Significance
Assembly-line polyketide synthases (PKSs) make many medicinally significant natural products. A better understanding of evolutionary mechanisms underlying polyketide diversification could open new avenues for PKS engineering and drug discovery. In the course of interrogating the role of gene conversion in assembly-line PKS evolution, we discovered not only that gene conversion is widespread in these PKSs but also that it is frequently associated with the presence of a genetic element, which we have designated GRINS (genetic repeats of intense nucleotide skew). Computational analysis suggests that the presence of GRINS may promote late-stage structural diversification of polyketide antibiotics. Our work sets the stage for further investigation of the underlying molecular mechanisms and evolutionary roles of these genetic elements.
Keywords: polyketide synthase, nucleotide skew, gene conversion
Abstract
Assembly-line polyketide synthases (PKSs) are large and complex enzymatic machineries with a multimodular architecture, typically encoded in bacterial genomes by biosynthetic gene clusters. Their modularity has led to an astounding diversity of biosynthesized molecules, many with medical relevance. Thus, understanding the mechanisms that drive PKS evolution is fundamental for both functional prediction of natural PKSs as well as for the engineering of novel PKSs. Here, we describe a repetitive genetic element in assembly-line PKS genes which appears to play a role in accelerating the diversification of closely related biosynthetic clusters. We named this element GRINS: genetic repeats of intense nucleotide skews. GRINS appear to recode PKS protein regions with a biased nucleotide composition and to promote gene conversion. GRINS are present in a large number of assembly-line PKS gene clusters and are particularly widespread in the actinobacterial genus Streptomyces. While the molecular mechanisms associated with GRINS appearance, dissemination, and maintenance are unknown, the presence of GRINS in a broad range of bacterial phyla and gene families indicates that these genetic elements could play a fundamental role in protein evolution.
Polyketide synthases (PKSs) are large enzymatic machines that synthesize structurally diverse natural products, many of which are used as antibiotics, immunosuppressants, anticancer agents, and other types of medicines. In bacteria, a substantial fraction of polyketides is synthesized by multimodular PKSs, where each module consists of a set of domains that collectively catalyze one round of elongation and chemical modification of the growing polyketide chain (1) (Fig. 1A). The homologous modules of each PKS operate in a defined assembly-line manner. The emergence of this multimodular architecture and its subsequent diversification has led to an astounding complexity and variety of polyketide natural products. However, the underlying evolutionary processes that drive the evolution of assembly-line PKSs are not well understood (2).
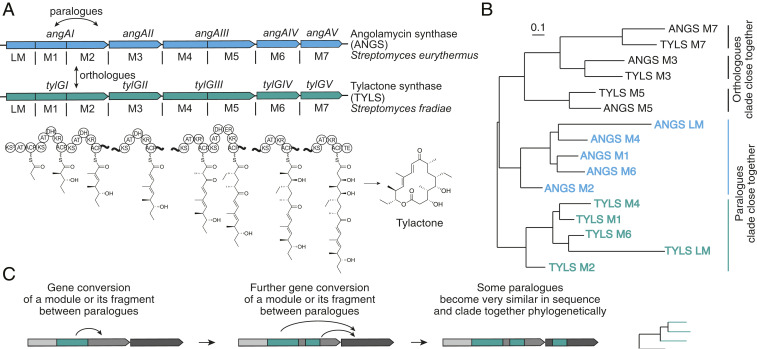
Fig. 1.
Gene conversion in assembly-line PKSs. (A) Typical architecture of an assembly-line PKS, exemplified by closely related angolamycin synthase (ANGS) and tylactone synthase (TYLS), which contain the same set of biosynthetic domains and produce the same polyketide, tylactone. Domains: KS, ketosynthase; KSQ, decarboxylative ketosynthase; AT, acyltransferase; DH, dehydratase; DHt, inactive dehydratase; ER, enoyl-reductase; KR, ketoreductase; KR°, ketoreductase-inactive epimerase; ACP, acyl carrier protein; TE, thioesterase. (B) Phylogenetic tree of protein sequences of angolamycin and tylactone synthase modules. Some orthologous modules clade together, as is expected for closely related PKSs that only recently diverged from a common ancestor through point mutations. However, in many cases paralogous modules clade more closely together, which can be explained by gene conversion between these modules. Sequence alignment performed using ClustalOmega (10); phylogenetic tree constructed using RAxML (30). (C) Gene conversion is a process in which a DNA sequence is nonreciprocally transferred from one homologous region to another and was implicated in the evolution of assembly-line PKSs (6, 7).
According to one model, present-day assembly-line PKSs mainly arose through successive duplications of a parent module, whereafter each prototypical multimodular PKS evolved into a family of distinct but functionally related contemporary PKSs. However, this model has several discordances. For instance, it predicts that orthologous modules of closely related PKSs should cluster together in phylogenetic trees, which is often not the case (Fig. 1B). An alternative model proposes that assembly-line PKSs arose through recombination between different modules in a mosaic-like manner (3), whereafter present-day PKSs evolved through a combination of point mutations, recombination, and gene conversion (2).
Gene conversion is a process in which a DNA sequence is nonreciprocally transferred from one homologous region to another, thereby homogenizing these homologous sequences (Fig. 1C). It is common in eukaryotic genomes, where it frequently occurs during mitosis, meiosis, and double-strand-break repair and has not only been implicated in the evolution of many gene families but also identified as the mechanism causing certain genetic diseases (4). In bacteria, gene conversion has been described only in a few systems, but its overall evolutionary role in prokaryotic genomes is not well understood (5). Gene conversion has also been implicated in assembly-line PKS evolution (6, 7), although its extent, role, and mechanism remain unclear.
