Significance
Understanding the cell origin and molecular mechanisms underlying rare genetic diseases provides invaluable insights into human biology and pathology of not only rare genetic diseases but also related common disorders. Autosomal dominant hyper immunoglobulin-E syndrome (AD-HIES) or Job syndrome is a genetic disease with skeletal defects resulting from mutations in the STAT3 gene. We have modeled the skeletal abnormality in Job Syndrome by deleting Stat3 using cell-specific Cre lines. We found that Stat3 is required in osteoblast lineage cells for skeletal development by maintaining Wnt/β-catenin signaling. Deletion of Stat3 resulted in impaired osteoblast differentiation due to reduced Wnt/β-catenin signaling with upregulated Sost expression. Our work provides a foundation for further understanding the principles whereby Stat3 governs bone formation and maintenance.
Keywords: Stat3, Job syndrome, Wnt/beta-catenin signaling, Sost, bone development
Abstract
Job syndrome is a rare genetic disorder caused by STAT3 mutations and primarily characterized by immune dysfunction along with comorbid skeleton developmental abnormalities including osteopenia, recurrent fracture of long bones, and scoliosis. So far, there is no definitive cure for the skeletal defects in Job syndrome, and treatments are limited to management of clinical symptoms only. Here, we have investigated the molecular mechanism whereby Stat3 regulates skeletal development and osteoblast differentiation. We showed that removing Stat3 function in the developing limb mesenchyme or osteoprogenitor cells in mice resulted in shortened and bow limbs with multiple fractures in long bones that resembled the skeleton symptoms in the Job Syndrome. However, Stat3 loss did not alter chondrocyte differentiation and hypertrophy in embryonic development, while osteoblast differentiation was severely reduced. Genome-wide transcriptome analyses as well as biochemical and histological studies showed that Stat3 loss resulted in down-regulation of Wnt/β-catenin signaling. Restoration of Wnt/β-catenin signaling by injecting BIO, a small molecule inhibitor of GSK3, or crossing with a Lrp5 gain of function (GOF) allele, rescued the bone reduction phenotypes due to Stat3 loss to a great extent. These studies uncover the essential functions of Stat3 in maintaining Wnt/β-catenin signaling in early mesenchymal or osteoprogenitor cells and provide evidence that bone defects in the Job Syndrome are likely caused by Wnt/β-catenin signaling reduction due to reduced STAT3 activities in bone development. Enhancing Wnt/β-catenin signaling could be a therapeutic approach to reduce bone symptoms of Job syndrome patients.
Job syndrome (Online Mendelian Inheritance in Man No. 147060), also known as autosomal dominant hyper immunoglobulin-E syndrome (AD-HIES), is caused by mutations in the STAT3 gene in more than two-thirds of cases (70%). Apart from severe primary immunodeficiencies, Job syndrome patients also show skeletal abnormalities such as long bone fractures, osteopenia, craniosynostosis, and scoliosis (1–5). STAT3 is a STAT family member of transcription factors that transduce signals of cytokines and growth factors from the activated receptors to the nucleus to activate transcription by binding to specific DNA motifs (6). In response to cytokines and growth factors such as interleukin-6 (IL-6), interleukin-10 (IL-10), leukemia inhibitory factor (LIF), and platelet derived growth factor (PDGF), STAT3 is phosphorylated and activated by a variety of tyrosine kinases such as Jaks, Src, EGF-R, and c-Met (7, 8). Activated STAT3 and other STAT proteins form either homo- or hetero-dimers and translocate into the nucleus to stimulate target gene expression by binding to specific DNA sequences (9). STAT3 signaling regulates a wide variety of cellular processes, including cell proliferation, survival, and differentiation during normal development and immune function as well as tumor growth, invasion, and cancer metastasis (10).
The STAT3 mutations associated with the AD-HIES/Job syndrome are primarily located in the DNA binding domain and Src homology (SH2) domain (4, 11, 12). These two domains are essential for the transcriptional activity of STAT3, and the mutations are shown to be either gain of function (GOF) or loss of function (LOF) (13, 14). Evidence has been provided that skeletal defects are more likely associated with dominant-negative or LOF mutations in STAT3 (15–17). It was shown that IL-6–mediated Stat3 activation in mice was required for osteoblast differentiation (18). In addition, loss of Stat3 in osteoblasts and osteocytes reduced load-driven bone formation and impairs the regulation of mitochondrial oxidative stress (19). Stat3 has also been reported to regulate the expression of Sox9 during embryonic cartilage development in mice (20), and Sox9 is a master transcription factor for chondrocyte differentiation and cartilage formation (21). Importantly, Sox9 is not expressed in osteoblast cells and is found to inhibit osteoblast differentiation (22, 23). Sox9 haploinsufficiency in campomelic dysplasia results in accelerated bone formation (22, 24), and Sox9 deletions in growth plate chondrocytes also result in premature osteoblast differentiation (23). Therefore, in contrast to the extensive studies of STAT3 functions in immune development, the cellular origin and molecular mechanisms underlying STAT3 function in bone formation during embryonic development are not clear, hampering therapeutic development to improve the skeletal phenotypes of Job syndrome patients. As osteoblasts are derived from perichondrium osteoblast progenitor cells and hypertrophic chondrocytes in embryonic development (25–27), to decipher the cell of origin in which Stat3 controls osteoblast differentiation, we have removed Stat3 in mesenchymal progenitor cells that give rise to both chondrocytes and osteoblasts, or osteoblast progenitor cells or hypertrophic chondrocytes, in the mouse embryos by deleting the SH2 domain of mouse Stat3 (28). Shortened and bowed limbs with multiple bone fractures similar to the skeletal defects observed in the Job Syndrome patients were found in mutants with Stat3 deficiency in early limb bud mesenchyme or osteoblast progenitors but not in hypertrophic chondrocytes. We found that the Stat3 LOF mutants showed drastically reduced bone formation, with no alteration in embryonic cartilage development. Our studies identify Stat3 as a critical regulator of Wnt/β-catenin signaling and suggest that up-regulating Wnt/β-catenin signaling might be a promising strategy to reduce the skeletal defects of Job syndrome patients.
Results
Loss of Stat3 in Early Limb Mesenchyme Resulted in Limb Defects Similar to Those in Job Syndrome Patients.
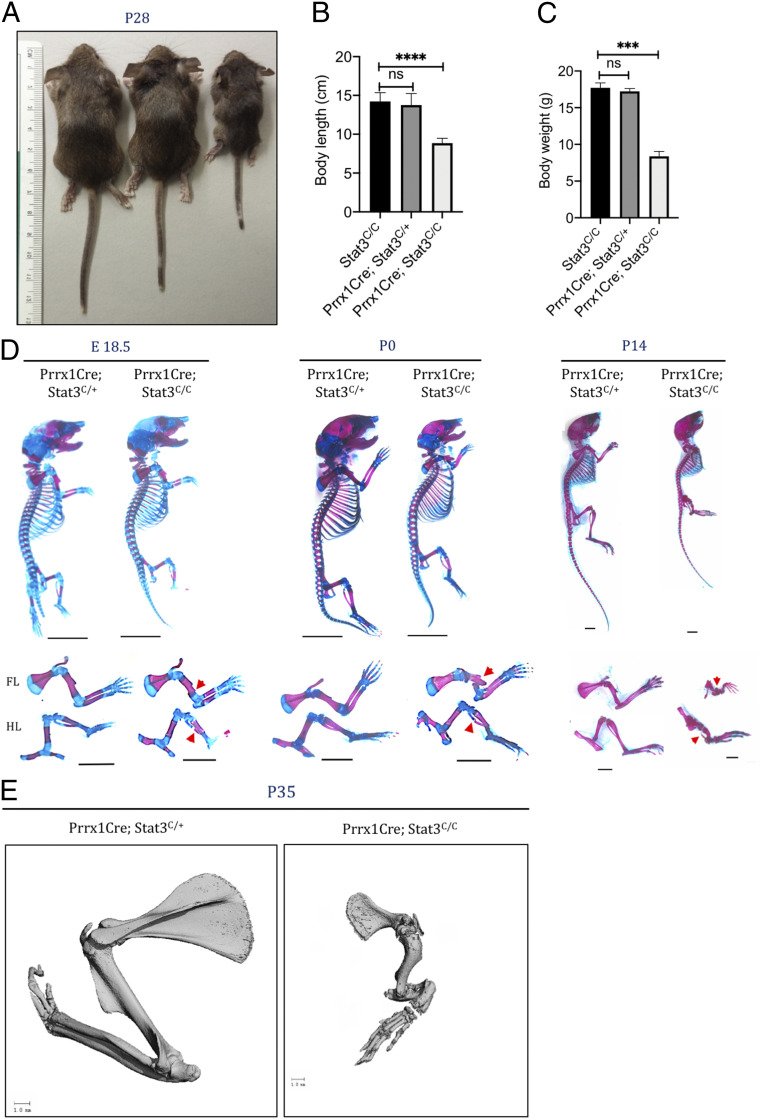
Stat3 plays important roles in many different cell types. To understand the cell of origin and the underlying molecular mechanism for the skeletal phenotypes in Job Syndrome, we determined whether the limb defects in the Job syndrome patients were caused by requirement of Stat3 in early mesenchymal progenitor cells. Stat3 is expressed ubiquitously in the developing limb (20). Importantly, we analyzed Stat3 activation by immunofluorescent (IF) staining of phospho-Stat3 (pStat3) and found it was strongly up-regulated in the differentiating osteoblasts and bone marrow (SI Appendix, Fig. S1A), suggesting that its activation may be required for bone formation. We therefore specifically removed Stat3 function from the limb bud mesenchyme cells early in development using the Prrx1Cre line (29) and a conditional Stat3 (Stat3C/C) mice with the floxed exon encoding the SH2 domain of Stat3 (28). Drastic reduction of pStat3 was found in the developing limb, indicating the efficiency of the Prrx1Cre line (SI Appendix, Fig. S1 A and E). The Prrx1Cre; Stat3C/C pups were born alive at Mendelian ratios but were readily distinguishable with their wild-type littermates due to their shorter and bowed limbs (SI Appendix, Fig. S1C and Fig. 1A). The Prrx1Cre; Stat3C/C mice showed multiple fractures in both forelimbs and hindlimbs starting from late embryonic development at E18.5 (Fig. 1 D and E). Forelimbs showed more severe defects as the Prrx1Cre induces more complete deletion in forelimbs (29). These mice could survive postnatally but were much smaller with reduced body length and weight (Fig. 1), whereas the Prrx1Cre; Stat3C/+ littermates were normal, healthy, and fertile, with slightly shortened tails compared to the wild-type controls, and used as control in all the analyses. The limb phenotypes of the Prrx1Cre; Stat3C/C mice were progressively more severe at later postnatal stages and similar to those found in the Job syndrome (Fig. 1 D and E). The manifestation of these skeletal phenotypes was similar in male and female mice. Taken together, Stat3 is required in the early limb mesenchyme for normal skeletal development. These data also indicate that loss of Stat3 function in mesenchymal cells contributes to skeletal defects in Job syndrome patients.
