Abstract
Klebsiella pneumoniae is a Gram-negative pathogen that has become a worldwide concern due to the emergence of multidrug-resistant isolates responsible for various invasive infectious diseases. Biofilm formation constitutes a major virulence factor for K. pneumoniae and relies on the expression of fimbrial adhesins and aggregation of bacterial cells on biotic or abiotic surfaces in a coordinated manner. During biofilm aggregation, bacterial cells communicate with each other through inter- or intra-species interactions mediated by signallng molecules, called autoinducers, in a mechanism known as quorum sensing (QS). In most Gram-negative bacteria, intra-species communication typically involves the LuxI/LuxR system: LuxI synthase produces N-acyl homoserine lactones (AHLs) as autoinducers and the LuxR transcription factor is their cognate receptor. However, K. pneumoniae does not produce AHL but encodes SdiA, an orphan LuxR-type receptor that responds to exogenous AHL molecules produced by other bacterial species. While SdiA regulates several cellular processes and the expression of virulence factors in many pathogens, the role of this regulator in K. pneumoniae remains unknown. In this study, we describe the characterization of sdiA mutant strain of K. pneumoniae. The sdiA mutant strain has increased biofilm formation, which correlates with the increased expression of type 1 fimbriae, thus revealing a repressive role of SdiA in fimbriae expression and bacterial cell adherence and aggregation. On the other hand, SdiA acts as a transcriptional activator of cell division machinery assembly in the septum, since cells lacking SdiA regulator exhibited a filamentary shape rather than the typical rod shape. We also show that K. pneumoniae cells lacking SdiA regulator present constant production of QS autoinducers at maximum levels, suggesting a putative role for SdiA in the regulation of AI-2 production. Taken together, our results demonstrate that SdiA regulates cell division and the expression of virulence factors such as fimbriae expression, biofilm formation, and production of QS autoinducers in K. pneumoniae.
Keywords: Klebsiella pneumonia, SdiA regulator, cell division, quorum sensing, type 1 fimbriae, biofilm
Introduction
Klebsiella pneumoniae is a Gram-negative bacterium responsible for various diseases that has become a worldwide concern due to the increase in cases of severe infections in the community (Keynan and Rubinstein, 2007; Lin et al., 2010; Holt et al., 2015). Much of K. pneumoniae pathogenicity comes from its ability to form biofilms (Li et al., 2014; Paczosa and Mecsas, 2016), which are microbial communities that grow attached to surfaces and typically surrounded by a matrix of self-produced extracellular polymeric substances (Hall-Stoodley et al., 2004). Biofilm formation by K. pneumoniae, which can occur on biotic and abiotic surfaces, represents a relevant mechanism to protect the bacterium from host immunity and antimicrobial agents (Li et al., 2014; Paczosa and Mecsas, 2016).
During biofilm aggregation, the bacterial cells communicate with each other through inter- or intra-species interactions mediated by a mechanism called quorum sensing (QS). By this process, bacteria produce and detect specific signaling molecules to coordinate gene expression according to the bacterial cell density (Bassler et al., 1993; Miller and Bassler, 2001). Signaling is mediated by chemical molecules, known as autoinducers (AI), that determine two main types of cell-cell communication: AI-1 and AI-2 QS regulatory systems.
AI-2 QS system allows intra- and inter-species communication and utilizes cyclic furanones compounds as AI-2 signaling molecules. AI-2 is synthesized by LuxS synthase, a key enzyme of the activated methyl cycle that converts S-ribosylhomocysteine to homocysteine and 4,5-dihydroxy-2,3-pentanedione, which spontaneously rearranges to form the AI-2 molecules (Schauder et al., 2001; Kendall and Sperandio, 2014). Highly conserved luxS gene homologues have been found in several Gram-negative and Gram-positive bacteria (Bassler et al., 1993; Miller and Bassler, 2001), including K. pneumoniae (Balestrino et al., 2005). The regulatory network for AI-2 metabolism in K. pneumoniae also relies on lsr (comprized of lsrACDBFG) and lsrRK operons, which are located adjacent to one another in the genome but are transcribed divergently (Xavier and Bassler, 2005b; Pereira et al., 2009; Pereira et al., 2013). The first four genes of the lsr operon (lsrACDB) encode an ATP-binding cassette (ABC) transporter system that uptakes the AI-2 molecules, while the remaining genes, lsrFG, are required for processing AI-2 molecules following internalization. Once inside the cell, AI-2 is phosphorylated by the cytoplasmic kinase LsrK, and the activated phospho-AI-2 molecule binds to the transcriptional repressor LsrR and inactivates it. In the absence of phospho-AI-2, LsrR represses the transcription of the lsr operon and regulates its own expression by repressing the lsrRK operon. While LsrR represses the expression of both lsrR and lsr operon, cyclic adenosine monophosphate (cAMP) complexed with the cAMP-receptor protein activates their expression (Wang et al., 2005). In K. pneumoniae, the operons lsr and lsrRK are up-regulated in mature biofilm (Guilhen et al., 2016). Besides, previous studies with K. pneumoniae luxS and lsrCD mutant strains revealed a regulatory role of AI-2 QS system on biofilm formation and lipopolysaccharide synthesis (Ng and Bassler, 2009; Tavio et al., 2010).
AI-1 QS system utilizes N-acyl-L-homoserine lactones (AHLs) as autoinducers and represent the major QS system used by Gram-negative bacteria for intra-species communication and to monitor their own population density (Engebrecht and Silverman, 1984; Ng and Bassler, 2009). In most bacteria, the AI-1 QS mediated by AHLs involves a typical system composed of LuxI and LuxR proteins: LuxI is the enzyme that synthesizes AHLs molecules and LuxR is the cognate receptor that acts as a transcriptional regulator in response to the binding of the AHL autoinducers (Engebrecht and Silverman, 1984; Ng and Bassler, 2009). A variety of AHLs molecules have been identified, each differing in length, oxidation state at β-position, and saturation degree of the N-acyl side chain. Intriguingly, some Gram-negative bacteria encode LuxR receptors but do not produce AHLs because they lack LuxI synthase. These LuxR-type receptors without their corresponding LuxI synthase are known as “solo” or “orphan” receptors (Fuqua, 2006; Patankar and González, 2009). For instance, bacteria from the genera Salmonella, Escherichia, and Klebsiella harbor no luxI gene homologues in their genome and, therefore, they are considered non-AHL producers (Michael et al., 2001). Nonetheless, these enteropathogens encode SdiA, an orphan LuxR-type receptor that senses and responds to AHLs synthesized by other species of bacteria (Ahmer, 2004; Janssens et al., 2007). Although most members of the Enterobacteriaceae family contains solo sdiA, species from the genus Pantoea and Erwinia harbor luxI homologs which represent descendants of the ancient LuxI protein paired with SdiA (Sabag-Daigle and Ahmer, 2012).
SdiA stands for “suppressor of cell division inhibition” and directly regulates gene expression by binding to regulatory elements, termed SdiA-box, located at the promoter region of the target genes (Yamamoto et al., 2001). Reports indicate that the nucleotide sequence of the SdiA-box consists of the sequence AAAA (with minor variations) at both ends, intercalated with a spacer sequence that can vary from 8, 10, to 18 nucleotides (Yamamoto et al., 2001; Shimada et al., 2014; Lu et al., 2017; Ma et al., 2020). SdiA regulates the transcription of the target genes by complexing with AHLs synthesized by other bacterial species (Michael et al., 2001; Smith and Ahmer, 2003; Ahmer, 2004), in response to synthetic AHLs (Shimada et al., 2014; Styles et al., 2020), or even in the absence of AHLs (Yamamoto et al., 2001; Dyszel et al., 2010; Shimada et al., 2014; Nguyen et al., 2015). Besides, non-AHL molecules have been identified as SdiA ligands, such as xylose (Yao et al., 2007) and the endogenous ligand 1-octanoyl-rac-glycerol (Nguyen et al., 2015). Indole has also been suggested to influence SdiA-mediated gene transcription (Lee et al., 2007; Lee et al., 2009), although this claim is contraditory, since some authors have reported that the effects of indole on Escherichia coli and Salmonella enterica is not mediated by sdiA (Sabag-Daigle et al., 2012; Kohli et al., 2018).
Acting as a transcriptional regulator, SdiA has been implicated in the regulation of several cellular processes including cell division (Sitnikov et al., 1996) and in the expression of virulence factors such as antibiotic resistance, motility and biofilm formation (Kanamaru et al., 2000; Sharma et al., 2010; Antunes et al., 2010; Tavio et al., 2010; Culler et al., 2018; Ma et al., 2020). Bacterial cell division relies on the ftsQAZ operon, which encodes the FtsQ, FtsA, and FtsZ proteins responsible for recruiting and assembling cell division machinery in the septum. Regulation of this operon is complex and involves multiple promoters and several transcriptional regulators (Joseleau-Petit et al., 1999). In E. coli, two promoters located upstream of ftsQ gene are responsible for independent transcriptional regulation of the full operon: P1 promoter is controlled by the stationary-phase Sigma factor RpoS, while P2 promoter is controlled by SdiA (Wang et al., 1991; Sitnikov et al., 1996). SdiA plays an important role during cell division because it acts as a positive regulator in the expression of ftsQAZ operon (Wang et al., 1991), and in vitro assays confirmed DNA-binding activity of purified SdiA to the ftsQ promoter (Shimada et al., 2014). Regarding virulence factors, SdiA regulates biofilm formation through regulation of fimbriae and curli genes (Culler et al., 2018). Studies have shown that fimbriae play an important role during biofilm formation by K. pneumoniae (Schroll et al., 2010; Alcántar-Curiel et al., 2013), but the role of SdiA in this process has not yet been addressed in this bacterium.
