013;23:406–420. doi: 10.1016/j.ccr.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 63.Baer R, et al. Pancreatic cell plasticity and cancer initiation induced by oncogenic Kras is completely dependent on wild-type PI 3-kinase p110α. Genes Dev. 2014;28:2621–2635. doi: 10.1101/gad.249409.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Albury TM, et al. Constitutively active Akt1 cooperates with KRas(G12D) to accelerate in vivo pancreatic tumor onset and progression. Neoplasia. 2015;17:175–182. doi: 10.1016/j.neo.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma J, et al. IGF-1 mediates PTEN suppression and enhances cell invasion and proliferation via activation of the IGF-1/PI3K/Akt signaling pathway in pancreatic cancer cells. J. Surg. Res. 2010;160:90–101. doi: 10.1016/j.jss.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 66.Zheng Y, et al. Insulin-like growth factor 1-induced enolase 2 deacetylation by HDAC3 promotes metastasis of pancreatic cancer. Signal Transduct. Target Ther. 2020;5:53. doi: 10.1038/s41392-020-0146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu X, Yu Y, Zong K, Lv P, Gu Y. Up-regulation of IGF2BP2 by multiple mechanisms in pancreatic cancer promotes cancer proliferation by activating the PI3K/Akt signaling pathway. J. Exp. Clin. Cancer Res. 2019;38:497. doi: 10.1186/s13046-019-1470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fu Y, et al. TMEM158 promotes pancreatic cancer aggressiveness by activation of TGFβ1 and PI3K/AKT signaling pathway. J. Cell Physiol. 2020;235:2761–2775. doi: 10.1002/jcp.29181. [DOI] [PubMed] [Google Scholar]
- 69.Jiang N, et al. Role of PI3K/AKT pathway in cancer: the framework of malignant behavior. Mol. Biol. Rep. 2020;47:4587–4629. doi: 10.1007/s11033-020-05435-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ren X, et al. lncRNA-PLACT1 sustains activation of NF-κB pathway through a positive feedback loop with IκBα/E2F1 axis in pancreatic cancer. Mol. Cancer. 2020;19:35. doi: 10.1186/s12943-020-01153-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khurana, N., Dodhiawala, P. B., Bulle, A. & Lim, K. H. Deciphering the role of innate immune NF-ĸB pathway in pancreatic cancer. Cancers12, 2675 (2020). [DOI] [PMC free article] [PubMed]
- 72.Prabhu L, Mundade R, Korc M, Loehrer PJ, Lu T. Critical role of NF-κB in pancreatic cancer. Oncotarget. 2014;5:10969–10975. doi: 10.18632/oncotarget.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol. 2018;18:309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 74.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 75.Sun Q, et al. Pin1 promotes pancreatic cancer progression and metastasis by activation of NF-κB-IL-18 feedback loop. Cell Prolif. 2020;53:e12816. doi: 10.1111/cpr.12816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou L, et al. MicroRNA-628-5p inhibits invasion and migration of human pancreatic ductal adenocarcinoma via suppression of the AKT/NF-kappa B pathway. J. Cell Physiol. 2020;235:8141–8154. doi: 10.1002/jcp.29468. [DOI] [PubMed] [Google Scholar]
- 77.Garg B, et al. NFκB in pancreatic stellate cells reduces infiltration of tumors by cytotoxic T cells and killing of cancer cells, via up-regulation of CXCL12. Gastroenterology. 2018;155:880–891.e888. doi: 10.1053/j.gastro.2018.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ratnam NM, et al. NF-κB regulates GDF-15 to suppress macrophage surveillance during early tumor development. J. Clin. Invest. 2017;127:3796–3809. doi: 10.1172/JCI91561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pramanik, K. C., Makena, M. R., Bhowmick, K. & Pandey, M. K. Advancement of NF-κB signaling pathway: a novel target in pancreatic cancer. Int. J. Mol. Sci. 19, 3890 (2018). [DOI] [PMC free article] [PubMed]
- 80.Song Y, Tang MY, Chen W, Wang Z, Wang SL. High JAK2 protein expression predicts a poor prognosis in patients with resectable pancreatic ductal adenocarcinoma. Dis. Markers. 2020;2020:7656031. doi: 10.1155/2020/7656031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: implications for cancer therapy. Nat. Rev. Cancer. 2016;16:131–144. doi: 10.1038/nrc.2016.14. [DOI] [PubMed] [Google Scholar]
- 82.Doi T, et al. The JAK/STAT pathway is involved in the upregulation of PD-L1 expression in pancreatic cancer cell lines. Oncol. Rep. 2017;37:1545–1554. doi: 10.3892/or.2017.5399. [DOI] [PubMed] [Google Scholar]
- 83.Lu C, Talukder A, Savage NM, Singh N, Liu K. JAK-STAT-mediated chronic inflammation impairs cytotoxic T lymphocyte activation to decrease anti-PD-1 immunotherapy efficacy in pancreatic cancer. Oncoimmunology. 2017;6:e1291106. doi: 10.1080/2162402X.2017.1291106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Biffi G, et al. IL1-induced JAK/STAT signaling is antagonized by TGFβ to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov. 2019;9:282–301. doi: 10.1158/2159-8290.CD-18-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mao W, Mai J, Peng H, Wan J, Sun T. YAP in pancreatic cancer: oncogenic role and therapeutic strategy. Theranostics. 2021;11:1753–1762. doi: 10.7150/thno.53438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Salcedo Allende MT, et al. Overexpression of Yes associated protein 1, an independent prognostic marker in patients with pancreatic ductal adenocarcinoma, correlated with liver metastasis and poor prognosis. Pancreas. 2017;46:913–920. doi: 10.1097/MPA.0000000000000867. [DOI] [PubMed] [Google Scholar]
- 87.Zhang W, et al. Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci. Signal. 2014;7:ra42. doi: 10.1126/scisignal.2005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gruber R, et al. YAP1 and TAZ control pancreatic cancer initiation in mice by direct up-regulation of JAK-STAT3 signaling. Gastroenterology. 2016;151:526–539. doi: 10.1053/j.gastro.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Q, et al. Fbxw7 deletion accelerates Kras(G12D)-driven pancreatic tumorigenesis via Yap accumulation. Neoplasia. 2016;18:666–673. doi: 10.1016/j.neo.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Murakami S, et al. A Yap-Myc-Sox2-p53 regulatory network dictates metabolic homeostasis and differentiation in Kras-driven pancreatic ductal adenocarcinomas. Dev. Cell. 2019;51:113–128.e119. doi: 10.1016/j.devcel.2019.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kapoor A, et al. Yap1 activation enables bypass of oncogenic kras addiction in pancreatic cancer. Cell. 2019;179:1239. doi: 10.1016/j.cell.2019.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tu, B. et al. YAP1 oncogene is a context-specific driver for pancreatic ductal adenocarcinoma. JCI Insight4, e130811 (2019). [DOI] [PMC free article] [PubMed]
- 93.Shao DD, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Murakami S, et al. Yes-associated protein mediates immune reprogramming in pancreatic ductal adenocarcinoma. Oncogene. 2017;36:1232–1244. doi: 10.1038/onc.2016.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu J, et al. Activation of the intrinsic fibroinflammatory program in adult pancreatic acinar cells triggered by Hippo signaling disruption. PLoS Biol. 2019;17:e3000418. doi: 10.1371/journal.pbio.3000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jiang Z, et al. Inhibiting YAP expression suppresses pancreatic cancer progression by disrupting tumor-stromal interactions. J. Exp. Clin. Cancer Res. 2018;37:69. doi: 10.1186/s13046-018-0740-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ram Makena, M. et al. Wnt/β-catenin signaling: the culprit in pancreatic carcinogenesis and therapeutic resistance. Int. J. Mol. Sci. 20, 4242 (2019). [DOI] [PMC free article] [PubMed]
- 98.Nakamoto M, Hisaoka M. Clinicopathological implications of wingless/int1 (WNT) signaling pathway in pancreatic ductal adenocarcinoma. J. UOEH. 2016;38:1–8. doi: 10.7888/juoeh.38.1. [DOI] [PubMed] [Google Scholar]
- 99.Morris JPT, Cano DA, Sekine S, Wang SC, Hebrok M. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J. Clin. Invest. 2010;120:508–520. doi: 10.1172/JCI40045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Manegold, P. et al. Differentiation therapy targeting the β-Catenin/CBP interaction in pancreatic cancer. Cancers10, 95 (2018). [DOI] [PMC free article] [PubMed]
- 101.Ercan G, Karlitepe A, Ozpolat B. Pancreatic cancer stem cells and therapeutic approaches. Anticancer Res. 2017;37:2761–2775. doi: 10.21873/anticanres.11628. [DOI] [PubMed] [Google Scholar]
