Significance
Based on “subfossil” skeletal remains it is known that multiple now-extinct giant lemur (primate) species with estimated body masses of up to ∼160 kg survived on Madagascar into the past millennium. In this study, we used ancient DNA methods to sequence the nuclear genome of one of these megafaunal lemurs, Megaladapis edwardsi (∼85 kg). With the power of the nuclear genome, we robustly resolved the phylogenetic relationship between Megaladapis and other lemurs, which had been a lingering uncertainty. We also identified multiple signatures of past positive natural selection across the M. edwardsi genome that support reconstructions of this taxon as a large-bodied, specialized folivore.
Keywords: paleogenomics, megafaunal extinction, phylogenomics, convergent evolution, dietary reconstruction
Abstract
No endemic Madagascar animal with body mass >10 kg survived a relatively recent wave of extinction on the island. From morphological and isotopic analyses of skeletal “subfossil” remains we can reconstruct some of the biology and behavioral ecology of giant lemurs (primates; up to ∼160 kg) and other extraordinary Malagasy megafauna that survived into the past millennium. Yet, much about the evolutionary biology of these now-extinct species remains unknown, along with persistent phylogenetic uncertainty in some cases. Thankfully, despite the challenges of DNA preservation in tropical and subtropical environments, technical advances have enabled the recovery of ancient DNA from some Malagasy subfossil specimens. Here, we present a nuclear genome sequence (∼2× coverage) for one of the largest extinct lemurs, the koala lemur Megaladapis edwardsi (∼85 kg). To support the testing of key phylogenetic and evolutionary hypotheses, we also generated high-coverage nuclear genomes for two extant lemurs, Eulemur rufifrons and Lepilemur mustelinus, and we aligned these sequences with previously published genomes for three other extant lemurs and 47 nonlemur vertebrates. Our phylogenetic results confirm that Megaladapis is most closely related to the extant Lemuridae (typified in our analysis by E. rufifrons) to the exclusion of L. mustelinus, which contradicts morphology-based phylogenies. Our evolutionary analyses identified significant convergent evolution between M. edwardsi and an extant folivore (a colobine monkey) and an herbivore (horse) in genes encoding proteins that function in plant toxin biodegradation and nutrient absorption. These results suggest that koala lemurs were highly adapted to a leaf-based diet, which may also explain their convergent craniodental morphology with the small-bodied folivore Lepilemur.
Madagascar is exceptionally biodiverse today. Yet, the island’s endemic diversity was even greater in the relatively recent past. Specifically, there is an extensive “subfossil” record of now-extinct Malagasy fauna, with some of these species persisting until at least ∼500 y B.P. (1). The Late Holocene extinction pattern in Madagascar resembles other “megafaunal extinction” patterns in that it is strikingly body-mass structured, with the majority of extinct subfossil taxa substantially larger than their surviving counterparts. For example, the average adult body mass of the largest of the ∼100 extant lemur (primates) species is 6.8 kg (2), well below that of the 17 described extinct subfossil lemur taxa, for which estimated adult body masses ranged from ∼11 kg to an incredible ∼160 kg (3).
Despite a tropical and subtropical environment in which nucleotide (nt) strands rapidly degrade, in a select subset of Malagasy subfossil samples, ancient DNA (aDNA) is sufficiently preserved for paleogenomic analysis (4–10). In our group’s previous study (6), we reconstructed complete or near-complete mitochondrial genomes from five subfossil lemur species, with population-level data in two cases. As part of that work, we identified one Megaladapis edwardsi (body mass ∼85 kg) (3, 11) sample with an especially high proportion of endogenous aDNA. We have subsequently performed additional rounds of extraction and sequencing of this sample to amass sufficient data for studying the M. edwardsi nuclear genome.
In this study, we analyzed the M. edwardsi nuclear genome to help reconstruct subfossil lemur behavioral ecology and evolutionary biology. Our approach included an unbiased search across the genome for Megaladapis-specific signatures of positive selection at the individual gene level. We also searched for striking patterns of genomic convergence with a set of biologically diverse extant mammals across sets of functionally annotated genes. The results from these analyses may serve to extend current hypotheses or to offer potentially unexpected insights into the evolutionary biology of Megaladapis.
Additionally, we aimed to resolve lingering uncertainty over Megaladapis phylogenetic relationships with other lemurs. At one point, a sister taxon relationship between Megaladapis and extant sportive lemurs (genus Lepilemur) was inferred based on craniodental similarities (3, 12). A different phylogeny was estimated, however, following the successful recovery of several hundred base pairs (bp) of the Megaladapis mitochondrial genome in several early aDNA studies (4, 5). Specifically, Megaladapis and the extant Lemuridae (genera Eulemur, Lemur, Varecia, Prolemur, and Hapalemur) formed a clade to the exclusion of Lepilemur. Our more recent aDNA study (6) resolved a similar phylogeny but with greater confidence (e.g., 87% bootstrap support) given the near-complete recovery of the Megaladapis mitochondrial genome (16,714 bp). Still, the mitochondrial genome is a single, nonrecombining locus; in certain cases, true species-level phylogenies are not reconstructed accurately from mitochondrial DNA alone (13). Most recently, Herrera and Dávalos (14) estimated a “total evidence” phylogeny by analyzing the combination of both morphological and genetic characters. Their result was dissimilar to each of the above phylogenies, instead supporting an early divergence of the Megaladapis lineage from all other non-Daubentonia (aye-aye) lemurs.
Because the nuclear genome is comprised of thousands of effectively independent markers of ancestry, we expected to achieve a more definitive phylogenetic result with our Megaladapis paleogenome sequence. To distinguish among competing phylogenetic hypotheses, we also needed to generate genome data for representatives of the extant Lemuridae and Lepilemur lineages, which we did for Eulemur rufifrons (red-fronted lemur) and Lepilemur mustelinus (greater sportive lemur), respectively. We aligned the three lemur genome sequences with those previously published for extant lemurs Daubentonia madagascariensis (aye-aye) (15), Microcebus murinus (gray mouse lemur) (16), and Propithecus diadema (diademed sifaka) (17), and with 47 nonlemur outgroup species, for phylogenetic and evolutionary analyses.
Results
We used a high-volume shotgun sequencing approach to reconstruct the M. edwardsi nuclear paleogenome. From the well-preserved M. edwardsi specimen (UA [Université d'Antananarivo] 5180; a mandible from Beloha Anavoha, extreme southern Madagascar; 1,475 ± 65 calibrated years B.P.) (1) identified in our previous study (6), we performed additional rounds of aDNA extraction (total extractions = 3), double-stranded library preparation (total libraries = 9), and massively parallel high-throughput “shotgun” sequencing (total = 15 lanes on Illumina HiSeq 2000 and 2500 with 75 bp paired-end reads) to amass sufficient sequence data (total = 313 gigabases) for studying the nuclear genome despite the still relatively low endogenous nuclear DNA content (6.35%; see Materials and Methods and SI Appendix, Figs. S1 and S2 and Table S1).
The size distribution (18, 19) and damage pattern (20–22) (potentially damaged nts were subsequently masked; see Materials and Methods) of putative M. edwardsi sequence reads were both characteristic of authentic aDNA (SI Appendix, Figs. S3 and S4). Furthermore, we estimated a low 1.2% modern human DNA contamination rate among putative M. edwardsi reads (see Materials and Methods and SI Appendix, Table S2), consistent with or below that reported in other paleogenomic studies (23, 24) and overall contributing negligibly to the nuclear gene sequence reconstructions we report in this paper.
The set of quality-filtered and damage-masked M. edwardsi sequence reads were aligned to a version of the human reference genome (hg19) masked to contain only Reference Sequence (RefSeq) gene exons ±100 bp. We used a conservative approach to reconstruct orthologous, single-copy M. edwardsi gene coding region sequences with 2× minimum sequence coverage per position (median proportion of sites reconstructed per gene = 0.29; SI Appendix, Fig. S5 and Table S3). Exon sequences reconstructed from shotgun sequence reads from the five modern lemur species (including two with data newly generated for this study) and a golden snub-nosed colobine monkey (Rhinopithecus roxellana) were also included in our analysis (25). All of these reconstructed sequences were integrated with a canonical gene exon alignment of 46 vertebrate species for an overall total alignment with 53 species.
Reconstructing a Nuclear Genome–Based Phylogeny of Extinct and Extant Lemurs.
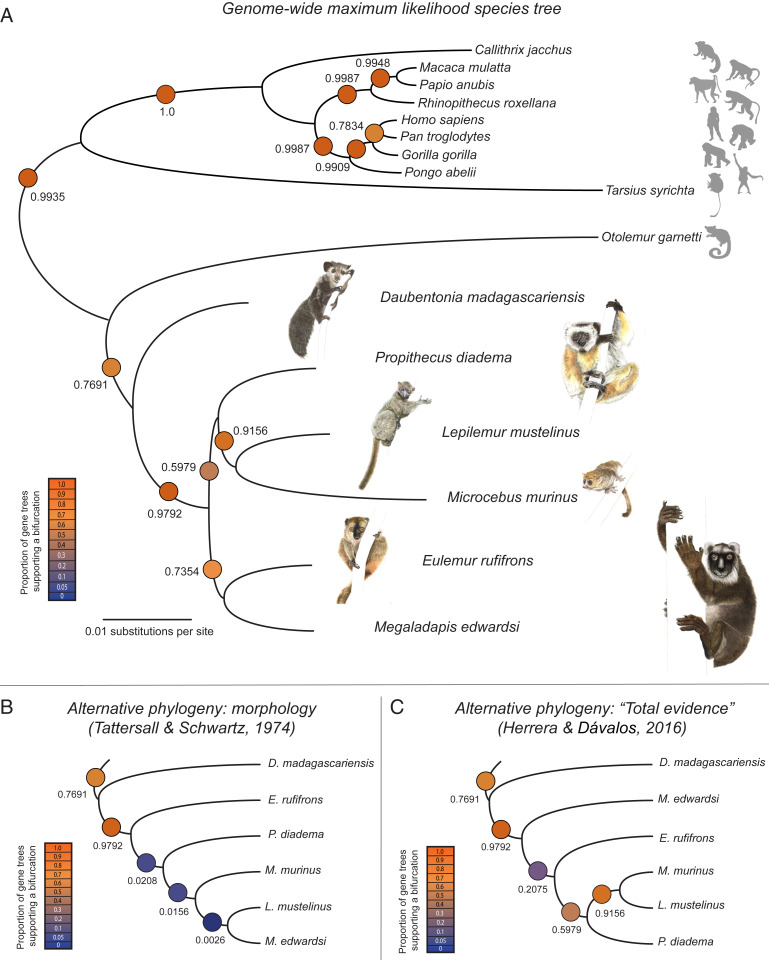
We used a genome-wide maximum likelihood approach (26) to estimate the phylogenetic placement of M. edwardsi among primates. We first considered alignment data from n = 896 genes for which at least 50% of Megaladapis sites were represented in our 2× minimum sequence coverage per position dataset (1.07 million bp in total) and estimated a single unrooted phylogeny from the concatenated alignment (Fig. 1A). This extinct and extant lemur phylogeny, estimated from concatenated nuclear genome sequences, matches the previously reconstructed mitochondrial genome–based phylogeny (6).