K. pneumoniae encodes several types of fimbriae. The most studied fimbriae are those of type 1, type 3 and the common pilus encoded by the fim, mrk, and ecp gene clusters, respectively (Alcántar-Curiel et al., 2013; Li et al., 2014). Type 3 fimbriae mediate adhesion to several types of cells and they are essential for biofilm formation on abiotic surfaces, thus playing a role in the development of infections in catheterized patients (Struve et al., 2009; Stahlhut et al., 2012; Li et al., 2014). On the other hand, type 1 fimbriae are essential for urinary tract infection (Struve et al., 2008). Due to the high affinity for mannose residues present on the bladder cells surface (Rozen and Skaletsky, 2000), type 1 fimbriae promote adhesion and invasion of epithelial bladder cells, leading to the formation of biofilm-like intracellular bacterial communities (Rosen et al., 2008). The expression of the fim cluster is regulated by a mechanism known as phase variation (van der Woude and Baumler, 2004). The phase variation involves the fimS element, a DNA fragment located immediately upstream to the fimA gene that harbors the promoter region of fim cluster and has the capacity to suffer inversion of its orientation. Thus, depending on the orientation of fimS, the expression of fim gene cluster is activated or inactivated and the bacterium can shift from a fimbriated (phase ON) to a non-fimbriated (phase OFF) phenotype. Two recombinases, encoded by the fimE and fimB genes located upstream to fimS, control the inversion of the fimS element (van der Woude and Baumler, 2004).
While the role of SdiA in regulating the expression of several virulence factors in many pathogens is well documented, little is known about the role of this regulator in K. pneumoniae. Therefore, the present work aimed to investigate the role of SdiA in Klebsiella pneumoniae pathogenicity by assessing biofilm formation, fimbriae expression and production of quorum sensing autoinducers on an sdiA mutant strain of K. pneumoniae.
Materials and Methods
Bacterial Strains and Culture Conditions
Klebsiella pneumoniae strain ATCC 10031 and an isogenic mutant strain deficient for sdiA gene were used throughout this study. Bacterial strains were routinely grown in Lysogeny Broth (LB; BD, United States) at 37°C with shaking at 200 rpm, and on LB agar under static cultures. Bacterial growth was monitored by measuring the optical density (O.D.) of the cultures at wavelength of 600 nm (O.D.600nm) using the GeneQuant Spectrophotometer (GE Healthcare). Antibiotics were added when appropriated at the following concentrations: ampicillin at 100 μg/mL, kanamycin at 25 μg/mL, chloramphenicol at 25 μg/mL, and erythromycin at 50 μg/mL.
For Reverse Transcription Quantitative Real-Time PCR (RT-qPCR) analyses and detection of autoinducers type 2, the strains were grown in LB medium with the addition of 2% glucose, since glucose stimulates K. pneumoniae to produce more AI-2 (Zhu et al., 2012).
To investigate the phenotypic effects of AHL on the K. pneumoniae strains, the assays carried out in this study were conducted using bacterial cells cultured in the absence or in the presence of 2 μM (final concentration) of the synthetic AHL N-Octanoyl-L-homoserine lactone (C8-HSL, Sigma-Aldrich). Previous studies have indicated that C8-HSL is an effective AHL autoinducer for both E. coli and Salmonella enterica SdiA (Michael et al., 2001; Yao et al., 2006; Lee et al., 2007; Kim et al., 2013; Shimada et al., 2014).
For indirect measurements of AI-2 molecules produced by the K. pneumoniae strains we used the reporter strain Vibrio campbellii MM32 (ATCC® BAA-1121TM). This strain is unable to produce AI-2 and to sense AHL due to mutations on luxS and luxN receptor genes, respectively. Autoinducer Bioassay medium (AB medium) was used to culture V. campbellii and also as the assay medium for AI-2 detection (Bassler et al., 1993), and consisted of 0.3 M NaCl, 0.05 M MgSO4 and 0.2% vitamin-free acid casamino, adjusted to pH 7.5 with KOH and sterilized by autoclaving. After reaching room temperature, 10 mL of 1 M potassium phosphate (pH 7.0), 10 mL of 0.1 M L-arginine and 20 mL of glycerol were added for each liter of the initial solution.
Generation of K. pneumoniae sdiA Mutant Strain
SdiA-deficient ATCC 10031 mutant strain was generated using TargeTron Gene Knockout System (Sigma-Aldrich), following a protocol previously standardized by us (Gomes et al., 2021). The TargeTron system produces an RNA-protein complex (RNP) that inserts a modified group II intron of Lactococcus lactis (L1. LtrB Intron) permanently on the coding region of the target gene. The knockout renders the mutant strain resistant to kanamycin antibiotic, because the group II intron RNA harbors a kanamycin resistance gene (kanR). A computer algorithm at Sigma-Aldrich TargeTron Design website was used to identify the most efficient target site on sdiA gene (Supplementary Table 1). The website also provided the nucleotide sequence of three primers (Supplementary Table 2) used to mutate (re-target) the intron by PCR reactions. The mutated 350 bp PCR fragment was cloned into the pACD4K-C vector provided by the manufacturer. Subsequently, the recombinant pACDK-4-C vector were transformed into E. coli DH5α strain to obtain clones. The pACD4K-C vector contains a T7 promoter to express the intron and RNP, and a source of T7 RNA Polymerase was provided by plasmid pAR1219. Therefore, competent ATCC 10031 cells were cotransformed with pAR1219 and recombinant pACDK-4-C, and incubated in LB broth containing ampicillin and chloramphenicol at 37°C for 18 h with shaking at 200 rpm. Next, a new incubation of ATCC 10031 cells was performed with fresh LB broth under the same conditions. When the O.D.600nm reached 0.2, the expression of RNP was induced with the addition of 0.1 M IPTG and incubation at 30°C for 30 min with shaking at 200 rpm. Then, the cells were centrifuged for 2 min at 10,000 g, resuspended in fresh LB broth, and incubated again at 30°C for 1 h. Colonies grown on agar plate supplemented with kanamycin were selected after 18 h of incubation at 37°C. Since gene knockout by TargeTron System is based on the insertion of kanR gene inside sdiA, the mutant strain of K. pneumoniae ATCC 10031 was renamed sdiA::kanR.
The complemented strain sdiA::kanRcomp was obtained by introducing the sdiA gene back into the mutant strain. For this, a DNA fragment comprising the entire coding region of sdiA plus the 3′ and 5′ flanking regions was PCR amplified using primers listed on Supplementary Table 3. The DNA fragment was inserted on pCRTM2.1 vector (InvitrogenTM) previously cloned with erythromycin-resistance gene. Chemically competent sdiA::kanR was transformed with the recombinant vector and plated on LB agar supplemented with 50 μg/mL erythromycin. Complemented strains were recovery by screening erythromycin-resistant colonies.
Growth Pattern and Optical Microscopy Analysis
The wild-type ATCC 10031, mutant sdiA::kanR, and complementary sdiA::kanRcomp strains were separately inoculated into LB medium and grown until saturation (overnight) at 37°C under shaking. The next day, the culture was diluted 1:100 in fresh LB and the bacterial growth was monitored every 15 min by measuring the O.D.600nm. Growth curves were constructed by plotting the O.D.600nm values against time. To investigate the morphology of the bacterial cells, 10 μL of each culture were harvested at the indicated O.D.600nm, stained with fuchsine and visualized under optical microscopy. Results were recorded under 1000 × magnification. Three independent cultures of each K. pneumoniae strain were conducted for the growth pattern and the optical microscopy analysis.
Biofilm Mass Assay and Pellicle Formation at the Air-Liquid Interface
Biofilm formation assays were carried out conducted in 96-well microtiter plates as described previously (Gomes et al., 2021). Saturated cultures of the bacterial strains were harvested by centrifugation and resuspended in LB broth to a final concentration of 106 cells/mL. 150 μL of each cell suspension were applied in 96-well microtiter plates containing 150 μL of LB supplemented or not with C8-HSL. The plates were incubated at 37°C under static conditions for 8, 24, 48, and 72 h of incubation. After each time, the medium was discarded, and the biofilm mass was gently rinsed with PBS. The wells were left to dry for 5 min and then stained with 0,1% of crystal violet for 15 min at room temperature. After staining, the crystal violet was discarded, the wells were rinsed 3 times with PBS and left to dry for 5 min. The biofilm-associated crystal violet was solubilized with 200 μL acetic acid (30%, v/v), and the absorbance of the acetic acid containing the eluted dye was measured at O.D.600nm with a Biotek Microplate reader. Biofilm formation assays were conducted in duplicates from three independent cultures of each K. pneumoniae strain.
We also investigated the formation of pellicle in air-liquid interface by the K. pneumoniae strains. This is a biofilm-like structure that requires a great organization due to the lack of solid surface for fixation. To investigate pellicle formation, the strains were inoculated overnight and subsequently grown in LB medium until O.D.600nm of 0.6. The cultures were diluted to obtain a final density of 5 × 106 CFU/mL. Six milliliters of each cell suspension were applied in glass tubes and incubated for 72 h under static conditions at 37°C. The formation of pellicles at the air-liquid interface in the glass tubes was recorded using a digital camera. Pellicle formation assays were performed in triplicate for each K. pneumoniae strain, and they were not conducted in presence of the autoinducer C8-HSL.
Phase Variation Assay of the fimS Element
The orientation of the fimS element, which contains the promoter region of the cluster fim, was investigated according to a phase variation assay previously described (Struve et al., 2008). K. pneumoniae strains were grown to O.D.600nm of 0.6 in LB broth at 37°C with shaking. Cells were harvested and the DNA were extracted with the Wizard® Genomic DNA Purification Kit (Promega). DNAs were then used to PCR amplify an 817 bp fragment containing the invertible fimS promoter element using primers CAS168 and CAS169 (Supplementary Table 3). The amplified fragments were cut with HinfI restriction enzyme and the pattern of the digested products was determined on 2% agarose gels stained with ethidium bromide. The HinfI restriction site is asymmetrically located on the fimS element, which results in different cleaved fragments depending on the orientation of the phase switch: a phase switched to the ON orientation results in fragments of 212 and 605 bp, whereas a phase switched to the OFF orientation results in fragments of 321 and 496 bp. The assays were performed from two independent cultures for each strain of K. pneumoniae.
Agglutination Assay
Yeast agglutination assays were performed to investigate the expression of type 1 fimbriae by the K. pneumoniae strains. The assays were conducted on Kline concavity slides as described previously, with minor modifications (Gomes et al., 2021). Overnight cultures of the bacterial strains were inoculated (1:100) into fresh LB broth supplemented or not with C8-HSL and cultured until they reached O.D.600nm of 0.6. Bacteria were then mixed with 5% (w/v) suspension of Saccharomyces cerevisiae cells (Sigma-Aldrich) prepared in PBS. The intensity of the agglutination was documented using a digital camera. The agglutination of the yeast cells is specifically mediated by type 1 fimbriae since these fimbriae have great affinity for mannose, a highly abundant residue on yeast cell-surface. Therefore, the assays were also performed in the presence of 5% D-(+)-Mannose (Sigma-Aldrich) to confirm if the agglutination was indeed mediated by the type 1 fimbriae. Assays were carried out in triplicate for each K. pneumoniae strain.
