Abstract
Evidence points epicardial adipose tissue (EAT) as an emerging cardiovascular risk marker. Whether genetic polymorphisms linked with atherosclerosis are associated with higher EAT is still unknown. We aim to assess the role of genetic burden of atherosclerosis and its association to EAT in a cohort of asymptomatic individuals without coronary disease. A total of 996 participants were prospectively enrolled in a single Portuguese center. EAT volume was measured by Cardiac Computed Tomography and participants were distributed into 2 groups, above and below median EAT. SNPs were genotyped and linked to their respective pathophysiological axes. A multiplicative genetic risk score (mGRS) was constructed, representing the genetic burden of the studied SNPs. To evaluate the association between genetics and EAT, we compared both groups by global mGRS, mGRS by functional axes, and SNPs individually. Individuals above-median EAT were older, had a higher body mass index (BMI) and higher prevalence of hypertension, metabolic syndrome, diabetes, and dyslipidemia. They presented higher GRS, that remained an independent predictor of higher EAT volumes. The group with more EAT consistently presented higher polymorphic burden across numerous pathways. After adjustment, age, BMI, and mGRS of each functional axis emerged as independently related to higher EAT volumes. Amongst the 33 SNPs, MTHFR677 polymorphism emerged as the only significant and independent predictor of higher EAT volumes. Patients with higher polymorphism burden for atherosclerosis present higher EAT volumes. We present the first study in a Portuguese population, evaluating the genetic profile of EAT through GWAS and GRS, casting further insight into this complicated matter.
Keywords: Epicardial adipose tissue, atherosclerosis, cardiovascular risk factors, genetic polymorphisms, Genetic Risk Score
Introduction
Epicardial adipose tissue (EAT) or epicardial fat, is often referred in the literature as a form of visceral fat located between the myocardial surface and the visceral layer of the pericardium, sharing a common embryologic origin with visceral fat deposits. 1 It should not be confused with pericardial fat, as they share different embryologic sources, blood supply, and represent a different percentage of total cardiac mass.1,2
EAT anatomical traits first raised the suspicion for a role in cardiovascular physiology and pathophysiology through its direct contact with the myocardium and coronary arteries as well as its shared microcirculation.1,3 However, only recently it has been recognized as a metabolically active tissue, producing several bioactive molecules, including pro-inflammatory cytokines. These could, via paracrine or vasocrine, be secreted to the myocardium, and the coronary arteries.3-5 EAT is now seen as an emerging marker of cardiovascular risk, linked to coronary artery disease, 6 atrial fibrillation, 7 and other cardiac disorders.8,9
Non-invasive quantification of EAT can be achieved through different cardiac imaging techniques such as echocardiography, magnetic resonance imaging (MRI), and cardiac computed tomography (CCT). 10 The higher accuracy, availability, and spatial resolution of the latter, make it a frequent choice for EAT quantification. However, a uniform standardized method for EAT quantification is still lacking, alongside normal range values for this variable, as these remain to be established.11,12
Increasing evidence derived from genome-wide association studies (GWAS) has identified numerous single-nucleotide polymorphisms (SNPs) across different biological pathways, associated with particular phenotypes, clinical traits, cardiovascular risk, and diseases. 13 Polygenic Risk Scores (PRS) are a useful way of conveying the burden of genetic variants and individual susceptibility.14,15 Despite the recent advances and interest regarding cardiogenesis, little is known about the role of genetics on embryonic heart progenitors and cardiovascular pathophysiology. Studies about the influence of individual and multiple genetic variants on epicardial fat volume remain scarce and limited.
This study aimed to address the underlying clinical characteristics, risk factors and genetic background responsible for higher volumes of epicardial fat and investigate whether Epicardial Adipose Tissue volume in healthy individuals is significantly and independently associated with the individual genetic load.
Methods
Patient selection
Subjects who underwent cardiac CT between 2013 and 2019 at Hospital Dr Nélio Mendonça (Funchal) were retrospectively recovered from the picture archiving and communication service (PACS). Our sample consisted of 996 asymptomatic volunteers (mean age 59 ± 8, 78% male), recruited from the control arm of the GENEMACOR study, an ongoing research study in Madeira Island with a prospective cohort (Supplemental Table S1). All GENEMACOR participants, aged between 30 and 65 years, are born and resident in Madeira for at least 2 generations.
Written informed consent was waived by all patients at the time of enrollment. The study was conducted according to the World Medical Association Declaration of Helsinki and approved by the Ethics committee and Institutional board of our Hospital.
Data was collected from all subjects in a standardized file comprising demographic, clinical characteristics, and traditional risk factors. Blood was collected for subsequent biochemical and genetic analysis, as described elsewhere. 16
Polygenic risk score (PRS)
SNPs were selected either from GWAS or candidate gene association studies.17,18 Entering criteria included genes with an Odds Ratio (OR) for CAD ⩾1 and, simultaneously, with a Minor Allele Frequency (MAF) ⩾2%.
Genotyping was performed by TaqMan allelic discrimination assay. A multiplicative genetic risk score (mGRS) was constructed and represents the genetic burden of the association of 33 polymorphisms studied (rs12190287 [TCF21], rs11556924 [ZC3HC1], rs599839 [PSRC1], rs12526453 [PHACTR1], rs17465637 [MIA3], rs6922269 [MTHFD1L], rs17228212 [SMAD3], rs964184 [ZNF259], rs3825807 [AMDAMTS7], rs4977574 [CDKN2B], rs1801282 [PPARG], rs17782313 [MC4R], rs266729 [ADIPOQ], rs3798220 [LPA], rs1326634 [SLC30A8], rs4402960 [IGF2BP2], rs705379 [PON1], rs20455 [KIF6], rs1884613 [HNF4A], rs8050136 [FTO], rs1376251 [TAS2R50], rs2114580 [PCSK9], rs618675 [GJA4], rs7903146 [TCF7L2], rs4340 [ACE], rs699 [AGT], rs5186 [AT1R], rs662 [PON192], rs1333049 [CDKN2B-AS1], rs7412/rs429358 [APOE], rs1801131 [MTHFR1298], rs1801133 [MTHFR677], rs854560 [PON55]). These variants were distributed in 5 main axes according to their participation in pathways leading to atherosclerosis or mechanisms of action involving effects on hypertension, lipid metabolism, oxidation, diabetes/obesity, and cellular (Supplemental Table S2).
