Significance
Receptors that separate ligand recognition and intracellular signaling into separate protein subunits are ubiquitous in immunity. These subunits mix and match to create combinatorial functional diversity. The transmembrane domains of these receptors assemble through the interaction between two acidic and one basic residue on different helices. Using computational methods to study the DAP12-NKG2C receptor complex, we identified a polar motif in which a singly-protonated Asp-Asp pair forms a carboxyl-carboxylate clamp that clasps a charged Lys side chain in the membrane. This local interaction allows dynamic variations in other regions of the helices that tolerate sequence diversity of the interacting subunits in this class of receptors, which signal through multisubunit clustering rather than propagation of rigid conformational changes through the membrane.
Keywords: immune receptor, transmembrane helix assembly, carboxyl-carboxylate pair, salt bridge, simulation
Abstract
Multicomponent immune receptors are essential complexes in which distinct ligand-recognition and signaling subunits are held together by interactions between acidic and basic residues of their transmembrane helices. A 2:1 acidic-to-basic motif in the transmembrane domains of the subunits is necessary and sufficient to assemble these receptor complexes. Here, we study a prototype for these receptors, a DAP12-NKG2C 2:1 heterotrimeric complex, in which the two DAP12 subunits each contribute a single transmembrane Asp residue, and the NKG2C subunit contributes a Lys to form the complex. DAP12 can also associate with 20 other subunits using a similar motif. Here, we use molecular-dynamics simulations to understand the basis for the high affinity and diversity of interactions in this group of receptors. Simulations of the transmembrane helices with differing protonation states of the Asp-Asp-Lys triad identified a structurally stable interaction in which a singly-protonated Asp-Asp pair forms a hydrogen-bonded carboxyl-carboxylate clamp that clasps onto a charged Lys side chain. This polar motif was also supported by density functional theory and a Protein Data Bank–wide search. In contrast, the helices are dynamic at sites distal to the stable carboxyl-carboxylate clamp motif. Such a locally stable but globally dynamic structure is well suited to accommodate the sequence and structural variations in the transmembrane helices of multicomponent receptors, which mix and match subunits to create combinatorial functional diversity from a limited number of subunits. It also supports a signaling mechanism based on multisubunit clustering rather than propagation of rigid conformational changes through the membrane.
Immune cells often employ multicomponent protein receptors that separate specific extracellular ligand recognition and intracellular downstream signaling functions into distinct subunits (1–3). Assembly of these single-pass protein subunits through helix–helix interactions of their transmembrane domains is required for receptor function. The ligand-binding subunit exhibits an elaborate extracellular domain that is adapted for ligand specificity, while the signaling subunit typically contains a pair of evolutionarily conserved intracellular signaling elements, immunoreceptor tyrosine-based activation motifs (ITAMs), that couple to downstream phosphorylation and signaling pathways (4). The details of receptor subunit assembly have not been fully worked out, obscuring the mechanism of signal transduction across the membrane.
The interaction between a pair of acidic aspartate residues on the signaling dimer and a basic residue (lysine or arginine) on the ligand-binding coreceptor are required for complex assembly within the otherwise nonpolar membrane (5–7). This 2:1 acidic-to-basic polar association motif is ubiquitous in multicomponent receptor families including Fc receptors (8), T-cell receptor–CD3 complexes (9–11), and the more than 20 receptors across most immune-cell types that associate with the signaling subunit DNAX-activation protein 12 (DAP12), a disulfide-linked homodimer with a minimal extracellular region (2, 12–14). DAP12–coreceptor complexes are noteworthy because malfunctions of DAP12 or its coreceptors have been implicated in an array of diseases (15–19). Notably, point mutations of DAP12-associated coreceptor TREM2 have recently been linked to Alzheimer’s disease (20, 21). Additionally, DAP12 must be able to assemble with coreceptors that are as diverse in sequence as they are in ligand-recognition capabilities (22). A molecular-level understanding of what enables this promiscuous yet specific association with so many ligand-binding coreceptors is limited. Therefore, DAP12–coreceptor association serves as an ideal model system for studying polar residue–induced assembly of 2:1 multicomponent receptors.
The sequence diversity of DAP12-associated receptors suggests that interaction stability of the polar triad association motif (Asp-Asp-Lys) is the primary driver of proper subunit assembly and therefore dictates signaling function in the receptor complex. This motif has the potential to exist in four protonation states: a fully ionized triad with an overall −1 charge (designated D−D−K+), a neutral triad in which the lysine and either of the two aspartates are ionized (D−D0K+), a second neutral state in which each residue is nonionized (D0D0K0), and the positively charged state due to ionization of the lysine (D0D0K+). The anionic D−D−K+ state has been widely accepted to drive complex assembly, but evidence for this protonation state is based solely on short, coarse-grained simulations in which resolution is limited or a relatively short (<60 ns) atomistic molecular-dynamics (MD) simulation in which the timescale prevented observation of conformational differences between protonation states (23–28). Given that the D−D−K+ state would result in an energetically unfavorable net charge in the center of the nonpolar bilayer, we felt that a reexamination of the ionization using multiple computational methods was warranted. We were particularly interested in comparing this fully charged state with the D−D0K+ and D0D0K0 states, which do not require burial of a charge near the center of the bilayer. The D−D0K+ appeared to be particularly favorable because aspartates on transmembrane peptides have elevated pKas that assist helix association, and such an elevation in pKa should allow facile protonation of one of the two carboxylate side chains (29–35). Moreover, there is an extensive literature on the role of carboxyl-carboxylate pairs (in which one of the two carboxylate groups is protonated and the other ionized) in stabilizing protein structures (36–39). Consequently, we hypothesized that the Asp-Asp-Lys interaction in these receptor complexes is stabilized by protonation that results in an overall neutral charge of the triad. We further wanted to understand how the presence and formation of this polar-residue network dictate the packing interface of the receptor helices and provide insights into how this conserved signaling-subunit is able to stably engage with so many diverse coreceptors.
Here, we used microsecond-long fully atomistic MD simulations to probe the conformational dynamics as a function of the polar-residue protonation state of the only known structure of DAP12 with a coreceptor, DAP12-NKG2C (Protein Data Bank [PDB] ID: 2L35), both at the polar-residue level and at the helix–helix level. The results suggest that effective assembly of multisubunit receptors is conformationally static at the local polar-residue level but conformationally dynamic at the level of the tertiary structure, thereby allowing a single conserved-signaling module, DAP12, to be simultaneously specific and promiscuous for lysine-containing coreceptors. This favorable polar arrangement is only possible with protonation of one of the aspartic acids on the DAP12 signaling subunit. Protonation ensures charge compensation and generates a carboxyl–carboxylate pair that conformationally stabilizes hydrogen bonding with the lysine residue on the coreceptor.