Fig. 1.
Stat3 is required for bone development. (A) Gross appearance of 4-wk-old Stat3C/C, Prrx1Cre: Stat3C/+, and Prrx1Cre: Stat3C/C (Stat3 knockout [KO]) mice (from Left to Right). (B and C) Quantification of body length (B) and weight (C) of 4-wk-old mice of the indicated genotypes. (D) Whole-mount Alizarin red and Alcian blue staining of Stat3 KO and littermate control mice at indicated ages. The forelimb (FL) and hindlimb (HL) were shown in the Lower. (Scale bars, 1 mm.) Bone fractures were indicated by red arrowheads. (E) Representative µCT images of FL of 5-wk-old littermate mice with the indicated genotypes. (Scale bars, 1 mm.) ***P < 0.001 and ****P < 0.0001 were considered significant; ns, not significant; the data are presented as mean ± SD.
Reduced Osteoblast Differentiation with Normal Cartilage Formation in the Prrx1Cre; Stat3C/C Embryos.
The smaller limb and body size in the Prrx1Cre; Stat3C/C mice led us to determine whether cartilage development was affected as cartilage formation precedes bone formation in endochondral ossification, and Stat3 has been implicated regulating cartilage development in the limb (20). Histological analysis of the developing long bones in the Prrx1Cre; Stat3C/C and littermate control embryos did not identify obvious defects in the chondrocyte morphology or cartilage structure at E16.5 and P0 (SI Appendix, Fig. S2 A and B). Furthermore, chondrocyte differentiation in the embryonic cartilage was analyzed by in situ hybridization for Col2a1, a chondrocyte-specific marker. Col2a1 expression was not altered in the Prrx1Cre; Stat3C/C cartilage as compared to the littermate control at E16.5 and P0 (SI Appendix, Fig. S2 A and B). Additionally, chondrocyte differentiation from mesenchymal progenitor cells in the developing limb was determined by in vitro micromass culture (SI Appendix, Fig. S2D). Limb bud cells were isolated from the E12.5 Stat3C/C embryos and transduced with adenovirus expressing GFP (Ad-GFP) or Cre recombinase (Ad-Cre). After 5-d culture under chondrogenic conditions, no difference in cartilage nodule formation could be found between the Ad-GFP– and Ad-Cre–infected samples (SI Appendix, Fig. S2C). We then determined chondrocyte hypertrophy in vivo by IF staining of Col10a1, a marker for hypertrophic chondrocytes. No alteration was found at E16.5 (SI Appendix, Fig. S2A), but at P0, while the nonhypertrophic chondrocytes marked by Col2a1 expression were similar, the Col10a1-marked hypertrophic chondrocyte region was expanded in the Prrx1Cre; Stat3C/C cartilage (SI Appendix, Fig. S2 A and B). This change was confirmed by histological analysis (SI Appendix, Fig. S2 B and C). These results suggest that replacement of hypertrophic chondrocytes by osteoblasts was delayed later in endochondral ossification. Taken together, loss of Stat3 did not reduce chondrocyte differentiation from mesenchymal progenitor cells but expanded the hypertrophic chondrocyte zone, which is consistent with previous findings (20).
We then asked whether Stat3 loss altered chondrocyte proliferation and/or survival. Sections of the developing long bones at E16.5 and P0 were processed for BrdU staining to detect proliferation and cleaved caspase 3 (Cl-Casp3) IF staining to detect apoptosis (SI Appendix, Fig. S1D). There was no difference in the number of BrdU+ chondrocyte between the Prrx1Cre; Stat3C/C and control littermates both at E16.5 and P0 (SI Appendix, Fig. S1D), indicating that Stat3 did not regulate chondrocyte proliferation in the embryonic cartilage in the limb. Interestingly, Stat3 loss led to a significant increase in Cl-Casp3+ cells only in the bone marrow at P0 (SI Appendix, Fig. S1D), indicating apoptotic cell death was increased in the absence of Stat3 specifically in the bone marrow. These data suggest that limb shortening in long bone development in the Prrx1Cre; Stat3C/C mice is not due to reduction in chondrocyte proliferation or differentiation. Furthermore, as osteoblast cells can be derived from hypertrophic chondrocytes (26), to determine whether Stat3 is specifically required in hypertrophic chondrocytes for bone formation, we removed Stat3 with the Col10a1Cre line (26). The Col10a1Cre; Stat3c/c embryos and newborn pups were normal as compared to their control littermates both morphologically and molecularly (SI Appendix, Fig. S3). These results indicate that Stat3 is not required for osteoblast differentiation from hypertrophic chondrocytes during embryonic development. As long bone growth was severely affected after birth (Fig. 1), and recently it was reported that there is a radical switch in clonality of growth plate chondrocytes shortly after birth (30), we then asked whether Stat3 is required in the growth plate to regulate both chondrocytes and their contribution to osteoblast differentiation by lineage tracing. We used the Col2a1CreER (31) and Rosa26-tdTomato (Jackson stock No. 007909) (Rosa26Tm) mice to label the chondrocytes with tdTomato (Td-Tm). We generated the Col2a1CreER; Stat3C/C; Rosa26Tm and the littermate control Col2a1CreER; Stat3C/+; Rosa26Tm mice. Tamoxifen (Tam) was injected at P2, and humerus was cryo-sectioned at P21 for analysis (SI Appendix, Fig. S4). We found that loss of Stat3 resulted in reduced chondrocyte number and chondrocyte columnar length in an environment of wild-type chondrocytes. Consistent with this, osteoblast cells derived from the Stat3-deficient chondrocytes were reduced. These results suggest that chondrocytes require Stat3 to expand for long bone growth and osteoblast differentiation in postnatal long bone development.