EAT quantification using CCT
EAT volume was independently measured by 1 operator with a quantitative semi-automated procedure using a postprocessing workstation—TeraRecon Aquarius Workstation (version 4.4.7, TeraRecon, Inc., San Mateo, CA, USA). We used a simplified method previously validated, performing a single slice measurement of EAT at the level of the left main coronary artery. This kind of evaluation proved to be an efficient and clinically practical measure, acting as a surrogate for total epicardial fat volume with prognostic relevance.19,20
Before imaging, metoprolol may have been used for heart rate control. The scans were performed using a 64-slice TOSHIBA Aquilion CXL CT scan.
A non-contrast-enhanced, prospective ECG-triggered image acquisition (collimation 3.0 mm × 4 mm; gantry rotation 1⁄2 250 ms; tube current 200 mA; tube voltage 120 kV) was performed at the 75% phase, and images were reconstructed using a slice thickness of 3 mm. We manually traced the pericardial outline at the level of the ostium of the left main coronary artery. The volume of tissue within the outlined boundary (ROI) with an attenuation of −250 to −30 HU was calculated (cm3). This process was repeated for each patient/CT (Figure 1). The same acquisitions were used for CAC score quantification using the Agatson score algorithm.
Figure 1.
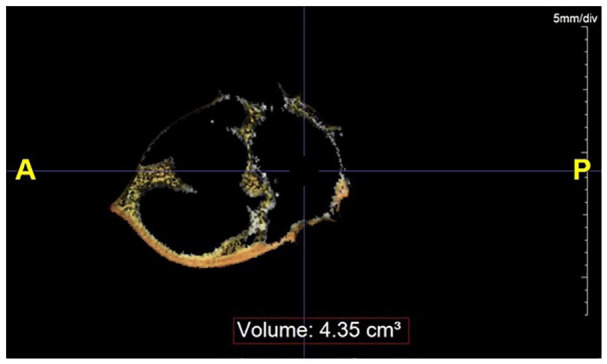
Epicardial adipose tissue (EAT) volume estimation from a single 3 mm slice at the level of the left main coronary, using a range of attenuation of −250 to −30 HU—TeraRecon Aquarius Workstation.
Statistical analysis
Due to the lack of a standardized cut-off to establish a comparison between high and low EAT volume, patients were divided into 2 groups above and below the median of EAT volume.
We compared both groups regarding clinical traits, risk factors, available biochemical parameters and their mean GRS. Normality of data distribution was assessed by the Shapiro–Wilk test. Normally distributed variables were reported as mean and SD, while non-normally distributed variables were reported as the median and interquartile range (IQR). Appropriate comparison tests were performed for parametric and nonparametric variables. Chi-square test compared categorical variables, while T-student or Mann–Whitney tests compared numerical and categorical ones.
As EAT-volume variable does not have a normal distribution, a non-parametric test (Spearman’s correlation) was used to evaluate the correlation between the EAT-volume and other variables of interest.
Finally, logistic regression was used to address the contribution and individual weight of genetics, where a model comprising the clinically relevant variables in the univariate analysis and potential confounders were addressed. To better illustrate and cover the impact of a genetic burden to EAT, we aimed to compare both groups by overall GRS (33 polymorphic variants) and by individual SNP. Afterwards, we evaluated GRS by genetic axis (including each axis of the studied SNPs belonging to main atherosclerosis pathways/axes: oxidation, renin-angiotensin system, diabetes/obesity, dyslipidemia, cell growth differentiation, and migration.
For calculating the additive (aGRS), we based on previous reports. Genotypes were coded “0” for no risk allele homozygous, “1” for heterozygous, and “2” for risk-allele homozygous and the additive score (aGRS) was generated by summing the number of risk alleles in each variant. For calculate multiplicative GRS (mGRS), each genotype was assigned the respective Odds Ratio (OR) and calculated the product of the ORs. In this work, only the multiplicative score was used.
A P-value of <.05 was deemed statistically significant. The statistical analysis was performed by statistical software IBM SPSS statistics, 25 (IBM, Corp).
Results
Description of the population characteristics, above and below EAT-volume median
The median of the EAT volume was 3.13 ± 1.26. A thorough and detailed description of the baseline characteristics of both groups is summarized in Table 1.
Table 1.
Baseline characteristics of the population.