- 102.Zhang Y, et al. Canonical wnt signaling is required for pancreatic carcinogenesis. Cancer Res. 2013;73:4909–4922. doi: 10.1158/0008-5472.CAN-12-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li H, et al. WD repeat-containing protein 1 maintains β-catenin activity to promote pancreatic cancer aggressiveness. Br. J. Cancer. 2020;123:1012–1023. doi: 10.1038/s41416-020-0929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jiang H, et al. Activation of the Wnt pathway through Wnt2 promotes metastasis in pancreatic cancer. Am. J. Cancer Res. 2014;4:537–544. [PMC free article] [PubMed] [Google Scholar]
- 105.Bo H, Gao L, Chen Y, Zhang J, Zhu M. Upregulation of the expression of Wnt5a promotes the proliferation of pancreatic cancer cells in vitro and in a nude mouse model. Mol. Med. Rep. 2016;13:1163–1171. doi: 10.3892/mmr.2015.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu X, et al. MUC1 regulates cyclin D1 gene expression through p120 catenin and β-catenin. Oncogenesis. 2014;3:e107. doi: 10.1038/oncsis.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhi X, et al. MUC4-induced nuclear translocation of β-catenin: a novel mechanism for growth, metastasis and angiogenesis in pancreatic cancer. Cancer Lett. 2014;346:104–113. doi: 10.1016/j.canlet.2013.12.021. [DOI] [PubMed] [Google Scholar]
- 108.Chartier C, et al. Therapeutic targeting of tumor-derived R-spondin attenuates β-catenin signaling and tumorigenesis in multiple cancer types. Cancer Res. 2016;76:713–723. doi: 10.1158/0008-5472.CAN-15-0561. [DOI] [PubMed] [Google Scholar]
- 109.Criscimanna A, et al. PanIN-specific regulation of Wnt signaling by HIF2α during early pancreatic tumorigenesis. Cancer Res. 2013;73:4781–4790. doi: 10.1158/0008-5472.CAN-13-0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang Q, et al. Hypoxia-inducible factor-2α promotes tumor progression and has crosstalk with Wnt/β-catenin signaling in pancreatic cancer. Mol. Cancer. 2017;16:119. doi: 10.1186/s12943-017-0689-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Beans C. News feature: targeting metastasis to halt cancer’s spread. Proc. Natl Acad. Sci. USA. 2018;115:12539–12543. doi: 10.1073/pnas.1818892115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hegde S, et al. Dendritic cell paucity leads to dysfunctional immune surveillance in pancreatic cancer. Cancer Cell. 2020;37:289–307.e289. doi: 10.1016/j.ccell.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Grünwald B, et al. Pancreatic premalignant lesions secrete tissue inhibitor of metalloproteinases-1, which activates hepatic stellate cells via CD63 signaling to create a premetastatic niche in the liver. Gastroenterology. 2016;151:1011–1024.e1017. doi: 10.1053/j.gastro.2016.07.043. [DOI] [PubMed] [Google Scholar]
- 114.Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–1558. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 115.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Guo X, et al. Circular RNA circBFAR promotes the progression of pancreatic ductal adenocarcinoma via the miR-34b-5p/MET/Akt axis. Mol. Cancer. 2020;19:83. doi: 10.1186/s12943-020-01196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cui L, et al. Small nucleolar noncoding RNA SNORA23, up-regulated in human pancreatic ductal adenocarcinoma, regulates expression of spectrin repeat-containing nuclear envelope 2 to promote growth and metastasis of xenograft tumors in mice. Gastroenterology. 2017;153:292–306.e292. doi: 10.1053/j.gastro.2017.03.050. [DOI] [PubMed] [Google Scholar]
- 119.Guo K, et al. The novel KLF4/MSI2 signaling pathway regulates growth and metastasis of pancreatic cancer. Clin. Cancer Res. 2017;23:687–696. doi: 10.1158/1078-0432.CCR-16-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.He P, Yang JW, Yang VW, Bialkowska AB. Krüppel-like factor 5, Increased in pancreatic ductal adenocarcinoma, promotes proliferation, acinar-to-ductal metaplasia, pancreatic intraepithelial neoplasia, and tumor growth in mice. Gastroenterology. 2018;154:1494–1508.e1413. doi: 10.1053/j.gastro.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gupta R, et al. KLF7 promotes pancreatic cancer growth and metastasis by up-regulating ISG expression and maintaining Golgi complex integrity. Proc. Natl Acad. Sci. USA. 2020;117:12341–12351. doi: 10.1073/pnas.2005156117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kong Y, et al. circNFIB1 inhibits lymphangiogenesis and lymphatic metastasis via the miR-486-5p/PIK3R1/VEGF-C axis in pancreatic cancer. Mol. Cancer. 2020;19:82. doi: 10.1186/s12943-020-01205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tiwari A, et al. Loss of HIF1A from pancreatic cancer cells increases expression of PPP1R1B and degradation of p53 to promote invasion and metastasis. Gastroenterology. 2020;159:1882–1897.e1885. doi: 10.1053/j.gastro.2020.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Roe JS, et al. Enhancer reprogramming promotes pancreatic cancer metastasis. Cell. 2017;170:875–888.e820. doi: 10.1016/j.cell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Argentiero A, et al. Bone metastasis as primary presentation of pancreatic ductal adenocarcinoma: a case report and literature review. Clin. Case Rep. 2019;7:1972–1976. doi: 10.1002/ccr3.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wen J, et al. The Eighth Edition of the American Joint Committee on Cancer Distant Metastases Stage Classification for Metastatic Pancreatic Neuroendocrine Tumors Might Be Feasible for Metastatic Pancreatic Ductal Adenocarcinomas. Neuroendocrinology. 2020;110:364–376. doi: 10.1159/000502382. [DOI] [PubMed] [Google Scholar]
- 127.Avula LR, Hagerty B, Alewine C. Molecular mediators of peritoneal metastasis in pancreatic cancer. Cancer Metastasis Rev. 2020;39:1223–1243. doi: 10.1007/s10555-020-09924-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lee JW, et al. Hepatocytes direct the formation of a pro-metastatic niche in the liver. Nature. 2019;567:249–252. doi: 10.1038/s41586-019-1004-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Maddipati R, Stanger BZ. Pancreatic cancer metastases harbor evidence of polyclonality. Cancer Discov. 2015;5:1086–1097. doi: 10.1158/2159-8290.CD-15-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tsai S, et al. A serum-induced transcriptome and serum cytokine signature obtained at diagnosis correlates with the development of early pancreatic ductal adenocarcinoma metastasis. Cancer Epidemiol. Biomark. Prev. 2019;28:680–689. doi: 10.1158/1055-9965.EPI-18-0813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pausch TM, et al. Metastasis-associated fibroblasts promote angiogenesis in metastasized pancreatic cancer via the CXCL8 and the CCL2 axes. Sci. Rep. 2020;10:5420. doi: 10.1038/s41598-020-62416-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lowder CY, et al. Clinical implications of extensive lymph node metastases for resected pancreatic cancer. Ann. Surg. Oncol. 2018;25:4004–4011. doi: 10.1245/s10434-018-6763-4. [DOI] [PubMed] [Google Scholar]
- 133.Nishiwada S, et al. A microRNA signature identifies pancreatic ductal adenocarcinoma patients at risk for lymph node metastases. Gastroenterology. 2020;159:562–574. doi: 10.1053/j.gastro.2020.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sun W, Ren Y, Lu Z, Zhao X. The potential roles of exosomes in pancreatic cancer initiation and metastasis. Mol. Cancer. 2020;19:135. doi: 10.1186/s12943-020-01255-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Costa-Silva B, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hoshino A, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhou P, et al. NMIIA promotes tumor growth and metastasis by activating the Wnt/β-catenin signaling pathway and EMT in pancreatic cancer. Oncogene. 2019;38:5500–5515. doi: 10.1038/s41388-019-0806-6. [DOI] [PubMed] [Google Scholar]
- 138.Krebs AM, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 2017;19:518–529. doi: 10.1038/ncb3513. [DOI] [PubMed] [Google Scholar]
- 139.Yamada S, et al. Epithelial-to-mesenchymal transition predicts prognosis of pancreatic cancer. Surgery. 2013;154:946–954. doi: 10.1016/j.surg.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 140.Ellenrieder V, et al. Transforming growth factor beta1 treatment leads to an epithelial-mesenchymal transdifferentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Res. 2001;61:4222–4228. [PubMed] [Google Scholar]