We recently proposed that extensive gene conversion between paralogous modules of assembly-line PKSs could explain why paralogous modules are often more similar to each other than orthologous ones (Fig. 1B) (2). In this work we sought to quantify the prevalence of gene conversion in assembly-line PKSs and investigate whether it might confer an evolutionary advantage. We discovered not only that gene conversion is widespread in assembly-line PKSs but also that it is frequently associated with the presence of a genetic element which recodes PKS genes and undergoes gene conversion. This association is particularly strong within Streptomyces bacteria, suggesting a major role in the diversification of assembly-line PKSs and possibly other gene families.
Results
Discovery of Genetic Repeats of Intense Nucleotide Skews in Assembly-Line PKS Genes.
To assess how common gene conversion is among assembly-line PKS genes, we selected 203 assembly-line PKSs that synthesize polyketides of known chemical structure and searched their encoding DNA for regions of unusually high (>80%) sequence identity to another region within the same PKS gene cluster. Almost all of these PKS gene clusters (93%) contained such regions, mostly 0.3 to 1.4 kb in size (SI Appendix, Table S1). Notably, these regions of high DNA sequence identity are often significantly longer than the stretches of DNA encoding the most highly conserved regions of PKS protein domains. For example, conserved regions of ketosynthase (KS) domains (>80% DNA sequence identity) are on average only 0.4 kb long. The duplicated regions that we detected often span entire domains, suggesting that gene conversion rather than protein sequence conservation is responsible for the high degree of sequence identity. A representative example of a PKS with duplicated regions is the tylactone synthase from Streptomyces fradiae (Figs. 1A and 2A).
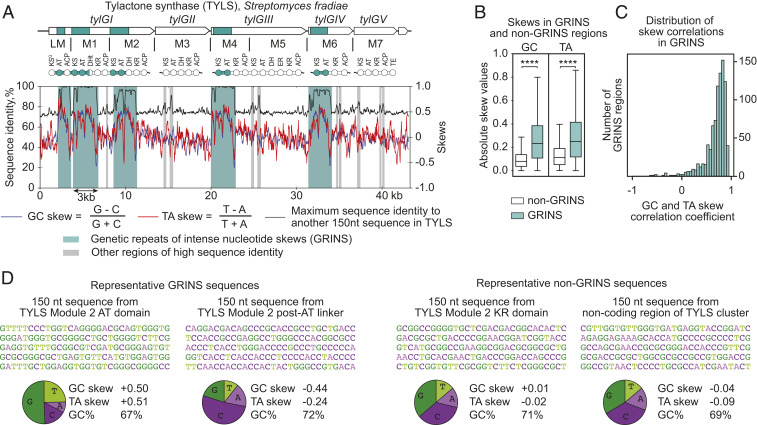
Fig. 2.
GRINS in assembly-line PKSs. (A) Tylactone synthase (TYLS) is a representative example of an assembly-line PKS containing GRINS. The GC and TA skews as well as the maximum sequence similarity to another window in tylactone synthase were calculated for a sliding window of 150 nt with a step of 30 nt. Duplicated regions were identified by high sequence similarity (black line) and are shaded. Several regions also have intense and correlated nucleotide skews (red and blue lines) and therefore are annotated as GRINS (shaded in green). (B) Absolute GC and TA skew values in GRINS and non-GRINS regions of 203 known assembly-line PKS clusters. ****Significant difference (t test, P < 0.0001). Values were calculated for a sliding window of 150 nt with a step of 30 nt along the PKS cluster. (C) Distribution of Pearson correlation coefficients between GC and TA skews in GRINS regions of 203 known assembly-line PKSs. (D) Two representative GRINS sequences and two representative non-GRINS sequences from tylactone synthase gene cluster and their nucleotide content.
Surprisingly, we observed that most duplicated regions have extreme nucleotide composition, characterized by intense and highly correlated GC and TA skews (Fig. 2A). GC skew is a measure of overabundance of G over C on the same DNA strand, while TA skew is a measure of overabundance of T over A. In bacteria, GC and TA skew values often show slight deviations from zero on the scale of the chromosome: typically, the leading strand is slightly positively skewed, and the lagging strand is slightly negatively skewed (8, 9).
The skews we observed in duplicated regions on assembly-line PKSs are qualitatively different and have not been reported in bacterial genomes so far. The fact that they are localized in duplicated regions of assembly-line PKSs that have presumably undergone gene conversion most likely reflects their biological relevance. We designate these genetic elements “GRINS”: genetic repeats of intense nucleotide skews. Even though most duplicated regions in PKS clusters have intense and highly correlated skews, not all of them do. We define GRINS as long (>500 bp) duplicated regions (>80% DNA sequence identity to another region within the same PKS) of intense nucleotide skews (means of absolute GC and TA skew values within the region >0.15). According to this definition, 64% of the 203 selected PKS clusters contain one or more GRINS (SI Appendix, Table S1).
Absolute GC and TA skew values in GRINS regions are more intense than non-GRINS regions within the same assembly-line PKS (P < 0.0001; Fig. 2B). Skews in GRINS often change abruptly and are correlated: A 150-nt window with a more intense GC skew usually has a more intense TA skew (Pearson r = 0.70, P < 0.0001); such correlation is not observed in non-GRINS regions (Pearson r = 0.09, P < 0.0001). Whereas GC and TA skews are positively correlated in most GRINS, anticorrelation was also observed (Fig. 2C). Skew values tend to vary quite abruptly; for example, a 150-nt region of the acyltransferase (AT) domain in module 2 of tylactone synthase is mostly encoded by G+T, whereas a 150-nt region of the post-AT linker in the same module is mostly encoded by C+A, even though these two sequences lie within 1 kb of each other (Fig. 2D).
