ABSTRACT
Alpha‐Klotho is a multi‐functional protein essential for maintenance of a myriad of cell functions. αKlotho is a single transmembrane protein with a large extracellular segment consisting of two domains (termed Kl1 and Kl2) which is shed into the extracellular fluid by proteolytic cleavage to furnish circulating soluble αKlotho. Based on cDNA sequence, an alternatively spliced mRNA is predicted to translate to a putative soluble αKlotho protein in mouse and human with only the Kl1 domain that represents a “spliced αKlotho Kl1” (spKl1) and is released from the cell without membrane targeting or cleavage. The existence of this protein remains in silico for two decades. We generated a novel antibody (anti‐spE15) against the 15 amino acid epitope (E15; VSPLTKPSVGLLLPH) which is not present in Kl1 or full‐length αKlotho and validated its specific reactivity against spKl1 in vitro. Using anti‐spE15 and two well‐established anti‐αKlotho monoclonal antibodies, we performed immunoblots, immunoprecipitation, and immunohistochemistry to investigate for expression of spKl1 in the mouse brain. We found anti‐spE15 labeling in mouse brain but were not able to see co‐labelling of Kl1 and spE15 epitopes on the same protein, which is the pre‐requisite for the existence of a spKl1 polypeptide, indicating that anti‐spE15 likely binds to another protein other than the putative spKl1. In isolated choroid plexus from mouse brain, we found strong staining with anti‐spE15, but did not find the spliced αKlotho transcript. We conclude that using reliable reagents and inclusion of proper controls, there is no evidence of the spKl1 protein in the mouse brain.
Keywords: alternative transcript, brain, Klotho, protein expression
1. INTRODUCTION
The murine klotho gene was cloned by serendipity over two decades ago when Kuro‐o and colleagues disrupted its 5’‐flanking region with the Na+/H+ exchanger‐1 cDNA during the creation of a murine model of hypertension. 1 , 2 A Klotho hypomorph with low levels of Klotho was thus generated with dramatic premature multi‐organ degeneration, earning Klotho the reputation as an anti‐aging protein. 1 , 3 Since its discovery, many advances have been made on multiple fronts. Klotho, now termed αKlotho, is part of a gene family with two more members, βKlotho and γKlotho. 4 , 5 βKlotho has critical roles in energy metabolism and bile acid regulation and the functional role of γKlotho is still elusive. 6 , 7 , 8 , 9 , 10 , 11 , 12 In contrast to the other members, αKlotho has an enormously wide range of functions beyond anti‐aging including mineral metabolism regulation, actions against oxidative damage, apoptosis, senescence, fibrosis, and hypertension. 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19
The predicted structure of αKlotho is a single‐pass transmembrane protein with a signal peptide, a large extracellular N‐terminus consisting of two homologous motifs called Kl1 and Kl2, a transmembrane region, and a very short cytoplasmic C‐terminus. 1 , 20 , 21 , 22 This structure is well preserved among human, mouse, and rat species. 1 , 20 , 21 , 22 The structure of the extracellular domain of both αKlotho and βKlotho were recently solved. 23 , 24 Transmembrane αKlotho serves as a co‐receptor for the bone‐derived mineral‐regulating hormone fibroblast growth factor FGF‐23, along with the FGF receptor (FGFR) family. 25 , 26 The extracellular region of transmembrane αKlotho is cleaved as full length soluble αKlotho, and released from the cell by secretases 27 , 28 , 29 , 30 into cerebrospinal fluid (CSF), blood, and urine 29 , 31 , 32 , 33 , 34 , 35 and acts on distal organs as an endocrine substance exerting a myriad of effects. 11 , 47 This model of αKlotho release requires plasma membrane targeting and proteolytic cleavage, which contribute to regulation of circulating αKlotho levels.
In addition to proteolytic cleavage, a second model of αKlotho release from cells was proposed based on complementary DNA reversed transcribed from αKlotho mRNA in mouse and human, where an alternatively spliced αKlotho transcript 20 , 21 , 22 , 48 is predicted to be translated to a putative αKlotho protein composed of Kl1 only and a short 15 amino acid peptide unique to the spliced protein, followed by a stop codon devoid of Kl2, transmembrane and intracellular domains. This constitutes a potential soluble Kl1 polypeptide, which we term called spliced αKlotho Kl1 (spKl1) protein with no transmembrane anchor and does not require cell surface membrane targeting and proteolytic shedding to be released from the cell (Figure 1A). The functions of full length soluble αKlotho can be reproduced by Kl1 in some cell systems, 48 , 49 , 50 , 51 a fact which supports the notion that the hypothetical spKl1 may be one form of fully functional secreted αKlotho, with a totally different mode of release from the cell completely independent of plasma membrane targeting and proteolytic cleavage. This is an important and highly fundamental paradigm for αKlotho biology but quite surprisingly, two decades after the discovery of this alternatively spliced transcript, spKl1 protein still mostly has a conjectural existence in silico. It is imperative to confirm or refute whether spKl1 actually exists in tissues and not just in transfected cultured cell lines.
FIGURE 1.
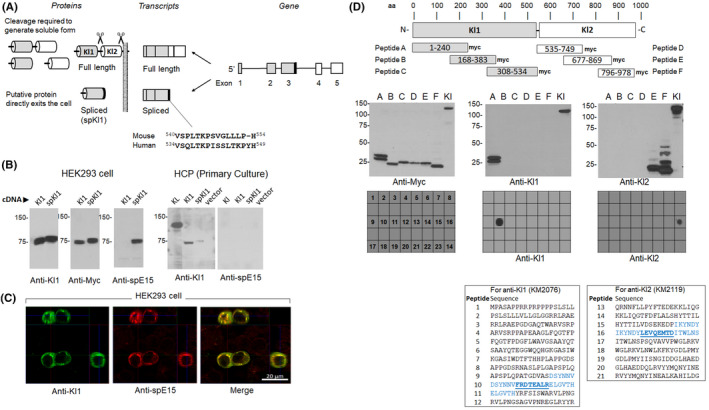
Model of spliced Kl1 αKlotho and characterization of antibodies. A. Model of spliced αKlotho transcript and the hypothetical spliced Kl1 αKlotho protein (spKl1). Fifteen amino acid for spKl1‐specific epitope (spE15) was used to generate antibody against spKl1. B. Left: myc‐tagged Kl1 and myc‐tagged spKl1 were expressed in HEK293 cells and immunoblotted with anti‐KL1, anti‐myc, and anti‐spE15 (raised against the 15 amino acid unique to spKl1). Right: primary culture of human choroid plexus cells (HCP) were transfected with full‐length αKlotho (KL), Kl1, spliced Kl1 (spKl1), and vector only, and cell lysate were immunoblotted with anti‐Kl1 or anti‐spE15. C. Immunocytochemistry of HEK293 cells transfected with spKl1 using anti‐Kl1 (left) or anti‐spE15 (middle). Merged image is on the right. D. Epitope mapping of rat anti‐KL1 and rat anti‐KL2. Top panel shows the location of the myc‐tagged peptides A to F in relation to full‐length αKlotho. Middle panel show immunoblots of peptides A to F, and full‐length αKlotho (Kl) by anti‐myc, anti‐Kl1 (KM2076), and anti‐Kl2 (KM2119) antibodies. The epitopes on polypeptides E and F were further refined by “peptide walking” using 21 overlapping short synthetic 20‐AA peptides by dot blot (bottom panel). The epitopes for anti‐KL1 is FRDTEALR and for anti‐KL2 is LEVQEMTD
A recent study used an antibody against a 15 amino acid epitope unique to spKl1 and not in Kl1 or full length αKlotho, and showed that spKl1 protein is expressed in the brain. 48 This represents the first study verifying the existence of a spKl1 polypeptide instead of a mere intriguing transcript, cDNA, and proposed polypeptide. If spKl1 is really destined for secretion, it should be detectable in tissues bearing its transcript, blood and even more likely CSF; unfortunately, such data are not available. In addition, specific labeling of spKl1 by the reagents was not demonstrated. 48 In contrast to the above study, Mencke and coworkers presented a dataset which supports a rather different model, 52 which indicates that the “spliced αKlotho mRNA” is destined for degradation by nonsense‐mediated mRNA decay, and thus unlikely to be translated into protein.
