Significance
Environmental factors can promote phenotypic variation through alterations in the epigenome and mediate adaptation of an organism to the environment. Observations suggest the adaptation of Poecilia mexicana fish to toxic, hydrogen sulfide–rich environments in southern Mexico may, in part, be promoted through epigenetic DNA methylation alterations that became generationally stable and are inherited to subsequent generations independent of the environment. Environmental epigenetics may provide an important mechanism mediating adaptation in this species. This is an observation that the epigenome is stably inherited generationally through the germline after the removal of an environmental stressor (i.e., hydrogen sulfide) from a wild population.
Keywords: epigenetic, inheritance, sulfidic environment, adaptation
Abstract
Environmental factors can promote phenotypic variation through alterations in the epigenome and facilitate adaptation of an organism to the environment. Although hydrogen sulfide is toxic to most organisms, the fish Poecilia mexicana has adapted to survive in environments with high levels that exceed toxicity thresholds by orders of magnitude. Epigenetic changes in response to this environmental stressor were examined by assessing DNA methylation alterations in red blood cells, which are nucleated in fish. Males and females were sampled from sulfidic and nonsulfidic natural environments; individuals were also propagated for two generations in a nonsulfidic laboratory environment. We compared epimutations between the sexes as well as field and laboratory populations. For both the wild-caught (F0) and the laboratory-reared (F2) fish, comparing the sulfidic and nonsulfidic populations revealed evidence for significant differential DNA methylation regions (DMRs). More importantly, there was over 80% overlap in DMRs across generations, suggesting that the DMRs have stable generational inheritance in the absence of the sulfidic environment. This is an example of epigenetic generational stability after the removal of an environmental stressor. The DMR-associated genes were related to sulfur toxicity and metabolic processes. These findings suggest that adaptation of P. mexicana to sulfidic environments in southern Mexico may, in part, be promoted through epigenetic DNA methylation alterations that become stable and are inherited by subsequent generations independent of the environment.
Toxicants are present in a wide variety of ecological contexts including natural and man-made, and environmental toxicants are becoming increasingly abundant due to anthropogenic activities. While many chemicals are categorized by their mutagenic effects, it has become increasingly clear that indirect modes of action are as important for short-term (e.g., phenotypic plasticity) and long-term (e.g., adaptive) effects (1–4). Responses to the toxicity of environmental chemicals have been shown, in part, to be mediated by epigenetics (4–6). For example, exposure to heavy metals, including cadmium and arsenic, has been associated with changes in DNA methylation in mammals (7–10). However, the findings are often complex and contradictory. In vitro cadmium exposure experiments in rat liver cells suggested initial inductions of DNA hypomethylation occurred, while prolonged exposure led to DNA hypermethylation (7). A wide variety of environmental compounds, such as the agricultural fungicide vinclozolin and the herbicide glyphosate, have been shown to modify the location and abundance of DNA methylation in rats (11–13). Moreover, vinclozolin and glyphosate promote the epigenetic transgenerational inheritance of DNA methylation states and disease susceptibility in the third generation following transient exposures (6, 11–13). The ability of these environmental toxicants to shape phenotypic variation through epigenetic changes that are stable across generations has potential evolutionary consequences that remain largely unexplored.
Natural systems with environmentally derived toxicants provide a framework in which it is possible to study the evolutionary effects of toxicants. By comparing populations adapted to a toxicant to nonadapted ancestral populations, we can determine the effect of the toxicant on the evolution of the populations. To make predictions about the outcomes from environmental exposure, it is necessary to have a model stressor with clearly defined and predictable effects. Hydrogen sulfide (H2S) is an ideal toxicant to study the role of epigenetics in responding to environmental toxicants, because H2S exposure has clear effects on sulfide processing and energy metabolism (14).
H2S is one of the most toxic inorganic gases for metazoan organisms. It occurs both naturally (e.g., in deep-sea hydrothermal vents, marine sediments, and sulfide springs) and as a by-product of pollution and industrial processes [e.g., in habitats impacted by farming, tanning, paper manufacturing, sewage treatment, oil refining, and gas exploration and refining (15, 16)]. For humans, with a recommended industrial daily exposure of 10 ppm (293 µM) (National Institute for Occupational Safety and Health, 2020), H2S can have detrimental health effects, including headache and eye irritation, even at low concentrations of 2.5 to 5 ppm (73 to 146 µM) (16), and exposure to high concentrations (>1,000 ppm/29,343 µM) can lead to instantaneous death (17). The primary reason for H2S’s high toxicity is that it directly inhibits cytochrome c oxidase, which is Complex IV of the mitochondrial electron transport chain (16, 18). The inhibition of cytochrome c oxidase by H2S results in a shutdown of the electron transport chain and cellular ATP generation (16, 18).
While H2S is toxic at high concentrations, H2S is produced at low concentrations endogenously as a product of cysteine catabolism and by intestinal bacteria (19, 20), and physiologically relevant concentrations are likely in the nanomolar range (21, 22). Due to the endogenous production of H2S, most organisms are able to detoxify low concentrations of H2S, and there are known sulfide detoxification enzymes present in most metazoans (23). Sulfide oxidation to thiosulfate is mediated by sulfide quinone oxidoreductase, a dioxygenase, and sulfur transferase (24). H2S is highly membrane permeable (25) and plays an important role as a cell-signaling molecule in the cardiovascular and nervous systems (26). For example, H2S is involved in the regulation of vasodilation and inflammation (27). The effect of H2S on epigenetic changes has not been investigated.
Very few organisms are able to tolerate exposure to high H2S concentrations (16, 28). However, some fish in the livebearing family Poeciliidae live in habitats with naturally occurring sustained and high concentrations of H2S (29–34). The current study focuses on the species Poecilia mexicana, which inhabits H2S-rich springs in southern Mexico (Fig. 1) (35). H2S in these springs ranges from 30 µM to over 1,000 µM, depending on the site (35). The concentrations fluctuate little across seasons and years (30, 36). There are also closely related populations of P. mexicana residing in nonsulfidic habitats adjacent to sulfidic springs. The populations are genetically differentiated but considered the same species (37–39). Wild-caught fish from sulfide spring populations exhibit significantly higher sulfide tolerance than fish from adjacent nonsulfidic populations (35). We have previously shown that P. mexicana from sulfidic populations can survive in high levels of H2S by constitutively expressing high levels of important H2S detoxification genes (40–42), and some sulfidic populations have evolved a resistant cytochrome c oxidase (43). Moreover, there is heritable variation in gene expression, especially in key genes related to H2S toxicity and detoxification (44), some of which may be driven by expression differences of relevant microRNAs (45). Despite these observations, a key gap in our knowledge is the effect of H2S exposure on epigenetics and whether expression changes due to H2S exposure and local adaptation are potentially mediated by DNA methylation changes. Fish are known to have an increased number of DNA methylation enzymes, DNA methyltransferases (DNMTs), and respond to environmental insults through DNA methylation (46).