Results
A Carboxyl-Carboxylate Pair Conformationally Stabilizes the 2:1 Polar-Residue Motif.
To interrogate the geometric and conformational stability of the (Asp-Asp-Lys) polar-residue network, we simulated the DAP12-NKG2C complex, starting from the solution NMR structure (7), in three different protonation states of the polar-residue triad (see Materials and Methods). In complex 1 (D−D−K+), all three residues were ionized leading to a 2:1 negative-to-positive charge ratio. In complex 2 (D0D−K+) and complex 3 (D−D0K+), the aspartate on one of the DAP12 monomers was protonated such that the charge ratio was 1:1. Finally, in complex 4 (D0D0K0), all polar groups were in their neutral protonation state. Three independent μs-long simulations were performed for each complex. To ensure structural stability in simulation, C-α root-mean-square deviations (RMSD) from the starting NMR structure were calculated for every complex. RMSD values plateaued within the first 50 ns of simulation and then remained relatively stable throughout the remainder of the microsecond simulation time, indicating equilibration in this time regime (SI Appendix, Fig. S1). Given that the simulations began with NMR structures, it is hence likely that we are evaluating relevant conformational ensembles on this timescale. After excluding the first 100 ns of each simulation, we performed principal component analysis (PCA) and cluster analysis of the polar-group side chains to understand the conformational variation as a function of protonation state (Fig. 1 and SI Appendix, Figs. S2 and S3). Structures for analysis were taken every 200 ps for a total of 15,000 structures per complex. Geometrically similar structures were grouped with a conservative RMSD cutoff of 0.5 Å (see Materials and Methods). In order to quantify the conformational stability of the polar triad as a function of protonation state, conformational clusters were obtained for each protonation state independently such that clusters from different protonation states were not related. For a comparison between the conformation of the largest cluster in each protonation state see SI Appendix, Fig. S3.
Fig. 1.
Polar-group conformational arrangement as a function of protonation state. (A) Complex 1 (D−D−K+), (B) complex 2 (D0D−K+), (C) complex 3 (D−D0K+), and (D) complex 4 (D0D0K0). For all complexes, the structural centroid of polar-residue (Asp-Asp-Lys) cluster 1 (Top) and the quantification of the proportion of time spent in each of the clusters highlighting the percent of time spent in cluster 1 are shown (Bottom). The dotted arrows denote the minimum aspartate-aspartate O-O distance.
Analysis of complex 1 (D−D−K+) indicated that the geometry of the polar groups populated multiple conformational states in the course of the simulation time (Fig. 1A). The top conformer for complex 1 was populated 39% of the time and frequently fluctuated to other conformations (SI Appendix, Fig. S4). An unfavorable electrostatic term to the short-range nonbonded potential energy between aspartate residues suggested that the conformational dynamics were driven by electrostatic repulsion between aspartates (SI Appendix, Fig. S5A). Indeed, the minimum aspartate-aspartate O-O distance was long and highly fluxional (3.9 ± 0.5 Å), and in two out of three simulations, a sodium ion bound the aspartates for over 40% of the time, consistent with uncompensated charge. Notably, a large influx of water molecules into the membrane (∼10 waters within 3.5 Å of the polar residues) disrupted critical hydrogen bonding interactions between aspartate residues and lysine (SI Appendix, Figs. S5 and S6). In sum, electrostatic repulsion between two negatively charged aspartates led to a dynamic and potentially less stable polar-residue triad arrangement.
In stark contrast to complex 1, the polar group arrangement was conformationally rigid when only one aspartate was protonated, as in the cases of complex 2 (D0D−K+) and complex 3 (D−D0K+). The predominant polar-residue conformers for these complexes were populated 83% and 70% of the time, respectively (Fig. 1 B and C). The aspartic acid formed a short ionic hydrogen bond with the aspartate, average O-O distance of 2.6 ± 0.1 Å, that persisted for greater than 97% of the simulation time (SI Appendix, Fig. S7 A and B). The syn-anti arrangement of the carboxyl-carboxylate clamp between DAP12 monomers was stabilized by association with lysine on the NKG2C helix. In the absence of the NKG2C helix, the carboxyl-carboxylate hydrogen bond was either broken or adopted an energetically unfavorable anti-anti arrangement (SI Appendix, Fig. S8). Persistent charge interactions and hydrogen bonding between lysine and the carboxyl-carboxylate clamp effectively stapled the complex together for 83% of the simulation time in the case of complex 2 (D0D−K+). Though three to five water molecules were always present in the proximity of this polar triad, the dynamic influx and orientation of these waters did not disrupt the conformational stability of the polar group and instead contributed to fulfilling otherwise unsatisfied hydrogen bonds with lysine and the deprotonated aspartate (SI Appendix, Figs. S9 and S10).
Finally, analysis of complex 4 (D0D0K0) demonstrated that assembly was the most conformationally dynamic when the polar residues in the triad were all in the neutral state. The top conformer for complex 4 was only populated 16% of the time (Fig. 1D), a dynamic arrangement due, in part, to a weaker carboxyl-carboxyl hydrogen bond between protonated aspartic-acid residues, which persisted for 59% of the simulation time (SI Appendix, Fig. S11A). Consistent with a weak hydrogen bond, a long and highly fluxional O-O distance of 3.4 ± 1.0 Å between aspartic-acid residues was observed (SI Appendix, Fig. S11B). Taken together, these data demonstrate that conformational stability of the polar triad can only be achieved if the charge is compensated and there is a strong, conformationally rigid interaction between the acidic residues facilitated by the carboxyl–carboxylate pair found in complex 2 (D0D−K+) and complex 3 (D−D0K+).
Polar-Triad Arrangement Imposed by Carboxyl-Carboxylate Pair Is Supported by Density Functional Theory Calculations and Database Search.