Indirect Detection of AI-2
The production of AI-2 molecules by the K. pneumoniae strains was indirectly measured using the reporter strain Vibrio campbellii MM32, as previously reported (Zhu et al., 2008). Initially, the strains were cultured overnight in LB broth containing 2% glucose. On the following day, the cultures were inoculated (1:100) into LB broth supplemented or not with C8-HSL, and samples were collected at O.D.600nm of 0.2, 0.4, 0.6, 0.8, and 1.0. Cell-free conditioned culture supernatant was obtained by centrifuging the cultures at 10,000 g and passing the supernatant through a Millipore membrane filter (pore size of 0.22 μm). V. campbellii MM32 was grown overnight at 30°C at 200 rpm in AB medium and then diluted 1:5000 in fresh AB medium. 180 μL of the MM32 diluted culture were distributed in 96-well microtiter plates, followed by the addition of 20 μL of the cell-free conditioned culture supernatant. The mixtures were incubated with shaking at 30°C, and the luminescence were measured every 15 h in the equipament GloMax® 96 Microplate Luminometer (Promega, Madison, WI, United States). Assays were carried out at least in triplicate for each K. pneumoniae strain. The results are expressed as arbitrary luminescence units and were obtained by dividing the light values measured on the experimental samples by the light values of the sterile LB culture medium.
RNA Extraction and Real-Time Quantitative PCR Analysis
K. pneumoniae cells were grown in LB broth at 37°C with shaking at 200 rpm until O.D.600nm of 0.2 and 0.6. Cell pellets were obtained after centrifugation, and total bacterial RNA were extracted by using the TRIzolTM MaxTM Bacterial RNA Isolation and MICROBEnrichTM kits (InvitrogenTM), following the manufacturer’s instructions. After treatment with DNAse, 1 μg of total RNA was reverse transcribed in cDNA using the High-Capacity cDNA Reverse Transcription kit (Applied BiosystemsTM), according to the manufacturer’s recommendations. Synthesized cDNA was used in RT-qPCR analyses, using primers listed in Supplementary Table 3.
Primers were designed using Primer3 version 0.4.0 web-program1 (Rozen and Skaletsky, 2000). Reactions were performed in triplicates on the Applied Biosystems® 7300 Real-Time PCR System equipment (ThermoFisher Scientific) with the PlatinumTM SYBRTM Green qPCR SuperMix-UDG kit (InvitrogenTM), following the manufacturer’s instructions. RT-qPCR results were normalized using rho as endogenous gene, which encodes the transcription termination factor Rho (Gomes et al., 2018). The relative expression levels of the genes were calculated using the 2–ΔΔCt relative quantification method (Livak and Schmittgen, 2001). GraphPad Prism 7.0 (GraphPad Software, Inc) was used for the statistical analyses.
Purification of K. pneumoniae SdiA Protein and Electrophoretic Mobility Shift Assays (EMSA)
The coding region of sdiA gene was amplified by PCR using K. pneumoniae genomic DNA as template and primers containing the restriction sites for NdeI and XhoI (Supplementary Table 3). After digestion with NdeI and XhoI, the amplicon was cloned into the expression vector pET28a(+) (Sigma-Aldrich) at the corresponding sites, and the resulting recombinant vector was transformed into E. coli BL21(DE3). The expression and purification of the histidine-tagged recombinant SdiA protein (His-SdiA) were conducted as described elsewhere (Gomes et al., 2018), with some modifications. Some reports have shown the expression of recombinant SdiA from culture medium supplemented with AHLs autoinducers (Yao et al., 2006; Abed et al., 2014). Since we aimed to conduct EMSA with the K. pneumoniae SdiA protein in its apo form (i.e., not complexed with autoinducers), we did not add AHLs in the culture medium, as has been done by others (Kanamaru et al., 2000; Yamamoto et al., 2001; Wu et al., 2008; Kim et al., 2013; Shimada et al., 2014; Nguyen et al., 2015; Lu et al., 2017). In brief, transformed BL21(DE3) was grown in 500 mL of LB medium at 37°C to an O.D.600nm of 0.4. At this moment, isopropyl-β-D-thiogalactopyranoside (IPTG, Sigma-Aldrich) was added to a final concentration of 1 mM and the culture was incubated for 4 h. After incubation, in-culture bacterial cell lysis was promoted by adding CelLyticTM Express 1 mL Tablets (Sigma-Aldrich), following the manufacturer’s instructions. The lysed cells were centrifuged at 16,000 g for 15 min to obtain a clarified supernatant. His-SdiA was purified from the clarified supernatant by affinity chromatography under native conditions using Ni-NTA Agarose matrix (Qiagen), following to the manufacturer’s protocol. Eluted fractions containing the recombinant SdiA were pooled and dialyzed overnight at 4°C on storage buffer (50 mM Tris-HCl pH 8.0, 2 mM DTT, 0.5 mM EDTA, and 10 % glycerol v/v). The concentration of the purified His-SdiA was determined by the Bradford method and the purity was verified by SDS-PAGE analysis.
EMSA was conducted using the purified His-SdiA protein and DNA probes containing the promoter region of the indicated genes. The probes were generated by PCR amplifications using the primers and conditions displayed on Supplementary Table 3. As a negative control, a 220 base pairs DNA fragment was obtained by PCR amplifying a recircularized pCRTM2.1 vector (InvitrogenTM) without insert using primers M13 (Supplementary Table 3). Reactions were performed using 2 or 10 ρmol of the recombinant His-SdiA protein, previously equilibrated for 15 min at 37°C in 40 μL of 1X binding buffer containing 10 mM Tris-HCl pH 8.0, 50 mM KCl, 1 mM DTT, 0.1 mM EDTA, 2.5 mM MgCl2, and 2 % glycerol. After pre-incubation, 50 ηg of the DNA probes were added and the reaction mixture were incubated for 30 min at 37°C. To investigate the effects of AHLs on the binding affinity of SdiA, EMSA were carried out in the absence and in the presence of 2 or 4 μM of C8-HSL. Samples were submitted to electrophoresis at either 2% (w/v) agarose gel at 80 volts for 60 min in 1X sodium borate buffer (5 mM) or native non-denaturing 6% bis-acrylamide gel at 80 volts for 2 h in 1X TBE buffer (89 mM Tris, 89 mM boric acid, and 2 mM EDTA). DNA probe-SdiA complexes formed were visualized under UV light after staining the gels with ethidium bromide solution (0.5 μg/mL). Images of the DNA bands were recorded with Molecular Imager® Gel DocTM XR System (Biorad) using the Image LabTM Software version 5.0 (Biorad).
Results
K. pneumoniae Depleted of SdiA Presents Impaired Cell Division and Abnormal Cell Morphology
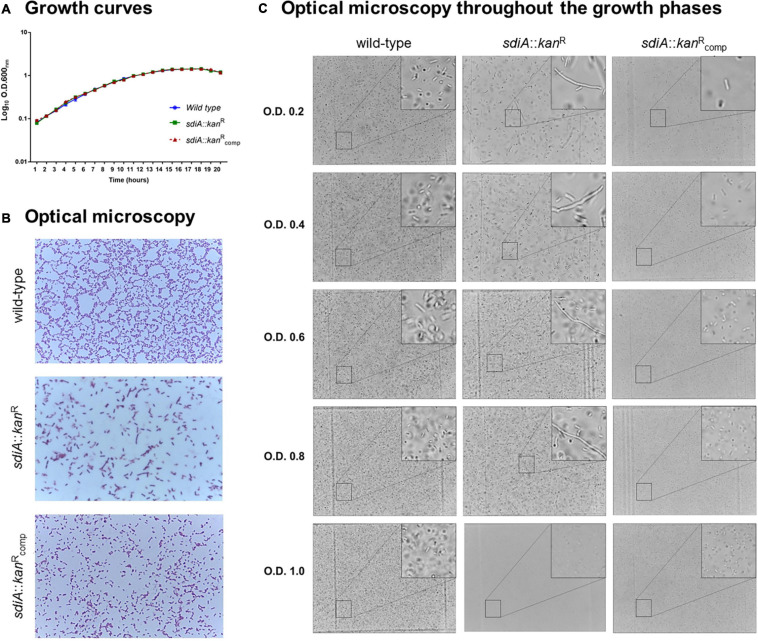
In order to investigate the role of SdiA in Klebsiella pneumoniae, a knockout sdiA mutant strain was generated and phenotypically characterized. Firstly, growth curves of the wild-type (ATCC 10031), mutant sdiA::kanR and complemented sdiA::kanRcomp strains were assessed. As shown in Figure 1A, no significant changes were observed in the growth curves of the mutant and the complemented strains in relation to the wild-type.
FIGURE 1.
Lack of SdiA affects cell division and leads to morphological alterations of K. pneumoniae cells. (A) Growth curves of the wild-type, mutant (sdiA::kanR) and complemented (sdiA::kanRcomp) strains are displayed as log scale of the optical density of the cultures of each strain measured at the indicated times. No differences in growth curves were observed among the strains. (B,C) Aliquots of the cultures of each strain were submitted to optical microscopy analyses to investigate the morphology of the bacterial cells. Disruption of sdiA led the mutant strain to exhibit a filamentous phenotype. The mutant strain recover the same morphology of the wild-type and complemented strains only at high cell densities (O.D.600 nm of 1.0).
However, disruption of sdiA led the mutant strain to assume a filamentous phenotype (Figure 1B), denoting a failure in bacterial cell division. On the other hand, the morphology of the complemented strain was similar to the wild-type. Analyzing the morphology of the strains throughout growth stages (Figure 1C), we observed that the mutant strain presents the filamentous phenotype at O.D.600nm of 0.2–0.8 and that the cell division of the mutant strain is recovered only at at high densities (O.D.600nm of 1.0). No changes in the morphology of the wild-type and the complemented strain were observed throughout the growth stages (Figure 1C).