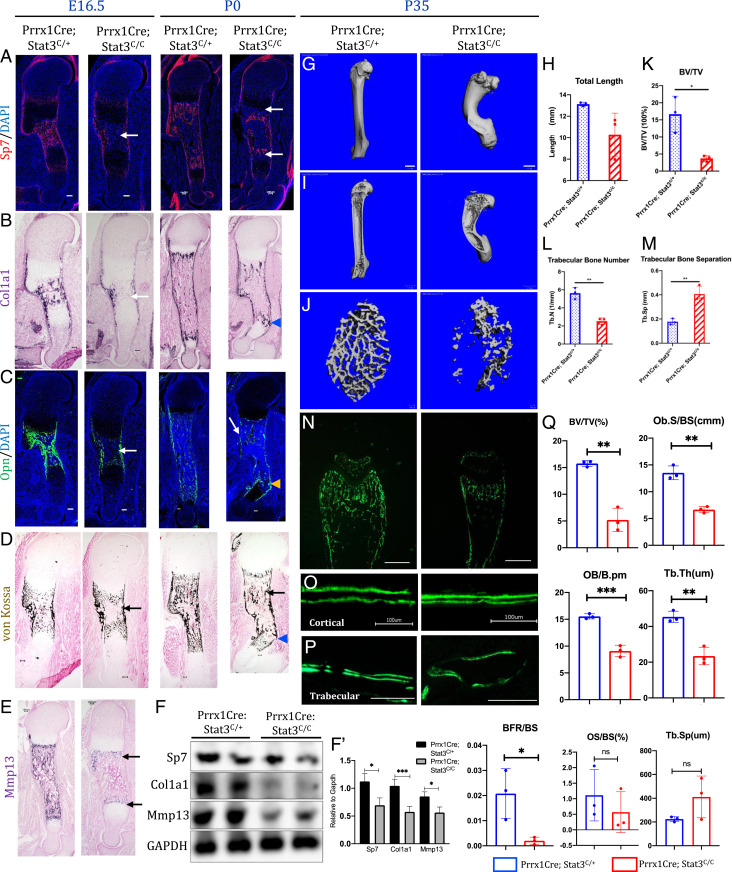
To determine whether the long bone defects in the Prrx1Cre; Stat3C/C mouse embryos were mainly caused by reduced osteoblast differentiation from mesenchymal progenitor cells, we analyzed osteoblast differentiation by IF staining and in situ hybridization (Fig. 2 A–C). The expression of Sp7 (Osterix), a master transcription factor required for early osteoblast lineage commitment, was reduced in the Prrx1Cre; Stat3C/C embryos. The number of Sp7+ cells was reduced in the Prrx1Cre; Stat3C/C long bone sections compared with the littermate control at E16.5 and P0 (Fig. 2A). In addition, expression of Col1a1, an osteoblast marker, and Osteopontin (Opn), a marker for mature osteoblasts, was both reduced at E16.5 and P0 in the developing long bone of the Prrx1Cre; Stat3C/C mutants compared to the control litter mates (Fig. 2 B and C). Consistent with reduced osteoblast differentiation and maturation, bone ossification indicated by von Kossa staining was reduced in the Prrx1Cre; Stat3C/C long bones (Fig. 2D). Furthermore, we found that expression of Mmp13, which is expressed in the terminal hypertrophic chondrocytes/osteoblasts and required for the replacement of hypertrophic chondrocytes by trabecular bone (32), was diminished in the Prrx1Cre; Stat3C/C sections at P0 (Fig. 2E), which explains the phenotype of expanded hypertrophic zone. Reduction in the expression of Sp7, Col1a1, and Mmp13 in the developing long bones of the Prrx1Cre; Stat3C/C embryos were further confirmed by Western blotting analysis (Fig. 2F). Therefore, fragile bones with fractures in the Prrx1Cre; Stat3C/C limbs were caused by reduced osteoblast differentiation and maturation during embryonic development. In addition, bone fracture and poor repair likely caused bone malformation and limb bowing.
Fig. 2.
Stat3 is required in limb mesenchymal cells for osteoblast differentiation and bone formation. (A–E) Representative images of Sp7 IF staining (A), Col1a1 in situ hybridization (B), Opn IF staining (C), von Kossa staining (D), and Mmp13 in situ hybridization (E) of the humerus sections from E16.5 littermate embryos or P0 littermate pups with indicated genotypes. The arrows indicate reduced osteoblast marker (protein/gene) expression or ossification. The arrowheads indicate bone fractures in the P0 mutant humerus. (Scale bars, 100 µm.) (F) Western blotting analyses of the humerus bone tissue lysates of the P0 pups with indicated genotypes. (F’) Quantification of the Western blotting results in (F). (G) Representative µCT images of femurs from 5-wk-old littermate mice with the indicated genotypes. (Scale bars, 1 mm.) (H) Quantification of humerus length (n = 3, mean ± SD). (I and J) Representative µCT images of the cortical (I) and trabecular (J) femur bones from 5-wk-old littermate mice. (Scale bars, 100 µm.) (K–M) Quantification of indicated parameters of µCT scanning. (N–P) Histomorphometric analysis of bone formation from 5-wk-old littermate mice of indicated genotypes. Representative images of double Calcein labeling in the distal femur heads (N), cortical bones (O), and trabecular bones (P) of the indicated genotypes. (Scale bars, 100 µm.) (Q) Quantification of indicated histomorphometric parameters of the distal femurs from 5-wk-old littermate mice of indicated genotypes. *P < 0.05, **P < 0.01, and ***P < 0.001 were considered significant; ns, not significant. The data are shown as means ± SD.
In the adult Prrx1Cre; Stat3C/C mice, bone mass and formation were also reduced compared to the control (Fig. 2 G–M). The femurs from 5-wk-old mice were analyzed by μCT scanning (Fig. 2 G–M). The femur was severely bowed and significantly shorter in the Prrx1Cre; Stat3C/C mice (Fig. 2 G–I). The trabecular bone was drastically reduced (Fig. 2 J–M), which was confirmed by quantification of bone parameters such as bone length, bone volume (BV)/trabecular volume, and trabecular number. To test whether reduced BV was due to reduced bone formation, bone histomorphometric analyses were performed by double calcein labeling (Fig. 2 N–Q). A strong reduction in calcein labeling was observed in the bone of the Prrx1Cre; Stat3C/C mice (Fig. 2N), and mineral acquisition rate in cortical (Fig. 2O) and trabecular (Fig. 2P) bones was also reduced as compared to the littermate control. These data showed that Stat3 is required for bone formation by controlling osteoblast differentiation and maturation during both embryonic development and postnatal life.
As osteoblast differentiation is coupled with osteoclast differentiation, we then determined whether Stat3 function in osteoblasts could indirectly regulate bone remodeling via osteoclast differentiation. Increased osteoclast activity leads to excessive loss of bone, leading to osteopenia and osteoporosis (33). Osteoclast cells were detected by tartrate-resistant acid phosphatase (TRAP) staining and quantified (SI Appendix, Fig. S5A). In the Prrx1Cre; Stat3C/C bone, the osteoclast cell surface normalized to bone surface was increased significantly compared to the control. Such increase in osteoclast cell number was associated with significant increase in the messenger RNA (mRNA) expression of both Receptor Activator of NF-κB Ligand (RANKL) and Osteoprotegrin (Opg) in the Prrx1Cre; Stat3C/C mice (SI Appendix, Fig. S5 B and C). The increased RANKL expression may have a dominant effect. However, interestingly, we also found many dead cells without nucleus in the Prrx1Cre; Stat3C/C bone (SI Appendix, Figs. S1D and S5A), suggesting that extensive osteoclast differentiation was induced by inflammatory responses to massive cell death due to Stat3 deletion in the Prrx1 lineage. Indeed, we found that macrophage and neutrophil numbers were up-regulated in regions where the dead cells were found (SI Appendix, Fig. S5D). These results show that reduced osteoblast differentiation/maturation, increased osteoblast cell death, and osteoclast differentiation together led to the reduction of bone mass in the Prrx1Cre; Stat3C/C mice.
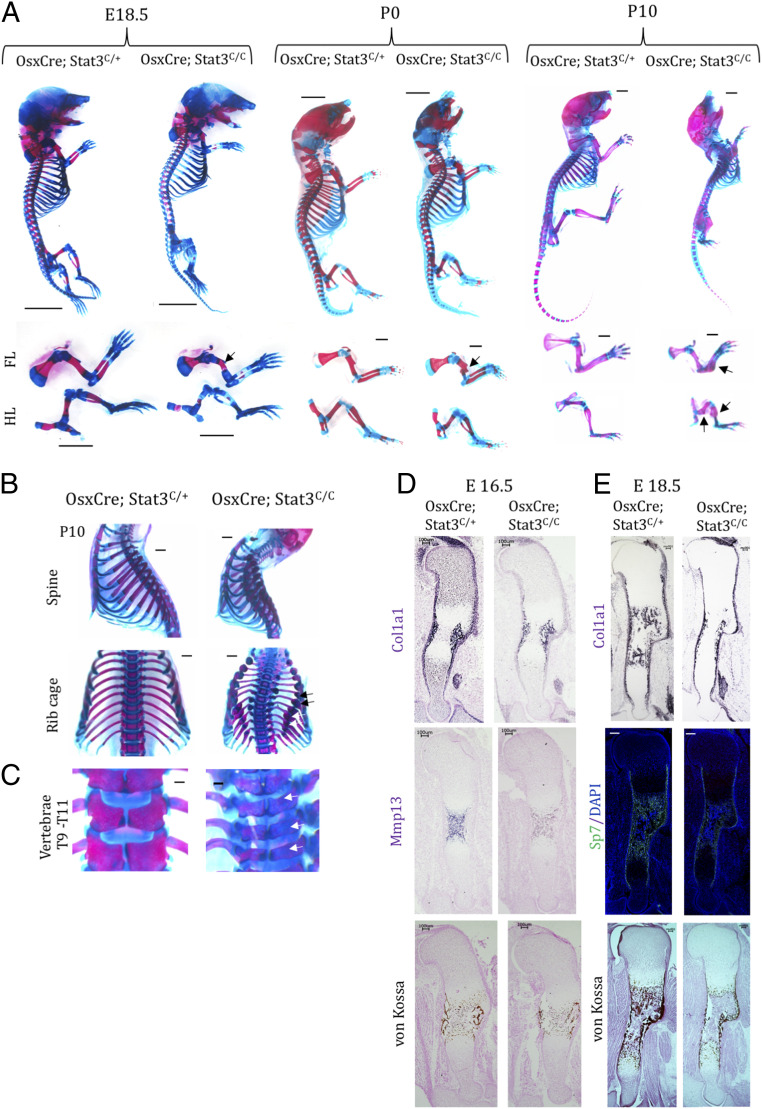
The requirement of Stat3 in bone formation in the developing skeleton of the Prrx1Cre; Stat3C/C mutant led us to test specifically whether Stat3 is required in the osteoblast lineage cells for bone formation. We employed the OsxCre line (34) to delete Stat3 in osteoblast lineage cells of the entire skeletal system. Interestingly, we observed broader skeletal phenotypes in the OsxCre; Stat3C/C mice compared to the Prrx1Cre; Stat3C/C mice (Fig. 3). Reduced bone formation was shown in the limb, skull, rib, and vertebral body (Fig. 3 A–C). The OsxCre; Stat3C/C mice died shortly after birth due to smaller and fractured ribcages (Fig. 3B). The OsxCre; Stat3C/C embryos showed similar long bone defects in the limbs compared to the Prrx1Cre; Stat3C/C embryos both morphologically (Fig. 3A) and molecularly (Fig. 3 D and E). As hypertrophic chondrocytes also express Osx but Stat3 is not required in hypertrophic chondrocytes for bone formation (SI Appendix, Fig. S3), these results further demonstrate that the skeletal defects in the OsxCre; Stat3C/C mouse were mainly caused by functional loss of Stat3 in the osteoblast lineage cells. Previous studies also showed that Stat3 is required in mature osteoblast or osteocytes to maintain homeotic bone formation as well as load-induced bone formation (19, 35).
