Significance
Antibody-dependent complement activation plays a major role in various pathophysiological processes in our body, including infection, inflammation, autoimmunity, and transplant rejection. In order to activate complement, antibodies should bind to target cells and recruit complement component C1. C1 is a large, multimolecular complex that consists of the antibody recognition protein C1q and a heterotetramer of proteases (C1r2s2). Although it is believed that interactions between C1 and IgGs are solely mediated by C1q, we here show that C1r2s2 proteases affect the capacity of C1q to form an avid complex with surface-bound IgG molecules. Furthermore, we demonstrate that C1q–IgG stability is influenced by IgG oligomerization and that promoting IgG oligomerization improves phagocytosis of the pathogenic bacterium Staphylococcus aureus.
Keywords: complement, C1, IgG subclasses, IgG hexamerization, Staphylococcus aureus
Abstract
Complement is an important effector mechanism for antibody-mediated clearance of infections and tumor cells. Upon binding to target cells, the antibody’s constant (Fc) domain recruits complement component C1 to initiate a proteolytic cascade that generates lytic pores and stimulates phagocytosis. The C1 complex (C1qr2s2) consists of the large recognition protein C1q and a heterotetramer of proteases C1r and C1s (C1r2s2). While interactions between C1 and IgG-Fc are believed to be mediated by the globular heads of C1q, we here find that C1r2s2 proteases affect the capacity of C1q to form an avid complex with surface-bound IgG molecules (on various 2,4-dinitrophenol [DNP]-coated surfaces and pathogenic Staphylococcus aureus). The extent to which C1r2s2 contributes to C1q–IgG stability strongly differs between human IgG subclasses. Using antibody engineering of monoclonal IgG, we reveal that hexamer-enhancing mutations improve C1q–IgG stability, both in the absence and presence of C1r2s2. In addition, hexamer-enhanced IgGs targeting S. aureus mediate improved complement-dependent phagocytosis by human neutrophils. Altogether, these molecular insights into complement binding to surface-bound IgGs could be important for optimal design of antibody therapies.
Antibodies are important mediators of the human complement response, which offers critical protection against microbial infections and damaged host cells (1). In order to initiate a complement response, an antibody molecule first needs to bind antigens on the target cell via its antigen-binding (Fab) domains (2–5). Subsequently, the antibody’s constant (Fc) domain recruits the first complement protein complex, C1, to the cell surface (SI Appendix, Fig. S1A). The large C1 complex (also denoted as C1qr2s2, 766 kDa) consists of the recognition protein C1q (410 kDa) and a heterotetramer of serine proteases C1r and C1s (denoted C1r2s2, 356 kDa) (SI Appendix, Fig. S1B). While C1q is responsible for antibody recognition, its attached proteases C1r2s2 induce the activation of downstream enzymatic complexes (i.e., C3 convertases [C4b2b (6)]) that catalyze the covalent deposition of C3-derived molecules (e.g., C3b and its degradation product iC3b) onto the target cell surface (SI Appendix, Fig. S1A) (7, 8). C3b opsonizes the target cell surface and can induce formation of lytic pores (membrane attack complex [MAC]) in the target cell membrane (9–11). In contrast to human cells and gram-negative bacteria, gram-positive bacteria are not susceptible to the MAC due to their thick cell wall (12). On these bacteria, efficient decoration with C3b and iC3b is essential for triggering effective phagocytic uptake of target cells via complement receptors (CR) expressed on phagocytes of which the integrin CR3 (also denoted CD11b/CD18) is considered most important (13, 14).
In recent years, our insights into IgG-dependent complement activation have increased significantly. A combination of structural, biophysical, and functional studies revealed that surface-bound IgG molecules (after Fab-mediated antigen binding) require organization into higher-ordered structures, namely hexamers, to induce complement activation most effectively (15–19). Hexamerized IgGs are being held together by noncovalent Fc–Fc interactions and form an optimal platform for C1q docking (SI Appendix, Fig. S1A). C1q has a “bunch of tulips–” like structure, consisting of six collagen arms that each end in a globular (gC1q) domain (SI Appendix, Fig. S1B) that binds the Fc region of an IgG. As the affinity of C1q for a single IgG is very weak (20, 21), avidity achieved through simultaneous binding of its globular domains to six oligomerized IgG molecules is paramount for a strong response (15, 17–19). Furthermore, it was found that IgG hexamerization could be manipulated by specific point mutations in the Fc–Fc contact region that enhance such oligomerization (15, 18, 22). While these hexamer-enhancing mutations in IgG potentiate the efficacy of MAC-dependent cytotoxicity on tumor cells and gram-negative bacteria (15, 23), their effect on complement-dependent phagocytosis is not known.
Because complement is an important effector mechanism to kill bacteria and tumor cells, development of complement-enhancing antibodies represents an attractive strategy for immune therapies (1, 24). Immunotherapy based on human monoclonal antibodies is not yet available for bacterial infections (25–28). Such developments are mainly hampered by the fact that little is known about the basic mechanisms of complement activation on bacterial cells. For instance, we do not understand why certain antibodies induce complement activation on bacteria and others do not. In this study, we set out to investigate how antibacterial IgGs induce an effective complement response. By surprise, we noticed that C1q–IgG stability differs between human IgG subclasses. More detailed molecular investigations revealed that C1r2s2 proteases are important for generating stable C1q–IgG complexes on various target surfaces. Furthermore, we demonstrate that C1q–IgG stability is influenced by antibody oligomerization. These molecular insights into C1q binding to surface-bound IgGs may pave the way for optimal design of antibody therapies.
Results
IgG-Mediated Complement Activation Does Not Always Correlate with Detection of C1q.
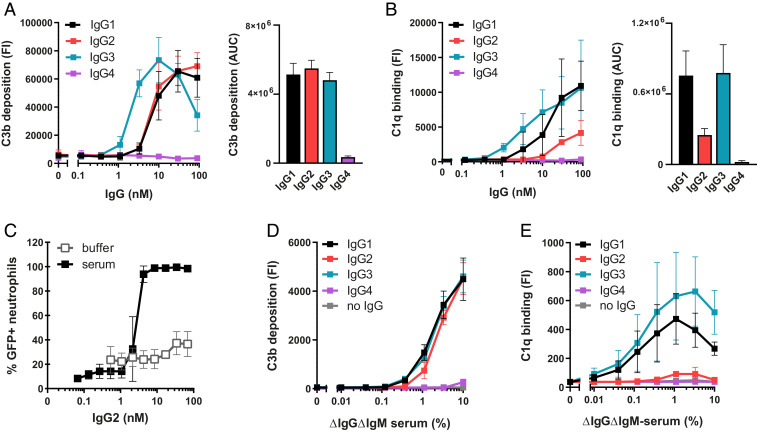
To enhance our understanding of complement activation by antibacterial IgGs, we studied complement activation by monoclonal antibodies against Staphylococcus aureus, an important gram-positive pathogen and the leading cause of hospital-acquired infections. We generated IgGs against wall teichoic acid (WTA), a highly abundant and immunogenic cell wall glycopolymer that comprises 40% of the staphylococcal cell wall (29–31). The variable domains of anti-WTA IgG1 4,497 (31) were cloned into HEK expression vectors encoding IgG1, IgG2, IgG3, and IgG4 Fc backbones. We included all IgG subclasses to obtain a better understanding of their variable complement effector functions (1, 32). After confirming that all IgG subclasses bound similarly to the bacterial surface (SI Appendix, Fig. S2), we examined complement activation by anti-WTA IgGs by incubating S. aureus with human serum as a complement source. To exclude involvement of naturally occurring anti-staphylococcal IgGs, we used serum that is depleted of natural IgG and IgM (ΔIgGΔIgM serum) (33). Complement activation was first quantified by measuring deposition of C3 cleavage products on the surface of S. aureus using flow cytometry (Fig. 1A). In line with recent results, IgG1 and IgG3 antibodies against WTA elicit effective C3b deposition on S. aureus (Fig. 1A) (34). Furthermore, IgG4 did not activate complement on S. aureus (Fig. 1A), which was expected because IgG4 has a reduced capability to interact with C1q (35–37), (38–41).
Fig. 1.
IgG-mediated complement activation does not always correlate with detection of C1q. (A and B) C3b deposition (A) and C1q binding (B) on anti-WTA IgG-labeled S. aureus Wood46 bacteria upon incubation with 5% or, respectively, 1% ΔIgGΔIgM serum as determined by flow cytometry. The data represent mean FI ± SD (Left) or mean area under the curve (AUC) ± SD (Right) of three independent experiments. AUC of C3b and C1q binding curves was determined after subtraction of a 2,500 (C3b) or 200 (C1q) baseline, respectively. (C) Phagocytosis of fluorescently labeled S. aureus Wood46 in either RPMI buffer or 1% ΔIgGΔIgM serum supplemented with anti-WTA IgG2 and human neutrophils. Bacterial uptake was quantified by flow cytometry and displayed as the percentage of GFP-positive neutrophils. The data represent mean ± SD of three independent experiments. (D and E) C3b deposition (D) and detection of C1q (E) on beads coated with 1 µg/mL DNP upon incubation with ΔIgGΔIgM serum and human monoclonal anti-DNP IgG (20 nM). Deposition of C3b and C1q molecules on the beads was determined by flow cytometry. The data represent geometric mean ± SD of three independent experiments.
Our particular interest was in IgG2, which is the predominant IgG subclass against WTA in a natural human immune response (30, 42). Although IgG2 is often described as a poor complement activator, we here observed that anti-WTA IgG2 effectively induced C3b deposition on S. aureus to a level that was comparable to anti-WTA IgG1 and IgG3 (Fig. 1A). These data support previous studies suggesting that IgG2 can activate complement when reacting with highly dense epitopes (42–45).