These two important papers presented opposing conclusions placing the existence of spKl1 from a previously “conjectural” state that lasted two decades, now into a “controversial” state. This fundamental model of spKl1 expression and release is in dire need to be confirmed or refuted after twenty years. To this end, we generated a new antibody, validated its ability to detect spKl1, and collected a body of data with the intent to investigate the existence of spKl1 in mouse brain.
2. METHODS
2.1. Experimental animals
All studies were conducted following the Guide for the Care and Use of Laboratory Animals by the National Institutes of Health and approved by the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center.
2.2. Custom antibody production and tissue preparation
A rabbit polyclonal antibody was raised against the amino acid unique to the putative spliced mouse Klotho (NH3‐CSPLTKPSVGLLLPH‐COOH), and affinity‐purified by a commercial vendor (YenZym Antibodies LLC).
Whole brain from 6‐month‐old C57BL/6J male mice was collected for homogenization and fixation. Brain tissue was homogenized using a Polytron (Kinematica, Lucerne) in lysis buffer containing 20 mM HEPES pH 7.4, NaCl and 1.2% Triton, with the addition of protease inhibitors (AEBSF, Aprotinin, Bestatin, E‐64, Leupeptin, Pepstatin A). For immunohistochemistry (IHC), the brain was fixed in 4% paraformaldehyde at 4°C overnight, and then washed with cold PBS. Four μm cryosections were prepared. Twenty microgram of protein from the brain lysate and from the transfected cell lysate were loaded onto gels.
2.3. Cell culture and transfection
Human HEK293 cells (ATTC) were cultured in six‐well plates with Dulbecco's Modified Eagle's medium (DMEM,) supplemented with 10% fetal bovine serum (Thermo Scientific, Rockford, IL) at 37°C, and 5% CO2. At 60–70% confluency, the cells were transfected with 2 μg of DNA mixed with Lipofectamine Plus (Thermo Scientific). The primary cell line, Human Choroid Plexus Epithelial cells (HCPEPiC) were cultured in Epithelial Cell Media (ScienCell Research laboratories), at 37°C, and 5% CO2. The cells were transfected as described above.
2.4. Plasmids
The human full‐length transmembrane α‐Klotho cDNA using the pcDNA3.1 vector (Addgene), was prepared by PCR amplification and ligation by inserting the primers in Table S1A. The six human Klotho peptides constructs were prepared by PCR amplification and ligation into the pEF1/Myc‐His(A) vector between BamHI and XbaI enzyme site (primers in Table S1B). The mouse full‐length soluble Klotho cDNA in pEF1/Myc‐His(A) was kindly provided by Dr. Makoto Kuro‐o (Jichi Medical University, Tochigi, Japan, and University of Texas Southwestern Medical Center, Dallas, TX USA). The mouse Kl1 plasmid was prepared by PCR amplification and ligation into the pEF1/Myc‐His(A) vector between EcoR I and XbaI sites using primers in Table S1C. Finally, the splice mouse Kl1 which has an extra 15 AA in C‐terminal was prepared by PCR amplification and ligation into the pEF1/Myc‐His(A) vector between EcoRI and XbaI enzyme site using the primers in Table S1D.
2.5. Immunoprecipitation and immunoblots
For immunoprecipitation (IP), 100 μg of brain tissue lysate or cell protein lysate was incubated with 3.0 μg of anti‐KL1 (KM2076, Trans Genic Inc, Japan), anti‐KL2 (KM2119, Trans Genic Inc), anti‐αklotho SB48 (formally known as SB106) 53 and anti‐splice Kl1 (spE15) (YenZym Antibodies LLC), or anti‐MTMR12 (Proteintech, Rosemont, IL), all diluted in 500 μl Krebs‐Ringer Buffer, in 1.5 ml conical low‐bind polypropylene tubes. The reaction occurred overnight rocking at 4°C, and on the next day, the antibody‐antigen complex was incubated with 50 μl of Protein A/G slurry (Thermo Scientific, Rockford, IL) for 2 h. followed by 3 washes with Krebs buffer, and eluted with 50 μl of 2X LDS sample buffer containing 100 mM DTT, and then boiled for 4 min. before gel electrophoresis. Transfected cells were harvested 36 h post‐transfection and lysed in buffer containing protease inhibitors. All protein samples were boiled for 5 min. in SDS sample buffer (Thermo Scientific) containing 100 mM DTT and subsequently loaded onto a 4–12% gradient Bis‐Tris gel (Thermo Scientific) for electrophoresis. For all immunoblots, 20 μg of total protein were loaded per lane, fractionated by SDS‐PAGE, and transferred overnight onto PVDF (Thermo Scientific), which was blocked in 5% non‐fat dried milk, followed by overnight incubation at 4°C with the appropriate antibodies: anti‐KL1 (KM2076), anti‐KL2 (KM2119) (both from Trans Genic Inc.); anti‐spliced Kl1 (anti‐spE15, Custom‐generated by YenZyme), anti‐Myc (Cell Signaling); anti‐myotubularin‐related protein‐12 (anti‐MTMR12, Proteintech). Signals were obtained using SuperSignal West Dura Chemiluminescent ECL substrate (Thermo Scientific).
2.6. Protein dot blot
Twenty peptides were synthesized by a commercial vendor (Pepset™, SynPhaseTM Lanterns, Mimotopes). The peptide lengths were 20 amino acids with an offset of 14 and an overlap of 6. The peptides were dissolved in 50% ETOH and diluted 100‐fold with TBS. Two μg of each peptide were applied to a PDVF membrane presoaked in TBS in a dot Blot apparatus (Bio‐Rad). The membrane was blocked with 5% milk for 1 h, and then incubated with anti‐KL1 or anti‐KL2 antibodies respectively. The conditions were otherwise identical to the immunoblots.
2.7. RNA extraction, reverse transcription‐polymerase chain reaction (RT‐PCR)
Total RNA was extracted using RNAeasy kit (Qiagen) from mouse brain tissues. Complementary DNA (cDNA) was generated with oligo‐dT primers using SuperScript III First Strand Synthesis System (Invitrogen) according to the manufacturer's protocol. PCR products were analyzed by 1.2% agarose ethidium bromide gel electrophoresis. Primers for RT‐PCR (98°C‐30 s; 98°C‐10 s.; 65°C‐30 s; 72°C‐1.5 min.) X35 cycles, and 72°C‐10 min., are shown in Table S2.