Fig. 3.
Loss of Stat3 in osteoprogenitor cells phenocopied the Prrx1Cre; Stat3 mice. (A) Whole-mount Alizarin red and Alcian blue staining of the indicated littermate mouse embryos or pups. The forelimb (FL) and hindlimb (HL) were shown in the Lower. Bone fracture is shown by arrows. (Scale bars, 1 mm.) (B) Enhanced curvature of spine and multiple fractures (arrows) in the ribcage of P10 mutant mice. (C) Poorly developed and less-mineralized spine bones of P10 mutant mice. The T9 to T11 vertebras of the littermate animals with indicated genotypes are shown. The arrows indicate the defects in vertebras. (Scale bars, 1 mm.) (D and E) Representative in situ hybridization images of indicated gene expression or Sp7::GFP florescent images in the humerus sections of E16.5 (D) and E18.5 (E) littermate embryos with indicated genotypes. (Scale bars, 100 µm.)
Stat3 Is Required to Promote Wnt/β-Catenin Signaling in the Developing Bone.
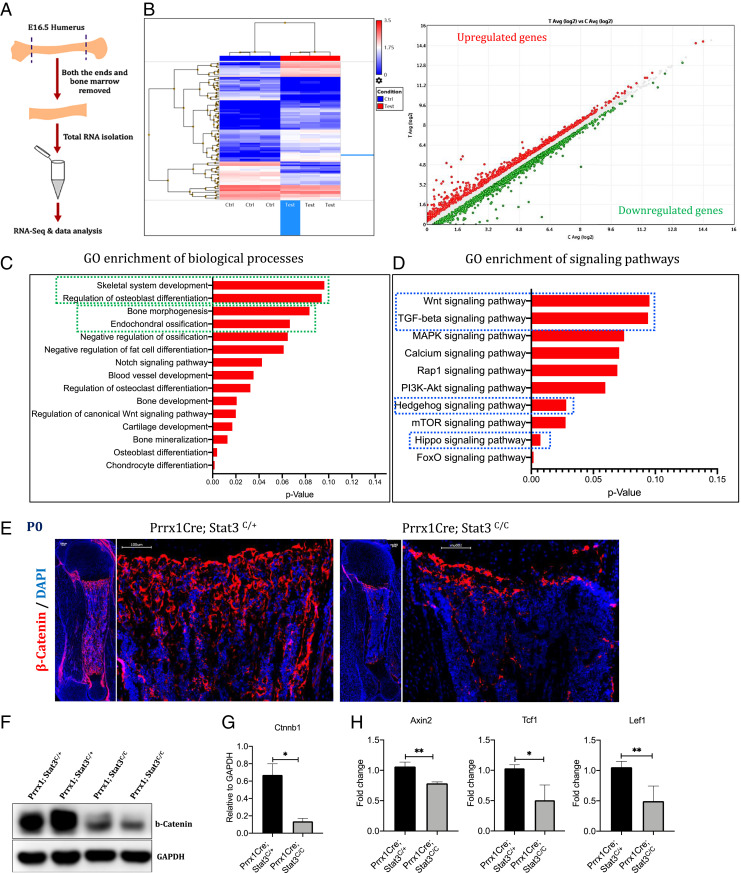
To investigate the molecular mechanism whereby Stat3 regulates osteoblast differentiation, we performed bulk RNA sequencing (RNA-seq) experiments to unbiasedly identify downstream pathways altered by Stat3 loss (Fig. 4). The developing humerus from the E16.5 embryos were isolated, and the cartilage tissues as well as the bone marrow were removed (Fig. 4A). The experiment was performed in triplicate, and profound gene expression changes were found (Fig. 4B). Analyses of gene ontology (GO) enrichment for biological processes showed that bone developmental processes were predominantly altered (Fig. 4C). In addition, GO enrichment for signaling pathways showed that major signaling pathways known to regulate bone development were altered by loss of Stat3, among which the Wnt signaling pathway is on the top of the list (Fig. 4D). As the canonical Wnt- or the Wnt/β-catenin–signaling pathway plays key roles in bone development, we focused our analysis on the Wnt/β-catenin signaling. We found that, indeed, the protein levels of β-catenin, a central signal transducer in the Wnt/β-catenin–signaling pathway, were reduced in the developing long bones of the newborn Prrx1Cre; Stat3C/C pups (Fig. 4 E and F). In addition, we found that expression of transcriptional target genes of the Wnt/β-catenin–signaling pathway such as Axin2, Lef1, and Tcf1 were reduced in the Prrx1Cre; Stat3C/C bone compared to the littermate control (Fig. 4G). These results indicate that Stat3 is required for bone formation and osteoblast differentiation by promoting Wnt/β-catenin signaling. However, we did not find protein interaction between Stat3 and β-catenin by coimmunoprecipitation assay of lysates from limb bud mesenchymal cells or bone samples (SI Appendix, Fig. S6A).
Fig. 4.
Stat3 deletion leads to reduction in Wnt/β-catenin signaling activity in the developing long bones. (A) Schematics of the procedure for isolating E16.5 humerus bones for the RNA-seq analysis. (B) Heatmap analysis of differentially expressed genes between Control and Prrx1Cre; Stat3c/c KO Samples. (C) GO enrichment analysis of the differentially expressed genes for biological processes revealed genes associated with skeleton development, osteoblast differentiation, and endochondral ossification (boxed in green). (D) GO enrichment analysis of down-regulated genes for signaling pathways revealed reduction in Wnt, TGF-β, Hedgehog, and Hippo signaling pathways (boxed in blue). (E) IF images of β-catenin expression in the humerus sections of littermate control and Stat3 KO P0 pups. (Scale bars, 100 µm.) (F) Western blotting analysis of β-catenin in the bone tissue lysates prepared from P0 pups with indicated genotypes. (G) Quantification of the Western blotting results in (F). (H) qRT-PCR analysis of Wnt/β-catenin signaling target genes in the humerus bone from the P0 littermate pups. *P < 0.05 and **P 0.005 were considered as significant; the data are presented as mean ± SD.
Consistent with the reduction of Wnt/β-catenin signaling in the developing long bone of the Prrx1Cre; Stat3C/C embryo, we found that expression of Sclerostin (Sost), which encodes a secreted inhibitor of Wnt/β-catenin signaling (36, 37), was precociously increased as compared to the control at E16.5 (SI Appendix, Fig. S6B). Sost is specifically expressed in the osteocytes, and we found that the increase in Sost expression was not due to cooresponding increase in osteocyte formation in the Prrx1Cre; Stat3C/C embryo, as the expression of other two osteocyte-specific genes Dmp1 and Phex was reduced, possibly due to reduced bone formation (SI Appendix, Fig. S6B). Increased Sost expression was also found in long bones at postnatal stages P0, P5, and P21 in the Prrx1Cre; Stat3C/C animals (SI Appendix, Fig. S6 C and D′). To further test the regulation of Wnt/β-catenin signaling by Stat3, we isolated the bone marrow stromal cells (BMSCs) from the Stat3C/C mice and cultured them in vitro as we have shown (38). When Stat3 was deleted by Ad-Cre, we found that osteoblast differentiation of BMSCs as indicated by alkaline phosphatase (ALP) staining was reduced (SI Appendix, Fig. S6E), and the expression of osteoblast markers (Col1a1, Sp7 (Osx), Alp, Runx2, and Ocn) and Wnt signaling target genes (Axin2, Tcf1, and Lef1) were reduced, and Sost expression was increased (SI Appendix, Fig. S6F). These results suggest that Stat3 may promote Wnt/β-catenin signaling by suppressing the expression of a Wnt inhibitor Sost.