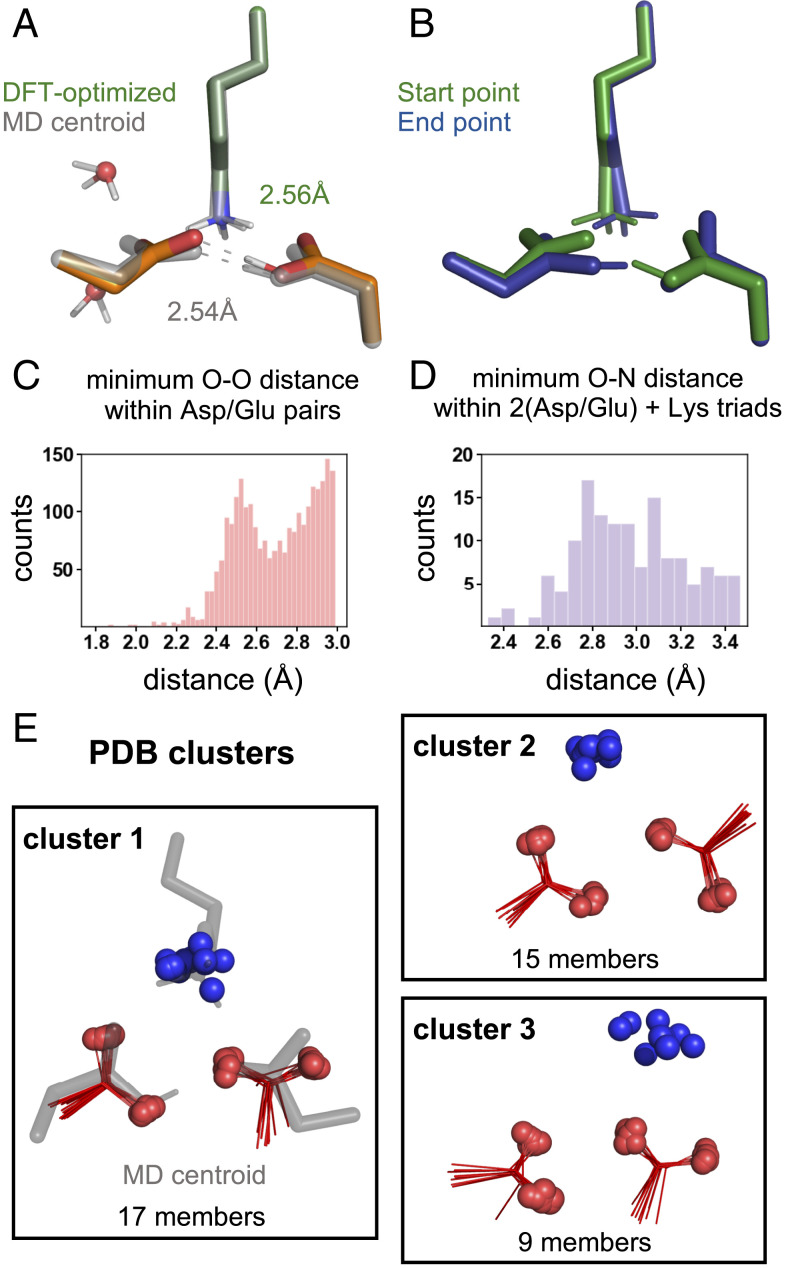
We next wanted to provide support for the polar-group configurations of complex 2 (D0D−K+) and complex 3 (D−D0K+) predicted from MD simulations. Because classical MD forcefields do not account for polarization and may lead to incorrect geometries, especially for charged moieties, we first performed density functional theory (DFT) calculations to determine if this was an energetically reasonable arrangement of the polar groups. We were particularly interested in interrogating the quantum-mechanical feasibility of the short carboxyl-carboxylate O-O distances that were responsible for the observed conformational stability. As a starting point, we used a model that contained the two aspartates, the lysine, a nearby threonine, and the two nearest waters with the positions extracted from the centroid of cluster 1 from complex 2 (SI Appendix, Density Functional Theory Methods). Holding the backbone atoms fixed and allowing the side chains to relax resulted in a structure that was very similar to that observed in MD. Indeed, the RMSD of 0.24 Å is well within our clustering cutoff of 0.5 Å (see SI Appendix for details). This suggests that any restrictions imposed by parameterization and fixed geometries in the force field were not responsible for the polar-group arrangement observed in these complexes. In particular, a short and strong carboxyl-carboxylate interaction was maintained in the relaxed structure (2.56 Å versus 2.54 ± 0.07 Å, Fig. 2A and SI Appendix, Fig. S12A).
Fig. 2.
Evaluation of the (Asp-Asp-Lys) geometry imposed by monoprotonation of the acidic pair. (A) Comparison of the DFT-optimized structure (green/orange) and centroid of cluster 1 from MD simulations of complex 2 (gray) highlighting the similarly short Asp-AspH hydrogen bond (AspH designates a protonated Asp). Overall RMSD of 0.24 Å of the clustered atoms (C-β and C-γ of the Asp/AspH and NZ of the lysine). (B) End points found by following the transition-state mode for proton transfer between the Asp and AspH, showing interconversion of complex 2 (start point, green) and complex 3 (end point, blue). Nonpolar protons, backbone atoms, and threonine in all DFT calculations are removed from the image for clarity. (C) Distribution of minimum carboxylate O-O distances from the PDB. (D) Distribution of minimum lysine-(carboxyl-carboxylate) N-O distances from the PDB. (E) Geometric cluster of lysine-(carboxyl-carboxylate) triads from the PDB that are in closest agreement with the complex-2 (D0D−K+) cluster-1 centroid from MD simulations.
Given the short nature of the carboxyl-carboxylate hydrogen-bonding interaction and that carboxyl-carboxylate pairs can have geometries in which the hydrogen is shared, we wanted to evaluate the energetic penalty for proton transfer between carboxylates. In this explicitly solvated model, proton transfer is uphill by ∼5 kcal/mol (SI Appendix, Fig. S13). However, in a model without explicit waters, we find a nearly barrierless proton transfer (ΔG‡ = +2 kcal/mol) that interconverts complex 2 (start point, Fig. 2B) and complex 3 (end point, Fig. 2B). These findings suggest that water reorganization may be the greatest barrier to proton transfer consistent with the nonpolar nature of the microenvironment. Nonetheless, these results strongly suggest that proton transfer is energetically feasible, and, therefore, all of the conformational states of complex 2 (D0D−K+) and complex 3 (D−D0K+) are likely simultaneously available to be explored during subunit assembly (SI Appendix, Figs. S14 and S15).