Fig. 1.
Phylogenetic analyses with the M. edwardsi nuclear genome sequence. (A) Phylogeny estimated from maximum likelihood analysis of a concatenated alignment of the n = 896 genes for which at least 50% of M. edwardsi sites were represented at minimum 2× sequence coverage (1.07 million bp in total). The heat map and printed values represent the proportions of strong phylogenetic signal–individual gene trees (a total of n = 771 genes with ≥90% mean bootstrap support and ≥20% of sites present across all lemurs in the study) supporting each bifurcation. Watercolor illustrations by Joel Borgerson. Silhouette images courtesy of PhyloPic (see Materials and Methods for attribution details). (B) The proportions of our strong phylogenetic signal–individual gene trees that support each bifurcation in a previously hypothesized phylogeny are inferred based on craniodental traits [Tattersall and Schwartz (12)]. (C) The proportions of our strong phylogenetic signal–individual gene trees that support each bifurcation in a previously published phylogeny are based on the analysis of a combined morphological and mtDNA dataset [Herrera and Dávalos (14)].
Second, we analyzed a larger database of n = 11,944 genes with aligned nts present across at least 20% of the sites per gene across all lemurs in our study (including M. edwardsi). For each of these genes, we estimated an independent phylogeny using the same model as above and performed 100 bootstrap replicates. For each of these gene trees, the mean level of bootstrap support across all branch bipartitions was calculated as a measure of gene tree phylogenetic signal (27). Among the 11,944 gene trees, the overall average mean bootstrap support value was 74.10% (SD = 12.65%; range = 7.69 to 98.85%; SI Appendix, Fig. S6).
We next considered the phylogenetic properties of the subset of individual gene trees with ≥90% mean bootstrap support. Of these n = 771 “strong phylogenetic signal” individual gene trees, 191 (25%) exactly matched the full species tree based on the concatenated gene sequences. The species tree was well supported at nearly every individual node (Fig. 1A). The placement of Megaladapis as a sister taxon to Eulemur was supported in 567 out of the 771 strong phylogenetic signal gene trees (74%). We used DiscoVista (28) to help confirm the relatively strong support for this topology (SI Appendix, Fig. S7).
We also explicitly examined the level of support for the two alternative, previously reported phylogenies involving Megaladapis. First, scholars have hypothesized common ancestry for Megaladapis and Lepilemur to the exclusion of other lemurs based on a shared set of derived craniodental traits (e.g., the absence of permanent upper incisors, premolar proportional size similarity, and an expanded articular facet on the mandibular condyle) between these two taxa (12, 29). Yet, a Megaladapis–Lepilemur sister taxon relationship was observed in only 2 of the 771 strong phylogenetic signal gene trees in our nuclear genome dataset (0.26%; Fig. 1B). Second, Herrera and Dávalos (14) combined genetic data [for M. edwardsi: sequences from two mitochondrial genes (6)] and morphological trait variables (for M. edwardsi: n = 169 traits) to reconstruct a “total evidence” phylogeny in which Megaladapis was placed as a sister taxon to a clade of all other non-Daubentonia lemurs. This bipartition was observed in 160 of the 771 strong phylogenetic signal gene trees (20.75%; Fig. 1C), the second-most observed result but a substantial ∼3.5-fold reduction compared to the Megaladapis–Eulemur sister taxon relationship (Fig. 1A).
Specific support for the position of M. edwardsi was further confirmed by comparison to its nearest neighbors. We counted the number of well-supported M. edwardsi and E. rufifrons bipartitions relative to the total number of other well-supported M. edwardsi groupings (equivalent to the Fig. 1A branch support values). Among the 771 gene trees with ≥ 90% mean bootstrap support, the M. edwardsi and E. rufifrons pairing itself is either supported or specifically conflicted by at least 90% of bootstrap replicates in 334 gene trees. Among these 334 Megaladapis placement-informative gene trees, the main topology is supported by 269 trees (80.5%), with 53 trees (15.9%) supporting the Herrera and Dávalos (14) topology, and the remaining 12 trees (3.6%) representing 8 other configurations.
Evolutionary Genomics.
The M. edwardsi nuclear genome sequence contains a wealth of information about the evolutionary biology of this extinct species. Reliably equating between-species nt differences to adaptive phenotypes is a considerable challenge regardless of genome quality (30, 31); in our case here, the challenge is compounded by stochastic patterns of paleogenomic sequence coverage. Still, even with incomplete data, the vast expanse of the nuclear genome provides abundant opportunities to identify potential signatures of past natural selection. Combined with inferences of likely gene functions and pathways based on studies conducted in other species, these results can contribute to our understandings of M. edwardsi phenotypic form, function, and genetically mediated behavior.
Nonsynonymous versus synonymous substitution rates.
One comparative evolutionary genomics approach is to compare the ratios of the rates (d) of nonsynonymous (N; amino acid–changing) to synonymous (S; not amino acid–changing) substitutions (dN/dS) across a gene. While not all synonymous mutations are completely neutral with respect to function and fitness (32), the fates of these mutations at least more closely reflect neutrality than those of nonsynonymous mutations. For the vast majority of genes in any interspecies comparison dN/dS << 1 because the majority of nonsynonymous mutations are detrimental to fitness and are typically removed from populations by purifying selection. However, in rare cases, the repeated emergence of strongly adaptive nonsynonymous mutations at different positions along the same gene, leading to repeated fixation by positive selection, can result in dN/dS >> 1.
We used a maximum likelihood–based method implemented in the program PAML (Phylogenetic Analysis Using Maximum Likelihood) (33, 34) to estimate dN/dS along each ancestral and terminal branch in our extant and extinct lemur genomic phylogeny. This analysis was restricted to the 3,342 genes with sufficient and high-quality sequence data for all lemurs and three outgroups (Homo sapiens, Pan troglodytes, and Gorilla gorilla; Materials and Methods). Because there are considerably fewer S than N sites per gene (e.g., in our dataset; 2.7 times fewer S sites overall), we further limited stochasticity in the dN/dS statistic by computing a single, per-lineage dSgenome value for use as the denominator in each gene-specific dN/dS calculation (15) (dN/dSgenome) for that lineage.
We considered the 53 genes (1.6% of 3,342) with Megaladapis lineage-specific dN/dSgenome > 1.5 to be the strongest positive selection candidates for this extinct subfossil lemur in our dataset (SI Appendix, Table S4). When we tested whether this set of genes was significantly enriched for any known biological functions or biochemical pathways, we found none following multiple test corrections (35). Still, included among these 53 candidates were several individual loci with potentially intriguing links to hypotheses concerning Megaladapis evolutionary biology and behavioral ecology.
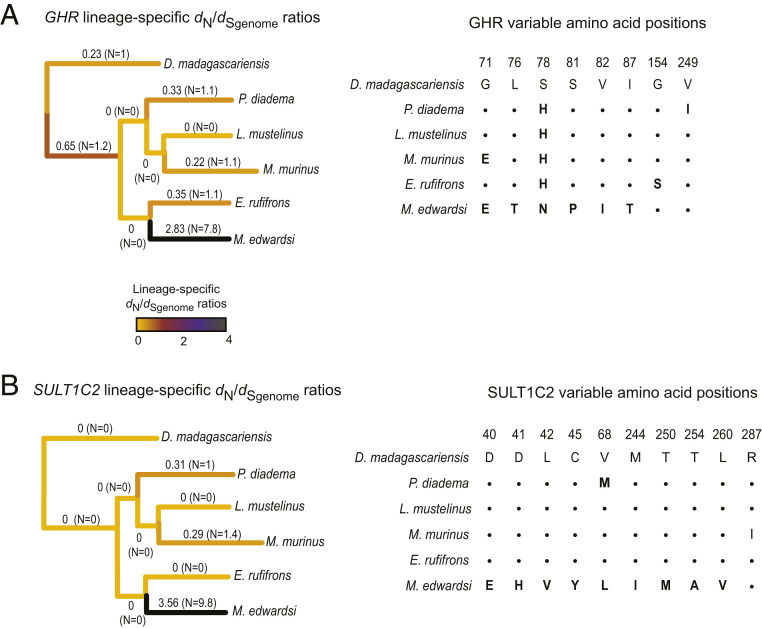
For example, M. edwardsi lineage dN/dSgenome = 2.83 for the growth hormone receptor (GHR) gene (n = 7.8; S = 2.0) whereas dN/dSgenome values for all other terminal and ancestral lemur branches range from 0.0 to 0.65 (Fig. 2A and SI Appendix, Fig. S8A). Biological activity of growth hormone (GH) is mediated by interaction with the GHR protein. Genetic changes in the GH/GHR pathway can result in marked body size phenotypes (36–38). Thus, the pattern of Megaladapis-specific positive selection in GHR marks this gene as a candidate contributor to the evolved gigantism in this lineage [estimated M. edwardsi body mass ∼85 kg (3, 11) versus maximum ∼6.8 kg for any extant lemur (2)].
Fig. 2.
Lineage-specific dN/dS ratios for GHR and SULT1C2. Using a maximum likelihood approach implemented in PAML, lineage-specific ratios of the rates (d) of nonsynonymous (N) versus synonymous (S) substitution along ancestral and terminal branches estimated with a maximum likelihood–based approach for (A) the growth hormone receptor (GHR) and (B) sulfotransferase 1C2 (SULT1C2) genes. For each branch, the dS denominator is based on the genome-wide synonymous substitution rate. dN/dSgenome estimates are recorded next to each branch and depicted by the heat map. The estimated number of N substitutions for each branch are reported within the parentheses. Branch lengths shown are based on those from Fig. 1A rather than these individual genes. For each gene, alignments of inferred amino acid residues for the encoded proteins are shown for all variable positions. Amino acid residues identical to those for D. madagascariensis are depicted with “.”, and amino acid position numbers are based on the human RefSeq (hg19/GRCh37).
For the sulfotransferase 1C2 (SULT1C2) gene, M. edwardsi lineage dN/dSgenome = 3.56 (n = 9.8; S = 3.6) compared to a range of 0 to 0.35 for all other branches (Fig. 2B and SI Appendix, Fig. S8B). SULT1C2 catalyzes reactions that detoxify xenobiotic compounds, including phenolics, to facilitate removal of potentially harmful metabolites from the body (39, 40). Phenolics are toxic compounds common in leafy plants (41). Based on craniodental and postcranial gross morphology, biomechanical analyses, dental microwear and topographic analyses, and biogeochemistry, M. edwardsi is inferred to have been highly folivorous (42–45). Thus, SULT1C2 nonsynonymous substitutions may have been part of a suite of adaptations to folivory in the Megaladapis lineage (see Convergent genomic evolution, below).