- 141.Sato M, et al. BACH1 promotes pancreatic cancer metastasis by repressing epithelial genes and enhancing epithelial-mesenchymal transition. Cancer Res. 2020;80:1279–1292. doi: 10.1158/0008-5472.CAN-18-4099. [DOI] [PubMed] [Google Scholar]
- 142.Niu N, et al. Loss of Setd2 promotes Kras-induced acinar-to-ductal metaplasia and epithelia-mesenchymal transition during pancreatic carcinogenesis. Gut. 2020;69:715–726. doi: 10.1136/gutjnl-2019-318362. [DOI] [PubMed] [Google Scholar]
- 143.Zheng X, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Satoh K, Hamada S, Shimosegawa T. Involvement of epithelial to mesenchymal transition in the development of pancreatic ductal adenocarcinoma. J. Gastroenterol. 2015;50:140–146. doi: 10.1007/s00535-014-0997-0. [DOI] [PubMed] [Google Scholar]
- 145.Fink DM, Steele MM, Hollingsworth MA. The lymphatic system and pancreatic cancer. Cancer Lett. 2016;381:217–236. doi: 10.1016/j.canlet.2015.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ray K. Pancreatic cancer: pancreatic cancer exosomes prime the liver for metastasis. Nat. Rev. Gastroenterol. Hepatol. 2015;12:371. doi: 10.1038/nrgastro.2015.93. [DOI] [PubMed] [Google Scholar]
- 147.Giovannetti E, et al. Never let it go: stopping key mechanisms underlying metastasis to fight pancreatic cancer. Semin. Cancer Biol. 2017;44:43–59. doi: 10.1016/j.semcancer.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 148.Griesmann H, et al. Pharmacological macrophage inhibition decreases metastasis formation in a genetic model of pancreatic cancer. Gut. 2017;66:1278–1285. doi: 10.1136/gutjnl-2015-310049. [DOI] [PubMed] [Google Scholar]
- 149.Huang C, et al. Interleukin 35 expression correlates with microvessel density in pancreatic ductal adenocarcinoma, recruits monocytes, and promotes growth and angiogenesis of xenograft tumors in mice. Gastroenterology. 2018;154:675–688. doi: 10.1053/j.gastro.2017.09.039. [DOI] [PubMed] [Google Scholar]
- 150.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 151.Sousa CM, Kimmelman AC. The complex landscape of pancreatic cancer metabolism. Carcinogenesis. 2014;35:1441–1450. doi: 10.1093/carcin/bgu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yang J, et al. The enhancement of glycolysis regulates pancreatic cancer metastasis. Cell Mol. Life Sci. 2020;77:305–321. doi: 10.1007/s00018-019-03278-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Halbrook CJ, Lyssiotis CA. Employing metabolism to improve the diagnosis and treatment of pancreatic cancer. Cancer Cell. 2017;31:5–19. doi: 10.1016/j.ccell.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 154.Karasinska JM, et al. Altered gene expression along the glycolysis-cholesterol synthesis axis is associated with outcome in pancreatic cancer. Clin. Cancer Res. 2020;26:135–146. doi: 10.1158/1078-0432.CCR-19-1543. [DOI] [PubMed] [Google Scholar]
- 155.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 156.Ying H, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bryant KL, Mancias JD, Kimmelman AC, Der CJ. KRAS: feeding pancreatic cancer proliferation. Trends Biochem. Sci. 2014;39:91–100. doi: 10.1016/j.tibs.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li X, et al. Mitochondria-translocated PGK1 functions as a protein kinase to coordinate glycolysis and the TCA cycle in tumorigenesis. Mol. Cell. 2016;61:705–719. doi: 10.1016/j.molcel.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Taparra K, et al. O-GlcNAcylation is required for mutant KRAS-induced lung tumorigenesis. J. Clin. Invest. 2018;128:4924–4937. doi: 10.1172/JCI94844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Santana-Codina N, et al. Oncogenic KRAS supports pancreatic cancer through regulation of nucleotide synthesis. Nat. Commun. 2018;9:4945. doi: 10.1038/s41467-018-07472-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Vernucci, E. et al. Metabolic alterations in pancreatic cancer progression. Cancers12, 2 (2019). [DOI] [PMC free article] [PubMed]
- 162.Nagarajan A, et al. Paraoxonase 2 facilitates pancreatic cancer growth and metastasis by stimulating GLUT1-mediated glucose transport. Mol. cell. 2017;67:685–701 e686. doi: 10.1016/j.molcel.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang C, et al. Tumour-associated mutant p53 drives the Warburg effect. Nat. Commun. 2013;4:2935. doi: 10.1038/ncomms3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang YP, et al. Arginine methylation of MDH1 by CARM1 inhibits glutamine metabolism and suppresses pancreatic cancer. Mol. Cell. 2016;64:673–687. doi: 10.1016/j.molcel.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 165.McDonald PC, et al. Regulation of pH by carbonic anhydrase 9 mediates survival of pancreatic cancer cells with activated KRAS in response to hypoxia. Gastroenterology. 2019;157:823–837. doi: 10.1053/j.gastro.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 166.Chaika NV, et al. MUC1 mucin stabilizes and activates hypoxia-inducible factor 1 alpha to regulate metabolism in pancreatic cancer. Proc. Natl Acad. Sci. USA. 2012;109:13787–13792. doi: 10.1073/pnas.1203339109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Shi M, et al. A novel KLF4/LDHA signaling pathway regulates aerobic glycolysis in and progression of pancreatic cancer. Clin. Cancer Res. 2014;20:4370–4380. doi: 10.1158/1078-0432.CCR-14-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Guillaumond F, et al. Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc. Natl Acad. Sci. USA. 2013;110:3919–3924. doi: 10.1073/pnas.1219555110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Xu R, et al. Reprogramming of amino acid metabolism in pancreatic cancer: recent advances and therapeutic strategies. Front. Oncol. 2020;10:572722. doi: 10.3389/fonc.2020.572722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kaira K, et al. Prognostic significance of L-type amino-acid transporter 1 expression in surgically resected pancreatic cancer. Br. J. Cancer. 2012;107:632–638. doi: 10.1038/bjc.2012.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J. Clin. Investig. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bernfeld E, Foster DA. Glutamine as an essential amino acid for KRas-driven cancer cells. Trends Endocrinol. Metab. 2019;30:357–368. doi: 10.1016/j.tem.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 173.Son J, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Biancur DE, et al. Compensatory metabolic networks in pancreatic cancers upon perturbation of glutamine metabolism. Nat. Commun. 2017;8:15965. doi: 10.1038/ncomms15965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Udupa S, et al. Upregulation of the glutaminase II pathway contributes to glutamate production upon glutaminase 1 inhibition in pancreatic cancer. Proteomics. 2019;19:e1800451. doi: 10.1002/pmic.201800451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lampson BL, et al. Targeting eNOS in pancreatic cancer. Cancer Res. 2012;72:4472–4482. doi: 10.1158/0008-5472.CAN-12-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wang H, Li QF, Chow HY, Choi SC, Leung YC. Arginine deprivation inhibits pancreatic cancer cell migration, invasion and EMT via the down regulation of Snail, Slug, Twist, and MMP1/9. J. Physiol. Biochem. 2020;76:73–83. doi: 10.1007/s13105-019-00716-1. [DOI] [PubMed] [Google Scholar]