2.8. Indirect immunofluorescence microscopy
Frozen brain sections were washed with PBS for 15 min, followed by 0.1% TritonX‐100 incubation for 5–10 min. the sections were incubated with a blocking solution (PBS, 3% BSA, 10% Donkey serum) for 40 min and then incubated at 4°C overnight with KM2076 antibody (2.5 μg/ml), MTMR antibody (2.0 μg/ml), or spE15 antibody (5 μg/ml). The next day, the sections were washed with PBS, and then incubated with AlexaFluor 488‐conjugated donkey anti‐rat IgG or AlexaFluor 488‐conjugated donkey anti‐rabbit IgG and Alexa Fluor 546‐conjugated donkey anti‐rabbit IgG (Invitrogen) respectively. After extensive washing with PBS, the sections were mounted and visualized under a Zeiss LSM880 confocal microscope. Transfected HEK293 cells were cultured on cover glass at 37°C for 24 h and were fixed in 4% paraformaldehyde at 4°C for overnight. The remaining of the staining procedure was identical as for the frozen brain sections.
3. RESULTS
3.1. Characterization of reagents
The configuration of the regular and alternatively spliced αKlotho transcripts and their cognate proteins are shown in Figure 1A. The first set of studies aimed to validate all our reagents. The new rabbit polyclonal affinity‐purified antibody named anti‐spE15, raised against the spliced epitope of 15 amino acids (spE15; VSPLTKPSVGLLLPH), reacted against the immunizing peptide and not the control peptide on dot blot (data not shown). We then confirmed its reactivity with the target epitope residing in the spKl1 protein. Soluble myc‐tagged Kl1 and spKl1 were transiently expressed in HEK293 cells and the cell lysates were probed with the anti‐spE15, anti‐Kl1, and anti‐myc antibodies. The anti‐spE15 antibody only recognized the cell lysates from cells expressing spKl1 (Figure 1B). The co‐staining for anti‐Kl1 and anti‐spE15 in transfected cells (Figure 1C) indicates that these epitopes colocalize on the transfected cells.
To fully characterize our antibodies, we epitope‐mapped the rat monoclonal anti‐αKlotho antibodies, anti‐KL1 (KM2076) and anti‐KL2 (KM2119). Six small recombinant proteins were synthesized (Table S1), with an overlap of approximately 80 amino acids on both ends and with a C‐terminal myc‐tag. These were: peptide A (AA 1–240), peptide B (AA 168–383), peptide C (AA 308–534), for Kl1 and peptide D (AA 535–749), peptide E (AA 677–869), and peptide F (796–978) for Kl2. The six peptides were blotted with anti‐myc, anti‐KL1 and anti‐KL2 antibodies. A strong band was positive for peptide A in the anti‐KL1 blot, and one for anti‐KL2 from peptide F (Figure 1D‐2nd panel). Next, we tested 24 shorter peptides based on positive reaction with peptides A & F. Only one single dot was exclusively positive for each antibody (Figure 1D‐3rd panel). Highlighted in bold, (Figure 1D 4th‐panel), are the two peptides selected as the cognate epitopes for KM2076 (SYNNVFRDTEALRELGVTH) and for KM2119 (IKYNDYLEVQEMTDITWLNS).
FIGURE 2.
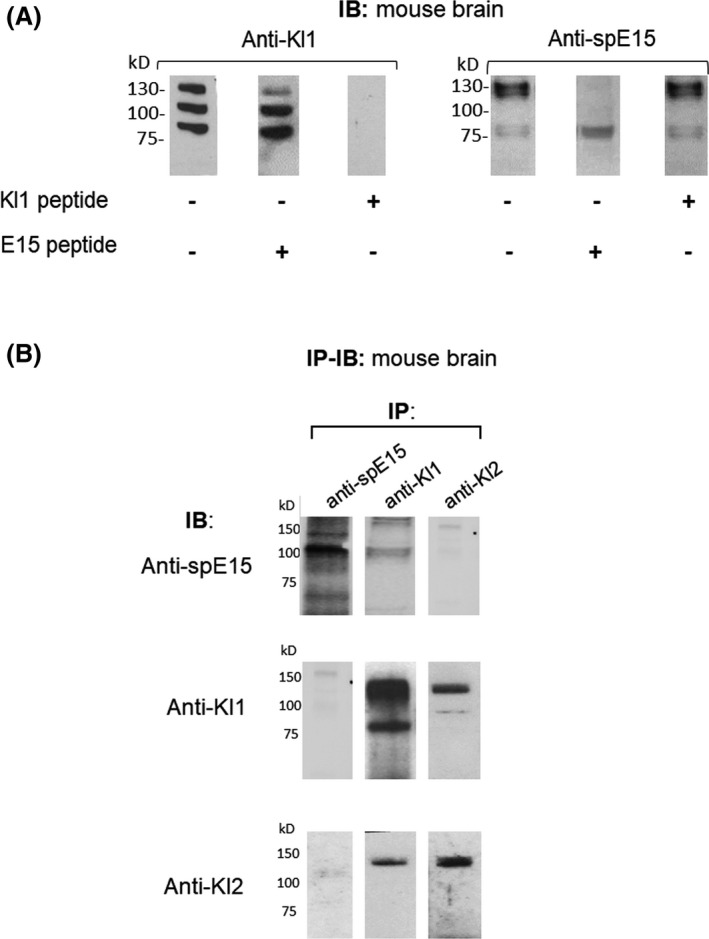
Immunoblot (IB) and immunoprecipitation (IP) of WT mouse brain. A, Whole WT mouse brain lysate was fractionated by SDS‐PAGE and immunoblotted with the antibodies listed below. On the left panel, anti‐KL1 was used as primary antibody for detecting full‐length αKlotho and KL1 proteins in the WT mouse brain. In the left blot, no competitor peptide was added to the primary antibody solution, whilst in the middle and right blots, E15 and the KL1 peptide [FDRTEALR] were added respectively. In the right panel, anti‐spE15 was used for IB, and again from left to right, either no peptide, E15 or KL1 peptides were added as competitors in this order respectively. B, Whole WT mouse brain lysate was immunoprecipitated and the pulled‐down immune complex was resolved by SDS‐PAGE and immunoblotted with the antibodies listed left side. Anti‐spE15, rat monoclonal anti‐KL1 and anti‐KL2 were used. The most left column of three blots shows the IP using only anti‐spE15, and IB anti‐spE15, anti‐KL1 and anti‐KL2 respectively. The middle column of three blots shows the IP using anti‐KL1, and IB, anti‐spE15, anti‐KL1 and anti‐KL2 respectively. In the most right column the three blots show the IB using anti‐KL2, and IP using anti‐spE15, anti‐KL1 and anti‐KL2 respectively
FIGURE 3.