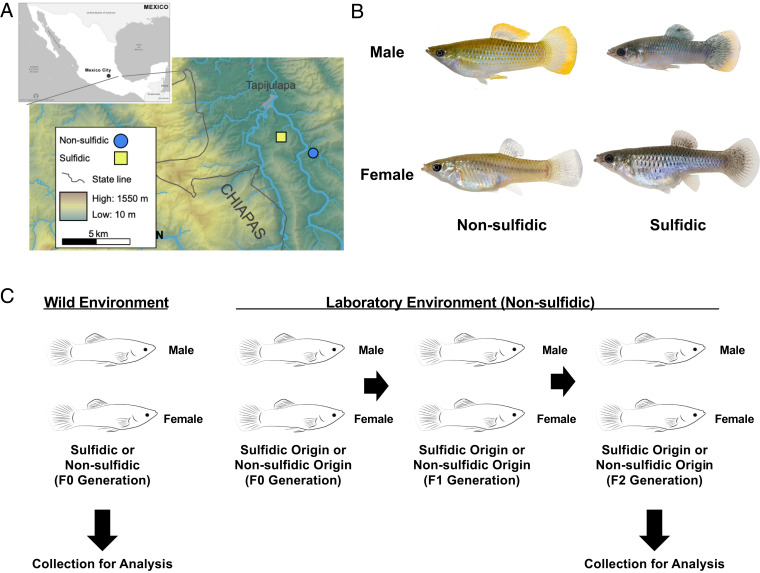
Fig. 1.
Collection site, P. mexicana fish morphology, and experimental design. (A) Collection sites in southern Mexico. Map from ref. 79, which is licensed under CC BY 3.0. (B) Images of P. mexicana adult male and female fish from sulfidic and nonsulfidic environments. (C) Experimental design.
The current study was designed to understand the long-term epigenetic changes that can occur in response to H2S adaptation. This study also sets the stage for future studies examining how short- and long-term environmental exposure may shape the epigenome and the role of epigenetic transgenerational inheritance in adaptation to H2S. The primary research objectives were to test whether 1) the abundance and distribution of differential DNA methylation regions (DMRs) differ between sulfidic and nonsulfidic populations for both sexes, when samples were derived from wild versus laboratory-reared individuals, and 2) the methylation differences between populations are consistent and related to living in H2S. We hypothesized that long-term H2S exposure in P. mexicana involved alterations in epigenetics (DNA methylation) and that those changes are stable over generations.
Results
Red blood cells (RBC) from male and female P. mexicana from H2S-rich habitats and nonsulfidic habitats in southern Mexico were collected to assess the environmental impacts of the sulfidic habitats on the fish epigenome (Fig. 1). The RBC in fish species contain nuclei, which allows DNA extraction from an easily purified single-cell type. Another set of P. mexicana from sulfidic and nonsulfidic habitats were collected as adults (pregnant females) in May 2013 and transported to a nonsulfidic laboratory environment and propagated for two generations in the nonsulfidic environment (Fig. 1C). Adult F2 generation laboratory fish harvested in August 2015 were used for analysis and comparisons (Fig. 1C). Genomic DNA from the nucleated RBC was isolated and used to identify differential DMRs between the sulfidic and nonsulfidic populations for males and females, separately. This experimental design allowed us to determine population differences in DMRs of wild fish populations and the stability of the epigenetic alterations in the laboratory propagated fish.
The RBC DNA was fragmented and the methylated DNA immunoprecipitated (MeDIP) with a methyl cytosine antibody to identify DMRs between the sulfidic and nonsulfidic populations. On average, similar numbers of DMRs were observed for the various comparisons between sulfidic and nonsulfidic populations (Table 1). Two-thirds of DMRs had one significant 100-base pair (bp) window, and the other third of DMRs exhibited multiple, significant 100-bp regions within the DMR (Table 2). The DMRs for each comparison are presented in SI Appendix, Tables S1–S4. For both wild male and female comparisons between sulfidic and nonsulfidic populations, ∼50% of the DMRs had an increase in DNA methylation in the sulfidic population, and the remainder of the DMRs had a decrease in methylation (SI Appendix, Fig. S1). Although a limitation of the study is the small number of fish analyzed, the robust and significant epigenetic data obtained support the observations presented.
Table 1.
DMR identification
| Comparison | All window | Multiple window |
| Sulfidic Male Wild (SMW) vs. Nonsulfidic Male Wild (NMW) | 1,049 | 440 |
| Sulfidic Female Wild (SFW) vs. Nonsulfidic Female Wild (NFW) | 1,461 | 606 |
| Sulfidic Male Laboratory (SML) vs. Nonsulfidic Male Laboratory (NML) | 1,619 | 641 |
| Sulfidic Female Laboratory (SFL) vs. Nonsulfidic Female Laboratory (NFL) | 1,451 | 544 |
The number of DMRs found in each comparison using P value threshold of P < 10−7. The All window (100 bp) column shows all DMRs. The Multiple window column shows the number of DMRs with multiple significant 100-bp windows within the DMR.
Table 2.
Number of DMRs with a given number of significant windows at p-value < 10−7
| Comparison | Number of significant windows | |||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ≥12 | |
| Sulfidic male wild (SMW) vs. nonsulfidic male wild (NMW) | 609 | 240 | 76 | 45 | 23 | 17 | 10 | 4 | 8 | 5 | 3 | 9 |
| Sulfidic female wild (SFW) vs. nonsulfidic female wild (NFW) | 855 | 331 | 119 | 61 | 37 | 17 | 12 | 3 | 9 | 2 | 4 | 10 |
| Sulfidic male laboratory (SML) vs. nonsulfidic male laboratory (NML) | 978 | 377 | 120 | 57 | 33 | 17 | 12 | 3 | 6 | 4 | 1 | 9 |
| Sulfidic female laboratory (SFL) vs. nonsulfidic female laboratory (NFL) | 907 | 329 | 110 | 44 | 18 | 12 | 6 | 6 | 3 | 2 | 2 | 12 |
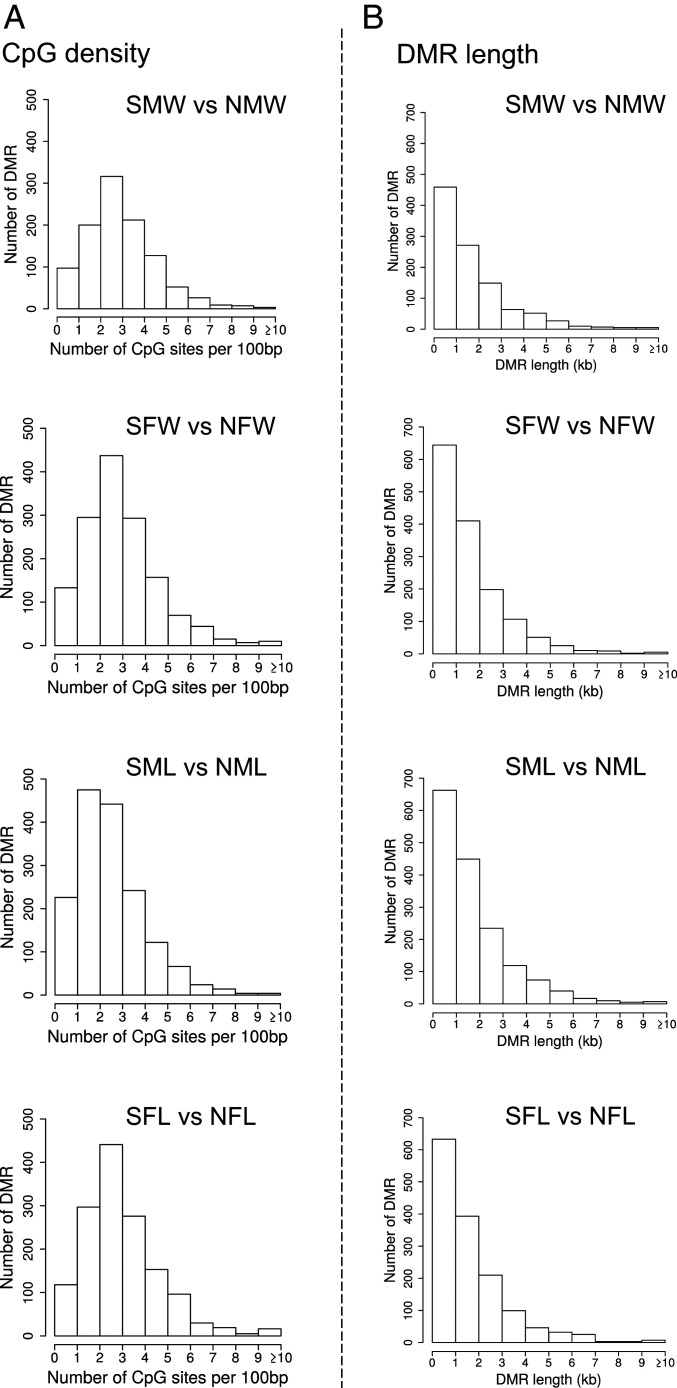
The genomic features associated with DMRs for each comparison were similar. The primary cytosine nucleotide followed by a guanine nucleotide (CpG) density was 1 to 6 CpG per 100 bp (Fig. 2). The average size of a DMR was around 1 kilobase (kb), with a range from 1 to 6 kb (Fig. 2). Therefore, the CpG density was low in 1 to 2 kb regions, similar to CpG deserts previously observed (47).