- 178.Goberdhan DC, Wilson C, Harris AL. Amino acid sensing by mTORC1: intracellular transporters mark the spot. Cell Metab. 2016;23:580–589. doi: 10.1016/j.cmet.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wang S, et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347:188–194. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wyant GA, et al. mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell. 2017;171:642–654.e612. doi: 10.1016/j.cell.2017.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Burke L, et al. The Janus-like role of proline metabolism in cancer. Cell Death Discov. 2020;6:104. doi: 10.1038/s41420-020-00341-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Olivares O, et al. Collagen-derived proline promotes pancreatic ductal adenocarcinoma cell survival under nutrient limited conditions. Nat. Commun. 2017;8:16031. doi: 10.1038/ncomms16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kottakis F, et al. LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature. 2016;539:390–395. doi: 10.1038/nature20132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mayers JR, et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science. 2016;353:1161–1165. doi: 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yue M, Jiang J, Gao P, Liu H, Qing G. Oncogenic MYC activates a feedforward regulatory loop promoting essential amino acid metabolism and tumorigenesis. Cell Rep. 2017;21:3819–3832. doi: 10.1016/j.celrep.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 186.Dong Y, Tu R, Liu H, Qing G. Regulation of cancer cell metabolism: oncogenic MYC in the driver’s seat. Signal Transduct. Target Ther. 2020;5:124. doi: 10.1038/s41392-020-00235-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mayers JR, et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat. Med. 2014;20:1193–1198. doi: 10.1038/nm.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Dey P, et al. Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer. Nature. 2017;542:119–123. doi: 10.1038/nature21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Röhrig F, Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer. 2016;16:732–749. doi: 10.1038/nrc.2016.89. [DOI] [PubMed] [Google Scholar]
- 190.Silvente-Poirot S, Poirot M. Cancer. Cholesterol and cancer, in the balance. Science. 2014;343:1445–1446. doi: 10.1126/science.1252787. [DOI] [PubMed] [Google Scholar]
- 191.Philip B, et al. A high-fat diet activates oncogenic Kras and COX2 to induce development of pancreatic ductal adenocarcinoma in mice. Gastroenterology. 2013;145:1449–1458. doi: 10.1053/j.gastro.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Guillaumond F, et al. Cholesterol uptake disruption, in association with chemotherapy, is a promising combined metabolic therapy for pancreatic adenocarcinoma. Proc. Natl Acad. Sci. USA. 2015;112:2473–2478. doi: 10.1073/pnas.1421601112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Tadros S, et al. De novo lipid synthesis facilitates gemcitabine resistance through endoplasmic reticulum stress in pancreatic cancer. Cancer Res. 2017;77:5503–5517. doi: 10.1158/0008-5472.CAN-16-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yu M, et al. Four types of fatty acids exert differential impact on pancreatic cancer growth. Cancer Lett. 2015;360:187–194. doi: 10.1016/j.canlet.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 195.Ding, Y. et al. Omega-3 fatty acids prevent early pancreatic carcinogenesis via repression of the AKT pathway. Nutrients10, 1289 (2018). [DOI] [PMC free article] [PubMed]
- 196.Snaebjornsson MT, Janaki-Raman S, Schulze A. Greasing the wheels of the cancer machine: the role of lipid metabolism in cancer. Cell Metab. 2020;31:62–76. doi: 10.1016/j.cmet.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 197.Ma Y, et al. Fatty acid oxidation: An emerging facet of metabolic transformation in cancer. Cancer Lett. 2018;435:92–100. doi: 10.1016/j.canlet.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.De Oliveira, M. P. & Liesa, M. The role of mitochondrial fat oxidation in cancer cell proliferation and survival. Cells9, 2600 (2020). [DOI] [PMC free article] [PubMed]
- 199.Lee, J. S. et al. ATP production relies on fatty acid oxidation rather than glycolysis in pancreatic ductal adenocarcinoma. Cancers12, 2477 (2020). [DOI] [PMC free article] [PubMed]
- 200.Rozeveld CN, Johnson KM, Zhang L, Razidlo GL. KRAS controls pancreatic cancer cell lipid metabolism and invasive potential through the lipase HSL. Cancer Res. 2020;80:4932–4945. doi: 10.1158/0008-5472.CAN-20-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Man J, Pajic M, Joshua AM. Fats and Mets, KRAS-driven lipid dysregulation affects metastatic potential in pancreatic cancer. Cancer Res. 2020;80:4886–4887. doi: 10.1158/0008-5472.CAN-20-3082. [DOI] [PubMed] [Google Scholar]
- 202.Gabitova-Cornell L, et al. Cholesterol pathway inhibition induces TGF-β signaling to promote basal differentiation in pancreatic cancer. Cancer Cell. 2020;38:567–583.e511. doi: 10.1016/j.ccell.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Freed-Pastor WA, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–258. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Moon SH, et al. p53 represses the mevalonate pathway to mediate tumor suppression. Cell. 2019;176:564–580.e519. doi: 10.1016/j.cell.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.McGregor GH, et al. Targeting the metabolic response to statin-mediated oxidative stress produces a synergistic antitumor response. Cancer Res. 2020;80:175–188. doi: 10.1158/0008-5472.CAN-19-0644. [DOI] [PubMed] [Google Scholar]
- 206.Oni, T. E. et al. SOAT1 promotes mevalonate pathway dependency in pancreatic cancer. J. Exp. Med.217, e20192389 (2020). [DOI] [PMC free article] [PubMed]
- 207.New M, et al. Molecular pathways controlling autophagy in pancreatic cancer. Front. Oncol. 2017;7:28. doi: 10.3389/fonc.2017.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Yang A, et al. Autophagy sustains pancreatic cancer growth through both cell-autonomous and nonautonomous mechanisms. Cancer Discov. 2018;8:276–287. doi: 10.1158/2159-8290.CD-17-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bryant KL, et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat. Med. 2019;25:628–640. doi: 10.1038/s41591-019-0368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kinsey CG, et al. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat. Med. 2019;25:620–627. doi: 10.1038/s41591-019-0367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kimmelman AC, White E. Autophagy and tumor metabolism. Cell Metab. 2017;25:1037–1043. doi: 10.1016/j.cmet.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Perera RM, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Yang S, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Santana-Codina N, Mancias JD, Kimmelman AC. The role of autophagy in cancer. Annu. Rev. Cancer Biol. 2017;1:19–39. doi: 10.1146/annurev-cancerbio-041816-122338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Yamamoto K, et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature. 2020;581:100–105. doi: 10.1038/s41586-020-2229-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Yamamoto K, Venida A, Perera RM, Kimmelman AC. Selective autophagy of MHC-I promotes immune evasion of pancreatic cancer. Autophagy. 2020;16:1524–1525. doi: 10.1080/15548627.2020.1769973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ansari D, Carvajo M, Bauden M, Andersson R. Pancreatic cancer stroma: controversies and current insights. Scand. J. Gastroenterol. 2017;52:641–646. doi: 10.1080/00365521.2017.1293726. [DOI] [PubMed] [Google Scholar]
- 218.Rhim AD, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ren B, et al. Tumor microenvironment participates in metastasis of pancreatic cancer. Mol. Cancer. 2018;17:108. doi: 10.1186/s12943-018-0858-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sherman MH. Stellate cells in tissue repair, inflammation, and cancer. Annu. Rev. Cell Dev. Biol. 2018;34:333–355. doi: 10.1146/annurev-cellbio-100617-062855. [DOI] [PubMed] [Google Scholar]
- 221.Fu Y, Liu S, Zeng S, Shen H. The critical roles of activated stellate cells-mediated paracrine signaling, metabolism and onco-immunology in pancreatic ductal adenocarcinoma. Mol. Cancer. 2018;17:62. doi: 10.1186/s12943-018-0815-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Farajzadeh Valilou S, Keshavarz-Fathi M, Silvestris N, Argentiero A, Rezaei N. The role of inflammatory cytokines and tumor associated macrophages (TAMs) in microenvironment of pancreatic cancer. Cytokine Growth Factor Rev. 2018;39:46–61. doi: 10.1016/j.cytogfr.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 223.Yoshikawa K, et al. Impact of tumor-associated macrophages on invasive ductal carcinoma of the pancreas head. Cancer Sci. 2012;103:2012–2020. doi: 10.1111/j.1349-7006.2012.02411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Jang JE, et al. Crosstalk between regulatory T cells and tumor-associated dendritic cells negates anti-tumor immunity in pancreatic cancer. Cell Rep. 2017;20:558–571. doi: 10.1016/j.celrep.2017.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Zhang Y, et al. Regulatory T-cell depletion alters the tumor microenvironment and accelerates pancreatic carcinogenesis. Cancer Discov. 2020;10:422–439. doi: 10.1158/2159-8290.CD-19-0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Thyagarajan, A. et al. Myeloid-derived suppressor cells and pancreatic cancer: implications in novel therapeutic approaches. Cancers11, 1627 (2019). [DOI] [PMC free article] [PubMed]
