Abstract
Mycoplasma hyopneumoniae causes swine respiratory disease worldwide. Due to the difficulty of isolating and cultivating M. hyopneumoniae, very few attenuated strains have been successfully isolated, which hampers the development of attenuated vaccines. In order to produce an attenuated M. hyopneumoniae strain, we used the highly virulent M. hyopneumoniae strain ES-2, which was serially passaged in vitro 200 times to produce the attenuated strain ES-2L, and its virulence was evidenced to be low in an animal experiment. In order to elucidate the mechanisms underlying virulence attenuation, we performed whole-genome sequencing of both strains and conducted comparative genomic analyses of strain ES-2 and its attenuated form ES-2L. Strain ES-2L showed three large fragment deletion regions including a total of 18 deleted genes, compared with strain ES-2. Analysis of single-nucleotide polymorphisms (SNPs) and indels indicated that 22 dels were located in 19 predicted coding sequences. In addition to these indels, 348 single-nucleotide variations (SNVs) were identified between strains ES-2L and ES-2. These SNVs mapped to 99 genes where they appeared to induce amino acid substitutions and translation stops. The deleted genes and SNVs may be associated with decreased virulence of strain ES-2L. Our work provides a foundation for further examining virulence factors of M. hyopneumoniae and for the development of attenuated vaccines.
Keywords: Mycoplasma hyopneumoniae, virulence, attenuation, in vitro passaging, sequence analysis
Introduction
Mycoplasma hyopneumoniae is a pathogen that colonizes the respiratory tract of pigs and can cause porcine enzootic pneumonia (EP), which is an important epidemic disease with very high morbidity and is difficult to eradicate (1–3). Even though mortality rates of EP are low, M. hyopneumoniae can colonize and destruct cilia barriers in pigs, which may result in complicated secondary bacterial and viral infections (4, 5). Therefore, it is essential to prevent or control this disease that causes considerable economic losses to the pig industry worldwide (6).
Vaccines play an important role to reduce the prevalence of EP (7, 8). Among them, available inactivated vaccines, based mainly on J strain, do not provide complete protection against lung lesions (9, 10). It has been recently demonstrated that M. hyopneumoniae can evade the host's immune protection by surface antigenic variation, inhibiting the complement activation pathway (11, 12). Moreover, more and more studies have revealed that mucosal immunity and cellular immunity play a major role in immune defense against M. hyopneumoniae (13, 14). For this reason, attenuated vaccines may thus be a promising alternative (15). Nevertheless, isolating and cultivating M. hyopneumoniae is notoriously difficult (16–18); thus, very few attenuated strains have been successfully isolated, which hampers the development of attenuated vaccines (19). Passaging of virulent strains in vitro may be a suitable method to produce attenuated strains, and in 2014, Zhang et al. reported an attenuated strain of Mycoplasma bovis produced by serial passaging (20). In addition, an attenuated M. hyopneumoniae strain 168L has been developed into a commercially available vaccine in China (21), but intrapulmonary injection is required for its usage, which limits its clinical application. In this context, it has enormous significance to develop a new M. hyopneumoniae attenuated strain.
Third-generation sequencing has been used increasingly to perform comparative genomics of pathogens (22), and recently, many novel virulence factors and pathogen–host interaction mechanisms of M. hyopneumoniae were elucidated using comparative genomics (23). Szczepanek et al. performed comparative genomics of a pathogenic strain of Mycoplasma gallisepticum and its attenuated form to identify putative virulence genes (24). However, only a few studies were conducted on the differences between pathogenic M. hyopneumoniae strains and attenuated forms using comparative genomics (23).
Mycoplasma hyopneumoniae strain ES-2 was isolated and identified and was considered a highly virulent strain through our preliminary work. In order to obtain an attenuated strain of M. hyopneumoniae, in the current study, strain ES-2 was serially passaged in vitro to produce the attenuated strain ES-2L, and its virulence was evaluated in an animal experiment. More comparative genomics was used to examine differences between the two strains on a genomic level. Our study provides a foundation for excavation of virulence factor and the development of attenuated vaccines against EP.
Materials and Methods
Mycoplasma Strains and Culture Conditions
Mycoplasma hyopneumoniae strain ES-2 (GenBank accession no. CP038641.1) was isolated from EP-affected pigs housed on a farm in Hubei province, China; and its genome was sequenced by State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University. The pathogen was cultured using modified Friis medium (25, 26) at 37°C in a 5% CO2-enriched atmosphere for 48–72 h, until the color of the medium changed from red to yellow (2, 27).
Virulence Attenuation of Strain ES-2 by in vitro Passaging
The strain ES-2 was cultivated in modified Friis liquid medium at 37°C with a 5% CO2-enriched atmosphere for 48–72 h and was then passaged to P2. Serial passaging cultivation was carried out as the above way. At passage P197, bacteria were plated on modified Friis solid medium and were cultured in a 5% CO2-enriched atmosphere at 37°C for approximately 7–10 days. Individual colony was selected using a low-power microscope and was then cultured in Friis liquid medium. After three cloning and purification steps, the strain produced from the last cloning step (P200) was termed M. hyopneumoniae strain ES-2L. During passaging, PCR assays and cell morphology examinations were performed to ensure no abnormality in the passaging process. Genomic DNA of the strain every 40 generations was extracted using a Mycoplasma gDNA Mini Kit (Biomiga, San Diego, CA, USA). The M. hyopneumoniae-specific P36 gene and 16SrRNA were amplified using specific primers (Supplementary File 1) with the following PCR program (28, 29): 95°C for 5 min, followed by 28 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C 1 min, 72°C for 10 min, and 16°C for 20 min. Cell morphology was observed by transmission electron microscopy.
Virulence Evaluation of ES-2 and Passaged Strains
Twenty healthy weaned cross-bred (Landrace × Large Yorkshire) pigs aged 25–28 days were purchased from a local farm in Hubei province, China, which was free of M. hyopneumoniae and porcine reproductive and respiratory syndrome virus (PRRSV). Blood samples were collected and were screened for M. hyopneumoniae and PRRSV antibodies using an enzyme-linked immunosorbent assay (IDEXX Co., Westbrook, ME, USA). Only animals that are diagnosed with negative antibodies for M. hyopneumoniae and PRRSV were used.