Fig. 2.
DMR genomic features for DMRs with a P value < 10−7. (A) The number of DMRs at different CpG densities for each of the pairwise comparisons. (B) The DMR lengths in kilobase (kb) for each of the pairwise comparisons. Sample names are: sulfidic female wild (SFW), nonsulfidic female wild (NFW), sulfidic male wild (SMW), nonsulfidic male wild (NMW), sulfidic female laboratory (SFL), nonsulfidic female laboratory (NFL), sulfidic male laboratory (SML), and nonsulfidic male laboratory (NML).
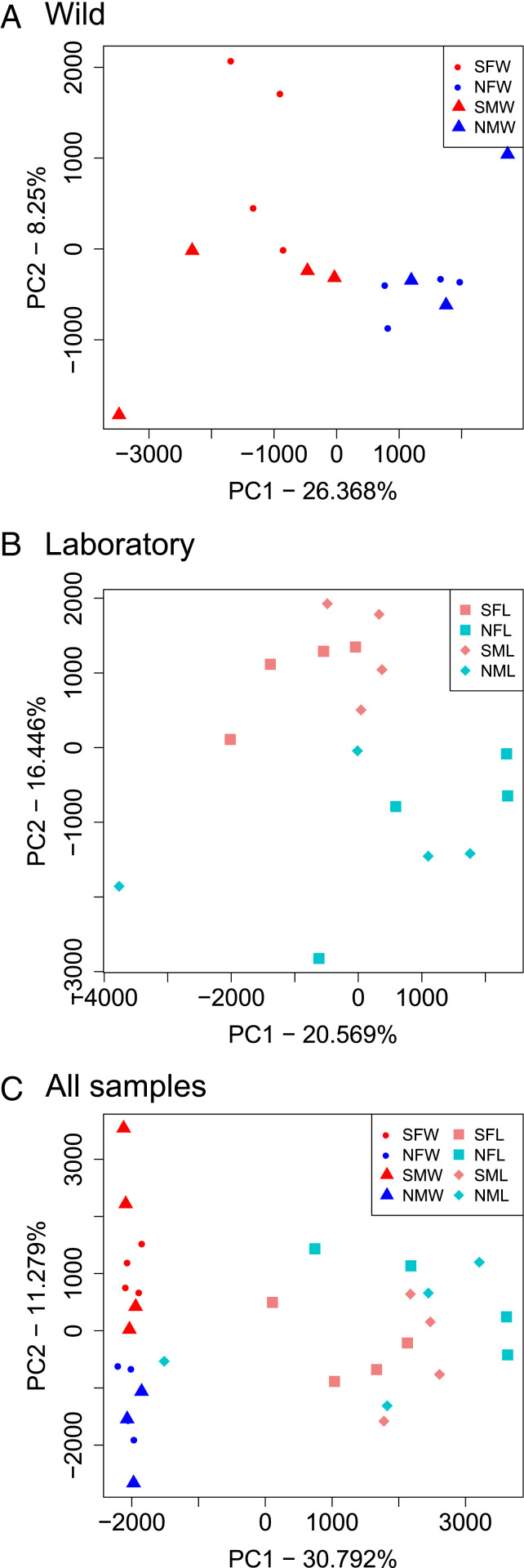
A principal component analysis (PCA) was performed to identify patterns in genome-wide methylation across all sample groups (Fig. 3). The reads per kilobase of transcript, per million mapped (RPKM) read depths at all genomic sites were used as the basis for the PCA. For samples collected in the wild, the sulfidic males and females clustered and were distinct from the nonsulfidic males and females (Fig. 3A). Therefore, the sulfidic habitat influences the PCA clustering for both sexes. For the laboratory samples from fish that were propagated in a nonsulfidic environment for multiple generations, the PCA clustering again showed that the individuals derived from the sulfidic population clustered distinctly from the nonsulfidic one (Fig. 3B). Hence, the samples were separated by habitat type in the PCA analysis for both the wild and laboratory populations. The PCA analysis with all samples from all comparisons further demonstrated that the wild samples clustered separately from the laboratory samples (Fig. 3C). Therefore, both the population of origin and the rearing environment (wild versus laboratory) impact methylation patterns.
Fig. 3.
PCA. (A) Wild samples. (B) Laboratory samples. (C) All samples. Sample labels are: sulfidic female wild (SFW), nonsulfidic female wild (NFW), sulfidic male wild (SMW), nonsulfidic male wild (NMW), sulfidic female laboratory (SFL), nonsulfidic female laboratory (NFL), sulfidic male laboratory (SML), and nonsulfidic male laboratory (NML).
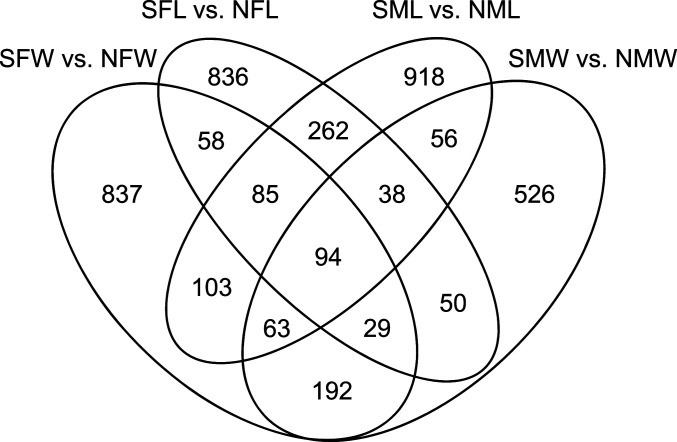
The majority of the DMRs were unique to a specific dataset at P < 10−7, with the wild or laboratory male and female comparisons having the highest level of overlap (Fig. 4). Interestingly, 94 DMRs overlapped between all four comparisons (SI Appendix, Table S5), and nearly 20% of DMRs were shared between laboratory and field conditions at P < 10−7. We also performed an extended DMR overlap that compared the DMRs with P < 10−7 for each comparison with the genomic windows in a second comparison at P < 0.05 (Table 3). DMRs in one analysis were considered present in the second analysis if any overlapping genomic window in the second analysis had a P value of 0.05 or less. A comparison of the male and female data sets within the wild or laboratory comparisons demonstrated greater than 90% overlap. Therefore, as indicated by the PCA (Fig. 3) and Venn diagram (Fig. 4), the overlaps are very high between the sexes for each laboratory or wild samples. Interestingly, the extended overlaps for the comparisons between laboratory and wild samples for both males and females were greater than 80% (Table 3). Therefore, even after two generations of maintaining fish under nonsulfidic laboratory conditions, the majority of DMRs documented in the laboratory overlapped with those documented in the wild with the extended overlap comparison. Although this extended overlap with multiple P values is not standard, it does reveal a high degree of overlap not observed in the high statistical threshold comparison.