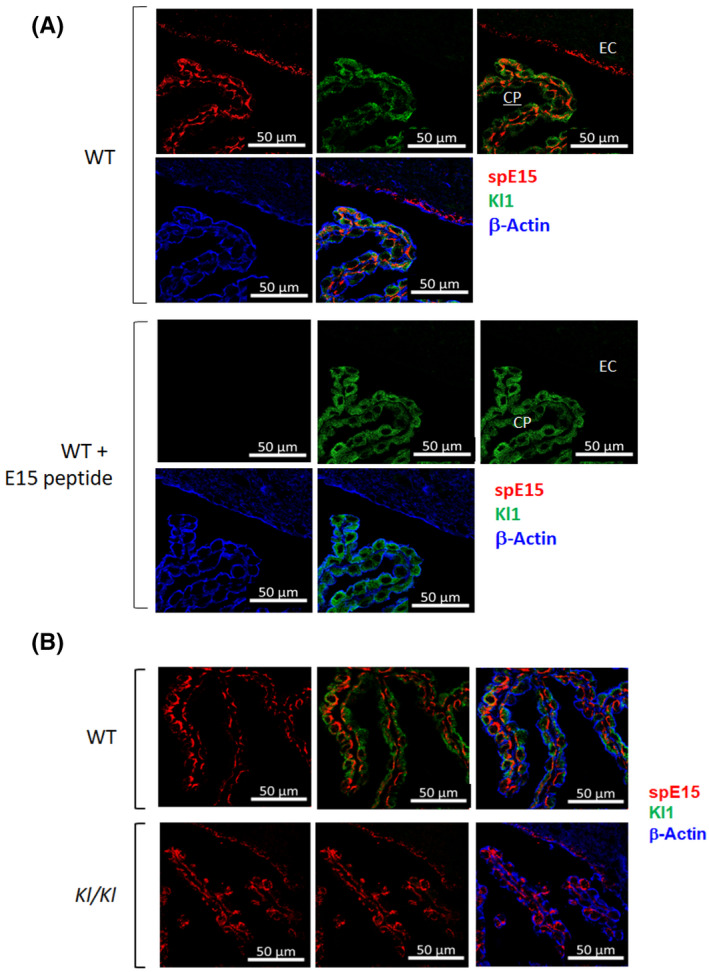
Immunohistochemistry of mouse brain Choroid‐plexus. Mouse brain sections were stained with anti‐KL1 (green) and anti‐spE15 (red). A, Wild type mouse brain sections was stained with anti‐spE15 (top panel) and with anti‐spE15 plus E15 peptide (bottom panel). B, (top) Wild type (top panel) and kl/kl (bottom panel) mouse brain sections was co‐stained with anti‐KL1 and anti‐spE15.
3.2. Expression of spKl1 protein in murine brain
Three methods were used to test for spKl1 protein expression in mouse brain using antibodies from Figure 1. First, by immunoblot: when we probed the brain with the anti‐spE15, several bands were visible, some of which disappeared with the addition of the spE15 peptide: except for the 75 kDa doublet bands. (Figure 2A right). This indicates that some of the proteins labelled by anti‐spE15 most likely harbor the 15 amino acids from spE15. In contrast, anti‐KL1 showed labelling of several bands in brain lysate, but when we pre‐incubated the antibody with the spE15 peptide, the same bands were still present, demonstrating that the spE15 peptide failed to compete with the protein detected by anti‐KL1. In contrast, Kl1 protein completely blocked all labelling by the anti‐Kl1 antibody (Figure 2A left).
Second, with the possibility that low quantity of protein loaded on the SDS‐PAGE gels for immunoblotting (IB) may explain the negative results, we additionally assessed the spKl1 and αKlotho protein expression by immunoprecipitation (IP) pull‐down of murine brain lysates (Figure 2B). In addition, sequential IP‐IB allows testing of the co‐existence two epitopes (spE15 and Kl1) on the same precipitated proteins, which is what one expects from the spliced spKl1 protein. We were not able to demonstrate the presence of spKl1 (~70 kDa) in the immune complex brought down by both anti‐spE15, anti‐KL1, anti‐KL2 (Figure 2B top row). IP with anti‐KL1 or anti‐KL2 brought down full‐length Klotho or the Kl1 polypeptide (Figure 2B middle and bottom rows).
Third, we looked for spKl1 protein expression by immunohistochemistry. While both anti‐spE15 and anti‐KL1 labelled the choroid plexus of normal WT mice very strongly, there was no co‐localization from the two antibodies (Figure 3A). The putative spKl1 protein should harbor both FRDTEALR in the Kl1 domain and VSPLTKPSVGLLLPH in the unique C‐tail region, thus mandating a co‐staining signal. The absence of co‐staining pattern indicates that the two epitopes reside on different proteins when examined in situ (Figure 3A). Furthermore, incubation of anti‐spE15 with the spE15 peptide eliminated anti‐spE15 staining, but not anti‐KL1 staining (Figure 3A). We next compared wild type to kl/kl hypomorphic mice. kl/kl mouse brain showed staining by anti‐spE15, which was surprisingly similar to the signal seen in the WT mouse brain (Figure 3B). There was no signal with anti‐KL1 antibody but clearly detectable signal with anti‐spE15 in kl/kl mice, further indicating that the antigen, which reacts with anti‐spE15 is not in αKlotho protein in mouse brain. Furthermore, when comparing bands in the spE15 IB, the WT and kl/kl mice brain showed very similar patterns with no band nearing 70 kDa (Figure S1A). We examined for αKlotho expression in the brain of the WT and kl/kl mice using anti‐KL1 and observed the absence of the 130 kDa band in kl/kl mouse brain lysate with anti‐KL1 antibody, but in sharp contrast, identical bands were detected with anti‐spE15, in both the WT and kl/kl mice brain lysate. In Figure S1B, we performed immunoprecipitation of both the WT and the kl/kl mouse brains using two different αKlotho antibodies, anti‐KL1 and the synthetic anti‐klotho SB48. 53 Both anti‐αKlotho antibodies clearly precipitated the full‐length αKlotho as well as an approximately ~75 kDa protein, possibly Kl1, only in the WT brain. These anti‐αKlotho antibodies were unable to precipitate a ~70 kDa protein from either the WT or kl/kl brains. When the immune complex was probed with anti‐spE15, we could not detect any difference between WT and kl/kl mouse brains. This is another evidence which further supports our other findings confirming the absence of a spKl1 protein in the WT and kl/kl mouse brains.
3.3. Concurrence of putative spKl1 protein and transcript
The data thus far raise doubts about the specificity of anti–spE15 for spKl1. We further tested whether the choroid plexus actually expresses the spliced αKlotho transcript. We dissected the region of the mouse choroid plexus of WT mouse brain that was most strongly stained by anti‐spE15 (Figure 4A), isolated RNA, reversely transcribed to cDNA, and tested with PCR using the primers indicated in Table S2 and Figure 4B. In the choroid plexus where the most intense staining was observed with anti‐spE15, we were not able to find the spliced transcript (Figure 4C). The αKlotho and Kl1 transcripts were in the mouse brains, but the spliced klotho (spkl1) transcript was not found. Only a very short spKl1 C‐terminal transcript was found (>900 bp; Figure 4C. lane 8).
FIGURE 4.