Pigs were assigned to five groups of four individuals each, which were designated as ES-2 (P1), P80, P120, ES-2L (P200), and negative control (NC) groups. ES-2 (P1), P80, P120, and ES-2L (P200) pigs were intratracheally injected with ES-2 (P1), P80, P120, and ES-2L (P200) strains, respectively, at 4.9 × 108 color changing unit (CCU). NC pigs were intratracheally injected with an equal volume of sterile Friis medium.
Clinical observations were recorded daily, particularly regarding coughing, which may suggest M. hyopneumoniae infection. On days 0, 1, 2, 3, 5, and 7 post-infection, rectal temperature was measured each day before feeding. Pigs were weighed on days 0 and 42 post-infection to calculate average daily weight gain.
On 42 days post-infection, all pigs were anesthetized and then exsanguinated. Lung tissue samples were collected and were fixed in formalin for hematoxylin and eosin (H&E) staining using conventional methods (30) and subjected to PCR assays. Pulmonary lesions were evaluated using the scoring method of Madec and Kobisch (31). Each lung lobe was scored as follows: no pathological lesion, 0 points; 1–25% of the area with lesions, 1 point; 26–50% of the area with lesions, 2 points; 51–75% of the area with lesions, 3 points; and 76–100% of the area with lesion, 4 points (total 28 points, seven lung lobes). For each animal, the lesion score (LS) was calculated from the sum of scores of seven pulmonary lobes and ranged between 0 and 28. This study was conducted after approval by the Animal Experiment Ethics Committee of the State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, China (Approval Number: HZAUSW-2019-014). Differences between groups were tested using t-tests. Statistical significance is reported at P < 0.05.
Genome Sequencing, Assembly, and Annotation of Strain ES-2
Genomic DNA of M. hyopneumoniae strain ES-2L was extracted as indicated above. Then, genomic libraries were sequenced using a combined Illumina NovaSeq (Illumina, San Diego, CA, USA) and Nanopore (Oxford Nanopore, Oxford, UK) sequencing approach (Nextomics, Wuhan, China). Genomes were assembled using Canu software version 1.8 with a combination of short and long reads, followed by error correction using Pilon software version 1.12. A circular chromosome with 2,300-fold average base coverage was produced and then was annotated using the best-placed reference protein set (GeneMarkS+) of the National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline version 3.3.
Comparative Genomics and Structure Variation Analysis
For comparative genomic analysis, a genome of M. hyopneumoniae primary strain ES-2 was downloaded from GenBank as a reference (accession no. CP038641.1, https://www.ncbi.nlm.nih.gov/nucleotide/CP038641.1). Clean NovaSeq reads were mapped to the reference genome using the Burrows–Wheeler Alignment tool, and structure variations were examined using the three software programs GATK Haplotype Caller version 4.0, FreeBayes version 1.3, and samtools mpileup version 1.8. A genome alignment analysis of strains ES-2 and ES-2L was conducted using the Artemis Comparison Tool.
Confirmation of Deleted Genes by PCR
Based on the above comparative genomics results, PCR assays were used to confirm whether the predicted genes were really deleted on the genome of strain ES-2. Primers for 18 genes were designed using Primer5 software and were commercially synthesized (TSINGKE Biological Technology, Beijing, China; Supplementary File 1). These genes were amplified by PCR with ES-2 and ES-2L genome templates, using the following program: 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 55°C for 45 s, and 72°C 1 min, 72°C for 10 min, and 16°C for 15 min.
Results
Identification of Strain ES-2L
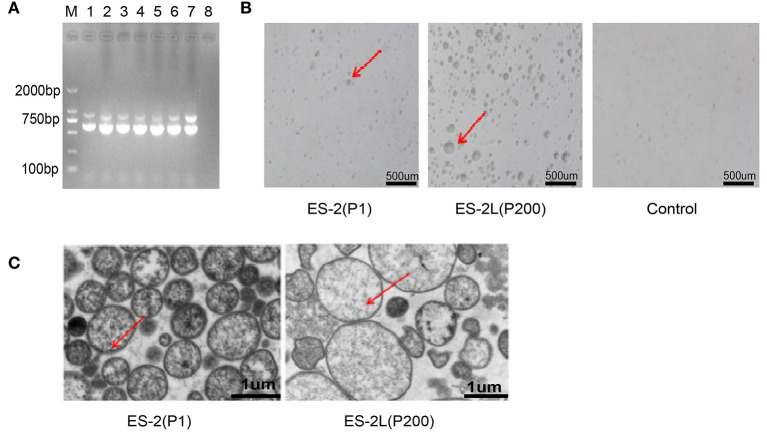
PCR assays indicated that M. hyopneumoniae-specific P36 (948 bp) and 16S RNA (627 bp) target genes could both be detected in ES-2 (P1), P80, P120, P160, and ES-2L (P200) (Figure 1A). Colonies of strain ES-2 and ES-2L on solid medium showed the morphological characteristics of rounded, distinct edges, and centers with granules (Figure 1B), which was accorded with biological characteristics of mycoplasma. In addition, transmission electron microscopy displayed that strain ES-2 had no cell walls, cell membranes were surrounded by capsules, and the cytoplasmic structure was loose. The morphological characteristics of strain ES-2L were similar to those of strain ES-2, but there were a few large-celled mutant individuals (Figure 1C).
Figure 1.
(A) Detection of Mycoplasma hyopneumoniae-specific P36 (948 bp) and 16S RNA (627 bp) genes for the different passage strains: M, DNA marker 2,000; lane 1, ES-2 (P1); lane 2, P40; lane 3, P80; lane 4, P120; lane 5, P160; lane 6, ES-2L (P200); lane 7, positive control; lane 8, negative control. (B) Morphological observation of strain ES-2L and ES-2 on solid medium using a low-power microscope; the colonies of strain ES-2 and ES-2L showed the characteristics of rounded, distinct edges and centers with granules, as indicated by the red arrow. (C) Morphological examination of strains ES-2L and ES-2 observed by transmission electron microscopy, indicating that strain ES-2 had no cell walls, cell membranes were surrounded by capsules, and the cytoplasmic structure was loose; characteristics of strain ES-2L were similar to those of strain ES-2, but there were a few large-celled mutant individuals, as indicated by the red arrow.