| Variables | EAT-volume; median = 3.13 ± 1.26 | P-value | |
|---|---|---|---|
| EAT volume | |||
| Above median | Below median | ||
| Age, years | 60.7 ± 7.7 | 56.5 ± 8.4 | <.0001 |
| Male gender, n (%) | 387 (77.7) | 393 (78.9) | .645 |
| Smoking habits, n (%) | 114 (22.9) | 111 (22.3) | .820 |
| Type 2 diabetes, n (%) | 85 (17.1) | 39 (7.8) | <.0001 |
| Dyslipidemia, n (%) | 366 (73.5) | 326 (65.5) | .006 |
| Physical inactivity, n (%) | 242 (48.6) | 187 (37.6) | <.0001 |
| Hypertension, n (%) | 303 (60.8) | 206 (41.4) | <.0001 |
| SBP, mmHg | 139.3 ± 18 | 132.8 ± 17.1 | <.0001 |
| DBP, mmHg | 86.2 ± 10.9 | 82 ± 10.3 | <.0001 |
| Alcohol, gr/day | 7.2 (34-167) | 1.1 (29.9-287) | .002 |
| Alcohol intake # , n (%) | 247 (49.6) | 204 (41) | .006 |
| BMI, kg/m2 | 29.8 ± 4.4 | 26.5 ± 3.8 | <.0001 |
| Fast glucose, mg/dl | 102 (93-113) | 97 (90-105) | <.0001 |
| Total cholesterol, mg/dl | 204 (180-232) | 204.5 (182-232) | .598 |
| Triglycerides, mg/dl | 130 (97-189) | 114 (84-167) | <.0001 |
| LDL, mg/dl | 125.1 (105.6-150) | 127.6 (106.6-149.2) | .370 |
| HDL, mg/dl | 47 (41-56) | 49.5 (42-59) | .006 |
| Apolipoprotein B, mg/dl | 93.4 (61.8-109.3) | 93.4 (58.8-114.2) | .435 |
| Lipoprotein (a), mg/dl | 14.1 (8.8-24.7) | 13.8 (9-26.2) | .735 |
| Fibrinogen, mg/dl | 374.5 (328-424) | 365.5 (318-405) | .014 |
| Homocysteine, mg/dl | 11.8 (10.2-13.6) | 11.4 (9.6-12.8) | <.0001 |
| Homocysteine >10 mg/dl, n (%) | 383 (76.9) | 348 (69.9) | .012 |
| hsCRP, mg/L | 2.7 (1.5-4.9) | 2.5 (1-2.9) | <.0001 |
| hsCRP > 3, n (%) | 187 (37.6) | 117 (23.5) | <.0001 |
| PWV, m/s | 8.5 ± 1.9 | 8.1 ± 1.5 | <.0001 |
| PWV >10 m/s, n (%) | 79 (15.9) | 44 (8.8) | .001 |
| Heart rate | 73.5 ± 11.5 | 70.1 ± 10.9 | <.0001 |
| Leucocytes, mg/dl | 6.7 (5.8-7.9) | 6.4 (5.4-7.5) | .005 |
| Hemoglobin, mg/dl | 14.8 (14.1-15.5) | 14.8 (14.1-15.5) | .873 |
| Metabolic syndrome, n (%) | 301 (60.4) | 170 (34.1) | <.0001 |
| Genetic Risk Score (GRS) | |||
| Multiplicative | 2.2 ± 2.1 | 1.5 ± 1.4 | <.0001 |
Abbreviations: BMI, body mass index; DBP, diastolic blood pressure; EAT, epicardial adipose tissue; HDL, high-density lipoprotein; hsCRP, high-sensitivity C-reactive protein; LDL, low-density lipoprotein; PWV, pulse wave velocity; SBP, systolic blood pressure.
Statistical significance for P < .05.
>40 g/week for women >60 g/week for men.
Patients presenting above-median EAT volume were older (60.7 ± 7.7 vs 56.5 ± 8.4, P < .0001), had a higher body mass index (BMI) (29.8 ± 4.4 vs 26.5 ± 3.8, P < .0001) and higher prevalence of hypertension (60.8% vs 41.4%, P < .0001), diabetes (17.1% vs 7.8%, P < .0001), dyslipidemia (73.5% vs 65.5%, P = .006) and higher alcohol daily consumption (7.2 vs 1.1 g, P = .002). No significant differences were found regarding gender or smoking habits. On physical examination, patients with high EAT registered a higher systolic and diastolic blood pressure and higher heart rates (P < .0001).
Regarding lab tests and biochemical variables, higher EAT volume patients had higher fasting glucose and triglycerides levels, lower HDL, and higher levels of homocysteine and high-sensitivity C-reactive protein (hsCRP). Fibrinogen, another inflammatory marker, was also significantly more elevated in the group above the EAT-median volume compared to below-median.
Metabolic Syndrome, as defined by the WHO criteria, 21 was more frequently present among patients with higher EAT volume in our study (60.4% vs 34.1%, P < .0001). Actually, a significant correlation between EAT volume and Metabolic Syndrome was observed (Figure 2), as well as with the previously mentioned inflammatory markers (Table 2).
Figure 2.
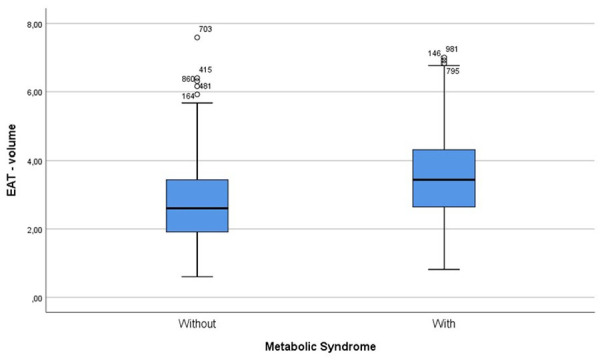
Distribution of EAT median values in metabolic syndrome.
Table 2.
Correlation between EAT volume and other clinical and biochemical variables.
| Variables | Spearman’s rho | P-value |
|---|---|---|
| hsCRP, mg/l | 0.237 | <.0001 |
| Homocysteine, mg/dl | 0.129 | <.0001 |
| Fibrinogen, mg/dl | 0.073 | <.0001 |
| SBP, mmHg | 0.224 | <.0001 |
| DBP, mmHg | 0.200 | <.0001 |
| Fast glucose, mg/dl | 0.264 | <.0001 |
| HDL, mg/dl | −0.107 | <.0001 |
| Triglycerides, mg/dl | 0.200 | <.0001 |
Abbreviations: DBP, diastolic blood pressure; EAT, epicardial adipose tissue; HDL, high-density lipoprotein; hsCRP, high-sensitivity C-reactive protein; SBP, systolic blood pressure.
Statistical significance for P < .05.
Mean Pulse Wave Velocity (PWV), a marker of vascular stiffness, was higher in the group above EAT median. Likewise, this group displayed higher percentages of PWV >10 m/seg.
The burden of genetics was elevated in the above-median EAT group, as shown by the higher mean global Genetic Risk Score—Figure 3.
Figure 3.