To further confirm that the geometry observed in complex 2 (D0D−K+) and complex 3 (D−D0K+) is both reasonable and frequently observed in protein structures, we analyzed lysine-(carboxyl-carboxylate) triads in the PDB (40) based on methods we developed in previous work (41) (SI Appendix, PDB Database Search Methods). We first explored the occurrence frequency and geometry of proteins with glutamate or aspartate carboxylate oxygens within a distance of 3.0 Å. Strikingly, a histogram of the minimum carboxylate O-O distances is bimodal, with a local maximum near 2.5 Å (Fig. 2C). This maximum is associated with carboxyl-carboxylate pairs based on previous surveys of the small-molecule crystallographic database (42), in which the hydrogens can be assigned explicitly, and is consistent with our MD and DFT results. Because of the two distinct populations in the distribution of carboxyl-carboxylate distances, we refined our dataset to only the population with the shortest O-O distances (<2.8 Å). Of those 1,583 carboxyl-carboxylate pairs, we observed 140 (9%) that form a salt bridge with the terminal side chain atom (NZ) of a lysine (i.e., N-O distance 3.5 Å). The distribution of minimum N-O distances observed for carboxyl-carboxylate pairs is similar to that previously seen for lysine-glutamate/aspartate salt bridges (43), showing a maximum near 2.8 Å (Fig. 2D). Importantly, this distribution is consistent with MD-derived N-O distances, which vary depending on the protonation state of the aspartate, 2.8 ± 0.1 Å for the deprotonated aspartate and 3.1 ± 0.4 Å for the protonated aspartate in the case of complex 2 (D0D−K+). Lastly, we grouped lysine-carboxyl-carboxylate triads into geometrically similar clusters with an RMSD cutoff of 0.6 Å (see Materials and Methods). Most interestingly, the top cluster showed a very close superposition with the geometry observed for the centroid of complex 2 (D0D−K+) (Fig. 2E and SI Appendix, Fig. S16). From these data, we conclude that the polar-triad geometry observed in the simulation is well represented in the examples of carboxyl-carboxylate ammonium triads found in the PDB.
Mutant Data Support a Biological Role for the Carboxyl-Carboxylate Pair.
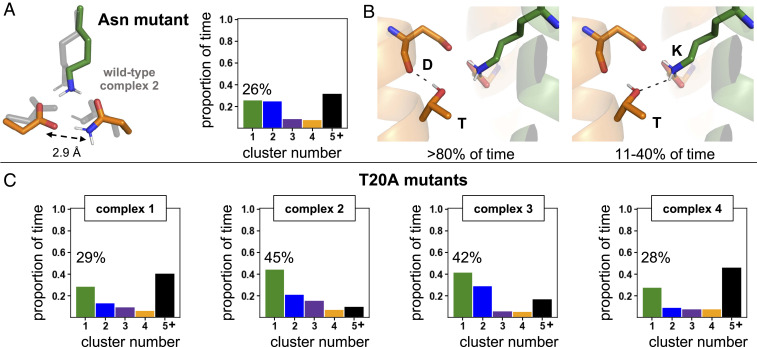
We wanted to understand the biological importance of a conformationally static arrangement of the polar residues. We simulated mutants that have been shown experimentally to have reduced complex assembly relative to the wild-type DAP12-NKG2C complex and interrogated the conformational dynamics of their polar triad. First, we simulated a conservative asparagine mutation (ND−K+), which was previously found to experimentally assemble with 27% of the efficiency of wild type (5). The top conformer of this mutant had a geometry and hydrogen-bonding network that closely resembled that of wild-type complex 2 (D0D−K+) but was observed only 26% of the simulation time (Fig. 3A and SI Appendix, Fig. S17A). A carboxamide-carboxylate bond with an average O-N distance of 2.9 ± 0.2 Å stabilized the DAP12 monomers for interaction with the coreceptor lysine. Analogous to DFT calculations performed on the wild type, we used the centroid of cluster 1 as the input structure and allowed the side chains to relax. This protocol again resulted in a structure in good agreement with the MD centroid (SI Appendix, Fig. S12C). Then, natural bond orbital (NBO) calculations were performed on this asparagine mutant and on wild-type complex 2 to provide insight into the relative strength of noncovalent interactions in the polar region. Consistent with other studies, we found that the Asn-Asp hydrogen bond was ∼20% weaker than the carboxyl-carboxylate hydrogen bond in the wild-type complex (SI Appendix, Fig. S12B). These data are consistent with previous computational evaluation of carboxylate/carboxamide substitutions (39) and in line with the broader finding that hydrogen-bonding distance and strength is strongly correlated with the pKa match between the donor and acceptor partner (44). Consistent with a longer and weaker interaction, the Asn-Asp hydrogen bond was broken in 49% of the MD simulation time (SI Appendix, Fig. S17 C and D). These combined data suggest that a short (2.6 ± 0.1 Å) and strong carboxyl-carboxylate hydrogen bond between wild-type DAP12 monomers is necessary for a conformationally stable interaction with the lysine on the NKG2C coreceptor and that even small deviations in the hydrogen-bond strength are conformationally destabilizing.
Fig. 3.
The role of mutation on polar-triad conformational stability. (A) Comparison of the top polar-residue conformer of the asparagine mutant (ND−K+) to the top conformer of wild-type complex 2 (D0D−K+) (Left) and a quantification of the proportion of time this mutant arrangement is present in the simulation (Right). (B) The two roles of threonine in complex assembly: threonine-aspartate side-chain-to-main-chain hydrogen bonding was present >80% of the simulation time (Left), and threonine-lysine hydrogen bonding was present 11 to 40% of the simulation time depending on ionization state (Right). (C) Quantification of the proportion of frames spent in each of the polar-residue (Asp-Asp-Lys) clusters for T20A mutants of complex 1 (D−D−K+), complex 2 (D0D−K+), complex 3 (D−D0K+), and complex 4 (D0D0K0). The percent of time spent in the top cluster is highlighted.