Clinical Observations of Virulence Evaluation Experiment
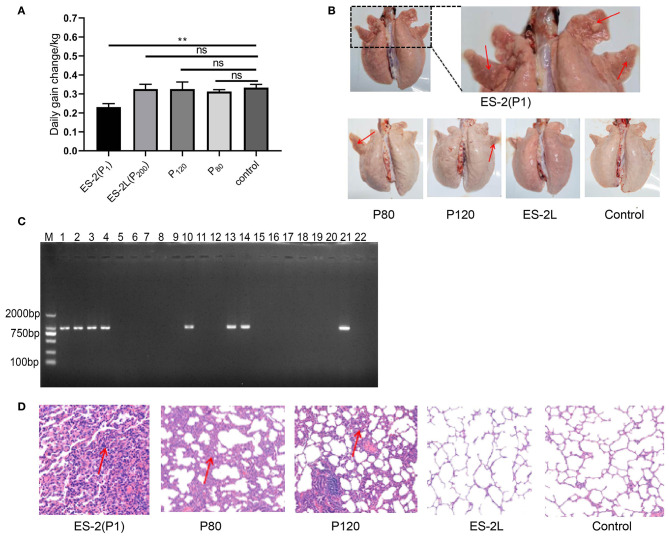
No consistent increase in body temperature was recorded in any of the treatment groups after infection. Further, two pigs in the P1 group appeared the symptom of severe cough along with the emaciated and depressed status on day 23 post-infection. Two pigs in the P80 group exhibited moderate cough symptom, and one pig in the P120 group showed mild cough. No coughing was observed in the ES-2L group throughout the experiment period. On the other hand, compared with the NC group, average daily weight gain in the P1 group was significantly reduced (Figure 2A), whereas no difference between the ES-2L group and the NC group was observed. The above results manifested that strain ES-2, not the attenuated strain ES-2L, could induce clinical symptoms of EP in animal infection experiment.
Figure 2.
(A) Change of average daily weight gain in each group; data are presented as the means ± SD (n = 4); **P < 0.01, Student's t-test. (B) Observation of lung tissue macroscopic lesions in each group; all pigs in the P1 group had typical “red consolidations” lesions of lung tissue (indicated by the red arrow). None of the pigs in the ES-2L group showed typical lesions. (C) Detection of P36 gene (948 bp) by PCR assay for pig's lungs of each group: M, DNA marker 2,000; lanes 1, 2, 3, and 4 correspond to P36 gene on ES-2 group; lanes 5, 6, 7, and 8 correspond to P36 gene on ES-2L group; lanes 9, 10, 11, and 12 correspond to P36 gene on P120 group; lanes 13, 14, 15, and 16 correspond to P36 gene on P80 group; lanes 17, 18, 19, and 20 correspond to P36 gene on control group; lane 21, positive control; lane 22, negative control. (D) Examination of lung tissue hematoxylin and eosin staining by microscopy at 100-fold magnification; the red arrow indicates that there were severe mononuclear cell and lymphocyte infiltration and alveolar consolidation in lung tissue.
Macroscopic and Microcosmic Pathological Observations
After dissection on 42 days post-infection, macroscopic lesions were examined. All pigs in the P1 group showed typical “red consolidation” lesions of the lungs (Figure 2B), with the average LS of 11 ± 1. Two pigs in the P80 group had moderate pneumonia lesions in the lungs, with the average LS of 7 ± 2; one pig in the P120 group displayed mild lesions of the lungs, and the average LS was 4 ± 1. None of the pigs in the ES-2L group showed typical lesions in the lungs (Figure 2B), and the average LS was 2 ± 1. In addition, the P36 gene could be detected by PCR assays in all pigs' lungs of the ES-2 group, in two pigs' lungs of the P80 group, in one pig's lungs of the P120 group, and in none of the ES-2L group (Figure 2C). For microcosmic pathological lesions, H&E staining of lung tissue indicated that there were severe mononuclear cell and lymphocyte infiltration and alveolar consolidation in the ES-2 group (Figure 2D). One pig in the P80 group exhibited lesions of severe inflammatory cell infiltration with widened alveolar stroma in the lungs, and one pig in the P120 group had also diseased regions of mild inflammatory cell infiltration in the lungs. No pathological lesions were observed in the ES-2L group (Figure 2D). These results demonstrated that strain ES-2 was virulent, and after serial passaging in vitro, its virulence was reduced, resulting in the attenuated strain ES-2L.
Genomic Characteristics of Strains ES-2L and ES-2
In total, 108,621 raw reads comprising 2,392,084,536 bases were produced from strain ES-2L (accession no CP058578.1, https://www.ncbi.nlm.nih.gov/nuccore/CP058578.1). Raw reads were filtered and optimized to assemble the first contig with a size of 918,900 bp [guanine–cytosine (GC) content 28.48%]. A total of 697 protein-encoding genes were predicted. The ES-2L genome was 37,614 bp smaller than that of ES-2 (956,514 bp). According to the collinearity analysis results, the ES-2L strain sequence indicated three large fragment deletion regions (Figure 3A), and these large fragment deletion regions indicated 18 gene deletions in ES-2L, compared with ES-2 (Table 1). Analysis of single-nucleotide polymorphism (SNP) and indels indicated that 22 dels were located in 19 predicted coding regions (Supplementary File 2). Except for these indels, 348 single-nucleotide variations (SNVs) were identified between ES-2L and ES-2 (Supplementary File 2). These SNVs mapped to 99 genes where they induce amino acid substitutions and translational stops.
Figure 3.