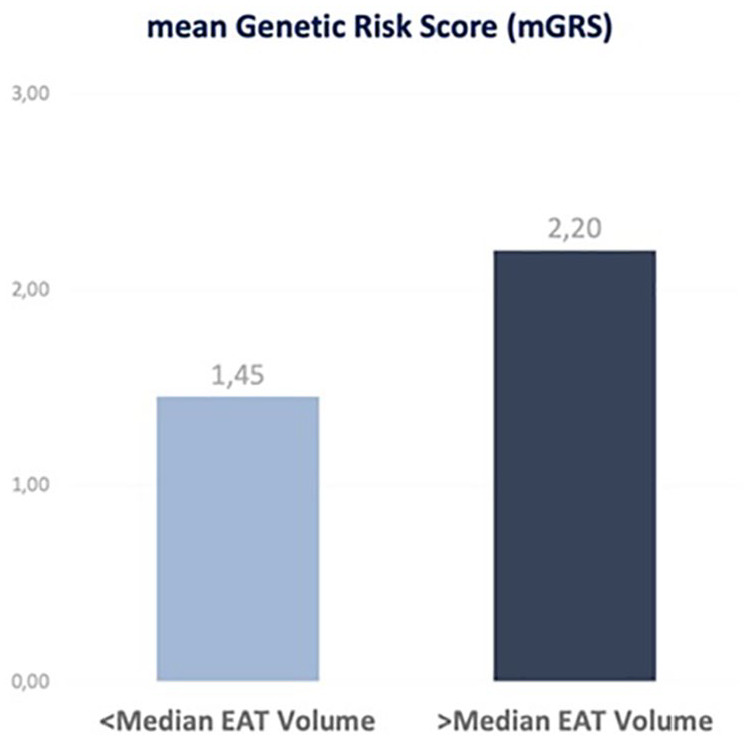
Comparison of the mGRS between the above- and below-median EAT groups.
Evaluation of total genetic load performance (global mGRS) and its association with EAT-Volume
After logistic regression and adjustment for confounders, mGRS remained an independent predictor for higher EAT volumes (OR 1.336, 95%CI 1.215-1.469; P < .0001, alongside with age and BMI (Table 3 and Figure 4).
Table 3.
Logistic regression for mGRS.
| Variables above EAT-volume median | OR | 95%CI | P-value |
|---|---|---|---|
| Age | 1.075 | 1.055-1.095 | <.0001 |
| BMI (kg/m²) | 1.250 | 1.201-1.300 | <.0001 |
| mGRS | 1.336 | 1.215-1.469 | <.0001 |
Abbreviations: BMI, body mass index; CI, confidence interval; EAT, epicardial adipose tissue; Mgrs, multiplicative genetic risk score; OR, odds ratio.
Statistically significant for P < .05.
Figure 4.
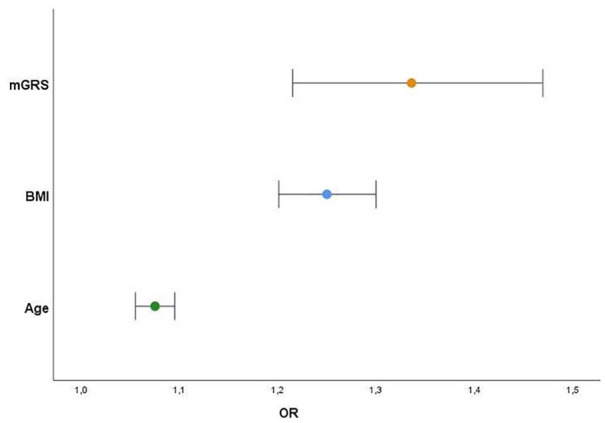
Variables independently associated with higher EAT volumes (above-median) and respective predictive risk (OR) (Sex, diabetes, dyslipidemia, hypertension, and physical inactivity [all P > .1]).
Genetic burden by physiopathological axes
In the analysis by genetic axes, subjects with more epicardial fat consistently presented a higher polymorphism burden (translated by a higher mGRS) across numerous axes/pathways: oxidation, renin-angiotensin system, cellular, diabetes/obesity, and dyslipidemia. After adjusting for confounders, the following have emerged as independently related to higher EAT volumes: mGRS from different functional axes, age, and BMI (Table 4 and Figure 5).
Table 4.
Logistic regression for variables associated with high EAT volumes.
| Variables above EAT-volume median | OR | 95%CI | P-value |
|---|---|---|---|
| mGRS oxidative axis | 2.131 | 1.324-3.429 | .002 |
| mGRS diabetes/obesity axis | 2.052 | 1.275-3.302 | .003 |
| mGRS RAAS axis | 1.886 | 1.082-3.287 | .025 |
| mGRS cellular axis | 1.475 | 1.202-1.808 | <.0001 |
| mGRS dyslipidemia axis | 1.314 | 1.026-1.685 | .031 |
| Age | 1.075 | 1.055-1.095 | <.0001 |
| BMI (kg/m²) | 1.250 | 1.201-1.300 | <.0001 |
Abbreviations: BMI, body mass index; CI, confidence interval; EAT, epicardial adipose tissue; Mgrs, multiplicative genetic risk score; OR, odds ratio.
Adjusted for confounders sex, dyslipidemia, hypertension, diabetes, physical inactivity (all > 0.1).
Figure 5.
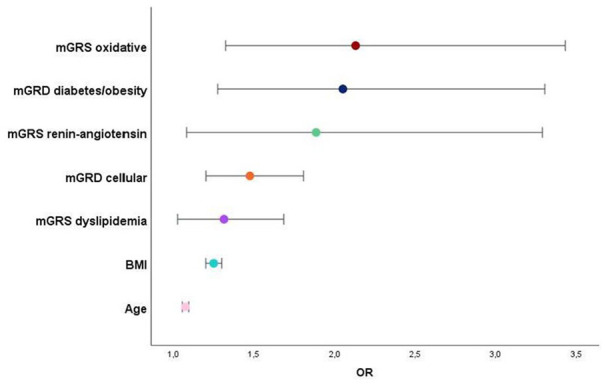
Variables independently associated with higher EAT volumes (above-median) and respective predictive risk (OR).
As expected, mGRS of the Oxidative axis cluster and Diabetes/Obesity axis presented the best risk prediction to higher EAT volume (Table 4).