When we took a closer look at different complement activation steps, we noticed an unexpected disparity between surface detection of C3b and C1q for IgG2. While recruitment of C1q is a prerequisite to initiate antibody-dependent complement activation, we observed that detection of C1q molecules on IgG2-coated bacteria was low compared to IgG1 and IgG3 (Fig. 1B). This was surprising because C3b deposition via these subclasses was similar (Fig. 1A). Using a monoclonal antibody that prevents C1q–IgG interactions (46), we showed that C3b deposition via IgG2 was driven by C1 (SI Appendix, Fig. S3). Also, inhibition of C1 via a bacterial protein that blocks C1r [BBK32 (47–49)] confirmed that C1 is required to deposit C3b onto IgG2-coated bacteria (SI Appendix, Fig. S3). Furthermore, we showed that C3b molecules deposited by anti-WTA IgG2 antibodies are functional. We studied phagocytosis of bacteria by human neutrophils, the primary mechanism for elimination of S. aureus (50). Although anti-WTA IgG2 did not potently induce Fc receptor–mediated phagocytosis of S. aureus [as expected from the predicted low affinity of IgG2 for FcγR (32)], we found that addition of complement strongly promoted the phagocytic uptake of IgG2-labeled S. aureus (Fig. 1C).
We wondered whether these findings could be translated to IgGs recognizing a different antigenic surface. To study this, we used an assay system in which beads are coupled with the model antigen 2,4-dinitrophenol (DNP) (51) and coated with human IgGs specific for DNP (SI Appendix, Fig. S4) (52). To mimic the highly abundant nature of WTA, beads were coated with saturating levels of DNP (quantified by measuring IgG binding using flow cytometry) (SI Appendix, Fig. S4B). In accordance with our findings on S. aureus, we found that anti-DNP antibodies of the IgG1, IgG2, and IgG3 subclasses all potently induced a complement response and deposit C3b molecules onto the surface of DNP-beads (Fig. 1D). Again, whereas C3b opsonization could be correlated with the presence of C1q on beads coated with IgG1 and IgG3, almost no C1q could be detected on the IgG2-coated surface (Fig. 1E).
In conclusion, on two independent surfaces, we showed that IgG1, IgG2, and IgG3 can all potently elicit C1-dependent deposition of C3b molecules. However, for IgG2, our data revealed an unexpected disparity between the detection of C1q and downstream deposition of C3b molecules.
C1r2s2 Proteases Enhance the Binding of C1q to Target-Bound IgG.
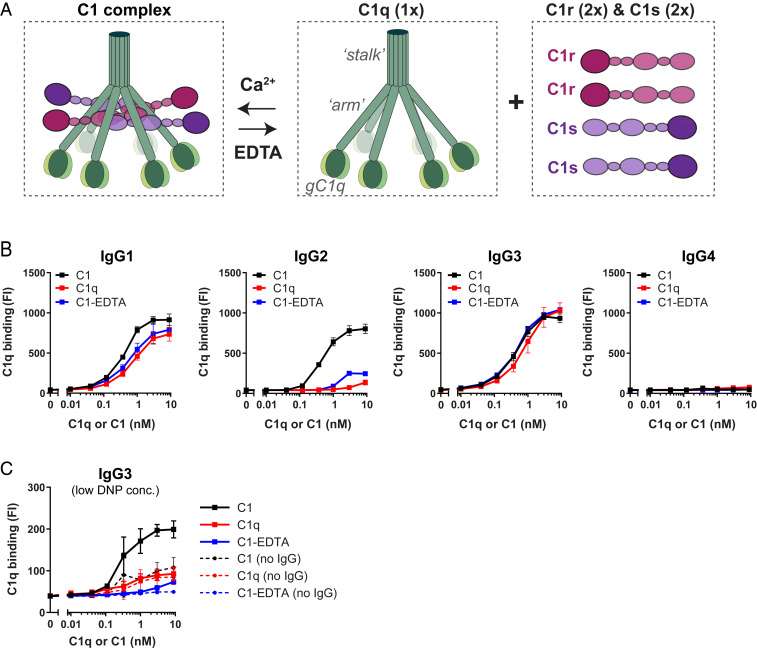
To better understand the above findings, we more closely examined the molecular interactions between C1q and target-bound IgGs by using purified C1 complexes. We included two forms of C1q in our analyses: 1) C1q in complex with C1r2s2 proteases (denoted C1), which is representative for circulating C1 complexes in human blood (53, 54) or 2) recognition molecule C1q without proteases (denoted C1q) (Fig. 2A); the absence of C1r and C1s in the C1q preparation was confirmed by mass spectrometry (SI Appendix, Fig. S5A). When we studied binding of different forms of C1q to anti-DNP antibodies on DNP-beads, we noticed a discrepancy between the binding of noncomplexed C1q molecules versus C1 (Fig. 2B and SI Appendix, Fig. S5B). Particularly in the case of IgG2, we observed that binding of C1 was more efficient than binding of noncomplexed C1q, as quantified by flow cytometric detection of surface-bound C1q (Fig. 2B). Western blotting was used to confirm that the different levels of C1q detected in flow cytometry actually represent different quantities of surface-bound C1q (SI Appendix, Fig. S5C). When C1 complexes were dissociated by ethylenediaminetetraacetic acid (EDTA), which disrupts the calcium-dependent attachment of C1r2s2 to C1q (55, 56), binding was similar to C1q alone (Fig. 2B). Reversely, reconstitution of noncomplexed C1q with purified C1r and C1s resulted in C1q binding similar as for purified C1 complex (SI Appendix, Fig. S5D). On IgG1-coated beads, we observed that C1r2s2 proteases had subtle effect on binding of C1q (Fig. 2B and SI Appendix, Fig. S5D). Furthermore, C1r2s2 proteases had a very limited effect on the binding of C1q to IgG3 (Fig. 2B and SI Appendix, Fig. S5D). Similar to these results on beads, we observed that EDTA reduced binding of purified C1 to S. aureus labeled with IgG1 and IgG2, while binding to IgG3 was much less affected (SI Appendix, Fig. S6). Altogether, these studies suggest that attached C1r2s2 proteases affect C1q–IgG interactions in a subclass-dependent manner. This finding is unexpected when considering that the direct interactions between C1 and IgG were shown to solely depend on the gC1q domains (16).
Fig. 2.
C1r2s2 proteases enhance the binding of C1q to target-bound IgG. (A) Schematic presentation of C1 consisting of the recognition molecule C1q in complex with a tetramer of proteases C1r and C1s (C1r2s2). C1q consists of six polypeptide chains that come together in an N-terminal stalk. At their C terminus, all six chains end in a globular domain (gC1q) that recognizes Fc domains of IgM and clustered IgGs. The C1r2s2 tetramer associates to the collagen arms of C1q via Ca2+-dependent interactions (56). The proteases dissociate from C1q in the presence of EDTA, a calcium chelator. (B) Binding of different forms of purified C1 to DNP beads (1 µg/mL DNP) coated with 20 nM anti-DNP IgG1-4. “C1” indicates the fully assembled C1 complex (= C1qr2s2), “C1q” is the recognition molecule C1q only, “C1-EDTA” sample consists of C1q, C1r, and C1s, but the proteases are not attached to C1q (because 10 mM EDTA disrupts the Ca2+-dependent association between proteases and C1q). (C) Binding of different forms of purified C1 to IgG3-labeled beads coated with 0.003 µg/mL (∼300-fold lower than in B). The dotted lines show aspecific binding of the C1q molecules in absence of IgG3. (B and C) Bound C1q was detected by polyclonal anti-C1q antibodies and flow cytometry. The data represent mean ± SD of three independent experiments.
Since the surface density of antigens was earlier proposed to be a critical parameter for IgG-mediated complement activation (57–59), we wondered whether C1r2s2 proteases also affect C1q–IgG interactions at lower antigen concentrations. In the bead system, we lowered the DNP concentration by ∼300-fold. At this lower DNP concentration, we found that IgG3 was the most potent subclass triggering complement activation in human serum (SI Appendix, Fig. S7) and that C3b deposition by IgG1 and IgG2 was inefficient. This corresponds with the idea that IgG3 more potently drives complement activation on low abundant antigens [presumably because it has a longer hinge region (21, 57, 58, 60)]. Upon studying binding of purified C1 complexes, we observed that C1r2s2 proteases can also affect C1q binding to IgG3 on beads with a lower DNP concentration. First, we observed that C1 bound more efficiently to IgG3 than noncomplexed C1q (Fig. 2C). Also, disruption of C1 with EDTA caused an almost complete reduction of C1q binding to IgG3 (Fig. 2C).
Altogether this suggests that C1r2s2 proteases can enhance binding of C1q to all IgG subclasses. However, the extent to which C1r2s2 proteases contribute to C1q binding depends both on the IgG subclass and antigen concentration.
C1r2s2 Proteases Enhance the Stability of Surface-Bound C1q–IgG Complexes.