- 227.Pergamo M, Miller G. Myeloid-derived suppressor cells and their role in pancreatic cancer. Cancer Gene Ther. 2017;24:100–105. doi: 10.1038/cgt.2016.65. [DOI] [PubMed] [Google Scholar]
- 228.von Ahrens D, Bhagat TD, Nagrath D, Maitra A, Verma A. The role of stromal cancer-associated fibroblasts in pancreatic cancer. J. Hematol. Oncol. 2017;10:76. doi: 10.1186/s13045-017-0448-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Pothula SP, et al. Key role of pancreatic stellate cells in pancreatic cancer. Cancer Lett. 2016;381:194–200. doi: 10.1016/j.canlet.2015.10.035. [DOI] [PubMed] [Google Scholar]
- 230.Bulle A, Lim KH. Beyond just a tight fortress: contribution of stroma to epithelial-mesenchymal transition in pancreatic cancer. Signal Transduct. Target Ther. 2020;5:249. doi: 10.1038/s41392-020-00341-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Leca J, et al. Cancer-associated fibroblast-derived annexin A6+ extracellular vesicles support pancreatic cancer aggressiveness. J. Clin. Invest. 2016;126:4140–4156. doi: 10.1172/JCI87734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Zhao H, et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife. 2016;5:e10250. doi: 10.7554/eLife.10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Richards KE, et al. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene. 2017;36:1770–1778. doi: 10.1038/onc.2016.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Lo, A. et al. Fibroblast activation protein augments progression and metastasis of pancreatic ductal adenocarcinoma. JCI Insight2, e92232 (2017). [DOI] [PMC free article] [PubMed]
- 235.Chen S, et al. MiR-21-mediated metabolic alteration of cancer-associated fibroblasts and its effect on pancreatic cancer cell behavior. Int. J. Biol. Sci. 2018;14:100–110. doi: 10.7150/ijbs.22555. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 236.Sousa CM, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536:479–483. doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Auciello FR, et al. A stromal lysolipid-autotaxin signaling axis promotes pancreatic tumor progression. Cancer Discov. 2019;9:617–627. doi: 10.1158/2159-8290.CD-18-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Shindo K, et al. Podoplanin expression in cancer-associated fibroblasts enhances tumor progression of invasive ductal carcinoma of the pancreas. Mol. Cancer. 2013;12:168. doi: 10.1186/1476-4598-12-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Feig C, et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc. Natl Acad. Sci. USA. 2013;110:20212–20217. doi: 10.1073/pnas.1320318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Hwang RF, et al. Inhibition of the hedgehog pathway targets the tumor-associated stroma in pancreatic cancer. Mol. Cancer Res. 2012;10:1147–1157. doi: 10.1158/1541-7786.MCR-12-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Laklai H, et al. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat. Med. 2016;22:497–505. doi: 10.1038/nm.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Tape CJ, et al. Oncogenic KRAS regulates tumor cell signaling via stromal reciprocation. Cell. 2016;165:910–920. doi: 10.1016/j.cell.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ho WJ, Jaffee EM, Zheng L. The tumour microenvironment in pancreatic cancer - clinical challenges and opportunities. Nat. Rev. Clin. Oncol. 2020;17:527–540. doi: 10.1038/s41571-020-0363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Vonderheide RH, Bayne LJ. Inflammatory networks and immune surveillance of pancreatic carcinoma. Curr. Opin. Immunol. 2013;25:200–205. doi: 10.1016/j.coi.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Meyer MA, et al. Breast and pancreatic cancer interrupt IRF8-dependent dendritic cell development to overcome immune surveillance. Nat. Commun. 2018;9:1250. doi: 10.1038/s41467-018-03600-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Tjomsland V, et al. The desmoplastic stroma plays an essential role in the accumulation and modulation of infiltrated immune cells in pancreatic adenocarcinoma. Clin. Dev. Immunol. 2011;2011:212810. doi: 10.1155/2011/212810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Mahajan UM, et al. Immune cell and stromal signature associated with progression-free survival of patients with resected pancreatic ductal adenocarcinoma. Gastroenterology. 2018;155:1625–1639.e1622. doi: 10.1053/j.gastro.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 248.Carstens JL, et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat. Commun. 2017;8:15095. doi: 10.1038/ncomms15095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, Bar-Sagi D. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell. 2012;21:836–847. doi: 10.1016/j.ccr.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Sivaram N, et al. Tumor-intrinsic PIK3CA represses tumor immunogenecity in a model of pancreatic cancer. J. Clin. Invest. 2019;129:3264–3276. doi: 10.1172/JCI123540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Hashimoto S, et al. ARF6 and AMAP1 are major targets of KRAS and TP53 mutations to promote invasion, PD-L1 dynamics, and immune evasion of pancreatic cancer. Proc. Natl Acad. Sci. USA. 2019;116:17450–17459. doi: 10.1073/pnas.1901765116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Wörmann SM, et al. Loss of P53 function activates JAK2-STAT3 signaling to promote pancreatic tumor growth, stroma modification, and gemcitabine resistance in mice and is associated with patient survival. Gastroenterology. 2016;151:180–193.e112. doi: 10.1053/j.gastro.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 253.Serrels A, et al. Nuclear FAK controls chemokine transcription, Tregs, and evasion of anti-tumor immunity. Cell. 2015;163:160–173. doi: 10.1016/j.cell.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Jiang H, et al. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat. Med. 2016;22:851–860. doi: 10.1038/nm.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Sodir NM, et al. Endogenous Myc maintains the tumor microenvironment. Genes Dev. 2011;25:907–916. doi: 10.1101/gad.2038411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Sodir NM, et al. MYC instructs and maintains pancreatic adenocarcinoma phenotype. Cancer Discov. 2020;10:588–607. doi: 10.1158/2159-8290.CD-19-0435. [DOI] [PubMed] [Google Scholar]
- 257.Muthalagu N, et al. Repression of the type I interferon pathway underlies MYC- and KRAS-dependent evasion of NK and B cells in pancreatic ductal adenocarcinoma. Cancer Discov. 2020;10:872–887. doi: 10.1158/2159-8290.CD-19-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Dovmark TH, Saccomano M, Hulikova A, Alves F, Swietach P. Connexin-43 channels are a pathway for discharging lactate from glycolytic pancreatic ductal adenocarcinoma cells. Oncogene. 2017;36:4538–4550. doi: 10.1038/onc.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Ye H, et al. Tumor-associated macrophages promote progression and the Warburg effect via CCL18/NF-kB/VCAM-1 pathway in pancreatic ductal adenocarcinoma. Cell Death Dis. 2018;9:453. doi: 10.1038/s41419-018-0486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Hutcheson J, et al. Immunologic and metabolic features of pancreatic ductal adenocarcinoma define prognostic subtypes of disease. Clin. Cancer Res. 2016;22:3606–3617. doi: 10.1158/1078-0432.CCR-15-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Penny HL, et al. Warburg metabolism in tumor-conditioned macrophages promotes metastasis in human pancreatic ductal adenocarcinoma. Oncoimmunology. 2016;5:e1191731. doi: 10.1080/2162402X.2016.1191731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wang X, et al. Cancer-FOXP3 directly activated CCL5 to recruit FOXP3(+)Treg cells in pancreatic ductal adenocarcinoma. Oncogene. 2017;36:3048–3058. doi: 10.1038/onc.2016.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wang X, et al. PD-L1 is a direct target of cancer-FOXP3 in pancreatic ductal adenocarcinoma (PDAC), and combined immunotherapy with antibodies against PD-L1 and CCL5 is effective in the treatment of PDAC. Signal Transduct. Target Ther. 2020;5:38. doi: 10.1038/s41392-020-0144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Zhu Y, et al. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 2014;74:5057–5069. doi: 10.1158/0008-5472.CAN-13-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Ijichi H, et al. Inhibiting Cxcr2 disrupts tumor-stromal interactions and improves survival in a mouse model of pancreatic ductal adenocarcinoma. J. Clin. Invest. 2011;121:4106–4117. doi: 10.1172/JCI42754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Vonderheide RH, Bear AS. Tumor-derived myeloid cell chemoattractants and T cell exclusion in pancreatic cancer. Front Immunol. 2020;11:605619. doi: 10.3389/fimmu.2020.605619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Sanford DE, et al. Inflammatory monocyte mobilization decreases patient survival in pancreatic cancer: a role for targeting the CCL2/CCR2 axis. Clin. Cancer Res. 2013;19:3404–3415. doi: 10.1158/1078-0432.CCR-13-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Hou P, et al. Tumor microenvironment remodeling enables bypass of oncogenic KRAS dependency in pancreatic cancer. Cancer Discov. 2020;10:1058–1077. doi: 10.1158/2159-8290.CD-19-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Zhang R, et al. CXCL5 overexpression predicts a poor prognosis in pancreatic ductal adenocarcinoma and is correlated with immune cell infiltration. J. Cancer. 2020;11:2371–2381. doi: 10.7150/jca.40517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Seifert L, et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature. 2016;532:245–249. doi: 10.1038/nature17403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Le DT, et al. Safety and survival with GVAX pancreas prime and Listeria monocytogenes-expressing mesothelin (CRS-207) boost vaccines for metastatic pancreatic cancer. J. Clin. Oncol. 2015;33:1325–1333. doi: 10.1200/JCO.2014.57.4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Oh MH, et al. Targeting glutamine metabolism enhances tumor-specific immunity by modulating suppressive myeloid cells. J. Clin. Invest. 2020;130:3865–3884. doi: 10.1172/JCI131859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Li J, et al. Tumor cell-intrinsic factors underlie heterogeneity of immune cell infiltration and response to immunotherapy. Immunity. 2018;49:178–193.e177. doi: 10.1016/j.immuni.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Zhang A, et al. Cancer-associated fibroblasts promote M2 polarization of macrophages in pancreatic ductal adenocarcinoma. Cancer Med. 2017;6:463–470. doi: 10.1002/cam4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Steins A, et al. High-grade mesenchymal pancreatic ductal adenocarcinoma drives stromal deactivation through CSF-1. EMBO Rep. 2020;21:e48780. doi: 10.15252/embr.201948780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Principe DR, et al. TGFβ signaling in the pancreatic tumor microenvironment promotes fibrosis and immune evasion to facilitate tumorigenesis. Cancer Res. 2016;76:2525–2539. doi: 10.1158/0008-5472.CAN-15-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Yin Z, et al. Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-β signaling pathway. J. Exp. Clin. Cancer Res. 2019;38:310. doi: 10.1186/s13046-019-1313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Lianyuan T, et al. Tumor associated neutrophils promote the metastasis of pancreatic ductal adenocarcinoma. Cancer Biol. Ther. 2020;21:937–945. doi: 10.1080/15384047.2020.1807250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Melzer, M. K. et al. An immunological glance on pancreatic ductal adenocarcinoma. Int. J. Mol. Sci. 21, 3345 (2020). [DOI] [PMC free article] [PubMed]
- 280.Martinez-Bosch, N., Vinaixa, J. & Navarro, P. Immune evasion in pancreatic cancer: from mechanisms to therapy. Cancers10, 6 (2018). [DOI] [PMC free article] [PubMed]
- 281.Kaur A, et al. A recombinant fragment of human surfactant protein d induces apoptosis in pancreatic cancer cell lines via Fas-mediated pathway. Front. Immunol. 2018;9:1126. doi: 10.3389/fimmu.2018.01126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 283.Jeune F, et al. Pancreatic cancer surgical management. Presse Med. 2019;48:e147–e158. doi: 10.1016/j.lpm.2019.02.027. [DOI] [PubMed] [Google Scholar]
- 284.Li HY, Cui ZM, Chen J, Guo XZ, Li YY. Pancreatic cancer: diagnosis and treatments. Tumour Biol. 2015;36:1375–1384. doi: 10.1007/s13277-015-3223-7. [DOI] [PubMed] [Google Scholar]
- 285.Zhang Y, et al. Tumor markers CA19-9, CA242 and CEA in the diagnosis of pancreatic cancer: a meta-analysis. Int. J. Clin. Exp. Med. 2015;8:11683–11691. [PMC free article] [PubMed] [Google Scholar]
- 286.van Manen, L. et al. Stage-specific value of carbohydrate antigen 19-9 and carcinoembryonic antigen serum levels on survival and recurrence in pancreatic cancer: a single center study and meta-analysis. Cancers12, 2970 (2020). [DOI] [PMC free article] [PubMed]
- 287.Daoud AZ, Mulholland EJ, Cole G, McCarthy HO. MicroRNAs in pancreatic cancer: biomarkers, prognostic, and therapeutic modulators. BMC Cancer. 2019;19:1130. doi: 10.1186/s12885-019-6284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Previdi MC, Carotenuto P, Zito D, Pandolfo R, Braconi C. Noncoding RNAs as novel biomarkers in pancreatic cancer: what do we know? Fut. Oncol. 2017;13:443–453. doi: 10.2217/fon-2016-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Cicenas, J. et al. KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 mutations in pancreatic cancer. Cancers9, 42 (2017). [DOI] [PMC free article] [PubMed]
- 290.Takai E, Yachida S. Circulating tumor DNA as a liquid biopsy target for detection of pancreatic cancer. World J. Gastroenterol. 2016;22:8480–8488. doi: 10.3748/wjg.v22.i38.8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Martini, V., Timme-Bronsert, S., Fichtner-Feigl, S., Hoeppner, J. & Kulemann, B. Circulating tumor cells in pancreatic cancer: current perspectives. Cancers11, 1659 (2019). [DOI] [PMC free article] [PubMed]
- 292.Nagrath S, Jack RM, Sahai V, Simeone DM. Opportunities and challenges for pancreatic circulating tumor cells. Gastroenterology. 2016;151:412–426. doi: 10.1053/j.gastro.2016.05.052. [DOI] [PubMed] [Google Scholar]
- 293.Heiler S, Wang Z, Zöller M. Pancreatic cancer stem cell markers and exosomes - the incentive push. World J. Gastroenterol. 2016;22:5971–6007. doi: 10.3748/wjg.v22.i26.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Zhao C, Gao F, Weng S, Liu Q. Pancreatic cancer and associated exosomes. Cancer Biomark. 2017;20:357–367. doi: 10.3233/CBM-170258. [DOI] [PubMed] [Google Scholar]
- 295.Lee JS, Park SS, Lee YK, Norton JA, Jeffrey SS. Liquid biopsy in pancreatic ductal adenocarcinoma: current status of circulating tumor cells and circulating tumor DNA. Mol. Oncol. 2019;13:1623–1650. doi: 10.1002/1878-0261.12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Riva F, et al. Clinical applications of circulating tumor DNA and circulating tumor cells in pancreatic cancer. Mol. Oncol. 2016;10:481–493. doi: 10.1016/j.molonc.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Gao Y, et al. Clinical significance of pancreatic circulating tumor cells using combined negative enrichment and immunostaining-fluorescence in situ hybridization. J. Exp. Clin. Cancer Res. 2016;35:66. doi: 10.1186/s13046-016-0340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Xu, Y. et al. Detection of circulating tumor cells using negative enrichment immunofluorescence and an in situ hybridization system in pancreatic cancer. Int. J. Mol. Sci. 18, 622 (2017). [DOI] [PMC free article] [PubMed]
- 299.Ankeny JS, et al. Circulating tumour cells as a biomarker for diagnosis and staging in pancreatic cancer. Br. J. Cancer. 2016;114:1367–1375. doi: 10.1038/bjc.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Effenberger KE, et al. Improved risk stratification by circulating tumor cell counts in pancreatic cancer. Clin. Cancer Res. 2018;24:2844–2850. doi: 10.1158/1078-0432.CCR-18-0120. [DOI] [PubMed] [Google Scholar]
- 301.Poruk KE, et al. Circulating tumor cell phenotype predicts recurrence and survival in pancreatic adenocarcinoma. Ann. Surg. 2016;264:1073–1081. doi: 10.1097/SLA.0000000000001600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Zhang Y, et al. Patterns of circulating tumor cells identified by CEP8, CK and CD45 in pancreatic cancer. Int. J. Cancer. 2015;136:1228–1233. doi: 10.1002/ijc.29070. [DOI] [PubMed] [Google Scholar]