Performance of the individually genetic variants
Of the 33 investigated variants, rs1333049 (CDKN2B-AS1) at 9p21 locus was the only significantly associated with EAT volume but only in the univariate analysis (Table 5).
Table 5.
Univariate analysis for SNPs associated with EAT volumes.
| rs1333049 (CDKN2B-AS1) | Above EAT-volume median | Below EAT-volume median | P-value |
|---|---|---|---|
| GG or GC | 336 (67.5) | 372 (74.7) | .012 |
| CC | 162 (32.5) | 126 (25.3) |
All other 32 genetic polymorphisms (P > .05).
Performing multivariate analysis with all 33 SNPs, environmental factors, and other confounding variables, only the variant rs1801133 of the MTHFR677 was significantly and independently associated with EAT-volume above the median. This gene variant with a critical role in regulating plasma homocysteine levels emerged as associated to higher EAT volumes in our population with an OR of approximately 1.4 (95%CI: 1.100-1.684, P = .005) together with Age and BMI (Table 6 and Figure 6).
Table 6.
Logistic regression for variables above EAT-volume median.
| Variables above EAT-volume median | OR | 95%CI | P-value |
|---|---|---|---|
| rs1801133 (MTHFR677) | 1.361 | 1.100-1.684 | .005 |
| Age | 1.075 | 1.056-1.095 | <.0001 |
| BMI (kg/m²) | 1.247 | 1.200-1.296 | <.0001 |
Abbreviations: BMI, body mass index; CI, confidence interval; EAT, epicardial adipose tissue; OR, odds ratio.
Sex, diabetes, dyslipidemia, hypertension, physical inactivity, and 32 other gene variants (all P > .1).
Figure 6.
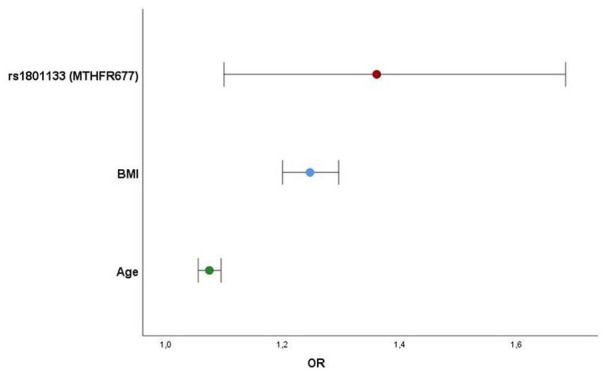
Variables independently associated with higher EAT volumes (above-median) and respective predictive risk (OR) (32 genetic variants, sex, dyslipidemia, hypertension, diabetes, physical inactivity [all P > .1]).
Discussion
Epicardial fat volume is probably the result of a complex interaction between environmental, genetic, and epigenetic factors that only recently we started to uncover.
In the present study with asymptomatic individuals, who have undergone non-contrast cardiac CT, we could see an excess of EAT volume in the group considered at risk of atherosclerosis. This group included the older participants with multiple traditional cardiovascular risk factors and higher levels of biochemical markers which could be related to subclinical atherosclerosis and cardiovascular risk. This fact comes in line with previous studies documenting such associations.22-24
EAT does not just reveal fat deposits: it is considered a biologically active structure representing an association between obesity and cardiovascular disease. 25
We must also account for the detail that some of the variables related to higher EAT volumes are also present in Metabolic Syndrome and its definition (dyslipidemia, diabetes, hypertension, and obesity), thus reinforcing the systemic and overlapping nature of EAT. In our study, the group with higher EAT volume had higher fasting glucose, triglycerides levels, and higher levels of homocysteine, fibrinogen, and high-sensitivity C-reactive protein (hsCRP). A positive correlation was also observed between EAT volume and the mentioned variables supporting a link with Metabolic Syndrome and inflammation. Fibrinogen, homocysteine, and CRP are inflammatory biomarkers, that when elevated, indicate the presence of inflammation, identifying individuals with a high risk for cardiovascular disorders. As we know, mediators of inflammation and oxidative stress play a role in the pathogenesis of cardiovascular disease and are upraised in people with obesity and Metabolic Syndrome.
Our findings are in line with previously published studies who pointed EAT as a risk factor for Metabolic Syndrome. As an integral part of visceral fat, EAT is a live and active metabolic tissue, producing adipokines responsible for adipose tissue inflammation, ectopic lipid deposits, and mitochondrial dysfunction. 26 Adipose tissue secretes many factors including leptin, adiponectin, tumor necrosis factor alpha (TNF-a), interleukin-6 (IL-6), among others, with a proven role in the pathophysiology of metabolic syndrome, systemic inflammation, and cardiovascular disease. 27 The rich expression of inflammatory mediators in epicardial depots as opposed to subcutaneous fat has been documented in previous studies. 4 Anthropomorphic measurements used to estimate visceral fat are imprecise and EAT volume assessed by imaging techniques has proven a solid surrogate of visceral fat volume and its effects, with a greater expression among patients with Metabolic Syndrome, as found by previous studies.28,29
This chronic inflammatory rich environment, will enhance lipolysis, release of free fatty acids, hypoxia, oxidative stress, and seems to be linked to metabolic disorders such as glucose intolerance, dyslipidemia and hypertension, ultimately implicated in the Metabolic Syndrome diagnosis.26,27,30 Further insight into the pathophysiology of inflammatory markers, visceral fat such as EAT depots and Metabolic Syndrome, although not the main focus of this article, may be found in recent reviews by Hamjane et al, 30 Dutheil et al, 27 and Villasante Frick and Iacobellis. 26
Age and BMI consistently emerged as independent predictors for EAT volume across the numerous analysis performed. Our results are similar to those of previously published works.8,22
Pulse wave velocity (PWV), a widely recognized marker of arterial stiffness, 23 was higher among the group with a higher volume of EAT. In the present study, 15.9% of the high-volume EAT group, showed a PWV >10 m/s compared to 8.9% in the group with EAT below the median. Similar results have been published, where EAT was independently associated with PWV in a diabetic and with established cardiovascular disease population. 31 Even though EAT can be a risk factor for the beginning of atherosclerotic development, its usefulness can be the identification of disease at subclinical stage, preventing future progression through preventive measures. Beyond its utility as a risk marker, EAT can be a flexible and modifiable risk factor. It can represent a therapeutic target for preventing plaque growth and cardiovascular events. Interventions like weight loss induced by diet, GLP-1 receptor agonists, or bariatric surgery modulating adipose tissue have demonstrated risk reduction and better prognosis. 32 However, further studies are needed in this complex new field.