Previous experimental studies have also shown that an alanine mutation of a threonine residue present one helical turn (i + 4 residues) away from each aspartate on DAP12 resulted in a 70% loss of assembly with NKG2C (7). To understand how this threonine-to-alanine (T20A) mutation on DAP12 influenced polar-triad conformational stability, we compared T20A mutants in each protonation state to wild-type complexes. Threonine performed two essential functions in wild-type assembly: 1) it formed a side-chain-to-main-chain hydrogen bond with the aspartate carbonyl oxygen, rigidifying that stretch of the DAP12 helix for greater than 80% of the simulation time and 2) it directly engaged the lysine residue through hydrogen bonding for 11 to 40% of the simulation time depending on protonation state (Fig. 3B and SI Appendix, Fig. S18). The T20A mutation conformationally destabilized the Asp-Asp-Lys polar triad (Fig. 3C and SI Appendix, Fig. S19A). Mutation in complex 2 (D0D−K+) and complex 3 (D−D0K+) resulted in a dramatic decrease in the population of the top conformer, with the T20A mutant of complex 2 (D0D−K+) exhibiting 45% representation compared to 83% in the wild type. This conformational destabilization was not as pronounced in complex 1 (D−D−K+), suggesting that the threonine residue in this protonation state would not negatively affect observed assembly, inconsistent with the experimental data. Consistent with this observation, the number of waters that formed hydrogen bonds with lysine increased with the T20A mutation only for complex 2 (D0D−K+) and complex 3 (D−D0K+) (SI Appendix, Fig. S19B). These T20A mutant data again demonstrate that local polar-group conformational stability correlates to experimentally observed assembly in the DAP12-NKG2C complex. Importantly, this and the asparagine-mutant data support the hypothesis that persistence of the polar groups in the specific arrangement necessary to stabilize DAP12-NKG2C complex assembly is only possible when the aspartate pair of wild-type DAP12 is monoprotonated to form a carboxyl-carboxylate clamp as in complex 2 (D0D−K+) and complex 3 (D−D0K+).
Complex Assembly Is Globally Dynamic.
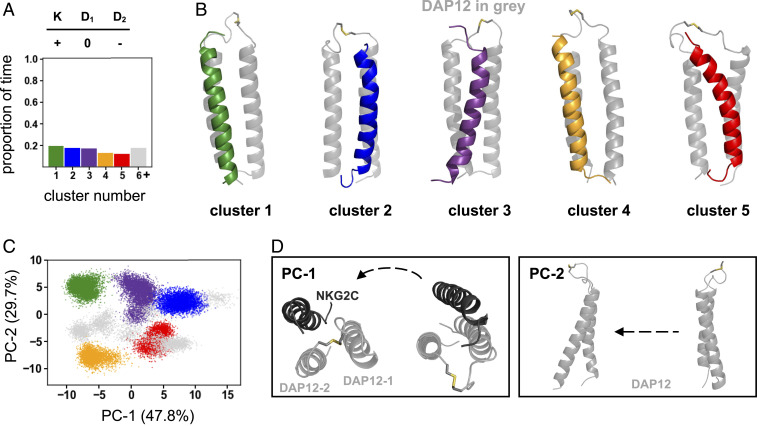
With substantial evidence in support of a charge-compensated polar-group arrangement, we sought to determine how local polar-group conformational stability influenced helix–helix conformational dynamics. We reasoned that helix-rearrangement dynamics would occur over longer timescales, and, in an effort to capture more conformational space, we performed two additional independent 1-μs–long simulations of complex 2 (D0D−K+) for a total of 5 μs. The local polar-triad arrangement of the 5 μs of simulations was in agreement with the arrangement found after 3 μs (SI Appendix, Fig. S20A), with a cluster 1 centroid RMSD of 0.13 Å. We then clustered simulations and performed PCA at the helix level based on the C-α RMSD of the 11 core residues of each helix, including the three polar residues in the triad, with a 1.5 Å RMSD cutoff (see Materials and Methods). Surprisingly, we found a conformationally dynamic arrangement of helices with multiple clusters (Fig. 4A). Visual inspection of aligned cluster centroids and RMSD analysis of helix pairs within the trimer suggested that the position of the NKG2C coreceptor helix relative to DAP12 varied considerably (Fig. 4B and SI Appendix, Fig. S20B). To corroborate this observation, we then used PCA to identify the greatest source of conformational variance. Principal component 1 (PC-1) accounted for 47.8% of variance and represented the translational motion of the NKG2C helix from close engagement with DAP12-1 to DAP12-2 (Fig. 4 C and D). PC-2 accounted for variance in the DAP12 homodimer crossing angle, but the biggest differentiating factor between clusters was the NKG2C coreceptor position. In sum, a local, fixed polar-group arrangement did not dictate the helix arrangement. Instead, the conformational arrangement of the polar groups in this complex was stably maintained even as the NKG2C position relative to the DAP12 homodimer varied such that the helical interface remained dynamic.
Fig. 4.
Global-helix dynamics of complex 2 (D0D−K+). (A) Quantification of the proportion of time DAP12-NKG2C complex spends in geometrically different 33-residue C-α clusters. (B) Structural centroids of the top five clusters. (C) Principal-component analysis of global-helix residues. Projections of structural coordinates along PC-1 and PC-2 are shown (points are colored according to the helical clustering analysis). (D) Visual representation of the global-helical motions along PC-1 and PC-2.
Discussion
Multicomponent receptor assembly that proceeds through a 2:1 acidic-to-basic polar-residue triad is conformationally static at the local polar-residue level only if the acidic-residue pair is monoprotonated. Atomistic MD simulations of the DAP12-NKG2C complex have demonstrated that, when both aspartates are deprotonated, multiple conformational states of the polar residues are populated due to electrostatic repulsion of the aspartates. Importantly, increased conformational dynamics of the polar residues correlate with reduced experimental complex assembly in asparagine and threonine mutants. There is no appreciable difference in polar-residue dynamics between the wild-type doubly-deprotonated state and these mutants, allowing us to establish this protonation state as unproductive for assembly. Protonation of one of the aspartates eliminates electrostatic repulsion and generates a stabilizing carboxyl-carboxylate clamp with a nearly invariant 2.6-Å hydrogen-bonding interaction that more than doubles the conformational stability of the polar triad. The carboxyl-carboxylate clamp engages with the coreceptor lysine in a conformationally persistent arrangement that was found to be energetically favorable through DFT and well precedented in soluble protein structures. The short carboxyl-carboxylate hydrogen bond between DAP12 monomers that staples this triad together is of particular interest because it carries the potential for added conformational stability. Given that the proton in the carboxyl-carboxylate pair has a low barrier for transfer between the two carboxylate groups, complex 2 (D0D−K+) and complex 3 (D−D0K+) can each be considered a limiting case, imposed by the classical force field, of what is likely a shared interaction. Therefore, both of the tautomeric states of complex 2 (D0D−K+) and complex 3 (D−D0K+) are likely available to be explored during subunit assembly. Conceivably, all residues in the triad could be in the neutral state if lysine (intrinsic pKa = 10.5) were deprotonated, but some coreceptors rely on arginine (intrinsic pKa = 12) for association, which is more likely to remain ionized. Therefore, we reasoned that lysine should also be charged, and the data from complex 4 (D0D0K0) corroborate our hypothesis that an ionized basic residue is necessary for receptor assembly.