Fig. 4.
DMR overlap for DMRs with a P value threshold of P < 10−7. Sample groups are: sulfidic female wild (SFW), nonsulfidic female wild (NFW), sulfidic male wild (SMW), nonsulfidic male wild (NMW), sulfidic female laboratory (SFL), nonsulfidic female laboratory (NFL), sulfidic male laboratory (SML), and nonsulfidic male laboratory (NML).
Table 3.
Extended DMR Overlap
| SFW versus NFW | SMW versus NMW | SFL versus NFL | SML versus NML | |
| SFW versus NFW | 1,372 (94%) | 1,207 (83%) | 1,205 (82%) | |
| SMW versus NMW | 984 (94%) | 871 (83%) | 860 (82%) | |
| SFL versus NFL | 1,192 (82%) | 1,155 (80%) | 1,324 (91%) | |
| SML versus NML | 1,356 (84%) | 1,291 (80%) | 1,496 (92%) |
The P < 10−7 left axis and P < 0.05 top axis with number of DMR overlap and percent of total for the specific comparison. Sample groups are: sulfidic female wild (SFW), nonsulfidic female wild (NFW), sulfidic male wild (SMW), nonsulfidic male wild (NMW), sulfidic female laboratory (SFL), nonsulfidic female laboratory (NFL), sulfidic male laboratory (SML), and nonsulfidic male laboratory (NML).
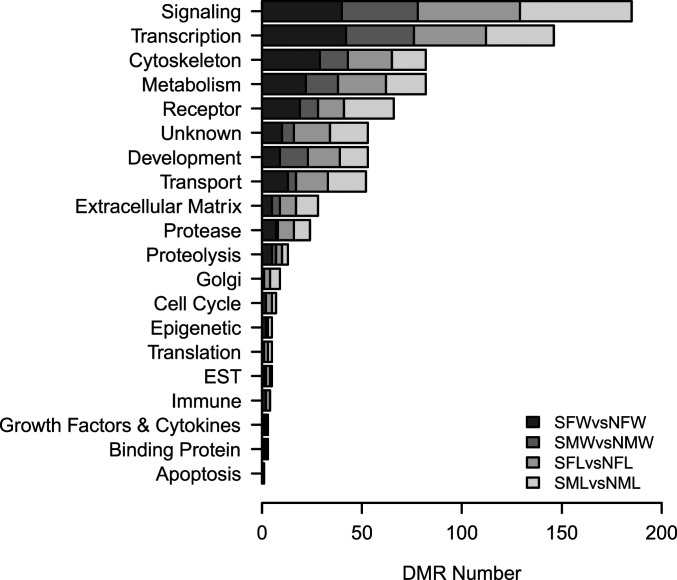
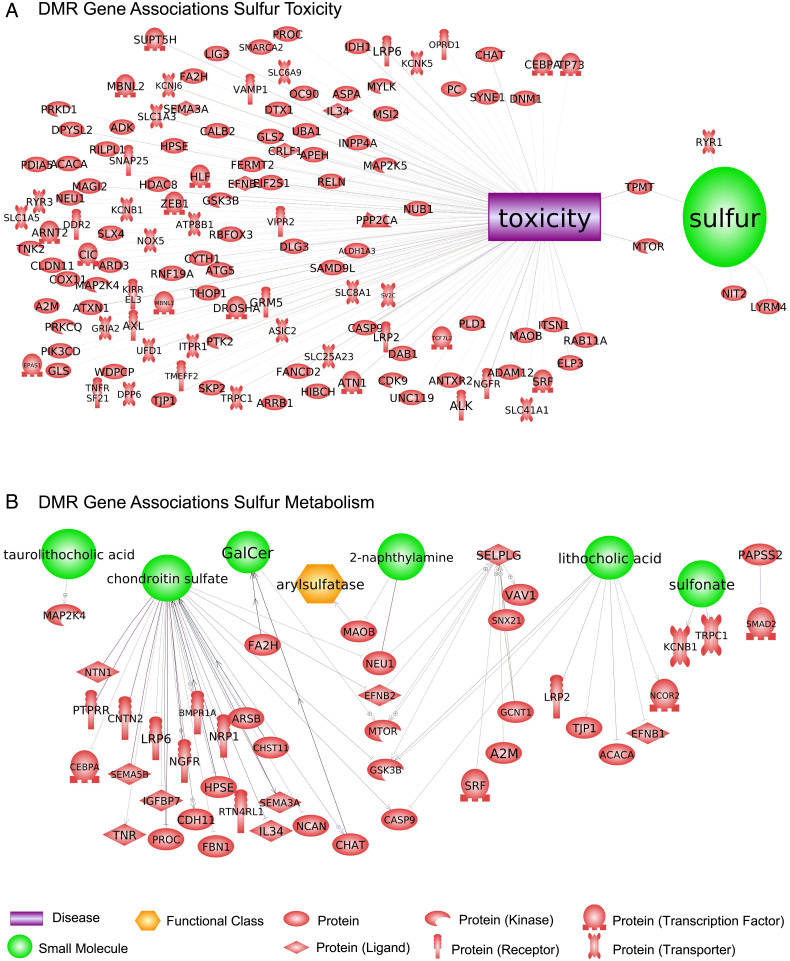
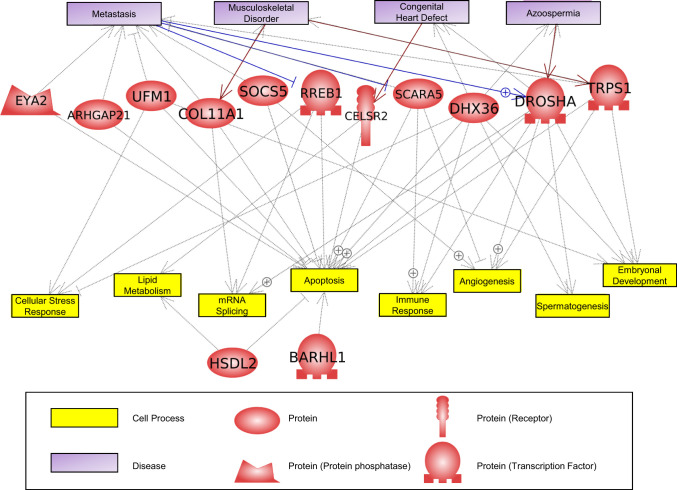
The DMR gene associations were investigated using DMRs identified within 10 kb of a gene so the promoter regions could be considered (SI Appendix, Tables S1–S4). The signaling, transcription, cytoskeleton, metabolism, and receptor functional categories were the most predominant DMR-associated genes in all comparisons (Fig. 5). There were consistent similarities in the DMR-associated gene numbers for all the categories. Analysis of the DMR-associated gene functions using a Pathway Studio program analysis identified a large number of genes related to sulfur toxicity, either directly or indirectly (Fig. 6A). A variety of related protein ligands, transcription factors, receptors, kinases, and phosphatases were present for the comparison data sets. Another relevant DMR-associated gene functional category involved sulfur metabolism (Fig. 6B). A number of different components of sulfur metabolism processes were listed, and all had connections to DMR-associated genes. The most predominant were chondroitin sulfate–associated genes that are a critical component of the extracellular matrix and proteoglycans. The final set of DMR-associated genes analyzed was the 94 overlapping DMRs between all comparisons (Fig. 4) with the DMR list and DMR-associated genes (SI Appendix, Table S5). These DMR-associated genes and correlated gene processes are presented in Fig. 7. Although this does not demonstrate direct links with sulfidic environments or toxicity, the pathways and gene families influenced are anticipated to be indirectly linked to the environmental exposure and potential adaptation.