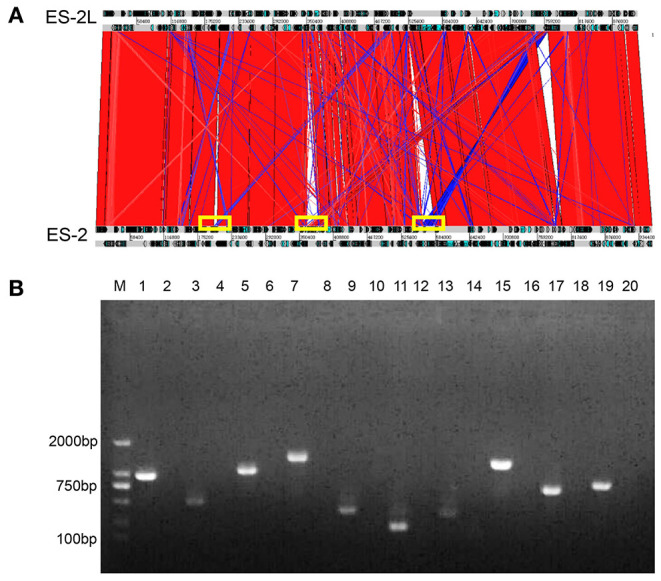
(A) Collinearity analysis results of strains ES-2L and ES-2; strain ES-2L showed three large fragment deletion regions indicated by the yellow box, compared with ES-2. (B) Confirmation of the deleted genes by PCR. M, DNA marker 2,000; lanes 1, 3, 5, 7, 9, 11, 13, 15, 17, and 19 correspond to genes E5E95_00935, E5E95_00940, E5E95_01845, E5E95_01850, E5E95_01855, E5E95_01860, E5E95_01905, E5E95_03755, E5E95_00670, and E5E95_00665 with strain ES-2 genome as a template, respectively. Lanes 2, 4, 6, 8, 10, 12, 14, 16, 18, and 20 indicate that these genes were amplified with strain ES-2L genome as template.
Table 1.
Eighteen genes in regions deleted from the genome of strain ES-2L.
| Variation type | Start site | Stop site | Size/bp | Including gene ID | Encoding protein |
|---|---|---|---|---|---|
| Deletion | 113,237 | 114,699 | 1,462 | E5E95_00505 | Serine protease |
| Deletion | 159,989 | 161,778 | 1,789 | E5E95_00670 | Hypothetical protein |
| E5E95_00665 | Hypothetical protein | ||||
| E5E95_00675 | Hypothetical protein | ||||
| Deletion | 203,386 | 208,867 | 5,481 | E5E95_00935 | Hypothetical protein |
| E5E95_00940 | Hypothetical protein | ||||
| Deletion | 261,435 | 262,128 | 693 | E5E95_01210 | Hypothetical protein |
| Deletion | 262,717 | 26,3417 | 700 | E5E95_01215 | Hypothetical protein |
| Deletion | 417,189 | 420,421 | 3,232 | E5E95_01850 | Hypothetical protein |
| E5E95_01845 | Hypothetical protein | ||||
| E5E95_01855 | Hypothetical protein | ||||
| E5E95_01860 | Alcohol dehydrogenase | ||||
| Deletion | 429,321 | 433,928 | 4,607 | E5E95_01905 | DNA gyrase subunit A |
| E5E95_01910 | Hypothetical protein | ||||
| E5E95_01915 | Serine protease | ||||
| Deletion | 558,155 | 560,135 | 1,980 | E5E95_02435 | IS1634 family transposase |
| E5E95_02440 | Site-specific DNA-methyltransferase | ||||
| Deletion | 925,798 | 927,220 | 1,422 | E5E95_03755 | DDE domain-containing protein |
Confirmation of Gene Deletion by PCR
Ten of 18 potentially deleted genes, E5E95_00935, E5E95_00940, E5E95_01845, E5E95_01850, E5E95_01855, E5E95_01860, E5E95_01905, E5E95_03755, E5E95_00670, and E5E95_00665, could be detected by PCR, with strain ES-2 genome as template. However, these genes could not be detected when using strain ES-2L genome as PCR template, which indicated that these 10 genes were in fact deleted in strain ES-2L, as confirmed by PCR (Figure 3B).
Discussion
In this study, the animal infection experiment demonstrated that strains ES-2L and ES-2 have significant difference in virulence. Especially, in terms of clinical symptoms of ES-2 challenged pigs, the ES-2 group developed cough symptoms 23 days after infection; nevertheless, other studies showed that coughing began 14 days after infection (32), which may be caused by the differences of source of the strain, infection dose, and experimental animals. Unlike other pathogens such as PRRSV, M. hyopneumoniae could not lead to continuous increase in body temperature after infection (33, 34). In the experiment, regardless of which group, there was no obvious increase in body temperature after infection until the end, which was consistent with other reports (34).
Serum IgG antibody response is not a reliable tool for assessing the infection status of an individual animal (35, 36). For our research, at 28 and 42 days post-infection, the ES-2 and ES-2L groups had a low seroconversion rate (50%), and only one pig in the P120 group was seropositive (results not shown), which could be explained by this mechanism that M. hyopneumoniae could weaken humoral immune response by inducing apoptosis of immune cells (37–39). Recently, intensive studies have revealed that IgA antibody level emerges in an uptrend after vaccination, which indicated that mucosal IgA responses play a crucial role in immune protection against M. hyopneumoniae (40); hence, it is necessary to detect IgA in the saliva and/or nose after infection in the following investigation.
Pigs infected with virulent M. hyopneumoniae strain can have clinical symptoms such as coughing and weight loss, and lung lesions characterized by dark red, firm consolidation in apical, cardiac, and intermediate lobes (6, 41, 42), but further confirmation by molecular diagnosis is still necessary (43). The lungs of healthy piglets infected with strain ES-2 showed typical “well-demarcated, red consolidations” (Figure 2B); especially, M. hyopneumoniae-specific gene P36 was detected in the cardiac lobe and apical lobe of the lungs. But gene P36 was not detected by PCR in the spleen, kidney, and liver of all ES-2 challenged pigs, which conflicted with another study that M. hyopneumoniae genomes have been detected by qPCR from inner organs, such as the liver, spleen, and kidneys in experimentally infected pigs (30). One possible explanation for the conflict between them was that the detection method adopted on our study was different from theirs. Taken together, these results demonstrated that strain ES-2 was a highly virulent and pathogenic strain, which was also consistent with the results of our previous studies; furthermore, its virulence decreased with the number of in vitro passages. Besides, pigs infected with strain ES-2L showed no typical lung lesions and cough symptoms, suggesting the lower virulence of this strain, which may thus be a vaccine candidate. However, further evaluation of its immunogenicity was necessary in the next exploration. Yu et al. obtained also similar macroscopic and microcosmic pathological observations by comparing the virulence of the M. hyopneumoniae highly virulent strain 168 and its attenuated strain 168L (32).