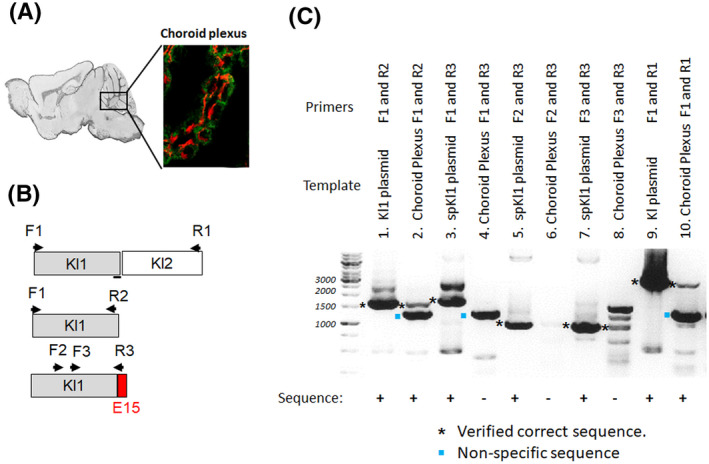
spE15 transcript is not present in mouse brain. A, Immunohistochemistry for spE15 in WT mouse choroid plexus. spE15; red, and KL1 in green. We Micro‐dissected choroid plexus from WT mouse brain were subjected to RT‐PCR using the different primers B. C, the primers (top panel), cDNA origin (middle panel) and RT‐PCR products in 1.2% agarose ethidium bromide gel (bottom panel). Lanes 2, 4, 6, 8, 10 showed strong transcript of Kl1, spKl1, and short spKl1, and very short transcripts of spKl1 and αKlotho respectively. Lanes 1, 3, 5, 7, 9, showed the transcripts in plasmids as PCR templates as positive control with. (* shows verified correct sequence.)
We next pose the logical question “what is anti‐spE15 staining if there is no evidence for spKl1 protein?” We searched for possible cross‐reacting proteins using the tripartite criteria: 1. containing primary sequence homology with the 15 amino acid epitope. 2. known to be strongly expressed (not necessarily exclusively) in the adult mouse brain. 3. predicted molecular weight and mobility around 75 kDa. As one dials up the stringency of the homology criteria, one can narrow it down to a few candidates. One protein standing out fulfilling all of the above criteria is myotubularin‐related‐protein or MTMR12 (Figure 5A). When probing mouse brain tissue for anti‐MTMR12 and anti‐spE15, both antibodies revealed a positive band of approximately ~75 kDa (Figure 5B). We then probed the immune complex brought down with anti‐spE15 and anti‐MTMR12 antibody and found the 75 kDa band was also present on both blots indicating a possible scenario for the precipitated proteins to be the same (Figure 5C). However, when we performed IHC on the brain sections with these two antibodies, we did not find co‐staining so anti‐MTMR12 and anti‐spE15 are not labelling the same protein (Figure 5C.)
FIGURE 5.
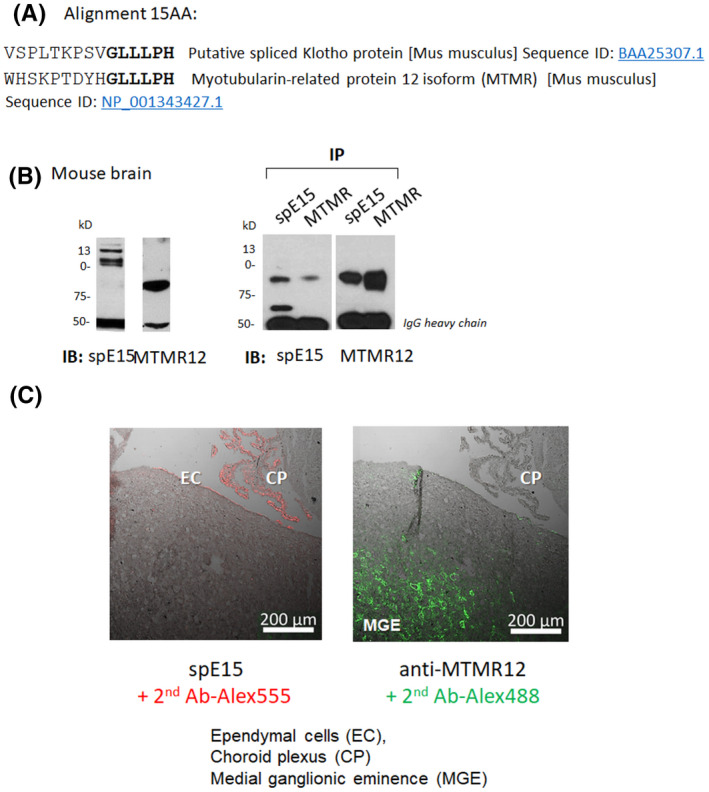
spE15 peptide “GLLLPH” is present also in myotubulin‐related protein‐12 isoform 2 protein (MTMR12). A, Amino acid alignment between splice Klotho isoform 15 AA sequence and MTMR12. B, Immunoblot for WT mouse brain lysate for anti‐spE15, and anti‐MTMR12 (left panel) and immunoprecipitation followed by immunoblot for spE15 and MTMR12 (right panel). In both MTMR12 immunoblots we detect a band nearing 75 kDa, and this same band appears in the IB for anti‐spE15. C. Immunohistochemistry on mouse WT brain sections using anti‐spE15 (left panel) and anti‐MTMR12 (right panel). CP: Choroid plexus; EC: Ependymal cells; MGE: medial ganglionic eminence. The anti‐spE15 stains in the choroid Plexus (CP) and the Ependymal cells (EC), while the MTMR12 stains in the Medial ganglionic eminence (MGE)
4. DISCUSSION
4.1. αKlotho locale and function in CNS
Brain expression of αKlotho commences in utero, increases steeply in the postnatal period, 54 and declines with age. 55 Previous studies found brain αKlotho expression to be mostly located to choroid plexus 56 , 57 , 58 and also in certain specific neurons. 54 , 59 Even during the short lifespan of the kl/kl mouse, severe and rapid onset of hippocampal‐dependent cognitive impairment was detected which can possibly a primary or secondary effect of αKlotho deficiency. 60 , 61 , 62 The kl/kl mice die shortly after cognitive decline is detectable 1 and the pre‐mortem multi‐organ failure can possibly contribute to secondary brain dysfunction. Clinical study showed low αKlotho in CSF of Alzheimer's disease compared to healthy senior individuals. 63 So normal expression of αKlotho in the brain is required for slowing of aging‐related CNS disease including Alzheimer's disease. 60 , 63 , 64
4.2. Implications of a spliced Klotho transcript and protein
The post‐translational processing and trafficking of αKlotho protein to reach the cell surface requires multiple steps of protein trafficking, and the eventual proteolytic cleavage and release of full length, Kl1, and Kl2 αKlotho fragments from the cell membrane. The generation of a soluble form of Kl1 devoid of the transmembrane domain theoretically bypasses the membrane targeting and proteolytic steps and represents an entirely different system of secretion. An unfortunate situation in the αKlotho field is that “soluble”, “secreted”, and “spliced” αKlotho have all been somehow abbreviated as “sKlotho” due to lack of uniform nomenclature. We want to exercise caution and use “spKl1” to denote the putative spliced Kl1 polypeptide. The spKl1 transcript is generated by alternative splicing of klotho exon 3 that can potentially produce a ~70 kDa protein that has all the primary amino acid sequence of Kl1. 20 To date, there are no reports of the detection of the Kl1 fragment in mouse or human blood. Based on the studies of αKlotho effect on tumor development, 51 , 65 Kl1 seems to possess most of the functional effects of full‐length αKlotho. Equipped with the full amino acid sequence of Kl1, spKl1 can theoretically be a functional equivalent of endocrine αKlotho protein, which does not require targeting to the membrane or cleavage, regardless of whether constitutive or regulated. Several studies have detected αKlotho in the cerebrospinal fluid (CSF) by ELISA 33 , 34 , 63 and by immunoblot. 35 Because there were no disclosures of the epitopes of their reagents or the size of the captured proteins, we actually cannot discern whether the ELISA’s are detecting full length or αKlotho fragments in the CSF.