Convergent genomic evolution.
The gene-by-gene dN/dS approach presented above (in Nonsynonymous versus synonymous substitution rates) provides limited opportunity to identify signatures of past positive selection, as detection requires a history of repeated fixation of nonsynonymous substitutions within a gene beyond the background synonymous substitution accumulation rate. This combination can be especially rare on relatively longer branches such as the Megaladapis terminal lineage [i.e., estimated 27.3 ± 4.2 My divergence from last common ancestor with Eulemur (6)], resulting in likely high false-negative rates relative to the true occurrence of past positive selection.
Therefore, we also used a convergent evolution-based approach to identify potential signatures of positive selection on the Megaladapis lineage. We scanned across biological functional categories (i.e., groups of genes linked by known function based on the Gene Ontology [GO] database) (46) to identify those functions with significantly higher proportions of convergent amino acid substitutions between Megaladapis and a distant species or clade relative to the genome-wide rate of convergence. We performed this analysis using amino acid alignments of 21,520 genes for 53 total species, comprised of the 6 lemurs in our study (including Megaladapis) along with 47 nonlemur vertebrates (SI Appendix, Fig. S9). In combination with extensive available knowledge for many of the extant species in our dataset, these results can be used to develop or extend hypotheses of Megaladapis evolutionary biology and behavioral ecology.
Specifically, for each possible comparison between Megaladapis and a distant taxon (either an individual species or a clade of species), we searched for codon positions with the following pattern of convergent evolution: Megaladapis and the distant comparison taxon shared the same predicted amino acid, while the sister species to Megaladapis (E. rufifrons) and the outgroup M. murinus (we also performed separate analyses with P. diadema as an outgroup; SI Appendix, Fig. S10) shared a different amino acid, and the sister and outgroup species to the comparative taxon likewise shared a different amino acid. For each gene we also counted the number of analyzable amino acid positions (see Materials and Methods). We then summed the numbers of convergent and analyzable sites across all genes represented in each GO term. For GO categories with ≥ 5 convergent amino acids, we tested whether the proportion of convergent sites was significantly different from expected based on the genome-wide ratio.
Using this approach, we performed 52 different comparisons between Megaladapis and a distant species/clade. Per comparison, we identified an average of 0.54 (SD = 0.90) GO categories significantly enriched for convergent amino acids at a low false discovery rate (FDR < 0.05). Within any particular comparison, significant functional categories were often nested within other significant categories, as expected given the structure of the GO database.
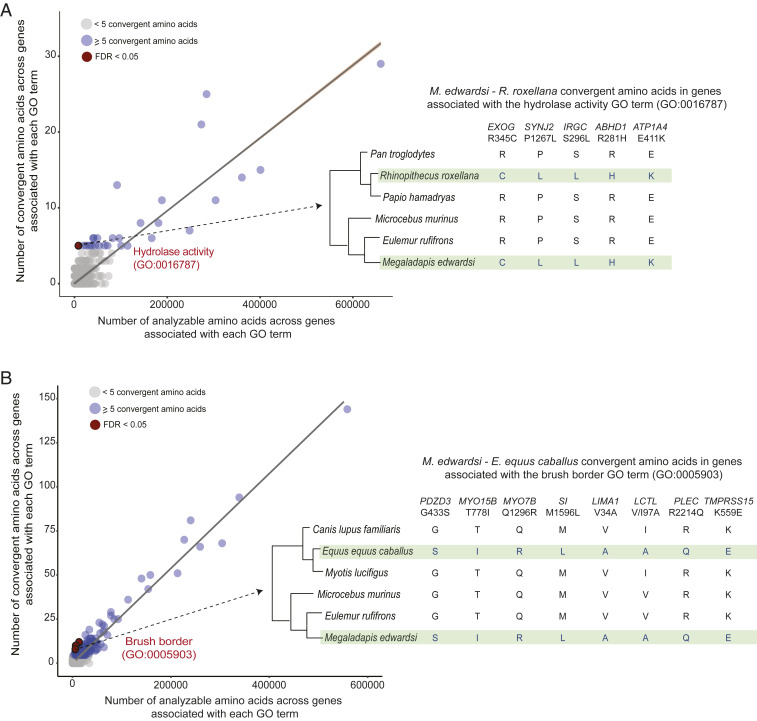
Included among the most striking convergent evolution results were several patterns that may reflect Megaladapis adaptations to folivory. For example, between M. edwardsi and the golden snub-nosed monkey (R. roxellana), a colobine primate with a lichen- and leaf-specialized diet, there were 5 total convergent amino acid positions across 5 different hydrolase activity genes (GO: 0016787; 8,535 total analyzable sites) versus an expectation of only 0.48 convergent sites (genome-wide convergent amino acids = 73; genome-wide analyzable positions = 1,273,496; Fisher’s exact test; P = 0.00018; FDR = 0.0054; Fig. 3A). Among the identified hydrolase activity genes were EXOG and ATP1A4, which encode proteins involved in the metabolism of xenobiotics, which is critically important for many folivores given their exposure to plant secondary compounds (41, 47). Moreover, while families of genes involved in xenobiotic metabolism have expanded via gene duplication in golden snub-nosed monkeys (25) and other herbivores (48, 49), in carnivores such genes are disproportionately pseudogenized (50).
Fig. 3.
Convergent amino acid evolution between M. edwardsi and extant herbivores. Results from scans to identify GO functional categories with unusual proportions (relative to genome-wide expectations) of inferred convergent amino acid positions between (A) M. edwardsi and the folivore R. roxellana and (B) M. edwardsi and the herbivore E. caballus. Convergent positions are those with identical residues between M. edwardsi and the comparison species but for which the sister and an outgroup species (for each of the comparison species) share a distinct amino acid residue (Right). (Left) The number of analyzable amino acid positions (aligned amino acids for all six species in the analysis plus identical residues in each sister species–outgroup pair) and convergent amino acid positions for each GO term. For terms with ≥5 convergent amino acids, we tested whether the proportion of convergent sites was significantly different from expected based on the genome-wide ratio and computed FDRs to account for the multiple tests. For two highlighted GO terms, all convergent amino acid positions between M. edwardsi and the comparison species along with gene name and position (based on the human RefSeq) are shown.
We also identified 8 total convergent amino acids across 8 different brush border genes (GO: 0005903) between Megaladapis and horse (Equus caballus) versus an expectation of 1.727 convergent sites (brush border gene analyzable sites = 5,787; genome-wide convergent positions = 316; genome-wide analyzable positions = 1,058,758; Fisher’s exact test; P = 0.00046; FDR = 0.0307; Fig. 3B). The brush border is the microvilli-covered surface of epithelial cells, for example the intestinal lining, which helps to facilitate the absorption and hydrolysis of nutrients (via brush border enzymes embedded in the microvilli) (51). Brush border genes with Megaladapis–horse convergent amino acids include LIMA1, which encodes an actin-binding protein with a role in cholesterol homeostasis (52), and the microvilli myosin-encoding gene MYO7B, which maintains brush border action (53). The digestive biology of these brush border proteins in horses is incompletely known, but the connection between the herbivorous diet of horses and the proposed specialized folivory of Megaladapis warrants further investigation into the functional impacts of these convergent amino acid changes (54–56).
A potential limitation of our analytical framework is that genes in certain GO categories might be relatively prone to amino acid convergence due to their intrinsic properties (e.g., different ancestral amino acid frequency distributions). Thus, the Megaladapis–Rhinopithecus GO: 0016787 and Megaladapis–Equus GO: 0005903 results highlighted above could theoretically reflect neutral patterns of convergence for these GO categories rather than histories of evolutionary adaptation (57). To consider this possibility, we conducted a post hoc analysis in which we identified the numbers of convergent and analyzable amino acid positions within each of the GO: 0016787 and GO: 0005903 categories and also genome-wide for all n = 1,856 possible taxonomic comparisons (excluding others involving Megaladapis) from our multispecies alignment. The Megaladapis–Rhinopithecus GO:0016787 category to genome-wide convergent evolution ratio was the most extreme (1/1,856) of all taxonomic comparisons for that GO term, with a Fisher’s exact test P value in the top 1.5% of comparisons (15/1,856). Meanwhile, the Megaladapis–Equus GO: 0005903 category to genome-wide ratio and P value were both in the top 0.3% (6/1,856 and 6/1,856) of all GO: 0005903 comparisons (SI Appendix, Fig. S11 and Table S5). These results support the notion that our primary findings are of potential evolutionary biology interest.
Discussion
For this study, we generated a nuclear genome sequence dataset for an extinct nonhominin primate species. Paleogenomic approaches have immense potential for helping to resolve phylogenetic relationships and for insights into the evolutionary biology of now-extinct taxa and ancestral clades (58). Our study follows the recent analysis of a nuclear genome sequence from a ∼5,800 y-old baboon (extant Papio ursinis) (59), the sequencing of a mitochondrial genome and seven nuclear genes from an extinct Caribbean monkey (Xenothrix mcgregori) (60), and prior mitochondrial DNA sequencing studies of multiple extinct subfossil lemur species (4–7). A promising outcome of our study and others is the recovery of paleogenomic data from tropical and subtropical taxa. We anticipate continued future expansion of aDNA work in biologically diverse tropical and subtropical habitats. In addition to paleogenomics, we are following continuing developments in the field of paleoproteomics (61) for similar insights from samples with inadequate aDNA preservation, including those considerably older. An exciting recent paper presenting and analyzing the enamel proteome of the extinct orangutan relative Gigantopithecus blacki demonstrated this point (62).
For the present study, we felt fortunate to generate M. edwardsi nuclear genome sequence data. Madagascar’s tropical and subtropical conditions severely challenge aDNA preservation. To date, our aDNA laboratory has screened multiple hundreds of extinct subfossil lemur samples (many had been collected previously for non-aDNA analyses). Yet we have considered endogenous DNA preservation sufficient in only two samples to attempt (at least with current technology) shotgun sequencing of the nuclear genome. The M. edwardsi sample UA 5180 studied here was the best preserved.
Phylogenetic Resolution of a Rapid Lemur Radiation with Incomplete Lineage Sorting.
Our ability to analyze sequence data from thousands of loci from across the M. edwardsi nuclear genome helped us to resolve ongoing extant–extinct lemur phylogenetic uncertainty, particularly the branching order of Lemuridae–Megaladapidae, Lepilemuridae–Cheriogalediae, and Indriidae. Prior analyses of mitochondrial DNA (mtDNA) sequence data from M. edwardsi and extant lemurs showed that Megaladapis and extant Lepilemur were likely not sister taxa (4, 6), as previously had been hypothesized based on morphological similarities (3, 12). Both of these phylogenetic reconstructions positioned Megaladapis distinctly from yet another, more recent phylogenetic analysis that was based on an extensive morphological plus mtDNA combined dataset (14).