In addition, our study found that in the process of continuous passage, the morphology of a small number of cells would undergo the changes to large-celled mutations observed by transmission electron microscopy (Figure 1C). These large-celled mutations contained more DNA compared with typical cell types in the population (44), which might contribute to form biofilms on abiotic surfaces (45), but their role in invading and adhering to host was currently unclear. As expected, the adaptability of strain ES-2L to the culture medium was better than that of strain ES-2, which may be attributed to serial passaging. Specifically, the growth titer of strain ES-2L could reach 1.0 × 1010 CCU/ml, which was much higher than that of strain ES-2. Also, the growth rate of strain ES-2L that only took 2 days to grow to the late-log phase was also considerably faster than that of strain ES-2 (results not shown). These phenomena allowed us to hypothesize that some mutated genes related to substance transport and metabolism, such as importing sugars, amino acids, peptides, and metal ions, should be the focus of our attention (46, 47). In line with our assumption, the results of SNP analysis depicted that there were a large number of mutated genes associated with microbial substance transport and metabolism processes, such as ABC transporter protein family.
In order to explain the mechanisms underlying attenuation of strain ES-2, we conducted comparative genomics of ES-2 and ES-2L. The ES-2L genome was 37,614 bp smaller than that of ES-2. During serial passaging, gene deletion may occur, which was observed in M. bovis regarding a deletion of a 14.2-kb fragment as assessed by comparative genomics of strain HB0801 and its attenuated form (47). Compared with strain ES-2, strain ES-2L showed several large fragment deletion regions that contained 18 genes (Table 1), and 10 of 18 genes were in fact deleted, as confirmed by PCR (Figure 3B). Unfortunately, the functions of most of these genes are unknown and thus require further examination. One of the deleted genes encodes alcohol dehydrogenase (AdhE; E5E95_01860), which is a key enzyme in the anaerobic fermentation pathway of ethanol and regulates ethanol generation in bacteria (48, 49). Under anaerobic conditions, the lack of AdhE may reduce ethanol synthesis, resulting in decreased hydrogen peroxide concentrations, which was closely associated with the toxicity of mycoplasma (50–52). Interestingly, we further analyzed the genome structure of strain ES-2L and ES-2 by Island Viewer 4 software, which indicated that there are two loss of genomic islands in the process of passaging. Genomic island was considered to be closely related to the horizontal gene transfer (53, 54). According to current researches, the genes encoded by genomic islands were associated with organism virulence, pathogenicity, metabolism, and drug resistance (55, 56). One of the genomic islands, encoding 10 genes, was located at start 920,852-bp site to end 943,294-bp site in the ES-2 genome. Two of the 10 genes, E5E95_03750 and E5E95_03795, encoded LppA family lipoprotein and transcription elongation factor GreA, respectively. Moreover, other researches have reported that LppA family lipoprotein and transcription elongation factor were two important virulence factors of M. hyopneumoniae, playing a significant role in the process of M. hyopneumoniae invading and adhering to the host (57–60). These findings suggested that the loss of genomic island might be related to the changes in strain ES-2 virulence and growth characteristics.
On the other hand, the results of SNP analysis showed that there were 348 SNVs in strain ES-2L, which may affect the functions of 99 genes. The adherence of M. hyopneumoniae to porcine ciliated respiratory cells plays a key step for infection. Liu et al. also exhibited through the comparative genomics results of M. hyopneumoniae strain 168 and 168L that there were a large number of SNVs, and these SNVs affected functional expression of some important adhesin gene, such as P97, P102, P46, and P65 (23). E5E95_00610 gene was annotated as adhesin protein by the NCBI Search database; it experienced two non-synonymous gene mutations (C–G and C–T) according to the results of SNP analysis (Supplementary File 2), which suggested that this gene may be a new potential virulence factor, although its biological significance needs further verification. More importantly, mycoplasmas can evade the killing of the host's humoral immunity by the MIB–MIP system that captures and cleaves immunoglobulin G (61–63). E5E95_03535 was an important virulence gene encoding putative immunoglobulin-blocking protein, which could bind to the immunoglobulin in the serum to cleave the immunoglobulin, playing a crucial role in evading the neutralization of serum antibodies. The protein undergoes three single-nucleotide mutations (C–T, G–C, and T–C) in the process of continuous passage (Supplementary File 2), which might affect its function. Taken together, gene deletion and SNVs affecting gene function may be responsible for the decrease in virulence of strain ES-2. However, there are still huge technical obstacles to construct gene-deleted strains (64, 65); we are currently unable to study the functions of specific gene through genetic manipulation methods. We look forward to greater progress in related technical operations for verifying the biological functions of these genes.
Conclusion
We successfully produced an attenuated M. hyopneumoniae strain by serial in vitro passaging, and lower virulence was confirmed in an animal experiment. Comparative genomics between strains ES-2 and its attenuated form ES-2L indicated numerous considerable differences on a genomic level. Our research provides a new perspective for the pathogenic mechanism of M. hyopneumoniae and the development of attenuated vaccines.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI GenBank; accession no.'s CP038641.1 and CP058578.1.
Ethics Statement
The animal study was reviewed and approved by Committee on the Ethics of Animal Experiments at the College of Huazhong Agricultural University.
Author Contributions
ZL performed experiments, analyses, and wrote the manuscript. ZL performed experiments with the assistance of YQ, XT, WL. YW, and YZhu performed analyses. CT, XW, YZha, and HC designed experiments and revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
HC and XT were employed by Wuhan Keqian Biology Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the science and technology major program of Hubei province (2020ABA016).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.696262/full#supplementary-material
Primers used for amplification.
Information of single nucleotide polymorphisms (SNP) and indels analysis.