4.3. Data regarding the existence of spliced αKlotho protein in the murine brain
The spliced transcript appears to exist in mouse and humans but not rat, 48 , 58 which is quite unusual but yet appears to be accepted at large. A second point of note is the low level of similarity between the mouse and human spKl1‐specific epitope (Figure 1A) when the murine‐human homology is usually 80% or greater in expressed proteins. The disparity between mouse (540VSPLTKPSVGLLLPH554) and human (534VSQLTKPISSLTKPYH549) strongly suggests that these predicted amino acid sequences may not be real. The report by Masso et al. was the first that attempted to address this question by generating an antibody specifically against spKl1. 48 While positive staining was reported on immunoblot, no IHC was done, and the possibility of cross reactivity of the antibody was not ruled out. The positive RT‐PCR can potentially be due to amplification of transcripts destined for degradation rather than translation (see below).
We unequivocally authenticated our reagents including their cognate epitopes and validated that anti‐spE15 unequivocally labels heterologously expressed spKl1 (Figure 1B). Our finding of anti‐spE15 reacting with a band on IB of mobility compatible with spKl1 (Figure 2A) is similar to and confirms the findings of Masso et al. 48 However, further and more detailed interrogation failed to support the conclusion that spKl1 is expressed in the brain. Five irrefutable findings are highlighted: 1. Anti‐KL1 and anti‐spE15 are not detecting proteins of the same mobility on SDS‐PAGE (Figure 2A); surely not the expected 70 kDa band. 2. Anti‐KL1 and anti‐spE15 do not precipitate the same immune‐complex (Figure 2B). 3. Anti‐KL1 and anti‐spE15 signals on IHC of the choroid plexus do not colocalize at all (Figure 3A). 4. The anti‐spE15 signal was unchanged while the anti‐KL1 signal was absent in the kl/kl hypomorphs (Figure 3B). 5. Regions of the choroid plexus that reacted positively with anti‐Kl1 and anti‐spE15 do not contain the spliced spKl1 transcript (Figure 3A). In concert, these findings are overwhelmingly strong in refuting the existence of a spKl1 polypeptide in the mouse brain choroid plexus. In supp. Figure 1A, we did not see any 70 kD band in the brain of WT and kl/kl mice that resembles the spKl1 from spKl1‐transfected cells. We used our anti‐αKlotho antibodies SB48 53 and anti‐KL1 to immunoprecipitate αKlotho from brain tissue of mice, (supp. Figure 1B) but were not able to detect any bands in the anti‐spE15 immunoblot as compared to the anti‐KL1 blot where we saw a band of approximately 75 kDa only visible in the WT and not kl/kl mouse brain. This same 75 kDa band was also pulled down using the anti‐KL1 antibody, which confirms the presence of KL1 in the brain of WT mice.
4.4. Data supporting that the transcript is not translated in mice brain
Mencke and coworkers examined the fate of the spKl1 transcript. 52 Premature stop codons, such as the ones present in spKl1 mRNA, can prime mRNAs for degradation by nonsense‐mediated mRNA decay (NMD). 66 NMD is coupled to translation and serves to rid mRNAs with premature translation stop codons. In mammalian cells, NMD is also linked to pre‐mRNA splicing, when the premature stop codon is 5ʹ‐upstream of an intron. It is proposed that the exon junction complex links splicing and NMD. When Mencke and coworkers disrupted NMD, they observed accumulation of spKl1, supportive of constitutive degradation of this transcript at normal basal states. 52 spKl1 is also associated with the NMD core factor UPF1 rather with polysomes, indicating that the spKl1 transcript is destined for αKlotho mRNA degradation rather than for a template of protein translation. 52 The results of Masso et al. 48 in fact can be reproduced by our laboratory but control experiments unambiguously indicated that anti‐spE15 is reacting to proteins other than Kl1 or spKl1.
4.5. Conclusion
Rather than denouncing negative data as uninspiring, it is in fact as true and as informative as positive data, If one subscribe to the rationale and philosophy of Sir Karl Popper, it is even more powerful and conclusive than positive data. 67 Approximately two decades after the conjecture that the Kl1 domain of αKlotho can be synthesized from an alternatively spliced transcript and released from cells as a soluble protein without membrane targeting and cell surface cleavage, definitive proof of its existence in tissues is still lacking, and available data are conflicting. Our antibody that was proven to react with the spliced αKlotho epitope produced in vitro, also label other proteins, and the lack of co‐labeling by anti‐spE15 and anti‐KL1, absence of the spKl1 transcript in cells positive for anti‐spE15 staining unambiguously, supports the model that the spliced Kl1 mRNA is not translated to protein, reinforcing as the prediction proposed by Mencke et al. Despite our efforts, we could not detect spliced Kl1 in the brain of adult mice. It remains possible that spKl1 protein can be expressed in the pre‐, post‐natal and senescent murine brain; which cannot be ruled by the current dataset. While αKlotho is undoubtedly critical for central nervous system function, effort should not be focused on the regulation or action of the “spliced Kl1 protein” which does not exist in mouse brain, rather on the traditional αKlotho protein.
AUTHOR CONTRIBUTIONS
Orson Moe contributed to study concept and design. Liping Li and Johanne Pastor contributed to analysis and interpretation of data. Ming‐Chang Hu and Johanne Pastor contributed to drafting of the manuscript. Johanne Pastor, Ming‐Chang Hu and Orson Moe contributed to critical revision of the manuscript for important intellectual content. Jianning Zhang contributed to statistical analysis. Orson Moe obtained funding. Orson Moe and Johanne Pastor contributed to study supervision. Experiments were performed by Liping Li, Johanne Pastor, Jianning Zhang and Taylor Davidson.
Supporting information
Fig S1
Supplementary Material
ACKNOWLEDGEMENT
The authors are supported by the National Institutes of Health (R01 DK091392 and R01 DK092461), the O’Brien Kidney Research Center (P30 DK‐079328), the Innovative Research Support and Endowed Professors Collaborative Research Support from the Charles Pak Foundation.
Liping Li and Johanne Pastor contributed equally.