Fig. 5.
DMR-associated gene categories and the number of DMRs for each comparison presented for each category. Sample groups are: sulfidic female wild (SFW), nonsulfidic female wild (NFW), sulfidic male wild (SMW), nonsulfidic male wild (NMW), sulfidic female laboratory (SFL), nonsulfidic female laboratory (NFL), sulfidic male laboratory (SML), and nonsulfidic male laboratory (NML).
Fig. 6.
DMR gene associations from Pathway Studio. (A) DMR gene associations with sulfur toxicity. (B) DMR gene associations with sulfur metabolism. Gene symbols are presented and the functional categories listed.
Fig. 7.
Overlapping 94 DMR P < 10−7 gene associations with cellular processes and pathologies from Pathway Studio. Gene symbols are presented and the functional categories listed.
Discussion
There is evidence that epigenetic modifications change in response to exposure to environmental toxicants (4, 7, 11) and that modified placement of epigenetic modifications are associated with phenotypic variation and disease (3). The analysis of methylation changes in populations adapted to a physiological stressor allows for the assessment of short-term responses to environmental stress as well as potential forecasting of long-term responses. Here, we used H2S-sensitive and H2S-tolerant fish, P. mexicana, living in naturally nonsulfidic and sulfidic environments, respectively, to elucidate the epigenetic changes in response to living in H2S-rich springs.
The current study investigates the epigenetic inheritance stability of putatively adaptive epigenetic alterations following the removal of the abiotic selective pressure. Although the P. mexicana genome (48) is not a chromosome-level assembly, sufficient information was available to assess the environmental influences on the DMRs, associated genomic features, and potential gene associations. The lack of a well-characterized genome will underestimate the DMR-gene associations, and future improved genomic information will likely improve and expand on the gene association information. The DMR size and locations in regions of low-density CpG (termed CpG deserts) found in this study are similar to environmentally responsive DMRs previously identified in other species (47, 49). Low-density CpG regions generally constitute over 90% of the genome for most species, while the high-density CpG islands generally constitute less than 5% of the genome (49). As shown in most species, from plants to mammals, CpG DNA methylation is predominant, with non-CpG methylation being negligible with no known functional significance currently identified (50). Although the MeDIP procedure used is biased to low-density CpG regions, it is the most efficient to examine the majority of the genome compared to other procedures, such as bisulfite sequencing, that can be biased to higher-density CpG analyses (47, 51). High-density CpG regions are predominantly restricted to 50% of promoters in mammals, which is less than 5% of the genome. While these regions are not detected efficiently with MeDIP sequencing, greater than 90% of the genome is detected with MeDIP sequencing (52).
The DMR-associated genes were investigated to provide insights into the potential impacts of the epigenetic alterations on the genome and physiology. Although a gene association is identified, this does not confirm a functional causal link of the epigenetic alteration and gene regulation. This is a limitation that will require future studies at the gene expression level to confirm. Due to the sulfidic-versus-nonsulfidic focus of the study, the initial analysis of the DMR-associated genes involved sulfur toxicity and metabolism. A large number of toxicity-associated genes that were either directly or indirectly correlated with sulfur that were associated with the DMRs identified including genes such as MTOR or TPMT have been shown to have direct impacts on sulfur toxicity (53–56). Sulfur metabolism genes were also associated with the DMRs identified. The chondroitin sulfate processes, which have been associated with sulfur exposures (57, 58), had the most represented associated genes. A number of cellular processes (e.g., apoptosis and angiogenesis) were also associated with the DMRs. Although these DMR-associated genes have links with sulfur toxicity and metabolism, further molecular work is needed to correlate these molecular processes and genes to adaptation of P. mexicana to H2S and the observed epigenetic inheritance.
An overlap of the DMRs between the comparisons and data sets demonstrated that the majority of DMRs were distinct for the comparison with DMRs at P < 10−7. The primary principal component in the PCA separated laboratory and wild samples, suggesting that there are largescale changes in DMRs when populations are brought into the laboratory for multiple generations. An approximately 20% overlap was observed between the male and female wild or laboratory DMR comparisons. To expand this analysis, an extended overlap was performed that compared the P < 10−7 DMR with the P < 0.05 DMRs for the other data sets. Although the extended overlap with a high and low statistical threshold is not standard, this allows for the identification of overlapping DMRs that may be marginally significant at any given P value threshold. The comparison showed a greater than 90% overlap between the male or female DMR comparisons for the wild or laboratory DMRs. Greater than 80% overlap was observed between the wild and laboratory comparisons; this level of overlap demonstrates that the majority of the sulfidic versus nonsulfidic DMRs identified were in common between the F0 generation wild populations and the F2 generation laboratory populations. Therefore, the majority of the methylation changes identified comparing wild individuals from sulfidic and nonsulfidic environments were found to be propagated for two generations in the laboratory in the absence of the sulfidic environmental exposure. These observations demonstrate an epigenetic inheritance of DMRs in the absence of the continued sulfidic environment. This is an observation of methylation inherited through the germline when an environmental stressor (i.e., H2S) is removed from a wild population and suggests that future studies on transgenerational inheritance beyond the F2 generation would be useful. This is consistent with previously described environmentally induced epigenetic transgenerational inheritance phenomena (4, 6).
Environmentally induced epigenetic inheritance of phenotypic variation and disease was first described in rats exposed to an agricultural fungicide vinclozolin (11) but now has been shown to occur in all species investigated from plants to humans (4, 6). Previous studies have primarily used the transient exposure of an individual to induce epigenetic alterations in the germline (sperm or egg) to promote the epigenetic transgenerational inheritance phenomenon (4, 6). The transmission of epigenetics between generations requires the involvement of the germline, as they are the only cells that can transmit molecular information between generations (4, 6). Direct exposure to an environmental factor can promote epigenetic alterations, and this can occur at any time during development. For example, the exposure of a gestating female mammal exposes the F0 generation mother, the F1 generation fetus, and germline that will generate the F2 generation individual (4, 59). P. mexicana is a livebearing fish that requires the same consideration. Therefore, the F3 generation is the first generation that has no direct exposure toxicity (59). For a nonlivebearing fish model, exposure of the F0 generation male or female will directly impact the F0 generation and the sperm or egg that will form the F1 generation; therefore, the F2 generation is the first generation (i.e., transgenerational) not having direct exposure toxicity (59). An example in fish involves the exposure of zebrafish to mercury that promoted neurodevelopmental abnormalities in zebrafish in the F2 generation offspring, which was the first epigenetic transgenerational inheritance observation in fish (60). However, livebearing fish are more like mammals in that the exposure of a gestating female will expose the germline that will generate the F2 generation. The current study observations suggest that an important future study in these livebearing fish is the investigation of DMRs in the F3 generation, the first with no direct exposure to H2S, which would be considered epigenetic transgenerational inheritance. Although the current study did not analyze the F3 generation in the nonsulfidic environment, propagation of the F2 generation in the laboratory environment demonstrates survival of the fish to the F3 generation and beyond. Future studies will need to compare the F1 generation of laboratory fish to determine whether there is a steady decrease in DMR shared with each subsequent generation or whether the changes are due to the initial transition to an aquarium environment and are stable and maintained at similar levels thereafter. It is also unknown whether fish from sulfidic habitats that have been raised in a nonsulfidic laboratory environment for multiple generations may have a higher survival than fish with nonsulfidic ancestry when transmitted back into a sulfidic environment. The question is whether an ancestral sulfidic environment adapted epigenetics is maintained, which may facilitate reintroduction into the sulfidic environment. This will be an important future experiment to consider for understanding how epimutations altered in an aquarium environment play a role in phenotypic variation and survival.