We then tried to test whether Stat3 directly regulates Sost expression. The only Stat3 binding site was found in the 3′ end of the Sost gene, where chromatin was weakly open, as indicated by the lower peak of the assay for transposase-accessible chromatin (ATAC) with high-throughput sequencing analyses of E14.5 embryonic limb bud tissue (SI Appendix, Fig. S7 A and B) (39). In contrast, strong ATAC signals was found in the enhancer/promoter of Socs3, a known transcription target of Stat3 (40) (SI Appendix, Fig. S7A). We then used chromatin immunoprecipitation–qPCR (ChIP-qPCR) to confirm whether Stat3 indeed binds to these sites (SI Appendix, Fig. S7C). Importantly, while the control experiments showed that Stat3 binds to its binding site in the enhancer/promoter of Socs3, Stat3 did not bind to the identified site in Sost in BMSCs (SI Appendix, Fig. S7C). In addition, CHIP-qPCR analysis of the epigenetic marker H3K27Ac, which marks active enhancer (41), showed that while the Stat3 binding sites in Socs3 gene were also bound strongly by H3K27Ac, the Stat3 site in the Sost gene only showed weak H3K27Ac binding (SI Appendix, Fig. S7C). These data suggest that Stat3 does not regulate Sost expression directly. We have also looked into the possibility whether Stat3 may directly regulate expression of Wnt target genes Axin2, Tcf1, and Lef1 (SI Appendix, Fig. S7D). While the Lef/Tcf binding sites were found in the promoter regions (42) with strong ATAC signals, Stat3 binding sites were only found in regions with lower ATAC peaks, suggesting that Stat3 may not regulate Wnt signaling by directly controlling Wnt target gene expression.
Enhanced Wnt/β-Catenin Signaling Restores Bone Formation in the Stat3 Mutants.
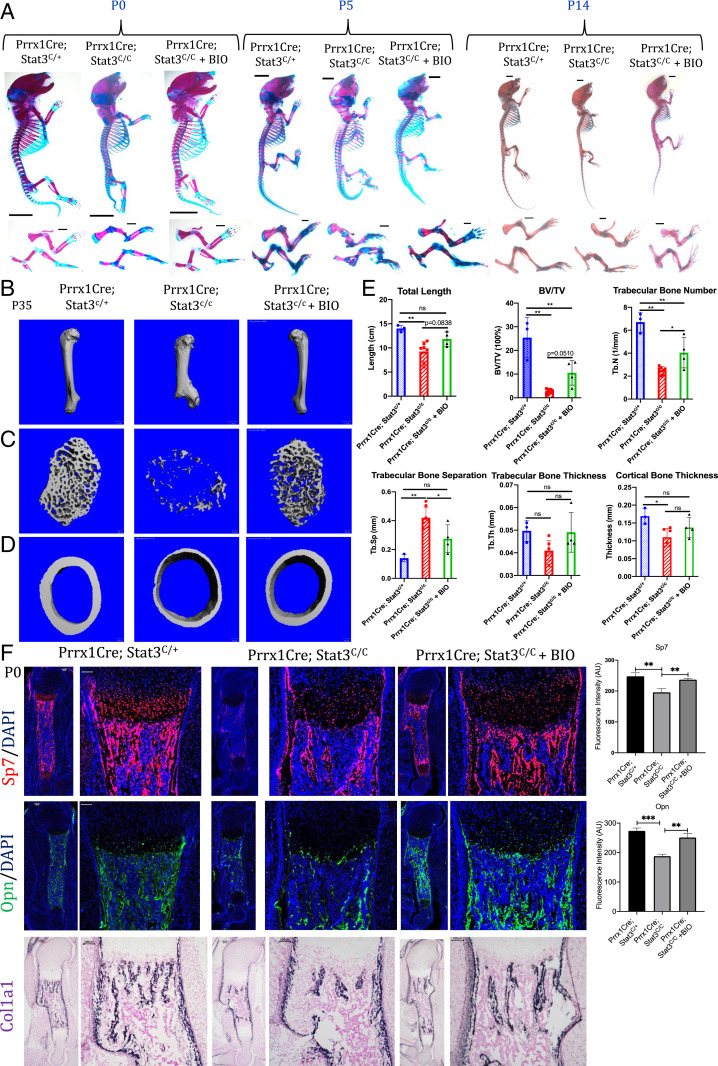
To determine whether Wnt/β-catenin signaling mediates the function of Stat3 in promoting osteoblast differentiation and bone formation, we decided to increase Wnt/β-catenin signaling first by treating the pregnant females with BIO, a natural product inhibitor of Gsk3-β (43, 44), to up-regulate Wnt/β-catenin signaling in the developing embryo. It is well-known that Gsk3-β inhibition leads to β-catenin stabilization and activation (45). We found that BIO treatment led to substantial rescue of the skeletal phenotypes and bone formation in the Prrx1Cre; Stat3C/C mice both embryonically and postnatally (Fig. 5A and SI Appendix, Fig. S8A). Restoration of trabecular and cortical bone mass was also found in the adult Prrx1Cre; Stat3C/C mice by μCT analyses (Fig. 5 B–E). However, likely due to the big variations and limited number of animals we could use in the analysis, we were not able to conclude that the difference between BIO-treated and nontreated mutant mice was statistically significant in all the parameters that were measured (Fig. 5E). However, at the molecular level, BIO treatment significantly up-regulated expression of Sp7, Opn, and Col1a1 at P0 (Fig. 5F and SI Appendix, Fig. S8B). Similar BIO treatment also rescued bone formation defects and fractures in the P0 and p28 OsxCre; Stat3C/C mice (SI Appendix, Fig. S8 C and D). It is likely that BIO treatment regimen needs to be optimized to observe more robust rescue. While 100% OsxCre; Stat3C/C embryos and pups at birth that were analyzed (n > 5) showed bone fractures and all pups died by P2 or P3, no fracture was observed at the time of birth in the OsxCre; Stat3C/C; Lrp5A/+ pups or the BIO-treated OsxCre; Stat3C/C pups, and these mice can survive much longer, though at later postnatal life, some long bones developed fractures. BIO treatment indeed increased bone mineralization of the Prrx1Cre; Stat3C/C mice, as shown by von Kossa staining and β-catenin expression (SI Appendix, Fig. S8 F and G). Treating the phenotypically normal heterozygous Prrx1Cre; Stat3C/+ mice with BIO did not cause baseline difference in bone formation (SI Appendix, Fig. S9).
Fig. 5.
Skeleton defects in Prrx1Cre; Stat3c/c mice were rescued by BIO treatment. (A) Whole-mount Alizarin red and Alcian blue staining of littermate mice of the indicated genotypes at P0, P5, and P14. The forelimb (FL) and hindlimb (HL) are shown in the Lower. (B–D) Representative µCT images of humerus (B) and cross-sections of the cortical (C) and trabecular (D) distal femur bones from 5-wk-old littermate mice of the indicated genotypes. (Scale bars, 1 mm in B and 100 µm in C and D.) (E) Quantification of humerus length (mean ± SD) and µCT parameters in femur bones from 5-wk-old littermate mice of the indicated genotypes. *P < 0.05, **P < 0.005, and ***P < 0.001. The data are shown as means ± SD. ns, not significant. (F) Representative images of fluorescent staining of Sp7, Opn, and Col1a1 in situ hybridization in the humerus sections of P0 littermate pups with indicated genotypes. (Scale bars, 100 µm.) Quantification was shown on the Right.
To test whether such rescue by BIO treatment was specific to the Wnt/β-catenin signaling activity, we use the Lrp5A/+ high bone mass mice (46) to genetically up-regulate Wnt/β-catenin signaling by generating the Prrx1Cre; Stat3C/C; Lrp5A/+ mice. We found that, again, bone formation was substantially rescued in the Prrx1Cre; Stat3C/C; Lrp5A/+ mice compared to the Prrx1Cre; Stat3C/C mice (Fig. 6A and SI Appendix, Fig. S8E). IF staining showed that at P0, Sp7, and β-catenin levels were largely restored (Fig. 6B). Mmp13 expression was also restored (Fig. 6C). Ossification indicated by von Kossa staining were also much improved to a level similar to that in the control (Fig. 6D). Taken together, these data show that Stat3 loss in mesenchymal progenitors or osteoblast lineage cells causes reduction in bone formation by reducing Wnt/β-catenin signaling. Our results also suggest that restoring Wnt/β-catenin signaling could be a therapeutic approach to improve bone formation in the Job syndrome patients.
Fig. 6.
The Lrp5 high bone mass (HBM) allele (Lrp5A/+) partially rescued the skeletal defects of the Prrx1Cre; Stat3c/c mice. (A) Whole-mount Alizarin red and Alcian blue staining of littermate mice of the indicated genotypes at P0 and P5. The forelimb (FL) and hindlimb (HL) were shown in the Lower. (Scale bars, 100 µm.) (B) IF images of β-catenin and Sp7 expression in the humerus sections of P0 pups of the indicated genotypes. Higher-magnification images are shown on the Right side. (Scale bars, 100 µm.) Quantification was shown on the Right. *P < 0.05 and **P < 0.005. (C) Representative IF images of Mmp13 in humerus sections from the indicated P5 mice. DAPI stained the nucleus. n = 3 biological replicates. (Scale bar, 50 μm.) (D) von Kossa staining of the humerus sections of P0 pups of the indicated genotypes. Higher-magnification images are shown on the Right side. (Scale bars, 100 µm.)