A monoprotonated acidic pair was suggested previously to be unlikely because of a threefold decrease in complex-assembly efficiency in the conservative mutation of one aspartate to asparagine (5). However, this change in stability corresponds to less than a single kilocalorie per mole, and a carboxamide-carboxylate pair is not a perfect mimic of the hydrogen bonding and electrostatic network of a carboxyl-carboxylate pair in the monoprotonated wild-type system. Moreover, aspartate to asparagine mutations have been previously shown to modestly decrease (2 to 3 °C) the thermostability of carboxyl-carboxylate pairs (45), a difference that could account for the similarly modest effect of an Asp-to-Asn mutation on assembly of the DAP12-NKG2C complex. In line with literature precedent, we find that, although asparagine shares many characteristics with aspartic acid, it is not a perfect mimic. The 2.6-Å carboxyl-carboxylate hydrogen bond conformationally stabilizes the DAP12 dimer interface, while a longer 2.9-Å carboxamide-carboxylate bond makes this point of attachment weaker and more fluxional in the asparagine mutant. This slight destabilization of the DAP12 homodimer interface turns out to be conformationally destabilizing for assembly with the lysine residue on the coreceptor. These subtle changes observed due to conservative mutations highlight the means by which the DAP12-signaling subunit ensures specific assembly with a coreceptor as long as the coreceptor contains a properly positioned lysine residue.
One notable feature of the 2:1 polar-residue motif is its residence in the nonpolar membrane. Because MD force fields represent the structures of both water-soluble and membrane-soluble proteins well, the geometry of the local atomic interactions in the polar triad observed in the simulation would be expected to hold across a wide range of environments. Indeed, the DFT analysis and the PDB search for this motif corroborate the conclusion that this geometry is locally optimized and largely invariant with its environment. Conversely, the energetics of polar-triad association, though locally favorable, will vary depending on the folding environment. Energetics of folding are based on the unfolded or reference state of a system. In the case of multicomponent receptor assembly, the “unfolded state” is composed of noninteracting, membrane-inserted helices with largely desolvated polar groups due to the nonpolar environment. Previous studies with model peptides have shown that aspartic acid in the membrane interior has an elevated pKa, which allows protonation near neutral pH (29–35). Folding would bring the interacting groups together without incurring the energetically costly dehydration of the polar side chains or protonation of one of the two carboxyl groups involved in the carboxyl-carboxylate clamp. By contrast, the formation of a (carboxyl-carboxylate)-lysine triad in water-soluble proteins would require a significant dehydration penalty as the chain folds from the hydrated random coil to its native structure (46). We suspect that this energetic advantage in the membrane is one of the reasons why this 2:1 motif is so prevalent across multicomponent receptor families. We further expect that (carboxyl-carboxylate)-lysine/arginine triads will be found across a variety of other membrane proteins as more structures become available. The precise geometries will likely vary based on the surrounding sequence, but the database search performed here should be generally helpful for identifying favorable geometries.
Interestingly, the local rigidity in the polar-group arrangement observed to be necessary for specific assembly did not carry forward to helix–helix arrangement and packing, which were conformationally dynamic. This dynamic arrangement of the helices is independent of the protonation state of the polar triad (SI Appendix, Fig. S20C). We propose that it is this locally static and globally dynamic arrangement that enables DAP12 to assemble specifically with so many sequence-diverse ligand-binding coreceptors. Because the global-complex arrangement is inherently dynamic without disrupting the polar-residue arrangement, it is better able to accommodate the sequence diversity. Additionally, the lack of a single structural conformation explains, in part, why detailed biophysical-structural characterization has been so challenging in these systems. Though a great degree of sequence tolerance is built into the association of DAP12 with a coreceptor, and specificity of assembly is centered on the polar-residue triad (5–7), the remaining transmembrane residues do play a role in specificity, which is why DAP12 cannot assemble with a coreceptor like NKG2D (47). The T20A mutant data clearly demonstrate the importance of distal residues on proper complex assembly, but additional studies are necessary to illuminate the nature of specificity-generating interactions in the rest of the transmembrane sequence.
The work presented here has clarified details of protein-subunit assembly structure and dynamics that define effectiveness in multicomponent receptors. These details help inform the mechanism of signal transduction across the membrane in this class of receptors, which is still unknown. The 2:1 acidic-to-basic polar-residue association motif within the membrane is found in receptors in which signaling is thought to be mediated by extracellular clustering of the ligand-binding subunits (1, 14, 48–50). This clustering hypothesis proposes that increased local concentrations of ITAM motifs dictate the efficiency of receptor phosphorylation and subsequent downstream signaling (51–53). This suggests that the goal of the transmembrane association motif is to staple the ligand-binding and signaling subunits in a stable conformation such that, when extracellular receptor clustering in response to a ligand occurs, the signaling subunits also cluster. For receptors that are hypothesized to signal through extracellular clustering, stable assembly with the signaling coreceptor, in this case, DAP12, is a necessary component for successful clustering of signaling motifs. The carboxyl-carboxylate clamp on DAP12 is strong enough to staple to lysine on a coreceptor and allows the two subunits to move in the membrane as one receptor. Additionally, the dynamic helix packing found for the DAP12-NKG2C receptor complex might contribute to this signaling-by-clustering hypothesis. The global dynamics observed appear to contradict a ligand-induced conformational-change mechanism. Instead, dynamic helix rearrangement could allow the ligand-binding subunit to freely adapt its position and orientation while still maintaining a stable point of attachment to the signaling subunit through the polar motif. This global conformational flexibility would be important for cluster-mediated signaling in order to allow the conformational changes necessary for clustering with other receptors in response to a ligand.
Conclusion
Overall, the findings reported here provide an atomic-level picture that helps explain the simultaneous specificity and promiscuity of DAP12 for lysine-containing receptors with very little sequence homology. DAP12 assembly with a coreceptor is mediated by a monoprotonated aspartate pair, not a doubly-deprotonated aspartate pair as was previously proposed. This carboxyl-carboxylate pair forms a short and strong hydrogen bond that mediates a conformationally static interaction with the coreceptor lysine. Specificity of interaction with the lysine results from both the conformational restriction and directionality imparted on the aspartate-aspartic acid pair by such a tight hydrogen bond as well as hydrogen bonding of lysine to a distal threonine. This local, fixed arrangement does not extend to the helix level, which is found to be highly flexible. Thus, the DAP12 signaling-module is able to promiscuously interact with many sequence-diverse coreceptors as long as they retain the lysine residue that imparts stability. These findings elucidate details of receptor assembly that proceeds by means of a 2:1 acidic-to-basic polar-residue motif, and they are widely applicable across receptor families because this motif is so pervasive. Further, this work provides evidence in support of cluster-mediated signaling in this class of receptors, which needs to be stable enough to ensure subunit assembly while also being dynamically adaptable to respond to a ligand.