Although the ability of environmental stressors to alter the epigenome of the individual exposed and impact the physiology of the individual is critical, the ability to alter the germline epigenome and propagate this transgenerationally to subsequent generations allows for an adaptive mechanism that can impact evolution (61). The phenotypic plasticity and molecular alterations in the epigenome and transcriptome to survive the sulfidic environment are essential to allow an organism to respond to its environment. The epimutations documented here may contribute to heritable variation in gene expression, which we have documented in detail in the field, the laboratory, and across multiple species (40, 41, 44, 62). The sulfidic and nonsulfidic populations of P. mexicana studied here are genetically differentiated, with a divergence time of ∼10,000 y ago and an average empirical FST of 0.11 (63), but considered the same species (37–39), and studying how these epigenetic changes play into adaptive changes will be a fruitful avenue of research. The population studied here is one of four pairs of sulfidic and nonsulfidic P. mexicana populations in tributaries of the Río Grijalva, and the replicated nature of the independently derived sulfidic populations provides an opportunity to test whether the findings in this drainage are consistent across multiple drainages. Future research will need to explore how genetic variation interacts with epimutations to shape phenotypic variation in this system. This project is a first step to evaluate epigenetic effects of toxicant exposure and sets the stage for future studies examining how long-term environmental exposure may shape the epigenome.
Materials and Methods
Samples.
Male and female P. mexicana were collected from H2S-rich habitats and nonsulfidic habitats in southern Mexico (Fig. 1) for the preparation of purified RBCs. Wild-caught samples were obtained from southern Mexico in proximate H2S-rich (sulfidic, n = 4 female, n = 4 male) (PSO, Latitude: 17.43843, Longitude: −92.77476) and nonsulfidic springs (n = 4 female, n = 3 male) (Bonita, Latitude: 17.42685, Longitude: −92.75213) in the Tacotalpa Drainage in southern Mexico. Another set of P. mexicana males and females from sulfidic or nonsulfidic habitats were collected and transported into a breeding population at Kansas State University and propagated for two generations (F2) in the nonsulfidic laboratory environment (Fig. 1C) (sulfidic, n = 4 female, n = 4 male) and (nonsulfidic, n = 4 female, n = 4 male). All laboratory animals were reared in nonsulfidic water, and n = 4 individuals were sampled per population. Adult fish for meDIP sequencing were collected in the wild in May 2015. For laboratory experiments, pregnant females were collected in May 2013, and adult F2 offspring were harvested in August 2015 for blood extractions. Of note, fish from sulfidic habitats are smaller than fish from the nonsulfidic habitats, and males are smaller than females.
DNA Extraction.
Fish were euthanized by cervical transection, and blood was immediately extracted by using microhematocrit capillary tubes (Fisher). Tubes were then spun down at 12,000 rpms for 3 min using a ZipCombo Centrifuge (LW Scientific) to separate blood cells from serum. After centrifugation, capillaries were broken to retain only RBCs. The portion of the capillary containing the RBCs was placed in a 1.5-mL Eppendorf tube for DNA extraction. DNA was extracted using the Qiagen DNeasy Blood & Tissue kit according to the manufacturer’s instructions.
MeDIP.
Extracted genomic DNA was used to isolate methylated DNA with a methyl-cytosine antibody precipitation procedure (MeDIP). The protocol is described in detail in ref. 64. A total of 5 μg total genomic DNA was sonicated using a Covaris sonicator. Sonicated DNA was diluted with TE (tris-ethylenediaminetetraacetic acid [EDTA]) buffer to 400 μL, heat denatured for 10 min at 95 °C, and cooled on ice for 10 min to create single-stranded DNA fragments. A total of 100 μL of 5× IP (immunoprecipitation) buffer and 5 μg of antibody (monoclonal mouse anti 5-methyl cytidine; Diagenode C15200006) were added to the fragmented single-stranded DNA. The mixture was incubated on a rotator overnight at 4 °C. Prewashed magnetic beads (50 μL, Dynabeads M-280 Sheep anti-Mouse IgG; Life Technologies 11201D) were added to the 500 μL of DNA–antibody mixture. The mixture was incubated for 2 h on a rotator at 4 °C, after which the samples were washed three times with 1× IP buffer. Washed samples were resuspended in 250 μL digestion buffer (5mM Tris pH 8, 10.mM EDTA, and 0.5% SDS [sodium dodecyl sulfate]) with 3.5 μL Proteinase K (20mg/mL) and incubated at 55° for three hours. A Phenol-Chloroform-Isoamyalcohol extraction was used to clean-up the DNA, and the DNA was resuspended in 20 μL of H2O. DNA concentration was measured in Qubit (Life Technologies) with the single-stranded (ss) DNA kit (Molecular Probes Q10212).
MeDIP Sequencing.
For library preparation, we used the NEBNext Ultra RNA Library Prep Kit for Illumina starting at step 1.4 of the manufacturer’s protocol to generate double-stranded DNA. After this step, the manufacturer’s protocol was followed. Each individual received a unique barcode. Enriched methylated DNA libraries were sequenced at the Washington State University Genomics Core in Spokane, WA, using an Illumina HiSeq 2500 with paired-end 50-bp reads.
Identifying and Analyzing Differentially Methylated Regions.
DMR identification and annotation followed previously published aapproaches (65, 66). The FastQC program (67) was used to assess data quality. Reads were trimmed to remove adapters and low-quality bases using Trimmomatic (68). The reads for each MeDIP sample were mapped to the P. mexciana (48) genome using Bowtie2 (69) with default parameter options. The mapped read files were then converted to sorted BAM (Binary Sequence Alignment/Map) files using SAMtools (70).