While the nucleotide composition of GRINS is unexpected per se, it is all the more surprising that they encode functional proteins. This unusual nucleotide composition of GRINS does not appear to restrict the functionality of the encoded protein. In part, this can be explained by the degeneracy of the genetic code: Both the intensity and the correlation of nucleotide skews are most marked in third codon positions, where most mutations are silent (SI Appendix, Fig. S2 A and B). Nonetheless, GRINS have a noticeable effect on the amino acid composition of PKSs; for example, amino acids encoded by GGN (Gly), GTN (Val), TGY (Cys), and TGG (Trp) are highly enriched in positively GC- and TA-skewed regions and depleted in negatively skewed regions, whereas amino acids encoded by CCN (Pro), ACN (Thr), CAY (His), and CAR (Gln) follow the opposite trend (SI Appendix, Fig. S2 C and D).
Potential Functional Implications of GRINS.
An illustrative example of the functional implications of GRINS can be found through sequence comparisons between tylactone synthase and six homologous assembly-line PKSs synthesizing closely related 16-membered lactones that comprise the macrocyclic cores of antibacterial agents with similar mechanisms of action (Fig. 3A and SI Appendix, Fig. S3). These octamodular PKSs share overall pairwise protein sequence similarity >60% [ClustalOmega alignment (10)], clearly indicative of common ancestry. However, each PKS harbors one or more unique GRINS (each labeled in a different color in Fig. 3A and SI Appendix, Fig. S3). Tylactone and angolamycin synthases have five copies of one GRINS each, while the mycinamicin synthase has four GRINS copies, which not only differ from tylactone and angolamycin GRINS by sequence but are also located in a different set of modules. The chalcomycin, niddamycin, midecamycin, and spiramycin PKS gene clusters have two types of GRINS each. While GRINS copies within a PKS are almost identical (75 to 95% DNA identity), they show only modest (45 to 75%) DNA sequence identity between homologous PKSs. Such differences in DNA sequence identity suggest that GRINS are products of relatively recent evolutionary activity in each PKS. The only exceptions are midecamycin and spiramycin synthases, where GRINS are very similar not only within but also between PKSs (40 to 90% DNA identity). It is possible that these two closely related PKSs (89% protein similarity) shared most GRINS before divergence, and later three additional GRINS copies appeared in M4, M5, and M6 of the midecamycin PKS gene cluster.
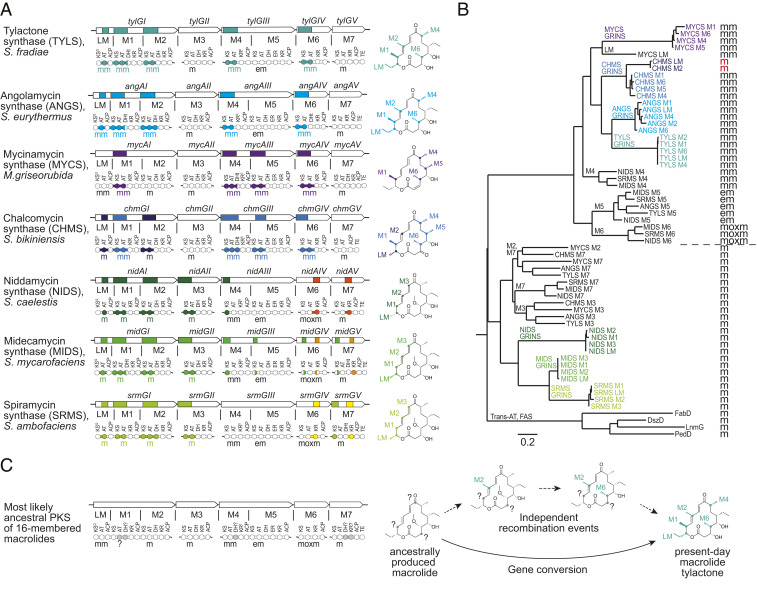
Fig. 3.
Evolutionary role of GRINS in assembly-line PKSs. (A) Seven homologous PKSs. For each module, the extender unit specificity of its AT domain is specified: mm, methylmalonyl extender; m, malonyl; em, ethylmalonyl; moxm, methoxymalonyl. Each color shows a different group of GRINS within a PKS (>80% DNA sequence identity within group) and their locations. Molecules biosynthesized by these PKSs are shown to the right. Each color shows the differences in chemical structure, all of which can be attributed to the different extender unit incorporated by the GRINS-encoded AT domains. (B) Phylogenetic tree of AT domains (Left) and their extender unit specificity (Right). Fatty acid synthase (FAS) and trans-AT PKS AT sequences were used as an outgroup. Sequence alignment performed using ClustalOmega (10); phylogenetic tree constructed using RAxML (30). (C) Proposed role of GRINS in assembly-line PKS evolution. Extender unit specificity of AT domains devoid of GRINS allows reconstruction of the most likely ancestral PKS and its product (Left), whose AT domain specificity differs in multiple modules from modern-day PKSs such as tylactone synthase. These differences would likely have involved several AT domain recombination events and consequently a long evolutionary path, if they occurred independently. Through gene conversion, GRINS allow concerted changes in multiple PKS modules, resulting in a more rapid polyketide diversification path.