Materials and Methods
All MD simulations used the DAP12-NKG2C NMR structure (PDB ID: 2L35) (7). The structure was embedded in a POPC lipid membrane and solvated in 150 mM NaCl using CHARMM-GUI (54). All production runs were performed in the isothermal–isobaric (NPT) ensemble using the Amber ff14SB protein force field and the lipid17 lipid force field with the Amber Graphics Processing Unit (GPU) engine (55–57). Changes to the ionization state of the polar residues and mutations were made in the Amber LEaP program (58). Three independent 1.1-μs simulations were performed per ionization or mutation state starting from the same initial position but different starting velocities. For analysis, trajectories were combined for a total of 3 μs per ionization state (after discarding the first 100 ns of each simulation). Two additional simulations were performed for complex 2 for analysis of global helix dynamics.
For DFT calculations, the input positions were taken from the centroid for the top cluster from the complex of interest. The inputs were completed by appropriately terminating the N- and carboxyl-termini of each residue. Then, optimization, frequency, and transition state calculations were done with the ORCA software package (version 4.2) (59). NBO calculations were performed with the Gaussian09 package (60, 61).
For the PDB search, we used a nonredundant dataset of 13,026 protein-crystal structures curated by PISCES (62) on September 14, 2020, with ≤30% sequence identity, R factor of ≤0.3, and resolution of ≤2.5 Å. All structures were rerefined by PDB-REDO (63). We searched for lysine-(carboxyl-carboxylate) triads, and identical triads within oligomers belonging to the same structure were eliminated.
All clustering analysis was performed using a hierarchical agglomerative average-linkage approach (64) with a 0.5 Å and 1.5 Å cutoff for the local polar-group and global-helix analysis of MD simulations, respectively. A 0.6 Å cutoff was used for clustering of PDB structures.
Further details on all MD, DFT, database search, and analysis methods can be found in the SI Appendix, Methods.
Supplementary Material
Acknowledgments
We thank Kevin J. Metcalf, Paola Bisignano, and Andrew Natale for useful discussions. This work was supported by NIH Grant K12GM081266 (L.-K.F.), and NIH Grant F32 GM139379 (M.J.C.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026318118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Call M. E., Wucherpfennig K. W., Common themes in the assembly and architecture of activating immune receptors. Nat. Rev. Immunol. 7, 841–850 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Lanier L. L., DAP10- and DAP12-associated receptors in innate immunity. Immunol. Rev. 227, 150–160 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lanier L. L., Turning on natural killer cells. J. Exp. Med. 191, 1259–1262 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Humphrey M. B., Lanier L. L., Nakamura M. C., Role of ITAM-containing adapter proteins and their receptors in the immune system and bone. Immunol. Rev. 208, 50–65 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Feng J., Garrity D., Call M. E., Moffett H., Wucherpfennig K. W., Convergence on a distinctive assembly mechanism by unrelated families of activating immune receptors. Immunity 22, 427–438 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng J., Call M. E., Wucherpfennig K. W., The assembly of diverse immune receptors is focused on a polar membrane-embedded interaction site. PLoS Biol. 4, e142 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Call M. E., Wucherpfennig K. W., Chou J. J., The structural basis for intramembrane assembly of an activating immunoreceptor complex. Nat. Immunol. 11, 1023–1029 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blázquez-Moreno A., et al., Transmembrane features governing Fc receptor CD16A assembly with CD16A signaling adaptor molecules. Proc. Natl. Acad. Sci. U.S.A. 114, E5645–E5654 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manolios N., Bonifacino J. S., Klausner R. D., Transmembrane helical interactions and the assembly of the T cell receptor complex. Science 249, 274–277 (1990). [DOI] [PubMed] [Google Scholar]
- 10.Call M. E., Pyrdol J., Wiedmann M., Wucherpfennig K. W., The organizing principle in the formation of the T cell receptor-CD3 complex. Cell 111, 967–979 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Call M. E., et al., The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell 127, 355–368 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanier L. L., Corliss B. C., Wu J., Leong C., Phillips J. H., Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature 391, 703–707 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Lanier L. L., Corliss B., Wu J., Phillips J. H., Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity 8, 693–701 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Turnbull I. R., Colonna M., Activating and inhibitory functions of DAP12. Nat. Rev. Immunol. 7, 155–161 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Paloneva J., et al., Loss-of-function mutations in TYROBP (DAP12) result in a presenile dementia with bone cysts. Nat. Genet. 25, 357–361 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Paloneva J., et al., Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am. J. Hum. Genet. 71, 656–662 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paloneva J., et al., DAP12/TREM2 deficiency results in impaired osteoclast differentiation and osteoporotic features. J. Exp. Med. 198, 669–675 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen S. T., et al., CLEC5A is critical for dengue-virus-induced lethal disease. Nature 453, 672–676 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Thomas R., et al., NKG2C deletion is a risk factor of HIV infection. AIDS Res. Hum. Retroviruses 28, 844–851 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulrich J. D., Ulland T. K., Colonna M., Holtzman D. M., Elucidating the role of TREM2 in Alzheimer’s disease. Neuron 94, 237–248 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Yeh F. L., Hansen D. V., Sheng M., TREM2, microglia, and neurodegenerative diseases. Trends Mol. Med. 23, 512–533 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Lanier L. L., Bakker A. B., The ITAM-bearing transmembrane adaptor DAP12 in lymphoid and myeloid cell function. Immunol. Today 21, 611–614 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Cheng X., Im W., NMR observable-based structure refinement of DAP12-NKG2C activating immunoreceptor complex in explicit membranes. Biophys. J. 102, L27–L29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma S., Juffer A. H., An atomistic model for assembly of transmembrane domain of T cell receptor complex. J. Am. Chem. Soc. 135, 2188–2197 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Sun H., Chu H., Fu T., Shen H., Li G., Theoretical elucidation of the origin for assembly of the DAP12 dimer with only one NKG2C in the lipid membrane. J. Phys. Chem. B 117, 4789–4797 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Wei P., Zheng B. K., Guo P. R., Kawakami T., Luo S. Z., The association of polar residues in the DAP12 homodimer: TOXCAT and molecular dynamics simulation studies. Biophys. J. 104, 1435–1444 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei P., et al., Molecular dynamic simulation of the self-assembly of DAP12-NKG2C activating immunoreceptor complex. PLoS One 9, e105560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dube N., Marzinek J. K., Glen R. C., Bond P. J., The structural basis for membrane assembly of immunoreceptor signalling complexes. J. Mol. Model. 