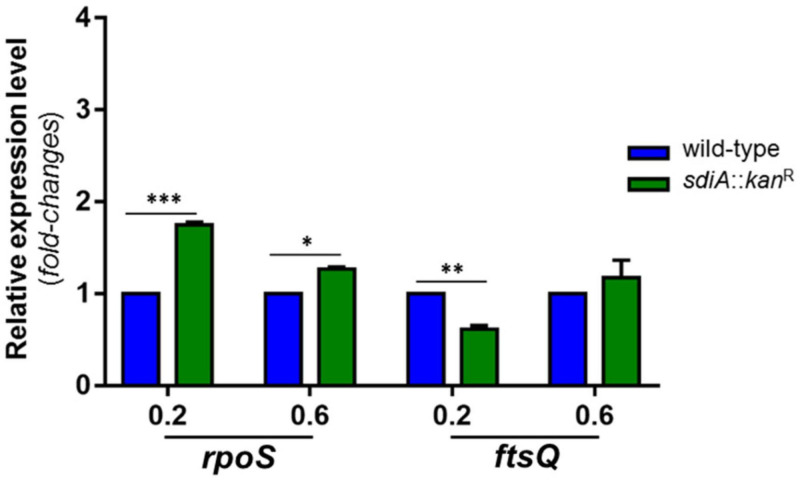
Since the mutant strain exhibited a filamentary shape in a manner dependent on the growth phase, we decided to investigate the expression pattern of ftsQ, from the ftsQAZ cell division gene cluster, and the rpoS gene, which encodes the stationary-phase Sigma factor RpoS. As displayed in Figure 2, the expression levels of rpoS in the mutant strain was almost twofold higher than the level of the wild-type strain at O.D.600nm of 0.2, and slightly up-regulated at O.D.600nm of 0.6. On the other hand, the expression levels of ftsQ in the mutant strain was slightly down-regulated at O.D.600nm of 0.2 and unchanged at O.D.600nm of 0.6, when compared to the wild-type strain.
FIGURE 2.
K. pneumoniae depleted of SdiA exhibits up-regulation of rpoS (stationary-phase Sigma factor RpoS) and down-regulation of ftsQ (gene cluster fts of cell division). The expression of rpoS and ftsQ genes was analyzed by RT-qPCR on the wild-type and mutant K. pneumoniae strains cultured at O.D.600 nm of 0.2 and 0.6. Error bars represent the standard deviation. Statistically significant differences between the strains were analyzed by multiple t-test (*p < 0.05; **p < 0.01; ***p < 0.001).
Lack of SdiA Increases Biofilm Formation and Yeast Cells Agglutination, and Leads to Down-Regulation of the Type 3 Fimbriae and Up-Regulation of Type 1 Fimbriae Expression
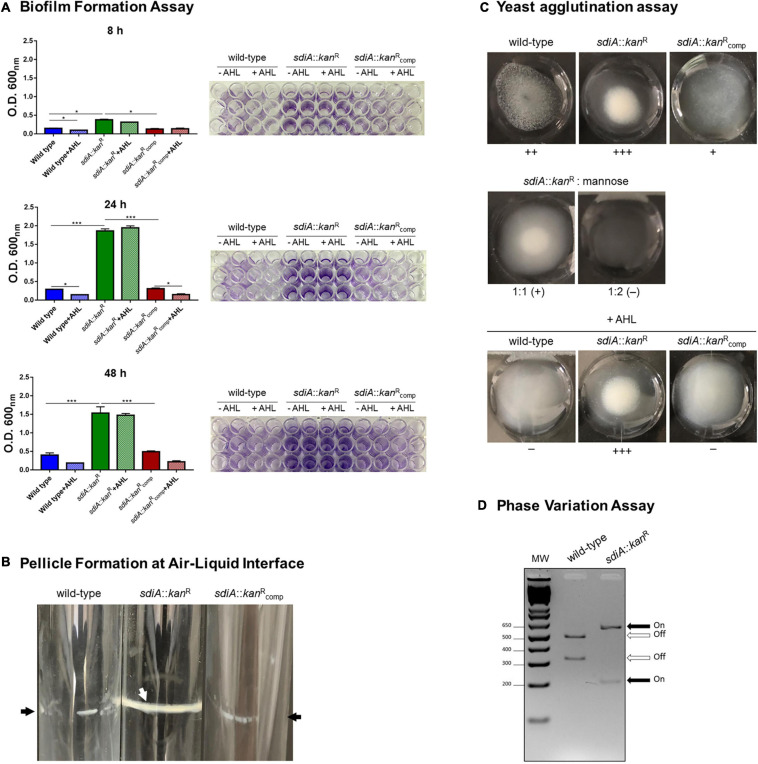
The role of SdiA as a regulator of biofilm formation is well recognized in many pathogens, but it is still uncertain in K. pneumoniae. To assess whether SdiA is also involved in biofilm formation by K. pneumoniae, the ability of the sdiA::kanR mutant strain to form biofilms was compared to the wild-type and complemented strains. As shown in Figure 3A, the biofilm formation by sdiA::kanR was significantly superior than the wild-type after 8, 24, and 48 h of incubation, while the complemented sdiA::kanRcomp strain restored the pattern originally exhibited by the wild-type. The addition of the AHL C8-HSL had no effect on biofilm formation by the mutant strain, but reduced the biofilm formed by the wild-type and the complemented strains. Both wild-type and sdiA::kanR strains were able to form a pellicle in the air-liquid interface (Figure 3B), but the mutant exhibited a thicker pellicle than the wild-type strain. Moreover, the phenotype was fully re-established on the complemented mutant strain.
FIGURE 3.
K. pneumoniae deficient in SdiA presents enhanced biofilm formation, yeast cells agglutination, and production of type 1 fimbriae. (A) The biofilm formation assays showed that the mutant strain (sdiA::kanR) forms more biofilm than the wild-type and complemented (sdiA::kanRcomp) strains. The addition of the AHL C8-HSL reduced the biofilm formed by the wild-type and the complemented strains at the indicated incubation times but did not affect biofilm formation by the mutant strain. For this assay, the strains were incubated at 37 °C in 96-well microtiter plates under static condition for 8, 24, and 48 h, and then stained with crystal violet. The biofilm-associated crystal violet was solubilized and measured at O.D.600 nm. Error bars represent the standard deviation. Statistically significant differences between the strains were analyzed using ANOVA (*p < 0.05; ***p < 0.001). The 96-well plates of each biofilm formation assay are shown to the right of the respective graphs. (B) The pellicle formation at the air-liquid interface assays showed that K. pneumoniae mutant strain presents thicker pellicle formation when compared to the wild-type and complemented strains. For this assay, strains were cultivated for 72 h at 37°C under shaking conditions. Black arrows indicate thin pellicle structure formation by the wild-type and complemented strain, while the white arrow indicates the formation of thick pellicle by the mutant strain. The images are from an individual representative experiment. (C) The yeast agglutination assays revealed that the mutant strain agglutinated yeast cells with more intensity than the wild-type strain. The absence of agglutination of yeast cells by the mutant strain in the presence of mannose confirms that the agglutination was mediated specifically by type 1 fimbriae. The addition of AHL reduced the agglutination by the wild-type and the complemented strains but had no effect on the agglutination of the yeast cells by the mutant strain. For yeast agglutination assays, cultures of the bacterial strains at O.D.600 nm of 0.6 were mixed with Saccharomyces cerevisiae cells, and the intensity of the agglutination was digitally documented. The images are from an individual representative experiment. (D) The phase variation assays showed the promoter region of the fim gene cluster at ON orientation in the mutant strain and at OFF orientation in the wild-type strain. For this assay, the fimS element, which contains the promoter region of the cluster fim, was PCR amplified using DNA extracted from the wild-type and mutant strains cultured in LB broth to O.D.600 nm of 0.6. The amplicons were cut with HinfI and the digested fragments were resolved by electrophoresis on agarose gels. The fimS element at ON orientation results in fragments of 212 and 605 bp, whereas at OFF orientation results in fragments of 321 and 496 bp. The image is from an individual representative experiment.
Since fimbriae are considered as important mediators of bacterial adhesion and the loss of sdiA has resulted in more intense biofilm formation, we sought to investigate the production of fimbriae by the K. pneumoniae strains. Firstly, we compared the ability of the K. pneumoniae strains to agglutinate yeast cells. As displayed in Figure 3C, the mutant strain sdiA::kanR was able to agglutinate yeast cells with more intensity than the wild-type, while the complemented strain partially recovered the phenotype exhibited by the wild-type. The addition of mannose abolished the agglutination of the yeast cells by the mutant strain, thus confirming that the agglutination was indeed mediated by type 1 fimbriae. The addition of AHL had no effect on the agglutination of the yeast cells by the mutant strain, but reduced the agglutination by the wild-type and the complemented strains.
Yeast agglutination is indicative of type 1 fimbriae expression because this type of fimbriae has a great affinity for mannose-containing receptors present on the surface of the yeast cells. To confirm that type 1 fimbriae expression is induced on the mutant strain, we sought to investigate the phase variation of the fimS invertible element containing the promoter region of the fim gene cluster. The phase variation assay indicated that the mutant strain presents the fimS element in the ON orientation (Figure 3D).
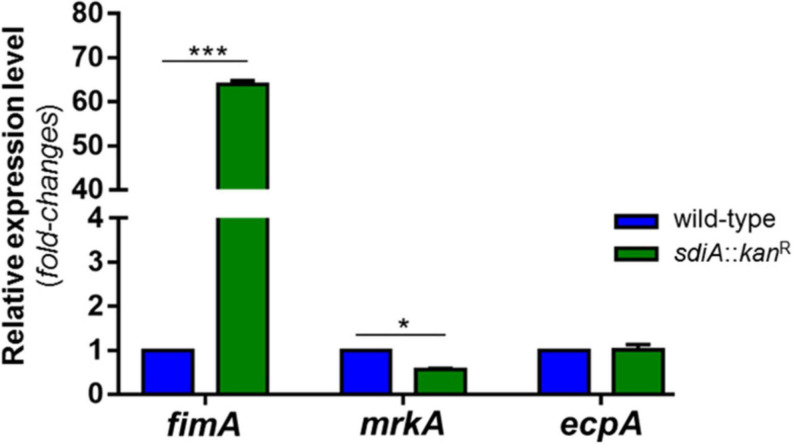
To finally confirm the up-regulation of type 1 fimbriae in the mutant strain, analyses of fimbrial genes expression were conducted in the wild-type and sdiA::kanR strains. As shown in Figure 4, the mutant strain exhibited significantly higher transcription levels of fimA (type 1 fimbriae) and slightly lower transcription of mrkA (type 3 fimbriae) than the wild-type strain. There was no difference statistically significant in the expression of ecpA (common pillus) between wild-type and mutant strains. These findings seem to indicate that the SdiA regulator modulates the expression of fimbriae in K. pneumoniae, by repressing the expression of type 1 fimbriae and inducing the expression of type 3 fimbriae.