Each of the seven homologous PKSs harbors a set of GRINS spanning the DNA encoding the ketosynthase–acyltransferase (KS-AT) fragment or portions thereof (Fig. 3A). AT domains with the same extender unit specificity are thought to originate from a common ancestor, in part because they comprise distinct malonyl- and methylmalonyl-specific clades in the phylogenetic tree (11, 12) (Fig. 3B). (The methylmalonyl-specific clade also contains AT domains specific to more atypical extender units such as ethylmalonyl and methoxymalonyl substrates.) This implies that changes in AT specificity between closely related PKSs must either be due to recombination and/or gene conversion, rather than point mutations. Gene conversion has previously been implicated in the evolution of extender unit specificity of AT domains (6). Analysis of the DNA encoding AT domains from these seven macrolide synthases reveals the presence of GRINS in every AT domain with divergent extender unit specificity (Fig. 3A). For example, in five PKSs where AT domains of module 5 (M5) do not harbor GRINS these ATs have specificity for ethylmalonyl-CoA–derived extender units. However, M5 AT domains in mycinamicin and chalcomycin synthases are encoded by GRINS and are specific for methylmalonyl-CoA, thus replacing an ethyl substituent by a methyl at the corresponding skeletal carbon atoms of mycinolide and chalcolactone, respectively (Fig. 3 A and B). In the mycinamicin synthase, the AT domain of M5 is encoded by a GRINS that is present in four almost identical copies within this PKS and leads to the incorporation of a methylmalonyl-CoA–derived extender unit by each of these modules (purple in Fig. 3A). Similarly, four almost identical copies of a GRINS are present in chalcomycin synthase and lead to the incorporation of a methylmalonyl-CoA–derived extender unit by the four corresponding modules (M1, M4, M5, and M6, light blue in Fig. 3A). In addition, chalcomycin synthase harbors another set of GRINS which result in the incorporation of a malonyl-CoA–derived extender unit by LM and M2 (dark blue in Fig. 3A). Surprisingly, phylogenetic reconstruction places these malonyl-specific AT domains of chalcomycin synthase in the middle of a methylmalonyl-specific AT clade, adjacent to the four methylmalonyl-specific AT domains of chalcomycin synthase (Fig. 3B, red). The DNA sequences of the AT domains of chalcomycin synthase are very similar not only within each set of GRINS (92 to 97% DNA identity in the set of four GRINS; 98% DNA identity in the set of two GRINS) but also between the two sets (80 to 81% DNA identity). This may be indicative of ongoing gene conversion between the two sets of GRINS in chalcomycin synthase, which is gradually increasing the sequence similarity of the AT domains of LM and M2 with those of M1, M4, M5, and M6. It is possible that if this process continues the specificity of the AT domains of LM and M2 will be altered, leading to the biosynthesis of a chemically distinct 16-membered macrolactone.
GRINS also appear in ketoreductase (KR), dehydratase (DH), and enoyl reductase (ER) domains, which reduce the growing polyketide chain. For example, in the family of homologous assembly-line PKSs that synthesize the antihelminthic macrolides milbemycin, avermectin, and nemadectin, differentially reduced positions of the polyketide backbones can be traced back to GRINS located in the reductive loops of corresponding modules (SI Appendix, Fig. S4). These chemical differences significantly affect the biological activity of the resulting compounds, highlighting the importance of GRINS from the perspective of structure/activity evolution within this family of natural products.
The distinct correlation between GRINS locations in PKS genes and differences in polyketide molecules suggests that GRINS might cause the structural diversification of these molecules. By changing the substrate specificity of AT domains or the composition of reductive loops, gene conversion of GRINS could be accelerating the divergence of biosynthesized polyketides between closely related PKSs.
Tracing Evolutionary Lineage through GRINS.
The distribution of GRINS in closely related assembly-line PKSs presents an opportunity to attempt tracing their evolutionary lineage. For example, orthologous AT domains of the PKSs shown in Fig. 3 that do not harbor GRINS are the most likely to resemble their common ancestor. By comparing the specificity of AT domains lacking GRINS it is possible to reconstruct the most likely biosynthetic product of the ancestral octamodular PKS (Fig. 3C). However, this approach does not allow us to infer the ancestral specificity of the extender unit incorporated by module 1, because each PKS harbors a GRINS in the AT domain of this module. Compared to the hypothesized ancestral PKS, all present-day 16-membered macrolide synthases have functionally diverged in at least one domain. We hypothesize the evolutionary benefit of gene conversion stems in part from its ability to simultaneously introduce more than one functionally relevant change in a PKS.
Distribution of GRINS among Assembly-Line PKSs.
By 2018, the sequences deposited into the NCBI database included 3,551 nonredundant assembly-line PKSs (2). Only 10% of these PKSs are functionally characterized, while the remaining PKSs are “orphan” in that their biosynthetic products are unknown. Approximately half of these orphan PKSs are homologous (>50% sequence similarity) to at least one known PKS. We reasoned that by searching this catalog for PKSs containing GRINS insights could be derived into the prevalence of GRINS and their role in PKS diversification.
We observed GRINS in 37% of assembly-line PKSs. Based on module architecture, assembly-line PKSs are separated into cis-AT PKSs (where the AT domain is covalently fused to the KS domain) and trans-AT PKSs (where the AT domain is expressed as a stand-alone protein). GRINS are far more common in cis-AT PKSs than in trans-AT PKSs (Fig. 4A); this is consistent with at least two aspects of our mechanistic and structural understanding of cis-AT vs. trans-AT PKSs. First, all modules of a trans-AT PKS are usually serviced by one copy of a malonyl-CoA specific AT domain (13), which prevents gene conversion between ATs and eliminates any benefit from such exchanges. Second, KS domains of trans-AT PKSs typically show higher specificity for their growing polyketide substrates (14), implying that small structural changes provided by GRINS would be less tolerated.