References
- 1.Betlach AM, Fano E, VanderWaal K, Pieters M. Effect of multiple vaccinations on transmission and degree of Mycoplasma hyopneumoniae infection in gilts. Vaccine. (2021) 39:767–74. 10.1016/j.vaccine.2020.10.096 [DOI] [PubMed] [Google Scholar]
- 2.Galvao Ferrarini M, Mucha SG, Parrot D, Meiffrein G, Ruggiero Bachega JF, Comte G, et al. Hydrogen peroxide production and myo-inositol metabolism as important traits for virulence of Mycoplasma hyopneumoniae. Mol Microbiol. (2018) 108:683–96. 10.1111/mmi.13957 [DOI] [PubMed] [Google Scholar]
- 3.Maes D, Segales J, Meyns T, Sibila M, Pieters M, Haesebrouck F. Control of Mycoplasma hyopneumoniae infections in pigs. Vet Microbiol. (2008) 126:297–309. 10.1016/j.vetmic.2007.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fablet C, Marois-Créhan C, Simon G, Grasland B, Jestin A, Kobisch M, et al. Infectious agents associated with respiratory diseases in 125 farrow-to-finish pig herds: a cross-sectional study. Vet Microbiol. (2012) 157:152–63. 10.1016/j.vetmic.2011.12.015 [DOI] [PubMed] [Google Scholar]
- 5.Razin S, Yogev D, Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. (1998) 62:1094–156. 10.1128/MMBR.62.4.1094-1156.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Almeida HMS, Mechler-Dreibi ML, Sonálio K, Ferraz MES, Storino GY, Barbosa FO, et al. Cytokine expression and Mycoplasma hyopneumoniae burden in the development of lung lesions in experimentally inoculated pigs. Vet Microbiol. (2020) 244:108647. 10.1016/j.vetmic.2020.108647 [DOI] [PubMed] [Google Scholar]
- 7.Feng ZX, Wei YN, Li GL, Lu XM, Wan XF, Pharr GT, et al. Development and validation of an attenuated Mycoplasma hyopneumoniae aerosol vaccine. Vet Microbiol. (2013) 167:417–424. 10.1016/j.vetmic.2013.08.012 [DOI] [PubMed] [Google Scholar]
- 8.Garza-Moreno L, Segalés J, Pieters M, Romagosa A, Sibila M. Acclimation strategies in gilts to control Mycoplasma hyopneumoniae infection. Vet Microbiol. (2018) 219:23–9. 10.1016/j.vetmic.2018.04.005 [DOI] [PubMed] [Google Scholar]
- 9.Matthijs AMF, Auray G, Boyen F, Schoos A, Michiels A, García-Nicolás O, et al. Efficacy of three innovative bacterin vaccines against experimental infection with Mycoplasma hyopneumoniae. Vet Res. (2019) 50:91. 10.1186/s13567-019-0709-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen Y, Hu W, Wei Y, Feng Z, Yang Q. The immune mechanism of Mycoplasma hyopneumoniae 168 vaccine strain through dendritic cells. BMC Vet Res. (2017) 13:285. 10.1186/s12917-017-1194-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leal Zimmer FMA, Paes JA, Zaha A, Ferreira HB. Pathogenicity and virulence of Mycoplasma hyopneumoniae. Virulence. (2020) 11:1600–22. 10.1080/21505594.2020.1842659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu Y, Wang J, Han R, Wang L, Zhang L, Zhang AY, et al. Mycoplasma hyopneumoniae evades complement activation by binding to factor H via elongation factor thermo unstable (EF-Tu). Virulence. (2020) 11:1059–74. 10.1080/21505594.2020.1806664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen AY, Fry SR, Forbes-Faulkner J, Daggard G, Mukkur TKS. Evaluation of the immunogenicity of the P97R1 adhesin of Mycoplasma hyopneumoniae as a mucosal vaccine in mice. J Med Microbiol. (2006) 55:923–9. 10.1099/jmm.0.46088-0 [DOI] [PubMed] [Google Scholar]
- 14.Thacker EL, Thacker BJ, Kuhn M, Hawkins PA, Waters WR. Evaluation of local and systemic immune responses induced by intramuscular injection of a Mycoplasma hyopneumoniae bacterin to pigs. Am J Vet Res. (2000) 61:1384–9. 10.2460/ajvr.2000.61.1384 [DOI] [PubMed] [Google Scholar]
- 15.Villarreal I, Maes D, Meyns T, Gebruers F, Calus D, Pasmans F, et al. Infection with a low virulent Mycoplasma hyopneumoniae isolate does not protect piglets against subsequent infection with a highly virulent M. hyopneumoniae isolate. Vaccine. (2009) 27:1875–9. 10.1016/j.vaccine.2008.12.005 [DOI] [PubMed] [Google Scholar]
- 16.Cook BS, Beddow JG, Manso-Silván L, Maglennon GA, Rycroft AN. Selective medium for culture of Mycoplasma hyopneumoniae. Vet Microbiol. (2016) 195:158–64. 10.1016/j.vetmic.2016.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodwin RF, Hurrell JM. Further observations on the problem of isolating Mycoplasma suipneumoniae from field cases of enzootic pneumonia in pigs. J Hyg. (1970) 68:313–25. 10.1017/S002217240002876X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian Y, Xu Z, Wen Y, Yang M, Ning Y, Wang Z, et al. Development of an indirect ELISA for detection of anti-Mycoplasma hyopneumoniae IgG in naturally infected pathogen-induced convalescent sera. BMC Vet Res. (2021) 17:123. 10.1186/s12917-021-02828-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tao Y, Shu J, Chen J, Wu Y, He Y. A concise review of vaccines against Mycoplasma hyopneumoniae. Res Vet Sci. (2019) 123:144–52. 10.1016/j.rvsc.2019.01.007 [DOI] [PubMed] [Google Scholar]
- 20.Zhang R, Han X, Chen Y, Mustafa R, Qi J, Chen X, et al. Attenuated Mycoplasma bovis strains provide protection against virulent infection in calves. Vaccine. (2014) 32:3107–14. 10.1016/j.vaccine.2013.12.004 [DOI] [PubMed] [Google Scholar]
- 21.Xiong Q, Wei Y, Xie H, Feng Z, Gan Y, Wang C, et al. Effect of different adjuvant formulations on the immunogenicity and protective effect of a live Mycoplasma hyopneumoniae vaccine after intramuscular inoculation. Vaccine. (2014) 32:3445–51. 10.1016/j.vaccine.2014.03.071 [DOI] [PubMed] [Google Scholar]