FIGURE 4.
K. pneumoniae deprived of SdiA presents up-regulation of fimA (type 1 fimbriae) and down-regulation of mrkA (type 3 fimbriae). No change in the expression of ecpA (common pillus) was observed between the wild-type and mutant strains. The expression of the genes was analyzed by RT-qPCR on the wild-type and mutant K. pneumoniae strains cultured at O.D.600nm of 0.6. Error bars represent the standard deviation. Statistically significant differences between the strains were analyzed by multiple t-test (*p < 0.05; ***p < 0.001).
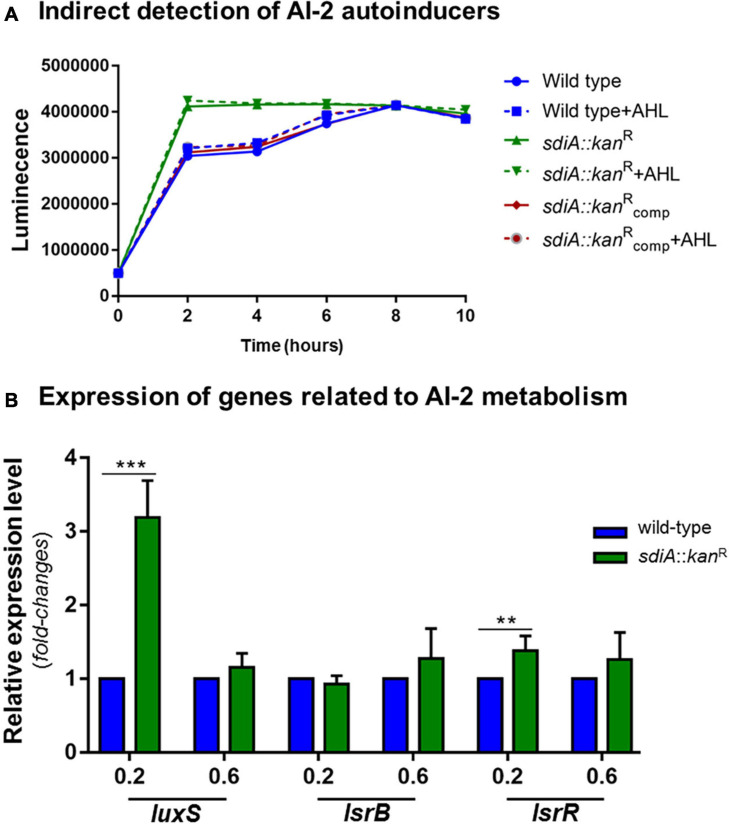
SdiA Deficient K. pneumoniae Strain Reaches Maximum Production of AI-2 Earlier
SdiA has been related to inter-species communication since this regulator senses and responds to AHLs synthesized by other bacteria species. To elucidate a possible relationship of SdiA with the inter-species communication mediated by the Autoinducers-2 (AI-2) QS signaling system, we used an indirect method based on the reporter strain V. campbellii to measure and compare the production of signal molecules AI-2 by the wild-type and the sdiA deficient mutant strains. According to Figure 5A, the mutant strain reaches maximum production of AI-2 after 2 h of culture, which corresponds to the lag phase of growth (O.D.600nm of 0.1) according to the growth curve of the strain (Figure 1A). On the other hand, the wild-type and the complemented strains took longer and reached the same level of AI-2 as the mutant strain only after 8 h of culture, which corresponds to the mid-log phase of growth (O.D.600nm of 0.6, Figure 1A). The addition of AHL had no apparent effect on the production of AI-2 molecules by the K. pneumoniae strains.
FIGURE 5.
K. pneumoniae cells lacking SdiA regulator reach maximum production of AI-2 QS signaling molecules earlier and present up-regulation of luxS and lrsR. (A) Cell-free supernatants were collected from wild-type and sdia::kanR mutant strains cultured at the indicated O.D. and tested for AI-2 production by indirect measuring the level of bioluminescence induced in the V. campbellii MM32 reporter strain. The mutant strain reaches maximum production of AI-2 after 2 h of culture, while the wild-type and complemented strains reached maximum production of AI-2 only after 8 h of culture. The addition of AHL had no effect on the production of AI-2 molecules by the K. pneumoniae strains. (B) The absence of SdiA causes up-regulation of luxS and lsrR genes, which encode the AI-2 synthase and the LsrR repressor, respectively. No change in the expression of lsrB (receptor of AI-2 molecules) was observed between the wild-type and mutant strains. Gene expression analysis were performed by RT-qPCR on the wild-type and mutant K. pneumoniae strains cultured at O.D.600 nm of 0.2 and 0.6. Error bars represent the standard deviation. Statistically significant differences between the strains were analyzed by multiple t-test (**p < 0.01; ***p < 0.001).
Based on these results, we proceeded to gene expression analyses of luxS, lsrB, and lsrR, genes related to AI-2 synthesis, uptake, and uptake regulation, respectively. As displayed in Figure 5B, the expression levels of luxS in the mutant strain was more than threefold higher than the wild-type strain at O.D.600nm of 0.2 and slightly induced at O.D.600nm of 0.6. No statistically significant difference was observed for the expression of lsrB between wild-type and mutant strains, whereas the expression levels of lsrR was slightly up-regulated at O.D.600nm of 0.2 and unchanged at O.D.600nm of 0.6 in the mutant strain, compared to that in the wild-type.
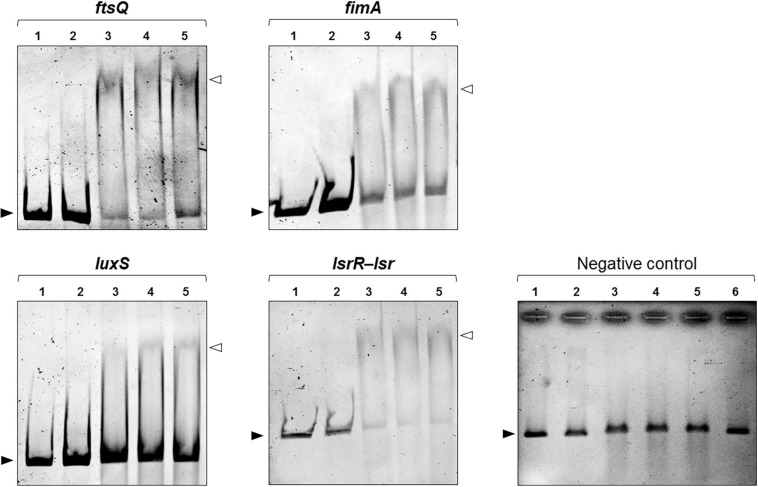
SdiA Binds to the Promoter Region of ftsQ, fimA, luxS, and lsrR-lsrA
The significant up-regulation of fimA and luxS in the mutant strain led us to examine whether SdiA exerts a direct role on the expression modulation of these genes by binding on their promoter region. Firstly, bioinformatic analyses were employed to identify putative SdiA-boxes on the promoter region of fimA, luxS and on the intergenic region between lsrR and lsrA. The promoter region of the ftsQAZ operon was included in the analyses as a positive control because it is well known by previously published studies that SdiA binds in the promoter region of the operon and controls the transcription of ftsQAZ genes (Wang et al., 1991; Sitnikov et al., 1996; Yamamoto et al., 2001; Shimada et al., 2014). SdiA-binding sequences that resembles to the consensus sequence of SdiA-box (5′-AAAA(N5–30)AAAA-3′) were found on the promoter region of fimA, luxS, lsrR-lsrA and ftsQAZ (Supplementary Table 4). Next, EMSA was performed using the recombinant His-SdiA protein from K. pneumoniae and DNA fragments containing the putative SdiA-boxes as probes. As shown in Figure 6, shifted bands corresponding to SdiA-DNA probe complexes were observed only with 10 ρmol of the recombinant His-SdiA protein. The addition of C8-HSL, both 2 and 4 μM, had no apparent effect on the shifting mobility of the bands. No shifted bands were observed when His-SdiA was incubated with the negative control probe. These results indicate the direct binding of K. pneumoniae SdiA to the promoter region of fimA, luxS, lsrR-lsrA and ftsQAZ, and that this binding occurs in an AHL-independent manner.
FIGURE 6.
DNA Electrophoretic Mobility Shift Assays (EMSA) revealed that SdiA binds to the promoter region of ftsQ, fimA, and the intergenic region between the lsrR and lsr operon. For EMSA, 50 ηg of the DNA probes (in all lanes) were incubated with 2 ρmoles (lane 2) or 10 ρmoles (lanes 3–5) of the recombinant His-SdiA protein for 30 min at 37°C. To investigate the effects of AHL on the binding affinity of SdiA, 2 μM (lane 4) or 4 μM (lane 5) of C8-HSL were added to the binding reaction. Mobility shifts (indicated by open arrowheads) are observed only at higher concentration of purified SdiA (10 ρmoles, lanes 3–5). These shifts represent the formation of SdiA-DNA probe complexes and confirm the binding activity of SdiA on the DNA probes. The addition of C8-HSL have no apparent effect on the shifting mobility of the bands (lanes 4 and 5 compared). Closed arrowheads indicate the free DNA probes. No shifted bands were observed when SdiA was incubated with the negative control DNA probe. Lane 6 represents the incubation of the negative DNA probe (50 ηg) with 4 μM of C8-HSL in the presence of 10 ρmoles of the purified SdiA.
Discussion
In many pathogens, the SdiA regulator modulates the expression of several virulence factors, such as adherence and motility (Kanamaru et al., 2000; Sharma et al., 2010), multidrug resistance (Tavio et al., 2010), biofilm formation (Culler et al., 2018) and acid tolerance (Ma et al., 2020). Concerning K. pneumoniae, little is known about the role of SdiA in the pathogenesis of this bacterium and, to date, no member of the SdiA regulon had been described in K. pneumoniae. In the present study, we provided new insights into the role of SdiA in the expression of virulence factors by K. pneumoniae through characterization of a strain depleted from sdiA gene. We also investigated the presence of putative SdiA binding sites within the promoter region of genes responsible for the synthesis of type 1 fimbriae, bacterial cell division, and the metabolism of type 2 autoinducers.