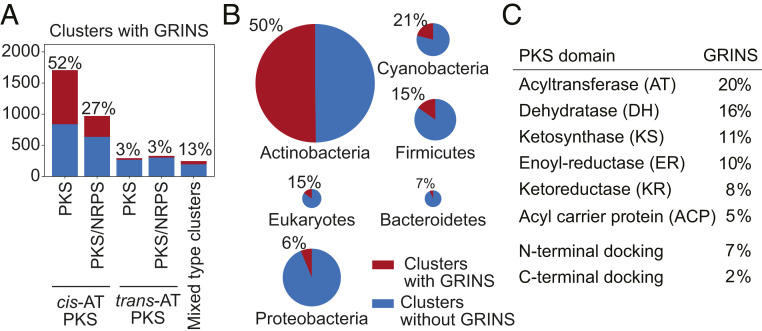
Fig. 4.
Distribution of GRINS in 3,551 assembly-line PKS clusters. (A) Distribution of GRINS among different cluster types. (B) Distribution of GRINS in clusters from different taxons. (C) Distribution of GRINS among different PKS domains.
PKSs often collaborate with another family of assembly-line enzymes: nonribosomal polypeptide synthetases (NRPSs). Predictably, GRINS are more common in PKS–NRPS hybrids with cis-AT PKSs than in hybrids with trans-AT PKSs (Fig. 4A). When GRINS were detected in such hybrid clusters they were most commonly found in DNA encoding PKS modules, although some GRINS elements were also observed in DNA encoding NRPS modules.
GRINS are also nonhomogeneously distributed among assembly-line PKSs from different bacterial phyla: They are common in actinobacteria, less so in cyanobacteria and firmicutes, and rare in bacteroidetes and proteobacteria (Fig. 4B). In part, it could be due to the fact that cis-AT PKSs are widespread in actinobacteria and cyanobacteria, while trans-AT PKSs are typically found in firmicutes and bacteroidetes (2). However, it does not explain the low abundance of GRINS in proteobacteria, both in cis- and trans-AT PKSs. It is possible that other factors contribute to the heterogeneous distribution of GRINS among bacterial phyla, such as the niches they inhabit and the biological activity of molecules they produce. For instance, GRINS may provide a greater evolutionary advantage to PKSs that produce antibiotics than to those producing pigments.
Last, but not least, we also detected GRINS in assembly-line PKSs of amoeba and choanoflagellates. The evolutionary history of eukaryotic assembly-line PKSs is unclear. While it is possible that these PKSs (along with their GRINS) were acquired from a bacterium, the fact that relatively intense skews persist even in third-codon positions of these PKSs suggests that the mechanisms responsible for their maintenance could be present in eukaryotic genomes.
Distribution of GRINS among Assembly-Line PKS Domains.
GRINS occur in virtually all domains that comprise PKS modules (Fig. 4C), including docking domains, methyltransferases, beta-branching ACP domains, and others (SI Appendix, Table S5). Consistent with their possible role in accelerating the appearance of small adjustments to the chemical structure of the product, GRINS are most common in AT domains overlapping ∼20% of all AT domains in assembly-line PKSs. GRINS could also potentially change the reduction level of the backbone, which is consistent with their distribution in the avermectin synthase and backed up by their relatively frequent occurrence in DH, ER, and KR domains. The functional implications of GRINS in 11% KS and 5% ACP domains are less clear. It is known that KS domains show a certain degree of specificity for their upstream and downstream ACPs, as well as for the incoming polyketide chain. Gene conversion of KS and ACP would change their specificity for partner domains, thereby promoting changes to the product structure. An example of structural diversification through recombination of KS and upstream ACP domains was described in four homologous PKSs that biosynthesize large aminopolyols (15). Our analysis of these PKSs revealed numerous GRINS (SI Appendix, Table S1), suggesting that GRINS occurring in KS and ACP domains may play a role in the structural diversification of their polyketide products.
Phylogenetic Distribution of GRINS.
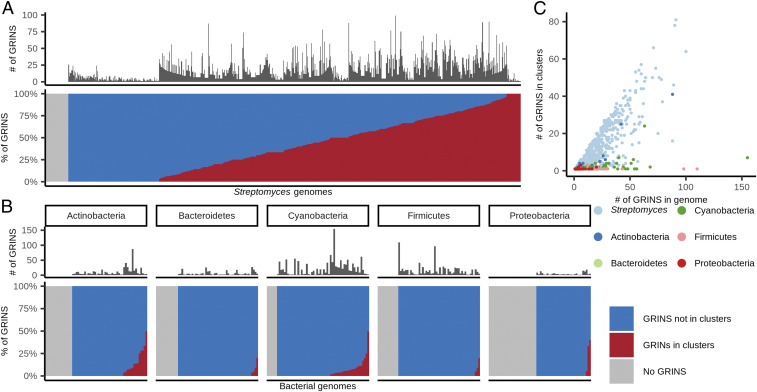
Up to this point our analysis focused on assembly-line PKSs. However, these sequences are not representative of bacterial diversity. In order to limit ascertainment bias and to get a more accurate view of GRINS in bacterial evolution, we analyzed 803 high-quality and nonredundant Streptomyces genomes. For each genome we predicted both GRINS and biosynthetic gene clusters (Materials and Methods). We found that most (95%) Streptomyces species harbor at least one GRINS element and that most of those genomes (80%) harbor at least one GRINS overlapping a biosynthetic gene cluster (Fig. 5A). In order to determine if this prevalence is evolutionarily meaningful, we also predicted GRINS and biosynthetic gene clusters in 300 high-quality genomes each representing a unique bacterial genus, evenly split among five different bacterial phyla (Materials and Methods and SI Appendix, Table S6). In this diverse group of bacteria we found a lower proportion of genomes harboring GRINS (53 to 90%) when compared with Streptomyces. Even when GRINS are present, a smaller proportion of them overlap with biosynthetic gene clusters (6 to 42%; Fig. 5 B and C). Finally, we tested for an enrichment of GRINS within biosynthetic gene clusters by comparing the fraction of each genome that encodes such clusters with the fraction of GRINS that overlap with the same clusters in each genome. We found that an average of 12% genomic content in each Streptomyces encodes biosynthetic gene clusters and that these cluster overlap 40% of GRINS, implicating a highly significant 3.28-fold enrichment (P = 2.8e-80, two-sided Wilcoxon rank sum test). Among the genomes in the five phyla tested, only actinobacteria showed some level of enrichment (1.66-fold), but it was not significant (P = 0.17). The other four phyla all had a significant depletion (P < 0.05) of GRINS among biosynthetic gene clusters.