A sister taxon relationship between Megaladapis and extant Lemuridae (represented in our study by E. rufifrons) was robustly supported in our nuclear genome–based analysis (Fig. 1A) relative to alternative phylogenies (Fig. 1 B and C). This result is consistent with the prior phylogenetic reconstructions based on mtDNA sequences only. Our nuclear phylogeny does not support a close relationship of the Megaladapidae and the Lepilemuridae; instead, the latter is the sister to the Cheirogaleidae, and the Lepilemurid–Cheirogaleid clade is the sister to a clade comprising the Archaeolemuridae, Indriidae, and Paleopropithecidae.
We propose two nonmutually exclusive explanations for the past phylogenetic inference discrepancies. First, based on patterns of dental microwear (42–44), dental topography (45), craniodental features (12, 29), infraorbital foramen size (IOF) (63), and isotopic data (64–66) with further support from our evolutionary genomic results, Megaladapis was likely a specialized folivore. Meanwhile, the diets of sportive lemurs (Lepilemur spp.) are also highly folivorous (67–69). Megaladapis–Lepilemur morphological similarities may thus represent convergent biological adaptations to similar behavioral ecology, rather than shared inheritance from a common ancestor. For example, the absence of upper incisors in both taxa could represent convergent adaptation to folivory in the context of a plesiomorphic lower toothcomb. Such processes could affect phylogenetic analyses based on morphological features.
Second, a rapid early diversification of lemur lineages (other than Daubentonia) occurred ∼34 Mya (6). Potentially, this rapid radiation was triggered by the Eocene–Oligocene extinction event, a period of dramatic climate shift (global cooling) and flora/fauna turnover (forest reduction and niche fragmentation) (6, 70, 71). Alternatively, based on recent African fossil evidence, there may have been two separate lemur colonizations of Madagascar—one, by an ancestor exclusive to the Daubentonia lineage and another by an ancestor of all non-Daubentonia lemurs (which could have occurred at ∼34 Mya during the Cenozoic) (72). Regardless, rapidpt radiations like this likely complicate lemur phylogenetic reconstructions.
Specifically, within a closely timed radiation, a proportion of ancestral genetic variants may remain polymorphic across multiple lineages through the duration of splitting events, to only subsequently become fixed—potentially with a fixation pattern that is not representative of species-level relationships. This “incomplete lineage sorting” process (73, 74) can lead to conflicting locus-to-locus phylogenetic signals, thereby resulting in a minority of gene trees differing from the overall species tree.
Incongruences due to incomplete lineage sorting are not uncommon among primates. For example, this phenomenon has been well-documented for humans, chimpanzees, and gorillas. Across the autosomal nuclear genome of these species, ∼30% of alignments support incongruent branching orders—(chimpanzee, (human, gorilla)) or (human, (chimpanzee, gorilla))—instead of the true species order (gorilla, (human, chimpanzee)) (75). Indeed, this finding is replicated by our own gene tree/species tree phylogenetic analysis, with results from 604 of 771 genes (78.3%) supporting the true (gorilla, (human, chimpanzee)) phylogeny (Fig. 1A) versus results from 78/771 genes (10.12%) supporting (chimpanzee, (human gorilla)) and 89/771 (11.54%) supporting (human, (gorilla, chimpanzee)) incongruent branching orders.
Using the same set of genes, we observed a similar signature of incomplete lineage sorting among lemur clades involving Megaladapis. Specifically, the ((Megaladapis, Eulemur), all other non-Daubentonia lemurs) typology was supported by 567 of the 771 gene trees with strong phylogenetic signal (73.5%; Fig. 1A). The second-most common branching order involving Megaladapis (Daubentonia, (Megaladapis, all other lemurs)) was supported by 160/771 (20.8%) of gene trees (Fig. 1C). This signature of incomplete lineage sorting strongly supports the notion of a rapid radiation among non-Daubentonia lemurs on Madagascar, possibly immediately following either a mass extinction event (6, 76) or a non-Daubentonia lemur colonization of the island (72).
Evolutionary Genomic Reconstruction of Megaladapis as a Large-Bodied Specialized Folivore.
We approached our evolutionary genomic analyses with care, and we suggest cautious interpretation of the results. The Megaladapis lineage branch length is relatively long, making it difficult to identify individual genes with histories of positive selection based on the detection of excessive nonsynonymous substitution fixation rates, especially with the stochastic sequence coverage of our dataset (we limited this analysis to sites with ≥2× coverage). Still, our set of candidate genes with dN/dS-based signatures of positive selection on the Megaladapis lineage included the growth hormone receptor (GHR), a finding of interest given the large reconstructed body size of M. edwardsi (∼85 kg) (3, 11), one of the “giant” extinct subfossil lemurs. Yet, we did not observe an enrichment for body size or growth-related functional pathways among the overall candidate gene set. Given that body size variation is often highly polygenic (77–79), the absence of such an enrichment is not unexpected.
We have more confidence in the connection of several evolutionary genomic results to potential Megaladapis diet-related adaptations. Specifically, we identified enrichments for convergent amino acid evolution between M. edwardsi and the golden snub-nosed monkey (a folivore) across genes with hydrolase activity functions and between M. edwardsi and horse (a grazing herbivore) across genes with brush border functions. Hydrolases help to break down plant secondary compounds (47), while brush border microvilli play crucial roles in nutrient absorption and hydrolysis in the gut (53, 56). Additionally, our set of candidate genes with dN/dS-based signatures of positive selection on the Megaladapis lineage included SULT1C2, which encodes an enzyme involved in the detoxification of toxic phenolic compounds common in leafy plants (39, 41). Interestingly, a recent genomic study of sifakas (genus Propithecus), extant lemurs with partially folivorous diets, also reported signatures of convergent evolution or potential parallel adaptation with nonlemur folivores in pathways and genes involved in nutrient absorption (including intestinal microvilli morphology) and xenobiotic metabolism (80). In the future, the biology of the Megaladapis molecular changes could be examined via colobine monkey and horse in vivo or in vitro studies or other functional evolutionary genomics approaches (81), as appropriate.
Our evolutionary genomic findings support developing reconstructions of Megaladapis as a specialized folivore. Specifically, molar microwear patterns are important proxies for inference of the diet of an individual animal in the weeks prior to its death; shearing tough foods, such as leaves, often produces scratches on the tooth surface, while consuming fruits with hard pericarps or hard seeds may lead to punctures or pits (44). M. edwardsi microwear patterns feature scratches characteristic of leaf eaters (42, 43), with similarities to extant primate folivores, including Presbytis entellus (42) and Lepilemur petteri (with a habitat in Madagascar overlapping that of M. edwardsi) (43). Furthermore, M. edwardsi dental topography features including low molar occlusal surface complexity and high “Dirichlet normal energy” (a measurement that roughly captures crown profile “relief” or, more precisely, changes in the direction of occlusal surface tangents) suggest adaptation for efficiency in shearing leaves (45).
Stable isotope data are also consistent with the reconstruction of a folivorous diet for Megaladapis. Specifically, carbon isotope ratios can help differentiate the relative contributions of C3 plants, C4 plants, and stem/leaf succulents, as well as nonphotosynthetic plant tissues (e.g., fruits, flowers) in the diets of extinct species (64, 65). The spiny thicket habitat in southern and southwest Madagascar is dominated by succulents with patches of C4 grasslands and C3 trees, yet M. edwardsi carbon isotope ratios ubiquitously suggest a C3-based, herbivorous leafy diet (64).
Several M. edwardsi craniodental traits suggest adaptations to a “browsing-via-plucking” mode of leaf eating, including the loss of upper incisors, ventrally flexed nasal bones, posteriorly expanded temporomandibular joint surfaces for compressive mastication, and a postcanine diastema (12, 29, 42, 82). Similar to koalas, M. edwardsi has a caudally positioned foramen magnum and limited midface projection relative to length, interpreted to facilitate greater head movement to facilitate direct foraging on leaves (12, 82).
Finally, variation in infraorbital foramen size is an osteological proxy for “maxillary mechanoreception” (i.e., how mammals use their “snouts” to acquire and process foods) and dietary inference, at least among primates: frugivores tend to have larger infraorbital foramen size areas relative to folivores and insectivores, perhaps reflecting adaptations for selecting and evaluating fruit (83). The relative infraorbital foramen size area of M. edwardsi is significantly smaller than that of any frugivorous extant lemur and is instead more similar to relatively more-strict extant lemur folivores (e.g., Lepilemur) (63).
Conclusion
Overall, our work highlights both the challenges and exciting prospects of nonhuman primate paleogenomics. Many nonhuman primates live in the tropics or subtropics, which can be challenging environments for aDNA recovery and analysis. In this study, we had the opportunity to focus a concerted shotgun sequencing effort on a particular M. edwardsi sample with higher-than-typical levels of aDNA preservation for a subfossil specimen from Madagascar. In the future, improved methods to extract DNA from tropical and subtropical samples (84) alongside further technological innovations may facilitate future recoveries of additional nuclear genome sequences from other extinct lemurs or nonlemur primates. For now, we are excited to have been able to analyze M. edwardsi nuclear genome sequences for insights into the evolutionary biology and behavioral ecology of this extinct subfossil lemur and to robustly resolve its phylogenetic relationship with other lemurs.
Materials and Methods
Sample Preparation and Sequencing of the M. edwardsi Nuclear Genome.
DNA extraction.
The UA 5180 mandible was sampled under a collaborative agreement with the Department of Paleontology and Biological Anthropology at the University of Antananarivo, Madagascar. All ancient materials were processed in dedicated sterile facilities with positive pressure at the Pennsylvania State University, with physically separate post-PCR processing facilities. As part of a previous study (6), we identified an M. edwardsi mandible, UA 5180, from the site of Beloha Anavoha, southern Madagascar (6), with sufficient endogenous DNA quality and quantity for a whole-nuclear genome shotgun sequencing effort. The UA 5180 specimen was directly AMS (Accelerator Mass Spectrometry) 14C dated (CAMS 142541) as part of a previous study (1). We have used a recently-updated calibration curve (SHCal20) (85) to recalibrate (86) the 14C age (1,640 ± 30) to 1,475 ± 65 cal y B.P. For this study, we prepared eight additional DNA extractions from the UA 5180 using an established protocol for animal hard tissue (87) and following our previously described subsampling strategy (6). Negative controls were included with every extraction and library preparation and assessed with gel electrophoresis, with no evidence of contamination. While the negative controls were not sequenced, we did estimate the proportion of human DNA contamination in each sequenced library (see Authenticity of M. edwardsi Genomic Data, below).
Library preparation.