- 303.Bissolati M, et al. Portal vein-circulating tumor cells predict liver metastases in patients with resectable pancreatic cancer. Tumour Biol. 2015;36:991–996. doi: 10.1007/s13277-014-2716-0. [DOI] [PubMed] [Google Scholar]
- 304.Tien YW, et al. A high circulating tumor cell count in portal vein predicts liver metastasis from periampullary or pancreatic cancer: a high portal venous CTC count predicts liver metastases. Medicine. 2016;95:e3407. doi: 10.1097/MD.0000000000003407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Chang MC, et al. Clinical significance of circulating tumor microemboli as a prognostic marker in patients with pancreatic ductal adenocarcinoma. Clin. Chem. 2016;62:505–513. doi: 10.1373/clinchem.2015.248260. [DOI] [PubMed] [Google Scholar]
- 306.Bettegowda C, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014;6:224ra224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Cohen JD, et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc. Natl Acad. Sci. USA. 2017;114:10202–10207. doi: 10.1073/pnas.1704961114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Kinugasa H, et al. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer. 2015;121:2271–2280. doi: 10.1002/cncr.29364. [DOI] [PubMed] [Google Scholar]
- 309.Perets R, et al. Mutant KRAS circulating tumor DNA is an accurate tool for pancreatic cancer monitoring. Oncologist. 2018;23:566–572. doi: 10.1634/theoncologist.2017-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Pietrasz D, et al. Plasma circulating tumor DNA in pancreatic cancer patients is a prognostic marker. Clin. Cancer Res. 2017;23:116–123. doi: 10.1158/1078-0432.CCR-16-0806. [DOI] [PubMed] [Google Scholar]
- 311.Creemers A, et al. Clinical value of ctDNA in upper-GI cancers: a systematic review and meta-analysis. Biochim. Biophys. Acta Rev. Cancer. 2017;1868:394–403. doi: 10.1016/j.bbcan.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 312.Hadano N, et al. Prognostic value of circulating tumour DNA in patients undergoing curative resection for pancreatic cancer. Br. J. Cancer. 2016;115:59–65. doi: 10.1038/bjc.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Singh N, Gupta S, Pandey RM, Chauhan SS, Saraya A. High levels of cell-free circulating nucleic acids in pancreatic cancer are associated with vascular encasement, metastasis and poor survival. Cancer Invest. 2015;33:78–85. doi: 10.3109/07357907.2014.1001894. [DOI] [PubMed] [Google Scholar]
- 314.Mashouri L, et al. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer. 2019;18:75. doi: 10.1186/s12943-019-0991-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Whiteside TL. Tumor-derived exosomes and their role in cancer progression. Adv. Clin. Chem. 2016;74:103–141. doi: 10.1016/bs.acc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Zhu L, et al. Isolation and characterization of exosomes for cancer research. J. Hematol. Oncol. 2020;13:152. doi: 10.1186/s13045-020-00987-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Melo SA, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Lewis JM, et al. Integrated analysis of exosomal protein biomarkers on alternating current electrokinetic chips enables rapid detection of pancreatic cancer in patient blood. ACS Nano. 2018;12:3311–3320. doi: 10.1021/acsnano.7b08199. [DOI] [PubMed] [Google Scholar]
- 319.Théry C. Cancer: diagnosis by extracellular vesicles. Nature. 2015;523:161–162. doi: 10.1038/nature14626. [DOI] [PubMed] [Google Scholar]
- 320.Lai X, et al. A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett. 2017;393:86–93. doi: 10.1016/j.canlet.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Liang, K. et al. Nanoplasmonic quantification of tumor-derived extracellular vesicles in plasma microsamples for diagnosis and treatment monitoring. Nat. Biomed. Eng.1, 0021 (2017). [DOI] [PMC free article] [PubMed]
- 322.Allenson K, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann. Oncol. 2017;28:741–747. doi: 10.1093/annonc/mdx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.An M, et al. Quantitative proteomic analysis of serum exosomes from patients with locally advanced pancreatic cancer undergoing chemoradiotherapy. J. Proteome Res. 2017;16:1763–1772. doi: 10.1021/acs.jproteome.7b00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Yang, K. S. et al. Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy. Sci. Transl. Med.9, eaal3226 (2017). [DOI] [PMC free article] [PubMed]
- 325.Lim, M., Kim, C. J., Sunkara, V., Kim, M. H. & Cho, Y. K. Liquid biopsy in lung cancer: clinical applications of circulating biomarkers (CTCs and ctDNA). Micromachines9, 100 (2018). [DOI] [PMC free article] [PubMed]
- 326.Cheng DT, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J. Mol. Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Razavi P, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 2019;25:1928–1937. doi: 10.1038/s41591-019-0652-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Zehir A, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Samstein RM, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019;51:202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Law HC, et al. The proteomic landscape of pancreatic ductal adenocarcinoma liver metastases identifies molecular subtypes and associations with clinical response. Clin. Cancer Res. 2020;26:1065–1076. doi: 10.1158/1078-0432.CCR-19-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Aung KL, et al. Genomics-driven precision medicine for advanced pancreatic cancer: early results from the COMPASS Trial. Clin. Cancer Res. 2018;24:1344–1354. doi: 10.1158/1078-0432.CCR-17-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Hayashi A, et al. A unifying paradigm for transcriptional heterogeneity and squamous features in pancreatic ductal adenocarcinoma. Nat. Cancer. 2020;1:59–74. doi: 10.1038/s43018-019-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Er JL, et al. Identification of inhibitors synergizing gemcitabine sensitivity in the squamous subtype of pancreatic ductal adenocarcinoma (PDAC) Apoptosis. 2018;23:343–355. doi: 10.1007/s10495-018-1459-6. [DOI] [PubMed] [Google Scholar]
- 334.Kalimuthu SN, et al. Morphological classification of pancreatic ductal adenocarcinoma that predicts molecular subtypes and correlates with clinical outcome. Gut. 2020;69:317–328. doi: 10.1136/gutjnl-2019-318217. [DOI] [PubMed] [Google Scholar]
- 335.Somerville TDD, et al. TP63-mediated enhancer reprogramming drives the squamous subtype of pancreatic ductal adenocarcinoma. Cell Rep. 2018;25:1741–1755.e1747. doi: 10.1016/j.celrep.2018.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Roa-Peña L, et al. Keratin 17 identifies the most lethal molecular subtype of pancreatic cancer. Sci. Rep. 2019;9:11239. doi: 10.1038/s41598-019-47519-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Kaissis, G. A. et al. Image-based molecular phenotyping of pancreatic ductal adenocarcinoma. J. Clin. Med. 9, 724 (2020). [DOI] [PMC free article] [PubMed]
- 338.Aguirre AJ, et al. Real-time genomic characterization of advanced pancreatic cancer to enable precision medicine. Cancer Discov. 2018;8:1096–1111. doi: 10.1158/2159-8290.CD-18-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Collisson EA, Bailey P, Chang DK, Biankin AV. Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2019;16:207–220. doi: 10.1038/s41575-019-0109-y. [DOI] [PubMed] [Google Scholar]
- 340.Aparicio S, Hidalgo M, Kung AL. Examining the utility of patient-derived xenograft mouse models. Nat. Rev. Cancer. 2015;15:311–316. doi: 10.1038/nrc3944. [DOI] [PubMed] [Google Scholar]
- 341.Huang L, et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat. Med. 2015;21:1364–1371. doi: 10.1038/nm.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Vlachogiannis G, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Hingorani SR, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/S1535-6108(03)00309-X. [DOI] [PubMed] [Google Scholar]
- 344.Olive KP, Tuveson DA. The use of targeted mouse models for preclinical testing of novel cancer therapeutics. Clin. Cancer Res. 2006;12:5277–5287. doi: 10.1158/1078-0432.CCR-06-0436. [DOI] [PubMed] [Google Scholar]