Discussion
Mutations in human STAT3, either LOF or GOF in transcriptional activities, are linked to the genetic cause of AD-HIES/Job syndrome. Here, we show that removing Stat3 function in early embryonic mesenchymal progenitor cells or osteoblast lineage cells led to severe skeletal malformation and reduced bone formation, similar to the skeletal defects in the Job syndrome. Some STAT3 mutations have been experimentally determined for the reduction in transcriptional activity as well as poor phosphorylation after being stimulated by cytokines or growth factors (47). The skeletal defects in autosomal AD-HIES/Job syndrome are therefore likely due to STAT3 haploinsufficiency or dominant-negative effects. We identified promoting Wnt/β-catenin signaling as an important function of Stat3 during osteoblast differentiation, possibly by suppressing Sost expression in the forming long bones. Enhancing Wnt/β-catenin signaling pharmacologically or genetically can rescue the bone phenotypes of the Stat3 mutants, suggesting that the skeletal defects of Job syndrome due to Stat3 LOF can be improved by enhancing Wnt/β-catenin signaling. The Lrp5A allele that partially rescued the bone defect of Stat3 mutants is GOF and resistant to Sost (48). Our results are consistent with a previous observation that Sost expression was up-regulated by Stat3 loss in osteocytes (35). In bone development, Stat3 can be activated by the LIF (49) and other growth factors such as PDGFs (50), all of which have been shown to promote osteoblast differentiation.
It is important to note that stronger pStat3 staining was detected in differentiating osteoblasts than in chondrocytes or mesenchymal progenitor cells, and loss of Stat3 in early limb mesenchymal progenitors that give rise to both cartilage and bone or in osteoblast lineage cells results in similar phenotypes of bone reduction. Indeed, no cartilage formation or chondrocyte differentiation defect was observed in the Prrx1Cre; Stat3C/C embryos. These results are consistent with a previous study in Zebrafish (51). Knocking-down Stat3 in Zebrafish resulted in normal cartilage formation with defects in the spine and immune system. Stat3 has recently been found to regulate cartilage development through Sox9 expression (20, 52). Such requirement appears to be location (in the embryonic rib cartilage) (52) or timing dependent (after birth in the TCre; Stat3C/C bone) (20). While Stat3 regulates Sox9 expression, the requirement of Stat3 in osteoblast linage cells that do not express Sox9 indicate that Stat3 has distinct targets to control bone development. The osteoblast cells in the developing endochondral bone are derived from Osx+ cells, a fraction of Prrx1+ cells. In this study, we provide strong evidence that similar bone defects were observed in Prrx1Cre- and OsxCre-driven Stat3 mutants, in which BIO treatment or Lrp5A expression also had similar effects. The cell of origin for Stat3 function in embryonic bone development is the Osx+ osteoprogenitor, and Wnt/β-catenin downstream targets should be similarly down-regulated in the OsxCre model. Given the intimate interaction between perichondrial osteoblast progenitor cells and the chondrocytes in the growth plates (53–56), our results also suggest that reduced bone formation due to Stat3 loss may also contribute to later cartilage defects. Multiple modes of Stat3 and β-catenin interactions have been reported in different context (57–60). Interestingly, Stat3 activation has been reported to increase β-catenin by repressing Gsk3-β and the SWI/SNF gene Arid1b driving neurofibroma initiation (61). Related to our finding, a STAT3-miR-92a-DKK1 pathway has been found to activate β-catenin in promoting malignant progression of ovarian cancer (59). Here, we show that in the developing long bone, Stat3 is activated and promotes Wnt/β-catenin signaling, possibly by repressing Sost expression. It will be interesting to further test whether Stat3 also acts through microRNA or other noncoding RNA to suppress Sost expression.
In the mosaic lineage-tracing experiments (SI Appendix, Fig. S4), the defects of Stat3−/− cells were likely compensated by the large amount of wild-type cells, which may explain why there was no obvious difference of growth plate heights between the control and mutant mice. Furthermore, as Stat3 plays important roles regulating cell proliferation and survival (62, 63), mosaic loss of Stat3 may render the mutant cells disadvantageous during cell competition, a phenomenon that cells with lower translation rates or lower levels of proteins involved in signal transduction, polarity, and cellular growth can survive in a homogenous environment but are killed when surrounded by cells of higher fitness (64). Lastly, being a key mediator of LIF-dependent signaling, Stat3 controls the embryonic stem cell maintenance and pluripotency (65–67). Stat3 has also been found to regulate maintenance and proliferation of other stem cells (68, 69). It is possible that loss of Stat3 reduced chondrocyte stemness, as Col2a1CreER has been shown to label chondrocyte stem cells in early postnatal mice (30), causing reduction in mutant chondrocyte numbers and column length. In addition, as Col2A1CreER also labels mesenchymal stem cells in the bone marrow (70), reduced osteoblast differentiation in the Col2a1CreER; Stat3C/C; Rosa26Tm mice (SI Appendix, Fig. S4) may also reflect the requirement of Stat3 in mesenchymal stem cells in the bone marrow, as we have shown in the in vitro experiments (SI Appendix, Fig. S6 E and F). It is possible that the functional mechanism of Stat3 in the postnatal chondrocytes and osteoblast progenitors or mesenchymal stem cells is distinct. One scenario could be that the role of Stat3 in chondrocytes is Wnt-signaling independent, while its role in mesenchymal stem cells or osteoblast progenitor cells is mediated at least in part by Wnt/β-catenin signaling. This may explain the observed poorer rescue of the Stat3 mutant long bone growth by BIO or Lrp5A expression later in postnatal life.
In osteoblast lineage, it appears that Stat3 is required in distinct differentiation stages. While deleting Stat3 in the osteoprogenitors and early osteoblasts using the OsxCre resulted in severely reduced bone formation with bone fractures and lethality shortly after birth (Fig. 3), removing Stat3 in early or mature osteoblast with the Col3.6-Cre or Col2.3-Cre, respectively, resulted in similar but less severe bone formation defects (19). Noticeable at an age of 3 to 4 wk, 10% of these Stat3-deficient mice were extremely small with a spine deformity. The other mutant mice can survive to older ages and showed reduced bone mass and shorter femurs at 18 wk of age (19). When Stat3 was deleted in more-mature osteoblasts/osteocytes using the Dmp1Cre, however, the mutant mice were indistinguishable from wild-type littermates, with no difference in body weight, bone mineral content, or bone mineral density at 18 wk of age. More-detailed analyses showed Stat3 deficiency in osteocytes did not affect bone mineral accrual but reduced bone formation in adult mice (35).Therefore, Stat3 plays more important roles for bone formation in osteoprogenitor cells and earlier osteoblast cells. Taken together, our studies show that modeling the skeletal phenotypes of Job syndrome in mice provides insights into the cell origin (osteoblast) and molecular mechanism (Wnt/β-catenin signaling) underlying the skeletal defects.
Materials and Methods
Mouse Lines Used.
All mouse studies were conducted according to the protocols approved by the Harvard Medical School Institutional Animal Care and Use Committee (IACUC). The mice strains were either previously reported or purchased from the Jackson Laboratories are as follows: Stat3C/C mice were generated by inserting loxP sites in the introns 17 and 20 as reported earlier (28). Prrx1Cre (stock No. 005584), Osx1-GFP::Cre (OsxCre, stock No. 006361), Rosa26-tdTomato (Jackson stock No. 007909) (71), FVB-Tg(Col2a1-cre/ERT)KA3Smac/J (Jackson stock No. 006774), and the Lrp5-GOFA214V-neo mice were reported earlier (46). The Col10a1Cre mice were generated by Kathy Cheah’s laboratory Department of Biochemistry, University of Hong Kong, China (26). In postnatal studies, sex-matched littermate mice were analyzed. Representative data from analyses of a minimum of three control and mutant littermates in each experiment are shown.
Gene Expression Analysis by In Situ Hybridization.
In situ hybridization was performed using digoxygenin-labeled antisense probes on either 5-μm thick cryosections or on paraffin sections as described before (72). The probe sequences were described previously (73).
IF Staining.
Embryos and early postnatal tissues were fixed overnight in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) and processed for either cryostat or paraffin sections. Sections were rehydrated, permeabilized with PBT (1× PBS + 0.1% Tween 20), and were blocked in 10% donkey/goat serum in PBT. Immunohistochemistry or IF staining were performed using standard methods, and details of primary and secondary antibodies are provided in SI Appendix, Table S1. Sections were mounted in mounting medium containing nuclear stain DAPI from Vector laboratories (Cat. No. H-1200). The fluorescence intensity of IF images was quantified using Image J software as described earlier (74).
Skeletal Preparation and μCT Scanning Analyses.
The protocol for Alcian blue staining for cartilage and Alizarin red staining for mineralized tissues was described before (75). μCT scanning of postnatal long bones was conducted using a SCANCO μCT 35 according to standard procedures and data were analyzed using software from the manufacturer.
von Kossa and Masson’s Trichrome Staining.
For von Kossa staining, paraffin sections were incubated with 1% silver nitrate solution under a 60-W lamp for 1 h. Slides were washed three times in distilled water and incubated with 5% solution of sodium thiosulfate for 5 min. Slides were washed three times in distilled water and counterstained with 0.1% nuclear fast red. Slides were rinsed three times in distilled water before mounted in mounting medium. Masson’s trichrome staining was performed on paraffin sections by refixing in Bouin’s solution for 1 h at 56 °C followed by staining sequentially with Weigert’s iron hematoxylin solution, Biebrich scarlet-acid fuchsin solution, and aniline blue solution (76).