25, 277 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Gratkowski H., Lear J. D., DeGrado W. F., Polar side chains drive the association of model transmembrane peptides. Proc. Natl. Acad. Sci. U.S.A. 98, 880–885 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou F. X., Merianos H. J., Brunger A. T., Engelman D. M., Polar residues drive association of polyleucine transmembrane helices. Proc. Natl. Acad. Sci. U.S.A. 98, 2250–2255 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caputo G. A., London E., Cumulative effects of amino acid substitutions and hydrophobic mismatch upon the transmembrane stability and conformation of hydrophobic α-helices. Biochemistry 42, 3275–3285 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Caputo G. A., London E., Position and ionization state of Asp in the core of membrane-inserted α helices control both the equilibrium between transmembrane and nontransmembrane helix topography and transmembrane helix positioning. Biochemistry 43, 8794–8806 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Senes A., Engel D. E., DeGrado W. F., Folding of helical membrane proteins: The role of polar, GxxxG-like and proline motifs. Curr. Opin. Struct. Biol. 14, 465–479 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Moore D. T., Berger B. W., DeGrado W. F., Protein-protein interactions in the membrane: Sequence, structural, and biological motifs. Structure 16, 991–1001 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shahidullah K., Krishnakumar S. S., London E., The effect of hydrophilic substitutions and anionic lipids upon the transverse positioning of the transmembrane helix of the ErbB2 (neu) protein incorporated into model membrane vesicles. J. Mol. Biol. 396, 209–220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langkilde A., et al., Short strong hydrogen bonds in proteins: A case study of rhamnogalacturonan acetylesterase. Acta Crystallogr. D Biol. Crystallogr. D64, 851–863 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawyer L., James M. N., Carboxyl-carboxylate interactions in proteins. Nature 295, 79–80 (1982). [DOI] [PubMed] [Google Scholar]
- 38.Flocco M. M., Mowbray S. L., Strange bedfellows: Interactions between acidic side-chains in proteins. J. Mol. Biol. 254, 96–105 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Wohlfahrt G., Analysis of pH-dependent elements in proteins: Geometry and properties of pairs of hydrogen-bonded carboxylic acid side-chains. Proteins 58, 396–406 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Berman H. M., et al., The protein data bank. Nucleic Acids Res. 28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan S. K., et al., Modulating integrin alphaIIbbeta3 activity through mutagenesis of allosterically regulated intersubunit contacts. Biochemistry 58, 3251–3259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’Ascenzo L., Auffinger P., A comprehensive classification and nomenclature of carboxyl-carboxyl(ate) supramolecular motifs and related catemers: Implications for biomolecular systems. Acta Crystallogr. B Struct. Sci. Cryst. Eng. Mater. 71, 164–175 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donald J. E., Kulp D. W., DeGrado W. F., Salt bridges: Geometrically specific, designable interactions. Proteins 79, 898–915 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Remer L. C., Jensen J. H., Toward a general theory of hydrogen bonding: The short, strong hydrogen bond [HOH···OH]. J. Phys. Chem. A 104, 9266–9275 (2000). [Google Scholar]
- 45.Lin J., Pozharski E., Wilson M. A., Short carboxylic acid-carboxylate hydrogen bonds can have fully localized protons. Biochemistry 56, 391–402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeGrado W. F., Gratkowski H., Lear J. D., How do helix-helix interactions help determine the folds of membrane proteins? Perspectives from the study of homo-oligomeric helical bundles. Protein Sci. 12, 647–665 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu J., Cherwinski H., Spies T., Phillips J. H., Lanier L. L., DAP10 and DAP12 form distinct, but functionally cooperative, receptor complexes in natural killer cells. J. Exp. Med. 192, 1059–1068 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Germain R. N., T-cell signaling: The importance of receptor clustering. Curr. Biol. 7, R640–R644 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Duchemin A. M., Ernst L. K., Anderson C. L., Clustering of the high affinity Fc receptor for immunoglobulin G (Fc gamma RI) results in phosphorylation of its associated gamma-chain. J. Biol. Chem. 269, 12111–12117 (1994). [PubMed] [Google Scholar]
- 50.Ma Y., et al., Clustering of the zeta-chain can initiate T cell receptor signaling. Int. J. Mol. Sci. 21, 3498 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Underhill D. M., Goodridge H. S., The many faces of ITAMs. Trends Immunol. 28, 66–73 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Love P. E., Shores E. W., ITAM multiplicity and thymocyte selection: How low can you go? Immunity 12, 591–597 (2000). [DOI] [PubMed] [Google Scholar]
- 53.James J. R., Tuning ITAM multiplicity on T cell receptors can control potency and selectivity to ligand density. Sci. Signal. 11, eaan1088 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jo S., Kim T., Im W., Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One 2, e880 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maier J. A., et al., ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dickson C. J., et al., Lipid14: The Amber lipid force field. J. Chem. Theory Comput. 10, 865–879 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salomon-Ferrer R., Götz A. W., Poole D., Le Grand S., Walker R. C., Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 9, 3878–3888 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Case D. A., et al., AMBER 2016 (University of California, San Francisco, 2016). [Google Scholar]
- 59.Neese F., Software update: The ORCA program system, version 4.0. WIREs Comput. Mol. Sci. 8, e1327 (2018). [Google Scholar]
- 60.Glendening E. D., Reed A. E., Carpenter J. E., Weinhold F., NBO Version 3.1 (Gaussian Inc., Pittsburgh, 2003). [Google Scholar]
- 61.Frisch M. J.et al., Gaussian 09, Revision E.01 (Gaussian, Inc., Wallingford, CT, 2016). [Google Scholar]
- 62.Wang G., Dunbrack R. L. Jr, PISCES: A protein sequence culling server. Bioinformatics 19, 1589–1591 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Joosten R. P., Long F., Murshudov G. N., Perrakis A., The PDB_REDO server for macromolecular structure model optimization. IUCrJ 1, 213–220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shao J., Tanner S. W., Thompson N., Cheatham T. E., Clustering molecular dynamics trajectories: 1. Characterizing the performance of different clustering algorithms. J. Chem. Theory Comput. 3, 2312–2334 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.