- 345.Hingorani SR, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 346.Aguirre AJ, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Bardeesy N, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc. Natl Acad. Sci. USA. 2006;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Drosos Y, et al. ATM-deficiency increases genomic instability and metastatic potential in a mouse model of pancreatic cancer. Sci. Rep. 2017;7:11144. doi: 10.1038/s41598-017-11661-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Russell R, et al. Loss of ATM accelerates pancreatic cancer formation and epithelial-mesenchymal transition. Nat. Commun. 2015;6:7677. doi: 10.1038/ncomms8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Siveke JT, et al. Concomitant pancreatic activation of Kras(G12D) and Tgfa results in cystic papillary neoplasms reminiscent of human IPMN. Cancer Cell. 2007;12:266–279. doi: 10.1016/j.ccr.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 351.Bardeesy N, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Kojima K, et al. Inactivation of Smad4 accelerates Kras(G12D)-mediated pancreatic neoplasia. Cancer Res. 2007;67:8121–8130. doi: 10.1158/0008-5472.CAN-06-4167. [DOI] [PubMed] [Google Scholar]
- 353.Gopinathan A, Morton JP, Jodrell DI, Sansom OJ. GEMMs as preclinical models for testing pancreatic cancer therapies. Dis. Model. Mech. 2015;8:1185–1200. doi: 10.1242/dmm.021055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Francia G, Cruz-Munoz W, Man S, Xu P, Kerbel RS. Mouse models of advanced spontaneous metastasis for experimental therapeutics. Nat. Rev. Cancer. 2011;11:135–141. doi: 10.1038/nrc3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Wolff RA, Varadhachary GR, Evans DB. Adjuvant therapy for adenocarcinoma of the pancreas: analysis of reported trials and recommendations for future progress. Ann. Surg. Oncol. 2008;15:2773–2786. doi: 10.1245/s10434-008-0002-3. [DOI] [PubMed] [Google Scholar]
- 356.Fenocchio, E. et al. Is there a standard adjuvant therapy for resected pancreatic cancer? Cancers11, 0 (2019). [DOI] [PMC free article] [PubMed]
- 357.Merkow RP, et al. Postoperative complications reduce adjuvant chemotherapy use in resectable pancreatic cancer. Ann. Surg. 2014;260:372–377. doi: 10.1097/SLA.0000000000000378. [DOI] [PubMed] [Google Scholar]
- 358.Dhir M, et al. Neoadjuvant treatment of pancreatic adenocarcinoma: a systematic review and meta-analysis of 5520 patients. World J. Surg. Oncol. 2017;15:183. doi: 10.1186/s12957-017-1240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Kim S, et al. Comparative effectiveness of nab-paclitaxel plus gemcitabine vs FOLFIRINOX in metastatic pancreatic cancer: a retrospective nationwide chart review in the United States. Adv. Ther. 2018;35:1564–1577. doi: 10.1007/s12325-018-0784-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Sohal DP, Mangu PB, Laheru D. Metastatic pancreatic cancer: American Society of Clinical Oncology Clinical Practice Guideline Summary. J. Oncol. Pract. 2017;13:261–264. doi: 10.1200/JOP.2016.017368. [DOI] [PubMed] [Google Scholar]
- 361.Sohal DPS, et al. Metastatic pancreatic cancer: ASCO Clinical Practice Guideline Update. J. Clin. Oncol. 2018;36:2545–2556. doi: 10.1200/JCO.2018.78.9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Wang-Gillam A, et al. Nanoliposomal irinotecan with fluorouracil and folinic acid in metastatic pancreatic cancer after previous gemcitabine-based therapy (NAPOLI-1): a global, randomised, open-label, phase 3 trial. Lancet. 2016;387:545–557. doi: 10.1016/S0140-6736(15)00986-1. [DOI] [PubMed] [Google Scholar]
- 363.Ueno M, et al. A randomized phase II study of S-1 plus oral leucovorin versus S-1 monotherapy in patients with gemcitabine-refractory advanced pancreatic cancer. Ann. Oncol. 2016;27:502–508. doi: 10.1093/annonc/mdv603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.McCormick F. KRAS as a therapeutic target. Clin. Cancer Res. 2015;21:1797–1801. doi: 10.1158/1078-0432.CCR-14-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Kamerkar S, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Doebele RC, et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1-2 trials. Lancet Oncol. 2020;21:271–282. doi: 10.1016/S1470-2045(19)30691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Drilon A, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N. Engl. J. Med. 2018;378:731–739. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Lin JJ, Riely GJ, Shaw AT. Targeting ALK: precision medicine takes on drug resistance. Cancer Discov. 2017;7:137–155. doi: 10.1158/2159-8290.CD-16-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Singhi AD, et al. Identification of targetable ALK rearrangements in pancreatic ductal adenocarcinoma. J. Natl Compr. Cancer Netw. 2017;15:555–562. doi: 10.6004/jnccn.2017.0058. [DOI] [PubMed] [Google Scholar]
- 370.Kaufman B, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J. Clin. Oncol. 2015;33:244–250. doi: 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Royal RE, et al. Phase 2 trial of single agent Ipilimumab (anti-CTLA-4) for locally advanced or metastatic pancreatic adenocarcinoma. J. Immunother. 2010;33:828–833. doi: 10.1097/CJI.0b013e3181eec14c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Brahmer JR, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Marabelle A, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair-deficient cancer: results from the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2020;38:1–10. doi: 10.1200/JCO.19.02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Toole BP, Slomiany MG. Hyaluronan: a constitutive regulator of chemoresistance and malignancy in cancer cells. Semin. Cancer Biol. 2008;18:244–250. doi: 10.1016/j.semcancer.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Hingorani SR, et al. HALO 202: randomized phase II study of PEGPH20 plus nab-paclitaxel/gemcitabine versus nab-paclitaxel/gemcitabine in patients with untreated, metastatic pancreatic ductal adenocarcinoma. J. Clin. Oncol. 2018;36:359–366. doi: 10.1200/JCO.2017.74.9564. [DOI] [PubMed] [Google Scholar]
- 376.Van Cutsem E, et al. Randomized phase III trial of pegvorhyaluronidase alfa with nab-paclitaxel plus gemcitabine for patients with hyaluronan-high metastatic pancreatic adenocarcinoma. J. Clin. Oncol. 2020;38:3185–3194. doi: 10.1200/JCO.20.00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Alistar A, et al. Safety and tolerability of the first-in-class agent CPI-613 in combination with modified FOLFIRINOX in patients with metastatic pancreatic cancer: a single-centre, open-label, dose-escalation, phase 1 trial. Lancet Oncol. 2017;18:770–778. doi: 10.1016/S1470-2045(17)30314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Philip PA, et al. A phase III open-label trial to evaluate efficacy and safety of CPI-613 plus modified FOLFIRINOX (mFFX) versus FOLFIRINOX (FFX) in patients with metastatic adenocarcinoma of the pancreas. Fut. Oncol. 2019;15:3189–3196. doi: 10.2217/fon-2019-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Wolpin BM, et al. Phase II and pharmacodynamic study of autophagy inhibition using hydroxychloroquine in patients with metastatic pancreatic adenocarcinoma. Oncologist. 2014;19:637–638. doi: 10.1634/theoncologist.2014-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Bennewith KL, et al. The role of tumor cell-derived connective tissue growth factor (CTGF/CCN2) in pancreatic tumor growth. Cancer Res. 2009;69:775–784. doi: 10.1158/0008-5472.CAN-08-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Neesse A, et al. CTGF antagonism with mAb FG-3019 enhances chemotherapy response without increasing drug delivery in murine ductal pancreas cancer. Proc. Natl Acad. Sci. USA. 2013;110:12325–12330. doi: 10.1073/pnas.1300415110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Sonbol MB, et al. CanStem111P trial: a Phase III study of napabucasin plus nab-paclitaxel with gemcitabine. Fut. Oncol. 2019;15:1295–1302. doi: 10.2217/fon-2018-0903. [DOI] [PubMed] [Google Scholar]
- 383.Li C, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 384.Gurlevik E, et al. Administration of gemcitabine after pancreatic tumor resection in mice induces an antitumor immune response mediated by natural killer cells. Gastroenterology. 2016;151:338–350.e337. doi: 10.1053/j.gastro.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 385.Izeradjene K, et al. Kras(G12D) and Smad4/Dpc4 haploinsufficiency cooperate to induce mucinous cystic neoplasms and invasive adenocarcinoma of the pancreas. Cancer Cell. 2007;11:229–243. doi: 10.1016/j.ccr.2007.01.017. [DOI] [PubMed] [Google Scholar]