- 22.Liu W, Fang L, Li M, Li S, Guo S, Luo R, et al. Comparative genomics of Mycoplasma: analysis of conserved essential genes and diversity of the pan-genome. PloS ONE. (2012) 7:e35698. 10.1371/journal.pone.0035698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W, Xiao S, Li M, Guo S, Li S, Luo R, et al. Comparative genomic analyses of Mycoplasma hyopneumoniae pathogenic 168 strain and its high-passaged attenuated strain. BMC Genomics. (2013) 14:80. 10.1186/1471-2164-14-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szczepanek SM, Tulman ER, Gorton TS, Liao X, Lu Z, Zinski J, et al. Comparative genomic analyses of attenuated strains of Mycoplasma gallisepticum. Infect Immun. (2010) 78:1760–1771. 10.1128/IAI.01172-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calus D, Maes D, Vranckx K, Villareal I, Pasmans F, Haesebrouck F. Validation of ATP luminometry for rapid and accurate titration of Mycoplasma hyopneumoniae in Friis medium and a comparison with the color changing units assay. J Microbiol Methods. (2010) 83:335–40. 10.1016/j.mimet.2010.09.001 [DOI] [PubMed] [Google Scholar]
- 26.Friis N. A selective medium for Mycoplasma suipneumoniae. Acta Vet Scand. (1971) 12:454–6. 10.1186/BF03547746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friis NF. Some recommendations concerning primary isolation of Mycoplasma suipneumoniae and Mycoplasma flocculare a survey. Nord Vet Med. (1975) 27:337–9. [PubMed] [Google Scholar]
- 28.Caron J, Ouardani M, Dea S. Diagnosis and differentiation of Mycoplasma hyopneumoniae and Mycoplasma hyorhinis infections in pigs by PCR amplification of the p36 and p46 genes. J Clin Microbiol. (2000) 38:1390–6. 10.1128/JCM.38.4.1390-1396.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mattsson JG, Bergström K, Wallgren P, Johansson KE. Detection of Mycoplasma hyopneumoniae in nose swabs from pigs by in vitro amplification of the 16S rRNA gene. J Clin Microbiol. (1995) 33:893–7. 10.1128/JCM.33.4.893-897.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woolley LK, Fell S, Gonsalves JR, Walker MJ, Djordjevic SP, Jenkins C, et al. Evaluation of clinical, histological and immunological changes and qPCR detection of Mycoplasma hyopneumoniae in tissues during the early stages of mycoplasmal pneumonia in pigs after experimental challenge with two field isolates. Vet Microbiol. (2012) 161:186–95. 10.1016/j.vetmic.2012.07.025 [DOI] [PubMed] [Google Scholar]
- 31.Xiong Q, Wei Y, Feng Z, Gan Y, Liu Z, Liu M, et al. Protective efficacy of a live attenuated Mycoplasma hyopneumoniae vaccine with an ISCOM-matrix adjuvant in pigs. Vet J (Lond.). (2014) 199:268–74. 10.1016/j.tvjl.2013.11.001 [DOI] [PubMed] [Google Scholar]
- 32.Yu Y, Liu M, Hua L, Qiu M, Zhang W, Wei Y, et al. Fructose-1,6-bisphosphate aldolase encoded by a core gene of Mycoplasma hyopneumoniae contributes to host cell adhesion. Vet Res. (2018) 49:114. 10.1186/s13567-018-0610-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vicca J, Maes D, Thermote L, Peeters J, Haesebrouck F, de Kruif A. Patterns of Mycoplasma hyopneumoniae infections in Belgian farrow-to-finish pig herds with diverging disease-course. J Vet Med B Infect Dis Vet Public Health. (2002) 49:349–53. 10.1046/j.1439-0450.2002.00579.x [DOI] [PubMed] [Google Scholar]
- 34.Vicca J, Stakenborg T, Maes D, Butaye P, Peeters J, de Kruif A, et al. Evaluation of virulence of Mycoplasma hyopneumoniae field isolates. Vet Microbiol. (2003) 97:177–90. 10.1016/j.vetmic.2003.08.008 [DOI] [PubMed] [Google Scholar]
- 35.Deiters U, Mühlradt PF. Mycoplasmal lipopeptide MALP-2 induces the chemoattractant proteins macrophage inflammatory protein 1alpha (MIP-1alpha), monocyte chemoattractant protein 1, and MIP-2 and promotes leukocyte infiltration in mice. Infect Immun. (1999) 67:3390–8. 10.1128/IAI.67.7.3390-3398.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchioro SB, Maes D, Flahou B, Pasmans F, Del Pozo Sacristán R, Vranckx K, et al. Local and systemic immune responses in pigs intramuscularly injected with an inactivated Mycoplasma hyopneumoniae vaccine. Vaccine. (2013) 31:1305–11. 10.1016/j.vaccine.2012.12.068 [DOI] [PubMed] [Google Scholar]
- 37.Fourour S, Marois-Créhan C, Martelet L, Fablet C, Kempf I, Gottschalk M, et al. Intra-species and inter-species differences in cytokine production by porcine antigen-presenting cells stimulated by Mycoplasma hyopneumoniae, M. hyorhinis, M. flocculare. Pathogens. (2019) 8:34. 10.3390/pathogens8010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Messier S, Ross RF, Paul PS. Humoral and cellular immune responses of pigs inoculated with Mycoplasma hyopneumoniae. Am J Vet Res. (1990) 51:52–8. [PubMed] [Google Scholar]
- 39.Redondo E, Masot AJ, Fernández A, Gázquez A. Histopathological and immunohistochemical findings in the lungs of pigs infected experimentally with Mycoplasma hyopneumoniae. J Comp Pathol. (2009) 140:260–70. 10.1016/j.jcpa.2008.12.008 [DOI] [PubMed] [Google Scholar]
- 40.Maes D, Boyen F, Devriendt B, Kuhnert P, Summerfield A, Haesebrouck F. Perspectives for improvement of Mycoplasma hyopneumoniae vaccines in pigs. Vet Res. (2021) 52:67. 10.1186/s13567-021-00941-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pallarés FJ, Añón JA, Rodríguez-Gómez IM, Gómez-Laguna J, Fabré R, Sánchez-Carvajal JM, et al. Prevalence of mycoplasma-like lung lesions in pigs from commercial farms from Spain and Portugal. Porcine Health Manag. (2021) 7:26. 10.1186/s40813-021-00204-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarradell J, Andrada M, Ramírez AS, Fernández A, Gómez-Villamandos JC, Jover A, et al. A morphologic and immunohistochemical study of the bronchus-associated lymphoid tissue of pigs naturally infected with Mycoplasma hyopneumoniae. Vet Pathol. (2003) 40:395–404. 10.1354/vp.40-4-395 [DOI] [PubMed] [Google Scholar]