Differential coverage between sulfidic and nonsulfidic populations was calculated using the MEDIPS R package (71). P value from edgeR (72) was used to determine the significance of the difference between the two groups for each 100-bp genomic window. Windows with an edgeR P value less than a specified threshold (P < 10−7) were considered the initial start of the DMR. DMR edges were extended until no genomic window with a P value less than 0.1 remained within 1,000 bp of the DMR. The extended DMR overlap compared the DMRs, with at least one 100-bp window with P < 10−7 from one comparison, with the genomic windows (100-bp regions) in a second comparison. Windows that had a P value <0.05 in the second comparison were considered overlapping. The Ensembl database (73), accessed with the biomaRt R package (74), was used to annotate DMRs. Genes that were overlapping a DMR, including 10 kb on either side of the DMR, were input into a Kyoto Encyclopedia of Genes and Genomes pathway search (75, 76) to identify relevant associated pathways. The DMR-associated genes were sorted into functional groups using information provided by the DAVID (Database for Annotation, Visualization and Integrated Discovery) (77) and Panther (78) databases incorporated into an internal curated database (https://www.skinner.wsu.edu/). DMR-associated gene correlations present in published literature were further analyzed using Pathway Studio software (version 12.2.1.2: Database of functional relationships and pathways of mammalian proteins; Elsevier). R code computational tools are available at GitHub (https://github.com/skinnerlab/MeDIP-seq) and https://www.skinner.wsu.edu.
Ethical Statement.
Procedures for all experiments were approved by the Institutional Animal Care and Use Committee at Kansas State University (protocol no. 3418). Field work was approved by the Mexican government (fieldwork permit DGOPA.00093.120110.-0018).
Supplementary Material
Acknowledgments
We thank the field crew for assistance with sampling. We also appreciate Centro de Investigación e Innovación para la Enseñanza y Aprendizaje and Universidad Juárez Autónoma de Tabasco for their hospitality and support over many years of research. J.L.K. would like to thank her writing group and the Cornejo and Kelley laboratory groups for feedback on the manuscript. We acknowledge the technical assistance of Dr. Eric Nilsson for gene association analysis and critically reviewing the manuscript. We acknowledge Ms. Amanda Quilty for editorial assistance and Ms. Heather Johnson for assistance in preparation of the manuscript. This work was funded by grants from the NSF (IOS-1121832, IOS-1463720, IOS-1557860, IOS-1557795, IOS-1931650, and IOS-1931657) and the US Army Research Office (W911NF-15-1-0175) to M.T. and J.L.K. as well as Templeton Foundation support to M.K.S.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014929118/-/DCSupplemental.
Data Availability
Sequencing data, code, and internal databases data have been deposited in https://github.com/skinnerlab/MeDIP-seq as well as https://www.skinner.wsu.edu (GSE157730).
References
- 1.Reamon-Buettner S. M., Mutschler V., Borlak J., The next innovation cycle in toxicogenomics: Environmental epigenetics. Mutat. Res. 659, 158–165 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Dolinoy D. C., Weidman J. R., Jirtle R. L., Epigenetic gene regulation: Linking early developmental environment to adult disease. Reprod. Toxicol. 23, 297–307 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Portela A., Esteller M., Epigenetic modifications and human disease. Nat. Biotechnol. 28, 1057–1068 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Jirtle R. L., Skinner M. K., Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 8, 253–262 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baccarelli A., Bollati V., Epigenetics and environmental chemicals. Curr. Opin. Pediatr. 21, 243–251 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nilsson E., Sadler-Riggleman I., Skinner M. K., Environmentally induced epigenetic transgenerational Inheritance of disease. Environ. Epigenet. 4, dvy016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takiguchi M., Achanzar W. E., Qu W., Li G., Waalkes M. P., Effects of cadmium on DNA-(Cytosine-5) methyltransferase activity and DNA methylation status during cadmium-induced cellular transformation. Exp. Cell Res. 286, 355–365 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Zhong C. X., Mass M. J., Both hypomethylation and hypermethylation of DNA associated with arsenite exposure in cultures of human cells identified by methylation-sensitive arbitrarily-primed PCR. Toxicol. Lett. 122, 223–234 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Chanda S., et al., DNA hypermethylation of promoter of gene p53 and p16 in arsenic-exposed people with and without malignancy. Toxicol. Sci. 89, 431–437 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Zhao C. Q., Young M. R., Diwan B. A., Coogan T. P., Waalkes M. P., Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc. Natl. Acad. Sci. U.S.A. 94, 10907–10912 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anway M. D., Cupp A. S., Uzumcu M., Skinner M. K., Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308, 1466–1469 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerrero-Bosagna C., et al., Epigenetic transgenerational inheritance of vinclozolin induced mouse adult onset disease and associated sperm epigenome biomarkers. Reprod. Toxicol. 34, 694–707 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubsad D., et al., Assessment of glyphosate induced epigenetic transgenerational inheritance of pathologies and sperm epimutations: Generational toxicology. Sci. Rep. 9, 6372 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tobler M., et al., The evolutionary ecology of animals inhabiting hydrogen sulfide–rich environments. Annu. Rev. Ecol. Evol. Syst. 47, 239–262 (2016). [Google Scholar]
- 15.National Research Council , Hydrogen Sulfide (University Park Press, Baltimore, 1979). [Google Scholar]
- 16.Reiffenstein R. J., Hulbert W. C., Roth S. H., Toxicology of hydrogen sulfide. Annu. Rev. Pharmacol. Toxicol. 32, 109–134 (1992). [DOI] [PubMed] [Google Scholar]
- 17.Ellenhorn M. J., Ordog G. J., Schonwald S., Ellenhorn’s Medical Toxicology: Diagnosis and Treatment of Human Poisoning (Lippincott Williams & Wilkins, 1997). [Google Scholar]
- 18.Cooper C. E., Brown G. C., The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: Chemical mechanism and physiological significance. J. Bioenerg. Biomembr. 40, 533–539 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Kimura H., Hydrogen sulfide as a neuromodulator. Mol. Neurobiol. 26, 13–19 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Stipanuk M. H., Sulfur amino acid metabolism: Pathways for production and removal of homocysteine and cysteine. Annu. Rev. Nutr. 24, 539–577 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Furne J., Saeed A., Levitt M. D., Whole tissue hydrogen sulfide concentrations are orders of magnitude lower than presently accepted values. Am. J. Physiol. Regul. Integr. Comp. Physiol. 295, R1479–R1485 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Levitt M. D., Abdel-Rehim M. S., Furne J., Free and acid-labile hydrogen sulfide concentrations in mouse tissues: Anomalously high free hydrogen sulfide in aortic tissue. Antioxid. Redox Signal. 15, 373–378 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Hildebrandt T. M., Grieshaber M. K., Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS J. 275, 3352–3361 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Lagoutte E., et al., Oxidation of hydrogen sulfide remains a priority in mammalian cells and causes reverse electron transfer in colonocytes. Biochim. Biophys. Acta 1797, 1500–1511 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Mathai J. C., et al., No facilitator required for membrane transport of hydrogen sulfide. Proc. Natl. Acad. Sci. U.S.A. 106, 16633–16638 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L., Rose P., Moore P. K., Hydrogen sulfide and cell signaling. Annu. Rev. Pharmacol. Toxicol. 51, 169–187 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Bhatia M., Hydrogen sulfide as a vasodilator. IUBMB Life 57, 603–606 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Bagarinao T., Sulfide as an environmental factor and toxicant: Tolerance and adaptations in aquatic organisms. Aquat. Toxicol. 24, 21–62 (1992). [Google Scholar]
- 29.Tobler M., Hastings L., Convergent patterns of body shape differentiation in four different clades of poeciliid fishes inhabiting sulfide springs. Evol. Biol. 38, 412–421 (2011). [Google Scholar]
- 30.Tobler M., Plath M., “Living in extreme habitats” in Ecology and Evolution of Poeciliid Fishes, Evans J., Pilastro A., Schlupp I., Eds. (University of Chicago Press, Chicago, 2011), pp. 120–127. [Google Scholar]
- 31.Greenway R., et al., Patterns of macroinvertebrate and fish diversity in freshwater sulphide springs. Diversity 6, 597–632 (2014). [Google Scholar]