First, we compared the growth pattern of the sdiA mutant strain with the wild-type and the complemented strains. Although no changes in bacterial growth were observed among the strains, K. pneumoniae cells lacking SdiA regulator presented a filamentary shape rather than the typical rod shape, revealing a failure in cell division by the mutant cells. Bacterial cell division relies on the ftsQAZ operon, which encodes essential cell division proteins. Regulation of this operon is complex and involves multiple promoters and several transcriptional regulators (Joseleau-Petit et al., 1999). Two promoter regions located upstream of ftsQAZ drive the transcription of the entire operon: P1 promoter is controlled by the stationary-phase Sigma factor RpoS and P2 promoter by SdiA (Joseleau-Petit et al., 1999). In E. coli, SdiA plays an important role in cell division because positively regulates the transcription of the ftsQAZ operon (Wang et al., 1991; Sitnikov et al., 1996; Shimada et al., 2014). In our study, K. pneumoniae cells depleted of the SdiA regulator presented down-regulation of the ftsQ gene, and EMSA analyses confirmed DNA-binding activity of the purified K. pneumoniae SdiA to the ftsQAZ promoter. These results suggest that the filamentary shape of the mutant strains seems to be due to the down-regulation of the ftsQAZ operon with consequent failure in septum division, and that SdiA acts as a transcriptional activator of the operon also in K. pneumoniae. We also observed a growth-phase dependent effect of SdiA, since the rod-shaped pattern was restored only at the stationary phase of growth. Since ftsQAZ expression mediated by SdiA shows cell density dependence and that Sigma Factor RpoS and SdiA act in a coordinated manner to guarantee ftsQAZ expression (Sitnikov et al., 1996), the restored rod-shaped pattern observed at stationary phase of growth may be attributed to RpoS. Although RpoS is responsible for gene expression activation when cells enter the stationary phase, the up-regulation of rpoS observed at the mutant strain throughout the phases of growth seems to compensate for the absence of sdiA.
Subsequently, we investigated how the lack of SdiA influences biofilm formation by K. pneumoniae. We observed greater biofilm and pellicle formation at air-liquid interface by the sdiA mutant strain as compared to the wild-type strain. Biofilm formation in the presence of exogenous AHL rendered distinct results: while no effect was observed for the mutant strain, the wild-type formed less biofilm. These results are in agreement with studies that reported an increase in biofilm formation and pellicle formation by E. coli sdiA mutant strains (Lee et al., 2007; Sharma et al., 2010; Culler et al., 2018). Similarly to our findings, these authors also noticed no change in biofilm formation by the mutant strain in the presence of AHL, whereas the wild-type strain has the formation of biofilm inhibited. Our results indicate that SdiA responds to AHL and represses biofilm formation in K. pneumoniae.
Expression of fimbriae is the first essential step in biofilm formation by K. pneumoniae and previous reports by others indicate that SdiA exerts its effects on biofilm formation by regulating the expression of fimbrial genes (Culler et al., 2018). Recently, Shimada and colleagues reported a direct link between SdiA regulator and expression of type 1 fimbriae encoded by fim gene cluster (Shimada et al., 2014). They showed that SdiA binds the promoter region of fim cluster, although the precise binding site has not been determined. These authors (Shimada et al., 2014) and others (Janssens et al., 2007) also described a decrease in fimA transcription on the wild-type E. coli strain in the presence of AHL. Although the current study has not analyzed gene expression in the presence of AHL, we observed that cells lacking SdiA were able to agglutinate yeast cells with greater intensity than the wild-type strain, and that this agglutination is due specifically to the production of type 1 fimbriae. Likewise observed in biofilm formation assays, no effect on yeast agglutination by the mutant strain was observed when exogenous AHL was added, while the wild-type had its ability to agglutinate yeast cells reduced. RT-qPCR analyses showed up-regulation of fimA – the gene encoding the major structural subunit of type 1 fimbriae – in the mutant strain. Corroborating this result, phase variation analyses revealed that the fimS element, which contains the promoter region of the type 1 fim gene cluster, is oriented at ON position in the mutant strain. Furthermore, EMSA analyses confirmed direct binding of SdiA to an SdiA-box located in the immediate vicinity of the fimA initiation codon. All results together suggest that SdiA has a repressive role in the expression of type 1 fimbriae in K. pneumoniae, and that the lack of this regulator resulted in a hyperfimbriated phenotype that rendered the mutant strain with greater ability to form biofilm and to agglutinate yeast cells. Thus far, a regulatory role of the QS system on attachment and biofilm maturation by K. pneumoniae had been described before, but involving type 2 QS signaling molecules (Balestrino et al., 2005; De Araujo et al., 2010). Here we show that SdiA, the LuxR-type receptor of the AI-1 QS system, has a suppressive role in bacterial adherence and biofilm aggregation by K. pneumoniae as well.
It has been reported that AHL autoinducers function as a folding switch for the regulation of some LuxR homologues (Zhu and Winans, 2001; Schuster et al., 2004; Urbanowski et al., 2004; Lee et al., 2006). In these cases, the receptors only assume their active functional structure and with higher DNA-binding activity when complexed with their cognate AHLs ligands. For these LuxR receptors, AHLs are required for proper folding of the protein, stabilizing it, and preventing it from degradation; in vitro purification of these LuxR receptors in the active soluble form requires the presence of AHL in the culture medium used for bacterial expression. Interestingly, AHL molecules elicited phenotypic changes in the wild-type K. pneumoniae strain, but they had no apparent effect on the DNA-binding affinity of the recombinant SdiA protein, as revealed by EMSA analyses. Moreover, the ability of the purified SdiA to bind to the promoter region of the target genes indicates that the recombinant protein is correctly folded, even though it was expressed in a culture medium without supplementation with AHLs. These results suggest that AHL may not be a folding switch for K. pneumoniae SdiA, and the proper folding of SdiA in the absence of AHL during in vitro expression may be due to the binding of a yet unknown endogenous non-AHL ligands such as 1-octanoyl-rac-glycerol (Nguyen et al., 2015), in a mechanism that has already been suggested for other bacterial species (Sabag-Daigle et al., 2015).
In this study, we observed DNA-binding activity of K. pneumoniae SdiA in an AHL-independent manner. Although previous reports have shown the higher affinity of SdiA with DNA when complexed with AHLs (Nguyen et al., 2015), our results are in agreement with other reports that show DNA-binding activity of SdiA even in the absence of ligands (Yamamoto et al., 2001; Dyszel et al., 2010; Shimada et al., 2014; Nguyen et al., 2015). For instance, Kim et al. (2013) reported no effect of AHL to the binding activity of SdiA toward the promoter region of ftsQAZ operon, and that the AHLs increase the transcriptional activity of SdiA by promoting protein stability rather than by affecting the DNA-binding affinity of the protein. However, it is important to remind that in our study, EMSA was performed using the synthetic AHL N-Octanoyl-L-homoserine lactone (C8-HSL). Given the variety of AHL molecules according to the degree of oxidation and saturation and the length of the N-acyl side chains, further studies with other AHLs are needed to explore in more detail whether and how these autoinducers can modulate the DNA-binding activity of SdiA in K. pneumoniae.
The production of AI-2 signaling molecules was indirectly measured in the wild-type and mutant K. pneumoniae strains by performing bioluminescence assays with the AI-2 reporter strain Vibrio campbellii. We observed that when a functional SdiA is present, the production of AI-2 increases as the phases of bacterial growth progress. This increase in AI-2 production is consistent with the role of these signaling molecules as monitors of cell population density (Xavier and Bassler, 2005a; Rutherford and Bassler, 2012; Kendall and Sperandio, 2014). Our results are in accordance with a previous report indicating maximal accumulation of AI-2 by K. pneumoniae in the late-exponential phase (Balestrino et al., 2005). Intriguingly, K. pneumoniae cells without SdiA regulator show constant production of AI-2 molecules at maximum levels, regardless of the growth phases of the bacteria. Bacteria control population density by sensing and responding to AI-1 and AI-2 QS signaling molecules. The activation of the AI-2 QS system on the mutant strain, as revealed by the maximum production of AI-2 molecules, seems to indicate an attempt by the mutant strain to compensate for the loss of cell density control due to the SdiA absence.
Traditionally, intra- and inter-species communications in Gram-negative bacteria are attributed to QS systems mediated by type 1 and type 2 autoinducers, respectively. How these two QS regulatory systems are connected is controversial and remains poorly understood. While some authors report that sdiA and luxS work independently (Surette and Bassler, 1999), others suggest that SdiA plays a role in regulating AI-2 uptake and processing (Smith et al., 2010). For instance, DeLisa and coauthors observed that exogenous AI-2 slightly activates the transcription of sdiA in an E. coli luxS mutant strain (DeLisa et al., 2001), although it cannot be excluded that this activation is due to metabolic changes in the methyl cycle in the luxS mutant. More recently, Zhou and colleagues suggested that AI-1 and AI-2 QS systems might be linked in E. coli through a synergistic action of SdiA and YdiV to regulate the intracellular concentration cAMP (Zhou et al., 2008). According to these authors, SdiA activates ydiV expression by binding on its promoter region in the presence of exogenous AI-1 signaling molecules. YdiV, an EAL domain protein, regulates the production of cAMP that, in turn, positively regulates the expression of lsrR and the lsr operon. In the present study, we observed up-regulation of luxS in the mutant strain throughout the growth stages and slightly induction of lsrR at the initial phase of growth. We also observed the DNA-binding activity of SdiA on the luxS promoter region and the intergenic region between lsrR and lsr operon. Although the results presented here seem to indicate a possible direct role of SdiA in the synthesis, uptake, and processing of AI-2 molecules, more studies need to be performed to further strengthen this hypothesis and to better understand how SdiA plays a role in the interaction of AI-1 and AI-2 QS systems in K. pneumoniae.