We prepared a total of nine double-stranded libraries with barcoded adapters from specimen UA 5180 suitable for Illumina massively parallel sequencing platforms following the Meyer and Kircher protocol (88). We used 50 μL template as input for the initial blunt-end repair step without the enzymatic removal of uracil residues and abasic sites. The postreaction purification steps were carried out using the Qiagen MinElute PCR Purification kit after blunt-end repair and carboxyl-coated magnetic beads (Solid Phase Reversible Immobilization or SPRI) for the adapter ligation and fill-in steps. The final elution volume of 20 μL in TET (Tris EDTA-Tween-20) was then used as a template for the indexing reaction. Libraries were barcoded using a single unique P7 index primer where “xxxxxxx” represents the specific barcode for a library (200 nM, 5′-CAAGCAGAAGACGGCATACGAGATxxxxxxxGTGACTGGAGTTCAGACGTGT-3′) was added to each library (n = 9) with a universal IS4 forward primer (200 nM, 5′- AATGATAACGGCGACCGAGATCTACACTCTTTCCCTACACGACGCTCTT-3′) (88) in a 50 μL reaction that also included PCR buffer, 2 mM MgSO4, 200 μM dNTPs, and 2.5 U Platinum Taq High Fidelity DNA Polymerase (Thermo Scientific) prepared in the aDNA facility. Amplification of these libraries was performed under cycling conditions of a 5 min denaturation at 94°C; 24 cycles of 20 s at 94°C, 15 s at 60°C, and 20 s at 68°C; with a final extension of 5 min at 60°C. SPRI beads were used for postreaction clean up with elution in 15 μL TET (Tris EDTA-Tween-20) buffer.
Sequencing.
These nine uniquely indexed libraries were subject to multiple sequence runs at the Pennsylvania State Huck Institutes Genomics Core Facility and at the Schuster laboratory at Pennsylvania State University (Illumina HiSeq 2000 and 2500, 75-bp paired-end reads), generating a total of 2,067,295,157 paired-end reads and 313 giga bp of sequence data across 15 total lanes. Each lane contained only one library; some libraries were sequenced across multiple lanes. These sequences have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA), Accession no. SRP136389 (SRA BioProject no. PRJNA445550).
Bioinformatic processing of sequence data.
From the raw reads, the forward and reverse adapter sequences (introduced as part of the library preparation protocol) were trimmed, and overlapping paired-end reads were merged using the MergeReadsFastQ_cc script (89) with default settings, using a minimum 11-nt overlap and a phred quality score of 20 for merged sites. Unmerged reads were not used for downstream analyses due to limited yield. Exact duplicates from the trimmed and merged reads were removed premapping (https://github.com/smmarciniak/Megaladapis_nuc/blob/main/Pre-processing/rmdup.pl), resulting in the removal of 64.9% of all merged reads (SI Appendix, Table S1). Since PCR amplification and sequencing of the same original DNA fragment may also create duplicate reads that are not identical to each other (i.e., due to PCR or sequencing errors), we used perl to collapse such reads (identified as having the same start and end sequences) within each separate library to the single read with the best sum of FASTQ quality scores after alignment with lastZ (outlined in Sequence Read Alignments to Human Exons, below) (https://github.com/smmarciniak/Megaladapis_nuc/blob/main/fastq_filter.pl).
Authenticity of M. edwardsi Genomic Data.
To assess the authenticity of the M. edwardsi ancient nuclear genome sequence data, we considered the fragment length distributions (FLD) of all sequenced libraries and the nt damage pattern of the mapped reads, and we also estimated the proportion of human DNA contamination.
The FLD for each of the sequenced M. edwardsi libraries (n = 9; with 5 of the libraries sequenced twice) was composed of abundant short DNA fragments, which is characteristic of ancient specimens (19, 90) (SI Appendix, Figs. S3 and S4).
The authenticity of our M. edwardsi data are supported by the fragment size distribution (SI Appendix, Fig. S5A), base frequency fragmentation prior to read starts (SI Appendix, Fig. S5B), and elevated rates of C > T and G > A mismatches as expected at read ends (up to 20%) (SI Appendix, Fig. S5C). To further characterize DNA damage and degradation, we focused on DNA nt mismatches detectable in double- and single-stranded overhangs (δD and δs, respectively) that due to cytosine deamination are typically overrepresented in paleogenome samples in the 5′ termini as cytosine to thymine (C > T) mismatches (guanine to adenine or G > A on the complementary 3′ strand) (21, 90). We used mapDamage 2.0 (91) to quantify postmortem damage signals in the alignment of M. edwardsi reads to the hg19 hard-masked exon reference (UCSC Genome Browser) (92) from our genomic analyses (outlined in Sequence Read Alignments to Human Exons, below). Through mapDamage analysis, we estimated the probability of cytosine deamination (21) as δs = 0.73 to 73% of cytosine residues in single-stranded overhangs have been affected by deamination. To characterize the temporal rate of this chemical damage, we calculated a cytosine deamination rate of 8.55 × 10−3 site−1 year−1, placing deamination in the expected range for bone at a site with an annual mean temperature of 23.42 °C (19). The probability of a nt terminating an overhang was inferred using mapDamage (91) at λ = 0.26 (mean overhang length 3.4 nt). Therefore, the first nine nt within the end of a given fragment are expected to contain 95% of misincorporated deoxy-uracil residues under the geometric distribution. Accordingly, for our analyses of the M. edwardsi nuclear genome sequence, for all sequence reads we hard masked (i.e., replaced with “N”) sites potentially affected by cytosine deamination (5′ T residues and 3′ A residues) (93) within nine nt of fragment ends accordingly.
To estimate the level of human DNA contamination in our dataset, we aligned 45 million raw sequence reads sampled from across the multiple aDNA libraries to both the M. edwardsi mtDNA reference genome sequence (NC_026088.1) (6) and a human mitochondrial genome sequence (haplotype H6A1; EU256375.1) using bwa (Burrows-Wheeler Aligner) aln (94) with seeding disabled (−I 16500) and default mapping parameters (−n 0.01 and −o 2), filtering for a minimum read length of 20 nt and minimum mapping quality of 20 (SI Appendix, Table S2). We assigned 4,930 nonredundant reads to the M. edwardsi reference mtDNA, yielding 22× coverage of the complete mitochondrial genome, versus only 85 reads that mapped to the human reference mtDNA (1.7% of the total mapped reads). Of those 85 human-mapped reads, 25 were in regions strongly conserved across primates including M. edwardsi. The remaining 60 reads mapped uniquely to the human reference genome. Across each of the 13 M. edwardsi sequence libraries, an average of 126,194 reads (range 27,467 to 698,966) mapped to the M. edwardsi mtDNA reference genome sequence and an average of 457 reads (range 19 to 1,118) mapped to the human mtDNA sequence (an average of 330 reads mapped uniquely, ranging from 5 to 798). We thus estimate ∼1.2% contamination of our M. edwardsi sequence data with modern human DNA across all libraries with an average of 1.0% contamination per library (range 0.002 to 2.5%), which is consistent with or below reported human contamination rates in other studies (23, 24) and contributing negligibly to our gene sequence reconstructions, especially given our minimum 2× sequence coverage requirements.
We roughly estimated the proportion of endogenous M. edwardsi DNA in our sequencing libraries to be 6.35% by computing the number of sequence reads following merging and removal of identical sequence duplicates that were mapped to hg19 exons and flanks (see Sequence Read Alignments to Human Exons, below; n = 4,824,118) times 27.197 (given that these targets comprise ∼3% of the nuclear genome), all divided by the total number of all reads sequenced (n = 2,067,295,157).
Sample Preparation and Sequencing of Modern Lemur Genomes.
DNA extraction.
Ear punches were obtained from wild-caught lemurs, L. mustelinus (Weasel sportive lemur, TVY7.125 from Runhua, Madagascar) and E. rufifrons (RANO5.15 from Ranomafana and ISA2.23 from Isalo), with capture and sampling procedures approved by the Institutional Animal Care and Use Committee of Omaha's Henry Doorly Zoo and Aquarium (#12-101). Collection and export permits were obtained from Madagascar National Parks and the Ministère de l'Environnement, de l'Ecologie et des Forêts (MEEF) of Madagascar. The samples were imported under requisite Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) permits from the US Fish and Wildlife Service. Genomic DNA was extracted from these blood and tissue samples using a standard phenol/chloroform method from wild-caught individuals [as performed in Kistler et al. (6)] at Pennsylvania State University.
Library preparation and sequencing.
The L. mustelinus specimen underwent double-stranded library preparation and indexing following the protocols described above (in Sample Preparation and Sequencing of the M. edwardsi Nuclear Genome) for M. edwardsi. The single library generated was shotgun sequenced at the University of California Los Angeles Genomics Center on an Illumina HiSeq 2500 (100-bp paired end) (SI Appendix, Table S1). These sequence reads have been deposited in the NCBI SRA, Accession no. SRP136389 (SRA BioProject no. PRJNA445550). Libraries were prepared for the two E. rufifrons specimens with the TruSeq PCR-free library preparation kit, with subsequent whole-genome sequencing performed at the HudsonAlpha Institute for Biotechnology (Genomic Services Lab) on the Illumina HiSeq X Ten (150-bp paired end, one sample per lane) (SI Appendix, Table S1). The E. rufifrons sequence reads have been deposited in the NCBI SRA, Accession no. SRP136389 (SRA BioProject no. PRJNA445550).
Existing genomic data.
Previously published primate whole-genome sequence read data were used for D. madagascariensis (SRA043766.1) (15), P. diadema (PRJNA317769) (17), M. murinus (PRJNA285159) (16), and R. roxellana, (PRJNA230020) (25).
Sequence Read Alignments to Human Exons.
With the sequence read length and coverage restrictions of our aDNA data, it was not possible to construct a de novo assembly of the M. edwardsi nuclear genome. Thus, it was necessary to align our sequence reads to an existing reference genome sequence. We focused on exons, which tend to be relatively conserved across species, thereby aiding mapping and alignment efforts between lemur sequence reads and the human reference genome. We prepared an hg19 reference genome with NCBI RefSeq annotations (95), with hard masking so that only the RefSeq exons and 100-nt flanks to either side of each exonic region were available as alignment targets. The inclusion of the 100-nt flanks helps minimize loss of data from exon ends.
The modern lemur and colobine monkey sequence read data were mapped against this modified hg19 reference using bwa mem (96) with slightly relaxed mismatch penalty (option –B 2). The bwa mem algorithm has higher tolerance for divergent RefSeqs given a suitable minimum read length (≥70 bp) (i.e., 2% error for a 100-bp alignment) (96). The resulting SAM (Sequence Alignment Map) files were converted to BAM (Binary Alignment Map) format using SAMtools (97) and then used to generate exon consensus sequences using SAMtools mpileup (default settings).