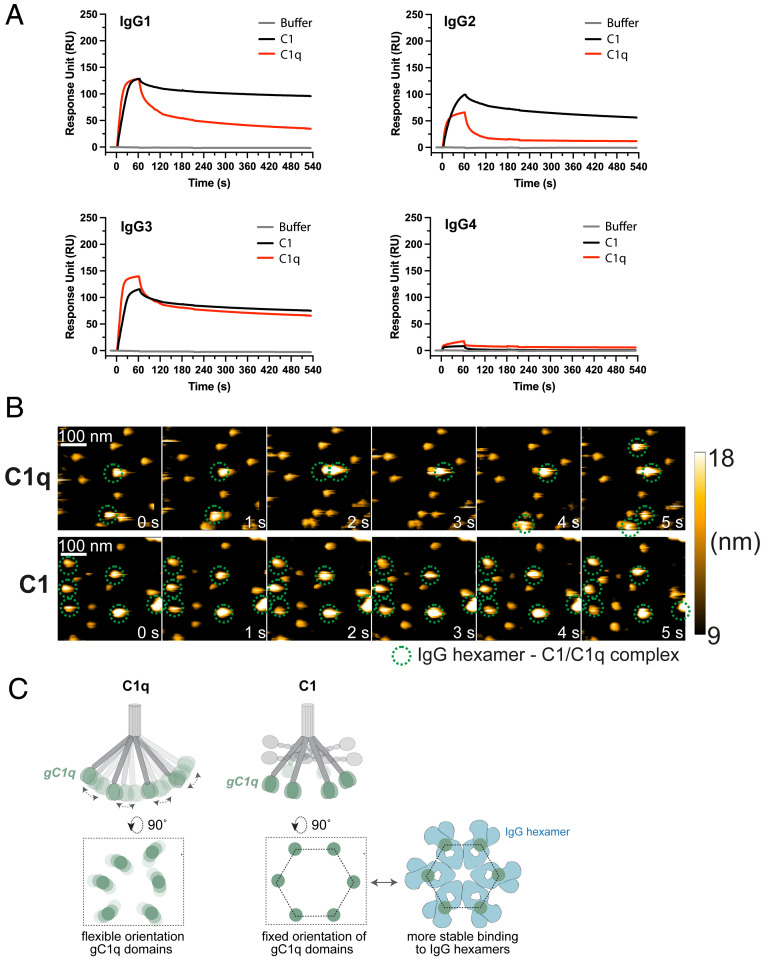
To corroborate these results, we employed surface plasmon resonance (SPR) as an orthogonal technique to confirm the binding profiles and obtain dynamic information about the formation and stability of C1q–IgG complexes (Fig. 3A and SI Appendix, Fig. S8). Using a flat DNP-labeled surface, prepared by coupling DNP-PEG-NHS to an activated carboxyl sensor chip (SI Appendix, Fig. S8A), monoclonal anti-DNP antibodies could be captured at high density and stability (SI Appendix, Fig. S8B). No binding was observed on a MeO-PEG-NHS surface that was used as a reference (SI Appendix, Fig. S8C). By injecting either C1q or C1 for 60 s, the assembly (during injection) and stability of C1q–IgG/C1–IgG complexes (during the dissociation period) could be monitored over time (SI Appendix, Fig. S8D). SPR analysis indeed confirmed that C1r2s2 proteases affect the stability of C1q–IgG complexes, especially for IgG1 and IgG2 (Fig. 3A). Whereas both C1q and C1 bound to the IgG1- and IgG2-coated sensor chips during protein injection, C1 dissociated much slower than C1q after the injection was stopped, which suggests that C1 forms more stable interactions with the antibody-coated surface. Additionally, we observed a weaker association of C1q in absence of C1r2s2 to IgG2-coated chips during the injection (Fig. 3A). In line with our studies on IgG3-labeled beads, C1r2s2 proteases did not notably affect C1q binding to IgG3-coated chips. However, SPR experiments showed that C1r2s2 proteases had a more-pronounced impact on binding of C1q when the IgG3 capturing concentration was lowered fivefold (SI Appendix, Fig. S8E). As expected, neither C1q nor C1 showed detectable binding to IgG4.
Fig. 3.
C1r2s2 proteases enhance the stability of surface-bound C1q–IgG complexes. (A) SPR experiment showing binding of purified C1 or C1q to sensor chips with immobilized DNP coated with 20 nM anti-DNP IgG. C1 or C1q was injected for 60 s to allow association, after which the injection was stopped and dissociation was monitored. Representative of two independent experiments. SPR responses were normalized to account for the molecular weight difference between C1 (766 kDa) and C1q (410 kDa). RU, response units. (B) HS-AFM image sequence of anti-DNP IgG1-RGY in complex with C1q in the presence of C1q in solution (Upper; taken from Movie S1) and C1 in the absence of C1 in solution (Lower; taken from Movie S2). The depicted height scale is relative to the membrane surface. The heights of the respective complexes were 12.2 ± 0.6 (anti-DNP IgG1-RGY) (17), 20.2 ± 1.5 (anti-DNP IgG1-RGY + C1q), and 18.5 ± 1.7 nm (anti-DNP IgG1-RGY + C1). (C) gC1q domains mediate binding to IgG. In C1q, the gC1q domains are flexible. We hypothesize that in C1 the associated C1r2s2 proteases fix the collagen arms and thereby orient the gC1q domains in a hexagon-like platform that favors binding to (hexameric) IgG clusters.
To directly visualize C1q–IgG complexes, we performed high-speed atomic force microscopy (HS-AFM) experiments. To enable reliable identification of C1q–IgG complexes in HS-AFM, we employed the anti-DNP triple mutant (IgG1-E345R, E430G, and S440Y, denoted IgG1-RGY), which was shown to efficiently associate into IgG1 hexamers in solution (15, 19). DNP-labeled supported lipid bilayers (SLBs) were therefore preincubated with anti-DNP IgG1-RGY (17). Then, C1q or C1 was added and the resulting complexes were visualized via HS-AFM (Fig. 3B and Movies S1 and S2). While the complexes of C1 that bound to the IgG1-RGY hexamers were not significantly disturbed by the minimal forces exerted by the HS-AFM tip, C1q alone was frequently removed from the IgG1-RGY hexamers as a result of the tip–sample interaction under the same experimental settings.
Altogether, the above data suggest that attached C1r2s2 proteases improve the stability of C1q–IgG complexes on target surfaces. We propose that C1r2s2 proteases affect the conformation of C1q in a way that facilitates stable docking to surface-bound IgGs. Earlier studies reported that the solution structure of C1q shows a high degree of flexibility (61–63). While the six collagen arms firmly bundle via disulphide bonds in the upper stalk, such interactions are lacking below the stalk and enable a rather flexible arrangement of the gC1q domains (Fig. 3C). A recent in situ three-dimensional structure of C1qr2s2, bound to surface-antigen–bound IgM, showed that the C1r2s2 proteases are packed inside the C1q molecule as two antiparallel C1rs heterodimers (64). Each C1rs dimer binds three of the six C1q collagen helices, which limits the flexibility of the collagen arms and results in a near-hexagonal arrangement of the six gC1q domains. Based on recent observations that IgG hexamers are the ideal docking platform for C1q (15, 17), we propose that this hexagon-like arrangement of the six gC1q domains induced by the attachment of C1r2s2 to C1q favors multivalent, high-avidity binding of C1q with clustered IgG (Fig. 3C).
Removal of C1r2s2 Proteases from Surface-Bound C1–IgG Complexes can Result in C1q Dissociation.
Next, we wondered how the observed differences in stability between C1–IgG and C1q–IgG could explain why we observed a discrepancy between C1q and C3b detection on IgG2-coated surfaces in serum (Fig. 1). Because C1q in human serum circulates as a complex with C1r2s2 proteases, we assume that IgG2-coated S. aureus and DNP-beads (1 µg/mL DNP) can recruit C1 and activate complement. However, since human serum contains an inhibitor that removes C1r2s2 proteases from C1q, we hypothesized that subsequent removal of proteases results in dissociation of C1q–IgG complexes. To test this, we incubated IgG-covered beads with purified C1 and, after washing, incubated the C1-bound beads with human C1-esterase inhibitor (C1-INH) (65). C1-INH is a human serpin that inactivates the proteases by forming a covalent bond with the catalytic site of both C1r and C1s. As expected, incubation of C1–IgG complexes on beads with C1-INH led to removal of C1r2s2 proteases from C1q on IgG1-, IgG2-, and IgG3-covered beads as evidenced by the detection of C1-INH−C1r and C1-INH−C1s complexes in the sample supernatant using Western blotting (SI Appendix, Fig. S9). In line with our hypothesis, we found that the dissociation of C1r2s2 by C1-INH had differential effects on the stability of the bead-bound C1q–IgG complexes (Fig. 4A). Whereas dissociation of C1r2s2 proteases by C1-INH did not affect C1q–IgG3 complexes, it caused a strong (80%) reduction of C1q binding to IgG2 beads and a 10% reduction on IgG1 beads (Fig. 4A). Similar results were obtained when C1r2s2 proteases were removed from C1–IgG complexes using EDTA (Fig. 4A).
Fig. 4.
Removal of C1r2s2 proteases from surface-bound C1–IgG complexes can result in C1q dissociation. (A) Detection of C1q on IgG-coated DNP beads that were first labeled with purified C1 and subsequently incubated with 10 mM EDTA or 200 nM C1-INH to remove C1r and C1s proteases. The data represent geometric mean ± SD of three independent experiments. Unpaired Student’s t test (buffer versus EDTA; buffer versus C1-INH); *P < 0.05, all other conditions not significant. (B) Schematic cartoon of our hypothesis that C1r2s2 dissociation by C1-INH can result in C1q dislodgement depending on the stability of the C1q–IgG complex. Our data suggest that removal of the C1r and C1s proteases from surface-bound C1 by C1-INH can result in two situations: 1) C1q dissociates from the surface-bound IgGs in the case the remaining C1q–IgG complexes are unstable (e.g., for C1q–IgG2 complexes) or 2) C1q remains bound since it has formed a stable interaction with the surface-bound IgGs.
Altogether, these data demonstrate that C1-INH can dislodge C1q from IgG-coated surfaces by removing C1r2s2 from surface-bound C1–IgG complexes. While C1-INH binds and removes C1r2s2 regardless of the IgG subclass, subsequent C1q dissociation depends on the stability of C1q–IgG complexes (Fig. 4B). It seems likely that the dislodgement of C1q as result of C1r2s2 dissociation explains why we could not detect C1q on IgG2 beads in a serum environment (Fig. 1E). The observed deposition of C3b molecules on IgG2 beads in serum (Fig. 1D) furthermore suggests that C1 inhibitory mechanisms lag behind on C1-mediated cleavage of complement proteins.