Cell Death Assay.
Cleaved caspase-3 IF was performed using standard procedures, and primary antibody was purchased from CST (Cat. No. 9664).
Micromass Culture.
Limb bud cells for micromass analyses were isolated from E11.5 embryos as reported earlier (77).
qRT-PCR.
Total RNA from mouse humerus and femur bone tissue devoid of bone marrow and cartilage was prepared using the TRIZOL reagent (Life Technologies) or RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocols. Complementary DNA (cDNA) was synthesized from total RNA (1 to 2 μg) using SuperScript II Reverse Transcriptase with random primers (Life Technologies). qRT-PCR were performed using SYBR Select Master Mix on StepOnePlus thermal cycler from Applied Biosystems. Gene expression levels were analyzed relative to β-actin or GAPDH. The primer sequences of the genes are given in the SI Appendix, Table S1.
RNA-Seq.
Total RNA was isolated from the shaft of the humerus of E16.5 embryos after removing both the ends and flushing out the marrow. Three Prrx1Cre; Stat3C/+ (Ctrl) and three Prrx1Cre; Stat3C/C (Test) embryos were used for total RNA isolation. The libraries were constructed using Ion AmpliSeq Transcriptome Mouse Gene Expression Panel, Chef-Ready Kit according to the manufacturer’s protocols. The library qualities and quantification were estimated using Bio-Analyzer 2100. Equal amount of six barcoded libraries were pooled together on an equimolar basis and sequenced using the Ion 550 Chip Kit. Genes with a fold change 1.2 were considered as differentially expressed genes and analyzed using the DAVID Bioinformatics Resources 6.8 (78). The RNA-seq data have been deposited to Gene Expression Omnibus (GEO) under accession number GSE159184.
Immunoprecipitation and Immunoblotting.
Bone tissues or cells were lysed using either lysis buffer (20 mM Tris [pH 7.4], 150 mM NaCl, 1% Triton X-100, 1 mM ethylenediamine tetraacetic acid (EDTA), 1 mM ethylene glycol tetraacetic acid (EGTA), 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate), or radioimmunoprecipitation assay (RIPA) buffer (Santa Cruz Biotechnology), supplemented with protease inhibitor mixture (Roche). Immunoprecipitated complex or total cell lysates were analyzed using Western blotting. Western blotting analyses were conducted using standard procedures. Densitometric analysis of Western blot data were done using Image J software as described earlier (79).
Small Molecule Treatment.
BIO (Calbiochem, CAS No. 667463-62-9) was prepared as described previously (43, 80). BIO was injected to pregnant females by intraperitoneal injection at a concentration of 2 μM every day from E15.5, and the postnatal mice were injected every other day. Equivalent volumes of vehicle were injected to control animals.
Antibodies Used.
The details of the antibodies used in this study are provided in SI Appendix, Table S1.
BMSC Isolation and Culture in Osteogenic Media.
BMSCs were flushed out from tibia and femur bone marrow cavity of Stat3c/c mice. Then, the cells were centrifuged and seeded with Alpha-MEM, 20% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin. Prior to reaching confluence, cells were infected with either Ad-Cre or Ad-GFP. Upon reaching confluence, cells were switched to osteogenic media (Alpha-MEM, 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, 10−4 M L-ascorbic acid 2-phosphate, and 10 mM β-glycerol phosphate) and cultured for the indicated time points (81).
ChIP Assay.
ChIP was performed using ChIP-Grade Protein G Enzymatic Kit (Biolegend, 699904) according to the manufacturer’s instructions. Briefly, BMSC cells were cross-linked with 1% formaldehyde for 10 min and stopped by 0.125M glycine in culture medium at room temperature. Chromatin was enzymatically digested for 5 min at 37 °C. Antibodies used in the ChIP experiment: Stat3 (Cell signaling technology, 1:100), H3K27ac (Abcam ab4729, 1µg), and IgG antibody (Biolegend, provided in the ChIP kit).
Adenovirus Treatment.
The Cre recombinase or GFP adenovirus (∼1012 pfu/mL) was diluted 1:2,000 to infect cells in vitro. After 4 h, fresh medium was added. After 24 h, the medium was changed.
von Kossa Staining.
Sections were hydrated and washed in distilled water, then stained with 5% silver nitrate under a 60-W lamp for 1 h. Sections were washed in distilled water three times in 5% sodium thiosulfate for 5 min and rinsed in water.
ALP Staining and Quantification.
For ALP staining, cells were fixed in 4% PFA for 20 min, washed with PBS three times, then stained with 1-Step nitro blue tetrazolium (NBT)/5-bromo-4-chloro-3-indolyl phosphate (BCIP) (Thermo, 34042) for 30 min and washed by PBS. There were three biological replicates in each group.
Statistical Analysis.
The data were analyzed using GraphPad Prism 7 (GraphPad Software). Data from at least three or more independent experimental groups were used for quantification. Statistical analysis between groups was performed by two-tailed Student’s t test to determine significance when only two groups were compared. One-way ANOVA test was used for μCT data. The P values of less than 0.05 and 0.01 were considered significant. Error bars represents the SD of the mean unless otherwise mentioned.
Supplementary Material
Acknowledgments
We thank the Y.Y. laboratory members for stimulating discussions. The work in the Y.Y. laboratory is supported by NIH grants from the National Institute of Dental and Craniofacial Research (NIDCR) (Grant DE025866), the National Institute of Arthritis and Musculoskeletal and Skin (NIAMS) (Grant AR070877), and the National Cancer Institute (NCI) (Grant CA222571) to Y.Y.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. B.O.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020100118/-/DCSupplemental.
Data Availability
RNA-seq data have been deposited in GEO (GSE159184). All other study data are included in the article and/or SI Appendix.
References
- 1.Brestel E. P., Klingberg W. G., Veltri R. W., Dorn J. S., Osteogenesis imperfecta tarda in a child with hyper-IgE syndrome. Am. J. Dis. Child. 136, 774–776 (1982). [DOI] [PubMed] [Google Scholar]
- 2.Kirchner S. G., Sivit C. J., Wright P. F., Hyperimmunoglobulinemia E syndrome: Association with osteoporosis and recurrent fractures. Radiology 156, 362 (1985). [DOI] [PubMed] [Google Scholar]
- 3.Höger P. H., Boltshauser E., Hitzig W. H., Craniosynostosis in hyper-IgE-syndrome. Eur. J. Pediatr. 144, 414–417 (1985). [DOI] [PubMed] [Google Scholar]
- 4.Grimbacher B., et al., Hyper-IgE syndrome with recurrent infections–An autosomal dominant multisystem disorder. N. Engl. J. Med. 340, 692–702 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Hafsi W., Yarrarapu S. N. S., “Job syndrome” in StatPearls (StatPearls Publishing, Treasure Island, FL, 2021). [PubMed] [Google Scholar]
- 6.Fu X. Y., A transcription factor with SH2 and SH3 domains is directly activated by an interferon alpha-induced cytoplasmic protein tyrosine kinase(s). Cell 70, 323–335 (1992). [DOI] [PubMed] [Google Scholar]
- 7.Song L., Turkson J., Karras J. G., Jove R., Haura E. B., Activation of Stat3 by receptor tyrosine kinases and cytokines regulates survival in human non-small cell carcinoma cells. Oncogene 22, 4150–4165 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Rawlings J. S., Rosler K. M., Harrison D. A., The JAK/STAT signaling pathway. J. Cell Sci. 117, 1281–1283 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Darnell J. E. Jr, STATs and gene regulation. Science 277, 1630–1635 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Levy D. E., Lee C. K., What does Stat3 do? J. Clin. Invest. 109, 1143–1148 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holland S. M., et al., STAT3 mutations in the hyper-IgE syndrome. N. Engl. J. Med. 357, 1608–1619 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Mertens C., Haripal B., Klinge S., Darnell J. E., Mutations in the linker domain affect phospho-STAT3 function and suggest targets for interrupting STAT3 activity. Proc. Natl. Acad. Sci. U.S.A. 112, 14811–14816 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woellner C., et al., Mutations in STAT3 and diagnostic guidelines for hyper-IgE syndrome. J. Allergy Clin. Immunol. 125, 424–432.e8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milner J. D., et al., Early-onset lymphoproliferation and autoimmunity caused by germline STAT3 gain-of-function mutations. Blood 125, 591–599 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minegishi Y., et al., Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature 448, 1058–1062 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Flanagan S. E., et al., Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nat. Genet. 46, 812–814 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forbes L. R., Milner J., Haddad E., Signal transducer and activator of transcription 3: A year in review. Curr. Opin. Hematol. 23, 23–27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itoh S., et al., A critical role for interleukin-6 family-mediated Stat3 activation in osteoblast differentiation and bone formation. Bone 39, 505–512 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Zhou H., et al., Osteoblast/osteocyte-specific inactivation of Stat3 decreases load-driven bone formation and accumulates reactive oxygen species. Bone 49, 404–411 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Hall M. D., Murray C. A., Valdez M. J., Perantoni A. O., Mesoderm-specific Stat3 deletion affects expression of Sox9 yielding Sox9-dependent phenotypes. PLoS Genet. 13, e1006610 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bi W., Deng J. M., Zhang Z., Behringer R. R., de Crombrugghe B., Sox9 is required for cartilage formation. Nat. Genet. 