For M. edwardsi, given the shorter read lengths of this dataset, we instead used a lastZ (98) alignment procedure. We chose lastZ for this step with the Megaladapis read data because the bwa aln algorithm (94) strongly penalizes divergence, and in our experience bwa mem (99) does not work as effectively with shorter read lengths as lastZ. Thus, while lastZ is computationally inefficient, there may be occasions when its complementary use could contribute positively, depending on DNA quality and genetic divergence to the RefSeq. The workflow involved parsing the target sequence into overlapping fragments that were then compared iteratively to the query sequences (individually and sequentially) and filtered by score to remove alignment blocks that did not meet the specified criteria (98). An extension matrix (Dryad) based on an aye-aye–human whole-genome alignment (15) was used to align curated M. edwardsi reads to the prepared hg19 reference, which functioned to modify the scoring scheme to reject or continue along a query sequence to reflect homology with variably diverged sequences. The following command line options were used: format = general:name1,zstart1,end1,text1,name2,strand2,zstart2,end2,text2,nucs2,quals2,identity,coverage,continuity –ambiguous = iupac. The alignment output uses lastZ’s “general format” (e.g., one line per alignment block) and reports the aligned pair of sequences as well as the number of mismatches for that pair (98). International Union of Pure and Applied Chemistry ambiguity codes for nts (e.g., N, B, D, H, K, M, R, S, V, W, Y) were treated as completely ambiguous and scored as zero when these substitutions were present (98).
For reads with more than one viable mapping location (lastZ returns all hits rather than a heuristically optimal hit), we calculated the mean identity (percentage of aligned bases matching the target or query), coverage (percentage of the alignment blocks that cover the entire target or query), and continuity (percentage of the alignment blocks that are not gaps) of each location. Given a 5% minimum difference in this mean between the best and second-best hit, we retained the top mapping location. Reads with two or more very similar scores (i.e., less than 5% difference between hg19 exons matching the same region of M. edwardsi) on this metric were discarded. The lastZ file was sorted according to genomic coordinates and then any remaining PCR duplicates with matching start and end position coordinates were discarded by retaining the single read among all matches with the greatest sum of FASTQ quality scores (https://github.com/smmarciniak/Megaladapis_nuc/blob/main/fastq_filter.pl).
We generated a simple positional pileup file from the damage-masked Megaladapis read alignment, and we summarized exonic nt positions in the “known canonical” reference gene set from the hg19 assembly (http://hgdownload.soe.ucsc.edu/goldenPath/hg19/multiz46way/) (92). We summarized only sites with a strict consensus among Megaladapis reads (leading to higher confidence in authenticity with some loss of heterozygous sites), and we enforced strict positionality to maintain the reading frame of the human reference exons, ignoring indels observed in read data. We likewise generated exon consensus sequences in the modern lemur and colobine monkey read alignments to the hg19 exonic reference, using SAMtools mpileup (default settings) (97) to summarize positional nts. From the pileup files, we summarized exon sequences matching the hg19 “known canonical” gene set enforcing a minimum 2× sequence coverage excluding masked sites, spliced exons to match the full transcripts, and added our M. edwardsi, modern lemur, and colobine monkey gene sequences with the remaining sequences, resulting in a 53-way multispecies alignment that we used in our analyses. Because our exon extraction and the 46-way alignment were already forced into the human reading frame, further multiple alignment of gene sequences was not needed.
Phylogenetic Analyses.
We used a genome-wide maximum likelihood approach to estimate the phylogenetic placement of M. edwardsi. First, a concatenated gene alignment was constructed, comprised of 15 primate species at RefSeq gene loci (described in Sequence Read Alignments to Human Exons, above) where at least 50% of Megaladapis sites were represented at minimum 2× coverage after damage masking (n = 896 loci; 1.07 Mbp or megabase pairs). Using RAxML (Randomized Axelerated Maximum Likelihood) (26), we estimated a single unrooted phylogeny from the concatenated alignment without partitioning under the GTR (General Time Reversible) GAMMA model (assuming variable nt frequency changes that are independent for each type of nt) (100).
Independent phylogenies were also estimated from each gene with at least 20% of sites covered across all lemur sequences (n = 11,944) using the same GTR GAMMA model, with 100 bootstrap replicates for each gene tree. Mean bootstrap support across all bipartitions was calculated as a measure of gene tree phylogenetic signal [following Salichos and Rokas (27)], with resulting values ranging from 7.69 to 98.85% (median 76.38%; SI Appendix, Fig. S7). As described previously (27, 101), internode certainty (IC) among gene trees with strong phylogenetic signal provides a robust validation of gene tree support for species tree branching order. We therefore also calculated IC across the concatenated gene tree among bootstrap consensus gene trees with at least 90% mean bootstrap consensus support (n = 771 loci) using RAxML (26).
Discordance among the 771 strong signal gene trees with the species tree was visualized with the “Relative Frequency” analysis in DiscoVista version 1.0 (28). The frequency of three potential topologies (or bipartitions) are inferred based on the focal internal branches of the species tree with the main topology (in red) and alternative topologies (in blue) (SI Appendix, Figure S7) (28).
For each of the n = 771 gene trees with at least 90% bootstrap support, we also conducted a nearest neighbor analysis, studying only the n = 334 gene trees in which the bipartition defining the smallest M. edwardsi clade is supported by at least 90% of bootstrap replicates. For example, in a gene tree, the smallest group created by a bipartition to contain M. edwardsi describes its phylogenetic position with regards to the various phylogenetics hypotheses being tested. If the smallest group containing M. edwardsi contains only M. edwardsi and E. rufifrons, that gene tree supports our main tree topology. If the bootstrap support for that bipartition in that gene tree was at least 90%, then it was included in this nearest neighbor analysis. Conversely, if the smallest group containing M. edwardsi in a particular gene tree is (M. edwardsi, E. rufifrons, L. mustelinus, P. diadema, M. murinus), then M. edwardsi must be the basal member of this group, and if the bootstrap value for that bipartition in that tree was at least 90%, then the Herrera and Dávalos (14) topology is supported.
PhyloPic silhouettes were used in Fig. 1A, with the following attributions: Callithrix jacchus, Papio anubis, R. roxellana, G. gorilla, Tarsius syrichta (Carlito syrichta), and Otolemur garnetti (Galagonidae) all under Public Domain Dedication 1.0 license, https://creativecommons.org/publicdomain/zero/1.0/. Macaca mulatta and Homo sapiens sapiens under Public Domain Mark 1.0 license https://creativecommons.org/publicdomain/mark/1.0/. Credit to T. Michael Keesey (vectorization) and Tony Hisgett (photography) for the P. troglodytes image, under license https://creativecommons.org/licenses/by/3.0/ (modified opacity). Credit to Gareth Monger for the Pongo abelii image, https://creativecommons.org/licenses/by/3.0/(modified opacity).
dN/dS Analyses.
We used the codeml function of PAML (33) to estimate lineage-specific dN/dS ratios across the phylogeny of the six lemurs in our study plus three nonlemur primates (H. sapiens, G. gorilla, and P. troglodytes). Prior to analysis, all sequences were checked for codon completeness across all nine species and any premature stop codons; violating codons were masked with “N”s (https://github.com/RBankoff/PAML_Scripts/) in accordance with the input requirements of codeml. We restricted our analysis to the set of n = 3,342 genes with 1) ≥100 intact Megaladapis codons in our ≥2× sequence coverage data, 2) ≥100 N sites present and aligned across all nine species in this analysis, and 3) Megaladapis lineage dS values not more than 2 SD greater than the genome-wide average dS value.
Based on the PAML codeml results, dN/dS ratios were calculated in two ways: first based on the synonymous substitution rate for an individual gene (dN/dS), and second, a dN/dSgenome ratio based on the genome-wide estimate of dS (15). The genome-wide estimate is calculated from the total number of synonymous substitutions across all genes divided by the total number of synonymous sites genome wide (15). Inferring a dN/dSgenome ratio is valuable for branches where the synonymous substitutions may be low or zero for an individual gene (15).
We used the GO database within g:Profiler (https://biit.cs.ut.ee/gprofiler/gost) (35) to identify any functional category enrichment among the set of genes with M. edwardsi lineage dN/dSgenome values >1.5, using all 3,342 genes analyzed as background.
Genome-Wide Convergent Amino Acid Evolution Analyses.
We translated our multispecies (n = 53) gene sequence alignments (for n = 21,520 genes) into amino acid sequences. We then queried each possible individual species (n = 35) and clade (n = 17) comparison with M. edwardsi [using ETE3 (102) to navigate the tree], recording the numbers of analyzable amino acid sites and convergent amino acids for each gene.
A convergent amino acid was defined as having the following properties: 1) both M. edwardsi and the distant comparison species/clade to which M. edwardi was being compared shared the same amino acid at that position; 2) the sister species to Megaladapis (E. rufifrons) and a member of the outgroup clade to the M. edwardsi and E. rufifrons clade (either M. murinus or P. diadema) shared the same amino acid with each other but not Megaladapis; and 3) the sister species (or member of the sister clade of species) to the distant comparison species/clade and a member of the outgroup clade to the distant comparison-sister species clade also shared the same amino acid with each other but not with the comparison species/clade.
A site was counted as “analyzable” if the following conditions were met: 1) at that site, amino acid information was available for all six of the species involved in the particular analysis; 2) E. rufifrons and the outgroup species (e.g., M. murinus) had identical amino acids to each other at that position; and 3) the sister and outgroup representatives for the distant comparison clade also had identical amino acids to each other at that position.
For the results presented in the main text and figures, we used M. murinus as the representative of the outgroup clade to the M. edwardsi–E. rufifrons grouping because there was a greater number of sites with inferred amino acids for M. murinus than for P. diadema. We repeated all analyses using P. diadema to confirm consistency.
For each comparison, we summed the numbers of convergent and analyzable sites across all genes represented in each GO term (46). For GO categories with ≥5 convergent amino acids, we tested whether the proportion of convergent sites was significantly different from expected based on the genome-wide ratio. Specifically, for each qualifying category, we used a Fisher’s exact test to compare the ratio of convergent to analyzable amino acid positions within that GO category to this ratio for all genes in the genome apart from those included in that category. We computed FDR (103) from the resulting P values to account for the multiple tests. The GO database release used was 2019-04-17, and the code for this analysis is available at https://github.com/MehreenRuhi/conv with the results deposited to Dryad (http://datadryad.org/stash/dataset/doi:10.5061/dryad.5qfttdz3c).
We also conducted a post hoc analysis that focused specifically on two GO categories (hydrolase activity GO: 0016787 and brush border GO: 0005903). For this analysis, we recorded the numbers of analyzable amino acid sites and convergent amino acids for each gene in the genome for each possible individual species and clade comparison across the entire tree of n = 53 species (n = 1,855 possible taxonomic comparisons excluding those involving Megaladapis). We counted the number of genome-wide and GO: 0016787 and GO: 0005903 convergent and analyzable sites for each comparison and computed odds ratios and P values as described in the preceding paragraph. We then compared the respective Megaladapis–Rhinopithecus (for GO: 0016787) and Megaladapis–Equus (for GO: 0005903) results to those of all other taxonomic comparisons.