Enhanced IgG Oligomerization Stabilizes C1q–IgG Interactions.
Finally, we determined how C1q–IgG interactions are influenced by IgG oligomerization. Recent studies demonstrated that oligomerization of target-bound IgG into hexamers can be enhanced by specific point mutations that strengthen Fc–Fc contacts (15, 18). In contrast to the RGY mutation used above, these so-called HexaBody mutations do not enhance hexamer formation in solution but specifically enhance hexamerization on target surfaces. Here, we modified anti-DNP IgGs using hexabody mutations E430G or E345K (18) and verified that these mutations did not affect IgG binding to beads (SI Appendix, Fig. S10 A and B). Upon studying interactions with purified forms of C1/C1q, we observed that both Fc mutations strongly improved binding of noncomplexed C1q (i.e., C1q or C1-EDTA) to IgG1 and IgG2 on DNP beads (Fig. 5A and SI Appendix, Fig. S10C). For IgG3, hexabody mutations did not affect binding of noncomplexed C1q (Fig. 5A and SI Appendix, Fig. S10C). SPR experiments corroborated the results in the bead assay, showing that both the E430G and E345K mutation increase complex stability of IgG1 and IgG2 with C1q and have little effect on the association of C1q with IgG3 (SI Appendix, Fig. S11). Similar results were obtained for S. aureus, where introduction of E430G into anti-WTA IgG strongly enhanced binding of noncomplexed C1q to IgG1 and IgG2 while not affecting IgG3 (SI Appendix, Fig. S12 A and B). Interestingly, we observed that hexabody mutations did not have a strong impact on the binding of fully assembled C1 to IgG1 and IgG2 on DNP-coated beads (Fig. 5B) or SPR chips (SI Appendix, Fig. S11). On S. aureus, we observed that the E430G mutation induced a subtle improvement of C1 binding (SI Appendix, Fig. S12C). These data suggest that unstable C1q–IgG interactions between noncomplexed C1q and IgG can be overcome by promoting formation of high-avidity multimeric IgG platforms on the surface. In situations where C1q–IgG complexes are already stable (e.g., for C1 binding to IgG1, IgG2, or IgG3 or C1q binding to IgG3), hexabody mutations do not or only slightly improve C1q–IgG binding. The observation that Fc mutation E430G enhances C1q binding to IgG1 and IgG2 in human ΔIgGΔIgM serum [both on beads (Fig. 5C) and S. aureus (SI Appendix, Fig. S13)] suggests that hexabody mutations also improve the ability of IgGs to retain C1q on target surfaces in a serum environment.
Fig. 5.
Enhanced IgG oligomerization stabilizes C1q–IgG interactions. (A and B) Binding of purified C1q (A) or C1 (B) to DNP beads (1 µg/mL DNP) labeled with 20 nM anti-DNP IgG, either wild-type or containing hexamer-enhancing mutations E430G or E345K. (C) Detection of C1q on DNP-beads (1 µg/mL DNP) after incubation with 1% ΔIgGΔIgM serum supplemented with wild-type or mutated (E430G) anti-DNP IgG. (A–C) C1q was detected by flow cytometry. The data represent geometric mean ± SD of three independent experiments.
We also examined whether enhanced hexamerization could affect complement binding and activation via IgG4, the IgG subclass that is considered incapable of reacting with C1q (39). Previous studies showed that the Ser at position 331 in the heavy chain of IgG4 is critical for determining its inability to bind C1q (38, 66). Structural modeling has furthermore shown that the two Fab arms of IgG4 obstruct its C1q binding site and that the short IgG4 hinge region allows only limited flexibility (35–37). As the IgG4 used in our study also contains the essential Ser331 residue, we expected no binding to C1q or C1. However, when we introduced the E430G or E345K mutation in anti-DNP IgG4, we observed dose-dependent binding of fully assembled C1 but not C1q (Fig. 5 A and B and SI Appendix, Fig. S10C). Also on S. aureus (SI Appendix, Fig. S12) and in SPR assays (SI Appendix, Fig. S11), we observed that enhancing Fc–Fc interactions in IgG4 enabled binding to the fully assembled C1 and negligible binding to noncomplexed C1q. On S. aureus in human serum, we could detect only low levels of C1q when introducing the hexabody mutation E430G in IgG4 (SI Appendix, Fig. S13B). This observation is not unexpected considering our earlier hypothesis that C1r2s2 removal by C1-INH results in dislodgement of C1q when C1q–IgG interactions are unstable. Taken together, these data suggest that the poor interaction of surface-bound IgG4 with C1q can be overcome in the presence of both hexabody mutations and C1r2s2 proteases, which confirms a role for the proteases in C1q binding.
Introduction of Hexamer-Enhancing Mutation E430G in anti-WTA Enhance Complement-Dependent Phagocytosis of S. aureus.
Having demonstrated that enhanced IgG hexamerization improves the stability of C1q–IgG complexes on S. aureus, we wondered whether hexabody mutations also have an impact on the downstream complement effector mechanisms. In previous studies, we and others have shown that phagocytosis of S. aureus, which is eminent for human immune defense against these bacteria (67), critically depends on the labeling of S. aureus with complement-derived opsonins (68–70). While antibacterial IgGs may trigger phagocytosis of S. aureus via Fc receptors, additional opsonization of the bacterial surface with C3b and iC3b enhances the efficacy of particle uptake via engagement of CR. To study the effect of hexamer-enhancing mutations on phagocytosis, we first determined the efficiency by which anti-WTA mutant IgGs triggered covalent deposition of opsonic C3b on S. aureus (Fig. 6A). Upon incubation of S. aureus with anti-WTA IgGs and ΔIgGΔIgM serum, we observed that the E430G mutation enhanced C3b deposition mediated by IgG1 and IgG2 but not IgG3 (Fig. 6A). Next, we determined whether enhanced IgG oligomerization on S. aureus improved the phagocytosis of bacteria by human neutrophils in serum (Fig. 6B). In full correspondence with the observed improvements at the level of C1q (SI Appendix, Fig. S13B) and C3b (Fig. 6A), we observed that introduction of the E430G mutation in anti-WTA IgG1 and IgG2 but not IgG3 enhanced the phagocytic uptake of fluorescent S. aureus in human serum. Consistent with the finding that anti-WTA IgG4-E430G antibodies can induce C3b deposition, we observed that this antibody could induce phagocytosis of S. aureus (Fig. 6B). As expected, the E430G mutation did not affect the IgG-dependent phagocytosis via Fc receptors in the absence of complement (SI Appendix, Fig. S14). Altogether these data demonstrate that Fc engineering of anti–S. aureus IgGs can be a useful strategy to improve a complement response against this important pathogen and subsequent uptake by phagocytes.
Fig. 6.
Introduction of hexamer-enhancing mutation E430G in anti-WTA can enhance complement-dependent phagocytosis of S. aureus. (A) C3b deposition on S. aureus Wood46 after incubation of bacteria in 5% ΔIgGΔIgM serum supplemented with anti-WTA IgG (wild-type or E430G mutant). The data represent mean ± SD of three independent experiments. (B) Phagocytosis in the absence and presence of complement. Phagocytosis of fluorescently labeled S. aureus Wood46 in either RPMI buffer or 1% ΔIgGΔIgM serum supplemented with anti-WTA IgG (wild-type or E430G mutant) and human neutrophils. Bacterial uptake was quantified by flow cytometry and displayed as the number of GFP-positive neutrophils relative to IgG1 wild type. The data represent relative mean ± SD of three independent experiments. Phagocytosis data shown for wild-type IgG2 (buffer and serum condition) are identical to the data shown in Fig. 1C.
Discussion
The classical pathway of complement activation is a major contributor to pathophysiological processes in our body, including infection, inflammation, autoimmunity, and transplant rejection. Here, we demonstrate in great detail how the initiating step of this pathway, namely the binding of the large C1 complex to the surface of IgG-labeled target cells, is influenced by its attached proteases C1r2s2 and antibody oligomerization.
When compared to the growing body of structural information on C1q and its associated proteases, our knowledge about the dynamic interactions between C1q and surface-bound IgGs is still limited. In this study, we combined biophysical approaches with recombinant antibody engineering to study C1q–IgG interactions on target surfaces. Our findings reveal that C1r2s2 proteases affect the capacity of C1q to form an avid complex with clustered surface-bound IgG molecules in a subclass-dependent manner. The C1q molecule contains six collagen arms that each can bind an Ig-Fc domain. Although the six C1q arms are kept together by disulphide bonds between the collagen-like regions in the upper stalk, the collagen arms below the stalk are not held together and thus create a flexible structure in solution (61–63). Although a modifying effect of C1r2s2 proteases on the flexibility of C1q was previously indicated by biophysical studies (61–63), it was not known how this would affect C1q binding to surface-bound IgG. Okada et al. (1985) proposed a potential role for C1r2s2 proteases in C1q binding to polyclonal rabbit IgG, but the mechanism and relevance for human IgG remained unclear (71). Since our data now show that C1r2s2 are important for generating stable C1q–IgG complexes on surfaces, we propose that the associated proteases limit the conformational flexibility of the C1q arms. The recently published structure of C1 bound to IgM identified the contact points of C1rC1s within the collagen helices of C1q (64). It is conceivable that these C1rC1s–collagen contacts reduce the flexibility of the collagen arms and stabilize a conformation that facilitates multivalent binding of C1q to antibody clusters on the surface (Fig. 3C). At the same time, the formation of IgG hexamers on target surfaces has been shown to greatly stabilize the interaction with C1q. In our assays, mutations that enhance IgG oligomerization on a surface overcome the need for proteases to establish stable C1q–IgG complexes, likely because six antibodies can also form a stable, high-avidity complex with six C1q arms. This idea corroborates the finding that formation of hexamers in solution (e.g., of IgGs combining several hexamer-enhancing mutations) can enhance C1q–IgG binding in the absence of C1r2s2 (19). Hexamer-enhancing mutations in IgG2 and IgG1 also increased C1 binding to S. aureus, indicating that even when C1r2s2 is associated, efficient IgG oligomerization further stabilizes C1–IgG complexes. Interestingly, the finding that IgG4-E430G enabled binding to fully assembled C1 but less strongly to noncomplexed C1q could suggest that IgG4-E430G does not efficiently form hexamers. In all, we hypothesize that differences between human IgG subclasses to bind C1q (in the absence of proteases) could be linked to their ability to form hexameric platforms. However, the binding affinity of IgG subclasses for gC1q may also play a role. It remains to be determined whether our experiments on beads and gram-positive bacteria (both rigid surfaces) can be translated to highly fluidic eukaryotic or gram-negative membranes. Overall, our findings suggest that interactions between C1q and IgGs are influenced both by the C1r2s2 proteases and the inherent capacity of IgGs to form oligomers.