Genome-wide association studies (GWAS) have allowed us to uncover the frequent polygenic nature of common complex disorders. 13 However, conveying genetic load conferred by combined set risk variants can be challenging. The creation of Genetic Risk Scores (GRS) is an emerging method that attempts to establish a correlation between an association of single nucleotide polymorphisms (SNPs) and clinical phenotypes, evolving from research into clinical practice. 15
The contribution of a genetic burden to higher epicardial fat volumes was evident in all analysis performed in the present study: First, by the higher global mean GRS (the result of the interaction of 33 different SNPs) in patients with higher epicardial fat volumes. Afterwards, by the mean GRS composed of polymorphisms of specific axis or biological pathways (oxidation, renin-angiotensin system, cellular, diabetes/obesity, and dyslipidemia pathways). This aspect consistently proves how a higher number of different mutations influenced EAT volume, an unparallel finding in the literature available so far. Most studies, focused mainly on alterations in the expression of the EAT genes on in vivo samples from CAD patients undergoing cardiac surgery, are for ethical reasons limited, resulting in a small number of participants.33,34 Vacca et al, 34 have suggested a role for miRNA, small non-coding RNAs modulating gene expression, in the crosstalk between epicardial fat and coronary arteries, thus to CAD. In the present study, the 2 pathophysiological axes englobing variants associated with inflammation/oxidation and diabetes/obesity presented higher EAT association and should be better investigated. In the same line, findings showed recently by Tan et al, 35 suggested, in analysis with enrichment and new bioinformatic technologies that pro-inflammatory and immunological genes were up-regulated in the EAT CAD patients and could act as regulators of CAD pathological process.
We believe that incorporating GWAS information and Genetic Risk Scores in the characterization and study of epicardial adipose tissue is another step forward in the understanding of this complex genetic substrate. Previously, a genome-wide association analysis including 5487 individuals of European ancestry from the Framingham Heart Study (FHS) and of the Multi-Ethnic Study of Atherosclerosis (MESA) identified a unique locus 10198628 near TRIB2 gene, associated to visceral fat adjusted for BMI. 36 Among the 33 genes we analyzed, only the MTHFRC677T polymorphism was identified as an independent predictor of high EAT volume. MTHFR C/T is a gene with a critical role in regulating plasma homocysteine levels. It codifies for the enzyme methylenetetrahydrofolate reductase (MTHFR), responsible for homocysteine degradation and recycling. Inherited mutations in the gene that make the MTHFR enzyme can lead to a loss of enzymatic function and higher homocysteine levels. 37 Connecting homocysteine to lipid metabolism has proven to be a difficult task, but recent work by Momin et al 38 has shown that homocysteine is independently associated with hypertriglyceridemia and low HDL-C levels, shedding light on how it might affect lipid metabolism and fat depots. Balcioğlu et al, 39 have also discovered a close connection between EAT thickness and plasma homocysteine in patients with Metabolic Syndrome, consistent with our findings. However, association does not necessarily imply causality and more studies will be needed to address this matter as this link between homocysteine and EAT remains to be further evaluated.
Strengths and limitations
As far as we concern, this is the first study which proposes to correlate the genetic burden information with the EAT-volume in the Portuguese population. Our sample is representative of a homogeneous European population due to Madeira Island’ isolation over a long time without genetic admixtures.
However, the current methodology behind the construction of a polygenic risk score (PRS), assumes a similar genetic ancestry to large GWAS studies, only widely reflecting European descent. Their applicability across different populations could be limited by lower reproducibility and accuracy.14,40,41 Therefore, our results cannot be generalized to other communities.
Although GRS is a useful way of reflecting the genetic load conferred by a combined set of risk variants, doubts remain regarding its clinical implementation, interpretation, and discriminative ability in the lower-risk general population (of which our sample is representative of). 14 Besides, we must not forget that our score encompassed only 33 variants, although quite significant for CAD. Larger samples with genetic scores containing rare but very weighty variants may open up other paths and unveil key genes and pathways affected in epicardial adipose tissue. These could help us to explore EAT’s role in CAD from new and in-depth perspectives.
As EAT is a very metabolically active tissue, new evidence suggests that this activity and degree of inflammation may be gauged by the attenuation and density of fat (Hu), reinforcing these measurements alongside volume.42,43 We focused solely on volume, measuring it through a simplified method which previously proved to correlate with total EAT volume.19,20 We recognize it may be desirable to quantify the total volume of EAT on consecutive slices acquired through complete cardiac coverage or even self-automated software.
Finally, the impact on hard clinical outcomes is not assessed in the present study, but this should be the aim of future investigations into the role of EAT.
Conclusion
Epicardial adipose tissue has attracted a lot of interest in the last years as an essential player in the pathophysiology of cardiovascular disease. To better understand this risk marker and to make it an appealing therapeutic target in the future, we must first go to its roots, recognizing the contribution of environmental and genetic traits to its form and role. We present one of the few studies in the literature and the first in a Portuguese population, evaluating the genetic profile of EAT through GWAS and GRS, casting further insight into this complicated matter.