Supplementary Material
Acknowledgments
We thank the Laboratoire de Primatologie et de Paléontologie des Vertébrés and the Mention Anthropobiologie et Développement Durable at the University of Antananarivo for permission to sample the UA 5180 M. edwardsi specimen and the Madagascar National Parks and the MEEF for permission to collect the modern lemur samples included in the study. Different components of this work were supported by the Pennsylvania State University College of the Liberal Arts, the Pennsylvania State University Huck Institutes of the Life Sciences, and by grants from the NSF (BCS-1317163 to G.H.P., BCS-1554834 to G.H.P., BCS-1750598 to L.R.G., and BCS-1749676 to B.E.C.) and from the Ahmanson Foundation (to E.E.L.). We thank Webb Miller for bioinformatic discussions and contributions. Joel Borgerson created the watercolor illustrations of extant and extinct lemurs shown in Fig. 1. Computations for this research were performed on the Pennsylvania State University’s Institute for Computational and Data Sciences’ supercomputing cluster. This content is solely the responsibility of the authors and does not necessarily represent the views of the Institute for Computational and Data Sciences.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. N.J.R. is a guest editor invited by the Editorial Board.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022117118/-/DCSupplemental.
Data Availability
All sequence data newly generated for this study have been deposited to the NCBI SRA for M. edwardsi (PRJNA445550), E. rufifrons (PRJNA445550), and L. mustelinus (PRJNA445550). Extant/extinct lemur mpileup exon files, masked/unmasked 2× M. edwardsi sequences integrated with the UCSC species and extant lemurs alignment data sets, gene alignments used in gene tree phylogeny estimation (input and output files), dN/dS input nt files, and resulting output table, functional enrichment output tables, genomic convergence output, and post hoc results (supplementary tables) have been deposited to the Dryad Digital Repository (http://datadryad.org/stash/dataset/doi:10.5061/dryad.5qfttdz3c) (104). Code for the removal of duplicate reads, dN/dS, and genomic convergence analyses used in this manuscript have been made available through the following github repositories: https:/github.com/smmarciniak/Megaladapis_nuc, https://github.com/RBankoff/PAML_Scripts/, and https://github.com/MehreenRuhi/conv. All other study data are included in the article and/or SI Appendix.
References
- 1.Crowley B. E., A refined chronology of prehistoric Madagascar and the demise of the megafauna. Quat. Sci. Rev. 29, 2591–2603 (2010). [Google Scholar]
- 2.Powzyk J. A., Mowry C. B., Dietary and feeding differences between sympatric Propithecus diadema diadema and Indri indri. Int. J. Primatol. 24, 1143–1162 (2003). [Google Scholar]
- 3.Godfrey L. R., Jungers W. L., Burney D. A., “Subfossil lemurs of Madagascar” in Cenozoic Mammals of Africa, Werdelin L., Sanders W. J., Eds. (University of California Press, 2010), pp. 351–367. [Google Scholar]
- 4.Karanth K. P., Delefosse T., Rakotosamimanana B., Parsons T. J., Yoder A. D., Ancient DNA from giant extinct lemurs confirms single origin of Malagasy primates. Proc. Natl. Acad. Sci. U.S.A. 102, 5090–5095 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orlando L., et al., DNA from extinct giant lemurs links archaeolemurids to extant indriids. BMC Evol. Biol. 8, 121 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kistler L., et al., Comparative and population mitogenomic analyses of Madagascar’s extinct, giant ‘subfossil’ lemurs. J. Hum. Evol. 79, 45–54 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Yoder A. D., Rakotosamimanana B., Parsons T. J., “Ancient DNA in subfossil lemurs: Methodological challenges and their solutions” in New Directions in Lemur Studies, Rasaminanana H., Rakotosamimanana B., Goodman S., Ganzhorn J., Eds. (Plenum Press, 1999), pp. 1–17. [Google Scholar]
- 8.Grealy A., et al., Eggshell palaeogenomics: Palaeognath evolutionary history revealed through ancient nuclear and mitochondrial DNA from Madagascan elephant bird (Aepyornis sp.) eggshell. Mol. Phylogenet. Evol. 109, 151–163 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Grealy A., et al., Tropical ancient DNA from bulk archaeological fish bone reveals the subsistence practices of a historic coastal community in southwest Madagascar. J. Archaeol. Sci. 75, 82–88 (2016). [Google Scholar]
- 10.Hekkala E., et al., Paleogenomics illuminates the evolutionary history of the extinct Holocene “horned” crocodile of Madagascar, Voay robustus. Commun. Biol. 4, 505 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jungers W. L., Demes B., Godfrey L. R., “How big were the ‘giant’ extinct lemurs of Madagascar?” in Elwyn Simons: A Search for Origins, Fleagle J. G., Gilbert C., Eds. (Springer, New York, 2008), pp. 343–360. [Google Scholar]
- 12.Tattersall I., Schwartz J. H., “Craniodental morphology and the systematics of the Malagasy lemurs" (Volume 52, part 3, Anthropological Papers of the American Museum of Natural History, New York, 1974).
- 13.Rubinoff D., Holland B. S., Between two extremes: Mitochondrial DNA is neither the panacea nor the nemesis of phylogenetic and taxonomic inference. Syst. Biol. 54, 952–961 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Herrera J. P., Dávalos L. M., Phylogeny and divergence times of lemurs inferred with recent and ancient fossils in the tree. Syst. Biol. 65, 772–791 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Perry G. H., et al., A genome sequence resource for the aye-aye (Daubentonia madagascariensis), a nocturnal lemur from Madagascar. Genome Biol. Evol. 4, 126–135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsen P. A., et al., Hybrid de novo genome assembly and centromere characterization of the gray mouse lemur (Microcebus murinus). BMC Biol. 15, 110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bankoff R. J., et al., Testing convergent evolution in auditory processing genes between echolocating mammals and the aye-aye, a percussive-foraging primate. Genome Biol. Evol. 9, 1978–1989 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allentoft M. E., et al., The half-life of DNA in bone: Measuring decay kinetics in 158 dated fossils. Proc. Biol. Sci. 279, 4724–4733 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kistler L., Ware R., Smith O., Collins M., Allaby R. G., A new model for ancient DNA decay based on paleogenomic meta-analysis. Nucleic Acids Res. 45, 6310–6320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Höss M., Jaruga P., Zastawny T. H., Dizdaroglu M., Pääbo S., DNA damage and DNA sequence retrieval from ancient tissues. Nucleic Acids Res. 24, 1304–1307 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Briggs A. W., et al., Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl. Acad. Sci. U.S.A. 104, 14616–14621 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ginolhac A., Rasmussen M., Gilbert M. T. P., Willerslev E., Orlando L., mapDamage: Testing for damage patterns in ancient DNA sequences. Bioinformatics 27, 2153–2155 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Allentoft M. E., et al., Population genomics of Bronze age Eurasia. Nature 522, 167–172 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Burbano H. A., et al., Targeted investigation of the Neandertal genome by array-based sequence capture. Science 328, 723–725 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X., et al., Whole-genome sequencing of the snub-nosed monkey provides insights into folivory and evolutionary history. Nat. Genet. 46, 1303–1310 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salichos L., Rokas A., Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 497, 327–331 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Sayyari E., Whitfield J. B., Mirarab S., DiscoVista: Interpretable visualizations of gene tree discordance. Mol. Phylogenet. Evol. 122, 110–115 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Wall C. E., The expanded mandibular condyle of the Megaladapidae. Am. J. Phys. Anthropol. 103, 263–276 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Varki A., Altheide T. K., Comparing the human and chimpanzee genomes: Searching for needles in a haystack. Genome Res. 15, 1746–1758 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Preuss T. M., Human brain evolution: From gene discovery to phenotype discovery. Proc. Natl. Acad. Sci. U.S.A. 109 (suppl. 1), 10709–10716 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chamary J. V., Parmley J. L., Hurst L. D., Hearing silence: Non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 7, 98–108 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Yang Z., PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Reimand J., Kull M., Peterson H., Hansen J., Vilo J., g:Profiler-a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35, W193–W200 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pagani S., Radetti G., Meazza C., Bozzola M., Analysis of growth hormone receptor gene expression in tall and short stature children. J. Pediatr. Endocrinol. Metab. 30, 427–430 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Fontanesi L., et al., Identification of polymorphisms in the rabbit growth hormone receptor (GHR) gene and association with finishing weight in a commercial meat rabbit line. Anim. Biotechnol. 27, 77–83 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Hinrichs A., et al., Growth hormone receptor-deficient pigs resemble the pathophysiology of human Laron syndrome and reveal altered activation of signaling cascades in the liver. Mol. Metab. 11, 113–128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Runge-Morris M., Kocarek T. A., Expression of the sulfotransferase 1C family: Implications for xenobiotic toxicity. Drug Metab. Rev. 45, 450–459 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Marto N., Morello J., Monteiro E. C., Pereira S. A., Implications of sulfotransferase activity in interindividual variability in drug response: Clinical perspective on current knowledge. Drug Metab. Rev. 49, 357–371 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Cork S. J., Foley W. J., “Digestive and metabolic strategies of arboreal mammalian folivores in relation to chemical defenses in temperate and tropical forests” in Plant Defenses Against Mammalian Herbivory, Palo R. T., Robbins C. T., Eds. (CRC Press, 1991), pp. 134–166. [Google Scholar]
- 42.Rafferty K. L., Teaford M. F., Jungers W. L., Molar microwear of subfossil lemurs: Improving the resolution of dietary inferences. J. Hum. Evol. 43, 645–657 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Scott J. R., et al., Dental microwear texture analysis of two families of subfossil lemurs from Madagascar. J. Hum. Evol. 56, 405–416 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Godfrey L. R., et al., Dental use wear in extinct lemurs: Evidence of diet and niche differentiation. J. Hum. Evol. 47, 145–169 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Godfrey L. R., Winchester J. M., King S. J., Boyer D. M., Jernvall J., Dental topography indicates ecological contraction of lemur communities. Am. J. Phys. Anthropol. 148, 215–227 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Ashburner M.et al.; The Gene Ontology Consortium , Gene ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hodgson E., Das P. C., Cho T. M., Rose R. L., “Phase 1 metabolism of toxicants and metabolic interactions” in Molecular and Biochemical Toxicology, Smart R. C., Hodgson E., Eds. (John Wiley & Sons, Fourth Edition, 2008), pp. 173–203. [Google Scholar]