Our study also highlights that IgG subclasses differ in their capacity to form stable complexes with “empty” C1q molecules. While the in vivo relevance may not be immediately apparent since fully assembled C1qr2s2 complexes act as initiators of complement activation, plasma regulators such as C1-INH are able to remove C1r/C1s proteases and generate “empty” C1q. Particularly in the case of IgG1 and IgG2, we observed a marked reduction of complex stability with free C1q when compared to assembled C1. Whether C1q may dissociate or remain bound to an antigenic surface upon protease removal may therefore be context specific and depend on IgG subclass. Differences in the ability to bind empty C1q between subclasses may have several implications for immunity. Firstly, the biological functions of C1q reach beyond its traditional role in complement activation (72), as C1q is a known ligand of cell surface receptors on innate and adaptive immune cells. These receptors either recognize the globular [e.g., DC-SIGN (72, 73)] or collagenous domains of C1q [e.g., calreticulin/CD91 (74)] and induce various immunologic functions, including phagocytosis and apoptotic cell clearance. We hypothesize that antibodies with the capacity to retain “empty” C1q can promote such receptor interactions more efficiently. Secondly, stable binding of C1q to IgGs may interfere with alternative immunoglobulin functions such as the interaction with Fc receptors. Indeed, there is evidence from literature that C1q blocks IgG–FcγR interactions on viruses, both in vitro and in vivo. When studying mechanisms of antibody-dependent enhancement (ADE) of viral infection, Mehlhop et al., for example, showed that ADE of flavivirus infection is impaired by C1q in a complement-independent manner (75, 76). They demonstrated that C1q but not C3 in human serum blocked viral uptake in FcγR-expressing cells. Intriguingly, while IgG1, IgG2, and IgG3 against West Nile Virus all induced ADE in vitro, C1q-dependent inhibition was only observed for IgG3 and IgG1 but not IgG2. This finding is consistent with our observation that IgG2 is a poor partner for empty C1q.
Our findings are also relevant for the correct interpretation of functional antibodies in in vitro assays frequently used in research and diagnostics. C1q binding is often used as an important parameter to determine the presence of complement-fixing antibodies. For instance, in transplantation, the serum of organ recipients is tested for the presence of donor-specific complement-binding anti-HLA antibodies (DSA) (which could induce antibody-mediated rejection) by measuring binding of purified C1q (77–82). Based on our findings, direct C1q binding is an unreliable measure for downstream complement activation, as complement-binding IgGs that bind fully assembled C1 but not noncomplexed C1q are not detected. Indeed, there are various studies in which patients with C1q-negative DSA still showed downstream complement activation (i.e., C4d or C3d deposition) and/or developed antibody-mediated rejection (78, 80, 82–84). Moreover, it should be noted that detection of IgG-bound C1q may be an unreliable readout for complement activation, especially when C1q binding is analyzed in a serum environment. In our study, this was most evident for wild-type IgG2 and IgG4-E430G antibodies, which could potently trigger C1-mediated complement activation (and phagocytosis), while we could hardly detect C1q on the surface. Other groups have also demonstrated an apparent “paradox” between C1q binding and downstream complement activation. For example, Bindon et al. studied complement activation on antibody-coated erythrocytes and observed that a direct correlation between C1q binding and cell lysis could only be shown for some Ig subclasses (85, 86). We have now pinpointed a differential instability of IgG–C1q complexes in human serum as a likely cause of this discrepancy. Our systematic investigation of C1q versus C1 binding to all IgG subclasses in well-defined, purified systems and in human serum was key in obtaining this insight.
Our data also shed light on the mechanisms by which C1-INH inactivates C1, which are not yet fully defined. In 1998, Chen and Boackle already showed that purified C1-INH, besides removing C1r2s2, can dislodge C1q molecules from an IgG-coated surface. Although they used pooled human IgG in their C1-INH studies, they suggested that the IgG subclasses might be differentially affected by C1-INH (87, 88). Our data now provide a better insight into the subclass-specific effects of C1-INH. They show that C1-INH inhibits the C1r2s2 proteases in surface-bound C1–IgG independent of the IgG subclass, but that it mediates C1q dissociation only in conditions where C1q–IgG complexes are not stable. This suggests that protease dissociation by C1-INH can result in either 1) complete removal of both C1q and C1r2s2 from the IgG-coated surface or 2) removal of only C1r2s2 while C1q remains stably bound to the IgG platform (Fig. 4B). Although C1-INH can inactivate C1r and C1s in solution too, it is important to note that we here only investigated the effect of C1-INH on C1 that is activated on an IgG-labeled surface.
Finally, understanding how C1q molecules form a stable interaction with surface-bound IgGs is crucial for future design of therapeutic antibodies. Antibodies against bacteria and tumor cells cannot directly neutralize an infection but require activation of the immune system. For gram-positive bacteria, the functionality of antibodies in human immune protection critically depends on the antibody’s capacity to induce phagocytic killing (67, 89), either directly or via complement. Here, we show that Fc engineering could be a useful strategy to combat gram-positive bacteria, as both enhancement of C1q and C1 binding to S. aureus can improve the opsonization and phagocytosis of these bacteria in human serum. Recent studies on tumor cells and Neisseria gonorrhoeae have already shown that enhanced hexamerization of IgG by Fc engineering can potentiate the capacity of monoclonal antibodies to induce complement-mediated lysis via MAC pores (15, 18, 19, 23). Here, we demonstrate that the same mutations can be used to improve complement-dependent phagocytosis. Furthermore, since complement activation also appears crucial to induce protective antibodies against viral infections such as HIV or SARS-CoV-2 (90, 91), designing effective complement-triggering antibodies may also be valuable for therapeutic development of antiviral antibodies (24). In addition to its role in complement activation, increasing evidence suggests that C1q also enhances the efficacy of antibodies in the absence of other complement proteins. For instance, C1q may enhance the neutralizing activity of certain antiviral antibodies (75). Furthermore, it was shown that C1q can act as a structural component in potentiating the efficacy of antitumor antibodies by inducing outside-in signaling of death receptors on tumor cells (92).
Materials and Methods
More details on the materials and methods used in this study are provided in SI Appendix, Materials and Methods.
IgG and Complement Binding to DNP-Coated Beads.
Magnetic Dynabeads M-270 Streptavidin (Invitrogen) (∼6 to 7 × 105 beads per sample) were washed in PBS-TH (phosphate-buffered saline [PBS], pH 7.4, 0.05% (vol/vol) Tween, 0.5% human serum albumin [HSA]) and incubated with 0.003 or 1 μg/mL biotinylated 2,4-dinitrophenol (DNP-PEG2-GSGSGSGK(Biotin)-NH2; 1,186 Da; synthesized by Pepscan Therapeutics B.V.) in 0.1 mL/sample PBS-TH for 30 min at 4 °C, shaking (±700 rpm). Next, DNP-coated beads were washed once in PBS-TH and incubated in 0.05 mL/sample PBS-TH with 20 nM anti-DNP IgG or a threefold serial dilution of anti-DNP IgG (starting from 90 nM) for 30 min at 4 °C, shaking (±700 rpm). Subsequent incubations mentioned below were performed in 0.025 mL/sample PBS-TH for 30 min at 4 °C, shaking (±700 rpm), unless otherwise stated. After each incubation, beads were washed three times with PBS-TH or VBS-TH ([veronal buffered saline] pH 7.4, 0.25 mM MgCl2, 0.5 mM CaCl2, 0.05% (vol/vol) Tween, 0.5% HSA) dependent on the buffer used in the subsequent incubation step.
For detection of IgG binding, beads were incubated with 1 µg/mL goat anti-human kappa-AlexaFluor647 (Southern Biotech, 2060-31). Next, beads were washed and fixed in 0.15 mL/sample 1% paraformaldehyde in PBS-TH.
For C1q binding experiments, beads were incubated in 0.025 mL/sample VBS-TH with a threefold serial dilution (starting at 10 nM) of C1 or C1q for 30 min at 37 °C, shaking (±700 rpm). To determine binding of “C1-EDTA,” C1 was incubated in VBS-THE (VBS-TH, 10 mM EDTA) instead. To study the effect of C1-INH and EDTA on C1q binding, beads were first incubated with 0.025 mL/sample 3 nM C1 and then with 0.025 mL/sample VBS-THE or 0.025 mL/sample 200 nM C1-INH in VBS-TH for 30 min at 37 °C, shaking (±700 rpm). For C1q detection, beads were incubated with 5 μg/mL fluorescein isothiocyanate (FITC)-conjugated rabbit anti-human C1q (Dako, F0254).