REFERENCES
- 1. Kuro‐o M, Matsumura Y, Aizawa H, et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature. 1997;390(6655):45‐51. [DOI] [PubMed] [Google Scholar]
- 2. Life‐extending hormone found in mice research may apply to all mammals. Health News. 2005;11(10):2. [PubMed] [Google Scholar]
- 3. Kurosu H, Yamamoto M, Clark JD, et al. Suppression of aging in mice by the hormone Klotho. Science. 2005;309(5742):1829‐1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ito S, Kinoshita S, Shiraishi N, et al. Molecular cloning and expression analyses of mouse betaklotho, which encodes a novel Klotho family protein. Mech Dev. 2000;98(1–2):115‐119. [DOI] [PubMed] [Google Scholar]
- 5. Ito S, Fujimori T, Hayashizaki Y, Nabeshima Y. Identification of a novel mouse membrane‐bound family 1 glycosidase‐like protein, which carries an atypical active site structure. Biochim Biophys Acta. 2002;1576(3):341‐345. [DOI] [PubMed] [Google Scholar]
- 6. Samms RJ, Cheng CC, Kharitonenkov A, Gimeno RE, Adams AC. Overexpression of beta‐Klotho in adipose tissue sensitizes male mice to endogenous FGF21 and provides protection from diet‐induced obesity. Endocrinology. 2016;157(4):1467‐1480. [DOI] [PubMed] [Google Scholar]
- 7. Lan T, Morgan DA, Rahmouni K, et al. FGF19, FGF21, and an FGFR1/beta‐Klotho‐activating antibody act on the nervous system to regulate body weight and glycemia. Cell Metab. 2017;26(5):709‐718 e703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Misrahi M. beta‐Klotho sustains postnatal GnRH biology and spins the thread of puberty. EMBO Mol Med. 2017;9(10):1334‐1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Somm E, Henry H, Bruce SJ, et al. beta‐Klotho deficiency protects against obesity through a crosstalk between liver, microbiota, and brown adipose tissue. JCI Insight. 2017;2(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen J, Hu J, Liu H, et al. FGF21 protects the blood‐brain barrier by upregulating PPARgamma via FGFR1/beta‐klotho after traumatic brain injury. J Neurotrauma. 2018;35(17):2091‐2103. [DOI] [PubMed] [Google Scholar]
- 11. Kuro OM. The Klotho proteins in health and disease. Nat Rev Nephrol. 2019;15(1):27‐44. [DOI] [PubMed] [Google Scholar]
- 12. Hu MC, Shiizaki K, Kuro‐o M, Moe OW. Fibroblast growth factor 23 and Klotho: physiology and pathophysiology of an endocrine network of mineral metabolism. Annu Rev Physiol. 2013;75:503‐533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lau WL, Leaf EM, Hu MC, et al. Vitamin D receptor agonists increase klotho and osteopontin while decreasing aortic calcification in mice with chronic kidney disease fed a high phosphate diet. Kidney Int. 2012;82(12):1261‐1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haussler MR, Whitfield GK, Kaneko I, et al. The role of vitamin D in the FGF23, klotho, and phosphate bone‐kidney endocrine axis. Rev Endocr Metab Disord. 2012;13(1):57‐69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoshida T, Fujimori T, Nabeshima Y. Mediation of unusually high concentrations of 1,25‐dihydroxyvitamin D in homozygous klotho mutant mice by increased expression of renal 1alpha‐hydroxylase gene. Endocrinology. 2002;143(2):683‐689. [DOI] [PubMed] [Google Scholar]
- 16. Doi S, Zou Y, Togao O, et al. Klotho inhibits transforming growth factor‐beta1 (TGF‐beta1) signaling and suppresses renal fibrosis and cancer metastasis in mice. J Biol Chem. 2011;286(10):8655‐8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kuro‐o M. Klotho, phosphate and FGF‐23 in ageing and disturbed mineral metabolism. Nat Rev Nephrol. 2013;9(11):650‐660. [DOI] [PubMed] [Google Scholar]
- 18. Yamamoto M, Clark JD, Pastor JV, et al. Regulation of oxidative stress by the anti‐aging hormone klotho. J Biol Chem. 2005;280(45):38029‐38034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu MC, Shi M, Moe OW. Role of alphaKlotho and FGF23 in regulation of type II Na‐dependent phosphate co‐transporters. Pflugers Arch European Journal of Physiology. 2019;471: 99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shiraki‐Iida T, Aizawa H, Matsumura Y, et al. Structure of the mouse klotho gene and its two transcripts encoding membrane and secreted protein. FEBS Lett. 1998;424(1–2):6‐10. [DOI] [PubMed] [Google Scholar]
- 21. Ohyama Y, Kurabayashi M, Masuda H, et al. Molecular cloning of rat klotho cDNA: markedly decreased expression of klotho by acute inflammatory stress. Biochem Biophys Res Commun. 1998;251(3):920‐925. [DOI] [PubMed] [Google Scholar]
- 22. Matsumura Y, Aizawa H, Shiraki‐Iida T, Nagai R, Kuro‐o M, Nabeshima Y. Identification of the human klotho gene and its two transcripts encoding membrane and secreted klotho protein. Biochem Biophys Res Commun. 1998;242(3):626‐630. [DOI] [PubMed] [Google Scholar]
- 23. Lee S, Choi J, Mohanty J, et al. Structures of beta‐klotho reveal a ‘zip code'‐like mechanism for endocrine FGF signalling. Nature. 2018;553(7689):501‐505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen G, Liu Y, Goetz R, et al. alpha‐Klotho is a non‐enzymatic molecular scaffold for FGF23 hormone signalling. Nature. 2018;553(7689):461‐466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Urakawa I, Yamazaki Y, Shimada T, et al. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature. 2006;444(7120):770‐774. [DOI] [PubMed] [Google Scholar]
- 26. Kurosu H, Ogawa Y, Miyoshi M, et al. Regulation of fibroblast growth factor‐23 signaling by klotho. J Biol Chem. 2006;281(10):6120‐6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu MC, Shi M, Zhang J, et al. Renal Production, Uptake, and Handling of Circulating alphaKlotho. J Am Soc Nephrol. 2016;27(1):79‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Loon EPM, Pulskens WP, van der Hagen EAE, et al. Shedding of klotho by ADAMs in the kidney. Am J Physiol Renal Physiol. 2015;309(4):F359‐368. [DOI] [PubMed] [Google Scholar]
- 29. Chen C‐D, Tung TY, Liang J, et al. Identification of cleavage sites leading to the shed form of the anti‐aging protein klotho. Biochemistry. 2014;53(34):5579‐5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bloch L, Sineshchekova O, Reichenbach D, et al. Klotho is a substrate for alpha‐, beta‐ and gamma‐secretase. FEBS Lett. 2009;583(19):3221‐3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hu MC, Shi M, Zhang J, et al. Klotho deficiency causes vascular calcification in chronic kidney disease. J Am Soc Nephrol. 2011;22(1):124‐136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hu MC, Shi M, Zhang J, et al. Klotho: a novel phosphaturic substance acting as an autocrine enzyme in the renal proximal tubule. FASEB J. 2010;24(9):3438‐3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kunert SK, Hartmann H, Haffner D, Leifheit‐Nestler M. Klotho and fibroblast growth factor 23 in cerebrospinal fluid in children. J Bone Miner Metab. 2017;35(2):215‐226. [DOI] [PubMed] [Google Scholar]