Indeed, the regulatory circuits that integrate and command both AI-1 and AI-2 QS systems are unknown and likely complex. Although LuxR regulators have been originally associated with intra-species signaling, SdiA is a LuxR-type regulator involved not only in inter-species signaling (Michael et al., 2001; Dyszel et al., 2010; Lu et al., 2017) but also in interkingdom communication and according to environmental cues (Smith et al., 2008; Ghosh et al., 2009; Hughes et al., 2010; Wang et al., 2020). SdiA is an orphan LuxR-type regulator encoded by bacteria that do not produce their own AHL and, as such, do not detect endogenous AHL but autoinducers produced by other bacteria. Structural analyses indicate that the ligand-binding domain of SdiA is wide and open enough to accommodate a variety of AHLs molecules, and it can sense and respond to a variety of ligands that can be not only exogenous or synthetic AHL, but also non-AHL chemical compounds (Kim et al., 2013; Nguyen et al., 2015; Styles et al., 2020). This ability to respond to a range of molecules and environmental signals is consistent with the possible role of SdiA as a master modulator for both intra- and inter-species communication.
In summary, K. pneumoniae encodes SdiA, an orphan LuxR-type QS regulator since K. pneumoniae does not produce its own AHL autoinducers. Nonetheless, SdiA recognizes and responds to AHL produced by other species, indicating some level of inter-species cell-cell communication mediated by SdiA. We herein showed the role of the SdiA regulator in the pathogenesis of K. pneumoniae by controlling fimbriae expression, biofilm formation, and production of QS autoinducers. We also determined for the first time the SdiA binding sites within the promoter region of type 1 fimbrial gene cluster fim, the ftsQAZ cell division gene cluster, and the luxS and lsrA-lsrR, genes related to the synthesis and processing of AI-2 molecules in K. pneumoniae. As SdiA detects and responds to AHL produced by other species, we suppose that the modulation of these virulence factors may be orchestrated in a coordinated manner via SdiA-mediated inter-species communication.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
LFCF and TP conceived and designed the experiments. TP, AEIG, NMGS, and LA executed the experiments and the analysis. MD and HV contributed with reagents, materials, and analysis tools. LFCF and TP wrote the manuscript. MD and HV assisted with critical revision of the manuscript and LFCF coordinated its revision. All authors contributed to the manuscript revision and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was funded by the São Paulo Research Foundation (FAPESP), Grants #2015/26524-1 and #2018/26203-9 to LFCF. TP, AEIG, NMGS, and LA were supported by scholarships from the Coordination for the Improvement of Higher Education Personnel (CAPES, Ministry of Education of Brazil). The authors gratefully acknowledge Casa de Nossa Senhora da Paz - Ação Social Franciscana (CNSP-ASF) for the financial support for the publication fees of this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.597735/full#supplementary-material
References
- Abed N., Grépinet O., Canepa S., Hurtado-Escobar G. A., Guichard N., Wiedemann A., et al. (2014). Direct regulation of the pefI-srgC operon encoding the Rck invasin by the quorum-sensing regulator SdiA in Salmonella Typhimurium. Mol. Microbiol. 94 254–271. 10.1111/mmi.12738 [DOI] [PubMed] [Google Scholar]
- Ahmer B. M. M. (2004). Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol. Microbiol. 52 933–945. 10.1111/j.1365-2958.2004.04054.x [DOI] [PubMed] [Google Scholar]
- Alcántar-Curiel M. D., Blackburn D., Saldaña Z., Gayosso-Vázquez C., Iovine N. M., De la Cruz M. A., et al. (2013). Multi-functional analysis of Klebsiella pneumoniae fimbrial types in adherence and biofilm formation. Virulence 4 129–138. 10.4161/viru.22974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes L. C. M., Ferreira R. B. R., Buckner M. M. C., Finlay B. B. (2010). Quorum sensing in bacterial virulence. Microbiology 156 2271–2282. 10.1099/mic.0.038794-0 [DOI] [PubMed] [Google Scholar]
- Balestrino D., Haagensen J. A., Rich C., Forestier C. (2005). Characterization of type 2 quorum sensing in Klebsiella pneumoniae and relationship with biofilm formation. J. Bacteriol. 187 2870–2880. 10.1128/JB.187.8.2870-2880.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler B. L., Wright M., Showalter R. E., Silverman M. R. (1993). Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9 773–786. 10.1111/j.1365-2958.1993.tb01737.x [DOI] [PubMed] [Google Scholar]
- Culler F. H., Couto C. F. S., Higa S. J., Ruiz M. R., Yang J. M., Bueris V., et al. (2018). Role of SdiA on biofilm formation by atypical enteropathogenic Escherichia coli. Genes 9:253. 10.3390/genes9050253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Araujo C., Balestrino D., Roth L., Charbonnel N., Forestier C. (2010). Quorum sensing affects biofilm formation through lipopolysaccharide synthesis in Klebsiella pneumoniae. Res. Microbiol. 161 595–603. 10.1016/j.resmic.2010.05.014 [DOI] [PubMed] [Google Scholar]
- DeLisa M. P., Wu C. F., Wang L., Valdes J. J., Bentley W. E. (2001). DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183 5239–5247. 10.1128/jb.183.18.5239-5247.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyszel J. L., Soares J. A., Swearingen M. C., Lindsay A., Smith J. N., Ahmer B. M. (2010). E. coli K-12 and EHEC genes regulated by SdiA. PLoS One 5:e8946. 10.1371/journal.pone.0008946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J., Silverman M. (1984). Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. U. S. A. 81 4154–4258. 10.1073/pnas.81.13.4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C. (2006). The QscR quorum-sensing regulon of Pseudomonas aeruginosa: an orphan claims its identity. J. Bacteriol. 188 3169–3171. 10.1128/JB.188.9.3169-3171.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D., Roy K., Williamson K. E., Srinivasiah S., Wommack K. E., Radosevich M. (2009). Acyl-homoserine lactones can induce virus production in lysogenic bacteria: an alternative paradigm for prophage induction. Appl. Environ. Microbiol. 75 7142–7152. 10.1128/AEM.00950-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes ÉI., Pacheco T., Santos C.d.S.d, Pereira J. A., Ribeiro M. L., Darrieux M., et al. (2021). Functional insights from Kpfr, a new transcriptional regulator of fimbrial expression that is crucial for Klebsiella pneumoniae pathogenicity. Front. Microbiol. 11:601921. 10.3389/fmicb.2020.601921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes A. ÉI., Stuchi L. P., Siqueira N. M. G., Henrique J. B., Vicentini R., Ribeiro M. L., et al. (2018). Selection and validation of reference genes for gene expression studies in Klebsiella pneumoniae using Reverse Transcription Quantitative real-time PCR. Sci. Rep. 8:9001. 10.1038/s41598-018-27420-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilhen C., Charbonnel N., Parisot N., Gueguen N., Iltis A., Forestier C., et al. (2016). Transcriptional profiling of Klebsiella pneumoniae defines signatures for planktonic, sessile and biofilm-dispersed cells. BMC Genom. 17:237. 10.1186/s12864-016-2557-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L., Costerton J. W., Stoodley P. (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2 95–108. 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- Holt K. E., Wertheim H., Zadoks R. N., Baker S., Whitehouse C. A., Dance D., et al. (2015). Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. U. S. A. 112 E3574–E3581. 10.1073/pnas.1501049112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D. T., Terekhova D. A., Liou L., Hovde C. J., Sahl J. W., Patankar A. V., et al. (2010). Chemical sensing in mammalian host-bacterial commensal associations. Proc. Natl. Acad. Sci. U. S. A. 107 9831–9836. 10.1073/pnas.1002551107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens J. C. A., Metzger K., Daniels R., Ptacek D., Verhoeven T., Habel L. W., et al. (2007). Synthesis of N-acyl homoserine lactone analogues reveals strong activators of SdiA, the Salmonella enterica serovar Typhimurium LuxR homologue. Appl. Environ. Microbiol. 73:535. 10.1128/AEM.01451-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseleau-Petit D., Vinella D., D’Ari R. (1999). Metabolic alarms and cell division in Escherichia coli. J. Bacteriol. 181 9–14. 10.1128/JB.181.1.9-14.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru K., Tatsuno I., Tobe T., Sasakawa C. (2000). SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 38 805–816. 10.1046/j.1365-2958.2000.02171.x [DOI] [PubMed] [Google Scholar]
- Kendall M., Sperandio V. (2014). Cell-to-cell signaling in Escherichia coli and Salmonella. EcoSal Plus 6 1–15. 10.1128/ecosalplus.ESP-0002-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keynan Y., Rubinstein E. (2007). The changing face of Klebsiella pneumoniae infections in the community. Int. J. Antimicrob. Agents 30 385–389. 10.1016/j.ijantimicag.2007.06.019 [DOI] [PubMed] [Google Scholar]
- Kim T., Duong T., Wu C. A., Choi J., Lan N., Kang S. W., et al. (2013). Structural insights into the molecular mechanism of Escherichia coli SdiA, a quorum-sensing receptor Acta Crystallogr. D Biol. Crystallogr. 70 694–707. 10.1107/S1399004713032355 [DOI] [PubMed] [Google Scholar]
- Kohli N., Crisp Z., Riordan R., Li M., Alaniz R. C., Jayaraman A. (2018). The microbiota metabolite indole inhibits Salmonella virulence: involvement of the PhoPQ two-component system. PLoS One 13:e0190613. 10.1371/journal.pone.0190613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Lequette Y., Greenberg E. P. (2006). Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol. Microbiol. 59 602–609. 10.1111/j.1365-2958.2005.04960.x [DOI] [PubMed] [Google Scholar]
- Lee J., Jayaraman A., Wood T. K. (2007). Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 7:42. 10.1186/1471-2180-7-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Maeda T., Hong S. H., Wood T. (2009). Reconfiguring the quorum-sensing regulator SdiA of Escherichia coli to control biofilm formation via indole and N-acylhomoserine lactones. Appl. Environ. Microbiol. 75 1703–1716. 10.1128/AEM.02081-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Zhao Y., Liu C., Chen Z., Zhou D. (2014). Molecular pathogenesis of Klebsiella pneumoniae. Fut. Microbiol. 9 1071–1081. 10.2217/fmb.14.48 [DOI] [PubMed] [Google Scholar]