To determine IgG, C1q, and C3b deposition to beads in human serum, DNP-coated beads (∼6 to 7 × 105 beads per sample) were incubated in 0.05 mL/sample VBS-TH with 20 nM anti-DNP IgG and a threefold serial dilution of ΔIgGΔIgM serum (starting from 10%) for 30 min at 37 °C, shaking (±700 rpm). Otherwise, DNP-coated beads were incubated in VBS-TH with a threefold serial dilution of anti-DNP IgG (starting from 90 nM) and 1% ΔIgGΔIgM serum for 30 min at 37 °C, shaking (±700 rpm).
For IgG, C1q, and C3b detection, beads were washed and incubated in 0.025 mL/sample PBS-TH with a detection antibody for either IgG, C1q, or C3b (1:300 FITC-conjugated goat anti-C3 IgG, De Beer Medicals, 1,100). After incubation with detection antibodies, bead samples were washed and fixed in 0.15 mL/sample 1% paraformaldehyde in PBS-TH and binding of IgG, C1q, or C3b to the beads was determined by flow cytometry (BD FACSVerse). Data were analyzed based on single-bead population using FlowJo software and presented as fluorescence intensity (FI) means ± SD or geometric means ± SD of at least three independent experiments.
IgG and Complement Binding to S. aureus.
Depending on the experiment, S. aureus Wood46 (with low expression of staphylococcal protein A [Spa]) or S. aureus Newman Δspa/sbi (knock-out of Protein A and the second immunoglobulin-binding protein [Sbi]) (93) was or was not fluorescently labeled by transformation with a green fluorescent protein (GFP)-expressing plasmid (94). Bacteria were grown until log phase in Todd Hewitt Broth medium, washed, and frozen at −20 °C until use. To prepare samples, bacteria were thawed and diluted to optical density (OD) = 0.1 in RPMI-H (Roswell Park Memorial Institute medium [RPMI], 0.05% HSA). All subsequent incubations mentioned below were performed in RPMI-H at 4 °C, shaking (±700 rpm), unless stated otherwise. After each incubation, bacteria were washed once in RPMI-H by centrifugation.
For detection of IgG binding, 0.025 mL bacteria was incubated with 0.025 mL threefold serial dilution anti-WTA IgG (starting from 90 nM, final concentration). Next, bacteria were incubated in 0.025 mL/sample RPMI-H with 1 µg/mL goat anti-human kappa-AlexaFluor647.
For C1q binding experiments, 0.025 mL bacteria was incubated with 0.025 mL 50 nM anti-WTA IgG (final concentration). Bacteria were next incubated in 0.025 mL/sample RPMI-H with a threefold serial dilution of C1 or C1q (starting at 30 nM) for 30 min at 37 °C, shaking (±700 rpm). To determine binding of “C1-EDTA,” C1 was incubated in RPMI-HE (RPMI-H, 10 mM EDTA) instead. For C1q detection, bacteria were incubated with 5 μg/mL FITC-conjugated rabbit anti-human C1q.
To determine IgG, C1q, and C3b binding to bacteria in serum, 0.050 mL bacteria were mixed with 0.025 mL threefold serial dilution anti-WTA IgG (starting from 90 nM, final concentration) and 0.025 mL 1% (IgG or C1q detection) or 5% (C3b detection) ΔIgGΔIgM serum (final concentration) and incubated 30 min at 37 °C, shaking (±700 rpm). For IgG, C1q, or C3b detection, bacteria were next incubated with detection antibody for IgG, C1q, or C3b (1:300 FITC-conjugated goat anti-C3 IgG).
To determine C3b deposition on bacteria in serum in the presence of C1-blocking molecules, 0.020 mL bacteria were mixed with 0.030 mL RPMI-H with a threefold serial dilution anti-WTA IgG (starting from 67 nM, final concentration), 1% ΔIgGΔIgM serum (final concentration), and either monoclonal anti-C1q antibody [HB8327 a4b11 (46), ATCC, 10 µg/mL final concentration] or BBK32 (kindly provided by Brandon Garcia, Greenville, NC, 10 µg/mL final concentration). Bacteria were incubated 20 min at 37 °C, shaking (±700 rpm). For C3b detection, bacteria were next incubated with FITC-conjugated goat anti-C3 IgG (1:300).
After incubation with detection antibodies, bacteria were washed and fixed in 0.15 mL/sample 1% paraformaldehyde in RPMI-H and analyzed using flow cytometry (BD FACSVerse). Data were analyzed by FlowJo software and presented as FI means ± SD or geometric means ± SD of at least three independent experiments.
Phagocytosis.
Human neutrophils were isolated from blood of healthy donors by the Ficoll-Histopaque gradient method (95). Donor samples were de-identified prior to use. Phagocytosis was performed in a round-bottom 96-well plate. GFP-labeled Wood46 bacteria (20 µL 3.75 × 107 cells/mL) were mixed with twofold serial dilutions of anti-WTA IgG in RPMI-H or 1% ΔIgGΔIgM serum (20 µL volume) for 15 min at 37 °C for opsonization. Subsequently, neutrophils (10 µL 7.5 × 106 cells/mL) were added, giving a 10:1 bacteria-to-cell ratio and incubated for 15 min at 37 °C on a shaker (750 rpm) in a final volume of 50 µL. The reaction was stopped with 1% ice-cold paraformaldehyde, and neutrophil-associated fluorescent bacteria were analyzed by flow cytometry by scatter gating on neutrophils. Phagocytosis was defined by the percentage of cells with a positive fluorescent signal (% GFP-positive cells) of all neutrophils representing the overall phagocytosis efficacy.
SPR Studies.
All SPR experiments were performed at 25 °C using a Biacore T200 instrument (Cytiva, GE Healthcare) equipped with a flat carboxymethyldextran sensor chip (Xantec). For DNP surface preparations, flow cells were activated for 7 min with a 1:1 mixture of 0.1 M N-hydroxysuccinimide and 0.1 M (3-(N,N-dimethylamino)propyl-N-ethylcarbodiimide) at a flow rate of 10 µL/min, followed by a 7-min injection of 0.1 M ethylenediamine in 0.1 M sodium borate pH 8.5. DNP-NH-PEG (4)-NHS (Iris Biotech) was reacted at a concentration of 5 mM in PBS-T with the aminated surface to reach a coupling density of 250 response units (RU). As a reference surface, MeO-PEG (4)-NHS (20 mM) was coupled to a density of 200 RU as described for the DNP surface. Binding analyses were performed in HBST++ (10 mM Hepes, 150 mM NaCl, 0.5 mM CaCl2, 0.25 mM MgCl2, 0.005% Tween 20, pH 7.4) at a flow rate of 10 µL/min if not stated otherwise. Anti-DNP antibodies at concentrations of 4 and 20 nM were prepared in HBST++ and injected for 120 s over the DNP and control surfaces. Subsequently, C1 or C1q (20 nM) were injected for 60 s to associate on top of the antibodies and dissociation was observed for 300 s. The surfaces were regenerated with a 60-s injection of 10 mM glycine pH 2.0 at a flow rate of 20 µL/min. Data were collected at a rate of 1 Hz and analyzed with Scrubber (version 2.0c; BioLogic). Reference signals from the control surface (MeO) were subtracted from the DNP surface signals. The responses were normalized for analyte size contribution by dividing the data points of each analyte by the molecular weight of C1 (766 kDa) or C1q (410 kDa), respectively, and multiplying them by 100. The data were exported to Prism (version 8.1.2.; Graphpad).
DNP-Labeled Liposomes.
DNP-labeled liposomes consisting of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE), and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[6-[(2,4-dinitrophenyl)amino]hexanoyl] (DNP-cap-DPPE) were used to generate SLBs on mica and SiO2 substrates. The lipids were purchased from Avanti Polar Lipids, mixed at a ratio of DPPC:DPPE:DNP-cap-DPPE = 90:5:5 (molar ratio) and dissolved in a 2:1 mixture of chloroform and methanol. After the solvents were rotary evaporated for 30 min, the lipids were again dissolved in chloroform, which was then rotary evaporated for 30 min. Drying was completed at a high vacuum pump for 2 h. The lipids were dissolved in 500 µL Milli-Q H2O while immersed in a water bath at 60 °C, flooded with argon, and sonicated for 3 min at 60 °C to create small unilamellar vesicles. These were diluted to 2 mg/mL in buffer No. 1 (10 mM Hepes, 150 mM NaCl, 2 mM CaCl2, pH 7.4) and frozen for storage using liquid N2.
HS-AFM.
HS-AFM (96, 97) (RIBM) was conducted in tapping mode at room temperature (RT) in buffer, with free amplitudes of 1.5 to 2.5 nm and amplitude set points larger than 90%. Silicon nitride cantilevers with electron beam–deposited tips (USC-F1.2-k0.15, Nanoworld AG), nominal spring constants of 0.15 N m−1, resonance frequencies around 500 kHz, and a quality factor of approximately 2 in liquids were used. Imaging was performed in buffer No. 1. All IgGs were diluted and incubated in the same buffer.
DNP-labeled SLBs for HS-AFM were prepared on muscovite mica. The liposomes were incubated on the freshly cleaved surface (500 µg/mL in buffer No. 1), placed in a humidity chamber to prevent evaporation, and heated to 60 °C for 30 min. Then, the temperature was gradually cooled down to RT within 30 min, followed by exchanging the solution with buffer No. 1. After 10 min of equilibration at RT and 15 more buffer exchanges, the SLB was ready for imaging. In order to passivate any exposed mica, SLBs were incubated with 333 nM IgG1-b12 (irrelevant human IgG1 control antibody against HIV-1 gp120) (98) for 10 min before the molecules of interest were added. Anti-DNP IgG1-RGY (20 µg/mL) was incubated for 5 min followed by several buffer exchanges to remove unbound IgGs from solution. After that, C1q (20 µg/mL) or C1 (15 µg/mL) was added for 10 min, followed by another buffer exchange. In case of C1q, imaging was performed in the presence of C1q in solution to enable rebinding during the experiment.