- 32.Riesch R., Plath M., Schlupp I., Tobler M., Brian Langerhans R., Colonisation of toxic environments drives predictable life-history evolution in livebearing fishes (Poeciliidae). Ecol. Lett. 17, 65–71 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Tobler M., Kelley J. L., Plath M., Riesch R., Extreme environments and the origins of biodiversity: Adaptation and speciation in sulphide spring fishes. Mol. Ecol. 27, 843–859 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Greenway R., et al., Convergent evolution of conserved mitochondrial pathways underlies repeated adaptation to extreme environments. Proc. Natl. Acad. Sci. USA 117, 16424–16430 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tobler M., et al., Evolution in extreme environments: Replicated phenotypic differentiation in livebearing fish inhabiting sulfidic springs. Evolution 65, 2213–2228 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Tobler M., et al., Toxic hydrogen sulfide and dark caves: Phenotypic and genetic divergence across two abiotic environmental gradients in Poecilia mexicana. Evolution 62, 2643–2659 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Plath M., et al., Genetic differentiation and selection against migrants in evolutionarily replicated extreme environments. Evolution 67, 2647–2661 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Tobler M., Riesch R., Tobler C. M., Schulz-Mirbach T., Plath M., Natural and sexual selection against immigrants maintains differentiation among micro-allopatric populations. J. Evol. Biol. 22, 2298–2304 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Plath M., et al., Complementary effect of natural and sexual selection against immigrants maintains differentiation between locally adapted fish. Naturwissenschaften 97, 769–774 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Kelley J. L., et al., Mechanisms underlying adaptation to life in hydrogen sulfide-rich environments. Mol. Biol. Evol. 33, 1419–1434 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tobler M., Henpita C., Bassett B., Kelley J. L., Shaw J. H., H2S exposure elicits differential expression of candidate genes in fish adapted to sulfidic and non-sulfidic environments. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 175, 7–14 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Greenway R., et al., Convergent evolution of conserved mitochondrial pathways underlies repeated adaptation to extreme environments. Proc. Natl. Acad. Sci. U.S.A. 117, 16424–16430 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfenninger M., et al., Parallel evolution of cox genes in H2S-tolerant fish as key adaptation to a toxic environment. Nat. Commun. 5, 3873 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Passow C. N., et al., The roles of plasticity and evolutionary change in shaping gene expression variation in natural populations of extremophile fish. Mol. Ecol. 26, 6384–6399 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Kelley J. L., et al., microRNA expression variation as a potential molecular mechanism contributing to adaptation to hydrogen sulphide. J. Evol. Biol. 10.1111/jeb.13727 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Cavalieri V., Spinelli G., Environmental epigenetics in zebrafish. Epigenetics Chromatin 10, 46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skinner M. K., Guerrero-Bosagna C., Role of CpG deserts in the epigenetic transgenerational inheritance of differential DNA methylation regions. BMC Genomics 15, 692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warren W. C., et al., Clonal polymorphism and high heterozygosity in the celibate genome of the Amazon molly. Nat. Ecol. Evol. 2, 669–679 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skinner M. K., Manikkam M., Haque M. M., Zhang B., Savenkova M. I., Epigenetic transgenerational inheritance of somatic transcriptomes and epigenetic control regions. Genome Biol. 13, R91 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Law J. A., Jacobsen S. E., Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nair S. S., et al., Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics 6, 34–44 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Boers R., et al., Genome-wide DNA methylation profiling using the methylation-dependent restriction enzyme LpnPI. Genome Res. 28, 88–99 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu Y., et al., Organosulfur compounds induce cytoprotective autophagy against apoptosis by inhibiting mTOR phosphorylation activity in macrophages. Acta Biochim. Biophys. Sin. (Shanghai) 50, 1085–1093 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Yan S., et al., Dietary sulfur amino acids affect jejunal cell proliferation and functions by affecting antioxidant capacity, Wnt/β-catenin, and the mechanistic target of rapamycin signaling pathways in weaning piglets. J. Anim. Sci. 96, 5124–5133 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mokmak W., Tongsima S., Jenwitheesuk E., Molecular dynamics simulation of a human thiopurine S-methyltransferase complexed with 6-mercaptopurine model. Bioinformation 4, 59–62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cannon L. M., Butler F. N., Wan W., Zhou Z. S., A stereospecific colorimetric assay for (S,S)-adenosylmethionine quantification based on thiopurine methyltransferase-catalyzed thiol methylation. Anal. Biochem. 308, 358–363 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Fujiwara Y., Tsumura N., Yamamoto C., Kaji T., Differential effects of cadmium on proteoglycan synthesis of arterial smooth muscle cells: Increase in small dermatan sulfate proteoglycans, biglycan and decorin, in the extracellular matrix at low cell density. Toxicology 170, 89–101 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Parcell S., Sulfur in human nutrition and applications in medicine. Altern. Med. Rev. 7, 22–44 (2002). [PubMed] [Google Scholar]
- 59.Skinner M. K., What is an epigenetic transgenerational phenotype? F3 or F2. Reprod. Toxicol. 25, 2–6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carvan M. J., et al., Mercury-induced epigenetic transgenerational inheritance of abnormal neurobehavior is correlated with sperm epimutations in zebrafish. PLoS One 12, e0176155 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skinner M. K., Environmental epigenetics and a unified theory of the molecular aspects of evolution: A Neo-Lamarckian concept that facilitates Neo-darwinian evolution. Genome Biol. Evol. 7, 1296–1302 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Greep R., VanDyke H., Chow B., Gonadotropins of the swine pituitary: Various effects of purified thylakentrin (FSH) and pure metakentrin (ICSH). Endocrinology 30, 635 (1942). [Google Scholar]
- 63.Brown A. P., Arias-Rodriguez L., Yee M. C., Tobler M., Kelley J. L., Concordant changes in gene expression and nucleotides underlie independent adaptation to hydrogen-sulfide-rich environments. Genome Biol. Evol. 10, 2867–2881 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nilsson E., et al., Vinclozolin induced epigenetic transgenerational inheritance of pathologies and sperm epimutation biomarkers for specific diseases. PLoS One 13, e0202662 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McBirney M., et al., Atrazine induced epigenetic transgenerational inheritance of disease, lean phenotype and sperm epimutation pathology biomarkers. PLoS One 12, e0184306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ben Maamar M., et al., Alterations in sperm DNA methylation, non-coding RNA expression, and histone retention mediate vinclozolin-induced epigenetic transgenerational inheritance of disease. Environ Epigenet 4, dvy010 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fast QC, Version 0.11.9. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 1 December 2019.
- 68.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H.et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lienhard M., Grimm C., Morkel M., Herwig R., Chavez L., MEDIPS: Genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 30, 284–286 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cunningham F., et al., Ensembl 2015. Nucleic Acids Res. 43, D662–D669 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Durinck S., Spellman P. T., Birney E., Huber W., Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kanehisa M., Goto S., KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kanehisa M., et al., Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 78.Mi H., Muruganujan A., Casagrande J. T., Thomas P. D., Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mexico Capital Map Black and White. http://www.mapsopensource.com/mexico-capital-map-black-and-white.html. Accessed 30 June 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data, code, and internal databases data have been deposited in https://github.com/skinnerlab/MeDIP-seq as well as https://www.skinner.wsu.edu (GSE157730).