Fig. 5.
GRINS in bacterial genomes. (A) Distribution of GRINS in 803 Streptomyces high-quality nonredundant genomes. (Upper) Each bar/sliver is a genome; the dark bars show the number of detected GRINS in the corresponding genome. (Lower) The fraction of GRINS that overlap with biosynthetic gene clusters (red) versus those that do not overlap with such clusters (blue). Genomes without GRINS appear gray in this plot. (B) Distribution of GRINS among 300 bacterial genera from five phyla. The plot is similar to A, Lower, but each genome represents a unique genus from the corresponding phylum. (C) Streptomyces-specific enrichment of GRINS in biosynthetic gene clusters. Each point is a genome, and color indicates the taxonomic group of each genome. Only for Streptomyces bacteria does the number of GRINS in biosynthetic gene clusters (y axis) increase with the number of GRINS in a genome (x axis).
We also searched for overrepresentation of GRINS among other types of genes. We found that GRINS occur frequently in genes involved in replication, recombination, and repair. Transposases were the most commonly represented in four of the five phyla. Notably, 77% of cyanobacterial GRINS overlapped with a transposase (SI Appendix, Fig. S7), which presumably explains why GRINS are rare in cyanobacterial biosynthetic gene clusters despite these organisms having the highest average number of such clusters (Fig. 5B). Other mobile genetic elements such as integrases and phages were also represented with no significant differences between phyla (SI Appendix, Fig. S7). Finally, endonucleases overlapped with 15% of actinobacterial GRINS and were practically absent among GRINS from other taxa (SI Appendix, Fig. S7).
Taken together, these results imply that, while GRINS elements are widely distributed across bacterial lineages, their association with biosynthetic gene clusters is highly specific for the Streptomyces genus. As such, their role in PKS evolution might be particularly relevant within this lineage. Streptomyces have linear and relatively unstable chromosomes, which undergo frequent deletions and other recombination events (16). In addition, they lack homologs of the RecBCD or RecFOR pathways, instead relying on the less conventional AdnAB pathway for homologous recombination (17, 18). The resolution of Holliday junctions formed during recombination relies not only on the activity of RuvA and RuvG homologs but also on the yet-unknown alternative factors (19). Considering these unusual features of Streptomyces, we hypothesized that proteins involved in replication, recombination, or repair could be responsible for the high number of GRINS elements within this genus. To test this hypothesis, we performed phylogenetic profiling to detect gene copy number or amino acid sequence variants that associate with GRINS number, while controlling for phylogenetic relationships and genome quality (Materials and Methods). We found that a number of orthologous groups were quantitatively associated with the number of GRINS in Streptomyces genomes. Most notably, the RuvA helicase gene (OG:05KA4 in the eggNOG database) is negatively associated with GRINS number (SI Appendix, Fig. S8A). RuvA is known to bind Holliday junctions formed during DNA recombination and to promote branch migration of these junctions, which in turn can lead to gene conversion (20). Further analysis of the ruvA gene revealed that two sequence variants among identifiable RuvA orthologs are also strongly associated with differences in GRINS number (SI Appendix, Fig. S8 B and C). More work is needed to confirm if these sequence differences are indeed directly responsible for observed differences in distributions frequency, but our associations suggest that GRINS in Streptomyces are the product of complex pathways.
Discussion
GRINS are an unprecedented example of genetic elements that recode homologous proteins with a biased nucleotide composition. This process of gene conversion presumably facilitates evolution of the target proteins. Even though the underlying mechanisms of the origin and persistence of GRINS remain unknown, our observations have led us to formulate certain hypotheses.
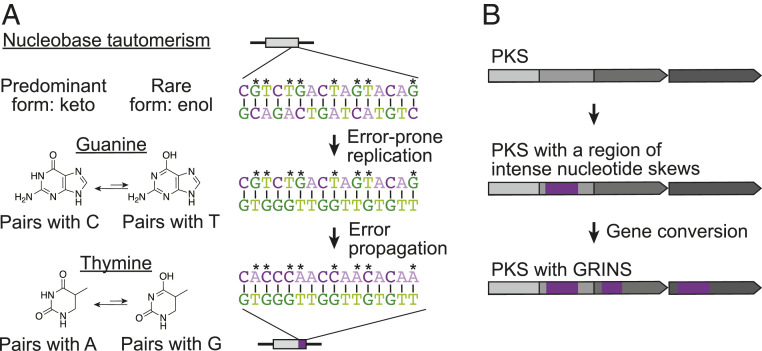
First, because of the wide distribution of GRINS among bacterial phyla and their presence in eukaryotes, the mechanisms involved in generating nucleotide skews must be relatively ubiquitous. It is possible that they involve multifactorial processes resulting in replication strand bias, such as cytosine deamination on the single-stranded DNA of the lagging strand (21). However, they do not explain why both intense positive and negative skews are observed on the same strand within GRINS elements. Mechanisms to account for the high intensity GC and TA skews observed in GRINS include, for example, error-prone replication due to the presence of tautomeric bases within the DNA (Fig. 6A). There is growing evidence that tautomeric bases exist in nucleic acids (22). Because G and T undergo keto/enol tautomerism whereas C and A undergo amino/imino tautomerism, the ability of an error-prone polymerase to stabilize one type of isomer or another could explain the strong association of GC and TA skews.