In the end, we believe our work contributes with new evidence in this field: (1) patients with a higher genetic polymorphic burden for atherosclerosis and CAD present a higher level of epicardial fat; (2) analyzing genetic variants by pathophysiological axes according to the assigned function, the genetic cellular axis (cell cycle, cell proliferation, apoptosis, and vascular smooth muscle cells migration) is associated significantly to EAT volume together with the already expected diabetes/obesity, dyslipidemia, and oxidative/inflammatory axis; (3) rs1801133 of the MTHFR677 was significantly and independently associated with EAT-volume above the median. This gene variant, with a critical role in regulating plasma homocysteine levels, emerged in our population as associated with higher EAT volumes with an odds ratio higher than age and body mass index; (4) after logistic regression and adjustment for confounders, GRS remained an independent predictor for higher EAT volumes, proving its contribution even beyond traditional risk factors; (5) Pulse wave velocity (PWV), a widely recognized marker of arterial stiffness, was higher among the group with higher EAT volume, in asymptomatic and healthy individuals representative of our everyday population, and not just in diabetes or with established cardiovascular disease patients, as proven before in the literature.
Even though EAT can be a risk factor for the beginning of atherosclerotic development, its usefulness can be the identification of disease at a subclinical stage, preventing future progression through preventive and even therapeutic measures.
Supplemental Material
Supplemental material, sj-zip-1-cic-10.1177_11795468211029244 for Epicardial Adipose Tissue: The Genetics Behind an Emerging Cardiovascular Risk Marker by João Adriano Sousa, Maria Isabel Mendonça, Marco Serrão, Sofia Borges, Eva Henriques, Sónia Freitas, Margarida Tentem, Marina Santos, Pedro Freitas, António Ferreira, Graça Guerra, António Drumond and Roberto Palma Reis in Clinical Medicine Insights: Cardiology
Footnotes
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: J. Sousa, P. Freitas and A. Ferreira conceived of the presented idea; P. Freitas and A. Ferreira mentored and assisted J. Sousa through the post-processing of CT scans and collecting data; M. Serrao and M. Temtem acquired and conceived the CT scans and were responsible for the revision of the manuscript; J. Sousa collected data and drafted the manuscript, alongside MI. Mendonça and S. Borges; E. Henriques and S. Freitas performed the statistical analysis and assisted in the creation of tables, figures and graphs; M. Santos, M. Temtem, G. Guerra and A. Drumond helped collecting data; MI Mendonca and RP Reis revised and supervised the findings of this work. All authors discussed the results and contributed to the final manuscript.
ORCID iD: Maria Isabel Mendonça  https://orcid.org/0000-0001-5450-5213
https://orcid.org/0000-0001-5450-5213
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Nagy E, Jermendy AL, Merkely B, Maurovich-Horvat P. Clinical importance of epicardial adipose tissue. Arch Med Sci. 2017;13:864-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Iacobellis G. Epicardial and pericardial fat: close, but very different. Obesity. 2009;17:625; author reply 626-627. [DOI] [PubMed] [Google Scholar]
- 3. Sacks HS, Fain JN. Human epicardial adipose tissue: a review. Am Heart J. 2007;153:907-917. [DOI] [PubMed] [Google Scholar]
- 4. Mazurek T, Zhang L, Zalewski A, et al. Human epicardial adipose tissue is a source of inflammatory mediators. Circulation. 2003;108:2460-2466. [DOI] [PubMed] [Google Scholar]
- 5. Talman AH, Psaltis PJ, Cameron JD, Meredith IT, Seneviratne SK, Wong DT. Epicardial adipose tissue: far more than a fat depot. Cardiovasc Diagn Ther. 2014;4:416-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nerlekar N, Brown AJ, Muthalaly RG, et al. Association of epicardial adipose tissue and high-risk plaque characteristics: a systematic review and meta-analysis. J Am Heart Assoc. 2017;6:e006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou M, Wang H, Chen J, Zhao L. Epicardial adipose tissue and atrial fibrillation: possible mechanisms, potential therapies, and future directions. Pacing Clin Electrophysiol. 2020;43:133-145. [DOI] [PubMed] [Google Scholar]
- 8. Bertaso AG, Bertol D, Duncan BB, Foppa M. Epicardial fat: definition, measurements and systematic review of main outcomes. Arq Bras Cardiol. 2013;101:e18-e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koepp KE, Obokata M, Reddy YNV, Olson TP, Borlaug BA. Hemodynamic and functional impact of epicardial adipose tissue in heart failure with preserved ejection fraction. JACC Heart Fail. 2020;8:657-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davidovich D, Gastaldelli A, Sicari R. Imaging cardiac fat. Eur Heart J Cardiovasc Imaging. 2013;14:625-630. [DOI] [PubMed] [Google Scholar]
- 11. Nerlekar N, Baey YW, Brown AJ, et al. Poor correlation, reproducibility, and agreement between volumetric versus linear epicardial adipose tissue measurement: a 3D computed tomography versus 2D echocardiography comparison. JACC Cardiovasc Imaging. 2018;11:1035-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Milanese G, Silva M, Bruno L, et al. Quantification of epicardial fat with cardiac CT angiography and association with cardiovascular risk factors in symptomatic patients: from the ALTER-BIO (Alternative Cardiovascular Bio-Imaging markers) registry. Diagn Interv Radiol. 2019;25:35-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tam V, Patel N, Turcotte M, Bosse Y, Pare G, Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20:467-484. [DOI] [PubMed] [Google Scholar]
- 14. Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 2020;12:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Igo RP, Jr, Kinzy TG, Cooke Bailey JN. Genetic risk scores. Curr Protoc Hum Genet. 2019;104:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pereira A, Mendonca MI, Sousa AC, et al. Genetic risk score and cardiovascular mortality in a southern european population with coronary artery disease. Int J Clin Pract. 2017;71:1-8. [DOI] [PubMed] [Google Scholar]