- 43.Vangroenweghe F, Willems E, Malášek J, Thas O, Maes D. Use of trachea-bronchial swab qPCR testing to confirm Mycoplasma hyopneumoniae seropositivity in an SPF breeding herd. Porcine Health Manag. (2018) 4:12. 10.1186/s40813-018-0088-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raymond BBA, Jenkins C, Turnbull L, Whitchurch CB, Djordjevic SP. Extracellular DNA release from the genome-reduced pathogen Mycoplasma hyopneumoniae is essential for biofilm formation on abiotic surfaces. Sci. Rep. (2018) 8:10373. 10.1038/s41598-018-28678-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen H, Yu S, Hu M, Han X, Chen D, Qiu X, et al. Identification of biofilm formation by Mycoplasma gallisepticum. Vet Microbiol. (2012) 161:96–103. 10.1016/j.vetmic.2012.07.013 [DOI] [PubMed] [Google Scholar]
- 46.Higgins CF, Hiles ID, Salmond GP, Gill DR, Downie JA, Evans IJ, et al. A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature. (1986) 323:448–50. 10.1038/323448a0 [DOI] [PubMed] [Google Scholar]
- 47.Rasheed MA, Qi J, Zhu X, Chenfei H, Menghwar H, Khan FA, et al. Comparative genomics of Mycoplasma bovis strains reveals that decreased virulence with increasing passages might correlate with potential virulence-related factors. Front Cell Infect Microbiol. (2017) 7:177. 10.3389/fcimb.2017.00177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arndt A, Eikmanns BJ. The alcohol dehydrogenase gene adhA in Corynebacterium glutamicum is subject to carbon catabolite repression. J Bacteriol. (2007) 189:7408–16. 10.1128/JB.00791-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pei J, Zhou Q, Jiang Y, Le Y, Li H, Shao W, et al. Thermoanaerobacter spp. control ethanol pathway via transcriptional regulation and versatility of key enzymes. Metab. Eng. (2010) 12:420–8. 10.1016/j.ymben.2010.06.001 [DOI] [PubMed] [Google Scholar]
- 50.Ferrarini MG, Siqueira FM, Mucha SG, Palama TL, Jobard É, Elena-Herrmann B, et al. Insights on the virulence of swine respiratory tract mycoplasmas through genome-scale metabolic modeling. BMC Genomics. (2016) 17:353. 10.1186/s12864-016-2644-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hames C, Halbedel S, Hoppert M, Frey J, Stülke J. Glycerol metabolism is important for cytotoxicity of Mycoplasma pneumoniae. J Bacteriol. (2009) 191:747–53. 10.1128/JB.01103-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinlan CL, Goncalves RL, Hey-Mogensen M, Yadava N, Bunik VI, Brand MD. The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I. J Biol Chem. (2014) 289:8312–25. 10.1074/jbc.M113.545301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. (2005) 3:679–87. 10.1038/nrmicro1204 [DOI] [PubMed] [Google Scholar]
- 54.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. (2000) 405:299–304. 10.1038/35012500 [DOI] [PubMed] [Google Scholar]
- 55.Guo FB, Wei W. Prediction of genomic islands in three bacterial pathogens of pneumonia. Int J Mol Sci. (2012) 13:3134–44. 10.3390/ijms13033134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hentschel U, Steinert M, Hacker J. Common molecular mechanisms of symbiosis and pathogenesis. Trends Microbiol. (2000) 8:226–31. 10.1016/S0966-842X(00)01758-3 [DOI] [PubMed] [Google Scholar]
- 57.Bai F, Ni B, Liu M, Feng Z, Xiong Q, Xiao S, et al. Mycoplasma hyopneumoniae-derived lipid-associated membrane proteins induce apoptosis in porcine alveolar macrophage via increasing nitric oxide production, oxidative stress, caspase-3 activation. Vet Immunol Immunopathol. (2013) 155:155–61. 10.1016/j.vetimm.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 58.Jiang F, He J, Navarro-Alvarez N, Xu J, Li X, Li P, et al. Elongation factor Tu and heat shock protein 70 are membrane-associated proteins from Mycoplasma ovipneumoniae capable of inducing strong immune response in mice. PLoS ONE. (2016) 11:e0161170. 10.1371/journal.pone.0161170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leal Zimmer F, Paludo GP, Moura H, Barr JR, Ferreira HB. Differential secretome profiling of a swine tracheal cell line infected with mycoplasmas of the swine respiratory tract. J Proteomics. (2019) 192:147–59. 10.1016/j.jprot.2018.08.018 [DOI] [PubMed] [Google Scholar]
- 60.Yu Y, Wang H, Wang J, Feng Z, Wu M, Liu B, et al. Elongation factor thermo unstable (EF-Tu) moonlights as an adhesin on the surface of Mycoplasma hyopneumoniae by binding to fibronectin. Front Microbiol. (2018) 9:974. 10.3389/fmicb.2018.00974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arfi Y, Minder L, Di Primo C, Le Roy A, Ebel C, Coquet L, et al. MIB-MIP is a mycoplasma system that captures and cleaves immunoglobulin G. Proc Natl Acad Sci USA. (2016) 113:5406–5411. 10.1073/pnas.1600546113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grover RK, Zhu X, Nieusma T, Jones T, Boreo I, MacLeod AS, et al. A structurally distinct human mycoplasma protein that generically blocks antigen-antibody union. Science. (2014) 343:656–61. 10.1126/science.1246135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nottelet P, Bataille L, Gourgues G, Anger R, Lartigue C, Sirand-Pugnet P, et al. (2021) The mycoplasma surface proteins MIB and MIP promote the dissociation of the antibody-antigen interaction. (2021) Sci Adv. 7:eabf2403. 10.1126/sciadv.abf2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maglennon GA, Cook BS, Deeney AS, Bossé JT, Peters SE, Langford PR, et al. Transposon mutagenesis in Mycoplasma hyopneumoniae using a novel mariner-based system for generating random mutations. Vet Res. (2013) 44:124. 10.1186/1297-9716-44-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maglennon GA, Cook BS, Matthews D, Deeney AS, Bossé JT, Langford PR, et al. Development of a self-replicating plasmid system for Mycoplasma hyopneumoniae. Vet Res. (2013) 44:63. 10.1186/1297-9716-44-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for amplification.
Information of single nucleotide polymorphisms (SNP) and indels analysis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI GenBank; accession no.'s CP038641.1 and CP058578.1.