Fig. 6.
Possible mechanistic origins of GRINS. (A) Nucleotide skews in GRINS could be generated through nucleobase tautomerism. Error-prone replication of minor tautomeric forms of G and T could result in C→T or A→G transitions in the daughter strand followed by appearance of correlated GC and TA skews after the next round of replication. Analogously, amino/imino tautomerism of C and A nucleotides would lead to opposite skews. (B) We propose that once a region of intense nucleotide skews is formed within a PKS it is more likely to undergo gene conversion between homologous module regions, thereby giving rise to GRINS.
Second, we propose that a skew in the DNA encoding all or part of one PKS module leads to recoding of a neighboring homolog at a higher frequency (Fig. 6B). Sequences with a high G+C bias are known to recombine more frequently. We hypothesize that additional nucleotide composition bias would make them more likely to undergo gene conversion due to their decreased sequence complexity, thereby giving rise to GRINS.
Third, we expect that present-day GRINS are active elements that are observed at different stages of a “life cycle” that involves accumulation of nucleotide skews, propagation through gene conversion, and erosion of sequence identity through random mutation. Examples shown in Fig. 3 most likely correspond to GRINS around the peak of their activity: High sequence similarity between GRINS copies suggests relatively recent gene conversion or strong purifying selection. However, we also observed regions of intense nucleotide skews present in only one copy, which could represent either an initial substrate for further gene conversion or an ancient GRINS element that is in the process of being lost. In many PKSs, GRINS sequences are highly similar but not identical, most likely because the skews are counterbalanced by both natural selection and mutation. Further understanding of the mechanisms involved in GRINS appearance, dissemination, and dissipation will provide more insight into the dynamics of these genetic elements.
It is also possible that a better understanding of GRINS chemical biology could spawn new methods for engineering PKSs. For example, in the course of targeted domain replacement efforts in the rapamycin and tylosin gene clusters Wlodek et al. identified unanticipated recombinants that produced new molecules, often at high titers (23). Our bioinformatic analysis indicates that all of the reported recombination events occurred between GRINS elements, highlighting the potential of deliberately leveraging GRINS for engineering other assembly-line PKSs to yield diverse “unnatural” natural products.
Materials and Methods
GRINS Detection in PKS Clusters.
The list of 203 known assembly-line PKS gene clusters was compiled by choosing PKSs for which sequence is available in the NCBI database, the polyketide product is known, and a biosynthetic mechanism has been proposed in the literature (SI Appendix, Table S1). Among these, the list and sequences of known trans-AT PKS clusters was kindly provided by E. J. Helfrich as reported in ref. 24. The list of 3,551 nonredundant assembly-line PKSs was reported previously in ref. 2.
First, we identified regions of high sequence identity in each PKS gene cluster. The DNA sequence was split into 150-nt-long fragments using a sliding window with a step of 30 nt. Pairwise alignments between all nonoverlapping 150-nt-long regions were performed using the Biopython pairwise2 module, and for each region the highest score of DNA sequence identity was kept. Regions of high sequence identity were defined as >500-bp regions in which the average DNA sequence identity over five adjacent windows is above 80%. Next, among these regions we identified GRINS as those that have mean absolute GC and TA skew intensities >0.15. All scripts are available at https://github.com/surh/grins.
Genome Selection.
For Streptomyces species, all genomes published in the NCBI database as of February 2020 were downloaded and submitted to CheckM (25). Only genomes predicted to be >95% complete, <5% contaminated, and assigned to the Streptomycetaceae family by CheckM were kept. These genomes were dereplicated by calculating their genomic average nucleotide identity (gANI) via FastANI (26) and clustered at 95% gANI. From each resulting cluster, a random representative was chosen, resulting in 803 genomes. For the analysis of the bacterial phyla Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, and Proteobacteria, representative genomes from Escherichia coli, Haemophilus influenzae, Pseudomonas aeruginosa, Mycobacterium tuberculosis, Mycobacterium leprae, and Synechocystis spp. were manually chosen, and the list of all RefSeq genomes from the phyla above was downloaded. This list was used to generate a random list of 70 additional RefSeq genomes from each phylum, ensuring that only one representative of any given genus was chosen. The selected 350 genomes were downloaded and processed with CheckM. Genomes with less than 95% completeness or more than 5% contamination were discarded. Of the remaining genomes, we randomly down-sampled to 60 genomes per phylum, ensuring the presence of the six manually selected genomes, resulting in a final set of 300 genomes (SI Appendix, Table S6).
GRINS Detection in Genomes.
We developed a pipeline to detect GRINS from assembled genomes. Briefly, the pipeline first partitions each genome into partially overlapping 150-bp windows with a step size of 30 bp. Then, BOWTIE2 (27) is used to map each window back to the genome using global (–end-to-end) and sensitive (–sensitive) alignment. Similar results were obtained with –very-sensitive and local alignments. After removing self-alignments, duplicated regions were identified and filtered by size, keeping only duplicated regions >500 bp. Next, among these regions we identified GRINS as those that have mean absolute GC and TA skew intensities >0.15. All scripts are available at https://github.com/surh/grins.
Genome Annotations.
In order to consistently identify biosynthetic gene clusters and other gene families, each assembled genome was annotated from scratch. First, antiSMASH5.0 (28) was run on each genome to detect biosynthetic gene clusters. We used Prodigal through antiSMASH (–genefinding-tool prodigal) in order to obtain CDS predictions, and we annotated all the resulting CDS sequences with the eggNOG mapper utility from the eggNOG4.5 database (29). All scripts are available at https://github.com/surh/grins.
Phylogenetic Profiling.