- Lin W. H., Wang M. C., Tseng C. C., Ko W. C., Wu A. B., Zheng P. X., et al. (2010). Clinical and microbiological characteristics of Klebsiella pneumoniae isolates causing community-acquired urinary tract infections. Infection 38 459–464. 10.1007/s15010-010-0049-5 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu Y., Zeng J., Wu B. E. S., Wang L., Cai R. (2017). Quorum sensing N-acyl homoserine lactones-SdiA suppresses Escherichia coli-Pseudomonas aeruginosa conjugation through inhibiting traI expression. Front. Cell. Infect. Microbiol. 7:7. 10.3389/fcimb.2017.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Zhang S., Xu Z., Li H., Xiao Q., Qiu F., et al. (2020). SdiA improves the acid tolerance of E. coli by regulating GadW and GadY expression. Front. Microbiol. 11:1078. 10.3389/fmicb.2020.01078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael B., Smith J. N., Swift S., Heffron F., Ahmer B. M. (2001). SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183 5733–5742. 10.1128/jb.183.19.5733-5742.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. B., Bassler B. L. (2001). Quorum Sensing in Bacteria. Annu. Rev. Microbiol. 55 165–199. 10.1146/annurev.micro.55.1.165 [DOI] [PubMed] [Google Scholar]
- Ng W.-L., Bassler B. L. (2009). Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43 197–222. 10.1146/annurev-genet-102108-134304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Y., Nguyen N. X., Rogers J. L., Liao J., MacMillan J. B., Jiang Y., et al. (2015). Structural and mechanistic roles of novel chemical ligands on the Sdia quorum-sensing transcription regulator. mBio 6 :e02429–14. 10.1128/mBio.02429-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paczosa M. K., Mecsas J. (2016). Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80 629–661. 10.1128/MMBR.00078-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patankar A. V., González J. E. (2009). Orphan LuxR regulators of quorum sensing. FEMS Microbiol. Rev. 33 739–756. 10.1111/j.1574-6976.2009.00163.x [DOI] [PubMed] [Google Scholar]
- Pereira C. S., de Regt A. K., Brito P. H., Miller S. T., Xavier K. B. (2009). Identification of functional LsrB-like autoinducer-2 receptors. J. Bacteriol. 191 6975–6987. 10.1128/JB.00976-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira C. S., Thompson J. A., Xavier K. B. (2013). AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 37 156–181. 10.1111/j.1574-6976.2012.00345.x [DOI] [PubMed] [Google Scholar]
- Rosen D. A., Pinkner J. S., Jones J. M., Walker J. N., Clegg S., Hultgren S. J. (2008). Utilization of an intracellular bacterial community pathway in Klebsiella pneumoniae urinary tract infection and the effects of FimK on type 1 pilus expression. Infect. Immun. 76 3337–3345. 10.1128/IAI.00090-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132 365–386. [DOI] [PubMed] [Google Scholar]
- Rutherford S. T., Bassler B. L. (2012). Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2:a012427. 10.1101/cshperspect.a012427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabag-Daigle A., Ahmer B. M. M. (2012). ExpI and PhzI are descendants of the long lost cognate signal synthase for SdiA. PLoS One 7:e47720. 10.1371/journal.pone.0047720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabag-Daigle A., Soares J. A., Smith J. N., Elmasry M. E., Ahmer B. M. M. (2012). The acyl homoserine lactone receptor. SdiA, of Escherichia coli and Salmonella enterica Serovar Typhimurium does not respond to indole. Appl. Environ. Microbiol. 78 5424–5431. 10.1128/AEM.00046-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabag-Daigle A., Dyszel J. L., Gonzalez J. F., Ali M. M., Ahmer B. M. M. (2015). Identification of sdiA-regulated genes in a mouse commensal strain of Enterobacter cloacae. Front. Cell. Infect. Microbiol. 5:47. 10.3389/fcimb.2015.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder S., Shokat K., Surette M. G., Bassler B. L. (2001). The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41 463–476. 10.1046/j.1365-2958.2001.02532.x [DOI] [PubMed] [Google Scholar]
- Schroll C., Barken K. B., Krogfelt K. A., Struve C. (2010). Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 10:179. 10.1186/1471-2180-10-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Urbanowski M. L., Greenberg E. P. (2004). Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. U. S. A. 101 15833–15839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V. K., Bearson S. M. D., Bearson B. L. (2010). Evaluation of the effects of sdiA, a luxR homologue, on adherence and motility of Escherichia coli O157:H7. Microbiology 156 1303–1312. 10.1099/mic.0.034330-0 [DOI] [PubMed] [Google Scholar]
- Shimada T., Shimada K., Matsui M., Kitai Y., Igarashi J., Suga H., et al. (2014). Roles of cell division control factor SdiA: recognition of quorum sensing signals and modulation of transcription regulation targets. Genes Cells 19 405–418. 10.1111/gtc.12139 [DOI] [PubMed] [Google Scholar]
- Sitnikov D. M., Schineller J. B., Baldwin T. O. (1996). Control of cell division in Escherichia coli: regulation of transcription of ftsQA involves both rpoS and SdiA-mediated autoinduction. Proc. Natl. Acad. Sci. U.S.A. 93 336–341. 10.1073/pnas.93.1.336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. N., Ahmer B. M. M. (2003). Detection of other microbial species by Salmonella: expression of the SdiA regulon. J. Bacteriol. 185 1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. N., Dyszel J. L., Soares J. A., Ellermeier C. D., Altier C., Lawhon S. D., et al. (2008). SdiA, an N-acylhomoserine lactone receptor, becomes active during the transit of Salmonella enterica through the gastrointestinal tract of turtles. PLoS One 3:e2826. 10.1371/journal.pone.0002826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. L., Fratamico P. M., Yan X. (2010). Eavesdropping by bacteria: the role of SdiA in Escherichia coli and Salmonella enterica serovar Typhimurium quorum sensing. Foodborne Pathog. Dis. 8 169–178. 10.1089/fpd.2010.0651 [DOI] [PubMed] [Google Scholar]
- Stahlhut S. G., Struve C., Krogfelt K. A., Reisner A. (2012). Biofilm formation of Klebsiella pneumoniae on urethral catheters requires either type 1 or type 3 fimbriae. FEMS Immunol. Med. Microbiol. 65 350–359. 10.1111/j.1574-695X.2012.00965.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struve C., Bojer M., Krogfelt K. A. (2008). Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect. Immun. 76 4055–4065. 10.1128/IAI.00494-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struve C., Bojer M., Krogfelt K. A. (2009). Identification of a conserved chromosomal region encoding Klebsiella pneumoniae type 1 and type 3 fimbriae and assessment of the role of fimbriae in pathogenicity. Infect. Immun. 77 5016–5024. 10.1128/IAI.00585-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styles M. J., Early S. A., Tucholski T., West K. H. J., Ge Y., Blackwell H. E. (2020). Chemical control of quorum sensing in E. coli: identification of small molecule modulators of Sdia and mechanistic characterization of a covalent inhibitor. ACS Infect. Dis. 6 3092–3103. 10.1021/acsinfecdis.0c00654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette M. G., Bassler B. L. (1999). Regulation of autoinducer production in Salmonella typhimurium. Mol. Microbiol. 31 585–595. 10.1046/j.1365-2958.1999.01199.x [DOI] [PubMed] [Google Scholar]
- Tavio M. M., Aquili V. D., Poveda J. B., Antunes N. T., Sanchez-Cespedes J., Vila J. (2010). Quorum-sensing regulator sdiA and marA overexpression is involved in in vitro-selected multidrug resistance of Escherichia coli. J. Antimicrob. Chemother. 65 1178–1186. 10.1093/jac/dkq112 [DOI] [PubMed] [Google Scholar]
- Urbanowski M. L., Lostroh C. P., Greenberg E. P. (2004). Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein. J. Bacteriol. 186 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude M. W., Baumler A. J. (2004). Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17 581–611. 10.1128/CMR.17.3.581-611.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. D., de Boer P. A., Rothfield L. I. (1991). A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. Embo J. 10 3363–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hashimoto Y., Tsao C.-Y., Valdes J. J., Bentley W. E. (2005). Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J. Bacteriol. 187 2066–2076. 10.1128/JB.187.6.2066-2076.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Payne G. F., Bentley W. E. (2020). Quorum sensing communication: molecularly connecting cells, their neighbors, and even devices. Annu. Rev. Chem. Biomol. Eng. 11 447–468. 10.1146/annurev-chembioeng-101519-124728 [DOI] [PubMed] [Google Scholar]
- Wu C., Lokanath N. K., Kim D. Y., Nguyen L. D. N., Kim K. K. (2008). Crystallization and preliminary X-ray studies of SdiA from Escherichia coli Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64 19–21. 10.1107/S1744309107059696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier K. B., Bassler B. L. (2005b). Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187 238–248. 10.1128/JB.187.1.238-248.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier K. B., Bassler B. L. (2005a). Interference with AI-2-mediated bacterial cell-cell communication. Nature 437 750–753. 10.1038/nature03960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Yata K., Fujita N., Ishihama A. (2001). Novel mode of transcription regulation by SdiA, an Escherichia coli homologue of the quorum-sensing regulator. Mol. Microbiol. 41 1187–1198. 10.1046/j.1365-2958.2001.02585.x [DOI] [PubMed] [Google Scholar]
- Yao Y., Martinez-Yamout M. A., Dickerson T. J., Brogan A. P., Wright P. E., Dyson H. J. (2006). Structure of the Escherichia coli quorum sensing protein SdiA: activation of the folding switch by acyl homoserine lactones. J. Mol. Biol. 355 262–273. 10.1016/j.jmb.2005.10.041 [DOI] [PubMed] [Google Scholar]
- Yao Y., Dickerson T. J., Hixon M. S., Dyson H. J. (2007). NMR detection of adventitious xylose binding to the quorum-sensing protein SdiA of Escherichia coli. Bioorg. Med. Chem. Lett. 17 6202–6205. 10.1016/j.bmcl.2007.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Meng X., Sun B. (2008). An EAL domain protein and cyclic AMP contribute to the interaction between the two quorum sensing systems in Escherichia coli. Cell Res. 18 937–948. 10.1038/cr.2008.67 [DOI] [PubMed] [Google Scholar]
- Zhu J., Winans S. C. (2001). The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl. Acad. Sci. U. S. A. 98 1507–1512. 10.1073/pnas.98.4.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Shen Y. L., Wei D. Z., Zhu J. W. (2008). Inhibition of quorum sensing in Serratia marcescens H30 by molecular regulation. Curr. Microbiol. 56 645–650. 10.1007/s00284-008-9140-x [DOI] [PubMed] [Google Scholar]
- Zhu H., Liu H. J., Ning S. J., Gao Y. L. (2012). The response of type 2 quorum sensing in Klebsiella pneumoniae to a fluctuating culture environment. DNA Cell Biol. 31 455–459. 10.1089/dna.2011.1375 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.