Statistical Analysis.
Statistical analysis was performed with Prism software (version 8.0.1; GraphPad). All data are presented as means ± SD or geometric means ± SD from three independent experiments unless otherwise stated.
Supplementary Material
Acknowledgments
We kindly thank Dr. Rob de Jong, Prof. Albert Heck, Dr. Annette Stemerding, Dr. Lubka Roemenia, and Dr. Leendert Trouw for scientific advice. We thank Dr. Brandon Garcia for providing BBK32. This work was supported by an European Research Council (ERC) Starting Grant (Grant No. 639209-ComBact to S.H.M.R.) and ERC Advanced Grant (Grant No. 233229-Coco to P.G.), the Utrecht University Molecular Immunology Hub, and the Swiss NSF (Grant No. 31003A_176104 to D.R.). J.P. acknowledges support by the European Fund for Regional Development (Regio 13), the Federal State of Upper Austria, and the Austrian Science Fund (FWF, Grant No. P33958 and P34164).
Footnotes
Competing interest statement: A.K., J.A.G.v.S., P.W.H.I.P., K.P.M.v.K., F.J.B., J. Schuurman, and S.H.M.R. are coinventors on a patent describing antibody therapies against S. aureus.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102787118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Lu L. L., Suscovich T. J., Fortune S. M., Alter G., Beyond binding: Antibody effector functions in infectious diseases. Nat. Rev. Immunol. 18, 46–61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walport M. J., Complement. First of two parts. N. Engl. J. Med. 344, 1058–1066 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Gros P., Milder F. J., Janssen B. J. C., Complement driven by conformational changes. Nat. Rev. Immunol. 8, 48–58 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Papanastasiou M., et al., Structural implications for the formation and function of the complement effector protein iC3b. J. Immunol. 198, 3326–3335 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ricklin D., Hajishengallis G., Yang K., Lambris J. D., Complement: A key system for immune surveillance and homeostasis. Nat. Immunol. 11, 785–797 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohlson S. S., Garred P., Kemper C., Tenner A. J., Complement nomenclature—Deconvoluted. Front. Immunol. 10, 1308 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mortensen S., et al., Structural basis for the function of complement component C4 within the classical and lectin pathways of complement. J. Immunol. 194, 5488–5496 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Law S. K. A., Dodds A. W., The internal thioester and the covalent binding properties of the complement proteins C3 and C4. Protein Sci. 6, 263–274 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heesterbeek D. A., et al., Bacterial killing by complement requires membrane attack complex formation via surface-bound C5 convertases. EMBO J. 38, e99852 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan B. P., Boyd C., Bubeck D., Molecular cell biology of complement membrane attack. Semin. Cell Dev. Biol. 72, 124–132 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Tomlinson S., Complement defense mechanisms. Curr. Opin. Immunol. 5, 83–89 (1993). [DOI] [PubMed] [Google Scholar]
- 12.Berends E. T. M., et al., Distinct localization of the complement C5b-9 complex on Gram-positive bacteria. Cell. Microbiol. 15, 1955–1968 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Dunkelberger J. R., Song W.-C., Complement and its role in innate and adaptive immune responses. Cell Res. 20, 34–50 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Stuart L. M., Ezekowitz R. A. B., Phagocytosis. Immunity 22, 539–550 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Diebolder C. A., et al., Complement is activated by IgG hexamers assembled at the cell surface. Science 343, 1260–1263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ugurlar D., et al., Structures of C1-IgG1 provide insights into how danger pattern recognition activates complement. Science 359, 794–797 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Strasser J., et al., Unraveling the macromolecular pathways of IgG oligomerization and complement activation on antigenic surfaces. Nano Lett. 19, 4787–4796 (2019). [DOI] [PubMed] [Google Scholar]
- 18.de Jong R. N., et al., A novel platform for the potentiation of therapeutic antibodies based on antigen-dependent formation of IgG hexamers at the cell surface. PLoS Biol. 14, e1002344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang G., et al., Molecular basis of assembly and activation of complement component C1 in complex with immunoglobulin G1 and antigen. Mol. Cell 63, 135–145 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Hughes-Jones N. C., Gardner B., Reaction between the isolated globular sub-units of the complement component C1q and IgG-complexes. Mol. Immunol. 16, 697–701 (1979). [DOI] [PubMed] [Google Scholar]
- 21.Feinstein A., Richardson N., Taussig M. I., Immunoglobulin flexibility in complement activation. Immunol. Today 7, 169–174 (1986). [DOI] [PubMed] [Google Scholar]
- 22.Strasser J., et al., Weak fragment crystallizable (Fc) domain interactions drive the dynamic assembly of IgG oligomers upon antigen recognition. ACS Nano 14, 2739–2750 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Gulati S., et al., Complement alone drives efficacy of a chimeric antigonococcal monoclonal antibody. PLoS Biol. 17, e3000323 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurtovic L., Beeson J. G., Complement factors in COVID-19 therapeutics and vaccines. Trends Immunol. 42, 94–103 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irani V., et al., Molecular properties of human IgG subclasses and their implications for designing therapeutic monoclonal antibodies against infectious diseases. Mol. Immunol. 67, 171–182 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Laxminarayan R., et al., Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 13, 1057–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Sause W. E., Buckley P. T., Strohl W. R., Lynch A. S., Torres V. J., Antibody-based biologics and their promise to combat Staphylococcus aureus infections. Trends Pharmacol. Sci. 37, 231–241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogers L. M., Veeramani S., Weiner G. J., Complement in monoclonal antibody therapy of cancer. Immunol. Res. 59, 203–210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown S., Santa Maria J. P., Walker S., Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 67, 313–336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Dalen R., Peschel A., van Sorge N. M., Wall teichoic acid in Staphylococcus aureus host interaction. Trends Microbiol. 28, 985–998 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Fong R., et al., Structural investigation of human S. aureus- targeting antibodies that bind wall teichoic acid. MAbs 10, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vidarsson G., Dekkers G., Rispens T., IgG subclasses and allotypes: From structure to effector functions. Front. Immunol. 5, 520 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zwarthoff S. A., Magnoni S., Aerts P. C., van Kessel K. P. M., Rooijakkers S. H. M., Method for depletion of IgG and IgM from human serum as naive complement source. Methods Mol. Biol. 2227, 21–32 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Cruz A. R., et al., Staphylococcal protein A inhibits complement activation by interfering with IgG hexamer formation. Proc. Natl. Acad. Sci. U.S.A. 118, e2016772118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu Y., et al., Solution conformation of wild-type and mutant IgG3 and IgG4 immunoglobulins using crystallohydrodynamics: Possible implications for complement activation. Biophys. J. 93, 3733–3744 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abe Y., Gor J., Bracewell D. G., Perkins S. J., Dalby P. A., Masking of the Fc region in human IgG4 by constrained X-ray scattering modelling: Implications for antibody function and therapy. Biochem. J. 432, 101–114 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Rayner L. E., et al., The Fab conformations in the solution structure of human immunoglobulin G4 (IgG4) restrict access to its Fc region. J. Biol. Chem. 289, 20740–20756 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tao M. H., Smith R. I., Morrison S. L., Structural features of human immunoglobulin G that determine isotype-specific differences in complement activation. J. Exp. Med. 178, 661–667 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruggemann M., Comparison of the effector functions of human immunoglobulins using a matched set of chimeric antibodies. J. Exp. Med. 166, 1351–1361 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lilienthal G. M., et al., Potential of murine IgG1 and Human IgG4 to inhibit the classical complement and Fcγ receptor activation pathways. Front. Immunol. 9, 958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kretschmer A., Schwanbeck R., Valerius T., Rösner T., Antibody isotypes for tumor immunotherapy. Transfus. Med. Hemother. 44, 320–326 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jung D.-J., et al., Specific serum Ig recognizing staphylococcal wall teichoic acid induces complement-mediated opsonophagocytosis against Staphylococcus aureus. J. Immunol. 189, 4951–4959 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Barrett D. J., Ayoub E. M., IgG2 subclass restriction of antibody to pneumococcal polysaccharides. Clin. Exp. Immunol. 63, 127–134 (1986). [PMC free article] [PubMed] [Google Scholar]
- 44.Ferrante A., Beard L. J., Feldman R. G., IgG subclass distribution of antibodies to bacterial and viral antigens. Pediatr. Infect. Dis. J. 9, S16–S24 (1990). [PubMed] [Google Scholar]
- 45.Siber G. R., Schur P. H., Aisenberg A. C., Weitzman S. A., Schiffman G., Correlation between serum IgG-2 concentrations and the antibody response to bacterial polysaccharide antigens. N. Engl. J. Med. 303, 178–182 (1980). [DOI] [PubMed] [Google Scholar]
- 46.Reckel R. P., Harris J. L., Wellerson R. Jr, Shaw S. M., Kaplan P. M., “Method for detecting immune complexes in serum.” US Patent 4595654A (1986).