- 34. Emami Aleagha MS, Siroos B, Ahmadi M, et al. Decreased concentration of Klotho in the cerebrospinal fluid of patients with relapsing‐remitting multiple sclerosis. J Neuroimmunol. 2015;281:5‐8. [DOI] [PubMed] [Google Scholar]
- 35. Imura A, Iwano A, Tohyama O, et al. Secreted Klotho protein in sera and CSF: implication for post‐translational cleavage in release of Klotho protein from cell membrane. FEBS Lett. 2004;565(1–3):143‐147. [DOI] [PubMed] [Google Scholar]
- 36. Neyra JA, Hu MC. Potential application of klotho in human chronic kidney disease. Bone. 2017;100:41‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lu X, Hu MC. Klotho/FGF23 axis in chronic kidney disease and cardiovascular disease. Kidney Dis (Basel). 2017;3(1):15‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bian A, Neyra JA, Zhan M, Hu MC. Klotho, stem cells, and aging. Clin Interv Aging. 2015;10:1233‐1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hu MC, Kuro‐o M, Moe OW. alphaKlotho and vascular calcification: an evolving paradigm. Curr Opin Nephrol Hypertens. 2014;23(4):331‐339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hum JM, O'Bryan L, Smith RC, White KE. Novel functions of circulating Klotho. Bone. 2017;100:36‐40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xie J, Wu YL, Huang CL. Deficiency of soluble alpha‐Klotho as an independent cause of uremic cardiomyopathy. Vitam Horm. 2016;101:311‐330. [DOI] [PubMed] [Google Scholar]
- 42. Rubinek T, Wolf I. The role of Alpha‐Klotho as a universal tumor suppressor. Vitam Horm. 2016;101:197‐214. [DOI] [PubMed] [Google Scholar]
- 43. Donate‐Correa J, Martín‐Núñez E, Delgado NP, et al. Implications of Fibroblast growth factor/Klotho system in glucose metabolism and diabetes. Cytokine Growth Factor Rev. 2016;28:71‐77. [DOI] [PubMed] [Google Scholar]
- 44. Bian A, Xing C, Hu MC. Alpha Klotho and phosphate homeostasis. J Endocrinol Invest. 2014;37(11):1121‐1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hu MC, Kuro‐o M, Moe OW. Renal and extrarenal actions of Klotho. Semin Nephrol. 2013;33(2):118‐129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hu MC, Moe OW. Klotho as a potential biomarker and therapy for acute kidney injury. Nat Rev Nephrol. 2012;8(7):423‐429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang CL, Moe OW. Klotho: a novel regulator of calcium and phosphorus homeostasis. Pflugers Arch. 2011;462(2):185‐193. [DOI] [PubMed] [Google Scholar]
- 48. Masso A, Sanchez A, Gimenez‐Llort L, et al. Secreted and transmembrane alphaKlotho isoforms have different spatio‐temporal profiles in the brain during aging and alzheimer's disease progression. PLoS One. 2015;10(11):e0143623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu H, Fergusson MM, Castilho RM, et al. Augmented Wnt signaling in a mammalian model of accelerated aging. Science. 2007;317(5839):803‐806. [DOI] [PubMed] [Google Scholar]
- 50. Chateau MT, Araiz C, Descamps S, Galas S. Klotho interferes with a novel FGF‐signalling pathway and insulin/Igf‐like signalling to improve longevity and stress resistance in Caenorhabditis elegans. Aging (Albany NY). 2010;2(9):567‐581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Abramovitz L, Rubinek T, Ligumsky H, et al. KL1 internal repeat mediates klotho tumor suppressor activities and inhibits bFGF and IGF‐I signaling in pancreatic cancer. Clin Cancer Res. 2011;17(13):4254‐4266. [DOI] [PubMed] [Google Scholar]
- 52. Mencke R, Harms G, Moser J, et al. Human alternative Klotho mRNA is a nonsense‐mediated mRNA decay target inefficiently spliced in renal disease. JCI Insight. 2017;2(20):e94375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Barker SL, Pastor J, Carranza D, et al. The demonstration of alphaKlotho deficiency in human chronic kidney disease with a novel synthetic antibody. Nephrol Dial Transplant. 2015;30(2):223‐233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Clinton SM, Glover ME, Maltare A, et al. Expression of klotho mRNA and protein in rat brain parenchyma from early postnatal development into adulthood. Brain Res. 2013;1527:1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Duce JA, Podvin S, Hollander W, Kipling D, Rosene DL, Abraham CR. Gene profile analysis implicates Klotho as an important contributor to aging changes in brain white matter of the rhesus monkey. Glia. 2008;56(1):106‐117. [DOI] [PubMed] [Google Scholar]
- 56. Li SA, Watanabe M, Yamada H, Nagai A, Kinuta M, Takei K. Immunohistochemical localization of Klotho protein in brain, kidney, and reproductive organs of mice. Cell Struct Funct. 2004;29(4):91‐99. [DOI] [PubMed] [Google Scholar]
- 57. Kato Y, Arakawa E, Kinoshita S, et al. Establishment of the anti‐Klotho monoclonal antibodies and detection of Klotho protein in kidneys. Biochem Biophys Res Commun. 2000;267(2):597‐602. [DOI] [PubMed] [Google Scholar]
- 58. Zhu L, Stein LR, Kim D, et al. Klotho controls the brain‐immune system interface in the choroid plexus. Proc Natl Acad Sci U S A. 2018;115(48):E11388‐E11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. German DC, Khobahy I, Pastor J, Kuro OM, Liu X. Nuclear localization of Klotho in brain: an anti‐aging protein. Neurobiol Aging. 2012;33(7):1483.e25‐1483.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shiozaki M, Yoshimura K, Shibata M, et al. Morphological and biochemical signs of age‐related neurodegenerative changes in klotho mutant mice. Neuroscience. 2008;152(4):924‐941. [DOI] [PubMed] [Google Scholar]
- 61. Uchida A, Komiya Y, Tashiro T, et al. Neurofilaments of Klotho, the mutant mouse prematurely displaying symptoms resembling human aging. J Neurosci Res. 2001;64(4):364‐370. [DOI] [PubMed] [Google Scholar]
- 62. Nagai T, Yamada K, Kim H‐C, et al. Cognition impairment in the genetic model of aging klotho gene mutant mice: a role of oxidative stress. FASEB J. 2003;17(1):50‐52. [DOI] [PubMed] [Google Scholar]
- 63. Semba RD, Moghekar AR, Hu J, et al. Klotho in the cerebrospinal fluid of adults with and without Alzheimer's disease. Neurosci Lett. 2014;558:37‐40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cararo‐Lopes MM, Mazucanti CHY, Scavone C, Kawamoto EM, Berwick DC. The relevance of alpha‐KLOTHO to the central nervous system: some key questions. Ageing Res Rev. 2017;36:137‐148. [DOI] [PubMed] [Google Scholar]
- 65. Ligumsky H, Rubinek T, Merenbakh‐Lamin K, et al. Tumor suppressor activity of Klotho in breast cancer is revealed by structure‐function analysis. Mol Cancer Res. 2015;13(10):1398‐1407. [DOI] [PubMed] [Google Scholar]
- 66. Brogna S, Wen J. Nonsense‐mediated mRNA decay (NMD) mechanisms. Nat Struct Mol Biol. 2009;16(2):107‐113. [DOI] [PubMed] [Google Scholar]
- 67. Popper K. The logic of scientific discovery. London and New York: Routledge Classics; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Supplementary Material