22, 85–89 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Zhou G., et al., Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 19004–19009 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dy P., et al., Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev. Cell 22, 597–609 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bi W., et al., Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc. Natl. Acad. Sci. U.S.A. 98, 6698–6703 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bianco P., Cancedda F. D., Riminucci M., Cancedda R., Bone formation via cartilage models: The “borderline” chondrocyte. Matrix Biol. 17, 185–192 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Yang L., Tsang K. Y., Tang H. C., Chan D., Cheah K. S., Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc. Natl. Acad. Sci. U.S.A. 111, 12097–12102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou X., et al., Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet. 10, e1004820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welte T., et al., STAT3 deletion during hematopoiesis causes Crohn’s disease-like pathogenesis and lethality: A critical role of STAT3 in innate immunity. Proc. Natl. Acad. Sci. U.S.A. 100, 1879–1884 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Logan M., et al., Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33, 77–80 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Newton P. T., et al., A radical switch in clonality reveals a stem cell niche in the epiphyseal growth plate. Nature 567, 234–238 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Nakamura E., Nguyen M. T., Mackem S., Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev. Dyn. 235, 2603–2612 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Stickens D., et al., Altered endochondral bone development in matrix metalloproteinase 13-deficient mice. Development 131, 5883–5895 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charles J. F., Aliprantis A. O., Osteoclasts: More than ‘bone eaters’. Trends Mol. Med. 20, 449–459 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodda S. J., McMahon A. P., Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 133, 3231–3244 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Corry K. A., et al., Stat3 in osteocytes mediates osteogenic response to loading. Bone Rep. 11, 100218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellies D. L., et al., Bone density ligand, Sclerostin, directly interacts with LRP5 but not LRP5G171V to modulate Wnt activity. J. Bone Miner. Res. 21, 1738–1749 (2006). [DOI] [PubMed] [Google Scholar]
- 37.ten Dijke P., Krause C., de Gorter D. J., Löwik C. W., van Bezooijen R. L., Osteocyte-derived sclerostin inhibits bone formation: Its role in bone morphogenetic protein and Wnt signaling. J. Bone Joint Surg. Am. 90, 31–35 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Zhou T., et al., Piezo1/2 mediate mechanotransduction essential for bone formation through concerted activation of NFAT-YAP1-ß-catenin. eLife 9, e52779 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorkin D. U., et al., An atlas of dynamic chromatin landscapes in mouse fetal development. Nature 583, 744–751 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Starr R., et al., A family of cytokine-inducible inhibitors of signalling. Nature 387, 917–921 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Creyghton M. P., et al., Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doumpas N., et al., TCF/LEF dependent and independent transcriptional regulation of Wnt/β-catenin target genes. EMBO J. 38, e98873 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato N., Meijer L., Skaltsounis L., Greengard P., Brivanlou A. H., Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 10, 55–63 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Tseng A. S., Engel F. B., Keating M. T., The GSK-3 inhibitor BIO promotes proliferation in mammalian cardiomyocytes. Chem. Biol. 13, 957–963 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Nusse R., Clevers H., Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Cui Y., et al., Lrp5 functions in bone to regulate bone mass. Nat. Med. 17, 684–691 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaimowitz N. S., et al., A novel STAT3 mutation in a Qatari patient with hyper-IgE syndrome. Front Pediatr. 7, 130 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niziolek P. J., et al., High bone mass-causing mutant LRP5 receptors are resistant to endogenous inhibitors in vivo. J. Bone Miner. Res. 30, 1822–1830 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poulton I. J., McGregor N. E., Pompolo S., Walker E. C., Sims N. A., Contrasting roles of leukemia inhibitory factor in murine bone development and remodeling involve region-specific changes in vascularization. J. Bone Miner. Res. 27, 586–595 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Wang Y. Z., et al., Activation of Stat3 preassembled with platelet-derived growth factor beta receptors requires Src kinase activity. Oncogene 19, 2075–2085 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Shuting Xiong J. W., et al., Loss of stat3 function leads to spine malformation and immune disorder in zebrafish. Sci. Bull. (Beijing) 62, 185–196 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Mochizuki Y., et al., Combinatorial CRISPR/Cas9 approach to elucidate a far-upstream enhancer complex for tissue-specific Sox9 expression. Dev. Cell 46, 794–806.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Long F., Linsenmayer T. F., Regulation of growth region cartilage proliferation and differentiation by perichondrium. Development 125, 1067–1073 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Vortkamp A., et al., Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science 273, 613–622 (1996). [DOI] [PubMed] [Google Scholar]
- 55.Alvarez J., Horton J., Sohn P., Serra R., The perichondrium plays an important role in mediating the effects of TGF-beta1 on endochondral bone formation. Dev. Dyn. 221, 311–321 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Ohbayashi N., et al., FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 16, 870–879 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fragoso M. A., et al., The Wnt/β-catenin pathway cross-talks with STAT3 signaling to regulate survival of retinal pigment epithelium cells. PLoS One 7, e46892 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ibrahem S., et al., STAT3 paradoxically stimulates β-catenin expression but inhibits β-catenin function. Int. J. Exp. Pathol. 95, 392–400 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen M. W., et al., The STAT3-miRNA-92-wnt signaling pathway regulates spheroid formation and malignant progression in ovarian cancer. Cancer Res. 77, 1955–1967 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Shin M., et al., STAT3 potentiates SIAH-1 mediated proteasomal degradation of β-catenin in human embryonic kidney cells. Mol. Cells 39, 821–826 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu J., et al., Insertional mutagenesis identifies a STAT3/arid1b/β-catenin pathway driving neurofibroma initiation. Cell Rep. 14, 1979–1990 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin L., et al., STAT3 is necessary for proliferation and survival in colon cancer-initiating cells. Cancer Res. 71, 7226–7237 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Y., Sepich D. S., Solnica-Krezel L., Stat3/Cdc25a-dependent cell proliferation promotes embryonic axis extension during zebrafish gastrulation. PLoS Genet. 13, e1006564 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amoyel M., Bach E. A., Cell competition: How to eliminate your neighbours. Development 141, 988–1000 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burdon T., Chambers I., Stracey C., Niwa H., Smith A., Signaling mechanisms regulating self-renewal and differentiation of pluripotent embryonic stem cells. Cells Tissues Organs 165, 131–143 (1999). [DOI] [PubMed] [Google Scholar]
- 66.Matsuda T., et al., STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 18, 4261–4269 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martello G., Bertone P., Smith A., Identification of the missing pluripotency mediator downstream of leukaemia inhibitory factor. EMBO J. 32, 2561–2574 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sherry M. M., Reeves A., Wu J. K., Cochran B. H., STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells 27, 2383–2392 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yuan Y., et al., STAT3 stimulates adipogenic stem cell proliferation and cooperates with HMGA2 during the early stage of differentiation to promote adipogenesis. Biochem. Biophys. Res. Commun. 482, 1360–1366 (2017). [DOI] [PubMed] [Google Scholar]
- 70.Ono N., Ono W., Nagasawa T., Kronenberg H. M., A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat. Cell Biol. 16, 1157–1167 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Madisen L., et al., A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prashar P., Yadav P. S., Samarjeet F., Bandyopadhyay A., Microarray meta-analysis identifies evolutionarily conserved BMP signaling targets in developing long bones. Dev. Biol. 389, 192–207 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Khan S. K., et al., Induced Gnas R201H expression from the endogenous Gnas locus causes fibrous dysplasia by up-regulating Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. U.S.A. 115, E418–E427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jensen E. C., Quantitative analysis of histological staining and fluorescence using ImageJ. Anat. Rec. (Hoboken) 296, 378–381 (2013). [DOI] [PubMed] [Google Scholar]
- 75.Rigueur D., Lyons K. M., Whole-mount skeletal staining. Methods Mol. Biol. 1130, 113–121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Asonova S. N., Migalkin N. S., [Use of Masson’s trichrome method for staining decalcified bone tissue]. Arkh. Patol. 58, 66–67 (1996). [PubMed] [Google Scholar]
- 77.Paulsen D. F., Solursh M., Microtiter micromass cultures of limb-bud mesenchymal cells. In Vitro Cell. Dev. Biol. 24, 138–147 (1988). [DOI] [PubMed] [Google Scholar]
- 78.Jiao X., et al., DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics 28, 1805–1806 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gallo-Oller G., Ordoñez R., Dotor J., A new background subtraction method for Western blot densitometry band quantification through image analysis software. J. Immunol. Methods 457, 1–5 (2018). [DOI] [PubMed] [Google Scholar]
- 80.Gunn W. G., Krause U., Lee N., Gregory C. A., Pharmaceutical inhibition of glycogen synthetase kinase-3β reduces multiple myeloma-induced bone disease in a novel murine plasmacytoma xenograft model. Blood 117, 1641–1651 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Regard J. B., et al., Activation of Hedgehog signaling by loss of GNAS causes heterotopic ossification. Nat. Med. 19, 1505–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in GEO (GSE159184). All other study data are included in the article and/or SI Appendix.