- 17. McCarthy JJ, Parker A, Salem R, et al. Large scale association analysis for identification of genes underlying premature coronary heart disease: cumulative perspective from analysis of 111 candidate genes. J Med Genet. 2004;41:334-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McPherson R. Genome-wide association studies of cardiovascular disease in European and Non-European populations. Curr Genet Med Rep. 2014;2:1-12. [Google Scholar]
- 19. Tran T, Small G, Cocker M, Yam Y, Chow BJ. A single slice measure of epicardial adipose tissue can serve as an indirect measure of total epicardial adipose tissue burden and is associated with obstructive coronary artery disease. Eur Heart J Cardiovasc Imaging. 2014;15:423-430. [DOI] [PubMed] [Google Scholar]
- 20. Oyama N, Goto D, Ito YM, et al. Single-slice epicardial fat area measurement: do we need to measure the total epicardial fat volume? Jpn J Radiol. 2011;29:104-109. [DOI] [PubMed] [Google Scholar]
- 21. Parikh RM, Mohan V. Changing definitions of metabolic syndrome. Indian J Endocrinol Metab. 2012;16:7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rabkin SW. The relationship between epicardial fat and indices of obesity and the metabolic syndrome: a systematic review and meta-analysis. Metab Syndr Relat Disord. 2014;12:31-42. [DOI] [PubMed] [Google Scholar]
- 23. Christensen RH, Hansen CS, von Scholten BJ, et al. Epicardial and pericardial adipose tissues are associated with reduced diastolic and systolic function in type 2 diabetes. Diabetes Obes Metab. 2019;21:2006-2011. [DOI] [PubMed] [Google Scholar]
- 24. Li Y, Liu B, Li Y, et al. Epicardial fat tissue in patients with diabetes mellitus: a systematic review and meta-analysis. Cardiovasc Diabetol. 2019;18:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tadros TM, Massaro JM, Rosito GA, et al. Pericardial fat volume correlates with inflammatory markers: the Framingham Heart Study. Obesity. 2010;18:1039-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Villasante Fricke AC, Iacobellis G. Epicardial adipose tissue: clinical biomarker of cardio-metabolic risk. Int J Mol Sci. 2019;20:5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dutheil F, Gordon BA, Naughton G, et al. Cardiovascular risk of adipokines: a review. J Int Med Res. 2018;46:2082-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iacobellis G, Assael F, Ribaudo MC, et al. Epicardial fat from echocardiography: a new method for visceral adipose tissue prediction. Obes Res. 2003;11:304-310. [DOI] [PubMed] [Google Scholar]
- 29. Iacobellis G, Ribaudo MC, Assael F, et al. Echocardiographic epicardial adipose tissue is related to anthropometric and clinical parameters of metabolic syndrome: a new indicator of cardiovascular risk. J Clin Endocrinol Metab. 2003;88:5163-5168. [DOI] [PubMed] [Google Scholar]
- 30. Hamjane N, Benyahya F, Nourouti NG, Mechita MB, Barakat A. Cardiovascular diseases and metabolic abnormalities associated with obesity: what is the role of inflammatory responses? A systematic review. Microvasc Res. 2020;131:104023. [DOI] [PubMed] [Google Scholar]
- 31. Al-Talabany S, Mordi I, Graeme Houston J, et al. Epicardial adipose tissue is related to arterial stiffness and inflammation in patients with cardiovascular disease and type 2 diabetes. BMC Cardiovasc Disord. 2018;18:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sahasrabuddhe AV, Pitale SU, Sivanesan SD, Deshpande PK, Deshpande SP, Daiwile A. Pathogenic gene expression of epicardial adipose tissue in patients with coronary artery disease. Indian J Med Res. 2020;151:554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chechi K, Vijay J, Voisine P, et al. UCP1 expression-associated gene signatures of human epicardial adipose tissue. JCI Insight. 2019;4:e123618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vacca M, Di Eusanio M, Cariello M, et al. Integrative miRNA and whole-genome analyses of epicardial adipose tissue in patients with coronary atherosclerosis. Cardiovasc Res. 2016;109:228-239. [DOI] [PubMed] [Google Scholar]
- 35. Tan L, Xu Q, Wang Q, Shi R, Zhang G. Identification of key genes and pathways affected in epicardial adipose tissue from patients with coronary artery disease by integrated bioinformatics analysis. PeerJ. 2020;8:e8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fox CS, White CC, Lohman K, et al. Genome-wide association of pericardial fat identifies a unique locus for ectopic fat. PLoS Genet. 2012;8:e1002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moll S, Varga EA. Homocysteine and MTHFR Mutations. Circulation. 2015;132:e6-e9. [DOI] [PubMed] [Google Scholar]
- 38. Momin M, Jia J, Fan F, et al. Relationship between plasma homocysteine level and lipid profiles in a community-based Chinese population. Lipids Health Dis. 2017;16:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Balcioğlu AS, Durakoğlugil ME, Ciçek D, Bal UA, Boyaci B, Müderrisoğlu H. Epicardial adipose tissue thickness and plasma homocysteine in patients with metabolic syndrome and normal coronary arteries. Diabetol Metab Syndr. 2014;6:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peterson RE, Kuchenbaecker K, Walters RK, et al. Genome-wide association studies in ancestrally diverse populations: opportunities, methods, pitfalls, and recommendations. Cell. 2019;179:589-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morales J, Welter D, Bowler EH, et al. A standardized framework for representation of ancestry data in genomics studies, with application to the NHGRI-EBI GWAS Catalog. Genome Biol. 2018;19:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nerlekar N, Thakur U, Lin A, et al. The natural history of epicardial adipose tissue volume and attenuation: a long-term prospective cohort follow-up study. Sci Rep. 2020;10:7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Goeller M, Achenbach S, Marwan M, et al. Epicardial adipose tissue density and volume are related to subclinical atherosclerosis, inflammation and major adverse cardiac events in asymptomatic subjects. J Cardiovasc Comput Tomogr. 2018;12:67-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-zip-1-cic-10.1177_11795468211029244 for Epicardial Adipose Tissue: The Genetics Behind an Emerging Cardiovascular Risk Marker by João Adriano Sousa, Maria Isabel Mendonça, Marco Serrão, Sofia Borges, Eva Henriques, Sónia Freitas, Margarida Tentem, Marina Santos, Pedro Freitas, António Ferreira, Graça Guerra, António Drumond and Roberto Palma Reis in Clinical Medicine Insights: Cardiology