- 47.Garcia B. L., Zwarthoff S. A., Rooijakkers S. H. M., Geisbrecht B. V., Novel evasion mechanisms of the classical complement pathway. J. Immunol. 197, 2051–2060 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia B. L., Zhi H., Wager B., Höök M., Skare J. T., Borrelia burgdorferi BBK32 inhibits the classical pathway by blocking activation of the C1 complement complex. PLoS Pathog. 12, e1005404 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie J., et al., Structural determination of the complement inhibitory domain of Borrelia burgdorferi BBK32 provides insight into classical pathway complement evasion by Lyme disease spirochetes. PLoS Pathog. 15, e1007659 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amulic B., Cazalet C., Hayes G. L., Metzler K. D., Zychlinsky A., Neutrophil function: From mechanisms to disease. Annu. Rev. Immunol. 30, 459–489 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Zwarthoff S. A., et al., Functional characterization of alternative and classical pathway C3/C5 convertase activity and inhibition using purified models. Front. Immunol. 9, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gonzalez M., Structural analysis of IgG2A monoclonal antibodies in relation to complement deposition and renal immune complex deposition. Mol. Immunol. 40, 307–317 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Watanabe S., et al., Serum C1q as a novel biomarker of sarcopenia in older adults. FASEB J. 29, 1003–1010 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Naito A. T., et al., Complement C1q activates canonical Wnt signaling and promotes aging-related phenotypes. Cell 149, 1298–1313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Colomb M. G., Arlaud G. J., Villiers C. L., Fothergill L. A., Activation of C1 [and discussion]. Philos. Trans. R. Soc. Lond. B Biol. Sci. 306, 283–292 (1984). [DOI] [PubMed] [Google Scholar]
- 56.Bally I., et al., Identification of the C1q-binding sites of human C1r and C1s. A refined three-dimensional model of the C1 complex of complement. J. Biol. Chem. 284, 19340–19348 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cooper L. J., et al., Role of heavy chain constant domains in antibody-antigen interaction. Apparent specificity differences among streptococcal IgG antibodies expressing identical variable domains. J. Immunol. 150, 2231–2242 (1993). [PubMed] [Google Scholar]
- 58.Giuntini S., Reason D. C., Granoff D. M., Combined roles of human IgG subclass, alternative complement pathway activation, and epitope density in the bactericidal activity of antibodies to meningococcal factor H binding protein. Infect. Immun. 80, 187–194 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wright J. K., Tschopp J., Jaton J. C., Preparation and characterization of chemically defined oligomers of rabbit immunoglobulin G molecules for the complement binding studies. Biochem. J. 187, 767–774 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cooper L. J. N., Robertson D., Granzow R., Greenspan N. S., Variable domain-identical antibodies exhibit IgG subclass-related differences in affinity and kinetic constants as determined by surface plasmon resonance. Mol. Immunol. 31, 577–584 (1994). [DOI] [PubMed] [Google Scholar]
- 61.Shelton E., Yonemasu K., Stroud R. M., Ultrastructure of the human complement component, Clq. Proc. Natl. Acad. Sci. U.S.A. 69, 65–68 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walker K. T., et al., Non-linearity of the collagen triple helix in solution and implications for collagen function. Biochem. J. 474, 2203–2217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poon P. H., Schumaker V. N., Phillips M. L., Strang C. J., Huxley H. E., Conformation and restricted segmental flexibility of C1, the first component of human complement. J. Mol. Biol. 168, 563–577 (1983). [DOI] [PubMed] [Google Scholar]
- 64.Sharp T. H., et al., Insights into IgM-mediated complement activation based on in situ structures of IgM-C1-C4b. Proc. Natl. Acad. Sci. U.S.A. 136, 201901841 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Windfuhr J. P., Alsenz J., Loos M., The critical concentration of C1-esterase inhibitor (C1-INH) in human serum preventing auto-activation of the first component of complement (C1). Mol. Immunol. 42, 657–663 (2005). [DOI] [PubMed] [Google Scholar]
- 66.Brekke O. H., Michaelsen T. E., Sandlie I., The structural requirements for complement activation by IgG: Does it hinge on the hinge? Immunol. Today 16, 85–90 (1995). [DOI] [PubMed] [Google Scholar]
- 67.van Kessel K. P. M., Bestebroer J., van Strijp J. A. G., Neutrophil-mediated phagocytosis of Staphylococcus aureus. Front. Immunol. 5, 467 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ko Y. P., et al., Phagocytosis escape by a Staphylococcus aureus protein that connects complement and coagulation proteins at the bacterial surface. PLoS Pathog. 9, e1003816 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rooijakkers S. H. M., et al., Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat. Immunol. 6, 920–927 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Rooijakkers S. H. M., Van Wamel W. J. B., Ruyken M., Van Kessel K. P. M., Van Strijp J. A. G., Anti-opsonic properties of staphylokinase. Microbes Infect. 7, 476–484 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Okada M., Udaka K., Utsumi S., Co-operative interaction of subcomponents of the first component of complement with IgG: A functional defect of dimeric Facb from rabbit IgG. Mol. Immunol. 22, 1399–1406 (1985). [DOI] [PubMed] [Google Scholar]
- 72.Ghebrehiwet B., Hosszu K. K., Valentino A., Peerschke E. I. B., The C1q family of proteins: Insights into the emerging non-traditional functions. Front. Immunol. 3, 1–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hosszu K. K., et al., DC-SIGN, C1q, and gC1qR form a trimolecular receptor complex on the surface of monocyte-derived immature dendritic cells. Blood 120, 1228–1236 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ogden C. A., et al., C1q and mannose binding lectin engagement of cell surface calreticulin and CD91 initiates macropinocytosis and uptake of apoptotic cells. J. Exp. Med. 194, 781–795 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mehlhop E., et al., Complement protein C1q reduces the stoichiometric threshold for antibody-mediated neutralization of West Nile virus. Cell Host Microbe 6, 381–391 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mehlhop E., et al., Complement protein C1q inhibits antibody-dependent enhancement of flavivirus infection in an IgG subclass-specific manner. Cell Host Microbe 2, 417–426 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen G., Sequeira F., Tyan D. B., Novel C1q assay reveals a clinically relevant subset of human leukocyte antigen antibodies independent of immunoglobulin G strength on single antigen beads. Hum. Immunol. 72, 849–858 (2011). [DOI] [PubMed] [Google Scholar]
- 78.Loupy A., et al., Complement-binding anti-HLA antibodies and kidney-allograft survival. N. Engl. J. Med. 369, 1215–1226 (2013). [DOI] [PubMed] [Google Scholar]
- 79.Loupy A., Lefaucheur C., Antibody-mediated rejection of solid-organ allografts. N. Engl. J. Med. 379, 1150–1160 (2018). [DOI] [PubMed] [Google Scholar]
- 80.Malheiro J., et al., Detection of complement-binding donor-specific antibodies, not IgG-antibody strength nor C4d status, at antibody-mediated rejection diagnosis is an independent predictor of kidney graft failure. Transplantation 102, 1943–1954 (2018). [DOI] [PubMed] [Google Scholar]
- 81.Thurman J. M., Panzer S. E., Le Quintrec M., The role of complement in antibody mediated transplant rejection. Mol. Immunol. 112, 240–246 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Viglietti D., et al., Complement-binding anti-HLA antibodies are independent predictors of response to treatment in kidney recipients with antibody-mediated rejection. Kidney Int. 94, 773–787 (2018). [DOI] [PubMed] [Google Scholar]
- 83.Sicard A., et al., Detection of C3d-binding donor-specific anti-HLA antibodies at diagnosis of humoral rejection predicts renal graft loss. J. Am. Soc. Nephrol. 26, 457–467 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yabu J. M., et al., C1q-fixing human leukocyte antigen antibodies are specific for predicting transplant glomerulopathy and late graft failure after kidney transplantation. Transplantation 91, 342–347 (2011). [DOI] [PubMed] [Google Scholar]
- 85.Bindon C. I., Hale G., Brüggemann M., Waldmann H., Human monoclonal IgG isotypes differ in complement activating function at the level of C4 as well as C1q. J. Exp. Med. 168, 127–142 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bindon C. I., Hale G., Waldmann H., Complement activation by immunoglobulin does not depend solely on C1q binding. Eur. J. Immunol. 20, 277–281 (1990). [DOI] [PubMed] [Google Scholar]
- 87.Chen C. H., Lam C. F., Boackle R. J., C1 inhibitor removes the entire C1qr2s2 complex from anti-C1Q monoclonal antibodies with low binding affinities. Immunology 95, 648–654 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen C.-H., Boackle R. J., A newly discovered function for C1 inhibitor, removal of the entire C1qr2s2Complex from immobilized human IgG subclasses. Clin. Immunol. Immunopathol. 87, 68–74 (1998). [DOI] [PubMed] [Google Scholar]
- 89.Weber S. S., Oxenius A., “Antibody-dependent cellular phagocytosis and its impact on pathogen control” in Antibody Fc, Ackerman M. E., Nimmerjahn F., Eds. (Elsevier, 2014), pp. 29–47. [Google Scholar]
- 90.Richardson S. I., et al., HIV-specific Fc effector function early in infection predicts the development of broadly neutralizing antibodies. PLoS Pathog. 14, e1006987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Atyeo C., et al., Distinct early serological signatures track with SARS-CoV-2 survival. Immunity 53, 524–532.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Overdijk M. B., et al., Dual epitope targeting and enhanced hexamerization by DR5 antibodies as a novel approach to induce potent antitumor activity through DR5 agonism. Mol. Cancer Ther. 19, 2126–2138 (2020). [DOI] [PubMed] [Google Scholar]
- 93.Sibbald M. J. J. B., et al., Synthetic effects of secG and secY2 mutations on exoproteome biogenesis in Staphylococcus aureus. J. Bacteriol. 192, 3788–3800 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kuipers A., et al., The Staphylococcus aureus polysaccharide capsule and Efb-dependent fibrinogen shield act in concert to protect against phagocytosis. Microbiology 162, 1185–1194 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bestebroer J., et al., Staphylococcal superantigen-like 5 binds PSGL-1 and inhibits P-selectin-mediated neutrophil rolling. Blood 109, 2936–2943 (2007). [DOI] [PubMed] [Google Scholar]
- 96.Ando T., et al., A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl. Acad. Sci. U.S.A. 98, 12468–12472 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Preiner J., et al., IgGs are made for walking on bacterial and viral surfaces. Nat. Commun. 5, 1–8 (2014). [DOI] [PubMed] [Google Scholar]
- 98.Burton D. R., et al., Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266, 1024–1027 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.