We took a linear modeling approach to detect associations between orthologous group (OG) copy number and number of GRINS per genome. We used the results from the eggNOG annotation to assign each CDS in the 803 Streptomyces genomes to its most likely orthologous group (bestOG). We then performed principal component analysis in the copy number matrix derived from these annotations. We included the 10 top principal components in a linear model that associated the log copy number as a predictor variable and the log number of GRINS as a dependent variable. We also included covariates for genome size and number of contigs in the assembled genomes (log(n_grins) ∼ log(OG copy number) + PCs + genome_size + n_contigs). The resulting P values were adjusted for multiple testing with the Benjamini–Hochberg method. Finally, for selected genes (e.g., RuvA; SI Appendix, Fig. S8) we generated multiple sequence alignments of all genes in the orthologous group (i.e., including paralogues and orthologs). We then performed phylogenetic reconstruction via RAxML (30) and removed paralogues by eliminating the multicopy genes that had the most substitutions within each genome. With the resulting list of orthologs we reused our phylogenetic profiling linear model (with the same controlling covariates) but using every variable site as predictor variable. All scripts are available at https://github.com/surh/grins.
Supplementary Material
Acknowledgments
This research was supported, in part, by NIH Grant R01 GM087934, FRM Fellowship SPE20170336834 (to A.N.), and the Simons Foundation fellowship of the Life Sciences Research Foundation (to S.H.P.).
Footnotes
The authors declare no competing interest.
See QnAs, e2109638118, in vol. 118, issue 26.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100751118/-/DCSupplemental.
Data Availability
All scripts are available in GitHub at https://github.com/surh/grins. All sequence data are publicly available, and accessions are listed in SI Appendix.
References
- 1.Robbins T., Liu Y.-C., Cane D. E., Khosla C., Structure and mechanism of assembly line polyketide synthases. Curr. Opin. Struct. Biol. 41, 10–18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nivina A., Yuet K. P., Hsu J., Khosla C., Evolution and diversity of assembly-line polyketide synthases. Chem. Rev. 119, 12524–12547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen T., et al., Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat. Biotechnol. 26, 225–233 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Chen J.-M., Cooper D. N., Chuzhanova N., Férec C., Patrinos G. P., Gene conversion: Mechanisms, evolution and human disease. Nat. Rev. Genet. 8, 762–775 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Santoyo G., Romero D., Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol. Rev. 29, 169–183 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Zucko J., Long P. F., Hranueli D., Cullum J., Horizontal gene transfer and gene conversion drive evolution of modular polyketide synthases. J. Ind. Microbiol. Biotechnol. 39, 1541–1547 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Medema M. H., Cimermancic P., Sali A., Takano E., Fischbach M. A., A systematic computational analysis of biosynthetic gene cluster evolution: Lessons for engineering biosynthesis. PLoS Comput. Biol. 10, e1004016 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rocha E. P. C., The replication-related organization of bacterial genomes. Microbiology (Reading) 150, 1609–1627 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Grigoriev A., Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 26, 2286–2290 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sievers F., et al., Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haydock S. F., et al., Divergent sequence motifs correlated with the substrate specificity of (methyl)malonyl-CoA:acyl carrier protein transacylase domains in modular polyketide synthases. FEBS Lett. 374, 246–248 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Jenke-Kodama H., Sandmann A., Müller R., Dittmann E., Evolutionary implications of bacterial polyketide synthases. Mol. Biol. Evol. 22, 2027–2039 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Helfrich E. J. N., Piel J., Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 33, 231–316 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Jenner M., et al., Acyl-chain elongation drives ketosynthase substrate selectivity in trans-acyltransferase polyketide synthases. Angew. Chem. Int. Ed. Engl. 54, 1817–1821 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Zhang L., et al., Characterization of giant modular PKSs provides insight into genetic mechanism for structural diversification of aminopolyol polyketides. Angew. Chem. Int. Ed. Engl. 56, 1740–1745 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Chen C. W., Huang C.-H., Lee H.-H., Tsai H.-H., Kirby R., Once the circle has been broken: Dynamics and evolution of Streptomyces chromosomes. Trends Genet. 18, 522–529 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Zhang L., et al., The adnAB locus, encoding a putative helicase-nuclease activity, is essential in Streptomyces. J. Bacteriol. 196, 2701–2708 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoff G., Bertrand C., Piotrowski E., Thibessard A., Leblond P., Genome plasticity is governed by double strand break DNA repair in Streptomyces. Sci. Rep. 8, 5272 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoff G., Bertrand C., Piotrowski E., Thibessard A., Leblond P., Implication of RuvABC and RecG in homologous recombination in Streptomyces ambofaciens. Res. Microbiol. 168, 26–35 (2017). [DOI] [PubMed] [Google Scholar]
- 20.West S. C., Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 31, 213–244 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Rocha E. P. C., Touchon M., Feil E. J., Similar compositional biases are caused by very different mutational effects. Genome Res. 16, 1537–1547 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimsey I. J., Petzold K., Sathyamoorthy B., Stein Z. W., Al-Hashimi H. M., Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes. Nature 519, 315–320 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wlodek A., et al., Diversity oriented biosynthesis via accelerated evolution of modular gene clusters. Nat. Commun. 8, 1206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helfrich E. J. N., et al., Automated structure prediction of trans-acyltransferase polyketide synthase products. Nat. Chem. Biol. 15, 813–821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W., CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain C., Rodriguez-R L. M., Phillippy A. M., Konstantinidis K. T., Aluru S., High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114–5118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25–R10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blin K., et al., antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huerta-Cepas J., et al., eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All scripts are available in GitHub at https://github.com/surh/grins. All sequence data are publicly available, and accessions are listed in SI Appendix.