- 48.Zhu F., Moural T. W., Nelson D. R., Palli S. R., A specialist herbivore pest adaptation to xenobiotics through up-regulation of multiple Cytochrome P450s. Sci. Rep. 6, 20421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dearing M. D., Foley W. J., McLean S., The influence of plant secondary metabolites on the nutritional ecology of herbivorous terrestrial vertebrates. Annu. Rev. Ecol. Evol. Syst. 36, 169–189 (2005). [Google Scholar]
- 50.Hecker N., Sharma V., Hiller M., Convergent gene losses illuminate metabolic and physiological changes in herbivores and carnivores. Proc. Natl. Acad. Sci. U.S.A. 116, 3036–3041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hooton D., Lentle R., Monro J., Wickham M., Simpson R., “The secretion and action of brush border enzymes in the mammalian small intestine” in Reviews of Physiology, Biochemistry and Pharmacology, Nilius B., et al., Eds. (Springer Verlag, 2015), pp. 59–118. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y. Y., et al., A LIMA1 variant promotes low plasma LDL cholesterol and decreases intestinal cholesterol absorption. Science 360, 1087–1092 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Crawley S. W., Mooseker M. S., Tyska M. J., Shaping the intestinal brush border. J. Cell Biol. 207, 441–451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dicks L. M. T., Botha M., Dicks E., Botes M., The equine gastro-intestinal tract: An overview of the microbiota, disease and treatment. Livest. Sci. 160, 69–81 (2014). [Google Scholar]
- 55.Richards N., Choct M., Hinch G. N., Rowe J. B., Examination of the use of exogenous α-amylase and amyloglucosidase to enhance starch digestion in the small intestine of the horse. Anim. Feed Sci. Technol. 114, 295–305 (2004). [Google Scholar]
- 56.Dyer J., et al., Molecular characterisation of carbohydrate digestion and absorption in equine small intestine. Equine Vet. J. 34, 349–358 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Zou Z., Zhang J., Are convergent and parallel amino acid substitutions in protein evolution more prevalent than neutral expectations? Mol. Biol. Evol. 32, 2085–2096 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Veeramah K. R., “Primate paleogenomics” in Paleogenomics: Genome-Scale Analysis of Ancient DNA, Lindqvist C., Rajora O. P., Eds. (Springer, 2019), pp. 353–374. [Google Scholar]
- 59.Mathieson I., et al., An ancient baboon genome demonstrates long-term population continuity in Southern Africa. Genome Biol. Evol. 12, 407–412 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woods R., Turvey S. T., Brace S., MacPhee R. D. E., Barnes I., Ancient DNA of the extinct Jamaican monkey Xenothrix reveals extreme insular change within a morphologically conservative radiation. Proc. Natl. Acad. Sci. U.S.A. 115, 12769–12774 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Welker F., Palaeoproteomics for human evolution studies. Quat. Sci. Rev. 190, 137–147 (2018). [Google Scholar]
- 62.Welker F., et al., Enamel proteome shows that Gigantopithecus was an early diverging pongine. Nature 576, 262–265 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muchlinski M. N., Godfrey L. R., Muldoon K. M., Tongasoa L., Evidence for dietary niche separation based on infraorbital foramen size variation among subfossil lemurs. Folia Primatol. (Basel) 81, 330–345 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Crowley B. E., Godfrey L. R., Irwin M. T., A glance to the past: Subfossils, stable isotopes, seed dispersal, and lemur species loss in southern Madagascar. Am. J. Primatol. 73, 25–37 (2011). [DOI] [PubMed] [Google Scholar]
- 65.Crowley B. E., Godfrey L. R., Why all those spines?: Anachronistic defences in the Didiereoideae against now extinct lemurs. S. Afr. J. Sci. 109, 1–7 (2013). [Google Scholar]
- 66.Crowley B. E., Godfrey L. R., Strontium isotopes support small home ranges for extinct lemurs. Front. Ecol. Evol. 7, 490 (2019). [Google Scholar]
- 67.Ganzhorn J. U., et al., Selection of food and ranging behaviour in a sexually monomorphic folivorous lemur: Lepilemur ruficaudatus. J. Zool. (Lond.) 263, 393–399 (2004). [Google Scholar]
- 68.Dröscher I., Rothman J. M., Ganzhorn J. U., Kappeler P. M., Nutritional consequences of folivory in a small-bodied lemur (Lepilemur leucopus): Effects of season and reproduction on nutrient balancing. Am. J. Phys. Anthropol. 160, 197–207 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Hladik C. M., Charles-Dominique P., “The behaviour and ecology of the sportive lemur (Lepilemur mustelinus) in relation to its dietary peculiarities” in Prosimian Biology, Martin R. D., Doyle G. A., Walker A. C., Eds. (Duckworth, London, 1974), pp. 25–37. [Google Scholar]
- 70.Lear C. H., Elderfield H., Wilson P. A., Cenozoic deep-Sea temperatures and global ice volumes from Mg/Ca in benthic foraminiferal calcite. Science 287, 269–272 (2000). [DOI] [PubMed] [Google Scholar]
- 71.Coxall H. K., Wilson P. A., Pälike H., Lear C. H., Backman J., Rapid stepwise onset of Antarctic glaciation and deeper calcite compensation in the Pacific Ocean. Nature 433, 53–57 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Gunnell G. F., et al., Fossil lemurs from Egypt and Kenya suggest an African origin for Madagascar’s aye-aye. Nat. Commun. 9, 3193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Galtier N., Daubin V., Dealing with incongruence in phylogenomic analyses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 4023–4029 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pamilo P., Nei M., Relationships between gene trees and species trees. Mol. Biol. Evol. 5, 568–583 (1988). [DOI] [PubMed] [Google Scholar]
- 75.Hobolth A., Christensen O. F., Mailund T., Schierup M. H., Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model. PLoS Genet. 3, e7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Godfrey L. R., et al., Mid-Cenozoic climate change, extinction, and faunal turnover in Madagascar, and their bearing on the evolution of lemurs. BMC Evol. Biol. 20, 97 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wood A. R.et al.; Electronic Medical Records and Genomics (eMEMERGEGE) Consortium; MIGen Consortium; PAGEGE Consortium; LifeLines Cohort Study , Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46, 1173–1186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bouwman A. C., et al., Meta-analysis of genome-wide association studies for cattle stature identifies common genes that regulate body size in mammals. Nat. Genet. 50, 362–367 (2018). [DOI] [PubMed] [Google Scholar]
- 79.Silva C. N. S., et al., Insights into the genetic architecture of morphological traits in two passerine bird species. Heredity 119, 197–205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guevara E. E., et al., Comparative genomic analysis of sifakas (Propithecus) reveals selection for folivory and high heterozygosity despite endangered status. Sci. Adv. 7, eabd2274 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grogan K. E., Perry G. H., Studying human and nonhuman primate evolutionary biology with powerful in vitro and in vivo functional genomics tools. Evol. Anthropol. 29, 143–158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tattersall I., The functional significance of airorhynchy in Megaladapis. Folia Primatol. (Basel) 18, 20–26 (1972). [DOI] [PubMed] [Google Scholar]
- 83.Muchlinski M. N., A comparative analysis of vibrissa count and infraorbital foramen area in primates and other mammals. J. Hum. Evol. 58, 447–473 (2010). [DOI] [PubMed] [Google Scholar]
- 84.Nieves-Colón M. A., et al., Comparison of two ancient DNA extraction protocols for skeletal remains from tropical environments. Am. J. Phys. Anthropol. 166, 824–836 (2018). [DOI] [PubMed] [Google Scholar]
- 85.Hogg A. G., et al., SHCal20 Southern Hemisphere calibration, 0–55,000 Years cal BP. Radiocarbon 62, 759–778 (2020). [Google Scholar]
- 86.Stuiver M., Reimer P. J., Reimer R. W., CALIB 8.2. http://calib.org/calib/calib.html. Accessed 5 October 2020.
- 87.Rohland N., “DNA extraction of ancient animal hard tissue samples via adsorption to silica particles” in Ancient DNA: Methods and Protocols, Shapiro B., Hofreiter M., Eds. (Humana Press, 2012), pp. 21–28. [DOI] [PubMed] [Google Scholar]
- 88.Meyer M., Kircher M., Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, pdb.prot5448 (2010). [DOI] [PubMed] [Google Scholar]
- 89.Kircher M., “Analysis of high-throughput ancient DNA sequencing data” in Ancient DNA: Methods and Protocols, Shapiro B., Hofreiter M., Eds. (Humana Press, 2012), pp. 197–228. [DOI] [PubMed] [Google Scholar]
- 90.Dabney J., Meyer M., Pääbo S., Ancient DNA damage. Cold Spring Harb. Perspect. Biol. 5, a012567 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jónsson H., Ginolhac A., Schubert M., Johnson P. L. F., Orlando L., mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kent W. J., et al., The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kennett D. J., et al., Archaeogenomic evidence reveals prehistoric matrilineal dynasty. Nat. Commun. 8, 14115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.O’Leary N. A., et al., Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li H., Durbin R., Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li H.et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harris R. S., Improved Pairwise Alignment of Genomic DNA (The Pennsylvania State University, 2007). [Google Scholar]
- 99.Li H., Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint] (2013). https://arxiv.org/abs/1303.3997v2 (Accessed 7 May 2021).
- 100.Nei M., Kumar S., Molecular Evolution and Phylogenetics (Oxford University Press, 2000). [Google Scholar]
- 101.Salichos L., Stamatakis A., Rokas A., Novel information theory-based measures for quantifying incongruence among phylogenetic trees. Mol. Biol. Evol. 31, 1261–1271 (2014). [DOI] [PubMed] [Google Scholar]
- 102.Huerta-Cepas J., Serra F., Bork P., ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- 104.Marciniak S., et al., Data from: Evolutionary and phylogenetic insights from a nuclear genome sequence of the extinct, giant ‘subfossil’ koala lemur Megaladapis edwardsi. Dryad Digital Repository. 10.5061/dryad.5qfttdz3c. Deposited 21 October 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data newly generated for this study have been deposited to the NCBI SRA for M. edwardsi (PRJNA445550), E. rufifrons (PRJNA445550), and L. mustelinus (PRJNA445550). Extant/extinct lemur mpileup exon files, masked/unmasked 2× M. edwardsi sequences integrated with the UCSC species and extant lemurs alignment data sets, gene alignments used in gene tree phylogeny estimation (input and output files), dN/dS input nt files, and resulting output table, functional enrichment output tables, genomic convergence output, and post hoc results (supplementary tables) have been deposited to the Dryad Digital Repository (http://datadryad.org/stash/dataset/doi:10.5061/dryad.5qfttdz3c) (104). Code for the removal of duplicate reads, dN/dS, and genomic convergence analyses used in this manuscript have been made available through the following github repositories: https:/github.com/smmarciniak/Megaladapis_nuc, https://github.com/RBankoff/PAML_Scripts/, and https://github.com/MehreenRuhi/conv. All other study data are included in the article and/or SI Appendix.